Summary
Bacterial colonization of the rhizosphere is critical for the establishment of plant–bacteria interactions that represent a key determinant of plant health and productivity. Plants influence bacterial colonization primarily through modulating the composition of their root exudates and mounting an innate immune response. The outcome is a horizontal filtering of bacteria from the surrounding soil, resulting in a gradient of reduced bacterial diversity coupled with a higher degree of bacterial specialization towards the root. Bacteria–bacteria interactions (BBIs) are also prevalent in the rhizosphere, influencing bacterial persistence and root colonization through metabolic exchanges, secretion of antimicrobial compounds and other processes. Traditionally, bacterial colonization has been examined under sterile laboratory conditions that mitigate the influence of BBIs. Using simplified synthetic bacterial communities combined with microfluidic imaging platforms and transposon mutagenesis screening approaches, we are now able to begin unravelling the molecular mechanisms at play during the early stages of root colonization. This review explores the current state of knowledge regarding bacterial root colonization and identifies key tools for future exploration.
Introduction
Soil provides a diverse habitat for billions of individual microorganisms, many of which form complex interactions with plants spanning the continuum of ecological outcomes from beneficial to pathogenic (Bardgett and Van Der Putten, 2014). To attract beneficial microbes from nutrient‐poor bulk soil, plants exude up to 20% of their photosynthate into the rhizosphere (soil–root interface), providing carbon for microbial growth and proliferation (Estabrook and Yoder, 1998). Some individuals form more intimate associations with plants, colonizing the rhizoplane (root surface) as epiphytes or endosphere (space between root cells) as endophytes (Fig. 1A) (Bulgarelli et al., 2013; Reinhold‐Hurek et al., 2015; Tkacz et al., 2015). Epiphytic and endophytic lifestyles allow microorganisms to remain anchored in a nutrient‐rich environment and facilitate the development of beneficial plant–bacteria interactions (PBIs), thus providing a key advantage over a free‐living lifestyle.
Fig. 1.
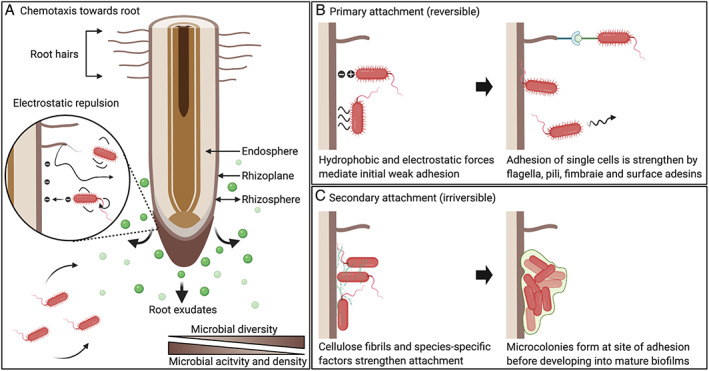
Bacterial colonization of plant roots is a multistep process. A. Plants secrete photosynthetically fixed carbon into the rhizosphere forming chemical gradients, which chemotactically attract motile bacteria from the soil towards the root surface. Flagella and pili propel bacteria, allowing them to overcome any electrostatic repulsion at the root surface. B. Primary attachment results in weak reversible binding of single cells to the root surface. This is initially mediated by hydrophobic and electrostatic interactions and subsequently strengthened by proteinaceous appendages and species‐specific surface adhesins. C. Secondary attachment leads to strong irreversible binding of bacteria to the root surface, promoting microcolony formation at the initial site of attachment. This process is mediated by the production of cellulose fibrils and other species‐specific factors including polysaccharides extracellular proteins. Created with BioRender.com
Root‐associated microbiota positively influence plant health and productivity through various mechanisms including enhancing nutrient acquisition, priming of plant defences and control of plant pathogens (Berendsen et al., 2012; Philippot et al., 2013; Wei et al., 2015; Trivedi et al., 2020). In recent years, metagenome studies have identified a wealth of microorganisms inhabiting the various root niches and revealed bacteria to be the most prevalent form of root‐associated microbiota. Despite the vast bacterial diversity present in soil, bacteria from four phyla, Actinobacteria, Bacteroidetes, Firmicutes and Proteobacteria, account for the major fraction of the root microbiome (Uroz et al., 2010; Bulgarelli et al., 2012; Lundberg et al., 2012). However, taxonomic composition varies widely at the genus and species levels due to unique selective pressures imposed by host genotype, sub‐localisation and abiotic environmental factors (Turner et al., 2013a; Turner et al., 2013b; Schlaeppi et al., 2014; Lebeis et al., 2015; Tkacz et al., 2020).
Exploitation of the root microbiome is often touted for its enormous potential to substitute environmentally deleterious agrochemicals such as fertilizers and pesticides that are crucial for current agricultural productivity (Busby et al., 2017). As such, there is escalating interest regarding the mechanistic characterization of beneficial PBIs. Genomics and multi‐omics approaches have facilitated the identification of many bacterial genes shared across phylogenetically diverse bacterial taxa involved in adaption to root niches, such as those required for colonization and bacteria–bacteria interactions (BBIs) (Levy et al., 2018). However, the processes driving bacterial assembly in root niches at the community level remain elusive since studies have predominantly validated the role of individual genes through analysis of loss‐of‐function mutations on sterile root systems. One exception is the recent application of simplified synthetic communities (SynComs) of bacteria, which have begun to shed light on the influence of individual bacteria and plant‐derived metabolites during the colonization of root niches. In this review we explore the current knowledge of bacterial root colonization from chemotaxis towards the rhizosphere, through to attachment on the root surface (Table 1) and highlight the tools available to aid future characterization of bacterial assembly in root niches.
Table 1.
Genes shown to affect colonization of plant roots.
| Colonization stage | Category | Gene(s) | Strain(s) | Comments | Reference(s) |
|---|---|---|---|---|---|
| Chemotaxis towards the root | Chemotaxis | cheA | P. fluorescens | cheA mutants demonstrate reduced competitive tomato‐root‐tip colonization when inoculated 1:1 with wild‐type | de Weert et al. (2002) |
| che1 gene clusters | R. leguminosarum | Che1, which encodes the conserved set of cheAWYRB homologues, facilitates root colonization and promotes competitive nodulation of peas | Miller et al. (2007) | ||
| mcpA, mcpB, tlpA, tlpB, mcpC, tlpC, yvaQ, yoaH, yfmS, heamAT | B. subtilis | Deletion of all 10 chemoreceptors results in reduced colonization of Arabidopsis roots relative to wild type | Allard‐Massicotte et al. (2016) | ||
| cheY1, cheY2 | A. caulinodans | Regulate chemotaxis, competitive root colonization and competitive nodulation of Sesbania rostrata | Liu et al. (2020) | ||
| Movement towards/over the root and potential role in primary attachment as adhesins | Flagella | fliC, fleQ, fleS | P. fluorescens | Single mutants are non‐motile. Colonize alfalfa roots when inoculated alone but impaired in colonization when co‐inoculated with wild type. These genes have been found in every pseudomonad analysed | Capdevila et al. (2004) |
| flhD, flhC | S. enterica | Double mutant is non‐motile as defective in flagella synthesis and fails to effectively colonize Arabidopsis roots or invade lateral root junctions | Cooley et al. (2003) | ||
| flmA, flmB | A. brasilense | Mutation in either gene results in non‐motile cells due to altered flagellum assembly. Mutations also affect cells competitive ability to attach to maize roots | Rossi et al. (2016) | ||
| Type IV pili | pilA, pilB | Azoarcus sp. BH72 | Single and double mutants failed to colonize rice roots or infect the root epidermal cells | Dörr et al. (1998) | |
| Primary attachment | Major outer membrane proteins (MOMPs) | oprF | Specific to Pseudomonas genus | Purified OprF strongly and selectively binds wheat, barley, maize and sunflower roots, but not leaves. Pseudomonas fluorescens oprF mutants show reduced attachment to cucumber and tomato roots 1 h post‐inoculation | De Mot and Vanderleyden (1991); Alvarez Crespo and Valverde (2009) |
| omaA | A. brasilense | Purified OmaA has a stronger binding affinity for cereal roots relative to legume and tomato roots | Burdman et al. (2001) | ||
| Unipolar polysaccharide (UPP) | gmsA | R. leguminosarum | Determines glucomannan synthesis which under acidic conditions mediates reversible polar attachment of single cells to pea and vetch roots by binding plant lectins | Laus et al. (2006) | |
| Secondary attachment and/or microcolony and biofilm formation | uppABCDEF | A. tumefaciens | Encodes UPP similar to glucomannan but mediates irreversible polar attachment to plant tissue and abiotic surfaces | Tomlinson and Fuqua (2009); Xu et al. (2012) | |
| Cellulose fibrils | celA | R. leguminosarum | Encodes cellulose synthase. Mutation does not alter attachment to root hairs but prevents cap formation under acidic and alkaline conditions. Mutants were also able to form biofilms in vitro (on glass) but not on root hairs | Williams et al. (2008) | |
| A. tumefaciens | Mutants attach to carrot tissue culture cells but unable to form aggregates and easily removed by washing plant tissue | Matthysse (1983) | |||
| Extracellular proteins | lapA |
Specific to P. fluorescens P. putida |
Surface adhesin and biofilm matrix component. Drives transition from reversible to irreversible attachment. Pseudomonas putida lapA mutants are impaired in competitive colonization of corn roots | Espinosa‐Urgel et al. (2000); Hinsa et al. (2003); Yousef‐Coronado et al. (2008); Gjermansen et al. (2010); Duque et al. (2013) | |
| lapF | Specific to P. putida | Mediates cell–cell interactions during biofilm development. Mutants impaired in micrology formation and biofilm development. Also impaired in individual and competitive colonization for corn and alfalfa roots | Martinez‐Gil et al. (2010) | ||
| rapA1 | Confined to R. leguminosarum and R. etli | Overexpression of rapA1 in R. leguminosarum bv. trifolii increases attachment to red clover roots fivefold | Mongiardini et al. (2008) | ||
| Important for attachment to plant roots but not to abiotic surfaces | LPS | rffB |
Rhizobium sp. IRBG74 >99% identity to A. tumefaciens rffB |
Mutants impaired in dTDP‐rhamnose synthesis resulting in altered LPS. Show reduced ability to colonize legume and rice roots, which negatively affects nodulation and endophytic colonization respectively. Mutants not affected in attachment to polypropylene plates so plant attachment specific trait | Mitra et al. (2016) |
| rmlD | A. brasilense | Disruption of dTDP‐rhamnose biosynthesis modifies LPS core and increased EPS production resulting in impaired attachment to maize roots and reduced colonization | Jofré et al. (2004) | ||
| rfbB, rfbC | H. seropedicae | Mutants lack rhamnose‐containing LPS and show 100‐fold reduction in attachment to maize roots relative to wild type. Also impaired in endophytic colonization. No difference in attachment to glass fibre suggesting recognition of rhamnose‐containing LPS is important for colonization of hosts | Balsanelli et al. (2010) | ||
| Root‐hair attachment | EPS | pssA | R. leguminosarum | pssA regulates EPS biosynthesis, with mutants showing reduced attachment to root hairs and impaired cap formation. Attachment to the epidermis was still observed | Williams et al. (2008) |
Chemotaxis towards the root
Plant photosynthates secreted into the rhizosphere form gradients that are perceived by bacteria occupying the surrounding soil, resulting in activation of chemosensory pathways and movement of motile bacteria towards the root (Fig. 1A). Bacterial motility is primarily mediated by proteinaceous appendages termed flagella that protrude from the cell surface or through type IV pili (Alexandre, 2015). Genes homologous to known components of the chemosensory and flagella pathways have been identified in most sequenced bacterial genomes suggesting that chemotaxis provides a selective advantage, particularly in nutrient scarce environments such as soil (Armitage, 1999; Szurmant and Ordal, 2004; Wadhams and Armitage, 2004).
