Abstract
Targeted kinase inhibitors improve the prognosis of lung cancer patients with ALK alterations (ALK+). However, due to the emergence of acquired resistance and varied clinical trajectories, early detection of disease progression is warranted to guide patient management and therapy decisions. We utilized 343 longitudinal plasma DNA samples from 43 ALK+ NSCLC patients receiving ALK-directed therapies to determine molecular progression based on matched panel-based targeted next-generation sequencing (tNGS), and shallow whole-genome sequencing (sWGS). ALK-related alterations were detected in 22 out of 43 (51%) patients. Among 343 longitudinal plasma samples analyzed, 174 (51%) were ctDNA-positive. ALK variant and fusion kinetics generally reflected the disease course. Evidence for early molecular progression was observed in 19 patients (44%). Detection of ctDNA at therapy baseline indicated shorter times to progression compared to cases without mutations at baseline. In patients who succumbed to the disease, ctDNA levels were highly elevated towards the end of life. Our results demonstrate the potential utility of these NGS assays in the clinical management of ALK+ NSCLC.
Subject terms: Non-small-cell lung cancer, Cancer genomics
Introduction
About 3–7% of non-small-cell lung cancer (NSCLC) belong to a molecular subgroup defined by the presence of ALK (anaplastic lymphoma kinase) rearrangements1. Since its discovery, ALK-rearranged (ALK+) NSCLC has been a model disease for targeted therapies using tyrosine kinase inhibitors (TKIs) that potently attenuate the function of ALK2–5. While initial responses to selective TKIs are durable, ALK+ tumors eventually and inevitably develop resistance to targeted therapy. For example, the first FDA/EMA-approved ALK inhibitor crizotinib elicited improved objective response rates and progression-free survival (PFS) in randomized phase III trials compared to chemotherapy, but disease relapse was nonetheless observed within 1 year of treatment6. Consequently, significant insights into the molecular underpinnings of TKI therapy resistance have been gained. At the same time, increasingly potent and selective next-generation ALK TKIs have also been developed and approved for clinical use7–10. Although sequential therapy using next-generation ALK inhibitors improves PFS and mitigates brain metastases11, ALK+ tumors continue to adapt and develop alternative resistance mechanisms against these drugs.
The dynamic adaptability of ALK+ tumors against ALK inhibitors requires a personalized approach to disease monitoring for accurate and timely clinical patient management. Next-generation sequencing (NGS)-based analysis of tissue DNA is an established and a highly robust method for baseline genotypic evaluations12,13. However, while longitudinal tissue biopsies could identify emerging resistance mutations, this strategy is limited by a number of factors: (a) single-site tumor biopsies would most likely fail to capture the full spectrum of genomic alterations of a molecularly heterogeneous tumor14; (b) multiple tumor re-biopsies pose procedural risks, and sometimes are not feasible15; and (c) tissue sample preparation such as formalin fixation could lead to false-positive results in molecular assays due to high levels of base transitions15,16. Therefore, liquid biopsy technologies are currently emerging as minimally invasive and easily accessible alternatives to tissue biopsies. Peripheral blood plasma naturally harbors molecular components that can be analyzed and attributed to certain pathologies. Among these, the circulating tumor DNA (ctDNA) compartment provides highly sensitive and insightful genetic information for evaluating tumor heterogeneity and clonal evolution. In addition to being a surrogate for localized tissue biopsy, ctDNA has the potential to capture the complete genomic profile of the primary tumor and metastases at expansive time points without spatial bias. Detection of such exhaustive genetic tumor profiles is particularly relevant for disease monitoring of patients undergoing treatment, where tumor subclones could emerge due to selective pressures introduced by various therapeutic challenges.
Previously, overall ctDNA levels have been utilized as independent biomarkers of disease progression or therapy response in multiple cancer entities including metastatic breast cancer17, melanoma18, metastatic gastrointestinal cancer19, metastatic bladder cancer20, and lung cancer21. ctDNA detection has also been explored for prediction of disease recurrence and minimal residual disease22–24. In ALK+ NSCLC, liquid biopsies have successfully been utilized for ALK resistance mutation profiling, presenting important implications for therapy decisions21,25,26.
We first reported the potential clinical utility of a combination of cfDNA assays for longitudinal monitoring of ALK+ NSCLC under TKI therapy27. We performed matched capture-based high coverage targeted NGS (tNGS) using a commercially available panel comprised of genes optimized for longitudinal tumor burden monitoring in lung and colorectal cancers. Simultaneously, we applied the trimmed median absolute deviation from the copy number neutral state (t-MAD) score using shallow whole-genome sequencing (sWGS) to quantify global copy number changes derived from ctDNA28. The information derived from these ctDNA assays revealed the complex mutational kinetics of ALK+ tumors across therapy lines, as well as the increasing genomic instability of the tumor after sequential TKI treatments. Here, we extend the number of longitudinal plasma samples from our initial ALK+ NSCLC cohort and for the first time, using tNGS and sWGS data, identify threshold values for changes in variant allele frequencies (VAFs) and t-MAD scores that potentially indicate disease progression earlier than current conventional modalities. With additional sampling points, we show that a combined NGS assay approach can reveal molecular progression that precedes radiographic assessment, particularly in patients with detectable ctDNA. Furthermore, our study cohort demonstrates the potential of targeted sequencing in baseline risk stratification of patients, and corroborates previous data about the negative prognostic impact of TP53 alterations detected during therapy.
Results
Patient characteristics
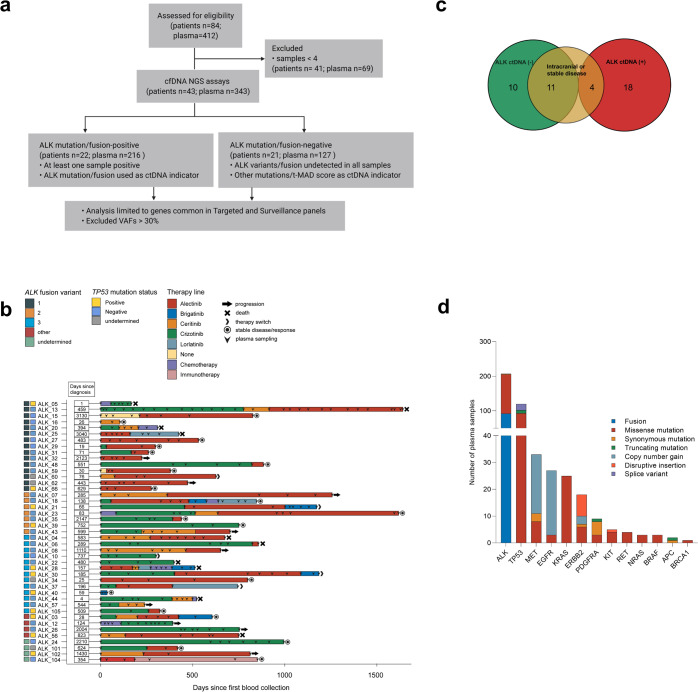
In total, 43 metastatic ALK+ patients with corresponding 343 longitudinal plasma samples were considered for NGS analysis (Fig. 1a, see Methods). The median age of the study population was 57 years (range 39–80), with almost equal distribution of males and females (53% males). Seventy eight percent of patients were never/light-smokers (<10 pack-years, Table 1). Based on tissue biopsies, EML4-ALK variant 1 (V1, E13:A20) was identified in 35% (15/43), V2 (E20:A20) in 16% (7/43), V3 (E6:A20) in 30% (13/43), and other variants in 9% (4/43) of tumors. Other cases of ALK fusion were detected with tissue RNA-NGS at initial diagnosis. TP53 mutation were detectable in 11/39 (28%) patients with available tissue DNA at diagnosis. The therapy regimens that patients underwent at each sampling time point are shown in Fig. 1b. The first-generation TKI crizotinib was administered to 25/43 patients. Second-generation TKIs (ceritinib, alectinib, brigatinib) were administered to 36/43 patients, and third-generation TKI lorlatinib was given to 4/43 patients.
