Abstract
The diverse applications of genetically modified cells and organisms require more precise and efficient genome-editing tool such as clustered regularly interspaced short palindromic repeats/CRISPR-associated protein (CRISPR/Cas). The CRISPR/Cas system was originally discovered in bacteria as a part of adaptive-immune system with multiple types. Its engineered versions involve multiple host DNA-repair pathways in order to perform genome editing in host cells. However, it is still challenging to get maximum genome-editing efficiency with fewer or no off-targets. Here, we focused on factors affecting the genome-editing efficiency and precision of CRISPR/Cas system along with its defense-mechanism, orthologues, and applications.
Keywords: CRISPR/cas system, classification, host DNA repair, genome-editing efficiency, epigenome
1 Introduction
The manipulation of DNA molecules to study genes and their applications in the field of biotechnology became possible through the development of recombinant DNA technology in the 1970s. With advancements in genome engineering, it has become possible to edit the target genome at the systematic level under natural cellular conditions. The function of specific genes or regulatory elements can be studied by insertion, deletion, or modification of the associated DNA sequences. The larger scale network of genes or proteins can be interrogated by multiplex genome editing which helps in the understanding of complex polygenic disorders. The cellular organization and architecture of the genomic material and its associated functions are revealed by manipulating chromatin and transcriptional regulation. The precise manipulation enables reconstruction of biological systems with enhanced or better features, e.g., genetically modified microbes, animals, and plants. This targeted modification can be used in human gene therapy to correct harmful genetic mutations. The successful execution of these processes holds immense promise to transform various fields, such as medicine, biotechnology, and basic science.
It is difficult to manipulate billions of DNA bases in the eukaryotic genome. The first breakthrough came in terms of homologous recombination (HR) based transgene integration at the target site, but efficiency was quite low (1 in 106–109 cells) (Capecchi, 1989). This process was significantly increased by introducing site-specific DNA double-strand breaks (DSBs) (Rudin et al., 1989; Bibikova et al., 2001). However, in the absence of a repair template, DSBs result in insertion and deletion mutations (indels) via the error-prone DNA repair pathway non-homologous end joining (NHEJ) (Bibikova et al., 2002). In this regard, four classes of programmable DNA-binding proteins have been engineered, including meganucleases from microbial mobile genetic elements (Smith et al., 2006), zinc finger nucleases (ZFNs) (Urnov et al., 2005), transcription activator-like effector nucleases (TALENs) (Moscou and Bogdanove, 2009; Miller et al., 2010), and the RNA-guided DNA endonuclease Cas (Jinek et al., 2012a; Gasiunas et al., 2012). Of these, the Cas enzyme derived from the microbial adaptive immune system CRISPR (clustered regularly interspaced short palindromic repeats) is considered the most efficient, advanced, and user-friendly.
The CRISPR/Cas system can be used to target any part of the human genome associated with protospacer adjacent motif (PAM) sequence by using short guide RNA, which follows Watson–Crick base pairing to recognize the target sequence. The CRISPR story began in 1987 when a set of 29 nucleotides was found downstream of the iap gene in Escherichia coli, a product which caused the conversion of alkaline phosphatase (Ishino et al., 1987). There were five 32 nucleotide long non-repetitive sequences intervening in these 29 nucleotide repeats, a phenomenon opposite to most repetitive elements that usually take the shape of tandem repeats like those of TALE repeat monomers. During the next 10 years, genome sequencing of various bacterial and archaeal strains confirmed the presence of additional such repeat elements (Mojica et al., 2000), which were eventually named CRISPR (Jansen et al., 2002). In addition, various well-conserved CRISPR-associated (cas) gene clusters were discovered adjacent to the repeat elements that led to initial classification of the CRISPR system into three main types (types I–III) (Jansen et al., 2002; Haft et al., 2005; Makarova et al., 2011). In types I and III of the CRISPR system, multiple Cas proteins recognize and destroy target nucleic acids, while type II consists of lesser number of these proteins (Brouns et al., 2008; Hale et al., 2009).
In 2005, phage-associated and extrachromosomal origins of the intervening spacer sequences between the successive direct repeats were confirmed through a systematic analysis (Bolotin et al., 2005; Mojica et al., 2005; Pourcel et al., 2005). Further studies demonstrated that CRISPR loci can be transcribed (Tang et al., 2002) and those viruses cannot infect the host cell which have their relevant spacer sequence integrated into the host genome (detailed mechanisms are shown in Figure 1) (Mojica et al., 2005). These studies speculated the immune memory and defensive nature of CRISPR arrays against invading bacteriophages (Mojica et al., 2005; Pourcel et al., 2005). The immune nature of CRISPR loci proposed a challenging puzzle regarding the working mechanism of spacers which led to several hypotheses, including that spacers cleave target DNA at Watson–Crick base pairing region by directing Cas enzymes (Bolotin et al., 2005) or that spacers behave like RNA guides to cleave viral transcripts in a mechanism similar to that of RNAi (Makarova et al., 2006).
FIGURE 1.
Detailed mechanism of CRISPR immunity in bacteria. (A) Protospacer acquisition: (I) Type I naive adaptation involves nuclease/helicase RecBCD in Gram-negative organisms (or AddAB in Gram-positive organisms) to generate substrate products for the Cas1-Cas2 complex. (II) Type I nuclease-inactive Cas3 (inCas3)-primed mechanism involves a conformational change in the Cse1 subunit of Cascade after imperfect recognition of the target region that recruits inCas3. The Cas3 moves along the target strand to find the suitable site where it recruits the Cas1-Cas2 complex for spacer acquisition. (III) The type I Cas3 mechanism allows Cascade to recognize foreign DNA in a PAM-dependent manner and to recruit Cas3 to generate substrate products for the Cas1-Cas2 complex. (IV) Type I Cas3-independent mechanism employs the inherent ability of the Cas1-Cas2 complex to recognize the target in a PAM-dependent manner. (V) Type II adaptation utilizes the PAM-interacting domain (PID) of Cas9 (loaded with tracrRNA) to guide the Cas1-Cas2 complex (along with accessory protein Csn2) for retrieving protospacers. (B) Spacer integration into the CRISPR array: (I) The Cas1-Cas2 complex guides the protospacer (3′-OH) to execute 1st nucleophilic attack at the leader end (L) of the first repeat (R). This bends the repeat DNA to help protospacer in executing 2nd nucleophilic attack at the spacer end (S) of the same repeat. As a result of these cleavage-ligation reactions, a double-stranded protospacer is bound to single-stranded repeat sequences via its 3′ ends. The gaps are filled and repaired via polymerases and ligases. (II) The type I system allows integration host factor (IHF) to bend DNA after binding to the conserved sequence in the leader region, which allows the Cas1-Cas2 complex to interact with the leader and IHF to perform the cleavage-ligation reaction. In the type II system, recognition of the leader anchoring sequence (LAS) by Cas1 is sufficient to execute polarized integration of the spacer. (C) Sequential phage interference: CRISPR immunity consists of three steps. The acquisition step involves integration of the new spacer (red) between the two repeat elements (black) with the help of the Cas1-Cas2 complex. crRNA synthesis step involves the transcription of the CRISPR array from the leader sequence (white) to make pre-crRNA which is further converted into a series of crRNAs. During the interference step, crRNA is assembled with the Cas protein (blue) to make the effector complex that targets and cleaves the complementary sequence in the genome of the attacking phage.
The first experimental proof for a natural, nucleic acid based-adaptive immune role of the type II CRISPR system was revealed while working with Streptococcus thermophiles, a bacterial strain used in the dairy industry (Barrangou et al., 2007). A series of studies revealed the functional mechanism of adaptive immunity conferred by all three types of CRISPR loci (Figure 1). In the type I CRISPR system of E. coli, spacers containing small crRNAs are produced by transcription of CRISPR arrays in which the spacer region guides the Cas protein for its nuclease activity (Brouns et al., 2008). In the type III-A CRISPR system of Staphylococcus epidermidis, Cas enzymes block plasmid conjugation by targeting DNA rather than RNA (Marraffini and Sontheimer, 2008). However, a different type III system (type III-B) in Pyrococcus furiosus also revealed the RNA cleaving ability of crRNA (Hale et al., 2009; Hale et al., 2012). The importance of PAMs was revealed by analyzing the circumvention in CRISPR interference because of a mutation in the PAM region (Bolotin et al., 2005; Deveau et al., 2008). However, the type III system requires mismatches between the target DNA and the 5′ end of crRNA for plasmid interference (Marraffini and Sontheimer, 2010).
Until now, the CRISPR/Cas system has been classified into six types (type I–VI) based on their signature genes which are grouped into two main classes depending upon the nature of the effector complexes (Figure 2) (Makarova et al., 2011; Makarova et al., 2015; Shmakov et al., 2015). The types included in the class I system (types I, III, and IV) are composed of effector complexes with multiple subunits while those included in the class II system (II, V, and VI) are composed of effector complexes with a single subunit (Makarova et al., 2015; Shmakov et al., 2015). The first discovered and most studied types include types I–III (as mentioned above), while types IV–VI were discovered afterwards (Makarova et al., 2015; Makarova and Koonin, 2015; Shmakov et al., 2015). In the type I system, Cas3 is the signature protein that cleaves the target DNA, recognized by the multiprotein-crRNA complex Cascade (CRISPR associated complex for antiviral defense), with the help of its helicase and nuclease domains. The type II system uses its signature protein, Cas9, for interference. The type III system assembles its signature protein, Cas10, into a cascade-like interfering complex to find and destroy the target. The uncharacterized protein of the type IV system, Csf1, is suggested to be the part of the cascade-like complex, however these systems often exist alone as cas genes without any allied CRISPR array (Makarova and Koonin, 2015). A single Cas9-like nuclease in the type V system might be either Cpf1, C2c1, or C2c3 depending upon the subtype (Zetsche et al., 2015a; Shmakov et al., 2015). The type VI system has a single large protein, C2c2, with two HEPN (higher eukaryotes and prokaryotes nucleotide binding) RNase domains (Shmakov et al., 2015).
FIGURE 2.
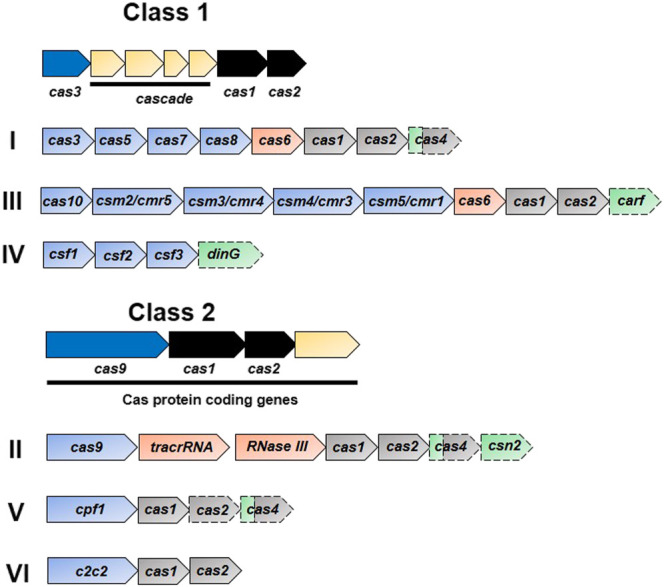
Functional organization of CRISPR systems. The CRISPR/Cas system has two main classes depending upon the nature of the effector nuclease complexes: class 1 has a multi-protein complex while there is single main protein in class 2. Each class is further subdivided into three main types based upon the signature and complementary genes. Representative operons specific to each type are shown in the figure. Dispensable elements are represented by a dashed outline, while the two colors for cas4 indicate the involvement of gene product in two stages. Genes involved in the interference, crRNA-synthesis, adaptation, and accessory role are represented in blue, orange, black, and green, respectively.
2 Era of Genome Editing
Application of the CRISPR tool in genome editing began after discovery of the basic components of the native type II CRISPR system. Cleavage of the target DNA in S. thermophiles is mediated by only the Cas9 enzyme among all members of the cas gene cluster (Figure 2) (Garneau et al., 2010). Later, noncoding trans-activating crRNA (tracrRNA) was discovered as a key component involved in generating and processing the crRNA, which facilitates RNA-guided targeting of the Cas9 enzyme after hybridization with the crRNA (Deltcheva et al., 2011). This hybrid of tracrRNA and crRNA combines with Cas9 and endogenously expressed RNase III to process transcripts of the CRISPR array into mature crRNA (Deltcheva et al., 2011). These studies revealed the minimum essential components (Cas9, crRNA, and tracrRNA) required to work in the type II CRISPR nuclease system. Because of the genome editing ability of ZFNs and TALENs, Cas9 endonuclease was also thought to be exploited in the same way which started a new race afterwards.
