Abstract
Radiotherapy is an effective anticancer treatment, but combinations with targeted agents that maximize efficacy while sparing normal tissue are needed. Here, we assess the radiopotentiation profiles of DNA damage response inhibitors (DDRi) olaparib (PARP1/2), ceralasertib (ATR), adavosertib (WEE1), AZD0156 (ATM), and KU-60648 (DNA-PK). We performed a radiotherapy combination screen and assessed how drug concentration and cellular DDR deficiencies influence the radiopotentiation ability of DDRi. We pre-selected six lung cancer cell lines with different genetic/signaling aberrations (including mutations in TP53 and ATM) and assessed multiple concentrations of DDRi in combination with a fixed radiotherapy dose by clonogenic assay. The effective concentration of DDRi in radiotherapy combinations is lower than that required for single-agent efficacy. This has the potential to be exploited further in the context of DDR deficiencies to increase therapeutic index and we demonstrate that low concentrations of AZD0156 preferentially sensitized p53-deficient cells. Moreover, testing multiple concentrations of DDRi in radiotherapy combinations indicated that olaparib, ceralasertib, and adavosertib have a desirable safety profile showing moderate increases in radiotherapy dose enhancement with increasing inhibitor concentration. Small increases in concentration of AZD0156 and particularly KU-60648, however, result in steep increases in dose enhancement. Radiopotentiation profiling can inform on effective drug doses required for radiosensitization in relation to biomarkers, providing an opportunity to increase therapeutic index. Moreover, multiple concentration testing demonstrates a relationship between drug concentration and radiotherapy effect that provides valuable insights that, with future in vivo validation, can guide dose-escalation strategies in clinical trials.
Introduction
Radiotherapy remains a major treatment modality in cancer, with approximately 50% of patients receiving radiotherapy treatment. Treatment success, however, is still limited by toxicities, which impact the quality of life of a patient. There is an urgent requirement for combinations with agents that increase the therapeutic window by further sensitizing tumor but not the normal tissue (1).
Recently, potent and targeted inhibitors of the DDR have entered the clinic in multiple disease settings. These include olaparib (PARP1/2), ceralasertib (ATR), adavosertib (WEE1), AZD0156 (ATM), and agents targeting DNA-PK (2–7). Preclinical data and their cellular function suggest that these inhibitors may potentiate radiotherapy in tumors by driving higher levels of DNA damage such as DNA double-strand breaks (DSB) and increased cell kill specifically through inactivation of various aspects of the DDR (5, 8–18). Importantly, to succeed clinically, dosing of these agents must be carefully informed to maximize therapeutic index.
Assessment of efficacy and tolerability of radiotherapy combinations is challenging, and the lack of high-quality preclinical studies of radiotherapy combinations was recently reported as posing a risk to successful implementation of inhibitors in radiotherapy combination trials (19, 20). Moreover, preclinical studies rarely test multiple concentrations/doses of targeted agents in combination with radiotherapy, because systemic small-molecule drug development in oncology is typically driven by the assumption that higher doses provide greater potential for antitumor efficacy (21). Thus, phase I clinical trials establish the maximum-tolerated dose (MTD) that can be recommended for further testing in phase II. However, when two treatment modalities are combined to increase efficacy, combination of both MTDs may not be necessary and is best avoided, particularly if clinical adverse events of the two agents overlap. Consequently, establishing the minimally effective dose (MED) of an inhibitor, that preferentially provides combination activity with radiotherapy in tumors, by, for example, exploiting a tumor-specific DDR deficiency could drive a significant increase in the therapeutic index. Indeed, use of biomarkers to select for tumors most likely to respond to targeted agents in radiotherapy combinations has the potential to significantly reduce the MED required, ultimately driving the best therapeutic ratio (10). This is particularly relevant to radiotherapy combinations, where irreversible radiotherapy toxicity may occur late as opposed to toxicity to systemic treatment that is usually more acute. Surprisingly, despite patient-selection strategies revolutionizing cancer care with targeted agents in the past two decades, there has been little focus on the identification of biomarkers to improve the effectiveness of radiotherapy combinations.
We performed detailed head-to-head radiopotentiation profiling of multiple DNA-damage response inhibitors (DDRi) for early clinical development in a lung cancer setting. We adhered to recently published guidelines to meet the standards required for these data to be robust and reproducible with the view to potentially inform the design of follow-up in vivo experiments that may influence radiotherapy combination trials (20). We devised a screening methodology that, although limited to a small number of representative lung cancer cell lines, allowed us to assess the influence of drug concentration and genetic/signaling DDR deficiencies on the radiopotentiation ability of multiple DDRi by long-term clonogenic survival assays, the gold-standard for assessing cell reproductive death after ionizing radiotherapy.
We demonstrate that radiopotentiation profiling performed in this way can provide important insights into MEDs required for radiosensitization in relation to genetic/cellular signaling aberrations. Moreover, assessment of the relationship between drug concentration and radiotherapy effect highlights important considerations to be made before embarking on combining DDRi with radiotherapy. These insights could be used to trigger more detailed in vitro and in vivo follow-up studies to help inform clinical trial design to improve therapeutic index and clinical outcome.
Materials and Methods
Compounds
All inhibitors were synthesized at AstraZeneca (2, 4, 6, 22, 23) and diluted in dimethylsulfoxide (DMSO; Sigma-Aldrich 276855) to a concentration of 10 mmol/L (olaparib, adavosertib, ceralasertib, AZD0156) or 5 mmol/L (KU-60648) and stored under nitrogen.
Cell lines
HOP-92 cells were sourced from the NCI Division of Cancer Treatment and Diagnosis (DCTD). All other human cell lines were sourced from the ATCC. A549 TP53 knock-out isogenic cells were generated by CRISPR-Cas9 technology. Short guide (sg)RNAs-targeting TP53 on exon 5 (TGACTGCTTGTAGATGGCCA or GAGCGCTGCTCAGATAGCGA) were designed and cloned into a vector containing Cas9 and a GFP cassette (azPGE02-Cas9-T2A-GFP). A549 cells were subsequently transfected with the vector using Lipofectamine 3000 (Thermo Fisher Scientific). After 48 hours, GFP-positive cells were single-cell sorted and individual clones sequenced by PCR to confirm loss of their wild-type (TP53) allele before expansion to obtain isogenic cell lines.
Cell lines were cultured in RPMI (Life Technologies 21875091; NCI-H460, NCI-H1299, NCI-H1703, HOP-92 and NCI-H23), DMEM (Life Technologies 21885–025; A549) or Ham's F12 (Life Technologies 11330057; A549 TP53 KO) supplemented with 10% FCS (Life Technologies 10270–106) and 2 mmol/L Glutamax (ThermoFisher Scientific; 35050061). All cell lines were incubated at 37°C and 5% CO2 and passaged up to a maximum of 15 passages. All cell lines were authenticated by the AstraZeneca cell bank using short tandem repeat analysis using CellCheck (IDEXX Bioanalytics) and validated free of Mycoplasma contamination using the STAT-Myco assay (IDEXX Bioanalytics).
Radiotherapy
Cells were exposed to single-radiotherapy doses (0.2–6 Gy) using a Faxitron benchtop irradiator set to 150kV and 5mA. Control cells were sham-irradiated.
Clonogenic assay
Cells were plated at a low density (500 cells for A549, NCI-H460 and NCI-H1299, 2,000 for HOP-92, 2,500 for NCI-H1703, and 3,000 for NCI-H23) into 6-well cell culture plates (Corning; VWR 734–1599) in a total of 2–3 mL growth medium. The following day proliferating cells were inspected to ensure that there were no more than 20% cell doublets to guarantee that colonies originated from single cell clones. Thereafter, cells were treated with the indicated drug concentrations or vehicle control (DMSO) for 1-hour before irradiation. All drug and vehicle treatments were performed at a final concentration of 0.1% DMSO. Cells were continuously treated or drug washed out 24 hours after radiotherapy. Cells were incubated until control colonies had grown to a size of approximately 100–200 cells, typically after 7–12 days (7–8 days for A549, A549 TP53 KO cells, NCI-H460 and NCI-H1299, 11 days for NCI-H1703 and HOP-92 and 11–12 days for NCI-H23). Medium was aspirated and cells carefully washed once in PBS before fixing and staining in Coomassie Brilliant Blue stain (Sigma-Aldrich B8522) supplemented with 25% methanol (VWR 20837.320) and 5% acetic acid (Sigma-Aldrich 537020) for a minimum of 15 minutes. Stain was thereafter aspirated and cells washed twice in distilled water and air-dried. Colonies were counted on a GelCount platform using a manually developed bespoke detection assay for each cell line. Single-agent IC50 values were determined from non-irradiated controls using GraphPad Prism software (v8.0.1). Clonogenic survival curves were generated by normalizing to untreated DMSO controls of the respective radiotherapy treatment or non-irradiated controls.
