Abstract
Severe early-onset obesity is mainly attributed to single gene variations of the hypothalamic leptin-melanocortin system, which is critical for controlling the balance between appetite and energy expenditure. Adenylate cyclase 3 (ADCY3), a transmembrane enzyme localized in primary neuronal cilia, is a key genetic candidate, which appears to have an essential role in regulating body weight. The present study aimed to identify ADCY3 genetic variants in severely obese young patients of Greek-Cypriot origin by genomic sequencing. Apart from previously reported variants, the novel and probably pathogenic variant c.349T>A, causing a p.Leu117Met substitution within one of the two pseudo-symmetric halves of the transmembrane part of the protein, was reported. Molecular modelling analysis used to delineate bonding interactions within the mutated protein structure strongly suggested a change in interactive forces and energy levels affecting the pseudo-twofold symmetry of the transmembrane domain of the protein and probably its catalytic function. These results support the involvement of ADCY3 in the pathology of the disease and point towards the requirement of defining protein function and evaluating the clinical significance of the detected variants.
Keywords: ADCY3, variants, obesity, body mass index, molecular modelling
Introduction
Obesity is a global pandemic demonstrating increasing international dimensions (1,2). According to a recent pooled analysis of 2,416 population-based measurement studies involving 128.9 million subjects, the worldwide prevalence of overweight adults increased from 21.5 to 38.9% over the last 40 years (3-5). This complex disease frequently starts early in childhood and continues through adulthood. It appears that obesity in both children and adolescents has exhibited marked increases through the years, with 38 million patients under the age of five years diagnosed as overweight or obese in 2019 (6-8).
Obesity is unquestionably a disease multifactorial in origin. Its phenotypic manifestations and metabolic complications are ascribed to various genetic, environmental and epigenetic factors (9). Early-onset severe childhood obesity, a rare disorder caused predominantly by a variation of a single gene, is described as monogenic obesity. Single gene variations in the hypothalamic leptin-melanocortin pathway have been reported to affect the balance between appetite and energy expenditure from a young age. Such genes are leptin, leptin receptor, melanocortin-4 receptor (MC4R), proopiomelanocortin and prohormone convertase 1/3 genes (10-12). Of course, there are other monogenic syndromic forms of childhood obesity, which are considered separate entities studied by clinical geneticists. On the contrary, common obesity is thought to be affected by numerous genes (polygenic) triggered by behavioral and environmental factors. Genome-wide association studies (GWAS) have reported variations in the four above-mentioned major genes of the hypothalamic leptin-melanocortic pathway or other loci implicated either in common or monogenic obesity (11,12). In addition, obesity susceptibility loci associated with alterations in body mass index (BMI) in both children and adults have been also identified by GWAS in targeted or population studies (10-12).
Adenylate cyclase 3 (ADCY3) is a candidate gene gaining increasing attention. ADCY3 loss-of-function mutations have been associated with early-onset childhood obesity (13). ADCY3 is located on chromosome 2p23.3, consisting of 21 exons covering the positions 24,819,169-24,920,237 bp (GRCh38.p13) (14). Its product, an enzyme characterized by a pseudo-symmetric structure of two transmembrane and two cytoplasmic domains, is a member of the adenylyl cyclase family and catalyses the synthesis of 3′,5′-cyclic adenosine monophosphate (cAMP) from adenosine triphosphate upon G-protein signaling (14). Research has been performed in an effort to elucidate its role in metabolic processes, including energy metabolism, adipogenesis and changes in BMI (15-18). Its genetic variability has also been studied in numerous populations of various geographical areas (19). The present study aimed to determine the occurrence of variations in the ADCY3 gene in 33 Greek-Cypriot patients diagnosed with early-onset severe obesity (as from the age of 3 years).
Materials and methods
Patient inclusion criteria
A total of 33 unrelated patients were included in the present study, comprising 18 females and 15 males aged 15-20 years. All patients were recruited from the Archbishop Makarios III Hospital (Nicosia, Cyprus), the Paedi Center for Specialized Paediatrics (Nicosia, Cyprus) and the Aretaeio Hospital (Nicosia, Cyprus). All patients were severely obese with BMI > +2.5 standard deviation score (SDS) at the time of genetic testing and were diagnosed with early-onset obesity defined as BMI > +2 SDS from the age of 3 years onwards. All patients had a 6-12 month follow-up in a pediatric endocrinology clinic. According to their medical records, patients were not diagnosed with any other underlying medical conditions or complex syndromes. A total of 51 age-matched non-obese individuals (40 females and 11 males) of Greek-Cypriot origin were included as a control group in the present study. The project was approved by the Cyprus National Ethics Committee (Nicosia, Cyprus) and informed consent was given by patients/parents prior to any genetic testing.
Genomic sequencing and in silico analysis
Total genomic DNA samples were isolated from peripheral whole blood using the Gentra Puregene Blood kit (Qiagen GmbH). DNA sequencing was performed with 100 ng genomic DNA, which was amplified using primers designed by Primer3 software ver. 0.4.0 (http://frodo.wi.mit.edu/). PCR mixtures were prepared using the Taq DNA Polymerase kit (Qiagen GmbH); they had a final volume of 20 µl and contained 2 µl PCR buffer (10X), 2 µl Q Solution (5X), 2 µl dNTPs (2 mM), 0.3 µl of each primer (10 µM), 0.2 µl Taq polymerase (5 U/µl) and 100 ng genomic DNA. Amplification was performed with an initial denaturing temperature at 95°C for 5 min, followed by 30 cycles of denaturation (95°C, 45 sec), annealing (57°C, 60 sec) and extension (72°C, 45 sec), with a final extension at 72°C for 5 min. The ADCY3 gene primers covered all 21 exons (listed in Table SI). The PCR products were analysed on an Applied Biosystems 3130xl Genetic Analyzer and the results were analysed using Sequencing Analysis R 5.3 software (Applied Biosystems; Thermo Fisher Scientific, Inc.). The χ2 test of independence and Fisher's exact test (for low counts) were used to determine any association between the presence of variants and sex or BMI SDS. For the association between the presence of variants and BMI SDS, patients were separated according to their BMI SDS into three groups: +2.00-+2.99 SDS, +3.00-+3.99 SDS and ≥ +4.00 SDS. P≤0.05 was considered to indicate statistical significance. In silico prediction on protein function of the pathogenicity effect of the novel amino acid substitution was performed using three different protein functionality prediction tools: PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), SIFT (https://sift.bii.a-star.edu.sg/www/SIFT_seq_submit2.html) and Mutation Taster (http://www.mutationtaster.org/). The PolyPhen-2 method was based on sequence homology (values >0.908 were considered probably damaging, values >0.446 and ≤0.908 were considered possibly damaging). The SIFT method was based on sequence homology and the physico-chemical properties of the amino acids (values <0.05 were considered deleterious whereas values ≥0.05 were considered tolerated). Mutation Taster was used to determine the position of a splice site change relative to intron/exon borders. A confidence score >0.3 was indicative of a gain of a completely new splice site. Linkage disequilibrium analysis for the variants was performed in all available populations using the Ensemble Linkage Disequilibrium Calculator (https://grch37.ensembl.org/Homo_sapiens/Tools/LD?db=core).
