Abstract
Up to 80% of people meeting DSM-IV definitions for schizophrenia will exhibit difficulties with sleep along with a breakdown in circadian entrainment and rhythmicity. The changes to the sleep and circadian systems in this population are thought to be interdependent, as evidenced by the frequent use of the combined term “sleep and circadian rhythm disruption” or “SCRD” to describe their occurrence. To understand links between sleep and circadian problems in the schizophrenia population, we analyzed the duration and rhythmicity of sleep behavior in mice lacking function of the immediate early gene early growth response 3 (Egr3−/−). EGR3 has been associated with schizophrenia risk in humans, and Egr3-deficient mice display various features of schizophrenia that are responsive to antipsychotic treatment. While Egr3−/− mice slept less than their wild type (WT) littermates, they showed no evidence of circadian disorganization; in fact, circadian rhythms of activity were more robust in these mice compared with WT, as measured by time series analysis and the relative amplitude index of Van Someren’s suite of non-parametric circadian rhythm analyses. Differences in circadian robustness were maintained when the animals were transferred to several weeks of housing under constant darkness or constant light. Together, our results suggest that Egr3−/− mice retain control over the circadian timekeeping of sleep and wake, while showing impaired sleep. The findings are compatible with those from a surprising array of mouse models of schizophrenia and raise the possibility that SCRD may be 2 separate disorders in the schizophrenia population requiring different treatment strategies.
Keywords: schizophrenia, Egr3, mouse model, circadian, sleep monitoring, piezoelectric
Schizophrenia is a severe mental illness with a lifetime prevalence of approximately 1% (Eaton et al., 2011). The condition cuts across Western and Eastern cultures and is equally common in men and women, providing no simple clues as to its genetic origins, despite high rates of heritability in many families (Sartorius et al., 1986). Symptom clusters can vary from one person with schizophrenia to the next and can fluctuate within the same individual over time (Richard et al., 1996). Diagnostic criteria involve a combination of abnormalities within several psychopathological domains, including “positive symptoms” of perceptual disturbance (e.g., hallucinations and delusions), “negative symptoms” consisting of reductions in the range and intensity of emotional expression (e.g., avolition, anhedonia, and alogia), and “cognitive symptoms,” including disorganized thinking and poor executive function (reviewed in Chan, 2017). The range of symptoms expressed among individuals diagnosed with schizophrenia may arise from differences in genetic background and environmental experience during both prenatal development and life (Arnedo et al., 2015; Brugger and Howes, 2017).
Despite the heterogeneity observed at the genetic, neurological, and clinical levels, upwards of 80% of people meeting DSM-IV (Diagnostic and Statistical Manual of Mental Disorders, fourth edition) definitions for schizophrenia exhibit significant difficulties with sleep and altered circadian function (Wulff et al., 2010). Characteristic changes in sleep architecture and continuity are noted in many patients using polysomnography, with meta-analyses identifying longer latency and lower amounts of slow-wave sleep, decreased onset latency of rapid-eye movement sleep, reductions in total sleep time and efficiency, and more overnight awakenings (Chouinard et al., 2004; Cohrs, 2008; Monti and Monti, 2004). Circadian assessments also point to a conserved profile of changes. In particular, phase shifts in melatonin—or the systematic breakdown of circadian rhythms in melatonin secretion or entrainment—have been reported in individuals with schizophrenia (Bromundt et al., 2011; Monti et al., 2013 and references therein; Rao et al., 1994; Wirz-Justice et al., 1997; Wulff et al., 2012). Available evidence suggests that these sleep-circadian disruptions (SCRDs) are not secondary factors related to antipsychotic treatment, which has sedative effects, or irregular daily routines that lack defined schedules of light exposure and social interaction. SCRDs tend to precede diagnosis and are equally visible in medicated and medication-free patients, as well as patients residing in hospital facilities or living as outpatients without a regular work schedule (Bromundt et al., 2011; Yang and Winkelman, 2006). Results of intervention studies suggest a more central role for sleep-circadian function in the emergence and treatment of schizophrenia. Such studies have shown that cognitive-behavioral therapy to improve sleep quality subsequently reduces the positive and negative symptoms of schizophrenia (Hofstetter et al., 2005; Myers et al., 2011). This was found in existing patient groups, as well as prospectively in large samples of the general young-adult population within an age demographic where schizophrenia symptoms first emerge (Freeman et al., 2017).
EGR3 is a member of the early growth response (EGR) family of immediate early gene (IEG) transcription factors. The fact that both Egr3 and Egr1 are activated in the SCN following nighttime light exposure led to the hypothesis that these genes operate in a pathway mediating circadian entrainment to LD cycles (Dziema et al., 2003; Morris et al., 1998; O’Donovan et al., 1999). Several EGR family genes have also been implicated in schizophrenia susceptibility (Marballi and Gallitano, 2018). EGR3, the most prominent of these genes, is associated with schizophrenia across multiple populations (Huentelman et al., 2015; Kim et al., 2010; Yamada et al., 2007; Zhang et al., 2012) and is expressed at reduced levels in patient brains (Mexal et al., 2005; Yamada et al., 2007). Although EGR3 is not at a locus currently significant at the genome-wide level, it interacts with EGR-family proteins EGR1 and its binding protein, NGFIA-binding protein 2 (NAB2; also known as EGR-1-binding protein 2), which map to loci identified in the Psychiatric Genomics Consortium genome-wide association study that identified 108 loci for schizophrenia. In fact, EGR1 and NAB2 both contain the “index” single nucleotide polymorphism (most significant SNP) at their respective loci (Schizophrenia Working Group, 2014).
There is a growing consensus that psychiatric illnesses, including schizophrenia, will be more fully understood when changes to the sleep and circadian system are taken into account (Wulff et al., 2010). Because of the complexity of studying schizophrenia in humans, links between SCRDs and schizophrenia-relevant endophenotypes are most easily made in rodents, which can be genetically or pharmacologically engineered. To date, genetically modified mouse models that misexpress genes linked to schizophrenia risk capture a range of various SCRDs (Bhardwaj et al., 2015; Oliver et al., 2012; Pritchett et al., 2015). In mice overexpressing human DISC1 (disrupted in schizophrenia 1), for example, the circadian system remains grossly normal but sleep homeostasis is impaired; DISC1 mice stay awake longer than control littermates and demonstrate less slow-wave recovery sleep after 2 h of sleep deprivation (Jaaro-Peled et al., 2016).
