Summary
RNAs adopt various conformations to perform different functions in cells. Incapable of acquiring intermediates, the key initiations of ligand recognition in the adenine riboswitch have not been characterized. In this work, stopped-flow fluorescence was used to track structural switches in the full-length adenine riboswitch in real time. We used PLOR (position-selective labeling of RNA) to incorporate fluorophores into desired positions in the RNA. The switching sequence P1 responded to adenine more rapidly than helix P4 and the binding pocket, followed by stabilization of the binding pocket, P4, and annealing of P1. Moreover, a transient intermediate consisting of an unwound P1 was detected during adenine binding. These events were observed in both the WT riboswitch and a functional mutant. The findings provide insight into the conformational changes of the riboswitch RNA triggered by a ligand.
Subject areas: Biological sciences, Biochemistry, Molecular biology, Structural biology
Graphical abstract

Highlights
-
•
Real-time tracking of the adenine riboswitch at nucleotide resolution
-
•
A transient conformation with unwound P1 is identified in the adenine riboswitch
-
•
Helix P1 responds to ligand quicker than the binding pocket or expression platform
Biological sciences; Biochemistry; Molecular biology; Structural biology
Introduction
Riboswitches are a class of cis-regulatory RNA elements located within 5′-untranslated regions (5′-UTR) of mRNA and control the expression of downstream genes by interacting with their specific ligands (Mandal and Breaker, 2004b; Micura and Höbartner, 2020; Serganov and Nudler, 2013). A full-length riboswitch consists of an aptamer domain and expression platform. The aptamer domain recognizes a specific ligand among numerous metabolites in cells. The recognition, in turn, alters the structure of the expression platform, thereby controlling gene expression. In general, aptamer domains with available crystal structures have been studied widely. For example, the aptamer of the adenine riboswitch, which is ∼70 nucleotides (nt), has been characterized by various methods (Broft et al., 2020; Dalgarno et al., 2013; Greenleaf et al., 2008; Neupane et al., 2011; Noeske et al., 2005; Serganov et al., 2004; St-Pierre et al., 2021), and Wang group captured a holo (ligand-bound), two apo (ligand-free), and an intermediate conformation with a re-arranged binding pocket and identified the stability of P1 was strengthened after ligand binding by mix-and-inject XFEL serial crystallography (Stagno et al., 2017). However, the full-length adenine riboswitch (termed the adenine riboswitch in subsequent text), which is ∼120 nt in length, is poorly understood, and only a few studies have examined the dynamic switch of the full-length riboswitch (Frieda and Block, 2012; Reining et al., 2013; Warhaut et al., 2017; Tomezsko et al., 2020; Tian et al., 2018). Block group investigated the folding hierarchy of the adenine riboswitch through optical trapping method, in which the structural regions of the RNA responded to ligand successively, and the P1 helix was the last region to be stabilized by ligand binding (Greenleaf et al., 2008; Frieda and Block, 2012). Schwalbe group proposed a three-state model (two apo and one holo) for the adenine riboswitch based on NMR data, indicating that multiple conformations of the adenine riboswitch co-exist in solution (Reining et al., 2013). Rouskin group postulated populations for three conformations of the adenine riboswitch by DMS-MaPseq and DREEM methods (Tomezsko et al., 2020). Das group studied allosteric details of the adenine riboswitch by a four-dimensional expansion of chemical mapping method, and suggested additional structural heterogeneities may be general in riboswitch RNAs (Tian et al., 2018). These studies contributed greatly to our understanding of the dynamic nature of the adenine riboswitch in solution.
There is no doubt that intermediate states provide valuable information for understanding a reactive molecule (Manz et al., 2017; Tian et al., 2018; Warhaut et al., 2017). However, it is difficult to obtain intermediate states because of their transitory and unstable nature. NMR, FRET, and stopped-flow methods are the popular techniques used for studying RNA dynamics, but they have restrictions on detecting intermediates. Decent NMR signals are usually from concentrated and stable structures instead of the transitory intermediates. For smFRET, its dead time and time interval in data collection are usually in sub-seconds, hence this method is not suitable for tracking short-lived intermediates either. Stopped-flow method is powerful in monitoring quick changes because its dead time can be as low as about 1 ms, but it generally requires nmoles of fluorophore-labeled samples, and preparing nmoles of site-specific long RNAs (>100 nt) are very challenging for routine strategies.
In this report, we applied stopped-flow fluorescence to track ligand-dependent switching of the adenine riboswitch at single-nucleotide resolution. The popular stopped-flow method can be used to monitor kinetics as rapid as milliseconds (Förster et al., 2012; Nayak et al., 2012). PLOR (position-selective labeling of RNA) was applied to incorporate Cy3 dye or AP (2-aminopurine) into specific positions in the adenine riboswitch (∼120 nt) to facilitate fluorescence experiments. PLOR is a solid-liquid phase hybrid method with the unique advantage of position-specific labeling of long RNAs with great flexibility and reasonable yields (Liu et al., 2015, 2018). Various sites distributed at the adenine riboswitch were tracked by stopped-flow and steady-fluorescence experiments. Based on our results, helix P1 sensed adenine faster than the binding pocket and helix P4 and reached equilibrium following the binding pocket and P4. Moreover, a transient conformation with an open P1 helix was detected. The “breathing” P1 helix may enable the ligand to interact with the binding pocket and trigger a switch of the expression platform in the adenine riboswitch.
