Abstract
Pulsed-field gel electrophoresis, ribotyping, and fingerprinting analysis of 22 invasive isolates of multidrug-resistant (MDR) pneumococci from Korea showed that 59 to 82% were genetically related. DNA sequencing of the PBP 2B gene showed relatively uniform alterations in nucleotides (5.4 to 7.8%) and amino acids (3.0 to 4.3%), while Asn-276→Lys, Arg-285→Cys and Ser-305→Phe substitutions were unique to Korean MDR strains, suggesting the spread of a few epidemic clones of resistant pneumococci within Korea.
During the past three decades, the resistance of Streptococcus pneumoniae to penicillin, other β-lactams, and non-β-lactam agents has been rapidly increasing in many parts of the world (1, 12). To investigate the molecular characteristics of multidrug-resistant (MDR) S. pneumoniae isolates in Korea, where the prevalence of penicillin resistance was 80% (14), we performed pulsed-field gel electrophoresis (PFGE), ribotyping, fingerprinting of penicillin-binding protein (PBP) genes, and DNA sequencing of PBP 2B genes.
(Part of this data was presented at the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy in Toronto, Canada, September 28 to October 1, 1997.)
Bacterial isolates.
Isolates of pneumococci (a total of 22: 13 of serogroup 23, 6 of serogroup 19, and 3 of serogroup 6) were selected if they showed reduced susceptibility to three or more classes of antibiotics and were epidemiologically unrelated. Isolates were invasive pathogens which had caused community-acquired diseases such as bacteremia, pneumonia, meningitis, and peritonitis from 1989 to 1995 in Korea. MICs of penicillin, cefotaxime, ceftriaxone, cefaclor, cefuroxime, ampicillin, imipenem, tetracycline, erythromycin, and chloramphenicol were determined by the agar dilution method (10). MICs for these antimicrobials were interpreted according to the 1999 National Committee for Clinical Laboratory Standards breakpoints (11).
PFGE.
Isolates were subjected to PFGE analysis as previously described (8, 15). One penicillin-susceptible R6 strain and the internationally epidemic serotype 23F Spanish clone were also tested. The DNA was digested by SmaI or ApaI, and the fragments were resolved by PFGE with the CHEF-Mapper System (Bio-Rad Laboratories, Richmond, Calif.). The DNA fragment patterns generated by PFGE were interpreted according to recent criteria (16).
Ribotyping.
Ribotyping was performed with the restriction enzyme PvuII by using a [α-32P]dCTP-labeled gene probe from Escherichia coli 16S-plus-23S RNA. Strains that differed by more than two bands were considered to be different clones, while single band differences were interpreted as subtypes (2).
PCR fingerprinting of PBP genes.
pbp 1a, 2b, and 2x genes were amplified from chromosomal DNA by PCR as previously described (6, 15). Briefly, amplification of the gene encoding PBP 2B yielded a 1.5-kb product that included the region encoding the transpeptidase domain of the enzyme. The PBP 2X gene product was a 1.9-kb fragment, whereas the gene encoding PBP 1A was amplified as a 2.4-kb product. Gene fingerprinting was performed as previously described (6) by using HinfI or MseI plus DdeI as restriction enzymes. pBR322 DNA digested with HpaII and labeled with [α-32P]dCTP was used as a size marker. A dendrogram was generated based on the visible fingerprinting patterns.
DNA sequencing of pbp genes.
The 1.5-kb transpeptidase-encoding region (TER) of the PBP 2B gene was amplified and directly sequenced by using the ABI PRISM Big Dye Terminator cycle sequencing kit (Perkin-Elmer). The primer sequences for the amplification were as follows: upstream primer, 5′-GAT CCT CTA AAT GAT TCT CAG GTG GCT GTT-3′ and downstream primer, 5′-CA ATT AGC TTA GCA ATA GGT GTT GG-3′. The sequence data for strain R6 appears in the EMBL, GenBank, and DDBJ nucleotide sequence data libraries under the accession no. X 16022 (5).
