Abstract
The Pharmacogenomics Knowledgebase (PharmGKB) is an integrated online knowledge resource for the understanding of how genetic variation contributes to variation in drug response. Our focus includes not only pharmacogenomic information useful for clinical implementation (e.g., drug dosing guidelines and annotated drug labels), but also information to catalyze scientific research and drug discovery (e.g., variant-drug annotations and drug-centered pathways). As of April 2021, the annotated content of PharmGKB spans 715 drugs, 1761 genes, 227 diseases, 165 clinical guidelines, and 784 drug labels. We have manually curated data from more than 9000 published papers to generate the content of PharmGKB. Recently, we have also implemented an automated natural language processing (NLP) tool to broaden our coverage of the pharmacogenomic literature. This article contains a basic protocol describing how to navigate the PharmGKB website to retrieve information on how genes and genetic variations affect drug efficacy and toxicity. It also includes a protocol on how to use PharmGKB to facilitate interpretation of findings for a pharmacogenomic variant genotype or metabolizer phenotype. PharmGKB is freely available at http://www.pharmgkb.org.
Keywords: drug response, genetic variation, knowledge base, pharmacogenetics, pharmacogenomics
INTRODUCTION
Pharmacogenomics (PGx) is the study of how genetic variation contributes to variation in drug response. Driven by technology advancements in the post-genomic era, pharmacogenomics research has the potential to optimize drug efficacy and minimize toxicity. It bridges the gap between the scientific discoveries and clinical applications, and offers the exciting promise of precision medicine. The Pharmacogenomics Knowledgebase (PharmGKB; http://www.pharmgkb.org) was established in 2000 to collect, curate, and disseminate knowledge about pharmacogenomics from a number of sources, including the scientific literature, drug labels, and clinical guidelines (Klein & Altman, 2004; Whirl-Carrillo et al., 2012). It is the central knowledge repository for pharmacogenomics information including drug dosing guidelines, drug label annotations, clinical and variant annotations, drug-centered pathways, pharmacogene summaries, and relationships among genes, drugs, and diseases.
PharmGKB serves diverse user groups from both the clinical and scientific communities. It provides comprehensive and integrated drug, gene, variant, and disease information to pharmacologists, clinical investigators, biologists, and informaticians, as well as general public. The PharmGKB homepage has been designed in a way that highlights the primary interests of most users. All PharmGKB data is now freely available through a Creative Commons license, with no user registration required to view our content. PharmGKB is also an excellent educational portal for any person who is new to pharmacogenomics. We provide short “guided page tour” and training exercises (https://api.pharmgkb.org/v1/download/file/attachment/PharmGKB_Training_Exercises.zip) on how to use the website. We also provide lecture materials, tutorials, and useful links intended to help users familiarize themselves with the fundamental concepts of pharmacogenomics research and how the information is used clinically.
The protocols in this article describe how to use PharmGKB to browse and understand pharmacogenomic knowledge. Basic Protocol 1 walks a user through the PharmGKB homepage, helping users familiarize themselves with the content of PharmGKB and how to conduct basic searches. Basic Protocol 2 describes how to use PharmGKB to facilitate interpretation of pharmacogenomic variant genotypes or metabolizer phenotypes.
BASIC PROTOCOL 1
NAVIGATING THE HOMEPAGE OF PharmGKB AND SEARCHING BY DRUG
This protocol will introduce the basic techniques used for searching and browsing the content on the PharmGKB. It starts with a quick tour of the PharmGKB home page. Then, it walks through an example, searching by the drug “clopidogrel” and find the associated genes, variants, prescribing information, pathways and annotations.
Necessary Resources
Hardware
Computer with an Internet connection
Software
An up-to-date Web browser (such as Google Chrome, Firefox, or Safari)
Files
No input files required
-
Open the PharmGKB homepage at http://www.pharmgkb.org in a web browser.
The PharmGKB homepage is the common entry point for all users. It has been designed to highlight the types of information that are most sought after by diverse groups of users, providing short cuts to drugs, pathways, dosing guideline annotations, and drug label annotations (Fig. 1). The principal content of PharmGKB is listed in Table 1. The main search bar sits at the top of the page where a user can enter drugs, genes, variants, or combinations of these terms for a Google-type query. Prominently displayed in the center of the homepage are clickable icons that provides short cuts to drugs, pathways, dosing guidelines, and drug label annotations, as well as the summary statistics for them within the PharmGKB knowledge base. Right below the icons is a diagram where someone new to pharmacogenomics can click on “What is Pharmacogenomics?” (https://www.pharmgkb.org/whatIsPharmacogenomics) to learn about basic genetics and pharmacogenomics concepts, or “Learn more about PharmGKB” (https://www.pharmgkb.org/whatIsPharmgkb) to learn about various features at PharmGKB. There is also a link to an interactive “Page tour” (https://www.pharmgkb.org/?tour=true) at the top of the homepage under “Help.”
-
Click on the icon “Drug Label Annotations” in the left hand of the homepage to browse the list of labels annotated with a pharmacogenomics summary (Fig. 2).
As drug labels began to include information about how to adjust drug prescribing based on a person’s genotype or metabolizer phenotype, PharmGKB correspondingly annotated international labels from the U.S. Food and Drug Administration (FDA), European Medicines Agency (EMA), Swiss Agency of Therapeutic Products (Swissmedic), Pharmaceuticals and Medical Devices Agency, Japan (PMDA,) and Health Canada (Santé Canada) (HCSC). Figure 2 provides an overview of PharmGKB’s annotation of drug labels. By showing these international drug labels side by side, it gives a comparable summary on how different agencies provide PGx information within their labels. If the label provides a dose adjustment based on gene/protein/chromosomal variants or phenotypes, the label annotation is tagged with a “Dosing info” tag. For example, the FDA label for aripiprazole (https://www.pharmgkb.org/labelAnnotation/PA166104839) is tagged with “Dosing info” because the label states that CYP2D6 poor metabolizers taking aripiprazole should have their dose reduced by 50%. If the label states that a drug is either indicated or contraindicated for a particular set of patients based on gene/protein/chromosomal variants or phenotypes, the “Alternative Drug” tag is applied to the label annotation (e.g., FDA label for abacavir, https://www.pharmgkb.org/labelAnnotation/PA166104833, is tagged this way because the label states that abacavir is contraindicated in patients with the HLA-B*5701 allele due to risk for hypersensitivity reactions.)
-
Type the drug name clopidogrel in the search box, then click Enter.
Drugs can be searched for by either typing the drug’s generic name or trade name in the search box, or, by clicking on the “annotated drugs” icon from the home page and then browsing through the alphabetically sorted drug list. The search function of PharmGKB uses an autocomplete service to make suggestions as the user types. If no result is returned, try a synonym or partial name. Both alternative names and other symbols that might have been used in the literature for drugs or genes are included in PharmGKB.
-
Open the clopidogrel drug page (Fig. 3).
PharmGKB drug, gene, disease and variant pages are structured similarly, with the navigation pane on the left. The “overview” tab includes information on chemical structure of the drug, alternative names used, and metabolites, as well as details such as molecular properties and short summaries of pharmacokinetics (PK), pharmacodynamics (PD), and pharmacogenomics (PGx) for drugs that have been well-studied in pharmacogenomics. Additional tabs include prescribing info, drug label annotations, clinical and variant annotations, literature, pathways, automated annotations, links, and downloads. The user can click on each section on the left to retrieve specific information for the drug of interest.
-
Click on the “Prescribing Info” tab from the navigation pane on the left to access information regarding drug dose or therapy adjustments based on genetic information for clopidogrel (Fig. 4).
The prescribing information may come from clinical guidelines for how to adjust treatment of certain medications based on a person’s genetic information, drug labels that have prescribing recommendations based on a particular genotype/metabolizer phenotype, and Rx annotations from publications.
-
Under Source “Clinical Pharmacogenomics Implementation Consortium” (CPIC), click the “Read Now” button to access dosing guidelines for clopidogrel published by CPIC (Fig. 5).
PharmGKB annotates clinical dosing guidelines published by professional societies such as the Clinical Pharmacogenetics Implementation Consortium (CPIC), the Royal Dutch Association for the Advancement of Pharmacy–Pharmacogenetics Working Group (DPWG), the Canadian Pharmacogenomic Network for Drug Safety (CPNDS), or other professional society. The dosing guidelines are curated manually by PharmGKB curators to highlight the prescribing recommendations based on a patient’s genotype or metabolizer phenotype, with excerpts from the guidelines and links to the underlying evidence and the associated genes and drugs. In addition to the information directly provided by the guidelines, PharmGKB provides an interactive allele picker that allows specific alleles/genotypes to be selected from pull-down menus, and then provides the resulting inferred phenotype (e.g., ultra-metabolizer), associated dosing advice, and strength of the recommendation. PharmGKB has also created video explanations of a number of CPIC guidelines, embedded from a dedicated YouTube channel.
-
Click on the “Drug Label Annotations” tab from the navigation pane on the left to access label annotations from regulatory agencies around the world (Fig. 6). Click on the “View Drug Label Annotation Legend” to see a detailed description of the sources, and definition of PGx levels and tags. Click on “Read now” next to “U.S. Food and drug Administration” to access the individual label annotations from the FDA.
The “Drug Label Annotations” tab lists annotations on medication labels that contain PGx information. The label annotations include a brief summary of the PGx in the label, an excerpt from the label, and a downloadable highlighted label PDF file. The annotated labels are also tagged with “PGx Level” (“Testing required,” “Testing Recommended,” “Actionable PGx,” and “Informative PGx”; https://www.pharmgkb.org/page/drugLabelLegend#pgx-level) to indicate the PharmGKB interpretation of the level of action implied in each label. The “Prescribing” section of the annotation captures guidance from the label for patients with a particular genotype or metabolizer phenotype. For U.S. FDA labels, the annotation also includes information on whether the label is on the FDA’s Table of Pharmacogenomic Biomarkers in Drug Labeling (https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling). We recently also added a section a section on information from the Table of Pharmacogenetic Associations (https://www.fda.gov/medical-devices/precision-medicine/table-pharmacogenetic-associations) curated by the FDA. Furthermore, we have developed a new landing page (https://www.pharmgkb.org/fdaLabelAnnotations) specifically for FDA-approved drug label annotations that can be sorted and filtered by different criteria in the column headings, e.g., by genes, drugs, PGx levels, or the prescribing tags.
-
Click on the “Clinical Annotations” tab from the navigation pane on the left to access clinical annotations for clopidogrel. The clinical annotation table can be sorted and/or filtered by evidence level, variant, gene, type, or phenotype. Click on “Read Now” to access the clinical annotations for CYP2C19 *1, *2, *3, *4, *5, *6, *8 and clopidogrel response (Fig. 7)
For more details on clinical annotation, see Basic Protocol 2.
Clinical annotations summarize all of PharmGKB’s annotations of published literature for the relationship between a particular genetic variant and a medication. They are given a rating by PharmGKB depending on how much published evidence there is for a relationship found in PharmGKB, and the consistency and quality of that evidence. Clinical annotations are based on variant annotations in PharmGKB, as well as prescribing information from dosing guidelines and FDA labels. Clinical annotations are also tagged with the relevant drug, phenotype, and broad phenotypic categories of toxicity, dosage, efficacy, PD, or metabolism/PK.
-
Click on the “Variant Annotations” tab from the navigation pane on the left to access variant annotations for clopidogrel. The variant annotation table can be sorted and/or filtered by variant, molecules, p-value, study size, ethnicities, etc. Click on “24762860” under column “PMID” to see the list of variant annotations for that specific article.
Variant annotations are summaries of a genetic association between a single variant and a specific drug response (metabolism, efficacy, dose or toxicity) from a single publication. PharmGKB curators routinely scan pharmacogenomic literature, curating these associations and adding the information to PharmGKB. A single paper can by annotated with multiple variant annotations if multiple variants and/or phenotypes are investigated in the study. Sometimes variant annotations also document cases where no association is found between a variant and drug from the literature. It’s important to capture negative findings, as they also contribute to the overall evaluation of the evidence base between a variant and drug response phenotype. Clinical annotations are built upon summaries of variant annotations along with other variant-specific prescribing information. The associations that are less consistent across different studies are likely to be assigned a lower level of evidence. All variant annotations include a summary sentence constructed from standardized vocabularies. Curators can add additional notes or description of the findings as free text to appear below the summary sentence. Variant annotations are also tagged with a broad phenotypic category of toxicity, dosage, efficacy, PD, or metabolism/PK, and often include additional parameters about study size, cohort description, statistical significance, and ethnicity information. At the bottom of the variant annotation page, the history section describes when the annotation was created and if or when the annotation was changed or edited.