Not surprisingly, inactivation of bacterial chemotaxis or motility renders bacteria deficient for colonization of the rhizoplane (Ames and Bergman, 1981; Bauer and Caetano‐Anollés, 1990; de Weert et al., 2002; Allard‐Massicotte et al., 2016). This is illustrated in experiments where all the Bacillus subtilis chemoreceptors were genetically inactivated leading to a significant reduction in colonization of Arabidopsis thaliana roots 4 h post‐inoculation (Allard‐Massicotte et al., 2016). Chemotaxis and motility also play fundamental roles in the formation of Rhizobium‐legume symbioses. Rhizobia are motile alpha‐proteobacteria that infect legume root nodules and fix atmospheric di‐nitrogen (N2) into ammonia for plant utilization in return for carbon (Poole et al., 2018). Successful establishment of nodule symbiosis requires a highly specific molecular dialogue between the two partners. Beginning in the rhizosphere, exudation of chemoattractants by legumes draw rhizobia towards the root hairs which act as the entry point for nodule infection (Armitage et al., 1988; Barbour et al., 1991; Dharmatilake and Bauer, 1992). Mutation of chemosensory components impairs competitiveness for root colonization and nodule occupancy in Rhizobium leguminosarum (Miller et al., 2007) and Azorhizobium caulinodans (Liu et al., 2020). Although these mutant bacteria remain motile, they are unable to sense chemical gradients towards the root or possibly root hairs where most nodule infections are initiated.
Root attachment
Attachment of bacteria to the rhizoplane marks the first physical step in many PBIs, anchoring bacteria in the nutrient‐rich environment of the rhizosphere and securing a prime location for the subsequent development of more intimate associations. The molecular mechanisms underlying root attachment have been best defined in the agriculturally important bacterial genera: Rhizobium, Pseudomonas, Azospirillum, Agrobacterium and Salmonella (Wheatley and Poole, 2018). These proteobacteria share a common biphasic mechanism consisting of two phases: primary attachment, characterized by reversible binding of bacteria to the root surface, followed by secondary attachment which results in their irreversible adhesion.
Primary attachment
Primary attachment involves weak, non‐specific and reversible binding mediated by hydrophobic and electrostatic interactions between cells and adjacent surface molecules on the root (van Loosdrecht et al., 1987; Kendall and Roberts, 2015). Despite the benefits of root attachment only a small proportion of inoculated isogenic bacteria, typically representing 0.4%–3.5% of the population, actually attach to roots in controlled conditions (Rodríguez‐Navarro et al., 2007). This is primarily due to electrostatic repulsion, which occurs between the negatively charged bacterial cell envelope and root surface (Berne et al., 2015). To overcome these repulsive forces, bacteria use flagella and pili to propel themselves towards the root surface (Fig. 1A). Following these initial interactions adhesins present on the cell surface mediate a tighter but still reversible association with the root (Wheatley and Poole, 2018) (Fig. 1B). Bacterial adhesins involved in primary attachment include proteinaceous appendages (flagella, pili, fimbriae), surface proteins and polysaccharides (exo‐ and capsular polysaccharides).
Numerous studies have demonstrated the role of flagella and pili as adhesins, enabling bacteria to not only move to the root but also to attach and migrate across the root surface. Flagella‐defective mutants of A. brasilense fail to attach to wheat or maize roots, whereas purified polar flagella bind to wheat roots (Croes et al., 1993; Rossi et al., 2016). Likewise, P. fluorescens and S. enterica flagella‐deficient mutants are unable to competitively colonize alfalfa roots or invade Arabidopsis lateral root junctions respectively (Cooley et al., 2003; Capdevila et al., 2004). In the plant pathogen P. aeruginosa (Hahn, 1997; O'Toole and Kolter, 1998) and N2 fixing endophyte Azoarcus sp. BH72 type IV pili act as adhesins (Dörr et al., 1998). However, it is often difficult to elucidate whether adhesion or motility facilitated by the flagella and pili is necessary for primary attachment. Fimbriae also take part in primary attachment, but unlike flagella and pili they do not have an active role in motility. Fimbriae appear to be common primary attachment factors among rhizobacteria (Vesper and Bauer, 1986; Vesper, 1987; Tan et al., 2016), which contain a high proportion of hydrophobic amino acid residues thereby contributing to cell surface hydrophobicity and influencing attachment (Rosenberg and Kjelleberg, 1986; Donlan, 2002).
Considering that bacterial mutants lacking flagella, pili and fimbriae are still able to attach to the root surface (Tan et al., 2016), it is likely that other species‐specific factors with adhesive properties such as polysaccharides and surface proteins play a key role in primary attachment. For example, in A. brasilense and P. fluorescens various major outer membrane proteins (MOMPs) have been implicated in root adhesion and cellular aggregation (De Mot and Vanderleyden, 1991; Burdman et al., 2001; Alvarez Crespo and Valverde, 2009). These MOMPs are exposed on the outer side of the bacterial cell and function by interacting with the surface domains of proteins and polysaccharides located on the root exterior.
Secondary attachment
The second phase of attachment involves strong irreversible binding of the bacteria to the root surface, mediated by the synthesis of extracellular cellulose fibrils and species‐specific secondary attachment factors. Biosynthesis, secretion or exposure of these cellulose fibrils and secondary attachment factors is typically induced after successful primary attachment (Matthysse, 1983; Ausmees et al., 1999; Martinez‐Gil et al., 2010; Monteiro et al., 2012). Secondary attachment culminates in the formation of a bacterial microcolony on the root (Fig. 1C) and ensures that bacteria remain on the rhizoplane. For many bacteria, this is essential for subsequent endophytic colonization (Kandel et al., 2017).
Cellulose fibrils, which often extrude from multiple points over the bacterial cell surface, appear to be universal secondary attachment factors among proteobacteria (Thompson et al., 2018). These fibrils bind tightly to one another, thereby promoting the formation of bacterial aggregates on the rhizoplane. In Rhizobium, cellulose fibrils assist bacterial accumulation at the site of infection by tightly adhering rhizobial cells on root hair tips (Smit et al., 1987; Williams et al., 2008). Similarly, in Agrobacterium cellulose fibrils anchor bacteria at the site of primary attachment promoting tumour formation (Matthysse, 1983). Although attachment is a critical early step in Rhizobium infection and Agrobacterium pathogenesis, the role of cellulose fibrils in attachment is not essential for the establishment of these PBIs. Rhizobium and Agrobacterium mutants deficient in cellulose fibrils are still able to induce nodulation and tumour formation respectively (Matthysse, 1983; Smit et al., 1987). Nonetheless, it cannot be excluded that cellulose fibrils are important for attachment and development of these PBIs under field conditions since Agrobacterium mutants lacking cellulose fibrils are easily removed from the rhizoplane by washing and require inoculation of higher cell densities to induce tumour formation (Minnemeyer et al., 1991).
In addition to the conserved factors described above, species‐specific factors play a key role in secondary attachment. These factors include extracellular proteins and polysaccharides that permit accumulation of bacteria at the site of primary attachment (Rodríguez‐Navarro et al., 2007). In P. fluorescens and P. putida the large adhesin protein LapA defines the transition from reversible polar attachment of single cells to their irreversible adhesion (Hinsa et al., 2003). LapA is a Ca2+ binding protein secreted from the bacterial cell through ATP binding cassette transporters, which loosely associates with the bacterial cell surface ready to mediate surface interactions. Pseudomonas fluorescens lapA mutants attach to abiotic surfaces at levels comparable to the wild type 1‐h post‐inoculation (hpi) but after 5‐hpi show a significant reduction in attachment and are defective for biofilm formation, suggesting that LapA is not involved in the initial primary attachment of P. fluorescens to roots. In contrast, P. putida lapA mutants show reduced attachment to abiotic surfaces and corn seeds and are defective for biofilm formation at 1‐hpi implying that LapA plays a role in the initial adhesion of P. putida (Espinosa‐Urgel et al., 2000; Yousef‐Coronado et al., 2008; Duque et al., 2013). Moreover, P. putida lapA mutants are at a competitive disadvantage for colonization of corn roots when in competition with the wild type strain. In R. leguminosarum and R. etli, Rhizobium‐adhering proteins are important species‐specific primary attachment factors also thought to bind Ca2+ (Ramey et al., 2004). RapA1 is a secreted Ca2+‐binding protein that localizes on the extracellular surface at the cell poles and is predicted to promote aggregation through binding of exopolysaccharides (EPS) or capsular polysaccharide (Russo et al., 2006). Overexpression of rapA1 in R. leguminosarum enhances the number of bacteria attached to host legume roots by up to fivefold (Mongiardini et al., 2008).
