Abstract
Introduction
Identifying families with an underlying inherited cancer predisposition is a major goal of cancer prevention efforts. Mendelian risk models have been developed to better predict the risk associated with a pathogenic variant of developing breast/ovarian cancer (with BRCAPRO) and the risk of developing pancreatic cancer (PANCPRO). Given that pathogenic variants involving BRCA2 and BRCA1 predispose to all three of these cancers, we developed a joint risk model to capture shared susceptibility.
Methods
We expanded the existing framework for PANCPRO and BRCAPRO to jointly model risk of pancreatic, breast, and ovarian cancer and validated this new model, BRCAPANCPRO on three data sets each reflecting the common target populations.
Results
BRCAPANCPRO outperformed the prior BRCAPRO and PANCPRO models and yielded good discrimination for differentiating BRCA1 and BRCA2 carriers from non-carriers (AUCs 0.79, 95% CI: 0.73–0.84 and 0.70, 95% CI: 0.60–0.80) in families seen in high-risk clinics and pancreatic cancer family registries, respectively. In addition, BRCAPANCPRO was reasonably well calibrated for predicting future risk of pancreatic cancer (observed-to-expected (O/E) ratio = 0.81 [0.69, 0.94]).
Discussion
The BRCAPANCPRO model provides improved risk assessment over our previous risk models, particularly for pedigrees with a co-occurrence of pancreatic cancer and breast and/or ovarian cancer.
Subject terms: Cancer models, Risk factors
Introduction
Cancer family history is an established predictor of an individual’s personal cancer risk and whether he or she carries a cancer predisposition gene. Mendelian risk models have been shown to provide clinically useful, accurate individual-level risk assessment for the management of patients with hereditary cancer syndromes [1–4]. We have previously developed models for hereditary breast and ovarian cancer (BRCAPRO [1, 2]) and for hereditary pancreatic cancer (PANCPRO [3]). The BRCAPRO model provides individual estimates for the probability of carrying a deleterious mutation in BRCA1 and/or BRCA2, based on an individual’s family history of breast and/or ovarian cancer. Parameters for this model include the population carrier frequency for pathogenic variants in these genes as well as the associated penetrance. Based on the framework developed for BRCAPRO, we developed the PANCPRO risk model for familial pancreatic cancer that is based on segregation modelling for pancreatic cancer [3, 5]. Because the segregation model combines the effect of multiple dominant moderate-penetrance susceptibility loci into a single high-risk “locus”, the individual effects of pathogenic variants in BRCA1 or BRCA2, which increase the risk of pancreatic cancer in addition to breast and ovarian cancer, are not individually modelled. To overcome this limitation, we integrated our existing BRCAPRO and PANCPRO models into a single risk model for families with breast, ovarian, and pancreatic cancers. This integrated model, BRCAPANCPRO, was then validated in three independent data sets: two data sets based on ascertainment through a pancreatic cancer proband and one data set from a high-risk breast cancer clinic.
Methods
Model development
The BRCAPANCPRO model was derived from the pre-existing BRCAPRO [1, 2] and PANCPRO [3] models, which were each built under a general Mendelian risk prediction approach and have been described and validated previously [3, 6–8]. Both BRCAPRO and PANCPRO are available in the BayesMendel R package [9]. Briefly, BRCAPRO takes information on family history of breast and/or ovarian cancers and provides the probability that an individual carries a pathogenic variant in the BRCA1 or BRCA2 genes and, if unaffected with cancer, their future risk of developing breast or ovarian cancer. Similarly, the PANCPRO model takes information on family history of pancreatic cancer and provides the probability an individual carries a deleterious mutation in a pancreatic cancer susceptibility gene, modelled from segregation analysis [5] and, if unaffected, their future risk of developing pancreatic cancer. This modelled gene, PANC, represents the combined effect of pathogenic variants in multiple genes with similar inheritance patterns and penetrance (i.e. dominant moderate-risk genes). Thus, the “PANC” locus represents the portion of pancreatic cancer due to dominant susceptibility genes, both those that have been localised and those that remain to be localised. For the BRCAPANCPRO model, we subtracted the carrier probability of BRCA2 and BRCA1 from the population allele frequency for PANC in the PANCPRO model. Carrier probabilities for BRCA1 and BRCA2 are obtained from BRCAPRO in the BayesMendel R package version 2.1-8. The penetrance (age-specific cancer risk) of pancreatic cancer for PANC carriers remained the same as in the original PANCPRO model. The penetrance of breast and ovarian cancer for PANC carriers was set to be equivalent to that of non-BRCA carriers in the BRCAPRO model. The penetrance of breast and ovarian cancer in BRCA1 and BRCA2 carriers remained the same as in BRCAPRO. The penetrance of pancreatic cancer among BRCA1 and BRCA2 carriers was estimated by increasing the age-specific probabilities of developing pancreatic cancer in Surveillance, Epidemiology, and End Results [10] by the odd ratio (OR) reported in the published literature, specifically for BRCA1 carriers (OR: 2.58, 95% confidence interval (CI): 1.54, 4.05) and for BRCA2 carriers (OR: 6.20, 95% CI: 4.62, 8.17) [11]. Sensitivity analysis was conducted using alternative estimates based on studies conducted in the Breast Cancer Family Registry; however, model performance was lower (results not shown) [12] BRCAPANCPRO also retains the fully functionality of BRCAPRO by allowing for input of breast and ovarian pathological markers oestrogen receptor (ER), progesterone receptor (PR), HER2, cytokeratin 14 (CK14), and CK5/6 and history of bilateral mastectomy or oophorectomy.
