Abstract
A particular recurrent clonal type of Escherichia coli O157 has been isolated from multiple clinical, veterinary, food, and environmental sources throughout Scotland since 1989. Significant genotypic variation was detected among isolates from distinct outbreaks, with the presence or absence of single fragments being sufficient to delineate outbreak groups within the clonal type.
Since its first description in 1982, Escherichia coli O157:H7 has caused many major general outbreaks of infection worldwide including those in Japan (ca. 10,000 cases and 5 deaths) (7) and Scotland (496 cases and 20 deaths) (2).
Pulsed-field gel electrophoresis (PFGE) has been applied in investigations of several outbreaks of E. coli O157 infection (1, 2, 3). Systematic application of PFGE as a determinative method for E. coli O157 in the Scottish Reference Laboratory (SRL) (11) has, for the first time, enabled identification of recurrent clonal types defined according to the criteria of Orskov and Orskov (15). The most significant of these, termed the “West Lothian” (WL) clonal type, has been isolated from multiple veterinary (cattle and sheep), food (raw and cooked meat), environmental (dairy machinery and milk tanker), and clinical (>400 patients) sources throughout Scotland over the last 5 years. It has been responsible for several major outbreaks of infection, separate both temporally and geographically, including the largest milk-borne outbreak in the world (West Lothian, Scotland, 1994) and the most severe outbreak worldwide, the central Scotland outbreak (November 1996), which caused the deaths of 20 people.
The aim of this study was to further characterize isolates of the clonal type by enhanced fingerprinting in order to distinguish them.
Sources of bacterial isolates.
During the course of three major outbreaks caused by the putative recurrent clonal type, in excess of 600 isolates were analyzed by phenotypic and genotypic methods. The outbreaks occurred as follows: West Lothian, 1994, 100 cases; Highland, 1994, 8 cases; central Scotland, 1996, 496 cases. Isolates with numbers 318 to 340 inclusive were associated with the West Lothian outbreak, isolates with numbers 522 to 594 inclusive were associated with the Highland outbreak, and isolates with numbers 1476 to 1717 inclusive were associated with the central Scotland outbreak. In addition to isolates from the three outbreak groups, other representative phage type 2 isolates received by SRL (1992 to November 1996 [onset of central Scotland outbreak]) and November 1996 to March 1998) were similarly analyzed.
Standard typing and subtyping.
Isolates were characterized biochemically and serotyped as E. coli O157:H7 by standard microbiological methods. Phage typing was performed with the study group by the method of Khakria et al. (10). The supplementary phages were also included (R. Khakria, M. Mulvey, R. Ahmed, D. Woodward, and W. Johnson, Abstr. 3rd International Symposium and Workshop on Shiga Toxin (Verocytotoxin)-Producing Escherichia coli Infections, abstr. V124/I, 1997). All isolates were tested with primers specific for the genes encoding Verotoxin type 1 (VT1) and VT2 as described previously (18). All outbreak isolates were E. coli O157:H7, phage type 2, VT1 negative, and VT2 positive. The supplementary phages did not further discriminate the outbreak groups. Phage type 2 is common in Scotland, comprising 58.3% of the annual SRL sample load as a result of the central Scotland outbreak (1996–1997) and 18.5% of the annual sample load in 1997–1998 (21).
Enhanced strain fingerprinting.
Enhanced strain fingerprinting by several genotypic techniques was performed with a selected study group of 53 isolates from the three outbreaks caused by the phage type 2 WL clonal type. The isolates analyzed included an isolate from milk and a cattle isolate associated with the West Lothian outbreak and a food sample associated with the central Scotland outbreak.