Some species‐specific adhesins such as EPS and lipopolysaccharides (LPS) play a role in both the primary and secondary attachment of diverse bacterial species. EPS is a major cell surface component composed of carbohydrate polymers, which promote cellular aggregation and irreversible binding to the root surface by forming bridges between bacterial cells (Burdman et al., 2000). The structure of these polymers varies considerably between bacterial species, altering the electrostatic, hydrophobic and steric properties of the cell surface and in turn affecting attachment. In R. leguminosarum, mutation of the EPS biosynthesis regulator pssA results in reduced attachment to root hairs and impaired aggregation at root hair tips (Williams et al., 2008). However, attachment to the root epidermis was still observed. LPSs constitute a major component of the Gram‐negative outer membrane and are composed of large tripartite glycolipids with a hydrophobic portion called lipid A, a hydrophilic core oligosaccharide, and the hydrophilic O‐antigen side chain (Bertani and Ruiz, 2018). LPS plays a critical role in the establishment of effective associations between several plant growth–promoting bacteria and their hosts. Rhizobium mutants deficient in the production of the monosaccharide rhamnose, an integral component of the LPS O‐antigen, displayed reduced colonization of rice and Sesbania rostrata roots, nullifying plant growth promotion and impairing nodulation respectively (Mitra et al., 2016). Disruption of rhamnose biosynthesis also alters LPS composition in A. brasilense, Herbaspirillum seropedicae and A. caulinodans resulting in reduced colonization of the host plants (Jofré et al., 2004; Balsanelli et al., 2010) and for A. caulinodans, ineffective symbiosis with S. rostrata (Gao et al., 2001). Interestingly, several of these LPS mutants were not impaired in attachment to plastic and glass surfaces, indicating that LPS or a component of it is not required for general attachment (Balsanelli et al., 2010; Mitra et al., 2016).
Environmental factors influence root attachment
Attachment can be influenced by environmental factors such as soil pH, divalent cations (Ca2+ and Mg2+) and water availability (Caetano‐Anollés et al., 1989; Howieson et al., 1993). The effect of pH on attachment has been well characterized in R. leguminosarum. Under acidic conditions the unipolar polysaccharide (UPP) glucomannan mediates localized polar attachment of R. leguminosarum to pea and vetch root hairs through binding to plant lectins (Laus et al., 2006). The gene locus encoding glucomannan is conserved among Rhizobium and Agrobacterium where a UPP similar to glucomannan has been shown to mediate irreversible polar attachment to plant tissue (Tomlinson and Fuqua, 2009; Xu et al., 2012). Under alkaline conditions root lectins are solubilized preventing glucomannan from mediating attachment. It has been proposed that an extracellular Ca2+ binding protein termed ‘rhicadhesin’ may mediate attachment under alkaline conditions. This rhicadhesin is predicted to bind the bacterial cell wall via a Ca2+ that is thought to dissociate under acidic pH conditions; however, there is currently little evidence pertaining to the identity of this hypothetical protein (Matthysse, 2014; Thompson et al., 2018). In fact, evidence for the existence of rhicadhesin is based entirely on a single set of experiments where crude preparations of rhizobial membrane proteins inhibited attachment of various rhizobia to pea roots (Smit et al., 1989; Smit et al., 1991). Thus, it remains unclear as to whether a pure rhicadhesin protein facilitates attachment or whether the concerted action of several proteins is required for this process.
Bacterial biofilms
Following attachment, microcolonies develop into mature biofilms on the root surface. Biofilm formation is a key determinant of successful root colonization and is a common strategy employed by many soil bacteria. Biofilms provide a physical barrier against detrimental external stimulus such as the diffusion of antimicrobial compounds from the host plant or other microbiome members. They also protect bacteria from environmental stresses including changes in pH, osmotic stress and UV radiation (Davey and O'Toole, 2000). Fundamentally, biofilms consist of dynamic heterogeneous communities of bacterial cells embedded in a matrix of EPS which aids in adherence to the root surface and ensures cells remain proximal to one another (Branda et al., 2005; Flemming and Wingender, 2010). Within the biofilm, individual microcolonies are separated by water channels that facilitate diffusion of nutrients, oxygen, antimicrobial compounds and even DNA via horizontal gene transfer (Donlan, 2002; Flemming and Wingender, 2010); hence biofilms also play a significant role in the functioning of BBIs. Large adhesins play a fundamental role in biofilm formation by mediating cell–cell interactions in both Gram‐negative and Gram‐positive bacteria. In P. putida, lapF mutants are unable to form microcolonies at the initial site of attachment and display reduced colonization of corn and alfalfa roots when inoculated individually and co‐inoculated with the wild type (Martinez‐Gil et al., 2010). This suggests that LapF plays a role in both root colonization and the development of mature biofilms in P. putida. LapA has also been implicated in mediating cell–cell as well as cell–surface interactions during biofilm development in P. putida (Gjermansen et al., 2010). No orthologues of lapA or lapF have been identified in P. syringae, P. mendocina, P. stutzeri or P. aeruginosa strains suggesting that although both pathogenic and non‐pathogenic strains of Pseudomonas attach to, colonize and form biofilms on surfaces, the mechanisms by which they do this differ (Duque et al., 2013). Other large adhesins such as Bap and Esp have been shown to mediate surface colonization and biofilm formation in the Gram‐positive bacteria Staphylococcus and Enterococcus respectively (Cucarella et al., 2001; Toledo‐Arana et al., 2001). These proteins along with other adhesins have a similar structural organization to LapA and LapF and are widespread throughout prokaryotes suggesting that a similar mechanism for biofilm formation exists (Lasa and Penadés, 2006; Yousef and Espinosa‐Urgel, 2007).
Plant–bacteria interactions influence colonization
Plant roots grow among diverse bacterial communities with up to 104 bacterial species and 109 bacterial cells per gram of soil (Daniel, 2005). Not surprisingly, the reservoir of bacteria in the surrounding soil is a crucial factor influencing root‐associated microbiome structure, as is illustrated by microbiome analyses of 27 inbred field‐grown maize lines planted at five geographically distinct locations across the United States (Peiffer et al., 2013). Albeit, plants do have some control over the composition of their root‐associated microbiome through modulation of their root exudate composition and mounting of an innate immune response (Lebeis et al., 2015; Reinhold‐Hurek et al., 2015; Sasse et al., 2018). As such, there are clear taxonomic differences between microbiota associated with the root and surrounding soil (Peiffer et al., 2013; Bulgarelli et al., 2015; Edwards et al., 2015; de Souza et al., 2016; Cheng et al., 2020). In Medicago truncatula, the rhizosphere and rhizoplane community structures become distinct after only 1 week of growth in soil. The community structure of both environments becomes more robustly established by the second week and remains stable for a minimum of 3 weeks thereafter (Tkacz et al., 2020). Crucially, microbiome structure varies across different plant species and even among genotypes within a single species (Kuske et al., 2002; Aira et al., 2010).
Plant root exudates contain a wide range of primary metabolites including carbohydrates, organic acids and amino acids, which preferentially stimulate bacterial growth thereby shaping microbiome assembly (Sasse et al., 2018). In addition, secondary metabolites present in root exudates can have antimicrobial activity against pathogens (Olanrewaju et al., 2019), act as signals for the establishment of root symbioses (Abdel‐Lateif et al., 2012) and have profound effects on the bacterial transcriptome (Ramachandran et al., 2011; Carvalhais et al., 2013). Root exudate composition varies between plant species and is dynamically influenced by developmental stage, environmental conditions and the structure of root‐associated microbiome (Sasse et al., 2018). Root colonization by specific microbial communities affects the chemical composition of root exudates through a systemic root‐to‐root signalling mechanism termed systemically induced root exudation of metabolites (SIREM) (Korenblum et al., 2020). In tomato split root assays, inoculation of B. subtilis onto one side of the root induced a systemic signal that results in increased secretion of acyl sugars from the uninoculated side of the root. Transportation of these signals through shoots to uncolonised areas of the root can modulate colonization and assembly of SIREM‐specific microbial communities.
The evidence supporting plant immune signalling as a regulator of root‐associated microbiome structure is less compelling but nevertheless important (Yu et al., 2019). Mutants of A. thaliana impaired in salicylic acid (SA) mediated defence displayed distinct microbiome composition relative to wild‐type plants (Lebeis et al., 2015). However, SA‐dependent signalling was shown to have the largest impact on endophytic community structure with rhizosphere microbiome structure less affected. In wheat, activation of the jasmonic acid (JA) signalling pathway through application of exogenous JA was found to reduce the diversity of the endophytic microbiome but not rhizosphere microbiome (Liu et al., 2017). In contrast, two Arabidopsis mutants disrupted in JA‐dependent signalling showed distinct rhizosphere microbiome structures relative to the wild type, though this could be associated with the fact that their root exudation profiles were also affected (Carvalhais et al., 2015). Additionally, aboveground activation of the immune system by plant pathogens and insects has been shown to alter the rhizosphere microbiome structure of several plant species (Dudenhöffer et al., 2016; Kong et al., 2016; Berendsen et al., 2018; Yuan et al., 2018). Again, alteration of rhizosphere microbiome structure may be linked to alteration of root exudate composition in response to activation of the immune system (Yuan et al., 2018). These results indicate that the immune system can influence epiphytic colonization, but colonization of the rhizosphere is indirectly influenced through the modulation of root exudates in response to the mounting of an innate immune defence.
Bacteria–bacteria interactions influence colonization
Interactions among bacterial communities can be complex, involving both cooperation and competition (Deines and Bosch, 2016). Cooperative interactions include processes such as metabolite exchanges (Zelezniak et al., 2015), whilst production of antimicrobial toxins and deployment of mechanical weapons are examples of competitive interactions (Granato et al., 2019). Notably, most experiments characterizing factors required for bacterial colonization have done so with single strains under sterile laboratory conditions. A major challenge in understanding bacterial assembly in root niches is to move from single‐species studies to those that encompass entire communities to comprehend how individuals interact with each other and their host plant. Due to the enormous complexity of root‐associated microbiomes it is extremely challenging to experimentally characterize the molecular mechanisms underlying PBIs and BBIs, and their effects on plant health in natural systems. To facilitate this, synthetic communities (SynComs) have been used to investigate the role of individual species and plant‐derived metabolites during colonization of root niches (Niu et al., 2017; Voges et al., 2019). Importantly, SynCom assembly is affected by bacterial interactions (Mee et al., 2014), host genotype (Bodenhausen et al., 2014; Lebeis et al., 2015) and niche specificities (Bai et al., 2015). The same factors are known to affect the assembly of natural microbial communities, validating the use of SynComs as simplified microbiomes.
The power of using SynComs to study PBIs and BBIs was recently demonstrated with a simplified community of seven bacterial strains isolated from maize roots (Niu et al., 2017). When inoculated together, all members colonized to form a stable community on maize roots that reduced the prevalence of seedling blight by delaying colonization of the fungal pathogen Fusarium verticillioides. Removal of one specific SynCom member led to the failure of the remaining members to form a stable community, resulting in competitive dominance by a single strain which in isolation was less capable of warding off the pathogen. This study highlights the significance of BBIs during colonization of root niches and demonstrates that fundamental ecological principles, such as the role of keystone species, can be preserved within simplified SynComs. Thus far, the selection of bacterial strains used in SynComs has been guided by studies that have analysed the species composition of the specific plant‐associated microbial community when grown in soil. Moving forward, it could be of great benefit to identify a SynCom that is stable among different plant species. This would provide a simple system for probing specific population‐level determinants of colonization that are common to diverse plants or unique to individual plant species. The prospect of a ‘universal SynCom’ is not unfeasible since a similar set of plant‐associated bacteria is seen across diverse plant species (Müller et al., 2016).