We have previously characterized the schizophrenia-like behavioral profile of mice that lack Egr3 function, and the unique behavioral response of these animals to antipsychotic treatment (Gallitano-Mendel, Izumi, et al., 2007; Gallitano-Mendel, Wozniak, et al., 2008; Williams et al., 2012). In addition to common mouse schizophrenia endophenotypes (e.g., Oliver and Davies, 2009), Egr3-deficient (−/−) mice show attenuated delta-band responses to 6-h sleep deprivation resembling the diminished homeostatic sleep drive seen in DISC1 mice (Grønli et al., 2016). To determine if the sleep abnormalities in Egr3−/− mice were possibly related to changes in circadian function, the present study examined 2 cohorts of wildtype (WT) and Egr3−/− littermates using PiezoSleep. This non-invasive system detects sleep and circadian patterns of arousal with a high degree of accuracy (~90%) relative to electroencephalogram/electromyogram recordings (see Supplementary Materials for methodological details concerning the PiezoSleep Mouse Behavior System, Signal Solutions, Lexington, KY; Donohue et al., 2008; Mang et al., 2014).
Our first experiment tracked the wake-sleep behavior of female WT and Egr3−/− mice housed within the PiezoSleep chambers for 1 week under a 14–10 LD cycle (lights-on at 0600 PST defined as ZT 0; 200 lux, cool white fluorescent light). Here, Egr3-deficient mice slept less than WT littermates (average daily sleep, WT, n = 6, 820 ± 29 min; Egr3−/−, n = 7, 743 ± 20 min; average daily wake percentage distribution and break-down of sleep duration in the light v. the dark phases are shown in Figure 1). However, relative to WT, the animals exhibited more robust circadian patterns of wake activity, as quantified by the amplitude of the 24-h periodicity of a Fast Fourier Transform (FFT) across hourly intervals of the recording (Mann-Whitney U = 6, n1, WT = 6, n2, Egr3−/− = 7, P = 0.035; WT median = 0.049, Egr3−/− median = 0.074). These initial data suggested that manipulations of Egr3, like those of DISC1, produce a dichotomy in sleep and circadian function: loss of Egr3 interferes with sleep duration and sleep homeostasis (i.e., the accumulation of sleep need; Grønli et al., 2016) but does not negatively influence circadian timekeeping.
Figure 1.
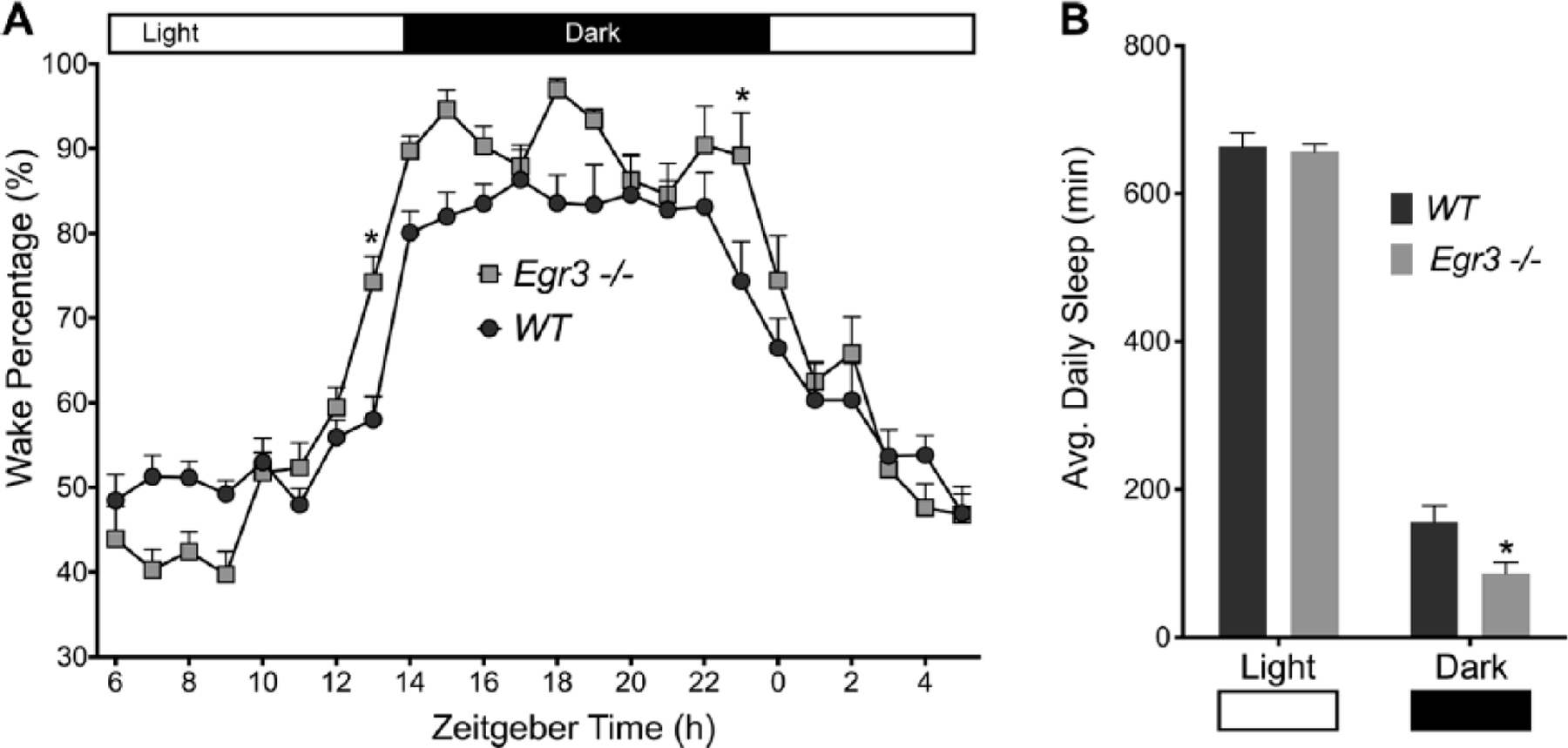
Sleep summary. (A) Wake percentage (%) was calculated across a 7-day period and then averaged to create a 24-h profile. Wake-Sleep behavior changed across the day in both WT and Egr3−/− mice, F(23, 253) = 62.34, P < 0.0001. However, Egr3−/− mice slept less than their WT littermates, F(1, 11) = 5.035, P = 0.0464. The negative effect of Egr3 deficiency on sleep varied within the 24-h cycle, F(23, 253) = 2.989, P < 0.0001; differences were most profound during L/D transitions at zeitgeber time 0 and 14 (Sidak’s multiple comparisons test, *P < 0.05). (B) Total sleep time (min) was lower in Egr3−/− mice v. WT controls when data were broken-down between the light and dark phases of the LD cycle (F(1, 11) = 5.035, P = 0.0464); though, as expected, both groups slept more during the light phase, F(1, 11) = 1162, P <0.0001. Data are presented as mean ± SEM.
