Abstract
The emergence of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), which is the etiologic agent of the coronavirus disease 2019 (COVID-19) pandemic, has dominated all aspects of life since of 2020. Research studies on the virus and exploration of therapeutic and preventive strategies has been moving at rapid rates to control the pandemic. In the field of bioinformatics or computational and structural biology, recent research strategies have used multiple disciplines to compile large datasets to uncover statistical correlations and significance, visualize and model proteins, perform molecular dynamics simulations, and employ the help of artificial intelligence and machine learning to harness computational processing power to further the research on COVID-19, including drug screening, drug design, vaccine development, prognosis prediction, and outbreak prediction. These recent developments should help us better understand the viral disease and develop the much-needed therapies and strategies for the management of COVID-19.
Keywords: COVID-19, SARS-CoV-2, Drug screening, Drug design, Vaccine development, Disease prediction, Artificial intelligence, Machine learning
1. Introduction
The coronavirus disease 2019 (COVID-19) is caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), which is thought to have a zoonotic origin and started as an outbreak in Wuhan, China, that later turned into a global pandemic. As of July 2021, the virus has led to>184 million people infected and over 3.9 million deaths globally, despite periodic controls [1]. This number is way higher than a combined death toll from the previous coronavirus outbreaks of the 2002 SARS and 2016 MERS (Middle East Respiratory Syndrome) of less than 2000 [2], [3]. SARS-CoV-2 is a single-stranded, enveloped virus that possesses a positive-sense RNA genome of roughly 29.9 kb in length, encoding the spike (S), envelope (E), membrane (M), and nucleocapsid (N) structural proteins and multiple other nonstructural proteins [4], [5], [6]. The virus’ life cycle initiates from binding to host angiotensin converting enzyme 2 (ACE2) by S protein, followed by viral genome release into cytoplasm. Precursor polyprotein is further auto-cleaved into various structural and non-structural proteins via papain-like protease (nsp3) and main protease (Mpro or 3CLpro; nsp5). Viral assembly takes place in ER-Golgi intermediate compartment, and nascent virions are released after S protein glycosylation in Golgi apparatus [7]. Patients with COVID-19 usually experience fever, cough, and dyspnea; however, some patients may be asymptomatic, while some others may develop fulminant disease and require intensive care [8].
Significant research efforts have been invested in COVID-19 research, which generated several vaccine and drug candidates [9], [10]. However, full immunization coverage and therapeutic efficacy evaluation in real-world situation remains an issue, which necessitates the continuous development of optimal therapeutic and prophylactic strategies for better management of COVID-19. Such development can be accelerated using bioinformatics, which has been rapidly evolving in recent years and is capable of tackling issues at a scale that previously would not have been feasible. This includes computational and structural biology, which is a relatively new frontier, but has the ability to ‘decode’ pathogens and hosts based on their genomic sequences, thus allowing researchers to predict and accelerate their understanding of the pathogen and also explore various strategies to help curb its spread. This aspect is critical to public health and has since garnered importance over the last decades with increasing number of emerging and re-emerging viral infections (such as influenza, Ebola, and Zika). The technology is further made powerful with the fast-paced development of computational technology, particularly in artificial intelligence (AI) and machine learning, that now has begun to see increasing applications in biology, medicine, and public health, and revolutionizing the way we approach a disease. Recently, it has been widely used for drug screening, vaccine/drug design and prediction of disease to tackle the COVID-19 pandemic. In this review, we summarize how computational and structural biology and AI platforms have been applied in the current pandemic.
2. Virtual drug screening
2.1. Identification of novel drugs
When the COVID-19 pandemic hit, one of the biggest concerns was finding an active antiviral. Structure-based in silico screening allows screening libraries of pharmacologically active compounds with documented activities to confer insight on how they may dictate interactions with host or viral proteins [11], [12]. Recently, computational models using molecular docking screening followed by absorption, distribution, metabolism, excretion, and toxicity (ADMET) analysis and molecular dynamics simulations have been widely utilized to identify compounds that potentially target SARS-CoV-2 proteins. Compounds identified include potential SARS-CoV-2 S receptor-binding domain (RBD)-specific terpenes NPACT01552, NPACT01557, and NPACT00631 [13], Mpro inhibitors tinosponone [14], ChEMBL275592, montelukast, ChEMBL288347 [15], quercetin-3-O-rhamnoside [16], and biflavone amentoflavone [17], RNA-dependent RNA polymerase (RdRp) inhibitors Galidesivir and the two drug-like compounds CID123624208 and CID11687749 [18]. Such method could also be utilized for high-throughput screening. A study screening plant secondary metabolites suggested flavonoid glycosides, biflavonoids, ellagitannins, anthocyanidins, and triterpenes to be potential TMPRSS2, SARS-CoV-2 S, Mpro and RdRp inhibitors [19]. Of note, one of the top-ranked triterpenoid saponins glycyrrhizic acid (glycyrrhizin) has demonstrated antiviral activities against SARS-CoV [20] and SARS-CoV-2 [21] in vitro and are being evaluated in clinical trials [22]. Another study incorporated molecular docking with machine learning to further expedite the screening procedure and identified six potential Mpro inhibitors from over 2000 natural compounds [23].
