Abstract
Background
Currently, immunotherapy is widely used for breast cancer (BC) patients, and tumor mutation burden (TMB) is regarded as a valuable independent predictor of response to immunotherapy. However, specific gene mutations and their relationship with TMB and tumor-infiltrating immune cells in BC are not fully understood.
Methods
Comprehensive bioinformatic analyses were performed using data from The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC) datasets. Survival curves were analyzed via Kaplan-Meier analysis. Univariate and multivariate Cox regression analyses were used for prognosis analysis. Gene set enrichment analysis (GSEA) was performed to explore regulatory mechanisms and functions. The CIBERSORT algorithm was used to calculate the tumor-infiltrating immune cell fractions.
Results
We analyzed somatic mutation data of BC from TCGA and ICGC datasets and found that 19 frequently mutated genes were reported in both cohorts, namely, SPTA1, TTN, MUC17, MAP3K1, CDH1, FAT3, SYNE1, FLG, HMCN1, RYR2 (ryanodine receptor 2), GATA3, MUC4, PIK3CA, KMT2C, TP53, PTEN, ZFHX4, MUC16, and USH2A. Among them, we observed that RYR2 mutation was significantly associated with higher TMB and better clinical prognosis. Moreover, GSEA revealed that RYR2 mutation-enriched signaling pathways were related to immune-associated pathways. Furthermore, based on the CIBERSORT algorithm, we found that RYR2 mutation enhanced the antitumor immune response by enriching CD8+ T cells, activated memory CD4+ T cells, and M1 macrophages.
Conclusion
RYR2 is frequently mutated in BC, and its mutation is related to increased TMB and promotes antitumor immunity; thus, RYR2 may serve as a valuable biomarker to predict the immune response.
1. Introduction
Breast cancer (BC) is the most common malignancy in women worldwide, with approximately 2.1 million new cases in 2018 [1]. Although current comprehensive treatments for BC, including surgery, chemotherapy, radiotherapy, endocrine therapy, and targeted drug therapy, have significantly improved the survival rate of patients, some patients still develop tumor recurrence due to drug resistance and eventually die [2]. In recent years, the discovery of immune checkpoints has opened a new era in the treatment of malignant tumors [3]. In clinical practice, immune checkpoint inhibition targeting programmed cell death receptor 1 (PD-1) and its ligand PD-L1 has been applied to a variety of solid tumors and significantly improved the overall survival (OS) of patients [4]. Currently, the American Food and Drug Administration (FDA) has approved atezolizumab (an inhibitor of PD-L1) in combination with albumin paclitaxel for first-line treatment of advanced triple-negative breast cancer (TNBC). However, BC is a “cold” tumor in immunotherapy [5]. In terms of overall efficacy, BC patients do not benefit as much from immunotherapy as patients with other solid tumors, such as non-small-cell lung cancer and melanoma [6]. Thus, the use of effective immune markers to screen out BC patients who are more likely to benefit from immunotherapy is an important research direction. The most critical problem for BC immunotherapy is how to choose the appropriate population and reasonable predictors of immunotherapy efficacy to prolong the survival time of patients and improve their quality of life.
The PD-1/PD-L1 signaling pathway is the most intensively studied pathway in immunotherapy [7]. Tumor cells achieve immune escape using the PD-1/PD-L1 signaling pathway to evade immune monitoring. Both tumor cells and immune cells can overexpress PD-L1. When PD-1 on the surface of T cells binds to PD-L1, the immune response of T cells to tumor cells is inhibited, enabling tumor cells to escape being killed by the immune system [8, 9]. PD-1 or PD-L1 antibodies can block the binding between PD-1 and PD-L1 and reactivate the immune response of T cells to tumor cells [8]. Thus, high PD-L1 expression is considered a good indicator for tumor immunotherapy. A meta-analysis of advanced BC showed that PD-L1 not only is related to the prognosis of advanced BC patients but also is a biomarker for screening the appropriate population for immunotherapy [10]. Although the expression level of PD-L1 in tumors can be clinically assessed by immunohistochemistry, PD-L1 expression in tumors is heterogeneous and affected by previous chemotherapy and radiotherapy [11, 12]. In addition, the methods for PD-L1 detection and the critical value of positive PD-L1 expression are not standardized [13]. Thus, the use of PD-L1 as a biomarker for immunotherapy sensitivity is still limited.
Tumor mutation burden (TMB) is the total number of nonsynonymous somatic cell mutations in tumor cells [14]. Nonsynonymous mutations can produce neoantigens recognized by the host immune system, thus triggering removal by the immune system and inducing the host immune response to scavenge tumor cells [15]. Therefore, tumor cells with higher levels of TMB may be more easily recognized by the immune system, which in turn triggers a stronger immune response to checkpoint inhibitors [16]. It has been confirmed that high TMB is associated with high tumor immunogenicity in different types of tumors, and the clinical response and survival rate of high TMB tumors, such as melanoma, lung cancer, and colorectal cancer, are significantly increased [17]. In a clinical trial, TMB was more significantly correlated with response rates than the expression level of PD-L1 [18]. These results suggest that TMB can be used as a tumor predictor of clinical benefit and a prognostic factor and that it has the potential to play a key role in predicting the efficacy of immune checkpoint inhibitors. A recent study analyzed clinical research data on PD-1 inhibitors and PD-L1 inhibitors in 27 solid tumor types, including BC, and found that TMB was significantly positively correlated with the objective response rate to immunotherapy [19]. TMB expression was significantly different among different gene mutation subtypes of BC. TMB expression was highest in TNBC patients, followed by HER2-positive patients, while TMB expression was lowest in patients with estrogen receptor- (ER-) positive, progesterone receptor- (PR-) positive, and human epidermal growth factor receptor-2- (HER2-) negative BC subtypes [20]. At present, whether TMB can directly predict the efficacy of immunotherapy for BC is still controversial, but some studies suggest that patients with a high TMB have a better prognosis when stratified by gene mutation and analyzed in combination with immune subtypes [21]. Therefore, high TMB-related gene mutation analysis may be a good predictor of immunotherapy efficacy, which can contribute to screening for the appropriate population for BC immunotherapy. However, specific gene mutations and their relationship with TMB and tumor-infiltrating immune cells in BC are not clear. Thus, this study is aimed at exploring whether TMB and prognosis-related genes are closely related to BC immunotherapy.