In 2011, it was revealed that the type II CRISPR locus derived from the Streptococcus thermophilus is able to perform CRISPR interference in Escherichia coli demonstrating transferability of this technology (Sapranauskas et al., 2011). The biochemical characterization of the Cas9 purified from Streptococcus pyogenes and S. thermophilus revealed that this enzyme cleaves target DNA after being guided by the crRNAs (Gasiunas et al., 2012). Moreover, in vitro cleavage of the target DNA is performed by a single guide RNA (sgRNA) which is formed by fusing the target DNA-specific crRNA and tracrRNA (Figure 2) (Jinek et al., 2012a).
By 2013, two simultaneous studies revealed the successful engineering of a type II system derived from S. pyogenes (Cong et al., 2013a; Mali et al., 2013b) and S. thermophiles (Cong et al., 2013a) to perform genome editing in mammalian cells. Homology directed repair (HDR) or NHEJ-mediated genome editing is stimulated in the mammalian cell genome after Cas9-mediated cleavage (details in Figure 3); this cleavage is directed by the heterologous expression of sgRNA (Cong et al., 2013a; Mali et al., 2013b) as well as a mature hybrid of crRNA-tracrRNA (Cong et al., 2013a). This system can also be used to target various genes simultaneously using multiple guide RNAs. Since then, the CRISPR/Cas9 system has been applied to various experimental models for genome editing by multiple laboratories (Sander and Joung, 2014).
FIGURE 3.
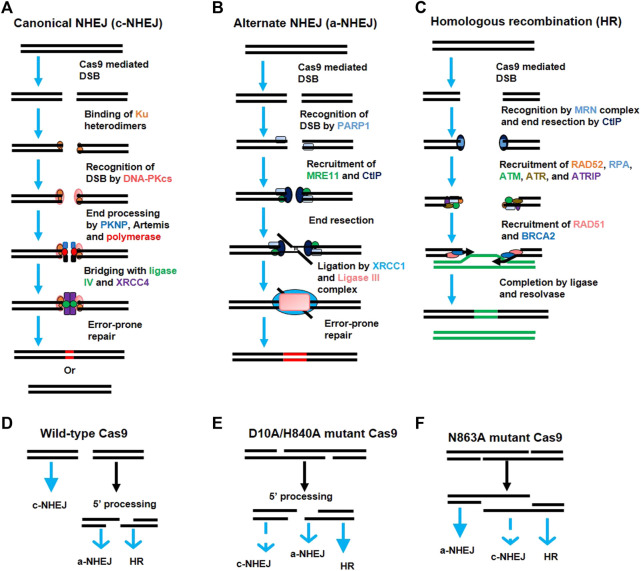
Activation of various DNA repair pathways after generation of double-strand breaks (DSBs) by Cas9. (A) The canonical non-homologous end joining (c-NHEJ) pathway. Broken DNA ends are recognized by the Ku heterodimer (Ku70 and Ku80), which recruits the DNA-PK catalytic subunit (DNA-PKcs). DNA-PKcs recruits various proteins like PKNP, Artemis, and polymerase for end processing. The ends are rejoined by ligase IV and XRCC4 with error-prone repair. (B) Alternate non-homologous end joining (a-NHEJ) pathway. The DSBs are recognized by the PARP1 protein which recruits MRE11 and CtlP for end resection. The internal microhomologies are associated with the larger deletions at junctions than that of c-NHEJ. Ligase III and XRCC1 ligate the strand ends. (C) The homologous recombination (HR) pathway. The DSBs are recognized by the MRE11-RAD50-NBS1 (MRN) complex which activates the DNA damage response via ATM kinase. The single-strand DNA (ssDNA) is formed after 5′ to 3′ end resection by CtlP which allows RAD52 and DNA replication protein A (RPA) to recognize the exposed ssDNA. This activates the Ataxia Telangiectasia and Rad3-related protein (ATR) to assist in HR repair. The RPA-coated ssDNA is replaced by the BRCA2 and RAD51 protein which performs strand invasion via searching for the homologous sequence. The junction is resolved and the ends are joined via resolvase and ligase. (D–F) Choice of DNA-repair pathway after DSBs. The predominance of a particular DNA-repair pathway (bold blue arrow) depends upon the type of lesion generated by a particular variant of Cas9.
By keeping the necessity of each of the three components (Cas9, crRNA, and tracrRNA) under consideration, researchers have reduced it to two components via making sgRNA (crRNA + tracrRNA). This conversion has made this tool more user-friendly for transcriptional control, genome editing, imaging, and RNA targeting. It has allowed it to be used in various types of cells and organisms ranging from stem cells and primary human T-cells to bacteria, fungi, plant, mice, and monkeys (Jiang and Marraffini, 2015; Sternberg and Doudna, 2015; Lin et al., 2017; Li et al., 2018). Cas9 has been used to produce various light-and chemical-inducible constructs for better spatiotemporal control as well as to employ orthologues of smaller sizes and different PAMs for easier packaging in adeno-associated virus vectors and broader targeting, respectively (Nihongaki et al., 2015a; Zetsche et al., 2015b; Davis et al., 2015; Polstein and Gersbach, 2015; Ran et al., 2015; Havlicek et al., 2017; Edraki et al., 2018; Shao et al., 2018).
Although other interference complexes have the potential to be used for genome manipulation, their multiple-subunit cascade composition makes them less suitable for genome editing unlike Cas9. However, their ability to bind stably has been employed for transcriptional silencing in E. coli (Rath et al., 2014). The Pyrococcus furiosus and Sulfolobus solfataricus derived Cmr system has been engineered to target various RNA substrates; however, targeting in mammalian cells has not been reported (Hale et al., 2009; Hale et al., 2012; Hale et al., 2014; Li et al., 2015a; Li et al., 2017). Three Cpf1 homologs have been validated in various cells for genome editing (Zetsche et al., 2015a; Kim et al., 2016; Zhang et al., 2017; Świat et al., 2017). The recognition of PAM different from those of Cas9 and the generation of staggered ends after cutting may facilitate the application of Cpf1 for genome editing by involving different DNA repair pathways. However, further investigation is needed to validate the off-targets and efficiency. The properties of various orthologues and engineered enzymes associated with CRISPR/Cas system are listed in Supplementary Table S1.
The genome editing process mediated by the CRISPR/Cas system depends upon various factors. Until now, many different ways have been adopted to increase genome-editing efficiency with minimized off-targets. This review describes such practical improvements so that researchers can choose the best conditions to achieve maximum on-target efficiency. The factors affecting various CRISPR applications are summarized in Figure 4 with the details mentioned below.
FIGURE 4.
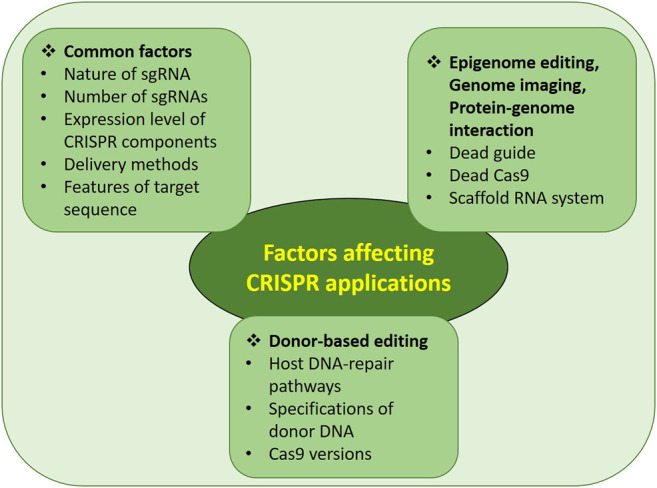
Factors affecting efficiency and specificity of CRISPR/Cas system.
3 Factors Affecting Most CRISPR Applications
3.1 Nature of sgRNA
3.1.1 Origin and GC Content
Genome editing efficiency using spacers varies depending upon the genomic region from where they were derived. Editing is highest for promoter regions followed by the exonic regions and intronic regions. This is because of the variability in retrieving spacers to the relevant target region, which further depends upon the variable GC content of the spacers (Labuhn et al., 2017), as different regions of the genome adopt various conformations based upon variation in their GC content (Pozzoli et al., 2008; Amit et al., 2012).
The GC content of the spacer region of designed sgRNA indirectly indicates the strength of the interaction between the spacer and protospacer sequences in various life domains that may affect the overall endonuclease activity of various Cas enzymes. In animals, spacers with average GC content tend to be more effective unlike those with unusually low or high GC content (Wang et al., 2014). Similarly, spacers in plants with GC content between 30 and 80% have been practically validated (Liang et al., 2016). However, lesser editing efficiency is observed in plants with 40% GC content as compared to those with higher GC content (Pan et al., 2016b). In mammalian cells, spacers with very low or very high GC content are less effective; however, 40–60% content is favorable for efficient genome editing (Doench et al., 2014; Wang et al., 2014; Liu et al., 2016c). Overall, spacers with more than 50% GC content often show high genome editing efficiency in microbes, animals, and plants (Jiang et al., 2013a; Jiang et al., 2013b; Feng et al., 2014; Wang et al., 2014; Zhang et al., 2014; Pan et al., 2016a). Inefficient sgRNAs can also be excluded by monitoring the GC content in the PAM distal region (4–13 nucleotides) (Labuhn et al., 2017). However, bulges arise between the sgRNA-DNA hybrid when GC content equals 70% (Lin et al., 2014c). DNA bulges formed due to mismatches 7–10 bp from PAM; thus, the 3′end or 5′end can result in mismatches, so these must be avoided (Lin et al., 2014b).
3.1.2 Nucleotide Preferences and Alterations
Nucleotide composition (purine or pyrimidine) of spacers may also affect Cas9 binding and nuclease activity. In animal models, it has been reported that the 3′ sequence of the spacer contributes to Cas9 binding preference, and purines are better to recruit the Cas9 enzyme than pyrimidines which eventually affects its nuclease activity (Wang et al., 2014). Doench et al., 2014) reported Cas9 preference for guanine and against cytosine at position 20; for cytosine and against guanine at position 16; for adenine in the middle and against cytosine at position three of the spacer sequence. Similarly, Xu et al. (2015a) revealed Cas9 preference for guanine at positions 19 and 20; against thymine from positions 17 to 20; for cytosine at position 18 (CRISPR/Cas9 complex DNA cleavage site); for adenines from positions nine to sixteen; and for guanines from positions four to seven. Moreover, PAM downstream nucleotides, unlike spacer upstream sequences, contribute to the editing efficiency of spacer (Doench et al., 2014; Xu et al., 2015a). The sequences rich in guanine can form noncanonical stable structures in vivo, called as G-quadruplexes (Huppert, 2008). Moreno-Mateos et al. (2015) injected in vitro transcribed guide sequences into zebrafish model and reported that sequences with more than eight guanines undergo G-quadruplex structures which are more efficient to edit due to their increased stability. However, this modification is not essential in case the guide is being expressed from constitutive promoter (like U6) after lentiviral transduction. On the other hand, these G-rich guides are not efficient in cultured mammalian cells (Malina et al., 2015). In contrast, no such nucleotide preference is observed in plants revealing a major difference in spacer designed to use in plants and animals (Liang et al., 2016).
Transcription from the U6 promoter by RNA polymerase III is inhibited when there is TTT stretch in the DNA, so guide RNAs with a UUU stretch (particularly in the seed region) are inefficient under the U6 promoter, and editing is more favorable when a stable duplex is formed between the target DNA and the guide RNA (Wong et al., 2015). Moreover, particular sequence motifs that affect the synthesis of the tracrRNA structures required for Cas9 interaction may also decrease guide activity. The presence of uridine at four nucleotide proximal to PAM makes it difficult for guide RNA to interact with Cas9 eventually resulting in lower activity (Wang et al., 2014; Hart et al., 2015).
To optimize sgRNA structure, researchers found that 1) substituting one of the T’s in four consecutive T’s with an A (to distort the string) shortly downstream of the spacer and 2) extending the sgRNA duplex region by five-nucleotides changes the sgRNA transcription rate and its structure, respectively, resulting in improved efficiency (Chen et al., 2013; Dang et al., 2015). This strategy also produced promising results with SaCas9 sgRNAs, signifying that this could be applied to other Cas9 orthologues to achieve maximum efficiency for different applications (Chen et al., 2016; Tabebordbar et al., 2016).
3.1.3 Secondary Structure
Ma et al. (2013) targeted the PVALB gene of humans with various sgRNAs and concluded that the absence of sgRNA secondary structures increases binding with the target gene, leading to more efficient genome editing. However, subsequent studies supported the existence of a secondary structure. Briefly, sgRNA consists of a crRNA sequence {guide/spacer [20 nucleotides (nt)] + repeat region [12 nt]} and a tracrRNA sequence [anti-repeat (14 nt)] and three stem loops (stem loops 1, 2, and 3). The fourth loop (called the RAR loop) is formed by the bases of repeat and anti-repeat regions that trigger RNase III-mediated processing of precursor CRISPR RNA (pre-crRNA). Stem loop 1 is important for formation of the Cas9-sgRNA-DNA functional complex while the other two loops provide stability to the complex and enhance in vivo function (Nishimasu et al., 2014). Liang et al. (2016) reported that all stem loops (except stem loop 1) must have an intact secondary structure for efficient genome editing.