2.2. Drug repurposing
Computational approaches such as network-based or expression-based algorithms and docking simulations have also been widely applied during the pandemic to identify candidates for drug repurposing [24], [25]. Incorporation of these methods with AI platforms may facilitate more efficient large-scale screening, and in vitro validation may further improve the platforms’ accuracy. For instance, Ke et al. constructed a deep neural network (DNN) platform to screen thousands of previously identified antivirals against SARS-CoV, influenza virus, and human immunodeficiency virus (HIV) or known 3CL pro inhibitors. The predicted drugs were then verified in vitro with a similar feline coronavirus, feline infectious peritonitis (FIP) virus, and reconfigured into the AI algorithm to refine future predictions [26]. Aside from antivirals, due to COVID-19 induced inflammatory response, databases were screened to locate clinical drugs with anti-inflammatory capabilities. For example, the Janus kinase (JAK) inhibitor baricitinib was predicted to be useful by BenevolentAI, a platform that combines Monte Carlo tree search (MCTS), neural networks, and symbolic AI [27], and was further verified for its anti-inflammatory and antiviral activities in vitro and in a small group of COVID-19 patients [28] with bigger clinical trials underway. AI can also be used to analyze how combinations of certain approved drugs affect their efficacy. IDentif.AI, a platform based on orthogonal array composite design (OACD), was utilized to identify a triple-drug combination of remdesivir, ritonavir, and lopinavir that increased antiviral efficacy by 6.5-fold compared to remdesivir alone in vitro [29]. While validation needs to be done in vivo, the applications of AI to predict synergistic effects can provide new platforms of developing treatment modalities.
2.3. Identification of druggable targets
Interestingly, computational analyses can be further adapted for identifying novel drug targets, such as host factors, in curbing the viral infection. For example, Gordon et al. established a high-throughput method to analyzed protein–protein interaction (PPI) between 26 SARS-CoV-2 viral proteins and host proteins that physically interact with them. Host factors extracted from PPIs of viral and human proteins will function as druggable targets for identifying candidates from approved, clinical, and preclinical drugs [30]. On the other hand, Riva et al. performed an in vitro high-throughput antiviral screening of more than 11000 compounds from the ReFRAME drug-repurposing library and evaluated the results with gene set enrichment analysis (GSEA) to determine drug targets and select compounds for further antiviral verification [31].
3. Drug design
3.1. Small molecules
Besides drug screening, computational analysis is also a powerful tool for designing small molecules or peptides targeting viral proteins. For example, Zhang et al. optimized α-ketoamide class Mpro inhibitors with additional functional groups by applying x-ray crystallography and molecular docking and validated with in vitro inhibition assay to determine the best candidates [32]. Similar approach is applied by Dai et al. to design novel Mpro inhibitors with a specific backbone [33]. Apart from small molecules, peptide-based inhibitors were developed to target viral proteins as well. A common strategy is to utilize the structure of human ACE2 and SARS-CoV-2 S RBD complex to design peptide inhibitors that contain critical ACE2 residues and are able to bind to the RBD, thereby blocking its interaction with ACE2 on host cells [34], [35], [36], [37].
3.2. Neutralizing antibodies
Another strategy often considered to tackle the disease is the identification and characterization of neutralizing antibodies. Incorporating in vitro neutralization assays and cryo-EM, several studies were able to identify neutralizing antibodies from convalescent plasma and reconstitute their antibody-S complexes for structural analyses [38], [39], [40], [41], [42], [43]. Knowing the structure of antibody-S complexes and critical residues for effective neutralization, Luan et al. were able to establish an automated workflow, using molecular docking simulation and free energy perturbation (FEP) method, to perform in silico mutagenesis and identify potential mutations that enhanced binding of neutralizing antibody to SARS-CoV-2 S [44]. Similarly, in a preprint, Boorla et al. were able to analyze solvent-exposed residues on the RBD and design potential antibody variable regions (Fv) with neutralizing properties [45].
4. Vaccine development strategies
While there were more than a few hundred vaccine candidates that started, only a handful of candidates have emerged as frontrunners [46], [47]. For this reason, reverse-vaccinology can be utilized to identify epitopes or immunogenic regions on SARS-CoV-2 proteins that can be targeted for vaccine design to reduce costs and provide another layer of verification to speed up vaccine research. To this end, immune-informatics approaches were applied to identify immunogenic T cell and B cell epitopes from SARS-CoV-2 viral proteins [48], [49]. Selected epitopes were reconstructed in silico and analyzed for their antigenicity, allergenicity, toxicity, physicochemical properties, and their binding stability to toll-like receptors (TLR), to identify the best vaccine constructs. An immune simulation was further carried out to predict the humoral and cellular immune responses after administering the vaccine candidates.