Molecular modeling
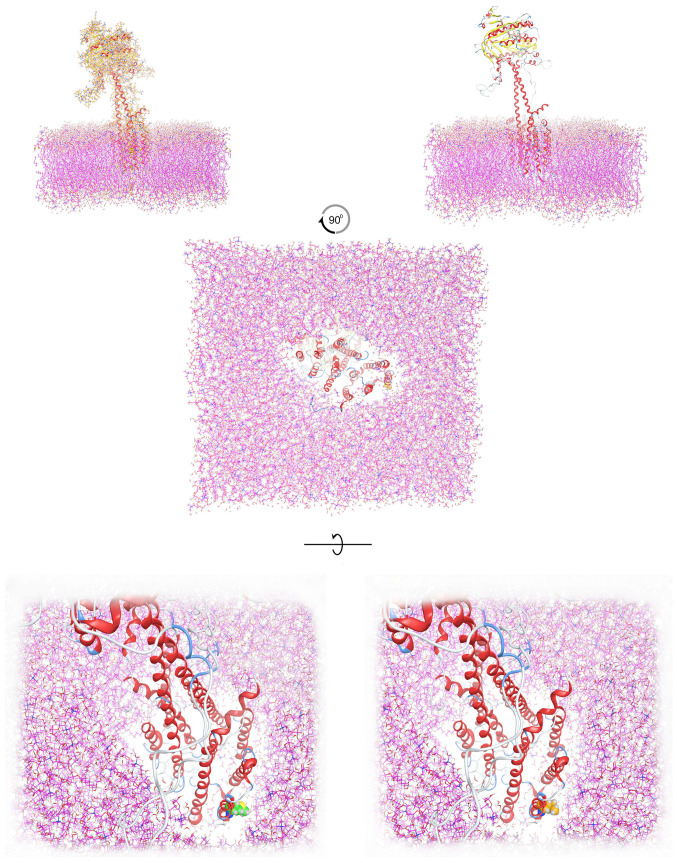
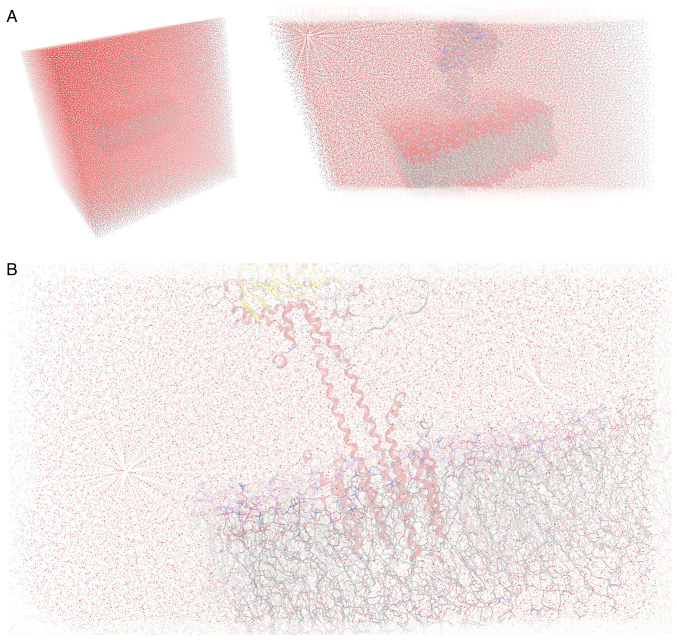
The homology modelling of ADCY3 was performed using Molecular Operating Environment (MOE; https://www.chemcomp.com/). The sequence of the human ADCY3 was used (accession no. 060266 in UniProtKB; https://www.uniprot.org/). The selection of template crystal structures for homology modelling was based on the primary sequence identity and the crystal resolution. Therefore, the selected template for the homology modelling was the structure of a membrane adenylyl cyclase bound to an activated stimulatory G protein (protein databank ID, 6R3Q). The primary alignment revealed a 74% coverage and a marginal 30% sequence identity, which however allowed for a conventional homology modelling experiment to be performed. The initial energy minimization for the ADCY3 model was performed in MOE using the CHARMM27 force-field up to a root mean square deviation gradient of 0.0001, in an effort to remove the geometrical strain. The ADCY3 model was subsequently solvated with simple point charge (SPC) water using the truncated octahedron box extending to 7Å from the model. Molecular dynamics simulation was performed at 300 K and 1 atm with a 2 fsecond step size for a total of 10 nanoseconds, using the NVT ensemble in a canonical environment (NVT stands for Number of atoms, Volume and Temperature that remain constant throughout the calculation). The ADCY3 model was also modelled in a full explicit phospholipid bilayer membrane protected by a layer of explicit SPC water molecules in a periodic system (Fig. 1). The results of the molecular dynamics simulation were collected into a database by MOE for further analysis (Fig. 2).
Figure 1.
Molecular modelling of ADCY3 in an explicit lipid bilayer, in an effort to depict the structural arrangement of ADCY3 in the membrane. The transmembrane and the outer membrane parts of ADCY3 are presented alongside the position of the amino acid position 117. Upper panel: Left, the wild-type ADCY3 model positioned in the lipid bilayer with all-atom representation; right, with ribbon representation. Middle panel: Bottom view of the positioned model of ADCY3 in the lipid bilayer. Lower panel: Left, bottom view of the wild-type ADCY3 model in the lipid bilayer (Leu117 shown in green space fill representation); right, bottom view of the mutant ADCY3 model (p.Leu117Met) in the lipid bilayer (Met117 displayed in orange space fill representation). ADCY3, adenylate cyclase 3.
Figure 2.
