Abstract
Aeromonas veronii (A. veronii) is a zoonotic pathogen. It causes clinically a variety of diseases such as dysentery, bacteremia, and meningitis, and brings huge losses to aquaculture. A. veronii has been documented as a multiple antibiotic resistant bacterium. Hfq (host factor for RNA bacteriophage Qβ replication) participates in the regulations of the virulence, adhesion, and nitrogen fixation, effecting on the growth, metabolism synthesis and stress resistance in bacteria. The deletion of hfq gene in A. veronii showed more sensitivity to trimethoprim, accompanying by the upregulations of purine metabolic genes and downregulations of efflux pump genes by transcriptomic data analysis. Coherently, the complementation of efflux pump-related genes acrA and acrB recovered the trimethoprim resistance in Δhfq. Besides, the accumulations of adenosine and guanosine were increased in Δhfq in metabonomic data. The strain Δhfq conferred more sensitive to trimethoprim after appending 1 mM guanosine to M9 medium, while wild type was not altered. These results demonstrated that Hfq mediated trimethoprim resistance by elevating efflux pump expression and degrading adenosine, and guanosine metabolites. Collectively, Hfq is a potential target to tackle trimethoprim resistance in A. veronii infection.
Keywords: Aeromonas veronii, Hfq, trimethoprim, antibiotic resistance, acrA/acrB, purine pathway
Introduction
Aeromonas veronii (A. veronii) is a rod-shaped gram-negative pathogen found in diseased grass fish, tilapia, and turtles. It can cause huge losses in the aquaculture industry but also infect humans (Liu et al., 2015; Wang et al., 2019). A. veronii performs multiple drug resistance to antibiotics such as ampicillin, kanamycin, streptomycin, and gentamycin, resulting in the increased risk of human diseases and the greater losses to the fishery (Liu et al., 2017; Wang et al., 2019; Zhang et al., 2019). Hfq is a relatively common molecular chaperone that interacts with small RNAs to mediate the binding of small RNA to mRNA and assists in the post-transcriptional regulation of bacterial genes. Hfq participates in several regulatory pathways as a global regulator (Kakoschke et al., 2016). Deletion of hfq reduces tolerance to harsh environments in Escherichia coli, Salmonella enterica, and Vibrio hollisae (Yamada et al., 2010; Hayashi-Nishino et al., 2012; Azam and Vanderpool, 2018). Hfq is involved in regulating the virulence of Aeromonas hydrophila, the adhesion of Vibrio alginolyticus, and the nitrogen fixation efficiency and plant interactions of Pseudomonas stutzeri (Hayashi-Nishino et al., 2012; Kakoschke et al., 2016; Azam and Vanderpool, 2018; Santiago-Frangos and Woodson, 2018).
Albeit there are few studies on Hfq-related drug resistance, Hfq is documented to affect multidrug resistance of E. coli (Yamada et al., 2010). In this study, the hfq knockout strain (Δhfq) of A. veronii conferred more sensitive to trimethoprim. Trimethoprim is a broad-spectrum antibacterial that inhibits the activity of dihydrofolate reductase and the synthesis of tetrahydrofolate (Stepanek et al., 2016). Tetrahydrofolate is a one-carbon unit donor that provides the raw materials needed to synthesize purine nucleotides and thymidine nucleotides in vivo. Tetrahydrofolate is concerned with the regulation of purine metabolism and is closely related to bacterial nucleic acid synthesis, energy metabolism, ion transport and signal transduction (Jinnah et al., 2013; Vazquez-Salazar et al., 2018).
In addition, the absence of hfq affects the expression of the efflux pump gene acrAB (Spaniol et al., 2015). AcrB belongs to the resistance-nodulation-cell division superfamily (RND). The substrates for the AcrAB-TolC efflux pump include a variety of antibiotics, detergents, bactericides, fuels, and free fatty acids (Li et al., 2015; Phetsang et al., 2016). The efflux pump can excrete trimethoprim, affecting the antibacterial effect of trimethoprim (Köhler et al., 1996; Podnecky et al., 2013). The efflux pump AcrAB accounts for a large proportion of the mechanisms of bacterial antibiotic resistance (Elsby et al., 2017; Zwama et al., 2018). Therefore, it is hypothesized that the sensitivity of A. veronii to trimethoprim may be related to the expression of the efflux pump. In summary, Hfq indirectly affects the sensitivity of trimethoprim by affecting purine metabolism and efflux pumping; these effects are important for further understanding of the molecular mechanisms of multidrug resistance.