AI and machine learning algorithms have also been developed to expedite reverse vaccinology. Software such as NEC Immune Profiler [50], the newly developed neural network-based ArdImmune Rank model [51], and the eXtreme Gradient Boosting (XGBoost)-based Vaxign-ML model [52], [53] have been used to identify immunogenic epitopes from the SARS-CoV-2 proteome. These approaches may provide significant help in designing multi-epitope chimeric vaccines with theoretically higher immunogenicity and assist the design of further biological experiments to examine the candidates.
5. Disease prediction
Aside from therapeutic development, machine learning has been widely explored to predict the severity or mortality of COVID-19. Proteomics and biochemical profile of blood and urine samples from patients with or without COVID-19, and with different severity and outcomes, were analyzed to determine prognostic biomarker combinations. Various machine learning models, such as regression analyses, XGBoost, random forest, Bayesian network, and support-vector machines (SVMs), have been used to select parameters that may predict mortality [54], [55], in-hospital mortality [56], [57], [58], [59], [60], and disease severity [61], [62]. A summary of the aforementioned examples and their evaluation matrices are listed in Table 1. Furthermore, lung lesion characterized by chest computed tomography (CT) scans were also proposed to predict disease progression [63], [64], [65]. An algorithm combining the imaging, clinical and biological attributes has been further constructed based on deep convolutional neural networks to generate a holistic forecast model, which has an area under curve (AUC) of 0.86 and 0.76 for predicting short-term and long-term mortality, respectively [66]. In addition, it is known that several variants of concern are reported to cause higher fatality rates [67], [68], [69], and it has been observed that the addition of viral clade or genetic information to demographic parameters (e.g. age and sex) could improve prediction model performance for severe outcomes [70], [71]. Nonetheless, increasing evidence are suggesting that, like other prediction models, external validation of these machine learning-based models is extremely crucial and should be performed prior to adopting them in clinical practice [72], [73].
Table 1.
Summary of machine learning models developed for disease prediction.
| Readout | Parameters | Algorithm | Sensitivity (Recall) | Specificity | Precision (PPV) | F1-score | Accuracy | AUROC | Test Cohort | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Mortality | 33 clinical parameters | Random forest | 85.71 % | 92.45% | – | – | 89.47% | 0.921 | No | [54] |
| Mortality | 45 proteins | Bayesian network | 92.68% | 86% | – | – | 89.01% | 0.953 | No | [54] |
| Mortality | CRP, BUN, serum calcium, serum albumin, lactic acid | SVM | 91% | 91% | 62.5% | – | – | 0.93 | No | [55] |
| In-hospital mortality | Age, lymphocyte, D-dimer, CRP, creatinine (ALDCC) | Logistic regression | 0.91 ± 0.03 | 0.78 ± 0.04 | 0.92 ± 0.03 | 0.92 ± 0.03 | 0.91 ± 0.03 | 0.992 | Yes | [56] |
| In-hospital mortality | Age, hs-CRP, lymphocyte, d-dimer | Logistic regression | 0.839 | 0.794 | – | – | – | 0.881 | Yes | [57] |
| In-hospital mortality | LDH, neutrophils, lymphocyte, hs-CRP, age (LNLCA) | Logistic regression | 92 ± 2.6% | 92 ± 3% | – | – | – | 0.991 | Yes | [58] |
| In-hospital mortality | PTA, urea, WBC, IL-2r, indirect bilirubin, myoglobin, FgDP | LASSO logistic regression | 98% | 91% | – | – | – | 0.997 | No | [59] |
| In-hospital mortality | Disease severity, age, hs-CRP, LDH, ferritin, IL-10 | Simple-tree XGBoost | >85% | – | >90% | >0.90 | >0.90 | 1.000 | Yes | [60] |
| Disease severity | 28 blood and urine parameters | SVM | – | – | – | – | 0.8148 | – | Yes | [61] |
| Disease severity | Different biomarker combinations | Penalized logistic regression | >82% | >71% | >87% | – | >85% | – | Yes | [62] |
BUN, blood urea nitrogen; CRP, c-reactive protein; FgDP, fibrinogen degradation products; hs-CRP, high-sensitivity C-reactive protein; IL-2r, interleukin-2 receptor; IL-10, interleukin-10; LASSO, least absolute shrinkage and selection operator; LDH, lactate dehydrogenase; MCHC, mean corpuscular hemoglobin concentration; PPV, positive predictive value; PTA, prothrombin; SVM, support vector machine; WBC, white blood cell activity; XGBoost, eXtreme Gradient Boosting.