Statistical analysis
A two-way ANOVA was performed using Repeated Measures ANOVA in GraphPad Prism software (v.8.0.1). Matched values were spread across rows and a Sidak's multiple comparison's test with a single pooled variance applied.
Generation of dose-enhancement factors
Dose-enhancement factors (DEF) were calculated from full radiotherapy dose–response curves in which cell densities were increased for higher radiotherapy dose exposures to compensate for cell kill. Surviving fractions (SF) were calculated by dividing the plating efficiency of treated samples by the plating efficiency of control samples. Radiotherapy doses resulting in 75% survival were calculated from linear quadratic (LQ) fits (SF = exp(−αD–βD2) with D = radiotherapy dose) for each drug concentration in each individual experiment. The ratio of radiotherapy doses causing 25% cell kill in the DMSO-treated samples over the radiotherapy dose causing 25% cell kill in drug-treated samples produced the DEF75 values.
To limit experimental burden and guide drug concentration selection for subsequent DEF validation experiments, “approximate” DEF values (aDEF) were generated in the initial concentration finding studies. This was done by overlaying extensive drug-concentration response data generated at one radiotherapy dose in the absence or presence of inhibitor onto previously obtained cell line-specific full radiotherapy dose–response curves (n = 2; Fig. 2B). Concentration response curves of each drug in each individual experiment were fitted using the LQ model as described above. These DEF values calculated in this procedure were classified as “approximate” as they rely on curve fit parameters derived from just two radiotherapy response data points for the individual drug concentration–radiotherapy combination treated survival data.
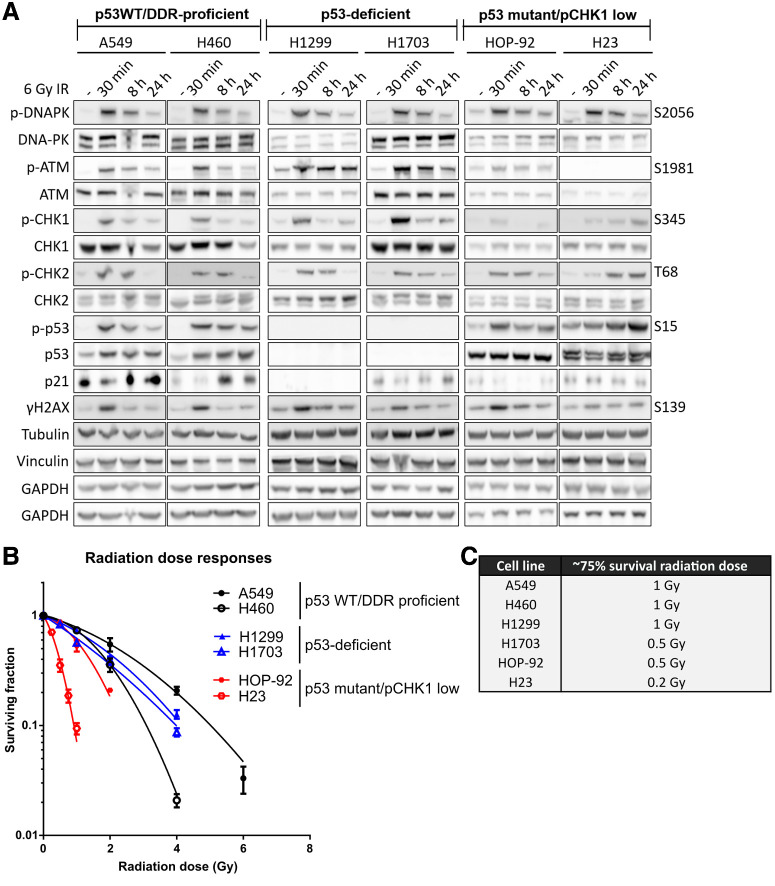
Figure 2.
Characterization of lung cancer cells. A, Cell lines were treated with a 6 Gy dose of radiotherapy and analyzed at the times indicated. Cell extracts were run across four separate gels and western blotted for total and phosphorylated (p-) proteins. Tubulin, GAPDH, and Vinculin served as total loading controls. B, Survival of cell lines after exposure to increasing doses of radiotherapy and as determined by clonogenic survival assays. “p53 WT/DDR proficient” cell lines are in black, “p53-deficient” cell lines in blue and “p53 mutant/pCHK1 low” cell lines are in red. Surviving fractions are the median of two independent experiments with error bars representing upper and lower limits. C, Radiotherapy doses resulting in approximately 75% cell survival in each cell line.
Target engagement assays
Cells were plated into 6-well plates in a total of 2 mL growth medium per well. At 60%–80% confluency cells were treated with the indicated concentrations of inhibitors or vehicle control (DMSO) for one hour before irradiation with 6 Gy. Cells were incubated for 30 minutes before medium was aspirated and cells washed 1x in PBS. Cells were lysed by scraping into 50–150 μL cell lysis buffer supplied in an HT PARP in vivo Pharmacodynamic Assay II kit (R&D systems 4520–096-K). Cell lysates were vortexed and following addition of 1% sodium dodecyl sulphate (SDS) boiled for 5 minutes at 80°C–100°C. Cellular DNA was degraded by incubation with DNAse I for 90 minutes. The concentration of protein was determined by Pierce BCA protein assay (ThermoFisher Scientific 23225) as per the manufacturer's instructions. Target engagement by DDRi was performed by analysis on standard SDS-PAGE following addition of sample-loading buffer and reducing agent (ThermoFisher Scientific NP0007 and NP0004). For western blotting, the following antibodies used were: rabbit anti–phospho-DNA-PK Ser-2056 (generated in-house), anti–DNA-PKcs (4215 or 12311), anti–phospho-CDK1 Tyr-15 (9111), anti–CDK1 POH-1 (9116), anti-CHK1 (2360), anti-phospho CHK1 Ser-345 (2341 or 2348), anti–phospho-CHK2 Thr-68 (2661), anti–phospho-p53 Ser-15 (9284 or 9286), anti–γH2AX Ser-139 (2577), anti-GAPDH (2118), and anti-WEE1 (4936) from Cell Signaling Technology, mouse anti-ATM (sc-23921), anti-ATR (sc-1887), anti-p53 (sc-126), and anti-p21 (sc-397) from Santa Cruz Biotechnology, anti–phospho-ATM Ser-1981 (ab81292), anti-CHK1 (ab40866), anti-ATM (ab78), and anti-KAP1 (ab10483) from Abcam, anti-CHK2 (#05–649), anti-γH2AX (#05–636), anti-H2AX (#07–627), and anti–phospho-ATM Ser-1981 (#MAB380:6) from Millipore, anti-CHK2 (2391) from ProScience, anti–phospho-KAP1 Ser-824 (IHC-00073) from Bethyl Laboratories, anti-p53 (AHO 0152) and anti-Vinculin (V9131) from Sigma-Aldrich. Bands were visualized on a G:BOX imaging platform (Syngene) using SuperSignal West Dura Substrate Extended Duration Substrate (ThermoFisher Scientific 34075) as per manufacturer's instructions. Target engagement by olaparib was measured by chemiluminescent ELISA (Trevigen HT PARP in vivo Pharmacodynamic Assay II, R&D systems 4520–096-K) to quantify polyADP-ribose as per the manufacturer's instructions.