2. Materials and Methods
2.1. Data Acquisition
A total of 1222 RNA expression profile samples of BC were downloaded from TCGA database (http://portal.gdc.cancer.gov/projects), including 1109 tumor samples and 113 normal samples. The clinical information of 1097 patients with BC was downloaded from TCGA database. After excluding those patients with missing clinical data, 980 samples were used for further analysis. The somatic mutation data of 877 United States patients from TCGA database and 508 European Union patients from the ICGC database (http://dcc.icgc.org/releases/current/Projects) were downloaded and extracted in Perl software so that it can be analyzed in R software.
2.2. Classification of BC Based on TMB
TMB was defined as the total number of somatic gene coding errors, gene insertions, gene deletions, and base substitutions detected per million bases. As described in previously published research, only mutations that cause changes in amino acids were counted [14].
2.3. Bioinformatic Analysis
As described in a previous study [22], R (v4.0.2) was used for bioinformatic analysis. The 30 genes with the highest mutation frequency in TGCA and IGGC databases were extracted with Perl. Somatic mutation data for both American and European Union BC samples were processed and visualized with the “GenVisR” package [23]. The R package “venn” was used to screen for the same mutated genes in both databases. The association between mutated genes and TMB was analyzed and visualized using the R package “ggpubr.” Due to the lack of survival information in the ICGC database, patients from TCGA database were sorted into two groups according to gene mutation states. Survival curves were analyzed and visualized using the R packages “survminer” and “survival.” Gene set enrichment analysis (GSEA) was performed with GSEA software (v4.1.0), and the conditions set included normalized enrichment score (NES) = 1000 and FDR p value < 0.05 [24]. The selected gene sets for GSEA contained “c5.go.bp.v7.2.symbols.gmt,” “c5.go.cc.v7.2.symbols.gmt,” and “c5.go.mf.v7.2.symbols.gmt.” The CIBERSORT algorithm was used to estimate the relative abundance of immune cell infiltration in patients with different RYR2 statuses [25]. The number of permutations was set to 1000, and a threshold p value of <0.05 was the criterion for the successful computation of a sample. Matrix data visualization was performed with the R package “corrplot.” Difference analysis of infiltrating immune cells between the RYR2-mutant and RYR2-wild-type groups was performed using the R package “limma” and visualized with the R package “vioplot.”
2.4. Statistical Analysis
Statistical analyses were performed with R software (version 4.0.2). Survival curves were analyzed using Kaplan-Meier survival analysis and a log-rank test. Univariate and multivariate Cox regression analyses were used for prognosis analysis. The correlation between mutant genes and TMB was analyzed with a Mann–Whitney U test. For all comparisons, a two-tailed p value < 0.05 was considered statistically significant.
3. Results
3.1. Landscape of Somatic Mutations in BC
We first analyzed the mutations of the top 30 genes with high mutation frequency in TCGA database (n = 877). As shown in Figure 1(a), the mutation spectrum of the top 30 genes was mainly missense mutations, and the five genes with the highest mutation frequencies were PIK3CA (36.7%), TP53 (35.8%), TTN (17.8%), CDH1 (13.0%), and GATA3 (11.3%). We then analyzed the genetic mutation profiles of BC patients in the European Union in the ICGC database (n = 508). The top 30 frequently mutated genes with high mutation frequency and the pattern of somatic mutation for the top 30 genes are illustrated in Figure 1(b), among which the five most frequently mutated genes were TP53 (35.6%), PIK3CA (29.9%), TTN (18.9%), GATA3 (8.1%), and MAP3K1 (7.5%).
Figure 1.
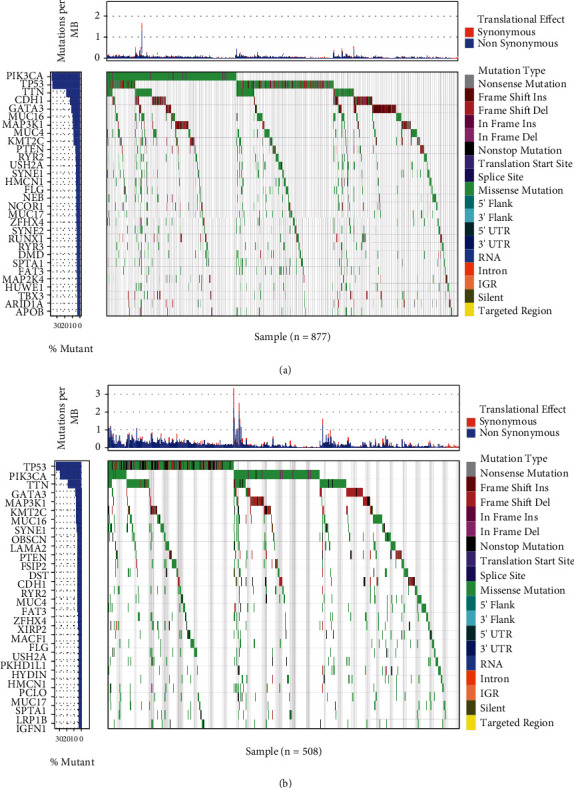
Overview of frequently mutated genes in BC. (a) Waterfall plot showing the top 30 mutated genes in TCGA BC cohort. Left panel: mutation frequency; right panel: different mutation types. (b) Waterfall plot showing the top 30 mutated genes in the ICGC BC cohort. Left panel: mutation frequency; right panel: different mutation types.
