Abstract
A major increase of bacterial resistance to colistin, a last-resort treatment for severe infections, was observed globally. Using colistin in livestock rearing is believed to be the ground of mobilized colistin resistance (mcr) gene circulation and is of crucial concern to public health. This study aimed to determine the frequency and virulence characteristics of colistin-resistant Gram-negative bacteria from the milk of mastitic cows and raw unpasteurized milk in Egypt. One hundred and seventeen strains belonging to Enterobacteriaceae (n = 90), Pseudomonas aeruginosa (n = 10), and Aeromonas hydrophila (n = 17) were screened for colistin resistance by antimicrobial susceptibility testing. The genetic characteristics of colistin-resistant strains were investigated for mcr-1–9 genes, phylogenetic groups, and virulence genes. Moreover, we evaluated four commonly used biocides in dairy farms for teat disinfection toward colistin-resistant strains. Multidrug-resistant (MDR) and extensive drug-resistant (XDR) phenotypes were detected in 82.91% (97/117) and 3.42% (4/117) of the isolates, respectively. Of the 117 tested isolates, 61 (52.14%) were colistin resistant (MIC >2 mg/L), distributed as 24/70 (34.29%) from clinical mastitis, 10/11 (90.91%) from subclinical mastitis, and 27/36 (75%) from raw milk. Of these 61 colistin-resistant isolates, 47 (19 from clinical mastitis, 8 from subclinical mastitis, and 20 from raw milk) harbored plasmid-borne mcr genes. The mcr-1 gene was identified in 31.91%, mcr-2 in 29.79%, mcr-3 in 34.04%, and each of mcr-4 and mcr-7 in 2.13% of the colistin-resistant isolates. Among these isolates, 42.55% (20/47) were E. coli, 21.28% (10/47) A. hydrophila, 19.12% (9/47) K. pneumoniae, and 17.02% (8/47) P. aeruginosa. This is the first report of mcr-3 and mcr-7 in P. aeruginosa. Conjugation experiments using the broth-mating technique showed successful transfer of colistin resistance to E. coli J53-recipient strain. Different combinations of virulence genes were observed among colistin-resistant isolates with almost all isolates harboring genes. Hydrogen peroxide has the best efficiency against all bacterial isolates even at a low concentration (10%). In conclusion, the dissemination of mobile colistin resistance mcr gene and its variants between MDR- and XDR-virulent Gram-negative isolates from dairy cattle confirms the spread of mcr genes at all levels; animals, humans, and environmental, and heralds the penetration of the last-resort antimicrobial against MDR bacteria. Consequently, a decision to ban colistin in food animals is urgently required to fight XDR and MDR bacteria.
Keywords: colistin, Gram-negative bacteria, mastitis, milk, mcr, virulence factors, multidrug-resistance
Introduction
Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa are the most common Gram-negative environmental pathogens in dairy farms. They could infect the mammary glands of dairy cows through the teat end and colonize the mammary tissue, causing mastitis (Scatamburlo et al., 2015; Gao et al., 2017). Although Aeromonas species are autochthonous to aquatic environments, Aeromonas hydrophila has been reported as a common contaminant in a diverse variety of foods such as milk and milk products (Sharma and Kumar, 2011). Contamination of milk with pathogenic Gram-negative bacteria is often derived directly from the udder excretion of infected animals or the farm environment, thus putting the consumers who consume unpasteurized milk at a high risk for developing foodborne diarrheal diseases (Garcia et al., 2019).
In Egypt, there are no regulations that control the use of antibiotics in animal husbandry for growth promotion or the prevention and treatment of bacterial diseases (WHO, 2013). The misuse of antibiotics in dairy production is a matter of concern for the effective antibiotic therapy of infected cows, food safety, and occupational exposure, and a potential threat to public health due to the selection of multidrug-resistant (MDR) bacteria and entry of antibiotic residues into bulk tank milk (Oliveira and Ruegg, 2014; Locatelli et al., 2019). The emergence of MDR and extensive drug-resistant (XDR) Gram-negative bacteria in livestock production increased the interest in the use of colistin (polymyxin E) as a last-resort antibiotic for the treatment of these pathogens (Kempf et al., 2016). Although colistin was prohibited in developed countries, it is still used in animal husbandry in Egyptian dairy farms (Lima Barbieri et al., 2017; Dandachi et al., 2019). Hence, the extensive use of colistin increases colistin-resistant Gram-negative bacteria. Also, it imposes a major public health concern (Lima Barbieri et al., 2017) due to the dissemination of colistin resistance genes (mcr) through mobilized plasmids among animal strains and subsequently transmitted to humans through the food chain or direct contact (McEachran et al., 2015; García et al., 2018; Hmede and Kassem, 2019). Nine plasmid-borne mcr-family genes (mcr-1–mcr-9) have been reported in over 40 countries from different continents across the globe; most of them were detected in several enterobacteria, including E. coli and K. pneumoniae (Luo et al., 2020). The mcr-1 gene has been detected in most continents, while other genes have only been found in a few countries (Ling et al., 2020). The mcr-1 gene has been reported previously in colistin-resistant E. coli isolated from raw milk (Coton et al., 2012; Hassen et al., 2019), mastitis bovine milk (Filioussis et al., 2019; Liu G. et al., 2020), and cheese (Coton et al., 2012; Hammad et al., 2019). A higher frequency of mcr-1 compared with other mcr genes has been reported in colistin-resistant E. coli (98.9%), K. pneumoniae (100%), and P. aeruginosa (100%) recovered from animal and human sources (Javed et al., 2020). The implementation of pre- and post-milking teat disinfection is a critical effective means of reducing the incidence of clinical and subclinical mastitis as well as new intramammary infections caused by Gram-negative environmental pathogens in dairy farms such as E. coli, K. pneumoniae, and P. aeruginosa (Tiwari, 2013; Kamal and Bayoumi, 2015; Böhm et al., 2017; Rowe et al., 2018). Choosing the ideal disinfectant with an accurate concentration is vital to control resistant bacteria in dairy farms. The resistant bacteria can survive at concentrations many folds below the minimum inhibitory concentration (MIC) of the biocidal agents (Hughes and Andersson, 2012). Notably, Gram-negative bacteria have a risk for promoting antibiotic resistance after exposure to sublethal concentrations of some disinfectants such as chlorhexidine (Kampf, 2018).
Most putative virulence determinants contributing to the pathogenicity of E. coli, K. pneumoniae, Aeromonas species, and P. aeruginosa are chromosomally encoded. However, previous studies have indicated the occurrence of some virulence factors on plasmids of E. coli (Rodriguez-Siek et al., 2005; Johnson et al., 2006; Mellata et al., 2010; Cyoia et al., 2015), K. pneumoniae (Dolejska et al., 2013; Chen et al., 2020), Aeromonas species (Brown et al., 1997), and P. aeruginosa (Morales-Espinosa et al., 2012). E. coli strains possess virulence genes that play a significant role in the survival and pathogenesis of the strain in the host through bacterial adhesion (ipfA), hemolysis (hlyF), iron acquisition (iroN and ireA), and increased serum survival and resistance to phagocytosis (iss and traT) (Sarowska et al., 2019; Adzitey et al., 2020). K. pneumoniae hypervirulent strains possess different genes that determine virulence and severity of infection in the host. These virulence genes include entB (enterobactin biosynthesis gene), allS (associated with allantoin metabolism), ybtS and irp (yersiniabactin biosynthesis), and mrkD and fimH (fimbrial adhesin that mediates binding to the extracellular matrix to form the biofilm) (Rosen et al., 2008; Wasfi et al., 2016; Ye et al., 2016; Russo and Marr, 2019). The most important A. hydrophila virulence factors include hemolysin A (hlyA), aerolysin (aer), cytotonic heat-stable enterotoxin (ast), cytotoxic enterotoxin (act), and lipases (lip) (Furmanek-Blaszk, 2014; Rather et al., 2014). P. aeruginosa produces extracellular products that contribute to its pathogenicity, such as protein exotoxin A (toxA), proteases (lasB), type III secretion system exoenzymes (exoU and exoS) (Casilag et al., 2016; Fadhil et al., 2016), and genes responsible for pyocyanin production (phzM) (Nowroozi et al., 2012). Genes encoding virulence and MDR/XDR are often found with mcr genes on plasmids in environmental isolates (Anyanwu et al., 2020). There are limited reports that studied the occurrence of plasmid-encoded virulence and colistin resistance genes (mcr) among colistin-resistant Gram-negative bacteria isolated from cow’s milk in our geographic region. Hence, the current study aimed to: (1) determine the frequency of colistin-resistant Gram-negative bacteria from the milk of mastitic cows as well as raw unpasteurized milk; (2) screen for mcr resistance determinants in bacterial isolates and their virulence characteristics; (3) evaluate four frequently used biocides in dairy farms for teat disinfection toward colistin-resistant isolates.
Materials and Methods
Gram-Negative Bacterial Strains
A total of 117 Gram-negative bacterial strains belonging to Enterobacteriaceae (n = 90), P. aeruginosa (n = 10), and A. hydrophila (n = 17) were included in this study. The bacterial isolates were recovered previously from milk samples of dairy cows showing mastitis as well as bulk tank raw milk during 2018–2020. Gram-negative isolates were selected based on the provided data from the Microbiology Department, Faculty of Veterinary Medicine, Zagazig University, Egypt. Enterobacteriaceae strains included non-O157 E. coli (n = 30), O157 E. coli (n = 12), K. pneumoniae (n = 18), Enterobacter species (n = 10), and Citrobacter species (n = 20). The isolated bacteria were sourced from different farms. All the isolates were independent and were not clonally duplicated from the same source. The presumptive bacterial isolates were confirmed adopting the standard microbiological procedures (Quinn et al., 1994). In brief, MacConkey’s, eosin methylene blue (Oxoid, Cambridge, UK) and HiCrome klebsiella-selective agar media (Himedia, Mumbai, India) were used for the cultivation of Enterobacteriaceae strains. Aeromonas species were grown on Rimler-Shotts medium (HiMedia, India) with a novobiocin (Oxoid, UK) supplement, while P. aeruginosa strains were cultivated on Pseudomonas cetrimide agar (Oxoid, UK). Serotyping of E. coli was applied using diagnostic polyvalent and monovalent O and H antisera (Denka Seiken Co., Tokyo, Japan). Genomic DNA was extracted from fresh bacterial cultures using the QIAamp DNA Mini kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Polymerase chain reaction (PCR)-based confirmation of Gram-negative bacteria was applied using genus- and species-specific primer sets depicted in Supplementary Table S1 .
Antimicrobial Susceptibility Testing
The antimicrobial susceptibilities of Gram-negative bacterial strains (n = 117) against a panel of 24 commonly used antimicrobial agents were determined using the Kirby-Bauer disk diffusion assay following the Clinical and Laboratory Standards Institute guidelines and interpretative criteria (CLSI, 2019). The tested antimicrobial discs (Oxoid, Cambridge, UK) were ampicillin (10 µg), amoxicillin-clavulanic acid (30 µg), piperacillin-tazobactam (40 µg), cefazolin (30 µg), cephalothin (30 µg), cefoxitin (30 µg), ceftriaxone (30 µg), cefotaxime (30 µg), ceftazidime (30 µg), cefepime (30 µg), imipenem (10 µg), nalidixic acid (30 µg), ciprofloxacin (5 µg), levofloxacin (5 µg), gentamicin (10 µg), tobramycin (10 µg), amikacin (30 µg), trimethoprim-sulphamethoxazole (25 µg), chloramphenicol (30 µg), tetracycline (30 µg), aztreonam (30 µg), tigecycline (15 µg), fosfomycin (50 µg), and colistin (25 µg). The minimum inhibitory concentration (MIC) of colistin (Sigma-Aldrich, Seelze, Germany) was determined against Gram-negative bacterial strains by broth microdilution technique following the relevant CLSI document (CLSI, 2019).
The multiple antibiotic resistance (MAR) indices were evaluated as documented elsewhere (Tambekar et al., 2006), while the MDR and XDR phenotypes were reported according to Magiorakos et al. (2012). E. coli ATCC 25922, P. aeruginosa ATCC 27853, and A. hydrophila ATCC 7966 were used as quality control strains.
Detection of Plasmid-Mediated Colistin Resistance Genes
Plasmid DNAs were extracted from colistin-resistant Gram-negative bacterial strains (E. coli, K. pneumoniae, P. aeruginosa, and A. hydrophila) using Plasmid DNA Miniprep Kits (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. PCR amplifications of colistin resistance genes (mcr-1 to mcr-9) from phenotypic colistin-resistant Gram-negative bacteria isolates were performed using oligonucleotide primer sequences and their annealing temperatures listed in Supplementary Table S1 . Conventional PCRs were performed using a programmable 2720 thermal cycler (Applied Biosystem, Waltham, MA, USA) in a total reaction volume of 25 μl containing 12.5 μL of EmeraldAmp Max PCR Master Mix (Takara, Japan), 1 μl of each primer (20 pmol) (Sigma-Aldrich, Co., St. Louis, MO, USA), 5 μl of template DNA, and 5.5 μl of nuclease-free water. The amplified products were visualized after 30 min of electrophoresis on a 2% agarose gel containing ethidium bromide.