-
Click on the “Pathways” tab to view all pathways associated with clopidogrel. Click on “Clopidogrel Pathway, pharmacokinetics (PK)” to view the simplified diagram of genes/proteins involved in the metabolism of clopidogrel (Fig. 8).
The interactive drug-centered pathways displayed on PharmGKB provide an overview of how genes are involved in the pharmacokinetics (PK) and pharmacodynamics (PD) of drugs. Our pharmacokinetics (PK) pathways describe candidate genes involved in the absorption, distribution, metabolism, and excretion of a given drug, while the pharmacodynamic (PD) pathways illustrate the physiological effects of the drug, its mechanism of action and possible side effects. The pathway diagrams use standard shapes and colors to represent genes, metabolites, drugs, and interactions. All genes and drugs on the pathway diagram are clickable. If the user clicks on these objects, the PharmGKB gene or drug page opens in a new browser window. Below the pathway picture is a “Description” of the pathway that describes the complex gene-drug relationships depicted in the pathway diagram. The pathway authors and the date of the most recent update are listed below the text of the description on the bottom of the pathway diagram. There is a section for components, related pathways, and downloads in the navigation pane on the left. If PharmGKB has both PK and PD pathways for a given drug, the user will find them under “Related pathways.” Under “Downloads,” the user can download pathway diagrams in both Adobe Illustrator and pdf formats. Pathway data can also be downloaded in tsv, bioPax, or GPML formats. In addition to being freely available on the website, PharmGKB VIPs and pathways are also typically featured in the journal “Pharmacogenetics and Genomics.”
-
Click on “Automated annotations” to see a list of annotations derived from a text-mining system that has scanned sentences in the literature (Fig. 9). The annotation highlights a chemical and a variation in a sentence that likely involves pharmacogenomic information.
In order to broaden our coverage of the ever-expanding pharmacogenomic literature, we have developed an automated NLP/text mining tool (PGxMine) to computationally extract possible variant-drug relationships from abstracts in PubMed or full-text articles in PubMed Central (Lever et al., 2020). These annotations use completely computational methods to find variants, drugs, and their associations, and have not been reviewed by a curator to check their accuracy. The sentences are extracted from abstracts in PubMed and full-text articles from the PubMed Central Open Access subset and Author Manuscript Collection.
-
Under the “Links & Downloads” tab, click on links to go to external databases where addition information on clopidogrel may be found.
PharmGKB has established bidirectional links with many leading gene, protein, and drug resources, such as NCBI PubChem, ChEBI, RxNorm, Entrez Gene, GeneCards, UniProtKB, and DrugBank, etc.
Figure 1.
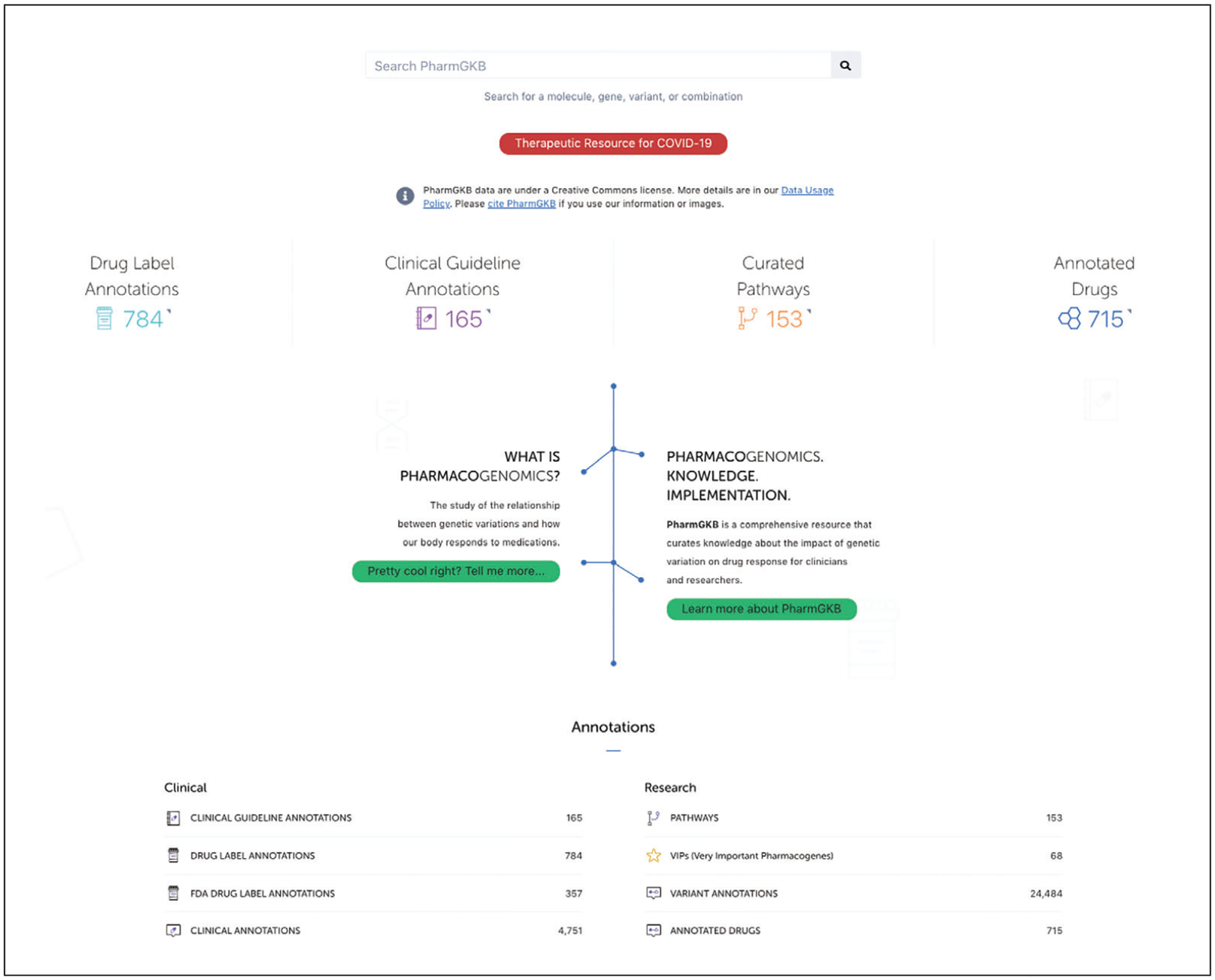
The PharmGKB home page (http://www.pharmgkb.org).
Table 1.
Summary of Content at PharmGKB
| Clinical pharmacogenomics | |
| Clinical guideline annotations | Clinical guidelines on how to adjust treatment of certain medications based on genetic information |
| Drug label annotations | Annotated drug labels containing PGx information from regulatory agencies around the world |
| Clinical annotations | Evidence-rated summaries of all the literature evidence for a particular genetic variant–drug association |
| Rx annotations | Summaries of individual publications that provide medication dosing or prescribing information based on genetic information |
| Research pharmacogenomics | |
| Variant annotations | Summaries of genetic variant–drug associations as reported in a single publication |
| Drug pathways | Illustrated diagrams of the pharmacokinetics or pharmacodynamics of a PGx-relevant drug and accompanying text describing the drug-gene interactions and pharmacogenomic findings |
| VIP PGx gene summaries | Written reviews of PGx-relevant genes, highlighting genomic organization, functional and clinical impact of genetic variations |
| PGx gene information tables | Gene resource tables created by CPIC and PharmGKB to summarize allele function, definition, frequency, and diplotype-phenotype translation |
Figure 2.
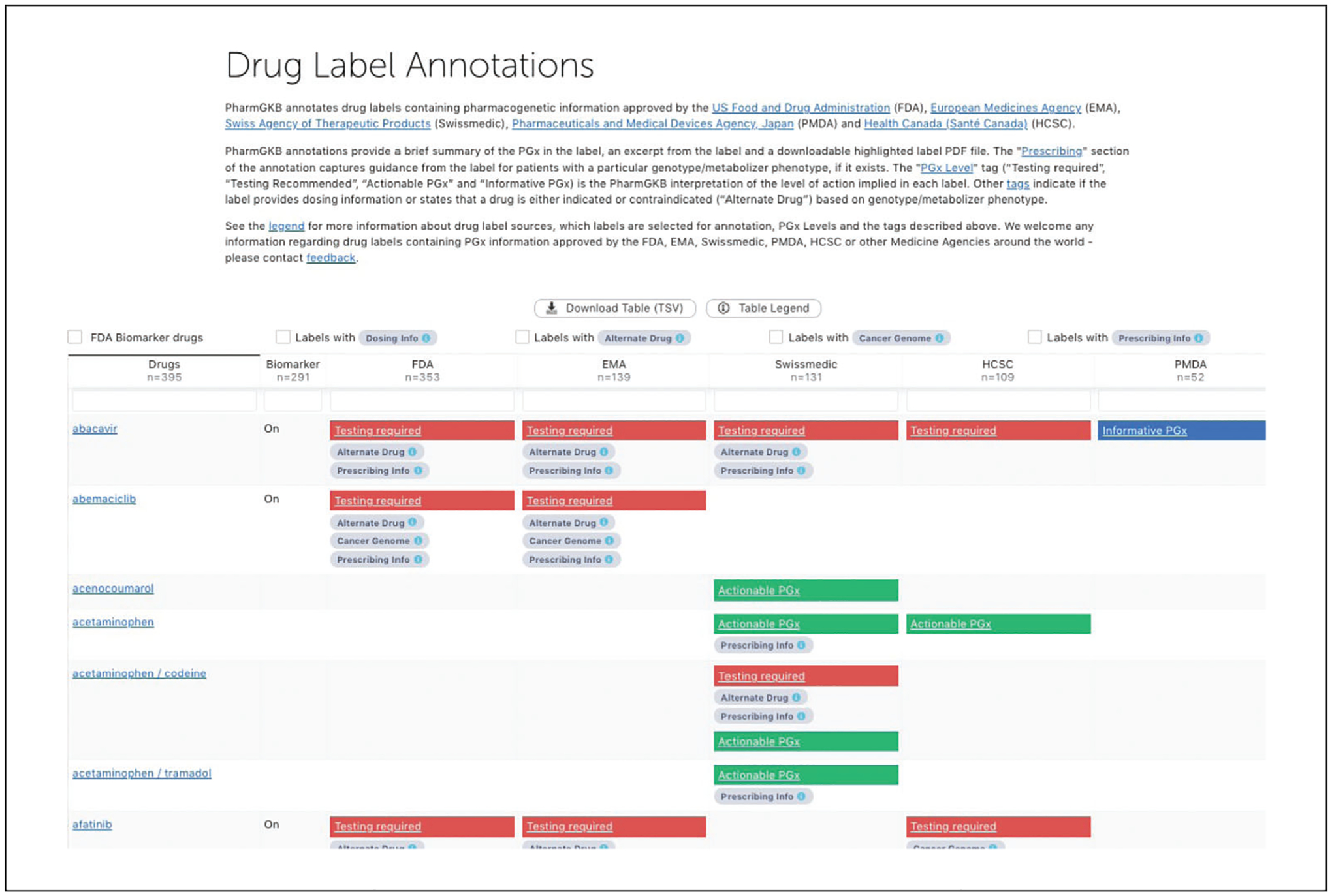
Overview of drug label annotations.
Figure 3.