Validation of study populations
To validate BRCAPANCPRO, we used data from three complementary data sources as detailed below. The Johns Hopkins IRB approved this study. Cohort and registry data participants at Johns Hopkins and participating sites provided informed consent at the enrolling site. The study was performed in accordance with the Declaration of Helsinki.
Hereditary breast and ovarian cancer: Johns Hopkins High Risk Clinic Cohort (JHHRCC)
Carrier probabilities were calculated in 319 families ascertained due to history of breast/ovarian cancer (286 had breast and/or ovarian cancer and 33 had breast/ovarian and a pancreatic cancer) using de-identified pedigree data and genetic testing results from patients undergoing genetic counselling at Johns Hopkins Hospital and data collected for clinial purposes. Data included cancer history (breast, ovarian, and pancreatic cancers, affection age) and current age. Of the 316 individuals who underwent testing, 34 were found to have pathogenic variants in BRCA2, 51 in BRCA1, and none in both genes. Individuals were included in the set to validate BRCA1/2 carrier probability estimates if they underwent genetic testing.
High-risk pancreatic cancer patients with genetic testing: Pancreatic Cancer Genetic Epidemiology (PACGENE) Consortium cohort
Carrier probabilities were calculated in 554 families ascertained due to pancreatic cancer in the proband (280 had pancreatic cancer but no breast/ovarian cancer and 365 had breast/ovarian cancer and pancreatic cancer), recruited from 5 sites participating in the PACGENE Consortium who underwent testing for BRCA1/2 as part of a previously published study [13, 14]. Of the 645 individuals who underwent testing, 22 were found to have pathogenic variants in BRCA2, 5 in BRCA1, and 1 in both genes. Individuals were included in the set to validate BRCA1/2 carrier probability estimates if they underwent genetic testing.
Validation of future pancreatic cancer risk: National Familial Pancreas Tumor Registry (NFPTR)
The NFPTR as Johns Hopkins is a registry of patients with pancreatic cancer and their family members. Families are ascertained through a patient undergoing care for their pancreatic cancer at Johns Hopkins Medicine or referred to the registry due to a history of pancreatic cancer. At enrollment, information on age, vital status, and cancer history is obtained for all first and selected (grandparent/avuncular) second-degree relatives of the pancreatic cancer patients. Families are contacted annually for updated health status as described in Wang et al. [3]. The observed incidence of pancreatic cancer in 26,329 pancreatic cancer-free individuals in 5095 families enrolled in NFPTR [15, 16] was compared to model-predicted risk of pancreatic cancer using the baseline family history for each individual during the follow-up period. Follow-up for each individual was from the date of enrollment in the registry, date of death, or date of last family health update or December 31, 2017. Individuals were included in the validation set if they met the following inclusion criteria: at least 1 pancreatic cancer in the family at baseline; aged 20–93 years; prospective follow-up data available. Of these individuals, 15,289 (58%) had family history of pancreatic cancer but no breast/ovarian cancer and 11,040 (42%) had family history breast/ovarian cancer and pancreatic cancer. These data were independent of the retrospective data on 287 families used in the segregation modelling, which provided the priors used for PANC in our model.
Validation of study design
BRCAPANCPRO was validated in two ways. First, carrier probabilities for BRCA1 and BRCA2 were assessed for model calibration and discrimination using the study data with germline testing for the BRCA1 and/or BRCA2 genes using data from the JHHRCC and PACGENE cohorts. Model calibration was evaluated as the ratio of the number of observed mutation carriers to the expected number, defined as the sum of carrier probabilities over the cohort. Model discrimination was evaluated with receiver operating characteristic (ROC) analysis. Second, future risk of pancreatic cancer was validated for all individuals in the NFPTR cohort using the observed incident cases of pancreatic cancer. Calibration was estimated by the ratio of the observed-to-expected (O/E) number of incident cases, and Hosmer–Lemeshow test evaluated goodness-of-fit of the model estimates, where P < 0.05 would suggest poor fit. Discrimination was evaluated with ROC analysis using the model-estimated annual risk of pancreatic cancer. CIs for all validation measures were estimated with 95% coverage using a bootstrap resampling approach with 10,000 simulations. The calibration of BRCAPANCPRO was compared to BRCAPRO and PANCPRO through paired t tests of the bootstrapped replicates. Improvement frequency (IF) was calculated as the proportion of bootstrap replicates where BRCAPANCPRO yielded better performance over BRCAPRO or PANCPRO.