Tools to investigate bacterial colonization of root niches
Traditional single‐isolate experimental approaches cannot unravel the population‐level dynamics of bacterial root colonization or the molecular mechanisms driving them. However, new tools have been developed for marking and monitoring bacterial strains. These include technologies such as engineered transposons for genomic integration of reporter genes (Schlechter et al., 2018) and microfluidic imaging platforms for dynamic mapping (Massalha et al., 2017; Aufrecht et al., 2018; Noirot‐Gros et al., 2020). Moreover, the development of strategies for genome‐wide transposon mutagenesis screens is providing unprecedented opportunities to identify population‐level genetic determinants of colonization (Cain et al., 2020). These technologies are discussed in the following subsections.
Marking bacteria to track and quantify colonization
Marking of bacterial strains with reporter genes exhibiting unique spectral or other visual properties has been widely used to study root colonization, enabling distinction between differentially labelled bacteria and roots. (Bloemberg et al., 2000; Stuurman et al., 2000; Lagendijk et al., 2010; Ramirez‐Mata et al., 2018). Reporter genes are commonly expressed in bacteria using broad‐host‐range plasmids, but they can also be integrated into the chromosome by several strategies including homologous recombination (Ledermann et al., 2015), CRISPR‐Cas9 (Wang et al., 2018) or transposase‐based systems (Schlechter et al., 2018). Stable integration into the chromosome has the advantage of reduced dosage effects and improved stability in the absence of selective pressure, both of which are useful for studies in the rhizosphere and rhizoplane. Transposon‐based systems for DNA integration vary in their host range and mechanism. Mini‐Tn5, for example, functions in a wide range of Gram‐negative bacteria where it randomly inserts into the genome (de Lorenzo et al., 1990; Reznikoff, 2008), whilst mini‐Tn7 integrates at specific attachment (attB Tn7 ) sites located downstream of the highly conserved chromosomal glmS gene (Bao et al., 1991; Craig, 1991). Importantly, mini‐Tn7 integration of reporter genes typically has no detrimental effect on bacterial growth or competitiveness (Enne et al., 2005). Moreover, attB Tn7 sites are prevalent in phylogenetically diverse species making it an attractive tool for differentially labelling diverse bacteria (Parks and Peters, 2007).
Marker genes such as gusA, celB and lacZ have traditionally been used to track bacteria on plants following incubation with a histochemical substrate, which the enzyme encoded by the reporter gene converts to a coloured product. (Sessitsch et al., 1998; Sánchez‐Cañizares and Palacios, 2013). Such reporters allow highly sensitive visualization of marked bacteria without the need for specialized equipment. However, in some instances, staining procedures result in significant cell death. Visualization of these markers is also affected by background activity in some bacteria and host plants (Sessitsch et al., 1998). More recently, fluorescent proteins have become the reporter of choice as they can be detected using non‐invasive methods allowing live cell imaging. There are many variants of fluorescent proteins that can be distinguished from one another based on their unique excitation and emissions wavelengths. By using fluorescent proteins with distinct spectral properties, a maximum of seven bacterial strains can be differentially marked and distinguished by confocal imaging (Schlechter et al., 2018). Fluorescent reporters can additionally be used for quantification of bacterial populations on the root surface by flow cytometry (Fig. 2A), which is a significantly more high‐throughput strategy than traditional culture‐dependent methods such as plate counting (Gamalero et al., 2004; Valdameri et al., 2015). Bioluminescent lux‐based reporters have also been used to measure bacterial attachment on whole‐root systems and for spatiotemporal mapping of root secretion (Pini et al., 2017). The primary benefit of using lux as a bioreporter over fluorescent proteins is improved sensitivity (Belkin, 2003). This is particularly useful when imaging bacteria during the early stages of root colonization, for example attachment at 1–2 h post‐inoculation of bacteria, since the number of cells on the root surface is relatively low (Parsons, 2019). However, light production via the lux proteins is energy‐intensive and can influence cell viability and competitiveness (Pini et al., 2017).
Fig. 2.
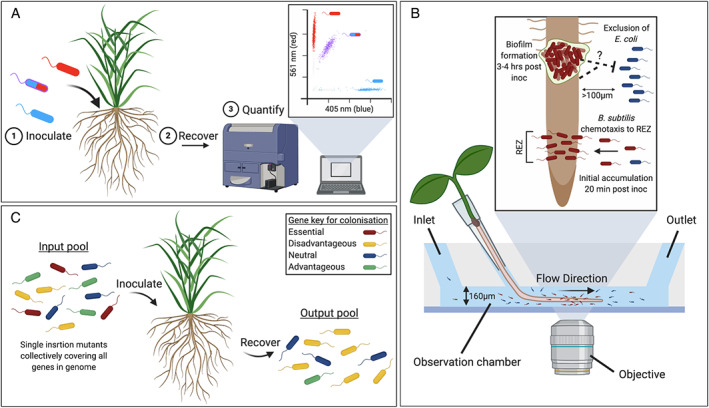
Tools to study bacterial root colonization. A. Flow cytometry: to track bacterial dynamics during colonisations bacterial species can be differentially marked with fluorescent proteins, e.g. red, blue or both, and quantified using flow cytometry. B. TRIS: a microfluidic device for real‐time visualization of bacterial–root interactions. The diagram shows a longitudinal section of a microfluidic channel containing root and bacterial cells (not drawn to scale). Seedlings are germinated through pipette tips into a channel to which bacteria can be introduced through the inlet. (Inset) a schematic of two bacteria, B. subtilis (red) and E. coli (blue) competing to attach to an A. thaliana root. TRIS showed that B. subtilis rapidly accumulates at the root elongation zone (REZ) within 20 min of bacterial inoculation, with subsequent aggregation occurring higher up the root (3–4 h post‐inoculation). Escherichia coli showed clear exclusion from the root, likely due to a diffusible element (represented by a dashed line) released by B. subtilis itself or the root when colonized by B. subtilis (Adapted from Massalha et al., 2017 ). C. Transposon mutagenesis screening: libraries containing single‐insertion transposon mutants that collectively cover all genes in the bacterial genome are inoculated onto a root system and recovered ‘X' days post‐inoculation. Comparison of input and output pools reveals whether a gene is essential (red), non‐essential (blue), advantageous (green) or disadvantages (yellow) for root colonization. Created with BioRender.com.
Competition between bacteria during colonization can alternatively be monitored by barcoding with oligonucleotides. In a recent example, 84 barcoded strains of R. leguminosarum were monitored for nodule occupancy of pea plants in co‐inoculation experiments (Mendoza‐Suárez et al., 2020). Sequencing of bacteria extracted from pea nodules was used to accurately determine the identity and relative abundance of rhizobial strains present in each nodule. In addition to carrying a unique barcode, each strain used in this study encoded a green fluorescent protein expressed from the promoter of the nifHDK (nitrogenase) operon, permitting crude quantification of N2 fixation. The ability to screen for elite competitiveness in large libraries of bacterial strains while simultaneously monitoring the expression of a biochemical marker will be revolutionary for the high‐throughput identification of agricultural inoculants.
Imaging bacterial root interactions
Real‐time imaging of bacterial root interactions is particularly challenging since they occur belowground and vary drastically in spatial scale. To overcome this, plants may be grown in rhizotrons or cultured on agar plates or similar (Schmidt et al., 2011); however, these strategies are not amenable to the high‐resolution imaging techniques required for dynamic mapping of bacterial root interactions. The use of microfluidic platforms combined with live imaging microscopy provides the controlled conditions necessary for continuous imaging of bacterial root interactions at the cellular and subcellular resolution over several days (Massalha et al., 2017; Aufrecht et al., 2018; Noirot‐Gros et al., 2020). The microfluidic device tracking root interactions system (TRIS) consists of nine independent chambers through which roots can simultaneously be grown (Massalha et al., 2017). Fluorescently labelled bacteria can then be introduced into these chambers and imaged with confocal microscopy. TRIS revealed that distinct chemotaxis of B. subtilis towards the root elongation zone (REZ) of Arabidopsis preceded colonization over the entire root length. This indicates that the REZ is a hotspot for initial bacterial root interactions, likely due to high concentrations of root exudates and that bacterial chemotaxis and motility towards these exudates is a prerequisite for root colonization (Fig. 2B). Modification of the original TRIS device into a two‐channel system divided by a semipermeable membrane, that allows free movement of solutes and bacteria whilst preventing the roots of two plants from touching, enables real‐time tracking of bacterial preference for root genotypes (Massalha et al., 2017). One caveat of TRIS and similar devices is that the chamber width limits its use to plants with root diameters less than 160 μm. Consequently, observation of bacterial root interactions is limited to plants with narrow roots such as Arabidopsis. One exception is the root‐microbe interaction (RMI) chip, which facilitates the growth of roots up to 800 μm wide and was successfully used to study bacterial root interactions with Aspen and Rice seedlings (Noirot‐Gros et al., 2020). Adaption of these microfluidics platforms to facilitate the growth of larger roots, such as RMI‐chip, will allow investigation of bacterial root interactions in agronomically relevant crops including cereals and legumes.
Microfluidics platforms may also be used to study cooperative or competitive interactions between differentially marked bacteria. TRIS revealed that co‐inoculation of B. subtilis and Escherichia coli resulted in the exclusion of E. coli from Arabidopsis roots, indicating an antagonistic compound is released from either B. subtilis or roots colonized by B. subtilis (Fig. 2B) (Massalha et al., 2017). Moving forward with these technologies, extending the number of co‐inoculated bacterial species beyond two will provide invaluable insights regarding the influence of BBIs on the early stages of root‐niche colonization.
Transposon screening to identify genetic candidates
Whilst quantification and imaging of bacterial root colonization is a powerful technique for looking at physiology, alternative approaches are required to identify genetic determinants involved in bacterial colonization of roots. Transposon insertion sequencing (TIS) is one such approach. TIS is a powerful technique whereby libraries of single‐insertion transposon mutants, which collectively saturate an organism's genome, are exposed to a specific condition and then analysed with next‐generation sequencing to simultaneously estimate the essentiality and/or fitness contribution of each gene in a bacterial genome. Comparison of gene mutation frequency in an ‘input pool’ relative to an ‘output pool’ following a challenge such as root colonization reveals whether a gene is essential, non‐essential, advantageous, or disadvantageous for growth and survival (Fig. 2C). There are several variations of this technique including INSeq and TnSeq, each following these same basic principles, allowing the determination of gene fitness at the genome‐scale across a variety of conditions (Cain et al., 2020).