Results
Ligand binding and gene regulation of add_FL were characterized
The adenine riboswitch used in this work, add_FL, is from Vibrio vulnificus and contains both the aptamer and expression platform. add_FL binds adenine specifically to control the translation of downstream adenosine deaminase (Mandal and Breaker, 2004a). The binding pocket located within the internal loop (green, Figure 1A) interacts directly with adenine by forming H-bonds (Figure S1). Helix P4 of add_FL (blue, Figure 1A) undergoes a structural change from a double-strand to single-strand conformation upon ligand binding, which releases the ribosome binding site (RBS) (magenta, Figure 1A) and is capable of recruiting the ribosome to translate the downstream gene. We performed ITC (isothermal titration calorimetry) and stopped-flow experiments to measure the binding affinity of add_FL toward adenine at room temperature (Figures 1B and 1C). ITC analysis showed that add_FL binds adenine with a dissociation constant (Kd) of 90.0 ± 0.96 nM, which is similar to the Kd of its aptamer (72.5 ± 0.77 nM) (Figure S2 and Table S1). This Kd shows that add_FL interacts with adenine strongly, and as expected, it is the aptamer domain instead of the expression platform that contributes mostly to ligand binding. AP has been reported to bind with the adenine riboswitch RNA with a comparable Kd as adenine (Mandal and Breaker, 2004a), and as a natural fluorescent analog of adenine, AP has been widely used in the studies of riboswitch RNA (Lemay et al., 2006; Mulhbacher and Lafontaine, 2007; Nozinovic et al., 2014). The fluorescence of AP quenched rapidly after mixing with add_FL in the stopped-flow experiments (Figure 1C). The quenching accelerated as the AP equivalents increased, and the association rate was determined to be 1.09 ± 0.01 × 105 M−1 s−1 (Figure S3 and Table S2). This rate is comparable with the rate constant reported elsewhere (Gilbert et al., 2006). In the reporter assays, the plasmid inserted with the adenine riboswitch and red fluorescent protein (RFP) was overexpressed in E. coli at 25°C, with a clear fluorescence increase in the presence of adenine (Figures 1D and S4). Our results are consistent with the well-known functions of the adenine riboswitch, i.e., binding adenine with high affinity and regulating gene expression.
Figure 1.
Characterization of ligand binding and gene regulation of the full-length adenine riboswitch, add_FL
(A) The well-accepted model of the adenine riboswitch. A ligand-binding incompetent conformation apoB and a ligand-binding conformation apoA co-exist in the absence of adenine. In the presence of adenine (orange), apoA switches to holo with the single-stranded RBS (magenta) available for ribosome binding. The binding pocket, helices P1, and P4 are shown in green, red, and blue, respectively.
(B) ITC thermogram of add_FL titrated against adenine. Adenine bound to 0.5 μM add_P1 with high affinity (Kd = 90.0 ± 0.96 nM).
(C) Stopped-flow fluorescence spectra of add_FL mixed with 2.5- to 20-fold AP. The binding rates of AP (2-aminopurine) to add_FL increased as higher amount of AP.
(D) Regulation of RFP expression by add_FL in E. coli in the presence of 0–1 mM adenine at room temperature. The expression level was adjusted by the internal control (EGFP expression) at different adenine concentrations.
Site-specific labeled add_FL was synthesized and characterized
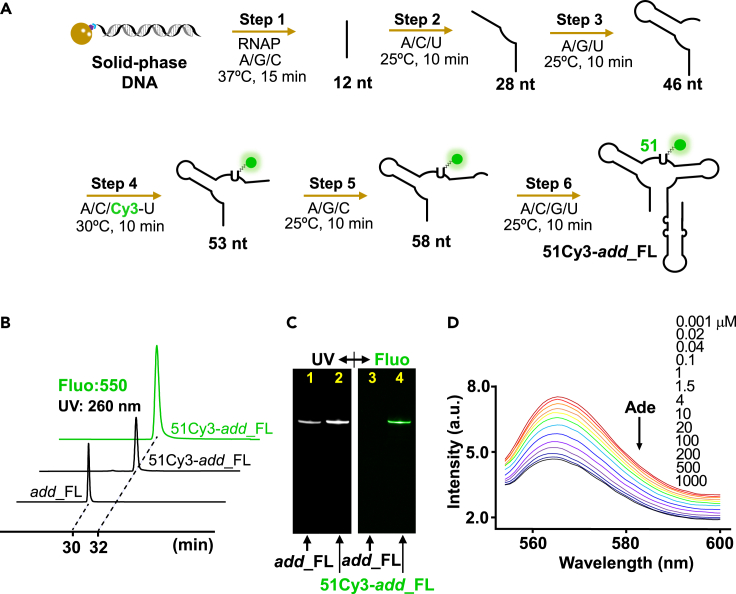
Environmental-sensitive dyes Cy3 and AP have been used to probe subtle structural changes in nucleic acids or proteins (Levitus and Ranjit, 2011; Morten et al., 2018; Rieder et al., 2007; Santiago-Frangos et al., 2016; Teplova et al., 2020). To study the kinetics of add_FL binding to adenine, we labeled various sites throughout the binding pocket, the switch helix P1 and P4. Site 51 of the binding pocket forms an H-bond with adenine (Figure S1). 51Cy3-add_FL is add_FL with Cy3 labeled at site 51, which was synthesized by the 6-step PLOR and purified by reversed-phase HPLC equipped with a C8 column (Figures 2A and 2B, Table S3). The observed single peak in HPLC or a sharp band in PAGE at UV and fluorescence excitation indicated the purity of 51Cy3-add_FL (Figures 2B and 2C). In addition, the elution time of 51Cy3-add_FL (middle and top curves) was ∼2 min longer than that of the unlabeled control, add_FL (bottom curve), which supported successful labeling of Cy3 to the RNA and the longer elution time was due to the hydrophobic nature of Cy3 (Figure 2B). Titration of adenine (0.001 μM–1 mM) to 0.2 μM 51Cy3-add_FL at room temperature resulted in a pronounced fluorescence decrease (42.8%), and the calculated KD was 5.37 ± 0.55 μM (Figures 2D and S5), which is similar to the reported KD measured using the aptamer domain (Stagno et al., 2017).
Figure 2.
Synthesis and characterization of 51Cy3-add_FL
(A) Diagram showing the 6-step PLOR synthesis of 51Cy3-add_FL. The DNA templates were immobilized on streptavidin-coated agarose beads (brown ball) in PLOR. Different NTPs were added at steps 1–6 to incorporate Cy3 (green ball) into site 51.
(B) HPLC spectra of purified add_FL and 51Cy3-add_FL with UV detection (black) and fluorescence excitation (green). The retention time of 51Cy3-add_FL was longer than the native add_FL because of the hydrophobic character of Cy3.
(C) Denaturing gel images of purified add_FL and 51Cy3-add_FL. The unlabeled RNA, add_FL was only observed under UV light (260 nm) (Lane 1). 51Cy3-add_FL was visualized by UV and fluorescence (λex, 530 nm) (lanes 2 and 4).