DNA Sequencing and Phylogenetic Analysis of mcr Genes
DNA sequencing of the PCR products was applied to validate their genetic identity and to verify any mutations in the mcr genes. The amplified DNA fragments were purified with the QIAquick PCR purification kit (Qiagen, Courtaboeuf, France). Sanger sequencing was performed in both directions using Bigdye Terminator V3.1 cycle sequencing kit (Perkin-Elmer, Inc. Waltham, MA, USA) in an applied Biosystems 3130 genetic analyzer (Hitachi, Tokyo, Japan). The nucleotide sequences were compared with those available in the National Center for Biotechnology Information (NCBI; www.ncbi.nlm.nih.gov). Nucleotide and deduced protein sequences were analyzed using the MEGA7 program (Kumar et al., 2016). To investigate the genetic relatedness and the evolutionary distance among Gram-negative strains harboring the mcr genes, a phylogenetic tree was constructed using the neighbor-joining (maximum composite likelihood) method. DNA sequences generated in the study were deposited into the GenBank database with accession numbers MW811398-MW811434 and MZ648218-MZ648227.
Conjugation Assay
The transmissibility of colistin-resistant mcr genes between donors (mcr-positive isolates) and the recipient bacteria (E. coli J53; Na azide-resistant) was evaluated by the conjugation assays using the broth mating technique (Wang et al., 2003). An equal ratio (1:1) of donor and recipient cells (0.5 ml of each) in a logarithmic phase were added to 4 ml of sterile Luria-Bertani (LB) broth (Sigma-Aldrich, USA) then incubated overnight at 37°C without shaking. The mating mixture was serially diluted then the transconjugants were cultured on LB agar (Sigma-Aldrich, USA) supplemented with colistin (2 mg/L) or Na azide (100 mg/L). MICs for the donors, recipients, and transconjugants were determined by the broth microdilution method following the CLSI guidelines (CLSI, 2019). Potential transconjugants were examined for the existence of mcr genes using PCR assays. The conjugation efficiency was calculated as the number of transconjugants per donor as previously documented (Wang et al., 2003).
PCR Amplification of Virulence-Associated Genes
PCR detection of virulence-related genes of E. coli (hlyF, ireA, iroN, iss, lpfA, and traT), K. pneumoniae (entB, alls, mrkD, fimH, ybtS, and irp1), P. aeruginosa (exoU, exoS, lasB, toxA, and phzM), and A. hydrophila (hlyA, aer, lip, ast, and act) was performed in conventional PCR assays using the oligonucleotide primer sequences presented in Supplementary Table S1 .
Disinfectant Susceptibility Testing
Gram-negative bacterial strains (three of each species exhibiting colistin resistance) were evaluated against four commonly used biocides in dairy farms for teat disinfection, namely chlorhexidine gluconate (0.5% in alcohol 70%; Pfizer, Manhattan, NY, USA), hydrogen peroxide (H2O2; 6%, w/v), iodine (0.5%, w/v), and alcohol (ethanol; 70% v/v) (El Nasr Pharmaceutical Chemicals Co., Cairo, Egypt). The disinfectants were diluted (10–90%) using sterile distilled water, in which the original concentrations were considered stock solutions. The agar well diffusion method was applied to evaluate their effectiveness against the suspected strains of equivalent turbidity to that of 0.5 McFarland standards. Diameters of inhibition zones from the well edge to the inhibition front equal to or larger than 15 mm were regarded as susceptible (Wanja et al., 2020). The MICs of teat disinfectants against bacterial strains were determined as described before (Baron and Feingold, 1990).
Bioinformatics and Data Analysis
Statistical analyses were performed using the software SAS (SAS Institute Inc., 2012). Results of conjugation efficiency of Gram-negative bacteria were graphed by a boxplot through GraphPad Software (version 8.0.1, GraphPad Software Inc., La Jolla, CA, USA). Resistance phenotypes and frequencies of virulence genes among isolates were compared using the Chi-squared (χ 2) test. P < 0.05 was considered significant. A heatmap with hierarchical clustering was generated to visualize the overall distribution of mcr variants and antimicrobial resistance phenotypes in colistin-resistant isolates using the “pheatmap” package in R software (version 3.4.2) (Kolde, 2019). To determine the shared mcr variants among colistin-resistant isolates from different sources, a Venn diagram was generated using the Venny 1.0 tool, https://bioinfogp.cnb.csic.es/tools/venny/index.html. Spearman correlations were used to provide an estimate for the association among various variables (resistance phenotypes, colistin resistance, and virulence genes). The correlation coefficients and their p-values were visualized using a correlation plot. The correlation analyses and visualization were done using R packages rcorr (https://hbiostat.org/R/Hmisc/) and corrplot (https://github.com/taiyun/corrplot). Genes or phenotypes that were present or absent in all analyzed subjects were not considered in these analyses.
Results
Resistance Phenotypes of Gram-Negative Bacteria Isolated From Mastitis and Raw Milk
Overall, the MDR phenotype was determined for 70 Enterobacteriaceae isolates including 37/42 (88.1%) E. coli (27/30 were non-O157 and 10/12 were O157 E. coli), 18/18 K. pneumoniae (100%), 10/10 Enterobacter species (100%), and 5/20 Citrobacter species (25%). All P. aeruginosa and A. hydrophila isolates were MDR with MAR indices ranged from 0.33 to 0.79. Three P. aeruginosa and one E. coli (O111:H4) exhibited an XDR phenotype as being resistant to at least one antibiotic of all tested antimicrobial classes but remained susceptible to two classes (carbapenems and monobactams in P. aeruginosa, carbapenems and glycylcyclines in E. coli).
E. coli isolates showed high resistance to ampicillin, cephalothin, cefazolin, ceftazidime (100% each), cefoxitin (97.62%), cefotaxime (95.23%), and amoxicillin-clavulanic acid (90.47%). In addition, over 50% of isolates were resistant to piperacillin-tazobactam (71.43%), colistin, and fosfomycin (57.14%, each).
All K. pneumoniae isolates were resistant to ampicillin, amoxicillin-clavulanic acid, fosfomycin, cefoxitin, cefazolin, and cephalothin. Meanwhile, 94.4% of the isolates were resistant to both ceftazidime and piperacillin-tazobactam, 88.89% were resistant to tigecycline, 77.78% to ceftriaxone, cefotaxime, and colistin, and 55.56% were resistant to sulfamethoxazole-trimethoprim.
A. hydrophila isolates were sensitive to amikacin, imipenem, ciprofloxacin, and levofloxacin (100% each). However, the highest resistance was for the tetracycline and cephalosporins groups (ceftriaxone, cefotaxime, ceftazidime, cefoxitin, and cephalothin (100% each)) followed by colistin (88.24%), nalidixic acid (76.47%), sulfamethoxazole-trimethoprim (70.59%), and cefazolin (58.8%).
P. aeruginosa isolates exhibited resistance to sulfamethoxazole-trimethoprim, amoxicillin-clavulanic acid, fosfomycin, cefoxitin, cefazolin, and cephalothin (100% each); tetracycline, chloramphenicol, and ampicillin (90% each); colistin and nalidixic acid (80% each); and ceftazidime, ciprofloxacin, and levofloxacin (60% each). However, the most active antimicrobial agents against P. aeruginosa isolates were imipenem (100%), aztreonam (90%), piperacillin-tazobactam, and ceftriaxone (80% each).
Cephalosporins (cefotaxime, ceftazidime, cefoxitin, cefazolin, and cephalothin), ampicillin, and amoxicillin-clavulanic acid were the least active antimicrobials against both Enterobacter and Citrobacter species as well as cefepime, sulfamethoxazole-trimethoprim, and tetracycline in Enterobacter species (100% resistance rate). Citrobacter species were completely sensitive to both sulfamethoxazole-trimethoprim and tetracycline. Meanwhile, both species were highly sensitive to imipenem, gentamicin, tobramycin, amikacin, ciprofloxacin, levofloxacin, colistin, piperacillin-tazobactam, and tigecycline (100% each).
Trends in Bacterial Resistance and MAR Indices
The overall resistance rates of Gram-negative isolates from clinical mastitis, subclinical mastitis, and raw milk to each tested antimicrobial were analyzed using χ 2 test. As revealed in Table 1 , a significant (p < 0.05) difference in resistance rates of isolates from clinical, subclinical mastitis, and raw milk was observed against ampicillin, cefazolin, ceftriaxone, nalidixic acid, tobramycin, fosfomycin, colistin, and tetracycline. All isolates from different sources were highly resistant to the first and second generations of cephalosporins especially cephalothin (100%). Furthermore, the MAR index of each antimicrobial was calculated to understand the level of antibiotic use. High MAR index (0.04) of ampicillin, amoxicillin-clavulanic acid, and the first and second generations of cephalosporins was observed. This implies the excessive use of these antimicrobials in the veterinary sector and human medicine. Meanwhile, imipenem achieved a lower MAR index.
Table 1.
Resistance rates of 117 Gram-negative bacteria from milk samples against the tested antimicrobial agents and the overall multiple antibiotic resistance (MAR) index.
| AMA | Clinical mastitis | Subclinical mastitis | Raw milk | MAR index a | p-value* | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli (n = 25) | K. pneumoniae (n = 8) | Enterobacter species (n = 10) | Citrobacter species (n = 20) | P. aeuriginosa (n = 7) | E. coli (n = 5) | K. pneumoniae (n = 3) | P. aeuriginosa (n = 3) | E. coli (n = 12) | K. pneumoniae (n = 7) | A. hydrophila (n = 17) | |||
| AM | 25 (100.00) | 8 (100.00) | 10 (100.00) | 20 (100.00) | 6 (85.71) | 5 (100.00) | 3 (100.00) | 3 (100.00) | 12 (100.00) | 7 (100.00) | 9 (52.94) | 0.04 | 0.0004 |
| AMC | 25 (100.00) | 8 (100.00) | 10(100.00) | 20 (100.00) | 7 (100.00) | 5 (100.00) | 3 (100.00) | 3 (100.00) | 8 (66.67) | 7 (100.00) | 8 (47.06) | 0.04 | 0.0000007 |
| CEF | 25 (100.00) | 8 (100.00) | 10 (100.00) | 20 (100.00) | 7 (100.00) | 5 (100.00) | 3 (100.00) | 3 (100.00) | 12 (100.00) | 7 (100.00) | 17 (100.00) | 0.04 | NA |
| CFZ | 25 (100.00) | 8 (100.00) | 10 (100.00) | 20 (100.00) | 7 (100.00) | 5 (100.00) | 3 (100.00) | 3 (100.00) | 12 (100.00) | 7 (100.00) | 10 (58.82) | 0.04 | 0.0002 |
| FOX | 24 (96.00) | 8 (100.00) | 10 (100.00) | 20 (100.00) | 7 (100.00) | 5 (100.00) | 3 (100.00) | 3 (100.00) | 12 (100.00) | 7 (100.00) | 17 (100.00) | 0.04 | 0.7128 |
| CAZ | 25 (100.00) | 8 (100.00) | 10 (100.00) | 20 (100.00) | 4 (57.14) | 5 (100.00) | 3 (100.00) | 2 (66.67) | 12 (100.00) | 6 (85.71) | 17 (100.00) | 0.04 | 0.6633 |
| CTX | 23 (92.00) | 5 (62.50) | 10 (100.00) | 20 (100.00) | 1 (14.28) | 5 (100.00) | 3 (100.00) | 1 (33.33) | 12 (100.00) | 6 (85.71) | 17 (100.00) | 0.04 | 0.1211 |
| CRO | 6 (24.00) | 6 (75.00) | 5 (50.00) | 15 (75.00) | 2 (28.57) | 0 (00.00) | 3 (100.00) | 0 (00.00) | 6 (50.00) | 6 (85.71) | 17 (100.00) | 0.02 | 0.00088 |
| FEB | 9 (36.00) | 2 (25.00) | 10 (100.00) | 0 (00.00) | 1 (14.28) | 2 (40.00) | 1 (33.33) | 1 (33.33) | 5 (41.67) | 4 (57.14) | 4 (23.53) | 0.01 | 0.8673 |
| IPM | 0 (00.00) | 1(12.50) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 0.00 | 0.7128 |
| NA | 5 (20.00) | 0 (00.00) | 0 (00.00) | 5 (25.00) | 5 (71.43) | 1 (20.00) | 0 (00.00) | 3 (100.00) | 5 (41.67) | 1 (14.28) | 13 (76.47) | 0.01 | 0.00466 |
| CIP | 4 (16.00) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 4 (57.14) | 0 (00.00) | 0 (00.00) | 2 (66.67) | 4 (33.33) | 2 (28.57) | 0 (00.00) | 0.006 | 0.7614 |
| LVX | 4 (16.00) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 4 (57.14) | 0 (00.00) | 0 (00.00) | 2 (66.67) | 4 (33.33) | 0 (00.00) | 0 (00.00) | 0.005 | 0.799 |
| TOB | 1 (4.00) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 3 (42.86) | 2 (40.00) | 2 (66.67) | 1 (33.33) | 0 (00.00) | 0 (00.00) | 4 (23.53) | 0.005 | 0.0005 |
| AK | 0 (00.00) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 3 (42.86) | 0 (00.00) | 0 (00.00) | 1 (33.33) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 0.001 | 0.2763 |
| CN | 3 (12.00) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 3 (42.86) | 0 (00.00) | 0 (00.00) | 1 (33.33) | 3 (25.00) | 0 (00.00) | 4 (23.53) | 0.005 | 0.2512 |
| SXT | 10 (40.00) | 6 (75.00) | 10 (100.00) | 0 (00.00) | 7 (100.00) | 0 (00.00) | 1 (33.33) | 3 (100.00) | 4 (33.33) | 3 (42.86) | 12 (70.59) | 0.02 | 0.623 |
| TGC | 11 (44.00) | 8 (100.00) | 0 (00.00) | 0 (00.00) | 3 (42.86) | 1 (20.00) | 3 (100.00) | 1 (33.33) | 2 (16.67) | 5 (71.43) | 6 (35.29) | 0.01 | 0.632 |
| ATM | 5 (20.00) | 1 (12.50) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 8 (47.06) | 0.005 | 0.068 |
| TPZ | 21(84.00) | 8 (100.00) | 0 (00.00) | 0 (00.00) | 0 (00.00) | 3 (60.00) | 3 (100.00) | 0 (00.00) | 5 (41.67) | 6 (85.71) | 1 (5.88) | 0.02 | 0.429 |
| CHL | 5 (20.00) | 2 (25.00) | 0 (00.00) | 0 (00.00) | 6 (85.71) | 0 (00.00) | 0 (00.00) | 3 (100.00) | 5 (41.67) | 1 (14.28) | 7 (41.17) | 0.01 | 0.1378 |
| FOF | 15 (60.00) | 8 (100.00) | 0 (00.00) | 0 (00.00) | 7 (100.00) | 5 (100.00) | 3 (100.00) | 3 (100.00) | 3 (25.00) | 7 (100.00) | 2 (11.76) | 0.02 | 0.0001 |
| CST | 15 (60.00) | 4 (50.00) | 0 (00.00) | 0 (00.00) | 5 (71.43) | 4 (80.00) | 3 (100.00) | 3 (100.00) | 5 (41.67) | 7 (100.00) | 15 (88.23) | 0.02 | 0.00001 |
| TET | 9 (36.00) | 4 (50.00) | 10 (100.00) | 0 (00.00) | 6 (85.71) | 2 (40.00) | 1 (33.33) | 3 (100.00) | 6 (50.00) | 2 (28.57) | 17 (100.00) | 0.02 | 0.0232 |
AMA, antimicrobial agent; AM, ampicillin; AMC, amoxicillin-clavulanic acid; CEF, cephalothin; CFZ, cefazolin; FOX, cefoxitin; CAZ, ceftazidime; CTX, cefotaxime; CRO, ceftriaxone; FEB, cefepime; IPM, imipenem; NA, nalidixic acid; CIP, ciprofloxacin; LVX, levofloxacin; TOB, tobramycin; AK, amikacin; CN, gentamicin; SXT, sulfamethoxazole-trimethoprim; TGC, tigecycline; ATM, aztreonam; TPZ, piperacillin-tazobactam; CHL, chloramphenicol; FOF, fosfomycin; CST, colistin; TET, tetracycline; MAR, multiple antibiotic resistance; NA, not applicable.