To scrutinize this dichotomy further, we subjected a second, independent cohort of Egr3−/− and WT littermate mice to a 3-mo evaluation period. The min-by-min waking activity of the animals was analyzed under entrained conditions (14:10 LD cycle) for 3 weeks, followed by 6 weeks of evaluation under free-running conditions—3 weeks each in constant darkness (DD) then constant light (LL; 200 lux, cool white fluorescent light) separated by 14 days of reentrainment (outlined in the Supplementary Materials and Methods).
Circadian (~24-h) trends in wake behavior were once again evident in Egr3−/− and WT mice (summarized in Figures 2, 3, and 4, with representative actograms in Figure 2A and Figure 4A). All of these animals free ran under constant conditions and reentrained to the LD cycle after extended time in DD. However, quantitative genotypic differences did emerge in several circadian measures and in overall wake-related activity. Most of these differences were observed irrespective of lighting condition. During the LD cycle, Egr3−/− mice displayed significantly stronger circadian oscillations of wake behavior as measured by the amplitude of 24-h periodicities in the Lomb-Scargle periodogram (LSP) and FFT of the time series data, and cosinor analysis (mean ± SEM, circadian quotient [cosinor amplitude/mesor], Egr3−/−, 0.96 ± 0.04, WT, 0.78 ± 0.04; t(12) > 3.25, P < 0.007) (Figure 2, B-C). The fact that the circadian quotient was larger in Egr3−/− than WT mice suggests that the stronger rhythms produced by Egr3 loss were not (simply) a byproduct of general hyperactivity; this measure normalizes the cosinor amplitude as a function of the mesor, a rhythms-adjusted mean based on the distribution of wake percentage values.
Figure 2.
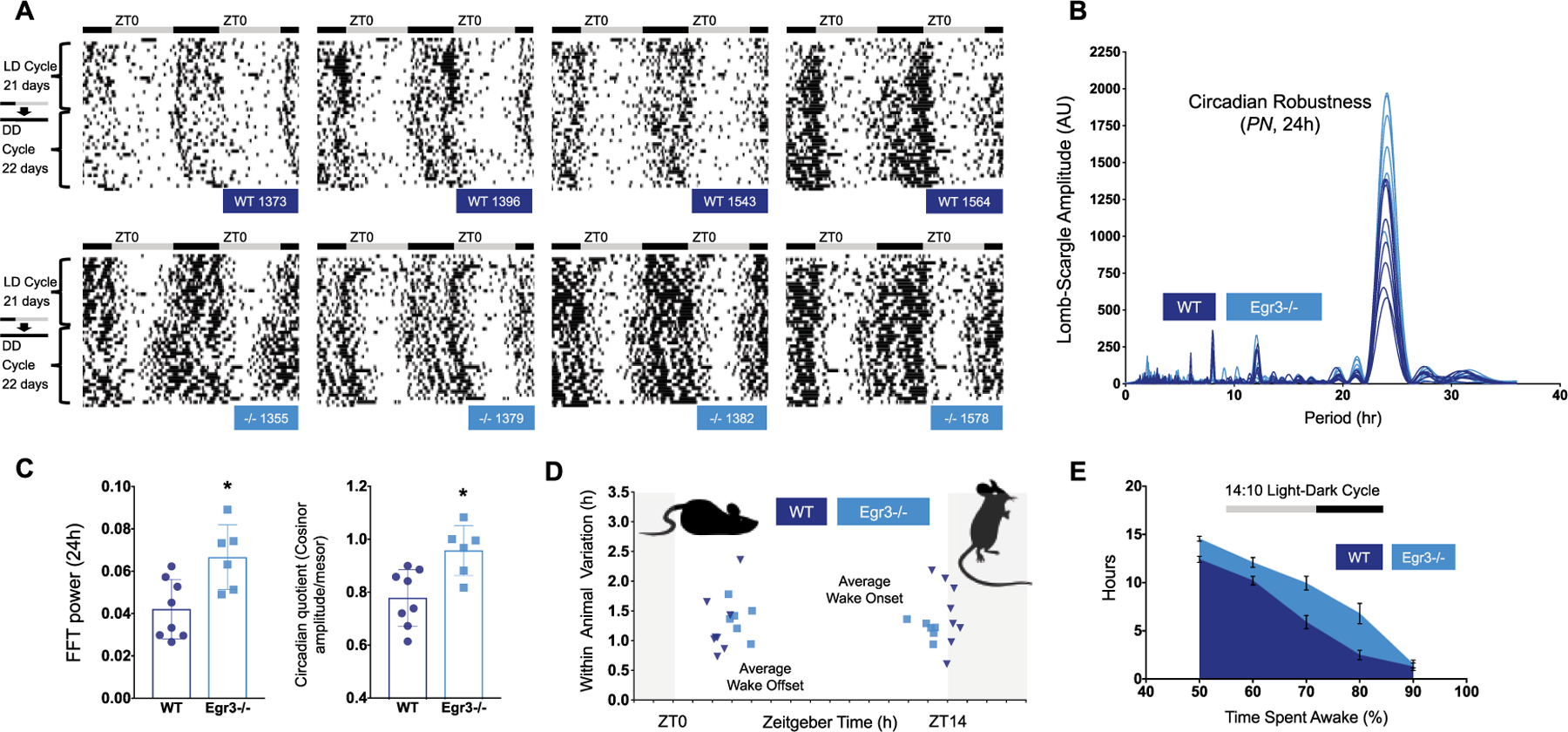
Summary of circadian function in LD. (A) Representative double-plotted actograms taken from WT (dark blue labels) and Egr3−/− littermates (cyan labels) housed in LD, then DD, over 6 weeks. Black bars indicate episodes where animals are scored >90% awake, smoothed over 25 min. For both sets of mice, sustained bouts of wake are consolidated to the dark phase of the LD cycle. In DD, this wake behavior freeruns. (B) The amplitudes of the major frequency components isolated by the Lomb-Scargle periodogram (LSP) are shown for each mouse under study in the second cohort. Spectral analysis indicates that 24-h patterns of wake behavior are more robust in Egr3−/− v. WT mice (mean ± SEM, LD-LSP24h, Egr3−/−, 1635 ± 147, WT, 1032 ± 116). (C) This disparity is noted in other assessments of circadian robustness as well, such as the amplitude of the FFT 24-h periodicity (FFT24h, Egr3−/−, 0.067 ± 0.006, WT, 0.042 ± 0.005) and the cosinor circadian quotient. All ts(12) > 3.1, ps < 0.009. (D) The rest period of Egr3−/− mice (cyan squares; 1 point = 1 animal) is truncated compared q=with WT littermates (dark blue triangles; 1 point = 1 animal) in a 14:10 LD cycle. Wake onsets are phase-advanced and offsets phase-delayed by 1 h (ts(12) > 3.20, ps < 0.008). (E) Total amount of time accrued (h) in which each minute had >50%, >60%, >70%, >80%, or >90% of its epochs defined as “wake” is plotted for each genotype (WT = dark blue; Egr3−/− = cyan). Egr3−/− mice log more waketime than WT animals in LD (repeated measures, 2-way ANOVA, main effect of genotype, F(1, 12) = 17.15, P < 0.002; main effect of wake category, F(4, 48) = 299.9, P < 0.0001).