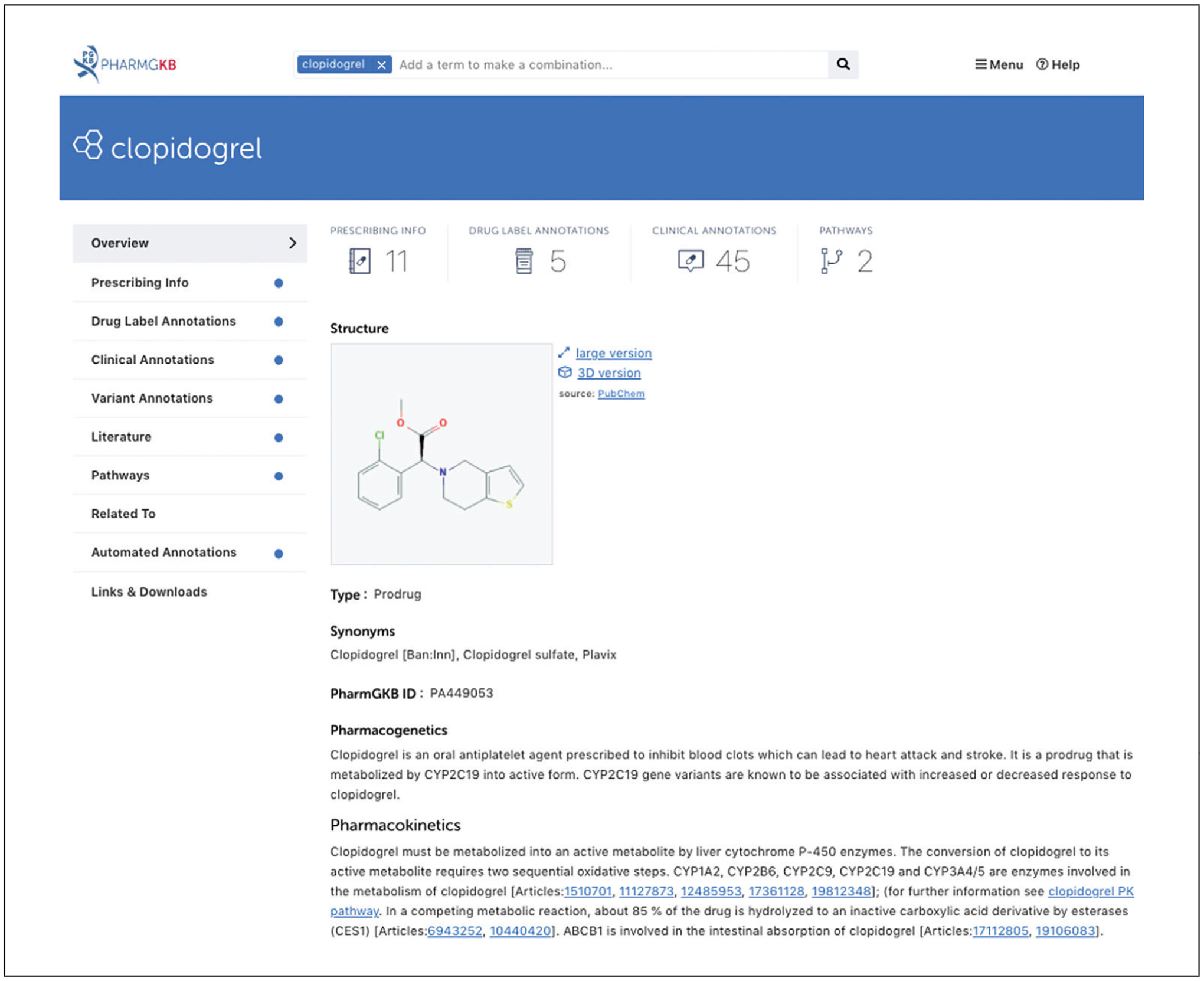
PharmGKB drug page for clopidogrel.
Figure 4.
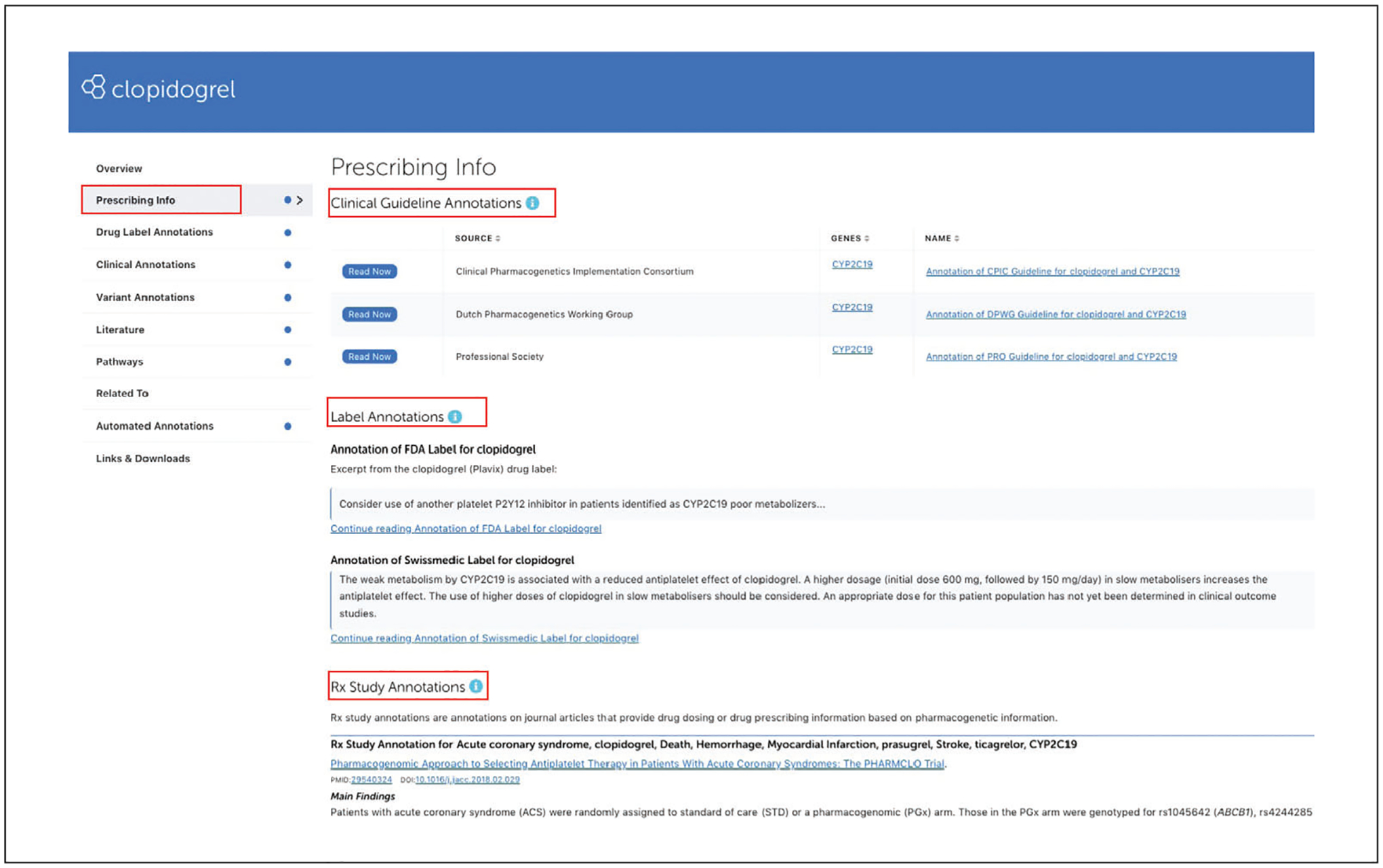
PharmGKB Prescribing info tab under the drug page for clopidogrel.
Figure 5.
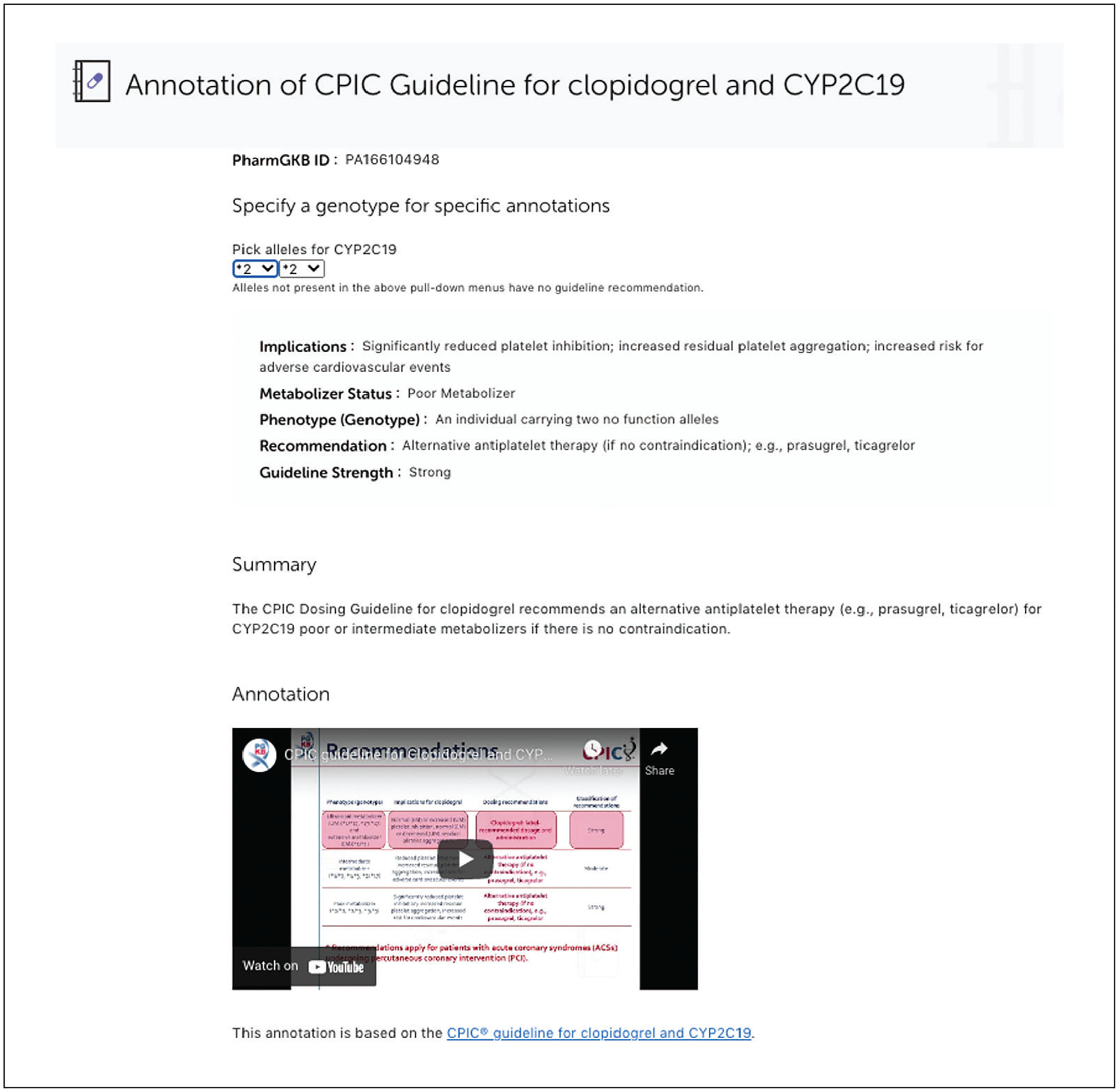
Example of PharmGKB clinical dosing guideline: The Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for clopidogrel and CYP2C19.
Figure 6.