3.1.4 Modified Versions
GGX20 modification of sgRNA involves addition of two additional mismatched guanines at the 5′ end of the designed guide RNA. Several studies have shown that this sgRNA modification enables them to differentiate the on-target site from other homologous sites differing by two or more nucleotides eventually increasing on-target specificity of sgRNA 10–100 times, but it also affects editing efficiency at the on-target and off-target locations (Cho et al., 2013; Fu et al., 2013; Cho et al., 2014; Sung et al., 2014; Kim et al., 2015). Moreover, only certain guides are compatible with such a modification, so there is a need to further validate this modification with different lengths and chemically modified sgRNAs.
Another specificity mediator of CRISPR/cas9 technology is the length of the designed sgRNA. Extending Spcas9 guide sequence from 20 nt to 30 nt to increase specificity results in its processing back to natural 20 nt length, so it is useless to increase length (Ran et al., 2013a). Short length (17 or 18 nt) spacer sequences, called truncated spacers or guides, have been reported to potentially increase Cas9 binding sensitivity to mismatches present within a smaller complementary sequence which causes more accurate but less active genome editing (Pattanayak et al., 2013; Fu et al., 2014). Even after eliminating off-target activity at many sites, truncated guides produce new off-target sites due to their shorter length (Fu et al., 2014; Tsai et al., 2015; Wyvekens et al., 2015; Slaymaker et al., 2016). Interestingly, these truncated guides show very low on-target efficiency when used with modified SpCas9 versions, such as eSpCas9 and SpCas9-HF (Kleinstiver et al., 2016; Slaymaker et al., 2016). Improvements in specificity need to be evaluated in other Cas9 orthologues with intact editing efficiency using these guide modifications (Hou et al., 2013; Friedland et al., 2015; Ran et al., 2015). Jinek et al. (2012b) detected efficient in vitro cleavage using truncated crRNA and tracrRNA but failed to detect cleavage at various loci previously modified using crRNA-tracrRNA duplexes with identical guide sequences (Cong et al., 2013b). Another study revealed that this cleavage difference is due to the length of tracrRNA sequence, as they used sgRNAs with tracrRNA tails extended to +67 and +85 nt to mediate cleavage at all previously tested target sites and found a five-fold increase in the level of indels than in the corresponding crRNA-tracrRNA duplex. They also observed increased expression of sgRNAs having longer tracrRNA sequences, predicting that higher sgRNA stability or expression is responsible for improved cleavage of the target sequence (Ran et al., 2013a).
In one study, full length 100 nt sgRNAs (crRNA + tracrRNA) were synthesized with three various chemical modifications of 2′-O-methyl (M), 2′-O-methyl 3′phosphorothioate (MS), or 2′-O-methyl 3′thioPACE (MSP) at the 5 and 3′ termini, which improved editing efficiency in CD34 + hematopoietic progenitor and stem cells, and human primary T-cells along with off-target activity at a few sites (Hendel et al., 2015). In another study, a 29 nt crRNA (scrRNA) phosphorothioate (PS) backbone was synthesized with chemical substitutions of 2′-fluoro (2′-F), 2′-O-methyl (2′-O-Me) and S-constrained ethyl (cEt) which increased their specificity but not on-target efficiency in human cells (compared to unmodified crRNA) due to its increased binding affinity to tracrRNA and metabolic stability (Rahdar et al., 2015). These contrasting results indicate that more exploration of chemically modified sgRNAs or only crRNAs is required to reach a conclusion.
3.1.5 Activity Scores of gRNA
The efficiency of guide RNA is difficult to predict due to its sequence dependency (Liu et al., 2016a), so a good option is to choose the best one from three tested gRNAs (Ran et al., 2013a) after confirmation by the T7E1 endonuclease assay (Larcher et al., 2014; Vouillot et al., 2015) and direct Sanger sequencing of the target sequence PCR products, particularly for knock-in experiments (Brinkman et al., 2014), DNA capillary electrophoresis of amplified products (Dahlem et al., 2012; Yang et al., 2015), or using fluorescent reporters (Kim et al., 2011). In many studies, guide efficiency correlating scores and sequence criteria have been identified which helps to reduce guide RNA number to test for genome editing. A list of online tools to predict the efficiency of sgRNA is shown in Supplementary Table S2. After predicting sgRNA using any of these tools, the highest score value should be obtained to increase efficiency and specificity.
3.2 Number of sgRNAs
The number of sgRNAs depends upon the particular application of the CRISPR/Cas system. Point mutation or knock-in can be achieved using single sgRNA along with wild-type Cas9. However, knock-in with single sgRNA and wild-type Cas9 results in many off-target effects that may destabilize genome integrity. Two sgRNAs have been used to cleave the flanking site of the target gene and replace it with a fluorescent marker to isolate the null allele in Drosophila melanogaster (Gratz et al., 2014) and Caenorhabditis elegans (Paix et al., 2015). The knock-out yield in mice can be increased to 95% by adopting a low dose triplet (three sgRNAs) rather than a high dose singlet (single sgRNA) strategy for the same target gene in oocytes (Sunagawa et al., 2016). Multiplex genome editing can be performed by simultaneously co-expressing multiple sgRNAs with Cas9. The recovery of homozygous mutants can be increased by targeting one gene with multiple sgRNAs in T0 tomato (Brooks et al., 2014) and rice plants (Xie et al., 2015; Wang et al., 2016). The doxycycline-inducible Cas9 system has been used in multiplex editing to efficiently and simultaneously delete lysine demethylase 5A, 5B, and 5C in vitro and in vivo (Cao et al., 2016).
3.3 Expression Level of CRISPR Components
The expression level of CRISPR components is associated with extension of culture period and individual expression level of sgRNA and Cas9. The proportion of mutated cells increases with extended culture time in soya bean somatic embryos (Jacobs et al., 2015) and rice callus infected with A. tumefaciens (Mikami et al., 2015). This is because of the acquisition of new mutations as well as proliferation of the existing mutants. However, the regenerative capacity of cells can be reduced by this method with an increased risk of producing chimeric plants (Xu et al., 2015b). The effect of expression level of sgRNA and Cas9 are explained below.
3.3.1 Expression Level of sgRNA
It depends upon the type of promoter and the host cell line. Most studies have employed the RNA polymerase III promoter (mostly constitutive and consisting of a few cellular promoters) for sgRNA expression which makes conditional or induced expression impossible (Orioli et al., 2012). Researchers have expressed sgRNA from artificial gene RGR which produces sgRNA mRNA with a ribozyme sequence at the flanking ends after transcription following cleavage and generation of a mature sgRNA with high in vitro and yeast genome editing (Gao and Zhao, 2014). Multiplex genome editing in human cells has been achieved using a cell-type specific promoter and the Csy-4 dependent method to form separate mature gRNAs from the same precursor mRNA to improve genome editing efficiency (Nissim et al., 2014). A synthetic hybrid promoter consisting of tRNA and RNA polymerase III has been employed in Yarrowia lipolytica to enhance gRNA expression, resulting in 100% transformants by inhibiting the NHEJ process (Schwartz et al., 2016).
The sgRNA expression level is one of the determinants for increased on-target efficiency of Cas9. Higher expression levels of sgRNA and the repair template increase genome editing in rice (Sun et al., 2016), tobacco (Baltes et al., 2014), S. cerevisiae (Stovicek et al., 2015), Yarrowia lipolytica (Schwartz et al., 2016), and mammals (Ran et al., 2013a). In contrast, higher sgRNA level limited genome editing efficiency in Arabidopsis (Ma et al., 2015b) and tomato (Pan et al., 2016a). Xie et al. (2015) reported that expression levels of Cas9 and sgRNA are lower in transgenic plants or the callus compared to the protoplast, which may affect editing efficiency. Ranganathan et al. (2014) reported less off-target activity than on-target activity in a human cell line when guide RNA was expressed from a weaker H1 promoter. These results indicate species-specific editing-efficiency based on the sgRNA expression level which needs to be further confirmed using more advanced approaches.
3.3.2 Expression Level of Cas9
Specificity and kinetics of gene editing can be affected by the level of Cas9 protein (Ran et al., 2013a; Pattanayak et al., 2013; Fu et al., 2014). For example, five-time drop in the level of Cas9 protein increased its specificity seven-times by affecting the on-target efficiency for just two-times (Ran et al., 2013a). Despite extensive application of CRISPR/Cas9 technology in plants for genome editing, there is variation in editing efficiencies, as the Cas9 expressing promoter, Cas9 codon optimization, and the positional effect can directly affect the Cas9 expression level and targeting efficiency (Wang et al., 2015a; Ma et al., 2015b; Yan et al., 2015; Mao et al., 2016). Constitutive Cas9 expression results in higher genome editing in mice (Platt et al., 2014), mammalian cells (Koike-Yusa et al., 2014), S. cerevisiae (DiCarlo et al., 2013), and rice (Mikami et al., 2015). In contrast to rice and some other higher plants, a higher Cas9 level reduces editing efficiency in mosaic Arabidopsis (Mao et al., 2013; Feng et al., 2014; Jiang et al., 2014; Ma et al., 2015b; Yan et al., 2015) and toxicity in Chlamydomonas reinhardtii (Shin et al., 2016) while no toxicity was observed in Aspergillus fumigatus with Cas9 constitutive expression (Fuller et al., 2015). Moreover, reduced search efficiency of Cas9 for heterochromatic regions indicates that access to DNA target site also contributes to specificity and efficiency (Knight et al., 2015).
Besides constitutive Cas9 expression, induced (chemical and light) and tissue-specific expression has also been evaluated in various studies to improve genome editing efficiency and specificity. In the case of CRISPR/Cas9 technology, chemically induced expression is achieved by using either an inducible promoter (e.g., doxycycline) or a Cas9 variant. Doxycycline-inducible Cas9 (iCas9) has been reported in vivo by genetic screening studies in humans (Wang et al., 2014); genome editing studies in mice (Dow et al., 2015); and human iPSCs (Zhu et al., 2015). This iCas9 system has also been reported in vitro for cell lineage specific reprogramming studies in a mouse cell line (Chakraborty et al., 2014); genomic loci imaging studies in human cell lines (Chen et al., 2013); reversible gene silencing studies in human iPSCs (Mandegar et al., 2016); multiplexed gene activations studies in cells and zygotes (Cheng et al., 2013); and biallelic gene knockout studies in hiPSCs (González et al., 2014). Cas9 variants have been developed to induce their activity in the presence of small cell-permeable 4-hydroxytamoxifen (4-HT) by combining Cas9 with either 4-HT responsive intein (Davis et al., 2015) or hormone-binding domain of the estrogen receptor (ERT2) (Liu et al., 2016b). Similarly, a light induced Cas9 system has been validated in mammalian cells to study: 1) transcriptional regulation using the p65-CRY2/dCas9-CIB1 construct (Nihongaki et al., 2015b) and the VP64-CRY2/CIB1-dCas9-CIB1 construct (Polstein and Gersbach, 2015) and 2) optogenetic control of genomic editing using the Cas9N713-pMag/nMagHigh1-Cas9C714 construct (named photoactivatable Cas9 (paCas9-1)) (Nihongaki et al., 2015a). The p65-CRY2/dCas9-CIB1 photoactivatable system constitutes sgRNAs and two fusion proteins. The first fusion protein constitutes dCas9 and CIB1; which acts like a genomic anchor probe and binds to target sequence with the help of sgRNAs. The second fusion protein constitutes transcriptional activator domain and photolyase homology region of CRY2 (CRY2PHR) which acts like an activator probe. The CIB1 and CRY2PHR are heterodimerized with the blue light irradiation and transcription is activated with the recruitment of activator domain to the target locus (Nihongaki et al., 2015b). Similarly, VP64-CRY2/CIB-dCas9-CIB1 construct constitutes light-inducible heterodimerizable proteins CRY2 and CIB1 attached to transactivation domain (VP64) and either C- or N-terminal catalytically inactive Cas9 (dCas9), respectively. This system is also directed to the target site with the help of sgRNAs and activates the transcription with blue light (Polstein and Gersbach, 2015). The Cas9N713-pMag/nMagHigh1-Cas9C714 construct constitutes photoinducible dimerization domains (named as Magnets) and split Cas9 fragments. Upon blue light irradiation, paCas9 induces genome editing by involving both homology-directed repair and nonhomologous end joining (Nihongaki et al., 2015a).
