Abstract
COVID-19 vaccination efficacy depends on serum levels of the neutralizing antibodies (NAs) specific to the receptor-binding domain of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein. Therefore, a high-throughput rapid assay capable of measuring the total SARS-CoV-2 NA level is urgently needed for COVID-19 serodiagnosis, convalescent plasma therapy, vaccine development, and assessment. Here, we developed a novel nanoplasmonic immunosorbent assay (NanoPISA) platform for one-step rapid quantification of SARS-CoV-2 NAs in clinical serum samples for high-throughput evaluation of COVID-19 vaccine effectiveness. The NanoPISA platform enhanced by the use of nanoporous hollow gold nanoparticle coupling was able to detect SARS-CoV-2 NAs with a limit of detection of 0.2 pM within 15 min without washing steps. The one-step NanoPISA for SARS-CoV-2 NA detection in clinical specimens yielded good results, comparable with those obtained in the gold-standard seroneutralization test and the surrogate virus-neutralizing enzyme-linked immunosorbent assay. Collectively, the one-step NanoPISA might be a rapid and high-throughput NA-quantification platform for evaluating the effectiveness of COVID-19 vaccines.
Keywords: COVID-19, SARS-CoV-2 neutralizing antibody, SARS-CoV-2 variant, Nanoplasmonic sensor, Gold nanoparticles
1. Introduction
The COVID-19 pandemic triggered by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) started in late 2019 (Chauhan et al., 2020; Gao et al., 2020). The rapid spread COVID-19 and exponential growth of the virus have caused health and economic havoc on a global scale, exceeding the limits of the existing healthcare and intensive care unit capacities (Muruato et al., 2020; Ranoa et al., 2020). Although the development of vaccines have effectively helped control the SARS-CoV-2 epidemic and limit morbidity and mortality, SARS-CoV-2 is evolving into new variants with the progression of transmission (Kannan et al., 2021; Shrotri et al., 2021). The current SARS-CoV-2 variants such as Delta, Delta Plus, and Lambda are more transmissible than preexisting variants, and they have rapidly become the dominant variants in several countries (CDC, 2021; Kannan et al., 2021). Whether the SARS-CoV-2 variants can re-infect people who have been vaccinated mainly depends on the long-term efficacy of vaccines and the level of neutralizing antibodies against SARS-CoV-2 variants (Chen et al., 2021, Kissler et al., 2020). Thus, quantitative detection of NAs will help to efficiently evaluate COVID-19 vaccine effectiveness and determine whether a COVID-19 vaccine booster shot is needed.
Spike (S) proteins are the major surface antigens expressed on the SARS-CoV-2 virus surface (Laurini et al., 2020; Wang et al., 2021). The receptor-binding domain of the SARS-CoV-2 S protein (S-RBD) can specifically attach to the host receptor angiotensin-converting enzyme 2 for viral entry into cells (Brufsky, 2020; Ge et al., 2013). Thus, S-RBD is among the most preferred targets for the creation of vaccines or therapeutics against SARS-CoV-2 (Du et al., 2009; Kang et al., 2021). The SARS-CoV-2 NAs produced in response to most vaccines effectively target the S-RBD of SARS-CoV-2 and block viral entry (Hoffmann et al., 2020). Different types of SARS-CoV-2 vaccines, including inactivated vaccines, nucleic acid vaccines, and adenovirus-based vector vaccines, have been successively approved for marketing worldwide (Chen et al., 2021; Dong et al., 2020; Krammer, 2020). The traditional and gold-standard method for evaluating vaccine effectiveness is the seroneutralization (SN) test, which is accurate but time-consuming and labor-intensive (Krammer, 2020). Generally, it takes 2–4 days to complete the SN test, which considerably impedes the evaluation of the effects of large-scale vaccination in a given population (Zettl et al., 2020). Thus, there is a pressing need for a simple, rapid, and high-throughput alternative method to evaluate the level of antibodies produced after vaccination (Kasetsirikul et al., 2020; Zettl et al., 2020).
Currently, efforts are being undertaken in both biomedical industry and basic academic research to identify new diagnostic solutions and develop point-of-care devices to detect SARS-CoV-2-specific antibodies. For example, the enzyme-linked immunosorbent assays (ELISAs) (Bundschuh et al., 2020; MacMullan et al., 2020), chemiluminescence assays (Gambino et al., 2020; Nuccetelli et al., 2020), and lateral flow immunoassays (LFIAs) (Huang et al., 2020) have recently been made available for the detection of SARS-CoV-2 antibodies. Although ELISA and chemiluminescence assays are accurate and sensitive, their time-consuming and complex multi-step operations complicate the application of these tests in a cost-effective and labor-saving manner. Moreover, LFIAs show superiority in point-of-care detection of SARS-CoV-2 antibodies because of their simple operation and rapid detection. In contrast, the conventional LFIA uses colloidal gold nanoparticles (GNPs) as the label probe and suffers from low throughput and sensitivity. A recently developed novel detection technology based on nanoplasmonic biosensors has allowed the scalability of detection schemes, low-cost operation, and high-throughput measurements, making it a highly promising option for point-of-care, rapid detection applications (Cetin and Topkaya, 2019; Hu et al., 2019; Soler et al., 2017). Additionally, owing to their excellent surface plasmon resonance (SPR) properties, GNPs have been widely used to enhance the detection sensitivity of nanoplasmonic biosensors (Belushkin et al., 2018, 2020). Previously, we reported a low-cost GNP-coupled nanoplasmonic sensor that allowed one-step rapid detection and quantification of the SARS-CoV-2 pseudovirus (Huang et al., 2021). Nanoporous hollow (NH) GNPs are core-shelled gold nanostructures with a hollow interior (Melancon et al., 2011; Xiong et al., 2018). In addition to their superior stability in the serum, NHGNPs have a larger surface area and stronger SPR than conventional colloidal GNPs, thereby supporting higher protein density and larger resonance coupling when binding to antibodies and then to the nanoplasmonic device surface (Li, Seo et al., 2018; Li, Y. et al., 2018; Wu et al., 2016). Therefore, we hypothesized that a combination of a nanoplasmonic nanocup sensor and NHGNPs might deliver a stronger SPR effect, which would, in turn, enable higher detection sensitivity.
