Abstract
This work describes the first epidemiological survey of Burkholderia cepacia involved in pulmonary infections among the Portuguese population with cystic fibrosis (CF) who attended the major CF treatment Center in Lisbon at Sta. Maria Hospital from 1995 to the end of 1997. The characterization of the genomic relatedness of the isolates was based on the analysis of their ribopatterns (with EcoRI) followed by construction of a ribotype-based phylogenetic tree. This study was complemented with macrorestriction fragment analysis by pulsed-field gel electrophoresis. After optimization of the solid growth medium, we found that exopolysaccharide (EPS) production by B. cepacia CF isolates is not as rare a phenomenon as was thought before; indeed, 70% of the isolates examined were EPS producers.
Burkholderia cepacia, originally described as a plant pathogen (3), has become an important opportunistic pathogen in patients with cystic fibrosis (CF) (10, 11, 14, 26) and an infrequent cause of nosocomial infection in patients without CF (13). There is increased evidence of transmission among patients with CF by social contact (9) The environment is also regarded as a potential source of strains capable of infecting these patients (4, 9). However, one risk factor for B. cepacia acquisition by patients with CF in the United States appeared to be hospitalization, and a recent hospital outbreak apparently involved patients both with and without CF (13). There are no clear data about the possible contribution of the polysaccharide produced extracellularly by specific B. cepacia isolates (1, 5, 6, 20) to the colonization and persistence of the species in the infected host, as was ascribed to alginate in the respiratory infection of patients with CF by Pseudomonas aeruginosa (10). Nevertheless, the mucoid colonial morphotype is thought to be relatively rare among B. cepacia CF isolates (10). However, the exopolysaccharides (EPSs) produced by several gram-negative bacterial species infecting plants or animals have been considered important virulence factors due to their contributions to the colonization and persistence of the producing microorganism in the infected host (19).
In Portugal, the first clearly identified B. cepacia isolate recovered from the sputum of a patient with CF attending the CF treatment center at Sta. Maria Hospital in Lisbon was found in 1992. The CF Center is attended by approximately 85% of the population with CF residing in the Lisbon area and by patients with CF living in the south of Portugal and in the Madeira and Azores Islands. From 9 of a total of 140 patients with CF registered at the CF Center between 1995 and the end of 1997, 23 isolates capable of growing on the selective Burkholderia Cepacia Selectatab medium (Mast Diagnostics, Merseyside, United Kingdom) were recovered at the Laboratory of Bacteriology of Sta. Maria Hospital. The isolates (Table 1) were obtained on different dates of isolation from bronchial secretions of different patients with CF, each identified by a letter (Table 1), after 3 days of incubation at 35°C followed by another day of incubation at room temperature in the selective medium Burkholderia Cepacia Selectatab. The four putative B. cepacia isolates obtained during 1996 were lost due to a prolonged storage period at the hospital and could not be examined in this study. To confirm that all 19 CF isolates obtained belonged to the species B. cepacia, the commercial systems API 20NE (Biomerieux, Marcy L'Etoile, France) and BIOLOG gram-negative (GN) (Biolog Inc., Hayward, Calif.) were used. The isolates identified as B. cepacia by the two systems were submitted to additional confirmation based on PCR amplification using the specific oligonucleotide primers CMG-16-1, C-16-21001, CMG-23-1, CM-16-2458, and G-23-2 proposed by Bauernfeind et al. (2). Further efforts to confirm the identification of the 19 isolates were undertaken at the Instituto Superior Técnico (IST) laboratory before comparing their genomic relatedness and their abilities to produce EPS, due to the increasing evidence of misidentification of B. cepacia by standard laboratory procedures (2). This analysis revealed that only the 16 IST isolates listed in Table 1 could be confirmed to be B. cepacia. They were stored at −70°C in 40% (wt/vol) glycerol, and when in use, the cultures were routinely maintained on Pseudomonas isolation agar (Difco, Detroit, Mich.) plates.
