Abstract
Bacteroides thetaiotaomicron is the second most frequently encountered species of the anaerobes isolated from clinical specimens. We developed a PCR-based assay for the rapid identification of B. thetaiotaomicron. Specific primers were based on shared amplicons of about 1.2 kb generated from B. thetaiotaomicron by randomly amplified polymorphic DNA. This 1.2-kb fragment was sequenced and then used to design a set of PCR amplification primers. This PCR generated an amplification product of 721 bp, which was unique to all 65 isolates of B. thetaiotaomicron tested. There was no amplification with isolates of other bacterial species. Restriction enzyme digestion of the amplification product and dot blot hybridization further verified the specificity of the assay. These results suggest that this PCR assay targets a nucleotide sequence that is strongly conserved in B. thetaiotaomicron. This simple and rapid PCR assay provides a rapid and accurate method for identification of B. thetaiotaomicron and shows promise for the detection of B. thetaiotaomicron in clinical samples.
Members of the Bacteroides fragilis group, which are bile-resistant, anaerobic, gram-negative rods, are opportunistic pathogens commonly recovered from patients with peritonitis, septicemia, and wound infections. Of the greatest clinical importance within this group are B. fragilis and B. thetaiotaomicron. B. thetaiotaomicron is the second most commonly encountered anaerobic gram-negative bacillus (1, 5; I. Brook, Letter, J. Antimicrob. Chemother. 25:473–474, 1990). Although it is part of the indigenous microflora of the gastrointestinal tract, it is also associated with infection, such as intraabdominal sepsis and bacteremia, and is resistant to many antimicrobial agents (1, 5; Brook, Letter). It has been reported that B. fragilis group bacteremia contributes significantly to morbidity and mortality (15). In addition, B. thetaiotaomicron has been reported to be more resistant to cephalosporins and clindamycin than B. fragilis (1, 5, 15, 16, 18). These clinical characteristics of B. thetaiotaomicron infection increase the need for rapid and accurate identification of the infection in clinical specimens and for immediate and effective management of infected patients.
Conventional biochemical identification of Bacteroides species is complicated and time-consuming. Species identification often requires 3 to 7 days and sometimes is inconclusive (19). The automated methods currently used are sometimes unreliable at identifying B. thetaiotaomicron clinical isolates or other species (2, 3). The phenotypic and biochemical similarity of closely related species makes it difficult to discriminate them. Precise identification is not always possible. This difficulty in correctly identifying the B. thetaiotaomicron strains may subsequently result in inappropriate antibiotic therapy and should be of concern to clinical laboratories. Thus, a more specific and rapid assay is needed for the identification of B. thetaiotaomicron. In recent years, PCR methods have been developed to identify a variety of microorganisms including anaerobes. Yamashita et al. used the glutamine synthetase gene as a target for amplification of B. fragilis (20). Kuwahara et al. reported the detection of B. fragilis by PCR amplification of the neuraminidase-encoding gene (11). Genotypic methods for the detection of B. thetaiotaomicron using PCR fingerprinting or PCR hybridization have been reported (4, 10). Kreader described the use of a PCR hybridization assay based on 16S rRNA sequences to identify B. thetaiotaomicron and other species from fecal extracts (10). However, it is difficult to adapt this test for use in clinical laboratories because of its complexity. The aim of the present study was to develop a simple and species-specific PCR assay for the identification of B. thetaiotaomicron.
Bacterial strains.
Twenty-seven reference strains obtained from the American Type Culture Collection, Rockville, Md., were used in this study. The PCR assay primers were based on the reference strain, B. thetaiotaomicron ATCC 29741. Other species representing 11 gram-positive and 15 gram-negative bacterial species were used as negative controls (Table 1). An additional 65 clinical isolates of B. thetaiotaomicron and 40 clinical isolates of Bacteroides species were also tested. All of the clinical isolates were collected from the Bacteriology Laboratory, National Taiwan University Hospital, a 2,000-bed teaching hospital in northern Taiwan. The Bacteroides species was identified by the Presumpto Plates method and sugar fermentation tests as previously described (19).