At present, it remains challenging to computationally predict the emergence of future clinically significant SARS-CoV-2 variants, but a couple of approaches have been developed to model the interaction between newly identified SARS-CoV-2 variants and their host and predict their infectivity. A computational pipeline “SpikePro”, consisting of three-step in silico mutagenesis experiments, calculates the stability of mutant spike protein, the binding affinity between mutant spike and human ACE2, and the binding affinity between mutant spike and neutralizing antibodies to predict viral fitness [74]. Another recently published work also established a neural network model that could predict binding affinity changes of spike mutations to human ACE2 [75]. Such tools may be helpful in screening emerging mutants/variants that are better adapted to humans and are potentially more infective.
6. Outbreak prediction
Finally, to better control the pandemic, machine learning has been investigated as a tool to predict the epidemic curve of COVID-19. Various algorithms such as long short-term memory (LSTM) network [76], [77], [78], [79], Grey Wolf Optimizer (GWO)-LSTM hybrid model [80], autoregressive integrated moving average (ARIMA) [81], [82], [83], XGboost [84], support vector regression (SVR) [85], [86], and genetic programming [87] were explored for their ability to forecast confirmed cases, recovered cases, and death in some of the most affected countries. With publicly available statistics, these models may be helpful in predicting COVID-19 transmission and may facilitate policy-making to prevent new outbreaks.
7. Discussion and perspectives
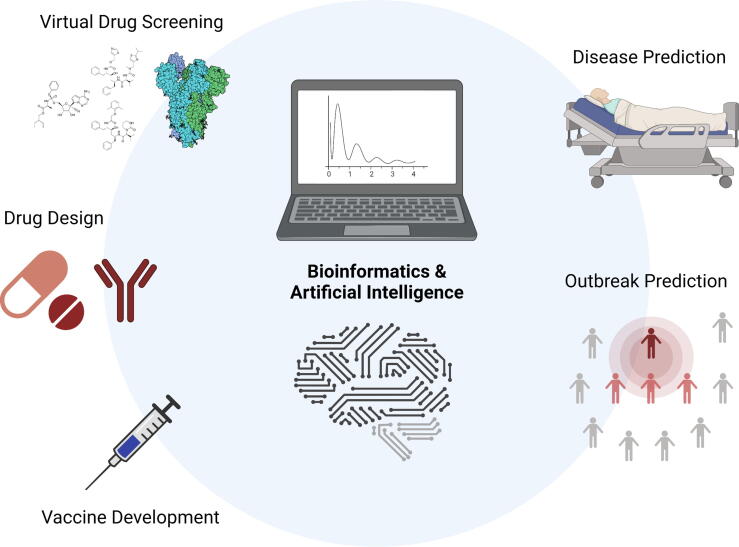
In summary, computational and structural biology with AI assistance has emerged as a new tool to tackle COVID-19 in prognosis and management of the disease (Fig. 1). However, due to the fact that these simulation models serve to provide candidates for preliminary selection, it is crucial that predictions generated from computational approaches be verified with biological confirmation, and to take into account the complex biological reactions [88]. Consequently, the accuracy of the computational models cannot be asserted merely based on simulation models. The representativeness of datasets utilized should also be carefully examined, since most of the prediction models rely on established databases or cohorts, and selection bias may magnify between different studies. Hence, it is vital to incorporate multiple datasets with diverse background to minimize the impact of selection bias. Nonetheless, results obtained from validation tests could be further used to optimize initial prediction models, thereby showing how AI can be utilized at multiple steps in various aspects. Indeed, computational research applied in the biology domain has emerged as a powerful technology to provide us with potential robust and efficient solutions in tackling challenging diseases including the COVID-19 pandemic. The rapid processing with AI may be especially beneficial when different variants are emerging worldwide, resulting in more cases, fatality, and decreased vaccine protection [89]. Disease prediction models may also become useful for identifying potential patients who are prone to post-acute symptoms or complications [90]. With accumulated experiences, the inclusion of AI-assisted computational and structural biology will likely continuously be refined and become a norm and critical parameter in future preparedness and rapid management of viral outbreaks and pandemic diseases.
Fig. 1.
Applications of computational and structural biology and artificial intelligence (AI) in the COVID-19 pandemic. Created with BioRender.com
CRediT authorship contribution statement
Ching-Hsuan Liu: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. Cheng-Hua Lu: Investigation, Writing – original draft. Liang-Tzung Lin: Conceptualization, Writing – original draft, Writing – review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors acknowledge Jonathan Ying Wang for help with the manuscript. LTL is supported by the Ministry of Science and Technology of Taiwan (MOST110-2320-B-038-041-MY3).