Materials and Methods
Strains and Culture
The strain information was listed in Table 1. The derivative A. veronii strains included wild-type, Δhfq, Δhfq:hfq, and Δhfq:acrAB. The strains were cultured in M9 minimal medium (M9) at 30°C, 150 r/min, supplemented with 50 μg/mL ampicillin. Δhfq represents the hfq knockout strain (Zhang et al., 2019). Δhfq:acrAB overexpresses the acrAB gene in the hfq knockout strain. The strain E. coli WM 3064 was applied to assist in the introduction of the plasmids into A. veronii by tri-parent conjugation (Simon et al., 1983; Ferrieres et al., 2010). For the culture of strain WM3064, 0.3 mM diaminopimelic acid was supplemented in LB at 37°C.
TABLE 1.
Strains and plasmids used in this paper.
| Strains or plasmids | Traits | Sources |
| E. coli WM3064 | Gene cloning strain | Liu et al., 2016 |
| Aeromonas veronii | Wild type strain | Liu et al., 2016 |
| Δhfq | hfq deletion mutant | Zhang et al., 2019 |
| Δhfq::hfq | hfq complement strain | Zhang et al., 2019 |
| Δhfq::acrAB | acrAB overexpression in hfqmutant strain | This paper |
| pBBR1MCS-2 | Gene cloning vector | Zhang et al., 2019 |
| pBBR acrAB | acrA/B overexpression vector | This paper |
Vector and Primers
The vector and primers were listed in Table 2. For the construction of the expression vector of the efflux pump-associated gene acrAB, the acrA and acrB genes were inserted into the plasmid pBBR1MCS-2, wherein the enzyme cleavage sites were SalI and EcoRI. The upstream and downstream primers required for the construction were acrA F 5′-ACGCGTCGACTTGGTATCGGCTGGGGATTG-3′ and acrB 5′-CCGGAATTCATGAGCGTCGGGAGAG-3′.
TABLE 2.
Primers used in this paper.
| Names of primers | Sequences (5′-3′) | Usage |
| WP_041202667.1-F | ATGGTCGCAGAGCTTGTC | Strain validation |
| WP_041202667.1-R | CAGCACAATAGAACACCAGAC | Strain validation |
| acrA SalI F | ACGCGTCGACTTGGTATCG GCTGGGGATTG | acrAB vector construction |
| acrB EcoRI R | CCGGAATTCATGAGCGTCGGGAGAG | acrAB vector construction |
| pBBR1MCS-2 F | GGCACCCCAGGCTTTACACT | Complement plasmid validation |
| pBBR1MCS-2 R | GATGTGCTGCAAGGCGATTAAG | Complement plasmid validation |
Minimum Inhibitory Concentration Test
Antibiotics were added to sterile 96-well plates at final concentrations of 64, 32, 16, 8, 4, 2, 1, 0.5, 0.25, and 0.125 μg/mL (Andrews, 2001). Then, 106 CFU broth was added to each well to a final volume of 200 μL. The 96-well plate was sealed with parafilm and cultured at 30°C with shaking at 150 r/min for 24 h. The experiment was repeated for 3 times.
Transcriptomic Analysis
The wild type and Δhfq strain were cultured in M9 medium containing 50 μg/mL ampicillin, cultured at 30°C and 150 r/min for 20 h, centrifuged to remove the culture medium, and washed with sterile PBS for transcriptomic analysis. The sequencing was carried out by BGI (Beijing Genomics Institution). The cells were collected and lysed, and the sample RNA was extracted with phenol-chloroform. The concentration and quality of the RNA samples were tested with the Agilent 2100. DNase I was used to remove double-stranded DNA, and a Ribo-Zero Magnetic Kit was used to remove ribosome RNA. Reverse transcription was performed with random primers and first strand cDNA as a template to synthesize the second strand. The linker sequence was attached to the 3′ end of the cDNA fragment. The cDNA sequence was amplified with a primer cocktail, and the purified product was sequenced on a HiSeq Xten (Illumina, San Diego, CA, United States) platform. The sequencing depth was chain-specific sequencing for 2 Gb of clean data. HISAT was attempted for genome assembly, potential coding sequence analysis and new transcript identification that may be present. The transcriptional differences between wild-type and hfq knockout were analyzed by Bowtie 2, and FPKM was used to normalize gene expression levels. Each gene expression was calculated using the Benjamini-Hochberg false discovery rate (FDR). The differential transcripts were tested for log-fold change, and the p value was corrected with FDR < 0.001. The differential genes were analyzed using GO classification, and disparity expression in the pathway was compared with the entire genomic background using hypergeometric analysis p ≤ 0.05 was a differential metabolic pathway. GEO accession number was GSE120603, and the URL of accession website was displayed as https://submit.ncbi.nlm.nih.gov/subs/sra/SUB6133286. The DESeq. 2 packages in R were applied to estimate the fold changes and perform other analysis. A. veronii TH0426 genome (Genomic Sequence: NZ_CP012504.1) was referenced for transcriptome analysis (Kang et al., 2016).
