Abstract
Mutations in the receptor binding domain (RBD) in SARS-CoV-2 are shown to enhance its replication, transmissibility, and binding to host cells. Recently, a new strain is reported in India that includes a mutation (T478K, and L452R) in the RBD, that is possibly increasing the infection rate. Here, using Molecular Mechanics (MM) and Monte Carlo (MC) sampling, we show that the double mutant variant of SARS-CoV-2 induced conformational change in ACE2-E37, which enhanced the electrostatic interactions by the formation of a salt-bridge with SARS-CoV-2-R403. In addition, we observed that the double mutated structure induced a significant change in the salt-bridge electrostatic interaction between RBD-T500 and ACE2-D355. Where that this interaction lost more than 70% of its value compared to its value in WT protein.
Keywords: SARS-CoV-2, Binding energy, Protein-Protein Interactions, Electrostatic Interactions, Monte Carlo
The Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) belongs to the large family of viruses that were reported to cause respiratory diseases in humans called Corona viruses (CoVs).[1] Coronaviruses family have caused two previous severe pandemics: SARS and MERS in 2003 and 2012.[1] SARS-CoV-2 is the virus that was reported to cause the novel COVID-19 pandemic. In December 2019, a cluster of severe unidentified pneumonia disease have been reported in Wuhan, China. In January 2020, COVID-19 outbreak emerged substantially and explosively worldwide till it was officially considered as a pandemic by the WHO. As of July 2021, the total number of confirmed COVID-19 cases have accumulated to over 197 million cases while the death toll was escalated to over 4 million deaths affecting more than 180 countries.[1], [2], [3], [4], [5], [6] The angiotensin converting enzyme 2 (ACE2) is the target protein in humans (hACE2) and many other species that mediates the cell entry via binding with the SARS-CoV-2 spike protein.[4], [7], [8], [9] Like all other viruses, SARS-CoV-2 exhibits changes all the time. Some changes may affect the viruses’ properties “severity, spreading rate, ...etc. while others may have no effect at all. It was recently reported that the mutations in the virus have led to an increase in the virus transmissibility due to change in the receptor-binding domain (RBD) or in the furin cleavage site.[10], [11], [12], [13] There are four variants of concern (VOC) so far; alpha, Beta, Gamma, and Delta have been emerged out of the original virus.[14] The Delta variant has mutations in the S protein that are suggested to affect the virus transmissibility alongside its susceptibility to be neutralized by the previous COVID-19 variants’ antibodies.[15], [16], [17] Those changes include two mutations in the RBD; Leucine at position 452 to Arginine (L452R) and Threonine at position 478 to Lysine (T478K).[16], [17] The mutation L452R have been reported to enhance the binding affinity to the host entry receptor ACE2, transmissibility, fitness, and infectivity and so improves the viral replication.[18], [19] Besides, L452R mutation recently reported to enhance the viral replication by increasing the S protein stability and viral infectivity and viral fusogenicity.[19], [20] The infectivity of the virus is mainly affected by different protein-protein interactions. [21] Mutations occur in the vital residues at the binding site would alter the adhesion of virus to host cells. [20] In this research, we study the effect of both of L452R and T478K variants in the RBD of the SARS-CoV2 on its binding to ACE2. The electrostatic interactions are known to be dominant over various protein-protein interactions. [15] Therefore, we investigate the role of electrostatic interactions on the energetics of binding. We herein used both of Molecular mechanics MM and Monte Carlo MC simulations to evaluate the interaction between RBD of S-protein and ACE2 at molecular level. We performed our calculations for the aforementioned mutations and compared our results with the wild type (Native) protein. All calculations were based on the protein structure retrieved from the protein data bank (PDB ID: 6m17). Firstly, the crystal structure was optimized using the python-based code openMM [22]. Followed by the generation of rotamers using MCCE [23], where that each rotatable bond was rotated by to, properly, sample the sidechains’ conformations. To build both variants L452R and T478K, the sidechains of L452 and T478 were replaced by sidechains of Arginine and Lysine, respectively, using MCCE. Finally, optimized protein structures with the most occupied conformers were used to calculate electrostatic interactions by Poisson Boltzmann equation. Where that DELPHI is used to calculate pairwise electrostatic interactions between conformers. Then, Boltzmann distribution for all conformers were computed at pH 7 by MCCE using MC sampling for the WT and mutated structures. The contribution of the electrostatic and the van der Waals to the interaction energies of SARS-CoV-2/ACE2 were calculated for single and double mutated structures as well as the WT protein. In contrary to T478K, the L452R variant is shown to enhance the total binding energy between RBD and ACE2 to be about 2.63 kcal/mol more negative than its value in WT protein. This value is shown to be enhanced even more in double mutated protein to be ∼7.29 kcal/mol more negative than its value in WT protein, see Table 1 .