To validate this hypothesis, we developed a novel nanostructure-coupled nanoplasmonic sensor platform for one-step rapid quantification of COVID-19 antibodies to evaluate vaccine effectiveness on a large scale (Scheme 1 ). Our low-cost nanoplasmonic sensor platform coupling with nanostructures can be used to detect total SARS-CoV-2 NAs produced by the human body. The nanoplasmonic immunosorbent assay (NanoPISA) achieved high-throughput detection without signal amplification and washing procedures within 15 min. The NHGNP-coupled NanoPISA platform was able to detect SARS-CoV-2 NAs in the serum with enhanced sensitivity and a limit of detection of 0.2 pM suggesting its potential efficacy for ultrasensitive detection of SARS-CoV-2 NAs to evaluate vaccine effectiveness on a large scale and to prevent the incidence of COVID-19 and other highly infectious diseases.
Scheme 1.
One-step rapid quantification of total SARS-CoV-2 NAs with the novel nanoparticle-coupled biosensor platform.
2. Materials and methods
2.1. Materials
Hexylsilane 6-mercapto-1-hexanol, L-cysteine, chloroauric acid, 1-ethyl-3-(dimethylaminopropyl) carbodiimide, N-hydroxysuccinimide, bovine serum albumin (BSA), ethanolamine, and phosphate-buffered saline (PBS) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Trisodium citrate dehydrate and NaBH4 were purchased from Nanjing Chemical Reagent Co., Ltd. (Nanjing, China). CoCl2 was purchased from Guangdong Guanghua Sci-Tech Co., Ltd. (Guangdong, China). SARS-CoV-2 S protein RBD and mouse anti-human IgG (catalog no. V90401) were purchased from Nanjing Genscript Co., Ltd. (Nanjing, China). Monoclonal antibodies against the SARS-COV-2 RBD (GHMA 105–1 and GHMA 105–2) were purchased from Goodhere Biological Technology Co., Ltd. (Hangzhou, China). Polyethylene glycol 2 W (PEG, MW, 20000) was purchased from Sinopharm Chemical Reagent Co., Ltd. All chemicals were used without further purification.
Serum samples were obtained from Hôpital Militaire d’Instruction Med V Rabat and stored at −80 °C until use. This study was approved by the Ethics Committee of the Science and Technology Department of Huazhong University of Science and Technology (certificate no. S1029).
2.2. Fabrication and characterization of the nanocup array sensor chip
The nanoplasmonic sensor chip was fabricated using the replica-molding technique with a mold. The original mold comprised a tapered nanopillar array with periodicity, height, and width of the nanocone of 400, 500, and 200 nm, respectively. The UV-curable polymer was evenly spread on the mold and placed on a polyethylene terephthalate sheet to produce the polymeric nanocup array structure, after which 9 nm of titanium and 70 nm of gold were subsequently deposited on the polymeric nanocup array in an electron beam evaporator. The sheet was then cut to 13 cm × 8.5 cm sections and glued to an open-bottom 96-well plate generated using a 3D printer (Objet30 Prime; Stratasys Ltd., Rehovot, Israel).
2.3. Surface functionalization
A seeded growth strategy for the chip surface modification was chosen based on the modified Turkevich/Frens reaction system reported elsewhere (Bundschuh et al., 2020; Bastús et al., 2011; Liu et al., 2012). A series of solutions of 1:1 L-cysteine and chloroauric acid (from 0.1 to 1.0 mM) in deionized water was evaluated. Briefly, 32 μL of L-cysteine and 40 μL of chloroauric acid of the same concentration were sequentially injected into each chip well and incubated for 30 min at room temperature. After cleaning the chip twice with deionized water, 50 μL of 8.0 μg/mL anti-human IgG in carbonate buffer solution (CBS, 1 × ) was added into the chip wells and incubated at 37 °C for 2 h. The chip wells were then rinsed twice with deionized water and blocked with 100 μL of 1% BSA in PBS for 30 min at 37 °C. Following the cleaning steps, the surface modified chip plate was stored at 4 °C until further use.
2.4. Preparation of GNPs and GNP-Labeled S-RBD protein
Colloidal GNPs were prepared using a sodium citrate-reduction method (Philip, 2008; Wang et al., 2014). Briefly, 300 mL of 1 mM HAuCl4 was heated to boiling. We then added 15 mL of 75 mM citrate, which resulted in a dark red solution, and continued the boiling process for another 15 min, followed by cooling to room temperature.
The labeling procedure for the S-RBD protein coupled to colloidal GNPs has been described previously (Huang et al., 2021). First, 1.5 mL of the colloidal GNP solution was adjusted to pH 7.4 using 0.1 M K2CO3 solution, after which 4.9 μL of 0.93 mg/mL S-RBD protein solution in PBS was added to the colloidal GNP solution and incubated for 15 min. After blocking with 22.5 μL of the blocking solution (10 mM PBS) containing PEG 2 W (10%, w/v) for 15 min, the colloidal GNP suspension was centrifuged at 7500 rpm for 22 min. The precipitate of the GNP-labeled S-RBD protein was resuspended in 530 μL of stabilizing buffer [20 mM Tris (pH 9.2), 0.3% sucrose, and 0.05% PEG 20000] and stored at 4 °C until use.
2.5. Preparation of NHGNPs and NHGNP-Labeled S-RBD protein
Ultrapure water (300 mL), 0.05 M sodium citrate (6 mL), and 0.4 M CoCl2 solution (300 μL) were consecutively added to a 500-mL round-bottomed flask. After stirring for 10 min in vacuo, 3 mL of 0.13 mM NaBH4 solution in water was added into the flask, followed by stirring for 15 min until the solution darkened. We then added 1.5 mL of 25 mM HAuCl4 solution to the flask and stirred overnight; thereafter, 300 μL of 20% polyvinylpyrrolidone solution was added to the reaction and stirred for 30 min. The final reaction solution was centrifuged for 15 min at 9000 rpm at 4 °C, the supernatant was discarded, and the precipitate was resuspended with ultrapure water and stored at 4 °C until use.
The morphology and size of the synthesized NHGNPs were characterized by transmission electron microscopy (TEM) (HT7700; Hitachi, Tokyo, Japan) using a charge-coupled device camera operated at an accelerating voltage of 100 kV. The NHGNP particle size distribution was determined by dynamic light scattering on a Zetasizer Nano-ZS90 analyzer (Malvern Instruments, Malvern, UK). The absorption spectra of NHGNPs, ranging from 400 to 900 nm, were obtained using a UV–vis spectrophotometer (cat # 8453; Agilent Technologies, Santa Clara, CA, USA) at 25 °C.
