Summary:
Long non-coding RNA (lncRNA) genes have well-established and important impacts on molecular and cellular functions. However, among the thousands of lncRNA genes, it is still a major challenge to identify the subset with disease or trait relevance. To systematically characterize these lncRNA genes, we used GTEx v8 genetic and multi-tissue transcriptomic data to profile the expression, genetic regulation, cellular contexts, and trait associations of 14,100 lncRNA genes across 49 tissues for 101 distinct complex genetic traits. Using these approaches, we identified 1,432 lncRNA gene-trait associations, 800 of which were not explained by stronger effects of neighboring protein-coding genes. This included associations between lncRNA quantitative trait loci and inflammatory bowel disease, type 1 and type 2 diabetes, and coronary artery disease, as well as rare variant associations to body-mass index.
Keywords: long non-coding RNA, lncRNA, expression quantitative trait loci, eQTL, co-expression, complex trait, disease, GTEx, colocalization
In Brief sentence:
A systematic analysis of NIH Genotype Tissue Expression (GTEx) project data provides insights into lncRNA gene expression patterns and functions, explores the impact of genetic variation on lncRNAs, and connects lncRNAs to complex traits and human disease.
Graphical Abstract

Introduction
Long non-coding RNA (lncRNA) genes are a prevalent and heterogeneous group of RNA molecules that lack protein-coding potential. They vary in their epigenetic marks, splicing and transcript structure (Amin et al., 2015; Hon et al., 2017; Melé et al., 2017; Quinn and Chang, 2016), and differ from protein-coding genes due to their lower expression, increased tissue-specificity, and greater variability in expression across individuals (Cabili et al., 2011; Djebali et al., 2012; Hon et al., 2017; Kornienko et al., 2016; Melé et al., 2015). Despite these differences, many lncRNA genes have important roles in gene regulation from epigenetic reprogramming to post-transcriptional regulation (Quinn and Chang, 2016; Wang and Chang, 2011). However, only a few of these lncRNAs have been connected to trait and disease outcomes, such as HOTAIR in cancer, BACE1-AS in Alzheimer’s disease, and PRNCR1 and PCGEM1 in prostate cancer (Faghihi et al., 2008; Gupta et al., 2010; Yang et al., 2013). Although the number of annotated lncRNA genes is increasing with more sensitive transcriptomic profiling in a wider range of contexts and across more individuals (Djebali et al., 2012; Hon et al., 2017; Iyer et al., 2015; Jiang et al., 2019), it remains a significant challenge to identify those lncRNAs with important functional consequences.
In this study, we used the Genotype-Tissue Expression (GTEx) project v8 data to profile genetic regulation of lncRNA genes across 49 human tissues. We combine multiple approaches, including expression quantitative trait loci (eQTL) analysis, gene expression outlier analysis, co-expression networks, and GWAS-QTL colocalization analysis, to identify putative functional lncRNA genes, define their cellular contexts, and systematically assess their relevance to diverse human traits. Together, this work increases the number of lncRNA genes with regulatory connections to human disease.
Results
Expression of lncRNA genes across multiple tissue transcriptomes
Across tissue transcriptomes, we observed expression of the majority (95%) of the 14,100 previously annotated lncRNA genes in at least one tissue (Figure S1A–B). Among these, 96% of our curated list of 954 lncRNA genes with prior established functions were expressed in at least one tissue (Table S1, Methods). We further stratified lncRNA genes into antisense and intergenic, as these two types make up the majority of lncRNA genes (at 5,220 and 7,433 genes, respectively), and observed expression of 96.5% and 94% of them, respectively.
There is previous evidence of the tissue-specificity of lncRNA gene expression, especially for intergenic lncRNA genes, which is also observed in GTEx data (Figure S1C)(Cabili et al., 2011; Djebali et al., 2012; Hon et al., 2017; Jiang et al., 2019; Melé et al., 2015). The tissue-specificity of lncRNA gene expression has been attributed to tissue-specific functions, non-specific transcription, and effects of thresholding on detecting lowly expressed genes. To extend these analyses and mitigate overestimates of tissue-specificity due to thresholding effects, we used an approach inspired by microarrays to identify tissue-specific lncRNA genes. For each of the 14,100 lncRNA genes, we defined a length-matched, non-genic region as close to the gene as possible; lncRNA genes were then called expressed in a given sample if the read count for the gene was significantly greater than the matched non-genic region (Figure S1D, Table S1, Methods). This resulted in lncRNA genes being detected in more tissues than a conventional TPM-based thresholding approach (Figure S1E), and produced a set of 316 tissue-specific lncRNA genes that were detected in only one broad tissue category (Figure 1A, Table S2, Methods). Tissue-specific lncRNA genes were most frequently expressed in testis, brain, blood, and skin tissues. The especially high numbers of genes preferentially expressed in the testis tissue may reflect “transcriptional scanning” during spermatogenesis (Xia et al., 2020).
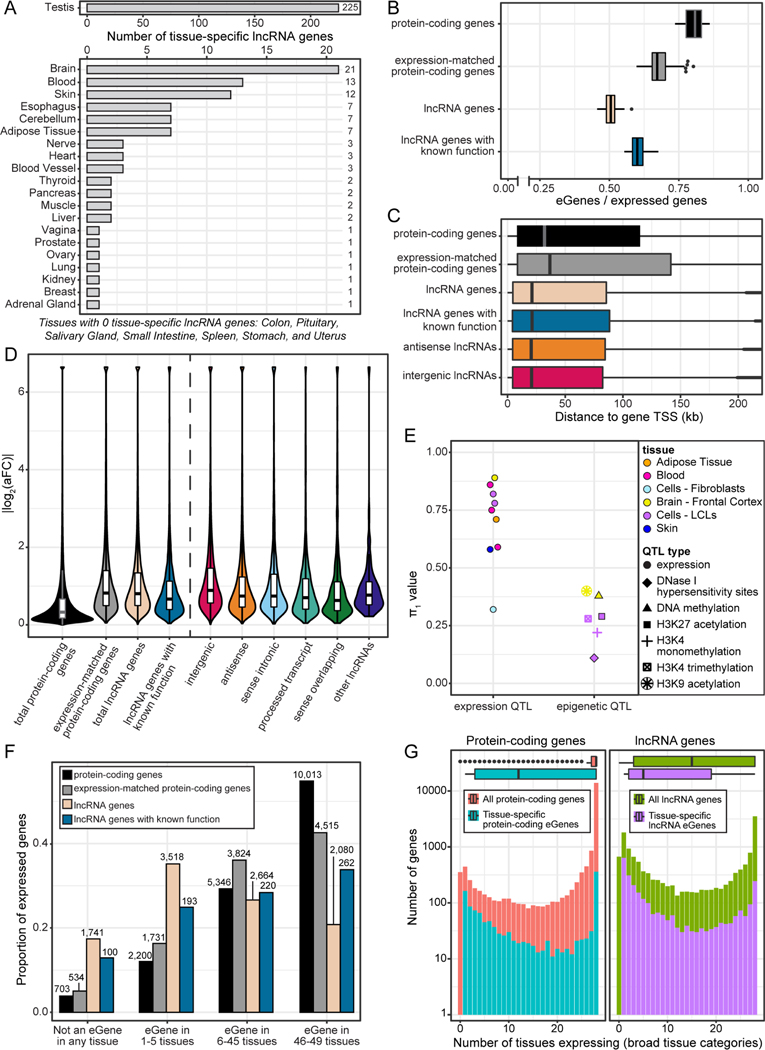
Figure 1.
Specificity of gene expression and presence of eQTLs across GTEx tissues.
(A) The numbers of lncRNA genes with tissue-specific expression across the broad tissue categories of GTEx.
(B) Proportion of expressed genes that were eGenes (MashR LFSR <0.05). Boxplots reflect the range of proportions across the 49 GTEx tissues.
(C) Distribution of distance between the eVariant and the gene’s transcription start site for the top eQTL for each gene in each tissue. The plot is truncated at 200kb for visibility, but the maximum outlier value was 1Mb. Gene group differences were significant (p <0.05/15, Wilcoxon test) between each lncRNA group and the protein-coding genes and the expression-matched coding genes, but not between lncRNA gene groups.
(D)Absolute effect size of the top eQTL for each gene in each tissue. Effect size was measured as log2(allelic fold-change). The dashed line separates the main comparison gene groups from the lncRNA gene types. Gene group differences were significant (p <0.05/23, Wilcoxon test) between all main gene groups (left of dashed line) except for expression-matched protein-coding genes vs. total lncRNA genes. Between lncRNA gene types, all differences were significant except for antisense vs. sense intronic and other lncRNAs, sense intronic vs. other lncRNAs, and processed transcript vs. sense overlapping and other lncRNAs.
(E) Summary of the π1 replication values between GTEx lncRNA gene eQTLs and other QTL studies.
(F) Proportion of each gene group that was an eGene in a certain number of tissues. Bar labels show the number of genes.
(G) The number of tissues expressing protein-coding genes (left) and lncRNA genes (right) at a threshold of ≥0.1 TPM in >20% of samples, compared to the subset of tissue-specific eGenes. Note the y-axis is on log scale.
For all boxplots, data represented are medians with first and third quartiles as boxes, and whiskers extending to 1.5 times the interquartile range. See also Figure S1, Tables S1–S2.
Most lncRNA gene expression is influenced by genetic variation
eQTL analysis provides a systematic approach to assess the regulatory impacts of genetic variants across the transcriptome. We found that 67.3% of all 14,100 annotated lncRNA genes were eGenes, meaning they had at least one variant significantly associated with their gene expression in at least one tissue (FDR <0.05)(The GTEx Consortium, 2020). Within each tissue, ~50% of expressed lncRNA genes were detected as eGenes, compared to ~80% of expressed protein-coding genes (Figure 1B). We observed a higher proportion of eGenes for the set of lncRNA genes with curated functions and, overall, comparable proportions across lncRNA types (Figure S1F). To assess whether lncRNA eGene discovery was limited by expression levels, we created an expression-matched protein-coding gene set within each tissue (Methods), and observed that a greater percentage (~67%) of these expression-matched protein-coding genes were eGenes (Figure 1B).
Alongside the lower abundance of lncRNA genes with eQTLs, we observed additional evidence that lncRNA genes have simpler regulatory mechanisms than protein-coding genes. First, the distance between the lead eQTL’s associated variant (eVariant) and its associated gene’s transcription start site (TSS) was shorter (median lncRNA eVariant-TSS distance = 21kb versus median protein-coding eVariant-TSS distance = 32kb; p <10−16, Wilcoxon test) (Figure 1C), suggesting that its regulatory mechanisms operated closer to the gene. Second, lncRNA eQTLs had higher effect sizes than protein-coding eQTLs (Figure 1D), suggesting fewer regulatory targets. This difference was partly explained by expression level, as we observed similarly high effect sizes for the expression-matched, protein-coding gene eQTLs. However, among lncRNA types, intergenic lncRNA genes had the highest eQTL effect sizes, despite having similar median expression across all tissues (median intergenic lncRNA TPM = 0.28, median lncRNA TPM = 0.34). Combined, these observations indicate that intergenic lncRNA genes have less complex regulation, supporting previous observations using massively-parallel reporter assays (Mattioli et al., 2019).
We assessed replication of GTEx lncRNA eQTLs against other QTL resources using the π1 value, an estimate of the proportion of true positive p-values. Replication of GTEx lncRNA eQTLs in other eQTL datasets for blood, EBV-transformed lymphocytes (LCLs), brain, adipose, and skin tissues had a median π1 of 0.75 (range = 0.32–0.89) (Methods, Figure 1E)(Buil et al., 2015; Gutierrez-Arcelus et al., 2013; Lepik et al., 2017; Ng et al., 2017; Võsa et al., 2018). To assess the replication of lncRNA eQTLs for other molecular phenotypes, we overlapped GTEx eQTLs with QTLs for epigenetic marks from both brain (frontal cortex)(Ng et al., 2017) and LCLs (Grubert et al., 2015). In the frontal cortex, where there were 1,612 lncRNA eGenes, there was a moderate overlap with QTLs for DNA methylation and H3K9 methylation (Figure 1E), illustrating the common coordination of non-coding RNA and epigenetic marks. In LCLs, where there were 932 lncRNA eGenes, overlaps with QTLs for DNase I-hypersensitivity sites, H3K27 acetylation, H3K4 monomethylation, and H3K4 trimethylation were lower (Figure 1E), which was predominantly related to differences in power and methods used for each study. Combined, these epigenetic QTL overlaps highlight many candidate regulatory connections influencing lncRNA expression.
Of the discovered lncRNA eGenes, we observed that 2,783 had evidence of tissue-specificity (Methods). This was more than was seen in protein-coding genes (Figure 1F), with testis, skin, blood, thyroid, and brain having the highest numbers (Table S2). Furthermore, we observed that this tissue-specificity was not solely driven by tissue-specific expression levels, as 15% of tissue-specific eGenes were expressed in all tissue categories (Figure 1G, S1G). Across the tissues, this ranged from 6% (testis) to 100% (uterus) of their tissue-specific eGenes being expressed in in all tissue categories (median = 24%), demonstrating tissue-specificity of lncRNA genetic regulatory effects.
Co-expression networks annotate cellular contexts of lncRNA genes
To improve our understanding of the cellular contexts of lncRNA genes, we performed weighted gene co-expression network analysis (WGCNA) (Langfelder and Horvath, 2008) within each GTEx tissue, and annotated these co-expression modules to specific cell types or compartments using existing single-cell datasets (Figure S2A–B, Data S1, Methods). On average, 80% of sufficiently expressed lncRNA genes were assigned to a module (Figure 2A, Methods). We noted that larger modules included higher proportions of lncRNA genes (Figure 2B); however, there were also some smaller modules mostly made up of lncRNA genes, which may represent hubs of lncRNA regulatory activity. For example, the adrenal gland, tibial artery, minor salivary gland, pancreas, and uterus tissues each had a small, unannotated co-expression module that was 75–89% lncRNA genes. There were 23 genes shared across these five tissues’ modules, 21 of which were lncRNA genes; these genes were widely spread across chromosomes, suggesting that this lncRNA-dominated group of co-expressed genes is not simply mediated by proximity, and might be performing a similar role within each of these tissues.
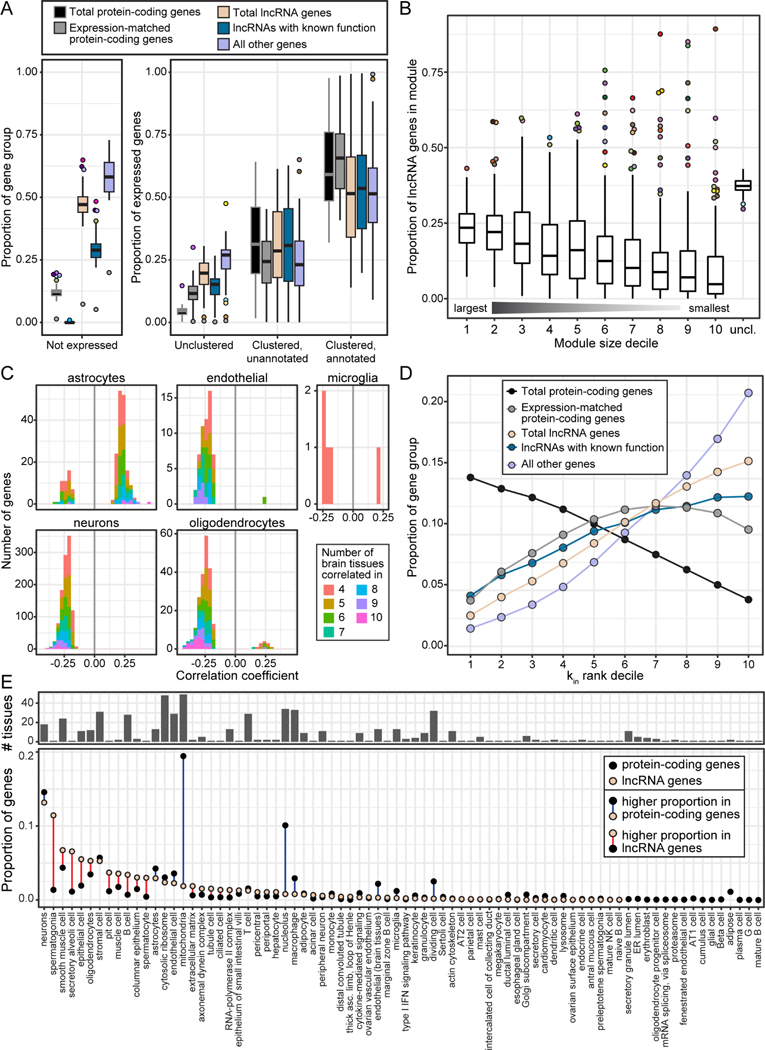
Figure 2.