Results
Characterization of a lung cancer cell line panel
For cross-comparison studies of radiopotentiation profiles of olaparib, ceralasertib, adavosertib, AZD0156 and KU-60648, we selected a panel of six non–small cell lung cancer (NSCLC) cell lines representing three NSCLC subtypes: adenocarcinoma (A549), squamous cell carcinoma (NCI-H1703), and large cell carcinoma (NCI-H460, NCI-H1299, HOP-92, and NCI-H23). We characterized their genetic aberrations using next-generation Cancer Cell Line Encyclopedia (CCLE) data (24), querying cancer genes as classified by the TCGA PanCancer Atlas project (25). Specifically, we focused on the RTK–RAS–MAPK, PI3K, and NRF2 pathways, which are typically altered in NSCLC, and on cell cycle and DDR genes that have the potential to influence their radiotherapy/DDRi combination response (Fig. 1; ref. 26). To accurately determine the functional status of the DDR across the cell line panel, we assessed the level of activation of DNA-PK, ATM, and ATR signaling in response to radiotherapy (Fig. 2). ATM is activated in response to DSBs and regulates processes, such as DSB repair by homologous recombination repair and non-homologous end joining (NHEJ), and also activates G1–S-phase checkpoints through CHK2 and the tumor-suppressor p53 (27). DNA-PK promotes NHEJ throughout the cell cycle, whereas ATR and its substrate CHK1 control intra-S, and S–G2 checkpoints to enable protection and restart of stalled replication forks, as well as a G2–M checkpoint to prevent DNA damage being taken into mitosis (27, 28).
Figure 1.
Molecular features of lung cancer cell lines. Lung cancer cell lines were classified using next-generation CCLE data. Genetic alterations in pathways typically altered in NSCLC, DDR, and cell cycle are reported. Each rectangle represents a specific cell line and percentages indicate the frequency at which genetic alterations in a specific gene (as specified by the legend) are present. NCI-H23 cells carry an additional ATM Q1919P mutation, which has not been annotated here due to uncertain significance to human disease (see text for details).
All cell lines within the panel were exposed to a 6 Gray (Gy) dose of radiotherapy and DNA-PK, ATM, and ATR signaling measured by western blotting. On the basis of their specific DDR aberrations, they were divided into three subgroups: p53 WT/DDR-proficient, p53-deficient, and p53 mutant/pCHK1 low (Fig. 2A).
In the “p53 WT/DDR-proficient” cell lines (A549 and NCI-H460), irradiation-induced autophosphorylation of DNA-PK (Ser-2056) and ATM (Ser-1981; Fig. 2A; refs. 29, 30). Downstream of ATM, CHK2 was phosphorylated (Thr-68), as well as p53 (Ser-15), which resulted in the stabilization of p21 indicative of functional p53 signaling (31). ATR signaling was also functional because we observed phosphorylation of its kinase target CHK1 (Ser-345; ref. 32). H2AX was phosphorylated within 30 minutes, indicative of ongoing DNA damage, and this was resolved within 8–24 hours coinciding with diminished ATM, ATR, and DNA-PK phosphorylation.
In the “p53-deficient” cell lines, DNA-PK, ATM/CHK2 and ATR/CHK1 signaling axes were activated within 30 minutes. However, p53 was not expressed and its target p21 was not stabilized. H2AX phosphorylation was observed at 30 minutes and resolved within 8 hours.
In the “p53 mutant/pCHK1 low” cell lines (HOP-92 and NCI-H23), DNA-PK was activated within 30 minutes. Activation of ATM was absent in NCI-H23 cells due to a genetic mutation (Q1919P, a homozygous missense mutation that leads to low expression levels of full-length ATM and thus impaired activation in response to IR; this mutation is not captured in Fig. 1 due to its uncertain significance to human health as annotated by ClinVar; refs. 33–35). Consequently, H2AX phosphorylation was low and CHK2 was only activated at the 8-hour time point, likely driven by alternative kinases. In HOP-92 cells, ATM is expressed, however, radiotherapy-induced ATM autophosphorylation was low. Nevertheless, there was no consequence on the downstream activation of CHK2. Interestingly, radiotherapy-induced CHK1 phosphorylation was low in both HOP-92 and NCI-H23 cells, indicative of defective ATR signaling. In addition, both cell lines harbor DNA-binding mutants of p53, and NCI-H23 has an additional mutation in the CDKN2A locus encoding the p16INK4A protein, suggesting that both cell lines likely harbor cell-cycle checkpoint defects. Although we grouped both HOP-92 and NCI-H23 cells as being “p53 mutant/pCHK1 low,” it is important to note that through lack of full-length ATM expression, NCI-H23 cells will likely be further differentiated from the “ATM low” HOP-92 cells, a distinction we deemed important to investigate.
Following characterization of the radiotherapy-induced DDR of the cell lines, we proceeded to determine their in vitro radiosensitivity, which largely depends on cell-cycle phase distributions and their ability to repair DNA damage. To generate statistically robust data, we wanted to determine a fixed radiotherapy dose that would only cause approximately 25% cell kill, ensuring a large enough assay window in which to assess the radiopotentiation ability of the DDRi. To this end, cells were exposed to multiple doses of radiotherapy and clonogenic survival assays performed (Fig. 2B).
Our data demonstrated that A549 cells were the most radioresistant among the cell line panel (Fig. 2B). Although NCI-H1299 and NCI-H1703 were expected to be more radioresistant due to their p53-deficient status, they were marginally more sensitive than A549 (36–38). Next, NCI-H460 cells were approximately 4–6-fold more sensitive to a 4 Gy dose than NCI-H1703 and NCI-H1299. They were, however, more resistant than HOP-92 cells, where clonogenic cell survival was undetectable after 4 Gy irradiation in our assay. This increased sensitivity to radiotherapy is consistent with their low ATM response (Fig. 2A). Finally, NCI-H23 cells were the most sensitive to radiotherapy, consistent with a deficiency in ATM signaling (Fig. 2A).
These data allowed selection of radiotherapy doses that resulted in approximately 25% cell kill across all cell lines for subsequent radiopotentiation profiling experiments. Specifically, these were 1 Gy in A549, NCI-H460, and NCI-H1299, 0.5 Gy in NCI-H1703 and HOP-92 cells and 0.2 Gy in NCI-H23 cells (Fig. 2C).
Concentrations required for single-agent efficacy are not required for radiosensitization
To assess and compare radiopotentiation provided by olaparib, ceralasertib, adavosertib, AZD0156 and KU-60648, with their single-agent activity, we performed clonogenic survival assays. Each cell line was treated with a 6-point concentration titration of inhibitor and then either irradiated with a single dose of radiotherapy causing approximately 25% cell kill or left non-irradiated (Fig. 2C). These data not only determined specific IC50 and IC90 concentrations (inhibitory concentrations resulting in 50% or 90% cell kill) of each inhibitor as a single agent (Fig. 3A), but also assessed how radiopotentiation depends on drug concentration and DDR proficiency.
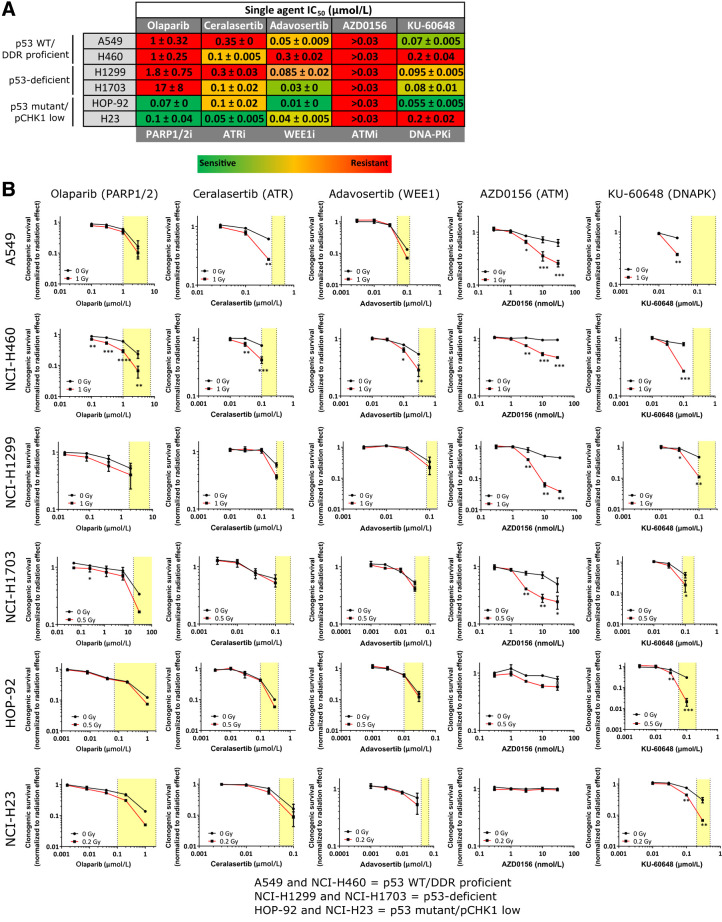
Figure 3.