TABLE 1.
Profiles of CF isolates used in the studya
| Patient or reference | Data of isolation | B. cepacia isolate | Ribopattern | EPS [g of total sugar · (g of cell protein)−1]b
|
|
|---|---|---|---|---|---|
| A | S | ||||
| A | August 95 | IST401 | 2 | 0.24 ± 0.09 | 4.30 ± 0.11 |
| A | September 95 | IST404 | 3 | ND | 0.60 ± 0.06 |
| B | April 95 | IST402 | 1 | ND | 0.80 ± 0.08 |
| B | April 95 | IST409 | 1 | ND | 1.1 ± 0.5 |
| C | February 95 | IST403 | 7 | ND | 0.10 ± 0.02 |
| D | December 95 | IST407 | 3 | ND | ND |
| E | December 95 | IST406 | 2 | 0.36 ± 0.02 | 2.5 ± 0.20 |
| F | January 95 | IST408 | 2 | 0.39 ± 0.01 | 2.20 ± 0.15 |
| G | April 95 | IST405 | 4 | ND | ND |
| G | June 95 | IST410 | 5 | ND | ND |
| G | February 95 | IST411 | 6 | ND | ND |
| H | January 97 | IST412 | 1 | ND | 0.50 ± 0.04 |
| H | March 97 | IST413 | 1 | ND | 0.80 ± 0.02 |
| H | May 97 | IST414 | 1 | ND | 0.80 ± 0.06 |
| H | July 97 | IST415 | 1 | ND | 0.80 ± 0.09 |
| I | September 97 | IST416 | 8 | ND | 0.40 ± 0.02 |
| Type strain | ATCC 25416 | 12 | ND | 4.00 ± 0.20 | |
| 11 | J2315 | 11 | ND | ND | |
| 23 | C1394 | 9 | ND | ND | |
| 27 | C1579 | 10 | ND | 0.50 ± 0.01 | |
CF patient hosts are identified by letters. Ribopatterns are shown in Fig. 3.
EPS produced (expressed as grams of total sugars present in the ethanol-precipitable material per gram of cell protein) after 5 days of incubation at 30°C on the surface of A or S solid medium by B. cepacia isolates. ND, not detectable.
B. cepacia prevalence rate and clinical course.
The value of the 3-year cumulative prevalence rate found during the period of surveillance (6.4%) did not suggest epidemic transmission of B. cepacia. This value is close to the prevalence rate observed in other surveillance studies carried out in other countries (11, 21, 22, 25), while the colonization rates in one large United Kingdom regional center, experiencing the spread of an epidemic strain, reached values close to the 40% prevalence experienced in a major North American CF center (9). Consistent with previous reports, the clinical courses of the nine Portuguese patients with CF infected with B. cepacia who were followed during this study were variable: patients A, D, and I exhibited stable pulmonary function after infection with B. cepacia; patient G died from the “cepacia syndrome”; patients B and H showed an increased deterioration of pulmonary function which was clearly associated with infection with the same B. cepacia isolate that continued to be isolated during 1998, and patients E and F died, although their deaths were considered unrelated to B. cepacia infection. Patient C moved to another geographical area, which prevented his clinical observation during the full duration of the present study.
Molecular typing.