Fig. 1. Overview of the ALK+ NSCLC cohort.
a Untargeted sWGS and targeted NGS assays were used to assess ctDNA in 43 ALK+ NSCLC patients corresponding to 343 longitudinal plasma samples. Targeted NGS analysis only included genes common to both the Avenio Targeted and Surveillance panels. In total, 51% (22 of 43) of patients had an ALK alteration in at least one liquid biopsy. b Timeline of plasma collection and therapy administration in the patient cohort. ALK fusion variants and TP53 status from tissue analysis at diagnosis are indicated by colored boxes. c Distribution of patients based on detected ALK alterations—including mutations and rearrangements. The group without detectable ALK alterations (ALK−) is enriched for patients with intracranial progression or stable disease. d Molecular alterations identified in 174 ctDNA (+) plasma samples based on tNGS.
Table 1.
Patient characteristics.
| ALK+ NSCLC patients analyzed in this study (n = 43) | ||
|---|---|---|
| Age, median (39–80) | 57 (10) | |
| Sex, % male | 53% | |
| Smoking status (% never smokers)a | 78% | |
| ECOG PS (%) at baseline | ||
| 0 | 26 | |
| 1 | 14 | |
| 2 | 1 | |
| No data | 2 | |
| Histologyb | ||
| adenocarcinoma | 42/43 | |
| ALK fusion variantc | ||
| EML4-ALK V3 | 13 | |
| EML4-ALK V1/V2 | 22 | |
| Other | 4 | |
| No data | 4 | |
| TP53 status at baseline, mutatedd | 11/39 | |
| ALK TKI, patient number | ||
| Crizotinib | 25 | |
| Ceritinib/alectinib/brigatinib | 36 | |
| Lorlatinib | 4 | |
| Chemotherapy | 7 | |
| Follow-up in months (median, [Q3-Q1]) | 37 (52–27) | |
| Number of samples analyzed per patient (mean [range]) | 8 (4–31) | |
| Percentage of cases with treatment-naive samples | 21% | |
| Number of samples at disease progression per patient, mean | 2.8 | |
| Number of TKI lines covered with liquid biopsy per patient, mean | 1.9 | |
SD standard deviation, EML4-ALK echinoderm microtubule-associated protein-like 4 (EML4) and anaplastic lymphoma kinase (ALK) fusion, PS performance status, TKI tyrosine kinase inhibitor.
aData available for 41/43 cases.
bOne patient had an ALK+ large-cell neuroendocrine lung carcinoma responsive to ALK inhibitors.
cData available for 39/43 cases; one case with E18A20, one with E9A20, one with K9A20 (KCL1), and one with K24A20 (KIF5B).
dData available for 39 cases by NGS of tissue biopsies at diagnosis of stage IV disease.
Genomic alteration landscape of ALK+NSCLC ctDNA
Among the patients under study, 22 (51%) had detectable ALK alterations (mutation or rearrangement) in at least one plasma sample based on tNGS profiling (Fig. 1a and Supplementary Table 1). A total of 36 ALK mutational events were detected, 34 of which were known or probable resistance mutations, while two were silent mutations. Of the remaining 21 patients, 11 (52%) had either intracranial progression (n = 6) or stable disease at all sampling points (n = 5), potentially explaining the low amount of ctDNA shedding and unmeasurable ALK alterations in plasma DNA (Fig. 1c). ctDNA was detected in a total of 174/343 (51%) plasma samples. Most of these were missense, synonymous, and truncating mutations, disruptive inframe insertions, as well as fusions and copy number changes in ALK and TP53 with 56% and 52% detection rate in ctDNA (+) plasma samples, respectively. Other genes prominently altered were MET, EGFR, KRAS, ERBB2, PDGFRA, KIT, RET, NRAS, BRAF, APC, and BRCA1 (Fig. 1d). Copy number gains were also detected in MET, EGFR, and ERBB2.
Dynamics of ALK rearrangement and resistance mutations during TKI therapy
We interrogated the concordance between ALK mutation or fusion abundance in ALK ctDNA+ (n = 22) patients and disease status. Emergence of at least one ALK mutation coincided with progressive disease in eight of these patients (Supplementary Table 2), while seven patients (ALK_06, 13, 18, 20, 25, 44, 62) showed increasing EML4-ALK fusion abundance upon progression on independent therapy lines (Supplementary Fig. 1). Five patients developed multiple secondary mutations (ALK_12, 13, 28, 44, 62), which included known TKI therapy resistance variants I1171N, I1171T, F1174L, F1174V, F1174C, L1196M, G1202R, and G1269A29. The median VAF of these ALK mutations was 0.18%. During the follow-up period, four patients (ALK_03, 13, 18, and 28) underwent subsequent treatment with a TKI that targeted the ALK mutation (Supplementary Fig. 1). In all cases, a reduction of the VAF of the mutation was observed after the change of therapy until the onset of further disease progression.
Prognostic utility of ctDNA
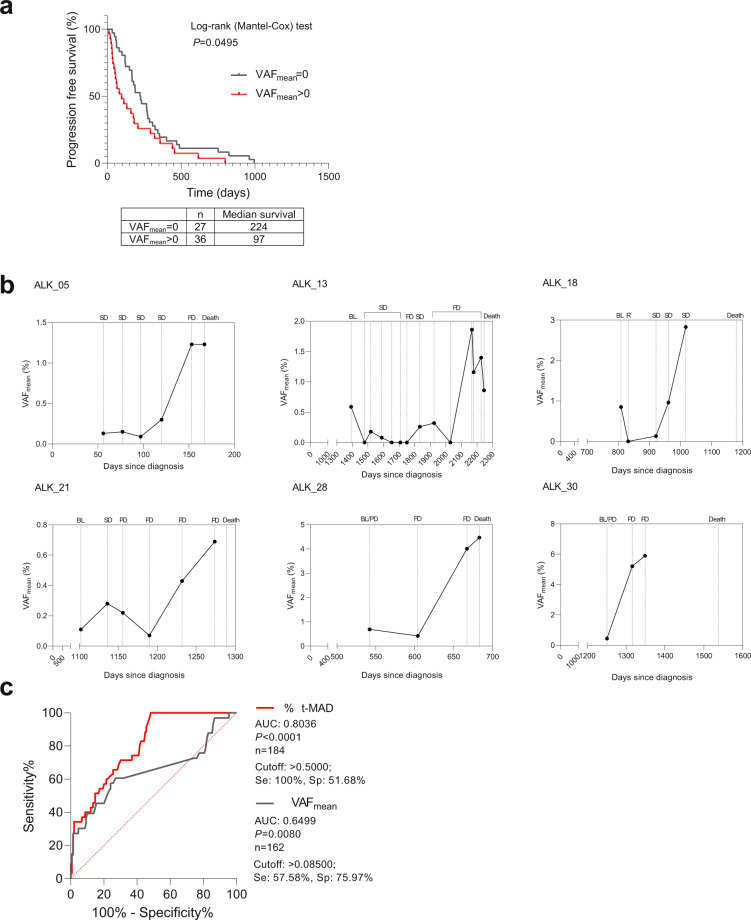
We analyzed the relevance of detecting variants at treatment initiation in relation to subsequent therapy durability, indicated by the time to progression from treatment baseline. We determined that upon independent evaluation of each therapy instance, detection of VAF (i.e., VAFmean > 0) at therapy baseline reflected significantly shorter time to progression compared to cases without detectable variants (median: 97 vs. 224 days) (P < 0.0495; Fig. 2a). Additionally, we observed elevated ctDNA levels as measured by VAFmean towards the end of life in 14 out of 16 patients who eventually succumbed to the illness. In six representative patients, the measured ctDNA levels were among the highest in samples prior to death (Fig. 2b).