Metabolomics Analysis
The non-target metabolomic and lipidomic detection platform (UHPLC-QTOF-MS) was applied to metabolomics for the detection of A. veronii samples. UHPLC-QTOF-MS included Ultra-Performance Liquid Chromatography 1290UHPLC (Agilent), ACQUITY UPLC BEH Amide column 1.7 μm, 2.1 × 100 mm (Waters) and High-Resolution Mass Spectrometry Triple TOF 6600 (AB Sciex). The original mass spectrum was converted to the mzXML format using Proteo Wizard software, and the peaks were identified using the R Programming Language package (Version 3.2) and self-built secondary mass spectrometry data. URL of accession website was displayed as www.ebi.ac.uk/metabolights/MTBLS1411.
Statistical Analysis
Statistical data were analyzed using the statistical Package for the Social Science (SPSS) version 20.0 (SPSS, Chicago, IL, United States) and GraphPad Prism version 8.0 (GraphPad, San Diego, CA, United States). The results are presented as the mean values of three independent experiments with standard deviation using one-way analysis of variance. p < 0.05 or 0.01 were represented as significant or extremely significant, respectively.
Results
Hfq Deletion Reduces Multiple Resistance to Antibiotics Including Trimethoprim
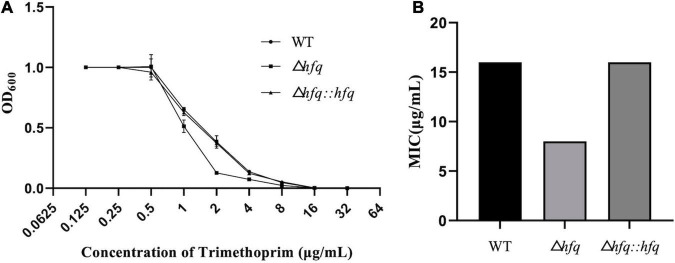
According to previous studies, A. veronii were resistant to gentamycin, kanamycin, streptomycin, and were sensitive to chloramphenicol, ciprofloxacin (Liu et al., 2018). Trimethoprim, as an antibiotic that inhibits folic acid metabolism, has a strong inhibitory effect on a variety of bacteria. Resistance to trimethoprim was found to be altered in the absence of hfq (Figure 1A). The mutant Δhfq was more sensitive to trimethoprim than wild type, which exhibited with a minimum inhibitory concentration (MIC) of 8 μg/mL in contrast to 16 μg/mL of wild type. The complemented strain attenuated the sensitivity of Δhfq to trimethoprim, and the MIC was the same as that of wild type (Figure 1B).
FIGURE 1.
Growth and Minimum inhibitory concentration (MIC) of trimethoprim. (A) The growth of three A. veronii strain: wildtype, Δhfq, and Δhfq:hfq. (B) The MIC of trimethoprim of wildtype, Δhfq, and Δhfq:hfq.
Upregulation of Purine Metabolic Gene Expression and Downregulation of Efflux Pump-Related Genes in Δhfq Strain