Labeling of the S-RBD protein with NHGNPs was performed as described previously, with minor modifications. Briefly, 9 μL of 0.93 mg/mL S-RBD solution in PBS was added to 0.2 mg/mL NHGNP solution and incubated for 15 min. We then added 15 μL of the blocking solution containing PEG 2 W (10%, w/v) and incubated for 15 min; subsequently, the solution was centrifuged at 7500 rpm for 20 min. The NHGNP precipitate was resuspended in 530 μL of the stabilizing buffer [20 mM Tris (pH 9.2), 0.3% sucrose, and 0.05% PEG 20000] and stored at 4 °C until use.
2.6. Antibody detection using the GNP-Coupled NanoPISA
To demonstrate the capabilities of the GNP-coupled NanoPISA platform, SARS-CoV-2 NAs were quantitatively detected using NanoSPR combination sensors. Specifically, 50 μL of SARS-CoV-2 NA solution of different concentrations (in 10 mM PBS and 1% BSA) was added to each individual well of the anti-human IgG-functionalized sensor chip plate. We then added 10 μL of a GNP-coupled S-RBD solution to each well, and spectra and dynamic interaction kinetics were recorded using a generic microplate reader (Biotek Epoch, USA) at the reported wavelengths. After 15 min, the final spectra of the solutions were recorded again.
Detection of SARS-CoV-2 NAs in human serum samples was performed using the same protocol; however, for sample preparation, the plasma or serum samples required 1:100 dilution with sample diluent buffer (10 mM PBS and 1% BSA) to produce samples with values within the dynamic range of the assay. Plasma samples were obtained by centrifuging blood samples at 2500 g and 4 °C for 15 min, after which the serum samples were stored at −80 °C.
3. Results and discussion
3.1. Theory of SARS-CoV-2 NA detection using NanoPISA
The technique of one-step, rapid, quantitative detection of SARS-CoV-2 NAs is based on a GNP-coupled nanoplasmonic sensor with an exceptional optical transmission effect. The coupling effect between GNPs and the nanocup array chip can significantly enhance detection sensitivity. NHGNPs have peculiar plasmonic properties and a large exposed surface area, which have garnered attention in the field of sensors for chemical and biological detection (Lang et al., 2010). Maximization of sensor sensitivity is achieved by the exceptionally high structural stability of ultrafine NHGNPs. In this system, we utilized GNP-coupled nanoplasmonic sensor chips to quantify SARS-CoV-2 NAs. The highly specific S-RBD, which is a key target of host NAs and an important factor in vaccine design, can be used to detect total NAs in human serum. The detection schematic is illustrated in Scheme 1. A mouse anti-human IgG solution was coated on a low-cost ultrasensitive biosensor integrated into a standard 96-well plate. Successive addition of serum positive for SARS-CoV-2 NAs and the (NH)GNP-labeled S-RBD protein results in the specific binding of target SARS-CoV-2 NAs in the serum to the (NH)GNP-labeled S-RBD, followed by capture by anti-human IgG on the 96-well chip surface, thereby forming sandwich immune-conjugate particles (protein–antibody–antibody). These immune-conjugate particles generate an SPR effect via the ultrasensitive plasmonic biosensor chip and change the OD at a specific wavelength in the microplate reader proportionate to the SARS-CoV-2 NA concentration in the serum.
3.2. Characterization of the nanoplasmonic sensor chip and NHGNPs
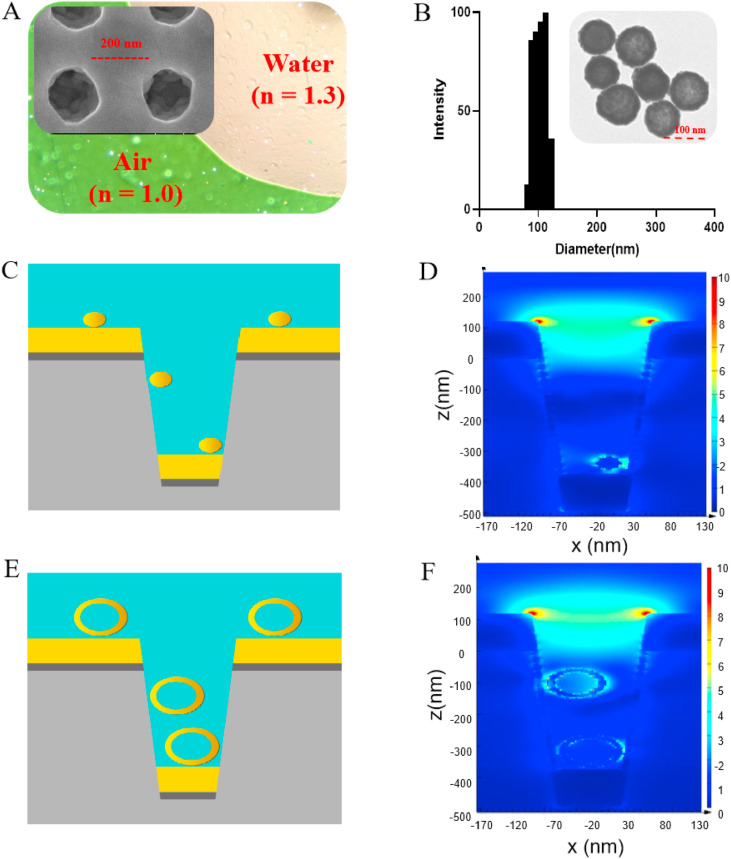
We fabricated the periodic nanocup array sensor chip on a polymer substrate (diameter, 200 nm; depth, 500 nm; and periodicity, 400 nm), in which the titanium and gold layers were 9- and 70-nm thick, respectively. The nanocup array exhibited high uniformity in scanning electron microscopy (SEM) (Fig. 1 A). The sensor chip surface showed distinct colors in the media with different refractive indices (RIs), including air (RI = 1.0) and water (RI = 1.3), suggesting that the sensor chip showed superior detection sensitivity (Fig. 1A). The characteristics of GNPs and NHGNPs were also investigated. For NHGNPs, the particle size was 107.2 nm with a polydispersity index of 0.163 (Fig. 1B). The synthesized NHGNPs had a negative charge (∼42.48 mV) on the surface that remained stable because of the electrostatic repulsion caused by the adsorption of citrate ions (Fig. S1). TEM images revealed that the morphology of the typical NHGNP was polyporous and hollow (Fig. 1B). Moreover, the solution of NHGNPs showed an effective absorption peak of ∼620 nm, which was similar to the resonance peak of the nanoplasmonic sensor chip (Fig. S2). In contrast, the absorption spectrum of GNPs exhibited a distinct SPR peak at ∼526 nm and the particles in TEM images showed dispersed spherical structure with an average size of ∼40 nm (Figs. S3–S5).