The genetic relatedness of the 16 B. cepacia isolates from Portuguese patients with CF was compared by ribotyping complemented with macrorestriction fragment analysis by pulsed-field gel electrophoresis (PFGE). The environmental type strain ATCC 25416 and the three highly transmissible epidemic strains J2315 (a representative of the highly epidemic Edinburgh-Toronto lineage [11]) and C1394 and C1579 (epidemic representatives of outbreaks of B. cepacia among patients with CF attending CF centers at Manchester [23] and Glasgow [27], United Kingdom, respectively) were used as reference strains. For ribotyping analysis, the isolation of total DNA, restriction with EcoRI (Gibco BRL Life Technologies, Gaithersburg, Md.), DNA blotting, and hybridization with the acetylaminofluorene-labeled 16S + 23S rRNA probe from E. coli (Eurogentec, Seraing, Belgium) were carried out as previously described (16). The sizes of the hybridization restriction fragments were estimated with DNA Simdex software from GenetX with RaoulI (Eurogentec) as the reference molecular size marker. In order to transform the restriction fragment length polymorphism (RFLP) data obtained from ribotyping for numerical analysis, each band of an RFLP profile obtained for each isolate under study was treated as a unit character and scored as 1 when present or 0 when absent across all isolates. Variations in staining intensity were not taken into account for the construction of the matrix. From the resulting binary data, a triangular similarity matrix was generated using the Dice similarity coefficient, SD (24). The construction of the dendrogram from the similarity matrix was performed by the UPGMA method (unweighted-pair group method using arithmetic means), which forms groups by successively pairing similar ribopatterns according to the magnitudes of their observed distances (24). The software package used was the program NTSYSpc version 2.02 (Exeter Software, Inc.). The cophenetic correlation, r, between the similarity and the cophenetic matrices was 0.968. The use of the Jaccard similarity coefficient or the change of the order in which isolate data were input into the software did not lead to significant changes in the final dendrogram. The UPGMA method was chosen because it has been shown to give the most accurate representation of data (18).
Chromosomal analysis by PFGE was carried out as described previously (16). Genomic DNA was prepared using approximately 5 × 108 cells (from an overnight culture of each strain under analysis) per ml of agarose (0.5% [wt/vol]) plugs. Digestions with the rare-cutting endonucleases AseI or AflII (New England Biolabs, Beverly, Mass.), were carried out by using 20 U of enzyme per one-third of a plug, according to the manufacturer's instructions. The plugs were loaded onto 1% (wt/vol) agarose gel prepared with the running buffer, 0.5× Tris-borate-EDTA buffer, pH 8.0. The macrorestriction fragments generated were separated by PFGE using the Gene Navigator (Pharmacia, Uppsala, Sweden) apparatus under appropriate conditions (180 V for 22 h with pulse times in the interpolation mode ranging from 2 to 50 s). Bacteriophage lambda concatamers (New England Biolabs) were used as size standards. After electrophoretic separation, the gels were stained with ethidium bromide and photographed under UV illumination.
