Abstract
OBJECTIVES:
This study is to explore the molecular mechanisms underlying how the lipid metabolic dysregulation is associated with systemic lupus erythematosus (SLE) pathogenesis.
METHODS:
B cells in peripheral blood from lupus patients and healthy controls were used for lipid bodipy analysis. B-cell specific IRE1α and SCD1 knockout mice were employed for studying the influence of IRE1α−SCD1/2 pathway on B-cell differentiation and auto-antibody production. The preclinical efficacy of IRE1α suppression in lupus treatment were elucidated in MRL/Lpr mice.
RESULTS:
We showed that supplementation of the monounsaturated fatty acid largely rescues plasma cell differentiation from IRE1α-null B cells, indicating that the frailer of IRE1α-null B-cell differentiation is due to a defect in monounsaturated fatty acid synthesis. IRE1α-XBP-1 activation is required for B cell expression of stearoyl-CoA desaturase 1 and 2 (SCD1 and SCD2), two critical enzymes catalyzing monounsaturated fatty acid synthesis. Mice with targeted Scd1 gene deletion largely phenocopies Ire1α-deficient mice with diminished B-cell differentiation into plasma cells. Importantly, IRE1α expression and Xbp-1 mRNA splicing in B-cells from lupus patients are significantly increased, which positively correlate with the expression of both Scd1 and Scd2 genes, as well as with the amount of lipid deposition in B cells. Either genetic or pharmacological IRE1α suppression protected mice from lupus pathogenesis.
CONCLUSION:
Our study reveals a molecular link of lipid metabolic dysregulation in lupus pathogenesis, demonstrates that the IRE1α-XBP-1 pathway controls plasma cell differentiation through SCD1/2-mediated monounsaturated fatty acid synthesis, and provides a rationale for targeting IRE1α and monounsaturated fatty acid synthesis in lupus treatment.
Keywords: monounsaturated fatty acid, Lupus, B cell differentiation, IRE1α, stearoyl-CoA desaturase 1
INTRODUCTION
One signature of systemic lupus erythematosus (SLE) is the production of self-reactive antibodies in lupus patients. The autoantibodies often deposit into tissues and organs that impairs their functions, such as the nephropathy, which leads to loss of the kidney functions. Since plasma cells, differentiated from B cells upon recognition of self-antigens, are the only resources of autoimmune antibodies, suppression of B-cell activation has been a focus for developing therapeutics for lupus as well as other Ab-mediated diseases [1–3]. Accumulated evidence suggests that the dysregulated lipid metabolism is involved in autoimmune diseases in human. Dramatic increases in free fatty acids, in particular the monounsaturated fatty acids oleic acid and palmitoleic acid, are positively correlated with SLE activity [4–7]. However, the direct connection between lipid metabolic dysregulation and SLE has remained largely unclear.
The immune responses can also be adversely affected by abnormalities in the unfolded protein response, which could potentially contribute to the development of autoimmunity [8, 9]. The transition of B cells into plasma cells provokes the unfolded protein response, as indicated by the Inositol-requiring enzyme 1α (IRE1α)-mediated mRNA splicing of the X-box binding protein 1 (Xbp-1), a transcription factor that promotes expression of ER chaperones [10]. IRE1α is an ER stress sensor that contains an ER-luminal sensor domain that recognizes unfolded proteins and a cytosolic kinase/RNase domain that regulates downstream effectors such as XBP-1 [11–14]. XBP-1s up-regulates the synthesis of lipids and chaperones, contributing to the ER expansion and increased Ig production in plasma cells [15–17]. As the only enzyme that catalyzes the XBP-1 splicing, it is not surprising that IRE1 is involved in regulating B cells differentiation [18–20]. The IRE1α RNase also selectively cleaves ER-bound mRNAs to alleviate ER protein load, a process known as the regulated IRE1-dependent decay (RIDD) [21]. Indeed, phosphorylation of IRE1α at S729 regulates RIDD in B cells and contributes to antibody production [18]. However, the exact molecular mechanisms underlying how IRE1α controls B-cell differentiation into antibody-secreting plasma cells are not fully defined.
Unexpectedly, we observed that addition of the monounsaturated fatty acid to the culture media fully rescues plasma cell differentiation from IRE1α-null B cells, indicating that the frailer of IRE1α-null B-cell differentiation is due to a defect in monounsaturated fatty acid synthesis. Importantly, analysis of B cells from lupus patients detected a dramatic increase in IRE1α-XBP-1 activation, which positively corelates both with the upregulation of stearoyl-CoA desaturase 1 and 2 (SCD1 and SCD2) expression and to the lipid accumulation in B cells. Similar to that of either Ire1α or Xbp-1, targeted deletion of Scd1 gene largely diminished B-cell differentiation into plasma cells. Ablation of IRE1α-XBP-1 pathway or treatment with IRE1α-specific inhibitor partially protected mice from the development of lupus-like autoimmunity. These studies define a previously unappreciated molecular mechanism underlying how IRE1α-XBP-1 pathway controls B cell immunity and provides a rationale for manipulation of the IRE1α-XBP-1 pathway in combating autoimmune diseases including lupus.
METHODS
Human sample collection and B cell isolation
This study was approved by the Ethics Committee at the Third Xiangya Hospital of Central South University (2019-S190). All the patients fulfilled the America College of Rheumatology 1982 criteria for SLE. Patients were clinically evaluated based on the Systemic Lupus Erythematosus Disease Activity Index 2000 (SLEDAI-2k) [22], and the patients with SLEDAI-2k cores ≥6, who were not under gone with immune suppressive treatment in particular with Rituximab, were included in the study. Fourteen patients with active SLE and eleven healthy subjects were enrolled in this study. All subjects involved gave informed written consent prior to study enrollment according to the Declaration of Helsinki. Whole venous blood samples were collected from SLE patients and healthy controls, the peripheral blood mononuclear cell (PBMC) were isolated by Ficoll-Hypaque density gradient centrifugation of EDTA-anticoagulated blood samples [23]. B lymphocytes were isolated from PBMC using CD19 microbeads (Miltenyi Biotec, Cat#: 130–050-301) according to the manufacturer’s instructions.
Mice
The Ire1αfl/fl [24] and CD19-Cre C57BL/6 mice[25] mice were backcrossed to MRL-MpJ-Faslpr /J strain (Stock Number: 000485. Jackson Laboratory) for at least 10 generations, respectively. All animals used in this study were maintained under specific pathogen-free conditions, and all experiments were approved by the Institutional Animal Care and Use Committees (IACUC) at Northwestern University. Detailed information is described in supplemental file.
Serum autoantibody analysis by ELISA and flow cytometry.
Serum titers of anti-nuclear antibodies and anti-dsDNA antibodies from Ire1αfl/fl and CD19-Cre/Ire1αfl/fl MRL.Faslpr mice were measured with the commercial ELISA kits (anti-ANA, Cat#: M-5210; anti-dsDNA, Cat#: M-5110; Alpha Diagnostic, San Antonio, TX). Absorbance at 450 nm was detected using a FilterMax F5 microplate reader (Molecular Devices). For the flow cytometry analysis, sera from Ire1αfl/fl and CD19-Cre Ire1αfl/fl MRL.Faslpr mice were diluted in 1% BSA, and incubated with prefixed and permeabilized mouse lymphoma EL4 cells on ice for 30 min, followed by staining with the appropriate fluorophore-conjugated anti-mouse Ig Abs, washed and analyzed on a FACSCanto II instrument (BD Biosciences).
Statistical Analysis
All data are indicated as mean±SD. Statistics were calculated by two-tailed student’s t test. The Gehan-Berslow-wilcoxon test was used for survival curves, and the Spearman rank test were applied for and correlations. * p<0.05, ** p<0.01, *** p<0.001, and **** p<0.0001.
RESULTS
Elevated lipid accumulation in B cells from SLE patients.