To improve spatial genome editing, tissue-specific promoters are being used in zebrafish (Ablain et al., 2015), drosophila (Xue et al., 2014b), mammalian cells (Yoshioka et al., 2015), and plants (Osakabe et al., 2016). Similarly, the mosaic effect in plants is reduced by using specialized promoters such as INCURVATA 2 promoter (Hyun et al., 2015), the meristem-specific YAO promoter (Yan et al., 2015), the germline-specific SPL promoter (Mao et al., 2016), the egg-cell-specific DD45 (Mao et al., 2016), and EC1.2 promoters (Wang et al., 2015b). For effective Cas9 expression in human iPSCs, EF1α promoter is proved stronger than CAG promoter (Matsui et al., 2014), while CMV and SV40 are not recommended due to transcriptional silencing (Hotta and Ellis, 2008). Programmable and multiplexed regulation of various gene networks in human cells is achieved by combining the RNA regulatory strategies with Cas9 based transcription factors (Nissim et al., 2014). For example, immune responses against transgene products can be minimized by using the mir-142-3p which can repress the expression of associated cellular transcripts by binding to their target sequences (Majowicz et al., 2013).
3.4 Delivery Methods
3.4.1 For Cas9 DNA
Cas9 is a sequence-specific endonuclease that is delivered to the host cell or organism as DNA, mRNA, or protein for a genome editing experiment. The first human genome editing was performed in 2013 in which all CRISPR components were delivered in the form of DNA plasmids and/or expression cassettes (Mali et al., 2013b). With advancements in the field, various vectors and methods have been attempted to achieve maximum editing efficiency and precision. For example, five-fold more HDR efficiency was achieved with a particle bombardment technique in rice (Sun et al., 2016) and maize (Svitashev et al., 2015) compared to Agrobacterium-mediated transformation. Enhanced genome editing was observed by 1) introducing components using adenovirus vector (AV) or adeno-associated vector (AAV) in mice (Senís et al., 2014; Swiech et al., 2015; Rodriguez et al., 2016) and a lentivirus into melanoma cell lines (Shalem et al., 2014) and 2) using the hydrodynamic injection technique in mice (Xue et al., 2014a). Mostly transfection methods such as electroporation or lipofectamine are used to transfer CRISPR components into host cell (Costa et al., 2007) but nucleofector is commonly used for human iPS cells genome editing (Ran et al., 2013b).
AAV vectors are common delivery tools due to safety and efficiency (Kaufmann et al., 2013; Kotterman and Schaffer, 2014; Kotterman et al., 2015). One problem is the size of the commonly used SpCas9 gene, which cannot be accommodated in wild type AAV, so smaller orthologues (Esvelt et al., 2013) or engineered AAV (Kotterman and Schaffer, 2014) are used as a solution. Moreover, orthologues have been developed to simultaneously perform multiplexed RNA-guided transcriptional repression, activation, and gene-editing (Esvelt et al., 2013). Similarly, AAV vector has shown some efficacy in multiple monogenic disorders including choroideremia (MacLaren et al., 2014), haemophilia B (Nathwani et al., 2011), and Leber’s congenital amaurosis type 2 (Hauswirth et al., 2008; Bennett et al., 2012). Lentiviral vectors have been used to study 1) drug resistant and cell viability genes via loss-of-function mutation in mouse and human cells (Koike-Yusa et al., 2014; Shalem et al., 2014; Wang et al., 2014; Zhou et al., 2014); 2) leukemia causing genes in mouse hematopoietic cells (Heckl et al., 2014); 3) the role of p53 and Pten in the formation of liver tumors using mouse 3T3 cells (Xue et al., 2014a) and 4) Kras gain-of-function mutation (KRAS G12D) in the Neuro-2a neuroblastoma cell line (Platt et al., 2014).
3.4.2 For Cas9 mRNA and Protein
Cas9 and sgRNA show persistent expression when delivered in the form of a cloned vector (Xue et al., 2014a; Yin et al., 2014; Yang et al., 2016); however, it might cause mutation with random integration of CRISPR components at on-target and off-target sites into the host genome (Cradick et al., 2013; Fu et al., 2013). The Cas9mRNA/sgRNA or Cas9protein/sgRNA is injected into the host embryo or zygote for specific genome editing. For example, the injection of sgRNA and Cas9mRNA causes 1) targeted mutagenesis in 88% of embryonically injected flies with a 33% transmittance rate (Bassett et al., 2013), 2) generation of one-step multiple allelic mutated mice via zygotic injection (Wang et al., 2013), 3) correction of mouse crygc allele (Wu et al., 2013) and dystrophin gene (Long et al., 2014) via zygotic injection, and 4) correction of human beta-hemoglobin gene via tripronuclear zygotic injection (Liang et al., 2015). Similarly, genome editing efficiency and specificity can be increased by 1) using the Cas9 protein and sgRNA delivered by electroporation (Kim et al., 2014), protein transduction (Ramakrishna et al., 2014), and lipofection (Zuris et al., 2015), 2) using the MS-modified sgRNA and Cas9 protein complex (Hendel et al., 2015), 3) synchronizing the delivery of ribonucleoprotein (RNP) with the stage of cell cycle in human cells (Lin et al., 2014a) and 4) using a solubilizing salt solution for the fluorescent Cas9/sgRNA complex in zebrafish embryos (Burger et al., 2016). The mixture of Cas9 mRNA and gRNA (Zhang et al., 2016b) or pre-assembled mixture of RNP (Woo et al., 2015; Svitashev et al., 2016; Liang et al., 2017) can be delivered to plant cells for efficient genome editing. These delivery methods are summarized in Supplementary Table S3.
3.5 Features of Target Sequence
3.5.1 GC Content
GC content is related to genome size in bacteria, but it is very complex in eukaryotes due to repetitive DNA segments and isochores. There is huge variation in GC content among microbes but less of a range in plants and animals, e.g., monocots and vertebrates have higher GC content than dicots and invertebrates, respectively. There is more GC content in animal chromosomes than that in plants but no correlation between chromosome size and GC content has been observed (Li and Du, 2014). In the case of CRISPR/Cas technology, target sequence GC content affects genome editing efficiency (Zhang et al., 2014; Ma et al., 2015b). For example, higher GC content (>70%) facilitates hybridization between the spacer and the protospacer, which may increase off-targets (Lin et al., 2014b; Tsai et al., 2015; Li et al., 2016). GC content lower than 30% also results in higher off-targets (Fu et al., 2013; Pattanayak et al., 2013). However, no off-targeting was observed with GC content of 57% in soybeans (Jacobs et al., 2015); 50% in Arabidopsis (Sauer et al., 2016); and 50–70% in rice (Ma et al., 2015b).
3.5.2 Stem Loop Structure
The genomic sites of the host target sequence that pair six continuous nucleotides with the sgRNA scaffold should not be selected because it results in the formation of a stem loop structure between the sgRNA scaffold and target sequence, which hinders perfect binding with the target sequence ultimately leading to less efficient genome editing (Ma et al., 2015b).
3.5.3 Chromatin State and Strand Preference
DNA exists in two chromatin states, i.e., euchromatin (less condensed) and heterochromatin (highly condensed); however, accessibility to the latter is a bit difficult. Thus, Cas9 targeting a DNA site in the highly condensed portion leads to decreased binding (Wu et al., 2014; Verkuijl and Rots, 2019), which may be due to less accessibility to the PAM sequence (Hinz et al., 2015). The heterochromatin state of the target sequence can reduce the diffusion of Cas9 (Knight et al., 2015) and CRISPR-Cas9 mediated mutagenesis by 7-fold (Kallimasioti-Pazi et al., 2018). The presence of epigenetic modifications or transcription factors may also affect Cas9 binding following genome editing. It has also been observed that sgRNAs targeting non-transcribed DNA strands are more effective compared to those targeting transcribed DNA strands (Wang et al., 2014). The cleavage efficiency of SpCas9 is independent of methylation of the target region (Ran et al., 2013a).
4 Factors Contributing to Donor-Based CRISPR Editing
4.1 Host DNA-Repair Pathways
The genome is continuously facing damages by various factors, ranging from metabolic byproducts to UV radiation. These factors affect the target genome by their own ways. One of them is the breakage of DNA phosphodiester backbone in either single strand or both strands (DSBs). These DSBs are mainly repaired by the NHEJ rather than HDR DNA-repair pathway (Figure 3). Integration of foreign DNA into the target genome by CRISPR/Cas technology is based on the principle of homologous recombination which is least prominent in the presence of NHEJ (as explained earlier); thus, either inhibiting NHEJ or stimulating HDR may lead to improved knock-in efficiency. To improve donor-based editing, various researchers have tried to modify the priorities of these naturally competing pathways. For example, the NHEJ pathway can be inhibited by inhibiting key components of the NHEJ pathway, such as DNA ligase IV, KU80, or KU70 in plants and animals (Chu et al., 2015; Maruyama et al., 2015; Endo et al., 2016). Stimulating the HDR pathway using RS-1 (HDR enhancer) also led to a two-to five-fold improved gene knock-in at various loci (Song et al., 2016). Yu et al. (2015) revealed that certain small chemical molecules can modulate the NHEJ and HDR pathways to improve genome editing. Moreover, NHEJ occurs throughout the cell cycle (Panier and Boulton, 2014) whereas HDR happens only during the G2 and S phases (Mali et al., 2013b; Wang et al., 2013; Yang et al., 2013), so transgenic Cas9 expression in meiotic cells led to high knock in efficiency due to higher HDR than NHEJ during gene drive experiments conducted in mosquitos (Gantz et al., 2015).
About 90% of DSBs generated in rapidly growing mammalian cells by ionizing radiation and Cas9 are repaired within 1 h (Metzger and Iliakis, 1991) and 15 h (Kim et al., 2014), respectively. This finding suggests a long lifetime (approximately 6 hour) of the Cas9-DNA complex. A close analysis of this complex revealed asymmetric cleavage and release of the 3′end of the non-targeted DNA strand by sgRNA. Richardson et al. (2016) achieved an improved knock-in efficiency in human cells using single stranded DNA (ssDNA) donors complementary to the first released strand. The donor-based knock-in can also be executed by the microhomology-mediated end joining (MMEJ) pathway (Figure 5) which is explained below.
FIGURE 5.
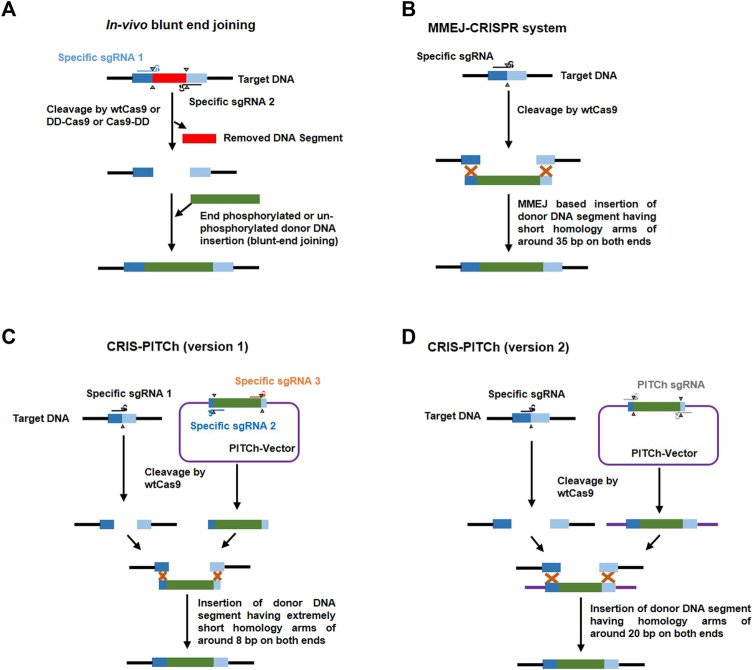
Representative methods in donor-based host genome editing. (A) In vivo blunt-end joining involves using two target specific sgRNAs (blue and black) to cleave target DNA [releasing a DNA segment (red)] with either wildtype cas9 (wtCas9) or Cas9 attached to the protein destabilization domain (DD) like FKBP12-L106P which destabilizes Cas9 in the absence of a stabilizing agent, such as Shield-1 (Clontech); it leads to precise knock-in of donor DNA (green) via non-homologous end joining (NHEJ). (B) The MMEJ-CRISPR system involves cleavage of target DNA using one specific sgRNA (black) and wtCas9; and integration of a donor DNA fragment (with around 35 bp homologous arms at both ends (dark and light blue) at the cut site through the process of microhomology-mediated end joining (MMEJ) during the M-early S phases of the cell cycle. (C) CRIS-PITCh version 1 involves three specific sgRNAs [one to cleave target DNA (black) and the other two (yellow and brown) to cleave the DNA sequence flanking outside the homologous sequence (8 bp) in the donor vector (PITCh-vector) which releases a donor DNA segment with homologous arms (dark and light blue)] and wtCas9 to precisely integrate any gene of interest without end trimming by proximal MMEJ. (D) CRIS-PITCh version 2 also involves three sgRNAs [one specific sgRNA (black) for host target genome cleavage and two general PITCh sgRNAs (grey) to cleave the DNA sequence flanking outside the homologous sequence (20 bp) in the donor vector (PITCh-vector) which releases a donor DNA segment with homologous arms (dark blue and light blue)] and wtCas9 to precisely integrate any gene of interest with end trimming by distal MMEJ.