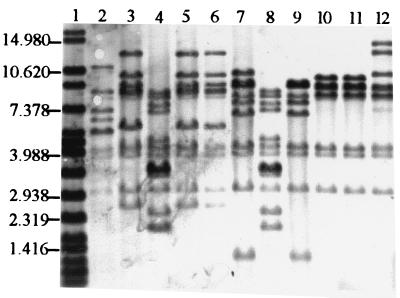
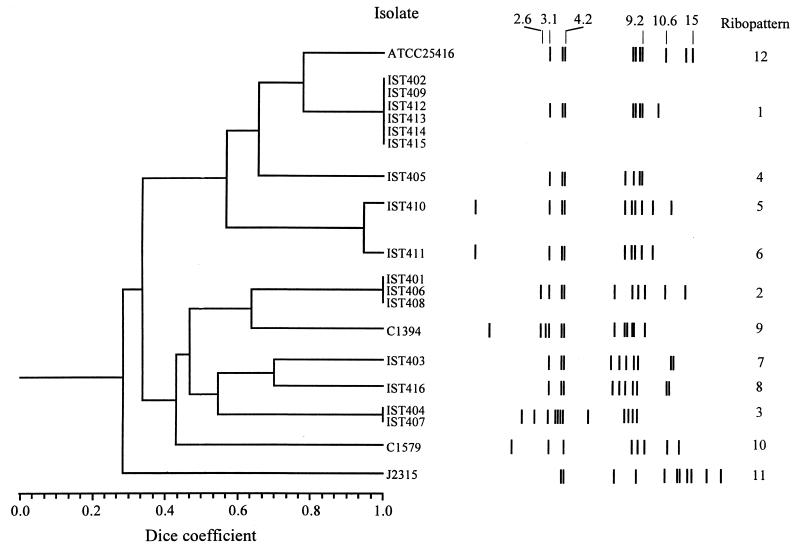
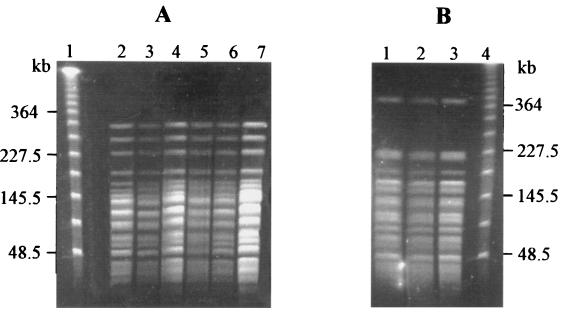
Eight distinct ribopatterns were found with the 16 Portuguese CF isolates examined. An example of the results obtained is given in Fig. 1, and the ribopatterns are schematically shown in Fig. 2. The ribopatterns obtained exhibited 7 to 12 bands, ranging from 1.25 to 27.0 kb. One band, approximately 4.2 kb, was common to all the isolates under study. Bands of approximately 4.0 and 3.1 kb were also consistently found for nearly all isolates, the exceptions being C1579 and J2315 (Fig. 1 and 2). The conserved 4.2-kb EcoRI band was also reported for all the B. cepacia isolates from patients at six North American CF centers (13). However, the other conserved EcoRI band in these North American B. cepacia isolates was reported to be 2.6 kb. The different size of the closer conserved band that was calculated in the present study (3.1 kb) may result from the different molecular size reference marker used. The genomic diversity of the isolates examined should be considered to be higher than the number that 8 ribopatterns for 16 isolates may suggest. Indeed, four of the isolates (IST412, IST413, IST414, and IST415), were serial isolates from the same patient, H (from January to July 1997), and exhibited the same ribopattern, 1; their RFLP-PFGE profiles with AseI and AflII were also indistinguishable (Fig. 3A and results not shown). Isolates IST404 and IST407, which gave rise to the same ribopattern (number 3), were obtained from patients A and D, but it was impossible to compare their RFLP-PFGE patterns because after repeated attempts these isolates produced lanes with smeared DNA, probably due to the degradation of the DNA by endogenous endonucleases. Additionally, three other isolates (IST401, IST406, and IST408) with the same ribopattern 2 were indistinguishable by RFLP-PFGE with both AseI (Fig. 3B) and AflII (results not shown); they were isolated from three different patients (A, E, and F). Because these patients were hospitalized in the Infectious Diseases Unit of Sta. Maria Hospital, although not simultaneously, during the temporary closure of the CF hospitalization unit for repairs, these typing results strongly suggest that the three isolates examined are the same strain that was acquired during hospitalization. Interestingly, this strain is genetically related to the CF isolate C1394 (65% similarity), a representative of a Manchester, United Kingdom, outbreak (23). It must be stressed that B. cepacia-positive patients are routinely segregated from B. cepacia-negative patients at Sta. Maria Hospital CF Center.
FIG. 1.
Example of the ribopatterns obtained with the B. cepacia isolates indicated below after digestion of total DNA with EcoRI and hybridization with the acetylaminofluorene-labeled 16S + 23S rRNA from E. coli. Lane 1, molecular size standard RaoulI; lanes 2 to 12, B. cepacia isolates IST403, IST406, IST407, IST401, IST408, IST410, IST404, IST411, IST409, IST402, and ATCC25416, respectively. Sizes in kilobases are given on the left.
FIG. 2.