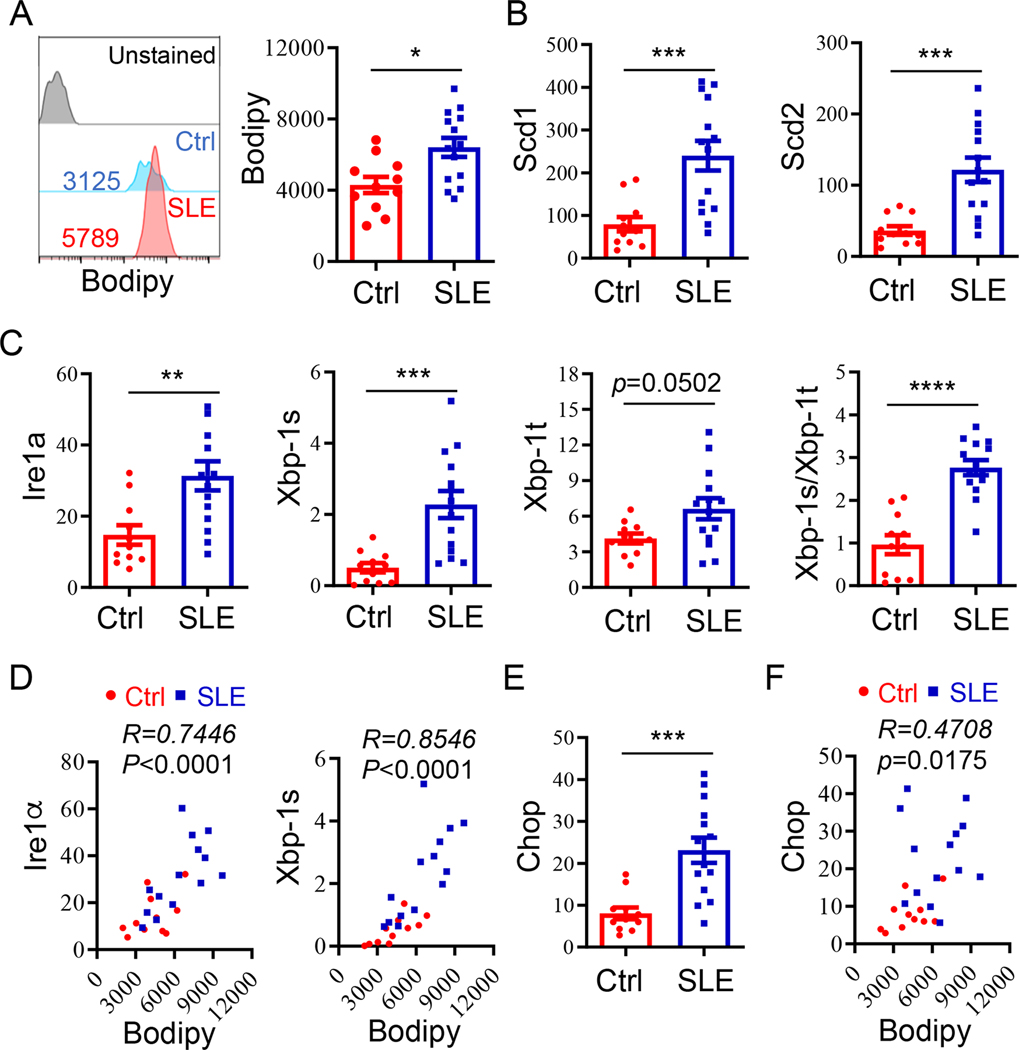
It has been well documented that dramatic increases in free fatty acids, in particular the monounsaturated fatty acids, are positively correlated with SLE activity [4–7, 26, 27]. We then speculated whether the lipid deposition is consequently accumulated in B cells from lupus patients. Bodipy analysis revealed a significant increase in the lipid volumes in B cells from lupus patients vs healthy controls (Fig. 1A). In contrast, while with a modest increase in lipid volumes in T lymphocytes from SLE patients, this increase was not statistically significant (Fig. s1). Further real-time RT-PCR analysis detected a drastic increase in the expression of Scd1 and Scd2, two critical enzymes for catalyzing the rate-limiting step in the formation of monounsaturated fatty acids [28], suggesting that the increased expression of Scd1 and Scd2 is involved in B-cell autoimmunity (Fig. 1B). We then analyzed whether the activation of IRE1α-XBP-1 pathway, which plays a critical role in lipid metabolic regulation [15, 28], is altered in lupus B cells. Indeed, a dramatic increase in the expression of Ire1α was detected in B cells from patients with active lupus compared with these from healthy controls. As a consequence, the levels of the spliced Xbp-1 (Xbp-1s) were significantly increased. In addition, we detected a modest increase in the total Xbp-1 (Xbp-1t) mRNA (Fig. 1C). More importantly, the ratios of Xbp-1s to total Xbp-1 transcripts, an indicator of IRE1α activation, were increased for almost 3-fold in B cells from lupus patients compared with that from healthy controlls (Fig. 1C). These results suggest that the elevated activation of the IRE1α-XBP-1 pathway in B cells is possibly involved in lupus pathogenesis. Therefore, B cells in SLE microenvironment exhibit a robust IRE1α-XBP-1 activation, which lead to more lipid accumulation in lupus B cells vs these from healthy controls through inducing Scd1 and Scd2 gene transcription. To support this notion, a strong positive correlation between IRE1α-XBP-1 activation with lipid accumulation levels was observed in B cells from lupus patients (Fig. 1D). In contrast, despite the expression of the ER stress responsive gene Chop was also increased, its correlation with B cell lipid levels is rather modest (Fig. 1E & F).
Fig. 1. Increased to lipid accumulation in B cell from SLE patients.
CD19+ B cells were isolated from PBMCs from patients with active SLE (SLEDAI ≥6; n =14) and healthy controls (n=11), respectively. (A) Intracellular lipid content in CD19+ B cell populations. (Left) Representative flow cytometric profile of lipid staining for B cells from patients with active SLE and healthy controls. (Right) Lipid quantification expressed as mean fluorescence intensity (MFI) of Bodipy 493/503 staining. (B) RT-qPCR analysis of expression levels of Scd1 and Scd2 in CD19+ B cells from PBMCs. (C) Expression of the indicated transcripts in B cell was determined by RT-qPCR. (D) Data plots show the correlation of intracellular lipid content with relative mRNA level of Ire1α and Xbp-1s. (E) RT-qPCR analysis of expression level of Chop in B cells from PBMCs. (F) Correlation between intracellular lipid content and Chop mRNA expression. Data are shown as mean±SD. Student’s t test (two-tailed) was used for statistical analysis (A-C & E), and the correlation coefficient R and P values were calculated by the Spearman rank correlation test (D & F). * p<0.05, ** p<0.01, *** p<0.001, and **** p<0.0001.
B cell-intrinsic IRE1α function is required for SLE progression and rapid mortality in mice.
To determine the possible pathogenic role of increased B-cell IRE1α activation in lupus disease, we backcrossed the Ire1αfl/fl/CD19-Cre mice, initially at the C57/B6 genetic background, with MRL.Faslpr mice, who develop spontaneous lupus that resembles human disease [29, 30], for at least 10 generations and generated the Ire1αfl/fl/CD19-Cre/MRL.Faslpr (Ire1ΔMRL.Faslpr) mice. The Cre recombinase expression under CD19 promoter, while was insufficient to delete the floxed Ire1α gene in naïve B cells, resulted in a sufficient Ire1α deletion upon 48-hour LPS stimulation (Fig. s2A & B). Similarly, while the Xbp-1s levels were indistinguishable in naïve B cells from WT and Ire1ΔMRL.Faslpr mice, its expression in activated B cells was largely diminished by IRE1α deletion (Fig. s2B). Further analysis revealed that neither the B-cell development nor its maturation were altered in Ire1ΔMRL.Faslpr mice (Fig. s2C), which is likely due to the insufficient Ire1α gene deletion during B cell development and maturation.