4.2 Specifications of Donor DNA
In CRISPR research so far, donor DNA has been used either in the form of a plasmid or single-stranded oligodeoxyribonucleotides (ssODNs), each having its own efficiency in particular target species. For example, using a donor in the form of ssODNs instead of a plasmid resulted in HDR efficiency from 10 to 80% in mice (Yang et al., 2014). In plants, ssODN donor showed ≈5% gene conversion (unidirectional interchange between similar sequences) efficiency in Arabidopsis (Sauer et al., 2016); a plasmid donor showed 9% gene replacement efficiency in Nicotiana benthamiana (Li et al., 2013); a ssODN donor (ssOligo2; 127 nt) showed 0.4% (4/1,000) mutation frequency in maize compared to 0.2% (2/1,000) using a plasmid donor (794 bp) (Svitashev et al., 2015); insertion frequency of ≈1 kb homologous arms containing the expression cassette was 4% in maize (Svitashev et al., 2015) and 4.6% in soybean (Li et al., 2015b); and the gene-replacement frequency in Arabidopsis was 0.8% using 733 and 825 bp homologous arms containing a 3.9 kb donor plasmid (Zhao et al., 2016). Moreover, increased editing efficiency and longer (100 bp) donor insertions have been achieved at ROSA loci of rats and mice using phosphorothioate-modified ssODNs (Renaud et al., 2016).
Normally, 700–1,000 bp length of a homologous sequence should be added to both sides of the donor cassette and the added sequence must be adjacent to but not include the sgRNA target site (Yusa, 2013). Modified systems have been developed for gene knock-in at the target region using donors with small homologous sequences or those without any such sequences. For example, Zhang et al. (2016a) introduced MMEJ of the donor construct into the host target genome and named it the MMEJ-CRISPR system. This system is active only during the M and early S phases when HR is inactive (Taleei and Nikjoo, 2013). They added approximately 35 bp of a homologous sequence to donor and achieved 95–100% accuracy in A. fumigatus and also demonstrated that this system is not dependent on the ku80 pathway (Figure 3). Sakuma et al. (2016) exploited the MMEJ phenomenon to introduce a more user-friendly donor based genome editing system known as CRIS-PITCH [two versions, i.e., CRIS-PITCh (v1) and CRIS-PITCh (v2)]. These PITCh vectors require a few days to construct and can successfully complete the knock-in process within 1 week in frog embryos and within 1.5 months in human cell lines. Geisinger et al. (2016) achieved highly precise genome editing in mouse and human cells via in vivo blunt-end cloning with the use of wt-Cas9 and Cas9 attached to the protein destabilization domain (FKBP12-L106P). Schematic illustrations of the MMEJ-CRISPR system, the CRIS-PITCH system and in vivo blunt end cloning are shown in Figure 5.
4.3 Cas9 Versions for Specificity Improvement
The target-specific sgRNAs may show some off-target effects that disturb the overall stability of the genome and knock-in of the donor cassette. This occurs because of the two endonuclease domains of the wild-type Cas9 enzyme; each cuts the opposite strand of the DNA resulting in DSBs. To minimize these hazardous effects, one of the domains is inactivated by a mutation (D10A or H840A) to form nickase Cas9 (nCas9). A nick is generated on opposite strands of two closely located target sites using pairs of sgRNAs and nCas9, which minimizes the off-targets by maintaining on-target cleavage efficiency (Cong et al., 2013a; Mali et al., 2013a). The paired Cas9 nickases recognize and cleave four 100 bp apart target sites on opposite strands (Ran et al., 2013a; Shen et al., 2014). Although some studies have reported fewer indel mutations using nCas9 with single sgRNA, paired sgRNAs with nCas9 have been observed with reduced off-targeting in human cells and mice (Sander and Joung, 2014). The gRNAs in the PAM-out orientation have more efficiency than those in the PAM-in orientation. Similarly, D10A nCas9 is more efficient than H840A nCas9 (Ran et al., 2013a).
The off-targets of monomeric nucleases can be minimized by using the dimerization-dependent RNA-guided FokI-dCas9 nucleases (RFNs) in which dCas9 is fused with FokI nuclease to form fCas9. This fCas9 uses a pair of sgRNAs targeting opposite strands of DNA with targets separated by 15–25 bp in a “PAM-out” orientation. This system shows a dramatic increase in specificity with comparable efficiency to that of nickase Cas9, which is two-thirds that of wild-type Cas9 (Guilinger et al., 2014; Tsai et al., 2014). Genome editing specificity can also be increased by combining two orthogonal approaches (independent approaches adopted to conclude via getting same or different results). For example, using tru-RFNs, made by combining truncated gRNAs (trugRNAs) with RFNs, results in increased specificity and efficiency in embryonic stem cells and human cancer cell lines (Wyvekens et al., 2015). Despite increased specificity, these methods are relatively complex and require more combination of guide RNAs that may have to be evaluated.
The precision in target recognition can also be enhanced by the fusion of programmable DNA-binding domain (pDBD) of other nucleases such as TALENs or ZFNs. For example, SpCas9MT3-ZFPTS2:TS3:TS4 was constructed by linking the mutated (R1335K) Cas9 (SpCas9MT3) with programmed zinc-finger protein (ZFPTS2:TS3:TS4), to target the closely spaced genomic DNA sites. The SpCas9MT3 version of Cas9 prefers NGN PAM while ZFPTS2:TS3:TS4 constitutes three different ZFPs to recognize sequence surrounding the guide RNA target sites 2, 3, and 4. This engineered Cas9 provides 150-fold increased specificity because of cooperative association between the two separate DNA binding events like the above-mentioned nickases and FokI (Bolukbasi et al., 2015). However, these strategies are less applicable because they require larger transgenes and additional components.
Recently, rational engineering of SpCas9 and SaCas9 has been done based on their crystal structures; the engineered versions showed enhanced specificity and thus named as eSpCas9 and eSaCas9, respectively. In eSpCas9 and eSaCas9, three and four positively charged residues of non-targeted DNA strand groove were neutralized by replacing them with alanine, respectively (Slaymaker et al., 2016). These mutations allow competitive rehybridization of DNA with the invasion of gRNA on target strand by weakening the protein binding on non-target strand. This increase in stringency between RNA and DNA matching dramatically reduced the genome-wide off-targeting. Surprisingly, on-target efficiency of eSpCas9 was comparable to that of wild-type SpCas9. A similar strategy was followed by another research group to design “high-fidelity SpCas9” (SpCas9-HF1) in which residues interacting with phosphate backbone of target DNA strand have been substituted with four alanine residues, unlike eSpCas9 (Kleinstiver et al., 2016). The on-target efficiency of SpCas9-HF slightly varies for tested guides as compared to that of eSpCas9; but, it is comparable (>70%) to that of wild-type (86%). Similarly, high-fidelity (HiFi) Cas9 variant was identified via unbiased bacterial screening approach in which a single point mutation (R691A) caused reduced off-targeting while maintaining higher on-target activity as a RNP complex (Vakulskas et al., 2018). Along with these, few more high-fidelity SpCas9 variants have been developed such as FeCas9 (Yin et al., 2019), evoCas9 (Casini et al., 2018), HypaCas9 (Chen et al., 2017), SpCas92Pro (Babu et al., 2019) LZ3 (Schmid-Burgk et al., 2020), and Sniper-Cas9 (Lee et al., 2018).
Another approach to engineer Cas9 is alteration in its PAM recognition sequence. This might increase the number of genome-wide targets as well as improve specificity by requiring less-abundant PAM or PAM with longer sequence across the genome. One strategy is to replace PAM-interacting domain (PID) of recipient with that of ortholog which recognizes a different PAM sequence. This has been done for Streptococcus thermophiles CRISPR-3 Cas9 (St3Cas9) and SpCas9 which retained their genome editing function (Fonfara et al., 2013; Nishimasu et al., 2014). Continuous phage-assisted evolution of SpCas9 generated a SpCas9 variant (xCas9) which can recognize broader range of PAM sequences including NG, GAT, GAA (Hu et al., 2018). Directed evolution has also been used to change the PAM specificity of SaCas9 (Kleinstiver et al., 2015a) and SpCas9 (Kleinstiver et al., 2015b). Remarkably, only four mutations brought about engineered “VRER SpCas9” with specificity for the PAM sequence “NCGC” which has 23 times less abundance in human genome than NGG. Directed evolution caused a point mutation (D1135E) in SpCas9 which increased its specificity for NGG over NAG PAM (Jiang et al., 2013a; Hsu et al., 2013; Kleinstiver et al., 2015b). In case of SaCas9, targeting range is increased by 2–4 fold after modifying its PAM from NNGRRT to NNNRRT via directed evolution (Kleinstiver et al., 2015a). Few more PAM-flexible variants have also been reported including SpCas9-NG (Nishimasu et al., 2018), SpG and SpRY (Walton et al., 2020). Overall, all these engineered versions of Cas9 improve their specificity to target without sacrificing their efficiency much.
5 Factors Contributing in Epigenome Editing, Genome Imaging, and Protein-Genome Interaction
5.1 Dead Guide
A type of guide sequence causing efficient binding of wild-type Cas9 to its target sequence without inducing cleavage is known as “dead guide (dRNA).” Researchers successfully synthesized these dRNAs by shortening the length of the normal guide to 14 or 15 nt and achieved good transcriptional control when co-transfected with wild-type Cas9 fused with a transcription-activating domain (Dahlman et al., 2015; Kiani et al., 2015). Moreover, orthogonal gene activation and knockout can be achieved by co-expressing wild-type Cas9 with dead guide (14 or 15 nt) and normal guide (20 nt), respectively, in the same cell (Dahlman et al., 2015). Although decreasing the length of guide can increase off-targets, the chances of transcriptional modulation at these sites may be low due to the presence of few off-targets within the transcriptional start site.
5.2 Dead or Deactivated Cas9 (dCas9)
The expression level of genes is naturally controlled by two main epigenetic marks, acetylation and methylation, executed by associated proteins (Javaid and Choi, 2017). dCas9 is generated by point mutation-mediated inactivation of both Cas9 catalytic domains, which does not affect its RNA-guided DNA binding ability (Jinek et al., 2012a). It can be used for epigenome editing (CRISPRa and CRISPRi systems) and genome-imaging. Moreover, various transcription factors (for example those involved in reprogramming) interact with target DNA in a specific manner (Yesudhas et al., 2017b). This interaction can be further validated by using the dCas9 version of the CRISPR endonucleases.
5.2.1 CRISPR Activation
CRISPRa is achieved by fusing the dCas9 enzyme to transcriptional activators, such as an ω subunit of RNA polymerase in bacterial cells (Bikard et al., 2013) and p65AD or VP64 in mammalian cells (Mali et al., 2013a; Gilbert et al., 2013; Konermann et al., 2013; Perez-Pinera et al., 2013). Various strategies have been adopted to enhance the efficiency of a CRISPRa system by recruiting multiple transcriptional activators: 1) In addition to multiple sgRNAs at a single promotor to recruit multiple activators (Cheng et al., 2013; Maeder et al., 2013; Chavez et al., 2015), strategies to recruit multiple activators to the dCas9 binding site have been developed (Tanenbaum et al., 2014; Chavez et al., 2015; Konermann et al., 2015). For example, the synergistic activation mediator system allows multiple activators to work synergistically by using both sgRNA and dCas9 as scaffolds (Konermann et al., 2015). In this system, sgRNA modified with two MS2 RNA aptamers is combined with dCas9-VP64. Each aptamer recruits a pair of similar RNA binding proteins, MCPs (MS2 bacteriophage coat proteins), which are bound with p65-and HSF1-activating domains (MCP-p65-HSF1) (Konermann et al., 2015; Nishimasu et al., 2015). This system has been applied for large-scale genome screening because of its increased efficiency (Konermann et al., 2015). 2) Multiple VP64 activators can be recruited to a single dCas9 binding site by combining the dCas9 system with multipeptide array, such as SunTag. For example, fusion of dCas9 to the polypeptide array (GCN4s) recruits multiple (10 or 24) copies of its cognate scFv (single-chain variable fragment), an engineered portion of the anti-GCN4 antibody. This scFv was further fused to VP64 which eventually led to strong upregulation of the target gene [chemokine (C-X-C motif) receptor 4 (CXCR4)] because of multiple VP64 copies per dCas9 (Tanenbaum et al., 2014). This system has also been used to reduce cell growth by upregulating the expression of CDKN1B (cyclin-dependent kinase inhibitor 1B) (Tanenbaum et al., 2014) and gain-of-function screening at the genome scale (Gilbert et al., 2014). 3) A tripartite activator system has been developed by fusing three various activators in tandem to dCas9 [VP64-p65-Rta (VPR)] which showed greater activation efficiency than dCas9-VP64 when used with multiple sgRNAs (Chavez et al., 2015). Reprogramming of target cells can be achieved by using multiple approaches (Anwar et al., 2016; Lee et al., 2017) and CRISPR activation system is one of them (Balboa et al., 2015). All of these systems mimic the intrinsic mechanism of gene activation by recruiting multiple activators at the target site (He and Weintraub, 1998; Govind et al., 2005).