Co-expression networks annotate cellular contexts of lncRNA genes.
(A) Summary of gene assignment to modules by gene group. The underlying boxplot shows the proportion of a gene group falling into that module status across tissues. Outlier point color indicates the tissue.
(B) Proportion of lncRNA genes in modules across all tissues, binned by module size. “uncl.” = unclustered genes.
(C) lncRNA genes with high confidence annotations in brain tissues, based on agreement of WGCNA annotations and correlation with CIBERSORTx estimated cell type proportions. The correlation coefficient is the median correlation across all relevant brain tissues between the estimated proportion of that cell type, and the lncRNA gene’s expression. The bar fill indicates the number of brain tissues in which the lncRNA gene’s expression level was significantly correlated with the estimated cell type proportion.
(D) Proportion of gene groups binned by intra-modular connectivity (kin) ranking. The most highly connected genes within their module are in the first kin rank decile, and the least connected genes within their module are in the tenth kin rank decile.
(E) Module annotations of genes in the top kin rank decile of their modules. The bottom panel shows the proportion of these highly-connected genes in each annotation group. The top panel shows the number of tissues in which a module is assigned that annotation term. In some cases, the association of many highly connected genes with a certain annotation term may reflect how common that module is across tissues: for example, there is at least one “mitochondria” module in all 49 tissues, which may result in the same hub genes for mitochondria being counted multiple times.
For all boxplots, data represented are medians with first and third quartiles as boxes, and whiskers extending to 1.5 times the interquartile range. See also Figures S2–S3, Data S1, Table S3.
We used these co-expression networks to refine the cellular contexts of tissue-shared and -specific lncRNA eGenes. We observed that, relative to all lncRNA genes, tissue-shared eGenes were enriched for the cell compartment annotations of mitochondria and cytosolic ribosomes, for the pathway annotation of type I interferon signaling pathway, and for the stomach cell type annotations of parietal cell and antral mucus cell (all OR >1.5 and p <0.05, Fisher’s test). In contrast, enriched module annotations for tissue-specific eGenes were dominated by sperm cell terms, which was consistent with many of these eGenes being specific for the testis tissue. Other enriched annotations in the tissue-specific eGenes included monocytes and megakaryocytes, keratinocytes, hepatocytes, and cardiomyocytes (OR >1.5 and p <0.05, Fisher’s test).
Since gene co-expression clustering in bulk tissues can be driven by cell composition, we further incorporated estimated cell proportions in the brain and blood tissues into co-expression analyses to identify the subset of lncRNA genes where both module and cell type proportion annotations were concordant (Methods). In the brain tissues, 1,554 lncRNA genes had matching annotation between the two methods, providing increased support for their cell type annotation (Figure 2C, Table S3). These genes were mostly annotated to the more common cell types such as neurons (1,069), oligodendrocytes (215), astrocytes (209), and endothelial cells (56) (Figure S3A–B). In blood, expression levels of 2,837 confident lncRNA blood cell type annotations were identified (Figure S3C–D, Table S3), most of which were annotated to monocytes (1,567) or T cells (1,193).
To identify additional cell type-relevant lncRNA genes, particularly in those tissues where single-cell data were not available, we used the WGCNA metric for within-module connectivity (kin) to identify lncRNA genes that were highly connected within specific annotated modules (Methods). We first observed that lncRNA genes were not as often highly connected compared to protein-coding genes, especially in larger co-expression modules (Figure 2D, S2C); part of this was due to a relationship between connectivity and expression level (Figure S2D). Relative to the total group of lncRNA genes, antisense lncRNA genes were enriched for being highly connected while intergenic lncRNA genes were depleted (antisense OR = 2.10, p <10−16; intergenic OR = 0.88, p = 0.002; Fisher’s test). Focusing on the most highly connected lncRNA and protein-coding genes (genes in the top kin decile in their assigned module), we identified the neuron module as a common annotation for both gene groups, and highly-connected lncRNA genes were additionally frequently assigned to early sperm cells, muscle cells, epithelium, and tissue-resident B cells (Figure 2E, S2E).
Intergenic lncRNA genes are enriched for having high allele-specific expression that is shared with their neighboring genes
Allele-specific expression, as measured by imbalance of expressed alleles in RNA-sequencing data, can provide an additional means to detect cis-acting regulatory variation. We observed 3,871 protein-coding, 1,138 intergenic lncRNA, and 863 antisense lncRNA genes with high ASE (mean gene Z score >3) (Table S4). Compared to protein-coding genes, both lncRNA gene groups were enriched for high ASE (intergenic OR = 1.67, p <10−16; antisense OR = 1.34, p = 3.8×10−11; Fisher’s test).
As some lncRNAs can operate on large genomic intervals that are detectable through allele-specific expression (ASE) patterns (e.g. XIST (Brown et al., 1991; Engreitz et al., 2013)), we sought to test local patterns of ASE for lncRNA genes. To assess this, we defined local sharing events, where ASE was present for both the lncRNA gene and adjacent genes within a 500-kb neighboring window (mean gene Z score >3 and mean neighbor Z score >3) (Figure 3A, Methods). In total, we identify 137 genes with high ASE-sharing that was consistent across tissues. Among these genes, there was an enrichment in intergenic lncRNA genes relative to protein-coding genes, while antisense lncRNA genes were slightly depleted for high ASE-sharing events (Figure 3B). We further identified 16 lncRNA genes (15 intergenic, 1 antisense) that were the only genes in their neighborhood to have a mean neighbor Z-score (>3). Although some of the genes around these lncRNAs showed ASE, they did not have high neighbor Z-scores, indicating the potential for these lncRNAs to have cis-acting impacts on their local regulatory regions (Figure 3C).
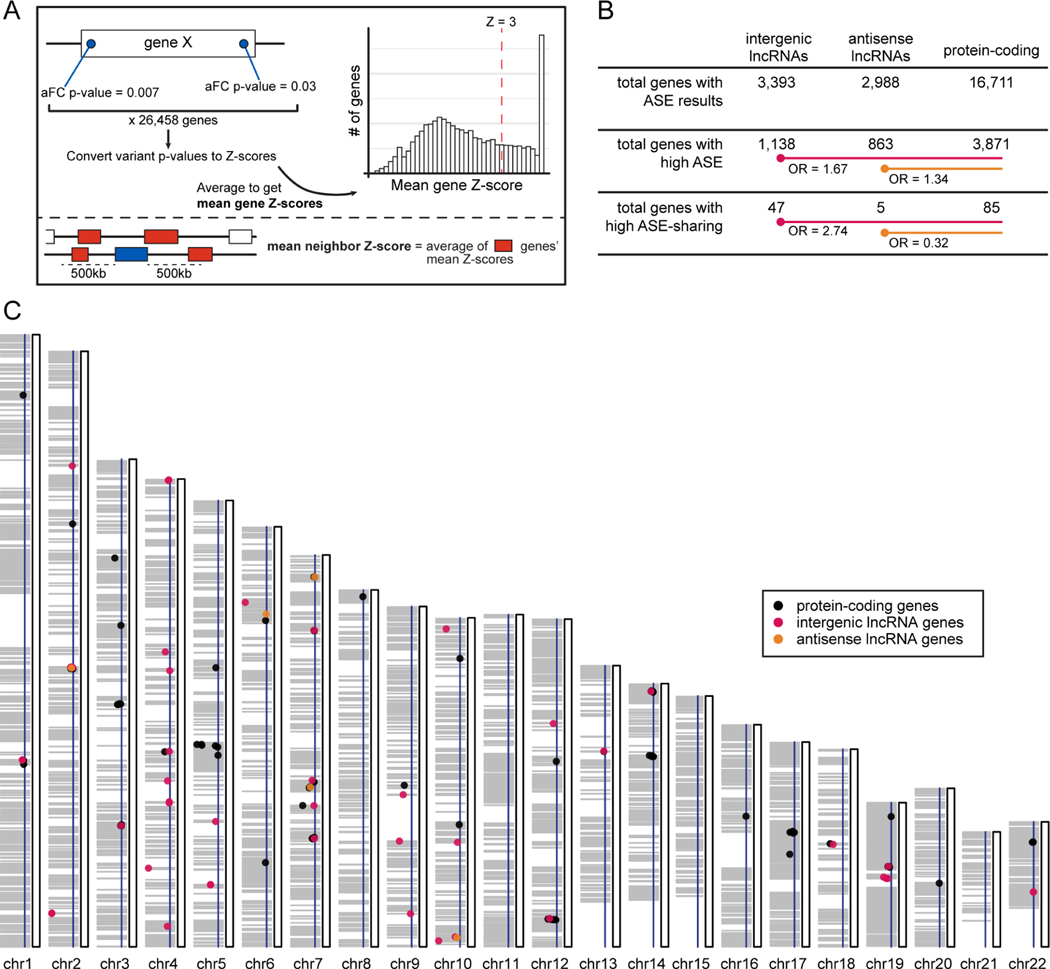
Figure 3.
Patterns in allele-specific expression (ASE) associated with lncRNA gene eQTLs.
(A) Scheme for calculating gene-level and neighbor gene-level ASE scores. aFC = allelic fold-change.
(B) ASE-sharing results for genes, grouped by gene type. Odds ratios (OR) were calculated for the lncRNA gene types relative to the protein-coding genes, with the background being total genes with ASE results. “High ASE” = mean gene Z score >3; “High ASE-sharing” = mean gene Z score >3 and mean neighbor Z score >3.
(C) Genome-wide distribution of high ASE-sharing genes. A dot’s horizontal position is its mean neighbor Z score, with a further left dot having a higher Z score and the blue horizontal line marking the Z=3 threshold. The grey shading illustrates stretches of the genome where, starting from a given gene with high ASE, at least one other gene within 500kb also has high ASE. See also Table S4.
Rare variation impacts intergenic lncRNA gene expression and complex traits
Rare genetic variation influences risk for both rare and common diseases (Keinan and Clark, 2012), and thousands of rare variants are present in each human genome (Auton et al., 2015; Bomba et al., 2017; Wright et al., 2018). We sought to define the properties of rarer genetic variants influencing lncRNA gene expression by applying an outlier enrichment approach (Ferraro et al., 2020). We first identified 1,563 intergenic lncRNA multi-tissue outlier events, with an outlier event being an individual-gene combination that was an outlier for the majority of that individual’s sampled tissues (|median Z score| >2) (Table S5, Methods). We then focused on the 1,119 outliers involving intergenic lncRNA genes with detectable expression in all tissues; for these widely expressed genes, a multi-tissue outlier individual is more likely a reflection of consistent transcriptional differences rather than spurious detection in a few tissues. These outlier events involved 497 unique intergenic lncRNA genes, and 545 unique individuals. There were 23 instances of shared multi-tissue outlier events between an intergenic lncRNA gene and a protein-coding gene within 10kb of each other; 14 of these outlier events involved a nearby rare variant, and three of these lncRNA/protein-coding gene outlier pairs occurred in more than one individual (Figure S4A–B). Relative to all tested genes, intergenic lncRNA genes were less likely to have any outlier individuals (Figure S4C). The majority of intergenic lncRNA outliers were over-expression outliers (Figure 4A). This may be partially due to lncRNA genes’ generally lower expression, since lowly expressed genes are more likely to fluctuate upward, and their under-expression is difficult to detect.
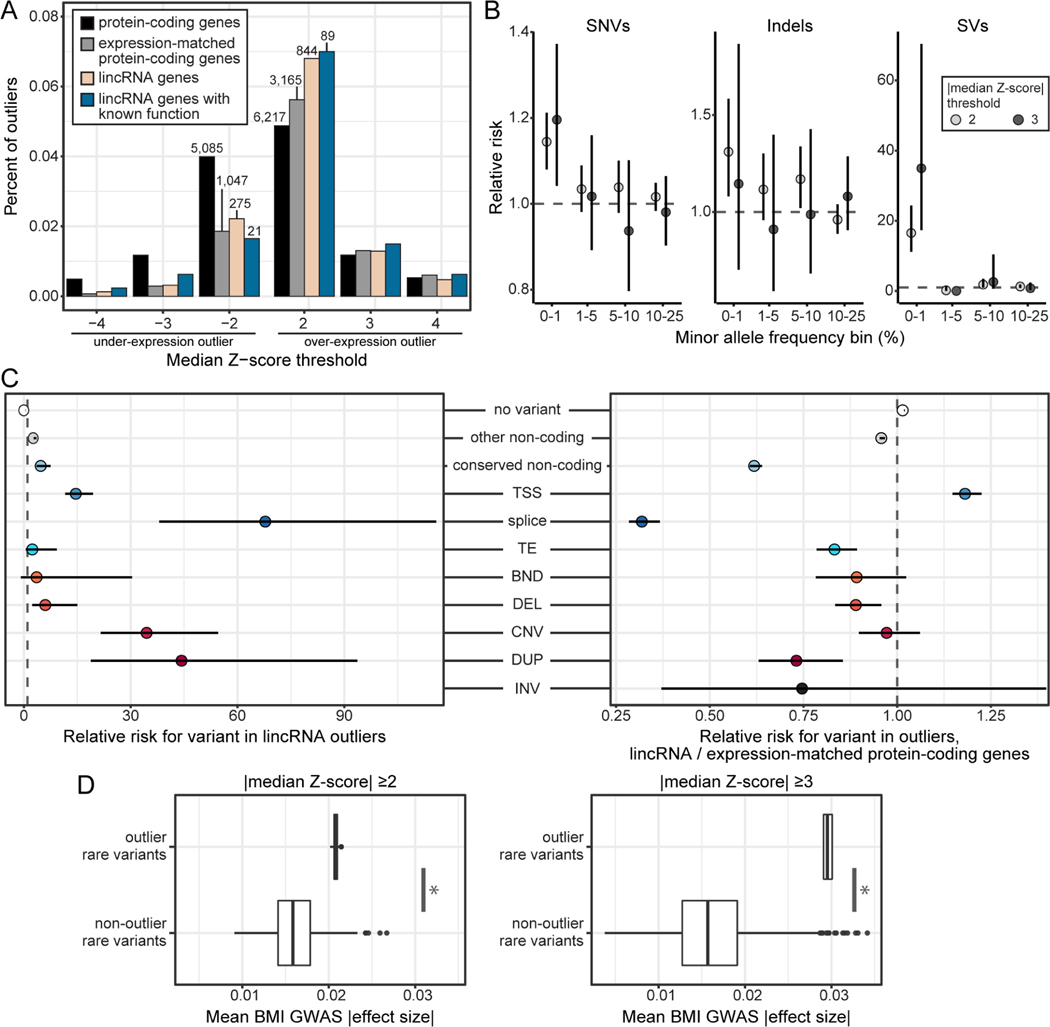
Figure 4.
Rare variation impacts intergenic lncRNA gene expression and complex traits.
(A) Percent of multi-tissue intergenic lncRNA gene outliers out of all gene-individual combinations tested. Labels indicate the number of outliers.
(B) Enrichment of variants within 10kb of the outlier gene in outlier individuals. Dots represent relative risk point estimate, with bars showing the 95% confidence intervals.
(C) Enrichment of rare variants (MAF <1%) within 10 kb of the outlier gene in outlier individuals. Left panel: the enrichment of rare variants in intergenic lncRNA outliers relative to non-outliers. Right panel: the enrichment values from the left relative to those same enrichment values for expression-matched protein-coding genes. Data are represented as in (B).
(D) The mean effect size in the UK Biobank GWAS for body-mass index of rare variants associated with intergenic lncRNA gene expression outlier events, compared to matched rare variants associated with non-outlier events. The heightened GWAS effect size of outlier-associated variants increases with gene expression outlier Z score (figure panels). * indicates p-value <0.05, Wilcoxon test. The boxplot represents medians with first and third quartiles as boxes, and whiskers extending to 1.5 times the interquartile range.
lincRNA: intergenic lncRNA, TSS: transcription splice site, TE: transposable element insertion, BND: breakend, DEL: deletion, CNV: copy number variation, DUP: duplication, INV: inversion. See also Figure S4, Table S5.