(D) Steady-state fluorescence of 51Cy3-add_FL measured in the presence of 0.001 μM–1 mM adenine. The fluorescence intensity decreased as adenine addition, and it dropped by 42.8% as the adenine concentration reached 1 mM.
Besides site 51, we individually labeled sites 18 and 86 in helix P1 and P4 with Cy3 by PLOR to generate 18Cy3-and 86Cy3-add_FL (Figures 3A and S5–S7 and Tables S4–S5). The fluorescence of 0.2 μM 18Cy3-and 86Cy3-add_FL decreased approximately 8.1% and 13.9%, respectively, as the adenine concentration reached 3 mM (Figure S5). Steady-fluorescence spectra reveal that the sites are allosteric with adenine, and structural changes around sites 18 and 86 are smaller than those at site 51, which is expected because site 51 interacts directly with adenine.
Figure 3.
Stopped-flow trajectories of Cy3-labeled add_FL with adenine
(A) The secondary structure of add_FL with labeled 18C (red), 51U (green), and 86U (blue).
(B–D) Stopped-flow curves of 0.5 μM 18Cy3- (B), 51Cy3- (C), and 86Cy3-add_FL (D) rapidly mixed with 1000-fold adenine in the presence of 1 mM Mg2+. The original and fitted traces in (B) and (D) are shown in dots and solid lines. The original and fitted traces in (C) are shown in black and green lines. The stopped-flow trajectory in (B) experienced a short and rapid fluorescence increase stage and then a decrease prior to the equilibrium.
(E) Normalized curves of 18Cy3- (red), 51Cy3- (green), and 86Cy3-add_FL (blue). The normalized curves in the first second were zoomed in the insert. The three sites, 18, 51, and 86 responded to ligand differently, and site 51 (green) reached equilibrium earlier than site 86 (blue) and site 18 (red).
(F–H) Steady-state fluorescence of 6 M urea titration to 0.2 μM 18Cy3- (F), 51Cy3- (G), and 86Cy3-add_FL (H) premixed with 1 mM MgCl2 and 1 mM adenine. All traces are an average of triplicate experiments.
Kinetic response of add_FL to adenine was differentiated regionally
Stopped-flow curves of all modified add_FL samples are shown in Figure 3. 18Cy3-add_FL experienced a steep-increasing stage over ∼0.1 s prior to the dropping stage after mixing with adenine (Figure 3B), and the data were fitted to single- and two-equilibrium curves, respectively. The calculated rate constants were 35.71 ± 3.18 s−1 for the increasing stage, 9.26 ± 1.36 and 0.33 ± 0.01 s−1 for the following stage (Figure S8 and Table S6). Fluorescence at site 51 showed the largest decrease after mixing with adenine, and data fitted better to three-equilibrium reactions rather than single- or double-equilibrium equations (Figures 3C and S9), indicating four conformations of add_FL exist under saturated adenine conditions (Table S6). The previous stopped-flow data of AP-labeled site 48 in the aptamer domain was fitted to a three-step kinetics model, which supports that at least four conformations of the adenine riboswitch co-exist upon ligand binding (Stagno et al., 2017). Although the sample size (aptamer domain vs. full-length), labeling sites (site 48 vs. 51), and labeling groups (AP vs. Cy3) are different, our stopped-flow results of the binding pocket are consistent with the previous research (Stagno et al., 2017). This also supports that the binding pocket reacts to ligand similarly in the presence or absence of the expression platform.
The rate constants for 51Cy3-add_FL binding adenine, k1obs, k2obs, and k3obs, were determined to be 8.62 ± 0.42, 2.35 ± 0.23, and 0.40 ± 0.05 s−1, respectively. The fluorescence of 86Cy3-add_FL decreased with adenine by the rate constants k1obs = 8.13 ± 1.42 s−1 and k2obs = 0.52 ± 0.15 s−1 (Figures 3D, S10 and Table S6). Comparing the rate constants between different fluorophore-labeled RNA samples, k1obs of 18Cy3-add_FL was clearly higher when compared with that of 51Cy3-and 86Cy3-add_FL, indicating that site 18 sensed adenine prior to sites 51 and 86. However, 18Cy3-add_FL reached equilibrium after ∼10 s (red, Figure 3E), which is slower than sites 51 (∼2 s, green) and 86 (∼6 s, blue, Figure 3E). We hypothesize that adenine promotes a fluctuation in helix P1 initially, which is followed by interaction with the binding pocket and unwinding of helix P4. Noteworthy, the stability of the binding pocket, in turn, contributes to the equilibrium of P1 and P4 and possibly the overall structure of the adenine riboswitch.
A P1-unwound intermediate was identified at add_FL
For site 18, a unique opposite change in fluorescence was detected (Figure 3B): a rapid increase followed by a decrease in fluorescence. The fluorescence changes indicated that P1 went through conformational changes oppositely in the two stages. The fluorescent intensities of cyanine dyes are environment-sensitive and affected complicatedly by many factors, including the local and global changes of nucleic acids (Morten et al., 2018). It is hard to speculate how structures change simply based on the rise or fall of fluorescence. Hence, we measured the fluorescent changes of individual site after urea denaturation to determine the correspondence between the changes in fluorescence and structures in the specific cases. Urea is a well-known denaturant to destabilize RNAs by breaking intra- and intermolecular interactions (Dalgarno et al., 2013). For site 51, the addition of urea caused an increase in fluorescence, probably because H-bonds were disrupted within the binding pocket and more fluorophores linked to site 51 were flexible and exposed to solvent (Figure 3G). The addition of urea caused a decrease in the fluorescence signal for site 86, which may arise from P4 dissociation (Figure 3H). This was in accordance with the fluorescence change brought to P4 by ligand addition. However, it was unforeseen that the fluorescent decrease of site 86 did not coincide with the fluorescent increase of site 51 after the addition of 6 M urea. One possible reason is that the effects of urea denaturation vary with the microstructural environments of different sites. Site 51 is located at an internal loop while P4 is located at the region connected 5′-helix and 3′-strand. Combined with the results of urea denaturation, the observed decrease in fluorescence intensity for site 51 may arise from stabilization of the internal loop due to the formation of an H-bond with adenine, and the fluorescent decrease for site 86 was because of the dissociation of helix P4 upon adenine binding as described earlier (Figures 3C and 3D). For site 18, urea caused P1 dissociation and a concomitant increase in the fluorescence signal (Figure 3F). Therefore, in Figure 3B, the trajectories can be attributed to the rapid uncoiling of P1 followed by restoration of the base pair at site 18.