Modelling of ADCY3 in the explicit lipid bilayer solvated in a water periodic box for the molecular dynamics simulations. (A) A cube-periodic box solvation system for the full molecular system (left) and the ADCY3 model positioned in the lipid bilayer (right). (B) Close-up view of the ADCY3 model and lipid bilayer solvated in the water periodic system during the molecular dynamics simulations. ADCY3, adenylate cyclase 3.
Results
Genetic findings and prediction analysis of the novel variation
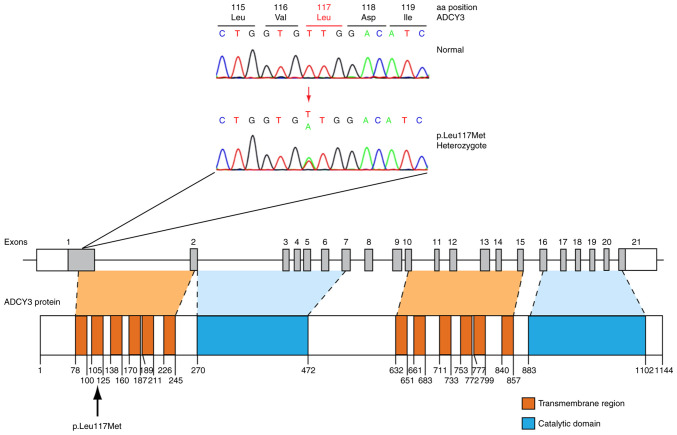
Sanger sequencing of ADCY3 revealed a total of five variants in patients (Table I), four of which were previously reported. A novel variant was identified in two patients (6%). Clinical and biochemical parameters for these patients are presented in Table SII. The novel variant involves a heterozygous c.349T>A change in exon 1 of the gene locus, leading to a missense p.Leu117Met substitution, as demonstrated in the sequence electropherogram presented in Fig. 3. Exon 1 codes for an α-helical transmembrane region of the protein (Fig. 3). A search in the widely used single nucleotide polymorphism (SNP) repositories such as the gnomAD V2.1.1 (accessed July 12, 2021; https://gnomad.broadinstitute.org/), 1000Genomes (accessed July 12, 2021; https://www.internationalgenome.org/) and TOPMED (accessed July 12, 2021; https://topmed.nhlbi.nih.gov/) verified that the novel p.Leu117Met variant was not previously reported. In silico prediction of the pathogenicity effect of the p.Leu117Met substitution was made using three different protein functionality prediction tools: PolyPhen-2, SIFT and Mutation Taster. PolyPhen-2 predicted p.Leu117Met to be 'possibly damaging' with a score of 0.646 (sensitivity: 0.80; specificity: 0.84). SIFT predicted substitution at position 117 from Leu to Met to 'affect protein function' with a score of 0.03. Finally, Mutation Taster predicted substitution at position 117 from Leu to Met as a 'polymorphism' resulting in splice changes with a confidence score of 0.91 for the predicted splice site. A gain of a completely new splice site was displayed if the confidence score of the newly created splice site was >0.3.
Table I.
Identification of previously reported variants in association to obesity and the novel adenylate cyclase 3 variant p.Leu117Met.
| SNP ID no. | Description nucleotide/amino acid | No. of homozygotes in patients (sex) | No. of heterozygotes in patients (sex) | MAF of patients (n=33) | MAF of controls (n=51) |
|---|---|---|---|---|---|
| Novel | c.349T>A/p.Leu117Met | 0 | 2 (M:2; F:0) | 0.03 | - |
| rs11676272 | c.319T>C/p.Ser107Pro | 10 (M:5; F:5) | 13 (M:5; F:8) | 0.50 | - |
| rs2241758 | c.1167C>G/p.Leu389= | 0 | 3 (M:1; F:2) | 0.05 | 0.06 |
| rs7604576 | c.2578-3T>C/NA | 9 (M:5; F:4) | 14 (M:6; F:8) | 0.48 | - |
| rs1127568 | c.2874A>G/p.Ser958= | 21 (M:12; F:9) | 9 (M:2; F:7) | 0.77 | 0.62 |
MAF, minor allele frequency; NA, not available; M, male; F, female.
Figure 3.
Identification of a probably pathogenic novel p.Leu117Met ADCY3 variant. The sequence electropherogram of the novel ADCY3 p.Leu117Met variant and the schematic representation of the ADCY3 gene and protein with the novel p.Leu117Met variant are displayed. ADCY3, adenylate cyclase 3.
The four previously reported variants include two synonymous (rs1127568 and rs2241758), one missense (rs11676272) and one intronic variant (rs7604576). The control group only revealed the two synonymous variations rs1127568 and rs2241758, located in exon 17 and exon 5, respectively. These SNPs have been previously identified in other populations as well. More specifically, rs1127568, which has a 0.8493 allele frequency based on gnomAD v2.1.1, was identified in a Chinese Han population sample in association to obesity (15), whereas rs2241758 (0.1068 allele frequency, based on gnomAD v2.1.1) was reported as a polymorphism in a Swedish population by Nordman et al (16). Both of the above-mentioned SNPs are synonymous, thus not causing any change of the amino acids in the protein structure. The frequency of these polymorphisms detected in the study population (Table I) was comparable to the results reported in gnomAD v2.1.1. Intronic variant rs7604576 and the missense variant rs11676272 were also among the SNPs reported in the Chinese Han population (15). It should be noted that both of these SNPs (rs7604576 and rs11676272) did not co-occur with the novel p.Leu117Met variant in any of the patients.
Linkage disequilibrium analysis for the variants was performed in all available populations. As expected, only neighboring variants rs1127568 and rs7604576 demonstrated a significant association (D=1.0, r2≥0.8).
The χ2 test of independence and Fisher's exact test were used to determine an association between variants and sex; however, no association was detected (P>0.05). A similar analysis revealed no association between the presence of variants and severity of obesity (measured as BMI SDS; P>0.05). However, it should be noted that the small sample size in the present study may have affected these results (Table SIII).
In light of recently reported findings suggesting the involvement of MC4R and ADCY3 in a common pathway towards obesity (20), the possible co-occurrence of MC4R variations in the patients was also investigated. However, Sanger sequencing of the MC4R locus in patients and controls revealed no variations within the MC4R locus (data not shown).