MAR index for each antimicrobial = total number of resistance scored/total number of antimicrobials tested × total number of isolates (Tambekar et al., 2006).
*Trends in bacterial resistance to the tested antimicrobials using χ2 test; p < 0.05 was considered significant.
Frequency of Colistin Resistance mcr Genes in MDR and XDR Gram-Negative Bacteria
In total, 61/117 tested isolates (52.14%), 24/70 (34.29%) from clinical mastitis, 10/11 (90.91%) from subclinical mastitis, and 27/36 (75%) from raw milk were colistin resistant, showing colistin MICs >2 mg/L. Among them, 19 (27.14%), 8 (72.73%), and 20 (55.56%) isolates from clinical, subclinical mastitis, and raw milk, respectively, were positive for plasmid-borne mcr genes.
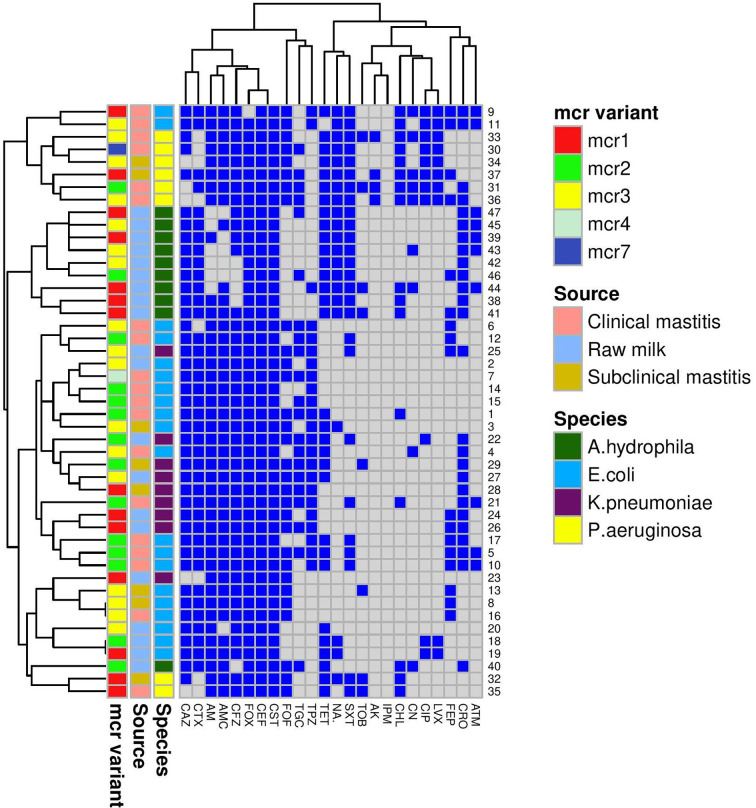
From the PCR and DNA sequencing results, the mcr-1 gene was identified in 31.91% (13 MDR and two XDR isolates), mcr-2 in 29.79% (13 MDR and one XDR), mcr-3 in 34.04% (15 MDR and one XDR), and each of mcr-4 and mcr-7 in 2.13% of the colistin-resistant Gram-negative isolates ( Table 2 ). Of these isolates, 42.55% (20/47) were E. coli, 21.28% (10/47) A. hydrophila, 19.12% (9/47) K. pneumoniae, and 17.02% (8/47) P. aeruginosa ( Figure 1 ).
Table 2.
Resistance phenotypes, virulence genes profiles, and plasmid-mediated mcr genes of 47 Gram-negative bacteria recovered from milk samples.
| Isolate No. | Species | Source | Antimicrobial resistance patterns | MAR index | MIC (mg/L) | mcr gene | Accession No. | Virulence gene |
|---|---|---|---|---|---|---|---|---|
| 1 | E. coli O111:H4 | Clinical mastitis | CFZ, FOX, AM, AMC, TPZ, CEF, CAZ, FOF, CTX, TET,CHL, TGC, CST | 0.54 | 128 | mcr-2 | MW811407 | hlyF, ireA, iss, lpfA |
| 2 | O26:H11 | Raw milk | CFZ, FOX, AM, AMC, TPZ, CEF, CAZ, FOF, CTX, CST | 0.42 | 8 | mcr-3 | MW811419 | hlyF, ireA, iroN, lpfA, traT |
| 3 | O111:H4 | Subclinical mastitis | CFZ, FOX, AM, AMC, TET, TPZ, CEF, CAZ, CST, FOF, CTX, NA | 0.5 | 32 | mcr-3 | MW811420 | hlyF, iroN, lpfA |
| 4 | O111:H4 | Clinical mastitis | CFZ, FOX, AM, AMC, TET, TPZ, CEF, CAZ, CTX, TGC, CST, CN, CRO | 0.54 | 64 | mcr-3 | MW811421 | ireA, iroN, iss, lpfA |
| 5 | O114:H21 | Clinical mastitis | CFZ, CRO, FOX, SXT, AM, AMC, TPZ, CEF, CAZ, TGC, CST, TET, ATM, FOF, FEP, CTX | 0.67 | 128 | mcr-2 | MW811408 | hlyF, ireA, iroN, lpfA, traT |
| 6 | O114:H21 | Clinical mastitis | CFZ, FEP, FOX, AM, AMC, TPZ, CEF, CAZ, FOF, TGC, CST | 0.46 | 64 | mcr-3 | MW811422 | hlyF, iroN, iss, lpfA, traT |
| 7 | O111:H4 | Clinical mastitis | CFZ, FOX, AM, AMC, TPZ, CEF, CAZ, CTX, TGC, CST, FOF | 0.46 | 16 | mcr-4 | MW811433 | hlyF, iroN, iss, lpfA, traT |
| 8 | O114:H4 | Subclinical mastitis | CFZ, FOX, AM, AMC, CEF, CAZ, CTX, FEP, FOF, CST | 0.42 | 16 | mcr-3 | MW811427 | hlyF, iroN, lpfA |
| 9 | O111:H4 | Clinical mastitis | CN, CFZ, TPZ, CEF, CRO, CAZ, CTX, FEP, NA, CIP, LVX, SXT, ATM, AM, AMC, CHL, TE, FOF, CST | 0.79 | 16 | mcr-1 | MW811398 | hlyF, ireA, iroN, iss, lpfA, traT |
| 10 | O114:H21 | Clinical mastitis | CFZ, TPZ, CEF, CAZ, CTX, CRO, FEP, FOX, SXT, AM, ATM, AMC, TET, CST, FOF | 0.63 | 16 | mcr-2 | MW811409 | hlyF, ireA, iroN, lpfA, traT |
| 11 | O111:H4 | Clinical mastitis | CFZ, CAZ, CEF, CRO, FEP, FOX, CTX, NA, CIP, LVX, SXT, ATM, AM, TPZ, AMC, CHL, CST, FOF | 0.75 | 64 | mcr-3 | MW811423 | hlyF, ireA, iroN, iss, lpfA |
| 12 | O26:H11 | Clinical mastitis | CFZ, CEF, FEP, FOX, CAZ, CTX, SXT, TGC, AM, TPZ, AMC, CST | 0.5 | 8 | mcr-2 | MW811410 | hlyF, iroN, iss, lpfA, traT |
| 13 | O111:H4 | Subclinical mastitis | CFZ, CEF, TOB, FOX, CAZ, CTX, FEP, AM, AMC, FOF, CST | 0.46 | 128 | mcr-3 | MW811424 | ireA, iroN, iss, lpfA, traT |
| 14 | O26:H11 | Clinical mastitis | CFZ, CEF, FOX, CAZ, CTX, AM, TPZ, AMC, CST | 0.38 | 16 | mcr-2 | MW811411 | hlyF, ireA, iroN, iss, lpfA, traT |
| 15 | O146:H- | Clinical mastitis | CFZ, CEF, FOX, CAZ, CTX, TGC, TPZ, AM, AMC, CST | 0.42 | 16 | mcr-2 | MW811412 | hlyF, iroN, iss, lpfA |
| 16 | O146:H- | Clinical mastitis | CFZ, CEF, FOX, CAZ, CTX, FEP, ATM, AMC, FOF, CST | 0.42 | 32 | mcr-3 | MW811425 | hlyF, ireA, iss, lpfA, traT |
| 17 | O157:H7 | Clinical mastitis | CFZ, CEF, FOX, CAZ, CTX, CRO, FEP, SXT, TPZ, AM, AMC, CST, TET | 0.54 | >128 | mcr-2 | MW811413 | hlyF, ireA, iss, lpfA, traT |
| 18 | O157:H7 | Raw milk | CFZ, CEF, FOX, CAZ, CTX, NA, CIP, LVX, AM, AMC, CST, TET | 0.5 | 64 | mcr-2 | MW811414 | ireA, iroN, iss, lpfA, traT |
| 19 | O157:H7 | Raw milk | CFZ, CEF, FOX, CAZ, CTX, NA, CIP, LVX, AM, AMC, CST, TET | 0.5 | >128 | mcr-1 | MW811399 | hlyF, iroN, iss, lpfA, traT |
| 20 | O157:H7 | Raw milk | CFZ, CEF, FOX, CAZ, CTX, AM, CST, TET | 0.33 | 128 | mcr-3 | MW811426 | hlyF, ireA, iroN, iss, lpfA |
| 21 | K. pneumoniae | Raw milk | CFZ, CEF, FOX, CAZ, CTX, CRO, SXT, TGC, ATM, TPZ, AM, AMC, CHL, FOF, CST | 0.63 | 8 | mcr-2 | MW811415 | entB, alls, mrkD, fimH, ybtS |
| 22 | K. pneumoniae | Raw milk | CFZ, CEF, FOX, CAZ, CTX, CRO, SXT, TGC, CIP, TPZ, AM, AMC, TET, FOF, CST | 0.63 | 128 | mcr-2 | MW811416 | entB, mrkD, fimH, ybtS, irp1 |
| 23 | K. pneumoniae | Raw milk | CFZ, CEF, FOX, ATM, AMC, FOF, CST | 0.29 | 4 | mcr-1 | MW811400 | entB, mrkD, ybtS, irp1 |
| 24 | K. pneumoniae | Raw milk | CFZ, CEF, FOX, CAZ, CTX, CRO, FEP, TPZ, AM, AMC, FOF, CST | 0.5 | 128 | mcr-1 | MW811401 | entB, alls, mrkD, fimH, irp1 |
| 25 | K. pneumoniae | Clinical mastitis | CFZ, CEF, FOX, CAZ, CTX, CRO, FEP, SXT, TGC, TPZ, AM, AMC, FOF, CST | 0.58 | 64 | mcr-3 | MW811428 | entB, alls, mrkD, fimH, irp1 |
| 26 | K. pneumoniae | Raw milk | CFZ, CEF, FOX, CAZ, CTX, CRO, FEP, TGC, TPZ, AM, AMC, FOF, CST | 0.54 | 64 | mcr-1 | MW811402 | entB, mrkD, fimH, ybtS, irp1 |
| 27 | K. pneumoniae | Raw milk | CFZ, CEF, FOX, CAZ, CTX, CRO, TGC, TPZ, AM, AMC, TET, FOF, CST | 0.54 | 8 | mcr-3 | MW811429 | entB, alls, mrkD, fimH, ybtS |
| 28 | K. pneumoniae | Subclinical mastitis | CFZ, CEF, FOX, CAZ, CTX, CRO, TGC, TPZ, AM, AMC, FOF, CST | 0.5 | 16 | mcr-1 | MW811403 | entB, fimH, ybtS, irp1 |
| 29 | K. pneumoniae | Subclinical mastitis | CFZ, CEF, FOX, CAZ, CTX, CRO, TOB, TGC, TPZ, AM, AMC, TET, FOF, CST | 0.58 | 32 | mcr-2 | MW811417 | entB, mrkD, fimH, ybtS, irp1 |
| 30 | P. aeuriginosa | Clinical mastitis | CEF, FOX, CAZ, CFZ, NA, CIP, LVX, SXT, TGC, AM, AMC, CHL, FOF, CST, TET | 0.63 | >128 | mcr-7 | MW811434 | toxA, exoS, lasB, exoU |