A contemporary intermediate was also identified in a mutant
The stability of P1 was related to gene regulation based on our reporter assays in E. coli (Figure 4B). The transitory uncoiling of P1 may facilitate the entrance of ligand, which is functionally important for the adenine riboswitch. To identify if such uncoiling commonly existed in functional mutants, we monitored the kinetics of add_P1. The mutant, add_P1, which maintained the WT base-pair mode in P1, was observed to regulate RFP expression in a similar fashion to that of the WT (Figures 4A and 4B). The possible impact of the mutations in add_P1 on P4 was evaluated by the mutant C117G, in which the base pair between sites 81 and 117 was supposed to be restored if present in the WT. No significant difference in functions between add_P1 and C117G was detected (Figure S4), indicating that the base pair between 81 and 117 was absent (Tian et al., 2018). Multiple mutations (A16G, A17C, UU79–80AA, and GAAG15–18CUUC) to destabilize P1 caused the riboswitch to lose function. These mutants reveal that the stability of helix P1 has a pronounced effect on gene regulation, as reported previously (Lemay et al., 2011). Other mutations in the internal loop (U22C) and P4 (U86C and A111C) affected RFP production, as expected (Figure 4B), in which U22C and A111C lost regulation because of disruption to the binding pocket or stabilized P4, whereas U86C destabilized P4 and hence increased RFP production.
Figure 4.
Characterization of the functional mutant, add_P1
(A) Secondary structure of add_P1 with mutations (boxed).
(B) E. coli reporter assays of WT, add_FL, and the mutants in the absence (blue columns) or presence of 0.5 mM adenine (orange columns) at room temperature. The production of the reporter, RFP was regulated by add_FL or its mutants.
(C) AFM greyscale images of add_FL and add_P1 in absence and presence of adenine. The overall shapes and sizes of add_FL and add_P1 were similar.
(D) Histogram of the radius and Z mean from 279 to 403 add_FL particles and from 289 to 325 particles of add_P1 in the absence and presence of adenine, respectively. Gaussian fitting was used to give the mean values shown on the top of each graph.
add_P1 and WT regulated RFP production similarly in bacterial culture experiments. ITC, stopped-flow, and AFM (atomic force microscopy) experiments were recorded to compare add_P1 and WT from ligand binding and structural viewpoints. ITC and stopped-flow experiments showed that the binding affinity of add_P1 and WT was similar with a Kd of 67.6 ± 0.53 vs. 90.0 ± 0.96 nM and association rates of 1.13 ± 0.02 × 105 vs. 1.09 ± 0.01 × 105 M−1 s−1, respectively (Figures S2 and S3). AFM was used to compare the structures of the mutant and WT (Ares et al., 2016, 2018; Ramakrishnan et al., 2021). WT and add_P1 showed no distinguishable differences in overall folding and both molecules adopted a more compact conformation in the presence of adenine, as judged by the more concentrated radius, Z-mean and S/V (Figures 4C, 4D, and S11). The S/V (surface area/volume) for WT and add_P1 were calculated to be 1.7 ± 0.3 vs. 1.6 ± 0.2 without adenine and 1.5 ± 0.2 vs. 1.5 ± 0.2 with adenine (Table S7). Overall, the mutations to add_P1 hardly impaired the structure and function of the adenine riboswitch. add_P1 was then used as an alternative molecule to identify whether the P1-uncoiled intermediate also existed in the functional mutant as in the WT of the adenine riboswitch.
We incorporated Cy3 or AP into the binding pocket (site 51), P1 (sites 76 and 78), and P4 (site 86) of add_P1 and measured the kinetics of these sites upon mixing with adenine by stopped-flow experiments (Figures 5 and S12–S16 and Tables S8–S11). In add_P1, fluorescence signals at site 78 exhibited two stages with opposing changes after mixing with adenine, and single- and two-equilibrium fitting were used for these two stages (Figures 5 and S16 and Table S12). Replacing Cy3 by AP and shifting the labeled position from 78 to 76, the uniquely reverse fluorescent change in one sample after mixing with ligand maintained although alleviated (Figure 5F). The results indicate that like the WT, the uncoiled P1 conformation is also present transiently in this functional mutant. Moreover, based on the response time and the rate constants of sites 51, 76, 78, and 86, we propose that P1 senses the ligand initially and undergoes uncoiling to facilitate the ligand reaching the binding pocket, and the rearrangement of the binding pocket then, in turn, stabilizes the P4 and P1 regions. Stabilization of P1 by the binding pocket is supported by MD simulations, in which removal of the ligand destabilizes the binding pocket, and in particular, P1 unwinds (Di Palma et al., 2013).
Figure 5.
Stopped-flow trajectories of Cy3- or AP-labeled add_P1 mixing with adenine
(A–D) Stopped-flow curves of 0.5 μM 51Cy3- (A), 78Cy3- (C), 86Cy3- (D), and 1 μM 76AP-add_P1 (B) upon rapid mixing with 1000-fold adenine in the presence of 1 mM Mg2+. The original and fitted traces for all RNAs are shown in dots and solid lines, respectively. The original and fitted trace in (A) is shown in black and green, respectively.
(E) Normalized curves of 51Cy3- (green), 78Cy3- (pink), and 86Cy3-add_P1 (blue). In add_P1, fluorescence signals at site 78 exhibited two stages with opposing changes after mixing with adenine. Site 51 reached equilibrium faster than the other two sites.
(F) Normalized curves of 76AP- (purple) and 78Cy3-add_P1 (pink). Both 76 and 78 exhibited two stages with opposing changes. The inserts in (E) and (F) were the zoomed fluorescence change of sites 76 and 78 in the first second.