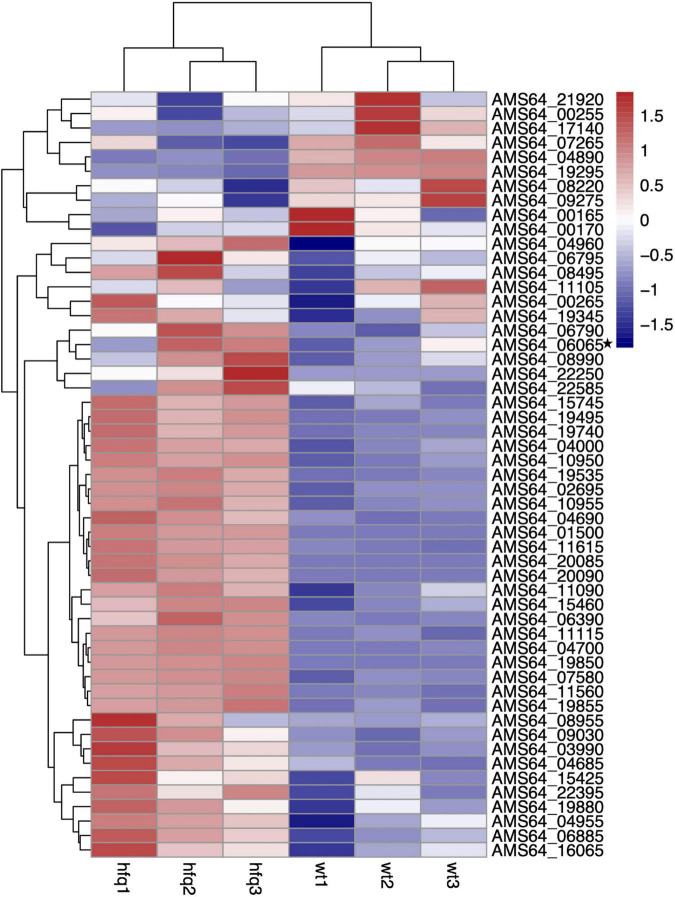
To understand the changes of drug resistance in Δhfq strains, transcriptome sequencing was used to compare metabolic pathways with significant variations in expression levels and to analyze their relationship with trimethoprim resistance. The clustering analysis revealed that many genes related to purine metabolism and efflux pump synthesis were expressed differently (Figure 2). Although the direct target of trimethoprim was dihydrofolate reductase, the transcriptions of dihydrofolate reductase were not significantly different between Δhfq and wild type (Figure 2 marked with star). However, the expression of 53 genes was affected in purine metabolism, which functioned as the downstream of folate metabolism (Figure 3). There were 21 genes marked with red were up-regulated which led to purine accumulation. Two genes marked with green, as purine consuming enzymes including xanthine nucleic acid transferase and hypoxanthine nucleic acid transferase, were significantly down-regulated.
FIGURE 2.
Transcriptomic data of Aeromonas veronii. Heat map of expression levels of genes involved in purine metabolism and efflux pump synthesis of wild-type and Δhfq strains.
FIGURE 3.
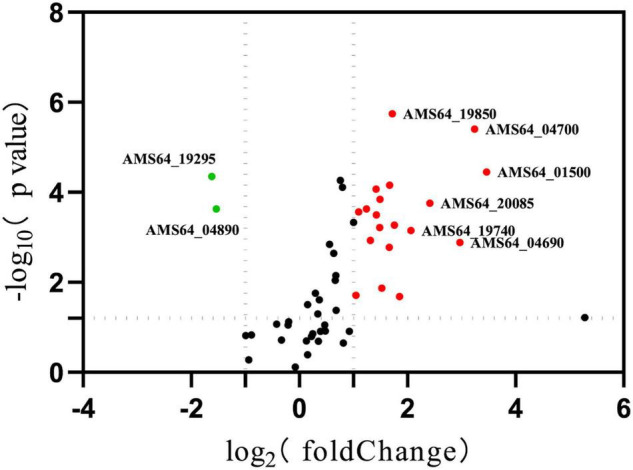
Expression of genes related to purine metabolism. Green represents down-regulated genes with significant differences. Red indicates up-regulated genes with a significant difference. *represents p < 0.05.
Metabolomics Analysis Displays That Δhfq Enhances Purine Metabolism
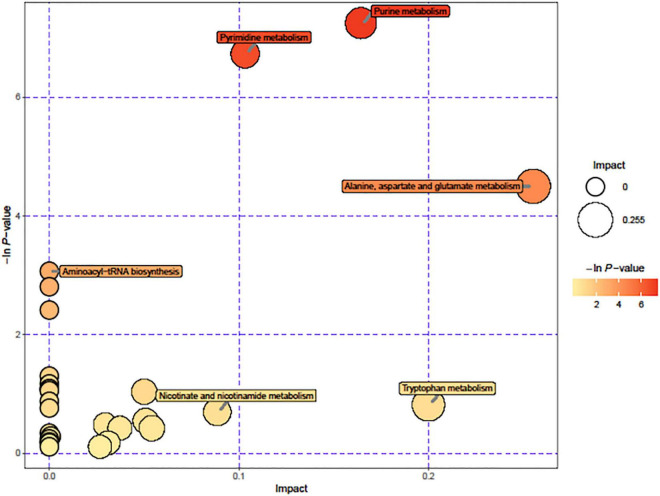
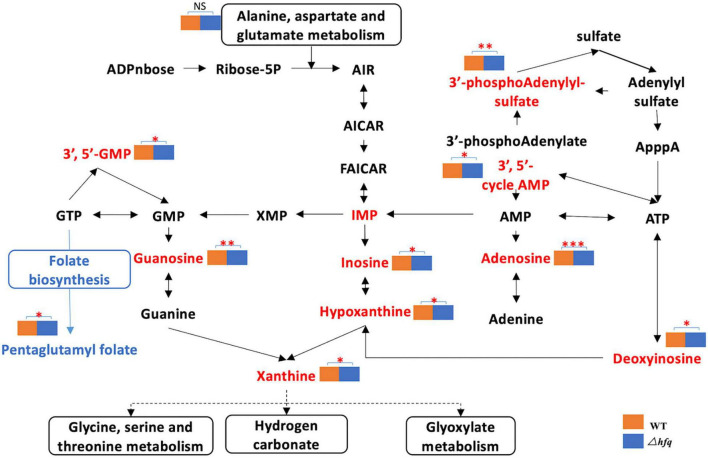
Due to the significant changes of transcription in metabolic pathways, variant metabolites of these pathways have been hypothesized to be responsible for trimethoprim resistance. The differential metabolites of wild-type and Δhfq were screened, analyzed, and classified into metabolic pathways (Figure 4). Purine metabolism, pyrimidine metabolism, and alanine, aspartate, and glutamic acid metabolism were greatly affected by the deletion of hfq, of which the effects on purine metabolism and pyrimidine metabolism were prominent. The deletion of Hfq incurred a significant increase of purine metabolites including adenosine, guanosine, and xanthine (Figure 5).