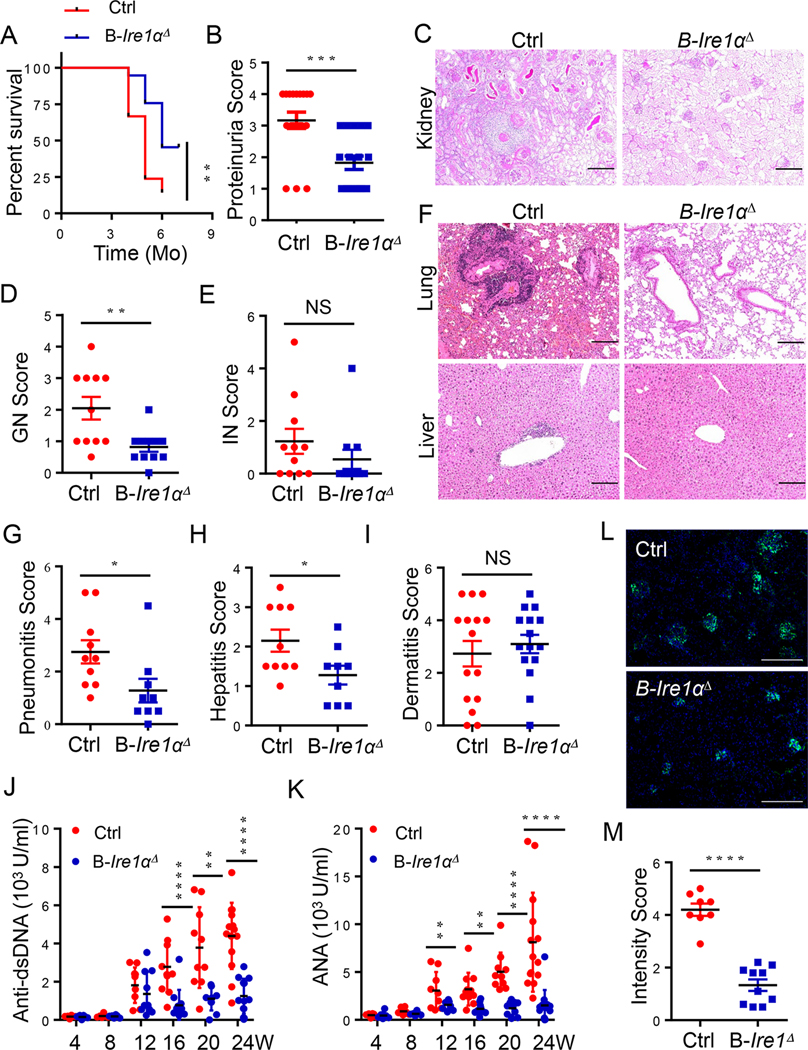
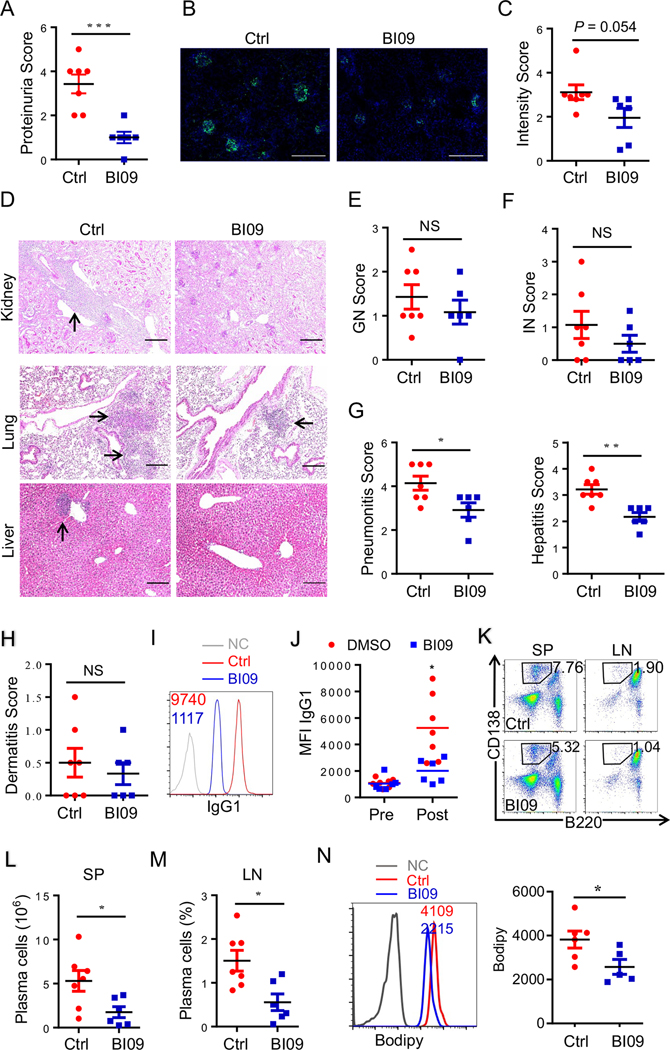
Importantly, the survival of lupus mice was dramatically improved by the B-cell IRE1α suppression when compared with their littermate controls (Fig. 2A). More than 50% of MRL.Faslpr mice with B-cell-specific Ire1α deletion survived for six months or more. In contrast, only12.5% of IRE1α-sufficient MRL.Faslpr mice were able to survive to the same age (Fig. 2A). The impaired kidney functions have been known as one of the important pathogenic factors responsible for lupus lethality [31]. Indeed, the proteinuria, a measure of kidney function, was markedly reduced in Ire1ΔMRL.Faslpr mice compared with their littermate controls (Fig. 2B). Further histopathological analysis detected that the decrease in proteinuria was associated with significantly less glomerulonephritis (GN) by IRE1α deletion, whereas interstitial nephritis was unaltered (Fig. 2C–E). A histological examination also demonstrated that a substantial reduction in lung and liver inflammation in mice with B-cell IRE1α deletion (Fig. 2F–H). However, dermatitis severity in Ire1ΔMRL.Faslpr and control mice was indistinguishable, demonstrating B-cell Ire1α ablation didn’t protect MRL.Faslpr mice from dermatitis (Fig. 2I). These data demonstrated that B-cell IRE1α expression is responsible for multi-organ pathology and rapid mortality in MRL.Faslpr mice.
Fig. 2. B cell-intrinsic IRE1α expression is responsible for SLE progression and rapid mortality in murine lupus.
(A) Survival of Cd19-Ire1αΔ mice and their littermate control mice (n=10). (B) Proteinuria scores of 16-week-old mice. (C-E) Representative low magnification images of PAS-stained kidneys sections (C), and scores of glomerular nephritis (D) and interstitial nephritis (E) were shown (n=11;). (F) Representative images of H&E–stained lung (top) and liver sections (bottom) illustrating perivascular infiltrates in target organs from 20-week-old mice. (G-I) Pneumonitis scores (G), Hepatitis scores (H), and Dermatitis scores (I) of control and Cd19-Ire1αΔ mice. (J &K) ELISA analysis of anti-dsDNA (J) and ANA (K) in serum from Cd19- Ire1αΔ mice and their litter controls. (L & M) Representative immunofluorescence histology staining of kidney glomeruli (L) and data analysis (M) of anti-IgG (Green) were shown (Red, n=8; blue, n=10). Significance determined by Gehan-Berslow-wilcoxon test (A) and an unpaired Student’s t-test (B, D, E, G-K, & M). * p<0.05, ** p<0.01, *** p<0.001, and **** p<0.0001. Scale bars, 200 μm (C, F & L).