TABLE 1.
Bacteria used as negative controls in this study
| Bacterial strain or isolate |
|---|
| Bacteroides ovatus ATCC 8483 and 6 clinical isolates |
| Bacteroides fragilis ATCC 25285 and 10 clinical isolates |
| Bacteroides distasonis ATCC 8503 and 5 clinical isolates |
| Bacteroides uniformis ATCC 8492 and 8 clinical isolates |
| Bacteroides vulgatus ATCC 8482 and 5 clinical isolates |
| Bacteroides caccae ATCC 43185 and 6 clinical isolates |
| Bacteroides stercoris ATCC 43183 |
| Clostridium perfringens ATCC 3624 |
| Clostridium difficile ATCC 9689 |
| Eubacterium lentum ATCC 43055 |
| Fusobacterium nucleatum ATCC 25586 |
| Fusobacterium varium ATCC 8501 |
| Prevotella intermedia ATCC 25611 |
| Peptostreptococcus anaerobius ATCC 27337 |
| Propionibacterium acnes ATCC 6919 |
| Veillonella parvula ATCC 10790 |
| Staphylococcus aureus ATCC 29213 |
| Staphylococcus epidermidis ATCC 14990 |
| Streptococcus bovis ATCC 9809 |
| Streptococcus anginosus ATCC 33397 |
| Streptococcus intermedius ATCC 27335 |
| Enterococcus faecalis ATCC 29212 |
| Escherichia coli ATCC 25922 |
| Aeromonas hydrophila ATCC 7965 |
| Acinetobacter baumanii ATCC 19606 |
| Pseudomonas aeruginosa ATCC 27853 |
RAPD fingerprint among Bacteroides species.
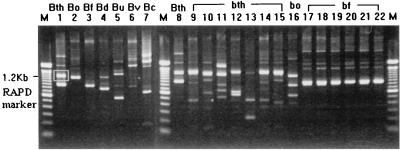
Randomly amplified polymorphic DNA (RAPD) patterns of tested strains were determined by means of arbitrarily primed PCR as described in our previous report (8). The arbitrary oligonucleotide primer OPH-9, 5′-TGTAGCTGGG-3′ (Operon Technologies, Alameda, Calif.), was used. Amplification products were analyzed by electrophoresis on a 1.5% agarose gel (FMC Bioproducts, Rockland, Me.). The RAPD fingerprint patterns obtained with the primer OPH-9 for B. thetaiotaomicron and other Bacteroides species are shown in Fig. 1. One shared band of about 1.2 kb was found in the RAPD fingerprints of all five of the B. thetaiotaomicron isolates tested. This fragment was absent from the RAPD fingerprints of the other six species (Fig. 1).
FIG. 1.
RAPD patterns generated from B. thetaiotaomicron and other Bacteroides species. RAPD patterns were obtained by AP-PCR amplification of genomic DNA with the primer OPA-09. Lanes M contain 100-bp DNA ladder (Gibco BRL). Lanes 1 and 8, B. thetaiotaomicron (Bth) ATCC 29741; lane 2, B. ovatus (Bo) ATCC 8483; lane 3, B. fragilis (Bf) ATCC 25285; lane 4, B. distasonis (Bd) ATCC 8503; lane 5, B. uniformis (Bu) ATCC 8492; lane 6, B. vulgatus (Bv) ATCC 8482; lane 7, B. caccae (Bc) ATCC 43185; lanes 9 to 15, B. thetaiotaomicron (bth) clinical isolates; lane 16, B. ovatus (bo) clinical isolate; lanes 17 to 22, B. fragilis (bf) clinical isolates. In lane 1, the 1.2-kb amplicon shared by B. thetaiotaomicron strains is indicated by the white box around the band.
Extraction and sequencing of DNA from a RAPD fingerprint band.