FIGURE 4.
Pathways affected by hfq knockout. The circle diameter indicates the degree of influence of the pathway, and the –ln (p value) and color indicate the significant difference in the number of genes regulated by the pathway for the entire genomic background.
FIGURE 5.
Metabolites of the purine metabolic pathway of Aeromonas veronii. The red font indicates the increased metabolites, and the blue color indicates the decreased metabolites. The metabolic pathway is framed by a solid line. * represents the difference between Δhfq and wild-type metabolites. * represents p < 0.05, ** represents p < 0.005, and *** represents p < 0.001.
The Accumulation of Purine Metabolites Enhances the Sensitivity of Δhfq to Trimethoprim
The purine metabolites, such as guanosine and adenosine, were significantly increased in Δhfq compared with wild type (Figure 3). To understand whether the accumulation of metabolites changed the trimethoprim sensitivity of hfq knockout, the downstream products such as 1 mM adenine, 1 mM guanine, and 1 mM ATP were added to the M9 medium to evaluate the MIC separately (Yang et al., 2019). The MIC of wild type was not altered when supplemented with 1 mM guanosine (Figure 6A), while that of Δhfq was decreased. There had little evident changes both in wild type and Δhfq after appending with 1 mM adenine or ATP. The above results suggested that the additional guanine enhanced the sensitivity of Δhfq to trimethoprim.
FIGURE 6.
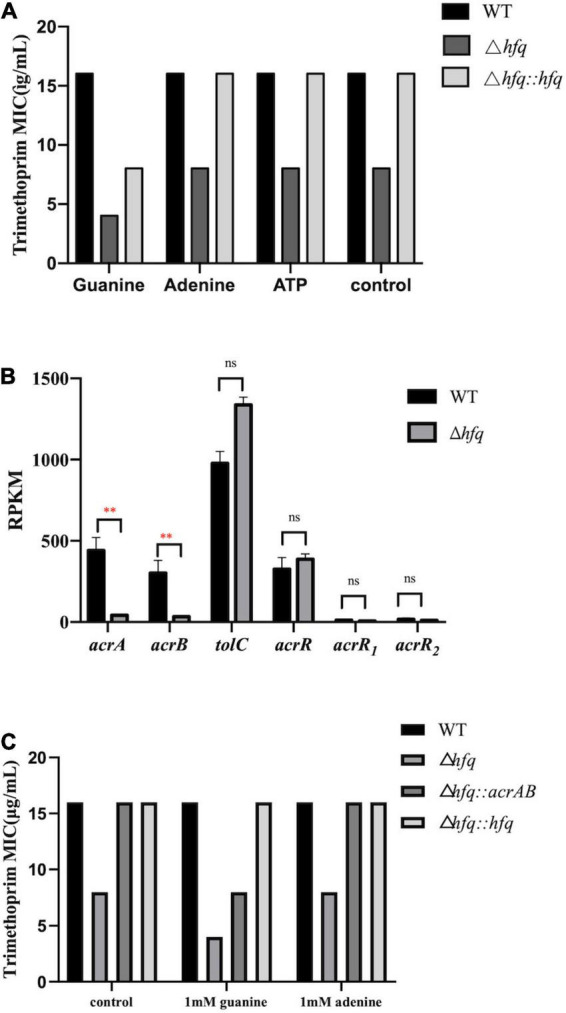
MIC of trimethoprim under conditions of exogenous purine metabolites and overexpressed AcrAB. (A) The concentrations of exogenous products are 1 mM for adenosine, guanosine, and adenosine triphosphate, and ATP. Equal volume of ddH2O was added as a control group (B). Expression of key genes in the efflux pump. The rpkm of three acrR copies was presented separately. (C) MIC of trimethoprim to Aeromonas veronii overexpressing acrAB. * represents p < 0.05, ** represents p < 0.01, and ns represents no significant difference.