Insertion Sequencing (INSeq) was recently used to identify bacterial genes important in the Rhizobium‐legume symbiosis at multiple stages of its development (Wheatley et al., 2020). To form a successful symbiosis, rhizobia must undergo several lifestyle changes. To investigate these, R. leguminosarum bv. viciae 3841 transposon libraries were inoculated onto its host legume Pisum sativum and recovered from four stages of the root‐nodule symbiosis: (i) free‐living growth in the rhizosphere, (ii) colonization of the root, (iii) nodule infection before differentiation into N2 fixing bacteroides and (iv) terminal differentiation into N2 fixing bacteroides (Fig. 3). While only 27 genes are assigned roles in the organization and regulation of N2 fixation, 593 genes were found to be required for the competitive ability to form a successful N2 fixing symbiosis. Of these, 146 were important for growth in the rhizosphere through to N2 fixing bacteroides, highlighting that competition in the rhizosphere is critical for establishing PBIs even in near‐isogenic populations.
Fig. 3.
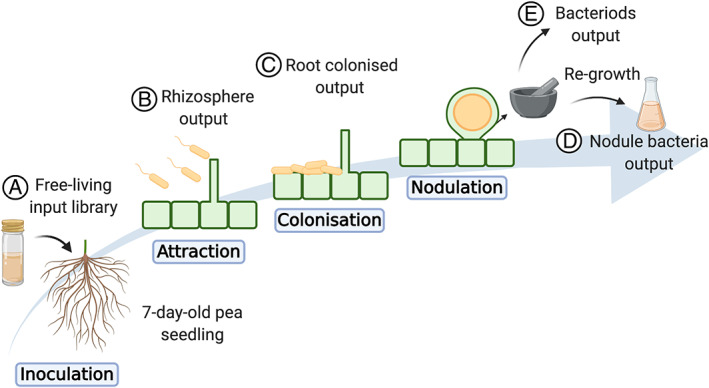
Lifestyle adaptations of Rhizobium from rhizosphere to symbiosis. Insertion sequencing was used to establish the role of Rhizobium leguminosarum bv. viciae 3841 (Rlv3841) genes at multiple stages of symbiosis with Pisum sativum. A. Rlv3841 transposon library was inoculated onto a 7‐day‐old pea seedling. Following inoculation bacteria were collected from four stages of symbiosis for analysis: (B) the rhizosphere (5 dpi), (C) the root (5 dpi), (D) nodule bacteria (28 dpi) and (E) N2 fixing bacteroides (28 dpi). Analysis of DNA purified from the input library and four output libraries enabled genome‐wide classification of gene fitness contributions at each stage. Created with BioRender.com.
A common limitation of most TIS techniques is that they require the mapping of transposon insertion locations for each mutant in the pool following exposure to each treatment. Random barcoded sequencing (RB‐TnSeq) is an extension of TIS techniques in which the transposable element contains a unique, but random, 20 nucleotide DNA ‘barcode’, so that each individual transposon mutant within a pool is identifiable by sequencing (Wetmore et al., 2015). After initial mapping of the transposon insertion site future experiments using the same mutant pool only require sequencing of the DNA barcodes in the input and output pools, saving considerable time and money and allowing multiplexing experiments where tens of strains can be simultaneously analysed. Such multiplexing experiments will be crucial for analysing how bacteria interact with the plant and one another during competitive root colonization. The power of RB‐TnSeq was recently demonstrated in a study that characterized mutant phenotypes of 32 diverse bacterial species in over 150 conditions to assign gene function en masse, resulting in the annotation of over 11 000 previously undefined protein‐coding genes (Price et al., 2018). Application of RB‐TnSeq to study P. simiae colonization of A. thaliana roots led to the identification of 115 genes required for optimal competitive colonization (Cole et al., 2017). These included genes with predicted roles in motility and carbon metabolism, and also 44 genes of unknown function. Undoubtedly, RB‐TnSeq experiments focussing on root colonization will continue to identify novel candidates involved in colonization. The next challenge will lie in deciphering their mechanism of action, particularly in the complex environment of the rhizosphere.
Concluding remarks
Bacterial colonization of plant roots is a sequential, multi‐step process that begins in the rhizosphere with chemotaxis towards the root, followed by attachment and subsequent biofilm formation. To date, most genetic determinants involved in bacterial colonization have been evaluated in single‐strain studies under sterile conditions. Here we have highlighted that both PBIs and BBIs can influence bacterial colonization but characterizing their molecular mechanisms has been hindered due to the enormous scale of complexity and diversity of root‐associated microbiota. With the advent of new DNA‐integration systems and fluorescent reporters, marked seven‐member bacterial SynComs have been successfully cultivated offering unprecedented advancement towards deciphering the underlying principles of bacterial assembly in root niches. Moving forward, genetic exploration of SynComs using transposon mutagenesis‐based screening approaches will be instrumental in unveiling the novel genetics at play. Overall, a more thorough understanding of PBIs and BBIs will advance our understanding of bacterial ecological processes and be invaluable to optimize the future development of biofertilisers for sustainable agriculture, whether this be through targeted selection or engineering of elite plant growth‐promoting bacteria.
Acknowledgements
This work was supported by the Biotechnology and Biological Sciences Research Council [Grant Nos. BB/M011224/1, BB/T001801/1] and the Royal Commission for the Exhibition of 1851 [Grant No. RF‐2019‐100238].
References
- Abdel‐Lateif, K. , Bogusz, D. , and Hocher, V. (2012) The role of flavonoids in the establishment of plant roots endosymbioses with arbuscular mycorrhiza fungi, rhizobia and Frankia bacteria. Plant Signal Behav 7: 636–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aira, M. , Gómez‐Brandón, M. , Lazcano, C. , Bååth, E. , and Domínguez, J. (2010) Plant genotype strongly modifies the structure and growth of maize rhizosphere microbial communities. Soil Biol Biochem 42: 2276–2281. [Google Scholar]
- Alexandre, G. (2015) Chemotaxis control of transient cell aggregation. J Bacteriol 197: 3230–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard‐Massicotte, R. , Tessier, L. , Lécuyer, F. , Lakshmanan, V. , Lucier, J.‐F. , Garneau, D. , et al. (2016) Bacillus subtilis early colonization of Arabidopsis thaliana roots involves multiple chemotaxis receptors. MBio 7: e01664‐01616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez Crespo, M.C. , and Valverde, C. (2009) A single mutation in the oprF mRNA leader confers strict translational control by the Gac/Rsm system in Pseudomonas fluorescens CHA0. Curr Microbiol 58: 182–188. [DOI] [PubMed] [Google Scholar]
- Ames, P. , and Bergman, K. (1981) Competitive advantage provided by bacterial motility in the formation of nodules by Rhizobium meliloti . J Bacteriol 148: 728–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage, J.P. (1999) Bacterial tactic responses. Adv Microb Physiol 41: 229–289. [DOI] [PubMed] [Google Scholar]
- Armitage, J.P. , Gallagher, A. , and Johnston, A.W.B. (1988) Comparison of the chemotactic behaviour of Rhizobium leguminosarum with and without the nodulation plasmid. Mol Microbiol 2: 743–748. [DOI] [PubMed] [Google Scholar]
- Aufrecht, J.A. , Timm, C.M. , Bible, A. , Morrell‐Falvey, J.L. , Pelletier, D.A. , Doktycz, M.J. , and Retterer, S.T. (2018) Quantifying the spatiotemporal dynamics of plant root colonization by beneficial bacteria in a microfluidic habitat. Adv Biosyst 2: 1800048. [Google Scholar]
- Ausmees, N. , Jonsson, H. , Höglund, S. , Ljunggren, H. , and Lindberg, M. (1999) Structural and putative regulatory genes involved in cellulose synthesis in Rhizobium leguminosarum bv. trifolii . Microbiology 145: 1253–1262. [DOI] [PubMed] [Google Scholar]
- Bai, Y. , Muller, D.B. , Srinivas, G. , Garrido‐Oter, R. , Potthoff, E. , Rott, M. , et al. (2015) Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528: 364–369. [DOI] [PubMed] [Google Scholar]
- Balsanelli, E. , Serrato, R.V. , De Baura, V.A. , Sassaki, G. , Yates, M.G. , Rigo, L.U. , et al. (2010) Herbaspirillum seropedicae rfbB and rfbC genes are required for maize colonization. Environ Microbiol 12: 2233–2244. [DOI] [PubMed] [Google Scholar]
- Bao, Y. , Lies, D.P. , Fu, H. , and Roberts, G.P. (1991) An improved Tn7‐based system for the single‐copy insertion of cloned genes into chromosomes of gram‐negative bacteria. Gene 109: 167–168. [DOI] [PubMed] [Google Scholar]
- Barbour, W.M. , Hattermann, D.R. , and Stacey, G. (1991) Chemotaxis of Bradyrhizobium japonicum to soybean exudates. Appl Environ Microbiol 57: 2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardgett, R.D. , and Van Der Putten, W.H. (2014) Belowground biodiversity and ecosystem functioning. Nature 515: 505–511. [DOI] [PubMed] [Google Scholar]
- Bauer, W.D. , and Caetano‐Anollés, G. (1990) Chemotaxis, induced gene expression and competitiveness in the rhizosphere. Plant Soil 129: 45–52. [Google Scholar]
- Belkin, S. (2003) Microbial whole‐cell sensing systems of environmental pollutants. Curr Opin Microbiol 6: 206–212. [DOI] [PubMed] [Google Scholar]
- Berendsen, R.L. , Pieterse, C.M. , and Bakker, P.A. (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17: 478–486. [DOI] [PubMed] [Google Scholar]
- Berendsen, R.L. , Vismans, G. , Yu, K. , Song, Y. , de Jonge, R. , Burgman, W.P. , et al. (2018) Disease‐induced assemblage of a plant‐beneficial bacterial consortium. ISME J 12: 1496–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berne, C. , Ducret, A. , Hardy, G.G. , and Brun, Y.V. (2015) Adhesins involved in attachment to abiotic surfaces by gram‐negative bacteria. Microbiol Spectr 3. 10.1128/microbiolspec.mb-0018-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani, B. , and Ruiz, N. (2018) Function and biogenesis of lipopolysaccharides. EcoSal Plus 8. 10.1128/ecosalplus.ESP-0001-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloemberg, G.V. , Wijfjes, A.H.M. , Lamers, G.E.M. , Stuurman, N. , and Lugtenberg, B.J.J. (2000) Simultaneous imaging of Pseudomonas fluorescens WCS365 populations expressing three different autofluorescent proteins in the rhizosphere: new perspectives for studying microbial communities. Mol Plant Microbe Interact 13: 1170–1176. [DOI] [PubMed] [Google Scholar]