| 31 | P. aeuriginosa | Clinical mastitis | AK, CN, TOB, CEF, CFZ, FOX, CTX, CRO, NA, CIP, LVX, SXT, AM, AMC, CHL, FOF, TGC, CST, TET | 0.79 | 128 | mcr-2 | MW811418 | toxA, exoS, lasB, exoU, phzM |
| 32 | P. aeuriginosa | Subclinical mastitis | TOB, CEF, FOX, CAZ, CFZ, NA, AM, AMC, SXT, CHL, FOF, TET, CST | 0.54 | 32 | mcr-1 | MW811404 | toxA, exoS, lasB |
| 33 | P. aeuriginosa | Clinical mastitis | AK, CN, TOB, CEF, FOX, CAZ, CFZ, SXT, NA, CIP, LVX, AM, AMC, CHL, FOF, TET, CST | 0.71 | 64 | mcr-3 | MW811430 | toxA, exoS, exoU |
| 34 | P. aeuriginosa | Subclinical mastitis | CEF, FOX, CFZ, NA, CIP, LVX, SXT, AM, AMC, CHL, FOF, TET, CST | 0.54 | 64 | mcr-3 | MW811431 | toxA, exoS, lasB |
| 35 | P. aeuriginosa | Clinical mastitis | TOB, CEF, FOX, CFZ, SXT, AM, AMC, CHL, FOF, TET, CST | 0.45 | 32 | mcr-1 | MW811405 | toxA,lasB, exoU |
| 36 | P. aeuriginosa | Clinical mastitis | AK, CN, CEF, FOX, CFZ, CRO, FEP, NA, CIP, LVX, SXT, TGC, AM, AMC, CHL, FOF, TET, CST | 0.75 | 16 | mcr-3 | MW811432 | toxA, exoS, lasB, exoU, phzM |
| 37 | P. aeuriginosa | Subclinical mastitis | AK, CN, CEF, FOX, CFZ, CAZ, CTX, FEP, NA, CIP, LVX, SXT, TGC, AM, AMC, CHL, FOF, TET, CST | 0.79 | >128 | mcr-1 | MW811406 | toxA, exoS, lasB, exoU |
| 38 | A. hydrophila | Raw milk | CAZ, CTX, CEF, CRO, FOX, NA, SXT, AM, AMC, CHL, CST, TET | 0.5 | 128 | mcr-1 | MZ648218 | aer |
| 39 | A. hydrophila | Raw milk | CFZ, CEF, FOX, CAZ, CTX, CRO, NA, SXT, ATM, AM, CST, TET | 0.5 | 16 | mcr-1 | MZ648219 | act |
| 40 | A. hydrophila | Raw milk | TOB, CEF, FOX, CAZ, CTX, CRO, TGC, AM, AMC, FOF, CHL, CST, TET | 0.54 | 32 | mcr-2 | MZ648224 | aer, lip |
| 41 | A. hydrophila | Raw milk | TOB, CEF, CFZ, FOX, CAZ, CTX, CRO, FEP, NA, SXT, AM, AMC, CHL, CST, TET | 0.63 | 64 | mcr-1 | MZ648220 | aer, lip |
| 42 | A. hydrophila | Raw milk | CEF, FOX, CAZ, CTX, CRO, NA, SXT, CST, TET | 0.38 | 64 | mcr-3 | MZ648226 | lip |
| 43 | A. hydrophila | Raw milk | CN, CEF, FOX, CAZ, CFZ, CTX, CRO, NA, SXT, ATM, CST, TET | 0.5 | 128 | mcr-3 | MZ648227 | aer |
| 44 | A. hydrophila | Raw milk | CN, TOB, CEF, FOX, CAZ, CTX, CRO, NA, SXT, ATM, TPZ, AMC, CHL, CST, TET | 0.63 | 128 | mcr-1 | MZ648221 | hlyA, aer |
| 45 | A. hydrophila | Raw milk | CEF, FOX, CAZ, CFZ, CTX, CRO, NA, SXT, ATM, AMC, CST, TET | 0.5 | 64 | mcr-1 | MZ648222 | act |
| 46 | A. hydrophila | Raw milk | CEF, FOX, CAZ, CTX, CRO, FEP, NA, SXT, TGC, CST, TET | 0.46 | 32 | mcr-2 | MZ648225 | aer |
| 47 | A. hydrophila | Raw milk | CEF, FOX, CAZ, CFZ, CTX, CRO, NA, SXT, TGC, ATM, CST, TET | 0.5 | >128 | mcr-1 | MZ648223 | aer, ast |
AM, ampicillin; AMC, amoxicillin-clavulanic acid; TPZ, piperacillin-tazobactam; CFZ, cefazolin; CEF, cephalothin; FOX, cefoxitin; CAZ, ceftazidime; CTX, cefotaxime; CRO, ceftriaxone; FEB, cefepime; IPM, imipenem; CN, gentamicin; TOB, tobramycin; AK, amikacin; NA, nalidixic acid; CIP, ciprofloxacin; LVX, levofloxacin; CST, colistin; TET, tetracycline; TGC, tigecycline; CHL, chloramphenicol; SXT, sulfamethoxazole-trimethoprim; ATM, aztreonam; FOF, fosfomycin. All isolates are multidrug resistant (MDR) except the bold ones are extensive drug resistant (XDR).
Figure 1.
A heatmap supported by a dendrogram for colistin-resistant Gram-negative isolates (n = 47) from bovine mastitis and raw milk showing their antimicrobial resistance phenotypes and the distribution of mcr genes. Mcr variant, source, and species are color-coded feature categories. AM, ampicillin; AMC, amoxicillin-clavulanic acid; TPZ, piperacillin-tazobactam; CFZ, cefazolin; CEF, cephalothin; FOX, cefoxitin; CAZ, ceftazidime; CTX, cefotaxime; CRO, ceftriaxone; FEB, cefepime; IPM, imipenem; CN, gentamicin; TOB, tobramycin; AK, amikacin; NA, nalidixic acid; CIP, ciprofloxacin; LVX, levofloxacin; CST, colistin; TET, tetracycline; TGC, tigecycline; CHL, chloramphenicol; SXT, sulfamethoxazole-trimethoprim; ATM, aztreonam; FOF, fosfomycin.
A Venn diagram ( Supplementary Figure S1 ) was generated to determine the shared mcr variants between colistin-resistant isolates from clinical mastitis, subclinical mastitis, and raw milk. E. coli mcr-4 and both mcr-7 and mcr-2 of P. aeruginosa were unique for clinical mastitis; whereas K. pneumonia mcr-3 and A. hydrophila mcr-1 to mcr-3 variants were exclusive for raw milk. Only E. coli mcr-3 and K. pneumonia mcr-2 were common among all sources.
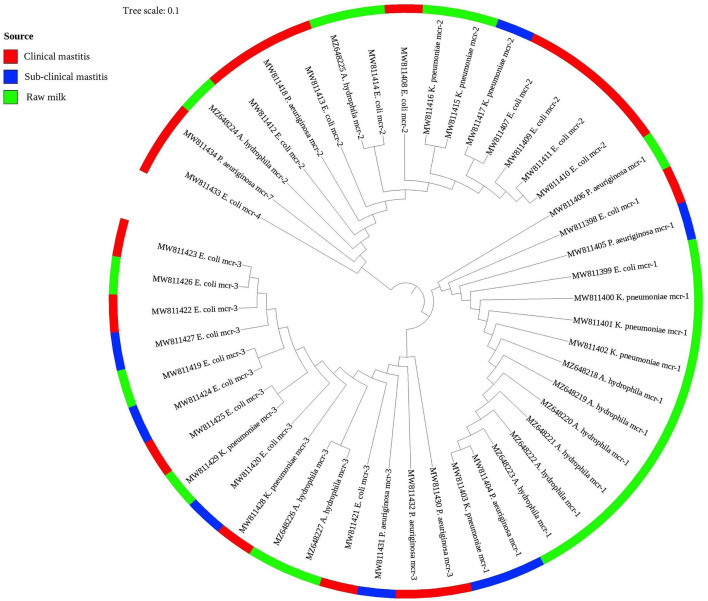
Phylogeny of Colistin Resistance mcr Variants
As presented in Figure 2 , the phylogenetic tree was constructed based on nucleotide sequences for all identified colistin resistance mcr variants (mcr-1 to mcr-4 and mcr-7) in 47 Gram-negative bacteria isolates of various species. The phylogenetic analysis revealed five potential clusters indicating sequence heterogeneity among the mcr variants. Despite the overall diversity of bacterial isolates, which were originating from different on-farm locations/sources, the phylogenetic tree demonstrated the presence of the same clonal lineages of each mcr variant separately. Nonetheless, two mcr variants (mcr-4, accession number MW811433, and mcr-7, accession number MW811434) were clonally related; whereas the latter shared the same lineage with another mcr-2 variant (accession number MW811412).
Figure 2.
A phylogenetic tree shows the genetic relationship between mcr gene sequences of colistin-resistant Gram-negative bacteria recovered from clinical, subclinical mastitis, and raw milk. The tree was constructed using a neighbor-joining method with a bootstrap value for 1,000 replicates.
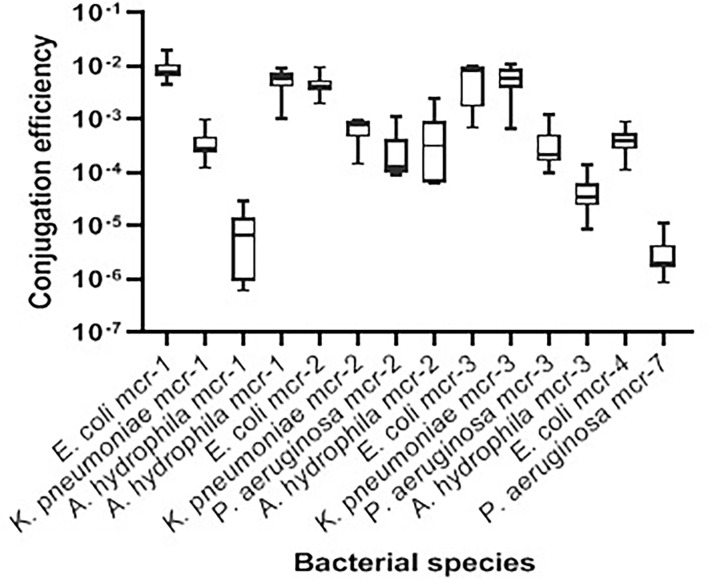
Transferability of mcr Genes
Transconjugation assays revealed that representative Gram-negative isolates harboring mcr-1, mcr-2, mcr-3, mcr-4, and mcr-7 were capable of transferring their genes to the E. coli J53-recipient strain. The conjugation efficiencies of these isolates ranged from 2.01 ± 0.816 × 10−6 to 9.50 ± 2.663 × 10−3 CFU/donor ( Table 3 and Figure 3 ). As expected, the recipient J53 strain did not grow on colistin and Na azide-incorporated media. The colistin MICs for transconjugants ranged from 4 to 64 µg/ml, demonstrating an increase of three to sevenfold relative to E. coli J53-recipient strain (MIC = 0.5 µg/ml) ( Table 3 ).
Table 3.