This 1.2-kb fragment was excised, purified, and sequenced. The selected amplicon was excised from the RAPD gel and was eluted by using the Gene-Clean kit (Bio 101, Inc., La Jolla, Calif.). The DNA fragment was cloned into the vector using the TA cloning kit and transformed into Escherichia coli. Following thermal cycling of the sequencing reactions with fluorescent-dye-labeled primers or terminators, the nucleotide sequence was determined on an Automated DNA Sequencer (ABI prism 377A DNA sequencer; Applied Biosystems Division, Perkin-Elmer Corp., Foster City, Calif.) The partial sequence of the 1.2-kb fragment is shown in Fig. 2. No significant homologies were found between the sequences of this fragment and those of other genes found in the GenBank database.
FIG. 2.
Partial sequences of the specific RAPD marker. The PCR primers are underlined.
Development of PCR.
The primer pair BTH-F (5′-TGGAGTTTTACTTTGAATGGAC-3′) and BTH-R (5′-CTGCCCTTTTACAATGGG-3′) based on the sequences of the 1.2-kb fragment were designed to generate a 721-bp product upon PCR amplification of DNA from B. thetaiotaomicron ATCC 29741 (Fig. 2). The amplification reaction mixtures contained 50 μl of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin, 1 U of Taq polymerase (Perkin-Elmer, Norwalk, Conn.), 200 μM each of the four deoxynucleotide triphosphates (dATP, dCTP, dGTP, and dTTP; Perkin-Elmer), 50 pmol of each primer, and 2 μl of DNA sample. PCR was performed in a DNA thermal cycler (MJ Research, Inc., Watertown, Mass.). A total of 35 cycles of PCR were done, with 1 cycle consisting of denaturation (94°C, 1 min), annealing (56°C, 1 min), and extension (72°C, 1 min) steps followed by a final extension step (72°C, 7 min). The PCR amplification products were analyzed by agarose gel electrophoresis in 1% agarose (FMC BioProducts) and stained with ethidium bromide. A visible band of the appropriate size (721 bp) was considered a positive reaction. The conserved sequence was further analyzed by digestion of the PCR products with the restriction enzymes HaeIII and HinfI (Gibco BRL, Gaithersburg, Md.). The DNA fragments were run on 1.8% agarose gels (FMC BioProducts).
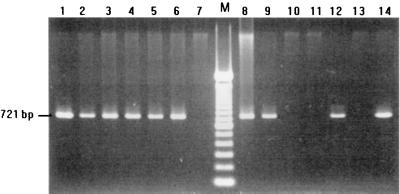
After PCR, the 721-bp DNA product was obtained only from isolates of B. thetaiotaomicron. No amplification was detected from other species (Fig. 3).
FIG. 3.
Specificity of PCR amplification. Specific amplification of the 721-bp DNA fragment was detected only in B. thetaiotaomicron isolates. Lane, M, 100-bp DNA ladder (Gibco BRL); lanes 1 to 6, 8, 9, 12, and 14, B. thetaiotaomicron isolates; lane 7, B. fragilis; lane 10, B. distasonis; lane 11, B. uniformis; lane 13, B. vulgatus.
Specificity and sensitivity of PCR assays.
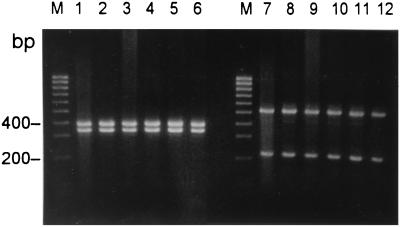
The specificity and sensitivity were further determined by testing a total of 65 isolates of B. thetaiotaomicron and 40 isolates of other species. All 65 clinical isolates identified as B. thetaiotaomicron by conventional methods yielded the 721-bp amplicon. Moreover, the PCR amplification products from B. thetaiotaomicron isolates all revealed identical HaeIII and HinfI restriction patterns (Fig. 4). No amplification products were detected with 40 isolates of all other species. The sensitivity of PCR was determined by testing with serial dilutions of the DNA sample. The level of sensitivity indicates that several copies of the target DNA are needed for detection.
FIG. 4.
PCR-restriction fragment length polymorphism analysis of B. thetaiotaomicron. The PCR products with primers BTH-F and BTH-R from B. thetaiotaomicron isolates were digested with restriction enzymes HaeIII (lanes 1 to 6) and HinfI (lanes 7 to 12). Lanes M contain 100-bp DNA ladder (Gibco BRL).