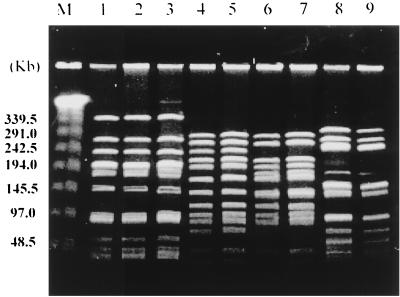
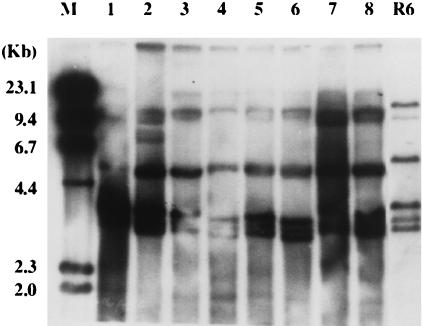
Phenotypic and genotypic characteristics of the isolates are presented in Table 1 and Table 2. Twenty-two pneumococcal isolates showed four major PFGE patterns after digestion with SmaI (Table 2). The most common PFGE pattern was type A, to which 82% (18 of 22) strains belonged (Fig. 1). The PFGE pattern of a serotype 23F strain from Spain was identical to the type A pattern. Isolates with different serotypes or antibiograms showed the same PFGE pattern. The PFGE pattern with ApaI was similar to that with SmaI digestion (data not shown). A total of seven different ribotypes were noted in 22 MDR strains. Thirteen of 22 isolates (59%) showed ribotype A or A subtype patterns (Fig. 2). Most strains with identical PFGE patterns belonged to the same ribotypes. Fingerprinting analysis of pbp 1a, 2x, and 2b genes digested with HinfI showed that 17 of 22 strains had identical fingerprinting patterns (data not shown). The number of fingerprint patterns of pbp 1a, 2x, and 2b genes was 5, 4, and 4, respectively. A dendrogram of similarities showed that clusters with greater than 90% similarities existed in 77, 77, and 82% of strains with pbp 1a, 2x, and 2b, respectively. Fingerprints with MseI and DdeI were the same as those with HinfI (data not shown). The PBP 2B TER of nine MDR pneumococcal strains showed very uniform alterations in nucleotide and amino acid sequences, ranging from 5.4 to 7.8% and from 3.0 to 4.3%, respectively. From 86.0 to 93.7% of nucleotide alterations and from 85.7 to 100% of amino acid alterations were noted in the hypervariable region which was located between bp 408 and 993, respectively. All isolates were identical to each other with respect to nucleotide and amino acid diversity between bp 652 (codon 218) and bp 915 (codon 305). A total of 30 amino acids in the TER were substituted in the Korean MDR strains, while 14 amino acid substitutions were shared by all strains. These amino acids alterations are as followings: Ser-218→Pro, Asn-228→Tyr, Thr-232→Lys, Gln-233→Leu, Gln-244→Glu, Thr-252→Ala, Leu-261→Ile, Asn-276→Lys, Ser-279→Thr, Glu-282→Gly, Arg-285→Cys, Ser-286→Ala, Thr-295→Ala, and Ser-305→Phe. Among these alterations, substitution of Lys for Asn-276, Cys for Arg-285, and Phe for Ser-305 were found in all Korean MDR strains but not in strains described in previous reports (4, 5, 13).
TABLE 1.
Antimicrobial susceptibility and serogroups of 22 isolates of MDR S. pneumoniae from Korea
| Strain no. | Isolation year | Specimen source | Serogroup | MIC of (μg/ml)a:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PC | CTX | CTR | CLO | CFX | AM | IMP | TC | EM | CM | ||||
| R6 | ≤0.03 | ≤0.03 | ≤0.03 | ≤1 | ≤0.03 | ≤0.03 | ≤0.03 | 0.5 | ≤0.06 | 4 | |||
| Sp23Fb | 1989 | 23 | 2 | 1 | 1 | 128 | 8 | 2 | 0.5 | 16 | ≤0.06 | 32 | |
| 1 | 1989 | Cerebrospinal fluid | 23 | 4 | 2 | 2 | 64 | 8 | 4 | 0.5 | 8 | 1 | 8 |