References
- 1.Center for Systems Science and Engineering at Johns Hopkins University. COVID-19 Dashboard [Online]. Available: <https://coronavirus.jhu.edu/map.html>; 2020 [accessed 03 September 2020].
- 2.Organization W.H. World Health Organization; Geneva: 2003. Consensus document on the epidemiology of severe acute respiratory syndrome (SARS) [Google Scholar]
- 3.Donnelly C.A., Malik M.R., Elkholy A., Cauchemez S., van Kerkhove M.D. Worldwide Reduction in MERS Cases and Deaths since 2016. Emerg Infect Dis. 2019;25:1758–1760. doi: 10.3201/eid2509.190143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim Biophys Acta, Mol Basis Dis. 2020;1866 doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2020;19(3):155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324(8):782. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 9.Dong Y., Dai T., Wei Y., Zhang L., Zheng M., Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct Target Ther. 2020;5:237. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C.-H., Lu C.-H., Wong S.H., Lin L.-T. Update on antiviral strategies against COVID-19: unmet needs and prospects. Front Immunol. 2021;11 doi: 10.3389/fimmu.2020.616595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choudhary S., Malik Y.S., Tomar S. Identification of SARS-CoV-2 cell entry inhibitors by drug repurposing using in silico structure-based virtual screening approach. Front Immunol. 2020;11:1664. doi: 10.3389/fimmu.2020.01664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panda P.K., Arul M.N., Patel P., Verma S.K., Luo W., Rubahn H.G., et al. Structure-based drug designing and immunoinformatics approach for SARS-CoV-2. Sci Adv. 2020;6:eabb8097. doi: 10.1126/sciadv.abb8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muhseen Z.T., Hameed A.R., Al-Hasani H.M.H., Tahir ul Qamar M., Li G. Promising terpenes as SARS-CoV-2 spike receptor-binding domain (RBD) attachment inhibitors to the human ACE2 receptor: Integrated computational approach. J Mol Liq. 2020;320:114493. doi: 10.1016/j.molliq.2020.114493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krupanidhi S., Abraham Peele K., Venkateswarulu T.C., Ayyagari V.S., Nazneen Bobby M., et al. Screening of phytochemical compounds of Tinospora cordifolia for their inhibitory activity on SARS-CoV-2: an in silico study. J Biomol Struct Dyn. 2020:1–5. doi: 10.1080/07391102.2020.1787226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abu-Saleh A.A.A., Awad I.E., Yadav A., Poirier R.A. Discovery of potent inhibitors for SARS-CoV-2's main protease by ligand-based/structure-based virtual screening, MD simulations, and binding energy calculations. Phys Chem Chem Phys. 2020;22(40):23099–23106. doi: 10.1039/d0cp04326e. [DOI] [PubMed] [Google Scholar]
- 16.Cherrak S.A., Merzouk H., Mokhtari-Soulimane N., Salahub D. Potential bioactive glycosylated flavonoids as SARS-CoV-2 main protease inhibitors: a molecular docking and simulation studies. PLoS ONE. 2020;15(10) doi: 10.1371/journal.pone.0240653. e0240653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryu Y.B., Jeong H.J., Kim J.H., Kim Y.M., Park J.Y., Kim D., et al. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CL(pro) inhibition. Bioorg Med Chem. 2010;18:7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aftab S.O., Ghouri M.Z., Masood M.U., Haider Z., Khan Z., Ahmad A., et al. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J Transl Med. 2020;18(1) doi: 10.1186/s12967-020-02439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puttaswamy H., Gowtham H.G., Ojha M.D., Yadav A., Choudhir G., Raguraman V., et al. In silico studies evidenced the role of structurally diverse plant secondary metabolites in reducing SARS-CoV-2 pathogenesis. Sci Rep. 2020;10:20584. doi: 10.1038/s41598-020-77602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361:2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van de Sand L., Bormann M., Alt M., Schipper L., Heilingloh C.S., Steinmann E., et al. Glycyrrhizin effectively inhibits SARS-CoV-2 replication by inhibiting the viral main protease. Viruses. 2021;13 doi: 10.3390/v13040609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murck H. Symptomatic Protective Action of Glycyrrhizin (Licorice) in COVID-19 Infection? Front Immunol. 2020;11:1239. doi: 10.3389/fimmu.2020.01239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Z., Yang L., Zhang X., Zhang Q., Yang Z., Liu Y., et al. Discovery of potential flavonoid inhibitors against COVID-19 3CL proteinase based on virtual screening strategy. Front Mol Biosci. 2020;7 doi: 10.3389/fmolb.2020.556481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parvathaneni V., Gupta V. Utilizing drug repurposing against COVID-19 - Efficacy, limitations, and challenges. Life Sci. 2020;259 doi: 10.1016/j.lfs.2020.118275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X., Guan Y. COVID-19 drug repurposing: a review of computational screening methods, clinical trials, and protein interaction assays. Med Res Rev. 2021;41(1):5–28. doi: 10.1002/med.