Dot blot assays.
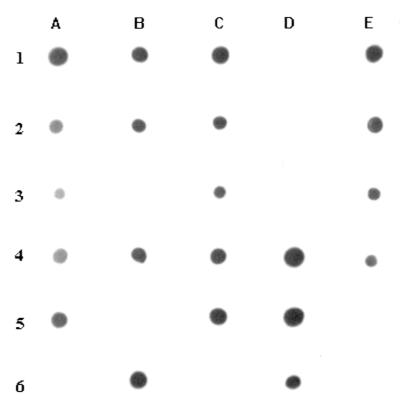
To confirm the specificity of this PCR assay, the 721-bp PCR product was labeled with digoxigenin and then used for hybridization to genomic DNA from various organisms. Probes were produced with the PCR method described above and simultaneously labeled by incorporation of digoxigenin-11-dUTP (Boehringer Mannheim, Mannheim, Germany). For each strain tested, 300 ng of chromosomal DNA was denatured by heating at 96°C for 10 min and spotted onto Hybond-N nylon membranes (Amersham). DNA was then fixed onto the filter by UV treatment at an intensity of 120 mJ/cm2 for 3 min on a UV Crosslinker. Both the prehybridization and hybridization temperatures were 61°C. All filters were prehybridized for 1 h in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Hybridization was performed overnight with heat-denatured probe. Detection was performed by using an antidigoxigenin antibody conjugated to alkaline phosphatase as a substrate according to the manufacturer's instructions. Only DNA from B. thetaiotaomicron showed a strong hybridization signal. No hybridization signal was detected by dot blot analysis of DNA from non-B. thetaiotaomicron strains (Fig. 5).
FIG. 5.
Dot blot hybridization showing the specificity of PCR amplification. All positive hybridization reactions (indicated by the presence of a hybridization signal) were obtained with B. thetaiotaomicron isolates. All negative reactions (no hybridization signal) were from other species as follows: B. ovatus (1D), B. uniformis (2D), B. distasonis (3B), B. fragilis (3D), B. vulgatus (5B), Staphylococcus aureus (5E), Streptococcus bovis (6A), Enterococcus faecalis (6C), and Escherichia coli (6E).
B. thetaiotaomicron isolates are commonly found with serious extraintestinal tract infections and are usually more resistant to antimicrobial agents than B. fragilis. Accurate identification of Bacteroides species is often problematic. Conventional identification protocols are usually laborious and time-consuming (19). It is sometimes difficult to differentiate among indole-positive Bacteroides species. For final identification, it was necessary to perform additional tests (production of acid from arabinose, trehalose, and xylane) to discriminate between B. fragilis, B. thetaiotaomicron, B. ovatus, and B. uniformis. It has been reported that misidentification of most Bacteroides species often involves B. thetaiotaomicron (2, 3). For such isolates, up to 48 h can be required for further identification assays. Phenotypic differentiation between B. thetaiotaomicron and B. ovatus is often difficult in a clinical laboratory setting (19). B. thetaiotaomicron is phylogenetically very close to B. ovatus. Although 16S rRNA has been widely used as a target for PCR primers or probes for the identification of microorganisms (6), the assay for B. thetaiotaomicron is less selective, since both B. thetaiotaomicron and B. ovatus were detected when the 16S rRNA probe of B. thetaiotaomicron was used (10). In fact, on the basis of 16S rRNA sequence comparison, these two species exhibited 96.9% sequence homology (13). Therefore, the development of rapid and sensitive DNA-based assays which are applicable for the direct detection of B. thetaiotaomicron may improve the rapidity and accuracy of the diagnosis of infections.