3.2. Gene Mutations Associated with TMB
We further screened out genes with high mutation frequency in both databases via a Venn diagram. Our results demonstrated that 19 frequently mutated genes reported in TCGA cohort were also reported in the ICGC cohort, namely, SPTA1, TTN, MUC17, MAP3K1, CDH1, FAT3, SYNE1, FLG, HMCN1, RYR2, GATA3, MUC4, PIK3CA, KMT2C, TP53, PTEN, ZFHX4, MUC16, and USH2A (Figure 2(a)). To further investigate the relationship between these highly mutated genes and TMB, the TMB value of each patient in TCGA database was calculated, and patients were assigned to the wild-type group or mutation group based on the 19 gene mutation statuses. The TMB score in BC ranged from 0.03 to 118.45 per Mb, with a median of 1.56 per Mb. As shown in Figure 2(b), among these high-frequency mutated genes in both databases, 14 of the 19 genes, including RYR2, with a mutation type had a higher TMB value than those genes with the wild type. Interestingly, patients with mutations in MAP3K1 showed a lower TMB, and there was no significant difference in TMB between the mutation group and the wild-type group of the other 4 genes (CDH1, GATA3, PIK3CA, and KMT2C) (Figure 2(b)).
Figure 2.
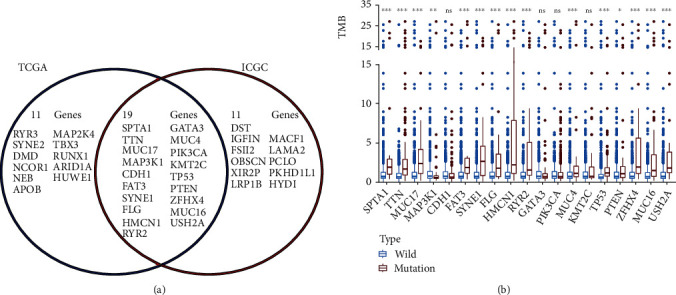
Gene mutations are correlated with TMB. (a) Venn diagram of the same frequently mutated genes in both TCGA and ICGC cohorts. (b) Mutated genes are correlated with a higher TMB. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ns: no significance.
3.3. RYR2 Mutation Associated with Prognosis
A previous study reported that higher TMB indicated favorable overall survival in BC patients [26]. To identify prognosis-related gene mutations, Kaplan-Meier analysis was performed on the TMB-related commonly mutated genes. The results demonstrated that only RYR2 mutation (HR = 0.140; 95% CI, 0.020–1.000; p = 0.021) was associated with a better prognosis (Figure 3). However, the RYR2 mutation did not remain statistically significant after considering age, sex, TNM classification, and TMB status in the Cox regression model (Figure 4).
Figure 3.
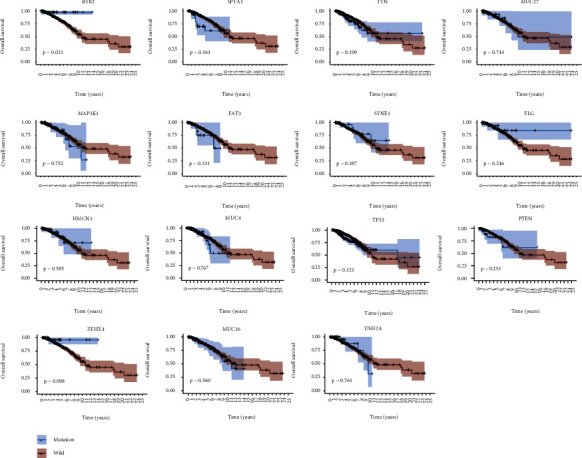
RYR2 mutation is associated with clinical prognosis. Kaplan-Meier curves of overall survival of the TMB-related mutated genes. The p value is marked in each plot.
Figure 4.
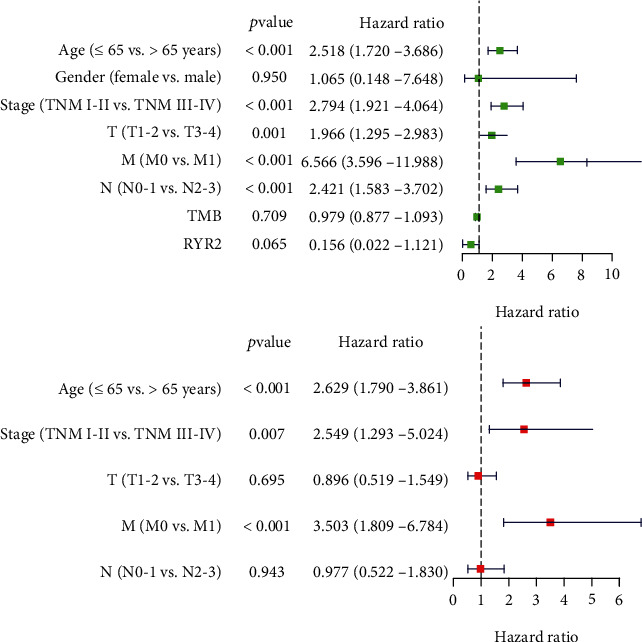
Univariate (a) and multivariate (b) overall survival analyses of BC patients using a Cox proportional hazards model.