Results
Clinical illustration
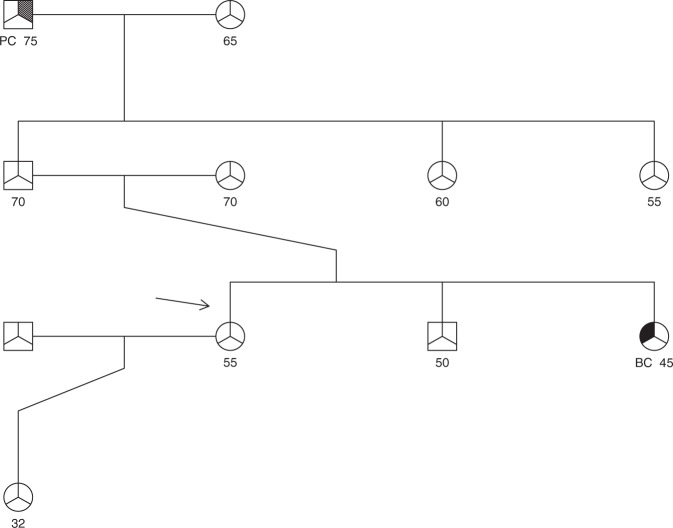
Table 1 illustrates how BRCAPANCPRO integrates information currently used in either BRCAPRO or PANCPRO to aid in clinical decision making. We show risk estimates for a hypothetical family and how the probability of carrying a pathogenic variant changes as different scenarios in the family history are applied (Fig. 1) for each of the three models: BRCAPRO, PANCPRO, and BRCAPANCPRO. As expected, the overall probability that the proband has a predisposition gene increases with the increasing number of family members and shifts to BRCA1/BRCA2 with an increased number of breast and ovarian cancers in the family.
Table 1.
Clinical illustration of BRCAPANCPRO for a hypothetical family shown in Fig. 1.
| Carrier probability (%) | Counselee’s absolute risk of developing cancer (%)a | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| By age 65 years | By age 85 years | ||||||||
| PANC | BRCA2 | BRCA1 | PC | BC | OC | PC | BC | OC | |
| As shown (counselee’s sister affected with breast cancer at 45 years and paternal grandfather with pancreatic cancer at 75 years) | 1.6 | 0.42 | 0.14 | 0.33 | 3.1 | 0.28 | 1.9 | 11.1 | 0.94 |
| As shown, and paternal aunt has pancreatic cancer at age 60 years | 13.6 | 0.49 | 0.11 | 1.3 | 3.1 | 0.29 | 5.2 | 11.2 | 0.96 |
| As shown, and paternal aunt has breast cancer at age 60 years | 1.6 | 1.4 | 0.34 | 0.34 | 3.4 | 0.41 | 1.9 | 11.7 | 1.4 |
| As shown, and paternal aunt has ovarian cancer at age 60 years | 1.4 | 4.2 | 2.7 | 0.36 | 4.5 | 1.2 | 2.2 | 14.2 | 3.6 |
| As shown, and paternal aunt has ovarian cancer at age 60 years and other paternal aunt has breast cancer at 55 years | 0.98 | 10.7 | 5.7 | 0.41 | 6.5 | 2.5 | 2.6 | 19 | 7.4 |
| As shown, and paternal aunt has pancreatic cancer at age 60 years and other paternal aunt has breast cancer at 55 years | 12.9 | 2 | 0.33 | 1.3 | 3.5 | 0.48 | 5.2 | 12.1 | 1.6 |
Diagnoses of breast, pancreas, and ovarian cancer are varied to show model behaviour. Rows correspond to variations on the pedigree.
PC pancreatic cancer, BC breast cancer, OC ovarian cancer.
aAssuming counselee has not developed any of these cancers before age 55 years.
Fig. 1. Hypothetical family.
Arrow indicates the counselee (female, age 55 years). PC pancreatic cancer, BC breast cancer.
Carrier probability validation
The JHHRCC is a high-risk breast and ovarian cancer cohort included in the validation to ensure that BRCAPANCPRO performed relatively similar to the existing BRCAPRO and did not produce unexpected results. BRCAPANCPRO’s overall discriminative ability calculated as the area under the corresponding ROC curve (AUC) for this cohort was 0.79 (95% CI: 0.73, 0.84) and BRCAPRO’s was 0.77 (95% CI: 0.70, 0.82). The IF for BRCAPANCPRO compared to BRCAPRO was 1.0 and similar (0.99 and 0.95) for BRCA1 and BRCA2, respectively. Other performance measures are in Table 2. The calibration for BRCAPANCPRO was O/E ratio 1.46 (95% CI: 1.22, 1.73) and for BRCAPRO was 1.42 (95% CI: 1.17, 1.70), P < 0.001. Carrier probabilities for individuals with a family history of breast/ovarian cancer had similar predictions in both the BRCAPRO and BRCAPANCPRO models, demonstrating that the addition of pancreatic cancer to the model did not compromise performance for these families (Supplementary eFig. 1A). However, individuals with a family history of both pancreatic cancer and breast/ovarian cancer had greater changes in the carrier probabilities.