Figure 3.
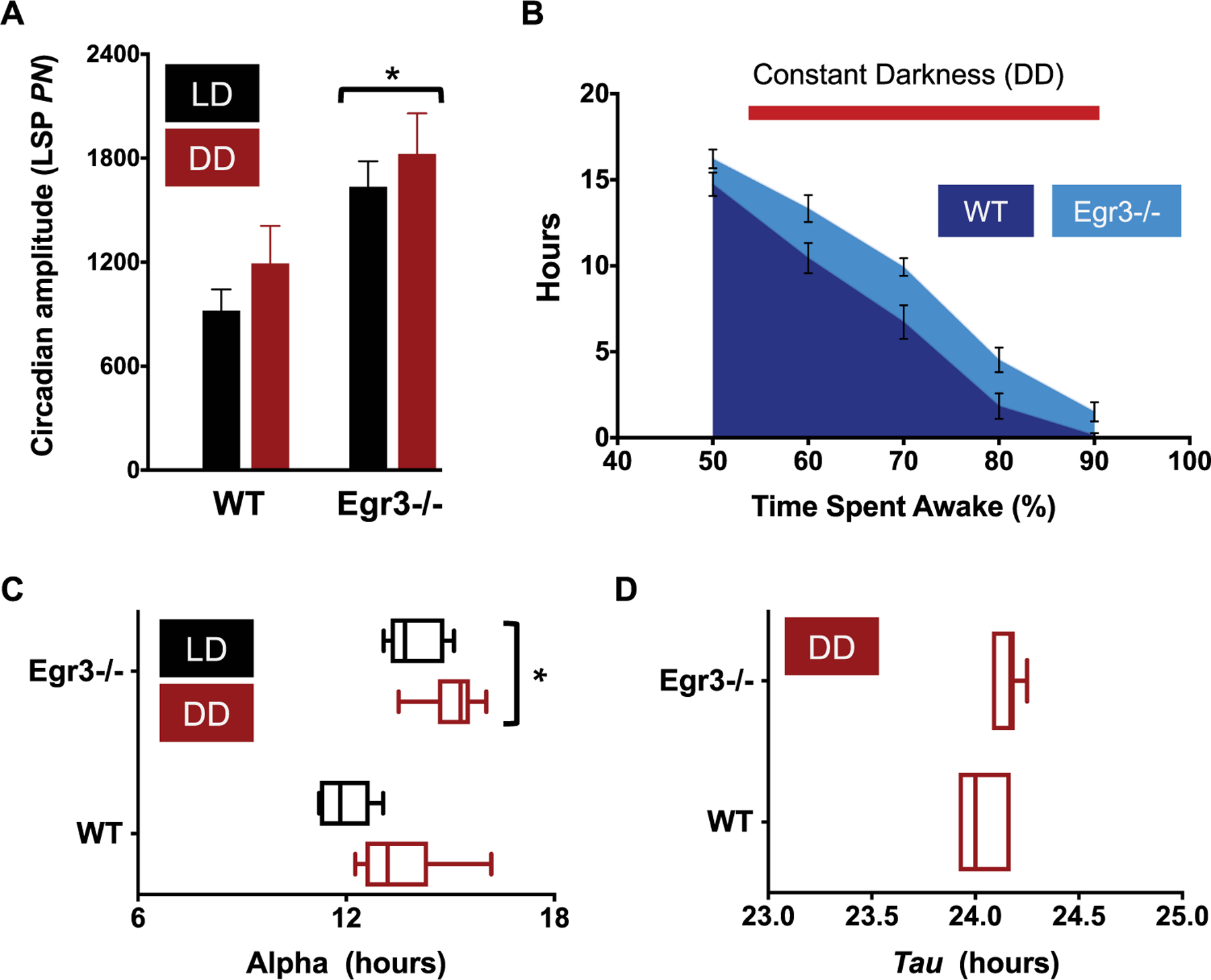
Summary of circadian function in DD. (A) Amplitude of the peak periodicity of the Lomb-Scargle periodogram (LSP; power normalized, PN, at tau) in LD (black bars) v. DD housing (red bars). Rhythms are more robust in Egr3−/− mice under both lighting regimens (repeated measures, 2-way ANOVA, main effect of genotype when comparing LD, DD, and LL lighting conditions, F(1, 10) = 8.722, P = 0.0145). (B) The total amount of time accrued, in hours, in which each minute had >50% to 90% of its epochs defined as “wake” is plotted for each genotype during their time in DD. Similar to their profile in LD, Egr3−/− mice log more high-percentage waketime than WT animals (main effect of genotype, F(1, 12) = 9.882, P = 0.0085; main effect of wake category, F(4, 48) = 234.5, P < 0.0001). (C) During DD, the active period is longer in Egr3−/− v. WT controls (Egr3−/−α DD, 15.1 ± 0.35 h; WTα DD, 13.5 ± 0.57 h; main effect of genotype when all lighting conditions are compared, F(1,10) = 40.49, P < 0.0001). (D) However, tau does not shorten in either group of mice.
Figure 4.