3.4. Identification of Enrichment Pathways for Patients with RYR2 Mutation
It is well known that TMB is an important indicator to judge the efficacy of tumor immunotherapy [17]. Thus, we next investigated whether RYR2 mutation-enriched signaling pathways are related to immunity. GSEA was performed, and the results showed that pathways were significantly enriched in the RYR2 mutant group, including antigen processing and presentation; antigen processing and presentation of peptide antigen via MHC class I/II, MHC class I/II protein binding, and NF-κB binding; regulation of response to IFN-γ; and response to IL-12. These results suggest that certain immune-related pathways are associated with RYR2 mutations in BC patients (Figure 5).
Figure 5.
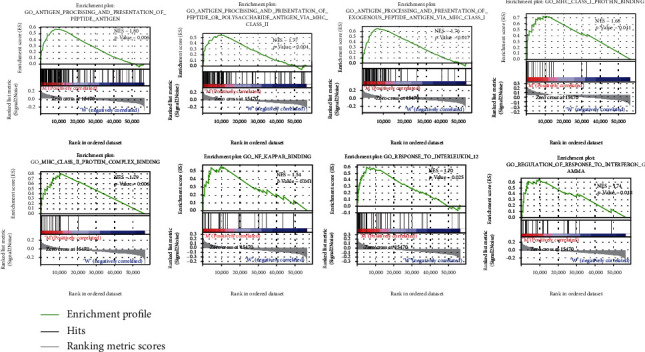
RYR2 mutation is associated with immune-related pathways. Gene enrichment plots showing that a series of immune-related pathways were enriched in the RYR2-mutant group. NES: normalized enrichment score. The p value is shown in each plot.
3.5. Tumor-Infiltrating Immune Cells Associated with RYR2 Mutation in BC
According to the CIBERSORT algorithm, we further assessed the association between RYR2 mutation and tumor-infiltrating immune cells in the BC microenvironment. Our results demonstrated that the composition of 22 immune cell types in each sample varied significantly, and the infiltrating immune cells were mainly T cells and macrophages in BC samples (Figure 6). In addition, we observed that CD8 T cells, native CD4 T cells (low infiltration in both the RYR2 mutant and wild-type groups), activated memory CD4 T cells, and M1 macrophages were more enriched in the RYR2 mutant-type group than in the wild-type group (Figure 7(a)). Furthermore, the correlation matrix results revealed that CD8 T cells had the strongest positive correlation with activated memory CD4 T cells and M1 macrophages but were negatively correlated with M2 macrophages (Figure 7(b)). Moreover, M1 macrophages were positively associated with activated memory CD4 T cells and CD8 T cells and negatively correlated with M2 macrophages (Figure 7(b)).
Figure 6.
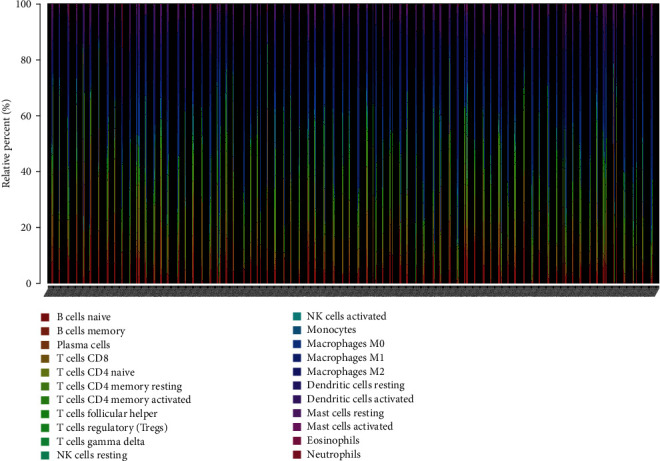
Distribution of tumor-infiltrating immune cells in BC samples. The stacked bar chart shows the distribution of 22 immune cells in each sample.
Figure 7.
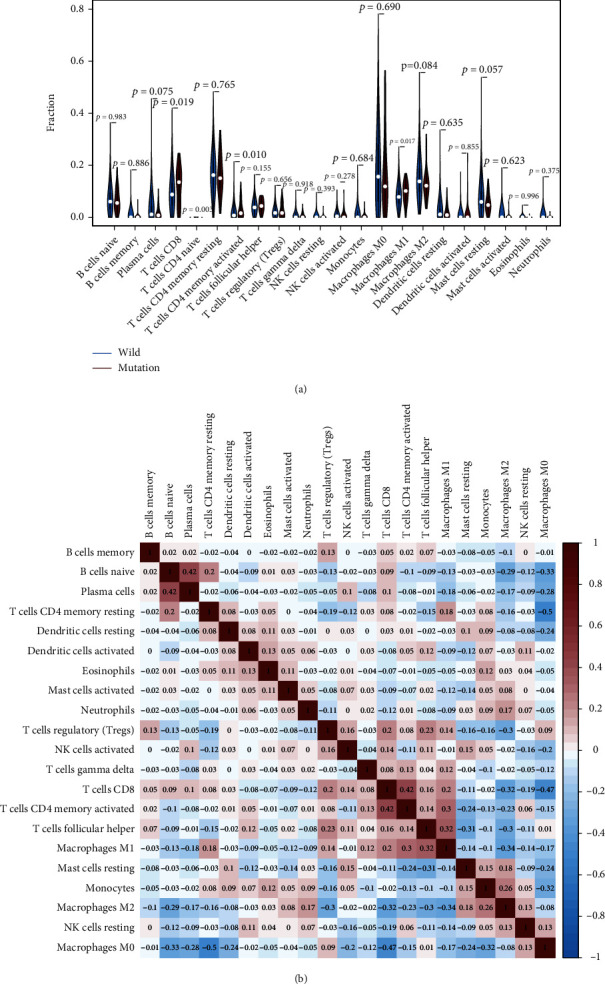
RYR2 mutation is associated with tumor-infiltrating immune cells. (a) Violin plot displaying the differentially infiltrated immune cells between the RYR2-mutant group and the RYR2-wild-type group. Blue represents the RYR2-wild-type group, and red represents the RYR2-mutant group. The p value is marked in the figure. (b) Correlation matrix of immune cell proportions. Red represents a positive correlation, and blue represents a negative correlation.