Fig. 1.
Characterization and finite-difference time-domain (FDTD) simulation of NHGNP-mediated enhancement of the nanoplasmonic sensor chip. (A) Transmission electron microscopy (TEM) image showing that air and water on the device surface exhibited different colors (green and pink, respectively). Scanning electron microscopy (SEM) image showing the replicated nanocup array (insert). (B) Size distribution and TEM image (insert) of the NHGNPs. (C) Simulation model of colloidal GNP coupled to the nanocup plasmonic sensor. (D) 3D-FDTD-simulated electric field distributions of the colloidal GNPs coupled to the nanocup sensor. (E) Simulation model of NHGNPs coupled to the nanocup plasmonic sensor. (F) 3D-FDTD-simulated electric field distributions of NHGNPs coupled to the nanocup sensor. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. FDTD simulations
Previous studies have reported that significant improvements in SPR performance could be achieved by coupling GNPs to nanoplasmonic nanocup arrays, and this was attempted to generate denser hot spots following the assembly of the plasmonic GNPs on the nanocup surface (Belushkin et al., 2018; Seo et al., 2016, 2018). GNPs (diameter, ∼40 nm) are generally recommended for use in biological-detection procedures owing to their sufficient stability and immune reactivity (OFarrell, 2015; Safenkova et al., 2012; Sharma et al., 2018). However, only large GNPs that fit in the nanocup can effectively help increase the intensity of the localized electric field and improve detection sensitivity (Muskens et al., 2007; Wu et al., 2012). Thus, NHGNPs with higher stability and a larger diameter (∼100 nm) were used to improve SPR effects on the nanocup sensor chip. We confirmed this using the FDTD method. As expected, a stronger electric field and absorption spectra were confined to the inner cavity of the nanocup following the assembly of the NHGNPs on the nanocup array than those obtained with colloidal GNPs (Figs. 1E, F, and S6).
3.4. Optimized conditions for Au nanosheet growth on the surface of the nanoplasmonic sensor chip
The deposition of GNPs on the metal film can cause a notable change in the plasmonic absorption wavelength because metal nanoparticles exhibit a strong localized SPR effect to the faint refractive index change of the surrounding materials (Baba et al., 2014; Inci et al., 2013; Kim et al., 2018). Therefore, we posited that the SPR effect of the nanoplasmonic sensor chip could be conveniently optimized by directly seeding the Au nanosheets on the gold surface with the nanocup array. At the same time, the efficiency of antibody immobilization can be improved by changing the roughness and charge of the chip surface. As a proof of concept, we screened a series of L-cysteine and chloroauric acid concentrations (0.1–1 mM) to determine the optimal chip modification condition. The number of Au nanosheets deposited on the chip surface gradually increased and the surface thickened as observed in the SEM images of the surface-modified sensor chip, and this was proportional to the concentrations of L-cysteine and chloroauric acid (Fig. 2 A). Moreover, the absorption peak at 590 nm of the absorption spectra of the surface-modified sensor chips decreased gradually with an increase in L-cysteine and chloroauric acid concentrations compared with that of original absorption spectra of all chip wells before modification (Figs. 2B and S7). To investigate the optical response of the Au nanosheet-modified chips, the absorption spectra of the chips were determined to be 1.333 for water and 1.339 for 5% sucrose. Fig. 2C shows the relative changes in OD at 610 and 590 nm before and after seeded growth on the surface of the sensor chip (blue bar), and the relative change in OD of the Au nanosheet-modified chips measured in different media (pink bar). With an increase in Au nanosheets, the optical response sensitivity increased gradually in the L-cysteine and chloroauric acid concentration range of 0–0.2 mM and then decreased significantly when L-cysteine and chloroauric acid concentrations were increased to 0.4–1.0 mM. On the basis of the above screening, both L-cysteine and chloroauric acid were used at a concentration of 0.2 mM in further analyses.
Fig. 2.
(A) SEM images of the nanoplasmonic sensor chip modified with L-cysteine and chloroauric acid at a range of concentrations (0.1–1 mM). (B) The absorption spectra of the nanoplasmonic sensor chip after modification with 1:1 L-cysteine and chloroauric acid at a range of concentrations (0.1–1 mM). (C) The relative change in OD at 610 and 590 nm before and after seeded growth on the surface of the sensor chip (a) and the relative change in OD of the Au nanosheet-modified chips were measured to be 1.333 for water and 1.339 for 5% sucrose (b).
3.5. Colloidal GNP-Coupled NanoPISA for quantitative determination of SARS-CoV-2 Nas
After immobilization of the mouse anti-human IgG on the surface of the Au nanosheet-modified biosensor chip, the GNP-coupled NanoPISA biosensor was effective for rapid and high-throughput (96 wells) antibody testing owing to its considerably shorter detection time (≤15 min in total) (Fig. 3 A). The original absorption spectra of the adjacent sensor modification steps exhibited an obvious change (Fig. 3B). We also observed that many GNPs bonded to the surface of the functionalized sensor chip in the SEM image of the tested chip (Fig. 3C). To validate the quantitative detection capability of SARS-CoV-2 NAs based on the colloidal GNP-coupled NanoPISA, 50 μL of solutions containing different concentrations of SARS-CoV-2 NAs (34–2176 pM) and 10 μL of a solution with the GNP-labeled S-RBD protein were successively added to each well of the anti-human IgG-modified sensor chip integrated with a microwell plate for detection using a generic microplate reader. The diluted serum solution without SARS-CoV-2 NAs was used as the control. As shown in Fig. 3D, the absorption spectra of solutions with various concentrations of SARS-CoV-2 NAs were different. The specific resonant wavelengths of the colloidal GNP-coupled nanocup array chip were 590 and 610 nm, with opposite variations observed at 590 and 610 nm with respect to the different concentrations of SARS-CoV-2 NAs. The real-time dynamic binding curves of the immune-conjugate particles at 610 nm are shown in Fig. 3E. The relative OD positively correlated with the SARS-CoV-2 NA concentration, whereas the dynamic curve of the control group showed almost no change. Furthermore, the difference between the relative OD change at 610 and 590 nm was obtained from the differential absorption spectra at 15 min. The standard curve of the relative OD value of the colloidal GNP-coupled NanoPISA relative to the concentration of SARS-CoV-2 NAs (34–2176 pM range) is shown in Fig. 3F. The standard curve was sigmoidal and was obtained using a four-parameter logistic regression model, with a correlation coefficient (R2) of 0.996. The limit of detection of the colloidal GNP-coupled NanoPISA for SARS-CoV-2 NA detection was estimated to be 20 pM.