Overexpression of acrAB Enhances the Tolerance to Trimethoprim
AcrAB-TolC is capable of actively transporting antibiotics (Li et al., 2015), and trimethoprim can be transported outside the cell membrane by an efflux pump of P. aeruginosa. According to the transcriptomic data, the mRNA levels of acrA and acrB in Δhfq were significantly reduced (by 9.30-fold and 9.34-fold) compared with wild type, but those of three copies of transcriptional repressor acrR, and that of component tolC (by 1.35-fold) were transcribed consistently (Figure 6B). The acrAB overexpression vector was constructed and transferred into the Δhfq strain. The overexpression strain Δhfq:acrAB showed an increased MIC and enhanced tolerance compared with Δhfq. Overexpression of acrAB reversed the loss of hfq, resulting in inefficient discharge of trimethoprim (Figure 6C).
Discussion
As a small chaperone protein, Hfq regulates gene expression by binding to sRNA and mRNA in response to external stress and environmental changes. Previous studies revealed that Hfq acts on a variety of membrane-associated protein genes, affecting bacterial growth, cell membrane formation, virulence, drug resistance, stress tolerance, and retention of retained bacteria (Hayashi-Nishino et al., 2012; Zhang et al., 2019).
Aeromonas veronii is highly resistant to ampicillin, kanamycin, gentamicin, streptomycin, and spectinomycin (Liu et al., 2016; Zhang et al., 2019). Previously the MIC of hfq knockout strain is significantly lower than that of wild type under the treatment of antibiotics (Zhang et al., 2019). As the substrate of nucleotide, the related genes and products of purine pathway showed significant differences in Hfq mutant strain (Figures 3, 5). But in fact, the productions of purines and nucleotides are affected by one carbon unit carrier tetrahydrofolate, and the latter is controlled by dihydrofolate reductase in turn (Paulsen et al., 2013). Since antibiotic trimethoprim targets dihydrofolate reductase specifically (Darrell et al., 1968; Bourne, 2014; Toulouse et al., 2020), trimethoprim is treated for Hfq knockout instead of other antibiotics. There are many mechanisms for resistance, of which efflux pump is important for multidrug resistance in bacteria (Elsby et al., 2017). The active transport function of the efflux pump is one of the main reasons for the decreased resistance to antibiotics (Abdali et al., 2017). The efflux pump AcrAB-TolC is an RND-type efflux pump that transports antibiotics through the inner membrane, periplasmic cavity, and outer membrane to the outside of the bacteria (Wang et al., 2017; Shi et al., 2019). The downregulations of the efflux pump-related genes acrA and acrB interfere with the assembly of the efflux pump, which reduce the ability of the efflux pump to bind and transport antibiotics and increase the sensitivity of the bacteria to trimethoprim (Cunrath et al., 2019). As a negative regulator of acrAB, the transcription level of acrR maintained a consistent in Δhfq, indicating that acrAB was regulated independently by Hfq rather than AcrR.
The enzymes of purine metabolism were enhanced in Δhfq strain, companying with the augmented productions of intermediate metabolites guanosine and adenosine. However, the quantities of downstream metabolites including glutamine, serine, threonine and glyoxylate were not significantly altered (Figure 3). In this study, metabolomics data showed that the deletion of hfq gene influenced on the basal metabolic pathways such as bacterial energy metabolism, hydrazine, and pyrimidine anabolism (Figures 4, 5).
Our experimental results demonstrated that Hfq affected the sensitivity of A. veronii to trimethoprim through different pathways. The downregulation of efflux pump system genes reduced the assembly of the efflux pump complex and decreased the ability of the cell to export trimethoprim. The transcriptional upregulation of many genes in purine metabolic pathway recruited the accumulation of metabolites, making A. veronii more sensitive to trimethoprim.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
ZL, XM, ML, DH, and DW contributed the conception and design of the study. DW, HL, YT, and HT performed the statistical analysis. DW and ZL drafted the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Funding
This work was supported by the grants from the National Natural Science Foundation of China Nos. 31772887 (to ZL) and 32060153 (to HL).