Fig. 2. Prognostic utility of ctDNA NGS assays.
a Mean variant allele frequency at baseline indicates therapy durability. Progression-free survival plot showing significantly longer therapy durability in instances when ctDNA is undetected (VAFmean = 0) compared to detection (VAFmean > 0) at treatment initiation. b VAF kinetics in representative patients showing elevated VAFmean towards end of life. SD stable disease, PD progressive disease, BL therapy baseline, R response. c Comparison of receiver operating characteristic curves (ROC) using ΔVAFmean (gray) or %Δt-MAD (red) in discriminating progressive and non-progressive disease states.
Evaluating the potential utility of changes in genome-wide copy number and VAFmean as indicators of disease progression
We asked whether the t-MAD score, as a measure of global copy number changes derived from ctDNA, could be an indicator of clinical progression. Since the range of absolute t-MAD scores for each patient varied due to the heterogeneity of clinical status and applied therapy lines, we utilized a relative measurement of t-MAD based on percentage of change (%Δt-MAD) to determine whether this score of genome-wide copy number aberration could indicate disease progression. We found that the relative change in t-MAD score at successive sampling points was associated with extracranial progressive disease coincident with therapy change (AUC: 0.8036 [0.7333–0.8740], P < 0.0001, Fig. 2c). The cutoff with maximum sensitivity (100%) and specificity (51.68%) was a t-MAD score increase of 0.5%.
Similarly, ctDNA levels based on tNGS were applied to assess and predict therapy response17,30–33. Here, we used the difference between the detectable VAFmean (ΔVAFmean) of consecutive sampling points to quantify the change in ctDNA abundance31. The ΔVAFmean also correlated significantly with clinical status (i.e., extracranial progressive disease coincident with therapy change), but with a lower AUC than %Δt-MAD (AUC: 0.6499 [0.5277–0.7720], P = 0.0080, Fig. 3b). The optimal ΔVAFmean cutoff was 0.0850, which showed a sensitivity of 57.58% and specificity of 75.97% (Fig. 2c).
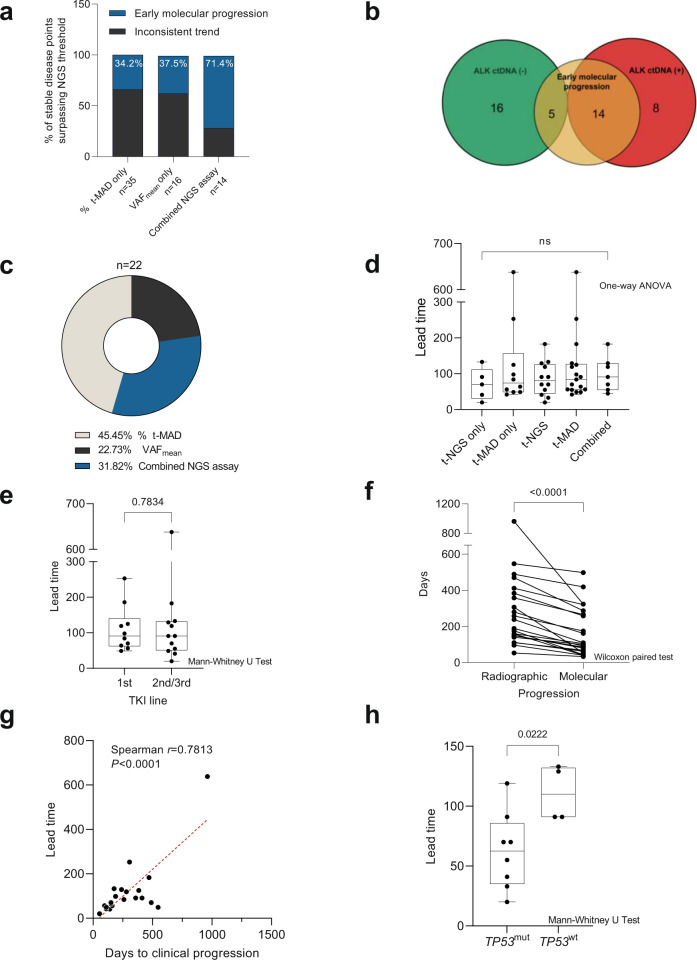
Fig. 3. Evaluation of the utility of ΔVAFmean and %Δt-MAD in identifying early molecular progression.
a Number of stable disease points that surpassed the numerical cutoffs per individual NGS assay (%Δt-MAD or ΔVAFmean) and in combination. Gray stacks indicate the percentage of points with inconclusive NGS assay values leading to clinical progression. Blue stacks represent points that showed sustained %Δt-MAD and/or ΔVAFmean increase leading to clinical progression, indicating early molecular progression. b Patients identified with early molecular progression were enriched in the ALK ctDNA (+) group. c Breakdown of 22 patients with early molecular progression based on NGS assays. d Comparison of called lead times based on NGS assays. e Comparison of called lead times based on therapy lines. f Comparison of cases with called lead time showing significant difference in days to radiographic progression versus molecular progression. g Length of lead time per therapy line was significantly correlated with duration of progression-free response to the respective treatment. h Lead times in cases with wild-type TP53 (TP53wt) were significantly longer compared to cases with mutated TP53 (TP53wt) detected by tNGS. The box plot show the the median, upper quartile, and lower quartile values. The whiskers indicate minimum and maximum values.
Molecular progression precedes clinical progression in a subset of ctDNA (+) patients
Next, we investigated whether ctDNA detection, as indicated by molecular progression, could predate and thus predict eventual clinical progression. Here, we used the ΔVAFmean and %Δt-MAD cutoff values defined from the ROC curve analysis of progressive disease points with therapy change as described above (i.e., ΔVAFmean = 0.0850, %Δt-MAD = 0.500%). Due to the heterogeneous therapy regimens across patients, we evaluated the time difference in days between early molecular and clinical progression (i.e., lead times) independently for each therapy line per patient. To identify early molecular progression, we examined sampling time points under treatment with stable disease according to RECIST v1.1. We identified 30 and 49 instances of such molecular progression using ΔVAFmean and %Δt-MAD, respectively, with 14 instances meeting both thresholds (Fig. 3a). Subsequently, we assigned a lead time if the initial increase in ctDNA was sustained at subsequent sampling points and eventually led to a progressive disease state. This second criterion validated 6 out of the 16 (37.5%) points based on ΔVAFmean alone and 12 out of 35 (34.2%) based on %Δt-MAD alone as true lead times. In the 14 cases where both NGS assay cutoffs were surpassed, 10 (71.4%) points were validated. These values indicated the higher sensitivity of %Δt-MAD compared to ΔVAFmean, while both showed similar specificity. Moreover, these results emphasized the potential of the combined NGS assays in increasing prediction confidence of early molecular progression. In total, we observed lead times in 19 out of 43 patients (44%), corresponding to 21 instances during TKI treatment, and one instance during chemotherapy. The median lead time to clinical progression was 88 days (range: 20–638 days). Fourteen of these 19 patients belonged to the group with detectable ALK alterations by tNGS (n = 22) as described above (Fig. 3b), indicating that calling lead times would be most relevant in patients or samples with high ctDNA content. Through this analysis, we also demonstrated the agreement and complementarity of tNGS (indicated by ΔVAFmean) and sWGS (indicated by %Δt-MAD) data in indicating molecular progression prior to standard radiographic assessment. Combined tNGS and sWGS data predicted lead times in 7 cases out of 22 (32%), tNGS data alone predicted lead times in five (23%) cases, and sWGS data alone was predictive in ten (45%) cases (Fig. 3c). No significant differences were found for the length of lead time predicted by either NGS assay, both independently and in combination (Fig. 3d). Lead time determination was also independent of therapy line (Fig. 3e). Time to molecular progression was significantly shorter in cases with lead time compared with clinical progression based on radiographic assessement (Fig. 3f). Lead times were longer for patients and/or therapy lines with longer duration of response (Fig. 3g). Out of 12 early molecular progression cases predicted by tNGS, 8 included TP53 mutations (TP53mut) in the variants monitored longitudinally. Upon lead time comparison of TP53wt versus TP53mut, we observed significantly shorter lead times in cases with TP53 mutation (Fig. 3h).