Conjugation efficiencies of some mcr-1-, mcr-2-, mcr-3-, mcr-4-, and mcr-7-positive isolates and colistin MIC values for transconjugants.
| Bacterial species | Code No. | mcr gene | Transferability | Transconjugant MIC (µg/ml)* | CFU/ml | |||
|---|---|---|---|---|---|---|---|---|
| Recipient (E. coli J53) | Donor | Transconjugant | Conjugation efficiency | |||||
| E. coli | 9 | mcr-1 | + | 16 | 2.62 × 108 | (3.14 ± 0.548) × 105 | (2.31 ± 0.142) × 103 | (7.36 ± 2.101) × 10−3 |
| K. pneumoniae | 26 | mcr-1 | + | 32 | (4.63 ± 0.856) × 106 | (1.36 ± 0.211) × 103 | (2.94 ± 0.621) × 10−4 | |
| P. aeruginosa | 37 | mcr-1 | + | 64 | (7.12 ± 0.891) × 107 | (5.70 ± 1.224) × 102 | (8.01 ± 2.663) × 10−6 | |
| A. hydrophila | 41 | mcr-1 | + | 64 | (5.53 ± 1.024) × 104 | (3.24 ± 0.882) × 102 | (5.86 ± 1.174) × 10−3 | |
| E. coli | 10 | mcr-2 | + | 16 | (1.69 ± 0.442) × 105 | (7.01 ± 1.663) × 102 | (4.15 ± 1.111) × 10−3 | |
| K. pneumoniae | 21 | mcr-2 | + | 8 | (5.46 ± 1.105) × 105 | (4.08 ± 1.022) × 102 | (7.47 ± 2.337) × 10−4 | |
| P. aeruginosa | 31 | mcr-2 | + | 32 | (3.91 ± 0.462) × 106 | (4.11 ± 1.121) × 102 | (1.05 ± 0.184) × 10−4 | |
| A. hydrophila | 40 | mcr-2 | + | 16 | (7.23 ± 1.335) × 105 | (2.64 ± 0.451) × 102 | (3.65 ± 0.326) × 10−4 | |
| E. coli | 3 | mcr-3 | + | 8 | (3.61 ± 0.710) × 105 | (3.43 ± 0.352) × 103 | (9.50 ± 2.663) × 10−3 | |
| K. pneumoniae | 27 | mcr-3 | + | 4 | (6.40 ± 0.633) × 106 | (4.26 ± 0.222) × 104 | (6.66 ± 1.812) × 10−3 | |
| P. aeruginosa | 34 | mcr-3 | + | 32 | (3.12 ± 0.223) × 107 | (6.47 ± 1.362) × 103 | (2.07 ± 0.634) × 10−4 | |
| A. hydrophila | 42 | mcr-3 | + | 32 | (6.46 ± 0.384) × 106 | (2.23 ± 0.331) × 102 | (3.45 ± 1.125) × 10−5 | |
| E. coli | 7 | mcr-4 | + | 16 | (4.15 ± 0.361) × 106 | (1.81 ± 0.114) × 103 | (4.36 ± 1.632) × 10−4 | |
| P. aeruginosa | 30 | mcr-7 | + | 64 | (5.46 ± 0.966) × 107 | (1.30 ± 0.314) × 102 | (2.01 ± 0.816) × 10−6 | |
*Colistin MIC value = 0.5 µg/ml.
Conjugation efficiency = number of transconjugants/number of donor cells.
CFU, colony-forming unit.
CFU values were retrieved from duplicate measurements from two independent experiments.
Figure 3.
Transconjugation frequencies of 14 mcr-harboring Gram-negative isolates from bovine milk. The data are presented as mean values ± standard deviations. Code numbers of isolates are shown in Table 2 .
Virulence Characteristics of Colistin-Resistant Gram-Negative Bacteria
The profiles of virulence-associated genes detected in 47 colistin-resistant isolates are depicted in Table 2 . Different combinations of virulence genes were observed among colistin-resistant isolates with almost all isolates harboring genes ranging from one to six. LpfA was the most frequently detected gene in the tested E. coli isolates (20/20; 100%) followed by hlyF (17/20; 85%), iroN (16/20; 80%), iss (15/20; 75%), ireA (13/20; 65%), and traT (12/20; 60%). The entB gene was detected in all nine colistin-resistant K. pneumoniae isolates examined; both mrkD and fimH were identified in 88.8%, ybtS and irp1 in 77.7%, and alls in 44.4% of K. pneumonia isolates. The frequencies of toxA, exoS, lasB, exoU, and phzM genes in eight colistin-resistant P. aeruginosa isolates were 100%, 87.5%, 87.5%, 75%, and 25%, respectively. The aer gene is the most prevalent (70%) in A. hydrophila isolates followed by lip (30%) and act (20%). Both ast and hlyA genes were detected in one A. hydrophila isolate (10%). There are significant differences in the frequencies of virulence genes among E. coli isolates (p = 0.04), A. hydrophila (p = 0.01), and P. aeruginosa (p = 0.005).
Correlations Between the Antimicrobial Resistance and Virulence Traits
The correlation plots among pairs of antimicrobial resistance phenotypes, colistin resistance genes (mcr), and virulence genes in E. coli, K. pneumoniae, P. aeruginosa, and A. hydrophila are displayed in Supplementary Figure S2 and Supplementary Tables S2A–H . Significant positive correlations were pronounced between fluoroquinolone resistance and the existence of mcr-1 gene in E. coli (r = 0.58–0.67; p < 0.001). On the other hand, a strong positive correlation was significantly detected between a fimH virulence gene and the resistance to certain antimicrobials; ceftazidime, cefotaxime, ceftriaxone, and piperacillin-tazobactam in K. pneumonia isolates (r = 1; p < 0.001). Correlations between virulence traits and antimicrobial resistance were obvious in P. aeruginosa and A. hydrophilus isolates. Concerning P. aeruginosa, non-significant positive correlations were found between the analyzed virulence genes and antimicrobial agents/genes including the last-resort ones. The correlations between exoU and phzM virulence genes and gentamicin (r = 0.58; p = 0.1), exoS and fluoroquinolones (r = 0.65; p = 0.07), exoU and tigecycline (r = 0.58; p = 0.1), phzM and mcr2 (r = 0.65; p = 0.07), and phzM and tigecycline (r = 0.58; p = 0.1) were mostly distinct. For A. hydrophila, the hlyA gene showed significant positive correlations with tobramycin (r = 0.67; p = 0.03) or piperacillin-tazobactam (r = 1; p < 0.001). Of interest, significant strong positive correlations were detected between the last resort antimicrobials; mcr-2 and tigecycline (r = 0.76; p = 0.01) and mcr-2 and fosfomycin (r = 0.67; p = 0.03).
Efficacy of Disinfectants on Colistin-Resistant Gram-Negative Bacteria
According to the agar well-diffusion results, certain disinfectants had a wide efficacy against the test bacteria. However, some disinfectants were ineffective even at high concentrations ( Supplementary Table S3 ). E. coli and K. pneumonia isolates were resistant to chlorohexdine gluconate at concentrations lower than 50% (MIC = 40%), whereas P. aeruginosa and A. hydrophila were sensitive to such disinfectant even at low concentrations (MIC = 10%). E. coli and K. pneumoniae were sensitive to iodine at high concentrations (MIC = 80%), while P. aeruginosa and A. hydrophila exhibited resistance at all tested concentrations. On the other hand, hydrogen peroxide had the best inhibitory effect against all tested bacterial isolates even at a low concentration (10%), except for E. coli, which was sensitive to H2O2 at a concentration of 50%. All tested bacteria were resistant to ethanol at all concentrations.
Discussion
The recent global dissemination of mcr genes has raised the alarm of colistin resistance as a serious and urgent threat to public health (WHO, 2019). Consequently, large numbers of studies were carried out to explore the distribution of mcr genes in different reservoirs. Herein, we studied colistin-resistant Gram-negative bacteria isolated from bovine milk concerning the mcr resistance determinants and virulence characteristics. Unfortunately, 52% of the tested isolates were colistin resistant. The high proportion of colistin-resistant bacteria and the diversity of mcr variants detected here indicates a lot of independent acquisitions of mcr genes by colistin-sensitive bacteria and confirm the hypothesis that animals and food chain may play a key role in mcr transmission (Luo et al., 2020). This is considering an alarming ratio since previous studies have detected colistin resistance in 37%, 40%, and 41% of bacterial isolates from pigs, poultry, and human clinical samples, respectively (Savin et al., 2020; Lay et al., 2021; Qadi et al., 2021). Of note, we identified these isolates in milk samples, which included various bacterial species. These findings could be attributed to the frequent usage of colistin in dairy farms that in turn could be responsible for the development of colistin resistance. This is consistent with a previous report of a Canadian dairy farm, which uses colistin to increase the milk yield although its usage is not officially licensed in veterinary medicine (Saini et al., 2012; Webb et al., 2017).
Colistin-resistant E. coli isolates exhibited resistance to cephalothin, cefazolin, ceftazidime, ampicillin, amoxicillin-clavulanic acid, cefoxitin, cefotaxime, piperacillin-tazobactam, and fosfomycin. Concurrently, nearly similar patterns were obtained recently for E. coli against several antibiotics in India, Croatia, and Egypt (Zdolec et al., 2016; Singh et al., 2018; Ramadan et al., 2021; Soliman et al., 2021). For K. pneumoniae isolates, the antibiotic resistance pattern was closely like the pattern of Brazilian MDR K. pneumoniae isolates (Ferreira et al., 2019). They showed resistance to cefazolin, cephalothin, cefoxitin, ceftazidime, cefotaxime, ceftriaxone, sulfamethoxazole-trimethoprim, and tigecycline. On the contrary, our colistin-resistant P. aeruginosa isolates showed resistance to most groups of antibiotics; however, previous colistin-resistant P. aeruginosa isolates from Iran showed only resistance to ticarcillin, ciprofloxacin, aztreonam, ceftazidime, gentamicin, imipenem, and piperacillin/tazobactam (Goli et al., 2016). Our A. hydrophila isolates have an antimicrobial resistance pattern much akin to the antibiotic resistance profiles of pathogenic aeromonads isolated from ornamental fish with resistance to gentamicin, cephalothin, cefoxitin, ceftazidime, cefotaxime, ceftriaxone, nalidixic acid, sulfamethoxazole-trimethoprim, aztreonam, and chloramphenicol (Saengsitthisak et al., 2020). Colistin-resistant isolates found in this study exhibited extreme emergence and spread of antimicrobial resistance and high virulence as well.
It is almost established that E. coli is deemed to be the top-most bacteria for the mcr genes by carrying mcr-1 up to mcr-5 with the chance for coexistence of more than one mcr gene (Yin et al., 2017; Hammerl et al., 2018). Similarly, colistin-resistant E. coli isolates in our study were found to carry mcr-1, mcr-2, mcr-3, or mcr-4 genes; 20% out of which were from raw milk and carried mcr-1, mcr-2, or mcr-3 genes. It is the first detection of mcr-carrying isolates in raw milk samples. A previous study has identified mcr-1 gene in one E. coli isolate from a cow suffering from subclinical mastitis in Egypt (Khalifa et al., 2016). Although the mcr-1 gene was detected frequently from different samples (Khalifa et al., 2016; Lima Barbieri et al., 2017; Ramadan et al., 2021), it is foremost found here in the raw milk samples. Marvelously, one E. coli isolate carrying mcr-1 gene obtained from hard cheese made from raw milk was reported in Egypt very lately (Ombarak et al., 2021), which in turn lightens the importance of raw milk quality control checks in Egypt, focusing on farm workers as well as the animal itself.
The prevalence of mcr-3 gene in our isolated E. coli was the dominant one, followed by the mcr-2 gene. Only one E. coli isolate was mcr-4 positive. However, in this study, the mcr-1, mcr-2, and mcr-3 genes were found in E. coli isolated from raw milk as well as clinical and subclinical mastitis samples; however, the only mcr-4-positive E. coli was isolated from a clinical mastitis sample. The variants mcr-2, mcr-3, and mcr-4 were identified formerly in E. coli isolates from Belgium, China, and Italy, respectively, and were isolated from calves and pigs (Xavier et al., 2016; Carattoli et al., 2017; Yin et al., 2017). All these data confirmed that E. coli is the chief bacteria carrying mcr genes from different samples and in different geographical areas.
Notably, all our colistin-resistant A. hydrophila were isolated from raw milk samples, and the predominant gene in them was mcr-1 followed by mcr-2 and mcr-3. In contrast, colistin-resistant A. hydrophila were found to harbor mcr-7 gene in cooked and raw meats and in environmental samples (Liu Y. et al., 2020). No previous reports until now showed the isolation of colistin-resistant A. hydrophila from raw milk samples. Our findings warrant that the source of raw milk contamination is most probably the farm workers rather than the animal source, particularly, that mcr-3 gene was previously identified in isolates from human rectal swabs (Shen et al., 2018).
Of the identified colistin-resistant isolates, 19.12% were K. pneumoniae and were carrying mcr-1, mcr-2, or mcr-3 genes. The three variants were identified in the isolates from raw milk samples, while mcr-1 was found in subclinical mastitis isolates and mcr-2 in both isolates from clinical and subclinical mastitis. It is almost agreed that the mcr-1 gene is one of the few and clear examples of animal origin of a resistance trait that could hit the entire human health system (Poirel and Nordmann, 2016). Appealingly, three mcr-1-positive K. pneumoniae isolates were isolated from raw milk samples ensuring the role of humans in contamination and the transmission from animals to humans.