Table 2.
BRCAPANCPRO and BRCAPRO validation measures for carrier probability of BRCA1/2 genes in JHHRCC and PACGENE families.
| BRCA1 or BRCA2 carriers (N) | Non-carriers (N) | AUC [95% CI] | O/E ratio Pa |
|||
|---|---|---|---|---|---|---|
| Overall | BRCA1 | BRCA2 | ||||
| High-risk breast/ovarian cohort (JHHRCC) | ||||||
| BRCAPANCPRO | 85 | 234 | 0.79 [0.73, 0.84] | 0.86 [0.81, 0.91] | 0.64 [0.53, 0.74] | 1.46 [1.22, 1.73] |
| P = 0.17 | ||||||
| BRCAPRO | 0.77 [0.70, 0.82] | 0.84 [0.78, 0.88] | 0.62 [0.51, 0.73] | 1.42 [1.17, 1.70] | ||
| P = 0.26 | ||||||
| High-risk pancreatic cancer cohort (PACGENE) | ||||||
| BRCAPANCPRO | 28 | 617 | 0.70 [0.60, 0.80] | 0.82 [0.55, 0.98] | 0.69 [0.55, 0.79] | 1.94 [1.26, 3.08] |
| P = 0.55 | ||||||
| BRCAPRO | 0.71 [0.58, 0.82] | 0.79 [0.48, 0.99] | 0.70 [0.58, 0.82] | 3.07 [1.88, 5.46] | ||
| P = 0.10 | ||||||
aP value for Hosmer–Lemeshow test for goodness of fit of the model prediction of carrier probability.
In the PACGENE cohort, which was enriched for pancreatic cancer, discrimination was similar for BRCAPANCPRO AUC of 0.70 (95%: 0.60, 0.80) and BRCAPRO AUC of 0.71 (95% CI: 0.58, 0.82). The IF suggested that these models perform similarly (IF = 0.46 overall). However, BRCAPANCPRO yielded a markedly improved calibration (O/E = 1.94, 95% CI: 1.26, 3.08) compared to BRCAPRO (O/E = 3.07, 95% CI: 1.88, 5.46, IF = 1, P < 0.001). When we examined the individual-level carrier predictions (Supplementary eFig. 1B), individuals with a family history of both pancreatic cancer and breast/ovarian cancer had higher carrier probabilities in the BRCAPANCPRO model compared to BRCAPRO, further demonstrating the value of the combined model. Individuals with a family history of pancreatic cancer had similar predictions in both the PANCPRO and BRCAPANCPRO models, demonstrating that the addition of breast/ovarian cancer to the model did not compromise performance.
Future risk of pancreatic cancer
In the NFPTR cohort, the observed incidence of pancreatic cancer compared to model predictions (O/E ratio) yielded a calibration of 0.82 (95% CI: 0.69, 0.94) for BRCAPANCPRO, an improvement over the original PANCPRO model, O/E ratio 0.73 (95% CI: 0.61, 0.84, IF = 1.0, P < 0.001, Supplementary eFig. 2). Calibration did not vary according to degree of relation to the proband but did change across subgroups defined by type of ascertainment and family history (Table 3). BRCAPANCPRO was better calibrated in individuals where there was a family history of pancreatic cancer and either breast or ovarian cancer (O/E ratio 0.95 [0.75, 1.16]), compared to those with pancreatic cancer only (O/E ratio 0.69 [0.54, 0.86], P < 0.001). Model discrimination was similar between BRCAPANCPRO (AUC = 0.743 [0.707, 0.778] and PANCPRO (AUC = 0.738 [0.702, 0.773]).
Table 3.
BRCAPANCPRO validation measures for future risk of pancreatic cancer in the NFPTR data.
| Incident cases (N) | Pancreatic cancer-free at follow-up (N) | O/E ratio [95% CI], Pc | ||
|---|---|---|---|---|
| BRCAPANCPRO | Original PANCPRO | |||
| Relation to proband | ||||
| All first degreea | 113 | 17,610 | 0.83 [0.67, 0.99], P = 0.79 | 0.73 [0.60, 0.88], P = 0.06 |
| >first degreeb | 46 | 8560 | 0.78 [0.57, 1.01], P = 0.84 | 0.71 [0.50, 0.93], P = 0.37 |
| Family history | ||||
| PC only | 72 | 15,217 | 0.69 [0.54, 0.86], P = 0.07 | 0.61 [0.48, 0.76], P < 0.001 |
| PC and BC/OC | 87 | 10,953 | 0.95 [0.75, 1.16], P = 0.99 | 0.85 [0.68, 1.04], P = 0.96 |
| Ascertainment method | ||||
| Clinic based | 47 | 12,199 | 1.06 [0.79, 1.40], P = 0.99 | 0.93 [0.69, 1.21], P = 0.99 |
| Referral | 112 | 13,971 | 0.74 [0.48, 0.87], P = 0.08 | 0.66 [0.53, 0.78], P < 0.001 |
| All individuals | 159 | 26,170 | 0.81 [0.69, 0.94], P = 0.38 | 0.72 [0.61, 0.84], P = 0.003 |
O/E observed to expected, CI confidence interval, N number.