Relevance of ALK resistance mutations in early molecular progression events
We observed that early molecular progression identified through increased ΔVAFmean were, in most cases, due to the cumulative contribution of known or probable ALK resistance mutations. Out of a total of 36 ALK mutations identified in our study, 20 overlapped with called lead times due to consistent and theshold-surpassing ΔVAFmean. From the remaining 16 mutations, 9 were detected at the time of radiologic progression (i.e., were not informative of lead time). The remaining seven instances represented mutations detected at the first sample analyzed for each patient (n = 3), which are likely clonal mutations present prior to therapy administration, and mutations arising under treatment (n = 4) whose associated ΔVAFmean did not reach the threshold. In addition, the VAFs of these mutations declined before the next instance of radiologic progression. For emergent non-ALK mutations (n = 23), which included alterations in BRAF, ERBB2, KRAS, MET, NRAS, RET, and TP53, 13 mutation events were detected at the time of radiologic progression. From the remaining 10 mutations, two contributed to early molecular progression detection (i.e., their emergence contributed to ΔVAFmean which exceeded the defined threshold), and 8 mutations emerged under treatment whose ΔVAFmean did not reach the threshold, and subsequently the VAFs of these mutations declined before the next instance of radiologic progression. Altogether, our data indicate that resistance mutations, particularly known ALK resistance mutations, can indicate early molecular progression in many cases. However, the detection of lead time is more reliable in instances where such novel mutations arise in conjunction with increased ΔVAFmean, as applied in this work. Otherwise, detection of isolated and low allele frequency mutations pose the risk of identifying false-positive pseudo-progression events34.
Longitudinal monitoring in representative ALK+ NSCLC patients
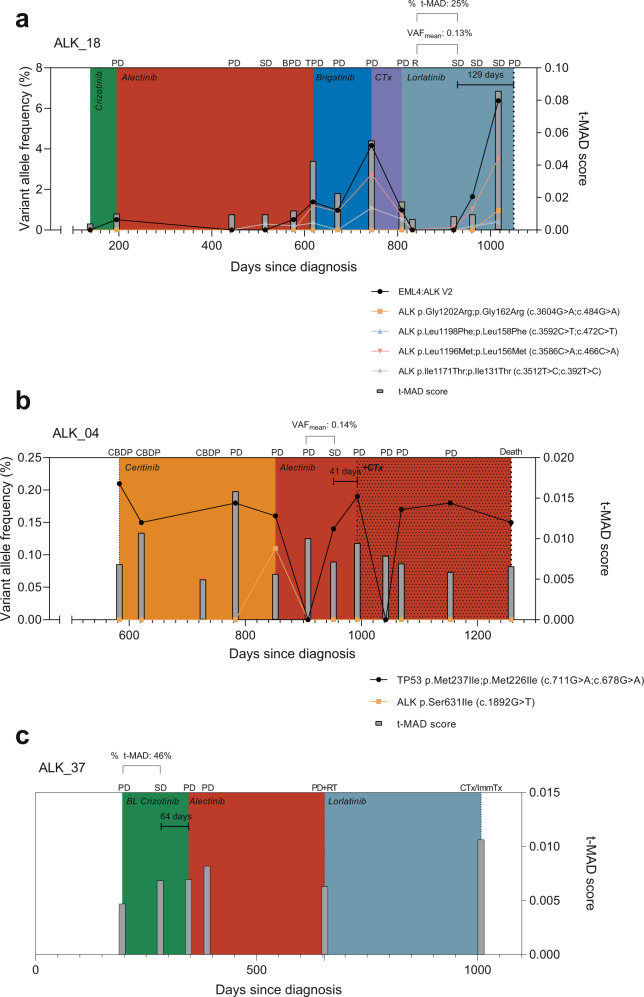
Representative cases of early molecular progression are illustrated in Fig. 4. Patient ALK_18 (Fig. 4a) depicts a case wherein both ΔVAFmean and %Δt-MAD were in agreement in detecting early molecular progression at a stable disease point. During lorlatinib therapy, ALK_18 initially exhibited response as reflected by decreasing VAF and t-MAD score 65 days post initiation of treatment. Early molecular progression was detected 112 days post therapy initiation while the disease was evaluated as stable, and two subsequent sampling points showed continued increase of NGS metrics (i.e., ΔVAFmean = 0.13%; %Δt-MAD = 25%) with stable disease until disease progression was observed. This point came 129 days after the onset of molecular progression. Patient ALK_04 (Fig. 4b) represents a case where ΔVAFmean was instrumental in identifying a lead time during alectinib therapy. After disease progression on ceritinib (852 days since diagnosis), the therapy was switched to alectinib in which stable disease was observed 100 days post administration. However, molecular analysis at this point already revealed ΔVAFmean of 0.14%, surpassing the threshold. Clinical progression was eventually observed 41 days later, supporting the result of ctDNA evaluation. Finally, patient ALK_37 (Fig. 4c) shows the utility of t-MAD monitoring particularly in the absence of detectable genetic alterations by tNGS. During crizotinib therapy, a %Δt-MAD increase of 46% was already apparent 86 days post treatment initiation, at which point the disease was still clinically stable. Disease progression occurred 64 days later, which led to a TKI therapy switch to alectinib. These case examples represent the 19 instances in our cohort where disease progression lead times were indicated by elevated levels of ctDNA through quantification of ΔVAFmean and %Δt-MAD.
Fig. 4. Representative cases illustrating early molecular progression as indicated by elevated ΔVAFmean and %Δt-MAD.
a Patient ALK_18 presented a case—during lorlatinib therapy—where both ΔVAFmean and %Δt-MAD were informative of the lead time. Early molecular progression was apparent 129 days prior to clinical progression, as indicated by ΔVAFmean of 0.13% and %Δt-MAD of 25%. Both values surpass the NGS metric thresholds identified in this study. b Patient ALK_04 depicts a case wherein ΔVAFmean indicated early molecular progression during alectinib therapy. A ΔVAFmean of 0.14% was calculated 41 days prior to clinical progression. c Patient ALK_37 emphasizes the relevance of untargeted NGS in ctDNA monitoring particularly in cases where genetic alterations remained undetected by tNGS. Here, %Δt-MAD of 46% was already apparent at a stable disease time point, 64 days prior to clinical progression. PD progressive disease, SD stable disease, BPD brain PD, TPD thoracic PD, R response, CBDP treatment continuation beyond disease progression, RT radiotherapy, CTx chemotherapy, ImmTx immunotherapy.
Discussion
Due to the relatively long duration of treatment and emergence of actionable acquired resistance, therapy monitoring is especially important for TKI-treated NSCLC patients35,36. Early detection of disease progression could facilitate significant improvement in patient outcomes, as approximately 25–30% of NSCLC patients failing ALK or EGFR TKI miss subsequent therapy or chemotherapy due to rapid clinical deterioration37,38. Previously, shallow whole-genome sequencing has been applied to estimate global copy number changes from ctDNA through computation of the t-MAD score28,39. We further showed that this metric was associated with shorter overall survival in a subset of our ALK+ NSCLC cohort27. Here, we systematically examine whether combined tNGS and sWGS could also enable the detection of acquired resistance and disease progression earlier than radiological imaging.