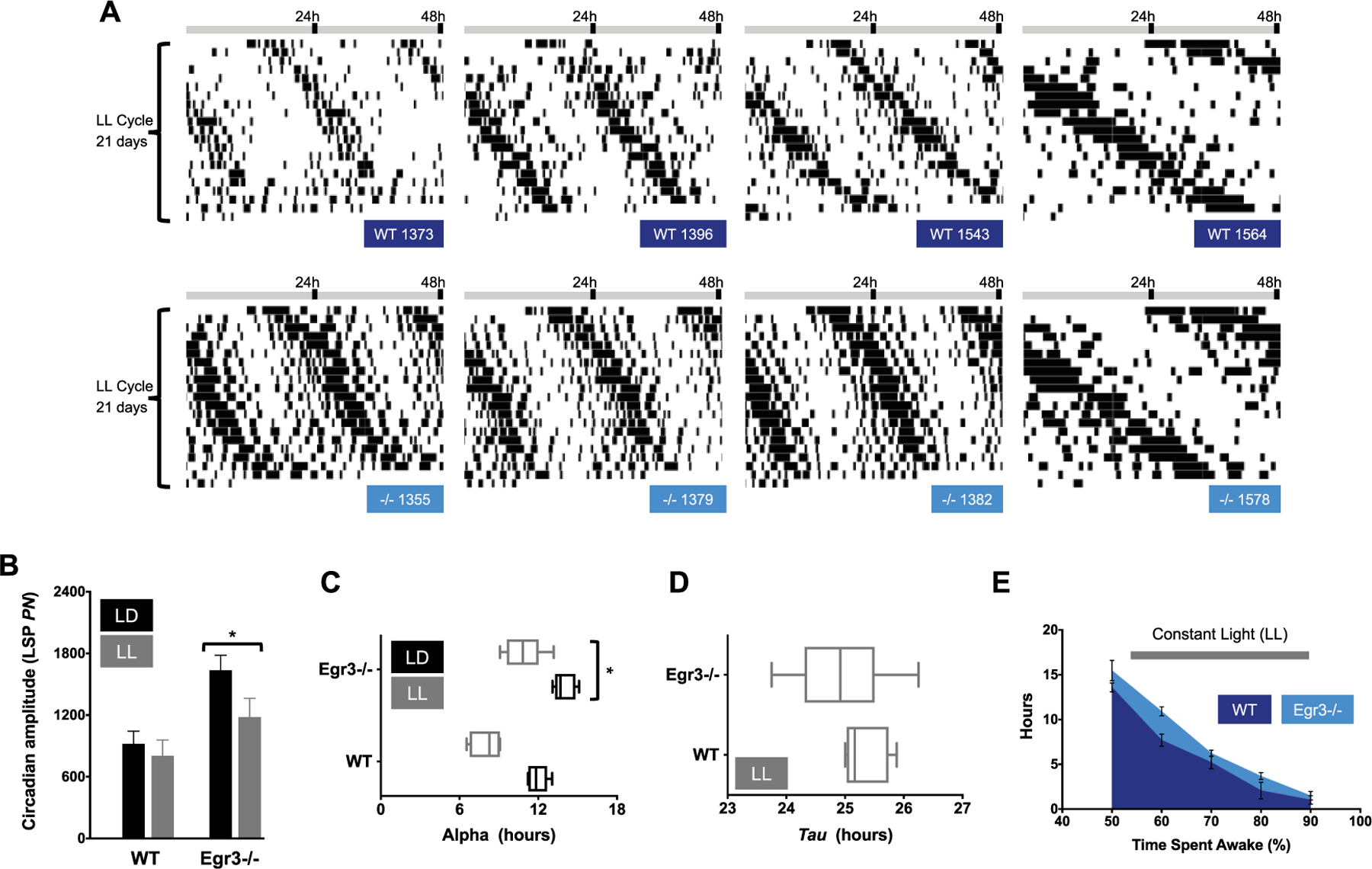
Summary of circadian function in LL. (A) Representative double-plotted actograms taken from WT (dark blue labels) and Egr3−/− littermates (cyan labels) housed in LL for 3 weeks. Wake behavior free runs under LL in mice of both genotypes. (B) Amplitude of the peak periodicity of the Lomb-Scargle periodogram (LSP; power normalized, PN, at tau) in LD (black bars) v. LL housing (gray bars). Rhythms are more robust in Egr3−/− mice under all light schedules tested in the current study (main effect of genotype, F(1, 10) = 8.722, P = 0.0145). Nevertheless, LL weakens rhythms without regard to Egr3 status (main effect of light schedule, F(2, 20) = 6.513, p = 0.0066; genotype × light schedule interaction, F(2, 20) = 0.757, P = 0.4820). (C) Length of the active period (alpha) is shortened in LL in mice of both genotypes, but more so in WT (main effect of lighting condition, F(2, 20) = 58.03, P < 0.0001; main effect of genotype, F(1, 10) = 40.49, P < 0.0001). (D) Tau slows with housing under LL in both Egr3−/− and WT mice. (E) The total amount of time accrued, in hours, in which each minute had >50% to 90% of its epochs defined as “wake” is plotted for each genotype. Even under LL, Egr3−/− mice log more high-percentage waketime than WT animals (repeated measures, 2-way ANOVA, main effect of genotype, F(1, 10) = 7.889, P = 0.0185; main effect of wake category, F(4, 40) = 175, P < 0.0001).
The amplitude and consistency of 24-h diurnal rhythms in humans can be quantified under entrained conditions in a subject’s home using a suite of non-parametric circadian rhythm analyses (NPCRA) developed by Van Someren (2011). NPCRA operationalizes amplitude and consistency with 3 measures: relative amplitude (RA), interdaily stability, and intradaily variability (Fernandez et al., 2017). The RA index represents the normalized difference in activity counts between the most active 10-h portion of the day (M10) and the least active 5-h portion (L5) (RA = (M10 – L5)/(M10 + L5)). Interdaily stability estimates how well an individual’s rest-activity pattern is replicated from one day to the next in a 24-h LD cycle, while intradaily variability estimates how well each day’s activity is consolidated (mathematical descriptions of these variables can be found in Van Someren et al., 1999).
Surveying these measures here, we found that Egr3−/− mice had higher RA indices than WT littermates, further suggesting that general hyperactivity does not factor into the enhanced circadian robustness of their wake behavior (Egr3−/−RA, 0.44 ± 0.01; WTRA, 0.35 ± 0.01; t(12) = 5.48, P < 0.0001). However, interdaily stability and intradaily variability were not significantly different, indicating that—despite this amplitude difference—mice of both genotypes were similarly able to consolidate waking activity to certain parts of the day and to maintain this organization across the entrainment period (interdaily stability, Egr3−/−, 0.32 ± 0.01, WT, 0.27 ± 0.02; intradaily variability, Egr3−/−, 1.30 ± 0.05, WT, 1.26 ± 0.08; ps > 0.05).