- Bodenhausen, N. , Bortfeld‐Miller, M. , Ackermann, M. , and Vorholt, J.A. (2014) A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLoS Genet 10: e1004283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda, S.S. , Vik, Å. , Friedman, L. , and Kolter, R. (2005) Biofilms: the matrix revisited. Trends Microbiol 13: 20–26. [DOI] [PubMed] [Google Scholar]
- Bulgarelli, D. , Garrido‐Oter, R. , Philipp, C.M. , Weiman, A. , Dröge, J. , Pan, Y. , et al. (2015) Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17: 392–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli, D. , Rott, M. , Schlaeppi, K. , Ver Loren van Themaat, E. , Ahmadinejad, N. , Assenza, F. , et al. (2012) Revealing structure and assembly cues for Arabidopsis root‐inhabiting bacterial microbiota. Nature 488: 91–95. [DOI] [PubMed] [Google Scholar]
- Bulgarelli, D. , Schlaeppi, K. , Spaepen, S. , Van Themaat, E.V.L. , and Schulze‐Lefert, P. (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64: 807–838. [DOI] [PubMed] [Google Scholar]
- Burdman, S. , Dulguerova, G. , Okon, Y. , and Jurkevitch, E. (2001) Purification of the major outer membrane protein of Azospirillum brasilense, its affinity to plant roots, and its involvement in cell aggregation. Mol Plant Microbe Interact 14: 555–561. [DOI] [PubMed] [Google Scholar]
- Burdman, S. , Okon, Y. , and Jurkevitch, E. (2000) Surface characteristics of Azospirillum brasilense in relation to cell aggregation and attachment to plant roots. Crit Rev Microbiol 26: 91–110. [DOI] [PubMed] [Google Scholar]
- Busby, P.E. , Soman, C. , Wagner, M.R. , Friesen, M.L. , Kremer, J. , Bennett, A. , et al. (2017) Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol 15: e2001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano‐Anollés, G. , Lagares, A. , and Favelukes, G. (1989) Adsorption of Rhizobium meliloti to alfalfa roots: dependence on divalent cations and pH. Plant Soil 117: 67–74. [Google Scholar]
- Cain, A.K. , Barquist, L. , Goodman, A.L. , Paulsen, I.T. , Parkhill, J. , and Van Opijnen, T. (2020) A decade of advances in transposon‐insertion sequencing. Nat Rev Genet 21: 526–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila, S. , Martínez‐Granero, F.M. , Sánchez‐Contreras, M. , Rivilla, R. , and Martín, M. (2004) Analysis of Pseudomonas fluorescens F113 genes implicated in flagellar filament synthesis and their role in competitive root colonization. Microbiology 150: 3889–3897. [DOI] [PubMed] [Google Scholar]
- Carvalhais, L.C. , Dennis, P.G. , Badri, D.V. , Kidd, B.N. , Vivanco, J.M. , and Schenk, P.M. (2015) Linking jasmonic acid signaling, root exudates, and rhizosphere microbiomes. Mol Plant Microbe Interact 28: 1049–1058. [DOI] [PubMed] [Google Scholar]
- Carvalhais, L.C. , Dennis, P.G. , Fan, B. , Fedoseyenko, D. , Kierul, K. , Becker, A. , et al. (2013) Linking plant nutritional status to plant‐microbe interactions. PLoS One 8: e68555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z. , Lei, S. , Li, Y. , Huang, W. , Ma, R. , Xiong, J. , et al. (2020) Revealing the variation and stability of bacterial communities in tomato rhizosphere microbiota. Microorganisms 8: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, B.J. , Feltcher, M.E. , Waters, R.J. , Wetmore, K.M. , Mucyn, T.S. , Ryan, E.M. , et al. (2017) Genome‐wide identification of bacterial plant colonization genes. PLoS Biol 15: e2002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley, M.B. , Miller, W.G. , and Mandrell, R.E. (2003) Colonization of Arabidopsis thaliana with Salmonella enterica and enterohemorrhagic Escherichia coli O157:H7 and competition by Enterobacter asburiae . Appl Environ Microbiol 69: 4915–4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, N.L. (1991) Tn7: a target site‐specific transposon. Mol Microbiol 5: 2569–2573. [DOI] [PubMed] [Google Scholar]
- Croes, C.L. , Moens, S. , van Bastelaere, E. , Vanderleyden, J. , and Michiels, K.W. (1993) The polar flagellum mediates Azospirillum brasilense adsorption to wheat roots. Microbiology 139: 2261–2269. [Google Scholar]
- Cucarella, C. , Solano, C. , Valle, J. , Amorena, B. , Lasa, I. , and Penadés, J.R. (2001) Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol 183: 2888–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel, R. (2005) The metagenomics of soil. Nat Rev Microbiol 3: 470–478. [DOI] [PubMed] [Google Scholar]
- Davey, M.E. , and O'Toole, G.A. (2000) Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol 64: 847–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo, V. , Herrero, M. , Jakubzik, U. , and Timmis, K.N. (1990) Mini‐Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram‐negative eubacteria. J Bacteriol 172: 6568–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mot, R. , and Vanderleyden, J. (1991) Purification of a root‐adhesive outer membrane protein of root‐colonizing Pseudomonas fluorescens . FEMS Microbiol Lett 81: 323–327. [Google Scholar]
- de Souza, R.S. , Okura, V.K. , Armanhi, J.S. , Jorrin, B. , Lozano, N. , da Silva, M.J. , et al. (2016) Unlocking the bacterial and fungal communities assemblages of sugarcane microbiome. Sci Rep 6: 28774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Weert, S. , Vermeiren, H. , Mulders, I.H.M. , Kuiper, I. , Hendrickx, N. , Bloemberg, G.V. , et al. (2002) Flagella‐driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens . Mol Plant Microbe Interact 15: 1173–1180. [DOI] [PubMed] [Google Scholar]
- Deines, P. , and Bosch, T.C.G. (2016) Transitioning from microbiome composition to microbial community interactions: the potential of the metaorganism hydra as an experimental model. Front Microbiol 7: 1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmatilake, A.J. , and Bauer, W.D. (1992) Chemotaxis of Rhizobium meliloti towards nodulation gene‐inducing compounds from alfalfa roots. Appl Environ Microbiol 58: 1153–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan, R.M. (2002) Biofilms: microbial life on surfaces. Emerg Infect Dis 8: 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr, J. , Hurek, T. , and Reinhold‐Hurek, B. (1998) Type IV pili are involved in plant‐microbe and fungus‐microbe interactions. Mol Microbiol 30: 7–17. [DOI] [PubMed] [Google Scholar]
- Dudenhöffer, J.‐H. , Scheu, S. , and Jousset, A. (2016) Systemic enrichment of antifungal traits in the rhizosphere microbiome after pathogen attack. J Ecol 104: 1566–1575. [Google Scholar]
- Duque, E. , de la Torre, J. , Bernal, P. , Molina‐Henares, M.A. , Alaminos, M. , Espinosa‐Urgel, M. , et al. (2013) Identification of reciprocal adhesion genes in pathogenic and non‐pathogenic Pseudomonas . Environ Microbiol 15: 36–48. [DOI] [PubMed] [Google Scholar]
- Edwards, J. , Johnson, C. , Santos‐Medellín, C. , Lurie, E. , Podishetty, N.K. , Bhatnagar, S. , et al. (2015) Structure, variation, and assembly of the root‐associated microbiomes of rice. Proc Natl Acad Sci U S A 112: E911–E920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enne, V.I. , Delsol, A.A. , Davis, G.R. , Hayward, S.L. , Roe, J.M. , and Bennett, P.M. (2005) Assessment of the fitness impacts on Escherichia coli of acquisition of antibiotic resistance genes encoded by different types of genetic element. J Antimicrob Chemother 56: 544–551. [DOI] [PubMed] [Google Scholar]
- Espinosa‐Urgel, M. , Salido, A. , and Ramos, J.‐L. (2000) Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J Bacteriol 182: 2363–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estabrook, E.M. , and Yoder, J.I. (1998) Plant‐plant communications: rhizosphere signaling between parasitic angiosperms and their hosts. Plant Physiol 116: 1–7. [Google Scholar]
- Flemming, H.‐C. , and Wingender, J. (2010) The biofilm matrix. Nat Rev Microbiol 8: 623–633. [DOI] [PubMed] [Google Scholar]
- Gamalero, E. , Lingua, G. , Giusy Capri, F. , Fusconi, A. , Berta, G. , and Lemanceau, P. (2004) Colonization pattern of primary tomato roots by Pseudomonas fluorescens A6RI characterized by dilution plating, flow cytometry, fluorescence, confocal and scanning electron microscopy. FEMS Microbiol Ecol 48: 79–87. [DOI] [PubMed] [Google Scholar]
- Gao, M. , D'Haeze, W. , De Rycke, R. , Wolucka, B. , and Holsters, M. (2001) Knockout of an azorhizobial dTDP‐L‐rhamnose synthase affects lipopolysaccharide and extracellular polysaccharide production and disables symbiosis with Sesbania rostrata . Mol Plant Microbe Interact 14: 857–866. [DOI] [PubMed] [Google Scholar]
- Gjermansen, M. , Nilsson, M. , Yang, L. , and Tolker‐Nielsen, T. (2010) Characterization of starvation‐induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol Microbiol 75: 815–826. [DOI] [PubMed] [Google Scholar]
- Granato, E.T. , Meiller‐Legrand, T.A. , and Foster, K.R. (2019) The evolution and ecology of bacterial warfare. Curr Biol 29: R521–R537. [DOI] [PubMed] [Google Scholar]
- Hahn, H.P. (1997) The type‐4 pilus is the major virulence‐associated adhesin of Pseudomonas aeruginosa – a review. Gene 192: 99–108. [DOI] [PubMed] [Google Scholar]
- Hinsa, S.M. , Espinosa‐Urgel, M. , Ramos, J.L. , and O'Toole, G.A. (2003) Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol 49: 905–918. [DOI] [PubMed] [Google Scholar]
- Howieson, J.G. , Robson, A.D. , and Ewing, M.A. (1993) External phosphate and calcium concentrations, and pH, but not the products of rhizobial nodulation genes, affect the attachment of Rhizobium meliloti to roots of annual medics. Soil Biol Biochem 25: 567–573. [Google Scholar]
- Jofré, E. , Lagares, A. , and Mori, G. (2004) Disruption of dTDP‐rhamnose biosynthesis modifies lipopolysaccharide core, exopolysaccharide production, and root colonization in Azospirillum brasilense . FEMS Microbiol Lett 231: 267–275. [DOI] [PubMed] [Google Scholar]