| 2 | 1989 | Cerebrospinal fluid | 23 | 4 | 2 | 2 | 64 | 8 | 4 | 1 | 8 | 1 | 8 |
| 3 | 1990 | Blood | 23 | 4 | 2 | 2 | 128 | 8 | 4 | 0.5 | 8 | 0.25 | 8 |
| 4 | 1990 | Sputum | 6 | 4 | 2 | 2 | 128 | 8 | 4 | 0.5 | 8 | 2 | 8 |
| 5 | 1990 | Ascites | 23 | 4 | 2 | 2 | 128 | 4 | 4 | 0.5 | 4 | 0.25 | 8 |
| 6 | 1991 | Ascites | 19 | 4 | 2 | 2 | 128 | 8 | 4 | 0.5 | 16 | 2 | 16 |
| 7 | 1991 | Sputum | 23 | 4 | 2 | 2 | 128 | 4 | 4 | 0.5 | 1 | 0.25 | 8 |
| 8 | 1991 | Blood | 23 | 4 | 2 | 2 | 128 | 8 | 4 | 0.5 | 0.5 | 2 | 8 |
| 9 | 1992 | Blood | 6 | 4 | 2 | 2 | 128 | 8 | 4 | 0.5 | 8 | 1 | 8 |
| 10 | 1992 | Blood | 23 | 4 | 2 | 2 | 128 | 8 | 4 | 0.5 | 1 | 0.25 | 8 |
| 11 | 1992 | Sputum | 23 | 4 | 2 | 2 | 128 | 4 | 4 | 0.5 | 0.5 | 1 | 16 |
| 12 | 1992 | Sputum | 19 | 4 | 2 | 2 | 128 | 8 | 4 | 0.5 | 16 | 0.5 | 16 |
| 13 | 1993 | Sputum | 19 | 4 | 2 | 2 | 128 | 8 | 4 | 0.5 | 2 | 2 | 8 |
| 14 | 1993 | Blood | 6 | 2 | 2 | 2 | 128 | 8 | 4 | 0.5 | 4 | 0.25 | 16 |
| 15 | 1993 | Blood | 19 | 4 | 2 | 2 | 128 | 8 | 8 | 0.5 | 4 | 0.25 | 16 |
| 16 | 1994 | Blood | 19 | 4 | 2 | 2 | 128 | 4 | 4 | 0.5 | 2 | 16 | 16 |
| 17 | 1994 | Sputum | 23 | 4 | 2 | 2 | 128 | 8 | 4 | 0.5 | 4 | 4 | 16 |
| 18 | 1995 | Sputum | 23 | 2 | 4 | 2 | 128 | 8 | 4 | 0.5 | 8 | 8 | 8 |
| 19 | 1995 | Sputum | 19 | 2 | 4 | 2 | 128 | 8 | 4 | 0.5 | 16 | 32 | 4 |
| 20 | 1995 | Sputum | 23 | 2 | 4 | 2 | 128 | 8 | 4 | 0.5 | 32 | 32 | 4 |
| 21 | 1995 | Sputum | 23 | 4 | 4 | 2 | 128 | 8 | 4 | 0.5 | 32 | 32 | 4 |
| 22 | 1995 | Sputum | 23 | 2 | 4 | 2 | 128 | 8 | 4 | 0.5 | 16 | 0.26 | 16 |
PC, penicillin; CTX, cefotaxime; CTR, ceftriaxone; CLO, cefaclor; CFX, cefuroxime; AM, ampicillin; IMP, imipenem; TC, tetracycline; EM, erythromycin; CM, chloramphenicol.
Sp23F, Spanish 23F clone.
TABLE 2.
Molecular characteristics of 22 MDR S. pneumoniae isolates from Korea
| Strain no. | PFGE pattern | Ribotype | Fingerprint profile of:
|
pbp 2b sequence
|
|||
|---|---|---|---|---|---|---|---|
| pbp 1a | pbp 2x | pbp 2b | Nucleotide variation (%) | Amino acid variation (%) | |||
| Sp 23Fa | A | NDb | I | I | I | 5.4 | 2.5 |
| 1 | A | 3 | I | I | I | ND | ND |
| 2 | A | 1 | I | I | I | ND | ND |
| 3 | A | 1 | I | I | I | 5.4 | 3.0 |
| 4 | A | 1 | I | I | I | ND | ND |
| 5 | A | 1 | I | I | I | ND | ND |
| 6 | A | 4 | I | I | I | ND | ND |
| 7 | A | 1 | I | I | I | ND | ND |
| 8 | A | 1 | I | I | I | 5.4 | 3.0 |
| 9 | A | 1 | I | I | I | 5.6 | 3.0 |
| 10 | A | 1ac | I | I | I | 5.4 | 3.0 |
| 11 | A | 1 | I | I | I | ND | ND |
| 12 | A | 6 | III | I | I | ND | ND |
| 13 | A | 1a | I | I | I | ND | ND |
| 14 | A | 1a | I | I | I | ND | ND |
| 15 | A | 1a | I | I | I | 5.6 | 3.0 |
| 16 | B | 7 | II | II | II | 5.4 | 3.0 |
| 17 | A | 1b | I | III | I | ND | ND |
| 18 | A | 1 | I | III | I | 5.6 | 3.4 |
| 19 | B | 4 | IV | IV | III | 6.8 | 3.4 |
| 20 | C | 5 | IV | IV | IV | 7.8 | 4.3 |
| 21 | D | 5a | V | I | IV | ND | ND |
| 22 | A | 2 | I | I | I | ND | ND |
Sp23 F, Spanish 23F clone.
