Abstract
Fecal pollution remains a challenge for water quality managers at Great Lakes and inland recreational beaches. The fecal indicator of choice at these beaches is typically Escherichia coli (E. coli), determined by culture-based methods that require over 18 h to obtain results. Researchers at the United States Environmental Protection Agency (EPA) have developed a rapid E. coli qPCR methodology (EPA Draft Method C) that can provide same-day results for improving public health protection with demonstrated sensitivity, specificity, and data acceptance criteria. However, limited information is currently available to compare the occurrence of E. coli determined by cultivation and by EPA Draft Method C (Method C). This study provides a large-scale data collection effort to compare the occurrence of E. coli determined by these alternative methods at more than 100 Michigan recreational beach and other sites using the complete set of quantitative data pairings and selected subsets of the data and sites meeting various eligibility requirements. Simple linear regression analyses of composite (pooled) data indicated a correlation between results of the E. coli monitoring approaches for each of the multi-site datasets as evidenced by Pearson R-squared values ranging from 0.452 to 0.641. Theoretical Method C threshold values, expressed as mean log10 target gene copies per reaction, that corresponded to an established E. coli culture method water quality standard of 300 MPN or CFU /100 mL varied only from 1.817 to 1.908 for the different datasets using this model. Different modeling and derivation approaches that incorporated within and between-site variability in the estimates also gave Method C threshold values in this range but only when relatively well-correlated datasets were used to minimize the error. A hypothetical exercise to evaluate the frequency of water impairments based on theoretical qPCR thresholds corresponding to the E. coli water quality standard for culture methods suggested that the methods may provide the same beach notification outcomes over 90% of the time with Method C results differing from culture method results that indicated acceptable and unacceptable water quality at overall rates of 1.9% and 6.6%, respectively. Results from this study provide useful information about the relationships between E. coli determined by culture and qPCR methods across many diverse freshwater sites and should facilitate efforts to implement qPCR-based E. coli detection for rapid recreational water quality monitoring on a large scale in the State of Michigan.
Keywords: E. coli, qPCR, EPA Draft Method C, Alternative Method, Ambient Surface Water, Recreational Water
1. Introduction
There is growing interest in the implementation of quantitative polymerase chain reaction (qPCR) methods for testing recreational water at beach sites for fecal contamination in the United States and Canada. The primary advantage of using qPCR methods over traditional culture-based methods for monitoring enterococci and Escherichia coli (E. coli) fecal indicator bacteria (FIB) at recreational beaches is their capability to provide water quality information within several hours, as opposed to the following day (Griffith and Weisberg, 2011). This important distinction allows for same day public notification of potentially unsafe waters that can vary from day to day (Boehm et al., 2002; Kim and Grant, 2004; Whitman and Nevers, 2004).
The United States Environmental Protection Agency (EPA) has established Beach Action Values (BAV) for recreational water testing of enterococci bacteria (U.S. EPA, 2012a) using EPA Method 1611 (U.S. EPA, 2012b) and, by extension, updated EPA methods 1609, 1611.1 and 1609.1 (U.S. EPA, 2013, U.S. EPA, 2015a, U.S. EPA, 2015b). These tools are already being used in a limited number of beach monitoring and public notification programs in the Great Lakes region (Byappanahalli et al., 2018; Dorevitch et al., 2017). While other factors such as the costs of implementing these tools are involved, the adoption of qPCR technology for enterococci in such programs has been slow in the Great Lakes and other inland regions because water quality standards and beach monitoring systems for most states are based on cultured E. coli rather than enterococci (U.S. EPA, 2003). Many of these authorities have been reluctant to change to both a different indicator organism and a different monitoring technology.
EPA has recently developed a qPCR method for E. coli that, like the EPA qPCR methods for enterococci, can estimate gene copy concentrations in recreational water samples in as few as three hours. This method for E. coli has been subject to an inter-laboratory performance study resulting in customized data acceptance criteria (Sivaganesan et al., 2019) and has been successfully tested with waters from several recreational sites (Aw et al., 2019). These efforts have led to the current development of EPA Draft Method C (hereafter referred to as Method C), a standardized procedure to assess E. coli levels in ambient freshwaters. While there has been significant progress towards the development of qPCR technology for E. coli, at present there are no epidemiological studies demonstrating a direct relationship between E. coli qPCR analysis results and the incidence of swimming-related illnesses—as has been demonstrated for enterococci qPCR methods (Wade et al., 2008, Wade et al., 2010). While direct evidence for a suitable correlation between E. coli qPCR and public health risk remains elusive, several studies have demonstrated positive relationships between qPCR and approved culture method results (Lavender and Kinzelman, 2009; Noble et al., 2010; Shrestha and Dorevitch, 2019).
Here, we describe a large-scale collaborative study between EPA and the Michigan Department of Environment, Great Lakes, and Energy (EGLE) designed to characterize the strength of the relationship between Method C and E. coli culture methods from almost 7000 water samples collected across 101 Great Lake Basin recreational water sites in Michigan.
Multiple statistical models and data eligibility requirements were considered to estimate the relationships between paired measurements within and among sites. In addition, a hypothetical exercise was conducted to evaluate the frequency of water impairments based on theoretical Method C qPCR threshold values developed from these relationships.
2. Materials and Methods
2.1. Water sample collection, processing, and distribution.
The state of Michigan has 1775 public and private beaches that have been monitored for public notification of recreational water quality by means of its BeachGuard system (https://www.egle.state.mi.us/beach/Default.aspx). Based on several factors, including past advisory rates and preferences of the participating local laboratories and beach managers, a total of 101 Great Lakes coastal and inland waterbody beach or river sites were selected for water sampling in this study. The ten local partner laboratories that collected samples and performed in-house analyses are listed (Table 1) and the selected beach or river sites are described in supplemental materials (Table S1). Water samples were collected during the summer months of 2016, 2017 and 2018 at varying frequencies and durations (Table S1), as determined by the local laboratories and beach managers. Map locations and descriptions of most of the beaches can also be seen at: https://arcg.is/r1eHW0.
Table 1.
Partner laboratories
| Laboratory | Location |
|---|---|
| Central Michigan District Health Dept., Assurance Water Laboratory | Gladwin, MI, 48624, USA |
| Ferris State University, Shimadzu Core Laboratory | Big Rapids, MI, 49307, USA |
| Grand Valley State University, Annis Water Resources Institute | Muskegon, MI, 49441, USA |
| Kalamazoo County Health and Community Services Laboratory | Kalamazoo, MI, 49001, USA |
| Lake Superior State University, Environmental Analysis Laboratory | Sault St. Marie, MI, 49783, USA |
| Marquette Area Wastewater Facility | Marquette, MI, 49855, USA |
| Oakland County Health Division Laboratory (OCHD) | Pontiac, MI, 48341, USA |
| Oakland University, HEART Laboratory | Rochester, MI, 48309, USA |
| United States Geological Survey, Upper Midwest Water Science Center | Lansing, MI, 48911, USA |
| Saginaw Valley State University, Dept. of Chemistry (SVSU) | University Center, MI, 48710, USA |
Except for three river sites included in the study, procedures for water sampling were consistent with EPA guidance (U.S. EPA, 2014a) and with EGLE’s EPA-approved quality assurance project plan (“Beach Program Quality Assurance Project Plan”) as briefly described below. Beach managers from the local health departments selected three monitoring points equally distributed along the beach sites and included areas of high bather use and potential pollution sources. When monitoring points varied from this scheme due to special circumstances, such as for river sites, variances were recorded. Samples were collected one foot below the surface in water with a depth of three to six feet or half-way between the surface and the bottom in areas where the water did not reach these depths. Samples were collected by filling either 500 mL or 1 L sterile sampling bottles and were transported to the respective local laboratories on ice in a cooler within 6 h.
Following their own established conventions, the ten partner laboratories listed in Table 1 either processed the individual leftmost, center and rightmost water samples from the three monitoring points at each site separately, or prepared composite samples composed of equal volumes of water from the three samples for processing (Table S1). Sample processing for Method C consisted of filtering 100 mL aliquots of the water samples through 0.4 μm pore size polycarbonate filters and transferring the filters into 2 mL, screw-capped microcentrifuge tubes containing 0.3 g of acid washed, 212–300 μm glass beads (Sigma-Aldrich, St. Louis, MO) as previously described (U.S. EPA, 2012b, U.S. EPA, 2013, U.S. EPA, 2015a, U.S. EPA, 2015b). All bead tubes with filters were stored in −80o C freezers until they were further processed for Method C analysis (Section 2.3). Two replicate filters were prepared in this manner by most of the laboratories. One filter from each water sample was saved for Method C analysis by the local laboratory collecting the sample and the other replicate filters, where available, were shipped overnight on dry ice to the EPA laboratory in Cincinnati, Ohio, where they were again stored at −80o C until Method C analysis was performed. All filters were held at −80o C for no longer than 12 months, and in most cases, less than six months before analysis. An additional aliquot of each water sample was immediately processed by the local laboratories for E. coli culture analysis (Section 2.2).
2.2. E. coli culture analyses.
In most instances, undiluted 100 mL aliquots of the water samples were directly analyzed using the Colilert® Quanti-Tray/2000 System (IDEXX Laboratories, Westbrook, ME) as per Standard Methods 9223B (Anonymous, 2017) and manufacturer instructions. Results were reported as most probable number (MPN) estimates of culturable E. coli within a quantitative range of 8 to 2419 MPN / 100 mL water sample volumes, except as noted below. In rare instances, where water samples were expected to contain high E. coli concentrations, they were diluted 10-fold prior to analysis and the MPN estimates were extrapolated to 100 mL sample volumes. A modified membrane filtration method with mTEC culture medium was used by one laboratory for E. coli enumeration as per EPA Method 1603 (U.S. EPA, 2014b) as noted in Table S1. Counts of colony forming units (CFU) determined by this membrane filtration culture method and MPN estimates obtained by the Colilert® culture method are both accepted by EPA.
2.3. DNA extraction and qPCR analyses.
While replicate filters were prepared by the partner laboratories from most of the water samples collected, as described in section 2.1, qPCR data from only one filter per water sample were used for data analysis in this study. DNA was extracted and analyzed from each of these filters by the procedures described below. Most of the qPCR data produced by these procedures were generated by the EPA laboratory. However, qPCR data for sampling lab SVSU samples from 2016 and OCHD samples from 2018, together representing about 24% of the total samples, were generated by the respective partner laboratories (Table S1) due to the unavailability of replicate filters or EPA analysis data. DNA extractions were in all cases performed as described by Sivaganesan et al. (2019). Briefly:
600 μL of SAE extraction buffer, containing 0.2 μg/mL of salmon DNA in Qiagen AE buffer, was added to each filter tube.
The tubes were sealed, bead milled at 5000 reciprocations/min for 60 s and centrifuged at 12,000 ×g for 1 min.
Supernatants were transferred to clean, low retention micro-centrifuge tubes and centrifuged for an additional 5 min.
Aliquots of the clarified supernatants were transferred to new microcentrifuge tubes for analysis. All DNA extracts were analyzed immediately after extraction.