Discussion
Detecting transient conformations is pivotal for uncovering dynamic biological processes and important in designing novel strategies for controlling riboswitches. For example, characterizing transient conformations can aid in the development of new antibiotics that selectively target transient conformations (Eubanks et al., 2019; Jones and Ferré-D’Amaré, 2017; Lund et al., 2020; Rode et al., 2018). No transient conformations of the full-length adenine riboswitch have been reported. The over 100-nt riboswitch RNA is too long to label using routine methods. Here, we combined PLOR and stopped-flow fluorescence to site-specific label and probe real-time kinetics of the adenine riboswitch and its mutant at nucleotide resolution. The labeled nucleotides at the internal loop and the switching helix were differentiated by allosteric regulation upon ligand addition. The functional importance of the stability and flexibility of P1 has remained unresolved (Buck et al., 2007; Frieda and Block, 2012; Lemay et al., 2006; Nozinovic et al., 2014; Stoddard et al., 2008). The stopped-flow data herein support the notion that the switching helix P1 of the adenine riboswitch exhibits distinct dynamics on adenine binding, and was observed to unwind transiently on conversion from the apo to holo conformation. This “breathing” helix has been observed in other riboswitches, such as the TPP riboswitch (Aboul-ela et al., 2015; Haller et al., 2013).
The short-lived P1 intermediate likely functions as a pathway for ligand entry into the RNA and regulates translation. Thus, P1 serves as a bridge between ligand-sensing and gene regulation. We have added the new intermediate to refine our understanding of the dynamic processes involved in adenine binding to the adenine riboswitch (Figure 6). In this “induced fit” model, P1 initially senses adenine and uncoils to facilitate ligand entry. The binding pocket then refolds to allow adenine binding, followed by helical dissociation of P4. After the internal loop is stabilized, P4 and P1 gradually reach their stable holo states. P1 returns to a helix conformation. P4, containing the RBS, is predominantly formed as a single strand and available for ribosome binding, switching on translation of the downstream gene, such as the RFP used in our tests in E. coli. Nucleotide-level conformational details on the ms–s timescale that are associated with ligand binding provide valuable insights that improve our understanding of the regulatory mechanism of riboswitches.
Figure 6.
Four-state kinetic model of the adenine riboswitch
apoB is a ligand-binding incompetent conformation, in which P1 and P2 form an alternative secondary structure in 5′ region. In apoA, P1 (red) and P4 (blue) with RBS (magenta) adopt base-paired helices and are unable to translate the downstream RFP (pink block) (“OFF” state). In the presence of adenine, P1 initially senses this substrate by gradual uncoiling of the helix, which favors entry of adenine into the binding pocket and dissociation of P4. This is the P1-uncoiled intermediate. Uncoiled P4 in the holo conformation enables ribosome (purple) recognition of the RBS for translation (“ON” state).
Significance
Allostery of riboswitch RNAs is important to understand their various functions in different environments. However, owing to the lack of intermediates, deep understanding of riboswitches is strictly limited. In our work, the structural changes of RNAs were probed at ms–s timescale, and a transient intermediate is detected. The unwound P1 in the intermediate is pivotal to uncover the dynamic process of ligand binding in the adenine riboswitch. What is more, the functional importance of the stability and flexibility of P1 is supported by in vivo experiments. The newly identified conformation serves as the missing link between ligand binding and gene regulation of the adenine riboswitch. The unique combination of PLOR with stopped-flow fluorescence used in this work can be extended to probe other RNAs in action, which can provide insight into structural changes of RNAs triggered by a ligand or a substrate.
Limitations of the study
This study combined the PLOR and stopped-flow fluorescence for real-time tracking the conformational change of full-length adenine riboswitch at nucleotide resolution, and an intermediate conformation was identified transiently in both the wild-type and a functional mutant. Additional investigation of mutants in vivo would be valuable to explore the functions of the intermediate. Furthermore, it would be interesting to identify if this intermediate state exists generally in the riboswitch RNAs with P1 helix.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Competent E. coli | Cowin Bio. | Cat. #CW0808S |
| Chemicals, peptides, and recombinant proteins | ||
| Adenine | Sangon Biotech | Cat. #A600013 |
| 2-aminopurine | Bide Pharmatech | Cat. #BD2694 |
| 2-Aminopurine riboside-5’-O-triphosphate | BioLog | Cat. #A124 |
| Cy3-UTP | ApexBio | Cat. #B8330 |
| Cy3-CTP | GE | Cat. #25801086 |
| Deposited data | ||
| Structure of V.vulnificus 71-mer add A-riboswitch | (Serganov et al., 2004) | PDB: 1Y26 |
| Structure of V.vulnificus adenine riboswitch aptamer | (Stagno et al., 2017) | PDB: 5E54, 5SWD, 5SWE |
| Recombinant DNA | ||
| Table S13 | N/A | Integrated DNA Technologies |
| Software and algorithms | ||
| Origin 8.5 | OriginLab | www.originlab.com |
| PyMol | Schrödinger | pymol.org |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Prof. Yu Liu (liuyu_sjtu@sjtu.edu.cn).
Materials availability
Plasmids generated in this study are available from the lead contact upon request.
Experimental model and subject details
Bacterial strain used in this work is listed in the key resources table. E. coli was grown in LB medium and minimal media at 20–37°C with appropriate antibiotics when required.
Method details
RNA preparation
All RNAs in this work were prepared in-house. The labeled RNA samples were prepared by PLOR, as reported (Liu et al., 2015, 2018). The unlabeled RNA samples were prepared by in vitro transcription with T7 polymerase. The crude products were purified by denaturing 12% PAGE (polyacrylamide gel electrophoresis) and exchanged to the desired buffer for further investigation (Liu et al., 2018). The reagent added for carrying out the individual steps of PLOR for sample preparation are shown in Tables S3–S5 and S8–S11. The constructs of the adenine riboswitch for PLOR, add_FL and add_P1, are shown in Figures 3A and 4A. The DNA templates used in PLOR containing 5'-biotin were generated by PCR and attached to the streptavidin-coated agarose beads, as described previously (Liu et al., 2018). The sequences of DNA templates are listed in Table S13. The fluorescent samples generated by PLOR were purified by reversed-phase HPLC using a C8 column (#EXL-122-2546U, Advanced Chromatography Technologies Ltd., UK) with multiple-wavelength detection (UV at 260 nm, fluorescence at 550 nm for Cy3 or 300 nm for AP) (Liu et al., 2018). The purified samples were then exchanged to a suitable buffer and stored at −80°C. Unless stated, all RNAs were incubated at 85°C for 5 min and then cooled to room temperature before use.