Intergenic lncRNA gene outlier events were enriched for the presence of nearby genetic variants, particularly for rare variants (minor allele frequency <1% in GTEx and gnomAD non-Finnish Europeans)(Karczewski et al., 2020) and especially for rare structural variants. This was assessed by relative risk (RR, the proportion of outlier individuals with variant / proportion of non-outlier individuals with variant); RRs were 1.14 for SNVs, 1.31 for small insertions or deletions (indels), and 16.52 for structural variants (SVs), with increasing enrichments at higher Z score thresholds (Figure 4B). In contrast to protein-coding genes, we observed that most of the enrichment was driven by the over-expression outliers (Figure S4D). Overall, 55% of the intergenic lncRNA outlier events in tested individuals were associated with a nearby rare variant.
We further stratified rare variant enrichments across different variant subclasses to refine identification of variant properties driving lncRNA outliers. Deletions, copy number variations (CNVs), and duplications were all specifically enriched in outlier individuals near their outlier genes (Figure 4C). Notably, splice variants were also strongly enriched near outlier genes (RR = 68.3, p <1016) – even more so than rare TSS variants (RR = 15.1, p <10−16). This is consistent with a similarly strong enrichment for rare splice variants observed near protein-coding gene outliers (Ferraro et al., 2020), as well as the strong enrichment of splice-related annotations for cis-eQTLs, including those that were distinct from splicing quantitative trait loci (sQTLs)(The GTEx Consortium, 2020). Although several different rare variant classes had strong enrichment near intergenic lncRNA outliers, these enrichments were not as strong as those for nearby expression-matched protein-coding gene outliers (Figure 4C). The exception was TSS variants, which had significantly stronger enrichment in lncRNA gene outliers (RR = 1.19, p <10−16); this is consistent with our findings from the eQTL analysis that lncRNA gene expression is regulated by simpler mechanisms than protein-coding genes.
Using the pool of rare variants identified from lncRNA outliers, we sought to test if these variants were enriched for subsequent impacts on complex traits. We identified 44 rare variants that were near lncRNA gene outliers, not near protein-coding gene outliers, and present in the UK Biobank. For comparison, we made a control, non-outlier pool of 3,173 rare variants that were near the same lncRNA genes but only present in individuals who were not outliers for those genes (|median Z score| <1). We observed that outlier-associated rare variants had higher effect sizes in the UK Biobank GWAS for body-mass index compared to non-outlier rare variants (p <0.05, Wilcoxon test) (Figure 4D). Although it may be expected that rare variants can have major impacts on a single gene’s expression, these analyses provide evidence that the rare variants associated with intergenic lncRNA gene expression influence common complex traits.
Colocalization of QTL and GWAS signals creates a catalog of trait-associated lncRNA genes
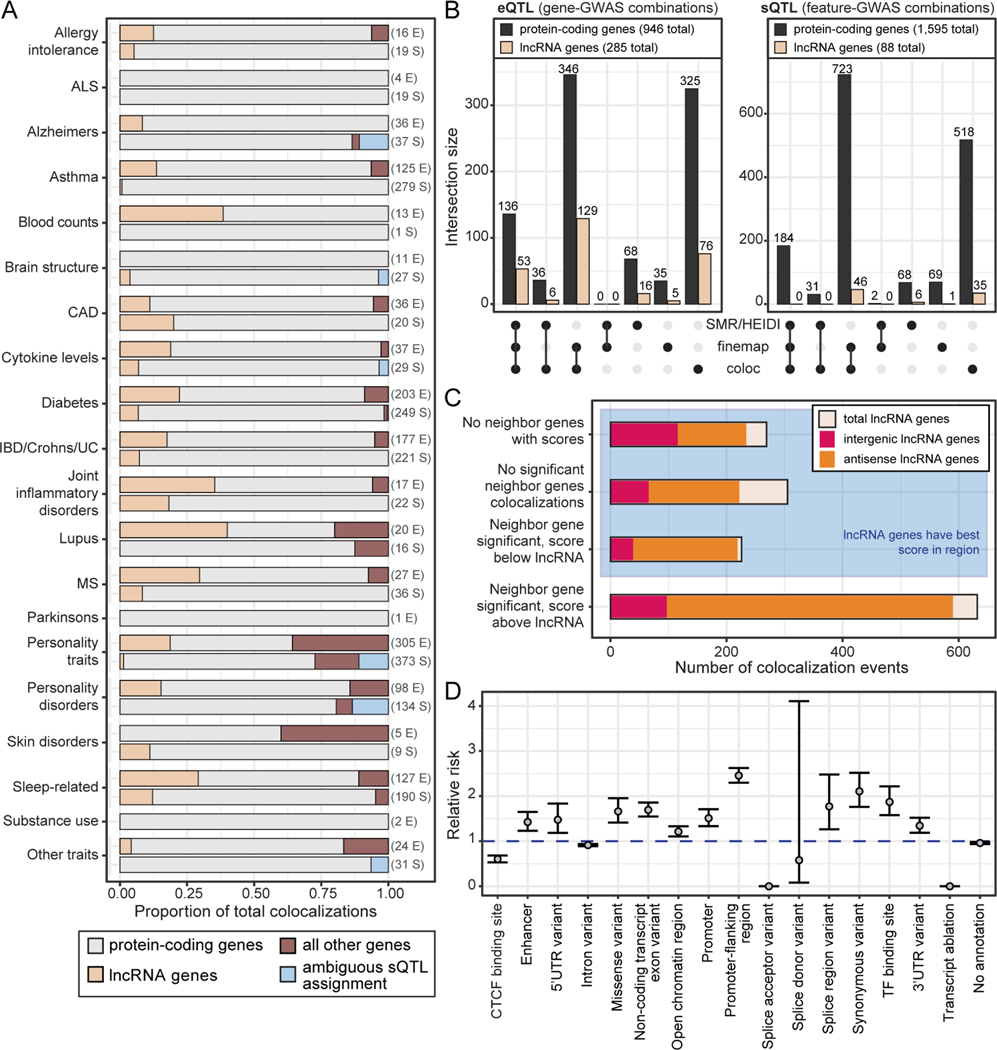
Discovering the trait and disease relevance of lncRNA genes has been a major challenge, with only a few examples robustly connected to phenotypic consequences (Wapinski and Chang, 2011). To address this challenge, we systematically assessed the roles of all lncRNA genes across a diverse set of traits through colocalization analyses. Following the standards of the recent GTEx GWAS analysis (Barbeira et al., 2021; The GTEx Consortium, 2020), we combine multiple colocalization approaches to improve performance: SMR+HEIDI (Zhu et al., 2016), FINEMAP+eCAVIAR (Benner et al., 2016; Hormozdiari et al., 2016) and coloc (Giambartolomei et al., 2014). By assessing 101 traits from 176 GWAS (Figure S5A, Table S6) for both expression and splicing QTLs, we provide a comprehensive evaluation of lncRNA genes’ roles in complex traits and disease.
We identified 1,432 significant lncRNA colocalization events, significantly expanding the lexicon of trait-associated lncRNA genes (Table S6). We defined a “colocalization event” as a robust relationship between a feature (such as a gene for eQTLs or a splice cluster for sQTLs) and a GWAS locus in a certain tissue. Together, these colocalization events encompassed 69 traits and 166 lncRNA features (119 genes, 47 splice clusters). As a point of reference, there were 9,167 significant protein-coding gene colocalization events, involving 82 traits and 1,096 unique features (416 genes, 680 splice clusters). Some trait categories had high proportions of lncRNA eQTL colocalization events, including lupus, multiple sclerosis, and blood cell counts (Figure 5A, S5B). For other traits, such as ALS, Parkinson’s disease, and substance use, no lncRNA colocalization events were observed for either QTL type. In these cases, there were few colocalized QTLs for any gene type; many of the GWAS within these trait categories had low numbers of candidate variants, which limited the number of QTL-GWAS colocalizations that our analyses could detect.
Figure 5.
GWAS-QTL colocalization identifies trait-associated lncRNA genes.
(A) Contribution of each gene type to significant colocalization events, collapsed across tissues (feature-GWAS combinations). GWAS were grouped on the y-axis by general trait categories. For each trait category, the top bar shows eQTL colocalizations and the bottom bar shows sQTL colocalizations. If a bar is missing from the plot, there were no colocalizations for that given trait category and QTL type. The numbers to the right of each bar are the total number of significant colocalization events (E: eQTL, S: sQTL).
(B) Number of significant colocalization events collapsed across tissues (feature-GWAS combinations) for each approach.
(C) Significant lncRNA colocalization events (feature-GWAS-tissue combinations) grouped by the colocalization status of protein-coding genes in the surrounding 1Mb range.
(D) Enrichment of variant annotation categories in the 95% credible sets of all significant lncRNA colocalization events discovered by FINEMAP. Enrichment was calculated relative to all GTEx variants that were not within the credible set and were within 400kb of an annotated gene. Dots represent relative risk point estimate, with bars showing the 95% confidence intervals.
For eQTL colocalization events, 19% of all unique lncRNA gene-GWAS discoveries were shared across all three colocalization approaches (Figure 5B). coloc yielded the highest number of eQTL colocalization discoveries (264), followed by FINEMAP+eCAVIAR (187), and SMR+HEIDI was the most conservative (75) (see Methods for thresholds of each colocalization approach). Compared to protein-coding genes, colocalizations with sQTLs for lncRNA genes were rare due to the low abundance of lncRNA sQTLs (Figure 5B).
Working with multi-tissue QTL data provides the unique opportunity to identify lncRNA genes with the strongest evidence for colocalization compared to any nearby protein-coding genes in the same or a different tissue. We first evaluated whether the lncRNA gene’s colocalization score was the greater than any protein-coding gene in its surrounding 1Mb range. Subsequently, we calculated a metric for how much better the lncRNA gene’s colocalization score was than that of adjacent genes (Table S6, Methods). We detected 574 (40%) lncRNA colocalization events (feature-GWAS-tissue combinations) that were surrounded by genes that either could not be tested for colocalization (for example, if they had no QTLs) or did not have a significant colocalization for the same GWAS in that same tissue (Figure 5C, S5C). An additional 226 (16%) events had neighboring genes with colocalizations that were significant, but with a weaker colocalization than the lncRNA gene, for a total of 800 lncRNA colocalization events with the strongest colocalization score in their 1 Mb region within a given tissue. Notably, we found that these included 120 unique lncRNA feature-GWAS combinations that met the more stringent requirement of having the strongest colocalization score in their 1 Mb region across all tissues (Table S6).
To identify putative causal variants in lncRNA gene-trait associations, we fine-mapped variants for all FINEMAP+eCAVIAR colocalization events where a lncRNA gene had the highest colocalization posterior probability (CLPP) in a 1Mb surrounding region (Table S6, Methods). We observed differences across annotation categories in these fine-mapped variants from lncRNA colocalization events relative to a background set of all GTEx variants either located in a gene or within 400kb of any gene: regulatory regions, such as enhancers and promoters, and non-coding exons were enriched in the set of fine-mapped variants, but these variants were also depleted for splice donor and acceptor sites, intron variants, and CTCF binding sites (Figure 5D).
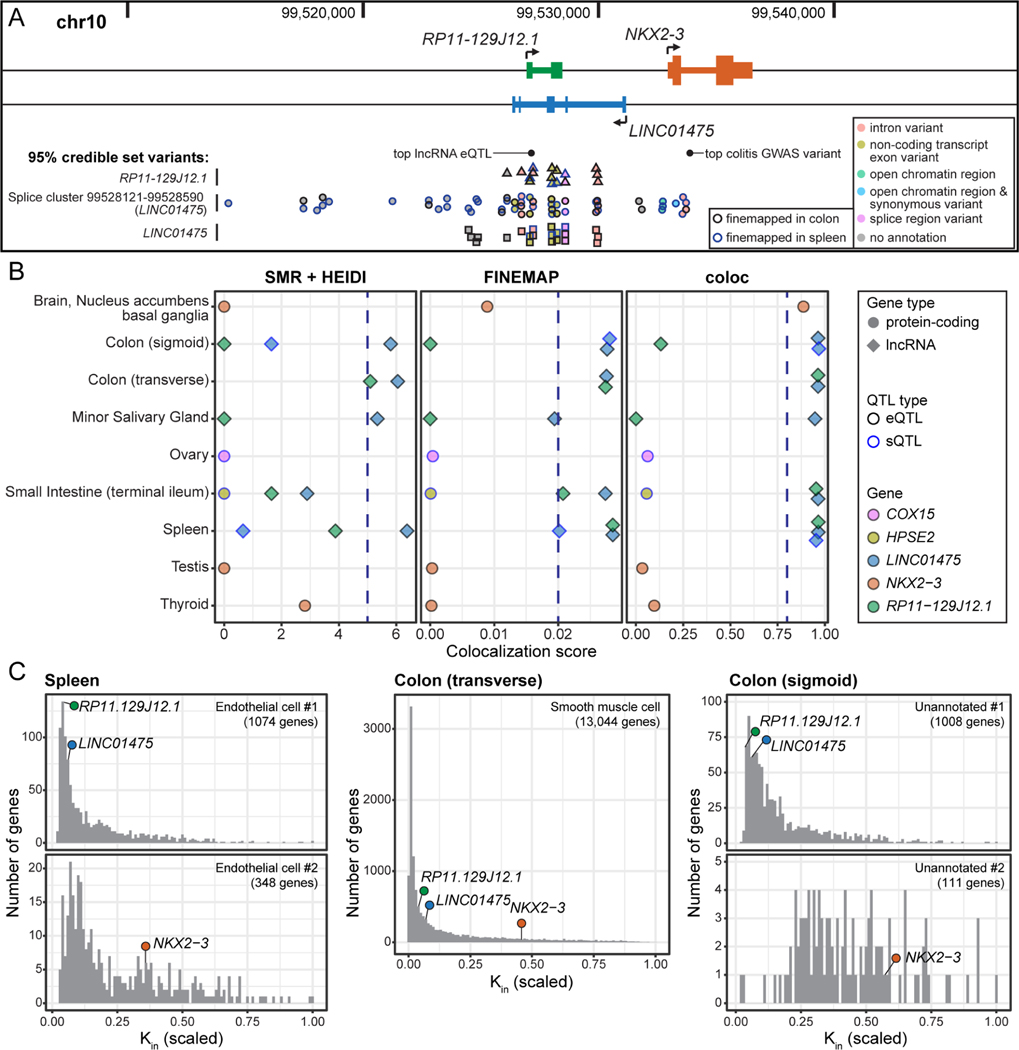
To demonstrate how our combined catalog of colocalization events and cell type annotations can generate hypotheses about lncRNA-mediated trait and disease pathways, we highlighted a colocalization between the lncRNA genes LINC01475 (ENSG00000257582) and RP11–129J12.1 (ENSG00000228778), and ulcerative colitis. The two genes overlap on opposite strands and are just upstream from the protein-coding gene NKX2–3 (Figure 6A). NKX2–3 has received attention related to the significant ulcerative colitis GWAS results in this genomic region (Lu et al., 2014). However, when looking at the colocalization scores, the two lncRNA genes have significant colocalizations with all three colocalization approaches across multiple tissues, whereas NKX2–3 only has a single significant colocalization for one method in one tissue (Figure 6B). Additionally, the tissues in which the lncRNA genes colocalize have relevance to ulcerative colitis: intestinal tissues (sigmoid colon, transverse colon, and small intestine), the spleen, which could be connected via immune system regulation, and the minor salivary gland (Muhvić-Urek et al., 2016). In contrast, the sole colocalization for NKX2–3 occurs in a single brain tissue (nucleus accumbens, basal ganglia). These patterns of colocalizations were also observed for GWAS from inflammatory bowel disease and Crohn’s disease, indicating that the regulatory pathway is involved in the development of both ulcerative colitis and Crohn’s disease.
Figure 6.
Exemplar significant colocalization of LINC01475 and RP11–129J12.1 with ulcerative colitis.
(A) Location of the lncRNA genes, the nearby protein-coding gene NKX2–3. Relevant variants are labeled, including the most significant ulcerative colitis GWAS variant, and the top eQTL for both lncRNA genes in the transverse colon, and the 95% credible sets for the FINEMAP colocalizations in spleen and colon tissues involving a RP11–129J12.1 eQTL (triangles), a LINC01475 sQTL (circles), and a LINC01475 eQTL (squares).
(B) Summary of colocalization scores for ulcerative colitis for the lncRNA genes and genes in the surrounding 1Mb; 14 genes had no score in any tissue, and are not shown. The thresholds for significant colocalization are indicated by the blue dashed line (Methods).
(C) Scaled intramodular connectivity (kin) of LINC01475, RP11–129J12.1, and NKX2–3 within their assigned modules in the gene co-expression networks for spleen, transverse colon, and sigmoid colon. Module annotation and size is indicated in the top right corner of each panel. See also Table S6.