Previously, an important challenge was the lack of a general consensus on how to measure ctDNA levels and response cutoffs. To date, multiple studies have used various ctDNA quantification methods including the maximum variant allele frequency40, the mean variant allele frequency31,41, and the total ctDNA concentration32. Similarly, response thresholds were typically established using a single time point after therapy baseline with arbitrary cutoff values18,32,42. Here, we employed ΔVAFmean and the %Δt-MAD scores because these metrics can capture ctDNA changes regardless of how many SNV and/or CNV are present in the baseline sample. The use of ultra-deep targeted sequencing with optimized gene panels enables efficient surveillance of alteration hotspots of high clinical impact. These somatic changes are then used for early screening, treatment monitoring, and detection of residual disease. In parallel, untargeted sWGS approaches to liquid biopsy have been successful in other cancer entities39,43,44 and have been shown to reflect tumor status, even in cases without detectable SNVs27. Our data show that ΔVAFmean offers better specificity than %Δt-MAD in discriminating progressive versus non-progressive disease. In contrast, %Δt-MAD showed increased sensitivity. These results suggest that ΔVAFmean could be a more relevant ctDNA metric for clinical utility in the advanced setting where specificity is a critical performance assay criterion and greater ctDNA shedding occurs44, whereas %Δt-MAD could be applied at early disease stages. Nonetheless, residual false-positive and false-negative calls of disease status based on ctDNA evaluations could still take place due to tumors that shed minimal amounts of ctDNA; due to cases with apparent disagreement between molecular progression and imaging evaluations; and due to clearance of tumors leading to ctDNA shedding during earliest cycles of treatment.
In our study population, VAFmean > 0 at therapy baseline predicted shorter therapy duration compared to cases without detected variants (VAFmean = 0). This parallels findings demonstrating that ctDNA at baseline indicates poor prognosis in various cancers18,27,45–47. Pretreatment VAF levels have also been shown to be prognostic of treatment response with immune checkpoint inhibitors in patients with advanced solid tumors31,48. Pretreatment VAFmean and maximum VAF were significantly inversely correlated with overall survival in NSCLC, urothelial cancer, microsatellite instable cancers, gastroesophageal cancer, and ovarian cancer31. Our results extend the potential of ctDNA detectability at therapy baseline as biomarkers for therapy response across various treatment lines in ALK+ NSCLC. Nonetheless, we recognize the marginal significance (P < 0.045) of our survival data. Variations in therapy sequence, therapy duration, and interpatient variabilities could be sources of confounding factors in the analysis. Each therapy line per patient was also treated as an independent case, discounting the potential effects of prior therapy.
A potential confounding factor in longitudinal monitoring lies in the assumption that visual evaluation of tumor radiographs accurately reflects tumor growth or progression49. In fact, there were sampling points attributed with clinically stable disease that showed consistently increasing levels of VAFmean and/or t-MAD, which could indicate onset of disease progression below the limit of detection of radiographic modalities. By specifying numerical cutoffs for both NGS assays through ROC analysis, we were able to systematically identify such early molecular progression events in 44% of patients, across all therapy lines. This suggests that in this subset of our study cohort, ctDNA analysis exhibits higher sensitivity in detecting tumor progression than visual staging. It is also worthwhile to note that in majority of cases, early emergence of known or probable ALK mutations accounted for increased ΔVAFmean that resulted in lead time identification. This emphasizes the sensitivity of ctDNA analysis in identifying potentially actionable mutations early in the disease course. From the tumor biology standpoint, such cases reflect the dynamic clonal evolution within the tumor that could potentially be missed by tissue re-biopsies. Nonetheless, there were also mutations—both ALK and non-ALK—that were inconsistent or sporadic throughout the longitudinal course. Using ΔVAFmean as a cumulative metric of ctDNA levels minimizes identification of such false-positive pseudo-molecular progression events that could have arisen from stochastic sampling issues50.
While both tNGS and sWGS were able to independently call early molecular progression events, we also present evidence that a combined evaluation increases the confidence in predicting lead times by approximately 50%, corroborating our previous observation27 that untargeted sWGS complements tNGS. Moreover, there was a positive correlation between the length of predicted lead time and durability of response to therapy. Therefore, molecular monitoring for earlier detection of disease progression could be even more useful for patients treated with newer ALK inhibitors, namely alectinib, brigatinib, or lorlatinib, whose progression-free survival extends well beyond two years, in contrast to 10–12 months under crizotinib8–10. This approach also limits the risk due to higher cumulative radiation exposure from regular CTs51. Interestingly, we further observed that lead times involving mutations in TP53 were shorter compared to TP53 wild types. This implies that the emergence of TP53 alterations during therapy course portends a swifter progression of disease, as previously noted in ALK+ NSCLC52–54. Ideally, early detection of molecular progression could prompt clinicians to halt ineffective treatments, avoiding potential side effects and added financial burden, and also facilitate an earlier therapy switch, while the patients are still capable of receiving next-line therapies38. Such a strategy could be particularly useful for cases with higher risk, such as those with TP53 mutations and/or the EML4-ALK variant 3 (refs. 53,55). This approach is complementary to monitoring of early ctDNA changes after therapy initiation, which have been shown to predict therapy durability44.
The main limitations of our study are related to its retrospective design, the relatively small patient number, and the lack of a separate validation cohort. Our findings should ideally be confirmed in a larger, prospective study with strictly defined sampling and imaging intervals. The variants by tNGS were also limited to the genes included in the panels utilized, thus other genetic alterations in ctDNA were not evaluated.
Our work demonstrates the applicability of a combined targeted and untargeted NGS analysis of cfDNA for longitudinal monitoring and identification of early molecular progression in ALK+ NSCLC. We established ΔVAFmean and %Δt-MAD as quantifiable metrics for ctDNA levels and determined thresholds to enable prediction of disease progression and identification of early molecular progression. Currently, the development of robust criteria assessing ctDNA dynamics that correlate with clinical status such as disease progression, therapy response, and therapy resistance remains challenging partly due to stochastic sampling issues50, and the spectrum of ctDNA shedding tendencies of different malignancies. Aside from ctDNA, it should also be noted that other cell-free markers have the capacity to detect progression and predict disease recurrence. In NSCLC, for example, miRNA signatures in the plasma have been shown to associate with higher risk for progression56,57, while a separate panel could predict survival in squamous cell carcinoma patients56. Serum carcinoembryonic antigen (CEA) is also associated with distinct cancer progression profiles and carries predictive information of risk recurrence in NSCLC58,59. Oncogenic mRNA markers in plasma have also shown clinical utility in NSCLC60,61. Thus, in addition to genomic alterations in ctDNA, future investigations of other plasma solutes including proteins, miRNAs, mRNAs, and the epigenomic properties of ctDNA (e.g., methylation, nucleosome positioning) will advance the potential of liquid biopsy in complementing imaging technologies in improving personalized patient management and efficient therapy decisions.
Methods
Patients and sample collection
The study was approved by the ethics committees of Heidelberg University (S-270/2001, S-296/2016) and Lübeck University (AZ 12-238). Written informed consent was obtained from all study participants. Newly diagnosed cases were screened for the presence of an ALK alteration in tissues by fluorescence in situ hybridization (FISH, ZytoLight SPEC ALK probe, ZytoVision GmbH, Bremerhaven, Germany) and reverse-transcription polymerase chain reaction until 2015, or by immunohistochemistry (D5F3 clone, Roche, Mannheim, Germany) and RNA-based next-generation sequencing (NGS, Thermo Fisher Lung Cancer Fusion Panel, Waltham, MA, USA) thereafter, as previously described12. Peripheral blood was prospectively drawn through venipuncture at each outpatient visit from 84 ALK+ patients with metastatic NSCLC under TKI therapy at Thoraxklinik Heidelberg and Lungenclinic Grosshansdorf, Germany62. In total, 412 longitudinal blood samples were collected and plasma was isolated within 1 h of blood draw, using the double spin method as previously described27. Samples were stored at −80 °C in the Lung Biobank Heidelberg/BMBH until further processing.