Fig. 3.
Label-free detection of total SARS-CoV-2 NAs using the colloidal GNP-coupled NanoPISA. (A) Schematic diagram of NanoPISA used to identify SARS-CoV-2 NAs along with colloidal GNP enhancement. (B) Typical original spectra before and after chip surface modification steps and detection with 2176 pM SARS-CoV-2 NAs based on the GNP-coupled NanoPISA sensors. (C) SEM images of the nanoplasmonic sensor chip surface captured binding of SARS-CoV-2 NAs (340 pM) with the GNP-labeled S-RBD protein. (D) Differential spectra of SARS-CoV-2 NAs (34–2176 pM) at 590 and 610 nm. (E) Dynamic binding curves of SARS-CoV-2 NAs (34–2176 pM) at 610 nm. (F) Standard curve for SARS-CoV-2 NAs (R2 = 0.996).
3.6. NHGNP-coupled NanoPISA for quantitative determination of SARS-CoV-2 NAs
The excellent colloidal stability of GNPs is important for their various applications in different fields. Unfortunately, the stability of colloidal GNPs can be easily affected by several external factors, including solution environment and protein concentration (Deng et al., 2020; Ye et al., 2010). However, NHGNPs with stronger SPR properties, larger surface area, and higher stability in aqueous solutions might exhibit higher detection sensitivity than colloidal GNPs. Based on the nanoplasmonic sensor chip plate, we developed an enhanced detection reagent with high stability using NHGNPs, which enabled a ∼100-fold increase in detection sensitivity (Fig. 4 A). Similarly, the original absorption spectra changed significantly before and after the chip modification steps, especially after the addition of enhanced NHGNP detection reagent (Fig. 4B). As observed in the SEM images of the tested chips with NHGNPs, the latter were bound to the interior of the nanocup (Fig. 4C). These results suggest that NHGNPs considerably changed the plasmonic absorption spectra in accordance with the above FDTD simulation results (Figs. 1E, F, and S6). We then applied this with the anti-human IgG-modified sensor surface and NHGNP-labeled S-RBD protein used for the rapid detection of SARS-CoV-2 NAs in a one-step method. In the typical differential absorption spectra of the NHGNP-couple sensor chip, signals appeared at 590 and 650 nm (Fig. 4D), with opposite variation values proportional to the different concentrations of SARS-CoV-2 NAs observed at 590 and 650 nm at 15 min. Additionally, dynamic curves of SARS-CoV-2 NAs at different concentrations and the control sample showed obvious differences within 5 min (Fig. 4E). As expected, the one-step NHGNP-coupled NanoPISA significantly improved the detection sensitivity of SARS-CoV-2 NAs at a concentration of 0.2 pM, which was 100-fold more sensitive than that obtained using the colloidal GNP-coupled NanoPISA. Furthermore, the standard curve of the difference between the relative OD values at 650 and 590 nm with respect to different concentrations of SARS-CoV-2 NAs was obtained from the differential spectra, resulting in an R2 value of 0.998 over the range of SARS-CoV-2 NA concentrations (2–400 pM) (Fig. 4F).
Fig. 4.
Label-free detection of total SARS-CoV-2 NAs following NHGNP enhancement of the NanoPISA platform. (A) Schematic diagram of the nanoplasmonic resonance sensor for the detection of SARS-CoV-2 NAs following NHGNP enhancement. (B) Typical original spectra before and after chip surface modification steps and detection of SARS-CoV-2 NAs (400 pM) based on the NHGNP-coupled NanoPISA sensors. (C) SEM images of the nanoplasmonic sensor chip surface showed the binding of SARS-CoV-2 NAs (400 pM) with NHGNP-labeled S-RBD protein. (D) Differential spectra of SARS-CoV-2 NAs at different concentrations (2–400 pM) at 600 and 650 nm. (E) Dynamic binding curves of SARS-CoV-2 NAs at different concentrations (2–400 pM). (F) Standard curve of SARS-CoV-2 NA detection by NHGNP-labeled S-RBD (R2 = 0.995; LOD = 0.2 pM).
Currently, a safe and effective COVID-19 vaccine is the best way to control and ultimately end the pandemic; therefore, the development of COVID-19 vaccines is progressing at an unprecedented speed. However, evaluation of vaccine effectiveness remains challenging. In this study, the one-step NanoPISA platform not only detected SARS-CoV-2 NAs with ultrahigh sensitivity and effectiveness but also significantly relieved the workload of medical personnel analyzing numerous serum samples from vaccinated people, consequently improving the efficiency of the evaluation process to determine vaccine effectiveness.
3.7. Accuracy and specificity Verification of NanoPISA for detecting SARS-CoV-2 NAs
Fig. S8 shows the comparison of the entire process of one-step NanoPISA with the multi-step ELISA along with the estimated time for each step in the detection of a single sample. SARS-CoV-2 antibodies were diluted in a buffer to a range of 66–4288 pM (the same concentration range used for ELISA); the sensor-calibration curve is shown in Fig. S9. Figs. S10 and 5A show the corresponding SARS-CoV-2 antibody standard curves measured by ELISA; they reveal a good correlation between NanoPISA and ELISA for detecting SARS-CoV-2 antibodies at concentrations of 66, 132, 264, 528, 1056, and 2112 pM (R2 = 0.995). To determine the accuracy of GNP-coupled NanoPISA for SARS-CoV-2 antibody detection, we analyzed three samples of the SARS-CoV-2 antibody in a buffer (132–2112 pM). As shown in Table 1 , the accuracy rate for the same sample was between 80% and 120%, indicating that both NanoPISA and ELISA showed good stability and accuracy. The use of the NHGNP-coupled nanoplasmonic sensor chip optimized the application of NanoPISA to relatively small sample volumes (∼50 μL), whereas the sensitivity was high and comparable to that of ELISA. Another major advantage of the NanoPISA technique is rapid detection of the SARS-CoV-2 antibody and with higher throughput than that afforded with ELISA. The high-throughput and detection speed of NanoPISA allowed all sample tests to be finished within 15 min, whereas it took >3 h to complete conventional ELISA.