4. Discussion
In our study, our data suggest that TMB and the prognosis-related gene RYR2 are closely related to the immune response of BC. These results support previous findings that these immune cells play a major role in the tumor microenvironment and enhance the immune response against tumor immune escape [27–29].
Intracellular calcium ions (Ca2+) play an important role in basic cellular physiology [30]. Accumulating evidence has shown that intracellular Ca2+ homeostasis is disrupted in tumor cells, and these changes are involved in genetic mutations, cancer cell proliferation, apoptosis, and migration [31–33]. RYR2 is a member of the RYR family and a major component of the Ca2+ pathway, which regulates the release of Ca2+ from the sarcoplasmic reticulum into the cytoplasm [34]. In an RYR2-positive prostate cancer cell line, RYR-related Ca2+ mobilization augments tumor cell apoptosis [35]. Moreover, RYR2 expression is correlated with poor prognosis in patients with thyroid carcinoma [36], and strong RYR2 upregulation was found in a BC cell line upon EGF-induced epithelial-to-mesenchymal transition [37]. In addition to RYR2 expression alteration, a high RYR2 mutation frequency is a common feature of numerous human malignancies. RYR2 somatic mutation is widely observed in cervical cancer patients, and it is speculated that RYR2 can be used as a target for cervical cancer treatment [38]. Cai et al. reported that RYR2 is a frequently mutated gene with predicted neoantigens presented by MHC class I and class II molecules and thus is a candidate for lung adenocarcinoma immunotherapy [39]. Schmitt et al. demonstrated that impaired RYR2 function due to somatic mutation is a common event in the pathogenesis of head and neck cancer [40]. For BC, RYR2 mutation was found to be correlated with a decreased BC risk restricted by PR, ER, and tumor stage [41]. In the present study, we found that RYR2 mutation was associated with a favorable prognosis and increased TMB in BC patients. TMB is the total number of somatic gene coding errors, gene insertions, gene deletions, and base substitutions detected per million bases [14]. High TMB in tumor cells produces more neoantigens that can be recognized by the host immune system and trigger the immune response [19, 42]. Thus, we speculate that RYR2 mutation with a high TMB in BC might drive the immune system to scavenge tumor cells.
The tumor microenvironment plays an important role in the occurrence and development of BC [43]. The immune microenvironment, composed of tumor-infiltrating lymphocytes, tumor-related macrophages, and other immune cells, is an important part of the tumor microenvironment [44]. Under the influence of different cellular activation mechanisms and cytokines, tumor-infiltrating immune cells produce different immune responses, which can directly reflect the local immune response of the tumor microenvironment [45]. The tumor-infiltrating immune cells in BC mainly derive from the tertiary lymphoid structure and are dominated by two opposing forces [46]. One is antitumor cells mainly composed of CD4+ Th1 cells, CD8+ cytotoxic T lymphocytes (CTLs), NK cells, M1 macrophages, and dendritic cells, while the other is capable of promoting tumor growth and includes CD4+ FOXP3+ T cells (Tregs), CD4+ Th2 cells, and M2 macrophages [47]. In our study, we found that samples with RYR2 mutation were more infiltrated by CD8 T cells, activated memory CD4 T cells, and M1 macrophages than samples with wild-type RYR2, indicating that RYR2 mutation may promote antitumor immunity in BC patients. This finding is in line with previous evidence indicating that RYR2 mutation and its transcriptomic signature are associated with a favorable outcome and immune infiltrates in basal-like tumors with high PD1 and PD-L1 expression [48].
It is generally believed that CD8+ T cells destroy tumor cells by binding to MHCI antigens [49], and the total number of CD8+ cells is positively correlated with tumor grade and better patient prognosis in BC [27]. Meanwhile, memory CD4+ T cells inhibit the outgrowth of tumor cells by promoting the proliferation of CD8+ cells [50]. The antitumor role of memory CD4+ T cells is also supported by previous evidence showing that increased disease-free survival of BC patients is directly related to an increase in activated memory CD4+ T cells [28]. Macrophages are an important type of immune cell in the tumor microenvironment and mainly include classically activated M1 macrophages and alternatively activated M2 macrophages [51]. Generally, the prooncogenic phenotype of M2-type macrophages promotes the occurrence and development of tumors by promoting tumor cell proliferation, inhibiting the tumor immune microenvironment, modulating matrix remodeling, and enhancing tumor angiogenesis [52–54]. In contrast, M1 macrophages are activated by Th1 cell factors; secrete inducible inductors, such as IL-12, iNOS, and TNF-α; and play a proinflammatory and antitumor role in the tumor microenvironment [29]. In our study, we demonstrated that the increased infiltrated immune cells in RYR2-mutant BC were all antitumorigenic immune cells, and CD8 T cells were positively correlated with activated memory CD4 T cells and M1 macrophages, while CD8 T cells, activated memory CD4 T cells, and M1 macrophages were negatively correlated with protumorigenic M2 macrophages. Thus, we suggest that the change in tumor-infiltrating immune cells induced by RYR2 contributes to antitumor immunity in BC.