Dendrogram showing the results of clustering analysis using UPGMA for the B. cepacia strains and isolates under study. The numbers in the horizontal axis indicate the percentage of similarity as determined with the Dice coefficient for the ribopatterns schematically represented on the left, with molecular sizes (in kilobases) arranged in a logarithmic scale.
FIG. 3.
Comparison of the AseI macrorestriction fragment patterns of the genomic DNAs from B. cepacia isolates separated by PFGE. (A) Lanes: 1, size standard of concatamerized phage λ DNA; 2, IST409; 3, IST412; 4, IST413; 5, IST414; 6, IST415; 7, IST402. (B) Lanes: 1, IST401; 2, IST406; 3, IST408; 4, size standard of concatamerized phage λ DNA.
A cluster analysis was undertaken of the ribopatterns generated during this study. Although we also had available the macrorestriction fragment profiles obtained by PFGE for all the Portuguese isolates examined in this study, due to the large number of bands generated, these profiles are difficult to analyze (Fig. 3A and B), and their cluster analysis was not considered. The inspection of the ribotype-based phylogenetic tree that was constructed suggests the possible acquisition of B. cepacia from environmental sources by one-third of the patients with CF examined during the period of surveillance. Indeed, the ribopattern of the environmental strain used as a reference, B. cepacia ATCC 25416, was very closely related (80% similarity) to the ribopattern generated by six Portuguese isolates that were sequentially isolated from the same two patients, B (IST402 and IST409) and H (IST412, IST413, IST414, and IST415), who resided in distinct geographical areas (on Madeira Island [B] and in the Lisbon area [H]) and who were never in contact or hospitalized. Isolates IST405, IST410, and IST411, with a similarity lower than 60%, were obtained (from February to June 1995) from patient G, who died of the cepacia syndrome. They gave rise to different, although related, ribotypes, particularly the isolates IST410 and IST411 (Fig. 3). Although it is possible that this patient harbored two or three different strains, we favor the hypothesis that the different ribotypes may result from genomic variations of the same colonizing strain. The results of this first epidemiological survey study of B. cepacia involved in pulmonary infections among the Portuguese population with CF did not reveal genomic relatedness between the Portuguese isolates and the Edinburgh-Toronto epidemic strain J2315 (10) or the epidemic Glasgow isolate C1579 (27).
EPS biosynthesis.
The mucoidy of B. cepacia isolates was assessed by comparing the morphologies of isolated colonies formed after incubation for 5 days at 30°C in the three different media examined, supplemented with 20 g of agar (Iberagar; Coina, Portugal) per liter. CDM and A media were described in the literature as leading to the production of EPS by a specific B. cepacia CF isolate (1, 20), and S medium was successfully used in the IST laboratory to overproduce the EPS gellan gum from Sphingomonas paucimobilis ATCC 31461 (8, 15, 17). Moreover, the use of S agar plates allowed the differentiation of the mucoid colonial morphotypes of S. paucimobilis variants, which produced mutated gellan gum in different yields (15, 17). CDM contained, in grams per liter, NaCl (0.175), KCl (0.224), (NH4)2SO4 (0.396), K2HPO4 (0.205), and glucose (10). Medium A contained, in grams per liter, only yeast extract (2) and glucose (10). S medium contained, in grams per liter, Na2HPO4 (10), KH2PO4 (3), K2SO4 (1), NaCl (1), MgSO4 · 7H2O (0.2), yeast extract (Difco) (1), Casamino Acids (Difco) (1), CaCl2 · 2H2O (0.01), FeSO4 · 7H2O (0.001), and glucose (20). Isolated colonies were obtained by spreading, after suitable dilution, 100 μl of liquid culture onto the surface of the solid growth medium. The liquid culture used to inoculate the agar plates resulted from overnight cultivation at 30°C with orbital agitation (250 revolutions · min−1) in Luria broth (Sigma) medium. EPS production by the different isolates was quantified after 5 days of incubation at 30°C of confluent cell growth in either A or S solid medium. The plates were scraped, the material was resuspended in 0.9% (wt/vol) NaCl by vortexing, and the cells were separated by centrifugation at 20,000 × g for 15 min. The EPS was precipitated from the cell-free supernatant by