Molecular modeling
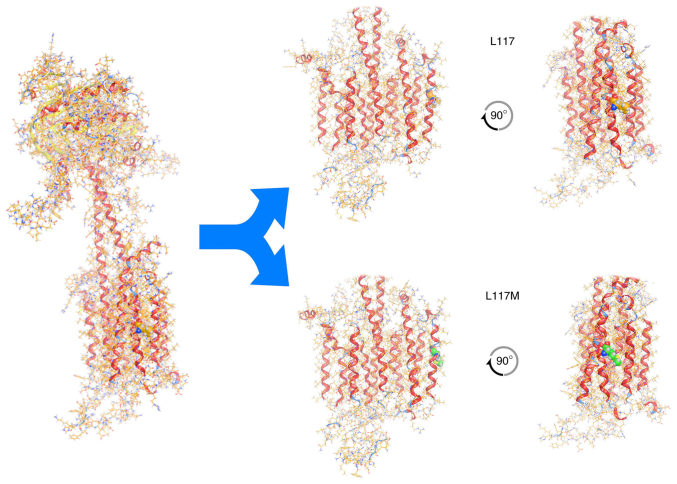
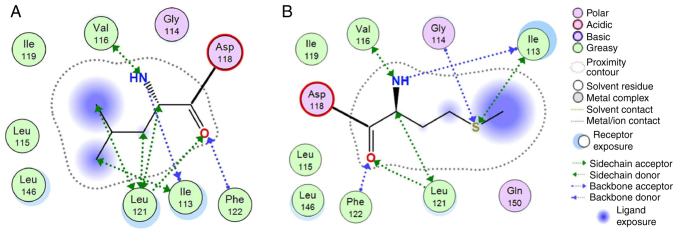
To investigate the possible effects of the novel p.Leu117Met variation on protein structure and function, the homology modelling of ADCY3 was performed using MOE. ADCY3 was modelled initially with the wild-type sequence and a Leu residue at position 117. The p.Leu117Met mutation was then induced and the model was energetically re-optimized using molecular dynamics (Fig. 4). The electrostatic potential and available conformational space were also calculated for both wild-type and mutant models. It was indicated, as expected, that even though Leu and Met have a similar size, the Met substitution introduces a sulphur atom and is therefore slightly bulkier than the Leu wild-type residue. More specifically, when superposing the two models and upon molecular dynamics optimization, the two interaction plots were deduced for both wild-type and mutant ADCY3 models (Fig. 5). Of note, while the wild-type Leu residue is only capable of establishing mild H-bonding interactions, the Met mutant substitution is establishing stronger interactions with adjacent residues. This is functionally profound; first, the different side chain of the Met residue, and furthermore, the huge differences in Van der Waals and interaction energies in the middle of the hydrophobic part of the lipid bilayer, may functionally and mechanistically explain the differences in structure that are able to lead to a significant shift in the functionality of ADCY3 and hence to pathology. The in silico calculated Van der Waals energy of the Leu to Met at position 117 was-1.7 and 31.1 respectively, which constitutes a significant change after the p.Leu117Met mutation (Table II). It is of utmost importance to focus on the structural and physicochemical significance of the Leu to Met substitution. Leu is a branched neutral hydrophobic amino acid, whilst Met contains the sulfur atom that alters its physicochemical properties and its potential atomic interaction. Even though the two amino acids exhibit small differences in terms of volume, charge and polarity, the sulfur of Met is reactive with electrophilic centers and is prone to oxidation. Leu to Met substitution decreases the stability of the protein structure with a higher entropic cost. The difference in their shape also affects the overall structure and, depending on the side chain's position, the substitution may cause steric effects. In addition, leucine's bulky side chain contributes to a greater hydrophobic stabilization and its substitution by the elongated sulfur-containing Met is able to destabilize the protein's structure.
Figure 4.
Molecular modelling of wild-type ADCY3 (on the left) and the ADCY3 variant c.349T>A/p.Leu117Met. Upper panel: The wild-type ADCY3 model with Leu117. Lower panel: The mutant ADCY3 model with the p.Leu117Met substitution. Note the position of the 117 amino acid (in spacefill representation) and the difference between wild-type and mutant. The latter induces a slight bend to the middle of the α-helix where it is located. ADCY3, adenylate cyclase 3.
Figure 5.
Molecular interactions 2D diagram. (A) 2D interaction diagram for the wild-type leucine residue at position 117 of the ADCY3 model. (B) 2D interaction diagram for the mutant methionine residue at position 117 of the ADCY3 model. Interactions are presented according to the conventions of the embedded interaction legend. ADCY3, adenylate cyclase 3.
Table II.
In silico calculated VdW energy of the L117 wild-type and the L117M mutated biological system (units are kcal/mol).
| System | Leucine 117
|
Methionine 117
|
||
|---|---|---|---|---|
| System energy | VdW | System energy | VdW | |
| Full system | 1263.102 | 115010.1 | 1298.466 | −133304 |
| Interaction | −1.714 | −1.730 | 32.309 | 31.149 |
VdW, Van der Waals.
Discussion
The genetic component of obesity is challenging to determine and monogenic forms of severe obesity starting at a young age remain elusive. Mendelian monogenic nonsyndromic obesity is associated with only 5% of obesity cases in the population (8). The present study focuses on the ADCY3 genetic profile of a small group of severely obese individuals diagnosed with obesity from the age of 3 years onwards. A small sample size in the present study may be a limitation and conclusive results may be affected. However, five ADCY3 gene variants (4 previously reported and one novel variant) were identified. Of the patients, 2 (6%) were heterozygous carriers of the novel p.Leu117Met variant in the ADCY3 gene. Taking into consideration the rarity of monogenic obesity, this is a noteworthy finding in a targeted group of patients diagnosed with severe obesity since childhood. Molecular modelling data of the mutated protein suggest significant changes in structure, as well as in the interaction with the surrounding lipid bilayer, indicating the possibility of functional changes.
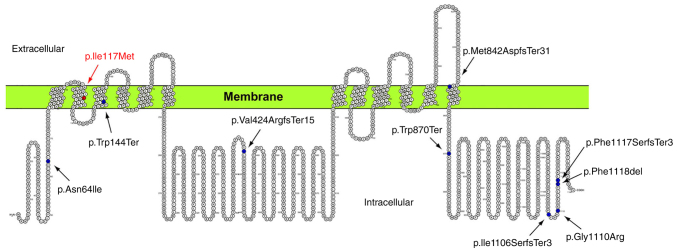
ADCY3 is a 128,960 Da integral protein, which consists of two transmembrane components of six-transmembrane helices each. These two transmembrane components alternate with two cytoplasmic domains, which contain the active site of the molecule at the N-terminus of the protein (Fig. 6). The amino acid Leu to Met change at position 117 of the protein is within an α-helical region embedded in the cellular membrane (105-125aa). Based on the molecular modelling data of the present study, it is suggested that the presence of the sulfur-carrying side chain of the Met residue alters the interaction with the surrounding environment. More specifically, the Met residue is capable of establishing stronger interactions with surrounding residues. Furthermore, interaction energies within the lipid bilayer of the membrane are increased. Although the enzyme's catalytic centers fall within a different region, the changes caused by the Leu to Met substitution may affect the interaction between the transmembrane clusters of the protein. According to previous studies, the interaction between these transmembrane regions of the protein appears to significantly affect the interaction of the catalytic domains (21). Hence, it is possible that the ADCY3 variant c.349T>A results in a reduction of ADCY3 activity.