Consistent with the finding in the first cohort—that sleep differences between WT and Egr3−/− mice were most distinct at the borders of the LD transitions occurring just after ZT0 and right before ZT14 of the LD schedule (Figure 1)—phase markers associated with wake onset and offset in the second cohort were significantly shifted so as to truncate the rest period in Egr3−/− mice (Figure 2D). The average onset of wake behavior in mice lacking Egr3 was phase-advanced by 1 h (Egr3−/− onset, ZT 13.0 ± 0.2; WT onset, ZT 14.1 ± 0.2), while the offset was phase-delayed by about 1 h (Egr3−/− offset, ZT 3.4 ± 0.2; WT offset, ZT 2.4 ± 0.2; Figure 2D). Other phase markers such as L5onset, which estimates the start of the least active 5-h stretch of the day, or M10onset, which estimates the start of the most active 10-h stretch of the day, did not distinguish animals with or without Egr3 expression (data not shown). However, although the variability of L5onset was not significantly different, that of M10onset was significantly lower in Egr3−/− mice (F-test of equality of variances, L5onset Variance: F(5,7) = 5.05, P = 0.0933; M10onset Variance: F(5,7) = 10.85, P = 0.0184). In keeping with this expansion of the wake period or alpha (Egr3−/−α LD, 13.9 ± 0.33 h; WTα LD, 12.0 ± 0.32 h), Egr3−/− mice showed greater wake percentage time under entrained conditions as compared with WT littermates (Figure 2E).
The increases in circadian amplitude (LSP24h), alpha, and wake percentage time that were noted in Egr3−/− mice under an LD schedule were still evident when the animals were transitioned to DD (Figure 3A-C). The free-running rhythm (tau) of nocturnal animals, such as mice, often accelerates under DD. However, neither WT nor their Egr3−/− littermates conformed to this trend, each revealing a free-running rhythm of wake behavior around 24 h (WTτ DD, 24.03 ± 0.05 h; Egr3−/−τ DD, 24.15 ± 0.03 h; Figure 3D). The lack of tau response might not be too surprising given that the background strain of the WT and Egr3−/− mice, C57BL/6, exhibits a relatively long mean tauDD greater than most of the values observed from other inbred strains (Schwartz and Zimmerman, 1990). The tauDD values in the current study might also have been influenced by the entrainment history of the animals (i.e., LD 14:10 instead of 12:12). Previous entrainment to long photoperiods can cause “after-effects” in rodents once they are released into DD, including lengthening of the free-running period to ≥24 h (Pittendrigh and Daan, 1976).
Under LL, circadian rhythms of wake behavior were severely disrupted in 2 WT mice but in none of the mice lacking Egr3. In subsequent analyses, these WT arrhythmic animals were removed from consideration of LL’s effects on activity. Among the remaining mice, daily recurring intervals of high-percentage and low-percentage wake were still evident (Fig. 4A). Egr3−/− mice continued to show more robust circadian patterns of wake behavior relative to WT under LL conditions as they had in LD and DD (Fig. 4B), and—once again—were awake for longer stretches of time (Egr3−/−α LL, 10.9 ± 0.59 h; WTα LL, 8.0 ± 0.44 h; Fig. 4C). The free-running rhythm of nocturnal animals, such as mice, often slows down in LL. Both Egr3−/− and WT mice conformed to this trend (WTτ LL, 25.32 ± 0.15 h; Egr3−/−τ LL, 24.93 ± 0.34 h; Fig. 4D). As documented throughout the study, Egr3−/− mice had higher-percentage waketime in LL relative to WT controls (Fig. 4E).
Together, our results suggest that Egr3−/− mice retain control over the circadian timekeeping of sleep/wake but exhibit several phenotypes that distinguish them from WT animals, including increased circadian amplitude, expanded alpha, and more wake-related behavior. These phenotypes are equally visible under entrained and free-running conditions and are noted in auxiliary analyses of wake-sleep bout duration in Egr3−/− v. WT mice (see Suppl. Figure 1). Why global loss of Egr3 should produce this particular cluster of sleep and circadian changes is not immediately clear, though one possible mechanism by which Egr3 influences sleep homeostasis (and thereby wake-related behavior) might be through regulation of one of its target genes, the serotonin (5-HT) 2A receptor (Htr2a). Sleep deprivation significantly increases Htr2a mRNA expression in the mouse cerebral cortex in an Egr3-dependent fashion (Maple et al., 2015) and, in human brain, significantly increases in vivo 5-HT2A receptor binding (Elmenhorst et al., 2012). The Egr3–5HT2A receptor signaling pathway is thought to promote recovery sleep after sleep deprivation (Grønli et al., 2016; Popa et al., 2005; Terao et al., 2003), and mice lacking 5-HT2A receptors reveal similar 24-h patterns of heightened arousal as we find in Egr3−/− mice (Popa et al., 2005). The reasons for the impact of Egr3 on the circadian system are even less clear but might relate to the mobilization of its expression in central pacemaker neurons following light exposure (Porterfield and Mintz, 2009). Whether Egr3 contributes to light’s phase-shifting or masking effects remains an open question, though our data raise this possibility given the early waking behavior of Egr3−/− mice an hour before lights-off in the LD schedule (i.e., weaker negative masking; see Oliver et al., 2012, for another example of this phenotype in a different schizophrenia mouse model). Future experiments will be necessary to determine how a lack of Egr3 expression in different brain regions might selectively replicate (and extend) different aspects of the Egr3−/− sleep-circadian profile and to what extent this regulation can be traced to specific clock genes (Franken et al., 2007).
Overall, our findings in the Egr3−/− mouse model are compatible with previous work in DISC1 mice (Jaaro-Peled et al., 2016), as well as Grm2/3 double knockouts (Pritchett et al., 2015), and add to the bourgeoning schizophrenia SCRD literature where there is a growing appreciation that—depending on the circumstances—mouse models of schizophrenia can show worse, normal, or better consolidation of daily waking activity (Bhardwaj et al., 2015; Oliver et al., 2012; Pritchett et al., 2015). Our data, along with the results from mice misexpressing other schizophrenia candidate alleles, suggest that the biological mechanisms contributing to sleep disruption in people with schizophrenia do not necessarily overlap with those responsible for the breakdown of circadian entrainment and rhythmicity. There is the possibility that different optimal treatment strategies may be required to sufficiently address the two (Franken et al., 2007). As we learn more about sleep in neurological conditions, sleep metrics become both a diagnostic and treatment target in the pursuit of precision and personalized medicine. The commonplace use of actigraphy could help identify at-risk patients.
Supplementary Material
Acknowledgments
The current work was supported by: NIH NIMH R01 MH097803 Award (ALG); the American Sleep Medicine Foundation, Focused Project Award (RKR); Science Foundation Arizona, Bisgrove Scholar Award (AMM, RKR, FF); the UA BIO5 Institute (FF); and the Arizona Department of Health Services (FF). The authors would like to thank Jordan L. Harrison for his assistance with the sleep apparatus.