Determination of sensitivity of cell lines to DDRi as single agent and in combination with radiotherapy. A, Table depicting IC50 concentrations of DDRi across cell lines as determined by clonogenic assay. Values are the average of two representative experiments with the standard error of the mean indicated. Lowest most sensitive IC50 for a particular drug are in green and highest most resistant in red. B, Clonogenic survival (normalized to untreated DMSO controls of the respective radiotherapy treatment) of cell lines in response to increasing concentrations of DDRi alone (black line; 0 Gy) or after radiotherapy at the indicated radiotherapy dose (red line). Yellow boxes indicate the single-agent IC50 to IC90 concentration range required to inhibit clonogenic survival in micromolar (μmol/L) or nanomolar (nmol/L) concentrations. Graphs represent the medians of two independent experiments performed in triplicate, with error bars representing upper and lower limits. Statistical significance was tested using a two-way ANOVA where *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We found varying single-agent sensitivities to each DDRi across the panel (Fig. 3A; Supplementary Fig. S1). NCI-H23 and HOP-92 cells—of the “p53 mutant/pCHK1 low” subgroup—were most sensitive to DDRi. Specifically, NCI-H23 cells were most sensitive to olaparib and ceralasertib, likely due to their ATM mutation (39), and HOP-92 cells were most sensitive to olaparib, adavosertib and KU-60648. Of note, most cell lines were sensitive to adavosertib with exception of NCI-H460. None of the cell lines showed single-agent sensitivity to AZD0156 at efficacious concentrations (16).
Next, we compared the effective single-agent concentration with the effective concentration required for radiosensitization. We performed clonogenic survival assays in cells treated with increasing inhibitor concentrations with or without radiotherapy (Fig. 3B). Strikingly, we found that where cells were robustly radiosensitized (indicated by asterisks signifying statistical significance by two-way ANOVA in Fig. 3B), inhibitor concentrations required for radiosensitization were consistently lower than those required for single-agent efficacy (Fig. 3A and B; Supplementary Fig. S1). For example, the IC50 and IC90 concentrations for ceralasertib in A549 cells were 350 and 650 nmol/L, respectively (Fig. 3B), but robust radiosensitization occurred at concentrations below the IC50 of 300 nmol/L. This was also the case for all the DDRi in NCI-H460 cells and with KU-60648 across all cell lines. Olaparib, ceralasertib, and adavosertib appeared to either not or only weakly sensitize the cell lines in the panel under these conditions (with exception of NCI-H460 cells and ceralasertib in A549 cells). We hypothesized that in vitro single-agent sensitivity may mask radiosensitization effects under continuous drug treatment. For example, adavosertib did not strongly radiosensitize any of the cell lines despite their inherent single-agent sensitivity (with the exception of NCI-H460 cells). To test this, we treated A549 cells either continuously or for only 24 hours with increasing concentrations of adavosertib in the presence or absence of 1 Gy radiotherapy (Supplementary Fig. S2A). A549 cells were not radiosensitized under either condition suggesting that the lack of radiosensitization was not, in fact, due to single-agent adavosertib sensitivity.
Next, we acknowledged that greater effect sizes (i.e., higher kill by greater radiotherapy doses) will be required to reveal statistically significant radiosensitization with milder radiosensitizers. We therefore chose a representative cell line from each subgroup and performed extended radiotherapy dose-response experiments to further investigate the capacity of these inhibitors to radiosensitize the cell lines (Supplementary Fig. S2B). DDR-proficient A549 cells were significantly radiosensitized with approximately IC50 concentrations of olaparib (1 μmol/L), ceralasertib (0.3 μmol/L), and adavosertib (0.03 μmol/L). p53-deficient NCI-H1299 cells were significantly radiosensitized with ≤IC50 concentrations of olaparib (0.4 μmol/L) and ceralasertib (0.3 μmol/L), but notably not a >IC50 concentration of adavosertib (0.3 μmol/L), and p53 mutant/pCHK1-low NCI-H23 cells were significantly radiosensitized with an approximately IC50 concentration of olaparib (0.2 μmol/L), but not ceralasertib (0.1 μmol/L) or adavosertib (0.03 μmol/L). These extended radiotherapy response curves reveal the potential strong radiopotentiation efficacy of some of these inhibitors at higher radiotherapy doses.
Because most cell lines were radiosensitized with DDRi concentrations that were lower or equal to the single-agent IC50 concentration, we tested the level of target engagement that could be achieved with these concentrations in radiotherapy combinations. We therefore pre-treated cells with single-agent IC50 or IC90 concentrations of each inhibitor required to inhibit colony formation, irradiated with 6 Gy and determined the level of target inhibition at these concentrations 30-minutes after irradiation. We assessed PARylation for olaparib, phospho-CHK1 (Ser-345) for ceralasertib (32), phospho-DNA-PK (Ser-2056) for KU-60648 (29), phospho-CDK1 (Tyr-15) for adavosertib (40) and phospho-ATM (Ser-1981) and phospho-KAP1 (Ser-824) for AZD0156 (30, 41).
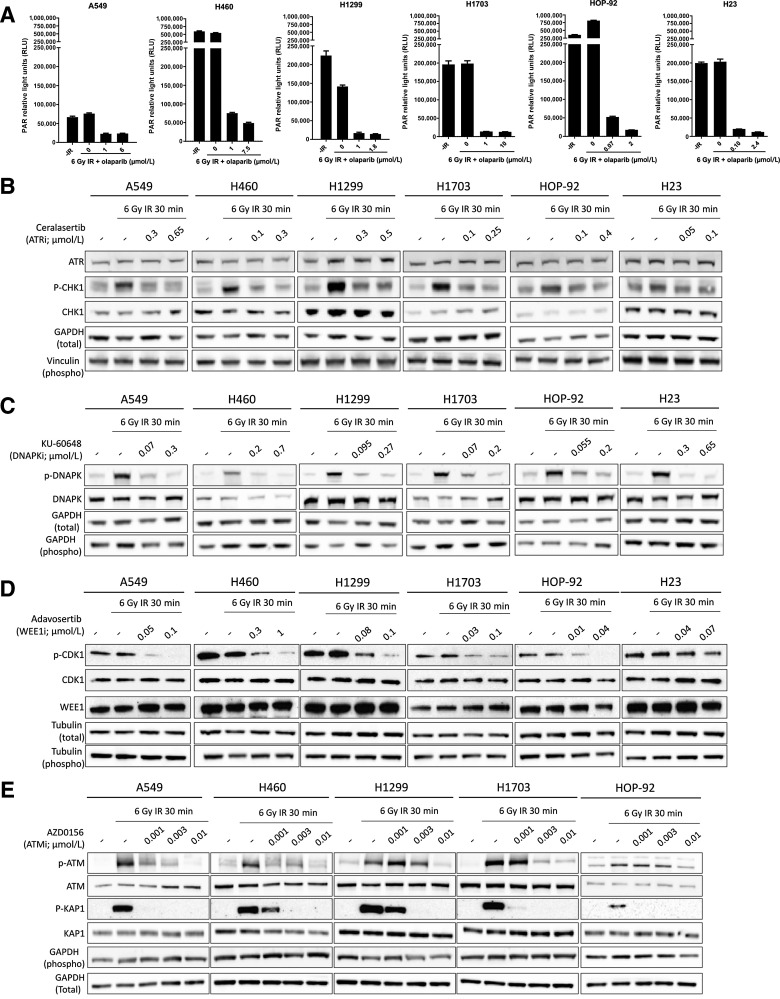
Under our assay conditions, baseline PARylation was highly variable across cell lines and radiotherapy–induced PARylation was only observed in HOP-92 cells, which are highly sensitive to single-agent olaparib (Fig. 4A). In HOP-92, PARylation was strongly inhibited using the IC50 concentration of olaparib (70 nmol/L), whereas inhibition with the IC90 concentration (2 μmol/L) led to complete inhibition. Similarly, in NCI-H460 cells the IC50 concentration (1 μmol/L) led to a marked inhibition of PARylation, with a slight further decrease following treatment with the IC90 (7.5 μmol/L) concentration (42). In all other cell lines, maximal inhibition of PARylation was seen at the IC50 concentrations of olaparib, with no further apparent decrease in PARylation at the IC90 concentration.