This research has some limitations. First, due to the lack of clinical data in the ICGC database, we could not determine whether RYR2 mutations are also associated with prognosis and tumor immunity in European Union patients. Second, only informatics analyses were conducted in this study, and further experimental validations are needed. Third, we did not classify the different BC subtypes in this study. However, the mutated genes and immune infiltrating cells in different subtypes may be different [20, 55].
In summary, antitumor immune cells are suppressed in the tumor microenvironment, leading to a decrease in and low activity of antitumor immune cells among tumor-infiltrated cells [27, 56]. This is also the root cause of immunotherapy failure. In our study, we combined somatic gene mutation and TMB analyses and found that RYR2 is frequently mutated in BC. RYR2 mutation was related to higher TMB and better patient prognosis. More importantly, RYR2 mutation induced an antitumor immune response. These findings reveal that RYR2 mutation could serve as a biomarker to predict the immune response in BC.
Acknowledgments
This work was supported by The General Project of Chongqing Natural Science Foundation, Chongqing Science and Technology Commission, China (grant number cstc2021jcyj-msxmX0153 (Linglong Peng)); the Key Medical Research Project of Chongqing Health Planning Commission, China (grant number 2017ZDXM012 (Jijian Wang)); The Free Exploring Basic Research Project of Science and Technology of Sichuan Province, China (grant number 2020YJ0186 (Yongfu Xiong)); and The Doctoral Research Startup Fund Project of North Sichuan Medical College, Sichuan, China (grant number CBY20-QD03 (He Zhou)).
Abbreviations
- BC:
Breast cancer
- CTLs:
Cytotoxic T lymphocytes
- ER:
Estrogen receptor
- FDA:
Food and Drug Administration
- GSEA:
Gene set enrichment analysis
- HER2:
Human epidermal growth factor receptor-2
- ICGC:
International Cancer Genome Consortium
- RYR2:
Ryanodine receptor 2
- OS:
Overall survival
- PD-1:
Programmed cell death receptor 1
- PD-L1:
Programmed cell death ligand 1
- PR:
Progesterone receptor
- TCGA:
The Cancer Genome Atlas
- TMB:
Tumor mutant burden
- TNBC:
Triple-negative breast cancer.
Contributor Information
Haitao Gu, Email: guhaitao_cqmu@163.com.
Jijian Wang, Email: wangjijian_cqmu@163.com.
Linglong Peng, Email: penglinglong_cqmu@cqmu.edu.cn.
Data Availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request. Data used included The Cancer Genome Atlas (TCGA, http://portal.gdc.cancer.gov/projects) and International Cancer Genome Consortium (ICGC, http://dcc.icgc.org/releases/current/Projects).
Consent
Consent is not applicable.
Conflicts of Interest
The authors declare that no competing interests exist.
Authors' Contributions
L. Peng, H. Gu, and J. Wang designed the research; Z. Xu and L. Xiang prepared the figures and drafted the manuscript; L. Peng, Z. Xu, L. Xiang, H. Gu, J. Wang, Y. Xiong, R. Wang, and H. Zhou analyzed the data; L. Peng, Z. Xu, and J. Wang contributed analytic tools and finalized the manuscript. All authors have read and approved the final manuscript.
Supplementary Materials
Supplementary File 1: detailed clinical information for this study.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA:a Cancer Journal for Clinicians . 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Waks A. G., Winer E. P. Breast cancer treatment: a review. Journal of the American Medical Association . 2019;321(3):288–300. doi: 10.1001/jama.2018.19323. [DOI] [PubMed] [Google Scholar]
- 3.Wei S. C., Duffy C. R., Allison J. P. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discovery . 2018;8(9):1069–1086. doi: 10.1158/2159-8290.CD-18-0367. [DOI] [PubMed] [Google Scholar]
- 4.Balar A. V., Weber J. S. PD-1 and PD-L1 antibodies in cancer: current status and future directions. Cancer Immunology Immunotherapy . 2017;66(5):551–564. doi: 10.1007/s00262-017-1954-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nanda R., Chow L. Q., Dees E. C., et al. Pembrolizumab in patients with advanced triple-negative breast cancer: phase Ib KEYNOTE-012 study. Journal of Clinical Oncology . 2016;34(21):2460–2467. doi: 10.1200/JCO.2015.64.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelderman S., Schumacher T. N., Haanen J. B. Acquired and intrinsic resistance in cancer immunotherapy. Molecular Oncology . 2014;8(6):1132–1139. doi: 10.1016/j.molonc.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solinas C., Gombos A., Latifyan S., Piccart-Gebhart M., Kok M., Buisseret L. Targeting immune checkpoints in breast cancer: an update of early results. ESMO Open . 2017;2(5, article e000255) doi: 10.1136/esmoopen-2017-000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Topalian S. L., Drake C. G., Pardoll D. M. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell . 2015;27(4):450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buqué A., Bloy N., Aranda F., et al. Trial watch: immunomodulatory monoclonal antibodies for oncological indications. Oncoimmunology . 2015;4(4, article e1008814) doi: 10.1080/2162402X.2015.1008814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M., Sun H., Zhao S., et al. Expression of PD-L1 and prognosis in breast cancer: a meta-analysis. Oncotarget . 2017;8(19):31347–31354. doi: 10.18632/oncotarget.15532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mansfield A. S., Aubry M. C., Moser J. C., et al. Temporal and spatial discordance of programmed cell death-ligand 1 expression and lymphocyte tumor infiltration between paired primary lesions and brain metastases in lung cancer. Annals of Oncology . 2016;27(10):1953–1958. doi: 10.1093/annonc/mdw289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng J., Hamanishi J., Matsumura N., et al. Chemotherapy induces programmed cell death-ligand 1 overexpression via the nuclear factor-κB to foster an immunosuppressive tumor microenvironment in ovarian cancer. Cancer Research . 2015;75(23):5034–5045. doi: 10.1158/0008-5472.CAN-14-3098. [DOI] [PubMed] [Google Scholar]