Table 1.
The interaction energies between SARS-CoV-2-RBD and ACE2 in WT, single mutated proteins L452R and T478K, and protein structure with T478K and L452R mutations combined.
| Coulomb | Van der Waals | Total | |
|---|---|---|---|
| T478K and L452R | -7.98 | -38.69 | -46.67 |
| L452R | -9.98 | -32.03 | -42.01 |
| WT | -6.64 | -32.74 | -39.38 |
| T478K | -6.09 | -32.79 | -38.88 |
| All values are in Kcal/mol | |||
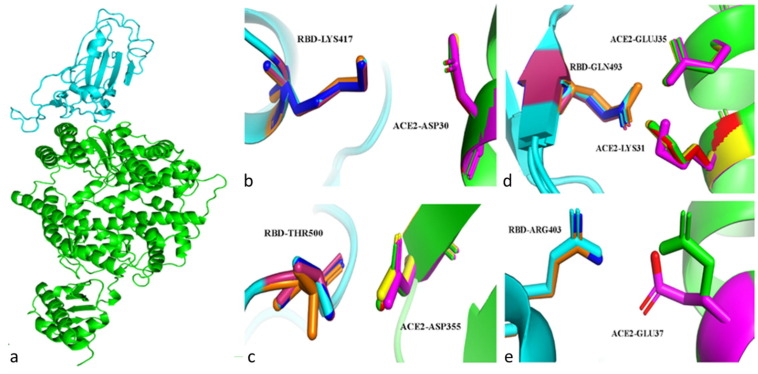
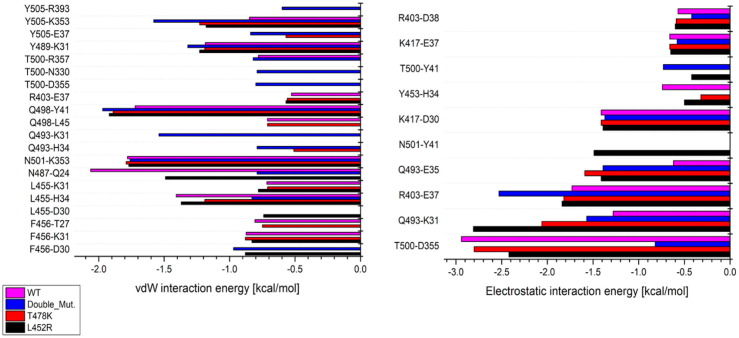
For WT protein, vdW interaction is shown to be maximum between RBD-N487 and ACE2-Q24 with ∼ -2.06 kcal/mol. Double mutation has noticeable effect on the vdW interaction between RBD-N487 and ACE2-Q24 by which interaction decreased by ∼ 1.27 Kcal/mol. Mutations induced vdW interactions with maximum values between RBD- Q498 and ACE2- Y41 with ∼ -1.97, -1.89 and -1.92 kcal/mol for double and single mutations T478K and L452R, respectively. Based on our calculations, the formation of RDB-R403/ACE2-D30, RBD-K417/ACE2-D30, RBD-K417/ACE2-E37, and RBD-R403/ACE2-D38 salt bridges were conserved among all structures with interaction energy of ∼ -0.4, -1.4, -0.6 and -0.4 kcal/mol on average, respectively. Upon mutations, the salt bridge between RBD-R403 and ACE2-E35 is shown to be formed with electrostatic energies of about -0.4 kcal/mol interaction energy. Single and double mutations in RBD are shown to enhance the electrostatic energy between RBD-Q493 and ACE2-K31, where that L452R mutation induced the maximum increase in the electrostatic interaction energy with about -1.53 kcal/mol compared to its value in WT protein. In WT and protein with single mutation T478K, Maximum electrostatic interactions were reported between RBD-T500 and ACE2-D355 with ∼ -2.94 and -2.8 kcal/mol, respectively. This interaction is shown to be, slightly, reduced by the single mutation L452R to be 0.52 kcal/mol more positive compared to WT protein. Upon double mutation, RBD-T500 and ACE2-D355 exhibit less attraction than that for WT by ∼ -2.12 kcal/mol. In contrary to single mutations, double mutation in RBD is shown to induce a significant conformational change in sidechain of ACE2-E37 residue, Figure 1 . Single mutations T478K and L452R induced an increase in the electrostatic interaction between RBD-R403 and ACE2-E37 with only ∼0.1 kcal/mol. The double mutation is shown to negatively shift the interaction energy for the salt bridge between RBD-R403 and ACE2-E37 to be ∼0.8 kcal/mol more negative than it is value in WT. Another salt bridge is formed between RBD-Q493 and ACE2-K31 with electrostatic interaction energy of ∼ -1.28 kcal /mol for WT, see Figure 2 .