ITC measurements
ITC experiments of the RNA samples were performed in a buffer containing 10 mM HEPES, 100 mM KCl, 10 mM MgCl2, pH 7.5, using a MicroCal ITC200 calorimeter (General Electric, USA). Two microliters of 350–450 μM adenine was injected into the cuvette containing 280 μL, 20–40 μM RNA with 90 s intervals between injections. The titration between the buffer and RNA was used as the background for ITC analysis. The background corrected ITC data were fitted by Origin ITC software (OriginLab, USA). Unless noted, all experiments were performed at room temperature.
Reporter assays of adenine riboswitch-controlled gene expression in E.coli
The sequence of the adenine riboswitch (or mutants) was inserted between enhanced green fluorescent protein (EGFP) and red fluorescent protein (RFP) genes in the vector pE1K to generate the vector pE1K_add. E. coli DH5α cells carrying the pE1K or pE1K_add were pre-incubated in LB media at 37°C for 8–10 h and then transferred to minimal media and cultured at 25°C until the OD600 reached ∼0.3. Culturing of E. coli was continued after the addition of 0, 0.01, 0.1, 0.5 and 1 mM adenine until the OD600 was 0.6–0.8. The fluorescence of EGFP (the internal control) and RFP was measured at 510 nm (λex = 480 nm) and 610 nm (λex = 580 nm), respectively, by a Cytation™ 5 multi-mode reader (BioTek, USA). All experiments were performed in triplicate. The expression level of RFP controlled by the adenine riboswitch was determined by:
| (Equation 1) |
whereR and E are the fluorescence of RFP and EGFP from pE1K_add, Rc and Ec are the fluorescence of RFP and EGFP from pE1K, and R0 and E0 are the fluorescent background of E. coli DH5α cells at 610 and 510 nm, respectively.
AFM(atomic force microscopy) experiments
The AFM experiments were performed in solution at 4°C using a Cypher VRS microscope (Asylum Research, Oxford Instruments). APS (1-(3-aminopropyl) silatrane)-treated mica surfaces were used to immobilized RNA and prepared as follows: freshly cleaved muscovite mica (Grade V1) (Ted Pella, Redding, CA) was coated with 0.17 mM APS for 30 min, rinsed with ultra-pure water and dried gently in filtered nitrogen. Ten microliters of 20 nM RNA was deposited on the APS-mica surface for ∼10 min followed by a wash step with 400 μL buffer (10 mM HEPES, 10 mM KCl, 1 mM MgCl2, 0 or 0.5 mM adenine ligand, pH 7.5). To avoid distortion caused by direct contact between the probe and molecular surfaces, we employed the amplitude modulated AC mode, commonly known as the tapping mode, to record images. All images were recorded using FASTSCAN-D-SS cantilevers (Bruker, CA), which have a nominal tip radius of 1 nm with a resonance frequency of ∼110 kHz and a spring constant of 0.25 N/m. Images were recorded at scan sizes of 500 x 500 nm2, 1024 x 1024 pixels (pixel resolution around 0.5 nm pixel–1) and a scanning rate of 0.98 Hz. The AFM images were processed using Gwyddion (version 2.56) as follows: horizontal scars correction to remove random scars, followed by plane leveling to correct any tilt and two-dimensional Fast Fourier Transform filtering to remove high-frequency noise. The digital resolution of the final images was increased to 4096 x 4096 pixels. Gaussian fitting was applied to the statistical analysis of the particle dimensions to extract the mean values using Mountains PIP (Image Metrology, France).
Fluorescence titration experiments
Steady-state fluorescence experiments were performed on an FLS 1000 photoluminescence spectrometer (Edinburgh Instruments Ltd., UK). For adenine titration experiments, 0.1 nM−3 mM adenine was titrated into a cuvette containing 200 μL of 0.2 μM Cy3-labeled or 0.5 μM AP-labeled RNAs in buffer (10 mM HEPES, 100 mM KCl, 1 mM MgCl2, pH 7.5). The mixture was pipetted up and down after adenine addition and equilibrated for ∼3 min before data collection. The total volume of the added adenine was less than 4 μL for one round of the experiment. For urea denaturation experiments, 6 M urea was titrated into a cuvette containing 200 μL of 0.2 μM Cy3-labeled or 0.5 μM AP-labeled RNAs in buffer (10 mM HEPES, 100 mM KCl, 1 mM MgCl2, 1 mM adenine, pH 7.5). The spectra were scanned from 555 to 600 nm (λex = 550 nm) and from 350 to 500 nm (λex = 300 nm) for Cy3-labeled and AP-labeled RNAs, respectively. The apparent dissociation constant KD was estimated by:
| (Equation 2) |
whereF0, F and Ff are the initial, the observed and the final fluorescence values, respectively, and [Ade] is the adenine concentration.
Stopped-flow fluorescence experiments
Stopped-flow fluorescence experiments were performed with a buffer of 10 mM HEPES, 100 mM KCl, 1 mM MgCl2, pH 7.5, mixed with an equal volume of the RNA solution with either AP (1.25–10 μM) or adenine (0.5 mM) using an SX20 stopped-flow spectrometer (Applied Photophysiscs Ltd., UK). 0.5 μM of unlabeled and Cy3-labeled RNA or 1.0 μM AP-labeled RNA were used in the stopped-flow fluorescence experiments. The stopped-flow trajectories of fluorescent reagents (Cy3-, AP-labeled RNA or AP) mixing with the buffer were collected as background. Data were collected with 550 nm excitation and a 570 nm long-pass filter for Cy3-labeled RNA, 300 nm excitation and a 360 nm long-pass filter for AP-labeled RNA or using AP as the ligand. The average data of at least three replicates with removal of background were fitted using a single-exponential Equation (3), biphasic rate Equation (4) or three-exponential Equation (5):
| (Equation 3) |
| (Equation 4) |
| (Equation 5) |
where F represents the detected fluorescence intensity at each time point t, A0 is the initial fluorescence, A1, A2 and A3 are constants, and k1obs, k2obs and k3obs are the apparent rate constants.