References
- Abdali N., Parks J. M., Haynes K. M., Chaney J. L., Green A. T., Wolloscheck D., et al. (2017). Reviving antibiotics: efflux pump inhibitors that interact with AcrA, a membrane fusion protein of the AcrAB-TolC multidrug efflux pump. ACS Infect. Dis. 3, 89–98. 10.1021/acsinfecdis.6b00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews J. M. (2001). Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 48(Suppl. 1) 5–16. 10.1093/jac/48.suppl_1.5 [DOI] [PubMed] [Google Scholar]
- Azam M. S., Vanderpool C. K. (2018). Translational regulation by bacterial small RNAs via an unusual Hfq-dependent mechanism. Nucleic Acids Res. 46 2585–2599. 10.1093/nar/gkx1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne C. R. (2014). Utility of the biosynthetic folate pathway for targets in antimicrobial discovery. Antibiotics (Basel). 3 1–28. 10.3390/antibiotics3010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunrath O., Meinel D. M., Maturana P., Fanous J., Buyck J. M., Saint Auguste P., et al. (2019). Quantitative contribution of efflux to multi-drug resistance of clinical Escherichia coli and Pseudomonas aeruginosa strains. EBioMedicine 41 479–487. 10.1016/j.ebiom.2019.02.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrell J. H., Garrod L. P., Waterworth P. M. (1968). Trimethoprim: laboratory and clinical studies. J. Clin. Pathol. 21 202–209. 10.1136/jcp.21.2.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsby R., Chidlaw S., Outteridge S., Pickering S., Radcliffe A., Sullivan R., et al. (2017). Mechanistic in vitro studies confirm that inhibition of the renal apical efflux transporter multidrug and toxin extrusion (MATE) 1, and not altered absorption, underlies the increased metformin exposure observed in clinical interactions with cimetidine, trimethoprim or pyrimethamine. Pharmacol. Res. Perspect. 5:e00357. 10.1002/prp2.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrieres L., Hemery G., Nham T., Guerout A.-M., Mazel D., Beloin C., et al. (2010). Silent mischief: bacteriophage mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J. Bacteriol. 192 6418–6427. 10.1128/jb.00621-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Nishino M., Fukushima A., Nishino K. (2012). Impact of hfq on the intrinsic drug resistance of salmonella enterica serovar typhimurium. Front. Microbiol. 3:205. 10.3389/fmicb.2012.00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinnah H. A., Sabina R. L., Van Den Berghe G. (2013). Metabolic disorders of purine metabolism affecting the nervous system. Handb. Clin. Neurol. 113 1827–1836. 10.1016/B978-0-444-59565-2.00052-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakoschke T. K., Kakoschke S. C., Zeuzem C., Bouabe H., Adler K., Heesemann J., et al. (2016). The RNA chaperone Hfq is essential for virulence and modulates the expression of four adhesins in Yersinia Enterocolitica. Sci. Rep. 6:29275. 10.1038/srep29275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Pan X., Xu Y., Siddiqui S. A., Wang C., Shan X., et al. (2016). Complete genome sequence of the fish pathogen Aeromonas veronii TH0426 with potential application in biosynthesis of pullulanase and chitinase. J. Biotechnol. 227 81–82. 10.1016/j.jbiotec.2016.04.009 [DOI] [PubMed] [Google Scholar]
- Köhler T., Kok M., Michea-Hamzehpour M., Plesiat P., Gotoh N., Nishino T., et al. (1996). Multidrug efflux in intrinsic resistance to trimethoprim and sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 40 2288–2290. 10.1128/aac.40.10.2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Z., Plesiat P., Nikaido H. (2015). The challenge of efflux-mediated antibiotic resistance in gram-negative bacteria. Clin. Microbiol. Rev. 28 337–418. 10.1128/CMR.00117-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Chen Y., Wang D., Tang Y., Tang H., Song H., et al. (2016). Genetic selection of peptide aptamers that interact and inhibit both small protein b and alternative ribosome-rescue factor a of Aeromonas veronii C4. Front. Microbiol. 7:1228. 10.3389/fmicb.2016.01228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Huang D., Hu X., Tang Y., Ma X., Yan R., et al. (2017). Targeting Inhibition of SmpB by peptide aptamer attenuates the virulence to protect zebrafish against Aeromonas veronii infection. Front. Microbiol. 8:1766. 10.3389/fmicb.2017.01766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Hu K., Tang Y., Li H., Tang H., Hu X., et al. (2018). SmpB down-regulates proton-motive force for the persister tolerance to aminoglycosides in Aeromonas veronii. Biochem. Biophys. Res. Commun. 507, 407–413. 10.1016/j.bbrc.2018.11.052 [DOI] [PubMed] [Google Scholar]