Table 1.
Accuracy of the proposed NanoPISA assay for SARS-CoV-2 antibody detection.
|
Concentration (pM) |
NanoPISA |
ELISA |
||
|---|---|---|---|---|
| Mean ± SD (pM) | Accuracy (%) | Mean ± SD (pM) | Accuracy (%) | |
| 132 | 133.2 ± 17.0 | 100.9% | 158.4 ± 5.9 | 120.0% |
| 528 | 493.2 ± 34.9 | 93.4% | 541.0 ± 28.0 | 102.5% |
| 2112 | 2019.1 ± 163.9 | 95.6% | 2097.2 ± 127.0 | 99.3% |
To further evaluate the potential cross-reactivity of the NanoPISA in SARS-CoV-2 NA detection, seven potential cross-reactant samples, namely, influenza A IgG, influenza B IgG, enterovirus IgG, adenovirus IgG, cytovirus IgG, Mycoplasma pneumoniae IgG (MP IgG), and SARS-CoV-2 virus N IgG, were tested. No false positive results were observed, indicating that the developed NanoPISA platform for SARS-CoV-2 NA detection has a good detection specificity (Fig. 5 B).
Fig. 5.
Validation of the detection performance of NanoPISA for SARS-CoV-2 NAs. (A) A correlation between NanoPISA and ELISA methods for the detection of SARS-CoV-2 antibody at concentrations of 66, 132, 264, 528, 1056, and 2112 pM. (B) The selectivity of the NanoPISA assay for seven potential cross-reactant samples.
3.8. Validation of NanoPISA using clinical samples
To confirm the diagnostic accuracy of the NanoPISA technique, we performed a scale-up test using clinical serum samples from Hôpital Militaire d’Instruction Med V Rabat, Morocco. The serum samples were obtained from 139 volunteers without SARS-CoV-2 infection before and after COVID-19 vaccination, and detection of SARS-CoV-2 NA level in each sample was performed as described previously and using only 50 μL of the diluted samples for each measurement. The measurements were performed in a blinded manner, where the actual NA concentrations were unknown. After the completion of the tests, the results collected from NanoPISA were compared with those of the surrogate virus-neutralizing ELISA, as well as those of the SN test (Fig. 6 ).
Fig. 6.
Determination of the cutoff value of NanoPISA for SARS-CoV-2 NA detection through ROC analysis. (A)The accuracy and precision of the NanoPISA platform for 139 clinical samples that were classified as positive or negative using the ELISA kit. (B) ROC curve analysis of the NanoPISA test results based on the data obtained using the ELISA kit. (C) The accuracy and precision of the NanoPISA platform for 68 clinical samples that were classified as positive or negative using the SN test. (B) ROC curve analysis of the NanoPISA test results based on the data obtained using the SN test. The comparison of two groups was followed by student's t-test (two-tailed). ****P < 0.001.
Similar to the detection principle of SARS-CoV-2 surrogate virus-neutralization ELISA kit, normal human serum was used as a negative control (NC) and the serum sample with an appropriate amount of SARS-CoV-2 NA standard (200 pM) as a positive control (PC) in the NanoPISA assay. A comparison of the NanoPISA results of the 139 clinical serum samples with those of the ELISA revealed a similar compliance rate of NA-positive samples and NA-negative samples (Fig. 6A and Table 2 ). The χ2 test used to compare the results showed a result for the kappa test of 0.878 (P < 0.001). Additionally, the sensitivity and specificity of NanoPISA detection were 95.7% and 89.1% respectively, as determined using the Receiver Operator Characteristic (ROC) curve analysis in Fig. 6B, and the cutoff value calculated using the ROC curve was 0.06 (8.8%*(△OD)PC - (△OD)NC). Moreover, the SARS-CoV-2 NA level in the 68 clinical serum samples showed a very strong concordance between the NanoPISA and SN test results. As shown in Fig. 6C and Table 2, the results of NanoPISA did not differ significantly from those of the SN test with a kappa coefficient of 0.792 (P < 0.001). From the corresponding ROC curve (Fig. 6D), a diagnostic cutoff value of 0.04 (5.3%*(△OD)PC - (△OD)NC) was assigned, which could yield 91.9% specificity and 80.7% sensitivity for the detection of SARS-CoV-2 NAs compared with the SN test (Table 3 ). These results demonstrated a high concordance between the results of NanoPISA and those from the two currently used immunoassays, suggesting robust NanoPISA performance. Thus, it could be applied to rapidly evaluate vaccine effectiveness on a large scale.
Table 2.
χ2 test comparing the results of NanoPISA with those of ELISA.
|
NanoPISA |
ELISA |
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 95 | 5 | 100 |
| Negative | 2 | 37 | 39 |
| Total | 97 | 42 | 139 |
Table 3.
χ2 test comparing the results of NanoPISA with those of the SN test.
|
NanoPISA |
SN test |
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 34 | 4 | 38 |
| Negative | 3 | 27 | 30 |
| Total | 37 | 31 | 68 |
4. Conclusions
Herein, we described a rapid and high-throughput nanoplasmonic sensor-based assay for detecting SARS-CoV-2 NAs. The surface of nanopore biosensor chip plate was pre-grown with Au nanosheets and immobilized with mouse anti-human IgG; then, serum samples and nanoparticle-labeled S-RBD were added into the chip well to allow direct measurement of SARS-CoV-2 NA level. The orientation of surface immobilized capture antibody of chip sensor was not only important to improve the immobilization efficiency of antibody, but also critical to increase the sensor's sensitivity. Therefore, in future studies, it is essential to establish more complete relationships of surface treatment and orientation of surface immobilized capture antibody for optimal detection performance. The unique nanoplasmonic mechanism of the NHGNP-coupled nanocup sensors enabled highly sensitive and rapid SARS-CoV-2 NA detection in one step without washing steps. Application of NanoPISA to clinical serum samples of 139 vaccinated people from two nations (China and Morocco) revealed a strong concordance with the results obtained using conventional ELISA and the SN tests. NanoPISA significantly shortened the total detection time from days and hours to ≤15 min and improved the testing capacity to simultaneous high-throughput detection of up to 96 samples. These results demonstrated that the NanoPISA platform is a low-cost, high-throughput device capable of rapidly evaluating vaccine effectiveness on a large scale, which would help vaccine development for SARS-CoV-2 variants and suppress the SARS-CoV-2 variants epidemic.