PFGE was performed in a CHEF DR-II apparatus (Bio-Rad, Hemel Hempstead, United Kingdom) by the method of Krause et al. (11). Cleavage of the agarose-embedded DNA was achieved with XbaI (Promega, Southampton, United Kingdom), NotI (Promega), SfiI (Promega), XhoI (Boehringer Mannheim, Lewes, United Kingdom), and AvrII (Boehringer Mannheim) according to the manufacturer's instructions. Run times and pulse times were optimized for each enzyme, as follows: for XbaI, pulse times of 37 to 47 s for 22 h or, alternatively, 15 to 50 s for 22 h; for NotI, pulse times of 10 to 30 s for 23 h; for SfiI, pulse times of 15 to 50 s for 22 h; for XhoI, pulse times of 1 to 15 s for 18 h; and for AvrII, pulse times of 5 to 50 s for 22 h, all with linear ramping. To assess variations in the small (<150-kb)-fragment profiles, inserts containing DNA cleaved with XbaI were run at pulse times of 6 to 30 s for 29 h or with AvrII at a pulse times of 6.75 to 26.29 s for 30.46 h.
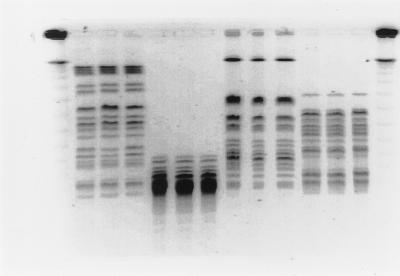
Systematic reference laboratory analyses have demonstrated that disparate isolates that share the same phage type (phage type 2) can be subdivided into at least 10 distinct pulsed-field groups by the standard PFGE method (XbaI digestion, pulse times of 37 to 47 s for 22 h) (1). During the course of the outbreaks SRL applied these run parameters in real-time analyses and found the macrorestriction patterns to be indistinguishable for isolates from each of the three outbreaks (Fig. 1). The patterns were stable during primary analysis and reproducible on subsequent repeated subculture. In this study the maximum period that elapsed between receipt of the isolate and the most recent subculture was 38 months.
FIG. 1.
PFGE profiles of genomic DNAs West Lothian, Highland, and central Scotland outbreak isolates cleaved with XbaI (lanes 2 to 4), XhoI (lanes 5 to 7), NotI (lanes 8 to 10), and SfiI (lanes 11 to 13). Lanes 1 and 14, bacteriophage lambda concatemer size markers. The unnumbered lanes correspond to lanes 1 to 14 from left to right, respectively.
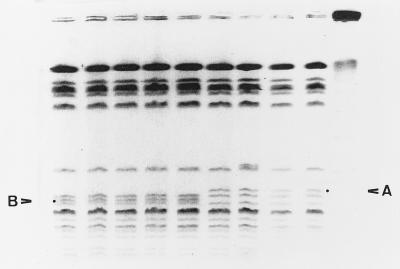
Indistinguishable profiles were also observed when genomic DNA was cleaved with XhoI, SfiI, and NotI (Fig. 1). When DNA was cleaved with AvrII and run with a pulse time of 5 to 50 s, differences were observed in the particular region of the macrorestriction profile where small fragments (<150 kb) are resolved. Electrophoretic conditions were altered (pulse times of 6.75 to 26.29 s for 30.46 h) to optimize separation of the small fragments in this region. It was then possible to distinguish the central Scotland outbreak isolates from other outbreak isolates by the presence of a triplet of fragments in the size range of 97 to 145 kb. Isolates from the Highland and West Lothian outbreaks also possessed a triplet, but the profiles varied by the absence of fragment A and the presence of fragment B (Fig. 2). The profile for isolate 1551 differed from the profiles for the rest of the central Scotland group by a single band (Fig. 2, lane 7).
FIG. 2.
PFGE profiles of AvrII-cleaved genomic DNAs from West Lothian (lanes 1 to 3), Highland (lanes 4 and 5), and central Scotland (lanes 6 to 9) outbreak isolates. Lane 10, bacteriophage lambda concatemer size markers. The fragment identifying central Scotland isolates is indicated by A. The fragment absent from central Scotland isolates but present in both West Lothian and Highland isolates is indicated by B. The unnumbered lanes correspond to lanes 1 to 10 from left to right, respectively.