Since our study was focused on longitudinal therapy monitoring, we limited the scope of our analysis only on patients with at least four evaluable plasma samples throughout their treatment course. Using these criteria, we excluded 41 patients (69 plasma samples). Ultimately, 43 metastatic ALK+ patients with corresponding 343 plasma samples were considered for NGS analysis (Fig. 1a). The characteristics of these patients are summarized in Table 1. Clinical data and the results of routine radiographic assessments (every 8–12 weeks) using chest/abdominal CT and brain MRI were collected through a review of patient records with a cutoff on September 15, 2020.
cfDNA isolation
The AVENIO cfDNA Isolation Kit (Roche Diagnostics) was used to isolate cfDNA from 2 mL of patient plasma, following the manufacturer’s protocol. The characteristic mononucleosomal molecular weight profile (160–200 bp) of the purified cfDNA was assessed using the Bioanalyzer 2100 High Sensitivity DNA Kit (Agilent Technologies). The cfDNA was quantified using the Qubit dsDNA High Sensitivity Kit (Thermo Fisher).
cfDNA NGS assays
Sequencing libraries for capture-based targeted sequencing were generated using the AVENIO ctDNA Library Preparation Kit with either the Targeted or Surveillance Panel (Roche Diagnostics), as previously described27. Equimolar 16-plex enriched library pools were sequenced on the Illumina NextSeq 550 platform with the High-Output Kit V2 (2 × 150 bp). The resulting raw BCL files were processed using the AVENIO ctDNA analysis software (Roche Diagnostics, version 2.0.0). The proprietary analysis pipeline applied by the software is adapted from the CAPP-Seq workflow with integrated digital suppression63,64. Called variants with variant allele frequencies (VAFs) ≥30% were classified as germline mutations and were excluded from subsequent analyses. Only genes common to both Targeted and Surveillance panels (Supplementary Table 3) were compared for all longitudinal assessments, with a VAF calling threshold of 0.01%, facilitated by an average of 4100× unique target coverage as previously reported27. For sWGS, sequencing libraries were prepared using the KAPA HyperPrep Kit with KAPA Dual-Indexed Adapters for Illumina platforms, as previously described27 using 1–2.5 ng cfDNA as input. Pooled multiplexes of 48–67 equimolar libraries were sequenced on the Illumina HiSeq 4000 platform (2 × 100 bp). To determine genome-wide copy number variations (CNVs) and calculate the t-MAD scores, raw sWGS sequencing data were processed and analyzed as previously described27. Briefly, automated sequence quality control and alignment were performed using the One Touch Pipeline65. Genome-wide copy number profiles and tumor fractions were estimated using ichorCNA implemented in R (version 3.3.1). T-MAD scores were computed using a 1-Mb bin size and without fragment size selection27.
Genomic alteration and t-MAD score determination
Somatic SNVs and indels detected by the AVENIO ctDNA analysis software were used to calculate the mean variant allele frequency (VAFmean) per sampling point. This value is the sum of all VAFs divided by the number of all mutations at each time point to account for polyclonality17. The change in mean VAF (ΔVAFmean) was the difference in VAFmean between two successive time points as shown by Eqs. (1) and (2).
| 1 |
where
| 2 |
Here, n represents the number of detected variants. If n = 0, or no variant was detected, VAFmean was set to 0. Positive ΔVAFmean values indicate increasing VAFmean. In parallel, the change in t-MAD (%Δt-MAD) was calculated, in percentage, as the difference between two successive t-MAD scores, divided by the t-MAD measured at the previous time point as shown by Eq. (3).
| 3 |
ΔVAFmean and %Δt-MAD thresholds for disease progression were derived using the respective receiver operating characteristic (ROC) curve across measurement time points with progressive disease accompanied by therapy change (PDtherapy change) as the event of interest. The annotation of PDtherapy change was verified manually based on radiological imaging according to RECIST criteria v1.1 (ref. 66), as well as information about patient treatment from the medical records. Area under the curve (AUC) values were generated using GraphPad Prism 9 (GraphPad software, La Jolla California USA), while cutoff values were identified using the Youden Index, which maximizes the sensitivity and specificity of the NGS metric (i.e., ΔVAFmean and %Δt-MAD)67. Seven patients (ALK_22, 37, 57, 59, 60, 66, 105) were excluded from ΔVAFmean measurements due to the absence of detectable variants in all plasma DNA samples. In total, 162 measurements of ΔVAFmean and 184 measurements of %Δt-MAD were used for ROC curve analyses (Supplementary Tables 4 and 5).
We defined early molecular progression as a change in either NGS metric (i.e., ΔVAFmean or %Δt-MAD) surpassing the threshold value defined from the ROC curve analysis, from a point where the tumor was radiographically stable according to RECIST to a later progression point. The lead time was defined as the number of days between the first observation of molecular progression until the point of an initial clinical progression as per radiographic assessment based on RECIST.
Statistical analysis and data visualization
Survival data were analyzed using the log-rank test. Lead time comparisons were performed using the Mann–Whitney U test or one-way ANOVA, as labeled in the graphs. Pairwise comparison of days to progression was performed using the Wilcoxon’s paired test. Since the primary aim of this study was to establish the relationship between ctDNA changes and disease progression, samples from different progression time points of the same patient were analyzed as independent, in order to account for the fact that several factors can influence liquid biopsy positivity differently in each sample of the same patient (e.g. progression site, progression rate, tumor volume, preceding treatment), and that these factors act similarly across different patients. Statistical analyses, including ROC curves, and relevant graphs were performed and generated, respectively, using GraphPad Prism 9.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This work was supported by the German Center for Lung Research (DZL), by the German Cancer Consortium (DKTK), by the Heidelberg Center for Personalized Oncology at the German Cancer Research Center (DKFZ-HIPO), and by Roche Sequencing Solutions (Pleasanton, CA, USA). The authors would like to thank Ingrid Heinzmann-Groth, Karin Schnorr-Teichert, and Saskia Oestringer of the Lung Biobank Heidelberg for assistance with patient sample collection. We also acknowledge the Genomics and Proteomics Core Facility at the German Cancer Research Center (DKFZ) for sample sequencing, and the Omics IT and Data Management Core Facility at the DKFZ for data management and processing. The authors thank Thomas Hielscher from the Division of Biostatistics at the DKFZ for support.
Author contributions
A.K.A.: conceptualization, methodology, data acquisition, formal analysis, writing—original draft, writing—review and editing, visualization; P.C.: conceptualization, methodology, data acquisition, formal analysis, writing—original draft, writing—review and editing, visualization, supervision; Z.Y.: methodology, formal analysis, visualization, writing—review and editing; S.B. and F.J.: methodology, writing—original draft, writing—review and editing; S.J.O.: data acquisition, writing—review and editing; M.R.: conceptualization, data acquisition, methodology, writing—review and editing; M.S.: methodology, data acquisition, formal analysis, writing—review and editing; M.M.: methodology, data acquisition, writing—review and editing; M.A.S.: data acquisition, methodology, writing—review and editing; S.D.: conceptualization, methodology, data acquisition, formal analysis, writing—review and editing; A.S.: methodology, data acquisition, supervision, writing—review and editing; M.T.: data acquisition, formal analysis, writing—review and editing; H.S.: conceptualization, methodology, writing—original draft, writing—review and editing, supervision, project administration, funding acquisition.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
All sequencing data supporting the findings in this study are deposited in the European Genome-phenome Archive (EGA) under accession number EGAS00001005327.