ND, not done.
1a indicates the subtype of the ribopattern which differed by only one band from original ribotype 1.
FIG. 1.
PFGE patterns of chromosomal DNA restriction fragments of MDR pneumococcal isolates from Korea digested with SmaI. The type A pattern (lanes 1 to 3) was the most common pattern among Korean MDR strains which was identical to that of the Spanish 23F clone (lane 1), while the type B pattern (lanes 4 to 5) was a pattern unique to Korean strains.
FIG. 2.
Ribotyping patterns of MDR pneumococcal isolates from Korea digested with PvuII. Lane M is the DNA marker. Ribotype 1 is noted in MDR strains in lanes 3 to 8 with subtypes of ribotype 1 in lanes 1 to 2, while that of a penicillin-susceptible R6 (lane 9) is different from those of MDR strains.
Korea has been considered to have the highest prevalences of penicillin and multidrug resistance in the world (3, 7, 14, 15). Data from one Korean hospital showed nonsusceptibility to penicillin rising from 29% in 1988 to 77% in 1993 (3). This rapid emergence of pneumococcal resistance in Korea might be due to the development and spread of resistant clones within Korea. Our previous work has documented the potential spread of the resistant clones within Korea and between different countries in Asia (14, 15). The current study clearly confirmed the genetic relatedness of MDR pneumococcal strains isolated from Korea during a 7-year period. The most common PFGE pattern of Korean strains was identical to the internationally epidemic Spanish 23F clone (25, 27). The international spread of this clone was first documented in the United States in 1989, when isolates of this clone were found in a Cleveland, Ohio, area day care center (9). Our data suggests that this clone was introduced into Korea in late 1980s, at about the same time that the spread of the 23F clone from Spain to the United States was recognized (9). Ribotyping and fingerprinting analysis of PBP genes also showed similar results with regard to the genetic relatedness of MDR strains. The present study used three different typing methods which showed that 12 out of 22 isolates (54.5%) had consistent genotypes by all three methods. Since these strains were isolated from different patients from various parts of South Korea during a 7-year period, the molecular characteristics of these strains by these methods strongly suggest that a few major clones have disseminated in South Korea during recent years. MDR isolates which seemed to originate from a common clone expressed different serogroups, which suggested that capsular transformation between serogroups 6, 19, and 23 occurs in vivo. Nucleotide sequence analysis of the pbp 2b gene revealed very uniform variation in nucleotide and amino acid sequences among the Korean strains. Among 14 common alterations in amino acid sequences in Korean MDR strains, Asn-276→Lys, Arg-285→Cys, and Ser-305→Phe substitutions were thought to be alterations unique to Korean strains since these mutations have not been documented in pneumococcal isolates from other countries and were not found in wild-type penicillin-susceptible strains from Korea (data not shown). Thr-252→Ala and Glu-282→Gly were also found in all Korean strains as in penicillin-resistant strains from other part of the world (13). According to the classification of the mutation patterns of the PBP 2B gene (4), mutations which were very similar to the class B mutation were found in seven Korean MDR strains (strains 3, 8, 9, 10, 15, 16, and 18) and the Spanish 23F clone, while two strains (strains 19 and 20) were found to have neither class A nor class B mutations. Class A mutation was not found in Korean strains. Sequencing data showed that a limited number of amino acid substitutions were shared by the wild-type Korean MDR pneumococcal strains in the transpeptidase domain of PBP 2B gene.
In conclusion, the present data show that widespread dissemination of a small number of MDR pneumococcus clones has occurred in Korea during the recent years. This might explain the rapid emergence of penicillin resistance among pneumococci in Korea in the 1990s.
Nucleotide sequence accession number.
Sequence data for the nine Korean isolates have been assigned GenBank accession numbers AF 180878 to AF 180886.
Acknowledgments
This work was supported by the Samsung Biomedical Research Institute (C-95-061-3) and a Samsung Medical Center research grant, Seoul, Korea.