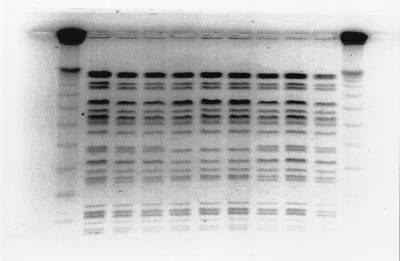
Following resolution of XbaI-cleaved fragments with parameters that optimize separation of fragments in the size range of 50 to 300 kb, a single band difference was observed in the isolates from the Highland outbreak (Fig. 3). The profiles for the isolates from the West Lothian and central Scotland outbreaks remained indistinguishable with the exception of that for isolate 340, whose profile was indistinguishable from those of the Highland outbreak isolates.
FIG. 3.
PFGE profiles of XbaI-cleaved genomic DNA from West Lothian (lanes 2 to 4), Highland (lanes 5 to 7), and central Scotland (lanes 8 to 10) outbreak isolates. Lanes 1 and 11, bacteriophage lambda concatemer size markers. The unnumbered lanes correspond to lanes 1 to 11 from left to right, respectively.
Similar PFGE analyses of a group of representative phage type 2 strains isolated either before or after the onset of the central Scotland outbreak demonstrated that the central Scotland outbreak clonal type, as identified by its macrorestriction profile, had been isolated on only one previous occasion in Scotland, from a single cluster of infection in a family in April 1995. From November 1996 to March 1998, however, the clonal type was isolated from 12 separate incidents (excluding the central Scotland outbreak) of infection from patients throughout Scotland.
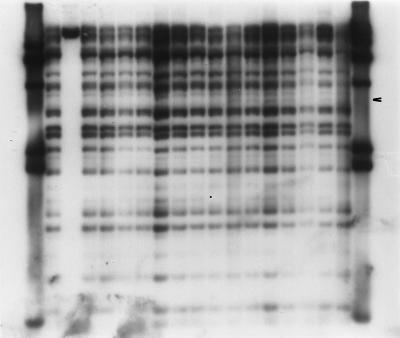
Bacteriophage lambda restriction fragment length polymorphism (RFLP) analysis was performed with purified genomic DNA (2 μg) cleaved with either PvuII or HindIII. The DNA fragments were separated, blotted, and probed with digoxigenin-labeled bacteriophage lambda (Bio-Rad) (16). Variations in RFLP profiles, which differentiated each of the three outbreak groups, were observed (Fig. 4). All central Scotland outbreak isolates possessed an extra band (denoted by an arrowhead in Fig. 4) not present in the other outbreak isolates.
FIG. 4.
Bacteriophage lambda RFLP analysis of PvuII-cleaved genomic DNA from West Lothian (lanes 2 to 10, and 18), Highland (lane 11), and central Scotland (lanes 12 to 17) outbreak isolates. Lanes 1 and 19, DNA molecular weight standards. The unnumbered lanes correspond to lanes 1 to 19 from left to right, respectively.
One particular fragment in the smaller size range was absent from all isolates from the Highland outbreak but was present in most isolates of the other outbreak groups (Fig. 4, lane 11; the missing band is denoted by a period). Two West Lothian isolates (isolates 325 and 340) were indistinguishable from the Highland isolates by this method. Isolate 325 could be differentiated from the Highland isolates by PFGE of XbaI-cleaved DNA, but isolate 340 could not. Similar experiments performed on HindIII-cleaved DNA demonstrated indistinguishable RFLP profiles for all isolates in all outbreak groups.