Similarly, dCas9 can be used to recruit epigenetic modifiers at a given locus to reshape the epigenome. For example, Neisseria meningitidis dcas9 (Nm dCas9) fused to histone demethylase LSD1 can reduce the expression of genes (which are controlled by the targeted enhancers) by decreasing the epigenetic marks H3K27ac and H3K4me2 near the targeted enhancer region (like Oct4 and Tbx3) (Kearns et al., 2015). Fusing the catalytic core domain of histone acetyltransferase p300 with dCas9 (Nm dCas9-p300 core and Sp dCas9-p300 core) can be used to activate the transcription of various endogenous genes by increasing the H3K27ac level at the targeted enhancer or promoter regions (Hilton et al., 2015).
5.2.2 CRISPR Interference
The dCas9 mediated regulation of gene expression is executed by recruiting various proteins and RNA factors at the target site (Gilbert et al., 2013; Qi et al., 2013). Transcriptional inhibition by the CRISPR machinery is termed CRISPRi and only dcas9 is sufficient in bacterial cells via steric hindrance of the transcriptional machinery (Bikard et al., 2013; Qi et al., 2013). However, it is not very effective in mammalian cells unless dCas9 is fused to a transcriptional repressor domain (such as KRAB of Kox1) (Gilbert et al., 2013; Konermann et al., 2013).
5.2.3 Genome Imaging
One of the important things in genome biology is to understand the correlation between linear genetic information imprinted on DNA and its three-dimensionally compact organization inside the cell nucleus because many studies have highlighted the impact of organization on the regulation of gene expression and cell differentiation (Lanctôt et al., 2007; Schneider and Grosschedl, 2007; Peric-Hupkes et al., 2010; Dixon et al., 2015). Because of the lack of a proper tool, it was difficult to visualize genomic dynamics in a sequence-specific manner. However, it has become possible with the sequence-specific binding of the dCas9 regardless of the genome architecture and epigenetic state. For example, fusing Streptococcus pyogenes dCas9 (Sp dCas9) to enhanced GFP can be used to visualize the repetitive and nonrepetitive genomic loci in living human cells (Chen et al., 2013). A similar approach was used to label the endogenous telomeres, pericentric regions, and centromeres (Anton et al., 2014). dCas9-based genome imaging has been expanded with various improvements. The SunTag peptide array with dCas9 was exploited to successfully amplify imaging strength (Tanenbaum et al., 2014). Multicolor genome imaging was achieved by individually tagging St1 dcas9, Nm dCas9, and Sp dCas9 with differently colored fluorescent proteins (FPs) and targeting each of them to different loci by the corresponding sgRNAs (Ma et al., 2015a). Fixed cells and tissues can be labeled by in vitro-assembled complexes of fluorescently labeled dCas9-sgRNA, a technique known as CASFISH (Deng et al., 2015). Using a single particle tracking method with fused dCas9 and the HaloTag system, it was found that there is a three-dimensional diffusion based analysis of genome by dCas9, with transient binding at off-targets and reduced efficiency to search at heterochromatic regions (Knight et al., 2015).
5.2.4 Protein-Genome Interaction
The sole or complex of different transcription factors interact with the target DNA by recognizing some consensus sequences (Yesudhas et al., 2017a; Yesudhas et al., 2019). Endogenous proteins interacting with a specific genome region can be found by using the CRISPR-based engineered DNA-binding molecule-mediated ChIP (enChIP) method. The loci specific proteins are pulled down by targeting the affinity-tagged dCas9 at that region using associated sgRNAs. This system has been successfully tested to analyze proteins interacting with an interferon-γ-responsive promoter (Fujita and Fujii, 2013; Fujita and Fujii, 2014).
5.3 Scaffold RNA System
The scRNA system is made by modifying sgRNA, instead of dCas9, to turn it into a scaffold to recruit various transcriptional regulators (Mali et al., 2013a; Konermann et al., 2015; Shechner et al., 2015; Zalatan et al., 2015). The orthogonal RNA aptamers are fused to the sgRNAs which recruit various RNA-binding proteins (RBPs) further fused to transcriptional repressors or activators. The multimodal gene regulation (i.e., simultaneous repression or activation) within the same cell can be achieved by coupling RNA aptamer-RBP pairs (such as PP7-PCP, MS2-MCP, and com-Com) to different sgRNAs and directing them to the target site with the help of dCas9 (Zalatan et al., 2015).
6 Applications of CRISPR/Cas System
Due to its flexibility, convenience and precision, CRISPR/Cas system is preferred over previously developed genome-editing tools (such as TALENs and ZFNs) for various applications (Figure 6). Gene targeting in embryonic stem cells by homologous recombination is used to modify animal genome which played a significant role in reverse genetics with reference to diseases. It has limited application due to lack of embryonic stem cells and long-time. Recently, precise germline-modifications of various model organisms have been achieved by CRISPR/Cas technology which has revolutionized the therapeutic industry (Hwang et al., 2013; Wang et al., 2013; Niu et al., 2014; Platt et al., 2014; Whitworth et al., 2014). Rapid, simple and scalable In vivo modification of target genes is achieved by microinjection of customizable sgRNA and Cas9-encoding mRNA into zebrafish embryos at one cell-stage (Chang et al., 2013; Hwang et al., 2013). The highly efficient biallelic mutations in mice is done by coinjecting sgRNAs targeting multiple genes and Cas9 mRNA into mouse zygotes (Wang et al., 2013). Targeting two sites of the same gene by respective sgRNA is used to generate mice with deleted mutations (Wang et al., 2013). Moreover, genetically modified knockin mutant mice has been generated by Cre-dependent Cas9 with simple injection of sgRNA (Platt et al., 2014). Along with mice, CRISPR/Cas technology has been used to genetically engineer other model organisms which include Caenrhabditis elegans (Friedland et al., 2013), Drosophila (Bassett et al., 2013; Gratz et al., 2013), Axolotl (Flowers et al., 2014), rat (Hu et al., 2013), Xenopus tropicalis (Blitz et al., 2013; Nakayama et al., 2013), and pig (Whitworth et al., 2014). Notably, this technology has also been used to modify the genome of cynomolgus monkey (Niu et al., 2014). The development of diverse model organism with various genomic modifications will help to develop therapeutic strategies for multiple human diseases.
FIGURE 6.
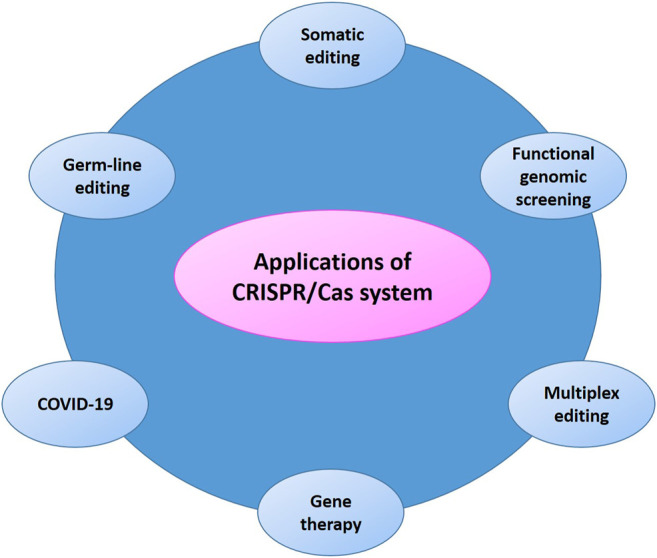
Applications of CRISPR/Cas system.
CRIPSR/Cas technology can also be used for somatic genome editing in various mouse models which is a tool for fast functional analysis of genes responsible for a particular disease (Xue et al., 2014a; Maddalo et al., 2014; Sánchez-Rivera et al., 2014). Hydrodynamic-based delivery of Cas9 and Pten targeting sgRNAs, with or without p53 mutation, in liver revealed the effects of p53 and PTEN knockouts (Xue et al., 2014a). The mouse model with Eml4-Alk-mediated lung cancer has been developed after Eml4-Alk inversion (chromosomal rearrangement) with CRISPR/Cas9 system (Maddalo et al., 2014). These tumors have typical molecular and histopathological characteristics of human ALK (+) non-small cell lung cancer with a sensitivity to ALK targeting inhibitors (Maddalo et al., 2014). This highlights the reliability of this technology to mutate oncogenes and tumor suppressor genes in somatic cells, which provides new approach for the development of respective disease models.
Functional genomic screening is a process to identify the function of a gene in a particular cellular process. Previously used RNA interference (RNAi) technique has limited applications because of multiple off-targets (Jackson et al., 2003; Chang et al., 2006; Adamson et al., 2012; Sigoillot et al., 2012). Moreover, RNAi cannot silence nuclear RNAs. On the other hand, CRISPR/Cas technology has a lot of success in loss-of-function screening at genomic scale (Findlay et al., 2014; Shalem et al., 2014; Wang et al., 2014). Most of the expected genes in DNA mismatch repair pathway have been identified by using lentiviral sgRNA library at genomic-scale (Wang et al., 2014). Genes responsible for resistance to late-stage melanoma drug, vemurafenib, and cell viability in pluripotent stem cells and cancer has been screened by using GeCKO (genome-scale CRISPR-Cas9 knockout) library (Shalem et al., 2014). Genes involved in cellular response to diphtheria and anthrax toxins have been screened by using CRISPR/Cas-based knockout library (Zhou et al., 2014). Functional screening of both trans-acting factors and cis-regulatory elements at high resolution in the genome can be done by linking CRISPR/Cas technology with multiplex HDR by using complex library of donor templates (Findlay et al., 2014). Two other powerful tools for functional screening are CRISPR-mediated activation (CRISPRa) and CRISPR-mediated repression (CRISPRi). Their libraries have been employed to map complex pathways by screening essential genes involved in tumor suppression, growth, differentiation, and sensitivity to various toxins (Mali et al., 2013a; Gilbert et al., 2013; Maeder et al., 2013; Perez-Pinera et al., 2013; Gilbert et al., 2014). The results promise the utilization of this tool to identify essential endogenous genes for various biological processes.
The precise and easy multiplexing ability of CRISPR/Cas system in human cells than other genome editing tools (such as ZFNs and TALENs) has made it popular. This feature is exploited for simultaneous editing of PVALB and EMX1 loci in human cells; this is done by expressing both targeting sgRNAs in a single expression cassette (Cong et al., 2013b). The simultaneous expression of two sgRNAs resulted in the deletion of 19-bp segment (Mali et al., 2013b). For multiplex engineering, vector systems for the expression of multiple sgRNAs simultaneously have been developed. For example, plasmid carrying expression cassettes of two to seven gRNAs showed editing efficiency from 4.3 to 37.8% (Sakuma et al., 2014). In another study, four gRNAs under different promoters are assembled in a single lentiviral vector via Golden Gate assembly. This construct is used to simultaneously edit one locus in each AAVS1 and HBG1 and two loci in IL1RN with editing efficiency of 4.8–18.4% in fibroblasts and 17.9–33.3% in HEK293T cells (Kabadi et al., 2014).
Genetic therapies are considered as powerful tool to cure monogenic diseases, cancers, HIV, or degenerative diseases. One of the exciting applications of CRISPR/Cas technology is its ability to cure genetic diseases. The dominant mutation of Crygc gene in cataracts mouse model has been corrected by simultaneous injection of mutant Crygc targeting sgRNA and Cas9 mRNA into zygotes (Wu et al., 2013). Subsequent study revealed 100% efficient production of offsprings with corrected phenotype by correcting Crygc gene in spermatogonial stem cells with CRISPR/Cas system (Wu et al., 2015). Development of muscular dystrophy in mutant mice has been prevented by correcting the dystrophin gene in germ line with coinjection of sgRNA, Cas9 and donor template into mouse zygotes (Schwank et al., 2013). The mutated locus of cystic fibrosis transmembrane conductor receptor (CFTR) in cultured intestinal stem cells derived from human cystic fibrosis patient has been correcting after homologous recombination with the help of CRISPR/Cas technology (Long et al., 2014).
Considering antiviral adaptive role of CRISPR/Cas system in bacteria, it can be used to eliminate genomes of pathogens from patients in order to cure them from various infectious disease. The new HIV infection is prevented by eliminating the HIV-1 genome from patients (Ebina et al., 2013; Hu et al., 2014). Upon transfecting the HIV-1 long terminal repeats (LTR)-specific sgRNA into human cells integrated with HIV-1 provirus, LTR target sites are cleaved and mutated which suppressed the expression of various viral genes. Additionally, it also eliminated the viral genes from chromosome of host cell (Ebina et al., 2013). Recently, the precise editing of HIV-1 genome with Cas9/sgRNAs has also been done which immunize the cells for the prevention of further infection (Hu et al., 2014).