- Kandel, S. , Joubert, P. , and Doty, S. (2017) Bacterial endophyte colonization and distribution within plants. Microorganisms 5: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall, K. , and Roberts, A.D. (2015) Van der Waals forces influencing adhesion of cells. Philos Trans R Soc Lond B Biol Sci 370: 20140078–20140078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, H.G. , Kim, B.K. , Song, G.C. , Lee, S. , and Ryu, C.‐M. (2016) Aboveground whitefly infestation‐mediated reshaping of the root microbiota. Front Microbiol 7: 1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenblum, E. , Dong, Y. , Szymanski, J. , Panda, S. , Jozwiak, A. , Massalha, H. , et al. (2020) Rhizosphere microbiome mediates systemic root metabolite exudation by root‐to‐root signaling. Proc Natl Acad Sci U S A 117: 3874–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuske, C.R. , Ticknor, L.O. , Miller, M.E. , Dunbar, J.M. , Davis, J.A. , Barns, S.M. , and Belnap, J. (2002) Comparison of soil bacterial communities in rhizospheres of three plant species and the interspaces in an arid grassland. Appl Environ Microbiol 68: 1854–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagendijk, E.L. , Validov, S. , Lamers, G.E. , de Weert, S. , and Bloemberg, G.V. (2010) Genetic tools for tagging gram‐negative bacteria with mCherry for visualization in vitro and in natural habitats, biofilm and pathogenicity studies. FEMS Microbiol Lett 305: 81–90. [DOI] [PubMed] [Google Scholar]
- Lasa, I. , and Penadés, J.R. (2006) Bap: a family of surface proteins involved in biofilm formation. Res Microbiol 157: 99–107. [DOI] [PubMed] [Google Scholar]
- Laus, M.C. , Logman, T.J. , Lamers, G.E. , Van Brussel, A.A.N. , Carlson, R.W. , and Kijne, J.W. (2006) A novel polar surface polysaccharide from Rhizobium leguminosarum binds host plant lectin. Mol Microbiol 59: 1704–1713. [DOI] [PubMed] [Google Scholar]
- Lebeis, S.L. , Paredes, S.H. , Lundberg, D.S. , Breakfield, N. , Gehring, J. , McDonald, M. , et al. (2015) Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349: 860–864. [DOI] [PubMed] [Google Scholar]
- Ledermann, R. , Bartsch, I. , Remus‐Emsermann, M.N. , Vorholt, J.A. , and Fischer, H.M. (2015) Stable fluorescent and enzymatic tagging of Bradyrhizobium diazoefficiens to analyze host‐plant infection and colonization. Mol Plant Microbe Interact 28: 959–967. [DOI] [PubMed] [Google Scholar]
- Levy, A. , Salas Gonzalez, I. , Mittelviefhaus, M. , Clingenpeel, S. , Herrera Paredes, S. , Miao, J. , et al. (2018) Genomic features of bacterial adaptation to plants. Nat Genet 50: 138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Carvalhais, L.C. , Schenk, P.M. , and Dennis, P.G. (2017) Effects of jasmonic acid signalling on the wheat microbiome differ between body sites. Sci Rep 7: 41766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Bai, X. , Li, Y. , Min, J. , Kong, Y. , and Hu, X. (2020) CheY1 and CheY2 of Azorhizobium caulinodans ORS571 regulate chemotaxis and competitive colonization with the host plant. Appl Environ Microbiol 86: e00599‐00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg, D.S. , Lebeis, S.L. , Paredes, S.H. , Yourstone, S. , Gehring, J. , Malfatti, S. , et al. (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488: 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez‐Gil, M. , Yousef‐Coronado, F. , and Espinosa‐Urgel, M. (2010) LapF, the second largest Pseudomonas putida protein, contributes to plant root colonization and determines biofilm architecture. Mol Microbiol 77: 549–561. [DOI] [PubMed] [Google Scholar]
- Massalha, H. , Korenblum, E. , Malitsky, S. , Shapiro, O.H. , and Aharoni, A. (2017) Live imaging of root‐bacteria interactions in a microfluidics setup. Proc Natl Acad Sci U S A 114: 4549–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse, A.G. (1983) Role of bacterial cellulose fibrils in Agrobacterium tumefaciens infection. J Bacteriol 154: 906–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse, A.G. (2014) Attachment of Agrobacterium to plant surfaces. Front Plant Sci 5: 252–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mee, M.T. , Collins, J.J. , Church, G.M. , and Wang, H.H. (2014) Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci U S A 111: E2149–E2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza‐Suárez, M.A. , Geddes, B.A. , Sánchez‐Cañizares, C. , Ramírez‐González, R.H. , Kirchhelle, C. , Jorrin, B. , and Poole, P.S. (2020) Optimizing Rhizobium‐legume symbioses by simultaneous measurement of rhizobial competitiveness and N2 fixation in nodules. Proc Natl Acad Sci U S A 117: 9822–9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, L.D. , Yost, C.K. , Hynes, M.F. , and Alexandre, G. (2007) The major chemotaxis gene cluster of Rhizobium leguminosarum bv. viciae is essential for competitive nodulation. Mol Microbiol 63: 348–362. [DOI] [PubMed] [Google Scholar]
- Minnemeyer, S.L. , Lightfoot, R. , and Matthysse, A.G. (1991) A semiquantitative bioassay for relative virulence of Agrobacterium tumefaciens strains on Bryophyllum daigremontiana . J Bacteriol 173: 7723–7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, S. , Mukherjee, A. , Wiley‐Kalil, A. , Das, S. , Owen, H. , Reddy, P.M. , et al. (2016) A rhamnose‐deficient lipopolysaccharide mutant of Rhizobium sp. IRBG74 is defective in root colonization and beneficial interactions with its flooding‐tolerant hosts Sesbania cannabina and wetland rice. J Exp Bot 67: 5869–5884. [DOI] [PubMed] [Google Scholar]
- Mongiardini, E.J. , Ausmees, N. , Pérez‐Giménez, J. , Julia Althabegoiti, M. , Ignacio Quelas, J. , López‐García, S.L. , and Lodeiro, A.R. (2008) The rhizobial adhesion protein RapA1 is involved in adsorption of rhizobia to plant roots but not in nodulation. FEMS Microbiol Ecol 65: 279–288. [DOI] [PubMed] [Google Scholar]
- Monteiro, R.A. , Balsanelli, E. , Tuleski, T. , Faoro, H. , Cruz, L.M. , Wassem, R. , et al. (2012) Genomic comparison of the endophyte Herbaspirillum seropedicae SmR1 and the phytopathogen Herbaspirillum rubrisubalbicansM1 by suppressive subtractive hybridization and partial genome sequencing. FEMS Microbiol Ecol 80: 441–451. [DOI] [PubMed] [Google Scholar]
- Müller, D.B. , Vogel, C. , Bai, Y. , and Vorholt, J.A. (2016) The plant microbiota: systems‐level insights and perspectives. Annu Rev Genet 50: 211–234. [DOI] [PubMed] [Google Scholar]
- Niu, B. , Paulson, J.N. , Zheng, X. , and Kolter, R. (2017) Simplified and representative bacterial community of maize roots. Proc Natl Acad Sci U S A 114: E2450–E2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirot‐Gros, M.‐F. , Shinde, S.V. , Akins, C. , Johnson, J.L. , Zerbs, S. , Wilton, R. , et al. (2020) Functional imaging of microbial interactions with tree roots using a microfluidics setup. Front Plant Sci 11: 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole, G.A. , and Kolter, R. (1998) Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30: 295–304. [DOI] [PubMed] [Google Scholar]
- Olanrewaju, O.S. , Ayangbenro, A.S. , Glick, B.R. , and Babalola, O.O. (2019) Plant health: feedback effect of root exudates‐rhizobiome interactions. Appl Microbiol Biotechnol 103: 1155–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks, A.R. , and Peters, J.E. (2007) Transposon Tn7 is widespread in diverse bacteria and forms genomic islands. J Bacteriol 189: 2170–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons, J. (2019) Characterising Root Attachment in Rhizobium‐Legume Symbioses, Oxford: University of Oxford. [Google Scholar]
- Peiffer, J.A. , Spor, A. , Koren, O. , Jin, Z. , Tringe, S.G. , Dangl, J.L. , et al. (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci U S A 110: 6548–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot, L. , Raaijmakers, J.M. , Lemanceau, P. , and van der Putten, W.H. (2013) Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11: 789–799. [DOI] [PubMed] [Google Scholar]
- Pini, F. , East, A.K. , Appia‐Ayme, C. , Tomek, J. , Karunakaran, R. , Mendoza‐Suarez, M. , et al. (2017) Bacterial biosensors for in vivo spatiotemporal mapping of root secretion. Plant Physiol 174: 1289–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole, P. , Ramachandran, V. , and Terpolilli, J. (2018) Rhizobia: from saprophytes to endosymbionts. Nat Rev Microbiol 16: 291–303. [DOI] [PubMed] [Google Scholar]
- Price, M.N. , Wetmore, K.M. , Waters, R.J. , Callaghan, M. , Ray, J. , Liu, H. , et al. (2018) Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557: 503–509. [DOI] [PubMed] [Google Scholar]
- Ramachandran, V.K. , East, A.K. , Karunakaran, R. , Downie, J.A. , and Poole, P.S. (2011) Adaptation of Rhizobium leguminosarum to pea, alfalfa and sugar beet rhizospheres investigated by comparative transcriptomics. Genome Biol 12: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey, B.E. , Koutsoudis, M. , Bodman, S.B.v. , and Fuqua, C. (2004) Biofilm formation in plant–microbe associations. Curr Opin Microbiol 7: 602–609. [DOI] [PubMed] [Google Scholar]
- Ramirez‐Mata, A. , Pacheco, M.R. , Moreno, S.J. , Xiqui‐Vazquez, M.L. , and Baca, B.E. (2018) Versatile use of Azospirillum brasilense strains tagged with egfp and mCherry genes for the visualization of biofilms associated with wheat roots. Microbiol Res 215: 155–163. [DOI] [PubMed] [Google Scholar]
- Reinhold‐Hurek, B. , Bünger, W. , Burbano, C.S. , Sabale, M. , and Hurek, T. (2015) Roots shaping their microbiome: global hotspots for microbial activity. Annu Rev Phytopathol 53: 403–424. [DOI] [PubMed] [Google Scholar]
- Reznikoff, W.S. (2008) Transposon Tn5. Annu Rev Genet 42: 269–286. [DOI] [PubMed] [Google Scholar]