On the other hand, no colistin-resistant P. aeruginosa were found in the raw milk samples. They were isolated only from clinical and subclinical mastitis and were carrying mcr-1, mcr-2, mcr-3, or mcr-7 genes. Although, we detected 37.5% and 12.5% of the colistin-resistant P. aeruginosa isolates carrying mcr-1 and mcr-2 genes; respectively, it was found lately in Egypt that colistin-resistant P. aeruginosa were negative for mcr-2 gene and 50% were positive for mcr-1 (Abd El-Baky et al., 2020). We also found that the mcr-3 variant was prevalent with 37.5% among our colistin-resistant P. aeruginosa isolates. Even, to our knowledge, there is no report detected on mcr-3 in P. aeruginosa isolates before. Moreover, this is the first-time reporting mcr-7 from Egyptian isolates, and it was identified in P. aeuriginosa from clinical mastitic milk. The first plasmid-mediated colistin resistance gene mcr-7 was detected in K. pneumoniae from chickens in China (Yang et al., 2018). Recently, the mcr-7 gene was detected in a water sample from an environment of an alligator (dos Santos et al., 2020). The detection of mcr-7 in our isolate from clinical mastitis confirms the dissemination of mcr gene and its variants at all levels; humans, animals, and environmental. This was confirmed here while application of the conjugation assay as the plasmid harboring mcr genes (mcr-1 to mcr-4, and mcr-7) successfully transferred colistin resistance to the recipient strain through a broth-mating assay, representing their capability to mobilize the mcr-genes between isolates (Migura-Garcia et al., 2020).
The virulome of our colistin-resistant isolates was screened in each species. Many of the identified virulence genes in our study were required for bacterial survival and had a role in causing mastitis (Tartor and El-Naenaeey, 2016; Gao et al., 2019; Guerra et al., 2020). This in turn raises the alarm that most of the colistin-resistant Gram-negative isolates are highly pathogenic. Here, the establishment of an interplay between antimicrobial resistance and virulence traits corresponded to each analyzed Gram-negative bacteria, revealing positive correlations between the existence of certain virulence genes and resistance to antimicrobial agents/genes. A previous study suggested that the mobile genetic elements of proficient pathogens favor the coselection of both resistance and virulence-associated traits (Hennequin and Robin, 2016).
The virulome, as well as the resistome patterns of our colistin-resistant isolates, confirmed the elevated pathogenicity and the widespread antibiotic resistance that correlated to these isolates. The phylogenetic analysis of the nucleotide sequences of our colistin resistance mcr variants showed three distinguishable clusters. Interestingly, each cluster has the same mcr variant, and this is in accordance with what was reported before about mcr-1, mcr-2, and mcr-3 (Zhang et al., 2018). The presence of each variant of the mcr gene in the same clonal lineage despite the different sources of isolation confirms the high spread of colistin resistance from one source to another. Each of the mcr-4 and mcr-7 variants was shown in a unique cluster. This was unsurprising since our study showed the first report of mcr-7 in P. aeruginosa. Similarly, mcr-4 was previously found in a unique phylogenetic group (Zhang et al., 2019). Our findings proved that each mcr gene has a distinct evolution and spread path producing a characteristic phenotypic colistin resistance.
All the above information has raised the alert for controlling the spread of colistin-resistant isolates. Thus, we decided to evaluate four commonly used biocides in the dairy farms against colistin-resistant isolates. In accordance with Lanjri et al. (2017), all isolates were sensitive to chlorhexidine with different concentrations. This is attributed to its cationic nature that helps in electrostatic interaction with the anionic group lipopolysaccharide in the cytoplasmic membrane of Gram-negative bacteria, resulting in modification of the membrane permeability and causing membrane disruption and cell death due to coagulation of the cytoplasm (Estrela et al., 2003). Another tested biocide was iodine; it showed an inhibitory effect on E. coli and K. pneumoniae at high concentrations and no inhibition effect on P. aeuriginosa and A. hydrophila at all tested concentrations. Iodine is proven to be considered a broad-spectrum, rapidly acting bactericidal, and effective against all mastitis-causing bacteria (Gleeson et al., 2009). It acts through its penetration into the proteins, nucleotides, and fatty acids of microorganisms causing cell death (Gupta et al., 2018), which makes the development of resistance unlikely. However, in the field, the presence of some residues in the environment like the organic matter of the teat skin or in the challenge suspension may have been reacting with the disinfectant and thus less germicidal activity may have been produced (Enger et al., 2015). These conditions permit some bacteria to be exposed too much to lower concentrations of the active ingredients consequently and thus increasing their resistance to the used disinfectants (Wales and Davies, 2015). These results were in agreement with what was reported earlier that a dip containing the highest free iodine provided a greater significantly superior reduction of pathogens compared with a lower concentration of iodine dip (Foret et al., 2005). Furthermore, all isolates showed a sensitivity to H2O2, and this had been confirmed previously. H2O2 produces oxidants as singlet oxygen, superoxide radicals, and hydroxyl radicals with high penetration power and not specific in the points of contact by attacking bacteria at any essential cell components, leading to a decrease in the probability of bacteria to create resistance with these compounds (Nilima, 2011; Benavides, 2014). This was in contrast with Enger et al. (2015) who reported that H2O2 was the least efficacious tested teat dip, and these differences may be due to the evolution of resistance by the gradual use of the disinfectant (Wales and Davies, 2015). However, different concentrations of ethanol showed no antimicrobial activity against all tested bacteria, and this is in agreement with what was proved before about ethanol effectiveness (Al-Dabbbagh et al., 2015). This ethanol tolerance mainly appears to result in large part from adaptive and evolutionary changes in cell membrane composition (Ingram, 1990).
Conclusion
Ultimately, the spread of mobile mcr genes between MDR and XDR virulent Gram-negative isolates from dairy cattle confirms the dissemination of the mcr gene and its variants at all levels: human, animal, and environmental. Consequently, a decision to stop using colistin in food animals is a principal demand to defy XDR and MDR emergence. Additionally, establishing a perfect quality control check system in Egypt, focusing on both farm workers and the animal itself is urgent.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .
Author Contributions
YT and NA contributed equally in the conception and design of the study and participated with RG, HE, EK, and AA in the application of classical microbiological techniques. YT and NA carried out the PCR assays, sequencing approaches, and participated with HR in sequence analysis. HR performed the bioinformatics and statistical analyses of the data. RG, HE, SE, EK, AA, SA, and HR conceived the study and participated in the design. YT, RG, NA, HE, SE, and AA wrote the initial draft of the manuscript. All authors revised the manuscript critically for important intellectual content and gave the final approval of the version to be published.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.761417/full#supplementary-material
| A Venn diagram showing the intersection between the mcr variants of colistin-resistant isolates from clinical, subclinical mastitis, and raw milk.
| Correlation plot between antimicrobial resistance phenotypes, colistin resistance genes (mcr) and virulence genes in E. coli, K. pneumoniae, P. aeruginosa, and A. hydrophila. In each plot, each cell represents the R-value (correlation coefficient) for each pair of features. The color and circle size indicate the value of correlation (red = negative and blue = positive correlation). The more intense the color, the higher the correlation value. Stars referring to significant correlation determined at a cutoff = 0.05. Insignificant correlations are left without stars.
References
- Abd El-Baky R. M., Masoud S. M., Mohamed D. S., Waly N. G., Shafik E. A., Mohareb D. A., et al. (2020). Prevalence and Some Possible Mechanisms of Colistin Resistance Among Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa . Infect. Drug Resist. 13, 323–332. doi: 10.2147/IDR.S238811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzitey F., Asante J., Kumalo H. M., Khan R. B., Somboro A. M., Amoako D. G. (2020). Genomic Investigation Intothe Virulome, Pathogenicity, Stress Response Factors, Clonal Lineages, and Phylogenetic Relationship of Escherichia coli Strains Isolated from Meat Sources in Ghana. Genes 11 (12), 1504. doi: 10.3390/genes11121504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Dabbbagh S. Y. A., Ali H. H., Khalil I. I., Hamada M. (2015). A Study of Some Antibiotics; Disinfectants and Antiseptics Efficacy Against Some Species of Pathogenic Bacteria. Assiut. Vet. Med. J. 61 (147), 210–217. [Google Scholar]
- Anyanwu M. U., Jaja I. F., Nwobi O. C. (2020). Occurrence and Characteristics of Mobile Colistin Resistance (Mcr) Gene-Containing Isolates From the Environment: A Review. Int. J. Environ. Res. Public Health 17, 1028. doi: 10.3390/ijerph17031028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron J. E., Feingold S. M. (1990). Bailey and Scott's Diagnostic Microbiology, 8th ed. The C.V. Mosby Company, Saint Louis, MO. [Google Scholar]
- Benavides M. L. (2014). “Must Know Facts About Teat Disinfectants and Their Role in the Prevention of Mastitis in Modern Dairy Herds,” in In Proceedings of the British Mastitis Conference, (2014) Sixways, Worcester, The Dairy Group, The University of Nottingham and DairyCo, 25–31. [Google Scholar]
- Böhm F., Wente N., Krömker V. (2017). The Efficacy of a Foaming Iodine-Based Pre-Milking Teat Disinfectant. Milk Sci. Int. - Milchwiss. 70 (2), 6–9. [Google Scholar]
- Brown R. L., Sanderson K., Kirov S. M. (1997). Plasmids and Aeromonas Virulence. Immunol. Microbiol. 17 (4), 217–223. doi: 10.1111/j.1574-695X.1997.tb01015 [DOI] [PubMed] [Google Scholar]
- Carattoli A., Villa L., Feudi C., Curcio L., Orsini S., Luppi A., et al. (2017). Novel Plasmid-Mediated Colistin Resistance Mcr-4 Gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium 2015 to 2016. Eurosurveillance 22 (31), 30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casilag F., Lorenz A., Krueger J., Klawonn F., Weiss S., Häussler S. (2016). The LasB Elastase of Pseudomonas aeruginosa Acts in Concert With Alkaline Protease AprA To Prevent Flagellin-Mediated Immune Recognition. Infect. Immun. 84 (1), 162. doi: 10.1128/IAI.00939-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Zhou J., Wu S., Yang Y., Yu D., Wang X., et al. (2020). Characterization of the IncX3 Plasmid Producing BlaNDM–7 From Klebsiella pneumoniae ST34. Front. Microbiol. 11, 1885. doi: 10.3389/fmicb.2020.01885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute – CLSI (2019). Performance Standard for Antimicrobial Susceptibility Testing. 29th ed. CLSI supplement M100. Wayne, PA: CLSI; [Google Scholar]
- Coton M., Delbes-Paus C., Irlinger F., Desmasures N., Le Fleche A., Stahl V., et al. (2012). Diversity and Assessment of Potential Risk Factors of Gram-Negative Isolates Associated With French Cheeses. Food Microbiol. 29 (1), 88–98. doi: 10.1016/j.fm.2011.08.020 [DOI] [PubMed] [Google Scholar]
- Cyoia P. S., Rodrigues G. R., Nishio E. K., Medeiros L. P., Koga V. L., Pereira A. P. D., et al. (2015). Presence of Virulence Genes and Pathogenicity Islands in Extraintestinal Pathogenic Escherichia coli Isolates Brazil. J. Infect. Dev. Ctries. 9 (10), 1068–1075. doi: 10.3855/jidc.6683 [DOI] [PubMed] [Google Scholar]