Funding
This work was supported by a grant from the National Key R & D Plan of China [grant number 2020YFC0861900 and 2019kfyXMPY002]; the National Natural Science Foundation of China [grant numbers 91959107 and 81972892]; the Science and Technology Innovation Project in Hubei Province [grant number 2021ACB001]; and Fundamental Research Funds for the Central Universities [grant number 2020kfyXGYJ111].
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
CRediT authorship contribution statement
Liping Huang: Conceptualization, Methodology, Investigation, Data curation, Visualization, and, Writing – original draft. Ying Li: Data acquisition, Investigation, and, Software. Changyou Luo: Data acquisition. Youqian Chen: Data acquisition. Nadia Touil: Clinical data acquisition. Hicham-El Annaz: Clinical data acquisition. Shaoqi Zeng: GNP-labeled S-RBD preparation. Tang Dang: Software. Jiawei Liang: Software. Wenjun Hu: Supervision. Hao Xu: Supervision, and, Funding acquisition. Jiasheng Tu: Investigation, Resources. Lin Wang: Supervision, Funding acquisition, and, Project administration, and. Yan Shen: Supervision, Writing – review & editing, and, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Liping Huang, Ying Li, Changyou Luo and Youqian Chen contributed equally to this work. The authors thank Xiaofeng Du, Hanlin Zhou, Hong Yang, and Hui Guo for their help with data acquisition. We also thank Editage (www.editage.cn) for English language editing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113868.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Baba A., Yoshida K.I.A., Tanaka D., Tamada K. SpringerPlus. 2014;3(1):1–10. doi: 10.1186/2193-1801-3-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastús N.G., Puntes J.C.V. 2011. Langmuir. [DOI] [PubMed] [Google Scholar]
- Belushkin A., Yesilkoy F., Altug H. ACS Nano. 2018;125:4453–4461. doi: 10.1021/acsnano.8b00519. [DOI] [PubMed] [Google Scholar]
- Belushkin A., Yesilkoy F., Gonzalez-Lopez J.J., Ruiz-Rodriguez J.C., Ferrer R., Fabrega A., Altug H. Small. 2020;163 doi: 10.1002/smll.201906108. [DOI] [PubMed] [Google Scholar]
- Brufsky A. J. Med. Virol. 2020;929:1386–1390. doi: 10.1002/jmv.25902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundschuh C., Egger M., Wiesinger K., Gabriel C., Clodi M., Mueller T., Dieplinger B. Clin. Chim. Acta. 2020;509:79–82. doi: 10.1016/j.cca.2020.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC, Centers for Disease Control and Prevention CDC . Centers for Disease Control and Prevention; 2021. CDC SARS-CoV-2 Variant Classifications and Definitions. [Google Scholar]
- Cetin A.E., Topkaya S.N. Biosens. Bioelectron. 2019;132:196–202. doi: 10.1016/j.bios.2019.02.047. [DOI] [PubMed] [Google Scholar]
- Chauhan G., Madou M., Kalra S., Chopra V., Ghosh D., Martinez-Chapa S. ACS Nano. 2020;147:7760–7782. doi: 10.1021/acsnano.0c04006. [DOI] [PubMed] [Google Scholar]
- Chen X., Azman Z., Sun A.S., Lu R., Zheng W., Zhou N., Wu J., Deng Q., Zhao X., Chen Z., Ge X., Yang S., Leung J., Yu D.T.H. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciab646. [DOI] [Google Scholar]
- Chen M., Yuan Y., Zhou Y., Deng Z., Zhao J., Feng F., Zou H., Sun C. Infect Dis Poverty. 2021;101:94. doi: 10.1186/s40249-021-00878-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H., Huang K., Fang Q., Lv Y., He S., Peng H., Xia X., Chen W. Sensor. Actuator. B Chem. 2020;311 [Google Scholar]
- Dong Y., Dai T., Wei Y., Zhang L., Zheng M., Zhou F. Signal Transduct Target Ther. 2020;51:237. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., He Y., Zhou Y., Liu S., Zheng B.-J., Jiang S. Nat. Rev. Microbiol. 2009;73:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambino C.M., Lo S.B., Colomba C., Giglio R.V., Agnello L., Bivona G., Ciaccio M. Biochem. Med. 2020;303 doi: 10.11613/BM.2020.030901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., Gao H., Ge X., Kan B., Hu Y., Liu J., Cai F., Jiang D., Yin Y., Qin C., Li J., Gong X., Lou X., Shi W., Wu D., Zhang H., Zhu L., Deng W., Li Y., Lu J., Li C., Wang X., Yin W., Zhang Y., Qin C. Science. 2020;3696499:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Li J., Yang X., Chmura A., Zhu G., Epstein J., Mazet J., Hu B., Zhang W., Peng C., Zhang Y., Luo C., Tan B., Wang N., Zhu Y., Crameri G., Zhang S., Wang L., Daszak P., Shi Z. Nature. 2013;5037477:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. Cell. 2020;1812:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Dang T., Li Z., Lei L., Wang G., Li Y., Xu H., Zhou Z., Liu G.L. J. Biomed. Nanotechnol. 2019;158:1724–1733. doi: 10.1166/jbn.2019.2769. [DOI] [PubMed] [Google Scholar]
- Huang C., Wen T., Shi F.-J., Zeng X.-Y., Jiao Y.-J. ACS Omega. 2020;521:12550–12556. doi: 10.1021/acsomega.0c01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Ding L., Zhou J., Chen S., Chen F., Zhao C., Xu J., Hu W., Ji J., Xu H., Liu G.L. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inci F., Wang O.T.S., Gurkan U.A., Tasoglu S., et al. ACS Nano. 2013;7(6):4733–4745. doi: 10.1021/nn3036232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.-F., Sun C., Zhuang Z., Yuan R.-Y., Zheng Q., Li J.-P., Zhou P.-P., Chen X.-C., Liu Z., Zhang X., Yu X.-H., Kong X.-W., Zhu Q.-Y., Zhong Q., Xu M., Zhong N.-S., Zeng Y.-X., Feng G.-K., Ke C., Zhao J.-C., Zeng M.-S. ACS Nano. 2021;152:2738–2752. doi: 10.1021/acsnano.0c08379. [DOI] [PubMed] [Google Scholar]