In the present study, we have developed a rapid PCR-based assay to improve the identification of B. thetaiotaomicron. Initially, RAPD analysis was performed to compare the patterns generated from Bacteroides species. A conserved fragment was observed with B. thetaiotaomicron isolates. Subsequently, the primers were designed based on the sequences of this fragment and a PCR assay was set up. RAPD analysis has been used mostly for intraspecies discrimination in epidemiological studies (8, 9). However, PCR assays based on the primers designed from conserved fragments generated by RAPD have been previously reported (7, 12, 14, 17). For some organisms, such as Prevotella, Porphyromonas, Legionella, and Candida species, this method has been successfully applied for species identification (7, 12, 14, 17). This strategy can be seen as a universal method for designing detection assays without the need for prior knowledge of the genetic characteristics of the target species.
The species specificity of this PCR assay was further tested on 65 clinical isolates identified as B. thetaiotaomicron by conventional methods and 11 species of gram-positive and 15 species of gram-negative clinically important bacteria tested. The specific 721-bp amplicon can be obtained only from all B. thetaiotaomicron isolates, not from other species. The sensitivity of this PCR assay indicates that only several copies of the target DNA are needed for detection. The sensitivity level should be sufficient for the direct detection of B. thetaiotaomicron in clinical specimens.
In order to ensure the specificity of amplification and also examine the intraspecies heterogeneity, restriction digestion was performed. All of the B. thetaiotaomicron clinical isolates displayed identical HaeIII and HinfI restriction patterns. Therefore, no intraspecies heterogeneity was found. The conservation of target sequences was also confirmed by dot blot hybridization. Under these conditions, only DNA from B. thetaiotaomicron showed a strong hybridization signal. No hybridization signal was detected by dot blot analysis of DNA from non-B. thetaiotaomicron strains. This assay shows promise for the detection of B. thetaiotaomicron from clinical specimens.
The PCR-based diagnostic assay described in this study targets a nucleotide sequence that is conserved in B. thetaiotaomicron. No significant homologies were found between the sequence of the 721-bp amplification product and those of other genes found in the GenBank database. No probe is needed in this assay. The simple and rapid PCR assay offers an alternative to currently used methods and may lead to the early diagnosis of B. thetaiotaomicron infections.
Nucleotide sequence accession number.
The partial sequence of the 1.2-kb fragment from the RAPD fingerprints of B. thetaiotaomicron isolates was deposited in GenBank and assigned accession no. AF182955.
REFERENCES
- 1.Appleman M D, Heseltine P N R, Cherubin C E. Epidemiology, antimicrobial susceptibility, pathology, and significance of Bacteroides fragilis group organisms isolated at Los Angeles County-University of Southern California Medical Center. Clin Infect Dis. 1991;13:12–18. doi: 10.1093/clinids/13.1.12. [DOI] [PubMed] [Google Scholar]
- 2.Arzese A, Minisini R, Botta G A. Evaluation of an automated system for identification of anaerobic bacteria. Eur J Clin Microbiol Infect Dis. 1994;13:135–141. doi: 10.1007/BF01982186. [DOI] [PubMed] [Google Scholar]
- 3.Cavallaro J J, Wiggs L S, Miller J M. Evaluation of the BBL Crystal Anaerobe Identification system. J Clin Microbiol. 1997;35:3186–3191. doi: 10.1128/jcm.35.12.3186-3191.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claros M C, Gerardo H, Citron D M, Goldstein E J C, Schonian G, Rodloff A C. Use of polymerase chain reaction fingerprinting to compare clinical isolates of Bacteroides fragilis and Bacteroides thetaiotaomicron from Germany and the United States. Clin Infect Dis. 1997;25(Suppl. 2):S295–S298. doi: 10.1086/516200. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein E J C. Anaerobic bacteremia. Clin Infect Dis. 1996;23(Suppl. 1):S97–S101. doi: 10.1093/clinids/23.supplement_1.s97. [DOI] [PubMed] [Google Scholar]