Competing interests
P.C. reports research funding from AstraZeneca, Novartis, Roche, and Takeda as well as advisory board and/or lecture fees from Boehringer Ingelheim, Chugai, Lilly, Pfizer, and Takeda, outside the submitted work. M.A.S. reports personal fees from the German Canter for Lung Research (DZL), outside the submitted work. S.D. reports speaker’s honoraria from Roche and research funding from Takeda, outside the submitted work. M.R. reports personal fees from Amgen, AstraZeneca, BMS, Boehringer Ingelheim, Lilly, Merck, MSD, Novartis, Pfizer, Roche, and Samsung, outside the submitted work. M.T. reports advisory board honoraria from Novartis, Lilly, BMS, MSD, Roche, Celgene, Takeda, AbbVie, Boehringer, speaker’s honoraria from Lilly, MSD, Takeda, research funding from AstraZeneca, BMS, Celgene, Novartis, Roche and travel grants from BMS, MSD, Novartis, Boehringer, outside the submitted work. A.S. reports advisory board honoraria from BMS, Bayer, AstraZeneca, Thermo Fisher, Novartis, Seattle Genomics speaker’s honoraria from BMS, Bayer, Illumina, AstraZeneca, Novartis, Thermo Fisher, MSD, Roche, as well as research funding from Chugai, Bayer and BMS, outside the submitted work. H.S. reports grants from Roche Sequencing Solutions, during the conduct of the study, and personal fees from Roche, outside the submitted work. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Arlou Kristina Angeles, Petros Christopoulos.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-021-00239-3.
References
- 1.Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat. Rev. Cancer. 2019;19:495–509. doi: 10.1038/s41568-019-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solomon BJ, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N. Engl. J. Med. 2014;371:2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 3.Kim DW, et al. Activity and safety of ceritinib in patients with ALK-rearranged non-small-cell lung cancer (ASCEND-1): updated results from the multicentre, open-label, phase 1 trial. Lancet Oncol. 2016;17:452–463. doi: 10.1016/S1470-2045(15)00614-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw AT, et al. Alectinib in ALK-positive, crizotinib-resistant, non-small-cell lung cancer: a single-group, multicentre, phase 2 trial. Lancet Oncol. 2016;17:234–242. doi: 10.1016/S1470-2045(15)00488-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim DW, et al. Brigatinib in patients with crizotinib-refractory anaplastic lymphoma kinase-positive non-small-cell lung cancer: a randomized, multicenter phase II trial. J. Clin. Oncol. 2017;35:2490–2498. doi: 10.1200/JCO.2016.71.5904. [DOI] [PubMed] [Google Scholar]
- 6.Chuang JC, Neal JW. Crizotinib as first line therapy for advanced ALK-positive non-small cell lung cancers. Transl. Lung Cancer Res. 2015;4:639–641. doi: 10.3978/j.issn.2218-6751.2015.03.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gainor JF, et al. Molecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK-rearranged lung cancer. Cancer Discov. 2016;6:1118–1133. doi: 10.1158/2159-8290.CD-16-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters S, et al. Alectinib versus crizotinib in untreated ALK-positive non-small-cell lung cancer. N. Engl. J. Med. 2017;377:829–838. doi: 10.1056/NEJMoa1704795. [DOI] [PubMed] [Google Scholar]
- 9.Camidge DR, et al. Brigatinib versus crizotinib in ALK-positive non-small-cell lung cancer. N. Engl. J. Med. 2018;379:2027–2039. doi: 10.1056/NEJMoa1810171. [DOI] [PubMed] [Google Scholar]
- 10.Shaw AT, et al. First-line lorlatinib or crizotinib in advanced ALK-positive lung cancer. N. Engl. J. Med. 2020;383:2018–2029. doi: 10.1056/NEJMoa2027187. [DOI] [PubMed] [Google Scholar]
- 11.Wang W, Sun X, Hui Z. Treatment optimization for brain metastasis from anaplastic lymphoma kinase rearrangement non-small-cell lung cancer. Oncol. Res. Treat. 2019;42:599–606. doi: 10.1159/000502755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volckmar AL, et al. Combined targeted DNA and RNA sequencing of advanced NSCLC in routine molecular diagnostics: analysis of the first 3,000 Heidelberg cases. Int. J. Cancer. 2019;145:649–661. doi: 10.1002/ijc.32133. [DOI] [PubMed] [Google Scholar]
- 13.Benayed R, et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden. Clin. Cancer Res. 2019;25:4712–4722. doi: 10.1158/1078-0432.CCR-19-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ilie M, Hofman P. Pros: can tissue biopsy be replaced by liquid biopsy? Transl. Lung Cancer Res. 2016;5:420–423. doi: 10.21037/tlcr.2016.08.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Rubis G, Rajeev Krishnan S, Bebawy M. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol. Sci. 2019;40:172–186. doi: 10.1016/j.tips.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Velimirovic, M. et al. Rising circulating tumor DNA as a molecular biomarker of early disease progression in metastatic breast cancer. JCO Precis. Oncol., 1246–1262 (2020). [DOI] [PubMed]
- 18.Lee JH, et al. Longitudinal monitoring of ctDNA in patients with melanoma and brain metastases treated with immune checkpoint inhibitors. Clin. Cancer Res. 2020;26:4064–4071. doi: 10.1158/1078-0432.CCR-19-3926. [DOI] [PubMed] [Google Scholar]
- 19.Parikh AR, et al. Serial ctDNA monitoring to predict response to systemic therapy in metastatic gastrointestinal cancers. Clin. Cancer Res. 2020;26:1877–1885. doi: 10.1158/1078-0432.CCR-19-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vandekerkhove G, et al. Plasma ctDNA is a tumor tissue surrogate and enables clinical-genomic stratification of metastatic bladder cancer. Nat. Commun. 2021;12:184. doi: 10.1038/s41467-020-20493-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang EW, et al. Association between circulating tumor DNA burden and disease burden in patients with ALK-positive lung cancer. Cancer. 2020;126:4473–4484. doi: 10.1002/cncr.33118. [DOI] [PubMed] [Google Scholar]
- 22.Tie J, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016;8:346ra392. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Murillas I, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med. 2015;7:302ra133. doi: 10.1126/scitranslmed.aab0021. [DOI] [PubMed] [Google Scholar]
- 24.Chaudhuri AA, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 2017;7:1394–1403. doi: 10.1158/2159-8290.CD-17-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dagogo-Jack, I. et al. Tracking the evolution of resistance to ALK tyrosine kinase inhibitors through longitudinal analysis of circulating tumor DNA. JCO Precis. Oncol. 2018, 10.1200/PO.17.00160 (2018). [DOI] [PMC free article] [PubMed]
- 26.Noe J, et al. ALK mutation status before and after alectinib treatment in locally advanced or metastatic ALK-positive NSCLC: pooled analysis of two prospective trials. J. Thorac. Oncol. 2020;15:601–608. doi: 10.1016/j.jtho.2019.10.015. [DOI] [PubMed] [Google Scholar]
- 27.Dietz S, et al. Longitudinal therapy monitoring of ALK-positive lung cancer by combined copy number and targeted mutation profiling of cell-free DNA. EBioMedicine. 2020;62:103103. doi: 10.1016/j.ebiom.2020.103103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mouliere, F. et al. Detection of cell-free DNA fragmentation and copy number alterations in cerebrospinal fluid from glioma patients. EMBO Mol. Med. 10, 10.15252/emmm.201809323 (2018). [DOI] [PMC free article] [PubMed]