the addition of 2 volumes of cold ethanol, air dried, and redissolved in distilled water. The total sugar content was assessed by the phenol-sulfuric acid method (7) using the EPS produced by isolate IST408 as a standard. For this purpose, the EPS produced by isolate IST408 was further dialyzed against distilled water at 4°C for 24 h and recovered by freeze-drying. The cell pellets obtained from each plate were washed once with 0.9% NaCl, and the protein content was quantified by the biuret method (12) using bovine serum albumin fraction V (Merck) as a standard. The results of EPS production were expressed as grams of total sugars per gram of protein and are the means of at least three independent cultivations and of three determinations of total sugar and protein contents in each independent sample. After 5 days of incubation at 30°C on solid CDM medium, no mucoid colonies were detected by visual inspection. Confirming this observation, we were unable to detect any ethanol-precipitable material from the cell-free supernatants. However, when grown on solid medium A, isolates IST401, IST406, and IST408 formed colonies that evidenced a very clear mucoid phenotype, while the other isolates maintained the nonmucoid appearance (results not shown). The indications of this visual observation were confirmed by quantitative analysis of EPS production (Table 1). The use of solid S medium to cultivate the whole collection of isolates allowed the identification of 12 Portuguese isolates (out of 16) giving rise to mucoid colonies after 5 days of incubation at 30°C. The quantification of the EPS produced by the isolates during growth in this solid medium confirmed the colony morphotypes (Table 1). Only the isolates IST405, IST407, IST410, and IST411 (4 out of 16) were consistently unable to produce EPS in any of the three media tested. Among the three CF epidemic isolates used as reference strains, only one was able to produce EPS in S medium while the environmental type strain ATCC 25416 produced high levels of EPS (Table 1). In general, the relative level of EPS produced on S plates by the different isolates was reproduced in S liquid culture carried out in shake flasks (results not shown). The structural analysis of the EPS produced by IST408 in A agar plates indicated that the polymer is very similar, if not identical, to the EPS produced by a French clinical isolate (5; P. Cescutti, M. Bosco, F. Picotti, J. A. Richau, J. H. Leitão, and I. Sá-Correia, Abstr. 10th Eur. Carbohydr. Symp., abstr. OB05, p. 85, 1999), and a detailed description will be published elsewhere. Moreover, the sugar composition of the EPS from B. cepacia IST408 (composed of glucose, mannose, rhamnose, galactose, and glucuronic acid in the molar ratio 1.0:1.0:1.0:3.0:1.0 [Cescutti et al., Abstr. 10th Eur. Carbohydr. Symp.]) is similar to the compositions of the EPSs produced by B. cepacia isolates from patients with CF in the United States (20) and in the United Kingdom (1).
Mucoid colonial morphotypes are considered rare in both environmental and clinical isolates of B. cepacia (10). However, although in solid CDM medium all the isolates were nonmucoid and in solid A medium only 3 of the 16 isolates examined were mucoid, 70% of the isolates were mucoid in solid S medium. It should be noted that the calculated percentage of mucoid isolates among the B. cepacia isolates from the Portuguese patients with CF (75% in S agar plates) could be overvalued. In fact, four of the mucoid isolates examined were serial isolates from the same patient, H, and exhibited the same ribopattern; three of them were indiscernible by RFLP-PFGE. Two other mucoid isolates are presumably the same strain obtained on different dates from patient B, and the strain acquired by three different patients, probably during their hospitalization, was also mucoid. Nevertheless, the results of the present study suggest that the concept that the mucoid phenotype is rarely found among B. cepacia CF isolates may have resulted from the use of culture media unsuitable to the clear expression of EPS biosynthesis in B. cepacia. They also indicate that EPS production by B. cepacia might not be as rare as initially thought and suggest that the B. cepacia EPS may indeed play a role in the colonization and persistence of B. cepacia in the lung in CF, as ascribed to alginate in CF infection by P. aeruginosa.
Acknowledgments
This work was supported by Fundação para a Ciência e a Tecnologia (FCT), by FEDER and the PRAXIS XXI program (grant PRAXIS/PSAU/P/SAU/59/96), and Ph.D. and M.Sc. scholarships to J.R. and M.C., respectively.
We gratefully acknowledge the supply of B. cepacia J2315, C1394, and C1579 by J. R. W. Govan (U. of Edinburgh Medical School, Edinburgh, United Kingdom).
REFERENCES
- 1.Allison D G, Goldsbrough M J. Polysaccharide production in Pseudomonas cepacia. J Basic Microbiol. 1994;34:3–10. doi: 10.1002/jobm.3620340102. [DOI] [PubMed] [Google Scholar]
- 2.Bauernfeind A, Schneider I, Jungwirth R, Roller C. Discrimination of Burkholderia gladioli from other Burkholderia species detectable in cystic fibrosis patients by PCR. J Clin Microbiol. 1998;36:2748–2751. doi: 10.1128/jcm.36.9.2748-2751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burkholder W H. Sour skin, a bacterial rot of onion bulbs. Phytopathology. 1950;50:115–117. [Google Scholar]
- 4.Butler S L, Doherty C J, Hughes J E, Nelson J W, Govan J R W. Burkholderia cepacia and cystic fibrosis: do natural environments present a potential hazard? J Clin Microbiol. 1995;33:1001–1004. doi: 10.1128/jcm.33.4.1001-1004.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cérantola S, Lemassu-Jacquier A, Montrozier H. Structural elucidation of a novel exopolysaccharide produced by a mucoid clinical isolate of Burkholderia cepacia. Characterization of a trisubstituted glucuronic acid in a heptasaccharide repeating unit. Eur J Biochem. 1999;260:373–383. doi: 10.1046/j.1432-1327.1999.00171.x. [DOI] [PubMed] [Google Scholar]
- 6.Cérantola S, Marty N, Montrozier H. Structural studies of the acidic exopolysaccharide produced by a mucoid strain of Burkholderia cepacia, isolated from cystic fibrosis. Carbohydr Res. 1996;14:285. doi: 10.1016/s0008-6215(96)90170-6. :59–67. [DOI] [PubMed] [Google Scholar]
- 7.Dubois M, Gilles K A, Hamilton J K, Rebers P A, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- 8.Fialho A M, Martins L O, Donval M-L, Leitão J H, Ridout M J, Jay A J, Morris V J, Correia I S. Structures and properties of gellan polymers produced by Sphingomonas paucimobilis ATCC 31461 from lactose compared with those produced from glucose and from cheese whey. Appl Environ Microbiol. 1999;65:2485–2491. doi: 10.1128/aem.65.6.2485-2491.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Govan J R W, Brown P H, Maddison J, Doherty C J, Nelson J W, Dodd M, Greening A P, Webb A K. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet. 1993;342:15–19. doi: 10.1016/0140-6736(93)91881-l. [DOI] [PubMed] [Google Scholar]
- 10.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Govan J R W, Hughes J E, Vandamme P. Burkholderia cepacia: medical, taxonomic and ecological issues. J Med Microbiol. 1996;45:395–407. doi: 10.1099/00222615-45-6-395. [DOI] [PubMed] [Google Scholar]
- 12.Herbert D, Phipps P J, Strange R E. Chemical analysis of microbial cells. In: Norris J R, Ribbons D W, editors. Methods in microbiology. 5B. London, United Kingdom: Academic Press; 1971. pp. 209–344. [Google Scholar]
- 13.Holmes A, Nolan R, Taylor R, Finley R, Riley M, Jiang R-Z, Steinbach S, Goldstein R. An epidemic of Burkholderia cepacia transmitted between patients with and without cystic fibrosis. J Infect Dis. 1999;179:1197–1205. doi: 10.1086/314699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isles A, Maclusky I, Corey M, Gold R, Prober C, Fleming P, Levison H. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J Pediatr. 1984;104:206–210. doi: 10.1016/s0022-3476(84)80993-2. [DOI] [PubMed] [Google Scholar]
- 15.Jay A J, Colquhoun I J, Ridout M J, Brownsey G J, Morris V J, Fialho A M, Leitão J H, Correia I S. Analysis of structure and function of gellans with different substitution patterns. Carbohydr Polym. 1998;35:179–188. [Google Scholar]
- 16.Leitão J H, Alvim T, Correia I S. Ribotyping of Pseudomonas aeruginosa isolates from patients and water springs and genome fingerprinting of variants concerning mucoidy. FEMS Immunol Med Microbiol. 1996;13:287–292. doi: 10.1111/j.1574-695X.1996.tb00253.x. [DOI] [PubMed] [Google Scholar]
- 17.Martins L O, Fialho A M, Rodrigues P L, Correia I S. Gellan gum production and activity of biosynthetic enzymes in Sphingomonas paucimobilis mucoid and non-mucoid variants. Biotechnol Appl Biochem. 1996;23:47–54. [Google Scholar]
- 18.Priest F, Austin B. Numerical taxonomy. In: Priest F, Austin B, editors. Modern bacterial taxonomy. 2nd ed. London, United Kingdom: Chapman & Hall; 1993. pp. 14–49. [Google Scholar]
- 19.Roberts I S. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 20.Sage A, Linker A, Evans L R, Lessie T G. Hexose phosphate metabolism and exopolysaccharide formation in Pseudomonas cepacia. Curr Microbiol. 1990;20:191–198. [Google Scholar]
- 21.Segonds C, Chabanon G, Couetdic G, Michel-Briand Y, Bingen E. Epidemiology of pulmonary colonization with Burkholderia cepacia in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis. 1996;15:841–842. doi: 10.1007/BF01701534. [DOI] [PubMed] [Google Scholar]
- 22.Shreve M R, Butler S, Kaplowitz H J, Rabin H R, Stokes D, Light M, Regelmann W E. Impact of microbiology practice on cumulative prevalence of respiratory tract bacteria in patients with cystic fibrosis. J Clin Microbiol. 1999;37:753–757. doi: 10.1128/jcm.37.3.753-757.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simpson I N, Finlay J, Winstanley D J, Dewhurst N, Nelson N, Butler S, Govan J R W. Multi-resistance isolates possessing characteristics of both Burkholderia (Pseudomonas) cepacia and Burkholderia gladioli from patients with cystic fibrosis. J Antimicrob Chemother. 1994;34:353–361. doi: 10.1093/jac/34.3.353. [DOI] [PubMed] [Google Scholar]
- 24.Sneath P H A, Sokal R R. Numerical taxonomy. San Francisco, Calif: W. H. Freeman and Co.; 1973. pp. 129–137. [Google Scholar]
- 25.Taccetti G, Campana S. Microbiologic data overview of Italian cystic fibrosis patients. Eur J Epidemiol. 1997;13:323–327. doi: 10.1023/a:1007373700089. [DOI] [PubMed] [Google Scholar]
- 26.Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Govan J R W. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 27.Witheford M L, Wilkinson J D, McColl J H, Conlon F M, Michie J R, Evans T J, Paton J Y. Outcome of Burkholderia (Pseudomonas) cepacia colonisation in children with cystic fibrosis following a hospital outbreak. Thorax. 1995;50:1194–1198. doi: 10.1136/thx.50.11.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]