aFirst-degree relatives include parents, siblings, and offspring.
bAbove first degree includes all second-degree relatives (grandparents, aunts, uncles, nieces, and nephews), third degree and above.
cP value for Hosmer–Lemeshow test for goodness of fit of the model prediction of future risk of pancreatic cancer.
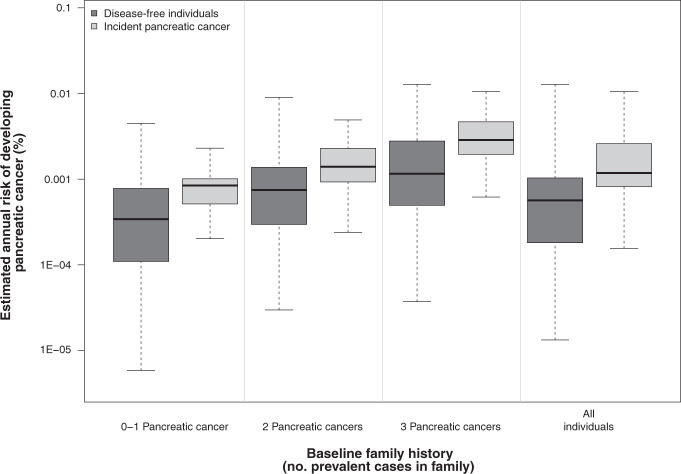
Risk estimates were higher among individuals who developed pancreatic cancer during follow-up compared to individuals who remained disease free (Fig. 2, Kolmogorov–Smirnov P < 0.001). This difference held in each stratum (P < 0.001 for 0 to 1, 2, and 3 or more affected relatives at baseline).
Fig. 2. Estimated risk of Pancreatic cancer.
Distribution of estimated average annual risk of developing pancreatic cancer for incident pancreatic cancer patients and disease-free individuals in BRCAPANCPRO according to the number of pancreatic cancers in the family at baseline.
Discussion
Identification of families at an increased risk of developing a hereditary cancer is an important step in reducing the burden of cancer in these families. Risk models, including the BayesMendel R Package discussed here, BOADICEA (CanRisk), and the Gail (BCRAT), are widely used as part of risk assessment for families with breast/ovarian cancer or pancreatic cancer [9, 17–19]. However, the overlap in the genes underlying these models is substantial with 2.5–7% of all newly diagnosed pancreatic cancers carrying a pathogenic variant in BRCA2 or BRCA1 [11, 20–22]. Other susceptibility genes also confer risk for both pancreatic and women’s cancers [23, 24]. The BOADICEA model implemented in (CanRisk) incorporates the effects of BRCA1, BRCA2, PALB2, CHEK2, and ATM and other information to calculate risk of breast and ovarian cancer. The BRCAPANCPRO model provides risk estimates for pancreatic cancer in addition to breast/ovarian cancer and includes additional genetic effects on pancreatic cancer risk via PANC.
Our BRCAPANCPRO model was designed to address this need and demonstrates improved performance in families with breast/ovarian and pancreatic cancers without compromising performance when there is only breast/ovarian cancer (BRCAPRO) or pancreatic cancer (PANCPRO) in the family. The inclusion of pancreatic cancer family history in the BRCAPANCPRO model does not sacrifice discrimination of BRCA1 or BRCA2 carriers from non-carriers, and the validation suggests improved calibration compared to the original BRCAPRO for families with hereditary breast or ovarian cancer and PANCPRO for families with hereditary pancreatic cancer. However, while the O/E ratios for BRCAPANCPRO were in most cases closer to 1.0 than those of prior models, there remained some underprediction of BRCA1 and BRCA2 carrier status in these families that underwent genetic testing. This is likely attributable to overrepresentation of breast, ovarian, or pancreatic cancer in the PACGENE and Johns Hopkins High Risk Cohort as a result of the ascertainment mechanisms. In our future risk validation, for families ascertained due to high risk, all models overestimated pancreatic cancer risk (O/E ratio <1.0); however, the extent of this overestimation was less for BRCAPANCPRO compared to PANCPRO (Table 3). In families unselected for cancer history who were instead clinically ascertained for a single proband, BRCAPANCPRO performed well (O/E 1.06 (95% CI 0.79–1.40), Table 3) indicating that overprediction of pancreatic cancer risk may be related to case ascertainment. However, genetic testing data are not widely available in these data, and thus we cannot directly validate carrier status estimates in these same data. Furthermore, since PANC incorporates genetic risk due to yet unmapped loci, it cannot be validated by evaluation of genetic testing results but can be evaluated by assessing the accuracy of future cancer risk estimates. Further studies may reveal whether these calibration differences are due to ascertainment efforts or inaccuracies in our choices for penetrance and prevalence.