- Dandachi I., Chaddad A., Hanna J., Matta J., Daoud Z. (2019). Understanding The Epidemiology of Multi-Drug Resistant Gram-Negative Bacilli in The Middle East Using AOne Health Approach. Front. Microbiol. 10, 1941. doi: 10.3389/fmicb.2019.01941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolejska M., Villa L., Dobiasova H., Fortini D., Feudi C., Carattoli A. (2013). Plasmid Content of Aclinically Relevant Klebsiella pneumoniae Clone From The Czech Republic Producing CTX-M-15 and Qnrb1 . Antimicrob. Agents Chemother. 57 (2), 1073–1076. doi: 10.1128/AAC.01886-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos L. D. R., Furlan J. P. R., Ramos M. S., Gallo I. F. L., de Freitas L. V. P., Stehling E. G. (2020). Co-Occurrence of Mcr-1, Mcr-3, Mcr-7 and Clinically Relevant Antimicrobial Resistance Genes in Environmental and Fecal Samples. Arch. Microbiol. 202 (7), 1795–1800. doi: 10.1007/s00203-020-01890-3 [DOI] [PubMed] [Google Scholar]
- Enger B. D., Fox L. K., Gay J. M., Johnson K. A. (2015). Reduction of Teat Skin Mastitis Pathogen Loads: Differences Between Strains, Dips, and Contact Times. J. Dairy Sci. 98 (2), 1354–1361. doi: 10.3168/jds.2014-8622 [DOI] [PubMed] [Google Scholar]
- Estrela C., Ribeiro R. G., Estrela C. R. A., Pécora J. D., Sousa-Neto M. D. (2003). Antimicrobial Effect of 2% Sodium Hypochlorite and 2% Chlorhexidine Tested by Different Methods. Braz. Dent. J. 14 (1), 58–62. doi: 10.1590/S0103-64402003000100011 [DOI] [PubMed] [Google Scholar]
- Fadhil L., Al-Marzoqi A., Al Taee Z. M., Shalan A. A. (2016). Molecular and Phenotypic Study of Virulence Genes in a Pathogenic Strain of Pseudomonas aeruginosa isolated From Various Clinical Origins by PCR: Profiles of Genes and Toxins. Res. J. Pharm. Biol. Chem. Sci. 7 (1), 590–5985. [Google Scholar]
- Ferreira R., da Silva B., Rezende G., Nakamura-Silva R., Pitondo-Silva A., Campanini E., et al. (2019). High Prevalence of Multidrug-Resistant Klebsiella pneumoniae Harboring Several Virulence and β-Lactamase Encoding Genes in a Brazilian Intensive Care Unit. Front. Microbiol. 9, 3198. doi: 10.3389/FMICB.2018.03198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filioussis G., Kachrimanidou M., Christodoulopoulos G., Kyritsi M., Hadjichristodoulou C., Adamopoulou M., et al. (2019). Short Communication: Bovine Mastitis Caused By A Multidrug-Resistant, Mcr-1-Positive (Colistin-Resistant), Extended-Spectrum Beta-Lactamase-Producing Escherichia coli Clone on A Greek Dairy Farm. J. Dairy Sci. 103 (1), 852–857. doi: 10.3168/jds.2019-17320 [DOI] [PubMed] [Google Scholar]
- Foret C. J., Corbellini C., Young S., Janowicz P. (2005). Efficacy of Two Iodine Teat Dips Based on Reduction of Naturally Occurring New Lntramammary Infections. J. Dairy Sci. 88 (1), 426–432. doi: 10.3168/jds.S0022-0302(05)72704-1 [DOI] [PubMed] [Google Scholar]
- Furmanek-Blaszk B. (2014). Phenotypic and Molecular Characteristics of an Aeromonas hydrophila Strain Isolated From the River Nile. Microbiol. Res. 169 (7–8), 547–552. doi: 10.1016/J.MICRES.2013.11.001 [DOI] [PubMed] [Google Scholar]
- Gao J., Barkema H. W., Zhang L., Liu G., Deng Z., Cai L., et al. (2017). Incidence of Clinical Mastitis and Distribution of Pathogens on Large Chinese Dairy Farms. J. Dairy Sci. 100 (6), 4797–4806. doi: 10.3168/JDS.2016-12334 [DOI] [PubMed] [Google Scholar]
- Gao J., Li S., Zhang J., Zhou Y., Xu S., Barkema H. W., et al. (2019). Prevalence of Potential Virulence Genes in Klebsiella spp. Isolated From Cows With Clinical Mastitis on Large Chinese Dairy Farms. Foodborne Pathog. Dis. 16 (12), 856–863. doi: 10.1089/fpd.2019.2657 [DOI] [PubMed] [Google Scholar]
- García V., García-Meniño I., Mora A., Flament-Simon S. C., Díaz-Jiménez D., Blanco J. E., et al. (2018). Co-Occurrence of Mcr-1, Mcr-4 and Mcr-5 Genes in Multidrug-Resistant ST10 Enterotoxigenic and Shiga Toxin-Producing Escherichia coli in Spain, (2006-2017). Int. J. Antimicrob. Agents 52 (1), 104–108. doi: 10.1016/J.IJANTIMICAG.2018.03.022 [DOI] [PubMed] [Google Scholar]
- Garcia S. N., Osburn B. I., Cullor J. S. (2019). A One Health Perspective on Dairy Production and Dairy Food Safety. One Heal. 7, 100086. doi: 10.1016/J.ONEHLT.2019.100086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson D., O’Brien B., Flynn J., O’Callaghan E., Galli F. (2009). Effect of Pre-Milking Teat Preparation Pocedures on the Microbial Count on Teats Prior to Clusters Application. Ir. Vet. J. 62 (7), 461–467. doi: 10.1186/2046-0481-62-7-461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goli H. R., Nahaei M. R., Rezaee M. A., Hasani A., Kafil H. S., Aghazadeh M. (2016). Emergence of Colistin Resistant Pseudomonas aeruginosa at Tabriz Hospitals, Iran. Iran J. Microbiol. 8 (1), 62–69. [PMC free article] [PubMed] [Google Scholar]
- Guerra S. T., Orsi H., Joaquim S. F., Guimarães F. F., Lopes B. C., Dalanezi F. M., et al. (2020). Short Communication: Investigation of Extra-Intestinal Pathogenic Escherichia coli Virulence Genes, Bacterial Motility, and Multidrug Resistance Pattern of Strains Isolated From Dairy Cows With Different Severity Scores of Clinical Mastitis. J. Dairy Sci. 103 (4), 3606–3614. doi: 10.3168/jds.2019-17477 [DOI] [PubMed] [Google Scholar]
- Gupta P., Bhatia M., Gupta P., Omar B. (2018). Emerging Biocide Resistance Among Multidrug-Resistant Bacteria: Myth or Reality? A Pilot Study. J. Pharm. Bioallied Sci. 10 (2), 96–101. doi: 10.4103/JPBS.JPBS_24_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad A. M., Hoffmann M., Gonzalez-Escalona N., Abbas N. H., Yao K., Koenig S., et al. (2019). Genomic Features of Colistin Resistant Escherichia coli ST69 Strain Harboring Mcr-1 on IncHI2 Plasmid From Raw Milk Cheese in Egypt. Infect. Genet. Evol. 73, 126–131. doi: 10.1016/j.meegid.2019.04.021 [DOI] [PubMed] [Google Scholar]
- Hammerl J. A., Borowiak M., Schmoger S., Shamoun D., Grobbel M., Malorny B., et al. (2018). Mcr-5 and a Novel Mcr-5.2 Variant in Escherichia coli Isolates From Food and Food-Producing Animals, Germany 2010 to 2017. J. Antimicrob. Chemother. 73 (5), 1433–1435. doi: 10.1093/jac/dky020 [DOI] [PubMed] [Google Scholar]
- Hassen B., Saloua B., Abbassi M. S., Ruiz-Ripa L., Mama O. M., Hassen A., et al. (2019). Mcr-1 Encoding Colistin Resistance in CTX-M-1/CTXM-15-Producing Escherichia coli Isolates of Bovine and Caprine Origins in Tunisia. First Report of CTX-M-15-St394/D E. coli from Goats. Comp. Immunol. Microbiol. Infect. Dis. 67, 101366. doi: 10.1016/j.cimid.2019.101366 [DOI] [PubMed] [Google Scholar]
- Hennequin C., Robin F. (2016). Correlation Between Antimicrobial Resistance and Virulence in Klebsiella pneumoniae . Eur. J. Clin. Microbiol. Infect. Dis. 35 (3), 333–341. doi: 10.1007/s10096-015-2559-7 [DOI] [PubMed] [Google Scholar]
- Hmede Z., Kassem I. I. (2019). First Report of the Plasmid-Borne Colistin Resistance Gene (Mcr-1) in Proteus Mirabilis Isolated From a Toddler in Non-Clinical Settings. IDCases 18, e00651. doi: 10.1016/J.IDCR.2019.E00651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D., Andersson D. I. (2012). Selection of Resistance at Lethal and Non-Lethal Antibiotic Concentrations. Curr. Opin. Microbiol. 15 (5), 555–560. doi: 10.1016/J.MIB.2012.07.005 [DOI] [PubMed] [Google Scholar]
- Ingram L. O. (1990). Ethanol Tolerance in Bacteria. Crit. Rev. Biotechnol. 9 (4), 305–319. doi: 10.3109/07388558909036741 [DOI] [PubMed] [Google Scholar]
- Javed H., Saleem S., Zafar A., Ghafoor A., Shahzad A. B., Ejaz H., et al. (2020). Emergence of Plasmid-Mediated Mcr Genes From Gram-Negative Bacteria at The Human-Animal Interface. Gut Pathog. 12 (1), 1–9. doi: 10.1186/s13099-020-00392-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T. J., Siek K. E., Johnson S. J., Nolan L. K. (2006). DNA Sequence of A ColV Plasmid and Prevalence of Selected Plasmid-Encoded Virulence Genes Among Avian Escherichia coli Strains. J. Bacteriol. 188 (2), 745–758. doi: 10.1128/JB.188.2.745-758.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal R., Bayoumi M. (2015). Efficacy of Premilking and Postmilking Teat Dipping as a Control of Subclinical Mastitis in Egyptian Dairy Cattle. Int. Food Res. J. 22 (2), 1037–1042. [Google Scholar]
- Kampf G. (2018). Biocidal Agents Used for Disinfection Can Enhance Antibiotic Resistance in Gram-Negative Species. Antibiotics 7 (4), 110. doi: 10.3390/ANTIBIOTICS7040110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf I., Jouy E., Chauvin C. (2016). Colistin Use and Colistin Resistance in Bacteria From Animals. Int. J. Antimicrob. Agents 48 (6), 598–606. doi: 10.1016/J.IJANTIMICAG.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Khalifa H. O., Ahmed A. M., Oreiby A. F., Eid A. M., Shimamoto T., Shimamoto T. (2016). Characterisation of the Plasmid-Mediated Colistin Resistance Gene Mcr-1 in Escherichia coli Isolated From Animals in Egypt. Int. J. Antimicrob. Agents 47 (5), 413–414. doi: 10.1016/j.ijantimicag.2016.02.011 [DOI] [PubMed] [Google Scholar]
- Kolde R. (2019). Pheatmap: Pretty Heatmaps. R Package Version 1.0. vol. 12, pp. 1–8. [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanjri S., Uwingabiye J., Frikh M., Abdellatifi L., Kasouati J., Maleb A., et al. (2017). In Vitro Evaluation of the Susceptibility of Acinetobacter baumannii Isolates to Antiseptics and Disinfectants: Comparison Between Clinical and Environmental Isolates. Antimicrob. Resist. Infect. Control 6 (1), 36. doi: 10.1186/s13756-017-0195-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay K. K., Jeamsripong S., Sunn K. P., Angkititrakul S., Prathan R., Srisanga S., et al. (2021). Colistin Resistance and ESBL Production in Salmonella and Escherichia coli From Pigs and Pork in the Thailand, Cambodia, Lao PDR, and Myanmar Border Area. Antibiotics 10 (6), 657. doi: 10.3390/ANTIBIOTICS10060657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima Barbieri N., Nielsen D. W., Wannemuehler Y., Cavender T., Hussein A., Yan S., et al. (2017). Mcr-1 Identified in Avian Pathogenic Escherichia coli (APEC). PloS One 12 (3), e0172997. doi: 10.1371/journal.pone.0172997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Z., Yin W., Shen Z., Wang Y., Shen J., Walsh T. R. (2020). Epidemiology of Mobile Colistin Resistance Genes Mcr-1 to Mcr-9 . J. Antimicrob. Chemother. 75 (11), 3087–3095. doi: 10.1093/jac/dkaa205 [DOI] [PubMed] [Google Scholar]
- Liu G., Ali T., Gao J., Rahman S. U., Yu D., Barkema H. W., et al. (2020). Co-Occurrence of Plasmid-Mediated Colistin Resistance (Mcr-1) and Extended-Spectrum β-Lactamase Encoding Genes in Escherichia coli From Bovine Mastitic Milk in China. Microb. Drug Resist. 26 (6), 685–696. doi: 10.1089/mdr.2019.0333 [DOI] [PubMed] [Google Scholar]
- Liu Y., Cui Y., Peng W., Huang B., Ma L., Zheng M., et al. (2020). Prevalence of Pathogens Harbouring Mobile Antimicrobial Resistance Genes and Virulence Factors in Retail Beef and Mutton. FEMS Microbiol. Lett. 367 (12), fnaa089. doi: 10.1093/femsle/fnaa089 [DOI] [PubMed] [Google Scholar]
- Locatelli C., Barberio A., Bonamico S., Casula A., Moroni P., Bronzo V. (2019). Identification of Multidrug-Resistant Escherichia coli From Bovine Clinical Mastitis Using a Ceftiofur-Supplemented Medium. Foodborne Pathog. Dis. 16 (8), 590–596. doi: 10.1089/FPD.2018.2598 [DOI] [PubMed] [Google Scholar]
- Luo Q., Wang Y., Xiao Y. (2020). Prevalence and Transmission of Mobilized Colistin Resistance (Mcr) Gene in Bacteria Common to Animals and Humans. Biosaf. Heal. 2 (2), 71–78. doi: 10.1016/j.bsheal.2020.05.001 [DOI] [Google Scholar]
- Magiorakos A., Srinivasan A., Carey R. B., Carmeli Y., Falagas M. E., Giske C. G., et al. (2012). Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Rsistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 18 (3), 268–281. doi: 10.1111/j.14690691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- McEachran A. D., Blackwell B. R., Hanson J. D., Wooten K. J., Mayer G. D., Cox S. B., et al. (2015). Antibiotics, Bacteria, and Antibiotic Resistance Genes: Aerial Transport From Cattle Feed Yards via Particulate Matter. Environ. Health Perspect. 123 (4), 337–343. doi: 10.1289/EHP.1408555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellata M., Ameiss K., Mo H., Curtiss R. (2010). Characterization of The Contribution to Virulence of Three Large Plasmids of Avian Pathogenic Escherichia coli χ7122 (O78: K80: H9). Infect. Immun. 78 (4), 1528–1541. doi: 10.1128/IAI.00981-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migura-Garcia L., González-López J. J., Martinez-Urtaza J., Aguirre Sánchez J. R., Moreno-Mingorance A., Perez de Rozas A., et al. (2020). Mcr-Colistin Resistance Genes Mobilized by IncX4, IncHI2, and IncI2 Plasmids in Escherichia coli of Pigs and White Stork in Spain. Front. Microbiol. 10, 3072. doi: 10.3389/fmicb.2019.03072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Espinosa R., Soberón-Chávez G., Delgado-Sapién G., Sandner-Miranda L., Méndez J. L., González-Valencia G., et al. (2012). Genetic and Phenotypic Characterization of A Pseudomonas aeruginosa Population With High Fequency of Genomic Islands. PloS One 7 (5), e37459. doi: 10.1371/journal.pone.0037459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilima S. (2011). Effect of Hydrogen Peroxide on E. coli and S. aureus . J. Pure Appl. Microbiol. 5, 875–878. [Google Scholar]