Within the cell type annotations, all three genes were assigned to the same “smooth muscle cell” module in the transverse colon co-expression network, while in the spleen co-expression network, NKX2–3 was assigned to one “endothelial cell” module, and the two lncRNA genes were assigned to a different “endothelial cell” module (Figure 6C). NKX2–3 is a homeobox gene that is key for the development of the spleen and the visceral mesoderm, which develops several essential cell types of the gastrointestinal tract including endothelial cells, immune cells, and - notably - smooth muscle cells. Knockout mouse studies have shown that loss of this gene affects spleen architecture, and lymphocyte maturation and homing (Pabst et al., 1999, 2000; Robles et al., 2016; Tarlinton et al., 2003; Vojkovics et al., 2018). When combined with the colocalization data, this suggests that the functions of these two lncRNAs, through regulation of NKX2–3, in both the colon and spleen influence ulcerative colitis susceptibility.
Discussion
Genetic studies of gene expression have significantly contributed to the identification of the molecular basis of diverse complex traits (Albert and Kruglyak, 2015). To date, the majority of this effort has focused on regulatory effects of non-coding sequences on protein-coding genes, instead of on non-coding genes such as lncRNA genes. Recent advances in population-scale transcriptomics across human tissues from GTEx (The GTEx Consortium, 2020) combined with growing resources from GWAS (Buniello et al., 2019) and population biobanks (Bycroft et al., 2018), now provide the means to systematically incorporate lncRNA genes into these endeavors. To this end, we have assessed lncRNA genes’ regulatory patterns and significantly expanded annotation of their potential roles in specific cellular contexts and across diverse complex traits and diseases. The multi-tissue aspect of the GTEx data allowed us to address a major challenge of identifying trait and disease associations that were specific to lncRNA genes and not instead driven by protein-coding genes in other tissue contexts.
The GTEx v8 data provides extensive annotation of genetic effects impacting lncRNA genes, identifying eQTLs for 67.3% of all 14,100 annotated lncRNA genes (The GTEx Consortium, 2020). However, a major challenge with multi-tissue eQTL data has been assessing the degree of tissue-specificity of genetic effects (Urbut et al., 2019). Our work demonstrated that tissue-specificity of lncRNA gene expression can be influenced by how specificity is defined and we add to evidence of more widespread lncRNA gene expression. Alongside this, we observe that lncRNA eQTLs can be tissue-specific even when the genes are expressed across all tissue types, with 8.8% of lncRNA tissue-specific eGenes expressed in all broad tissue categories. Combined, the GTEx catalog of lncRNA eQTLs greatly expands the annotation of genetic variants influencing lncRNA gene expression and highlights the role of genetic variation in contributing to tissue-specificity.
A complement to multi-tissue transcriptome data has been ongoing efforts to map cellular identities using single-cell sequencing techniques (Darmanis et al., 2015; Han et al., 2018; Regev et al., 2017). Combining these data now provides an opportunity to refine cell type annotations of lncRNA genes. We integrated co-expression analysis with single-cell gene expression reference maps and provide cell type and compartment annotations for 94.4% of lncRNA genes in at least one tissue. These data provide a resource for understanding the cellular contexts of lncRNA genetic effects and subsequently identifying their pathological cellular contexts in diverse diseases.
Both rare and common variants have the potential to impact complex traits and diseases. However, the involvement of genetic variants impacting lncRNA genes and contributing to complex disease remains difficult to ascertain. Examples of prominent rare variants impacting lncRNA genes in disease have included in prostate cancer (Walavalkar et al., 2020), HELLP syndrome (van Dijk et al., 2015), and limb malformation (Allou et al., 2021). By applying gene expression outlier analysis, we were able to identify rare variants that impact lncRNA genes and connect those effects to body-mass index, a highly polygenic trait. To systematically map lncRNA genes to complex traits and diseases, we applied colocalization analysis combining common GWAS, eQTL and sQTL genetic variants across 14,100 lncRNA genes, 101 traits, and 49 tissues using three approaches. We identified 800 lncRNA colocalization events in which there was no stronger protein-coding colocalization within 1 Mb; notably, this included 120 unique lncRNA-GWAS combinations in which no nearby protein-coding genes had a greater colocalization score in any tissue. These colocalization events represent robust connections between genetic variation, lncRNA gene expression, and complex traits. While dissecting the functional impacts of even a single lncRNA gene has been a major challenge, by combining these analyses with enhanced cell type classifications, we have generated a comprehensive catalog of trait-associated lncRNA genes and their cellular contexts.
Limitations of Study
Although this multi-tissue dataset allowed us to identify lncRNA genes with robust connection to cell types and complex diseases, targeted assessment of the cellular and organismal impacts of disease-associated lncRNAs in model systems would further confirm our findings. Additionally, developmental and environmental influences such as immune responsivity, behavior, and medication can impact gene expression and may have different regulatory genetic effects that were not well captured in the GTEx cohort, limiting our ability to catalog the impacts of disease risk variants in all potential contexts. Despite these limitations, these findings significantly extend the discovery of lncRNA genes with potential impacts on human traits and diseases.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Stephen Montgomery (smontgom@stanford.edu).
Materials Availability
Residual GTEx biospecimens have been banked, and are available as a resource for further studies (access can be requested on the GTEx Portal, at https://www.gtexportal.org/home/samplesPage).
Data and Code Availability
All GTEx protected data are available via dbGaP (accession phs000424.v8). Access to the raw sequence data is now provided through the AnVIL platform (https://gtexportal.org/home/protectedDataAccess). Public-access data, including QTL summary statistics and expression levels, are available on the GTEx Portal, as downloadable files and through multiple data visualizations and browsable tables (www.gtexportal.org), as well as in the UCSC and Ensembl browsers.
All components of the single tissue cis-QTL pipeline are available at https://github.com/broadinstitute/gtex-pipeline (https://doi.org/10.5281/zenodo.3727189), and analysis scripts are available at https://github.com/broadinstitute/gtex-v8 (https://doi.org/10.5281/zenodo.3930961). From the colocalization analyses, summary statistics and additional input files can be automatically downloaded and formatted consistently using the scripts available at https://github.com/mikegloudemans/gwas-download.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
GTEx subjects
All human donors were deceased, with informed consent obtained via next-of-kin consent for the collection and banking of de-identified tissue samples for scientific research. The research protocol was reviewed by Chesapeake Research Review Inc., Roswell Park Cancer Institute’s Office of Research Subject Protection, and the institutional review board of the University of Pennsylvania.
There were 838 donors (557 biological sex male, 281 biological sex female). Donors ranged in age from 20–70, with most enrolled donors being older individuals. For more details on donor characteristics and sample collection, see the GTEx v8 main paper (The GTEx Consortium, 2020).
METHOD DETAILS
Biospecimen collection
The biospecimen collection is described in detail in the GTEx v8 main paper (The GTEx Consortium, 2020). In brief, whole blood and skin samples were collected from each donor and shipped overnight to the GTEx Laboratory Data Analysis and Coordination Center (LDACC) at the Broad Institute. These samples were used for DNA genotyping (primarily from whole blood), RNA expression analysis, and culturing and transformation of fibroblast and lymphoblastoid cell lines, respectively. In addition to these samples, two adjacent aliquots were prepared from all other sampled tissues and preserved in PAXgene tissue kits, with ischemic time varying across the different tissue sites. Within each sample pair, one was embedded in paraffin (PFPE) for histopathological review and the second was shipped to the LDACC for processing and molecular analysis. Brains were collected from approximately one-third of the donors, and were shipped on ice to the brain bank at the University of Miami, where eleven brain sub-regions were sampled and flash-frozen. These samples were then shipped to the LDACC for processing and analysis.
A robust quality management program was established and implemented for data management, Standard Operating Procedure (SOP) development, and auditing of collections. Document control software was used to ensure all biospecimen collection sites used current versions of SOPs, and training was conducted prior to implementation of all new procedures. Supporting quality documents were developed to provide consistency and clarity to the program.
Molecular analyte extraction and QC
DNA and RNA extraction and sequencing details are provided in the GTEx v8 main paper (The GTEx Consortium, 2020). The same extraction protocols were used for all GTEx samples to avoid introduction of batch effects among samples, which were processed continually throughout the project. To control for variable RNA quality, RNA sequencing was only performed for samples with a RIN score of 5.5 or higher and with at least 500 ng of total RNA. The 49 tissues with ≥70 genotyped samples that were included in the QTL and other downstream analyses vary in their sample size (n=73 to 706), ischemic time, and RNA quality (RIN). Additionally, the donor age range varies by tissue; notably the brain samples were collected primarily from older individuals.
QUANTIFICATION AND STATISTICAL ANALYSIS
GTEx data
The v8 freeze of GTEx data includes whole genome sequencing (WGS) data from the whole blood of 838 post-mortem individuals, and RNA-sequencing (RNA-seq) data from 54 tissues. Each tissue has a different sample size for RNA-seq; we confined our analyses to the 49 tissues with N >70, for a total of 15,201 samples. For more details on data production, see the GTEx v8 main paper (The GTEx Consortium, 2020).
Poly(A) selection was performed prior to RNA-seq. RNA-seq libraries prepared by ribosomal RNA depletion and by poly(A) selection quantify similar numbers of lncRNA genes (Sultan et al., 2014). lncRNA genes unique to poly(A) selection tend to be antisense transcripts, whereas lncRNA genes unique to ribosomal RNA depletion tend to be intergenic or intronic lncRNA genes (Sultan et al., 2014). However, in the GTEx data, 96.5% of antisense lncRNA genes were detected in at least one tissue and 94% of intergenic lncRNA genes were detected in at least one tissue; additionally, there were no significant differences in median expression level of lncRNA types across tissues (data not shown). This indicates that poly(A) selection has not drastically skewed quantification of lncRNA types in the GTEx dataset. However, poly(A) selection does prevent the quantification of transcripts that are not polyadenylated, such as enhancer RNAs and excised introns (Li et al., 2016; Yang et al., 2011).
For the tissue-specific expression analyses in this paper, the expression data used were gene-level TPM quantifications produced by RNA-SeQC (DeLuca et al., 2012) following read alignment by STAR (Dobin et al., 2013) to the same GENCODE v26 collapsed single-transcript gene annotation from the GTEx v8 main paper (The GTEx Consortium, 2020), which are available on the GTEx Portal.
Defining gene groups
Four gene groups were compared throughout this paper: “total protein-coding genes”, “expression-matched protein-coding genes”, “total lncRNA genes”, and “lncRNA genes with known function”. The “total protein-coding genes” group includes any genes with the “protein_coding” biotype in the GTEx GENCODE v26 GTF (19,291 genes). The “total lncRNA genes” group includes any genes with a long non-coding gene biotype (“processed_transcript”, “non_coding”, “sense_intronic”, “sense_overlapping”, “antisense”, “lincRNA”, “macro_lncRNA”, “bidirectional_promoter_lncRNA”, “3prime_overlapping_ncRNA”) in the GTEx GENCODE v26 GTF (14,100 genes).
The “expression-matched protein-coding genes” group were identified through the pairmatch() function of the R package optmatch (Hansen and Klopfer, 2006), which takes a treatment group and a larger control reservoir and pairs treatment units to controls in a way that minimizes the sum of the discrepancies between these groups. Expression matching was done separately in each tissue, and matches were limited to the middle 50% of lncRNA genes expressed in that tissue (ranked by median TPM values). Limiting matching to the middle 50% of expressed lncRNA genes kept the group from being so large that sub-optimal matches were made just to ensure that each lncRNA gene had a match. Within each tissue, the treatment group was the middle 50% of expressed lncRNA genes, the larger control reservoir was all expressed protein-coding genes, and pairmatch() was run to minimize discrepancies in mean gene expression (TPM). Since expression matching was done within each tissue, the same lncRNA could be matched with different protein-coding genes in different tissues. The expression-matched protein-coding genes across all 49 tissues was a set of 11,178 genes. Whenever possible, the tissue-specific sets of expression-matched protein-coding genes were used, since a pair of genes that have similar expression profiles in one tissue will not necessarily be similar in a different tissue. The only time the entire union set is used at once is the tissue-specificity comparisons in Figure 1F and S1A,C,E.
The “lncRNA genes with known function” are a manually combined set of 954 genes from lncRNAdb (Quek et al., 2015), the HUGO gene nomenclature committee (HGNC, https://www.genenames.org/data/genegroup/#!/group/788), and recent work that identified functional lncRNA genes through splice-site-targeted CRISPR (Liu et al., 2018) and CRISPR interference (Liu et al., 2017) screens; plus 5 genes found in the literature that were not covered in these three sources. Genes from lncRNAdb and the HGNC were only included if they had at least one reference in which direct manipulation of the gene (e.g., knockdown or overexpression) had some effect on cellular phenotype or other genes’ expression.
Assessing tissue-specificity of gene expression
A set of tissue-specific lncRNA genes were defined based on having a significantly higher read count than a non-genic region of the same length, as inspired by microarrays and following an approach described by (Chen et al., 2015). First, the coordinates for all non-genic regions were identified by removing all GENCODE v26 exons from the genome, leaving only regions where there was no exonic sequence on either strand. An additional 100 bp was trimmed from both sides of intronic regions, and 1,000 bp from intergenic regions. Then, for each lncRNA gene, a length-matched non-genic region was mapped. This was done one exon at a time: the exon was shifted to the nearest right non-genic region; if it did not fit in that region, it was shifted to the nearest left non-genic region; and so on, bouncing between the next-nearest right and then the next-nearest left region until a non-genic region was found that the exon fit into. In some cases, this sacrificed exon order to select non-genic regions of the same length that were as close to the actual lncRNA gene as possible. Finally, the read counts of the lncRNA genes were compared to the read counts in their non-genic regions, using a paired one-sided Wilcoxon signed rank test (where n = number of samples for a given tissue, ranging from 85 to 803). Genes were called expressed in a tissue if the lncRNA gene count was significantly greater than its matched non-genic region, with a p-value <0.05.
The 49 GTEx tissues have been assigned by GTEx into 28 broad tissue categories (e.g. “Heart, Atrial Appendage” and “Heart, Left Ventricle” tissues are both in the broad tissue category “Heart”). A given gene was considered tissue-specific if the tissues in which it passed the expression test were all a part of the same broad tissue category, and if the median TPM of the genes in all other tissues was <0.1.
For Figure S1C, Tau scores were used as an additional assessment of tissue-specificity of gene expression (Yanai et al., 2005). Inputs to calculating Tau scores were log2(TPM + 1).
Identifying independent cis-eQTLs and cis-sQTLs by forward stepwise regression-backward selection
The same independent cis-eQTLs and cis-sQTLs mapped using FastQTL (Ongen et al., 2016) for the main GTEx paper were used in this paper, with expression data normalized by Probabilistic Estimation of Expression Residuals (PEER) (Stegle et al., 2012) and edgeR (Robinson et al., 2010) and splicing quantified by LeafCutter (Li et al., 2018). For full details of the methods used, see the GTEx v8 main paper (The GTEx Consortium, 2020). In each tissue, the variants tested were those within 1 Mb of the TSS of each gene and with minor allele frequencies ≥0.01, with the minor allele observed in at least 10 samples of that tissue. Independent eQTLs were used for Figure 1B–D and S1F, and as inputs for the colocalization analyses (see section below). For Figure 1D, the effect sizes of lead eQTLs for protein-coding and lncRNA genes were calculated as allelic fold-change (Mohammadi et al., 2017).
For Figure 1C,D, gene groups were compared by Wilcoxon tests, with the n of each gene group equal to the number all lead eQTLs across all tissues for genes in that gene group. Of the main gene groups compared, this ranged from 10,487 for “lncRNAs with known function” to 325,644 for “protein-coding genes”.
Identifying tissue-shared cis-eQTLs with mash
The mash (Urbut et al., 2019) results from the main GTEx paper were used (The GTEx Consortium, 2020). The output of mash is local false sign rate (LFSR), which is analogous to the false discovery rate, as well as a beta-value effect size. Variant-gene associations with LFSR <0.05 were considered significant. The mash output data were used in analyses of tissue-specificity of eQTLs, in Figure 1F,G, S1G, and Table S2.