- Liu Z., Liu P., Liu S., Song H., Tang H., Hu X. (2015). Small protein B upregulates sensor kinase bvgS expression in Aeromonas veronii. Front. Microbiol. 6:579. 10.3389/fmicb.2015.00579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen J. L., Viswanathan K., Wright D. L., Anderson A. C. (2013). Structural analysis of the active sites of dihydrofolate reductase from two species of Candida uncovers ligand-induced conformational changes shared among species. Bioorg. Med. Chem. Lett. 23 1279–1284. 10.1016/j.bmcl.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phetsang W., Pelingon R., Butler M. S., Kc S., Pitt M. E., Kaeslin G., et al. (2016). Fluorescent trimethoprim conjugate probes to assess drug accumulation in wild type and mutant Escherichia coli. ACS Infect. Dis. 2 688–701. 10.1021/acsinfecdis.6b00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podnecky N. L., Wuthiekanun V., Peacock S. J., Schweizer H. P. (2013). The BpeEF-OprC efflux pump is responsible for widespread trimethoprim resistance in clinical and environmental Burkholderia pseudomallei Isolates. Antimicrob. Agents Chemother. 57 4381–4386. 10.1128/aac.00660-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Frangos A., Woodson S. A. (2018). Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip. Rev. RNA 9:e1475. 10.1002/wrna.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Chen M., Yu Z., Bell J. M., Wang H., Forrester I., et al. (2019). In situ structure and assembly of the multidrug efflux pump AcrAB-TolC. Nat. Commun. 10:2635. 10.1038/s41467-019-10512-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R., Priefer U., Pühler A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Naturebiotechnology 1 784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- Spaniol V., Bernhard S., Aebi C. (2015). Moraxella catarrhalis AcrAB-OprM efflux pump contributes to antimicrobial resistance and is enhanced during cold shock response. Antimicrob. Agents Chemother. 59 1886–1894. 10.1128/AAC.03727-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanek J. J., Schäkermann S., Wenzel M., Prochnow P., Bandow J. E. (2016). Purine biosynthesis is the bottleneck in trimethoprim-treatedBacillus subtilis. Proteomics Clin. Appl. 10 1036–1048. 10.1002/prca.201600039 [DOI] [PubMed] [Google Scholar]
- Toulouse J. L., Shi G., Lemay-St-Denis C., Ebert M. C. C. J. C., Deon D., Gagnon M., et al. (2020). Dual-Target inhibitors of the folate pathway inhibit intrinsically trimethoprim-resistant DfrB dihydrofolate reductases. ACS Med. Chem. Lett. 11 2261–2267. 10.1021/acsmedchemlett.0c00393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Salazar A., Becerra A., Lazcano A. (2018). Evolutionary convergence in the biosyntheses of the imidazole moieties of histidine and purines. PLoS One 13:e0196349. 10.1371/journal.pone.0196349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Li H., Ma X., Tang Y., Tang H., Hu X., et al. (2019). Small RNA AvrA regulates IscR to increase the stress tolerances in SmpB deficiency of Aeromonas veronii. Front. Cell Infect. Microbiol. 9:142. 10.3389/fcimb.2019.00142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Fan G., Hryc C. F., Blaza J. N., Serysheva I. I., Schmid M. F., et al. (2017). An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. Elife 6:e24905. 10.7554/eLife.24905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada J., Yamasaki S., Hirakawa H., Hayashi-nishino M., Yamaguchi A., Nishino K. (2010). Impact of the RNA chaperone HFQ on multidrug resistance in Escherichia coli. J. Antimicrob. Chemother. 65 853–858. 10.1093/jac/dkq067 [DOI] [PubMed] [Google Scholar]
- Yang J. H., Wright S. N., Hamblin M., McCloskey D., Alcantar M. A., Schrübbers L., et al. (2019). A white-box machine learning approach for revealing antibiotic mechanisms of action. Cell 177 1649.e–1661.e. 10.1016/j.cell.2019.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Yu W., Tang Y., Li H., Ma X., Liu Z. (2019). RNA chaperone hfq mediates persistence to multiple antibiotics in Aeromonas veronii. Microb. Pathog. 132 124–128. 10.1016/j.micpath.2019.04.045 [DOI] [PubMed] [Google Scholar]
- Zwama M., Yamasaki S., Nakashima R., Sakurai K., Nishino K., Yamaguchi A. (2018). Multiple entry pathways within the efflux transporter AcrB contribute to multidrug recognition. Nat. Commun. 9:124. 10.1038/s41467-017-02493-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.