- Rodríguez‐Navarro, D.N. , Dardanelli, M.S. , and Ruíz‐Saínz, J.E. (2007) Attachment of bacteria to the roots of higher plants. FEMS Microbiol Lett 272: 127–136. [DOI] [PubMed] [Google Scholar]
- Rosenberg, M. , and Kjelleberg, S. (1986) Hydrophobic interactions: role in bacterial adhesion. In Advances in Microbial Ecology. Marshall, K.C. (ed). Springer US: Boston, MA, pp. 353–393. [Google Scholar]
- Rossi, F.A. , Medeot, D.B. , Liaudat, J.P. , Pistorio, M. , and Jofré, E. (2016) In Azospirillum brasilense, mutations in flmA or flmB genes affect polar flagellum assembly, surface polysaccharides, and attachment to maize roots. Microbiol Res 190: 55–62. [DOI] [PubMed] [Google Scholar]
- Russo, D.M. , Williams, A. , Edwards, A. , Posadas, D.M. , Finnie, C. , Dankert, M. , et al. (2006) Proteins exported via the PrsD‐PrsE type I secretion system and the acidic exopolysaccharide are involved in biofilm formation by Rhizobium leguminosarum . J Bacteriol 188: 4474–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Cañizares, C. , and Palacios, J. (2013) Construction of a marker system for the evaluation of competitiveness for legume nodulation in Rhizobium strains. J Microbiol Methods 92: 246–249. [DOI] [PubMed] [Google Scholar]
- Sasse, J. , Martinoia, E. , and Northen, T. (2018) Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci 23: 25–41. [DOI] [PubMed] [Google Scholar]
- Schlaeppi, K. , Dombrowski, N. , Oter, R.G. , Ver Loren Van Themaat, E. , and Schulze‐Lefert, P. (2014) Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci U S A 111: 585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlechter, R.O. , Jun, H. , Bernach, M. , Oso, S. , Boyd, E. , Muñoz‐Lintz, D.A. , et al. (2018) Chromatic bacteria – a broad host‐range plasmid and chromosomal insertion toolbox for fluorescent protein expression in bacteria. Front Microbiol 9: 3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, H. , Eickhorst, T. , and Tippkötter, R. (2011) Monitoring of root growth and redox conditions in paddy soil rhizotrons by redox electrodes and image analysis. Plant Soil 341: 221–232. [Google Scholar]
- Sessitsch, A. , Hardarson, G. , de Vos, W.M. , and Wilson, K.J. (1998) Use of marker genes in competition studies of Rhizobium . Plant Soil 204: 35–45. [Google Scholar]
- Smit, G. , Kijne, J.W. , and Lugtenberg, B.J. (1987) Involvement of both cellulose fibrils and a Ca2+−dependent adhesin in the attachment of Rhizobium leguminosarum to pea root hair tips. J Bacteriol 169: 4294–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, G. , Logman, T.J. , Boerrigter, M.E. , Kijne, J.W. , and Lugtenberg, B.J. (1989) Purification and partial characterization of the Rhizobium leguminosarum biovar viciae Ca2+−dependent adhesin, which mediates the first step in attachment of cells of the family Rhizobiaceae to plant root hair tips. J Bacteriol 171: 4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, G. , Tubbing, D.M.J. , Kijne, J.W. , and Lugtenberg, B.J.J. (1991) Role of Ca2+ in the activity of rhicadhesin from Rhizobium leguminosarum biovar viciae, which mediates the first step in attachment of Rhizobiaceae cells to plant root hair tips. Arch Microbiol 155: 278–283. [Google Scholar]
- Stuurman, N. , Bras, C.P. , Schlaman, H.R.M. , Wijfjes, A.H.M. , Bloemberg, G. , and Spaink, H.P. (2000) Use of green fluorescent protein color variants expressed on stable broad‐host‐range vectors to visualize rhizobia interacting with plants. Mol Plant Microbe Interact 13: 1163–1169. [DOI] [PubMed] [Google Scholar]
- Szurmant, H. , and Ordal, G.W. (2004) Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev 68: 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, M.S.F. , White, A.P. , Rahman, S. , and Dykes, G.A. (2016) Role of fimbriae, flagella and cellulose on the attachment of Salmonella Typhimurium ATCC 14028 to plant cell wall models. PLOS One 11: e0158311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, M.A. , Onyeziri, M.C. , and Fuqua, C. (2018) Function and regulation of Agrobacterium tumefaciens cell surface structures that promote attachment. Curr Top Microbiol Immunol 418: 143–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkacz, A. , Bestion, E. , Bo, Z. , Hortala, M. , and Poole, P.S. (2020) Influence of plant fraction, soil, and plant species on microbiota: a multikingdom comparison. MBio 11: e02785–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkacz, A. , Cheema, J. , Chandra, G. , Grant, A. , and Poole, P.S. (2015) Stability and succession of the rhizosphere microbiota depends upon plant type and soil composition. ISME J 9: 2349–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo‐Arana, A. , Valle, J. , Solano, C. , Arrizubieta, M.J. , Cucarella, C. , Lamata, M. , et al. (2001) The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl Environ Microbiol 67: 4538–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson, A.D. , and Fuqua, C. (2009) Mechanisms and regulation of polar surface attachment in Agrobacterium tumefaciens . Curr Opin Microbiol 12: 708–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi, P. , Leach, J.E. , Tringe, S.G. , Sa, T. , and Singh, B.K. (2020) Plant–microbiome interactions: from community assembly to plant health. Nat Rev Microbiol 18: 607–621. [DOI] [PubMed] [Google Scholar]
- Turner, T.R. , James, E.K. , and Poole, P.S. (2013a) The plant microbiome. Genome Biol 14: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, T.R. , Ramakrishnan, K. , Walshaw, J. , Heavens, D. , Alston, M. , Swarbreck, D. , et al. (2013b) Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J 7: 2248–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uroz, S. , Buee, M. , Murat, C. , Frey‐Klett, P. , and Martin, F. (2010) Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ Microbiol Rep 2: 281–288. [DOI] [PubMed] [Google Scholar]
- Valdameri, G. , Kokot, T.B. , Pedrosa, F.,.O. , and de Souza, E.M. (2015) Rapid quantification of rice root‐associated bacteria by flow cytometry. Lett Appl Microbiol 60: 237–241. [DOI] [PubMed] [Google Scholar]
- van Loosdrecht, M.C. , Lyklema, J. , Norde, W. , Schraa, G. , and Zehnder, A.J. (1987) The role of bacterial cell wall hydrophobicity in adhesion. Appl Environ Microbiol 53: 1893–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesper, S.J. (1987) Production of pili (fimbriae) by Pseudomonas fluorescens and correlation with attachment to corn roots. Appl Environ Microbiol 53: 1397–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesper, S.J. , and Bauer, W.D. (1986) Role of pili (fimbriae) in attachment of Bradyrhizobium japonicum to soybean roots. Appl Environ Microbiol 52: 134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges, M.J.E.E.E. , Bai, Y. , Schulze‐Lefert, P. , and Sattely, E.S. (2019) Plant‐derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc Natl Acad Sci U S A 116: 12558–12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams, G.H. , and Armitage, J.P. (2004) Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol 5: 1024–1037. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Hong, W. , Dong, S. , Zhang, Z.T. , Zhang, J. , Wang, L. , and Wang, Y. (2018) Genome engineering of Clostridium difficile using the CRISPR‐Cas9 system. Clin Microbiol Infect 24: 1095–1099. [DOI] [PubMed] [Google Scholar]
- Wei, Z. , Yang, T. , Friman, V.‐P. , Xu, Y. , Shen, Q. , and Jousset, A. (2015) Trophic network architecture of root‐associated bacterial communities determines pathogen invasion and plant health. Nat Commun 6: 8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore, K.M. , Price, M.N. , Waters, R.J. , Lamson, J.S. , He, J. , Hoover, C.A. , et al. (2015) Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar‐coded transposons. MBio 6: e00306‐00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley, R.M. , Ford, B.L. , Li, L. , Aroney, S.T.N. , Knights, H.E. , Ledermann, R. , et al. (2020) Lifestyle adaptations of Rhizobium from rhizosphere to symbiosis. Proc Natl Acad Sci U S A 117: 23823–23834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley, R.M. , and Poole, P.S. (2018) Mechanisms of bacterial attachment to roots. FEMS Microbiol Rev 42: 448–461. [DOI] [PubMed] [Google Scholar]
- Williams, A. , Wilkinson, A. , Krehenbrink, M. , Russo, D.M. , Zorreguieta, A. , and Downie, J.A. (2008) Glucomannan‐mediated attachment of Rhizobium leguminosarum to pea root hairs is required for competitive nodule infection. J Bacteriol 190: 4706–4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Kim, J. , Danhorn, T. , Merritt, P.M. , and Fuqua, C. (2012) Phosphorus limitation increases attachment in Agrobacterium tumefaciens and reveals a conditional functional redundancy in adhesin biosynthesis. Res Microbiol 163: 674–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef, F. , and Espinosa‐Urgel, M. (2007) In silico analysis of large microbial surface proteins. Res Microbiol 158: 545–550. [DOI] [PubMed] [Google Scholar]
- Yousef‐Coronado, F. , Travieso, M.L. , and Espinosa‐Urgel, M. (2008) Different, overlapping mechanisms for colonization of abiotic and plant surfaces by Pseudomonas putida . FEMS Microbiol Lett 288: 118–124. [DOI] [PubMed] [Google Scholar]
- Yu, K. , Pieterse, C.M.J. , Bakker, P.A.H.M. , and Berendsen, R.L. (2019) Beneficial microbes going underground of root immunity. Plant Cell Environ 42: 2860–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, J. , Zhao, J. , Wen, T. , Zhao, M. , Li, R. , Goossens, P. , et al. (2018) Root exudates drive the soil‐borne legacy of aboveground pathogen infection. Microbiome 6: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelezniak, A. , Andrejev, S. , Ponomarova, O. , Mende, D.R. , Bork, P. , and Patil, K.R. (2015) Metabolic dependencies drive species co‐occurrence in diverse microbial communities. Proc Natl Acad Sci U S A 112: 6449–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]