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ke Y.-Y., Peng T.-T., Yeh T.-K., Huang W.-Z., Chang S.-E., Wu S.-H., et al. Artificial intelligence approach fighting COVID-19 with repurposing drugs. Biomed J. 2020;43(4):355–362. doi: 10.1016/j.bj.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., et al. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. The Lancet. 2020;395:e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stebbing J., Krishnan V., de Bono S., Ottaviani S., Casalini G., Richardson P.J., et al. Mechanism of baricitinib supports artificial intelligence-predicted testing in COVID-19 patients. EMBO Mol Med. 2020;12 doi: 10.15252/emmm.202012697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blasiak A., Lim J.J., Seah S.G.K., Kee T., Remus A., Chye D.H., et al. IDentif.AI: Rapidly optimizing combination therapy design against severe Acute Respiratory Syndrome Coronavirus 2 (SARS-Cov-2) with digital drug development. Bioeng Transl Med. 2021;6 doi: 10.1002/btm2.10196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han Y., Král P. Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020;14:5143–5147. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao L., Goreshnik I., Coventry B., Case J.B., Miller L., Kozodoy L., et al. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science. 2020;370:426–431. doi: 10.1126/science.abd9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pomplun S., Jbara M., Quartararo A.J., Zhang G., Brown J.S., Lee Y.C., et al. De novo discovery of high-affinity peptide binders for the SARS-CoV-2 spike protein. ACS Cent Sci. 2021;7:156–163. doi: 10.1021/acscentsci.0c01309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang X., Pearce R., Zhang Y. De novo design of protein peptides to block association of the SARS-CoV-2 spike protein with human ACE2. Aging (Albany NY) 2020;12:11263–11276. doi: 10.18632/aging.103416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 39.Zhou D., Duyvesteyn H.M.E., Chen C.P., Huang C.G., Chen T.H., Shih S.R., et al. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat Struct Mol Biol. 2020;27:950–958. doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- 40.Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182(828–842) doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182(1):73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 43.Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A., et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luan B., Huynh T. In silico antibody mutagenesis for optimizing its binding to spike protein of severe acute respiratory syndrome coronavirus 2. J Phys Chem Lett. 2020;11:9781–9787. doi: 10.1021/acs.jpclett.0c02706. [DOI] [PubMed] [Google Scholar]
- 45.Boorla, VS, Chowdhury, R, Maranas, CD. 2020. [DOI] [PMC free article] [PubMed]
- 46.Le T.T., Cramer J.P., Chen R., Mayhew S. Evolution of the COVID-19 vaccine development landscape. Nat Rev Drug Discov. 2020;19(10):667–668. doi: 10.1038/d41573-020-00151-8. [DOI] [PubMed] [Google Scholar]
- 47.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586(7830):516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 48.Rahman M.S., Hoque M.N., Islam M.R., Akter S., Rubayet-Ul-Alam A.S.M., Siddique M.A., et al. Epitope-based chimeric peptide vaccine design against S, M and E proteins of SARS-CoV-2 etiologic agent of global pandemic COVID-19: an in silico approach. PeerJ. 2020;8 doi: 10.7717/peerj.9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rahman N., Ali F., Basharat Z., Shehroz M., Khan M.K., Jeandet P., et al. Vaccine design from the ensemble of surface glycoprotein epitopes of SARS-CoV-2: an immunoinformatics approach. Vaccines (Basel) 2020;8(3):423. doi: 10.3390/vaccines8030423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malone B., Simovski B., Moline C., Cheng J., Gheorghe M., Fontenelle H., et al. Artificial intelligence predicts the immunogenic landscape of SARS-CoV-2 leading to universal blueprints for vaccine designs. Sci Rep. 2020;10:22375. doi: 10.1038/s41598-020-78758-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mazzocco G., Niemiec I., Myronov A., Skoczylas P., Kaczmarczyk J., Sanecka-Duin A., et al. AI aided design of epitope-based vaccine for the induction of cellular immune responses against SARS-CoV-2. Front Genet. 2021;12 doi: 10.3389/fgene.2021.602196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ong E., Wong M.U., Huffman A., He Y. COVID-19 coronavirus vaccine design using reverse vaccinology and machine learning. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ong E., Wang H., Wong M.U., Seetharaman M., Valdez N., He Y. Vaxign-ML: supervised machine learning reverse vaccinology model for improved prediction of bacterial protective antigens. Bioinformatics. 