- 29.Lin JJ, Riely GJ, Shaw AT. Targeting ALK: precision medicine takes on drug resistance. Cancer Discov. 2017;7:137–155. doi: 10.1158/2159-8290.CD-16-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia N, et al. Serial monitoring of circulating tumor DNA in patients with metastatic colorectal cancer to predict the therapeutic response. Front. Genet. 2019;10:470. doi: 10.3389/fgene.2019.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q, et al. Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade. Cancer Discov. 2020;10:1842–1853. doi: 10.1158/2159-8290.CD-20-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nabet BY, et al. Noninvasive early identification of therapeutic benefit from immune checkpoint inhibition. Cell. 2020;183:363–376 e313. doi: 10.1016/j.cell.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moding EJ, et al. Circulating tumor DNA dynamics predict benefit from consolidation immunotherapy in locally advanced non-small-cell lung cancer. Nat. Cancer. 2020;1:176–183. doi: 10.1038/s43018-019-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ricciuti, B. et al. Early plasma circulating tumor DNA (ctDNA) changes predict response to first-line pembrolizumab-based therapy in non-small cell lung cancer (NSCLC). J. Immunother. Cancer9, 10.1136/jitc-2020-001504 (2021). [DOI] [PMC free article] [PubMed]
- 35.Schrank, Z. et al. Current molecular-targeted therapies in NSCLC and their mechanism of resistance. Cancers (Basel)10, 10.3390/cancers10070224 (2018). [DOI] [PMC free article] [PubMed]
- 36.Gristina, V. et al. The emerging therapeutic landscape of ALK inhibitors in non-small cell lung cancer. Pharmaceuticals (Basel)13, 10.3390/ph13120474 (2020). [DOI] [PMC free article] [PubMed]
- 37.Magios, N. et al. Real-world implementation of sequential targeted therapies for EGFR-mutated lung cancer. Ther. Adv. Med. Oncol.13, 1758835921996509, 10.1177/1758835921996509 (2021). [DOI] [PMC free article] [PubMed]
- 38.Elsayed, M. et al. Feasibility and challenges for sequential treatments in ALK-rearranged non-small-cell lung cancer. Front. Oncol.11, 10.3389/fonc.2021.670483 (2021). [DOI] [PMC free article] [PubMed]
- 39.Smith CG, et al. Comprehensive characterization of cell-free tumor DNA in plasma and urine of patients with renal tumors. Genome Med. 2020;12:23. doi: 10.1186/s13073-020-00723-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldberg SB, et al. Early assessment of lung cancer immunotherapy response via circulating tumor DNA. Clin. Cancer Res. 2018;24:1872–1880. doi: 10.1158/1078-0432.CCR-17-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ococks, E. et al. Longitudinal tracking of 97 esophageal adenocarcinomas using liquid biopsy sampling. Ann. Oncol., 10.1016/j.annonc.2020.12.010 (2021). [DOI] [PubMed]
- 42.Bratman SV, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat. Cancer. 2020;1:873–881. doi: 10.1038/s43018-020-0096-5. [DOI] [PubMed] [Google Scholar]
- 43.Mouliere, F. et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 10, 10.1126/scitranslmed.aat4921 (2018). [DOI] [PMC free article] [PubMed]
- 44.Davis AA, et al. Early assessment of molecular progression and response by whole-genome circulating tumor DNA in advanced solid tumors. Mol. Cancer Ther. 2020;19:1486–1496. doi: 10.1158/1535-7163.MCT-19-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacob, S. et al. The use of serial circulating tumor DNA to detect resistance alterations in progressive metastatic breast cancer. Clin. Cancer Res., 10.1158/1078-0432.CCR-20-1566 (2020). [DOI] [PubMed]
- 46.Cavallone L, et al. Prognostic and predictive value of circulating tumor DNA during neoadjuvant chemotherapy for triple negative breast cancer. Sci. Rep. 2020;10:14704. doi: 10.1038/s41598-020-71236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Christopoulos P, et al. Earlier extracranial progression and shorter survival in ALK- rearranged lung cancer with positive liquid rebiopsies. Transl Lung Cancer Res. 2021;10:2118–2131. doi: 10.21037/tlcr-21-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marsavela G, et al. Circulating tumor DNA predicts outcome from first-, but not second-line treatment and identifies melanoma patients who may benefit from combination immunotherapy. Clin. Cancer Res. 2020;26:5926–5933. doi: 10.1158/1078-0432.CCR-20-2251. [DOI] [PubMed] [Google Scholar]
- 49.Keup, C. et al. Longitudinal multi-parametric liquid biopsy approach identifies unique features of circulating tumor cell, extracellular vesicle, and cell-free DNA characterization for disease monitoring in metastatic breast cancer patients. Cells10, 10.3390/cells10020212 (2021). [DOI] [PMC free article] [PubMed]
- 50.Hrebien S, et al. Early ctDNA dynamics as a surrogate for progression-free survival in advanced breast cancer in the BEECH trial. Ann. Oncol. 2019;30:945–952. doi: 10.1093/annonc/mdz085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Volckmar AL, et al. A field guide for cancer diagnostics using cell-free DNA: from principles to practice and clinical applications. Gene Chromosome Cancer. 2018;57:123–139. doi: 10.1002/gcc.22517. [DOI] [PubMed] [Google Scholar]
- 52.VanderLaan PA, et al. Mutations in TP53, PIK3CA, PTEN and other genes in EGFR mutated. Lung Cancers. 2017;106:17–21. doi: 10.1016/j.lungcan.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Christopoulos, P. et al. Detection of TP53 mutations in tissue or liquid rebiopsies at progression identifies ALK(+) lung cancer patients with poor survival. Cancers11, 10.3390/cancers11010124 (2019). [DOI] [PMC free article] [PubMed]
- 54.Christopoulos P, et al. Identification of a highly lethal V3(+)TP53(+) subset in ALK(+) lung adenocarcinoma. Int. J. Cancer. 2019;144:190–199. doi: 10.1002/ijc.31893. [DOI] [PubMed] [Google Scholar]
- 55.Christopoulos P, Kirchner M, Endris V, Stenzinger A, Thomas M. EML4-ALK V3, treatment resistance, and survival: refining the diagnosis of ALK(+) NSCLC. J. Thorac. Dis. 2018;10:S1989–S1991. doi: 10.21037/jtd.2018.05.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanfiorenzo C, et al. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS ONE. 2013;8:e54596. doi: 10.1371/journal.pone.0054596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen Y, et al. microRNA expression profiles associated with survival, disease progression, and response to gefitinib in completely resected non-small-cell lung cancer with EGFR mutation. Med. Oncol. 2013;30:750. doi: 10.1007/s12032-013-0750-1. [DOI] [PubMed] [Google Scholar]
- 58.Grunnet M, Sorensen JB. Carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung Cancer. 2012;76:138–143. doi: 10.1016/j.lungcan.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 59.Kuo YS, et al. Association of divergent carcinoembryonic antigen patterns and lung cancer progression. Sci. Rep. 2020;10:2066. doi: 10.1038/s41598-020-59031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Castellanos-Rizaldos E, et al. Exosome-based detection of EGFR T790M in plasma from non-small cell lung cancer patients. Clin. Cancer Res. 2018;24:2944–2950. doi: 10.1158/1078-0432.CCR-17-3369. [DOI] [PubMed] [Google Scholar]
- 61.Krug AK, et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann. Oncol. 2018;29:2143. doi: 10.1093/annonc/mdy261. [DOI] [PubMed] [Google Scholar]
- 62.Wessels S, et al. Comprehensive serial biobanking in advanced NSCLC: feasibility, challenges and perspectives. Transl. Lung Cancer Res. 2020;9:1000–1014. doi: 10.21037/tlcr-20-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Newman AM, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Newman AM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 2016;34:547–555. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reisinger E, et al. OTP: an automatized system for managing and processing NGS data. J. Biotechnol. 2017;261:53–62. doi: 10.1016/j.jbiotec.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 66.Eisenhauer EA, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur. J. Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 67.Ruopp MD, Perkins NJ, Whitcomb BW, Schisterman EF. Youden Index and optimal cut-point estimated from observations affected by a lower limit of detection. Biom. J. 2008;50:419–430. doi: 10.1002/bimj.200710415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data supporting the findings in this study are deposited in the European Genome-phenome Archive (EGA) under accession number EGAS00001005327.