In 2018, first Phase 1 CRISPR clinical trial opened in the United States for cancer immunotherapy to edit autologous T cells against multiple tumors. During this, T lymphocytes collected from patients’ blood were engineered ex vivo to knockout α and β chains of endogenous T cell receptor (TCR). Later, NY-ESO-1 antigen specific TCR encoding gene was delivered to these cells via lentiviral transduction system (Baylis and McLeod, 2017). The first clinical trial in the United States to mediate gene disruption for therapeutic purpose were conducted for the patients with β-thalassemia and sickle-cell anemia, therapy named as CTX001. It involves CRISPR-mediated disruption of BCL 11 A gene in autologous progenitor and hematopoietic stem cells collected from peripheral blood (Basak et al., 2015; Basak and Sankaran, 2016). The first trial with in vivo delivery of CRISPR/Cas9 were conducted in leber congenital amaurosis patients in which a non-functional protein is produced due to the intronic IVS26 mutation, a therapy named as EDIT-101 (Maeder et al., 2019). Similarly, 32-base deletion was conducted in CCR5 gene (CCR5Δ32) of human embryos by using CRISPR/Cas9 and both embryos were implanted back into their mother (Cohen, 2019; Greely, 2019). In current state, germline gene editing is ethically unfavorable unless safety data of ongoing somatic CRISPR-based therapy clinical trials is obtained.
Recently, a novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) caused a global pandemic, coronavirus disease 2019 (COVID-19) (Sironi et al., 2020). The timely detection of SARS-CoV-2 and its cure is very important to prevent the death of the patients. A CRISPR Cas12-based assay has been developed to detect COVID-19 with 95% accuracy and turnaround time of around 40 min, named as SARS-CoV-2 DETECTR. The assay involves isothermal amplification of reverse transcribed RNA of SARS-CoV-2. Cas12 and guide RNAs against nucleoprotein and envelop genes are targeted and their cleavage is visualized by fluorescent reporter system (Broughton et al., 2020). In addition to detection, CRISPR may also provide therapeutic potential for COVID-19 patients. The PAC-MAN (Prophylactic Antiviral CRISPR in huMAN cells) utilizes Ruminococcus flavefaciens derived VI-D CRISPR-Cas13d variant which can degrade the SARS-CoV-2 RNA by simultaneously targeting multiple regions (Abbott et al., 2020). With these advancements, CRISPR/Cas machinery may serve as a virus-battling system during this pandemic.
7 Concluding Remarks
Not long ago, genome editing in humans was considered a hypothetical idea, but the CRISPR tool has provided a hopeful platform to achieve it. The CRISPR/Cas system is an emerging biotechnological tool for genome editing. In the presence of other competitive tools, such as like ZFNs and TALEN, what makes CRISPR better? The answer lies in the high cost-effectiveness, less laborious, specificity, and efficient compared to others. Since the application of CRISPR/Cas system into the genome editing, various experimental improvements have been achieved to enhance its specificity and efficiency (Table 1). Despite the swift progress in the CRISPR field, there are many fundamental unanswered mechanistic questions regarding spacer acquisition and discriminating between self and non-self among various CRISPR subtypes. The mechanism of crRNA biogenesis and interference is relatively well understood for certain subtypes, such as type I-E and type II-A. However, Type IV, V, and VI needs to be characterized further as some of them show mechanisms different from the traditional systems (Makarova and Koonin, 2015). In addition, many subtypes with possibly novel working mechanisms need to be discovered to enhance biotechnological application of the system.
TABLE 1.
Summary to enhance the efficiency of CRISPR-mediated genome and epigenome editing.
| Sr. no | Recommendation/Strategy |
|---|---|
| 1 | GC content of designed sgRNA must range from >30% to <70% |
| 2 | Target promoter region rather than exon or intron for gene disruption (if possible) |
| 3 | Prefer purine-rich spacer sequences (if possible) |
| 4 | Existence of secondary structure in sgRNA improves its processing and genome-editing capability |
| 5 | Truncating gRNA or adding extra guanines at its 5′end increases its specificity |
| 6 | Stabilize the gRNA with G-quadruplexe structure |
| 7 | Eliminate seed regions with UUU sequence |
| 8 | Chemically modify the gRNA |
| 9 | Substitute one of the nucleotides in the continuous stretch of four to six |
| 10 | Avoid constitutively higher expression level of sgRNA and Cas9 to prevent off-targeting |
| 11 | Select appropriate method to deliver CRISPR components |
| 12 | Avoid targeting heterochromatin region (if possible) |
| 13 | Inhibit NHEJ or stimulate HDR to increase the knock-in efficiency of transgene |
| 14 | Select appropriate type of donor-template and DNA-repair pathway |
| 15 | Use SpCas9MT-pDBD, paired nickase-Cas9 or dCas9-FokI to increase specificity |
| 16 | Prefer PAM-out orientation over PAM-in and D10A mutant over H840A |
| 17 | Use dead-guide or dead-Cas9 for epigenome editing |
| 18 | Recruit multiple activators by using modified guide and dCas9; combining dCas9 system with multipeptide array like SunTag; or using tripartite system to increase the efficiency of CRISPR activation |
| 19 | Modify dCas9 or sgRNA to recruit epigenetic modifiers at the target site |
There are still challenges related to off-targeting and less efficiency caused by Cas9 in the clinical and in vitro research venue. Moreover, dominating HDR over NHEJ to enhance the process of homology-based knock-in of transgenes remains a challenge (Chu et al., 2015; Maruyama et al., 2015). The Cas endonuclease can be delivered in various ways (Supplementary Table S3) in the form of DNA, mRNA, or protein to maximize the output in clinical applications; however, selecting an appropriate delivery method for a particular form is still a challenge (Lin et al., 2014a; Gori et al., 2015; Howes and Schofield, 2015; Zuris et al., 2015). Some of the advanced applications, such as CRISPR-enChIP, also face challenges like off-targeting (O'Geen et al., 2015). Due to its capability of genome editing in germlines, various social and ethical issues must be considered while editing not only in humans but also in other organisms (Rodriguez, 2016).
The precise and efficient genome editing are the main attributes of CRISPR/Cas technology; however, these might not remain same from cell to cell; cell to organism; or organism to organism. Based on research, various parameters and modifications have a significant role in improving the specificity and efficiency of genome editing. Briefly, number and nature of sgRNA; expression level and delivery of CRISPR components; features of target sequence; host DNA-repair pathways; and modified versions of Cas9 and guide RNA must be kept under consideration.
Other aspects of the CRISPR/Cas system do not fit the scope of this review. Along with providing adaptive-immunity (Figure 1), it also regulates genomic evolution, DNA repair, and group behavior (Westra et al., 2014; Ratner et al., 2015). The spatiotemporal regulation of the CRISPR/Cas system in response to the stress signal and phage infection requires further study to understand the complete mechanism (Bondy-Denomy and Davidson, 2014; Garrett et al., 2015; Patterson et al., 2015). Another active research area has been opened after identifying phage evasion of CRISPR-mediated immunity by various mechanisms, including mutational escape, anti-CRISPR proteins, and DNA modification (Deveau et al., 2008; Bondy-Denomy et al., 2013; Bondy-Denomy et al., 2015; Bryson et al., 2015; Paez-Espino et al., 2015). Further exploration of various organisms promises to discover new technologies like that of the CRISPR/Cas system.
Author Contributions
NJ conceptualized and wrote the original manuscript. SC reviewed and edited the draft.
Funding
This work was supported by the National Research Foundation of Korea (Grant No. NRF-2020R1F1A1071517, 2019M3D1A1078940 and 2019R1A6A1A11051471).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.761709/full#supplementary-material
Glossary
- CRISPR
clustered regularly interspaced short palindromic repeats
- Cas
CRISPR-associated protein
- HR
homologous recombination
- DSBs
double-strand breaks
- indels
insertion and deletions
- NHEJ
non-homologous end joining
- ZFNs
zinc finger nucleases
- TALENs
transcription activator-like effector nucleases
- PAM
protospacer adjacent motif
- Cascade
CRISPR associated complex for antiviral defense
- tracrRNA
trans-activating crRNA
- sgRNA
single guide RNA
- HDR
homology directed repair
- AV
adenovirus vector
- AAV
adeno-associated vector
- RNP
ribonucleoprotein
- ssDNA
single stranded DNA
- MMEJ
microhomology-mediated end joining
- ssODNs
single-stranded oligodeoxyribonucleotides
- RFNs
RNA-guided FokI-dCas9 nucleases
- trugRNAs
truncated gRNAs
- pDBD
programmable DNA-binding domain
- HiFi Cas9
high-fidelity Cas9 variant
- PID
PAM-interacting domain
- St3Cas9
Streptococcus thermophiles CRISPR-3 Cas9
- dRNA
dead guide
- dCas9
dead or deactivated Cas9
- CRISPRa
CRISPR Activation
- MCPs
MS2 bacteriophage coat proteins
- scFv
single-chain variable fragment
- CXCR4
chemokine (C-X-C motif) receptor 4
- CDKN1B
cyclin-dependent kinase inhibitor 1B
- Nm dCas9
Neisseria meningitidis dcas9
- Sp dCas9
Streptococcus pyogenes dCas9
- FPs
fluorescent proteins
- enCHIP
engineered DNA-binding molecule-mediated ChIP
- scRNA
scaffold RNA
- RBPs
RNA-binding proteins
- RNAi
RNA interference
- GeCKO
genome-scale CRISPR-Cas9 knockout
- CRISPRi
CRISPR-mediated repression
- CFTR
cystic fibrosis transmembrane conductor receptor
- LTR
long terminal repeats
- TCR
T cell receptor
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- COVID-19
coronavirus disease 2019
- PAC-MAN
Prophylactic Antiviral CRISPR in huMAN
References
- Abbott T. R., Dhamdhere G., Liu Y., Lin X., Goudy L., Zeng L., et al. (2020). Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell 181 (4), 865–876. e812. 10.1016/j.cell.2020.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablain J., Durand E. M., Yang S., Zhou Y., Zon L. I. (2015). A CRISPR/Cas9 Vector System for Tissue-specific Gene Disruption in Zebrafish. Developmental Cel. 32 (6), 756–764. 10.1016/j.devcel.2015.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson B., Smogorzewska A., Sigoillot F. D., King R. W., Elledge S. J. (2012). A Genome-wide Homologous Recombination Screen Identifies the RNA-Binding Protein RBMX as a Component of the DNA-Damage Response. Nat. Cel Biol 14 (3), 318–328. 10.1038/ncb2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit M., Donyo M., Hollander D., Goren A., Kim E., Gelfman S., et al. (2012). Differential GC Content between Exons and Introns Establishes Distinct Strategies of Splice-Site Recognition. Cel Rep. 1 (5), 543–556. 10.1016/j.celrep.2012.03.013 [DOI] [PubMed] [Google Scholar]
- Anton T., Bultmann S., Leonhardt H., Markaki Y. (2014). Visualization of Specific DNA Sequences in Living Mouse Embryonic Stem Cells with a Programmable Fluorescent CRISPR/Cas System. Nucleus 5 (2), 163–172. 10.4161/nucl.28488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar M. A., Kim S., Choi S. (2016). The Triumph of Chemically Enhanced Cellular Reprogramming: a Patent Review. Expert Opin. Ther. patents 26 (2), 265–280. 10.1517/13543776.2016.1118058 [DOI] [PubMed] [Google Scholar]