Assessing replication of GTEx lncRNA eQTLs
Replication datasets for lncRNA eQTLs were obtained for the following tissues: whole blood (Buil et al., 2015; Lepik et al., 2017; Võsa et al., 2018); EBV-transformed lymphoblastoid cell lines (Buil et al., 2015; Gutierrez-Arcelus et al., 2013); fibroblast cell lines (Gutierrez-Arcelus et al., 2013); brain frontal cortex tissue (Ng et al., 2017); adipose tissue (Buil et al., 2015); and skin (Buil et al., 2015). The eQTL results from Buil et al., Gutierrez-Arcelus et al., and Lepik et al. were obtained from the eQTL Catalogue (Kerimov et al., 2021). For each replication dataset, a set of one variant-gene pair per lncRNA gene was defined, where the variant had the lowest GTEx p-value and the variant-gene pair was also tested in the replication dataset. From this set of shared variant-gene pairs, π1 values were then calculated using the replication dataset’s p-values for those eQTL tests. The π1 value is calculated as π1 = 1 - π0, and π0 is the estimate of the proportion of null p-values as calculated by R package qvalue (Storey et al., 2020). Limitations in this overlap include differences in gene and variant annotation between the studies, expression thresholds for eQTL mapping, and biological differences between groups (for example, GENCORD samples were collected from newborns while GTEx samples were collected from post-mortem adults).
Assessing overlap of GTEx lncRNA eQTLs with other studies’ epigenetic QTLs
Datasets for other types of eQTLs (referred to generally as xQTLs) were obtained for the following tissues and QTL types: DNA methylation (DNAm) and H3K9 acetylation (H3K9Ac) QTLs in brain frontal cortex (Ng et al., 2017); and DNase I hypersensitive site (DHS), H3K27 acetylation (H3K27Ac), and H3K4 mono- and tri-methylation (H3K4Me1 and H3K4Me3) QTLs in EBV-transformed lymphoblastoid cell lines (Grubert et al., 2015). For each xQTL dataset, a set of one variant-gene pair per lncRNA gene was defined, where the variant was also tested in the xQTL dataset and the variant was the closest one to the xQTL peak. From this set of variant-gene pairs, π1 values were then calculated using the xQTL dataset’s p-values for the marker’s QTL tests. The π1 value is calculated as π1 = 1 - π0, and π0 is the estimate of the proportion of null p-values as calculated by R package qvalue (Storey et al., 2020). The range size for these xQTL overlaps varied by study and dataset: within 40kb of all histone marker QTLs, and 5kb of the DNAm and DHS QTLs.
Weighted gene correlation network analysis (WGCNA)
Transcript per million (TPM) values quantified by RNASeQC (DeLuca et al., 2012) were normalized on a per tissue basis using the variance-stabilized normalization (VSN) as implemented by the vsn package (Huber et al., 2002). Only genes which met an expression cutoff of at least 0.1 TPM in at least 20% of samples were included. The effects of gene expression batch, Hardy death type, and ischemic time were removed from normalized TPM values in each tissue using an empirical Bayes linear model implemented by the WGCNA package (Langfelder and Horvath, 2008). Latent factors were not removed from the expression data, because we found that doing so eliminated biological signals necessary for constructing the co-expression networks.
Adjacency matrices were computed using biweight mid-correlation and the default softthresholding power of 12. The adjacency matrix was transformed into a topological overlap matrix (TOM) and then subtracted from 1 to create a dissimilarity TOM suitable for hierarchical clustering.
Co-expression modules were identified using the dynamic tree-cutting approach provided by the WGCNA package (Langfelder and Horvath, 2008; Langfelder et al., 2008). The dissimilarity TOM was transformed into a Euclidean distance matrix and a hierarchical clustering tree was created from this matrix using average-linked hierarchical clustering. The hybrid dynamic tree-cutting algorithm was used with a minimum module size of 50 to prevent the creation of very small modules and a deepSplit parameter of 3 to favor more small modules over few large modules. The pamRespectsDendro parameter was set to true, which will force the partitioning around medoids (PAM) step to respect the hierarchical clustering tree when attempting to assign unclustered genes to modules or to merge very similar modules. This is more conservative than setting pamRespectsDendro to false and leads to more genes remaining unclustered.
Eigengenes were computed from the first principal component of the expression values of the genes assigned to each module. Modules whose eigengenes had a biweight mid-correlation greater than 0.8 were merged using the mergeCloseModules function to reduce the number of highly correlated modules. Module membership was estimated as the biweight midcorrelation between each gene and its module eigengene. Scaled intramodular connectivity (kin) was computed from intramodular connectivity for each gene by dividing its intramodular connectivity by the largest intramodular connectivity value in that module (scaled kin range = 0 to 1).
We observed that the number of co-expression modules defined for each tissue ranged from 8 (in cultured fibroblast cells) to 78 (in ovary), with the number of identified modules unrelated to the sample size of the tissue (Figure S2A). Of these modules, 18% (in ovary) to 81% (in stomach) were annotated (Figure S2B). This percentage was related to both the number of modules in a tissue, and the possible cell type gene sets established for that tissue. For most tissues, just over half of lncRNA genes met the expression requirements to be included in the co-expression networks (median included lncRNA genes across tissues = 53%; see Figure 2A). The proportion of included genes was higher for lncRNA genes with known functions (median = 71%).
Co-expression network module annotation via gene set enrichment
In large modules, gene set enrichment was limited to the top 500 genes as ranked by module membership. Enrichments for both cell compartments and cell types were assessed.
For cell compartment gene sets, only four terms were tested: nucleolus (GO:0005730), mitochondrion (GO:0005739), mitochondrial inner membrane (GO:0005743), and cytosolic ribosome (GO:0022626). This was because our aim was to eliminate spurious cell type enrichment driven by cell compartment rather than cell types. Enrichment was computed using the hypergeometric overlap test between the genes in the module and list of cell compartment genes provided by GO Cellular Compartment (Ashburner et al., 2000; The Gene Ontology Consortium, 2019). Enrichment p-values were adjusted for the number of modules and the number of terms in each tissue network using Bonferroni correction.
Enrichment for cell types across 20 tissues were computed using cell type specificity index (SI) values estimated by the pSI package (Dougherty et al., 2010; Xu et al., 2014) in the datasets described in the Methods section “Cell type annotation sources”. For each module, a linear model was fitted with a dummy variable indicating membership in the module or (or the top 500 genes in the module for large modules) as the predictor, and the SI values as the outcome. Only genes found in both the network and annotation were used in the model. Because a lower SI value indicates a higher cell type specificity, only models with a negative coefficient were considered a valid enrichment. All enrichment p-values were Bonferroni-corrected for the number of modules and the number of cell types in each dataset.
Final annotations were decided by combining the cell compartment and cell type annotations. Modules with strong enrichment for mitochondria or ribosomes were annotated for those compartments over cell types. Cell types present in multiple tissues from the annotation sources (such as resident immune cells, epithelial cells, or stromal cells) were accepted as annotations if there was agreement from multiple tissues. Tissue-specific cell types were only used as a module annotation if they were in the appropriate tissue. For tissues without a direct analog in the annotation sources, cell type annotations were assigned by looking across all available tissues in the annotation sources. If a module was annotated for a mix of cell types, it was submitted to Enrichr (Chen et al., 2013; Kuleshov et al., 2016) to assist in identifying other pathways. If this did not clarify the annotation, the module was called “unannotated”.
Cell type annotation sources
Blood cell type-specific expression data was obtained from a published dataset (GSE24759) (Novershtern et al., 2011). Raw CEL files were imported and normalized with RMA using the affy package (Gautier et al., 2004). Progenitor cells and cell types with a small number of samples were excluded, and some cell subtypes were aggregated. The final dataset used to estimate specificity index values was the averaged expression values for the samples corresponding to the following 13 cell types: naive CD4+ T-cell, memory CD4+ T-cell, naive CD8+ T-cell, memory CD8+ T-cell, naive B-cell, mature B-cell, mature NK cell, monocyte, myeloid dendritic cell, granulocyte (neutrophil), basophil, eosinophil, and megakaryocyte.
Cell type-specific expression for the central nervous system was obtained from a published single-cell RNA dataset (Darmanis et al., 2015). Fetal cell types were excluded and log counts were averaged for estimating specificity index values for six cell types: neuron, astrocyte, oligodendrocyte, oligodendrocyte progenitor cell (OPC), microglia and endothelial cell.
The remaining cell type specific expression data was obtained from the Mouse Cell Atlas (Han et al., 2018). Counts data was downloaded from Mouse Cell Atlas website (http://bis.zju.edu.cn/MCA/) and log normalized. All adult tissues with a matching tissue in GTEx were used except peripheral blood and brain and neonatal heart was also included because no adult heart sample was available. Some cell subtypes were collapsed, and averaged cell type matrices were computed for 18 tissues: heart, kidney, liver, lung, mammary gland (involution), mammary gland (lactation), mammary gland (pregnancy), mammary gland (virgin), muscle, ovary, skin, pancreas, prostate, small intestine, spleen, stomach, testis, and uterus.
Identifying lncRNA genes with high confidence cell type annotations in brain and blood tissues
For lncRNA genes assigned to cell type modules in brain and blood tissues, we assessed which of these genes’ annotation was further supported by correlation of the gene’s expression with that cell type’s estimated proportion. First, we estimated cell composition in all GTEx samples from brain tissues and whole blood using CIBERSORTx (Newman et al., 2019). The LM22 blood cell type reference (Newman et al., 2015) provided by CIBERSORTx was used for estimating blood cell composition, while a published single cell brain dataset (Darmanis et al., 2015) was used as a reference from brain regions. Default settings were used for creating the signature matrix for the brain reference and imputing cell fractions, with the recommended B-mode batch correction being used to normalize GTEx samples to the reference datasets.
Then, within each GTEx brain tissue and within GTEx whole blood, we performed a Pearson correlation test between each lncRNA gene’s expression and each cell type’s estimated proportion. The input gene expression data was log2(read counts + 1). The n for each test was the number of samples for the given brain or blood tissue, ranging from 139 to 755. Benjamini-Hochberg multiple test correction (Benjamini and Hochberg, 1995) was done across all tests within each tissue, and a significant correlation was an adjusted p-value <0.05.
Finally, we identified which lncRNA genes showed agreement in their WGCNA module annotations, and their correlation with CIBERSORTx-estimated cell type proportion. For brain, since there are ten different brain tissue types sampled in GTEx, we required agreement of WGCNA and CIBERSORTx-based annotation in multiple brain tissues. Specifically, to have high confidence cell type annotation, a lncRNA gene had to be significantly correlated with the same cell type in at least four brain tissues, and this correlation had to be in the same direction (all positive or all negative). In at least one of these brain tissues, the gene must be assigned to a WGCNA module annotated as that cell type. The gene must also not be assigned to a WGCNA module annotated as any other cell type in the other brain tissues (although a cell compartment annotation, such as “mitochondria”, would be acceptable). For blood, since there is only one tissue type (whole blood), we could not be as stringent as we were with gene annotations in brain tissue. Instead, a lncRNA gene had high confidence cell type annotation if it was significantly correlated with the same cell type to which it was annotated using WGCNA. The cell types estimated by CIBERSORTx for blood were more specific than the annotation categories used in WGCNA, so the two approaches just had to annotate the gene to similar cell types to be considered in agreement. For example, significant correlation with CIBERSORTx-estimated proportion of “T cells, CD4 naive” and WGCNA assignment to “T cell” module in whole blood was considered an agreement.
Allele-specific expression (ASE)
Autosomal ASE data were produced using GATK ASEReadCounter tool (Castel et al., 2015) and the WASP filtering strategy (van de Geijn et al., 2015) to remove read mapping bias, as described in the GTEx main paper (The GTEx Consortium, 2020) and the GTEx ASE companion paper (Castel et al., 2020). ASE sites were removed if they were in low-mappability regions (75-mer mappability with leq2 mismatches < 1), showed mapping bias in simulation (Panousis et al., 2014), or had no more reads supporting two alleles than would be expecting from sequencing noise alone, indicating potential genotyping error (FDR < 1%, see Castel et al., 2015 for description of test).
We used phASER-POP (Castel et al., 2016) to obtain p-values for regulatory effects of the unique set of top eQTL and sQTL in every gene in every tissue in GTEx. Inputs for phASER-POP were the WASP-corrected haplotype expression matrix, the read-backed phased VCF generated for the GTEx ASE companion paper (Castel et al., 2020), and gene-variant pairs corresponding to the top eQTL and sQTL for every gene in every tissue obtained from the eQTL and sQTL summary statistics generated for the GTEX main paper (The GTEx Consortium, 2020). We used the default setting of 10,000 bootstrap samples for estimating allelic fold change (aFC) p-values. Although this approach allows us to score and assign significance to the level of ASE as mediated by a given variant, a limitation of it was that only genes with either an eQTL or sQTL could be analyzed.
Since the aFC p-values were obtained via bootstrapping, some variants had p-values of 0. These values were changed to 1×10−4 (the equivalent to one bootstrap sample supporting the null) so that they could be converted to finite Z scores. Variant-level Z scores were calculated from aFC p-values, and averaged over the variants in each gene to produce gene-level Z scores. For a given gene, its mean neighbor Z score was calculated by averaging the gene-level Z scores for all genes within +/−500kb of the gene.
We defined ASE-sharing as all occurrences where a gene-level Z score and its corresponding mean neighbor Z score was greater than 3. We computed gene biotype enrichment for ASE-sharing using Fisher’s test, comparing the number of genes showing ASE-sharing in intergenic lncRNA genes and antisense lncRNA genes to protein coding genes, relative to the total number of genes in each class. The numbers of genes in each comparison are provided in Figure 3B.
ASE data can be noisy, since differences in read coverage along a gene results in variation in ASE measurements across the informative variants within that gene. Although we have mitigated this by aggregating ASE scores across the variants in a gene and by using stringent Z score thresholds to select a robust set of genes with high ASE, one might want to prioritize genes with low variation in their ASE measurements. In addition to reporting the mean Z score for a gene and its neighboring genes, we also provide the coefficients of variation for these two Z scores (Table S4).
Multi-tissue gene expression outlier discovery
We subset expression data in each tissue to genes with 6 reads and TPM > 0.1 in at least 20% of individuals. Within each tissue, the TPM values were log transformed, (log2(TPM + 2)), and scaled across individuals for each gene. We regressed out the effects of the first three genotype principal components, sex, and hidden factors discovered via PEER (Stegle et al., 2012), the number of which depends on sample size for that tissue and was consistent with the GTEx eQTL discovery pipeline (The GTEx Consortium, 2020): for tissues with less than 150 samples, we removed 15 PEER factors; less than 250 samples, 30 factors; less than 350, 45 factors; and 60 for the remaining tissues. We additionally corrected for the genotype of the strongest cis-eQTL per gene per tissue to magnify rare variant effects, which has been shown to improve nearby rare variant enrichments (Ferraro et al., 2020). We then re-scaled expression values across individuals within each gene to generate corrected Z scores per individual per gene per tissue.
For each gene-individual pair, if that individual has expression measurements in at least five tissues, we calculate a median Z score for that gene. We define outlier individuals as those with a |median Z score| greater than 2, and non-outliers as all other individuals for the same set of genes. We removed as global outliers 39 individuals for whom the proportion of tested genes were outliers at a threshold of |median Z score| > 3 exceeded 1.5 times the interquartile range of the distribution of proportion outlier genes per individual. For outlier analysis, we include all autosomal intergenic lncRNA and protein-coding genes. We focused much of our outlier analysis on widely-expressed intergenic lncRNA genes, which were genes with read counts significantly greater than their length-matched non-genic regions in all tissues.
To assess variant enrichments, we subset to 714 individuals who self-report with European ancestry, as allele frequencies are less comparable between continental populations. We retain all SNPs and indels that pass quality control in the GTEx VCF. Structural variants were called in a subset of these individuals as in Chiang et al., 2017 with GenomeSTRiP GSCNQUAL (Handsaker et al., 2015) set to limit the false discovery rate (FDR) for each variant type. GenomeSTRiP’s IntensityRankSumAnnotator was used to evaluate FDR based on available Illumna Human Omni 5M gene expression array data. GSCNQUAL was limited to ≥ 1 for GenomeSTRiP deletions and ≥ 8 for multi-allelic copy number variants, corresponding to an FDR of 10%. The GSCNQUAL cutoff for GenomeSTRiP duplications was set at ≥ 17, the point where the FDR plateaued at 15.1% and did not fluctuate more than ±1% for over 50 steps in increasing GSCNQUAL score. Additionally, the Mobile Element Locator Tool (MELT) (Gardner et al., 2017) version 2.1.4 was run using MELT-SPLIT to identify ALU, SVA, and LINE1 insertions into the test genomes. MELT calls that were categorized as “PASS” in the VCF info field, had an ASSESS score ≥ 3, and SR count ≥ 3 were retained.