- Kannan S.R., Spratt A.N., Cohen A.R., Naqvi S.H., Chand H.S., Quinn T.P., Lorson C.L., Byrareddy S.N., Singh K. J. Autoimmun. 2021;124 doi: 10.1016/j.jaut.2021.102715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasetsirikul S., Umer M., Soda N., Sreejith K.R., Shiddiky M.J.A., Nguyen N.-T. Analyst (London) 2020;14523:7680–7686. doi: 10.1039/d0an01609h. [DOI] [PubMed] [Google Scholar]
- Kim J., Shukla S.Y.O.S., Hong S.B., Heo N.S., et al. Biosens. Bioelectron. 2018;107:118–122. doi: 10.1016/j.bios.2018.02.019. [DOI] [PubMed] [Google Scholar]
- Kissler S.M., Tedijanto C., Goldstein E., Grad Y.H., Lipsitch M. Science. 2020;3686493:860–868. doi: 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. Nature. 2020;5867830:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Lang X.Y., Guo H., Chen L.Y., Kudo A., Yu J.S., Zhang W., Inoue A., Chen M.W. J. Phys. Chem. C. 2010;1146:2600–2603. [Google Scholar]
- Laurini E., Marson D., Aulic S., Fermeglia M., Pricl S. ACS Nano. 2020;149:11821–11830. doi: 10.1021/acsnano.0c04674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Wu Q., Ma P., Zhang Y., Song D., Wang X., Sun Y. Talanta. 2018;180:156–161. doi: 10.1016/j.talanta.2017.12.051. [DOI] [PubMed] [Google Scholar]
- Li Y., He D., Tu J., Wang R., Zu C., Chen Y., Yang W., Shi D., Webster T.J., Shen Y. Nanoscale. 2018;1018:8628–8641. doi: 10.1039/c7nr09083h. [DOI] [PubMed] [Google Scholar]
- Liu X., Xia H.X.H., Wang D. 2012. Langmuir. [DOI] [PubMed] [Google Scholar]
- MacMullan M.A., Ibrayeva A., Trettner K., Deming L., Das S., Tran F., Moreno J.R., Casian J.G., Chellamuthu P., Kraft J., Kozak K., Turner F.E., Slepnev V.I., Le Page L.M. 2020. Scientific Reports 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melancon M.P., Zhou M., Li C. Accounts Chem. Res. 2011;4410:947–956. doi: 10.1021/ar200022e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muruato A.E., Fontes-Garfias C.R., Ren P., Garcia-Blanco M.A., Menachery V.D., Xie X., Shi P.-Y. Nat. Commun. 2020;111 doi: 10.1038/s41467-020-17892-0. 4059-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskens O.L., Giannini V., Sánchez-Gil J.A., Gómez Rivas J. Nano Lett. 2007;79:2871–2875. doi: 10.1021/nl0715847. [DOI] [PubMed] [Google Scholar]
- Nuccetelli M., Pieri M., Grelli S., Ciotti M., Miano R., Andreoni M., Bernardini S. Cell Death Discovery. 2020;61 doi: 10.1038/s41420-020-0275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OFarrell B. Top. Companion Anim. Med. 2015;304:139–147. doi: 10.1053/j.tcam.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Philip D. Spectrochim. Acta Mol. Biomol. Spectrosc. 2008;711:80–85. doi: 10.1016/j.saa.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Ranoa D., Holland R., Alnaji F.G., Green K., Wang L., Brooke C., Burke M., Fan T., Hergenrother P.J. 2020. bioRxiv. [Google Scholar]
- Safenkova I., Zherdev A., Dzantiev B. Anal. Bioanal. Chem. 2012;4036:1595–1605. doi: 10.1007/s00216-012-5985-8. [DOI] [PubMed] [Google Scholar]
- Seo S., Zhou X., Liu G.L. Small. 2016;1225:3453–3462. doi: 10.1002/smll.201600451. [DOI] [PubMed] [Google Scholar]
- Seo S., Chang T.-W., Liu G.L. 2018. Scientific Reports 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C., Singh M., Upmanyu V., Chander V., Verma S., Chakrovarty S., Sharma G.K., Dhanze H., Singh P., Shrivastava S., Kumar J., Goswami T.K., Gupta V.K. Arch. Virol. 2018;1639:2359–2368. doi: 10.1007/s00705-018-3846-2. [DOI] [PubMed] [Google Scholar]
- Shrotri M., Navaratnam A.M.D., Nguyen V., Byrne T., Geismar C., Fragaszy E., Beale S., Fong W.L.E., Patel P., Kovar J., Hayward A.C., Aldridge R.W. Lancet. 2021;39810298:385–387. doi: 10.1016/S0140-6736(21)01642-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler M., Belushkin A., Cavallini A., Kebbi-Beghdadi C., Greub G., Altug H. Biosens. Bioelectron. 2017;94:560–567. doi: 10.1016/j.bios.2017.03.047. [DOI] [PubMed] [Google Scholar]
- Wang Y.T., Lu X.M., Zhu F., Zhao M. Bio Med. Mater. Eng. 2014;241:885–892. doi: 10.3233/BME-130882. [DOI] [PubMed] [Google Scholar]
- Wang C., Wang S., Chen Y., Zhao J., Han S., Zhao G., Kang J., Liu Y., Wang L., Wang X., Xu Y., Wang S., Huang Y., Wang J., Zhao J. ACS Nano. 2021;15:6340–6351. doi: 10.1021/acsnano.0c06836. [DOI] [PubMed] [Google Scholar]
- Wu H., Choi C., Cunningham B. Small. 2012;818:2878–2885. doi: 10.1002/smll.201200712. [DOI] [PubMed] [Google Scholar]
- Wu Q., Sun Y., Zhang D., Li S., Wang X., Song D. Biosens. Bioelectron. 2016;86:95–101. doi: 10.1016/j.bios.2016.06.035. [DOI] [PubMed] [Google Scholar]
- Xiong C., Lu W., Zhou M., Wen X., Li C. Cancer Nanotechnol. 2018;91:6. doi: 10.1186/s12645-018-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Zhou Y., Mo Z., Cheng W., Yang S., Wang X., Chen F. Talanta. 2010;813:792–798. doi: 10.1016/j.talanta.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Zettl F., Meister T.L., Vollmer T., Fischer B., Steinmann J., Krawczyk A., V'Kovski P., Todt D., Steinmann E., Pfaender S., Zimmer G. Vaccines (Basel) 2020;83:386. doi: 10.3390/vaccines8030386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.