For comparison of the fluorescence change under different experimental conditions, the stopped-flow trajectories could be normalized using the following Equation (6):
| (Equation 6) |
where F represents fluorescence intensity detected in stopped-flow analysis, Fmax and Fmin are the maximum and minimum fluorescence intensity during ligand binding. After normalizing, the range of fluorescence change is from 0 to 1. The fluorescence intensity is adjusted with the removal of background.
Quantification and statistical analysis
All of the statistical methods for each experiment can be found in method details section. The fluorescent data generated from stopped-flow analysis were processed using Origin 8.5. The average value of three replicate samples was expressed as mean ± standard deviation. A value of P < 0.05 was considered statistically significant.
Acknowledgments
The work was supported by the National Natural Science Foundation of China [31872628 and 32071300].
Author contributions
W.L. and Y.L. designed the experiments and wrote the manuscript. W.L. prepared RNA samples, collected and analyzed ITC, bacterial culture, fluorescence titration, and stopped-flow experiments. D.C. contributed to data analysis. J.D. collected and analyzed AFM.
Declaration of interests
The authors declare no competing interests.
Published: December 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103512.
Supplemental information
Data and code availability
Data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to analyze the data reported in this paper is available from the lead contact upon request.
References
- Aboul-ela F., Huang W., Abd Elrahman M., Boyapati V., Li P. Linking aptamer-ligand binding and expression platform folding in riboswitches: prospects for mechanistic modeling and design. Wiley Interdiscip. Rev. RNA. 2015;6:631–650. doi: 10.1002/wrna.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares P., Fuentes-Perez M.E., Herrero-Galan E., Valpuesta J.M., Gil A., Gomez-Herrero J., Moreno-Herrero F. High resolution atomic force microscopy of double-stranded RNA. Nanoscale. 2016;8:11818–11826. doi: 10.1039/c5nr07445b. [DOI] [PubMed] [Google Scholar]
- Ares P., Gomez-Herrero J., Moreno-Herrero F. High-resolution atomic force microscopy imaging of nucleic acids. Methods Mol. Biol. 2018;1814:3–17. doi: 10.1007/978-1-4939-8591-3_1. [DOI] [PubMed] [Google Scholar]
- Broft P., Dzatko S., Krafcikova M., Hansel-Hertsch R., Wacker A., Doetsch V., Trantirek L., Schwalbe H. In-cell NMR of functional riboswitch aptamers in eukaryotic cells. Angew. Chem. Int. Ed. 2020;60:865–872. doi: 10.1002/anie.202007184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck J., Fürtig B., Noeske J., Wöhnert J., Schwalbe H. Time-resolved NMR methods resolving ligand-induced RNA folding at atomic resolution. Proc. Natl. Acad. Sci. U S A. 2007;104:15699–15704. doi: 10.1073/pnas.0703182104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgarno P.A., Bordello J., Morris R., St-Pierre P., Dubé A., Samuel I.D., Lafontaine D.A., Penedo J.C. Single-molecule chemical denaturation of riboswitches. Nucleic Acids Res. 2013;41:4253–4265. doi: 10.1093/nar/gkt128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Palma F., Colizzi F., Bussi G. Ligand-induced stabilization of the aptamer terminal helix in the add adenine riboswitch. RNA. 2013;19:1517–1524. doi: 10.1261/rna.040493.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubanks C.S., Zhao B., Patwardhan N.N., Thompson R.D., Zhang Q., Hargrove A.E. Visualizing RNA conformational changes via pattern recognition of RNA by small molecules. J. Am. Chem. Soc. 2019;141:5692–5698. doi: 10.1021/jacs.8b09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster U., Weigand J.E., Trojanowski P., Suess B., Wachtveitl J. Conformational dynamics of the tetracycline-binding aptamer. Nucleic Acids Res. 2012;40:1807–1817. doi: 10.1093/nar/gkr835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieda K.L., Block S.M. Direct observation of cotranscriptional folding in an adenine riboswitch. Science. 2012;338:397–400. doi: 10.1126/science.1225722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert S.D., Stoddard C.D., Wise S.J., Batey R.T. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J. Mol. Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Greenleaf W.J., Frieda K.L., Foster D.A.N., Woodside M.T., Block S.M. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller A., Altman R.B., Soulière M.F., Blanchard S.C., Micura R. Folding and ligand recognition of the TPP riboswitch aptamer at single-molecule resolution. Proc. Natl. Acad. Sci. U S A. 2013;110:4188–4193. doi: 10.1073/pnas.1218062110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C.P., Ferré-D’Amaré A.R. Long-Range interactions in riboswitch control of gene expression. Annu. Rev. Biophys. 2017;46:455–481. doi: 10.1146/annurev-biophys-070816-034042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay J.F., Desnoyers G., Blouin S., Heppell B., Bastet L., St-Pierre P., Massé E., Lafontaine D.A. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS Genet. 2011;7:e1001278. doi: 10.1371/journal.pgen.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay J.F., Penedo J.C., Tremblay R., Lilley D.M., Lafontaine D.A. Folding of the adenine riboswitch. Chem. Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Levitus M., Ranjit S. Cyanine dyes in biophysical research: the photophysics of polymethine fluorescent dyes in biomolecular environments. Q. Rev. Biophys. 2011;44:123–151. doi: 10.1017/S0033583510000247. [DOI] [PubMed] [Google Scholar]