We define rare variants as those with < 1% frequency in GTEx, and for SNPs and indels, also < 1% frequency in non-Finnish Europeans from the gnomAD database (Karczewski et al., 2020). Remaining bins are defined by GTEx allele frequencies. We calculate relative risk as the proportion of outlier individuals with a variant of a given frequency within 10kb of the outlier gene or in the gene body over the proportion of non-outlier individuals with a variant of a given frequency within 10kb of or in the gene body of the same set of genes. Variant categories were annotated using Ensembl VEP (version 88) and each gene-individual pair was assigned to the most enriched variant category, regardless of the number of nearby rare variants.
Data to assess the GWAS effect size for body-mass index in outlier-associated rare variants was obtained from http://www.nealelab.is/uk-biobank. We first identified all intergenic lncRNA gene expression outliers with a nearby rare variant that was not observed in any control individuals (those with |median Z score| < 1), and that had no protein-coding gene expression outliers within 1Mb. At an outlier threshold of |median Z score| ≥2, this produced an outlier pool of 44 rare variants for 26 outlier intergenic lncRNA genes. We next identified a non-outlier pool of 3,173 rare variants from control individuals with |median Z score| <1 for these same genes. We then performed 1,000 permutations of randomly selecting one outlier variant and one non-outlier variant from each gene, and calculating the mean GWAS effect sizes in the two groups; these produce the distributions shown in Figure 4D and compared by Wilcoxon test.
GWAS sources and data preparation for colocalization analysis
We downloaded publicly available full-genome summary statistics from 176 papers (Table S6). We focused on studies of diseases and traits related to neurological and immune function, since these are contexts in which smaller scale lncRNA studies have found these genes to have compelling roles (Heward and Lindsay, 2014; Roberts et al., 2014). We re-formatted the GWAS statistics into a standardized tab-separated format for compatibility with our colocalization pipeline tools, and indexed them using the bgzip and tabix command line utilities using the above Github repository.
Selecting colocalization tests
To restrict the total number of intended colocalization tests to a computationally tractable number, we first performed a naive overlap test of the GWAS summary statistics and the GTEx sQTL and eQTL association summary statistics. For each GWAS, we selected all SNPs in any of our selected GWAS with a nominal association p-value < 1e-12, chosen as the least stringent threshold that was computationally tractable. We additionally required that selected SNPs be at least 1 Mb apart from all SNPs already selected from the same GWAS, to ensure independence of effects and different loci. For every selected GWAS SNP, we identified all eQTL or sQTL features (gene expression and splice junction usage, respectively) that had a QTL association p-value < 1e-5 at any SNP positioned within 10kb of the most significant GWAS SNP at the locus. We make no quantitative claims about the significance of these naïve overlaps, as many such overlaps could be expected by chance; these merely formed the set of loci to test for colocalization in subsequent steps.
The result from this step was a list of 1,153 unique lead SNPs from 176 GWAS, and 49 GTEx QTL tissues with a total of 13,804 individual QTL features. Each site to be tested for colocalization consisted of a SNP / GWAS trait / QTL feature / QTL tissue combination.
We then tested every resulting pair of GWAS locus and QTL feature in our set, using three gene prioritization or colocalization methods: SMR+HEIDI (Zhu et al., 2016), FINEMAP+eCAVIAR (Benner et al., 2016; Hormozdiari et al., 2016) and coloc (Giambartolomei et al., 2014). For each of these GWAS-QTL pairs, we then narrowed our summary statistics to the set of the SNPs tested for association with both the given GWAS trait and the given QTL trait, and removed all sites containing less than 50 SNPs after this filter.
Colocalization approaches: SMR + HEIDI
Summary data-based Mendelian Randomization (SMR) tests for association between a feature and trait using variant-feature and variant-trait association statistics in a two-sample Mendelian Randomization framework (Zhu et al., 2016). The HEIDI test, typically applied to significant SMR results, eliminates cases where the association is driven by linkage or proximity of independent causal variants rather than a shared causal variant.
We ran SMR using the default parameter settings to obtain an SMR p-value for each locus and a HEIDI p-value at each locus. For the remainder of the tested loci, we report the Bonferroni-adjusted SMR p-values. Significant colocalizations were those with the number of overlapping GWAS/eQTL SNPs within the tested window >50, an SMR Bonferroni-adjusted p-value <1e-05, and that did not show evidence of heterogeneity of estimated effects (i.e. did not show evidence of linkage) using the HEIDI test (HEIDI p-value ≥ 0.05). SMR+HEIDI colocalization scores were reported as -log10(SMR adjusted p-value).
Colocalization approaches: FINEMAP
FINEMAP is a variant association fine-mapping tool that identifies the set of causal variants with a predefined probability for a given GWAS or QTL (Benner et al., 2016). eCAVIAR then can be used to combine the FINEMAP outputs for a given GWAS and a given QTL, and compute the probability of their colocalization (Hormozdiari et al., 2016).
Using the full 1000 Genomes dataset from phase 3 (2,504 individuals) as a reference population (Auton et al., 2015), we estimated LD between all of these SNP pairs using PLINK (Purcell et al., 2007). We then ran FINEMAP independently on the GWAS and the QTL summary stats to obtain posterior probabilities of causality for each of the remaining SNPs. These probabilities were then combined to compute a colocalization posterior probability (CLPP) using the formula described in the eCAVIAR method. These are the values we report in the paper, with significant colocalizations having CLPP ≥ 0.02, with the number of overlapping GWAS/eQTL SNPs within the tested window >50. We ran FINEMAP using the default settings and assumed the existence of exactly one causal variant in both the GWAS and the QTL summary stats, in accordance with common practice. FINEMAP colocalization scores were the CLPP scores.
Colocalization approaches: coloc
coloc is a Bayesian approach that estimates the support for all possible hypotheses involving a given GWAS and a given QTL: H0, neither trait has a genetic association in the region; H1: only the GWAS has a genetic association in the region; H2: only the QTL has a genetic association in the region; H3: both the GWAS and QTL are associated, but with different causal variants; H4: both the GWAS and QTL are associated and share a single causal variant (Giambartolomei et al., 2014).
At a given locus, we estimated the allele frequencies for each variant appearing in both the GWAS and the QTL summary statistics. The allele frequencies were supplied along with the p-values as inputs to the coloc package (using the coloc.abf function to estimate posterior probability of colocalization). The default priors were used for the function. The summary statistics we reported for each locus are the PP4 value, which denotes the probability that both the eQTL and the GWAS have a variant at the locus with non-zero effect, and that they are the same causal variant. Significant colocalizations were those that had a PP4 ≥ 0.80, with the number of overlapping GWAS/eQTL SNPs within the tested window >50. coloc colocalization scores were the PP4 values.
Integrating and interpreting the colocalization results
The GTEx variant annotation, which was used when examining the 95% credible sets of variants from FINEMAP colocalizations, was compiled from Ensembl’s Variant Effect Predictor and Loss-Of-Function Transcript Effect Estimator (VEP v85), and the Ensembl Regulatory Build (The GTEx Consortium, 2020; Zerbino et al., 2015).
For the neighboring genes assessment, each significant lncRNA gene colocalization score was compared to the colocalization scores for the same GWAS, GTEx tissue, and colocalization approach of all protein-coding genes 500kb up or downstream of the gene. In addition to categorically evaluating whether the lncRNA gene’s colocalization score was the greatest in its 1Mb range, we also calculated a metric comparing a lncRNA gene’s score to its adjacent genes. First, the score differences were calculated for each colocalization event: this was (gene of interest’s colocalization score - greatest adjacent protein-coding gene’s colocalization score). Then, since each colocalization approach has different scoring scales, these score differences were converted into ranks. All colocalization events with a score difference of 0 (i.e. the gene of interest had the exact same colocalization score as a nearby protein-coding gene) were given the same rank. These ranks were then scaled by the total number of significant colocalizations discovered by that approach. Finally, each colocalization event was discovered by anywhere from one to three different approaches: we calculated an aggregated score by taking the mean of the scaled rank values for each approach that discovered this colocalization (GWAS-feature-tissue combination).
Supplementary Material
Data S1. Gene-module Assignments and Connectivity Results from Weighted Gene Co-expression Network Analysis (WGCNA), and Module Annotations, Related to Figures 2 and S2. Field descriptions: tissue: GTEx tissue; ensembl_gene_id: Ensembl gene ID; symbol: gene symbol; gene_type: gene type, as annotated by GENCODE v26; assigned_module: module name (colors, not related to module annotation); kTotal: connectivity to the whole network; kWithin: connectivity within assigned module; kOut: connectivity to genes outside of assigned module; kDiff: kWithin - kOut; kWithin_scaled: kWithin values, scaled by the largest kWithin of that module (1 = most connected gene, 0 = least connected gene); module_annotation: cell type, compartment, or pathway annotation of module; annotation_class: general category of module annotation; moduleMembership_assigned_module: correlation of the gene to its assigned module eigengene; assigned_module_size: number of genes in module; kWithin_scaled_ranking: ranking of the gene within its module based on kWithin score (with the top rank of 1 = most connected); kWithin_scaled_decile_rank: the decile group of kWithin_scaled_ranking (with a decile of 1 indicating the top most connected genes), used for Figures 2D, S2C.
Table S1. Test Results for Significant Expression Above Length-matched Non-genic Region for All lncRNA Genes, Related to Figures 1 and S1. Includes annotation of the 954 lncRNA genes selected for the “lncRNA genes with known function” gene group.
Table S2. List of lncRNA Genes with Tissue-specific Expression and Tissue-specific eGenes (MashR LFSR <0.05) Across the 28 Broad Tissue Categories of GTEx, Related to Figures 1 and S1.
Table S3. lncRNA Genes with Refined Cell Type Annotation in Brain and Blood Tissues Based on Corresponding WGCNA Module Annotations and Correlation Between the Gene’s Expression and CIBERSORTx-estimated Proportion of that Cell Type, Related to Figures 2 and S3.
Table S4. Gene-level and Neighbor-level Allele-specific Expression (ASE) Values, Related to Figure 3.
Table S5. Multi-tissue Outliers in Intergenic lncRNA Gene Expression, Related to Figures 4 and S4.
Table S6. Summary of Significant Colocalization Events Detected by At Least One Colocalization Approach, Related to Figures 5, 6, and S5. Tab 2 summarizes all unique seed-SNP/feature/GWAS/tissue colocalizations. Since seed SNP is also included, some of the 1,432 colocalization events described in text (unique feature/GWAS/tissue combinations) are duplicated. The seed SNP (field “ref_snp”) was the starting point for the analysis, and not necessarily the causal SNP; it provided in this table for completeness of information. Includes information about GWAS used in colocalization analyses. Tab 3 provides the colocalization results for the 120 lncRNA gene-GWAS associations that were the strongest in their 1Mb region across all GTEx tissues. Tab 4 lists the 95% credible set of variants for colocalizations identified by FINEMAP+eCAVIAR in which a lncRNA has the best colocalization score in its 1Mb region.
Figure S1. Gene Expression and eQTL Patterns of lncRNA and Protein-coding Genes, Related to Figure 1, and Tables S1 and S2.
(A) The number of tissues expressing each gene type, at an expression threshold of TPM >0.1 in >20% of samples. Bar labels show the number of genes.
(B) The proportion of each gene group expressed in each tissue, at a threshold of TPM >0.1 in >20% of samples. Data points reflect the proportion of a gene group expressed in a given tissue, with point color indicating the tissue. The overlaying boxplots represent the medians with first and third quartiles as boxes, and whiskers extending to 1.5 times the interquartile range. The most extreme values are labeled with the tissue name and number of genes expressed.
(C) Tissue-specificity of gene expression based on Tau scores. Since the testis tissue has such a distinctive gene expression profile and drove much of the observed tissue-specificity, Tau scores were calculated both including expression values from the testis tissue (left) and excluding the testis tissue (right). The boxplots represent the medians with first and third quartiles as boxes, and whiskers extending to 1.5 times the interquartile range.
(D) Illustration of how lncRNA genes were called expressed relative to background read counts. The cartoon (left) shows how matched non-genic regions were found for lncRNA genes. Note that non-genic regions could be intergenic or intronic, as long as they did not overlap with a gene on either strand; and that proximity to the original lncRNA gene was prioritized over maintaining “exon” order. The read count histogram (right) shows an example of a lncRNA gene with greater expression than its matched non-genic region.
(E) The number of tissues expressing each lncRNA gene type, based on testing read counts between the lncRNA genes and their matched non-genic regions. Bar labels show the number of genes.
(F) Proportion of expressed lncRNA genes that were also eGenes, separated by lncRNA gene type. Boxplots reflect the range of proportions across the 49 GTEx tissues, representing the medians with first and third quartiles as boxes, and whiskers extending to 1.5 times the interquartile range.
(G) For each set of broad tissue-specific eGenes (rows of heatmap), the shading indicates the proportion that were expressed in a given number of tissues (columns of heatmap) at a threshold of ≥0.1 TPM in >20% of samples. Both protein-coding and lncRNA tissue-specific eGenes are included in this heatmap. The labels indicate many of the tissue-specific eGenes were expressed in the corresponding number of tissues. There is no row for Breast tissue because it had no tissue-specific eGenes.
Figure S3. Identifying lncRNA Genes with High-confidence Cell Type Annotation in Blood and Brain Tissues Using CIBERSORTx, Related to Figure 2C and Table S3.
(A) Estimated cell proportions of the six different cell types calculated by CIBERSORTx (the panels of the plot) in each of the GTEx brain tissues (x-axis).
(B) Median TPM of lncRNA genes across brain tissues, based on whether their expression was significantly correlated with the CIBERSORTx estimated proportion of a certain cell type (and also coincided with WGCNA-based module annotation in that brain tissue). In all boxplots, the data represented are the medians with first and third quartiles as boxes, and whiskers extending to 1.5 times the interquartile range.
(C) Estimated cell proportions of the different blood cell types calculated by CIBERSORTx in blood. The data represented are the medians with first and third quartiles as boxes, and whiskers extending to 1.5 times the interquartile range.
(D) Number of genes correlated with a given cell type proportion, as estimated by CIBERSORTx.
Figure S2. Connecting Genes Through Weighted Gene Co-expression Network Analysis (WGCNA), Related to Figure 2 and Data S1.
(A) The number of co-expression modules identified in each tissue was not related to the sample size of the tissue. The fill color of points indicates tissue.
(B) The number of modules with a cell type or cell compartment annotation versus the number that were unannotated in each tissue. There was also one module of unclustered genes per tissue, which is not depicted.
(C) Proportion of gene groups binned by intra-modular connectivity (kin) ranking, in the largest modules (left; 1,876-16,840 genes) and smallest modules (right; 50-72 genes). The most highly connected genes within their module are in the first kin rank decile, and the least connected genes within their module are in the tenth kin rank decile.
(D) Median expression level (TPM) of protein-coding and lncRNA genes binned by their kin rank decile. On average, gene expression was higher in genes with greater intra-modular connectivity. Data represented are the medians with first and third quartiles as boxes, and whiskers extending to 1.5 times the interquartile range.
(E) Module annotations of the genes in the top kin rank decile of their modules. The box fill reflects the proportion of genes assigned to a module with that annotation. Since genes were assigned to modules in multiple tissues, the labels reflect gene-tissue combinations, not individual genes.
Figure S4. Outliers in Intergenic lncRNA Gene Expression, Related to Figure 4 and Table S5.
(A) Summary of lncRNA/protein-coding gene pairs that were within 10kb of each other and both outliers in the same individual.
(B) The distance between intergenic lncRNA genes and protein-coding genes that were called multi-tissue outliers in the same individual and were on the same chromosome. The dashed line indicates the threshold of 10kb for the lncRNA/protein-coding gene outlier pairs.
(C) Enrichment of outlier events in specific gene groups relative to all genes. Data represented are the relative risk ratios, with bars showing the 95% confidence interval.
(D) Presence of rare variants (MAF <1%) within 10kb of the outlier gene based on outlier status. The bold line separates outlier events with some nearby rare variant from those with no nearby rare variant (white fill).
lincRNA = intergenic lncRNA, TSS = transcription splice site, TE = transposable element insertion, BND = breakend, DEL = deletion, CNV = copy number variation, DUP = duplication, INV = inversion.
Figure S5. GWAS-QTL Colocalization Events Involving lncRNA Genes, Related to Figure 5 and Table S6.
(A) The number of specific traits covered by the general trait categories used in Figure 5A and Figure S5B. The green and blue bars reflect the number of tested traits with eQTL or sQTL colocalizations (respectively) with any gene, not just lncRNA genes.