- 13.Büttner R., Gosney J. R., Skov B. G., et al. Programmed death-ligand 1 immunohistochemistry testing: a review of analytical assays and clinical implementation in non-small-cell lung cancer. Journal of Clinical Oncology . 2017;35(34):3867–3876. doi: 10.1200/JCO.2017.74.7642. [DOI] [PubMed] [Google Scholar]
- 14.Chalmers Z. R., Connelly C. F., Fabrizio D., et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Medicine . 2017;9(1):p. 34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsushita H., Vesely M. D., Koboldt D. C., et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature . 2012;482(7385):400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hellmann M. D., Ciuleanu T. E., Pluzanski A., et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. The New England Journal of Medicine . 2018;378(22):2093–2104. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodman A. M., Kato S., Bazhenova L., et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Molecular Cancer Therapeutics . 2017;16(11):2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenberg J. E., Hoffman-Censits J., Powles T., et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. The Lancet . 2016;387(10031):1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yarchoan M., Hopkins A., Jaffee E. M. Tumor mutational burden and response rate to PD-1 inhibition. The New England Journal of Medicine . 2017;377(25):2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narang P., Chen M., Sharma A. A., Anderson K. S., Wilson M. A. The neoepitope landscape of breast cancer: implications for immunotherapy. BMC Cancer . 2019;19(1):p. 200. doi: 10.1186/s12885-019-5402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas A., Routh E. D., Pullikuth A., et al. Tumor mutational burden is a determinant of immune-mediated survival in breast cancer. Oncoimmunology . 2018;7(10, article e1490854) doi: 10.1080/2162402X.2018.1490854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu G., Pei L., Li Y., Gou X. EP300 mutation is associated with tumor mutation burden and promotes antitumor immunity in bladder cancer patients. Aging . 2020;12(3):2132–2141. doi: 10.18632/aging.102728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayakonda A., Lin D. C., Assenov Y., Plass C., Koeffler H. P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Research . 2018;28(11):1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A., Tamayo P., Mootha V. K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of The United States of America . 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman A. M., Liu C. L., Green M. R., et al. Robust enumeration of cell subsets from tissue expression profiles. Nature Methods . 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang F., Tang C., Gao X., Xu J. Identification of a six-gene signature associated with tumor mutation burden for predicting prognosis in patients with invasive breast carcinoma. Annals of Translational Medicine . 2020;8(7):p. 453. doi: 10.21037/atm.2020.04.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahmoud S. M., Paish E. C., Powe D. G., et al. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. Journal of Clinical Oncology . 2011;29(15):1949–1955. doi: 10.1200/JCO.2010.30.5037. [DOI] [PubMed] [Google Scholar]
- 28.Zhang S. C., Hu Z. Q., Long J. H., et al. Clinical implications of tumor-infiltrating immune cells in breast cancer. Journal of Cancer . 2019;10(24):6175–6184. doi: 10.7150/jca.35901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hao N. B., Lu M. H., Fan Y. H., Cao Y. L., Zhang Z. R., Yang S. M. Macrophages in tumor microenvironments and the progression of tumors. Clinical & Developmental Immunology . 2012;2012, article 948098:1–11. doi: 10.1155/2012/948098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berridge M. J., Lipp P., Bootman M. D. The versatility and universality of calcium signalling. Nature Reviews Molecular Cell Biology . 2000;1(1):11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 31.Cui C., Merritt R., Fu L., Pan Z. Targeting calcium signaling in cancer therapy. Acta Pharmaceutica Sinica B . 2017;7(1):3–17. doi: 10.1016/j.apsb.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kondratskyi A., Yassine M., Kondratska K., Skryma R., Slomianny C., Prevarskaya N. Calcium-permeable ion channels in control of autophagy and cancer. Frontiers in Physiology . 2013;4:p. 272. doi: 10.3389/fphys.2013.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu H., Zhang H., Jin F., et al. Elevated Orai1 expression mediates tumor-promoting intracellular Ca2+ oscillations in human esophageal squamous cell carcinoma. Oncotarget . 2014;5(11):3455–3471. doi: 10.18632/oncotarget.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding Z., Yuan J., Liang Y., et al. Ryanodine receptor type 2 plays a role in the development of cardiac fibrosis under mechanical stretch through TGFβ-1. International Heart Journal . 2017;58(6):957–961. doi: 10.1536/ihj.16-572. [DOI] [PubMed] [Google Scholar]