- Liu Y., Holmstrom E., Yu P., Tan K., Zuo X., Nesbitt D.J., Sousa R., Stagno J.R., Wang Y.X. Incorporation of isotopic, fluorescent, and heavy-atom-modified nucleotides into RNAs by position-selective labeling of RNA. Nat. Protoc. 2018;13:987–1005. doi: 10.1038/nprot.2018.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Holmstrom E., Zhang J., Yu P., Wang J., Dyba M.A., Chen D., Ying J., Lockett S., Nesbitt D.J., et al. Synthesis and applications of RNAs with position-selective labelling and mosaic composition. Nature. 2015;522:368–372. doi: 10.1038/nature14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund P.E., Chatterjee S., Daher M., Walter N.G. Protein unties the pseudoknot: S1-mediated unfolding of RNA higher order structure. Nucleic Acids Res. 2020;48:2107–2125. doi: 10.1093/nar/gkz1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M., Breaker R.R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- Mandal M., Breaker R.R. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- Manz C., Kobitski A.Y., Samanta A., Keller B.G., Jäschke A., Nienhaus G.U. Single-molecule FRET reveals the energy landscape of the full-length SAM-I riboswitch. Nat. Chem. Biol. 2017;13:1172–1178. doi: 10.1038/nchembio.2476. [DOI] [PubMed] [Google Scholar]
- Micura R., Höbartner C. Fundamental studies of functional nucleic acids: aptamers, riboswitches, ribozymes and DNAzymes. Chem. Soc. Rev. 2020;49:7331–7353. doi: 10.1039/d0cs00617c. [DOI] [PubMed] [Google Scholar]
- Morten M.J., Lopez S.G., Steinmark I.E., Rafferty A., Magennis S.W. Stacking-induced fluorescence increase reveals allosteric interactions through DNA. Nucleic Acids Res. 2018;46:11618–11626. doi: 10.1093/nar/gky887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulhbacher J., Lafontaine D.A. Ligand recognition determinants of guanine riboswitches. Nucleic Acids Res. 2007;35:5568–5580. doi: 10.1093/nar/gkm572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak R.K., Peersen O.B., Hall K.B., Van Orden A. Millisecond time-scale folding and unfolding of DNA hairpins using rapid-mixing stopped-flow kinetics. J. Am. Chem. Soc. 2012;134:2453–2456. doi: 10.1021/ja208490w. [DOI] [PubMed] [Google Scholar]
- Neupane K., Yu H., Foster D.A., Wang F., Woodside M.T. Single-molecule force spectroscopy of the add adenine riboswitch relates folding to regulatory mechanism. Nucleic Acids Res. 2011;39:7677–7687. doi: 10.1093/nar/gkr305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noeske J., Richter C., Grundl M.A., Nasiri H.R., Schwalbe H., Wöhnert J. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc. Natl. Acad. Sci. U S A. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozinovic S., Reining A., Kim Y.-B., Noeske J., Schlepckow K., Wöhnert J., Schwalbe H. The importance of helix P1 stability for structural pre-organization and ligand binding affinity of the adenine riboswitch aptamer domain. RNA Biol. 2014;11:655–666. doi: 10.4161/rna.29439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan S., Stagno J.R., Conrad C.E., Ding J., Yu P., Bhandari Y.R., Lee Y.T., Pauly G., Yefanov O., Wiedorn M.O., et al. Synchronous RNA conformational changes trigger ordered phase transitions in crystals. Nat.Commun. 2021;12:1762. doi: 10.1038/s41467-021-21838-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reining A., Nozinovic S., Schlepckow K., Buhr F., Fürtig B., Schwalbe H. Three-state mechanism couples ligand and temperature sensing in riboswitches. Nature. 2013;499:355–359. doi: 10.1038/nature12378. [DOI] [PubMed] [Google Scholar]
- Rieder R., Lang K., Graber D., Micura R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. Chembiochem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- Rode A.B., Endoh T., Sugimoto N. Crowding shifts the FMN recognition mechanism of riboswitch aptamer from conformational selection to induced fit. Angew.Chem. Int. Ed. 2018;57:6868–6872. doi: 10.1002/anie.201803052. [DOI] [PubMed] [Google Scholar]
- Santiago-Frangos A., Kavita K., Schu D.J., Gottesman S., Woodson S.A. C-terminal domain of the RNA chaperone Hfq drives sRNA competition and release of target RNA. Proc. Natl. Acad. Sci. U S A. 2016;113:E6089–E6096. doi: 10.1073/pnas.1613053113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A., Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A., Yuan Y.R., Pikovskaya O., Polonskaia A., Malinina L., Phan A.T., Hobartner C., Micura R., Breaker R.R., Patel D.J. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagno J.R., Liu Y., Bhandari Y.R., Conrad C.E., Panja S., Swain M., Fan L., Nelson G., Li C., Wendel D.R., et al. Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography. Nature. 2017;541:242–246. doi: 10.1038/nature20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard C.D., Gilbert S.D., Batey R.T. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA. 2008;14:675–684. doi: 10.1261/rna.736908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre P., Shaw E., Jacques S., Dalgarno P.A., Perez-Gonzalez C., Picard-Jean F., Penedo J.C., Lafontaine D.A. A structural intermediate pre-organizes the add adenine riboswitch for ligand recognition. Nucleic Acids Res. 2021;49:5891–5904. doi: 10.1093/nar/gkab307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M., Falschlunger C., Krasheninina O., Egger M., Ren A., Patel D.J., Micura R. Crucial roles of two hydrated Mg2+ ions in reaction catalysis of the pistol ribozyme. Angew.Chem. Int. Ed. 2020;59:2837–2843. doi: 10.1002/anie.201912522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S., Kladwang W., Das R. Allosteric mechanism of the V. vulnificus adenine riboswitch resolved by four-dimensional chemical mapping. eLife. 2018;7:29602. doi: 10.7554/eLife.29602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomezsko P.J., Corbin V.D.A., Gupta P., Swaminathan H., Glasgow M., Persad S., Edwards M.D., McIntosh L., Papenfuss A.T., Emery A., et al. Determination of RNA structural diversity and its role in HIV-1 RNA splicing. Nature. 2020;582:438–442. doi: 10.1038/s41586-020-2253-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warhaut S., Mertinkus K.R., Höllthaler P., Fürtig B., Heilemann M., Hengesbach M., Schwalbe H. Ligand-modulated folding of the full-length adenine riboswitch probed by NMR and single-molecule FRET spectroscopy. Nucleic Acids Res. 2017;45:5512–5522. doi: 10.1093/nar/gkx110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to analyze the data reported in this paper is available from the lead contact upon request.