Figure 6.
Membrane topology of the adenylate cyclase 3 transmembrane protein determined using the Protter tool (https://wlab.ethz.ch/protter/start/). Mutations located in the coding region are indicated, including the p.Ile117Met mutation identified in the present study.
According to the literature, ADCY3 has been studied extensively in a spectrum of processes affecting metabolic mechanisms and obesity development. There are 10 isoforms of ADCY3 reported and characterized in mammals. A total of 9 isoforms are membrane-bound proteins found to be expressed in various human tissues, including adipocyte tissues and the hypothalamus (22). ADCY3 is Ca2+-calmodulin sensitive and it appears to be linked to olfactory signal transduction (23,24). A study involving ADCY3 heterozygous null mice demonstrated increased visceral adiposity in the absence of hyperphagia and impaired insulin sensitivity, dyslipidemia, as well as increased plasma levels of proinflammatory cytokines. According to the same study, a high-fat diet decreased the expression of ADCY3 and genes involved in thermogenesis, fatty acid oxidation and insulin signaling, whereas it increased the expression of genes related to adipogenesis (25). In a different study using diabetic rats, which demonstrated a dysfunctional hypothalamic melanocortin system and attenuation of the hypothalamic glucose-sensing pathway, after performing a hypothalamic transcriptomes analysis, the authors identified ADCY3 among the genes with a role in hypothalamic regulation. In fact, their results support a key role of ADCY3 in preventing obesity (26).
ADCY3 is an enzyme which catalyzes the formation of cAMP, a second messenger involved in signal transduction processes in cells, such as the activation of kinases and the control of metabolic processes of carbohydrates and lipids. More specifically, cAMP appears to be involved in intracellular signaling of molecules such as glucagon-like peptide 1, ghrelin, orexins, α-melanocyte-stimulating hormone and leptin (27,28).
Numerous studies support the involvement of ADCY3 in metabolism regulation and BMI or obesity risk. For instance, a study reported on ADCY3-knockout mice demonstrating obesity, low locomotor activity, hyperphagia and leptin resistance (29). Furthermore, the involvement of ADCY3 in the regulation of glucose homeostasis was suggested by a study using Goto-Kakizaki (GK) rats with type 2 diabetes. In that study, its overexpression was observed in the pancreatic islets as well as the striatum and hypothalamus regions of the brain in the GK rats (30). The involvement of ADCY3 in energy metabolism regulation was reported in another study, which used mutagenized mice with dominantly inherited resistance to diet-induced obesity. These mice were genetically screened and were indicated to contain a p.Met279Leu substitution in ADCY3 (17).
ADCY3 variants in association with obesity were identified in various human population studies. All reported data are presented in Table III (15,16,31-34). More specifically, ADCY3 variants (rs2033655 and rs1968482) were reported to be associated with obesity in a population of Swedish males with or without type 2 diabetes in 2008 (16). Two years later, ADCY3 polymorphisms (rs1127568, rs7604576 and rs753529) were reported to be associated with obesity in adults but not in children of a Chinese Han population sample (15). It should be noted that one of these variants (rs7604576) was also detected in the patients of the present study (minor allele frequency, 0.48). In 2018, a study identified a correlation between loss-of-function mutations and onset obesity and type 2 diabetes in a small Greenlandic population (rs1331776405). Carriers in this cohort appeared to also have decreased RNA levels (34). In the same year, a similar study involving children with severe obesity of a Pakistani population also revealed ADCY3 variants, including that identified in the Greenlandic study (13). Most recently, ADCY3 variant p.Gly1110Arg was detected by targeted exome sequencing in a Finnish population with early-onset severe obesity (Table III) (33).
Table III.
Genetic variants identified in ADCY3 gene with association to obesity and BMI.
| A, Causative variations
| |||||||
|---|---|---|---|---|---|---|---|
| Nucleotide change | Amino-acid change | Genetic location (relevant to ADCY3 gene) | Type of variant | dbSNP number | NCBI Refseq | Clinical characteristic | (Refs.) |
| c.191A>T | p.Asn64Ile | Exon 1 | Missense | rs541941351 | NM_004036.5:c.191A>T | Obesity | (13) |
| c.349T>A | p.Leu117Met | Exon 1 | Missense | NA | NM_004036.5:c.349T>A | Obesity | Present study |
| c.431G>A | p.Trp144Ter | Exon 1 | Stop-gained | rs760195447 | NM_004036.5:c.431G>A | Obesity/type II diabetes | (34) |
| c.1072-1G>A | NA | Intron 4 | Splice acceptor | NA | NM_004036.5:c.1072-1G>A | Obesity/type II diabetes | (34) |
| c.1268del | p.Val424ArgfsTer15 | Exon 6 | Frameshift | rs754914420 | NM_004036.5:c.1268del | Obesity/type II diabetes | (13,34) |
| c.1805+2T>C | NA | Exon 10 | Splice donor | rs761428196 | NM_004036.5:c.1805+2T>C | Obesity/type II diabetes | (34) |
| c.2433-1G>A | NA | Intron 13 | Splice acceptor | rs1331776405 | NM_004036.5:c.2433-1G>A | Obesity | (34) |
| c.2524_2525del | p.Met842AspfsTer31 | Exon 15 | Frameshift | rs759512660 | NM_004036.5:c.2524_2525del | Obesity/type II diabetes | (34) |
| c.2578-1G>A | NA | Intron 15 | Splice acceptor | rs1553333167 | NM_004036.5:c.2578-1G>A | Obesity | (13) |
| c.2609G>A | p.Trp870Ter | Exon16 | Stop-gained | rs763802630 | NM_004036.5:c.2609G>A | Obesity/type II diabetes | (34) |
| c.3315del | p.Ile1106SerfsTer3 | Exon 21 | Frameshift | rs1553329804 | NM_004036.5:c.3315del | Obesity | (13) |
| c.3328G>C | p.Gly1110Arg | Exon 21 | Missense | rs1292057640 | NM_004036.5:c.3328G>C | Obesity | (33) |
| c.3348AG>A | p.Phe1117SerfsTer3 | Exon 21 | Frameshift | NA | NM_004036.5:c.3348AG>A | Obesity/type II diabetes | (34) |