The presence of high titers of auto-Abs is necessary for the development of kidney failure during lupus-like autoimmunity [32]. Therefore, we evaluated whether Ire1α-deficiency in B cells affected the production of auto-Abs. As reported, the autoreactive antibody levels were gradually increased with age in the sera from MRL.Faslpr mice [33]. Importantly, B-cell specific IRE1α suppression largely diminished the production of both anti-dsDNA and anti-nuclear antibodies (Fig. 2J & K). Further analysis validated a similar reduction in autoreactive IgG, IgG1, IgG2a, and IgA levels in sera from Ire1ΔMRL.Faslpr mice in particularly in later stages of the disease (Fig. s3). As a consequence, the autoreactive immune-complex deposition in kidney of MRL.Faslpr mice was largely abolished by B-cell IRE1α suppression (Fig. 2L & M). Collectively, our data indicate that B-cell IRE1α activation drives lupus pathogenicity and IRE1α suppression is a potential therapy to treat lupus disease.
Since plasma cells are the primary sources of autoantibodies, we analyzed the differentiation of B cells in IRE1α-sufficient and -deficient MRL.Faslpr mice. As expected, both the percentages and absolute numbers of B220intCD138+ plasma cells in spleen and in lymph node, were markedly decreased in an age-dependent manner in MRL.Faslpr mice by B-cell Ire1α ablation (Fig. s4A & B), indicating that B-cell specific IRE1α suppression protect MRL.Faslpr mice from lupus pathogenesis via suppressing plasma cell differentiation. To support this, we further demonstrated that Ire1α gene deletion largely abolished B-cell differentiation to plasma cells in vitro (Fig. s4C & D). In addition, lack of Ire1α expression in B cells displayed a time-dependent reduction in both viability and proliferation (Fig. 3C–F). Collectively, these data demonstrate a pivotal role for IRE1α expression in promoting B-cell immunity including survival, growth and plasma cell differentiation.
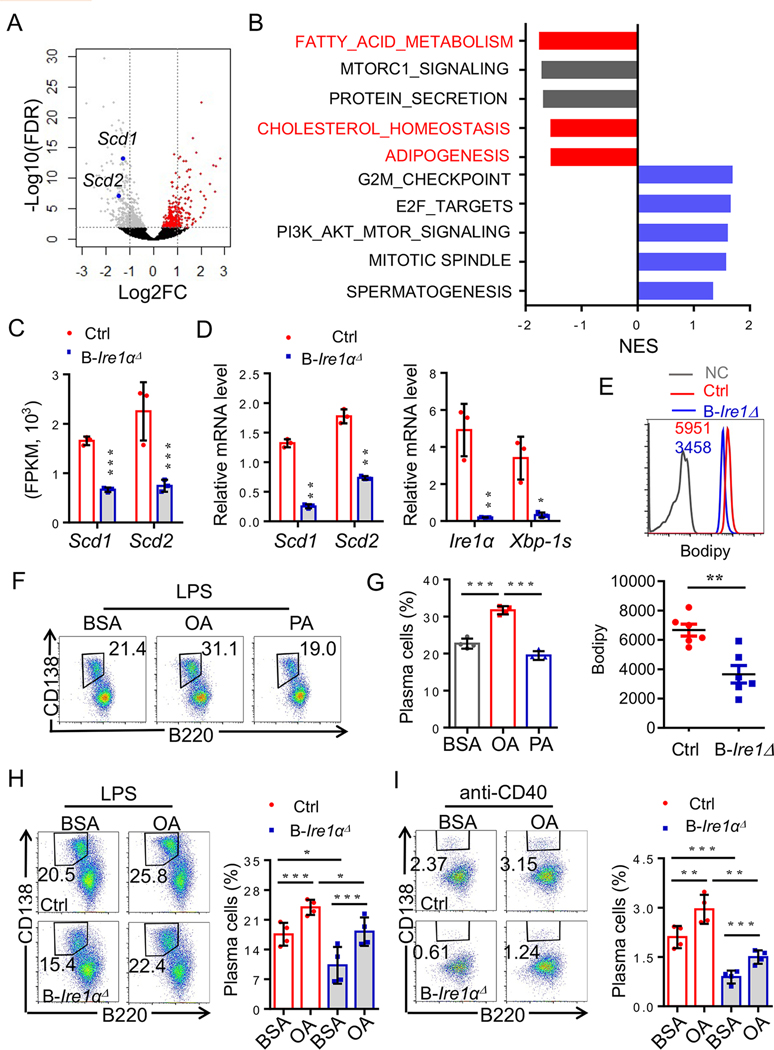
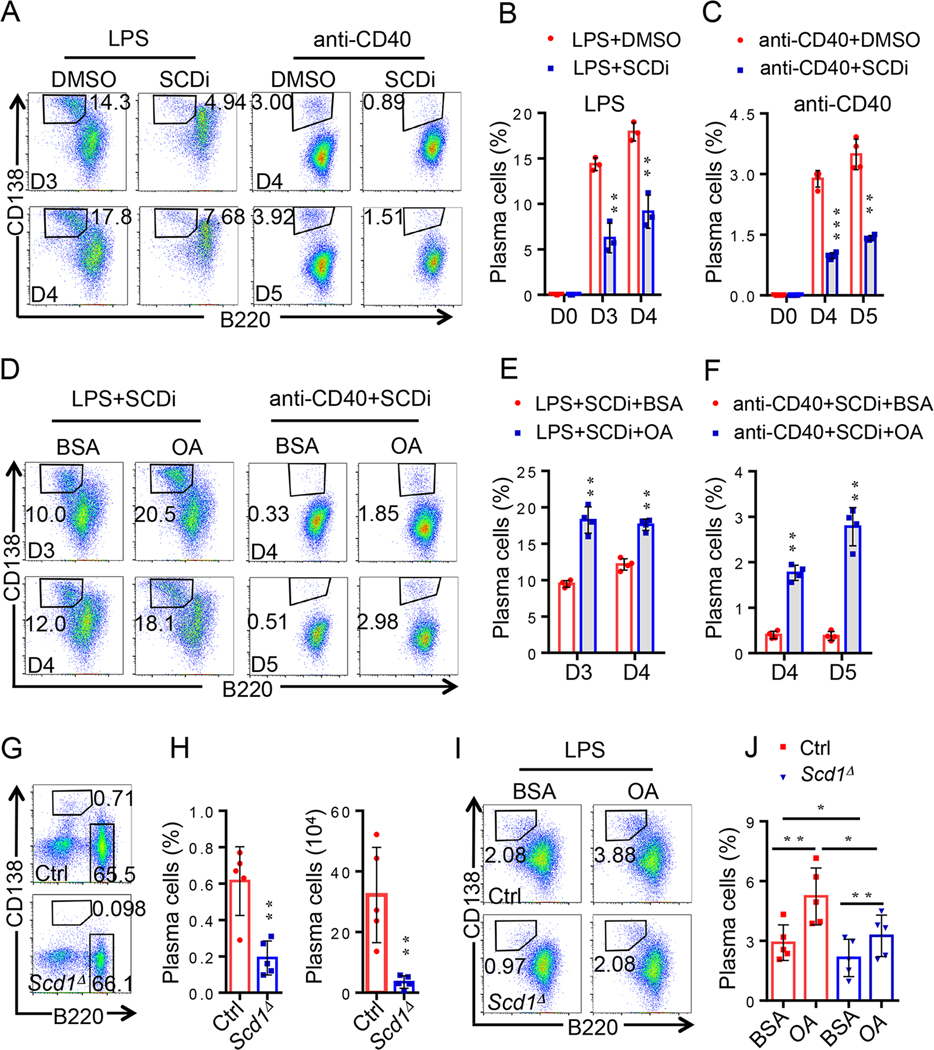
Fig. 3. Effects of the monounsaturated fatty acids on plasma cells differentiation from IRE1α–null B cells.