A limitation of the new BRCAPANCPRO model is that it incorporates cancer risk estimates from the existing literature, which limits generalisability to different populations according to race and ethnicity. However, validation studies of previous versions of BRCAPRO in Black and Hispanic populations have shown performance similar to that in European populations [8, 25]. For pancreatic cancer, there are only a few studies describing the prevalence of BRCA1 and BRCA2 pathogenic variants in patients of Chinese and Japanese ancestry but the data are even more limited for those of other ancestries [26]. The segregation model used to develop PANCPRO was based on families of primarily European Ancestry. Studies are ongoing to address this critical need. In contrast, several studies have examined the prevalence and associated penetrance of pathogenic variants in BRCA1 and BRCA2 in the various ancestral populations. Our model not only allows a user to specify both penetrance and prevalence estimates tailored to the proband’s race, ethnicity, and/or geographic region but also to the underlying rate in non-carriers. The underlying rate of cancer in the population can also impact model performance as shown in validation studies of BRCAPRO in Asian populations [27]. As cancer risk estimates improve, these can be readily incorporated into our model by replacing a user-specified input in the BayesMendel package. The BRCAPANPRO model is inherently limited by our imperfect knowledge of the genetic basis of pancreatic cancer as well as breast/ovarian cancer. While our combined BRCAPANCPRO model is a step forward by directly incorporating the effect of BRCA1 and BRCA2 on pancreatic cancer risk, additional pancreatic cancer susceptibility genes have been identified in the past several years, including ATM and PALB2 [24, 28]. In the future, the framework demonstrated here can readily be expanded to incorporate these effects directly vs indirectly via PANC. Finally given the lack of histological data on breast cancer cases in our validation set, we were unable to validate the contribution of pathological markers ER, PR, HER2, CK14, and CK5/6, in the context of our BRCAPANPRO model, as we had done previously for BRCAPRO [29].
Genetic testing guidelines have changed considerably over the past several years in part due to the decreasing cost of germline genetic testing, together with the improved therapeutic options for cancer patients with specific alterations. These include poly (ADP-ribose) polymerase inhibitors for BRCA-deficient breast, ovarian, and pancreatic cancers or checkpoint inhibitors for mismatch repair-deficient tumours [30–32]. While the United States has recently recommended that germline genetic testing be offered to all patients with pancreatic cancer, ovarian cancer, and young-onset breast cancer patients (<50 years) regardless of ancestry or family history, as well as breast cancer patients meeting certain family history criteria [33, 34], universal testing is not wide-spread in the rest of the world and risk models such as BRCAPANCPRO are widely used as a guide to help identify individuals who may benefit from germline genetic testing. Furthermore, patients, including >80% of those with a family history of pancreatic cancer [35], who undergo gene testing will not have a susceptibility gene mutation identified. Decision support tools such as risk models can help individuals with a familial history of cancer understand their future risk of cancer. Risk models can provide guidance on risk of pancreatic cancer among their relatives, which in turn could have an impact upon early detection screening decisions. BRCAPANCPRO is part of the R package BayesMendel and freely available for research use. Physician-facing interfaces are also available commercially.
The new BRCAPANCPRO model is an improved tool for families with a history of breast, ovarian, and/or pancreatic cancer as it leverages the overlap in risk for carriers of BRCA1, BRCA2, and the pancreatic cancer susceptibility gene to improve specification of cancer risk and subsequent personalisation of appropriate screening and follow-up for early detection.
Supplementary information
Acknowledgements
Deepest gratitude to the patients and families who participated in research studies contributing to this work.
Author contributions
ALB—study design, statistical analysis, drafting and revision of manuscript. EJC—statistical analysis, revision of manuscript. NP—data collection, statistical analysis, revision of manuscript. GMP, KGR, SG, AB, SS, MLC, AGS, MGG, RHH—data collection, revision of manuscript. GP—study design, supervision of data analysis, data collection, revision of manuscript, APK—study design, supervision of data analysis, data collection, provision of funding, drafting and revision of manuscript.
Funding
This work was supported by the Lustgarten Foundation and NCI RO1CA154823 (to APK), U01CA247283 (to APK), NCI P50 CA62924 (to APK), P30CA006973 (to Nelson), P30CA006516 (to Glimcher), R01 CA132829 (to SS), R01CA097075 (to GMP), Rolfe Pancreatic Cancer Foundation (to GMP), and the Sol Goldman Pancreatic Cancer Research Center.
Data availability
Data are available, through a collaborative agreement, upon request. BRCAPANCPRO is part of the R package BayesMendel and freely available for research use.