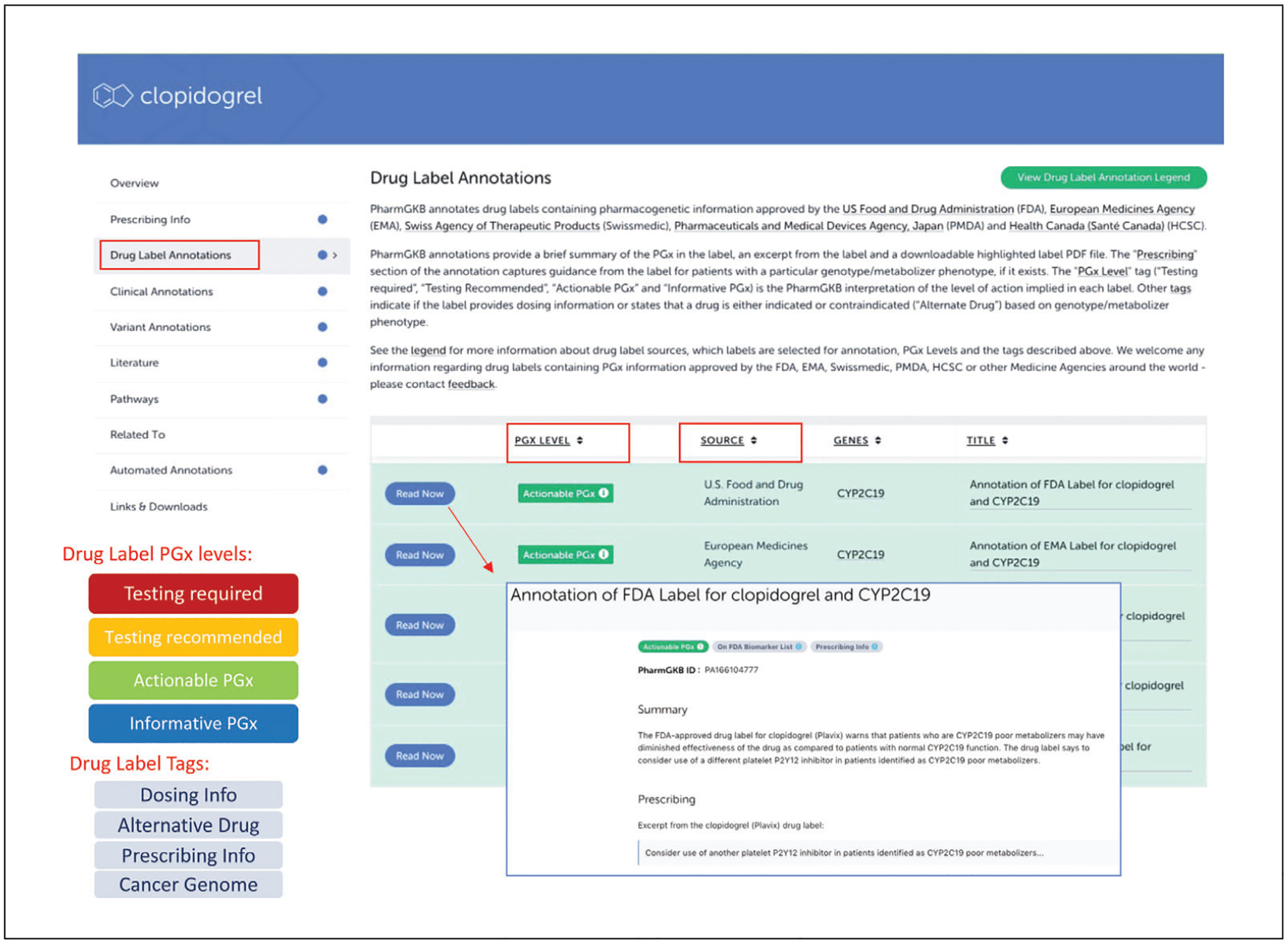
An overview of the annotated drug labels for clopidogrel on the PharmGKB.
Figure 7.
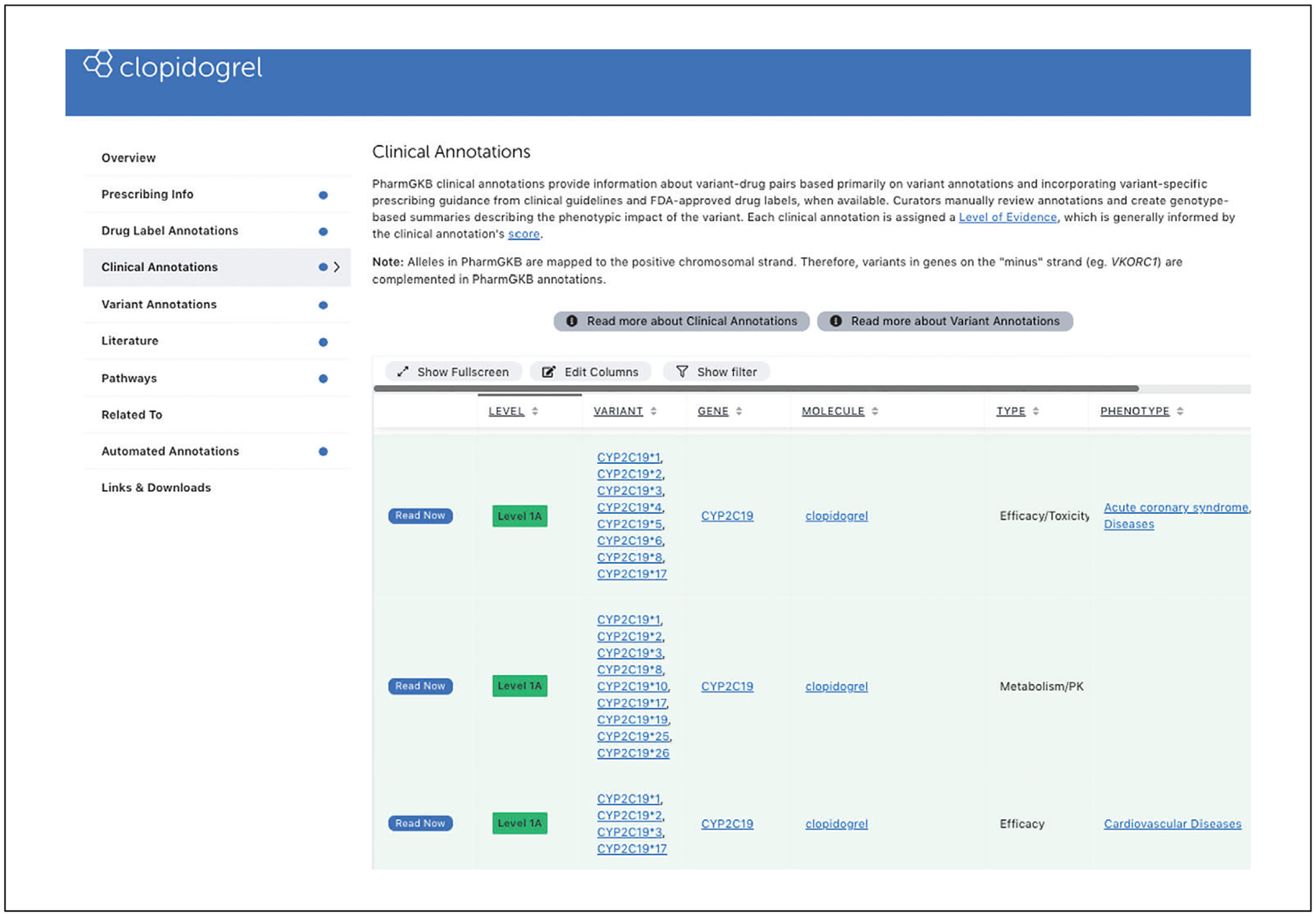
Example of PharmGKB clinical annotation for clopidogrel.
Figure 8.
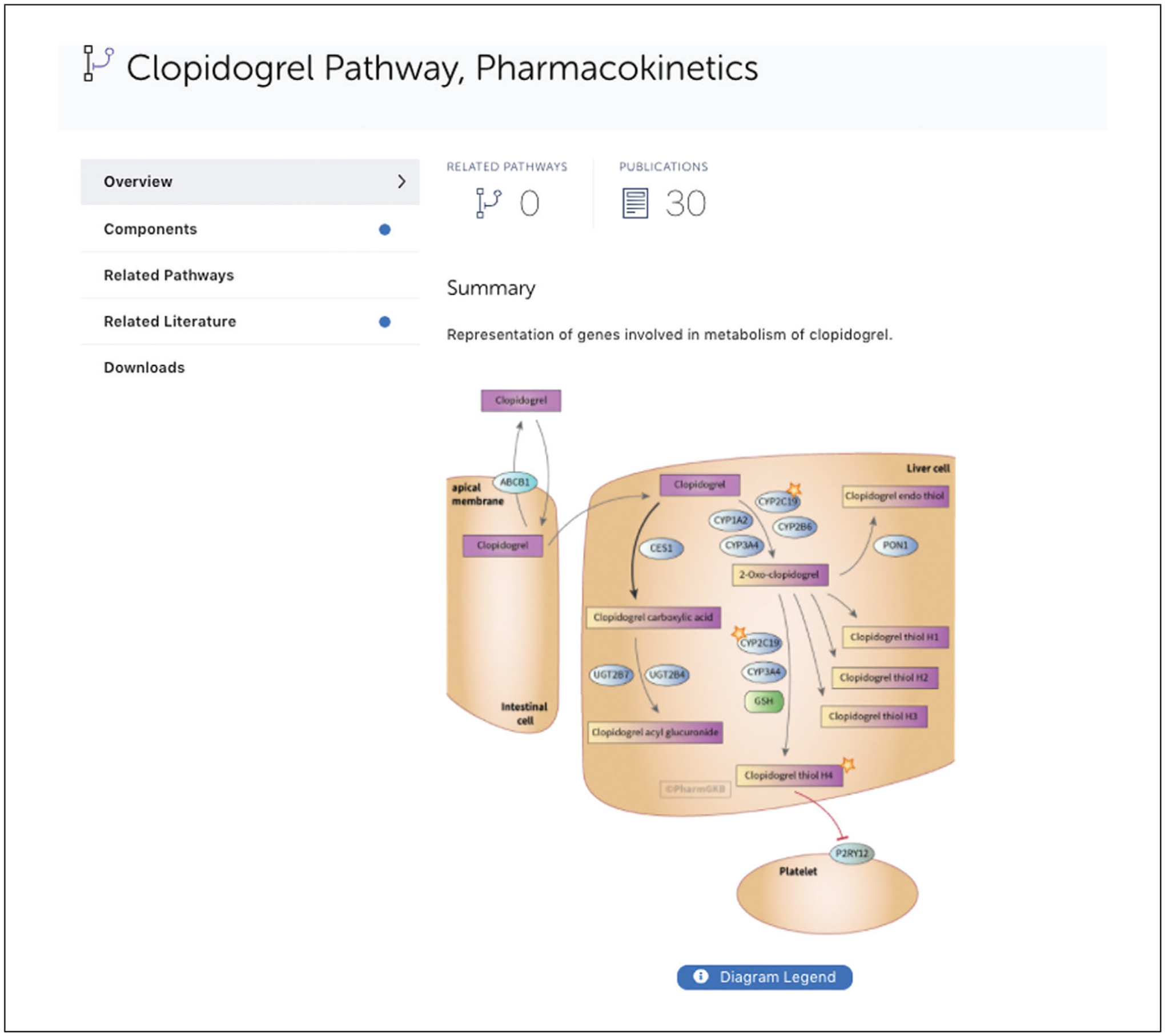
Example of PharmGKB drug-centered pathway for clopidogrel, pharmacokinetics.
Figure 9.