(A-C) Sorted B220intCD138hi plasma cells were analyzed by RNA sequencing (Red, n=3; blue, n=3). (A) Volcano plot comparing the P value of sorted B220intCD138hi plasma cells from Cd19-Ire1αΔ mice and their litter controls. Genes upregulated (red) and downregulated (blue). Scd1 and Scd2 were labeled in blue. (B) GSEA was performed, and significant downregulated and upregulated pathways were shown. (C) mRNA expression levels of Scd1 and Scd2 in plasma cells from the indicated mice. (D) RT-qPCR analysis of expression levels of Scd1, Scd2, Ire1α and Xbp-1s in Ire1α-deficient- and sufficient- plasma cells (n=3). (E) Lipid bodipy analysis of Ire1α-null B cells (n=6). (F-G) Representative flow cytometric profiles (F) and data plots (G) show frequencies of B220intCD138hi plasma cells under treatment with LPS plus BSA, or OA, or PA (n=4). (H-I) Representative FACS profiles and data analysis show OA largely rescued plasma cell differentiation from IRE1α – null B cell upon in vitro stimulation with either LPS (H) or anti-CD40 (I) (n=4). Statistical differences were tested using an unpaired unpaired Student’s t -test (two-tailed). * p<0.05, ** p<0.01, *** p<0.001.
Supplementation of the monounsaturated fatty acids largely rescues plasma cell differentiation from IRE1α-null B cells.
Since the increased activation of IRE1α-XBP-1 pathway positively correlates to both the expression in monounsaturated fatty acids synthetic genes, Scd1 and Scd2, as well as lipid accumulation in B cells from lupus patients (Fig. 1), we then hypothesized that IRE1α may promote plasma cell differentiation through facilitating B-cell lipid homeostasis. Indeed, we detected a significant reduction in the expression of lipid metabolic enzymes, including Scd1 and Scd2 through an unbiased genome-wide transcriptome analysis (Fig. 3A & C). Pathways analysis also confirmed the reduction in the expression of genes in multiple lipid metabolic pathways including fatty acids metabolism by IRE1α deletion in plasma cells (Fig. 3B). In contrast, the top upregulated genes in plasma cells by Ire1α deletion are involved in cell cycle progression and survival (Fig. 3B), further validating our discovery that IRE1α suppression attenuated the growth and promoted the death of plasma cells (Fig. s4D–F). Real-time RT-PCR analysis confirmed the expression of both Scd1 and Scd2 is largely inhibited in plasma cells by Ire1α suppression (Fig. 3D). A similar reduction in Scd1 and Scd2 gene expression was also detected in the activated IRE1α-null B cells (Fig. s5). As a consequence, the lipid volumes in B cells were dramatically decreased by Ire1α gene deletion (Fig. 3E). These results suggest that the reduction in the expression of lipid metabolic genes is possibly involved in the defect in plasma cell differentiation by IRE1α-null B cells.
If IRE1α-XBP-1 activation promotes B-cell differentiation through SCD1 and SCD2, we posed whether the SCD1 and SCD2 products, monounsaturated fatty acids, could facilitate plasma cell differentiation. Surprisingly, co-cultivation of B cells with oleic acid (OA) significantly enhanced B-cell differentiation into plasma cells in vitro (Fig. 3F & G). In contrast, addition of the saturated fatty acids, palmitic acid (PA) showed little effects (Fig. 3F & G). We then speculated whether OA administration rescues IRE1α-null B-cell differentiation into plasma cells. As expected, Ire1α gene suppression dramatically impaired B-cell differentiation into plasma cells upon in vitro stimulation (Fig. 3H & I). Notably, supplementation of OA largely rescued plasma cell differentiation from IRE1α-null B cells, clearly indicating that the impaired SCD1/2-medaited OA synthesis is responsible for the defect of IRE1α-null B-cell differentiation into plasma cells (Fig. 3H & I). These results reveal an IRE1α-XBP-1-SCD1/2 pathway essential for optimal plasma cell differentiation.
Activation of IRE1α-XBP-1 pathway promotes SCD1/2-mediated lipid accumulation during B-cell activation and plasma cell differentiation.
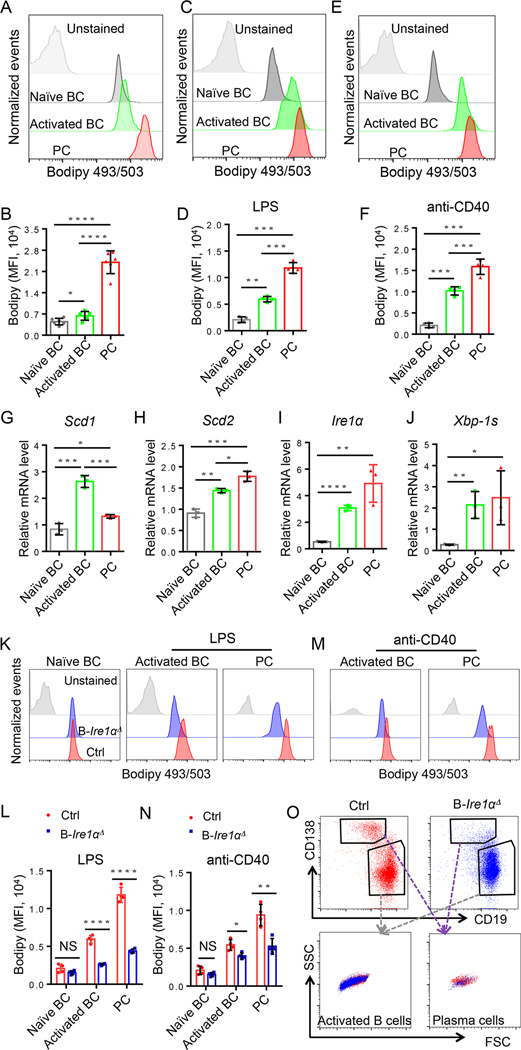
Our discovery that OA supplementation largely rescues IRE1α-null B-cell differentiation into plasma cells suggests that IRE1α controls plasma cell differentiation through promoting SCD1/2-mediated lipid synthesis. Indeed, analysis of the lipid levels in primary naïve and activated B cells and plasma cells revealed a gradual increase in lipid bodipy staining (Fig. 4A & B). This increase in lipid levels during B-cell activation and differentiation was further validated upon in vitro stimulation with either LPS (Fig. 4C & D) or anti-CD40 (Fig. 4E & F). Consistent with the increase in lipid levels, both Scd1 and Scd2 expressions were upregulated in activated B cells and plasma cells (Fig. 4G & H). Similarly, both the expression and activation of IRE1α were dramatically elevated during B-cell activation and differentiation (Fig. 4I & J). Together with the fact that Ire1α genetic deletion resulted in the reduced Scd1 and Scd2 expression in B cells, these results indicated that IRE1α-induced SCD1/2 expression is critical for B-cell activation and plasma cell differentiation. To support this, we further demonstrated that IRE1α suppression dramatically reduced the lipid levels in naïve, activated B cells and plasma cells (Fig. 4K–N). Importantly, Ire1α genetic suppression dramatically reduced the lipid Bodipy signal in activated B cells and plasma cells without affecting cell size as analyzed by their side and forward scatters (Fig. 4O). Collectively, our results suggesting that IRE1α regulates B-cell differentiation through SCD1/2-medited lipid synthesis.
Fig. 4. IRE1α activation promotes SCD1/2-mediated lipid accumulation during B cell activation and plasma cell differentiation.