Figure 1.
a. The SARS-CoV-2 RBD protein (cyan) with ACE2 protein (green). b-e. The interactions between selected residues at the RBD/ACE2 interface in WT protein and protein structures with single RBD mutations (L452R and T478K) and double mutation. The WT RBD and ACE2 are presented in cyan and green, respectively. RBDs of L452R, and T478K mutated structure are shown in Pink and Blue respectively, while ACE2 associated with these structures are shown in Yellow and Red, respectively. Finally, the double mutated structure is presented in Orange and Magenta for RBD and ACE2.
Figure 2.
Selected favorable vdW and Electrostatic interactions between residues at the RBD/ACE2 binding interface. The x-axis is the interaction energies (Energy in kcal/mol), while y-axis reflects the selected pairs of residues at the RBD/ACE2 interface. a) and b) Depiction of vdW and electrostatic interaction energies, respectively, associated with each structure (wild type (magenta), T478K mutated structure (red), L452R (black), and structures with double mutations (blue).
Herein, we showed that the binding affinity of SARS-CoV-2 to human ACE2 is higher in the double mutated structure than that in WT because of the significant change in the electrostatic and vdW interactions. Our data demonstrated a conformational change in the sidechain of E37 in ACE2 protein upon double mutation in the RBD protein, which induced a rise in the electrostatic interaction between this residue and R403 in RBD protein. Moreover, the salt-bridge electrostatic interaction decreased between RBD-T500 and ACE2-D355 to be 2.12 less than that in WT protein.
CRediT authorship contribution statement
Shaimaa S. Goher: . Fedaa Ali: Formal analysis. Muhamed Amin: Conceptualization, Supervision.
References
- 1.Zhu N.a., Zhang D., Wang W., Li X., Yang B.o., Song J., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karim, S. S. A.; de Oliveira, T. C or r e Sp Ondence New SARS-CoV-2 Variants — Clinical , Public Health , and Vaccine Implications. N Engl J Med2021, NEJMc2100362. [DOI] [PMC free article] [PubMed]
- 3.Wu F., Zhao S.u., Yu B., Chen Y.-M., Wang W., Song Z.-G., et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jt. WHO-China Study Team Rep. 2021;No. February:120. [Google Scholar]
- 6.Chaplin S. COVID-19: A Brief History and Treatments in Development. Prescriber. 2020;31(5):23–28. doi: 10.1002/psb.1843. [DOI] [Google Scholar]
- 7.Yan R., Zhang Y., Li Y., Ye F., Guo Y., Xia L.u., et al. Structural Basis for the Different States of the Spike Protein of SARS-CoV-2 in Complex with ACE2. Cell Res. 2021;31(6):717–719. doi: 10.1038/s41422-021-00490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z., Zhang Y., Liu K., Li Y., Lu Q., Wang Q., et al. The Molecular Basis for SARS-CoV-2 Binding to Dog ACE2. Nat. Commun. 2021;12(1) doi: 10.1038/s41467-021-24326-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ali F., Elserafy M., Alkordi M.H., Amin M. ACE2 Coding Variants in Different Populations and Their Potential Impact on SARS-CoV-2 Binding Affinity. Biochem. Biophys. Reports. 2020;24:100798. doi: 10.1016/j.bbrep.2020.100798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. SARS-CoV-2 Variants, Spike Mutations and Immune Escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ali F., Kasry A., Amin M. The New SARS-CoV-2 Strain Shows a Stronger Binding Affinity to ACE2 Due to N501Y Mutant. Med. Drug Discov. 2021;10:100086. doi: 10.1016/j.medidd.2021.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodcroft E.B., Domman D.B., Snyder D.J., Oguntuyo K., Van Diest M., Densmore K.H., et al. Emergence in Late 2020 of Multiple Lineages of SARS-CoV-2 Spike Protein Variants Affecting Amino Acid Position 677. medRxiv Prepr. Serv. Heal. Sci. 2021 doi: 10.1101/2021.02.12.21251658. [DOI] [Google Scholar]
- 13.Khailany R.A., Safdar M., Ozaslan M. Genomic Characterization of a Novel SARS-CoV-2. Gene Reports. 2020;19(1):1–5. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WHO. Tracking SARS-CoV-2 variants.