Figure 4.
Evaluation of target engagement of DDRi across cell lines. A, PAR relative light units (RLU) in no radiotherapy controls (-IR), or following a 6 Gy radiotherapy dose alone or in combination with an IC50 or IC90 concentration of olaparib (μmol/L) as indicated. 1 μmol/L was used as the maximum on-target concentration in in vitro assays. Hence, where IC50 values were higher than 1 μmol/L (NCI-H1299 and NCI-H1703), the level of PARylation inhibition of an IC50 concentration was compared with a 1μmol/L on-target concentration rather than the IC90 concentration. B–E, Cell lines were irradiated with 6 Gy in the presence or absence of an IC50 or IC90 concentration of ceralasertib (B), KU-60648 (C), adavosertib (D) and three concentrations of AZD0156 as indicated (E). Cell extracts were collected after 30 minutes and western blotted for total and phosphorylated (p-) proteins. GAPDH, Tubulin and Vinculin served as total loading controls for either phospho or total protein blots as indicated.
Similarly, for ceralasertib, adavosertib, and KU-60648, the level of target engagement was comparable between IC50 and IC90 concentrations (Fig. 4B–D). In the case of AZD0156—for which IC50 and IC90 concentrations could not be generated due to lack of single-agent sensitivity in the cell line panel—we tested 1, 3, and 10 nmol/L. Complete inhibition of phospho-ATM was typically observed using a 10 nmol/L concentration of AZD0156 across all cell lines, with varying degrees of inhibition at 1 or 3 nmol/L (Fig. 4E). Importantly, 3 nmol/L was sufficient to induce significant radiosensitization across all cell lines, with exception of HOP-92 cells, where radiosensitization was modest at the applied low dose of radiotherapy. This is consistent with HOP-92 cells having lower levels of radiotherapy–induced phospho-ATM in response to radiotherapy in comparison with other cell lines in the panel (Fig. 2A and 4E). AZD0156 also did not radiosensitize NCI-H23 cells, consistent with their ATM deficiency. This confirms the on-target specificity of the inhibitor-mediated radiosensitization effects across the other cell lines, consistent with a requirement for ATM in sensing radiotherapy–induced DNA damage (Figs. 2A and 3B).
These data therefore show that, when combined with radiotherapy, single-agent IC50 concentrations of olaparib, ceralasertib, adavosertib and KU-60648, as well as low concentrations of AZD0156, can sufficiently inhibit their respective target to drive radiosensitization in susceptible cell lines. In NCI-H460 cells, for example, the concentrations required for significant radiosensitization (Fig. 3B) were 0.1 μmol/L olaparib, 0.03 μmol/L ceralasertib, 0.1 μmol/L adavosertib, 3 nmol/L AZD0156 and 0.1 μmol/L KU-60648. Importantly, these concentrations are well below the clinical free Cmax concentrations achieved with clinical monotherapy doses of these inhibitors—typically 3.2 μmol/L for olaparib (300 mg steady-state), 5 μmol/L for ceralasertib (320 mg), and 542 nmol/L for adavosertib (300 mg QD 5/2) with AZD0156 not having reached the recommended Phase 2 dose and KU-60648 not being in clinical development.
Together, our data suggest that doses lower than those required for single-agent efficacy are sufficient to inhibit radiotherapy–induced activation of relevant drug targets when given concomitantly with radiotherapy, to provide effective radiosensitization.
The level of radiopotentiation depends on both the inhibitor target and tumor genetic/cellular DDR deficiencies
To compare the radiopotentiation ability of the DDRi across the cell line panel, we generated an approximate DEF (aDEF) at approximately 75% survival (aDEF75). This was done by overlaying the clonogenic survival data of each drug concentration in combination with radiotherapy onto previously generated radiotherapy dose–response curves (Fig. 2B; see Materials and Methods). We demonstrated the validity of this approach by confirming a crucial subset of aDEF75 values through determination of full radiotherapy dose–response curves. Specifically, two concentrations of each inhibitor were chosen, and radiotherapy dose–response curves were generated in one representative cell line per subgroup: NCI-H460 (p53 WT/DDR proficient), NCI-H1299 (p53-deficient), and NCI-H23 (p53 mutant/pCHK1 low). We did not generate DEFs for NCI-H23 and AZD0156 due to the absence of the drug target in this cell line. Crucially, we found that the DEF75 values calculated from full radiotherapy dose–response curves overlapped significantly with our previously generated aDEF75 values, giving us confidence in our screening approach (Supplementary Fig. S3).
We proceeded to plot all aDEF75 values against DDRi drug concentration to visualize the concentration-dependence profile of radiopotentiation of each inhibitor across the cell line panel (Fig. 5). Consistent with our statistical analysis in Fig. 3B, we found that AZD0156 and KU-60648 were more potent radiosensitizers than olaparib, ceralasertib or adavosertib, eliciting robust radiosensitization at the low radiotherapy doses used in the screen. They were also quick to reach the highest levels of radiosensitization within the tested drug concentration ranges. Olaparib, ceralasertib, and adavosertib behaved differently in that a doubling in DEF required an up to a 10-fold increase in drug concentration. These profiles highlight that the rate of increase in and degree of radiosensitization is defined by the inhibitor target and the genetics of each cell line. Olaparib robustly radiosensitized NCI-H460 cells (p53 WT/DDR proficient) whereas NCI-H23 (ATM-deficient) cells were only modestly sensitized under our screening conditions (but significantly in combination with higher radiotherapy doses; Fig. 3B; Supplementary Fig. S2B). Visualization of the concentration-dependency profile of radiotherapy response suggested that radiopotentiation was concentration-dependent in both cell lines, but that concentrations required for the same level of radiosensitization were approximately 10-fold higher in NCI-H460 cells in comparison with NCI-H23 cells (>1 vs. 0.2 μmol/L, respectively). Interestingly, whereas other cell lines were also mildly radiosensitized, the effect remained largely the same across concentrations of olaparib, consistent with our earlier findings that PARylation is fully inhibited with the IC50 in these cell lines (Fig. 4).
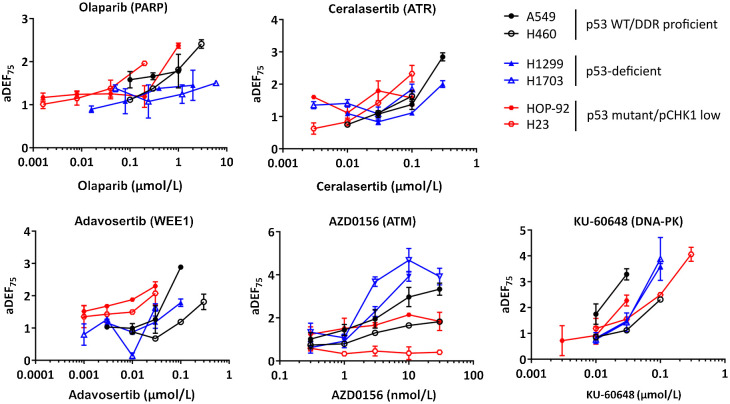
Figure 5.
The relationship between drug concentration and radiopotentiation in relation to genetic subtypes. Approximate radiotherapy DEFs at approximately 75% survival (aDEF75) of each inhibitor concentration determined by clonogenic assay across cell lines. aDEF75 values are the median of two independent experiments performed in triplicate, with error bars representing upper and lower limits.
Although concentration-dependency trends for ceralasertib were similar to olaparib with radiopotentiation being concentration-dependent in DDR-proficient A549, NCI-H460 and ATM-deficient NCI-H23 cells, there was not an equally strong differentiation at the level of drug concentration required to achieve radiopotentiation. This is consistent with ceralasertib not being able to significantly radiosensitize NCI-H23 cells at <IC50 concentrations (Supplementary Fig. S2B). Nevertheless, radiosensitization was frequently observed with concentrations around 100–300 nmol/L, well below the clinical Cmax of approximately 5 μmol/L. Interestingly, and similar to olaparib, ceralasertib-induced radiosensitization in p53-deficient cell lines showed weaker concentration dependency under the conditions tested.