| c.3348_3350CTT[2] | p.Phe1118del | Exon 21 | Deletion | rs750852737 | NM_004036.3:c.3348_3350CTT[2] | Obesity | (13) |
|
| |||||||
| B, Associated SNPs
| |||||||
| Nucleotide change | Amino-acid change | Genetic location (relevant to ADCY3 gene) | Type of variant | dbSNP number | NCBI Refseq | Clinical characteristic | (Refs.) |
|
| |||||||
| c.319T>C | p.Ser107Pro | Exon 1 | SNP | rs11676272 | NM_004036.5:c.319T>C | Linked to obesity | (36) |
| c.675+9866T>C | NA | Intron 1 | SNP | rs6545814 | NM_004036.5:c.675+9866T>C | Linked to BMI | (35) |
| c.675+14467G>A | NA | Intron 1 | SNP | rs6545809 | NM_004036.5:c.675+14467 G>A | Linked to BMI | (38) |
| c.676-21389G>A | NA | Intron 1 | SNP | rs7586879 | NM_004036.5:c.676-21389G>A | Linked to BMI | (42) |
| c.676-5504C>T/G | NA | Intron 1 | SNP | rs2033655 | NM_004036.5: c.676-5504C>T/G | Linked to obesity/type II diabetes | (16) |
| c.825+8581A>G | NA | Intron 2 | SNP | rs1968482 | NM_004036.5:c.825+8581A>G | Linked to BMI | (16) |
| c.1167C>G | p.Leu389= | Exon 5 | Synonymous (SNP) | rs2241758 | NM_004036.5:c.1167C>G | Obesity | (16) |
| c.2055+270C>T | NA | Intron 11 | SNP | rs753529 | NM_004036.3:c.2055+270C>T | Linked to obesity | (15) |
| c.2578-3T>C | NA | Intron 15 | SNP | rs7604576 | NM_004036.5:c.2578-3T>C | Linked to obesity | (15) |
| c.2874A>G | p.Ser958= | Exon 17 | Synonymous (SNP) | rs1127568 | NM_004036.5:c.2871A>G | Obesity | (15) |
| g.24927427A>G | NA | Intergenic | SNP | rs10182181 | NC_000002.12:g.24927427A>G | Linked to obesity/nutrition | (32) |
| g.24935139T>C | NA | Intergenic | SNP | rs713586 | NC_000002.12:g.24935139T>C | Linked to BMI | (43) |
ADCY3, adenylate cyclase 3; NCBI, National Center for Biotechnology Information; Refseq, reference sequence; NA, not available; SNP, single nucleotide polymorphism; BMI, body mass index.
Apart from population genetic studies, GWAS have also linked ADCY3 genetic polymorphisms to obesity susceptibility. The ADCY3 locus was indicated to be associated with BMI in the Genetic Investigation of ANthropometric Traits consortium meta-analysis (12) and also in a meta-analysis of East-Asian populations (rs6545814, rs11676272) (35). It should be noted that variant rs11676272, also detected in the patients of the present study, was reported in a GWAS of height-adjusted BMI in children. Of note, in this study, the authors discuss the possible effects on the second transmembrane-spanning α-helix of the M1 transmembrane cluster of the protein (same region where the novel p.Leu117Met of the present study was detected), thus possibly disturbing the interaction of M1 helix and M2 helix (36). ADCY3 was also identified in a GWAS meta-analysis of BMI across children aged 1 to 17 years from the Avon Longitudinal Study of Parents and Children and the Western Australian Pregnancy Cohort study (37). Similarly, an association to BMI was concluded in a GWAS concerning a Korean sample (38).
Adipogenesis is another process linked to ADCY3, as supported by animal studies performed by Tong et al (25) and Meng et al (26). A decrease in ADCY3 expression in the adipose tissue of obese patients was also reported in 2011 (18). However, recent findings concerning MC4R provided strong evidence regarding its possible relevance to the leptin-melanocortin pathway. More specifically, it was indicated that MC4R co-localizes with ADCY3 at the primary cilium of hypothalamic neurons and obesity-related MC4R mutations impaired ciliary localization. In addition, inhibition of ADCY3 signaling at the primary cilia of these neurons increased body weight (20). A previous study by our group reported a novel MC4R deletion in a pediatric patient with severe early-onset obesity (39). However, MC4R screening revealed no mutations in patients and healthy individuals of the present study.
Finally, epigenetic studies have also suggested a role of ADCY3 in human obesity, as DNA methylation and ADCY3 associations were identified in a study involving obese and healthy individuals (40). Similarly, a genome-wide DNA methylation quantitative trait locus analysis in human adipose tissue and genotype-DNA methylation associations were indicated to involve ADCY3 (41).
In conclusion, according to the literature discussed above, there appears to be strong evidence to suggest that ADCY3 is an important mediator of energy homeostasis with a possible role in the development of obesity. Although the lack of whole-exome sequencing or whole-genome sequencing may be considered a limitation of the present study, recent published data suggests that ADCY3 is considered a strong candidate among numerous loci previously reported to be associated with obesity (13,20,34). Thus, the ADCY3 locus has been the focus of the present study. The current findings support the association of ADCY3 with obesity and provide the first evidence that ADCY3 variations exist in the genetic spectrum of the Cypriot population. Further to identifying the novel, possibly pathogenic ADCY3 variant, causing an amino acid Leu to Met change at position 117 of the protein in young obese patients, the present study provided informative modelling data suggesting a change in the interaction of the two transmembrane halves of the enzyme, thus causing a disturbance in the pseudo-twofold symmetry of the transmembrane domain, a symmetry known to affect catalytic activity (21). Further biological assessment using cell lines or genetically modified animals may help to elucidate the role of the novel variant and other previously reported ADCY3 variants in genetic predisposition to obesity.
Supplementary Data
Acknowledgments
Not applicable.
Funding Statement
This research received funding from the AG Leventis Foundation (grant no. 3317312) and the RCB Bank Ltd. (grant no. 33173151).
Availability of data and materials
The data that support the findings of this study are openly available from the Zenodo repository at DOI (https://zenodo.org/record/5717389#.YZrIRM_RbX4).