Ethics approval and consent to participate
The Johns Hopkins IRB approved this study. Cohort and registry data participants at Johns Hopkins and participating sites provided informed consent at the enrolling site. The study was performed in accordance with the Declaration of Helsinki.
Consent to publish
Not applicable as no individual-level information is shared.
Competing interests
APK has previously consulted for MERCK and SS has consulted for Myriad Genetics.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01580-x.
References
- 1.Parmigiani G, Berry D, Aguilar O. Determining carrier probabilities for breast cancer-susceptibility genes BRCA1 and BRCA2. Am J Hum Genet. 1998;62:145–58. doi: 10.1086/301670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berry DA, Parmigiani G, Sanchez J, Schildkraut J, Winer E. Probability of carrying a mutation of breast-ovarian cancer gene BRCA1 based on family history. J Natl Cancer Inst. 1997;89:227–38. doi: 10.1093/jnci/89.3.227. [DOI] [PubMed] [Google Scholar]
- 3.Wang W, Chen S, Brune KA, Hruban RH, Parmigiani G, Klein AP. PancPRO: risk assessment for individuals with a family history of pancreatic cancer. J Clin Oncol. 2007;25:1417–22. doi: 10.1200/JCO.2006.09.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen S, Wang W, Lee S, Nafa K, Lee J, Romans K, et al. Prediction of germline mutations and cancer risk in the Lynch syndrome. JAMA. 2006;296:1479–87. doi: 10.1001/jama.296.12.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein AP, Beaty TH, Bailey-Wilson JE, Brune KA, Hruban RH, Petersen GM. Evidence for a major gene influencing risk of pancreatic cancer. Genet Epidemiol. 2002;23:133–49. doi: 10.1002/gepi.1102. [DOI] [PubMed] [Google Scholar]
- 6.Parmigiani G, Chen S, Iversen ES, Jr, Friebel TM, Finkelstein DM, Anton-Culver H, et al. Validity of models for predicting BRCA1 and BRCA2 mutations. Ann Intern Med. 2007;147:441–50. doi: 10.7326/0003-4819-147-7-200710020-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leonardi G, Marchi S, Falconi M, Zerbi A, Ussia V, de Bortoli N, et al. “PancPro” as a tool for selecting families eligible for pancreatic cancer screening: an Italian study of incident cases. Dig Liver Dis. 2012;44:585–8. doi: 10.1016/j.dld.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 8.Kurian AW, Gong GD, John EM, Miron A, Felberg A, Phipps AI, et al. Performance of prediction models for BRCA mutation carriage in three racial/ethnic groups: findings from the Northern California Breast Cancer Family Registry. Cancer Epidemiol Biomark Prev. 2009;18:1084–91. doi: 10.1158/1055-9965.EPI-08-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S, Wang W, Broman KW, Katki HA, Parmigiani G. BayesMendel: and R environment for Mendelian risk prediction. Stat Appl Genet Mol Biol. 2004;3:Article21. [DOI] [PMC free article] [PubMed]
- 10.SEER. Surveillance, Epidemiology, and End Results (SEER) Program Populations (1969-2018), National Cancer Institute, DCCPS, Surveillance Research Program, released December 2019. Bethesda, MD: National Cancer Institute, DCCPS, Surveillance Research Program, Surveillance Systems Branch; 2019.
- 11.Hu C, Hart SN, Polley EC, Gnanaolivu R, Shimelis H, Lee KY, et al. Association between inherited germline mutations in cancer predisposition genes and risk of pancreatic cancer. JAMA. 2018;319:2401–9. doi: 10.1001/jama.2018.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mocci E, Milne RL, Mendez-Villamil EY, Hopper JL, John EM, Andrulis IL, et al. Risk of pancreatic cancer in breast cancer families from the breast cancer family registry. Cancer Epidemiol Biomark Prev. 2013;22:803–11. doi: 10.1158/1055-9965.EPI-12-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petersen GM, de Andrade M, Goggins M, Hruban RH, Bondy M, Korczak JF, et al. Pancreatic cancer genetic epidemiology consortium. Cancer Epidemiol Biomark Prev. 2006;15:704–10. doi: 10.1158/1055-9965.EPI-05-0734. [DOI] [PubMed] [Google Scholar]
- 14.Zhen DB, Rabe KG, Gallinger S, Syngal S, Schwartz AG, Goggins MG, et al. BRCA1, BRCA2, PALB2, and CDKN2A mutations in familial pancreatic cancer: a PACGENE study. Genet Med. 2015;17:569–77. doi: 10.1038/gim.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brune KA, Lau B, Palmisano E, Canto M, Goggins MG, Hruban RH, et al. Importance of age of onset in pancreatic cancer kindreds. J Natl Cancer Inst. 2010;102:119–26. doi: 10.1093/jnci/djp466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein AP, Brune KA, Petersen GM, Goggins M, Tersmette AC, Offerhaus GJ, et al. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 2004;64:2634–8. doi: 10.1158/0008-5472.CAN-03-3823. [DOI] [PubMed] [Google Scholar]