Supplementary genotypic methods were applied to further distinguish isolates. Insertion sequence analysis (IS1) was performed with purified DNA (2 μg) cleaved with RsaI (Boehringer Mannheim). The DNA fragments were separated electrophoretically in 1% (wt/vol) agarose in 0.5× TBE (Tris-borate-EDTA) and were transferred to a positively charged nylon membrane (20). The IS1 insertion sequence was amplified (12) and the product was labeled with digoxigenin (DIG-High Prime; Boehringer Mannheim). High-stringency hybridization and detection of fragments were carried out according to the manufacturer's instructions. Hybridization demonstrated that all isolates possessed a single hybridizing fragment that was identical among the isolates. Identical results were obtained with different restriction enzymes (data not shown).
A range of DNA amplification-based fingerprinting methods was also applied. All amplifications were performed on a DNA Thermocycler 480 (Perkin-Elmer Biosystems, Warrington, United Kingdom), and reaction mixtures contained 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 1.25 U of Taq DNA polymerase, buffer (Bioline), 2 ng of purified bacterial genomic DNA per μl (17), and 0.25 μM primers.
Arbitrarily primed PCR (AP-PCR) was performed with two primers selected for their superior discriminatory abilities: 5′-GTGGATGCGA (14) and AB7-10 (5′-AAGAGGCCAG; Applied Biotechnologies, Epsom, United Kingdom) (L. J. Allison and F. M. Thomson-Carter, unpublished data). Thermocycler reaction conditions were an initial cycle of 94°C for 5 min, then 30 cycles of 94°C for 1 min, 30°C for 1 min, and 72°C for 2 min, and then an extension cycle at 72°C for 5 min.
Enterobacterial repetitive intergenic consensus (ERIC) PCR with the standard ERIC primers (13) was performed. Amplification conditions were an initial cycle of 94°C for 5 min and then 34 cycles of 94°C for 1 min, 37°C for 1 min, and 72°C for 2 min, followed by 72°C for 5 min.
Amplification of the 16S-23S spacer regions was performed with primers described previously (5). Amplification conditions were an initial cycle of 95°C for 1 min and then 30 cycles at 94°C for 1 min and 70°C for 2 min. A final extension cycle of 70°C for 5 min was performed. From the known genome sequence of E. coli K-12 (4), the expected size range for PCR products would be 963 to 1,066 bp; the sizes of the products observed in these experiments fell within this range, and the products were subsequently cleaved with either HaeIII or TaqI.
Amplified fragment length polymorphism (AFLP) analysis was performed with EcoRI and MseI or TaqI. Ligation of adapters, preselective and selective amplification, and labeling of the fragments were performed essentially as described previously (8). Fluorescently labeled fragments were separated on an ABI 377 XL automated DNA sequencer running Genescan application software.
None of the PCR-based assays (AP-PCR, ERIC PCR, 16S-23S spacer region RFLP analysis, and AFLP analysis) could discriminate the outbreak groups. Additionally, all WL clonal type isolates possessed copies of the normal and the variant VT2 gene (22), as determined by cleavage of the PCR-amplified B subunit with HaeIII.
The aim of the study described here was to differentiate isolates of a recurrent clonal type by enhanced DNA fingerprinting. The conventional method of subtyping of E. coli O157, bacteriophage typing, provided little useful information in investigation of the central Scotland outbreak since in excess of 58% of all bacterial isolates received by SRL during the period from 1 April 1996 to 31 March 1997 were phage type 2.
PFGE analyses with an extended panel of endonucleases demonstrated that the study group of isolates representing the three major outbreaks could be discriminated under strictly defined conditions by using XbaI and AvrII. Other restriction enzymes could not differentiate the clonal type. Different run parameters for XbaI-cleaved DNA distinguished Highland outbreak isolates from the other two groups, while AvrII distinguished the central Scotland outbreak group from the others. Two isolates (isolates 340 and 1551) did not share the indistinguishable profile for the outbreak isolates, with the profile for isolates 340 and 1551 differing from the profile for the outbreak isolates by one fragment. Isolate 340 from the West Lothian outbreak had a profile indistinguishable from that for the Highland isolates following cleavage with both XbaI and AvrII. The majority of isolates formed distinct identifiable outbreak groups that distinguished them from the remainder of the broader WL clonal type. Overreliance on one particular endonuclease should be avoided in PFGE analysis of E. coli O157.