2020;36:3185–3191. doi: 10.1093/bioinformatics/btaa119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sardar R., Sharma A., Gupta D. Machine learning assisted prediction of prognostic biomarkers associated With COVID-19, using clinical and proteomics data. Front Genet. 2021;12 doi: 10.3389/fgene.2021.636441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Booth A.L., Abels E., Mccaffrey P. Development of a prognostic model for mortality in COVID-19 infection using machine learning. Mod Pathol. 2020;34(3):522–531. doi: 10.1038/s41379-020-00700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rahman T., Al-Ishaq F.A., Al-Mohannadi F.S., Mubarak R.S., Al-Hitmi M.H., Islam K.R., et al. Mortality prediction utilizing blood biomarkers to predict the severity of COVID-19 using machine learning technique. Diagnostics (Basel) 2021;11 doi: 10.3390/diagnostics11091582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu C., Liu Z., Jiang Y., Shi O., Zhang X., Xu K., et al. Early prediction of mortality risk among patients with severe COVID-19, using machine learning. Int J Epidemiol. 2021;49:1918–1929. doi: 10.1093/ije/dyaa171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chowdhury M.E.H., Rahman T., Khandakar A., Al-Madeed S., Zughaier S.M., Doi S.A.R., et al. An Early warning tool for predicting mortality risk of COVID-19 patients using machine learning. Cognit Comput. 2021:1–16. doi: 10.1007/s12559-020-09812-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu G., Zhou S., Wang Y., Lv W., Wang S., Wang T., et al. A prediction model of outcome of SARS-CoV-2 pneumonia based on laboratory findings. Sci Rep. 2020;10:14042. doi: 10.1038/s41598-020-71114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guan X., Zhang B., Fu M., Li M., Yuan X., Zhu Y., et al. Clinical and inflammatory features based machine learning model for fatal risk prediction of hospitalized COVID-19 patients: results from a retrospective cohort study. Ann Med. 2021;53(1):257–266. doi: 10.1080/07853890.2020.1868564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yao H., Zhang N., Zhang R., Duan M., Xie T., Pan J., et al. Severity detection for the coronavirus disease 2019 (COVID-19) patients using a machine learning model based on the blood and urine tests. Front Cell Dev Biol. 2020;8:683. doi: 10.3389/fcell.2020.00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shu T., Ning W., Wu D., Xu J., Han Q., Huang M., et al. Plasma proteomics identify biomarkers and pathogenesis of COVID-19. Immunity. 2020;53(1108–1122) doi: 10.1016/j.immuni.2020.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu F., Zhang Q., Huang C., Shi C., Wang L., Shi N., et al. CT quantification of pneumonia lesions in early days predicts progression to severe illness in a cohort of COVID-19 patients. Theranostics. 2020;10(12):5613–5622. doi: 10.7150/thno.45985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y., Shang K., Bian W., He L., Fan Y., Ren T., et al. Prediction of disease progression in patients with COVID-19 by artificial intelligence assisted lesion quantification. Sci Rep. 2020;10:22083. doi: 10.1038/s41598-020-79097-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang M., Xia C., Huang L., Xu S., Qin C., Liu J., et al. Deep learning-based triage and analysis of lesion burden for COVID-19: a retrospective study with external validation. Lancet Digit Health. 2020;2:e506–e515. doi: 10.1016/S2589-7500(20)30199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chassagnon G., Vakalopoulou M., Battistella E., Christodoulidis S., Hoang-Thi T.N., Dangeard S., et al. AI-driven quantification, staging and outcome prediction of COVID-19 pneumonia. Med Image Anal. 2021;67 doi: 10.1016/j.media.2020.101860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Challen R., Brooks-Pollock E., Read J.M., Dyson L., Tsaneva-Atanasova K., Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. 2021;372 doi: 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davies N.G., Jarvis C.I., Group, C. C.-W., Edmunds W.J., Jewell N.P., Diaz-Ordaz K., et al. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593:270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grint D.J., Wing K., Williamson E., Mcdonald H.I., Bhaskaran K., Evans D., et al. Case fatality risk of the SARS-CoV-2 variant of concern B.1.1.7 in England, 16 November to 5 February. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.11.2100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Voss J.D., Skarzynski M., McAuley E.M., Maier E.J., Gibbons T., Fries A.C., et al. Variants in SARS-CoV-2 associated with mild or severe outcome. Evol Med Publ Health. 2021;9:267–275. doi: 10.1093/emph/eoab019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nakamichi K., Shen J.Z., Lee C.S., Lee A., Roberts E.A., Simonson P.D., et al. Hospitalization and mortality associated with SARS-CoV-2 viral clades in COVID-19. Sci Rep. 2021;11:4802. doi: 10.1038/s41598-021-82850-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barish M., Bolourani S., Lau L.F., Shah S., Zanos T.P. External validation demonstrates limited clinical utility of the interpretable mortality prediction model for patients with COVID-19. Nat Mach Intell. 2020;3:25–27. [Google Scholar]