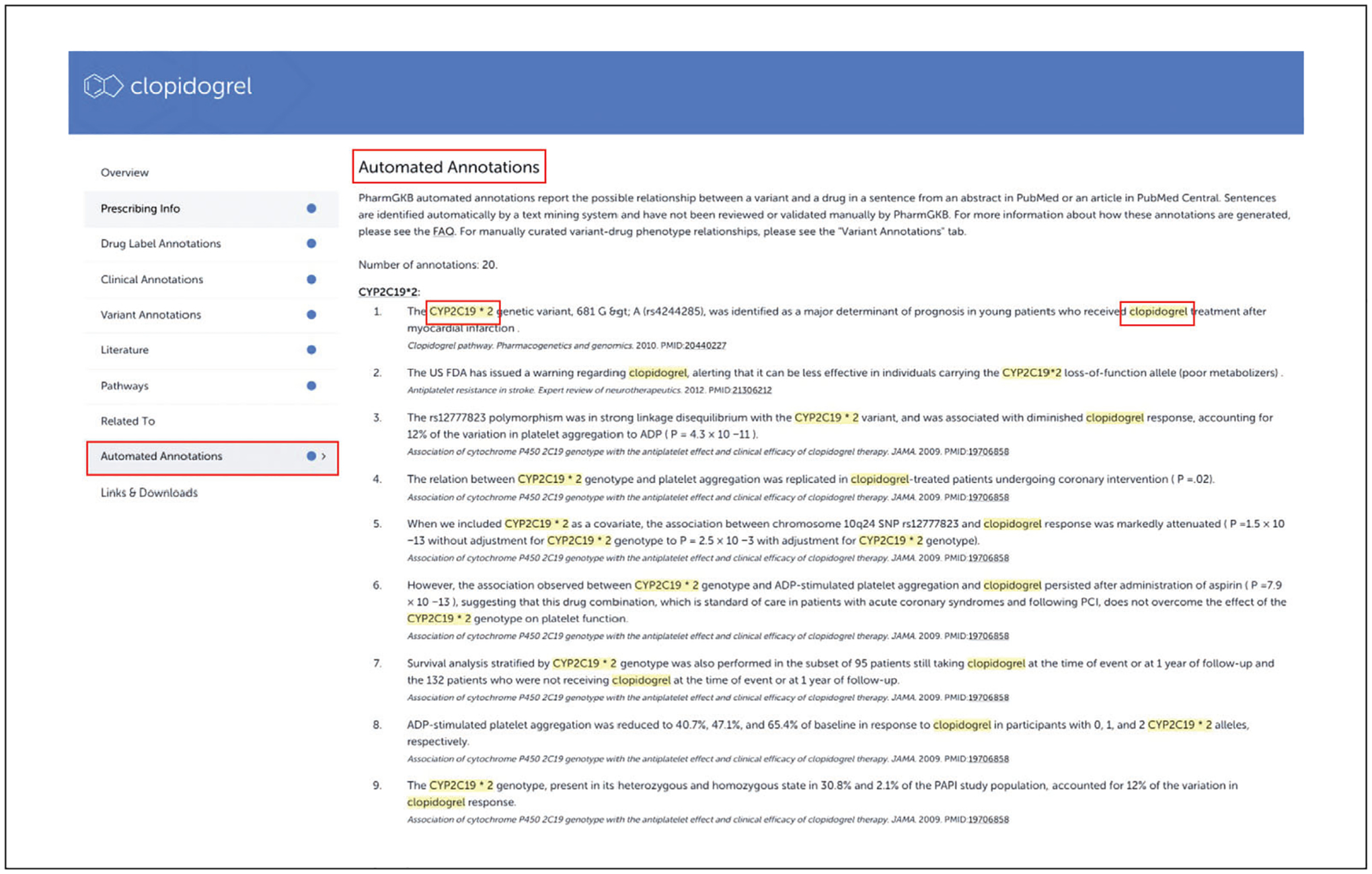
Automated annotations for clopidogrel using PGxMine.
BASIC PROTOCOL 2
SEARCHING BY VARIANT/HAPLOTYPE TO FACILITATE UNDERSTANDING OF THE IMPACT OF A GENETIC VARIATION ON DRUG RESPONSE
Direct-to-consumer genetic testing has become more widely available, and many people may have their genetic testing results in hand or have previously had their genome sequenced. The interpretation of genotyping results remains a challenge as more and more new evidence emerges. Many variants included in the tests on the market have varying levels of evidence to support their clinical relevance. Some may have an abundance of consistent clinical findings to support the claim, while others may either lack clinical data backing the claim or have disagreements about the role of the genetic variants contributing to a drug response phenotype. This protocol will introduce the basic techniques used for searching PharmGKB with a specific variant/haplotype to find out how it may impact the function of the gene and which drug therapies might be affected, as well as the level of confidence for a specific drug-variant association. We will use genotype calls on a specific SNP (rs4149056) and a diplotype call on a star allele (e.g., CYP2C9*3/*3) as examples. We will also demonstrate how to perform searches using combination of search terms.
Necessary Resources
Hardware
Computer with an internet connection
Software
An up-to-date Web browser including Google Chrome, Firefox, or Safari
Files
No input files required
Go to the PharmGKB homepage at http://www.pharmgkb.org in a web browser.
-
Enter variant ID rs41419056 in the search box. Open the variant page for “rs4149056.”
PharmGKB drug, gene, disease, and variant pages are structured similarly, with the navigation pane on the left. The “Overview” tab includes information on associated genes, aliases, and genome locations on human genome assembly GRCh38 and 37, as well allele frequencies.
Rs4149056 (c.T521C, p.V174A) is a nonsynonymous polymorphism in the SLCO1B1 gene, which encodes the organic anion–transporting polypeptide OATP1B1 that mediates the hepatic uptake of various drugs, including most statins and statin acids. This variant markedly decreases OATP1B1 function. Studies show that patients carrying the variant allele (C) tend to have higher circulating concentrations of drugs metabolized by OATP1B1 than patients carrying two reference alleles (TT). Many statins are substrates of OATP1B1, but to different degrees, which partially contributes to the varying responses to different statins in patients carrying the rs4149056 variant allele.
-
Click on the “Prescribing Info” tab from the navigation pane on the left to access dosing guidelines or drug labels that have specific prescribing information related to this variant.
The prescribing information may come from clinical guidelines for how to adjust treatment of certain medications based on a person’s genetic information, or drug labels that have prescribing recommendations based on a particular genotype/metabolizer phenotype. For “rs4149056,” you can find dosing guidance (https://www.pharmgkb.org/variant/PA166154579/guidelineAnnotation) based on rs4149056 from multiple clinical dosing guidelines (e.g., CPIC, DPWG and PRO) and drug labels for multiple statins.
-
Click on the “Clinical Annotations” tab from the navigation pane on the left to access clinical annotations associated with this variant. This table provides a good overall idea of the list of drugs that might be impacted by this genetic variation, whether the variation may influence the efficacy, toxicity or metabolism of the drug, and how strong the level of evidence is. The clinical annotation table can be sorted and/or filtered by evidence level, variant, gene, drug, or phenotype category. More information about the criteria for level of evidence can be accessed by clicking on the “i” next to the evidence level.
Click on the funnel icon “show filter” to enable the filter option for the clinical annotation table. To view the list of toxicity-related clinical annotations for this variant for statins, enter whole or part of word toxicity under column “type,” and statin under “Molecule” (Fig. 10).
Clinical annotations provide a summary for the association between a particular variant and drug, and are based on aggregated evidence from multiple variant annotations, as well as actionable clinical guidelines and drug labels with variant-specific recommendations. PharmGKB scientific curators manually review the evidence to create genotypeor haplotype-based summaries describing the phenotypic impact of the variant(s). Each clinical annotation is assigned a level of evidence (LOE) for the strength of these assertions. As new evidence emerges and is added to the clinical annotation, the level of evidence is reassessed. The level of evidence is clearly displayed, and users can click on the ‘i’ icon to be taken to the page explaining the clinical annotation levels of evidence. Clinical annotations are also tagged with the relevant drug, phenotype, and broad phenotypic categories of toxicity, dosage, efficacy, or metabolism/PK. A disclaimer is included in every clinical annotation description to highlight that other genetic and clinical factors may affect this association.
Clinical annotations fall into two primary categories: variant-level clinical annotations written on a single genetic locus (e.g., an rsID, as is the case for rs4149056), and gene-level clinical annotations written on haplotypes or star (*) alleles of a gene (e.g., CYP2C9*3, as demonstrated in step 9).
The clinical annotation search result for rs4149056 demonstrates that this variant is associated with statin-induced toxicity for multiple statins. However, the levels of evidence differ among them. The association with simvastatin and atorvastatin is currently at level 1A (score ≥80), with simvastatin associated with both CPIC and DPWG dosing guide lines and atorvastatin with the DPWG guidelines, suggesting a high level of evidence for the association. The association with other statins, e.g., pravastatin, fluvastatin, and rosuvastatin, is currently at level 3 (score -3-8), with conflicting evidence being reported from different studies, indicating a lower level of evidence for the association.
To find out more details about how we curate clinical annotations and the criteria for level of evidence, please check out https://www.pharmgkb.org/page/clinAnnLevels.
Click on the “Read Now” button to access the variant-level clinical annotation for rs4149056 and simvastatin-related toxicity (Fig. 11).
-
Pharmacogenomic variants in the cytochrome P450 genes are often referred to as * (star) alleles. To find pharmacogenomic information related to a particular genotype or diplotype on * alleles, enter the haplotype name CYP2C9*3 in the search box.
To find pharmacogenomic information related to a particular genotype or diplotype, users can either use the gene symbol or haplotype name in the search box to start.
-
Click on “Prescribing Info” tab in the left navigation pane to pull up drug-prescribing information related to CYP2C9*3.
Currently, there are 14 clinical dosing guideline annotations for gene CYP2C9 from CPIC, DPWG, and other professional societies. There are also FDA and EMA drug labels that specifically talk about possible changes of dose or therapy based on genotypes containing CYP2C9*3.
To find the metabolizer phenotype status for CYP2C9*3/*3, open a CPIC guideline annotation page for CYP2C9. PharmGKB enhances the CPIC guidelines by providing an interactive allele picker that allows specific alleles/genotypes to be selected from pull-down menus, and then provides the resulting inferred phenotype, associated dosing advice, and strength of the recommendation. Click on “Read Now” for “Annotation of CPIC Guideline for celecoxib, flurbiprofen, ibuprofen, lornoxicam and CYP2C9”. Enter “*3” “*3” under “Pick allele for CYP2C9”; this will retrieve inferred metabolizer status, as well as implications and recommendations for CYP2C9*3/*3 and NSAIDs drugs celecoxib, flurbiprofen, ibuprofen, and lornoxicam (Fig. 12).
Going back to the CYP2C9*3 page, click on “Clinical Annotation” in the left navigation pane to pull up the list of clinical annotations associated with CYP2C9*3. Click on the “Read Now” button to read gene-level clinical annotations written on haplotypes or star alleles of a gene (Fig. 13). Gene-level clinical annotations include phenotype descriptions for all alleles referenced by the supporting variant annotations. Allele functional status assigned by CPIC may also be displayed next to the alleles if available.
To conduct a combination search for a gene/variant AND a drug, first enter one term (e.g., VKORC1) in the search box and open that page. When on the page of the first term, enter the second term in the search box (e.g., warfarin) and hit Return. This will filter search results for items that contain both search terms. Please note that search terms on PharmGKB are primarily genes, variants, drugs, and diseases (or phenotypes). Click on “clinical annotation” to open a page containing all clinical annotations for the warfarin and VKORC1 combination (Fig. 14).
Figure 10.