Information on the specificity and sensitivity of the EC23S857 qPCR assay—which targets a segment of the multi-copy 23S ribosomal RNA genes of E. coli and is used in Method C—has been previously reported (Chern et al., 2011). Each DNA extract was analyzed in duplicate by the EC23S857 assay and by the Sketa22 assay (U.S. EPA, 2015a, U.S. EPA, 2015b) for the salmon DNA sample processing control (SPC) in the same instrument run. Analyses by each of the laboratories were performed as described by Sivaganesan et al. (2019). In brief: reaction mixtures for both assays contained 12.5 μL of Environmental Master Mix (Thermo Fisher Scientific, Microbiology Division, Lenexa, KS, #4396838), 2.5 μL of 2 mg/mL bovine serum albumin, 3 μL of primer-probe mix (for a final concentration of 1 μM of each primer and 80 nM of TaqMan® probe in the reactions), 2 μL of DNA-free water and 5 μL of the DNA extracts for a total reaction volume of 25 μL. However, EC23S857 assay reaction mixtures prepared by the EPA laboratory also contained about 100 copies of a multi-assay internal amplification control (IAC) DNA template and 80 nM of TaqMan® probe for this template (U.S. EPA, 2013, U.S. EPA, 2015a, U.S. EPA, 2015b). While incorporation of this multiplexed IAC assay has been recommended for Method C (Sivaganesan et al., 2019), it has not yet been incorporated into a standardized SOP for the method that is updated as needed each year for laboratories in the State of Michigan (supplementary material). All analyses were performed on StepOnePlus™ real-time PCR sequence detectors (Applied Biosystems, Foster City, CA). Thermal cycling protocols were 10 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 60 s at 56 °C.
DNA standards and control materials used for the qPCR analyses are described by Sivaganesan et al. (2019). In addition to five dilutions of the plasmid DNA standard IDTSMART-KAN: Std1_Xho1 with estimated copy numbers ranging from 22,699 to 11.8 per reaction (Sivaganesan et al., 2018), the EPA laboratory prepared a sixth serial dilution of this standard containing a nominal concentration of 5.9 copies per reaction. Analysis results of this additional standard were included in the generation of standard curves by the EPA lab to extend the method’s range of quantification in this study. Composite standard curves were prepared by the EPA laboratory—and by different analysts at the local Michigan laboratories as applicable—for each year of sample analyses using functionally equivalent prototypes of the Excel workbook presented by Lane et al. (2020a). Each of the EPA and local Michigan lab composite standard curves used in this study (cf. Curve IDs 17–20 and 5–7, 21, respectively, in Lane et al., 2020a) met all of the quality acceptance (QA) criteria currently incorporated in the workbooks as described by Lane et al. (2020a). QA criteria for positive and negative control samples analyzed with each batch of test samples, for test sample matrix interference, and for variability and lower limit of quantification (LLOQ) of test sample EC23S857 assay Cycle-threshold (Ct) measurements are described in Sivaganesan et al. (2019) and Aw et al. (2019) and were also followed in the workbooks with minor differences. These differences included: (1) the use of the multiplexed IAC assay with a previously established acceptance criterion (U.S. EPA, 2013, U.S. EPA, 2015a, U.S. EPA, 2015b) for all EPA sample analyses, (2) LLOQ acceptance criteria determined directly from each composite standard curve rather than from a single global value for all analyses (Lane et al., 2020a), and (3) no analyses of replicate test sample filters within the laboratories. Estimates of mean E. coli target gene quantities in the samples, reported as log10 copies per reaction (log10 copies/rxn), were calculated in the Excel workbooks using composite standard curves and a weighted linear regression model (Lane et al., 2020a) as summarized below:
where Usmp is the estimated mean test sample log10 copy number of the target gene, Ysmp is the mean sample E. coli Ct, Ssmp is the mean sample Sketa22 Ct, Scal is the mean calibrator Sketa22 Ct, αstd is the mean intercept estimate for the standard curve, and βstd is the mean slope estimate. All QA analyses and calculations of E. coli target gene quantity estimates in the test samples were automatically performed in the Excel workbooks.
2.4. Data eligibility conditions for statistical method comparisons.
Conditions used for excluding individual samples from datasets in the analyses were: (1) failure of Method C data to meet all QA criteria, (2) data from either Method C or the culture method were not available, or (3) data were not within the respective ranges of quantification of one or both methods. Selection conditions, sequentially imposed, for the cumulative exclusion of sites from the analyses were: (1) no site exclusions, (2) exclusion of sites having less than 10 samples with eligible paired data for both methods, (3) exclusion of sites having no exceedances of the Michigan water quality standard of 300 culturable E. coli / 100 mL, and (4) exclusion of sites not having a minimum Pearson R-squared value of 0.6 from linear regression analysis of the paired data. An additional eligibility factor considered was the inclusion or exclusion of outliers identified from linear regression analysis of the paired data by site, with outliers defined as: |studentized residual| > upper 95th percentile of the t distribution with n-3 degrees of freedom, where n is the sample size.
Sites with at least one exceedance were selected with the expectation that data from sites with no exceedances could (1) obscure the regression models due to their narrow ranges of concentration estimates from both methods and (2) would offer little additional information about comparative exceedance rates. The dataset eligibility condition of R-squared ≥0.6 was considered based on EPA guidance for establishing site-specific water quality criteria for alternative methods and indicators via demonstration of comparable results to those of an EPA-accepted method (U.S. EPA, 2014c).
2.5. Theoretical Method C threshold value development using simple linear regression.
Based on relationships between culture and the corresponding Method C distributions, theoretical Method C threshold values were developed. Simple linear regression was used to model Method C target gene copy concentration (Y) as a function of culturable MPN or CFU from selected individual sites and from composite (i.e., pooled) data from multiple sites based on the selection conditions described in Section 2.4. The fitted curves were then used to estimate Method C gene copy concentrations for given culture concentrations. The regression model is given by:
| (1) |
where, n is the total number of paired data, Yi is the log10 target gene copy/reaction concentration from Method C at the ith (log10 transformed) culture concentration, Xi, α and β are regression parameters and σ2 is the random error variance. In traditional (or Classical) methods, for a given culture value X0, the corresponding mean estimate of Y (e.g., Y0) can be estimated by:
| (2) |
where, and are least squares estimates of intercept α and slope β respectively. The standard deviation S0 for prediction of Y0 is given by:
| (3) |
where , i = 1,2….n.
This model was used to derive theoretical mean Method C threshold values corresponding to the Michigan culture standard of 300 E. coli / 100 mL ( ) for the selected site-specific and composite multi-site datasets.
2.6. Theoretical Method C threshold development using an averaged regression model
Equation (1) was modified to include the selected sites in the regression model given by:
| (4) |
for the jth selected site where is the total number of paired data, Yij is the log10 target gene copy/reaction concentration from Method C at the ijth (log10 transformed) culture concentration Xij, and σj 2 is the random error variance. First, as described in Section 2.5, least squares estimates for intercept α j and slope β j parameters were determined individually for each site j. Then, for a given (log transformed) culture method concentration X0, the mean estimate Y0, can be estimated by:
| (5) |
where and are the estimated intercept and slope parameters of the overall fitted curve.
Within-site variability and between-site variability can be pooled to estimate the standard error of prediction Y for given X. For a given site j, equation (3) can be modified to include the site number as subscript j, and the standard deviation S0j of the within-site variability is given by:
| (6) |
where , i = 1,2… nj.
Standard deviation of within-site variability Sw can be estimated as the square root of the average of all s and is given by:
Standard deviation of between-site variability Sb can be estimated as the standard deviation of all the predicted ’s, where
The estimated standard deviation ST for the multi-site prediction of Y0 for given X0 is given by:
| (7) |
This model was used to derive theoretical Method C threshold values from the selected multi-site datasets described in Section 2.4 corresponding to the Michigan culture standard of 300 E. coli / 100 mL or to the 83rd percentile of the Method C water quality distribution with the mean set to an EPA geometric mean criterion of 126 E. coli / 100 mL (Section 2.8).
2.7. Theoretical Method C threshold development using a Bayesian hierarchical regression model.
An alternative approach to the classical method to estimate an overall fitted curve is a Bayesian method using a Markov Chain Monte Carlo (MCMC) simulation. Equation (4) was modified to explicitly model the form of site-to-site variability in the intercept and slope parameters. The general form of the hierarchical Bayesian regression model is given by:
| (8) |
where the site-specific intercept α j and slope βj parameters are assumed to come from Normal distributions with means and have variances and , respectively. The following prior distributions were used to estimate the model parameters:
| (9) |
where, U stands for the standard Uniform distribution U(0,1) and var() and var(, respectively, are the estimated variances of the least squares estimates of intercept αj and slope βj. Note that the model described by equation (8) is like the master calibration model described in Sivaganesan et al., (2008).
For a given X0, the Posterior distribution of
| (10) |
is used to estimate the mean prediction and standard deviation of the prediction, where the intercept slope and random error variance parameters have the following normal distributions:
Random error variance accounts for within-site variability and is estimated as the average of the posterior mean estimates of all σj 2 = 1,2..m, defined by the model in Equation (8). Moreover, and account for site-to-site variability in intercept and slope parameters. Thus for a given X0, the mean and standard deviation of the posterior of Y0 defined by equation (10) provide the multi-site prediction and the corresponding standard deviation .
This model was used in a similar manner as the averaged regression model to derive theoretical Method C threshold values from selected multi-site datasets.
2.8. Theoretical Method C threshold derivations using a water quality distribution approach.
Using estimated Method C values corresponding to the EPA’s reported geometric mean (GM) water quality criterion of 126 culturable E. coli / 100ml in U.S. fresh waters (U.S. EPA, 2012a) and Method C standard deviations also determined at this GM value, the averaged and hierarchical Bayesian regression models were used to derive theoretical Method C threshold values corresponding to the 83rd percentile of the EPA water quality distribution by the procedures described in the respective sections for each model. The EPA’s reported BAV for recreational beach monitoring represent the 75th percentiles of the EPA’s water quality distributions for each of the EPA-approved methods in that publication as determined from their respective GMs and standard deviations (U.S. EPA, 2012a). However, we determined that the Michigan water quality standard of 300 culturable E. coli / 100 mL, used for beach advisories in that state, represents the 83rd percentile of the EPA water quality distribution for culturable E. coli.
2.9. Determining hypothetical frequencies and agreement of Method C and culture method threshold exceedances.
Hypothetical Method C threshold exceedances were compared with culture method exceedances based on results from the same sampling day. Samples with Method C log10 target gene copies/reaction estimates that exceeded the Method C threshold values from the different models and derivation approaches were tallied to determine the total number of exceedances for each corresponding approach and data set (i.e. site-specific or multi-site). Tallies of all samples that passed Method C data acceptance criteria, whether giving results that were above the LLOQ or not (Section 2.4), were used as the denominators for determining frequencies of exceedances. Similarly, all culture method results, whether considered quantitative or not, were used to determine frequencies of samples exceeding the Michigan standard for that method. This included culture method results that were both above and below the range of quantification. True negatives (TN) and true positives (TP) are identified in accordance to indications by the culture method, the currently accepted standard. Agreement of hypothetical exceedances of the different Method C thresholds with the “true” state of exceedance, as indicated by the culture method exceedance, was evaluated by the determination of false positive (identifying a non-exceedance as an exceedance) and false negative (identifying an exceedance as a non-exceedance) rates converted to percentages by the formulas:
False Positive rate (FPR) = FP/ (FP + TN)
False Negative rate (FNR) = FN/ (FN + TP)
where TP = total # of exceedances identified as exceedances by Method C; FP = # of non-exceedances identified as exceedances by Method C; TN = total # of non-exceedances identified as non-exceedances by Method C; and FN = # of exceedances identified as non-exceedances by Method C.