Radiosensitization with adavosertib was weak to absent in p53-deficient and p53 mutant/pCHK1 low cell lines and robust radiosensitization could also not be achieved at higher radiotherapy doses in NCI-H1299 or NCI-H23 cells (Fig. 3B; Supplementary Fig. S2B). However, adavosertib was able to significantly sensitize DDR proficient A549 and NCI-H460 cells (Fig. 3B; Supplementary Fig. S2B), particularly at higher concentrations.
Interestingly, although p53-deficient cell lines showed little response to radiotherapy combinations with olaparib, ceralasertib or adavosertib, we observed a trend for stronger concentration-dependent radiosensitization with AZD0156. This was supported by a mixed model two-way ANOVA analysis demonstrating a significant difference between cell lines and their DEFs across drug concentrations (P < 0.0001). p53-deficient NCI-H1703 cells, in particular, were sensitized with 3 nmol/L concentrations, well below the approximate free Cmax of 17 nmol/L interim calculated from a 60-mg clinical dose given in combination with olaparib.
KU-60648 strongly radiosensitized all cell lines within the panel with aDEF75 values nearly doubling in response to 3-fold increases in concentration, demonstrating a strong concentration–effect relationship between radiosensitization and drug concentration. Although cell line responses to KU-60648 varied within the panel, these could not be specifically aligned to subgroups pre-selected here.
In summary, evaluation of DEFs across inhibitor concentrations and cell lines with different genetic/signaling aberrations can provide insights into the concentration dependency of radiopotentiation and has the potential to identify biomarkers of increased sensitization.
Loss of p53 sensitizes A549 cells to radiosensitization with AZD0156
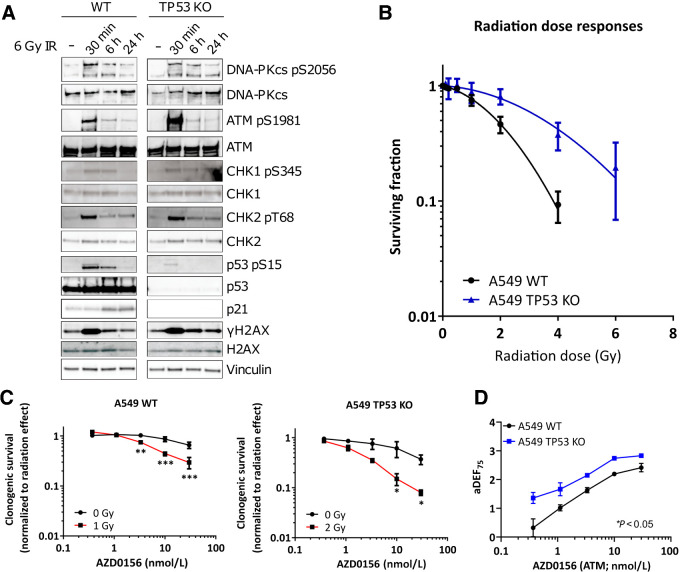
Having observed that p53-deficient cells may be preferentially sensitized by AZD0156, we proceeded to assess the direct consequence of functional loss of p53 on radiopotentiation by AZD0156 using an isogenic cell pair consisting of A549 wild-type (WT) controls and A549 TP53 knock-out (KO) cells. First, we confirmed loss of p53 expression in A549 TP53 KO cells by western blotting and characterized DDR signaling in response to radiotherapy (Fig. 6A). In A549 WT cells, DNA-PK, ATM/CHK2 and ATR/CHK1 signaling pathways were activated within 30 minutes of irradiation (Figs. 6A and 2A). H2AX was phosphorylated within 30 minutes and resolved within 24 hours. In A549 TP53 KO cells, DNA-PK, ATM/CHK2 and ATR/CHK1 signaling pathways were activated within 30 minutes, but loss of p53 expression resulted in p21 stabilization impairment and γH2AX was more sustained (Fig. 6A).
Figure 6.
Characterization of A549 WT and A549 TP53 KO isogenic cell pair. A, Cell lines were treated with a 6 Gy dose of radiotherapy and analyzed at the times indicated. Cell extracts were western blotted for total and phosphorylated (p-) proteins. Vinculin served as a total loading control. B, Clonogenic survival of cell lines after exposure to increasing doses of radiotherapy. C, Clonogenic survival (normalized to untreated DMSO controls of the respective radiotherapy treatment) of cell lines in response to concentrations indicated for AZD0156 alone (black line; 0 Gy) or after radiotherapy at the doses indicated (red line). D, Approximate radiotherapy DEFs at 75% (aDEF75) of each concentration of AZD0156 as determined in (C), with overall dose enhancement determined as significantly greater in TP53 KO cells. Graphs in B–D represent the medians of two independent experiments performed in triplicate, with error bars representing upper and lower limits. Statistical significance was tested using a two-way ANOVA and is indicated where *, P < 0.05; **, P < 0.01; or ***, P < 0.001.
Next, we compared the cell pair's in vitro radiosensitivity. Consistent with p53 loss, we observed that A549 TP53 KO cells were more radioresistant than A549 WT cells (Fig. 6B; refs. 36–38). For radiosensitization with AZD0156, WT cells were treated with 1 Gy and TP53 KO cells with 2 Gy to achieve comparable clonogenic survival of approximately 75%. Although both cell lines were significantly radiosensitized by AZD0156 (Fig. 6C), we observed that overall dose enhancement with AZD0156 was significantly greater following p53 deletion, confirming that p53 loss may indeed provide a vulnerability for radiosensitization with AZD0156 (P = 0.0235 by two-way ANOVA; Fig. 6D).
Discussion
To enhance the efficacy of radiotherapy, it will be crucial to identify combinations with targeted agents that can be dosed to preferentially sensitize tumor, thereby increasing therapeutic index and improving treatment outcomes. Preclinical testing of targeted agents in radiotherapy combinations are often limited by solely focusing on drug doses/concentrations identified as biologically effective in single-agent studies. They also rarely explore how (tumor-specific) biomarkers might influence the MED.
We therefore designed a study allowing radiopotentiation profiling by multiple concentration testing of a panel of targeted DDRi in combination with a fixed radiotherapy dose. This permitted identification of the effective drug concentrations required for radiosensitization and the relationship between drug concentration and radiotherapy effect across cell lines representing different molecular profiles. We demonstrate that where radiopotentiation was observed across our cell line panel, it typically occurred at concentrations lower than those required for single-agent efficacy, an observation that has been previously reported for olaparib in ovarian cancer cells irrespective of BRCA1 status (10). In addition, clinically, there is increasing evidence demonstrating that effective radiosensitization with olaparib occurs with doses as little as 25–50 mg BID, far below the clinical monotherapy dose of 300 mg BID (43, 44). Moreover, these preclinical data demonstrate that the concentrations required for radiopotentiation could be even lower in cell lines harboring existing DDR deficiencies. For example, our data suggested that p53-deficient cells were more readily sensitized by AZD0156, an observation confirmed by CRISPR-mediated knockdown. Indeed, this association with p53 status has been previously reported, warranting more mechanistic follow-up (15, 45, 46).
It was perhaps surprising that ceralasertib or adavosertib were not more radiosensitizing in p53-deficient or p53 mutant/pCHK1 low subgroups, which are inherently sensitive to these agents. Although we were able to confirm that ceralasertib was able to potently radiosensitize p53-deficient NCI-H1299 cells at low concentrations in combination with higher radiotherapy doses, this was not observed in ATM-deficient NCI-H23 cells. Furthermore, adavosertib did not significantly radiosensitize NCI-H1299 or NCI-H23 cells at ≤IC50 concentrations, even when assessed at higher radiotherapy doses. Our data therefore are consistent with a recent report by Parsels and colleagues (47) showing that 100 nmol/L (∼IC50) concentrations of adavosertib did not radiosensitize NCI-H23 cells with high doses of radiotherapy (up to 6 Gy) but significant radiosensitization was achieved with 300 nmol/L olaparib. Furthermore, this study demonstrated that adavosertib and olaparib did not significantly sensitize NCI-H1703 cells, also consistent with our study. Clearly, it will be important to appreciate the mechanism of action for each DDRi in different cancer cell backgrounds to provide a rationale for enhanced radiopotentiation to help inform on patient selection strategies to increase the therapeutic index. This has been exemplified by Parsels and colleagues (47) who went on to demonstrate that radiotherapy combined with WEE1 and PARP inhibition (through nucleotide depletion and PARP trapping) provides enhanced radiosensitization in KRAS mutant lung cancer cell lines through enhanced replication stress.