Footnotes
Conflict of Interest Statement
The author(s) have no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.[AQ: please confirm the conflict of interest statement]
Note
Supplementary material is available on the journal’s website at https://journals.sagepub.com/doi/suppl/10.1177/0748730418803802.
References
- Arnedo J, Svrakic DM, Del Val C, Romero-Zaliz R, Hernández-Cuervo H, Molecular Genetics of Schizophrenia Consortium, Fanous AH, Pato MT, Pato CN, de Erausquin GA, et al. (2015) Uncovering the hidden risk architecture of the schizophrenias: confirmation in three independent genome-wide association studies. Am J Psychiatry 172:139–153. [DOI] [PubMed] [Google Scholar]
- Bhardwaj SK, Stojkovic K, Kiessling S, Srivastava LK, and Cermakian N (2015) Constant light uncovers behavioral effects of a mutation in the schizophrenia risk gene Dtnbp1 in mice. Behav Brain Res 284:58–68. [DOI] [PubMed] [Google Scholar]
- Bromundt V, Koster M, Georgiev-Kill A, Opwis K, Wirz-Justice A, Stoppe G, and Cajochen C (2011) Sleep-wake cycles and cognitive functioning in schizophrenia. Br J Psychiatry 198:269–276. [DOI] [PubMed] [Google Scholar]
- Brugger SP, and Howes OD (2017) Heterogeneity and homogeneity of regional brain structure in schizophrenia: A meta-analysis. JAMA Psychiatry 74(11):1104–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan V (2017) Schizophrenia and psychosis. Child Adolesc Psychiatr Clin N Am 26:341–366. [DOI] [PubMed] [Google Scholar]
- Chouinard S, Poulin J, Stip E, and Godbout R (2004) Sleep in untreated patients with schizophrenia: a meta-analysis. Schizophr Bull 30:957–967. [DOI] [PubMed] [Google Scholar]
- Cohrs S (2008) Sleep disturbances in patients with schizophrenia: impact and effect of antipsychotics. CNS Drugs 22:939–962. [DOI] [PubMed] [Google Scholar]
- Donohue KD, Medonza DC, Crane ER, and O’Hara BF (2008) Assessment of a non-invasive high-throughput classifier for behaviours associated with sleep and wake in mice. Biomed Eng Online 7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziema H, Oatis B, Butcher GQ, Yates R, Hoyt KR, and Obrietan K (2003) The ERK/MAP kinase pathway couples light to immediate-early gene expression in the suprachiasmatic nucleus. Eur J Neurosci 17:1617–1627. [DOI] [PubMed] [Google Scholar]
- Eaton WW, Chen CY, and Bromet EJ (2011) Epidemiology of schizophrenia. In Textbook of Psychiatric Epidemiology, 3rd ed, Tsuang MT, Tohen M, Jones PB, eds, pp 263–287. [Google Scholar]
- Elmenhorst D, Kroll T, Matusch A, and Bauer A (2012) Sleep deprivation increases cerebral serotonin 2A receptor binding in humans. Sleep, 35:1615–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez F, Nyhuis CC, Anand P, Demara BI, Ruby NF, Spanò G, Clark C, and Edgin JO (2017). Young children with Down syndrome show normal development of circadian rhythms, but poor sleep efficiency: a cross-sectional study across the first 60 months of life. Sleep Med 33:134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken P, Thomason R, Heller HC, and O’Hara BF (2007) A non-circadian role for clock-genes in sleep homeostasis: a strain comparison. BMC Neurosci 8:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman D, Sheaves B, Goodwin GM, Yu LM, Nickless A, Harrison PJ, Emsley R, Luik AI, Foster RG, Wadekar V, et al. (2017) The effects of improving sleep on mental health (OASIS): a randomised controlled trial with mediation analysis. Lancet Psychiatry 4:749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallitano-Mendel A, Izumi Y, Tokuda K, Zorumski CF, Howell MP, Muglia LJ, Wozniak DF, and Milbrandt J (2007) The immediate early gene early growth response gene 3 mediates adaptation to stress and novelty. Neuroscience 148:633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallitano-Mendel A, Wozniak DF, Pehek EA, and Milbrandt J (2008) Mice lacking the immediate early gene Egr3 respond to the anti-aggressive effects of clozapine yet are relatively resistant to its sedating effects. Neuropsychopharmacology 33:1266–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grønli J, Clegern WC, Schmidt MA, Nemri RS, Rempe MJ, Gallitano AL, and Wisor JP (2016) Sleep homeostatic and waking behavioral phenotypes in Egr3-deficient mice associated with serotonin receptor 5-HT2 deficits. Sleep 39:2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstetter JR, Lysaker PH, and Mayeda AR (2005) Quality of sleep in patients with schizophrenia is associated with quality of life and coping. BMC Psychiatry 5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huentelman MJ, Muppana L, Corneveaux JJ, Dinu V, Pruzin JJ, Reiman R, Borish CN, De Both M, Ahmed A, Todorov A, et al. (2015) Association of SNPs in EGR3 and ARC with schizophrenia supports a biological pathway for schizophrenia risk. PLoS One 10:e0135076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaaro-Peled H, Altimus C, LeGates T, Cash-Padgett T, Zoubovsky S, Hikida T, Ishizuka K, Hattar S, Mongrain V, and Sawa A. (2016) Abnormal wake/sleep pattern in a novel gain-of-function model of DISC1. Neuroscience Res 112:63–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Song JY, Joo EJ, Lee KY, Ahn YM, and Kim YS (2010) EGR3 as a potential susceptibility gene for schizophrenia in Korea. Am J Med Genet B Neuropsychiatr Genet 153B:1355–1360. [DOI] [PubMed] [Google Scholar]
- Mang GM, Nicod J, Emmenegger Y, Donohue KD, O’Hara BF, and Franken P (2014) Evaluation of a piezoelectric system as an alternative to electroencephalogram/electromyogram recordings in mouse sleep studies. Sleep 37:1383–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maple AM, Zhao X, Elizalde DI, McBride AK, and Gallitano AL (2015) Htr2a expression responds rapidly to environmental stimuli in an Egr3-dependent manner. ACS Chem Neurosci 6:1137–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marballi KK, and Gallitano AL (2018) Immediate early genes anchor a biological pathway of proteins required for memory formation, long-term depression and risk for schizophrenia. Front Behav Neurosci 12:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mexal S, Frank M, Berger R, Adams CE, Ross RG, Freedman R, and Leonard S (2005) Differential modulation of gene expression in the NMDA postsynaptic density of schizophrenic and control smokers. Brain Res Mol Brain Res 139:317–332. [DOI] [PubMed] [Google Scholar]