Bacteriophage lambda RFLP analysis also differentiated the three outbreak groups. However, as with PFGE, certain isolates from different outbreaks were indistinguishable: West Lothian isolates 325 and 340 and Highland outbreak isolates. Isolate 340 could not be distinguished from Highland outbreak isolates by any method. If certain genetic events gave rise to the differences observed between the West Lothian and Highland outbreak groups, then conversely, their nonoccurrence or reversal in this particular isolate could lead to this observation.
A minor variation in PFGE profiles has previously been attributed to clonal turnover in individual patients shedding E. coli O157 for up to 62 days (9). In the present study, however, analyses were performed with multiple isolates from disparate sources while the outbreaks were ongoing.
The significance of the genomic differences elicited by PFGE and bacteriophage lambda RFLP analysis is not clear. Certainly, these are minor variations, at most a few small fragments, of a basically similar macrorestriction profile compared with, for example, the major differences observed between the PFGE profiles of phage types 2 and 49 (11). During the recent outbreaks in Japan analogous small-fragment differences were observed and found to be insignificant: on hybridization a minor fragment (75 kb) was found to be derived directly from a larger chromosomal fragment of the macrorestriction profile for the outbreak strain. Therefore, the discrepant fragment did not materially affect the grouping of the outbreak strains (7). However, the reasons why these observed differences arise remain as yet undetermined. The proposed interpretative criteria for PFGE profiles (20) do not include experimental evidence describing the means by which such variation occurs or the degree of variation seen, providing only a general outline of common genetic events and their potential effect on the PFGE profiles of various bacterial genera. It may be postulated from the evidence from this study that since the majority (80%) of genotypic analyses performed could not discriminate the outbreak groups, the isolates were genetically homogeneous. Overreliance on minor, arguably insignificant differences in genomic structure as evinced by the PFGE macrorestriction profiles should perhaps be resisted, particularly in light of the variation in genomic size apparent among certain isolates (6). Conversely, both PFGE and bacteriophage lambda RFLP analyses were able to separate members of the WL clonal type into discrete outbreak groups, indicating that whatever genetic alterations have occurred, although apparently minor, they can give rise to distinctive subsets of isolates. These variations may be exploited as epidemiological markers in the real-time investigation of outbreaks of E. coli O157 infection.
Acknowledgments
The expert technical assistance of the SRL staff is gratefully acknowledged. The continuing cooperation of colleagues in submission of isolates to SRL is gratefully acknowledged.
This study was funded by the Scottish Office Agriculture, Environment and Fisheries Department (project UAB/004/97) and the Acute Healthcare Research Commitee, Chief Scientist Office, Scottish Office Department of Health (project K/MRS/C50/2376). The Reference Laboratory for Campylobacter and Escherichia coli O157 is funded by the National Services Division, Scottish Office Department of Health.