(B) Contribution of each gene type to significant colocalization events, collapsed across tissues (feature-GWAS combinations) in a separate panel for each colocalization approach. GWAS were grouped on the y-axis by the general trait categories from Figure S5A. For each trait category, the top bar shows eQTL colocalizations and the bottom bar shows sQTL colocalizations. If a bar is missing from the plot, there were no colocalizations for that given trait category and QTL type. The numbers to the right of each bar are the total number of significant colocalization events.
(C) Significant lncRNA colocalization events (feature-GWAS-tissue combinations) grouped by the colocalization status of neighboring protein-coding genes. Each panel summarizes the results from one colocalization approach. Of the three colocalization approaches, SMR+HEIDI was the least favorable for finding lncRNA gene colocalizations with a strong, standalone signal, partially because this approach did not discover many lncRNA gene colocalizations to begin with. The numbers of events for each method will not sum up to the numbers in Figure 5C, since many colocalization methods were detected by multiple methods.
Highlights:
29% of lncRNA genes with eQTLs show tissue-specific genetic regulation
Co-expression networks and single-cell data provide annotations for 94% of lncRNAs
Rare variants near lncRNA expression outliers impact complex traits, like BMI
We identify 800 lncRNA-trait relationships not explained by protein-coding genes
Acknowledgments:
We thank the Montgomery and Kirkegaard labs for their feedback on this work. The GTEx Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health (NIH) and by the National Cancer Institute, the National Human Genome Research Institute (NHGRI), the National Heart, Lung, and Blood Institute (NHLBI), the National Institute on Drug Abuse (NIDA), the National Institute of Mental Health and the National Institute of Neurological Disorders and Stroke. We are thankful for support from a Stanford Graduate Fellowship & Bio-X Stanford Interdisciplinary Graduate Fellowship (O.M.de G.), National Science Foundation Graduate Research Fellowship (N.M.F.), NHLBI grant R01HL135313-01 (A.S.R.), NLM training grant 5T15LM007033-36 (T.Y.E.), grants HHSN268201000029C & 5U41HG009494 (F.A., K.G.A.), NIH grant R01GM122924 (S.E.C., T.L.), grant 1K99HG009916-01 (S.E.C.), a Marie-Skłodowska Curie fellowship H2020 Grant 706636 (S.K.-H.), NIH grant R01HG010067 (Y.P.), a Mr. and Mrs. Spencer T. Olin Fellowship for Women in Graduate Study (A.J.S.), grant R01MH109905 (A.B.), the Searle Scholar Program (A.B.), grant R01MH101822 (C.D.B.), NIH grants R01MH106842, R01HL142028, UM1HG008901, & R01GM124486 (T.L.), NIH grants R01MH107666 & P30DK020595 (H.K.I.), NIH grants R01HL109512, R01HL134817, R33HL120757, & R01HL139478 (T.Q.), the Chan Zuckerberg Foundation-Human Cell Atlas Initiative (T.Q.), Stanford University School of Medicine (K.K.), NIH grants R01MH101814 (NIH Common Fund; GTEx Program) (A.B., S.B.M), R01HG008150 (NHGRI; Non-Coding Variants Program) (A.B., S.B.M), R01AG066490, R01HL142015, U01HG009431, & U01HG009080 (S.B.M).
Declaration of Interests: F.A. is an inventor on a patent application related to TensorQTL; S.E.C. is a co-founder and chief technology officer at Variant Bio, and owns stock in Variant Bio; T.L. is on the scientific advisory board of Variant Bio, Goldfinch Bio, and GSK, and owns stock in Variant Bio; S.B.M. is on the scientific advisory board of MyOme Inc.
Footnotes
Secondary Author List:
Shankara Anand, Stacey Gabriel, Gad A. Getz, Aaron Graubert, Kane Hadley, Robert E. Handsaker, Katherine H. Huang, Xiao Li, Daniel G. MacArthur, Samuel R. Meier, Jared L. Nedzel, Duyen T. Nguyen, Ayellet V. Segrè, Ellen Todres, Brunilda Balliu, Rodrigo Bonazzola, Andrew Brown, Donald F. Conrad, Daniel J. Cotter, Nancy Cox, Sayantan Das, Emmanouil T. Dermitzakis, Jonah Einson, Barbara E. Engelhardt, Eleazar Eskin, Elise D. Flynn, Laure Fresard, Eric R. Gamazon, Diego Garrido-Martín, Nicole R. Gay, Roderic Guigó, Andrew R. Hamel, Yuan He, Paul J. Hoffman, Farhad Hormozdiari, Lei Hou, Brian Jo, Silva Kasela, Seva Kashin, Manolis Kellis, Alan Kwong, Xin Li, Yanyu Liang, Serghei Mangul, Pejman Mohammadi, Manuel Muñoz-Aguirre, Andrew B. Nobel, Meritxell Oliva, Yongjin Park, Princy Parsana, Ferran Reverter, John M. Rouhana, Chiara Sabatti, Ashis Saha, Matthew Stephens, Barbara E. Stranger, Nicole A. Teran, Ana Viñuela, Gao Wang, Fred Wright, Valentin Wucher, Yuxin Zou, Pedro G. Ferreira, Gen Li, Marta Melé, Esti Yeger-Lotem, Debra Bradbury, Tanya Krubit, Jeffrey A. McLean, Liqun Qi, Karna Robinson, Nancy V. Roche, Anna M. Smith, David E. Tabor, Anita Undale, Jason Bridge, Lori E. Brigham, Barbara A. Foster, Bryan M. Gillard, Richard Hasz, Marcus Hunter, Christopher Johns, Mark Johnson, Ellen Karasik, Gene Kopen, William F. Leinweber, Alisa McDonald, Michael T. Moser, Kevin Myer, Kimberley D. Ramsey, Brian Roe, Saboor Shad, Jeffrey A. Thomas, Gary Walters, Michael Washington, Joseph Wheeler, Scott D. Jewell, Daniel C. Rohrer, Dana R. Valley, David A Davis, Deborah C. Mash, Mary E. Barcus, Philip A. Branton, Leslie Sobin, Laura K. Barker, Heather M. Gardiner, Maghboeba Mosavel, Laura A. Siminoff, Paul Flicek, Maximilian Haeussler, Thomas Juettemann, W. James Kent, Christopher M. Lee, Conner C. Powell, Kate R. Rosenbloom, Magali Ruffier, Dan Sheppard, Kieron Taylor, Stephen J. Trevanion, Daniel R. Zerbino, Nathan S. Abell, Joshua Akey, Lin Chen, Kathryn Demanelis, Jennifer A. Doherty, Andrew P. Feinberg, Kasper D Hansen, Peter F. Hickey, Farzana Jasmine, Lihua Jiang, Rajinder Kaul, Muhammad G. Kibriya, Jin Billy Li, Qin Li, Shin Lin, Sandra E. Linder, Brandon L. Pierce, Lindsay F. Rizzardi, Andrew D. Skol, Kevin S. Smith, Michael Snyder, John Stamatoyannopoulos, Hua Tang, Meng Wang, Latarsha J. Carithers, Ping Guan, Susan E. Koester, A. Roger Little, Helen M. Moore, Concepcion R. Nierras, Abhi K. Rao, Jimmie B. Vaught, Simona Volpi
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albert FW, and Kruglyak L. (2015). The role of regulatory variation in complex traits and disease. Nature Reviews Genetics 16, 197–212. [DOI] [PubMed] [Google Scholar]
- Allou L, Balzano S, Magg A, Quinodoz M, Royer-Bertrand B, Schöpflin R, Chan W-L, Speck-Martins CE, Carvalho DR, Farage L, et al. (2021). Non-coding deletions identify Maenli lncRNA as a limb-specific En1 regulator. Nature 1–6. [DOI] [PubMed] [Google Scholar]
- Amin V, Harris RA, Onuchic V, Jackson AR, Charnecki T, Paithankar S, Subramanian SL, Riehle K, Coarfa C, and Milosavljevic A. (2015). Epigenomic footprints across 111 reference epigenomes reveal tissue-specific epigenetic regulation of lincRNAs. Nat Commun 6, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000). Gene Ontology: tool for the unification of biology. Nat Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Abecasis GR, Altshuler DM, Durbin RM, Abecasis GR, Bentley DR, Chakravarti A, Clark AG, Donnelly P, Eichler EE, et al. (2015). A global reference for human genetic variation. Nature 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeira AN, Bonazzola R, Gamazon ER, Liang Y, Park Y, Kim-Hellmuth S, Wang G, Jiang Z, Zhou D, Hormozdiari F, et al. (2021). Exploiting the GTEx resources to decipher the mechanisms at GWAS loci. Genome Biology 22, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y. (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological) 57, 289–300. [Google Scholar]
- Benner C, Spencer CCA, Havulinna AS, Salomaa V, Ripatti S, and Pirinen M. (2016). FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomba L, Walter K, and Soranzo N. (2017). The impact of rare and low-frequency genetic variants in common disease. Genome Biology 18, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, and Willard HF (1991). A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38–44. [DOI] [PubMed] [Google Scholar]
- Buil A, Brown AA, Lappalainen T, Viñuela A, Davies MN, Zheng H-F, Richards JB, Glass D, Small KS, Durbin R, et al. (2015). Gene-gene and gene-environment interactions detected by transcriptome sequence analysis in twins. Nature Genetics 47, 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, et al. (2019). The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47, D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, et al. (2018). The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, and Rinn JL (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Levy-Moonshine A, Mohammadi P, Banks E, and Lappalainen T. (2015). Tools and best practices for data processing in allelic expression analysis. Genome Biology 16, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Mohammadi P, Chung WK, Shen Y, and Lappalainen T. (2016). Rare variant phasing and haplotypic expression from RNA sequencing with phASER. Nature Communications 7, 12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Aguet F, Mohammadi P, Aguet F, Anand S, Ardlie KG, Gabriel S, Getz GA, Graubert A, Hadley K, et al. (2020). A vast resource of allelic expression data spanning human tissues. Genome Biology 21, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, and Ma’ayan A. (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H-IH, Liu Y, Zou Y, Lai Z, Sarkar D, Huang Y, and Chen Y. (2015). Differential expression analysis of RNA sequencing data by incorporating non-exonic mapped reads. BMC Genomics 16, S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Scott AJ, Davis JR, Tsang EK, Li X, Kim Y, Hadzic T, Damani FN, Ganel L, GTEx Consortium, et al. (2017). The impact of structural variation on human gene expression. Nature Genetics 49, 692–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Gephart MGH, Barres BA, and Quake SR (2015). A survey of human brain transcriptome diversity at the single cell level. PNAS 112, 7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire M-D, Williams C, Reich M, Winckler W, and Getz G. (2012). RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28, 1530–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk M, Visser A, Buabeng KML, Poutsma A, van der Schors RC, and Oudejans CBM (2015). Mutations within the LINC-HELLP non-coding RNA differentially bind ribosomal and RNA splicing complexes and negatively affect trophoblast differentiation. Human Molecular Genetics 24, 5475–5485. [DOI] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. (2012). Landscape of transcription in human cells. Nature 489, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty JD, Schmidt EF, Nakajima M, and Heintz N. (2010). Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res 38, 4218–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. (2013). The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341, 1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE St. Laurent G III, Kenny PJ, and Wahlestedt C. (2008). Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat Med 14, 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro NM, Strober BJ, Einson J, Abell NS, Aguet F, Barbeira AN, Brandt M, Bucan M, Castel SE, Davis JR, et al. (2020). Transcriptomic signatures across human tissues identify functional rare genetic variation. Science 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner EJ, Lam VK, Harris DN, Chuang NT, Scott EC, Pittard WS, Mills RE, Consortium, T. 1000 G.P., and Devine SE (2017). The Mobile Element Locator Tool (MELT): population-scale mobile element discovery and biology. Genome Res. 27, 1916–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, and Irizarry RA (2004). affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315. [DOI] [PubMed] [Google Scholar]
- van de Geijn B, McVicker G, Gilad Y, and Pritchard JK (2015). WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nature Methods 12, 1061–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, and Plagnol V. (2014). Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLoS Genet 10, e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubert F, Zaugg JB, Kasowski M, Ursu O, Spacek DV, Martin AR, Greenside P, Srivas R, Phanstiel DH, Pekowska A, et al. (2015). Genetic Control of Chromatin States in Humans Involves Local and Distal Chromosomal Interactions. Cell 162, 1051–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai M-C, Hung T, Argani P, Rinn JL, et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Arcelus M, Lappalainen T, Montgomery SB, Buil A, Ongen H, Yurovsky A, Bryois J, Giger T, Romano L, Planchon A, et al. (2013). Passive and active DNA methylation and the interplay with genetic variation in gene regulation. ELife 2, e00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F, et al. (2018). Mapping the Mouse Cell Atlas by Microwell-Seq. Cell 173, 1307. [DOI] [PubMed] [Google Scholar]
- Handsaker RE, Van Doren V, Berman JR, Genovese G, Kashin S, Boettger LM, and McCarroll SA (2015). Large multiallelic copy number variations in humans. Nature Genetics 47, 296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen BB, and Klopfer SO (2006). Optimal Full Matching and Related Designs via Network Flows. Journal of Computational and Graphical Statistics 15, 609–627. [Google Scholar]
- Heward JA, and Lindsay MA (2014). Long non-coding RNAs in the regulation of the immune response. Trends in Immunology 35, 408–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon C-C, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJL, Gough J, Denisenko E, Schmeier S, Poulsen TM, Severin J, et al. (2017). An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543, 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F, van de Bunt M, Segrè AV, Li X, Joo JWJ, Bilow M, Sul JH, Sankararaman S, Pasaniuc B, and Eskin E. (2016). Colocalization of GWAS and eQTL Signals Detects Target Genes. The American Journal of Human Genetics 99, 1245–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sültmann H, Poustka A, and Vingron M. (2002). Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18, S96–S104. [DOI] [PubMed] [Google Scholar]
- Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. (2015). The landscape of long noncoding RNAs in the human transcriptome. Nature Genetics 47, 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Cheng S-J, Ren L-C, Wang Q, Kang Y-J, Ding Y, Hou M, Yang X-X, Lin Y, Liang N, et al. (2019). An expanded landscape of human long noncoding RNA. Nucleic Acids Res 47, 7842–7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A, and Clark AG (2012). Recent Explosive Human Population Growth Has Resulted in an Excess of Rare Genetic Variants. Science 336, 740–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerimov N, Hayhurst JD, Peikova K, Manning JR, Walter P, Kolberg L, Samoviča M, Sakthivel MP, Kuzmin I, Trevanion SJ, et al. (2021). eQTL Catalogue: a compendium of uniformly processed human gene expression and splicing QTLs . BioRxiv 2020.01.29.924266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornienko AE, Dotter CP, Guenzl PM, Gisslinger H, Gisslinger B, Cleary C, Kralovics R, Pauler FM, and Barlow DP (2016). Long non-coding RNAs display higher natural expression variation than protein-coding genes in healthy humans. Genome Biology 17, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, and Horvath S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Zhang B, and Horvath S. (2008). Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24, 719–720. [DOI] [PubMed] [Google Scholar]
- Lepik K, Annilo T, Kukuškina V, eQTLGen Consortium, Kisand K, Kutalik Z, Peterson P, and Peterson H. (2017). C-reactive protein upregulates the whole blood expression of CD59 - an integrative analysis. PLOS Computational Biology 13, e1005766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, and Rosenfeld MG (2016). Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet 17, 207–223. [DOI] [PubMed] [Google Scholar]
- Li YI, Knowles DA, Humphrey J, Barbeira AN, Dickinson SP, Im HK, and Pritchard JK (2018). Annotation-free quantification of RNA splicing using LeafCutter. Nature Genetics 50, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, et al. (2017). CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Cao Z, Wang Y, Guo Y, Xu P, Yuan P, Liu Z, He Y, and Wei W. (2018). Genome-wide screening for functional long noncoding RNAs in human cells by Cas9 targeting of splice sites. Nature Biotechnology 36, 1203–1210. [DOI] [PubMed] [Google Scholar]
- Lu X, Tang L, Li K, Zheng J, Zhao P, Tao Y, and Li L-X (2014). Contribution of NKX2–3 Polymorphisms to Inflammatory Bowel Diseases: A Meta-Analysis of 35358 subjects. Scientific Reports 4, 3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli K, Volders P-J, Gerhardinger C, Lee JC, Maass PG, Melé M, and Rinn JL (2019). High-throughput functional analysis of lncRNA core promoters elucidates rules governing tissue specificity. Genome Res. 29, 344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, et al. (2015). The human transcriptome across tissues and individuals. Science 348, 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melé M, Mattioli K, Mallard W, Shechner DM, Gerhardinger C, and Rinn JL (2017). Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 27, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi P, Castel SE, Brown AA, and Lappalainen T. (2017). Quantifying the regulatory effect size of cis-acting genetic variation using allelic fold change. Genome Res. 27, 1872–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhvić-Urek M, Tomac-Stojmenović M, and Mijandrušić-Sinčić B. (2016). Oral pathology in inflammatory bowel disease. World J Gastroenterol 22, 5655–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, and Alizadeh AA (2015). Robust enumeration of cell subsets from tissue expression profiles. Nature Methods 12, 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, Khodadoust MS, Esfahani MS, Luca BA, Steiner D, et al. (2019). Determining cell type abundance and expression from bulk tissues with digital cytometry. Nature Biotechnology 37, 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng B, White CC, Klein H-U, Sieberts SK, McCabe C, Patrick E, Xu J, Yu L, Gaiteri C, Bennett DA, et al. (2017). An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nature Neuroscience 20, 1418–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. (2011). Densely Interconnected Transcriptional Circuits Control Cell States in Human Hematopoiesis. Cell 144, 296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongen H, Buil A, Brown AA, Dermitzakis ET, and Delaneau O. (2016). Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics 32, 1479–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst O, Zweigerdt R, and Arnold HH (1999). Targeted disruption of the homeobox transcription factor Nkx2–3 in mice results in postnatal lethality and abnormal development of small intestine and spleen. Development 126, 2215–2225. [DOI] [PubMed] [Google Scholar]