(A-B) In vivo intracellular lipid content in B cell populations of splenocytes from Ire1αfl/fl MRL.Faslpr mice (n=6). (A) Representative FACS analysis of lipid staining for B cells from the indicated populations. (B) Lipid quantification expressed of Bodipy 493/503 staining. (C-F) In vitro intracellular lipid content in B cell populations of splenocytes from Ire1αfl/fl MRL.Faslpr mice. Primary naïve B cells were isolated from splenocytes, and stimulated with LPS (1mg/ml) or anti-CD40 (4μg/ml) for 2 and 3 days, respectively (n=4). (G - J) RT-qPCR detection of expression levels of Scd1 (G), Scd2 (H), Ire1α (I), and Xbp-1s (J) in naïve B cells, activated B cells, and plasma cells (n=3). (K - N) Primary B cells were isolated from 4 to 5-week old of Cd19- Ire1αΔ mice and their littermate control mice and stimulated with LPS (K & L) or anti-CD40 (M & N) in vitro for 4 days (n=4). Representative FACS analysis (K & M) and data plots (L & N) of lipid staining for B cells from indicated sources. (O) Representative FACS analysis of cell size of activated B cell and plasma cells from Cd19- Ire1αΔ mice and their littermate controls. Significance determined by an unpaired Student’s t –test (two-tailed). * p<0.05, ** p<0.01, *** p<0.001, and **** p<0.0001.
Genetic and pharmacological SCD1 suppression impairs B-cell differentiation into plasma cells.
We first validated the critical functions of SCD1 in B-cell differentiation with a commercially available specific SCD inhibitor (SCDi) and tested its effects on plasma cell differentiation. As expected, SCDi treatment achieved a similar efficacy as that of Ire1α deletion by largely diminishing plasma cells differentiation in vitro (Fig. 5A–C). Similar to that of IRE1α-null B cells, addition of oleic acid largely rescued plasma cell differentiation from SCDi treatment (Fig. 5D–F). Therefore, SCD1-mediated synthesis of oleic acid is essential for the optimal B-cell differentiation into plasma cells.
Fig. 5. Genetic and pharmacological SCD1 suppression impairs B-cell differentiation into plasma cells.
(A-C) Representative flow cytometric profiles (A) and data plots (B & C) show frequencies of CD3-B220intCD138hi PCs under treatment with SCDi (n=3 or 4). Primary CD43- B cells were isolated from splenocytes, treated with SCDi (0.5μM) plus LPS (1mg/ml) for day 3 and day 4 (A & B) or with SCDi (0.5μm) plus anti-CD40 (4μg/ml) for day 4 and day 5 (A & C), respectively. (D-F) Representative flow cytometric profiles (D) and data plots (E & F) show OA partly rescued plasma cell differentiation from SCDi treatment upon in vitro stimulation with either LPS (D & E) or anti-CD40 (D & F) (n=4). (G & H) Representative flow cytometric profiles (G) and data plots (H) of CD3-B220intCD138hi PCs percentage and absolute number from Scd1-deficient B cells in vivo (n=5). (I & J) Representative flow cytometric profiles (I) and data plot (J) show OA partly rescued plasma cell differentiation from Scd1-null B cell upon in vitro stimulation with LPS (n=5). Significance determined by two-tailed Student’s t –test. * p<0.05, ** p<0.01, *** p<0.001, and **** p<0.0001.
To provide a proof-of-concept evidence to support our conclusion that SCD1 is required for plasma cell differentiation, we determined the impact of targeted Scd1 gene deletion on B-cell differentiation. Genetic deletion of Scd1 did not alter the frequency of B220+ B cells in the spleen of Scd1−/− mice. In contrast, both the absolute number and percentage of B220intCD138+ plasma cells in spleen were remarkably reduced in Scd1−/− mice when compared with their littermate controls (Fig. 5G & H), indicating that SCD1 functions are essential for maintaining the plasma cell pool in the steady state in mice. In addition, Scd1 gene deletion dramatically inhibited in vitro B-cell differentiation into plasma cells. More importantly, addition of OA could largely rescue the plasma cell differentiation from Scd1−/− B cells (Fig. 5I & J). These results clearly defined a critical role of SCD1-mediated monounsaturated fatty acids synthesis in B-cell differentiation into plasma cells.
IRE1α specific inhibitor as a potential therapeutic drug to treat lupus.
We then evaluated the preclinical efficacy of IRE1α-specific inhibitor BI09, which prevents its ability to splice Xbp-1 mRNA into the activated Xbp-1 transcription factor [34], in protecting MRL.Faslpr mice from lupus pathogenesis. We initiated the BI09 treatment of MRL.Faslpr mice from the age of 10 weeks, a time point after the disease onset in terms autoimmune antibody production. Unsurprisingly, a transient BI09 treatment of MRL.Faslpr mice largely protected them from lupus nephropathy because the levels of proteinuria were dramatically reduced comparing to those in untreated controls (Fig. 6A). Further immuno-histological staining detected a trend of reduction in the autoreactive antibody deposition in the kidney of the BI09-treated MRL.Faslpr mice with a p value of 0.054 (Fig. 6B & C), which is possibly due to the BI09 treatment, unlike the targeted genetic deletion as shown in Fig. 2, could only archive a partial suppression of IRE1α activity. Consistently, neither the reduction in kidney glomerulonephritis nor in interstitial nephritis reached to a statistically significant level (Fig. 6D–F). However, similar to B-cell specific IRE1α suppression, BI09 treatment significantly reduced the lymphocyte infiltration in the lung and liver without affect skin inflammation (Fig. 6D–H). These results indicate that IRE1α specific inhibitor BI09 protects MRL.Faslpr mice from autoreactive antibody-mediated lupus pathogenesis. To support this, the autoreactive antibody levels in the sera of BI09 treated mice were dramatically reduced (Fig. 6I & J). Flow cytometry analysis confirmed that BI09 treatment inhibited plasma cell differentiation in MRL.Faslpr mice (Fig. 6K–M). Consistent with our observation in Ire1α -null B cells, pharmacological IRE1α inhibition resulted in a dramatic reduction in B-cell lipid volumes (Fig. 6N & Fig. s6A). However, the autoimmune Ab levels were rebounded back 4-week after the termination of BI09 treatment (Fig. s6A), implying that the pharmacological IRE1α suppression, unlike that by genetic deletion, is transient in lupus treatment.
Fig. 6. IRE1α specific inhibitor BI09 protects mice from lupus pathogenesis.
(A) Data analysis of Proteinuria Score from lupus mice treated with BI09 and Control (Red, n=7; blue, n=6) for 3 weeks. (B & C) Representative images (B) and quantitative measures (C) of kidney glomeruli immunofluorescent staining of IgG1 from lupus mice treated with BI09 and Control DMSO. (D-G) Representative images of PAS- stained kidneys sections (D, top panels), its GN Score (E) and IN Score (F), as well as H&E–stained lung (middle panels) and liver (bottom panels) sections (D) and data analysis (G) of Pneumonitis Scores and Hepatitis Scores from lupus mice treated with BI09 and Control. (H) Data analysis of Dermatitis Scores from lupus mice treated with BI09 and Control. (I & J) Representative FACS image (I) and data plot (J) of IgG1 staining from serum samples from lupus mice treated with BI09 and control. (K-M) Representative flow cytometric profiles (K) and data plots (L & M) show and absolute numbers of CD3-B220intCD138hi PCs in splenocytes (K & L) and frequencies in lymph nodes (K & M) from lupus mice treated with BI09 and Control. (N) Lipid bodipy analyses of B cells from BI09 treated and control mice are shown. Significance determined by an unpaired Student’s t–test (two-tailed). * p<0.05, ** p<0.01, *** p<0.001, and **** p<0.0001. Scale bar, 200 μm.
We then validated the suppressive effects of BI09 on B-cell differentiation in vitro and observed that BI09 dose-dependently inhibited CD138+ plasma cell differentiation (Fig. s6B–D). This reduction in plasma cell differentiation by BI09 is likely due to a direct inhibition of IRE1α activation, because Xbp-1 mRNA splicing was largely inhibited by BI09 treatment (Fig. s6E). As a consequence, Scd1 and Scd2 mRNA in B cells was significantly inhibited (Fig. s6F & G). Collectively, our data indicate that both genetic and pharmacological IRE1α suppression block B-cell differentiation into plasma cells and consequently protected mice from B cell autoimmune pathogenesis.