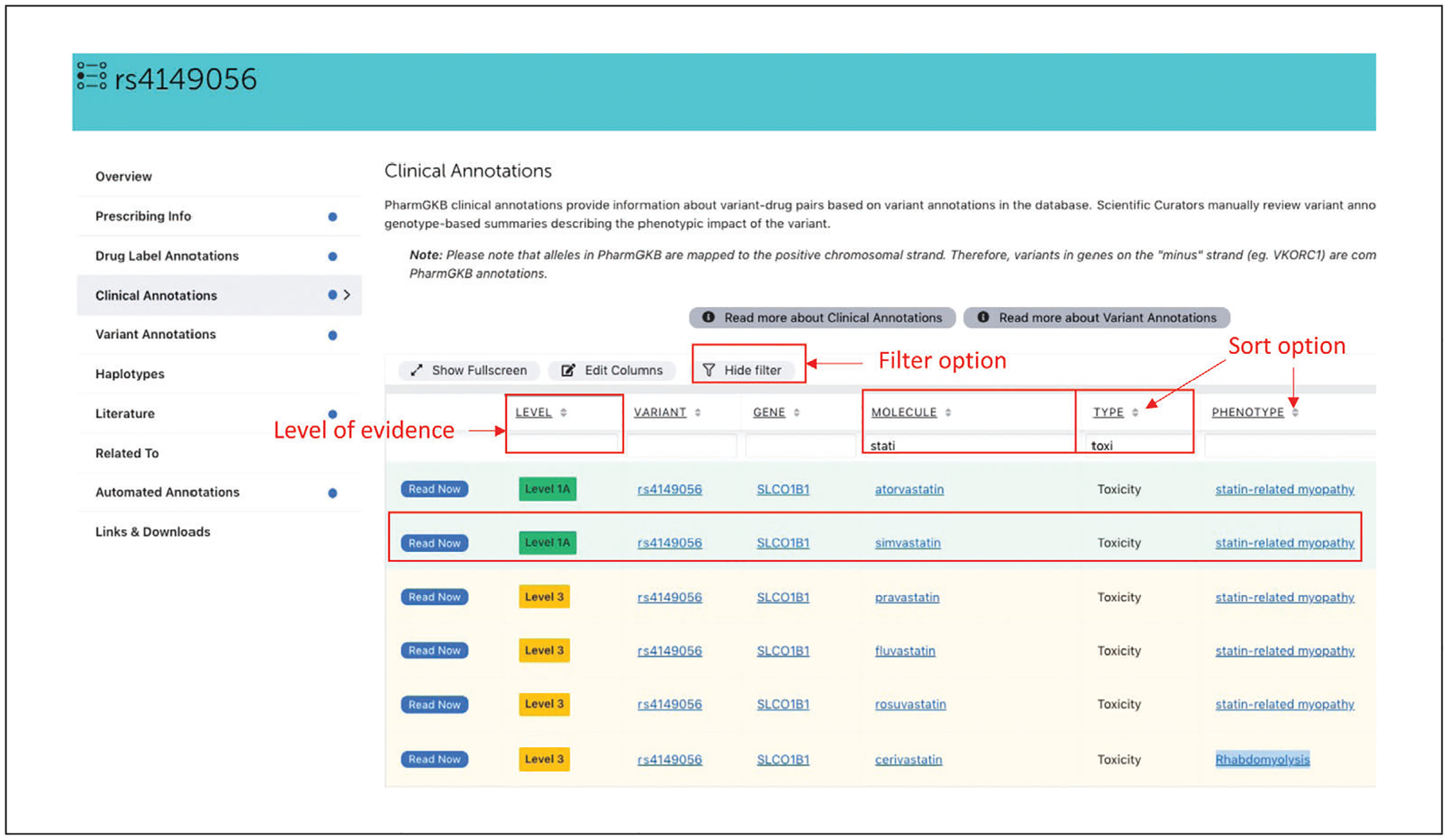
List of clinical annotations related to rs4149056 and statin-induced myopathy.
Figure 11.
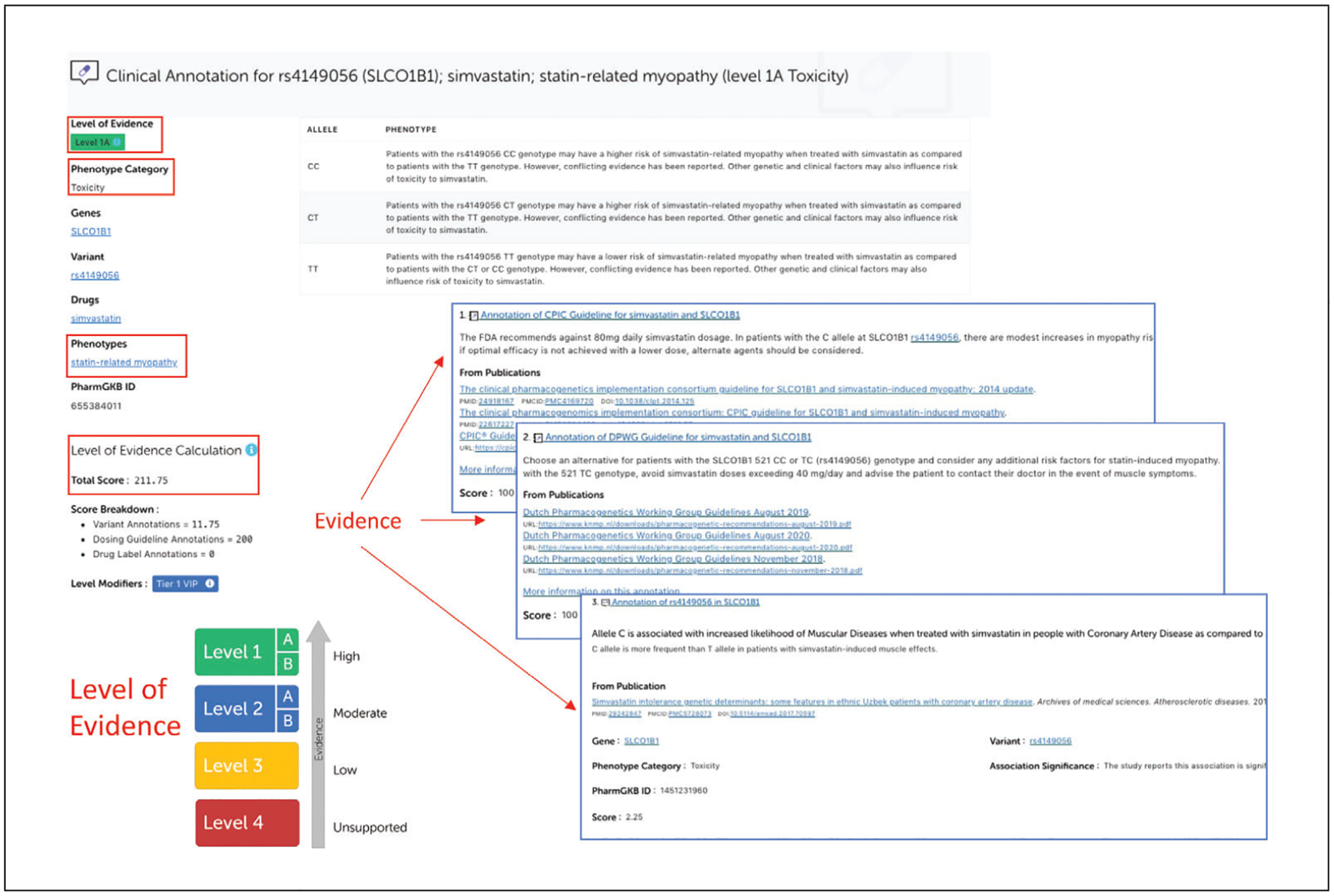
Example of variant-level clinical annotation for rs4149056 and simvastatin-induced myotoxcity.
Figure 12.
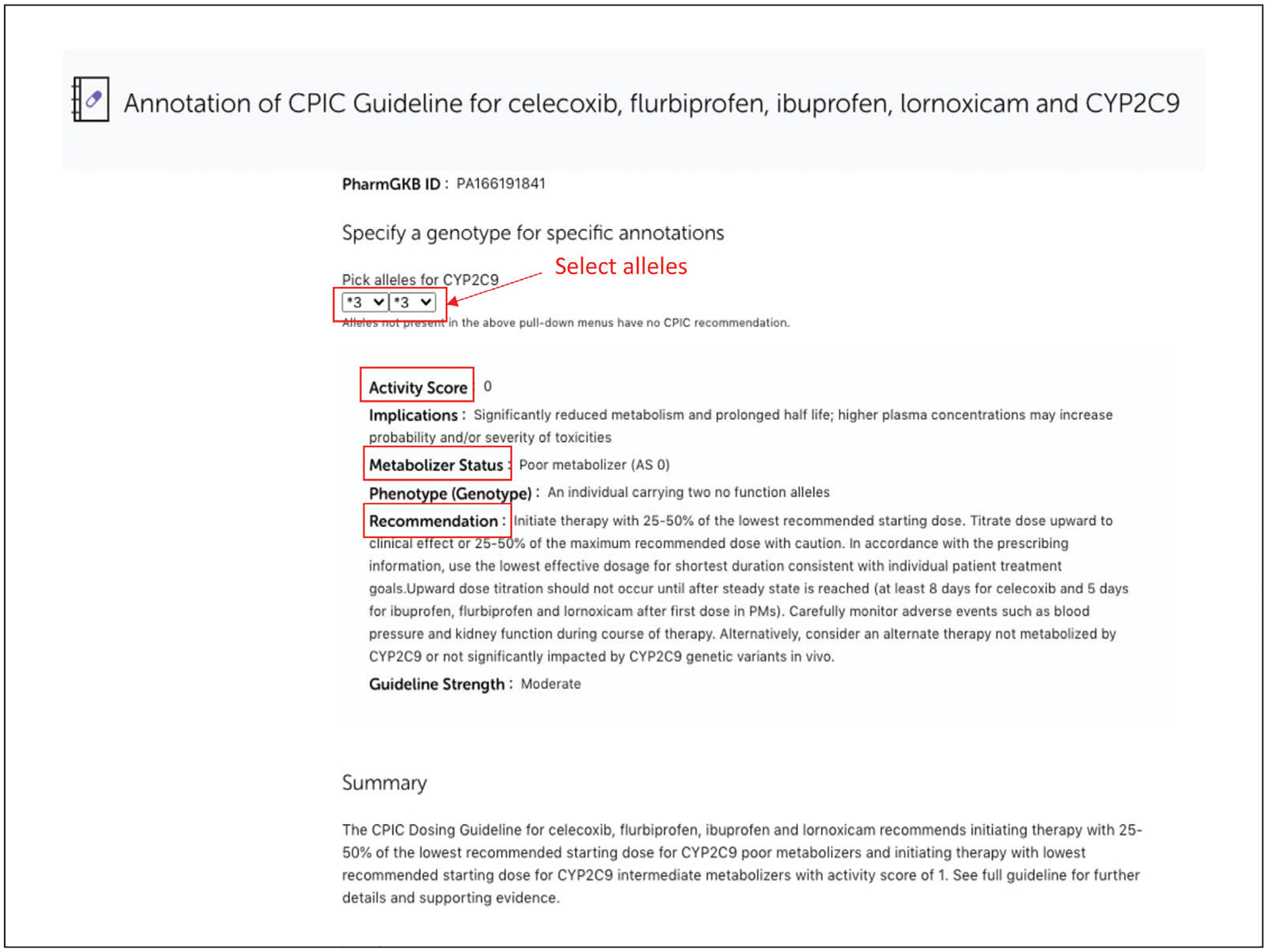
Example of PharmGKB interactive dosing table for a CPIC guideline annotation.
Figure 13.