The DNA-PK inhibitor KU-60648 produced the highest levels of radiosensitization across all cell lines tested. Although some cell lines appeared more readily radiosensitized than others, an expansion of these data with a clinically relevant DNA-PK inhibitor will likely be necessary to strengthen these observations and determine any genetic susceptibilities.
The level of target inhibition that could be achieved by single-agent IC50 concentrations of DDRi in radiotherapy combinations demonstrated that the drug target was already sufficiently inhibited with only a marginal further increase following treatment with an IC90 concentration. This observation was irrespective of genetic background and/or DDR proficiency of the cell lines, suggesting that IC50 concentrations of DDRi can potently inhibit radiotherapy–induced activation of a specific target to exert robust radiosensitization. This demonstrates that it is not only the level of target inhibition per se, but also the level of dependence of the cell on the drug target that drives the magnitude of sensitivity, further highlighting the importance of patient selection for DDRi radiotherapy combinations.
Testing multiple drug concentrations in radiotherapy combinations not only allows identification of the MED but it also allows assessment of the relationship between concentration and effect, that is, the level of dose enhancement one might expect following an increase in drug concentration. These data are important to consider when designing phase I dose-escalation trials. In particular, olaparib, ceralasertib, and adavosertib are mild radiosensitizers demonstrating only a moderate increase in DEF (in the range of approximately 1.3 to 2) with 3- or 5-fold increases in concentration. Thus, these inhibitors have a desirable safety profile and are more likely to spare normal tissue at MED, particularly if they can be targeted to tumors with an inherent susceptibility to the specific DDRi. Conversely, analysis of the concentration-dependence profile of KU-60648 demonstrated that 3-fold increases in concentration resulted in 2-fold increases in DEF with all cells within the panel responding similarly. Consistently, the potent and selective DNA-PK inhibitor, AZD7648, was recently also shown to achieve similar levels of radiopotentiation in both A549 and H1299 cells in vitro and in xenografts (17). Phase I clinical trials for an agent with this type of dose-dependence profile when combined with external beam radiotherapy will, therefore, need to be designed in a way that better anticipates high levels of potential toxicity from potent radiosensitization by choosing low starting doses and performing moderate and careful dose escalations. Clearly, it will also be important to identify biomarkers that render cells significantly more sensitive to radiotherapy combinations with DNA-PK inhibitors to increase therapeutic index. This will be particularly important because cells in healthy tissues rely on NHEJ to repair radiotherapy–induced DSBs.
AZD0156 was also a strong radiosensitizer but, in contrast with KU-60648, our data identified p53 deficiency as a potential biomarker, demonstrating that by targeting the right genetic background one can improve therapeutic index and avoid using higher concentrations of the inhibitor that could also “co-sensitize” normal tissue.
These data only provide the first step in assessing safety of DDRi radiotherapy combinations. To validate our findings, testing in carefully designed in vivo studies will be essential. Moreover, to truly assess therapeutic index, the inhibitors will also have to be vigorously tested in immune-competent models. The immune system plays a critical role not only in efficacy but also in normal tissue toxicities associated with radiotherapy treatment such as pneumonitis, fibrosis, esophagitis, and mucositis (48). Indeed, some key DDR proteins have been implicated in playing a functional role in the immune response, highlighting the importance of investigating the exact mechanisms underlying these roles in the context of radiotherapy combination efficacy and normal tissue toxicities (49–52).
In conclusion, we have demonstrated that multiple concentration testing can provide valuable insights into effective dose, concentration dependency and biomarkers of radiosensitization that can guide DDRi combinations with radiotherapy. When coupled with appropriate follow-up experiments in preclinical in vivo models, this has the potential to guide more effective approaches in the clinic.
Authors’ Disclosures
M.J. O'Connor reports employment and shareholder of AstraZeneca. S.J. Gill reports employment and shareholder of AstraZeneca. R.L. Lloyd reports employment with AstraZeneca at the time of conducting these studies. J. Cairns reports employment with AstraZeneca and may own stock in the company. A. Lau reports other support from Astrazeneca outside the submitted work; as well as reports employment with and shareholder in Astrazeneca. S.M. Galbraith reports other support from AstraZeneca PLC during the conduct of the study; as well as other support from BB Biotech outside the submitted work. No disclosures were reported by the other authors.
Acknowledgments
We thank the AstraZeneca Clinical and Quantitative Pharmacology team for providing clinical exposure data. We are also grateful to Dr. Josep Forment for providing comments on the article.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Contributions
M.J. O'Connor: Conceptualization, supervision, writing–review and editing. S.J. Gill: Formal analysis, validation, investigation, methodology, writing–original draft, writing–review and editing. P.W.G. Wijnhoven: Formal analysis, investigation, writing–review and editing. J.H.L. Fok: Formal analysis, investigation, writing–review and editing. R.L. Lloyd: Formal analysis, investigation. J. Cairns: Formal analysis. J. Armenia: Data curation, visualization. J. Nikkilä: Resources. A. Lau: Supervision. C.J. Bakkenist: Supervision. S.M. Galbraith: Conceptualization, supervision, writing–review and editing. C. Vens: Conceptualization, supervision, methodology, writing–review and editing.
References
- 1. Begg AC, Stewart FA, Vens C. Strategies to improve radiotherapy with targeted drugs. Nat Rev Cancer 2011;11:239–53. [DOI] [PubMed] [Google Scholar]
- 2. Foote KM, Nissink JWM, McGuire T, Turner P, Guichard S, Yates JWT, et al. Discovery and characterization of AZD6738, a potent inhibitor of ataxia telangiectasia mutated and rad3 related (ATR) kinase with application as an anticancer agent. J Med Chem 2018;61:9889–907. [DOI] [PubMed] [Google Scholar]
- 3. Hirai H, Iwasawa Y, Okada M, Arai T, Nishibata T, Kobayashi M, et al. Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol Cancer Ther 2009;8:2992–3000. [DOI] [PubMed] [Google Scholar]
- 4. Menear KA, Adcock C, Boulter R, Cockcroft XL, Copsey L, Cranston A, et al. 4-[3-(4-cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H-phthalazin- 1-one: a novel bioavailable inhibitor of poly(ADP-ribose) polymerase-1. J Med Chem 2008;51:6581–91. [DOI] [PubMed] [Google Scholar]
- 5. O'Connor MJ. Targeting the DNA damage response in cancer. Mol Cell 2015;60:547–60. [DOI] [PubMed] [Google Scholar]
- 6. Pike KG, Barlaam B, Cadogan E, Campbell A, Chen Y, Colclough N, et al. The identification of potent, selective, and orally available inhibitors of ataxia telangiectasia mutated (ATM) kinase: the discovery of AZD0156 (8-{6-[3-(Dimethylamino)propoxy]pyridin-3-yl}-3-methyl-1-(tetrahydro-2 H-pyran-4-yl)-1,3-dihydro-2 H-imidazo[4,5- c]quinolin-2-one). J Med Chem 2018;61:3823–41. [DOI] [PubMed] [Google Scholar]
- 7. U.S. National Institute of Health. n.d. May 17, 2017. ClinicalTrials.gov. Available from:www.clinicaltrials.gov. May 17, 2017. [Google Scholar]
- 8. Karnak D, Engelke CG, Parsels LA, Kausar T, Wei D, Robertson JR, et al. Combined inhibition of Wee1 and PARP1/2 for radiosensitization in pancreatic cancer. Clin Cancer Res 2014;20:5085–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Senra JM, Telfer BA, Cherry KE, McCrudden CM, Hirst DG, O'Connor MJ, et al. Inhibition of PARP-1 by olaparib (AZD2281) increases the radiosensitivity of a lung tumor xenograft. Mol Cancer Ther 2011;10:1949–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Verhagen CV, de Haan R, Hageman F, Oostendorp TP, Carli AL, O'Connor MJ, et al. Extent of radiosensitization by the PARP inhibitor olaparib depends on its dose, the radiation dose and the integrity of the homologous recombination pathway of tumor cells. Radiother Oncol 2015;116:358–65. [DOI] [PubMed] [Google Scholar]