3. Results
3.1. Data quality.
A total of 6,965 test samples from 101 sites were analyzed in the study. Among these samples, 291 were excluded by QA screening wherein 226 failed to pass Method C acceptance criteria for potential interferences by the sample matrices, as evidenced by SPC and/or IAC assay results. Acceptance criteria for positive and negative control samples were consistently met in a total of over 387 instrument runs conducted by the three labs contributing qPCR data to this study. Sixty-one samples had culture data that either failed QA criteria or were not reported and four samples failed both qPCR and culture QA criteria. Results for 20 samples from seven sites that were discovered to be incorrectly identified were also excluded. The largest contributor to the exclusion of sample data were results from 2,354 samples that were outside the range of quantification for one or both methods. Method C results that were excluded were due to Ct values above the LLOQ Ct values. LLOQ Ct values ranged from 36.10 to 36.78 for EPA composite curves prepared with six standards and from 35.07 to 36.01 for Michigan laboratory composite curves prepared with five standards. Colilert® values of < 8 MPN were excluded to maintain about the same upper bound of uncertainty as previously reported for qPCR results (Haugland et al. 2016) and to exclude a similar number of low concentration samples by each method. Five sites had no samples with quantitative data pairs and 13 additional sites had less than 10 samples with quantitative data pairs.
3.2. Relationships between E. coli culture and Method C across sites from a simple linear regression model.
Table 2 summarizes the relationships between Method C and culture results from quantitative data pairs as assessed by simple linear regression analysis of the pooled composite data. Results are presented utilizing different data eligibility conditions described in Section 2.4. As expected, the imposition of the site selection conditions and the exclusion of outliers increased the correlation between the methods results (R-squared values increased from 0.452 to 0.641) and decreased the standard deviations of resultant theoretical Method C threshold estimates corresponding to the water quality standard of 300 culturable E. coli / 100 ml from 0.417 to 0.323 log10 copies/reaction. However, the imposition of these conditions had only limited overall effects on the mean Method C threshold estimates (1.817 to 1.908 log10 copies/reaction).
Table 2.
Relationships between E. coli culture and Method C across sites utilizing different data eligibility conditions.
| Theoretical Threshold @ 300b | |||||||
|---|---|---|---|---|---|---|---|
| Selection criteria | N Sites | N Samples | Mean intercept | Mean slope | RSQa | Mean | Standard deviation |
| Quantitative results for both methods | 96 | 4305 | 0.142 | 0.710 | 0.452 | 1.902 | 0.416 |
| Quantitative results & statistical outliers removed | 96 | 4043 | 0.089 | 0.720 | 0.574 | 1.873 | 0.329 |
| Quantitative results, N ≥ 10 per site | 83 | 4229 | 0.151 | 0.705 | 0.439 | 1.900 | 0.417 |
| Quantitative results, N ≥ 10 per site, outliers removed | 83 | 3987 | 0.094 | 0.717 | 0.563 | 1.870 | 0.330 |
| Quantitative results, N ≥ 10 & culture ≥ 1 exceedance per site | 57 | 3127 | 0.095 | 0.731 | 0.486 | 1.908 | 0.412 |
| Quantitative results, N ≥ 10 & culture ≥ 1 exceedance per site, outliers removed | 57 | 2955 | 0.061 | 0.732 | 0.589 | 1.876 | 0.335 |
| Quantitative results, N ≥ 10, culture ≥ 1 exceedance & RSQ ≥ 0.6 per site | 21 | 920 | −0.073 | 0.763 | 0.623 | 1.817 | 0.351 |
| Quantitative results, N ≥ 10, culture ≥ 1 exceedance & RSQ ≥ 0.6 per site, outliers removed | 39 | 2092 | −0.005 | 0.756 | 0.641 | 1.867 | 0.323 |
R-squared
Mean Method C log10 copies/reaction estimates corresponding to 300 culturable E. coli MPN or CFU / 100 mL derived by simple linear regression model from composite data.
3.3. Site-specific relationships between culture and Method C.
Data from the 39 sites that met a minimum correlation requirement of R-squared ≥ 0.6 with ≥ 10 data pairs per site and statistical outliers removed were selected for further analyses starting with simple linear regression analyses of the relationships between the culture and qPCR methods at each of the sites. As indicated (Tables 1, 3, and S1), these sites represented both inland waterbodies and the Great Lakes from diverse regions of the state. Despite the consistency imposed by the minimum R-squared requirement for correlations between culture and Method C results, substantial variability was observed in the relationships between the methods at these sites as indicated by their slope and intercept values (Table 3) and illustrated by their fitted curves (Fig. 1). Differences were further demonstrated by the variability of the mean Method C values corresponding to the water quality standard of 300 culturable E. coli / 100 mL ranging from 1.331 to 2.291 log10 copies per reaction at the sites (Table 2). The 39 beaches with R-squared ≥ 0.6 also provided a useful dataset for further exploring the effects of site to site variability on theoretical Method C threshold determinations in subsequent analyses.
Table 3.
Theoretical site-specific relationships between Method C and culture method results.
| Site ID | Waterbody | N Samples | Intercept | Slope | RSQa | Mean Method C value @ 300 b |
|---|---|---|---|---|---|---|
| Ross | Ross Lake | 75 | −0.391 | 0.847 | 0.768 | 1.706 |
| Singing Bridge | Lake Huron (Saginaw Bay) | 74 | 0.305 | 0.483 | 0.696 | 1.502 |
| Orchard | Lake Michigan | 13 | −0.011 | 0.691 | 0.777 | 1.700 |
| Hoffmaster Campground | Lake Michigan | 34 | −0.371 | 0.888 | 0.839 | 1.830 |
| Meinert | Lake Michigan | 65 | −0.017 | 0.687 | 0.839 | 1.685 |
| New Buffalo | Lake Michigan | 48 | 0.248 | 0.519 | 0.685 | 1.535 |
| Warren | Lake Michigan | 55 | 0.105 | 0.600 | 0.754 | 1.593 |
| Brimley | Lake Superior | 39 | −0.094 | 0.731 | 0.660 | 1.717 |
| Sherman | Lake Superior | 32 | −0.360 | 0.922 | 0.758 | 1.925 |
| 1 | Adams Lake | 32 | 0.094 | 0.859 | 0.663 | 2.221 |
| 11 | Chalmers Lake | 51 | −0.071 | 0.928 | 0.644 | 2.226 |
| 114 | Orion Lake | 51 | −0.320 | 1.054 | 0.774 | 2.291 |
| 131 | Walled Lake | 53 | −0.024 | 0.716 | 0.704 | 1.749 |
| 137 | Cass Lake | 43 | 0.006 | 0.788 | 0.613 | 1.958 |
| 140 | Orchard Lake | 70 | −0.074 | 0.877 | 0.650 | 2.098 |
| 167 | Handsome Lake | 37 | 0.033 | 0.871 | 0.665 | 2.191 |
| 169 | Long Lake | 57 | −0.110 | 0.813 | 0.839 | 1.905 |
| 190 | Tipsico Lake | 58 | −0.257 | 0.990 | 0.940 | 2.196 |
| 207 | Walled Lake | 65 | −0.031 | 0.681 | 0.634 | 1.656 |
| 209 | Cass Lake | 50 | −0.033 | 0.821 | 0.661 | 2.000 |
| 214 | Eagle Lake | 69 | 0.162 | 0.746 | 0.646 | 2.009 |
| 268 | Woodhull Lake | 81 | −0.221 | 1.005 | 0.741 | 2.268 |
| 291 | Scotch Lake | 31 | −0.010 | 0.827 | 0.687 | 2.040 |
| 339 | Wolverine Lake | 52 | −0.368 | 0.931 | 0.624 | 1.939 |
| 38 | Commerce Lake | 26 | −0.278 | 1.014 | 0.837 | 2.234 |
| 64 | Stewart Lake | 20 | 0.079 | 0.741 | 0.610 | 1.915 |
| 74 | Duck Lake | 54 | −0.069 | 0.929 | 0.610 | 2.232 |
| 87 | Big Seven Lake | 63 | −0.038 | 0.757 | 0.636 | 1.838 |
| Holland | Lake Huron | 189 | 0.098 | 0.634 | 0.659 | 1.668 |
| Memorial | Lake St. Clair | 78 | 0.277 | 0.529 | 0.645 | 1.588 |
| Metro | Lake St. Clair | 139 | 0.013 | 0.671 | 0.767 | 1.676 |
| New Baltimore | Lake St. Clair | 63 | −0.172 | 0.607 | 0.685 | 1.331 |
| Bay City | Lake Huron (Saginaw Bay) | 31 | 0.265 | 0.700 | 0.786 | 1.998 |
| Bird Creek | Lake Huron | 50 | −0.053 | 0.705 | 0.753 | 1.695 |
| Brissette | Lake Huron (Saginaw Bay) | 19 | −0.037 | 0.706 | 0.738 | 1.713 |
| East Tawas | Lake Huron (Tawas Bay) | 19 | −0.355 | 0.876 | 0.904 | 1.816 |
| Gateway | Lake Huron (Tawas Bay) | 36 | 0.224 | 0.577 | 0.663 | 1.655 |
| Linwood | Lake Huron (Saginaw Bay) | 60 | 0.228 | 0.608 | 0.674 | 1.735 |
| VanEttan | Van Ettan Lake | 10 | −0.277 | 0.761 | 0.872 | 1.609 |
Peason R-squared
Mean Method C log10 copies/reaction estimate corresponding to 300 culturable E. coli / 100 mL from simple linear regression analysis
Figure 1.