We are very grateful to the members of the Asian Network for Surveillance of Resistant Pathogens for their active participation in the multinational surveillance study and technical assistance for this study and to Alexander Tomasz (Rockerfeller University) for generous donation of the Spanish 23F clone.
REFERENCES
- 1.Appelbaum P C. Antimicrobial resistance in Streptococcus pneumoniae: an overview. Clin Infect Dis. 1992;15:77–83. doi: 10.1093/clinids/15.1.77. [DOI] [PubMed] [Google Scholar]
- 2.Chetoui H, Delhalle E, Osterrieth P, Rousseaux D. Ribotyping for use in studying molecular epidemiology of Serratia marcescens: comparison with biotyping. J Clin Microbiol. 1995;33:2637–2642. doi: 10.1128/jcm.33.10.2637-2642.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chong Y, Lee K, Kwon O H, Henrichsen J. Capsular types and antimicrobial resistance of Streptococcus pneumoniae isolated in Korea. Eur J Clin Microbiol Infect Dis. 1995;14:528–531. doi: 10.1007/BF02113433. [DOI] [PubMed] [Google Scholar]
- 4.Dowson C G, Hutchison A, Brannigan J A, George R C, Hansman D, Linares J, Tomasz A, Maynard J, Spratt B G. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1989;86:8842–8846. doi: 10.1073/pnas.86.22.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dowson C G, Hutchison A, Spratt B G. Nucleotide sequence of the penicillin-binding protein 2B gene of Streptococcus pneumoniae strain R6. Nucleic Acids Res. 1989;17:7518. doi: 10.1093/nar/17.18.7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kell C M, Jordens J Z, Daniels M, Coffey T J, Bates J, Paul J, Gilks C, Spratt B G. Molecular epidemiology of penicillin-resistant pneumococci isolated in Nairobi, Kenya. Infect Immun. 1993;61:4382–4391. doi: 10.1128/iai.61.10.4382-4391.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee H-J, Park J-Y, Jang S-H, Kim J-H, Kim E-C, Choi K-W. High incidence of resistance to multiple antimicrobials in clinical isolates of Streptococcus pneumoniae from a university hospital in Korea. Clin Infect Dis. 1995;20:826–835. doi: 10.1093/clinids/20.4.826. [DOI] [PubMed] [Google Scholar]
- 8.Matushek M G, Bonten M J, Hayden M K. Rapid preparation of bacterial DNA for pulsed-field gel electrophoresis. J Clin Microbiol. 1996;34:2598–2600. doi: 10.1128/jcm.34.10.2598-2600.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munoz R, Coffey T J, Daniels M, Dowson C G, Laible G, Casal J, Hakenbeck R, Jacobs M, Musher J M, Spratt B G, Tomasz A. Intercontinental spread of a multiresistant clone of serotype 23F Streptococcus pneumoniae. J Infect Dis. 1991;164:302–306. doi: 10.1093/infdis/164.2.302. [DOI] [PubMed] [Google Scholar]
- 10.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th ed. 1997. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Villanova, Pa. [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing, 9th informational supplement. M100-S9. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 12.Schutze G E, Kaplan S L, Jacobs R F. Resistant pneumococcus: a worldwide problem. Infection. 1994;22:233–237. doi: 10.1007/BF01739904. [DOI] [PubMed] [Google Scholar]
- 13.Smith, A. M., and K. P. Klugman. 1995. Alterations in penicillin-binding protein 2B from penicillin-resistant wild-type strains of Streptococcus pneumoniae. 39:859–867. [DOI] [PMC free article] [PubMed]
- 14.Song J H, Lee N Y, Ichiyama S, Yoshida R, Hirakata Y, Wang F, Chongthaleong A, Aswapokee N, Chiu C-H, Lalitha M K, Thomas K, Perera J, Yee T T, Jamal F, Warsa U C, Vinh B X, Jacobs M R, Appelbaum P C the ANSORP Study Group. Spread of drug-resistant Streptococcus pneumoniae in Asian countries: Asian Network for Surveillance of Resistant Pathogens (ANSORP) study. Clin Infect Dis. 1999;28:1206–1211. doi: 10.1086/514783. [DOI] [PubMed] [Google Scholar]
- 15.Song J H, Yang J W, Peck K R, Kim S, Lee N Y, Jacobs M R, Appelbaum P C, Pai C H. Spread of multidrug-resistant Streptococcus pneumoniae in South Korea. Clin Infect Dis. 1997;25:747–749. doi: 10.1086/516945. [DOI] [PubMed] [Google Scholar]
- 16.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]