- Pabst O, Förster R, Lipp M, Engel H, and Arnold H-H (2000). NKX2.3 is required for MAdCAM-1 expression and homing of lymphocytes in spleen and mucosa-associated lymphoid tissue. The EMBO Journal 19, 2015–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panousis NI, Gutierrez-Arcelus M, Dermitzakis ET, and Lappalainen T. (2014). Allelic mapping bias in RNA-sequencing is not a major confounder in eQTL studies. Genome Biology 15, 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. (2007). PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quek XC, Thomson DW, Maag JLV, Bartonicek N, Signal B, Clark MB, Gloss BS, and Dinger ME (2015). lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res 43, D168–D173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JJ, and Chang HY (2016). Unique features of long non-coding RNA biogenesis and function. Nature Reviews Genetics 17, 47–62. [DOI] [PubMed] [Google Scholar]
- Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell P, Carninci P, Clatworthy M, et al. (2017). The Human Cell Atlas. ELife 6, e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TC, Morris KV, and Wood MJA (2014). The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos Trans R Soc Lond B Biol Sci 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles EF, Mena-Varas M, Barrio L, Merino-Cortes SV, Balogh P, Du M-Q, Akasaka T, Parker A, Roa S, Panizo C, et al. (2016). Homeobox NKX2–3 promotes marginal-zone lymphomagenesis by activating B-cell receptor signalling and shaping lymphocyte dynamics. Nat Commun 7, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegle O, Parts L, Piipari M, Winn J, and Durbin R. (2012). Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc 7, 500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Bass AJ, Dabney A, Robinson D, and Warnes G. (2020). qvalue: Q-value estimation for false discovery rate control. [Google Scholar]
- Sultan M, Amstislavskiy V, Risch T, Schuette M, Dökel S, Ralser M, Balzereit D, Lehrach H, and Yaspo M-L (2014). Influence of RNA extraction methods and library selection schemes on RNA-seq data. BMC Genomics 15, 675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarlinton D, Light A, Metcalf D, Harvey RP, and Robb L. (2003). Architectural Defects in the Spleens of Nkx2–3-Deficient Mice Are Intrinsic and Associated with Defects in Both B Cell Maturation and T Cell-Dependent Immune Responses. The Journal of Immunology 170, 4002–4010. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2019). The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47, D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium (2020). The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbut SM, Wang G, Carbonetto P, and Stephens M. (2019). Flexible statistical methods for estimating and testing effects in genomic studies with multiple conditions. Nature Genetics 51, 187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojkovics D, Kellermayer Z, Kajtár B, Roncador G, Vincze Á, and Balogh P. (2018). Nkx2–3—A Slippery Slope From Development Through Inflammation Toward Hematopoietic Malignancies. Biomark Insights 13, 1177271918757480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Võsa U, Claringbould A, Westra H-J, Bonder MJ, Deelen P, Zeng B, Kirsten H, Saha A, Kreuzhuber R, Kasela S, et al. (2018). Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. BioRxiv 447367. [Google Scholar]
- Walavalkar K, Saravanan B, Singh AK, Jayani RS, Nair A, Farooq U, Islam Z, Soota D, Mann R, Shivaprasad PV, et al. (2020). A rare variant of African ancestry activates 8q24 lncRNA hub by modulating cancer associated enhancer. Nature Communications 11, 3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, and Chang HY (2011). Molecular Mechanisms of Long Noncoding RNAs. Molecular Cell 43, 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O, and Chang HY (2011). Long noncoding RNAs and human disease. Trends in Cell Biology 21, 354–361. [DOI] [PubMed] [Google Scholar]
- Wright CF, FitzPatrick DR, and Firth HV (2018). Paediatric genomics: diagnosing rare disease in children. Nature Reviews Genetics 19, 253–268. [DOI] [PubMed] [Google Scholar]
- Xia B, Yan Y, Baron M, Wagner F, Barkley D, Chiodin M, Kim SY, Keefe DL, Alukal JP, Boeke JD, et al. (2020). Widespread Transcriptional Scanning in the Testis Modulates Gene Evolution Rates. Cell 180, 248–262.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Wells AB, O’Brien DR, Nehorai A, and Dougherty JD (2014). Cell Type-Specific Expression Analysis to Identify Putative Cellular Mechanisms for Neurogenetic Disorders. J. Neurosci 34, 1420–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R, Bar-Even A, Horn-Saban S, Safran M, Domany E, et al. (2005). Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21, 650–659. [DOI] [PubMed] [Google Scholar]
- Yang L, Duff MO, Graveley BR, Carmichael GG, and Chen L-L (2011). Genomewide characterization of non-polyadenylated RNAs. Genome Biol 12, R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al. (2013). lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 500, 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Wilder SP, Johnson N, Juettemann T, and Flicek PR (2015). The Ensembl Regulatory Build. Genome Biology 16, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, Montgomery GW, Goddard ME, Wray NR, Visscher PM, et al. (2016). Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nature Genetics 48, 481–487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Gene-module Assignments and Connectivity Results from Weighted Gene Co-expression Network Analysis (WGCNA), and Module Annotations, Related to Figures 2 and S2. Field descriptions: tissue: GTEx tissue; ensembl_gene_id: Ensembl gene ID; symbol: gene symbol; gene_type: gene type, as annotated by GENCODE v26; assigned_module: module name (colors, not related to module annotation); kTotal: connectivity to the whole network; kWithin: connectivity within assigned module; kOut: connectivity to genes outside of assigned module; kDiff: kWithin - kOut; kWithin_scaled: kWithin values, scaled by the largest kWithin of that module (1 = most connected gene, 0 = least connected gene); module_annotation: cell type, compartment, or pathway annotation of module; annotation_class: general category of module annotation; moduleMembership_assigned_module: correlation of the gene to its assigned module eigengene; assigned_module_size: number of genes in module; kWithin_scaled_ranking: ranking of the gene within its module based on kWithin score (with the top rank of 1 = most connected); kWithin_scaled_decile_rank: the decile group of kWithin_scaled_ranking (with a decile of 1 indicating the top most connected genes), used for Figures 2D, S2C.
Table S1. Test Results for Significant Expression Above Length-matched Non-genic Region for All lncRNA Genes, Related to Figures 1 and S1. Includes annotation of the 954 lncRNA genes selected for the “lncRNA genes with known function” gene group.
Table S2. List of lncRNA Genes with Tissue-specific Expression and Tissue-specific eGenes (MashR LFSR <0.05) Across the 28 Broad Tissue Categories of GTEx, Related to Figures 1 and S1.
Table S3. lncRNA Genes with Refined Cell Type Annotation in Brain and Blood Tissues Based on Corresponding WGCNA Module Annotations and Correlation Between the Gene’s Expression and CIBERSORTx-estimated Proportion of that Cell Type, Related to Figures 2 and S3.
Table S4. Gene-level and Neighbor-level Allele-specific Expression (ASE) Values, Related to Figure 3.
Table S5. Multi-tissue Outliers in Intergenic lncRNA Gene Expression, Related to Figures 4 and S4.
Table S6. Summary of Significant Colocalization Events Detected by At Least One Colocalization Approach, Related to Figures 5, 6, and S5. Tab 2 summarizes all unique seed-SNP/feature/GWAS/tissue colocalizations. Since seed SNP is also included, some of the 1,432 colocalization events described in text (unique feature/GWAS/tissue combinations) are duplicated. The seed SNP (field “ref_snp”) was the starting point for the analysis, and not necessarily the causal SNP; it provided in this table for completeness of information. Includes information about GWAS used in colocalization analyses. Tab 3 provides the colocalization results for the 120 lncRNA gene-GWAS associations that were the strongest in their 1Mb region across all GTEx tissues. Tab 4 lists the 95% credible set of variants for colocalizations identified by FINEMAP+eCAVIAR in which a lncRNA has the best colocalization score in its 1Mb region.
Figure S1. Gene Expression and eQTL Patterns of lncRNA and Protein-coding Genes, Related to Figure 1, and Tables S1 and S2.
(A) The number of tissues expressing each gene type, at an expression threshold of TPM >0.1 in >20% of samples. Bar labels show the number of genes.
(B) The proportion of each gene group expressed in each tissue, at a threshold of TPM >0.1 in >20% of samples. Data points reflect the proportion of a gene group expressed in a given tissue, with point color indicating the tissue. The overlaying boxplots represent the medians with first and third quartiles as boxes, and whiskers extending to 1.5 times the interquartile range. The most extreme values are labeled with the tissue name and number of genes expressed.
(C) Tissue-specificity of gene expression based on Tau scores. Since the testis tissue has such a distinctive gene expression profile and drove much of the observed tissue-specificity, Tau scores were calculated both including expression values from the testis tissue (left) and excluding the testis tissue (right). The boxplots represent the medians with first and third quartiles as boxes, and whiskers extending to 1.5 times the interquartile range.
(D) Illustration of how lncRNA genes were called expressed relative to background read counts. The cartoon (left) shows how matched non-genic regions were found for lncRNA genes. Note that non-genic regions could be intergenic or intronic, as long as they did not overlap with a gene on either strand; and that proximity to the original lncRNA gene was prioritized over maintaining “exon” order. The read count histogram (right) shows an example of a lncRNA gene with greater expression than its matched non-genic region.
(E) The number of tissues expressing each lncRNA gene type, based on testing read counts between the lncRNA genes and their matched non-genic regions. Bar labels show the number of genes.
(F) Proportion of expressed lncRNA genes that were also eGenes, separated by lncRNA gene type. Boxplots reflect the range of proportions across the 49 GTEx tissues, representing the medians with first and third quartiles as boxes, and whiskers extending to 1.5 times the interquartile range.
(G) For each set of broad tissue-specific eGenes (rows of heatmap), the shading indicates the proportion that were expressed in a given number of tissues (columns of heatmap) at a threshold of ≥0.1 TPM in >20% of samples. Both protein-coding and lncRNA tissue-specific eGenes are included in this heatmap. The labels indicate many of the tissue-specific eGenes were expressed in the corresponding number of tissues. There is no row for Breast tissue because it had no tissue-specific eGenes.
Figure S3. Identifying lncRNA Genes with High-confidence Cell Type Annotation in Blood and Brain Tissues Using CIBERSORTx, Related to Figure 2C and Table S3.
(A) Estimated cell proportions of the six different cell types calculated by CIBERSORTx (the panels of the plot) in each of the GTEx brain tissues (x-axis).
(B) Median TPM of lncRNA genes across brain tissues, based on whether their expression was significantly correlated with the CIBERSORTx estimated proportion of a certain cell type (and also coincided with WGCNA-based module annotation in that brain tissue). In all boxplots, the data represented are the medians with first and third quartiles as boxes, and whiskers extending to 1.5 times the interquartile range.
(C) Estimated cell proportions of the different blood cell types calculated by CIBERSORTx in blood. The data represented are the medians with first and third quartiles as boxes, and whiskers extending to 1.5 times the interquartile range.
(D) Number of genes correlated with a given cell type proportion, as estimated by CIBERSORTx.
Figure S2. Connecting Genes Through Weighted Gene Co-expression Network Analysis (WGCNA), Related to Figure 2 and Data S1.
(A) The number of co-expression modules identified in each tissue was not related to the sample size of the tissue. The fill color of points indicates tissue.
(B) The number of modules with a cell type or cell compartment annotation versus the number that were unannotated in each tissue. There was also one module of unclustered genes per tissue, which is not depicted.
(C) Proportion of gene groups binned by intra-modular connectivity (kin) ranking, in the largest modules (left; 1,876-16,840 genes) and smallest modules (right; 50-72 genes). The most highly connected genes within their module are in the first kin rank decile, and the least connected genes within their module are in the tenth kin rank decile.
(D) Median expression level (TPM) of protein-coding and lncRNA genes binned by their kin rank decile. On average, gene expression was higher in genes with greater intra-modular connectivity. Data represented are the medians with first and third quartiles as boxes, and whiskers extending to 1.5 times the interquartile range.
(E) Module annotations of the genes in the top kin rank decile of their modules. The box fill reflects the proportion of genes assigned to a module with that annotation. Since genes were assigned to modules in multiple tissues, the labels reflect gene-tissue combinations, not individual genes.
Figure S4. Outliers in Intergenic lncRNA Gene Expression, Related to Figure 4 and Table S5.
(A) Summary of lncRNA/protein-coding gene pairs that were within 10kb of each other and both outliers in the same individual.
(B) The distance between intergenic lncRNA genes and protein-coding genes that were called multi-tissue outliers in the same individual and were on the same chromosome. The dashed line indicates the threshold of 10kb for the lncRNA/protein-coding gene outlier pairs.
(C) Enrichment of outlier events in specific gene groups relative to all genes. Data represented are the relative risk ratios, with bars showing the 95% confidence interval.
(D) Presence of rare variants (MAF <1%) within 10kb of the outlier gene based on outlier status. The bold line separates outlier events with some nearby rare variant from those with no nearby rare variant (white fill).
lincRNA = intergenic lncRNA, TSS = transcription splice site, TE = transposable element insertion, BND = breakend, DEL = deletion, CNV = copy number variation, DUP = duplication, INV = inversion.
Figure S5. GWAS-QTL Colocalization Events Involving lncRNA Genes, Related to Figure 5 and Table S6.
(A) The number of specific traits covered by the general trait categories used in Figure 5A and Figure S5B. The green and blue bars reflect the number of tested traits with eQTL or sQTL colocalizations (respectively) with any gene, not just lncRNA genes.
(B) Contribution of each gene type to significant colocalization events, collapsed across tissues (feature-GWAS combinations) in a separate panel for each colocalization approach. GWAS were grouped on the y-axis by the general trait categories from Figure S5A. For each trait category, the top bar shows eQTL colocalizations and the bottom bar shows sQTL colocalizations. If a bar is missing from the plot, there were no colocalizations for that given trait category and QTL type. The numbers to the right of each bar are the total number of significant colocalization events.
(C) Significant lncRNA colocalization events (feature-GWAS-tissue combinations) grouped by the colocalization status of neighboring protein-coding genes. Each panel summarizes the results from one colocalization approach. Of the three colocalization approaches, SMR+HEIDI was the least favorable for finding lncRNA gene colocalizations with a strong, standalone signal, partially because this approach did not discover many lncRNA gene colocalizations to begin with. The numbers of events for each method will not sum up to the numbers in Figure 5C, since many colocalization methods were detected by multiple methods.
Data Availability Statement
All GTEx protected data are available via dbGaP (accession phs000424.v8). Access to the raw sequence data is now provided through the AnVIL platform (https://gtexportal.org/home/protectedDataAccess). Public-access data, including QTL summary statistics and expression levels, are available on the GTEx Portal, as downloadable files and through multiple data visualizations and browsable tables (www.gtexportal.org), as well as in the UCSC and Ensembl browsers.
All components of the single tissue cis-QTL pipeline are available at https://github.com/broadinstitute/gtex-pipeline (https://doi.org/10.5281/zenodo.3727189), and analysis scripts are available at https://github.com/broadinstitute/gtex-v8 (https://doi.org/10.5281/zenodo.3930961). From the colocalization analyses, summary statistics and additional input files can be automatically downloaded and formatted consistently using the scripts available at https://github.com/mikegloudemans/gwas-download.