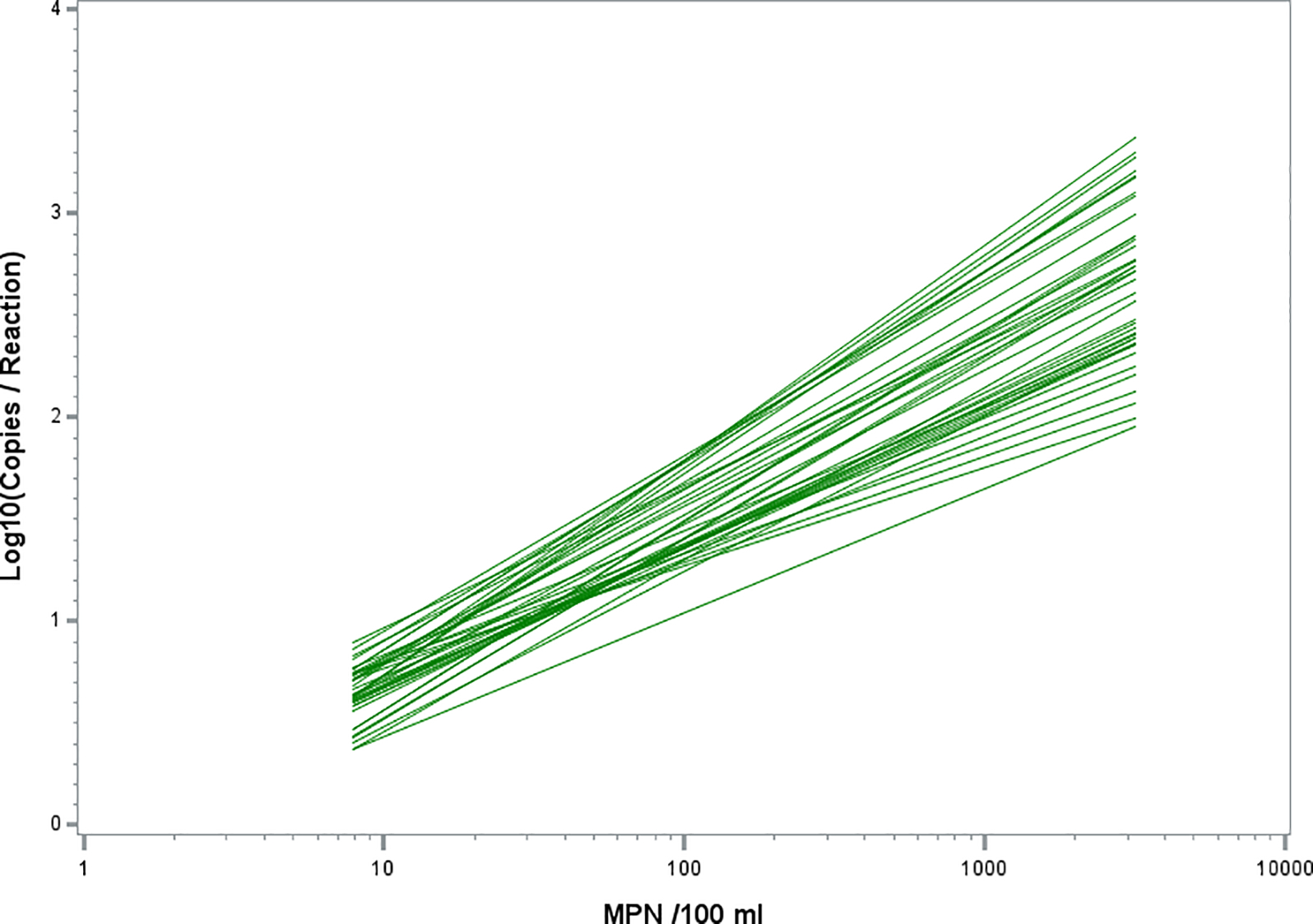
Best fit linear relationships between culture and Method C quantitative estimates for 39 individual sites with ≥ 1 culture exceedance, N ≥ 10 data pairs, R2 ≥ 0.6, and statistical outliers removed.
3.4. Across site relationships between culture and Method C assessed by average regression and Bayesian hierarchal regression models.
Average regression and Bayesian hierarchal regression models were used to further assess across site relationships between the methods under various data eligibility conditions and to predict corresponding theoretical Method C thresholds. Unlike simple linear regression analysis of the composite datasets, these two modeling approaches each accounted for the variability within and between the sites and allowed for the incorporation of this variability into the threshold estimates. Theoretical Method C thresholds were derived that either corresponded to the culture standard of 300 E. coli MPN or CFU / 100 mL or to the 83rd percentile of the Method C distribution.
For the averaged regression model, mean intercept and slope parameters were estimated for each of the sites meeting the different selection criteria independently. Averaged intercept and slope estimates were then used to estimate the mean log transformed DNA concentration Y300 at X0= log10(300). This estimate was the theoretical mean threshold value corresponding to the Michigan water quality standard of 300 culturable E. coli / 100 ml determined from this approach. Moreover, equation (7) was used to estimate the corresponding standard deviation ST_300, which incorporated within and between site variability. For the distribution derivation approach, the same equation was used to estimate Y126, at X0 = log10(126). Again, equation (7) was used to estimate the corresponding standard deviation ST_126, which incorporated within and between site variability. The 83rd percentile of a Normal distribution with the above-estimated mean and the standard deviation ST_126 was the theoretical Method C threshold value for this approach.
The Bayesian model was found not to be applicable for all datasets due to insufficient correlation of the methods results and was applied for comparison with the averaged model results for only the most highly correlated dataset from the 39 sites with R-squared ≥ 0.6. In this model, intercept and slope parameters of all included sites were assumed to come from Normal distributions. The standard deviations, σa and σb, of these Normal distributions accounted for site-to-site variability in intercept and slope parameters. The posterior mean of Y300 at X0= log10(300) was the theoretical EPA Method C threshold value corresponding to the Michigan water quality standard of 300 culturable E. coli / 100 ml. The posterior mean and standard deviation of Y126 at X0 =log10(126) were used to determine the theoretical EPA Method C threshold corresponding to the 83rd percentile.
The average regression and Bayesian modeling approaches provided more comprehensive information on the error of the theoretical multi-site threshold estimates than the simple linear regression model of composite data as evidenced by higher standard deviations of the mean estimates corresponding to 300 culturable E. coli / 100 mL for each of the selected datasets. Standard deviations from the averaged model ranged from 0.364 to 0.514 log10 copies/reaction (Table 4), as compared with 0.323 to 0.418 for the composite data model (Table 2). The Bayesian model produced a standard deviation of 0.387 for the 39-site dataset (Table 4). The mean theoretical threshold values corresponding to 300 culturable E. coli / 100 mL ranged from 1.828 to 1.899 log10 copies/reaction for the different datasets using the averaged model. A similar mean theoretical threshold value of 1.848 was obtained from the Bayesian model for the 39-site dataset.
Table 4.
Theoretical Method C threshold values from average regression and Bayesian hierarchal regression models employing three water quality definitions.
| Water Quality Definitiona | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Fitted Curve | (Normal) Distribution | ||||||||
| Dataset Option | Regression Model | RSQ b | Mean intercept | Mean slope β | Mean @300 | Std Dev @300 | Mean @126 | Std Dev @126 | 83rd percentile c |
| Quantitative results, N ≥ 10 per site | Averaged | 0.437 | 0.113 | 0.711 | 1.875 | 0.514 | 1.607 | 0.455 | 2.042 |
| Quantitative results, N ≥ 10 per site, outliers removed | Averaged | 0.559 | 0.091 | 0.701 | 1.828 | 0.422 | 1.563 | 0.364 | 1.912 |
| Quantitative results, N ≥ 10 & culture ≥ 1 exceedance per site | Averaged | 0.485 | 0.039 | 0.751 | 1.899 | 0.473 | 1.617 | 0.435 | 2.032 |
| Quantitative results, N ≥ 10 & culture ≥ 1 exceedance per site, outliers removed | Averaged | 0.586 | 0.032 | 0.733 | 1.847 | 0.397 | 1.571 | 0.356 | 1.911 |
| N ≥ 10, culture ≥ 1 exceedance, & RSQ ≥ 0.6 per site | Averaged | 0.622 | −0.115 | 0.789 | 1.838 | 0.399 | 1.541 | 0.365 | 1.890 |
| N ≥ 10, culture ≥ 1 exceedance, & RSQ ≥ 0.6 per site, outliers removed | Averaged | 0.640 | −0.049 | 0.772 | 1.863 | 0.364 | 1.572 | 0.332 | 1.889 |
| Regression Model | RSQ b | Mean intercept | Mean slope | Mean @300 | Std Dev @300 | Mean @126 | Std Dev @126 | 83rd percentile c | |
| N ≥ 10, culture ≥ 1 exceedance, & RSQ ≥ 0.6 per site, outliers removed | Bayesian | 0.639 | −0.021 | 0.754 | 1.848 | 0.387 | 1.564 | 0.362 | 1.909 |
Mean and standard deviation (Std Dev) values in log10 copies/reaction
Pearson R-squared
83rd percentile values can be considered to correspond to the Michigan water quality standard of 300 E. coli / 100 ml as described in Section 2.8
Theoretical Method C threshold values corresponding to the 83rd percentile, derived by the Normal distribution approach, ranged from 1.889 to 2.042 log10 copies/reaction using results from the different datasets and the averaged regression model. The threshold value for the 39-site dataset using the Bayesian model was 1.909 log10 copies/reaction. Standard deviations, determined for the mean theoretical Method C estimates corresponding to a culture method value of 126 E. coli MPN or CFU / 100 mL, ranged from 0.332 to 0.455 for the different datasets in this approach (Table 4).
3.5. Frequencies and variation of culture and hypothetical Method C threshold value exceedances.
The percentages of samples resulting in exceedances over the water quality standard of 300 culturable E. coli MPN or CFU / 100 mL, the percentages exceeding theoretical site-specific threshold values for Method C, and percentages exceeding an example theoretical multi-site threshold value for Method C were compared (Table 5). Site-specific threshold value exceedances are only shown for the 39 sites described in Section 3.3 and exceedances of the multi-site threshold value are only shown for the mean value developed from these sites using the averaged regression model (1.863 log10 gene copies/reaction). The overall percentage of the samples from all 101 sites that exceeded this multi-site threshold value was 11.39% (Table 5) and the results for all sites are illustrated in Figure 2. Overall exceedance rates of the theoretical multi-site Method C threshold values developed from each of the models and derivation approaches were highly similar, ranging from 10.41% to 11.68% (data not shown), and were greater than the average culture method exceedance rate of 6.65% in each case. Using same-day results of the culture methods as the standard, false positive and false negative Method C rates (FPR and FNR, respectively) for the 39 sites were 6% and 22%, respectively, using site-specific threshold values and about 7% and 30%, respectively, using the example multi-site Method C threshold value (Table 5). Overall disagreements in water quality decisions from culture and from Method C based on results of the entire 101 site dataset using this multi-site Method C threshold example would occur at a rate of 8.5%.
Table 5.
Water sample exceedances of E. coli culture method water quality standard, theoretical site-specific Method C threshold values, and an example, theoretical multi-site Method C threshold value.
| Site ID | N | E. coli culture method exceed | Site-Specific Method C exceed | TPa | FPb | FNc | TNd | FPRe | FNRf | Multi-site Method C exceed | TP | FP | FN | TN | FPR | FNR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ross | 87 | 28.74% | 25.29% | 18 | 4 | 7 | 58 | 6% | 28% | 17.24% | 14 | 1 | 11 | 61 | 2% | 44% |
| Singing Bridge | 91 | 29.67% | 29.67% | 18 | 9 | 9 | 55 | 14% | 33% | 3.30% | 3 | 0 | 24 | 64 | 0% | 89% |
| Orchard | 45 | 2.22% | 2.22% | 1 | 0 | 0 | 44 | 0% | 0% | 0.00% | 0 | 0 | 1 | 44 | 0% | 100% |
| Hoffmaster Campground | 68 | 5.88% | 4.41% | 3 | 0 | 1 | 64 | 0% | 25% | 4.41% | 3 | 0 | 1 | 64 | 0% | 25% |
| Meinert | 98 | 5.10% | 7.14% | 5 | 2 | 0 | 91 | 2% | 0% | 4.08% | 4 | 0 | 1 | 93 | 0% | 20% |
| New Buffalo | 73 | 8.22% | 9.59% | 3 | 4 | 3 | 63 | 6% | 50% | 1.37% | 1 | 0 | 5 | 67 | 0% | 83% |
| Warren | 71 | 14.08% | 19.72% | 8 | 6 | 2 | 55 | 10% | 20% | 11.27% | 6 | 2 | 4 | 59 | 3% | 40% |
| Brimley | 51 | 15.69% | 17.65% | 7 | 2 | 1 | 41 | 5% | 13% | 9.80% | 5 | 0 | 3 | 43 | 0% | 38% |
| Sherman | 48 | 6.25% | 6.25% | 3 | 0 | 0 | 45 | 0% | 0% | 6.25% | 3 | 0 | 0 | 45 | 0% | 0% |
| 1 | 60 | 3.33% | 8.33% | 2 | 3 | 0 | 55 | 5% | 0% | 13.33% | 2 | 6 | 0 | 52 | 10% | 0% |
| 11 | 93 | 5.38% | 9.68% | 4 | 5 | 1 | 83 | 6% | 20% | 27.96% | 5 | 21 | 0 | 67 | 24% | 0% |
| 114 | 93 | 8.60% | 10.75% | 6 | 4 | 2 | 81 | 5% | 25% | 12.90% | 7 | 5 | 1 | 80 | 6% | 13% |
| 131 | 90 | 4.44% | 4.44% | 4 | 0 | 0 | 86 | 0% | 0% | 4.44% | 4 | 0 | 0 | 86 | 0% | 0% |
| 137 | 100 | 9.00% | 18.00% | 6 | 12 | 3 | 79 | 13% | 33% | 18.00% | 6 | 12 | 3 | 79 | 13% | 33% |
| 140 | 93 | 1.08% | 4.30% | 0 | 4 | 1 | 88 | 4% | 100% | 8.60% | 0 | 8 | 1 | 84 | 9% | 100% |
| 167 | 86 | 3.49% | 8.14% | 3 | 4 | 0 | 79 | 5% | 0% | 10.47% | 3 | 6 | 0 | 77 | 7% | 0% |
| 169 | 88 | 2.27% | 3.41% | 2 | 1 | 0 | 85 | 1% | 0% | 3.41% | 2 | 1 | 0 | 85 | 1% | 0% |
| 190 | 81 | 3.70% | 3.70% | 3 | 0 | 0 | 78 | 0% | 0% | 4.94% | 3 | 1 | 0 | 77 | 1% | 0% |
| 207 | 93 | 1.08% | 3.23% | 0 | 3 | 1 | 89 | 3% | 100% | 1.08% | 0 | 1 | 1 | 91 | 1% | 100% |
| 209 | 92 | 9.78% | 20.65% | 9 | 10 | 0 | 73 | 12% | 0% | 23.91% | 9 | 13 | 0 | 70 | 16% | 0% |
| 214 | 223 | 30.94% | 34.53% | 52 | 25 | 17 | 129 | 16% | 25% | 43.50% | 58 | 39 | 11 | 115 | 25% | 16% |
| 268 | 93 | 10.75% | 18.28% | 8 | 9 | 2 | 74 | 11% | 20% | 34.41% | 10 | 22 | 0 | 61 | 27% | 0% |
| 291 | 158 | 25.95% | 27.22% | 31 | 12 | 10 | 105 | 10% | 24% | 35.44% | 37 | 19 | 4 | 98 | 16% | 10% |
| 339 | 86 | 4.65% | 6.98% | 3 | 3 | 1 | 79 | 4% | 25% | 8.14% | 3 | 4 | 1 | 78 | 5% | 25% |
| 38 | 84 | 3.57% | 3.57% | 3 | 0 | 0 | 81 | 0% | 0% | 7.14% | 3 | 3 | 0 | 78 | 4% | 0% |
| 64 | 90 | 3.33% | 8.89% | 3 | 5 | 0 | 82 | 6% | 0% | 10.00% | 3 | 6 | 0 | 81 | 7% | 0% |
| 74 | 83 | 2.41% | 8.43% | 2 | 5 | 0 | 76 | 6% | 0% | 18.07% | 2 | 13 | 0 | 68 | 16% | 0% |
| 87 | 86 | 1.16% | 6.98% | 1 | 5 | 0 | 80 | 6% | 0% | 6.98% | 1 | 5 | 0 | 80 | 6% | 0% |
| Holland | 60 | 5.00% | 10.00% | 3 | 3 | 0 | 54 | 5% | 0% | 6.67% | 2 | 2 | 1 | 55 | 4% | 33% |
| Memorial | 70 | 25.71% | 28.57% | 15 | 5 | 3 | 47 | 10% | 17% | 11.43% | 7 | 1 | 11 | 51 | 2% | 61% |
| Metro | 69 | 15.94% | 24.64% | 9 | 8 | 2 | 50 | 14% | 18% | 20.29% | 8 | 6 | 3 | 52 | 10% | 27% |
| New Baltimore | 77 | 6.49% | 9.09% | 4 | 3 | 1 | 69 | 4% | 20% | 2.60% | 2 | 0 | 3 | 72 | 0% | 60% |
| Bay City | 61 | 8.20% | 11.48% | 4 | 3 | 1 | 53 | 5% | 20% | 16.39% | 5 | 5 | 0 | 51 | 9% | 0% |
| Bird Creek | 68 | 14.71% | 19.12% | 8 | 5 | 2 | 53 | 9% | 20% | 16.18% | 8 | 3 | 2 | 55 | 5% | 20% |
| Brissette | 63 | 6.35% | 7.94% | 4 | 1 | 0 | 58 | 2% | 0% | 4.76% | 3 | 0 | 1 | 59 | 0% | 25% |
| East Tawas | 30 | 3.33% | 3.33% | 1 | 0 | 0 | 29 | 0% | 0% | 3.33% | 1 | 0 | 0 | 29 | 0% | 0% |
| Gateway | 58 | 5.17% | 5.17% | 1 | 2 | 2 | 53 | 4% | 67% | 0.00% | 0 | 0 | 3 | 55 | 0% | 100% |
| Linwood | 85 | 8.24% | 12.94% | 6 | 5 | 1 | 73 | 6% | 14% | 8.24% | 5 | 2 | 2 | 76 | 3% | 29% |
| Van Ettan | 28 | 17.86% | 10.71% | 3 | 0 | 2 | 23 | 0% | 40% | 7.14% | 2 | 0 | 3 | 23 | 0% | 60% |
| All 39 sites | 3,173 | 10.75% | 13.80% | 266 | 172 | 75 | 2,660 | 6% | 22% | 14.09% | 240 | 207 | 101 | 2,625 | 7% | 30% |
| All 101 Sites | 6,679 | 6.65% | 11.39% | 318 | 443 | 126 | 5,772 | 7% | 28% |
# of culture exceedances identified as exceedances by Method C
# of culture non-exceedances identified as exceedances by Method C
# of culture exceedances identified as non-exceedances by Method C
# of culture non-exceedances identified as non-exceedances by Method C
False Positive rate (FPR): FP/(FP + TN) * 100
False Negative rate (FNR): FN/(FN + TP) * 100
Figure 2.
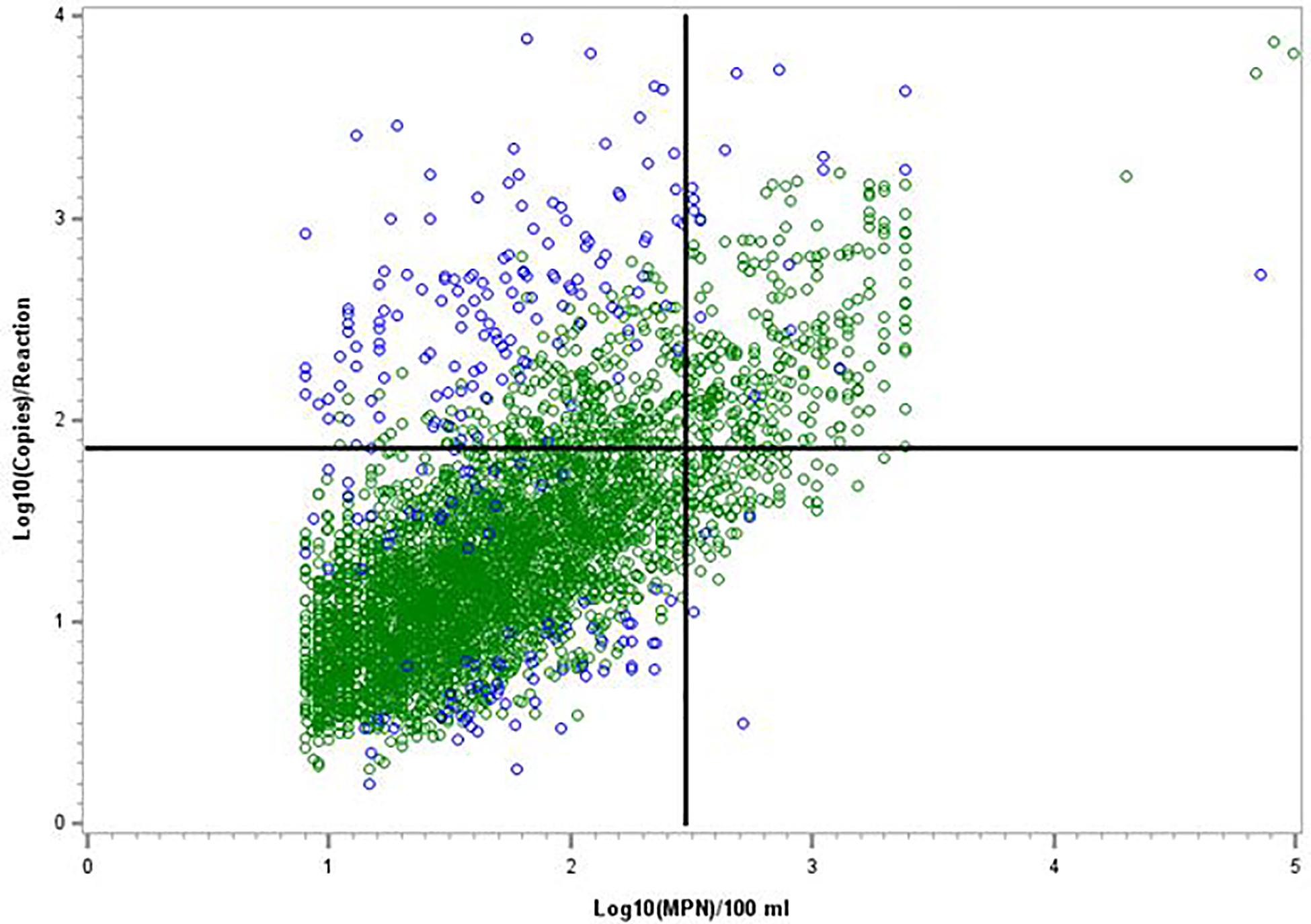
Comparison of E. coli culture method and Method C results with respective actual or theoretical threshold values. Vertical line indicates the culture method water quality standard of 300 E. coli MPN or CFU / 100 mL for beach notifications in Michigan. Horizontal line indicates a theoretical Method C beach notification threshold of 1.863 log10 copies/reaction that could be considered to correspond to the culture method standard. Data points are from 4305 samples with quantitative results for both methods (Table 2). Blue data points represent data pairs that were identified as statistical outliers in regression analyses of the paired datasets from each individual site.