- Nowroozi J., Sepahi A. A., Rashnonejad A. (2012). Pyocyanine Biosynthetic Genes in Clinical and Environmental Isolates of Pseudomonas aeruginosa and Detection of Pyocyanine’s Antimicrobial Effects With or Without Colloidal Silver Nanoparticles. Cell J. 14, 7. [PMC free article] [PubMed] [Google Scholar]
- Oliveira L., Ruegg P. L. (2014). Treatments of Clinical Mastitis Occurring in Cows on 51 Large Dairy Herds in Wisconsin. J. Dairy Sci. 97 (9), 5426–5436. doi: 10.3168/JDS.2013-7756 [DOI] [PubMed] [Google Scholar]
- Ombarak R. A., Awasthi S. P., Hatanaka N., Yamasaki S. (2021). Detection of Plasmid Mediated Colistin Resistance Mcr-1 Gene in ESBL Producing Escherichia coli Isolated From Raw Milk Hard Cheese in Egypt. Int. Dairy J. 117, 104986. doi: 10.1016/j.idairyj.2021.104986 [DOI] [Google Scholar]
- Poirel L., Nordmann P. (2016). Emerging Plasmid-Encoded Colistin Resistance: The Animal World as the Culprit? J. Antimicrob. Chemother. 71 (8), 2326–2327. doi: 10.1093/jac/dkw074 [DOI] [PubMed] [Google Scholar]
- Qadi M., Alhato S., Khayyat R., Elmanama A. A. (2021). Colistin Resistance Among Enterobacteriaceae Isolated From Clinical Samples in Gaza Strip. Can. J. Infect. Dis. Med. Microbiol. 2021:6. doi: 10.1155/2021/6634684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn P. J., Carter M. E., Markey B., Carter G. R. (1994). “Enterobacteriaceae,” in Clinical Veterinary Microbiology. Ed.s Quinn P. J., Carter M. E, Markey P. K., Carter G. R. (London: Wolfe Publishing; ), 209–236. Available at: https://www.scirp.org/(S(351jmbntvnsjt1aadkposzje))/reference/ReferencesPapers.aspx?ReferenceID=1778930. [Google Scholar]
- Ramadan H., Soliman A. M., Hiott L. M., Elbediwi M., Woodley T. A., Chattaway M. A., et al. (2021). Emergence of Multidrug-Resistant Escherichia coli Producing CTX-M, Mcr-1, and FosA in Retail Food From Egypt. Front. Cell. Infect. Microbiol. 11, 681588. doi: 10.3389/FCIMB.2021.681588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rather M., Willayat M., Wani S., Munshi Z., Hussain S. (2014). A Multiplex PCR for Detection of Enterotoxin Genes in Aeromonas Species Isolated From Foods of Animal Origin and Human Diarrhoeal Samples. J. Appl. Microbiol. 117 (6), 1721–1729. doi: 10.1111/JAM.12641 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Siek K. E., Giddings C. W., Doetkott C., Johnson T. J., Fakhr M. K., Nolan L. K. (2005). Comparison of Escherichia coli Isolates Implicated in Human Urinary Tract Infection and Avian Colibacillosis. Microbiology 151 (6), 2097–2110. doi: 10.1099/MIC.0.27499-0 [DOI] [PubMed] [Google Scholar]
- Rosen D., Pinkner J., Walker J., Elam J., Jones J., Hultgren S. (2008). Molecular Variations in Klebsiella pneumoniae and Escherichia coli FimH Affect Function and Pathogenesis in the Urinary Tract. Infect. Immun. 76 (7), 3346–3356. doi: 10.1128/IAI.00340-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe S., Tranter W., Laven R. (2018). Effect of Pre-Milking Teat Disinfection on Clinical Mastitis Incidence in a Dairy Herd in Northern Queensland, Australia. Aust. Vet. J. 96 (3), 69–75. doi: 10.1111/AVJ.12674 [DOI] [PubMed] [Google Scholar]
- Russo T. A., Marr C. M. (2019). Hypervirulent Klebsiella pneumoniae . Clin. Microbiol. Rev. 32 (3), e00001–e00019. doi: 10.1128/CMR.00001-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saengsitthisak B., Chaisri W., Punyapornwithaya V., Mektrirat R., Klayraung S., Bernard J., et al. (2020). Occurrence and Antimicrobial Susceptibility Profiles of Multidrug-Resistant Aeromonads Isolated From Freshwater Ornamental Fish in Chiang Mai Province. Pathog. (Basel Switzerland) 9 (11), 1–13. doi: 10.3390/PATHOGENS9110973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini V., McClure J. T., Léger D., Dufour S., Sheldon A. G., Scholl D. T., et al. (2012). Antimicrobial Use on Canadian Dairy Farms. J. Dairy Sci. 95 (3), 1209–1221. doi: 10.3168/jds.2011-4527 [DOI] [PubMed] [Google Scholar]
- Sarowska J., Futoma-Koloch B., Jama-Kmiecik A., Frej-Madrzak M., Ksiazczyk M., Bugla-Ploskonska G., et al. (2019). Virulence Factors, Prevalence and Potential Transmission of Extraintestinal Pathogenic Escherichia coli Isolated From Different Sources: Recent Reports. Gut Pathog. 11 (1), 1–16. doi: 10.1186/s13099-019-0290-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute Inc (2012). “SAS/STAT Statistics User’s Guide,” in Statistical Analytical System, 5th rev ed (Cary, NC, USA: SAS Institute Inc; ). [Google Scholar]
- Savin M., Bierbaum G., Blau K., Parcina M., Sib E., Smalla K., et al. (2020). Colistin-Resistant Enterobacteriaceae Isolated From Process Waters and Wastewater From German Poultry and Pig Slaughterhouses. Front. Microbiol. 11, 575391. doi: 10.3389/FMICB.2020.575391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scatamburlo T. M., Yamazi A. K., Cavicchioli V. Q., Pieri F. A., Nero L. A. (2015). Spoilage Potential of Pseudomonas Species Isolated from Goat Milk. J. Dairy Sci. 98 (2), 759–764. doi: 10.3168/JDS.2014-8747 [DOI] [PubMed] [Google Scholar]
- Sharma I., Kumar A. (2011). Occurrence of Enterotoxigenic Aeromonas Species in Foods of Animal Origin in North East India. Eur. Rev. Med. Pharmacol. Sci. 15 (8), 883–887. [PubMed] [Google Scholar]
- Shen Y., Xu C., Sun Q., Schwarz S., Ou Y., Yang L., et al. (2018). Prevalence and Genetic Analysis of Mcr-3-Positive Aeromonas Species From Humans, Retail Meat, and Environmental Water Samples. Antimicrob. Agents Chemother. 62 (9), e00404–e00418. doi: 10.1128/AAC.00404-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Chhabra D., Sikrodia R., Shukla S., Sharda R., Audarya S. (2018). Isolation of E. coli From Bovine Mastitis and Their Antibiotic Sensitivity Pattern. Int. J. Curr. Microbiol. Appl. Sci. 7 (10), 11–18. doi: 10.20546/ijcmas.2018.710.002 [DOI] [Google Scholar]
- Soliman A. M., Ramadan H., Zarad H., Sugawara Y., Yu L., Sugai M., et al. (2021). Co-Production of Tet(X7) Conferring High-Level Tigecycline Resistance, Fosfomycin FosA4 and Colistin Mcr-1.1 in Escherichia coli Strains From Chickens in Egypt. Antimicrob. Agents Chemother. 65 (6), e02084–e02020. doi: 10.1128/aac.02084-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambekar D., Dhanorkar D., Gulhane S., Khandelwal V., Dudhane M. (2006). Antibacterial Susceptibility of Some Urinary Tract Pathogens to Commonly Used Antibiotics. Afr. J. Biotechnol. 5 (17), 1562–1565. [Google Scholar]
- Tartor Y. H., El-Naenaeey E.-S. Y. (2016). RT-PCR Detection of Exotoxin Genes Expression in Multidrug Resistant Pseudomonas aeruginosa . Cell Mol. Biol. (Noisy-le-grand) 62, 56–62. [PubMed] [Google Scholar]
- Tiwari J. (2013). Trends In Therapeutic and Prevention Strategies for Management of Bovine Mastitis: An Overview. J. Vaccines Vaccin. 4, 176. doi: 10.4172/2157-7560.1000176 [DOI] [Google Scholar]
- Wales A. D., Davies R. H. (2015). Co-Selection of Resistance to Antibiotics, Biocides and Heavy Metals, and Its Relevance to Foodborne Pathogens. Antibiotics 4 (4), 567–604. doi: 10.3390/antibiotics4040567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Tran J. H., Jacoby G. A., Zhang Y., Wang F., Hooper D. C. (2003). Plasmid-Mediated Quinolone Resistance in Clinical Isolates of Escherichia coli From Shanghai, China. Antimicrob. Agents Chemother. 47 (7), 2242–2248. doi: 10.1128/AAC.47.7.2242-2248.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanja D. W., Mbuthia P. G., Waruiru R. M., Bebora L. C., Ngowi H. A., Nyaga P. N. (2020). Antibiotic and Disinfectant Susceptibility Patterns of Bacteria Isolated From Farmed Fish in Kirinyaga County, Kenya. Int. J. Microbiol. 2020, 8897338. doi: 10.1155/2020/8897338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasfi R., Elkhatib W. F., Ashour H. M. (2016). Molecular Typing and Virulence Analysis of Multidrug Resistant Klebsiella pneumoniae Clinical Isolates Recovered From Egyptian Hospitals. Sci. Rep. 6, 38929. doi: 10.1038/SREP38929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb H. E., Angulo F. J., Granier S. A., Scott H. M., Loneragan G. H. (2017). Illustrative Examples of Probable Transfer of Resistance Determinants From Food Animals to Humans: Streptothricins, Glycopeptides, and Colistin. F1000Res 6, 1805. doi: 10.12688/f1000research.12777.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2013). Report on the Consultative Meeting on Antimicrobial Resistance for Countries in the Eastern Mediterranean Region: From Policies to Action Sharm El Sheikh, Egypt. Available at: http://apps.who.int/iris/handle/10665/11.
- World Health Organization (2019). WHO List of Critically Important Antimicrobials for Human Medicine (WHO CIA List) (World Health Organization; ). Available at: https://apps.who.int/iris/handle/10665/325036. [Google Scholar]
- Xavier B. B., Lammens C., Ruhal R., Malhotra-Kumar S., Butaye P., Goossens H., et al. (2016). Identification of a Novel Plasmid-Mediated Colistin Resistance Gene, Mcr-2, in Escherichia coli, Belgium. Eurosurveill (Belgium) 21 (27). doi: 10.2807/1560-7917.ES.2016.21.27.30280 [DOI] [PubMed] [Google Scholar]
- Yang Y. Q., Li Y. X., Lei C. W., Zhang A. Y., Wang H. N. (2018). Novel Plasmid-Mediated Colistin Resistance Gene Mcr-7.1 in Klebsiella pneumoniae . J. Antimicrob. Chemother. 73 (7), 1791–1795. doi: 10.1093/jac/dky111 [DOI] [PubMed] [Google Scholar]
- Ye M., Tu J., Jiang J., Bi Y., You W., Zhang Y., et al. (2016). Clinical and Genomic Analysis of Liver Abscess-Causing Klebsiella pneumoniae Identifies New Liver Abscess-Associated Virulence Genes. Front. Cell. Infect. Microbiol. 6, 165. doi: 10.3389/FCIMB.2016.00165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Li H., Shen Y., Liu Z., Wang S., Shen Z., et al. (2017). Novel Plasmid-Mediated Colistin Resistance Gene Mcr-3 in Escherichia coli . MBio 8 (3), 543–560. doi: 10.1128/mBio.00543-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdolec N., Dobranić V., Butković I., Koturić A., Filipović I., Medvid V. (2016). Antimicrobial Susceptibility of Milk Bacteria From Healthy and Antimicrobial Susceptibility of Milk Bacteria From Healthy and Drug-Treated Cow Udder. Vet. Arh. 86 (2), 163–172. [Google Scholar]
- Zhang J., Chen L., Wang J., Yassin A. K., Butaye P., Kelly P., et al. (2018). Molecular Detection of Colistin Resistance Genes (mcr-1, mcr-2 and mcr-3) in Nasal/Oropharyngeal and Anal/Cloacal Swabs From Pigs and Poultry. Sci. Rep. 8 (1), 3705. doi: 10.1038/S41598-018-22084-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Hou M., Xu Y., Srinivas S., Huang M., Liu L., et al. (2019). Action and Mechanism of the Colistin Resistance Enzyme Mcr-4 . Commun. Biol. 2 (1), 1–14. doi: 10.1038/s42003-018-0278-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
| A Venn diagram showing the intersection between the mcr variants of colistin-resistant isolates from clinical, subclinical mastitis, and raw milk.
| Correlation plot between antimicrobial resistance phenotypes, colistin resistance genes (mcr) and virulence genes in E. coli, K. pneumoniae, P. aeruginosa, and A. hydrophila. In each plot, each cell represents the R-value (correlation coefficient) for each pair of features. The color and circle size indicate the value of correlation (red = negative and blue = positive correlation). The more intense the color, the higher the correlation value. Stars referring to significant correlation determined at a cutoff = 0.05. Insignificant correlations are left without stars.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .