Abstract
Early warning signals (EWSs) aim to predict changes in complex systems from phenomenological signals in time series data. These signals have recently been shown to precede the emergence of disease outbreaks, offering hope that policymakers can make predictive rather than reactive management decisions. Here, using a novel, sequential analysis in combination with daily COVID-19 case data across 24 countries, we suggest that composite EWSs consisting of variance, autocorrelation and skewness can predict nonlinear case increases, but that the predictive ability of these tools varies between waves based upon the degree of critical slowing down present. Our work suggests that in highly monitored disease time series such as COVID-19, EWSs offer the opportunity for policymakers to improve the accuracy of urgent intervention decisions but best characterize hypothesized critical transitions.
Keywords: bifurcation, coronavirus, critical transition, forecasting, monitoring, pandemic
1. Introduction
As with many natural systems, the emergence of infectious disease is often sudden and nonlinear, making it difficult for policymakers to identify and appropriately manage threats [1,2]. Balancing the health risks posed by novel diseases and the possible economic impact of imposing mitigation strategies is therefore a complicated process ultimately dependent on the timing of strategy implementation [3].
The severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2), which causes the coronavirus disease 2019 (COVID-19), exemplifies this challenge. There has been widespread criticism, in response to the harm the pandemic has inflicted, of the speed and severity of the national strategies to the virus threat [4,5]. To date, governments have imposed a spectrum of clinical (e.g. intensive care unit construction), non-pharmaceutical (e.g. large-scale lockdowns) and, most recently, vaccination-based strategies, with the timing of implementation dramatically influencing the case curve between legislative regions [6]. The optimum moment of action is unclear, however, with suggestions that, due to periods of cryptic transmission [7,8], strong non-pharmaceutical interventions (NPIs) two weeks earlier than performed may have halved cumulative deaths [9,10]. Identifying this cryptic window would therefore improve strategy decisions involving possible NPIs based upon the relevant COVID-19 situation.
Unfortunately, the causes of disease emergence and re-emergence often appear idiosyncratic [11], requiring the use of context-specific models [12,13] or risk assessments limited to initial emergence only [14]. These methods are powerful tools and have become keystones during the COVID-19 pandemic response, but are restricted by data availability [15,16] and potential for the lack of transparency [17]. Due to these difficulties, there have been suggestions to consider disease emergence as critical transitions [18] where a forcing pressure, such as host movement or pathogen evolution, drives the system towards a threshold. If considered in this manner, then a suite of alternative methods based upon the concept of critical slowing down (CSD) becomes applicable in the identification of transitions in disease and, potentially, COVID-19 dynamics.
CSD represents the compromised ability of a system to recover from perturbation as it approaches a threshold at which a small perturbation in state triggers a positive feedback loop and system shift [19]. From this phenomenon, various mechanism-independent and summary statistic indicators have been identified across a range of simulated [20,21] and empirical [22,23] studies. In disease systems, CSD was established as tracking a transcritical transition in the effective reproductive number, Reff, [24] or number of secondary infections an infectious individual causes. Below one, secondary infection is unlikely whereas above one, transmission is common and an outbreak occurs. The period where Reff increases towards one is represented by a region of ‘stuttering’ transmission [25] during which CSD also increases [24,26]. ‘Early warning signal’ (EWS) summary statistics based upon CSD will therefore precede the rapid increase in cases at the beginning of an outbreak [27].
EWSs have been shown to predict the emergence of diseases empirically and in simulations [24,28,29]. Variance and autocorrelation increased prior to malaria resurgence in Kenya [28] and before the initial emergence of COVID-19 in seven of the nine countries assessed [30]. In the latter, the exponential increase in cases resulted from the inherently high Reff of the SARS-CoV-2 pathogen (greater than 1) [31] rather than a transcritical transition. CSD is therefore not conceptually anticipated to be present [32]. However, wider research has suggested that CSD-based signals may be identifiable not only before a critical transition, but also prior to strong nonlinearity [20,33]; even in the absence of a defined bifurcation, EWSs may detect rapid case increases. This supposition has since been supported by simulated work showing EWSs increase during early stages of exponential and logistic change [34], implying a circumstance exploitable during disease monitoring to identify both exponential growth and transcritical transitions. For the successive COVID-19 re-emergences, however, NPIs restricting Reff [35] have theoretically introduced a period of stuttering transmission to cause CSD, and EWSs may act as expected. Thus, COVID-19 represents a unique opportunity to test the efficacy of EWSs in multiple sequential outbreaks by assessing the wave-like dynamics expressed in most countries [36].
Here, we test whether EWSs can predict sequential increases in COVID-19 daily cases, developing a novel methodology to detect and subset data into successive waves of infection. Using the suggested framework, we show evidence that EWSs can be identified prior to nonlinear COVID-19 case increases with the second wave best pre-empted, matching the theoretical predictions. Our results provide suggestions on how to use EWSs in a management scenario, where decisions must be made as data are collected, rather than post hoc.
2. Methods
(a) . Data availability
Daily COVID-19 case data were collected from the World Health Organization's dashboard (https://covid19.who.int/info/), spanning 30 January 2020 to 9 June 2021. We analysed positive, daily COVID-19 cases rather than cumulative cases as performed by other studies [30], thus allowing us to attempt the prediction of sequential COVID-19 outbreaks. Case data can be inaccurate from incomplete testing and cryptic cases [35], but we wanted to explore the viability of EWSs using the most universally collected and interpreted data type. The data were consequently analysed in its raw form with no detrending performed.
(b) . COVID-19 wave identification
To assess the increase and decline of COVID-19 cases and define nonlinear regions that can be considered ‘waves’, generalized additive models (GAMs) were iteratively fitted to daily cases in an ‘add-one-in’ fashion, using the R [37] package ‘mgcv’ [38] (see electronic supplementary material 1 for further details. The entire workflow is visualized in the electronic supplementary material 1, figure S1). The time series' inflection points, defined by the significant differing of the GAM smooth's first derivatives from zero, as assessed by 95% pointwise confidence intervals [39], were used to identify nonlinear regions of case increase/decrease and wave onset/subsidence.
(c) . Early warning signal calculation
The presence of EWSs was then calculated using the framework suggested by [40] and developed in [23,41]. Briefly, this approach differs from rolling window EWS methodologies [32,42] by assessing change in an expanding window via a composite metric consisting of multiple indicators. Here, we focussed on the two most used EWS indicators, variance (represented by the standard deviation, s.d.) and autocorrelation at first lag (acf) [43], as well as skewness (skew), following the initial authors [40]. Each indicator was normalized by subtracting its expanding mean from its calculated value at time t before dividing by its expanding standard deviation [23]. A composite metric was then constructed by summing all individual indicator values calculated per t. An EWS was considered present when the composite metric exceeded its expanding mean by 2σ [40]. The 2σ threshold was chosen based upon its equivalency to a 95% confidence interval and repeatedly favourable performance compared to other threshold levels [23,41].
As the expanding mean is the basis of assessment, a previous wave will often mask the appearance of second (electronic supplementary material 2, figure S2). Consequently, once a wave subsided, as assessed by GAM first derivatives, the data were cut and the EWS assessment-GAM fitting restarted, truncating the time series from the point of wave end. This resulted in a series of EWS and GAM assessments, each for a specific wave and independent from previous waves. Additionally, the expanding window approach is initially susceptible to false-positive signals due to the short time series length and high variability when few data points are supplied to the algorithm. To mitigate this, a seven-time step burn-in period was introduced to ‘train’ the signals.
Evidence suggests that a persistent signal for two time steps is sufficient to reduce the frequency of false positives [44] and so a ‘warning’ was acknowledged when signals were detected for two consecutive time steps. From the calculated indicators, we present both the individual indicator strengths over time as well as the difference between the time-of-first-detection and the estimated onset of nonlinearity in the UK and abroad. We also identify the superior indicator or combination of indicators for specifically pre-empting COVID-19 case increase.
3. Results
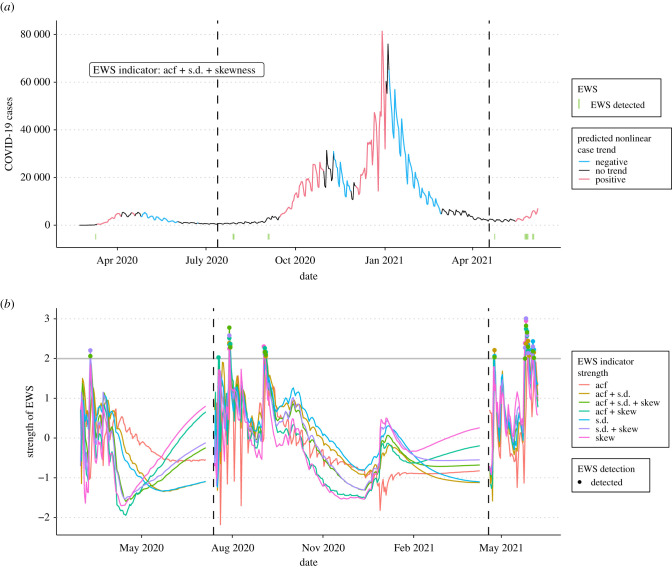
Using the UK as a case study, GAMs predicted three significantly increasing regions in daily COVID-19 cases. These regions correspond to the onset of Waves 1, 2 and, tentatively, 3 beginning on 9 March 2020, 15 September 2020 and 18 May 2021, respectively. From this prediction, two restarts of the EWS-GAM analysis were performed on 7 July 2020 and 12 April 2021 following the subsidence of each wave. Only ‘s.d. + skew’ and the composite metric exceeded the 2σ threshold at least once during each UK wave (figure 1). Under our definition of a ‘warning’, Wave 1 was un-detected, whereas Wave 2 was pre-empted by 48 days and indicators lagged Wave 3 by 10 days.
Figure 1.
Time series of (a) the daily UK COVD-19 cases and sequential GAMs' predicted trends, where green bars represent detected EWSs from the triple composite indicator ‘acf + s.d. + skew’. (b) Individual EWS indicator strengths, where coloured dots indicate time points exceeding the 2σ threshold (horizontal grey line). Dashed, vertical lines indicate the initialization of GAM and EWS reassessment.
Similar results were observed in 24 other countries, with GAMs predicting at least two significantly nonlinear increases in each country's daily COVID-19 cases (excluding Portugal). Nine countries also exhibited a third increase (electronic supplementary material 2, figure S3) and Japan alone exhibited a fourth increase. When warnings were averaged between countries, each sequential wave was more weakly pre-empted, although the first wave had the highest false-negative rate of all waves across all indicators (electronic supplementary material 2, table S1, μ ± s.e.: Wave 1 = 0.62 ± 0.02, Wave 2 = 0.27 ± 0.01, Wave 3 = 0.22 ± 0.01). The composite metric was the most robust indicator of both emergence and re-emergence (electronic supplementary material 2, table S1; figure 2) with a false negative rate of approximately 0.28 compared to approximately 0.37 for variance, approximately 0.43 for skewness and approximately 0.53 for autocorrelation. The composite's mean detection time did decrease from a 14.6 day pre-emption of COVID-19 emergence to 13.2 days for first re-emergence (Wave 2) and 5.0 day lag for second re-emergence (electronic supplementary material 2, table S1).
Figure 2.
The timeliness spread of individual EWS indicators across a range of countries, grouped by continent (‘other’ consists of Africa and the Middle East). Points represent the number of days that an EWS was detected prior to estimate nonlinear case increases. Failed detections are not plotted. EWS indicators include autocorrelation (acf), variance (s.d.) and skewness (skew).
European countries displayed the earliest mean pre-emption of the first wave when assessed using the composite metric (figure 2; +26.0 ± 2.8 days), but with a high false-negative rate (0.78) compared to Asia (0.2) which displayed the second earliest mean pre-emption (+19.0 ± 5.7 days). European countries also pre-empted the second wave earliest (+27.2 ± 2.1 days, 0.33 false negative) although no region pre-empted the third wave; other (Africa and the Middle East) exhibited the shortest lag (−0.5 ± 5.3 days, 0.00 false negative).
4. Discussion
These results show that EWSs based on the theory of CSD are sufficient to predict the emergence and re-emergence of COVID-19 in multiple countries, but that their ability is context dependent. Indicator ability is improved when time series are subset into individual waves unbiasedly estimated using an iterative GAM approach (figure 1; electronic supplementary material 2, figure S2) but, when EWSs are detected, they only appear for short periods before disappearing. This weakens the practicality of EWSs as a predictive tool for policymakers, as only a few warnings may appear prior to the nonlinear increase. Nevertheless, the observed degree of pre-emption supports EWSs use for characterizing both exponential growth at emergence and later periods of ‘stuttering’ transmission.
Time series variance consistently predicted nonlinear case increase, implying that the indicator is the most informative for characterizing disease outbreaks. This is congruent with previous disease [28,45] and ecological [23,43] findings. The relative weakness of autocorrelation was unexpected considering those studies also identified autocorrelation as a reliable indicator, yet the improvement of the triple composite EWS regardless highlights the benefit of the composite metric approach developed by [40] over individual indicators.
Although this study provides evidence for the partial success of EWS indicators during COVID-19 re-/emergence, their application is often hampered by the quality of data available [46]. Epidemiological data can be particularly problematic due to the reporting style of practitioners. Clinical testing effort changes over time [47] and case data can be aggregated into weekly or monthly counts with the exact date of infection of an individual unknown and many cases being cryptic due to a lack of symptoms [35]. While EWSs based upon disease incidence often display unique behaviour compared to the same indicators based upon disease prevalence [29], this study suggests EWSs can detect changes in daily data via an expanding window and is supported by other studies implementing the alternative rolling window approach [29,48]. COVID-19 case data from Europe does provide a best-case example for EWS assessment, due to the frequency of testing and defined periods of stationarity enforced by NPIs resulting in improved pre-emption compared to other regions (figure 2), but as many governments alter their approach towards future pandemics [49], the data quality may become universal.
The variable pre-emption between waves reported here is particularly important as, statistically, the lag between transition and disease emergence is not fully understood [27]. In this study, it is likely that second re-emergence is not pre-empted due to the shrinking stationary period between it and the previous wave, which in some circumstances may be only one to two weeks in length (e.g. Argentina, Colombia, India). This supports Dablander and colleagues' suggestion [32] that some ‘settling down’ of infection rates is required for EWSs to be persistent and pre-emptive, further weakening their generic usage. Similarly, first re-emergence displaying lowest false-negative EWS rates is consistent with critical theory; wave 2 is anticipated to be the only transcritical transition observed in the time series and so is the only wave thought to display CSD. We therefore suggest that while composite EWSs are not universally detectable prior to COVID-19 waves, they may supplement routine trend analysis in the current monitoring toolbox, alongside other possible early indicators (social media activity—[50], viral shedding in sewerage—[51]), as indicative of persistent nonlinear case growth rather than solely critical transitions.
In conclusion, we suggest that composite EWS indicators may provide useful predictive tools during monitoring, particularly where cases are maintained at low variability for extended periods. When waves rapidly follow one another, CSD-based tools can indicate strong nonlinearity, but these signals are most reliable when Reff is maintained sufficiently low that re-emergence results from a transcritical transition. These results support wider notions that EWSs are best suited as a ‘preliminary analysis' indicative of a system at risk and requiring intervention consideration. Although we advocate the use of sequential EWS assessment if used for disease monitoring, it is harder to apply this approach in multi-dimensional systems where a stationary period is likely unidentifiable. Ecosystems, for example, are constantly fluctuating in response to intrinsic or extrinsic drivers [52], with it unclear the minimum length of time series to definitively identify the system's trend [53]. Nonetheless, if periods of stationarity can be identified, we believe sequential assessment is necessary for EWS usage to prevent bias from historic transitions.
Acknowledgements
We are thankful to the GW4 + FRESH Centre for Doctoral Training in Freshwater Biosciences and Sustainability for their support of this project and to the four anonymous reviewers who greatly improved the quality and scope of work.
Data accessibility
All data are available from the World Health Organization's dashboard (https://covid19.who.int/info/). Code for the generation of all results and figures are available at https://doi.org/10.5281/zenodo.5556901 [54]. Additional supporting information and data are provided in the electronic supplementary material [55].
Authors' contributions
D.A.O. was involved in the conceptualization, methodology, investigation, formal analysis, writing-original draft preparation and visualization. C.F.C. was involved in the methodology, review and editing of the manuscript, and supervision. All authors agree to be held accountable for the content therein and approve the final version of the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This project was funded by a NERC GW4 + FRESH CDT PhD studentship awarded to D.A.O. (grant no. NE/R011524/1).
References
- 1.Howard CR, Fletcher NF. 2012. Emerging virus diseases: can we ever expect the unexpected? Emerging Microbes Infections 1, e46. ( 10.1038/emi.2012.47) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morens DM, Folkers GK, Fauci AS. 2004. The challenge of emerging and re-emerging infectious diseases. Nature 430, 242-249. ( 10.1038/nature02759) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson RM, Heesterbeek H, Klinkenberg D, Hollingsworth TD. 2020. How will country-based mitigation measures influence the course of the COVID-19 epidemic? Lancet 395, 931-934. ( 10.1016/S0140-6736(20)30567-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cairney P. 2021. The UK government's COVID-19 policy: assessing evidence-informed policy analysis in real time. Br. Politics 16, 90-116. ( 10.1057/s41293-020-00150-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devine D, Gaskell J, Jennings W, Stoker G. 2020. Trust and the coronavirus pandemic: what are the consequences of and for trust? An early review of the literature. Political Stud. Rev. 19, 274-285. ( 10.1177/1478929920948684) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prem K, et al. 2020. The effect of control strategies to reduce social mixing on outcomes of the COVID-19 epidemic in Wuhan, China: a modelling study. Lancet Public Health 5, e261-e270. ( 10.1016/S2468-2667(20)30073-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bedford T, et al. 2020. Cryptic transmission of SARS-CoV-2 in Washington state. Science 370, 571-575. ( 10.1126/science.abc0523) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li R, Pei S, Chen B, Song Y, Zhang T, Yang W, Shaman J. 2020. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science 368, 489-493. ( 10.1126/science.abb3221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ragonnet-Cronin M, et al. 2021. Genetic evidence for the association between COVID-19 epidemic severity and timing of non-pharmaceutical interventions. Nat. Commun. 12, 2188. ( 10.1038/s41467-021-22366-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hale T, Hale AJ, Kira B, Petherick A, Phillips T, Sridhar D, Thompson RN, Webster S, Angrist N. 2020. Global assessment of the relationship between government response measures and COVID-19 deaths. medRxiv, 2020.07.04.20145334. ( 10.1101/2020.07.04.20145334) [DOI] [PMC free article] [PubMed]
- 11.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. 2008. Global trends in emerging infectious diseases. Nature 451, 990-993. ( 10.1038/nature06536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y, Lee S, Chu C, Choe S, Hong S, Shin Y. 2016. The characteristics of Middle Eastern respiratory syndrome coronavirus transmission dynamics in South Korea. Osong Public Health Res. Perspect. 7, 49-55. ( 10.1016/j.phrp.2016.01.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertozzi AL, Franco E, Mohler G, Short MB, Sledge D. 2020. The challenges of modeling and forecasting the spread of COVID-19. Proc. Natl Acad. Sci. USA 117, 16 732-16 738. ( 10.1073/pnas.2006520117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson RN, Gilligan CA, Cunniffe NJ.. 2020. Will an outbreak exceed available resources for control? Estimating the risk from invading pathogens using practical definitions of a severe epidemic. J. R. Soc. Interface 17, 20200690. ( 10.1098/rsif.2020.0690) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmdahl I, Buckee C. 2020. Wrong but useful — what Covid-19 epidemiologic models can and cannot tell us. New Engl. J. Med. 383, 303-305. ( 10.1056/NEJMp2016822) [DOI] [PubMed] [Google Scholar]
- 16.Wood SN, Wit EC, Fasiolo M, Green PJ. 2021. COVID-19 and the difficulty of inferring epidemiological parameters from clinical data. Lancet Infect. Dis. 21, 27-28. ( 10.1016/S1473-3099(20)30437-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jalali MS, DiGennaro C, Sridhar D. 2020. Transparency assessment of COVID-19 models. Lancet Glob. Health 8, e1459-e1460. ( 10.1016/S2214-109X(20)30447-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drake JM, et al. 2019. The statistics of epidemic transitions. PLoS Comput. Biol. 15, e1006917. ( 10.1371/journal.pcbi.1006917) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheffer M, et al. 2009. Early-warning signals for critical transitions. Nature 461, 53-59. ( 10.1038/nature08227) [DOI] [PubMed] [Google Scholar]
- 20.Kéfi S, Dakos V, Scheffer M, Van Nes EH, Rietkerk M.. 2013. Early warning signals also precede non-catastrophic transitions. Oikos 122, 641-648. ( 10.1111/j.1600-0706.2012.20838.x) [DOI] [Google Scholar]
- 21.O'Regan SM, Drake JM. 2013. Theory of early warning signals of disease emergence and leading indicators of elimination. Theoret. Ecol. 6, 333-357. ( 10.1007/s12080-013-0185-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carpenter SR, et al. 2011. Early warnings of regime shifts: a whole-ecosystem experiment. Science 332, 1079. ( 10.1126/science.1203672) [DOI] [PubMed] [Google Scholar]
- 23.Clements CF, Ozgul A. 2016. Including trait-based early warning signals helps predict population collapse. Nat. Commun. 7, 10984. ( 10.1038/ncomms10984) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brett T, Ajelli M, Liu QH, Krauland MG, Grefenstette JJ, van Panhuis WG, Vespignani A, Drake JM, Rohani P.. 2020. Detecting critical slowing down in high-dimensional epidemiological systems. PLoS Comput. Biol. 16, e1007679. ( 10.1371/journal.pcbi.1007679) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blumberg S, Lloyd-Smith JO. 2013. Inference of R0 and transmission heterogeneity from the size distribution of stuttering chains. PLoS Comput. Biol. 9, e1002993. ( 10.1371/journal.pcbi.1002993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller PB, O'Dea EB, Rohani P, Drake JM. 2017. Forecasting infectious disease emergence subject to seasonal forcing. Theor. Biol. Med. Model. 14, 17. ( 10.1186/s12976-017-0063-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dibble CJ, O'Dea EB, Park AW, Drake JM. 2016. Waiting time to infectious disease emergence. J. R. Soc. Interface 13, 20160540. ( 10.1098/rsif.2016.0540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris MJ, Hay SI, Drake JM. 2020. Early warning signals of malaria resurgence in Kericho, Kenya. Biol. Lett. 16, 20190713. ( 10.1098/rsbl.2019.0713) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Southall E, Tildesley MJ, Dyson L. 2020. Prospects for detecting early warning signals in discrete event sequence data: application to epidemiological incidence data. PLoS Comput. Biol. 16, e1007836. ( 10.1371/journal.pcbi.1007836) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaur T, Sarkar S, Chowdhury S, Sinha SK, Jolly MK, Dutta PS. 2020. Anticipating the novel coronavirus disease (COVID-19) pandemic. Front. Public Health 8, 521. ( 10.3389/fpubh.2020.569669) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katul GG, Mrad A, Bonetti S, Manoli G, Parolari AJ. 2020. Global convergence of COVID-19 basic reproduction number and estimation from early-time SIR dynamics. PLoS ONE 15, e0239800. ( 10.1371/journal.pone.0239800) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dablander F, Heesterbeek H, Borsboom D, Drake JM. 2021. Overlapping time scales obscure early warning signals of the second COVID-19 wave. medRxiv, 2021.07.27.21261226. ( 10.1101/2021.07.27.21261226) [DOI] [PMC free article] [PubMed]
- 33.Dutta PS, Sharma Y, Abbott KC. 2018. Robustness of early warning signals for catastrophic and non-catastrophic transitions. Oikos 127, 1251-1263. ( 10.1111/oik.05172) [DOI] [Google Scholar]
- 34.Jäger G, Füllsack M. 2019. Systematically false positives in early warning signal analysis. PLoS ONE 14, e0211072. ( 10.1371/journal.pone.0211072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flaxman S, et al. 2020. Estimating the effects of non-pharmaceutical interventions on COVID-19 in Europe. Nature 584, 257-261. ( 10.1038/s41586-020-2405-7) [DOI] [PubMed] [Google Scholar]
- 36.Pedro SA, Ndjomatchoua FT, Jentsch P, Tchuenche JM, Anand M, Bauch CT. 2020. Conditions for a second wave of COVID-19 due to interactions between disease dynamics and social processes. Front. Phys. 8, 428. ( 10.3389/fphy.2020.574514) [DOI] [Google Scholar]
- 37.R Core Team. 2021. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. See https://www.R-project.org/. [Google Scholar]
- 38.Wood SN. 2011. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J. R. Stat. Soc. 73, 3-36. ( 10.1111/j.1467-9868.2010.00749.x) [DOI] [Google Scholar]
- 39.Simpson GL. 2021. gratia: graceful ‘ggplot’-based graphics and other functions for GAMs fitted using ‘mgcv’.
- 40.Drake JM, Griffen BD. 2010. Early warning signals of extinction in deteriorating environments. Nature 467, 456-459. ( 10.1038/nature09389) [DOI] [PubMed] [Google Scholar]
- 41.Clements CF, Blanchard JL, Nash KL, Hindell MA, Ozgul A. 2017. Body size shifts and early warning signals precede the historic collapse of whale stocks. Nat. Ecol. Evol. 1, 188. ( 10.1038/s41559-017-0188) [DOI] [PubMed] [Google Scholar]
- 42.Dakos V, et al. 2012. Methods for detecting early warnings of critical transitions in time series illustrated using simulated ecological data. PLoS ONE 7, e41010. ( 10.1371/journal.pone.0041010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dakos V, van Nes EH, D'Odorico P, Scheffer M. 2012. Robustness of variance and autocorrelation as indicators of critical slowing down. Ecology 93, 264-271. ( 10.1890/11-0889.1) [DOI] [PubMed] [Google Scholar]
- 44.Clements CF, McCarthy MA, Blanchard JL. 2019. Early warning signals of recovery in complex systems. Nat. Commun. 10, 1681. ( 10.1038/s41467-019-09684-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Regan SM, Lillie JW, Drake JM. 2016. Leading indicators of mosquito-borne disease elimination. Theoret. Ecol. 9, 269-286. ( 10.1007/s12080-015-0285-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clements CF, Drake JM, Griffiths JI, Ozgul A. 2015. Factors influencing the detectability of early warning signals of population collapse. Am. Nat. 186, 50-58. ( 10.1086/681573) [DOI] [PubMed] [Google Scholar]
- 47.Mercer TR, Salit M. 2021. Testing at scale during the COVID-19 pandemic. Nat. Rev. Genet. 22, 415-426. ( 10.1038/s41576-021-00360-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brett TS, O'Dea EB, Marty É, Miller PB, Park AW, Drake JM, Rohani P. 2018. Anticipating epidemic transitions with imperfect data. PLoS Comput. Biol. 14, e1006204. ( 10.1371/journal.pcbi.1006204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coccia M. 2021. The relation between length of lockdown, numbers of infected people and deaths of Covid-19, and economic growth of countries: lessons learned to cope with future pandemics similar to Covid-19 and to constrain the deterioration of economic system. Sci. Total Environ. 775, 145801. ( 10.1016/j.scitotenv.2021.145801) [DOI] [Google Scholar]
- 50.Yousefinaghani S, Dara R, Mubareka S, Sharif S. 2021. Prediction of COVID-19 waves using social media and Google search: a case study of the US and Canada. Front. Public Health 9, 359. ( 10.3389/fpubh.2021.656635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bibby K, Bivins A, Wu Z, North D. 2021. Making waves: plausible lead time for wastewater based epidemiology as an early warning system for COVID-19. Water Res. 202, 117438. ( 10.1016/j.watres.2021.117438) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dakos V, Carpenter SR, van Nes EH, Scheffer M.. 2015. Resilience indicators: prospects and limitations for early warnings of regime shifts. Phil. Trans. R. Soc. B 370, 20130263. ( 10.1098/rstb.2013.0263) [DOI] [Google Scholar]
- 53.Wauchope HS, Amano T, Sutherland WJ, Johnston A. 2019. When can we trust population trends? A method for quantifying the effects of sampling interval and duration. Methods Ecol. Evol. 10, 2067-2078. ( 10.1111/2041-210X.13302) [DOI] [Google Scholar]
- 54.O'Brien D, Clements C. 2021. Early warning signal reliability varies with COVID-19 waves. Zenodo ( 10.5281/ZENODO.5556901) [DOI] [PMC free article] [PubMed]
- 55.O'Brien DA, Clements CF. 2021. Early warning signal reliability varies with COVID-19 waves. Figshare. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- O'Brien D, Clements C. 2021. Early warning signal reliability varies with COVID-19 waves. Zenodo ( 10.5281/ZENODO.5556901) [DOI] [PMC free article] [PubMed]
- O'Brien DA, Clements CF. 2021. Early warning signal reliability varies with COVID-19 waves. Figshare. [DOI] [PMC free article] [PubMed]
Data Availability Statement
Daily COVID-19 case data were collected from the World Health Organization's dashboard (https://covid19.who.int/info/), spanning 30 January 2020 to 9 June 2021. We analysed positive, daily COVID-19 cases rather than cumulative cases as performed by other studies [30], thus allowing us to attempt the prediction of sequential COVID-19 outbreaks. Case data can be inaccurate from incomplete testing and cryptic cases [35], but we wanted to explore the viability of EWSs using the most universally collected and interpreted data type. The data were consequently analysed in its raw form with no detrending performed.
All data are available from the World Health Organization's dashboard (https://covid19.who.int/info/). Code for the generation of all results and figures are available at https://doi.org/10.5281/zenodo.5556901 [54]. Additional supporting information and data are provided in the electronic supplementary material [55].