4. Discussion
4.1. Data selection and analysis conditions.
To our knowledge, this study represents the first attempt to examine the relationships between results of an E. coli culture and E. coli qPCR method (Method C) for a wide range of sites representing different geographic regions and different types of freshwater waterbodies over an entire state. In the process, a potentially unprecedented volume of directly comparable quantitative data pairs from 4,305 samples and 96 sites were collected for these methods. A total of 226 or 3.2% of all 6965 samples that were analyzed failed Method C QA criteria due to presumptive sample matrix interference, which was consistent with previous studies using the same polymerase reagent in EPA Method 1609 for enterococci (Nappier et al., 2019). The use of several screening conditions for the selection of partial datasets from all the samples and sites analyzed in the study was an important step in this process. These screening conditions, as noted earlier, included removal of samples determined to be statistical outliers, exclusion of sites not having a minimum numbers of acceptable quantitative data pairs, exclusion of “clean” sites where no samples exceeded the state’s culture method water quality standard, and exclusion of sites not showing a minimum level of correlation between the methods results. This multi-tiered data selection approach allowed for exploration of the effects of varying numbers of different samples and sites on the relationships between the methods. The strength of the relationships was determined from linear regression analyses and resultant theoretical Method C threshold values. Several alternative data modeling and derivation approaches were employed to evaluate the consistency of the theoretical Method C threshold values determined from the different datasets. Most U.S. states and Canadian provinces have adopted different standards for E. coli culture methods based on guidance provided by the EPA for U.S. states (U.S. EPA, 2012a) and Health Canada for Canadian Provinces. (Health Canada, 2012). However, the E. coli culture standard for Michigan is 300 / 100 mL and corresponds to the 83rd percentile of the EPA water quality distribution. Therefore, since this study was specific to Michigan, we used these Michigan-based values as examples to derive corresponding Method C threshold values. Where similar datasets are available from states or provinces that utilize EPA-recommended or other water quality standards, the modeling and derivation methods described in this study can be applied with these standards.
4.2. Effects of data selection and analysis approaches on theoretical multi-site Method C threshold development.
Simple linear regression analyses of the composite datasets illustrated, as expected, that the imposition of various sample and site selection criteria improved the correlation between the qPCR and culture methods results—as indicated by increasing R-squared values. Consequently, the error or uncertainty of the Method C threshold estimates also decreased as evidenced by decreasing standard deviations. However, the mean Method C threshold values determined from the different datasets remained similar. The high variability of the different site-specific Method C thresholds was in contrast with the relative similarity of the multi-site thresholds obtained from the different datasets representing up to 83 different sites with n ≥ 10 data pairs. This raised the question of whether this simple modeling and derivation approach was adequate for developing a theoretical threshold value. Further analyses of the datasets incorporating site-to-site variability by the averaged regression modeling approach substantiated that the simple composite data regression model underestimated error, as evidenced by increases in the corresponding standard deviations. Comparison of the threshold estimates derived from the averaged model by the regression and distribution-based approaches for these datasets showed that the estimates from the two approaches increasingly diverged as the correlations within the datasets decreased and the error in the estimates increased. The threshold values obtained from the most highly correlated 39-site dataset showed the highest similarity between the two approaches and still consistently fell within the range of mean threshold estimates determined from the larger, less restricted datasets by the simple linear regression approach. These results suggest that inclusion of data analysis approaches that take into account the error in the estimates, such as the distribution-based approach, should be considered in the development of multi-site threshold values, at least to substantiate the values obtained by the simpler regression approach. Regression-based derivations only provide mean point estimates that do not directly incorporate this error but do have important advantages in terms of their relative simplicity and more easily understandable relationship to current water quantity standards. The distribution-based approach will incorporate this error into the determination of a threshold at any desired percentile but may be more difficult to intuitively relate to existing water quality standards that are often promulgated as numeric values (e.g., 300 E. coli / 100 mL in Michigan).
Our results further illustrated that the inclusion or exclusion of statistical outliers had an important effect on either the threshold estimates determined by the alternative derivation approaches or on the number of sites that could be included in the analyses. Method C threshold values from both derivation approaches remained relatively similar with each other with outliers either included or excluded, when a higher degree of correlation was maintained by imposing the minimum R-squared requirement. However, maintaining this similarity came at the expense of substantially reducing the number of sites represented in the analyses when outliers were included. The inclusion of outliers in the other, less restricted datasets substantially increased the error in the threshold estimates and increased the differences between estimates from the two derivation approaches. As a result of the higher error, estimates from the distribution-based approach increased for the datasets with outliers included while mean estimates from the regression approach remained relatively constant. Derivation of higher theoretical threshold values from relatively poorly correlated datasets by the distribution approach would have the effect of reducing beach advisory frequencies while the confidence in these decisions would decrease due to the higher error associated with these values.
The EPA has previously issued guidance in the form of a technical support material (TSM) publication for establishing water quality criteria for alternative methods and indicators based on the demonstration of comparable results to those of an EPA-accepted method (U.S. EPA, 2014c). However, this document was intended for application only at individual sites. One feature of the TSM guidance is that a minimum R-squared value of 0.6 is required to demonstrate adequate correlation between the results of the alternative and accepted methods for alternative method criteria development. Results from this study suggested that this ≥ 0.6 R-squared requirement provided a reasonable compromise between maintaining representation of data from a large number of different sites (e.g., 39) and maintaining confidence in theoretical multi-site threshold values obtained for the qPCR method. Excluding this criterion gave almost the same mean threshold value by the averaged regression approach from a substantially larger dataset representing 57 different sites (1.847 vs. 1.863 log10 copies/rxn, Table 4). Consistent with the slightly higher variability of this dataset (standard deviation 0.397 vs. 0.364 and R-squared 0.586 vs 0.622), it did produce a slightly larger discrepancy between estimated theoretical threshold values for Method C determined from the averaged regression and distribution based derivation approaches (1.847 vs. 1.911 log10 copies/rxn, respectively) with the latter value falling slightly outside the range established from all datasets by the simple linear regression approach.
4.3. Hypothetical comparisons of beach notification rates using culture methods and Method C.
Our results suggest that the implementation of Method C using the theoretical multi-site threshold values derived by simple regression from different datasets, including the entire unrestricted dataset of quantitative results from 96 sites, or using any of the values derived by alternative approaches from the selected 39-site dataset would result in an overall increase in threshold exceedances and hypothetical beach notifications of about 4% to 5%, compared to culture methods for all 101 sites included in the study. These relatively modest differences in overall beach notifications would contrast with the most extreme differences for individual sites. For example, the exceedance rate would increase by 20% to 25% at site #268 and decrease by 26% to 29% at the Singing Bridge site using the different Method C theoretical threshold values derived by the different approaches. Even using the site-specific threshold values for each of the 39 most highly correlated sites, the differences in Method C exceedances compared to culture methods would range from an 11% increase to a 7% decrease for Method C. Such site-specific differences in exceedances based on either multi-site or site-specific threshold values may be influenced by the differences in the correlation between culture and qPCR methods (e.g., R-squared values ranging from 0.61 to 0.94 for the reported 39 most highly correlated sites). A trend was also seen in the skewing of outlier results towards high Method C estimates which contributed to the higher overall exceedance rates by Method C (Figure 2). This observation was consistent with results from previous studies, suggesting that adjustments from the Sketa SPC assay tend to produce higher—and thus more conservative from a health protection standpoint—quantitative target organism estimates in samples that may be causing minor qPCR interferences (Haugland et al., 2016). However, as evidenced by the variability of the regression parameter values and site-specific threshold values for Method C, even at the 39 most highly correlated sites (Table 3), the greatest source of variability in exceedance differences between the methods among the individual sites using a multi-site threshold appeared to come from differences in the relationships between quantities of culturable cells and target gene copies at the different sites.
Results from this study further suggest that the assessment of threshold exceedances by the culture methods and by Method C using any of the theoretical threshold values developed from the different modeling and derivation approaches would differ for the same samples in some instances. Among the 39 selected sites, the false-positive rate (FPR, defined in section 2.9) for exceedances resulting from an example multi-site threshold value of 1.863 log10 gene copies/reaction was 7.3% using the exceedances of the currently accepted culture methods from same day samples as the standard. The false-negative rate (FNR) was 29.6%. These rates differed only slightly: 7.1% and 28.4%, respectively, for samples from all 101 study sites in the study. The false-negative rate for Method C would be of primary concern from a health protection standpoint, since such instances would suggest failures to advise the public of unacceptable water quality based on the presumed validity of the culture method decision. However, it has been suggested that care should be taken in interpreting the potential health implications of the terms “false positive” and “false negative” when comparing qPCR to culture method results in this manner (Raith et al., 2013). Using the same theoretical threshold value, Method C results from samples collected on the same day would differ from culture method results in indicating acceptable and unacceptable water quality at rates of 1.9% and 6.6%, respectively, based on all sample analyses in the study.
A more important consideration is that all accepted culture methods require at least overnight incubation of test samples and thus method results are not available until the following day. If the exceedance is due to a short-term event, the delay between sampling results from culture methods would result in an incorrect water quality decision based on the culture method result from the previous day. Nearly 70% of water quality criteria exceedances at California marine recreational beaches were single-day events (Leecaster and Weisberg, 2001). While such percentages could not be directly determined in this study due to the unavailability of consecutive day culture data, results of other freshwater recreational beach studies where overall frequencies of culture method water quality criteria exceedances were similar to those observed in this study have indicated that the overall rate of failures to provide warnings of unacceptable water quality the next day was 12.4% (66/530) and rates of incorrect warnings of unacceptable water quality was 12.6% (67/530) based on previous day E. coli culture results (Table 6).
Table 6.
Agreement of water quality decisions from previous day and next day E. coli culture method water sample analysis results.
| Site | N | Next day warning agreements a | Next day false warnings b | Next day warning failures c | Next day non-warning agreements d |
|---|---|---|---|---|---|
| West Beache | 183 | 7 | 15 | 15 | 146 |
| Zoo Beachf | 35 | 0 | 2 | 2 | 31 |
| Edgewater Beachg | 106 | 6 | 23 | 23 | 54 |
| Grant Park Beachg | 107 | 3 | 15 | 17 | 72 |
| Washington Park Beachg | 99 | 2 | 12 | 9 | 76 |
| All Sites | 530 | 18 | 67 | 66 | 379 |
# of exceedances from previous-day culture results that agreed with next-day culture exceedances. Exceedances based on EPA BAV of 235 E. coli MPN or CFU / 100 mL (U.S. EPA, 2012a).
# of exceedances from previous-day culture results where next-day culture results indicated a non-exceedance. Exceedances based on EPA BAV of 235 E. coli MPN or CFU / 100 mL.