- 35.Mariot P., Prevarskaya N., Roudbaraki M. M., et al. Evidence of functional ryanodine receptor involved in apoptosis of prostate cancer (LNCaP) cells. The Prostate . 2000;43(3):205–214. doi: 10.1002/(SICI)1097-0045(20000515)43:3<205::AID-PROS6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 36.Xu N., Zhang D., Chen J., He G., Gao L. Low expression of ryanodine receptor 2 is associated with poor prognosis in thyroid carcinoma. Oncology Letters . 2019;18(4):3605–3612. doi: 10.3892/ol.2019.10732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis F. M., Parsonage M. T., Cabot P. J., et al. Assessment of gene expression of intracellular calcium channels, pumps and exchangers with epidermal growth factor-induced epithelial-mesenchymal transition in a breast cancer cell line. Cancer Cell International . 2013;13(1):p. 76. doi: 10.1186/1475-2867-13-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Femi O. F. Genetic alterations and PIK3CA gene mutations and amplifications analysis in cervical cancer by racial groups in the United States. International Journal of Health Science . 2018;12(1):28–32. [PMC free article] [PubMed] [Google Scholar]
- 39.Cai W., Zhou D., Wu W., et al. MHC class II restricted neoantigen peptides predicted by clonal mutation analysis in lung adenocarcinoma patients: implications on prognostic immunological biomarker and vaccine design. BMC Genomics . 2018;19(1):p. 582. doi: 10.1186/s12864-018-4958-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitt K., Molfenter B., Laureano N. K., et al. Somatic mutations and promotor methylation of the ryanodine receptor 2 is a common event in the pathogenesis of head and neck cancer. International Journal of Cancer . 2019;145(12):3299–3310. doi: 10.1002/ijc.32481. [DOI] [PubMed] [Google Scholar]
- 41.Wei Y., Wang X., Zhang Z., et al. Impact of NR5A2 and RYR2 3'UTR polymorphisms on the risk of breast cancer in a Chinese Han population. Breast Cancer Research and Treatment . 2020;183(1):1–8. doi: 10.1007/s10549-020-05736-w. [DOI] [PubMed] [Google Scholar]
- 42.Gubin M. M., Artyomov M. N., Mardis E. R., Schreiber R. D. Tumor neoantigens: building a framework for personalized cancer immunotherapy. Journal of Clinical Investigation . 2015;125(9):3413–3421. doi: 10.1172/JCI80008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mittal S., Brown N. J., Holen I. The breast tumor microenvironment: role in cancer development, progression and response to therapy. Expert Reviews of Molecular Diagnostics . 2018;18(3):227–243. doi: 10.1080/14737159.2018.1439382. [DOI] [PubMed] [Google Scholar]
- 44.Junttila M. R., de Sauvage F. J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature . 2013;501(7467):346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 45.Renner K., Singer K., Koehl G. E., et al. Metabolic hallmarks of tumor and immune cells in the tumor microenvironment. Frontiers in Immunology . 2017;8:p. 248. doi: 10.3389/fimmu.2017.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee H. J., Park I. A., Song I. H., et al. Tertiary lymphoid structures: prognostic significance and relationship with tumour-infiltrating lymphocytes in triple-negative breast cancer. Journal of Clinical Pathology . 2016;69(5):422–430. doi: 10.1136/jclinpath-2015-203089. [DOI] [PubMed] [Google Scholar]
- 47.Emens L. A. Breast cancer immunobiology driving immunotherapy: vaccines and immune checkpoint blockade. Expert Review of Anticancer Therapy . 2012;12(12):1597–1611. doi: 10.1586/era.12.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cimas F. J., Manzano A., Baliu-Piqué M., et al. Genomic mapping identifies mutations in RYR2 and AHNAK as associated with favorable outcome in basal-like breast tumors expressing PD1/PD-L1. Cancers . 2020;12(8):p. 2243. doi: 10.3390/cancers12082243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farhood B., Najafi M., Mortezaee K. CD8+ cytotoxic T lymphocytes in cancer immunotherapy: a review. Journal of Cellular Physiology . 2019;234(6):8509–8521. doi: 10.1002/jcp.27782. [DOI] [PubMed] [Google Scholar]
- 50.Novy P., Quigley M., Huang X., Yang Y. CD4 T cells are required for CD8 T cell survival during both primary and memory recall responses. Journal of Immunology . 2007;179(12):8243–8251. doi: 10.4049/jimmunol.179.12.8243. [DOI] [PubMed] [Google Scholar]
- 51.Lee H. W., Choi H. J., Ha S. J., Lee K. T., Kwon Y. G. Recruitment of monocytes/macrophages in different tumor microenvironments. Biochimica ET Biophysica Acta . 2013;1835(2):170–179. doi: 10.1016/j.bbcan.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 52.Laoui D., Movahedi K., van Overmeire E., et al. Tumor-associated macrophages in breast cancer: distinct subsets, distinct functions. International Journal of Developmental Biology . 2011;55(7-8-9):861–867. doi: 10.1387/ijdb.113371dl. [DOI] [PubMed] [Google Scholar]
- 53.Chanmee T., Ontong P., Konno K., Itano N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers . 2014;6(3):1670–1690. doi: 10.3390/cancers6031670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sica A., Schioppa T., Mantovani A., Allavena P. Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anti-cancer therapy. European Journal of Cancer . 2006;42(6):717–727. doi: 10.1016/j.ejca.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 55.Solinas C., Carbognin L., De Silva P., Criscitiello C., Lambertini M. Tumor-infiltrating lymphocytes in breast cancer according to tumor subtype: current state of the art. Breast . 2017;35:142–150. doi: 10.1016/j.breast.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 56.Tang H., Qiao J., Fu Y. X. Immunotherapy and tumor microenvironment. Cancer Letters . 2016;370(1):85–90. doi: 10.1016/j.canlet.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1: detailed clinical information for this study.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request. Data used included The Cancer Genome Atlas (TCGA, http://portal.gdc.cancer.gov/projects) and International Cancer Genome Consortium (ICGC, http://dcc.icgc.org/releases/current/Projects).