REFERENCES
- 1.Allison L J, Stirrat A, Thomson-Carter F M. Genetic heterogeneity of Escherichia coli O157:H7 in Scotland and its utility in strain subtyping. Eur J Clin Microbiol Infect Dis. 1998;17:844–848. doi: 10.1007/s100960050204. [DOI] [PubMed] [Google Scholar]
- 2.Allison L J, Thomson-Carter F M. Reference laboratory investigation of a major outbreak of Escherichia coli O157:H7 infection. Enterohemorrhagic Escherichia coli. Fortschritte der Medizin Monographie 84. Munich, Germany: Urban und Vogel; 1997. [Google Scholar]
- 3.Barrett T J, Lior H, Green J H, Khakhria R, Wells J G, Bell B P, Greene K D, Lewis J, Griffin P M. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J Clin Microbiol. 1994;32:3013–3017. doi: 10.1128/jcm.32.12.3013-3017.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Gurtler V, Stanisich V A. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 6.Harsono K D, Kaspar C W, Luchansky J B. Comparison and genomic sizing of Escherichia coli O157:H7 isolates by pulsed-field gel electrophoresis. Appl Environ Microbiol. 1993;59:3141–3144. doi: 10.1128/aem.59.9.3141-3144.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Izumiya H, Terajima J, Wada A, Inagaki Y, Itoh K-I, Tamura K, Watanabe H. Molecular typing of enterohemorrhagic Escherichia coli O157:H7 isolates in Japan by using pulsed field gel electrophoresis. J Clin Microbiol. 1997;35:1675–1680. doi: 10.1128/jcm.35.7.1675-1680.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janssen P, Coopman R, Huys G, Swings J, Bleeker M, Vos P, Zabeau M, Kersters K. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology. 1996;142:1881–1893. doi: 10.1099/13500872-142-7-1881. [DOI] [PubMed] [Google Scholar]
- 9.Karch H, Russmann H, Schmidt H, Schwarzkopf A, Heesemann J. Long-term shedding and clonal turnover of enterohemorrhagic Escherichia coli O157 in diarrheal diseases. J Clin Microbiol. 1995;33:1602–1605. doi: 10.1128/jcm.33.6.1602-1605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khakria R, Duck D, Lior H. Extended phage-typing scheme for Escherichia coli O157:H7. Epidemiol Infect. 1990;105:511–520. doi: 10.1017/s0950268800048135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krause U, Thomson-Carter F M, Pennington T H. Molecular epidemiology of Escherichia coli O157:H7 by pulsed-field gel electrophoresis and comparison with that by bacteriophage typing. J Clin Microbiol. 1996;34:959–961. doi: 10.1128/jcm.34.4.959-961.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrence J G, Ochman H, Hart D L. The evolution of insertion sequences within enteric bacteria. Genetics. 1992;131:9–20. doi: 10.1093/genetics/131.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Louws F J, Fulbright D W, Stephens C T, De Bruijn F J. Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl Environ Microbiol. 1994;60:2286–2295. doi: 10.1128/aem.60.7.2286-2295.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madico G, Akopyants N S, Berg D E. Arbitrarily primed PCR DNA fingerprinting of Escherichia coli O157:H7 strains by using template from boiled cultures. J Clin Microbiol. 1995;33:1534–1536. doi: 10.1128/jcm.33.6.1534-1536.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orskov F, Orskov I. Summary of a workshop on the clone concept in the epidemiology, taxonomy and evolution of the Enterobacteriaceae and other bacteria. J Infect Dis. 1983;148:346–357. doi: 10.1093/infdis/148.2.346. [DOI] [PubMed] [Google Scholar]
- 16.Paros M, Tarr P I, Kim H, Besser T E, Hancock D D. A comparison of human and bovine Escherichia coli O157:H7 isolates by toxin genotype, plasmid profile, and bacteriophage lambda-restriction fragment length polymorphism profile. J Infect Dis. 1993;168:1300–1303. doi: 10.1093/infdis/168.5.1300. [DOI] [PubMed] [Google Scholar]
- 17.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 18.Pollard D K, Johnson W M, Lior H, Tyler S D, Rozee K R. Rapid and specific detection of verotoxin genes in Escherichia coli by the polymerase chain reaction. J Clin Microbiol. 1990;28:540–545. doi: 10.1128/jcm.28.3.540-545.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Tenover F, Arbeit R D, Goering R V, Michelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomson-Carter F M. Annual report. Scottish Reference Laboratory for Campylobacter and Escherichia coli O157. 1997. Aberdeen, Scotland. [Google Scholar]
- 22.Tyler S D, Johnson W M, Lior H, Wang G, Rozee K R. Identification of Verotoxin type 2 variant B subunit genes in Escherichia coli by the polymerase chain reaction and restriction fragment length polymorphism analysis. J Clin Microbiol. 1991;29:1339–1343. doi: 10.1128/jcm.29.7.1339-1343.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]