Authors' contributions
MT and NS examined the patients. MT gathered all clinical information. MP, PF and VN performed the database search and interpreted the genetic data. PF performed the genomic analysis experiments and processed genetic data. MP and PF performed data analysis. MP, MT and DV wrote the draft of the manuscript. DV worked on the molecular modelling data. LAP and CSM had substantial contribution to the design of the work and critically reviewed the manuscript. MP, NS, MT, DV, PF and VN confirm the authenticity of the raw data. All authors have read and given final approval of the version to be published.
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Written informed consent was obtained from the patients and the parents of minors that participated in the study. Bioethics approval was received from the Cyprus National Ethics Committee (Nicosia, Cyprus).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lobstein T, Baur L. Obesity in children and young people: A crisis in public health. Obes Rev. 2004;5(Suppl 1):S4–S104. doi: 10.1111/j.1467-789X.2004.00133.x. [DOI] [PubMed] [Google Scholar]
- 2.Nutrition Global Targets 2025. Geneva: WHO; 2018. http://www.who.int/nutrition/global-target-2025/en/. Accessed April 2, 2020. [Google Scholar]
- 3.NCD Risk Factor Collaboration (NCD-RisC) Worldwide trends in body-mass index, underweight, overweight, and obesity from 1975 to 2016: A pooled analysis of 2416 population-based measurement studies in 1289 million children, adolescents, and adults. Lancet. 2017;390:2627–2642. doi: 10.1016/S0140-6736(17)32129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberto CA, Swinburn B, Hawkes C, Huang TT, Costa SA, Ashe M, Zwicker L, Cawley JH, Brownell KD. Patchy progress on obesity prevention: Emerging examples, entrenched barriers, and new thinking. Lancet. 2015;385:2400–2409. doi: 10.1016/S0140-6736(14)61744-X. [DOI] [PubMed] [Google Scholar]
- 5.Templin T, Cravo Oliveira Hashiguchi T, Thomson B, Dieleman J, Bendavid E. The overweight and obesity transition from the wealthy to the poor in low- and middle-income countries: A survey of household data from 103 countries. PLoS Med. 2019;16:e1002968. doi: 10.1371/journal.pmed.1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.González-Muniesa P, Mártinez-González MA, Hu FB, Després JP, Matsuzawa Y, Loos RJF, Moreno LA, Bray GA, Martinez JA. Obesity. Nat Rev Dis Primers. 2017;3:17034. doi: 10.1038/nrdp.2017.34. [DOI] [PubMed] [Google Scholar]
- 7.Tremmel M, Gerdtham UG, Nilsson PM, Saha S. Economic burden of obesity: A systematic literature review. Int J Environ Res Public Health. 2017;14:435. doi: 10.3390/ijerph14040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.WHO fact sheet 311. https://www.who.int/en/news-room/fact-sheets/detail/obesity-and-overweight. Accessed April 2, 2020.
- 9.Sahoo K, Sahoo B, Choudhury AK, Sofi NY, Kumar R, Bhadoria AS. Childhood obesity: Causes and consequences. J Family Med Prim Care. 2015;4:187–192. doi: 10.4103/2249-4863.154628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, Inouye M, Freathy RM, Attwood AP, Beckmann JS, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Allen HL, Lindgren CM, Luan J, Mägi R, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saeed S, Bonnefond A, Tamanini F, Mirza MU, Manzoor J, Janjua QM, Din SM, Gaitan J, Milochau A, Durand E, et al. Loss-of-function mutations in ADCY3 cause monogenic severe obesity. Nat Genet. 2018;50:175–179. doi: 10.1038/s41588-017-0023-6. [DOI] [PubMed] [Google Scholar]
- 14.Ludwig MG, Seuwen K. Characterization of the human adenylyl cyclase gene family: cDNA, gene structure, and tissue distribution of the nine isoforms. J Recept Signal Transduct Res. 2002;22:79–110. doi: 10.1081/RRS-120014589. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Wu M, Zhu W, Shen J, Shi X, Yang J, Zhao Q, Ni C, Xu Y, Shen H, et al. Evaluation of the association between the AC3 genetic polymorphisms and obesity in a Chinese Han population. PLoS One. 2010;5:e13851. doi: 10.1371/journal.pone.0013851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nordman S, Abulaiti A, Hilding A, Långberg EC, Humphreys K, Ostenson CG, Efendic S, Gu HF. Genetic variation of the adenylyl cyclase 3 (AC3) locus and its influence on type 2 diabetes and obesity susceptibility in Swedish men. Int J Obes (Lond) 2008;32:407–412. doi: 10.1038/sj.ijo.0803742. [DOI] [PubMed] [Google Scholar]
- 17.Pitman JL, Wheeler MC, Lloyd DJ, Walker JR, Glynne RJ, Gekakis N. A gain-of-function mutation in adenylate cyclase 3 protects mice from diet-induced obesity. PLoS One. 2014;9:e110226. doi: 10.1371/journal.pone.0110226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurtado del Pozo C, Vesperinas-García G, Rubio MÁ, Corripio-Sánchez R, Torres-García AJ, Obregon MJ, Calvo RM. ChREBP expression in the liver, adipose tissue and differentiated preadipocytes in human obesity. Biochim Biophys Acta. 2011;1811:1194–1200. doi: 10.1016/j.bbalip.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 19.Wu L, Shen C, Seed Ahmed M, Östenson CG, Gu HF. Adenylate cyclase 3: A new target for anti-obesity drug development. Obes Rev. 2016;17:907–914. doi: 10.1111/obr.12430. [DOI] [PubMed] [Google Scholar]
- 20.Siljee JE, Wang Y, Bernard AA, Ersoy BA, Zhang S, Marley A, Von Zastrow M, Reiter JF, Vaisse C. Subcellular localization of MC4R with ADCY3 at neuronal primary cilia underlies a common pathway for genetic predisposition to obesity. Nat Genet. 2018;50:180–185. doi: 10.1038/s41588-017-0020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu C, Sorkin A, Cooper DM. Persistent interactions between the two transmembrane clusters dictate the targeting and functional assembly of adenylyl cyclase. Curr Biol. 2001;11:185–190. doi: 10.1016/S0960-9822(01)00044-6. [DOI] [PubMed] [Google Scholar]