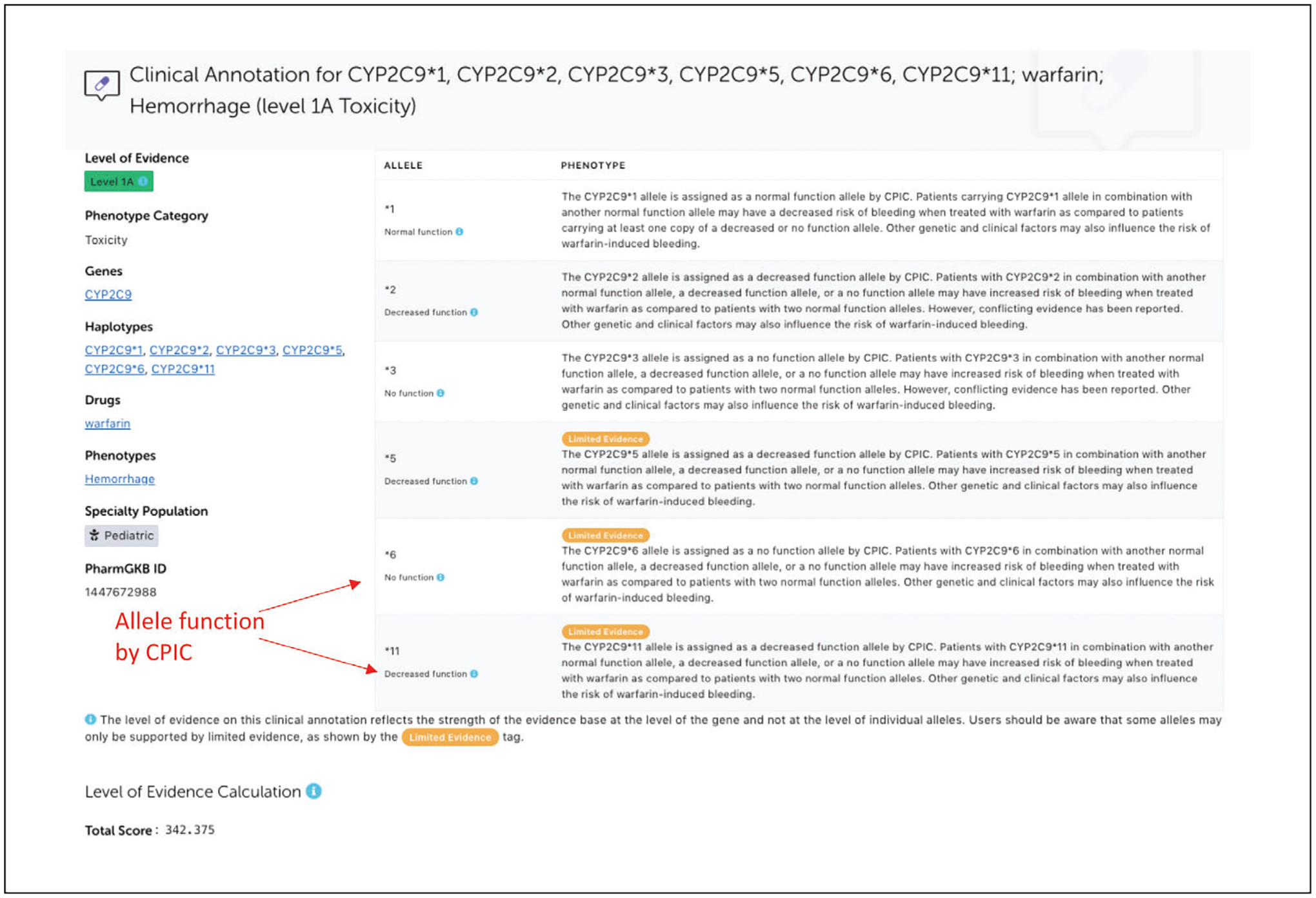
Example of gene-level clinical annotation for CYP2C9 and warfarin-related toxicity.
Figure 14.
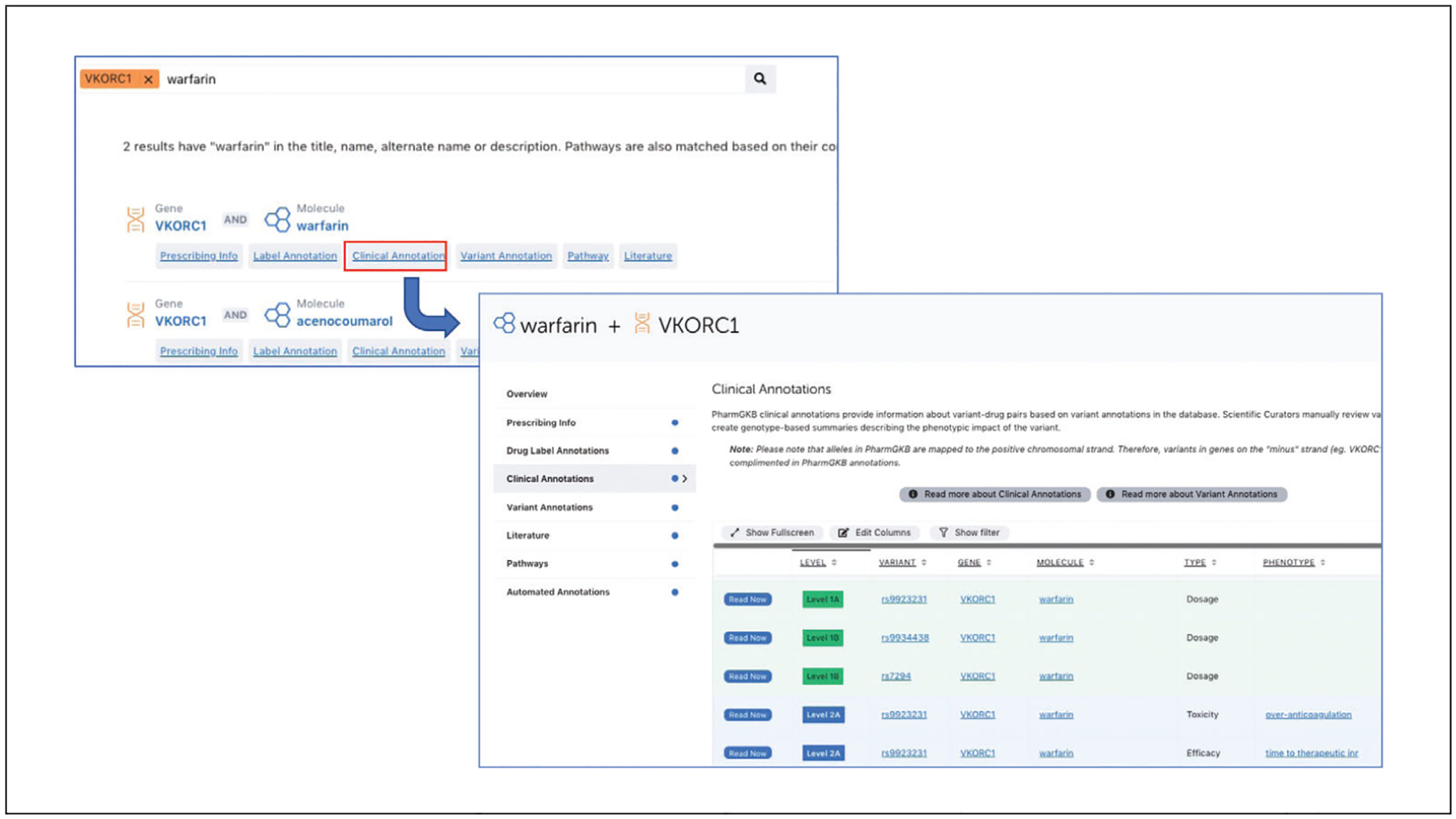
Combination search with a gene and a drug.
COMMENTARY
Background Information
PharmGKB began as the central data repository for the Pharmacogenetics Research Network (PGRN) and scientific community at large in 2000 (Giacomini et al., 2007; Long, 2007). It is designed to be a publicly available knowledge base with scientifically documented information connecting drug response phenotypes to genotypes. Over the past 20 years of development, PharmGKB has become a critical infrastructure for precision medicine. We have developed a comprehensive catalog of genes and genetic variations essential to drug response phenotypes. Our clinical and variant annotations are manually curated from peer-reviewed publications, linked to their evidence of support, and tagged with systematic ratings for the level of evidence. PharmGKB continuously summarizes important pharmacogenomic genes, drug-centered pathways, and associations between genes, drugs, and diseases. In addition to serving the research community, PharmGKB has greatly expanded PGx-related prescribing information to facilitate clinical implementation. As of April 2021, we have annotated 165 clinical dosing guidelines and 784 drug labels from the U.S., Europe, Switzerland, Japan, and Canada. PharmGKB also works with PGx consortia to examine important questions in pharmacogenomics. PharmGKB currently houses variant data associated with more than 1700 genes and 700 drugs, as well as greater than 9000 manually curated literature annotations, 153 drug-centered pathways of pharmacokinetics (PK) and pharmacodynam ics (PD), and 68 VIP gene summaries. Our comprehensive database makes it easier and faster for our diverse users to access key pharmacogenomic information without repeating searches in separate sources.
Manual expert curation has always been the gold standard in curating scientific literature. However, advances in technology have led to the exponential growth of literature, and it has been a huge challenge for manual curation to keep up with the rapid increase in information. There is an indisputable need for computational approaches to assist the curation process. At PharmGKB, we have been actively exploring natural language processing (NLP)/text mining approaches to help with the curation process to improve accuracy, coverage, and productivity. We have published multiple papers on identifying and extracting pharmacogenomic concepts and relationships from full text (Coulet, Shah, Garten, Musen, & Altman, 2010; Garten & Altman, 2009; Garten, Coulet, & Altman, 2010; Garten, Tatonetti, & Altman, 2010); the PharmGKB database has also been used repeatedly as the gold standard for evaluation of various text-mining tools in biomedical research (Guin et al., 2019; Mahmood et al., 2017; Monnin et al., 2019; Pakhomov et al., 2012; Ravikumar, Wagholikar, & Liu, 2014; Yang & Zhao, 2019). More recently, we have developed a supervised machine learning pipeline (PGxMine) to computationally extract possible variant-drug relationships from abstracts in PubMed or full-text articles in PubMed Central (Lever et al., 2020). PGxMine uses a text alignment process to identify mentions of drugs, genes, and variants in the text from data sources such as PubMed and PubTator Central. A supervised classifier is then trained and applied to all sentences accessible from PubMed abstracts and PubMed Central full-text papers to extract drug-variant associations. PGxMine identified more than 20,000 mentions of pharmacogenomic associations across 7170 papers and over 400 drugs. We have integrated the results of PGxMine into the PharmGKB knowledge base (as shown in Basic Protocol 1, step 11) and are currently developing tools to help triage papers for manual curation.
Despite challenges in clinical adoption, pharmacogenomics has been slowly incorporated in various clinical settings (Burt & Dhillon, 2013; Weng, Zhang, Peng, & Huang, 2013). To meet the increasing need of pharmacogenomics information from the clinical community, we have started developing a couple of domain-specific portals with specific clinical interests (e.g., cancer and COVID-19). Our “Cancer pharmacogenomics” portal (https://www.pharmgkb.org/page/cancerPgx) provides quick links to the important pharmacogenes for cancer research and cancer drug pathways illustrating the pharmacokinetics and mechanisms of action of the drug. It also lists cancer-related drug labels where pharmacogenomic biomarkers are used in diagnostic tests to select treatment or are predictive of response to treatment. Our portal for “Therapeutic Resource for COVID-19” (https://www.pharmgkb.org/page/COVID) contains annotation of therapeutic drugs from clinical trials and adjuvant therapies linking to associated genes and variants. This resource aims to assemble possible pharmacogenomics considerations that may impact drug choice for either the treatment of COVID-19 or the use of adjuvant therapies, including possible changes in drug metabolism or efficacy, as well as implied risks of side effects or drug-drug interactions. Given the rapidly changing landscape of COVID-19 research and several large national COVID-19 trials currently evaluating pharmacogenomics, we are constantly updating this resource as new information comes to our attention.
PharmGKB has forged strategic relationships with many key stakeholders of precision medicine, e.g., CPIC, ClinGen, ClinVar, PharmVar, ACMG, U-PGx, Emerge/Ignite, and the FDA. In August 2020, PharmGKB and CPIC have formalized a partnership with ClinGen to bring PGx expertise to a resource that defines the clinical relevance of genes and variants in the human genome. As of February 2021, 130 PGx genes curated by PharmGKB and/or CPIC are listed on the ClinGen website, with backlinks to the PharmGKB and CPIC websites for more detailed information. ClinGen displays all gene-drug pairs from PharmGKB with Level 1 and 2 clinical annotations, along with links to the relevant PharmGKB drug page and all PharmGKB clinical annotations for that gene-drug pair. We have also initiated the process of obtaining FDA recognition for PharmGKB level 1 and 2 clinical annotations under the FDA recognition of genomic databases.
Pharmacogenomics is a rapidly evolving field with many unmet challenges in translating the scientific findings in pharmacogenomics to clinical practice. However, the increasing understanding of how a person’s genetic makeup can influence his or her response to drugs provides the opportunity to improve the drug development process and deliver more effective and safer therapy for individual patients. As the largest PGx knowledge repository providing free and stable access to comprehensive information on the impacts of human genetic variation on drug responses, PharmGKB will continue to work closely with the PGx research and medical communities to catalyze scientific discoveries and clinical implementations, as well as empower public engagement with their genetic information.