- Monti JM, BaHammam AS, Pandi-Perumal SR, Bromundt V, Spence DW, Cardinali DP, and Brown GM (2013) Sleep and circadian rhythm dysregulation in schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry 43:209–216. [DOI] [PubMed] [Google Scholar]
- Monti JM, and Monti D (2004) Sleep in schizophrenia patients and the effects of antipsychotic drugs. Sleep Med Rev 4:263–276. [DOI] [PubMed] [Google Scholar]
- Morris ME, Viswanathan N, Kuhlman S, Davis FC, and Weitz CJ (1998) A screen for genes induced in the suprachiasmatic nucleus by light. Science 279:1544–1547. [DOI] [PubMed] [Google Scholar]
- Myers E, Startup H, and Freeman D (2011) Cognitive behavioural treatment of insomnia in individuals with persistent persecutory delusions: a pilot trial. J Behav Ther Exp Psychiatry 42:330–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan KJ, Tourtellotte WG, Millbrandt J, and Baraban JM (1999) The EGR family of transcription-regulatory factors: progress at the interface of molecular and systems neuroscience. Trends Neurosci 22:167–173. [DOI] [PubMed] [Google Scholar]
- Oliver PL, and Davies KE (2009) Interaction between environmental and genetic factors modulates schizophrenic endophenotypes in the Snap-25 mouse mutant blind-drunk. Hum Mol Genet 18:4576–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver PL, Sobczyk MV, Maywood ES, Edwards B, Lee S, Livieratos A, Oster H, Butler R, Godinho SI, Wulff K, et al. (2012) Disrupted circadian rhythms in a mouse model of schizophrenia. Curr Biol 22:314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh CS, and Daan S (1976) A functional analysis of circadian pacemakers in nocturnal rodents. J Comp Physiol 106:223–252. [Google Scholar]
- Popa D, Léna C, Fabre V, Prenat C, Gingrich J, Escourrou P, Hamon M, and Adrien J (2005) Contribution of 5-HT2 receptor subtypes to sleep-wakefulness and respiratory control, and functional adaptations in knock-out mice lacking 5-HT2A receptors. J Neurosci 25:11231–11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porterfield VM, and Mintz EM (2009) Temporal patterns of light-induced immediate-early gene expression in the suprachiasmatic nucleus. Neurosci Lett 463:70–73. [DOI] [PubMed] [Google Scholar]
- Pritchett D, Jagannath A, Brown LA, Tam SK, Hasan S, Gatti S, Harrison PJ, Bannerman DM, Foster RG, and Peirson SN (2015) Deletion of metabotropic glutamate receptors 2 and 3 (mGlu2 & mGlu3) in mice disrupts sleep and wheel-running activity and increases the sensitivity of the circadian system to light. PloS One 10:e0125523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao ML, Gross G, Strebel B, Halaris A, Huber G, Bräunig P, and Marler M (1994) Circadian rhythm of tryptophan, serotonin, melatonin, and pituitary hormones in schizophrenia. Biol Psychiatry 35:151–163. [DOI] [PubMed] [Google Scholar]
- Richard Y, Swann AC, and Burt DB (1996) Stability of diagnosis in schizophrenia. Am J Psychiatry 153:682–686. [DOI] [PubMed] [Google Scholar]
- Sartorius N, Jablensky A, Korten A, Ernberg G, Anker M, Cooper JE, and Day R (1986) Early manifestations and first-contact incidence of schizophrenia in different cultures: A preliminary report on the initial evaluation phase of the WHO Collaborative Study on Determinants of Outcome of Severe Mental Disorders. Psychol Med 16:909–928. [DOI] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014) Biological insights from 108 schizophrenia-associated genetic loci. Nature 511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz WJ, and Zimmerman P (1990) Circadian timekeeping in BALB/c and C57BL/6 inbred mouse strains. J Neurosci 10:3685–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terao A, Greco MA, Davis RW, Heller HC, and Kilduff TS (2003) Region-specific changes in immediate early gene expression in response to sleep deprivation and recovery sleep in the mouse brain. Neuroscience 20:1115–1124. [DOI] [PubMed] [Google Scholar]
- Van Someren EJ (2011) Actigraphic monitoring of sleep and circadian rhythms. Handb Clin Neurol 98:55–63. [DOI] [PubMed] [Google Scholar]
- Van Someren EJ, Swaab DF, Colenda CC, Cohen W, McCall WV, and Rosenquist PB (1999) Bright light therapy: improved sensitivity to its effects on rest-activity rhythms in Alzheimer patients by application of nonparametric methods. Chronobiol Int 16:505–518. [DOI] [PubMed] [Google Scholar]
- Williams AA, Ingram WM, Levine S, Resnik J, Kamel CM, Lish JR, Elizalde DI, Janowski SA, Shoker J, Kozlenkov A, et al. (2012) Reduced levels of serotonin 2A receptors underlie resistance of Egr3-deficient mice to locomotor suppression by clozapine. Neuropsychopharmacology 37:2285–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirz-Justice A, Cajochen C, and Nussbaum P (1997) A schizophrenic patient with an arrhythmic circadian rest–activity cycle. Psychiatry Res 73:83–90. [DOI] [PubMed] [Google Scholar]
- Wulff K, Dijk DJ, Middleton B, Foster RG, and Joyce EM (2012) Sleep and circadian rhythm disruption in schizophrenia. Br J Psychiatry 200:308–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff K, Gatti S, Wettstein JG, Foster RG (2010) Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease. Nature Rev Neurosci 11:589–599. [DOI] [PubMed] [Google Scholar]
- Yamada K, Gerber DJ, Iwayama Y, Ohnishi T, Ohba H, Toyota T, Aruga J, Minabe Y, Tonegawa S, and Yoshikawa T (2007) Genetic analysis of the calcineurin pathway identifies members of the EGR gene family, specifically EGR3, as potential susceptibility candidates in schizophrenia. Proc Natl Acad Sci U S A 104:2815–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, and Winkelman JW (2006) Clinical significance of sleep EEG abnormalities in chronic schizophrenia. Schizophrenia Res 82:251–260. [DOI] [PubMed] [Google Scholar]
- Zhang R, Lu S, Meng L, Min Z, Tian J, Valenzuela RK, Guo T, Tian L, Zhao W, and Ma J (2012) Genetic evidence for the association between the early growth response 3 (EGR3) gene and schizophrenia. PLoS One 7:e30237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