- 22.Cooper DM. Regulation and organization of adenylyl cyclases and cAMP. Biochem J. 2003;375:517–529. doi: 10.1042/bj20031061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valverde I, Vandermeers A, Anjaneyulu R, Malaisse WJ. Calmodulin activation of adenylate cyclase in pancreatic islets. Science. 1979;206:225–227. doi: 10.1126/science.225798. [DOI] [PubMed] [Google Scholar]
- 24.Wong ST, Trinh K, Hacker B, Chan GC, Lowe G, Gaggar A, Xia Z, Gold GH, Storm DR. Disruption of the type III adenylyl cyclase gene leads to peripheral and behavioral anosmia in transgenic mice. Neuron. 2000;27:487–497. doi: 10.1016/S0896-6273(00)00060-X. [DOI] [PubMed] [Google Scholar]
- 25.Tong T, Shen Y, Lee HW, Yu R, Park T. Adenylyl cyclase 3 haploinsufficiency confers susceptibility to diet-induced obesity and insulin resistance in mice. Sci Rep. 2016;6:34179. doi: 10.1038/srep34179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng Y, Guan Y, Zhang W, Wu YE, Jia H, Zhang Y, Zhang X, Du H, Wang X. RNA-seq analysis of the hypothalamic transcriptome reveals the networks regulating physiopathological progress in the diabetic GK rat. Sci Rep. 2016;6:34138. doi: 10.1038/srep34138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu TR, Yang Y, Ward R, Gao L, Liu Y. Orexin receptors: Multi-functional therapeutic targets for sleeping disorders, eating disorders, drug addiction, cancers and other physiological disorders. Cell Signal. 2013;25:2413–2423. doi: 10.1016/j.cellsig.2013.07.025. [DOI] [PubMed] [Google Scholar]
- 28.Wheeler MB, Lu M, Dillon JS, Leng XH, Chen C, Boyd AE., III Functional expression of the rat glucagon-like peptide-I receptor, evidence for coupling to both adenylyl cyclase and phospholipase-C. Endocrinology. 1993;133:57–62. doi: 10.1210/endo.133.1.8391428. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, Li V, Chan GC, Phan T, Nudelman AS, Xia Z, Storm DR. Adult type 3 adenylyl cyclase-deficient mice are obese. PLoS One. 2009;4:e6979. doi: 10.1371/journal.pone.0006979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seed Ahmed M, Kovoor A, Nordman S, Abu Seman N, Gu T, Efendic S, Brismar K, Östenson CG, Gu HF. Increased expression of adenylyl cyclase 3 in pancreatic islets and central nervous system of diabetic Goto-Kakizaki rats: A possible regulatory role in glucose homeostasis. Islets. 2012;4:343–348. doi: 10.4161/isl.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersen MK, Hansen T. Genetics of metabolic traits in Greenlanders: Lessons from an isolated population. J Intern Med. 2018;284:464–477. doi: 10.1111/joim.12814. [DOI] [PubMed] [Google Scholar]
- 32.Goni L, Riezu-Boj JI, Milagro FI, Corrales FJ, Ortiz L, Cuervo M, Martínez JA. Interaction between an ADCY3 genetic variant and two weight-lowering diets affecting body fatness and body composition outcomes depending on macronutrient distribution: A randomized trial. Nutrients. 2018;10:789. doi: 10.3390/nu10060789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loid P, Mustila T, Mäkitie RE, Viljakainen H, Kämpe A, Tossavainen P, Lipsanen-Nyman M, Pekkinen M, Mäkitie O. Rare variants in genes linked to appetite control and hypothalamic development in early-onset severe obesity. Front Endocrinol (Lausanne) 2020;11:81. doi: 10.3389/fendo.2020.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grarup N, Moltke I, Andersen MK, Dalby M, Vitting-Seerup K, Kern T, Mahendran Y, Jørsboe E, Larsen CVL, Dahl-Petersen IK, et al. Loss-of-function variants in ADCY3 increase risk of obesity and type 2 diabetes. Nat Genet. 2018;50:172–174. doi: 10.1038/s41588-017-0022-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen W, Cho YS, Zheng W, Dorajoo R, Kato N, Qi L, Chen CH, Delahanty RJ, Okada Y, Tabara Y, et al. Meta-analysis identifies common variants associated with body mass index in east Asians. Nat Genet. 2012;44:307–311. doi: 10.1038/ng.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stergiakouli E, Gaillard R, Tavaré JM, Balthasar N, Loos RJ, Taal HR, Evans DM, Rivadeneira F, St Pourcain B, Uitterlinden AG, et al. Genome-wide association study of height-adjusted BMI in childhood identifies functional variant in ADCY3. Obesity (Silver Spring) 2014;22:2252–2259. doi: 10.1002/oby.20840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warrington NM, Howe LD, Paternoster L, Kaakinen M, Herrala S, Huikari V, Wu YY, Kemp JP, Timpson NJ, St Pourcain B, et al. A genome-wide association study of body mass index across early life and childhood. Int J Epidemiol. 2015;44:700–712. doi: 10.1093/ije/dyv077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JS, Cheong HS, Shin HD. BMI prediction within a Korean population. PeerJ. 2017;5:e3510. doi: 10.7717/peerj.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neocleous V, Shammas C, Phelan MM, Fanis P, Pantelidou M, Skordis N, Mantzoros C, Phylactou LA, Toumba M. A novel MC4R deletion coexisting with FTO and MC1R gene variants, causes severe early onset obesity. Hormones (Athens) 2016;15:445–452. doi: 10.14310/horm.2002.1686. [DOI] [PubMed] [Google Scholar]
- 40.Voisin S, Almén MS, Zheleznyakova GY, Lundberg L, Zarei S, Castillo S, Eriksson FE, Nilsson EK, Blüher M, Böttcher Y, et al. Many obesity-associated SNPs strongly associate with DNA methylation changes at proximal promoters and enhancers. Genome Med. 2015;7:103. doi: 10.1186/s13073-015-0225-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volkov P, Olsson AH, Gillberg L, Jørgensen SW, Brøns C, Eriksson KF, Groop L, Jansson PA, Nilsson E, Rönn T, et al. A genome-wide mQTL analysis in human adipose tissue identifies genetic variants associated with DNA methylation, gene expression and metabolic traits. PLoS One. 2016;11:e0157776. doi: 10.1371/journal.pone.0157776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monda KL, Chen GK, Taylor KC, Palmer C, Edwards TL, Lange LA, Ng MC, Adeyemo AA, Allison MA, Bielak LF, et al. A meta-analysis identifies new loci associated with body mass index in individuals of African ancestry. Nat Genet. 2013;45:690–696. doi: 10.1038/ng.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available from the Zenodo repository at DOI (https://zenodo.org/record/5717389#.YZrIRM_RbX4).