- 6.Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guillot E, Mouton C. PCR-DNA probe assays for identification and detection of Prevotella intermedia sensu stricto and Prevotella nigrecens. J Clin Microbiol. 1997;35:1876–1882. doi: 10.1128/jcm.35.7.1876-1882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsueh P-R, Teng L-J, Ho S-W, Hsieh W-C, Luh K-T. Clinical and microbiological characteristics of Flavobacterium indologenes infections associated with indwelling devices. J Clin Microbiol. 1996;34:1908–1913. doi: 10.1128/jcm.34.8.1908-1913.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsueh P-R, Teng L-J, Yang P-C, Pan H-J, Chen Y-C, Wang L-H, Ho S-W, Luh K-T. Dissemination of two methicillin-resistant Staphylococcus aureus clones exhibiting negative Staphylase reactions in intensive care units. J Clin Microbiol. 1999;37:504–509. doi: 10.1128/jcm.37.3.504-509.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kreader C A. Design and evaluation of Bacteroides DNA probes for the specific detection of human fecal pollution. Appl Environ Microbiol. 1995;61:1171–1179. doi: 10.1128/aem.61.4.1171-1179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuwahara T, Akimoto S, Ugai H, Kamogashira T, Kinouchi T, Ohnishi Y. Detection of Bacteroides fragilis by PCR assay targeting the neuraminidase-encoding gene. Lett Appl Microbiol. 1996;22:361–365. doi: 10.1111/j.1472-765x.1996.tb01179.x. [DOI] [PubMed] [Google Scholar]
- 12.Menard C, Gosselin P, Duhaime J F, Mouton C. Polymerase chain reaction using arbitrary primer for the design and construction of a DNA probe specific for Porphyromonas gingivalis. Res Microbiol. 1994;145:595–602. doi: 10.1016/0923-2508(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 13.Paster B J, Dewhirst F E, Olsen I, Fraster G J. Phylogeny of Bacteroides, Prevotella, and Porphyromonas spp. and related bacteria. J Bacteriol. 1994;176:725–732. doi: 10.1128/jb.176.3.725-732.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Presti F L, Riffard S, Vandenesch F, Etienne J. Identification of Legionella species by random amplified polymorphic DNA profiles. J Clin Microbiol. 1998;36:3193–3197. doi: 10.1128/jcm.36.11.3193-3197.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Redondo M C, Arbo M D J, Grindlinger J, Snydman D R. Attributable mortality of bacteremia associated with the Bacteroides fragilis group. Clin Infect Dis. 1995;20:1492–1496. doi: 10.1093/clinids/20.6.1492. [DOI] [PubMed] [Google Scholar]
- 16.Snydman D R, McDermott L, Cuchural G J, Jr, Hecht D W, Iannini P B, Harrell L J, Jenkins S G, O'Keefe J P, Pierson C L, Rihs J D, Yu V L, Finegold S M, Gorbach S L. Analysis of trends in antimicrobial resistance patterns among clinical isolates of Bacteroides fragilis group species from 1990 to 1994. Clin Infect Dis. 1996;23(Suppl. 1):S54–S65. doi: 10.1093/clinids/23.supplement_1.s54. [DOI] [PubMed] [Google Scholar]
- 17.Steffan P, Vazquez J A, Boikov D, Xu C, Sobel J D, Akins R A. Identification of Candida species by randomly amplified polymorphic DNA fingerprinting of colony lysates. J Clin Microbiol. 1997;35:2031–2039. doi: 10.1128/jcm.35.8.2031-2039.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka-Bandoh K, Kato N, Watanabe K, Ueno K. Antibiotic susceptibility profiles of Bacteroides fragilis and Bacteroides thetaiotaomicron in Japan from 1990 to 1992. Clin Infect Dis. 1995;20(Suppl. 2):S352–S355. doi: 10.1093/clinids/20.supplement_2.s352. [DOI] [PubMed] [Google Scholar]
- 19.Whaley D N, Wiggs L S, Miller P H, Srivastava P U, Miller J M. Use of Presumpto Plates to identify anaerobic bacteria. J Clin Microbiol. 1995;33:1196–1202. doi: 10.1128/jcm.33.5.1196-1202.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamashita Y, Kohno S, Koga H, Tomono K, Kaku M. Detection of Bacteroides fragilis in clinical specimen by PCR. J Clin Microbiol. 1994;32:679–683. doi: 10.1128/jcm.32.3.679-683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]