# of non-exceedances from previous-day culture results where next-day culture results indicated an exceedance. Exceedances based on EPA BAV of 235 E. coli MPN or CFU / 100 mL.
# of non-exceedances from previous-day culture results that agreed with next-day culture exceedances. Exceedances based on EPA BAV of 235 E. coli MPN or CFU / 100 mL.
As would be expected, higher frequencies of disagreements between the culture and qPCR methods were observed for the 39 sites when using the example, theoretical multi-site Method C threshold value than when using their individual site-specific threshold values. However, disagreement rates for the individual sites still showed substantial variability regardless of whether a site-specific or multi-site theoretical threshold value was employed for Method C. Some of the seemingly great variability in disagreement rates among the individual sites may be attributable to the low numbers of exceedances by both the qPCR and culture methods at many of the sites which contributed to large fluctuations in the FPR and FNR values.
Previous studies have investigated the influences of different pollution sources and/or environmental factors on levels of both culturable FIB and their target genes detected by qPCR methods (Lane 2019; Lavender and Kinzelman, 2009; Molina et al., 2014; Telech et al., 2009). Collective observations from these studies indicate various environmental factors (e.g., rainfall, wind, turbidity) can differentially affect quantitative estimates by these methods. Thus, the relationships between the methods results can be highly site specific. A fundamental difference between culture and qPCR methods is that the former methods detect only viable target organisms, whereas the latter detect DNA from both viable and non-viable organisms. Consequently, the age and source of the fecal material impacting different sites might be expected to have an important effect on the relationships between results from these methods due to differences in the persistence of their analytes (Korajkic et al., 2014; Walters et al., 2009). Differences between the methods might also be expected for treated (e.g., disinfected wastewater effluents) versus untreated fecal sources due to the differential effects of these treatments on their analytes (Chern et al., 2014). Easily and commonly measured environmental data may be available from sanitary surveys and may help in understanding and predicting differences between culture and qPCR method results at sites when results from the methods, and resultant beach management decisions, differ substantially (Lavender and Kinzelman, 2009). The application of increasingly available molecular microbial source tracking technologies (Boehm et al., 2013, U.S. EPA, 2019a, 2019b) at these sites may also assist beach managers in assessing the health risks associated with different beach management decisions coming from the alternative methods. Given the important capability of qPCR methods to provide same day results and, based on examples from past EPA epidemiological studies using enterococci, the use of qPCR methods in place of the current culture methods may be advantageous.
4.4. Assumptions and limitations.
While numerous inland waterbody and Great Lakes sites from diverse regions of Michigan were included in this study, resulting in a seemingly unprecedented dataset, this research relied on multiple important assumptions that should be considered when interpreting results. One key set of assumptions are dataset eligibility conditions. While practices designed to ensure the exclusion of poor-quality data are widely accepted, factors such as the requirement for the inclusion or exclusion of outliers, inclusion of at least one sample that exceeds a culture-based water quality standard and a minimum R-squared threshold, among others, were selected in an effort to tailor datasets for recreational water quality monitoring scenarios. Here we explored multiple dataset eligibility permutations to characterize a range of possible outcomes, however, it was not practical to test all possible combinations. To address this limitation, the dataset will be publicly available allowing future researchers and water quality managers to evaluate any dataset eligibility scenario of interest. It is also important to note that datasets in this study were generated by several laboratories potentially introducing another source of variability in model estimates. A collaborative effort was necessary due to the large number of sites (n = 101) and samples (n = 6,965) considered. This collaborative approach was deemed acceptable based on the following: 1) future implementation of Method C would be conducted by multiple laboratories in practice; 2) all laboratories successfully met all QA criteria currently prescribed for Method C in all of their analyses in this study and 3) the majority of the qPCR data were generated by a single high capacity sample processing laboratory to help expedite testing and minimize influence of multiple laboratory variability.
Method C and culture results from ongoing recreational water sample analyses in subsequent years by laboratories in Michigan are expected to fill the role of validating the results reported in this study. Most of the Method C data utilized in our analyses came from a single EPA laboratory to facilitate consistency of results. Analyses of duplicate filters of the samples collected for this study by the partner Michigan laboratories have demonstrated a high level of consistency between their Method C results and those of the EPA laboratory (Lane et al., 2020b). Finally, it is important to recognize that there is still currently no direct scientific evidence demonstrating that the correlations between culture and qPCR method results, represented by the results of this study, correspond to a similar level of public health protection. Additional research is warranted to confirm this assumption using QMRA, epidemiology, or other approaches that can directly link quantitative E. coli estimates in water by EPA Method C to adverse health outcomes.
5. Conclusions
The Method C qPCR-based procedure can yield same day water quality results that should improve public health protection to recreational users. However, there remains limited information on the co-occurrence of E. coli measured by cultivation and EPA Draft Method C (Method C) necessary to assess the compatibility of these two approaches. This study reports a large-scale comparison of E. coli levels determined by culture and qPCR from 6,965 samples collected from 101 Michigan recreational water sites. A multi-tiered data selection approach was combined with multiple data modeling and derivation approaches to determine the correlation (Pearson R-Square value) between results of the methods across sites and to determine estimated theoretical Method C threshold values corresponding to the state’s culture-based water quality standard of 300 E. coli MPN or CFU / 100 mL.
Supplementary Material
Key findings include:
Despite differences in correlation between results of the methods, estimated mean theoretical Method C threshold values were similar for complete (unrestricted) and different selected subsets of Michigan water sample data using a simple linear regression model to analyze composite data relationships. Threshold values determined from alternative modeling and distribution-based derivation approaches that incorporated site-to-site variability were only in close agreement when error in the estimates was minimized by using relatively well-correlated datasets. Use of each of these data analysis approaches with a dataset from 39 sites meeting all of the data selection conditions with statistical outliers removed gave theoretical Method C threshold values that were within the range determined from the complete and other selected datasets by the simple linear regression model. Together, these analyses identified and substantiated a narrow range of theoretical multi-site Method C threshold values that could potentially be applied for monitoring recreational waters in Michigan.
Theoretical Method C threshold values determined from this study would hypothetically cause an overall increase of about 4% to 5% in beach notification frequencies over culture methods in Michigan. Differences in notification frequencies between culture method and Method C could vary substantially for some individual sites due to site-to-site variability in relationships between quantities of target gene copies estimated by Method C and viable cells measured by culture methods.
Hypothetical water quality decisions from Method C using an example theoretical threshold value would differ from culture methods at an overall rate of 8.5% for samples collected on the same day. Method C results would differ from culture method results in indicating acceptable and unacceptable water quality at overall rates of 1.9% and 6.6%, respectively. Previous studies have indicated that water quality decisions can differ at much higher rates when based on a previous day’s water sample analysis result from culture methods.
Results from this study suggest that a state-wide, multi-site threshold value may be feasible for Method C in Michigan despite site-to-site variability in its relationship to E. coli culture methods. Additional studies are warranted to determine factors responsible for site-to-site variability and to confirm if corresponding theoretical Method C thresholds postulated here represent an equivalent public health protection compared to established culture E. coli water quality standards.
Highlights.
A rapid E. coli qPCR method can provide same-day results for beach monitoring.
A large study compared the occurrence of E. coli by qPCR and culture in Michigan.
Results provide an indication of the degree of correlation between the methods.
Options for a beach notification threshold value for the PCR method are presented.
6. Acknowledgments
The authors wish to thank Drs. Shamima Akhter and John Wathen (U.S. EPA, Office of Water), and Drs. Eric Villegas, Larry Wymer and Orin Shanks (U.S. EPA, Office of Research and Development) for their helpful comments on this manuscript. Information has been subjected to U.S. EPA peer and administrative review and has been approved for external publication. The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
8. Acronyms
- BAV
Beach Action Value
- BNV
Beach Notification Value
- CFU
Colony Forming Units
- EGLE
Michigan Department of Environment, Great Lakes and Energy
- EPA
U.S. Environmental Protection Agency
- FN
False Negative
- FP
False Positive
- FNR
False Negative Rate
- FPR
False Positive Rate
- LLOQ
Lower Limit of Quantification
- MCMC
Markov Chain Monte Carlo
- OCHD
Oakland County Health Division
- QMRA
Quantitative Microbial Risk Assessment
- SVSU
Saginaw Valley State University
7. References
- Anonymous, 2017. 9223. Enzyme Substrate Coliform Test, In: Rice EW, Baird RB, Eaton AD (Eds.), Standard Methods for the Examination of Water and Wastewater. 23rd edition. Amer. Public Health Assoc./Amer. Water Works Assoc./Water Environment Federation, Washington, DC. [Google Scholar]
- Aw TG, Sivaganesan M, Briggs S, Dreelin E, Aslan A, Dorevitch S, Shrestha A, Isaacs N, Kinzelman J, Kleinheinz G, Noble R, Rediske R, Scull B, Rosenberg L, Weberman B, Sivy T, Southwell B, Siefring S, Oshima K, Haugland R, 2019. Evaluation of multiple laboratory performance and variability in analysis of Michigan recreational waters by an Escherichia coli qPCR method (Draft Method C). Water Res. 156, 465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm AB, Grant SB, Kim JH, Mowbray SL, McGee CD, Clark CD, Foley DM Wellman DE, 2002. Decadal and shorter period variability and surf zone water quality at Huntington Beach, California. Env. Sci. and Technol. 36, 3885–3892. [DOI] [PubMed] [Google Scholar]