- 15.Li, B.; Deng, A.; Li, K.; Hu, Y.; Li, Z.; Xiong, Q.; Liu, Z.; Guo, Q.; Zou, L.; Zhang, H.; et al. Viral Infection and Transmission in a Large Well-Traced Outbreak Caused by the Delta SARS-CoV-2 Variant. medRxiv2021, 2021.07.07.21260122. https://doi.org/10.1101/2021.07.07.21260122.
- 16.Yadav, P. D.; Mohandas, S.; Shete, A. M.; Nyayanit, D. A.; Gupta, N.; Patil, D. Y.; Sapkal, G. N.; Potdar, V.; Kadam, M.; Kumar, A.; et al. SARS CoV-2 Variant B.1.617.1 Is Highly Pathogenic in Hamsters than B.1 Variant. bioRxiv2021, 2021.05.05.442760. https://doi.org/10.1101/2021.05.05.442760.
- 17.Hoffmann, M.; Hofmann-Winkler, H.; Krüger, N.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Sidarovich, A.; Moldenhauer, A.-S.; Winkler, M. S.; Schulz, S.; et al. SARS-CoV-2 Variant B.1.617 Is Resistant to Bamlanivimab and Evades Antibodies Induced by Infection and Vaccination. bioRxiv2021, 2021.05.04.442663. https://doi.org/10.1101/2021.05.04.442663. [DOI] [PMC free article] [PubMed]
- 18.Ozono S., Zhang Y., Ode H., Sano K., Tan T.S., Imai K., et al. SARS-CoV-2 D614G Spike Mutation Increases Entry Efficiency with Enhanced ACE2-Binding Affinity. Nat. Commun. 2021;12(1) doi: 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tchesnokova, V.; Kulakesara, H.; Larson, L.; Bowers, V.; Rechkina, E.; Kisiela, D.; Sledneva, Y.; Choudhury, D.; Maslova, I.; Deng, K.; et al. Acquisition of the L452R Mutation in the ACE2-Binding Interface of Spike Protein Triggers Recent Massive Expansion of SARS-Cov-2 Variants. bioRxiv2021, 2021.02.22.432189. https://doi.org/10.1101/2021.02.22.432189. [DOI] [PMC free article] [PubMed]
- 20.Motozono C., Toyoda M., Zahradnik J., Saito A., Nasser H., Tan T.S., et al. SARS-CoV-2 Spike L452R Variant Evades Cellular Immunity and Increases Infectivity. Cell Host Microbe. 2021;29(7):1124–1136.e11. doi: 10.1016/j.chom.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norel R., Sheinerman F., Petrey D., Honig B. Electrostatic Contributions to Protein-Protein Interactions: Fast Energetic Filters for Docking and Their Physical Basis. Protein Sci. 2008;10(11):2147–2161. doi: 10.1110/ps.12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eastman P., Swails J., Chodera J.D., McGibbon R.T., Zhao Y., Beauchamp K.A., et al. OpenMM 7: Rapid Development of High Performance Algorithms for Molecular Dynamics. PLOS Comput. Biol. 2017;13(7):e1005659. doi: 10.1371/journal.pcbi.1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song Y., Mao J., Gunner M.R. MCCE2: Improving Protein PKa Calculations with Extensive Side Chain Rotamer Sampling. J. Comput. Chem. 2009;30(14):2231–2247. doi: 10.1002/jcc.21222. [DOI] [PMC free article] [PubMed] [Google Scholar]