- 73.Gupta R.K., Marks M., Samuels T.H.A., Luintel A., Rampling T., Chowdhury H., et al. Systematic evaluation and external validation of 22 prognostic models among hospitalised adults with COVID-19: an observational cohort study. Eur Respir J. 2020;56(6) doi: 10.1183/13993003.03498-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pucci F., Rooman M. Prediction and evolution of the molecular fitness of SARS-CoV-2 variants: introducing SpikePro. Viruses. 2021;13(5):935. doi: 10.3390/v13050935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen C., Boorla V.S., Banerjee D., Chowdhury R., Cavener V.S., Nissly R.H., et al. Computational prediction of the effect of amino acid changes on the binding affinity between SARS-CoV-2 spike RBD and human ACE2. Proc Natl Acad Sci U S A. 2021:118. doi: 10.1073/pnas.2106480118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kolozsvári L.R., Bérczes T., Hajdu A., Gesztelyi R., Tiba A., Varga I., et al. Predicting the epidemic curve of the coronavirus (SARS-CoV-2) disease (COVID-19) using artificial intelligence: an application on the first and second waves. Inform Med Unlocked. 2021;25:100691. doi: 10.1016/j.imu.2021.100691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hssayeni M.D., Chala A., Dev R., Xu L., Shaw J., Furht B., et al. The forecast of COVID-19 spread risk at the county level. J Big Data. 2021;8:99. doi: 10.1186/s40537-021-00491-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chimmula V.K.R., Zhang L. Time series forecasting of COVID-19 transmission in Canada using LSTM networks. Chaos, Solitons Fractals. 2020;135 doi: 10.1016/j.chaos.2020.109864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marzouk M., Elshaboury N., Abdel-Latif A., Azab S. Deep learning model for forecasting COVID-19 outbreak in Egypt. Process Saf Environ Prot. 2021;153:363–375. doi: 10.1016/j.psep.2021.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Prasanth S., Singh U., Kumar A., Tikkiwal V.A., Chong P.H.J. Forecasting spread of COVID-19 using google trends: a hybrid GWO-deep learning approach. Chaos, Solitons Fractals. 2021;142 doi: 10.1016/j.chaos.2020.110336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arunkumar K.E., Kalaga D.V., Sai Kumar C.M., Chilkoor G., Kawaji M., Brenza T.M. Forecasting the dynamics of cumulative COVID-19 cases (confirmed, recovered and deaths) for top-16 countries using statistical machine learning models: Auto-Regressive Integrated Moving Average (ARIMA) and Seasonal Auto-Regressive Integrated Moving Average (SARIMA) Appl Soft Comput. 2021;103:107161. doi: 10.1016/j.asoc.2021.107161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Singh R.K., Rani M., Bhagavathula A.S., Sah R., Rodriguez-Morales A.J., Kalita H., et al. Prediction of the COVID-19 pandemic for the top 15 affected countries: advanced autoregressive integrated moving average (ARIMA) model. JMIR Publ Health Surveill. 2020;6 doi: 10.2196/19115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ceylan Z. Estimation of COVID-19 prevalence in Italy, Spain, and France. Sci Total Environ. 2020;729 doi: 10.1016/j.scitotenv.2020.138817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mehta M., Julaiti J., Griffin P., Kumara S. Early stage machine learning-based prediction of US county vulnerability to the COVID-19 pandemic: machine learning approach. JMIR Public Health Surveill. 2020;6 doi: 10.2196/19446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ribeiro M., da Silva R.G., Mariani V.C., Coelho L.D.S. Short-term forecasting COVID-19 cumulative confirmed cases: perspectives for Brazil. Chaos, Solitons Fractals. 2020;135 doi: 10.1016/j.chaos.2020.109853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Parbat D., Chakraborty M. A python based support vector regression model for prediction of COVID19 cases in India. Chaos, Solitons Fractals. 2020;138 doi: 10.1016/j.chaos.2020.109942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Salgotra R., Gandomi M., Gandomi A.H. Time series analysis and forecast of the COVID-19 pandemic in india using genetic programming. Chaos, Solitons Fractals. 2020;138 doi: 10.1016/j.chaos.2020.109945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nussinov R., Tsai C.-J., Shehu A., Jang H. Computational structural biology: successes future directions, and challenges. Molecules. 2019;24(3):637. doi: 10.3390/molecules24030637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Abdool Karim S.S., De Oliveira T. New SARS-CoV-2 variants - clinical, public health, and vaccine implications. N Engl J Med. 2021;384:1866–1868. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nalbandian A., Sehgal K., Gupta A., Madhavan M.V., McGroder C., Stevens J.S., et al. Post-acute COVID-19 syndrome. Nat Med. 2021;27:601–615. doi: 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]