DISCUSSION
The current studies have defined a previously unappreciated molecular mechanism underlying how the IRE1α-XBP-1 pathway controls B cell differentiation and provide a rationale for IRE1α suppression in lupus therapy. First, IRE1α suppression blocks B-cell differentiation into plasma cells which can be rescued by supplementation of the monounsaturated fatty acids; Second, IRE1α activation positively regulates the transcription of monounsaturated fatty acid synthetic genes Scd1 and Scd2; Third, genetic and pharmacological suppression of SCD1 inhibits plasma cell differentiation. In addition, IRE1α suppression largely protects mice from lupus pathogenesis. Together with our discoveries that IRE1α-XBP-1 activation is elevated and positively correlated with both increased Scd1 and Scd2 expression and the lipid accumulation in B cells from lupus patients, our studies indicate that IRE1α is a potential therapeutic target to treat lupus and other B cell-mediated autoimmune diseases.
It has been well established that the transcription factor Xbp-1 is essential for B-cell differentiation into antibody producing plasma cells [20, 35]. As the only enzyme required for Xbp-1 mRNA splicing in B cells, it is not surprising that B cell-specific IRE1α suppressing largely diminished plasma cell differentiation and antibody secretion. XBP-1 promotes antibody production partially through induction of the production of IL-6, a cytokine that is critical for B-cell growth and plasma cell survival [36–38]. In addition, it is speculated that, in the absence of Xbp-1, cellular unfolded protein response is impaired and the inefficient processing and exportation of immunoglobulin results in an accumulation of unfolded protein and consequently cell death [39]. Surprisingly, we discovered that supplementation of monounsaturated fatty acids largely rescued plasma cell differentiation from B cells lacking IRE1α, indicating that lack of the monounsaturated fatty acid is, at least partially, responsible for the frailer of IRE1α-null B-cell differentiation into plasma cells. Indeed, as recently reported [28], we further confirmed that IRE1α is required for the optimal expression of two monounsaturated fatty acid synthetic enzymes, SCD1 and SCD2. As a proof of concept, genetic Scd1 suppression impaired B-cell differentiation into plasma cells. Therefore, the IRE1α-XBP-1 pathway appears to regulate B-cell differentiation to plasma cells through multiple mechanisms. Moreover, we have shown that Hrd1, an ER resident E3 ligase critical for degradation of misfolded proteins through the ubiquitin pathway and degradation, also catalyzes IRE1α degradation and functions as a negatively regulates IRE1α function to suppresses the ER stress induced cell apoptosis, which is involved in arthritis pathogenesis and intestinal homeostasis [40]. Interestingly, as an ER stress responsive gene, HRD1 has been shown as a direct target of Xbp-1 transcription factor, suggesting a feedback loop between IRE1α-XBP-1 pathway and HRD1-mediated ubiquitination. In addition to IRE1α, we have discovered HRD1 protects B cells from the activation-induced apoptosis by targeting the death receptor Fas [41]. It will be interesting to further elucidate whether, if yes how HRD1-mediated IRE1α degradation affect B-cell immunity.
Results from this study has defined a pathogenic role IRE1α-XBP-1 pathway in autoantibody producing plasma cell differentiation through SCD1, possibly as well as other SCD family members such as SCD2, mediated monounsaturated fatty acids synthesis and provided a rationale for manipulation of this pathway in lupus treatment. However, pharmacological IRE1α suppression could only achieve a transient efficacy because the autoreactive Ab levels were restored three weeks after BI09 treatment termination. In addition, we observed that, while BI09 treatment inhibited the production of autoimmune Abs and partially protected mice from proteinuria, neither the reduction in kidney glomerulonephritis nor in interstitial nephritis reached to a statistically significant level in the treated mice. Unlike B cell-specific targeted gene deletion, pharmacological IRE1α suppression with BI09 presumably inhibits all types of cells that express IRE1α. Indeed, suppression of T-cell IRE1α resulted in elevated Th2 immune response [42]. Obiedat et al reported that deletion of IRE1α or XBP1 was sufficient to promote the expression of NKG2D ligand, leading to the elevated killing of IRE1/XBP1 knockout target cells by NK cells [43]. Therefore, pharmacological IRE1α suppression is more complicated than B cell specific IRE1α gene deletion.
One of the most successful drugs developed by depleting B cells is the chimeric anti-human CD20 monoclonal Ab, Rituximab, which has been successfully used to treat lupus and other Ab-mediated autoimmune diseases such as RA [44, 45]. However, a recent review shows that, out of 71 patients, six deaths occurred in association with the Rituximab treatment. The paraneoplastic pemphigus, a disease characteristically resistant to conventional medication and with a high mortality rate, appears to be the number one cause of the death, as 4 out of the six deaths are with this disease. Infectious disease, both viral and bacterial infections, is often associated with Rituximab usage[45]. After binding of Rituximab to CD20+ cells, cells undergo apoptosis via direct effect, complement and antibody dependent cytotoxicity, and inhibition of cell proliferation [44]. Recovery of B cells begins 6–9 months after Rituximab treatment, with levels returning to normal one year later [46]. In contrast, our observations here suggest that IRE1α could be a better target for Ab-mediated disease therapy: (i) genetic suppression of IRE1α abolishes antigen-specific antibody production without reducing B cell numbers in mice. (ii) IRE1α suppression by its specific inhibitor BI09 suppresses plasma cell differentiation and protected mice from lupus pathogenesis in mice. These are important facts in drug development, because we could expect that, in the case of infection occurs during lupus treatment, termination of IRE1α inhibitor treatment allows an immediate B-cell function recovery to combat the pathogens. Moreover, a combination of Rituximab with an IRE1α inhibitor could produce a synergistic effect on lupus treatment. More importantly, while Rituximab suppresses autoantibody production by B-cell depletion, presumably it has no effects on suppressing inflammatory cytokine production by myeloid cells. Results from our previous publications [24] and data in the current studies demonstrate that IRE1α suppression has a “kill two birds with one stone” efficacy to inhibit autoimmune response of both myeloid and B cells.
Supplementary Material
Acknowledgements
We thank Fang lab members for critical reading of the manuscript and constructive suggestions during our research. This work was supported by National Institutes of Health (NIH) R01 grants AI079056, AI108634, AR006634 and CA232347 (to D.F.), NIH R01 DK 062388 and DK 118093 (to J.M.N.), NIH RO1 DK60635 (to Y.K.) as well as NIH R01 DK090313 (to K.Z.).
Funding supports:
This work was supported by National Institutes of Health R01 grants AI079056, AI108634, AR006634 and CA232347 (to D.F.), NIH R01 DK062388 and DK118093 (to J.M.N.), RO1DK60635 (to YK) and DK090313 (to K.Z.).
Footnotes
Conflict of interests: All authors have no conflict of interest for the study.