- 11. Barazzuol L, Jena R, Burnet NG, Meira LB, Jeynes JC, Kirkby KJ, et al. Evaluation of poly (ADP-ribose) polymerase inhibitor ABT-888 combined with radiotherapy and temozolomide in glioblastoma. Radiat Oncol 2013;8:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol 2011;7:428–30. [DOI] [PubMed] [Google Scholar]
- 13. Sarcar B, Kahali S, Prabhu AH, Shumway SD, Xu Y, Demuth T, et al. Targeting radiation-induced G(2) checkpoint activation with the Wee-1 inhibitor MK-1775 in glioblastoma cell lines. Mol Cancer Ther 2011;10:2405–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dillon MT, Barker HE, Pedersen M, Hafsi H, Bhide SA, Newbold KL, et al. Radiosensitization by the ATR inhibitor AZD6738 through generation of acentric micronuclei. Mol Cancer Ther 2017;16:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Durant ST, Zheng L, Wang Y, Chen K, Zhang L, Zhang T, et al. The brain-penetrant clinical ATM inhibitor AZD1390 radiosensitizes and improves survival of preclinical brain tumor models. Sci Adv 2018;4:eaat1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Riches LC, Trinidad AG, Hughes G, Jones GN, Hughes AM, Thomason AG, et al. Pharmacology of the ATM inhibitor AZD0156: potentiation of irradiation and olaparib responses preclinically. Mol Cancer Ther 2020;19:13–25. [DOI] [PubMed] [Google Scholar]
- 17. Fok JHL, Ramos-Montoya A, Vazquez-Chantada M, Wijnhoven PWG, Follia V, James N, et al. AZD7648 is a potent and selective DNA-PK inhibitor that enhances radiation, chemotherapy and olaparib activity. Nat Commun 2019;10:5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 2008;9:616–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Coleman CN, Higgins GS, Brown JM, Baumann M, Kirsch DG, Willers H, et al. Improving the predictive value of preclinical studies in support of radiotherapy clinical trials. Clin Cancer Res 2016;22:3138–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stone HB, Bernhard EJ, Coleman CN, Deye J, Capala J, Mitchell JB, et al. Preclinical data on efficacy of 10 drug-radiation combinations: evaluations, concerns, and recommendations. Transl Oncol 2016;9:46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sharma RA, Plummer R, Stock JK, Greenhalgh TA, Ataman O, Kelly S, et al. Clinical development of new drug-radiotherapy combinations. Nat Rev Clin Oncol 2016;13:627–42. [DOI] [PubMed] [Google Scholar]
- 22. Cano C, Saravanan K, Bailey C, Bardos J, Curtin NJ, Frigerio M, et al. 1-substituted (Dibenzo[b, d]thiophen-4-yl)-2-morpholino-4H-chromen-4-ones endowed with dual DNA-PK/PI3-K inhibitory activity. J Med Chem 2013;56:6386–401. [DOI] [PubMed] [Google Scholar]
- 23. Matheson CJ, Casalvieri KA, Backos DS, Reigan P. Development of potent pyrazolopyrimidinone-based WEE1 inhibitors with limited single-agent cytotoxicity for cancer therapy. ChemMedChem 2018;13:1681–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER III, et al. Next-generation characterization of the cancer cell line encyclopedia. Nature 2019;569:503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in the cancer genome atlas. Cell 2018;173:321–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Foundation Medicine. n.d. December 2020. FoundationOne CDx Technical Specifications. Available from:https://assets.ctfassets.net/w98cd481qyp0/YqqKHaqQmFeqc5ueQk48w/0a34fcdaa3a71dbe460cdcb01cebe8ad/F1CDx_Technical_Specifications_072020.pdf.December 2020. [Google Scholar]
- 27. Jones RM, Petermann E. Replication fork dynamics and the DNA damage response. Biochem J 2012;443:13–26. [DOI] [PubMed] [Google Scholar]
- 28. Saldivar JC, Hamperl S, Bocek MJ, Chung M, Bass TE, Cisneros-Soberanis F, et al. An intrinsic S/G2 checkpoint enforced by ATR. Science 2018;361:806–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K, et al. Cell-cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double-strand breaks. J Biol Chem 2005;280:14709–15. [DOI] [PubMed] [Google Scholar]
- 30. Kozlov SV, Graham ME, Peng C, Chen P, Robinson PJ, Lavin MF. Involvement of novel autophosphorylation sites in ATM activation. EMBO J 2006;25:3504–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a potential mediator of p53 tumor suppression. Cell 1993;75:817–25. [DOI] [PubMed] [Google Scholar]
- 32. Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol 2001;21:4129–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weber AM, Drobnitzky N, Devery AM, Bokobza SM, Adams RA, Maughan TS, et al. Phenotypic consequences of somatic mutations in the ataxia-telangiectasia mutated gene in non–small cell lung cancer. Oncotarget 2016;7:60807–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 2018;46:D1062-D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vendetti FP, Lau A, Schamus S, Conrads TP, O'Connor MJ, Bakkenist CJ. The orally active and bioavailable ATR kinase inhibitor AZD6738 potentiates the anti-tumor effects of cisplatin to resolve ATM-deficient non-small cell lung cancer in vivo. Oncotarget 2015;6:44289–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fei P, El-Deiry WS. P53 and radiation responses. Oncogene 2003;22:5774–83. [DOI] [PubMed] [Google Scholar]
- 37. Lee JM, Bernstein A. p53 mutations increase resistance to ionizing radiation. Proc Natl Acad Sci U S A 1993;90:5742–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ochi S, Nishiyama Y, Morita A. Development of p53-targeting drugs that increase radioresistance in normal tissues. J Med Invest 2019;66:219–23. [DOI] [PubMed] [Google Scholar]
- 39. Lloyd RL, Wijnhoven PWG, Ramos-Montoya A, Wilson Z, Illuzzi G, Falenta K, et al. Combined PARP and ATR inhibition potentiates genome instability and cell death in ATM-deficient cancer cells. Oncogene 2020;39:4869–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McGowan CH, Russell P. Human Wee1 kinase inhibits cell division by phosphorylating p34cdc2 exclusively on Tyr15. EMBO J 1993;12:75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol 2006;8:870–6. [DOI] [PubMed] [Google Scholar]
- 42. Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res 2012;72:5588–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. de Haan R, van Werkhoven E, van den Heuvel MM, Peulen HMU, Sonke GS, Elkhuizen P, et al. Study protocols of three parallel phase 1 trials combining radical radiotherapy with the PARP inhibitor olaparib. BMC Cancer 2019;19:901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Karam SD, Reddy K, Blatchford PJ, Waxweiler T, DeLouize AM, Oweida A, et al. Final report of a phase I trial of olaparib with cetuximab and radiation for heavy smoker patients with locally advanced head and neck cancer. Clin Cancer Res 2018;24:4949–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Biddlestone-Thorpe L, Sajjad M, Rosenberg E, Beckta JM, Valerie NC, Tokarz M, et al. ATM kinase inhibition preferentially sensitizes p53-mutant glioma to ionizing radiation. Clin Cancer Res 2013;19:3189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Takao N, Kato H, Mori R, Morrison C, Sonada E, Sun X, et al. Disruption of ATM in p53-null cells causes multiple functional abnormalities in cellular response to ionizing radiation. Oncogene 1999;18:7002–9. [DOI] [PubMed] [Google Scholar]
- 47. Parsels LA, Karnak D, Parsels JD, Zhang Q, Velez-Padilla J, Reichert ZR, et al. PARP1 trapping and DNA replication stress enhance radiosensitization with combined WEE1 and PARP Inhibitors. Mol Cancer Res 2018;16:222–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Prasanna PG, Stone HB, Wong RS, Capala J, Bernhard EJ, Vikram B, et al. Normal tissue protection for improving radiotherapy: where are the Gaps? Transl Cancer Res 2012;1:35–48. [PMC free article] [PubMed] [Google Scholar]
- 49. Chatzinikolaou G, Karakasilioti I, Garinis GA. DNA damage and innate immunity: links and trade-offs. Trends Immunol 2014;35:429–35. [DOI] [PubMed] [Google Scholar]
- 50. Miyamoto S. Nuclear initiated NF-kappaB signaling: NEMO and ATM take center stage. Cell Res 2011;21:116–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rosado MM, Bennici E, Novelli F, Pioli C. Beyond DNA repair, the immunological role of PARP-1 and its siblings. Immunology 2013;139:428–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Taccioli GE, Amatucci AG, Beamish HJ, Gell D, Xiang XH, Torres Arzayus MI, et al. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity 1998;9:355–66. [DOI] [PubMed] [Google Scholar]