- Boehm AB, Van De Werfhorst LC, Griffith JF, Holden PA, Jay JA, Shanks OC, Wang D, Weisberg SB, 2013. Performance of forty-one microbial source tracking methods: A twenty-seven lab evaluation study. Water Res. 47(18), 6812–6828. [DOI] [PubMed] [Google Scholar]
- Byappanahalli MN, Nevers MB, Shively DA, Spoljaric A, Otto C, 2018. Real-time water quality monitoring at a Great Lakes National Park. J. Environ. Qual. Spec. Sec. Microb. Water Qual. Monit. Model. 47(5), 1086–1093. [DOI] [PubMed] [Google Scholar]
- Chern EC, Siefring SC, Paar J, Doolittle M, Haugland RA, 2011. Comparison of quantitative PCR assays for Escherichia coli targeting ribosomal RNA and single copy genes. Lett. Appl. Microbiol. 52, 298–306. [DOI] [PubMed] [Google Scholar]
- Chern EC, Brenner KP, Wymer L, Haugland RA, 2014. Influence of wastewater disinfection on densities of culturable fecal indicator bacteria and genetic markers. J Water Health 12, 410–417. [DOI] [PubMed] [Google Scholar]
- Dorevitch S, Shrestha A, DeFlorio-Barker S, Breitenbach C, Heimler I, 2017. Monitoring urban beaches with qPCR vs. culture measures of fecal indicator bacteria: Implications for public notification. Environmental Health 16(1), 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JF, Weisberg SB, 2011. Challenges in implementing new technology for beach water quality monitoring: Lessons from a California Demonstration Project. Mar. Technol. Sci. J. 45, 65–73. [Google Scholar]
- Harwood VJ, Boehm AB, Sassoubre LM, Vijayavel K, Stewart JR, Fong TT, Caprais MP, Converse RR, Diston D, Ebdon J, Fuhrman JA, Gourmelon M, Gentry-Shields J, Griffith JF, Kashian DR, Noble RT, Taylor H, Wicki M, 2013. Performance of viruses and bacteriophages for fecal source determination in a multi-laboratory, comparative study. Water Res. 47(18), 6929–6943. [DOI] [PubMed] [Google Scholar]
- Haugland RA, Siefring S, Varma M, Oshima KH, Sivaganesan M, Cao Y, Raith M, Griffith J, Weisberg SB, Noble RT, Blackwood AD, Kinzelman J, Anan’eva T, Bushon RN, Harwood VJ, Gordon KV, Sinigalliano C, 2016. Multi-laboratory survey of qPCR enterococci analysis method performance in U.S. coastal and inland surface waters. J. Microbiol. Methods 123, 114–125. [DOI] [PubMed] [Google Scholar]
- Health Canada, 2012. Guidelines for Canadian Recreational Water Quality, Third Edition. Water, Air and Climate Change Bureau, Healthy Environments and Consumer Safety Branch, Health Canada, Ottawa, Ontario. [Google Scholar]
- Kim JH, Grant SB, 2004. Public mis-notification of coastal water quality: A probabilistic evaluation of posting errors at Huntington Beach, California. Environ Sci Technol. 38(9), 2497–504. [DOI] [PubMed] [Google Scholar]
- Korajkic A, McMinn BR, Harwood VJ, Shanks OC, Fout GS, Ashbolt NJ, 2013. Differential decay of enterococci and Escherichia coli originating from two fecal pollution sources. Appl Environ Microbiol 79, 2488–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis JR, Koch GG, 1977. The measurement of observer agreement for categorical data. Biometrics. 33(1), 159–174. [PubMed] [Google Scholar]
- Lane MJ, Frobish DJ, McNair JN, Rediske RR, 2019. Assessing the impact of environmental variables on E. Coli concentrations measured with Colilert® and Draft Method C (qPCR) at inland lake and Lake Michigan beaches within Muskegon County, MI. Chapter 3, Master’s Thesis, Grand Valley State University, Allendale, MI, U.S.A. [Google Scholar]
- Lane MJ, McNair JN, Rediske RR, Briggs S, Sivaganesan M, Haugland R, 2020a. Simplified analysis of measurement data from a rapid E. coli qPCR method (EPA Draft Method C) using a standardized Excel workbook. Water 12, 775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane MJ, Rediske R, McNair J, Briggs S, Rhodes G, Dreelin E, Sivy T, Flood M, Scull B, Szlag D, Southwell B, Isaacs N, Pike S, 2020b. A comparison of E. coli concentration estimates quantified by the U.S. EPA and a Michigan laboratory network using U.S. EPA Draft Method C. J. Microbiol. Methods, 10.1016/j.mimet.2020.106086. [DOI] [PubMed] [Google Scholar]
- Lavender JS, Kinzelman JL, 2009. A cross comparison of QPCR to agar-based or defined substrate test methods for the determination of Escherichia coli and enterococci in municipal water quality monitoring programs. Water Res. 43(19), 4967–4979. [DOI] [PubMed] [Google Scholar]
- Leecaster M, Weisberg SB, 2001. Effect of sampling frequency on shoreline microbiology assessments. Mar. Pollut. Bull. 42, 1150–1154. [DOI] [PubMed] [Google Scholar]
- Molina M, Hunter S, Cyterski M, Peed LA, Kelty CA, Sivaganesan M, Mooney T, Prieto L, Shanks OC, 2014. Factors affecting the presence of human-associated and fecal indicator real-time quantitative PCR genetic markers in urban-impacted recreational beaches. Water Res. 64, 196–208. [DOI] [PubMed] [Google Scholar]
- Nappier SP, Ichida A, Jaglo K, Haugland R, Jones KR, 2019. Advancements in mitigating interference in quantitative polymerase chain reaction (qPCR) for microbial water quality Monitoring. Sci. Total Environ. 671, 732–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble RT, Blackwood AD, Griffith JF, McGee CD, Weisberg SB 2010. Comparison of rapid quantitative PCR-based and conventional culture-based methods for enumeration of Enterococcus spp. and Escherichia coli in recreational waters. Appl. Environ. Microbiol. 76:7437–7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raith MR, Ebentier DL, Cao Y, Griffith JF, Weisberg SB, 2013. Factors affecting the relationship between quantitative polymerase chain reaction (qPCR) and culture-based enumeration of Enterococcus. J. Appl. Microbiol. 116, 737–746. [DOI] [PubMed] [Google Scholar]
- Shrestha A, Dorevitch S, 2019. Evaluation of rapid qPCR method for quantification of E. coli at non-point source impacted Lake Michigan beaches. Water Res. 156, 395–403. [DOI] [PubMed] [Google Scholar]
- Sivaganesan M, Seifring S, Varma M, Haugland RA, Shanks OC, 2008. A Bayesian method for calculating real-time quantitative PCR calibration curves using absolute plasmid DNA standards. BMC Bioinformatics 9, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaganesan M, Varma M, Siefring S, Haugland R, 2018. Quantification of plasmid DNA standards for U.S. EPA fecal indicator bacteria qPCR methods by droplet digital PCR analysis. J. Microbiol. Methods 152, 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaganesan M, Aw TG, Briggs S, Dreelin E, Aslan A, Dorevitch S, Shrestha A, Isaacs N, Kinzelman J, Kleinheinz JG, Noble R, Rediske R, Scull B, Rosenberg S, Weberman B, Sivy T, Southwell B, Siefring S, Oshima K, Haugland R, 2019. Standardized data quality acceptance criteria for a rapid E. coli qPCR method (Draft Method C) for water quality monitoring at recreational beaches. Water Res. 156, 456–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telech JW, Brenner KP, Haugland R, Sams E, Dufour AP, Wymer L, Wade TJ, 2009. Modeling Enterococcus densities measured by quantitative polymerase chain reaction and membrane filtration using environmental conditions at four Great Lakes beaches. Water Res. 43(19), 4947–4955. [DOI] [PubMed] [Google Scholar]
- U.S. EPA, 2002. National Beach Guidance and Required Performance Criteria for Grants. EPA-823-B-02–004. U.S. Environmental Protection Agency, Office of Water, Washington, D.C. [Google Scholar]
- U.S. EPA, 2003. Bacterial Water Quality Standards for Recreational Waters (Freshwater and Marine Waters) Status Report. EPA-823-R-03–008. U.S. Environmental Protection Agency, Office of Water, Washington, D.C. [Google Scholar]
- U.S. EPA, 2012a. Recreational Water Quality Criteria. EPA-820-F-12–058. U.S. Environmental Protection Agency, Office of Water, Washington, D.C. [Google Scholar]
- U.S. EPA, 2012b. Method 1611: Enterococci in water by TaqMan® quantitative polymerase chain reaction (qPCR) assay. EPA-821-R-12–008. U.S. Environmental Protection Agency, Office of Water, Washington, D.C. [Google Scholar]
- U.S. EPA, 2013. Method 1609: Enterococci in water by TaqMan® quantitative polymerase chain reaction (qPCR) assay with internal amplification control (IAC) assay. EPA-820-R-13–005. U.S. Environmental Protection Agency, Office of Water, Washington, D.C. [Google Scholar]
- U.S. EPA, 2014a. National Beach Guidance and Required Performance Criteria for Grants, 2014 Edition. EPA-823-B-14–001. U.S. Environmental Protection Agency, Office of Water, Washington, D.C. [Google Scholar]
- U.S. EPA, 2014b. Method 1603: Escherichia coli (E. coli) in water by membrane filtration using modified membrane-thermotolerant Escherichia coli agar (Modified mTEC). EPA 821-R-14–010. U.S. Environmental Protection Agency, Office of Water, Washington, D.C. [Google Scholar]
- U.S. EPA, 2014c. Site-Specific Alternative Recreational Criteria Technical Support Materials for Alternative Indicators and Methods. EPA-820-R-14–011. U.S. Environmental Protection Agency, Office of Water, Washington, D.C. [Google Scholar]
- U.S. EPA, 2015a. Method 1609.1: Enterococci in water by TaqMan® quantitative polymerase chain reaction (qPCR) assay with internal amplification control (IAC) assay. EPA-820-R-15–009. U.S. Environmental Protection Agency, Office of Water, Washington, D.C. [Google Scholar]
- U.S. EPA, 2015b. Method 1611.1: Enterococci in water by TaqMan® quantitative polymerase chain reaction (qPCR) assay. EPA-820-R-12–008. U.S. Environmental Protection Agency, Office of Water, Washington, D.C. [Google Scholar]
- U.S. EPA, 2019a. Method 1696: Characterization of human fecal pollution in water by HF183/BacR287 TaqMan® quantitative Polymerase Chain Reaction (qPCR) assay. EPA-821-R-19–002. U.S. Environmental Protection Agency, Office of Water, Washington, D.C. [Google Scholar]
- U.S. EPA, 2019b. Method 1697: Characterization of human fecal pollution in water by HumM2 TaqMan® quantitative Polymerase Chain Reaction (qPCR) assay. EPA-821-R-19–003. U.S. Environmental Protection Agency, Office of Water, Washington, D.C. [Google Scholar]
- Wade TJ, Calderon RL, Brenner KP, Sams E, Beach M, Haugland R, Wymer LJ, Dufour AP, 2008. High sensitivity of children to swimming-associated gastrointestinal illness: Results using a rapid assay of recreational water quality. Epidemiology 19(3), 375–383. [DOI] [PubMed] [Google Scholar]
- Wade TJ, Sams E, Brenner KP, Haugland R, Chern E, Beach M, Wymer L, Rankin CC, Love D, Li Q, Noble R, Dufour AP, 2010. Rapidly measured indicators of recreational water quality and swimming-associated illness at marine beaches: A prospective cohort study. Environ. Health 9, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters SP, Yamahara KM, Boehm AB, 2009. Persistence of nucleic acid markers of health-relevant organisms in seawater microcosms: Implications for their use in assessing risk in recreational waters. Water Res 43, 4929–4939. [DOI] [PubMed] [Google Scholar]
- Wanjugi P, Sivaganesan M, Korajkic A, McMinn B, Kelty CA, Rhodes E, Cyterski M, Zepp R, Oshima K, Stachler E, Kinzelman J, Kurdas SR, Citriglia M, Hsu FC, Acrey B, Shanks OC, 2018. Incidence of somatic and F+ coliphage in Great Lake Basin recreational waters. Water Res. 140, 200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman RL, Nevers MB, 2004. Escherichia coli sampling reliability at a frequently closed Chicago Beach: Monitoring and management implications. Environ. Sci. Technol. 38(16), 4241–4246. [DOI] [PubMed] [Google Scholar]
- Wymer LJ, Brenner KP, Martinson JW, Stutts WR Schaub SA, Dufour AP, 2005. The EMPACT Beaches Project. Results and Recommendations from a Study on the Microbiological Monitoring of Recreational Waters. EPA 600/R-04/023. U.S. Environmental Protection Agency, Office of Research and Development, Washington DC. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