References
- 1.Weide R, et al. , Successful long-term treatment of systemic lupus erythematosus with rituximab maintenance therapy. Lupus, 2003. 12(10): p. 779–82. [DOI] [PubMed] [Google Scholar]
- 2.Leandro MJ, et al. , An open study of B lymphocyte depletion in systemic lupus erythematosus. Arthritis Rheum, 2002. 46(10): p. 2673–7. [DOI] [PubMed] [Google Scholar]
- 3.Perrotta S, et al. , Anti-CD20 monoclonal antibody (Rituximab) for life-threatening autoimmune haemolytic anaemia in a patient with systemic lupus erythematosus. Br J Haematol, 2002. 116(2): p. 465–7. [PubMed] [Google Scholar]
- 4.Shin TH, et al. , Analysis of the free fatty acid metabolome in the plasma of patients with systemic lupus erythematosus and fever. Metabolomics, 2017. 14(1): p. 14. [DOI] [PubMed] [Google Scholar]
- 5.Wu T, et al. , Metabolic disturbances associated with systemic lupus erythematosus. PLoS One, 2012. 7(6): p. e37210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez-Carrio J, et al. , Intestinal Dysbiosis Is Associated with Altered Short-Chain Fatty Acids and Serum-Free Fatty Acids in Systemic Lupus Erythematosus. Front Immunol, 2017. 8: p. 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ormseth MJ, et al. , Free fatty acids are associated with metabolic syndrome and insulin resistance but not inflammation in systemic lupus erythematosus. Lupus, 2013. 22(1): p. 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, et al. , Deficiency of IRE1 and PERK signal pathways in systemic lupus erythematosus. Am J Med Sci, 2014. 348(6): p. 465–73. [DOI] [PubMed] [Google Scholar]
- 9.Todd DJ, Lee AH, and Glimcher LH, The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol, 2008. 8(9): p. 663–74. [DOI] [PubMed] [Google Scholar]
- 10.Gass JN, Gifford NM, and Brewer JW, Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J Biol Chem, 2002. 277(50): p. 49047–54. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y and Brandizzi F, IRE1: ER stress sensor and cell fate executor. Trends Cell Biol, 2013. 23(11): p. 547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen X, et al. , Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell, 2001. 107(7): p. 893–903. [DOI] [PubMed] [Google Scholar]
- 13.Korennykh AV, et al. , The unfolded protein response signals through high-order assembly of Ire1. Nature, 2009. 457(7230): p. 687–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calfon M, et al. , IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature, 2002. 415(6867): p. 92–6. [DOI] [PubMed] [Google Scholar]
- 15.Cubillos-Ruiz JR, et al. , ER Stress Sensor XBP1 Controls Anti-tumor Immunity by Disrupting Dendritic Cell Homeostasis. Cell, 2015. 161(7): p. 1527–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee AH, Iwakoshi NN, and Glimcher LH, XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol, 2003. 23(21): p. 7448–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGehee AM, et al. , XBP-1-deficient plasmablasts show normal protein folding but altered glycosylation and lipid synthesis. J Immunol, 2009. 183(6): p. 3690–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang CH, et al. , Phosphorylation of IRE1 at S729 regulates RIDD in B cells and antibody production after immunization. J Cell Biol, 2018. 217(5): p. 1739–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang K, et al. , The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest, 2005. 115(2): p. 268–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reimold AM, et al. , Plasma cell differentiation requires the transcription factor XBP-1. Nature, 2001. 412(6844): p. 300–7. [DOI] [PubMed] [Google Scholar]
- 21.Hollien J and Weissman JS, Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science, 2006. 313(5783): p. 104–7. [DOI] [PubMed] [Google Scholar]
- 22.Gladman DD, Ibanez D, and Urowitz MB, Systemic lupus erythematosus disease activity index 2000. J Rheumatol, 2002. 29(2): p. 288–91. [PubMed] [Google Scholar]
- 23.Zhu YY, et al. , The largely normal response to Toll-like receptor 7 and 9 stimulation and the enhanced expression of SIGIRR by B cells in systemic lupus erythematosus. PLoS ONE [Electronic Resource], 2012. 7(8): p. e44131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu Q, et al. , Toll-like receptor-mediated IRE1alpha activation as a therapeutic target for inflammatory arthritis. EMBO J, 2013. 32(18): p. 2477–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rickert RC, Roes J, and Rajewsky K, B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res, 1997. 25(6): p. 1317–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang N and Perl A, Metabolism as a Target for Modulation in Autoimmune Diseases. Trends Immunol, 2018. 39(7): p. 562–576. [DOI] [PubMed] [Google Scholar]
- 27.Lo MS and Tsokos GC, Recent developments in systemic lupus erythematosus pathogenesis and applications for therapy. Curr Opin Rheumatol, 2018. 30(2): p. 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie H, et al. , IRE1alpha RNase-dependent lipid homeostasis promotes survival in Myc-transformed cancers. J Clin Invest, 2018. 128(4): p. 1300–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rubtsova K, et al. , B cells expressing the transcription factor T-bet drive lupus-like autoimmunity. J Clin Invest, 2017. 127(4): p. 1392–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papadimitraki ED, et al. , Expansion of toll-like receptor 9-expressing B cells in active systemic lupus erythematosus: implications for the induction and maintenance of the autoimmune process. Arthritis Rheum, 2006. 54(11): p. 3601–11. [DOI] [PubMed] [Google Scholar]
- 31.Yap DY, et al. , Survival analysis and causes of mortality in patients with lupus nephritis. Nephrol Dial Transplant, 2012. 27(8): p. 3248–54. [DOI] [PubMed] [Google Scholar]
- 32.Davidson A, What is damaging the kidney in lupus nephritis? Nat Rev Rheumatol, 2016. 12(3): p. 143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinoshita K, et al. , Costimulation by B7–1 and B7–2 is required for autoimmune disease in MRL-Faslpr mice. J Immunol, 2000. 164(11): p. 6046–56. [DOI] [PubMed] [Google Scholar]
- 34.Tang CH, et al. , Inhibition of ER stress-associated IRE-1/XBP-1 pathway reduces leukemic cell survival. J Clin Invest, 2014. 124(6): p. 2585–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Todd DJ, et al. , XBP1 governs late events in plasma cell differentiation and is not required for antigen-specific memory B cell development. J Exp Med, 2009. 206(10): p. 2151–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suematsu S, et al. , IgG1 plasmacytosis in interleukin 6 transgenic mice. Proc Natl Acad Sci U S A, 1989. 86(19): p. 7547–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jego G, Bataille R, and Pellat-Deceunynck C, Interleukin-6 is a growth factor for nonmalignant human plasmablasts. Blood, 2001. 97(6): p. 1817–22. [DOI] [PubMed] [Google Scholar]
- 38.Iwakoshi NN, et al. , Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol, 2003. 4(4): p. 321–9. [DOI] [PubMed] [Google Scholar]
- 39.Iurlaro R and Munoz-Pinedo C, Cell death induced by endoplasmic reticulum stress. FEBS J, 2016. 283(14): p. 2640–52. [DOI] [PubMed] [Google Scholar]
- 40.Sun S, et al. , IRE1alpha is an endogenous substrate of endoplasmic-reticulum-associated degradation. Nat Cell Biol, 2015. 17(12): p. 1546–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong S, et al. , Endoplasmic reticulum-resident E3 ubiquitin ligase Hrd1 controls B-cell immunity through degradation of the death receptor CD95/Fas. Proc Natl Acad Sci U S A, 2016. 113(37): p. 10394–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Metcalfe S and Sikora K, A new marker for testicular cancer. Br J Cancer, 1985. 52(1): p. 127–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Obiedat A, et al. , Transcription of the NKG2D ligand MICA is suppressed by the IRE1/XBP1 pathway of the unfolded protein response through the regulation of E2F1. FASEB J, 2019. 33(3): p. 3481–3495. [DOI] [PubMed] [Google Scholar]
- 44.Kazkaz H and Isenberg D, Anti B cell therapy (rituximab) in the treatment of autoimmune diseases. Curr Opin Pharmacol, 2004. 4(4): p. 398–402. [DOI] [PubMed] [Google Scholar]
- 45.Peterson JD and Chan LS, Effectiveness and side effects of anti-CD20 therapy for autoantibody-mediated blistering skin diseases: A comprehensive survey of 71 consecutive patients from the Initial use to 2007. Ther Clin Risk Manag, 2009. 5(1): p. 1–7. [PMC free article] [PubMed] [Google Scholar]
- 46.Arin MJ, et al. , Anti-CD20 monoclonal antibody (rituximab) in the treatment of pemphigus. Br J Dermatol, 2005. 153(3): p. 620–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.