Critical Parameters and Troubleshooting
PharmGKB is designed to be a valuable resource for both expert researchers in the pharmacogenomics field, as well as for novice users and the general public. PharmGKB’s homepage prominently displays the information that our users are looking for most frequently with a distinct icon system to represent different data types and knowledge. Typical searches conducted at PharmGKB are for information about drugs, genes, variants, diseases, and pathways. Searches are normally conducted using the general search box. If too many search results are returned, users can narrow the search with more specific terms or add another search term (e.g., variant + drug name) in a combination search, as demonstrated in Basic Protocol 2. If the user encounters difficulties in finding information of interest, using alternative names or partial names, or loosening of the search criteria is suggested. Please note that not all genes, chemicals, and variants in PharmGKB are associated with annotations. If a gene, chemical, or variant is not annotated in PharmGKB, it indicates that we currently have no pharmacogenomic information linked to that object. Users can also use bulk downloadable files under the download tab to filter the data to see exactly which genes, chemicals, and variants have been annotated in PharmGKB. If users have difficulties with finding information on PharmGKB, the best way to resolve the problem is to send questions and concerns to feed-back@pharmgkb.org. Our scientific staff will typically respond to the inquiry within 48 hr.
Understanding Results
The protocols described in this article are designed to give users a broad overview of the content and capability of PharmGKB. In the basic protocols, we demonstrate how to use a simple query on a drug or a variant to retrieve relevant annotations on how the genetic variant might influence response to the drug. It is worth noting that this protocol did not cover every aspect of the PharmGKB search capability. A more detailed instruction on how to perform refined searches in PharmGKB can be accessed at https://www.pharmgkb.org/page/searchingPharmgkb.
Primary pharmacogenomics literature is the cornerstone of our curation efforts. PharmGKB curators routinely scan a set of high-impact pharmacogenomics journals and annotate drug-variant associations to form variant annotations. A variant annotation is a single sentence that describes a single finding from a single publication. PharmGKB curators will also add information about the study to the variant annotation (e.g., study type, sample size, ethnicity, and statistical significance). Please note that findings are reported exactly as they are presented in the original paper; we do not carry out any data interpretation on variant annotations. Variant annotations also document published reports of no association found between a variant and drug. Variant annotations are systematically scored in a five-step process based on study attributes and aggregated into clinical annotations which are assigned a level of evidence (LOE) based on a sum of scores from all supporting evidence. All clinical annotations will have at least one variant annotation as supporting evidence, and may also include variant-specific prescribing information from clinical dosing guidelines and FDA drug labels. Each piece of evidence will receive a score based on multiple factors. The score for each clinical annotation is based on the sum of the scores for each supporting variant annotation, guideline, and drug label. This score informs the assignment of an LOE to the clinical annotation, and indicates the relative strength of the supporting evidence for the assertion. This systematic and objective process ensures consistent levels of evidence assignment based on curated literature across curators, and enables adjusted scoring and LOE assignment as new evidence is added to clinical annotations. Sometimes conflicting findings are reported for the same drug-variant associations; these are noted in our clinical annotation when we summarize the findings, and the inconstant evidence will decrease the overall score for the clinical annotation. The scoring of the variant annotations is not a judgement of study quality; it is a metric used by PharmGKB when aggregating variant annotations to create and update clinical annotations.
Clinically relevant/actionable pharmacogenomic information is also curated from drug labels and dosing guidelines. PharmGKB curates dosing guidelines by extracting guideline excerpts for gene-drug pairs and linking them to the underlying evidence and the associated genes and drugs. Drug labels are annotated with the drug name, gene name, a short summary sentence, a longer description that includes excerpts from the label, and, for U.S. labels, whether or not the label is on the FDA’s Biomarker table (see Internet Resources) or is mentioned in the FDA’s Table of Pharmacogenetic Associations (also see Internet Resources). Occasionally, there are differences in the prescribing guidance from different clinical guidelines, or from different regulatory agencies around world. PharmGKB presents the information from various sources but leaves it to the user to decide what information to implement and how to prescribe medications. PharmGKB itself does not make clinical recommendations. We also do not advocate for the use of recommendations from one organization over another or recommend specific pharmacogenetic tests or testing companies. It is the responsibility of the user to assess all of the available guidance and come to their own conclusions.
PharmGKB is committed to adhere to the FAIR principles: Findable, Accessible, Interoperable, and Reusable (“FAIR principles”; see Internet Resources). These principles ensure that PharmGKB data is usable in a machine-readable format by other groups now and in the future. We import vocabularies, chemical structures, allele frequencies, and gene and variant location information from leading genomic and drug resources (e.g., NCBI, dbSNP, HGNC, Ensemble, gnomAD, PubChem etc.). We annotate data with standard vocabularies and ontologies (e.g., MeSH and RxNorm) that enable us to meet domain-relevant community standards. To support findability and interoperability, PharmGKB makes data available via API in JSON-LD, a lightweight Linked Data format that supports the creation of a network of standards-based, machine-readable data across websites. The PharmGKB API and data are freely available under a Creative Commons Attribution ShareAlike 4.0 International License and do–not require user registration. Any reuse or redistribution of our data should be made available under the same terms of this license. More information about our Data Usage Policy can be found at https://pharmgkb.org/page/dataUsagePolicy and about the Creative Commons license at https://creativecommons.org/licenses/by-sa/4.0/.
Time Considerations
The search function on PharmGKB normally returns results within a few seconds.
Acknowledgments
PharmGKB is supported by the NIH/NHGRI/NICHD (U24 HG010615) and managed at Stanford University. The authors thank the entire PharmGKB team (https://www.pharmgkb.org/about) that have contributed to the development of PharmGKB.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Data Availability Statement
The data that support the findings of this study are openly available in PharmGKB at http://www.pharmgkb.org.
Literature Cited
- Burt T, & Dhillon S (2013). Pharmacogenomics in early-phase clinical development. Pharmacogenomics, 14(9), 1085–1097. doi: 10.2217/pgs.13.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulet A, Shah NH, Garten Y, Musen M, & Altman RB (2010). Using text to build semantic networks for pharmacogenomics. Journal of Biomedical Informatics, 43(6), 1009–1019. doi: 10.1016/j.jbi.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garten Y, & Altman RB (2009). Pharmspresso: A text mining tool for extraction of pharmacogenomic concepts and relationships from full text. BMC Bioinformatics, 10(Suppl 2), S6. doi: 10.1186/1471-2105-10-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garten Y, Coulet A, & Altman RB (2010). Recent progress in automatically extracting information from the pharmacogenomic literature. Pharmacogenomics, 11(10), 1467–1489. doi: 10.2217/pgs.10.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garten Y, Tatonetti NP, & Altman RB (2010). Improving the prediction of pharmacogenes using text-derived drug-gene relationships. Pacific Symposium on Biocomputing, 305–314. doi: 10.1142/9789814295291_0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomini KM, Brett CM, Altman RB, Benowitz NL, Dolan ME, Flockhart DA, … Pharmacogenetics Research Network. (2007). The pharmacogenetics research network: From SNP discovery to clinical drug response. Clinical Pharmacology and Therapeutics, 81(3), 328–345. doi: 10.1038/sj.clpt.6100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guin D, Rani J, Singh P, Grover S, Bora S, Talwar P, … Kukreti R (2019). Global text mining and development of pharmacogenomic knowledge resource for precision medicine. Frontiers in Pharmacology, 10, 839. doi: 10.3389/fphar.2019.00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein TE, & Altman RB (2004). PharmGKB: The pharmacogenetics and pharmacogenomics knowledge base. Pharmacogenomics Journal, 4(1), 1. doi: 10.1038/sj.tpj.6500230. [DOI] [PubMed] [Google Scholar]
- Lever J, Barbarino JM, Gong L, Huddart R, Sangkuhl K, Whaley R, … Altman RB (2020). PGxMine: Text mining for curation of PharmGKB. Pacific Symposium on Biocomputing, 25, 611–622. [PMC free article] [PubMed] [Google Scholar]
- Long RM (2007). Planning for a national effort to enable and accelerate discoveries in pharmacogenetics: The NIH Pharmacogenetics Research Network. Clinical Pharmacology and Therapeutics, 81(3), 450–454. doi: 10.1038/sj.clpt.6100099. [DOI] [PubMed] [Google Scholar]
- Mahmood A, Rao S, McGarvey P, Wu C, Madhavan S, & Vijay-Shanker K (2017). eGARD: Extracting associations between genomic anomalies and drug responses from text. PloS One, 12(12), e0189663. doi: 10.1371/journal.pone.0189663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnin P, Legrand J, Husson G, Ringot P, Tchechmedjiev A, Jonquet C, … Coulet A (2019). PGxO and PGxLOD: A reconciliation of pharmacogenomic knowledge of various provenances, enabling further comparison. BMC Bioinformatics, 20(Suppl 4), 139. doi: 10.1186/s12859-019-2693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakhomov S, McInnes BT, Lamba J, Liu Y, Melton GB, Ghodke Y, … Birnbaum AK (2012). Using PharmGKB to train text mining approaches for identifying potential gene targets for pharmacogenomic studies. Journal of Biomedical Informatics, 45(5), 862–869. doi: 10.1016/j.jbi.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar KE, Wagholikar KB, & Liu H (2014). Towards pathway curation through literature mining–a case study using PharmGKB. Pacific Symposium on Biocomputing, 352–363. Available at https://www.ncbi.nlm.nih.gov/pubmed/24297561. [PMC free article] [PubMed] [Google Scholar]
- Weng L, Zhang L, Peng Y, & Huang RS (2013). Pharmacogenetics and pharmacogenomics: A bridge to individualized cancer therapy. Pharmacogenomics, 14(3), 315–324. doi: 10.2217/pgs.12.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, … Klein TE (2012). Pharmacogenomics knowledge for personalized medicine. Clinical Pharmacology and Therapeutics, 92(4), 414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CC, & Zhao M (2019). Mining heterogeneous network for drug repositioning using phenotypic information extracted from social media and pharmaceutical databases. Artificial Intelligence in Medicine, 96, 80–92. doi: 10.1016/j.artmed.2019.03.003. [DOI] [PubMed] [Google Scholar]
Internet Resources
- https://www.pharmgkb.org/; PharmGKB: The PharmGKB is a pharmacogenomics knowledge resource that encompasses clinical information including clinical guidelines and drug labels, potentially clinically actionable gene-drug associations, and genotypephenotype relationships.
- https://cpicpgx.org/; CPIC: The Clinical Pharmacogenetics Implementation Consortium (CPIC®) is an international consortium of individual volunteers and a small dedicated staff who are interested in facilitating use of pharmacogenetic tests for patient care. CPIC creates and curates freely available, peer-reviewed, evidence-based, updatable, and detailed gene/drug clinical practice guidelines.
- https://www.knmp.nl/patientenzorg/medicatiebewaking/farmacogenetica/pharmacogenetics-1/pharmacogenetics.; DPWG: The Dutch Pharmacogenetics Working Group (DPWG) aims to develop pharmacogenetics-based therapeutic (dose) recommendations and to assist drug prescribers and pharmacists by integrating the recommendations into computerized systems for drug prescription and automated medication surveillance.
- https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling.; FDA table of PGx biomarkers in drug labeling: This table lists therapeutic products from Drugs@FDA with pharmacogenomic information found in the drug labeling.
- https://www.fda.gov/medical-devices/precision-medicine/table-pharmacogenetic-associations.; FDA table of pharmacogenetic associations: The FDA curated a table that lists pharmacogenetic associations that they have evaluated and believe there is sufficient scientific evidence to suggest that subgroups of patients with certain genetic variants, or genetic variant-inferred phenotypes (i.e., affected subgroup in the table below), are likely to have altered drug metabolism, and in certain cases, differential therapeutic effects, including differences in risks of adverse events.
- https://www.pharmvar.org/; PharmVar: The Pharmacogene Variation (PharmVar) Consortium is a central repository for pharmacogene (PGx) variation that focuses on haplotype structure and allelic variation.
- https://www.go-fair.org/fair-principles/; FAIR principles (2021).
- https://www.fda.gov/medical-devices/precision-medicine/table-pharmacogenetic-associations.; Table of Pharmacogenetic Associations (FDA).
- https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling.; Table of Pharmacogenomic Biomarkers in Drug Labeling (FDA).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in PharmGKB at http://www.pharmgkb.org.