- 17.Gail MH, Brinton LA, Byar DP, Corle DK, Green SB, Schairer C, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81:1879–86. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 18.Antoniou AC, Cunningham AP, Peto J, Evans DG, Lalloo F, Narod SA, et al. The BOADICEA model of genetic susceptibility to breast and ovarian cancers: updates and extensions. Br J Cancer. 2008;98:1457–66. doi: 10.1038/sj.bjc.6604305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carver T, Hartley S, Lee A, Cunningham AP, Archer S, Babb de Villiers C, et al. CanRisk tool-a web interface for the prediction of breast and ovarian cancer risk and the likelihood of carrying genetic pathogenic variants. Cancer Epidemiol Biomark Prev. 2021;30:469–73. doi: 10.1158/1055-9965.EPI-20-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shindo K, Yu J, Suenaga M, Fesharakizadeh S, Cho C, Macgregor-Das A, et al. Deleterious germline mutations in patients with apparently sporadic pancreatic adenocarcinoma. J Clin Oncol. 2017;35:3382–90. [DOI] [PMC free article] [PubMed]
- 21.Yurgelun MB, Chittenden AB, Morales-Oyarvide V, Rubinson DA, Dunne RF, Kozak MM, et al. Germline cancer susceptibility gene variants, somatic second hits, and survival outcomes in patients with resected pancreatic cancer. Genet Med. 2019;21:213–23. doi: 10.1038/s41436-018-0009-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holter S, Borgida A, Dodd A, Grant R, Semotiuk K, Hedley D, et al. Germline BRCA mutations in a large clinic-based cohort of patients with pancreatic adenocarcinoma. J Clin Oncol. 2015;33:3124–9. doi: 10.1200/JCO.2014.59.7401. [DOI] [PubMed] [Google Scholar]
- 23.Tischkowitz MD, Sabbaghian N, Hamel N, Borgida A, Rosner C, Taherian N, et al. Analysis of the gene coding for the BRCA2-interacting protein PALB2 in familial and sporadic pancreatic cancer. Gastroenterology. 2009;137:1183–6. doi: 10.1053/j.gastro.2009.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones S, Hruban RH, Kamiyama M, Borges M, Zhang X, Parsons DW, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huo D, Senie RT, Daly M, Buys SS, Cummings S, Ogutha J, et al. Prediction of BRCA mutations using the BRCAPRO model in clinic-based African American, Hispanic, and other minority families in the United States. J Clin Oncol. 2009;27:1184–90. doi: 10.1200/JCO.2008.17.5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takai E, Yachida S, Shimizu K, Furuse J, Kubo E, Ohmoto A, et al. Germline mutations in Japanese familial pancreatic cancer patients. Oncotarget. 2016;7:74227–35. doi: 10.18632/oncotarget.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S, Blackford AL, Parmigiani G. Tailoring BRCAPRO to Asian-Americans. J Clin Oncol. 2009;27:642–3. doi: 10.1200/JCO.2008.20.6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts NJ, Jiao Y, Yu J, Kopelovich L, Petersen GM, Bondy ML, et al. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov. 2012;2:41–46. doi: 10.1158/2159-8290.CD-11-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tai YC, Chen S, Parmigiani G, Klein AP. Incorporating tumor immunohistochemical markers in BRCA1 and BRCA2 carrier prediction. Breast Cancer Res. 2008;10:401. doi: 10.1186/bcr1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med. 2019;381:317–27. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–13. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Domchek SM, Aghajanian C, Shapira-Frommer R, Schmutzler RK, Audeh MW, Friedlander M, et al. Efficacy and safety of olaparib monotherapy in germline BRCA1/2 mutation carriers with advanced ovarian cancer and three or more lines of prior therapy. Gynecol Oncol. 2016;140:199–203. doi: 10.1016/j.ygyno.2015.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoffel EM, McKernin SE, Brand R, Canto M, Goggins M, Moravek C, et al. Evaluating susceptibility to pancreatic cancer: ASCO provisional clinical opinion. J Clin Oncol. 2019;37:153–64. doi: 10.1200/JCO.18.01489. [DOI] [PubMed] [Google Scholar]
- 34.Daly MB, Pilarski R, Yurgelun MB, Berry MP, Buys SS, Dickson P, et al. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 1.2020. J Natl Compr Canc Netw. 2020;18:380–91. doi: 10.6004/jnccn.2020.0017. [DOI] [PubMed] [Google Scholar]
- 35.Roberts NJ, Norris AL, Petersen GM, Bondy ML, Brand R, Gallinger S, et al. Whole genome sequencing defines the genetic heterogeneity of familial pancreatic cancer. Cancer Discov. 2016;6:166–75. doi: 10.1158/2159-8290.CD-15-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available, through a collaborative agreement, upon request. BRCAPANCPRO is part of the R package BayesMendel and freely available for research use.