- Babu K., Amrani N., Jiang W., Yogesha S. D., Nguyen R., Qin P. Z., et al. (2019). Bridge helix of Cas9 Modulates Target DNA Cleavage and Mismatch Tolerance. Biochemistry 58 (14), 1905–1917. 10.1021/acs.biochem.8b01241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa D., Weltner J., Eurola S., Trokovic R., Wartiovaara K., Otonkoski T. (2015). Conditionally Stabilized dCas9 Activator for Controlling Gene Expression in Human Cell Reprogramming and Differentiation. Stem Cel. Rep. 5 (3), 448–459. 10.1016/j.stemcr.2015.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes N. J., Gil-Humanes J., Cermak T., Atkins P. A., Voytas D. F. (2014). DNA Replicons for Plant Genome Engineering. The Plant Cell 26 (1), 151–163. 10.1105/tpc.113.119792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., et al. (2007). CRISPR Provides Acquired Resistance against Viruses in Prokaryotes. Science 315 (5819), 1709–1712. 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- Basak A., Hancarova M., Ulirsch J. C., Balci T. B., Trkova M., Pelisek M., et al. (2015). BCL11A Deletions Result in Fetal Hemoglobin Persistence and Neurodevelopmental Alterations. J. Clin. Invest. 125 (6), 2363–2368. 10.1172/jci81163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak A., Sankaran V. G. (2016). Regulation of the Fetal Hemoglobin Silencing Factor BCL11A. Ann. N.Y. Acad. Sci. 1368 (1), 25–30. 10.1111/nyas.13024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett A. R., Tibbit C., Ponting C. P., Liu J.-L. (2013). Highly Efficient Targeted Mutagenesis of Drosophila with the CRISPR/Cas9 System. Cel Rep. 4 (1), 220–228. 10.1016/j.celrep.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylis F., McLeod M. (2017). First-in-human Phase 1 CRISPR Gene Editing Cancer Trials: Are We Ready? Curr. Gene Ther. 17 (4), 309–319. 10.2174/1566523217666171121165935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett J., Ashtari M., Wellman J., Marshall K. A., Cyckowski L. L., Chung D. C., et al. (2012). AAV2 Gene Therapy Readministration in Three Adults with Congenital Blindness. Sci. Transl Med. 4 (120), 120ra15–120ra115. 10.1126/scitranslmed.3002865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M., Carroll D., Segal D. J., Trautman J. K., Smith J., Kim Y.-G., et al. (2001). Stimulation of Homologous Recombination through Targeted Cleavage by Chimeric Nucleases. Mol. Cel Biol 21 (1), 289–297. 10.1128/mcb.21.1.289-297.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M., Golic M., Golic K. G., Carroll D. (2002). Targeted Chromosomal Cleavage and Mutagenesis in Drosophila Using Zinc-finger Nucleases. Genetics 161 (3), 1169–1175. 10.1093/genetics/161.3.1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., Marraffini L. A. (2013). Programmable Repression and Activation of Bacterial Gene Expression Using an Engineered CRISPR-Cas System. Nucleic Acids Res. 41 (15), 7429–7437. 10.1093/nar/gkt520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitz I. L., Biesinger J., Xie X., Cho K. W. Y. (2013). Biallelic Genome Modification in F0Xenopus Tropicalisembryos Using the CRISPR/Cas System. genesis 51 (12), 827–834. 10.1002/dvg.22719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A., Quinquis B., Sorokin A., Ehrlich S. D. (2005). Clustered Regularly Interspaced Short Palindrome Repeats (CRISPRs) Have Spacers of Extrachromosomal Origin. Microbiology 151 (8), 2551–2561. 10.1099/mic.0.28048-0 [DOI] [PubMed] [Google Scholar]
- Bolukbasi M. F., Gupta A., Oikemus S., Derr A. G., Garber M., Brodsky M. H., et al. (2015). DNA-binding-domain Fusions Enhance the Targeting Range and Precision of Cas9. Nat. Methods 12 (12), 1150–1156. 10.1038/nmeth.3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J., Davidson A. R. (2014). To Acquire or Resist: the Complex Biological Effects of CRISPR-Cas Systems. Trends Microbiology 22 (4), 218–225. 10.1016/j.tim.2014.01.007 [DOI] [PubMed] [Google Scholar]
- Bondy-Denomy J., Garcia B., Strum S., Du M., Rollins M. F., Hidalgo-Reyes Y., et al. (2015). Multiple Mechanisms for CRISPR-Cas Inhibition by Anti-CRISPR Proteins. Nature 526 (7571), 136–139. 10.1038/nature15254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J., Pawluk A., Maxwell K. L., Davidson A. R. (2013). Bacteriophage Genes that Inactivate the CRISPR/Cas Bacterial Immune System. Nature 493 (7432), 429–432. 10.1038/nature11723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman E. K., Chen T., Amendola M., van Steensel B. (2014). Easy Quantitative Assessment of Genome Editing by Sequence Trace Decomposition. Nucleic Acids Res. 42 (22), e168. 10.1093/nar/gku936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks C., Nekrasov V., Lippman Z. B., Van Eck J. (2014). Efficient Gene Editing in Tomato in the First Generation Using the Clustered Regularly Interspaced Short Palindromic repeats/CRISPR-Associated9 System. Plant Physiol. 166 (3), 1292–1297. 10.1104/pp.114.247577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J. P., Deng X., Yu G., Fasching C. L., Servellita V., Singh J., et al. (2020). CRISPR-Cas12-based Detection of SARS-CoV-2. Nat. Biotechnol. 38 (7), 870–874. 10.1038/s41587-020-0513-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns S. J. J., Jore M. M., Lundgren M., Westra E. R., Slijkhuis R. J. H., Snijders A. P. L., et al. (2008). Small CRISPR RNAs Guide Antiviral Defense in Prokaryotes. Science 321 (5891), 960–964. 10.1126/science.1159689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson A. L., Hwang Y., Sherrill-Mix S., Wu G. D., Lewis J. D., Black L., et al. (2015). Covalent Modification of Bacteriophage T4 DNA Inhibits CRISPR-Cas9. MBio 6 (3), e00648–00615. 10.1128/mBio.00648-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger A., Lindsay H., Felker A., Hess C., Anders C., Chiavacci E., et al. (2016). Maximizing Mutagenesis with Solubilized CRISPR-Cas9 Ribonucleoprotein Complexes. Development 143 (11), 2025–2037. 10.1242/dev.134809 [DOI] [PubMed] [Google Scholar]
- Cao J., Wu L., Zhang S. M., Lu M., Cheung W. K., Cai W., et al. (2016). An Easy and Efficient Inducible CRISPR/Cas9 Platform with Improved Specificity for Multiple Gene Targeting. Nucleic Acids Res. 44 (19), e149. 10.1093/nar/gkw660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi M. R. (1989). Altering the Genome by Homologous Recombination. Science 244 (4910), 1288–1292. 10.1126/science.2660260 [DOI] [PubMed] [Google Scholar]
- Casini A., Olivieri M., Petris G., Montagna C., Reginato G., Maule G., et al. (2018). A Highly Specific SpCas9 Variant Is Identified by In Vivo Screening in Yeast. Nat. Biotechnol. 36 (3), 265–271. 10.1038/nbt.4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S., Ji H., Kabadi A. M., Gersbach C. A., Christoforou N., Leong K. W. (2014). A CRISPR/Cas9-based System for Reprogramming Cell Lineage Specification. Stem Cel. Rep. 3 (6), 940–947. 10.1016/j.stemcr.2014.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K., Elledge S. J., Hannon G. J. (2006). Lessons from Nature: microRNA-Based shRNA Libraries. Nat. Methods 3 (9), 707–714. 10.1038/nmeth923 [DOI] [PubMed] [Google Scholar]
- Chang N., Sun C., Gao L., Zhu D., Xu X., Zhu X., et al. (2013). Genome Editing with RNA-Guided Cas9 Nuclease in Zebrafish Embryos. Cell Res 23 (4), 465–472. 10.1038/cr.2013.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A., Scheiman J., Vora S., Pruitt B. W., Tuttle M., P R Iyer E., et al. (2015). Highly Efficient Cas9-Mediated Transcriptional Programming. Nat. Methods 12 (4), 326–328. 10.1038/nmeth.3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Gilbert L. A., Cimini B. A., Schnitzbauer J., Zhang W., Li G.-W., et al. (2013). Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell 155 (7), 1479–1491. 10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Hu J., Almeida R., Liu H., Balakrishnan S., Covill-Cooke C., et al. (2016). Expanding the CRISPR Imaging Toolset with Staphylococcus aureus Cas9 for Simultaneous Imaging of Multiple Genomic Loci. Nucleic Acids Res. 44, 75. 10.1093/nar/gkv1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. S., Dagdas Y. S., Kleinstiver B. P., Welch M. M., Sousa A. A., Harrington L. B., et al. (2017). Enhanced Proofreading Governs CRISPR-Cas9 Targeting Accuracy. Nature 550 (7676), 407–410. 10.1038/nature24268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A. W., Wang H., Yang H., Shi L., Katz Y., Theunissen T. W., et al. (2013). Multiplexed Activation of Endogenous Genes by CRISPR-On, an RNA-Guided Transcriptional Activator System. Cel Res 23 (10), 1163–1171. 10.1038/cr.2013.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Kim S., Kim J. M., Kim J.-S. (2013). Targeted Genome Engineering in Human Cells with the Cas9 RNA-Guided Endonuclease. Nat. Biotechnol. 31 (3), 230–232. 10.1038/nbt.2507 [DOI] [PubMed] [Google Scholar]
- Cho S. W., Kim S., Kim Y., Kweon J., Kim H. S., Bae S., et al. (2014). Analysis of Off-Target Effects of CRISPR/Cas-derived RNA-Guided Endonucleases and Nickases. Genome Res. 24 (1), 132–141. 10.1101/gr.162339.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu V. T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., et al. (2015). Increasing the Efficiency of Homology-Directed Repair for CRISPR-Cas9-Induced Precise Gene Editing in Mammalian Cells. Nat. Biotechnol. 33 (5), 543–548. 10.1038/nbt.3198 [DOI] [PubMed] [Google Scholar]
- Cohen J. (2019). Did CRISPR Help—Or Harm—The First-Ever Gene-Edited Babies. Science 21. [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., et al. (2013a). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339, 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., et al. (2013b). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339 (6121), 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M., Dottori M., Sourris K., Jamshidi P., Hatzistavrou T., Davis R., et al. (2007). A Method for Genetic Modification of Human Embryonic Stem Cells Using Electroporation. Nat. Protoc. 2 (4), 792–796. 10.1038/nprot.2007.105 [DOI] [PubMed] [Google Scholar]
- Cradick T. J., Fine E. J., Antico C. J., Bao G. (2013). CRISPR/Cas9 Systems Targeting β-globin and CCR5 Genes Have Substantial Off-Target Activity. Nucleic Acids Res. 41 (20), 9584–9592. 10.1093/nar/gkt714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlem T. J., Hoshijima K., Jurynec M. J., Gunther D., Starker C. G., Locke A. S., et al. (2012). Simple Methods for Generating and Detecting Locus-specific Mutations Induced with TALENs in the Zebrafish Genome. Plos Genet. 8 (8), e1002861. 10.1371/journal.pgen.1002861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman J. E., Abudayyeh O. O., Joung J., Gootenberg J. S., Zhang F., Konermann S. (2015). Orthogonal Gene Knockout and Activation with a Catalytically Active Cas9 Nuclease. Nat. Biotechnol. 10.1038/nbt.3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y., Jia G., Choi J., Ma H., Anaya E., Ye C., et al. (2015). Optimizing sgRNA Structure to Improve CRISPR-Cas9 Knockout Efficiency. Genome Biol. 16 (1), 280. 10.1186/s13059-015-0846-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis K. M., Pattanayak V., Thompson D. B., Zuris J. A., Liu D. R. (2015). Small Molecule-Triggered Cas9 Protein with Improved Genome-Editing Specificity. Nat. Chem. Biol. 11 (5), 316–318. 10.1038/nchembio.1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E., Chylinski K., Sharma C. M., Gonzales K., Chao Y., Pirzada Z. A., et al. (2011). CRISPR RNA Maturation by Trans-encoded Small RNA and Host Factor RNase III. Nature 471 (7340), 602–607. 10.1038/nature09886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Shi X., Tjian R., Lionnet T., Singer R. H. (2015). CASFISH: CRISPR/Cas9-mediated In Situ Labeling of Genomic Loci in Fixed Cells. Proc. Natl. Acad. Sci. USA 112 (38), 11870–11875. 10.1073/pnas.1515692112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H., Barrangou R., Garneau J. E., Labonté J., Fremaux C., Boyaval P., et al. (2008). Phage Response to CRISPR-Encoded Resistance in Streptococcus Thermophilus. J. Bacteriol. 190 (4), 1390–1400. 10.1128/jb.01412-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo J. E., Norville J. E., Mali P., Rios X., Aach J., Church G. M. (2013). Genome Engineering in Saccharomyces cerevisiae Using CRISPR-Cas Systems. Nucleic Acids Res. 41, 4336–4343. 10.1093/nar/gkt135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J. R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J. E., Lee A. Y., et al. (2015). Chromatin Architecture Reorganization during Stem Cell Differentiation. Nature 518 (7539), 331–336. 10.1038/nature14222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J. G., Hartenian E., Graham D. B., Tothova Z., Hegde M., Smith I., et al. (2014). Rational Design of Highly Active sgRNAs for CRISPR-Cas9-Mediated Gene Inactivation. Nat. Biotechnol. 32 (12), 1262–1267. 10.1038/nbt.3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow L. E., Fisher J., O'rourke K. P., Muley A., Kastenhuber E. R., Livshits G., et al. (2015). Inducible In Vivo Genome Editing with CRISPR-Cas9. Nat. Biotechnol. 33 (4), 390–394. 10.1038/nbt.3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina H., Misawa N., Kanemura Y., Koyanagi Y. (2013). Harnessing the CRISPR/Cas9 System to Disrupt Latent HIV-1 Provirus. Sci. Rep. 3, 2510. 10.1038/srep02510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edraki A., Mir A., Ibraheim R., Gainetdinov I., Yoon Y., Song C.-Q., et al. (2018). A Compact, High-Accuracy Cas9 with a Dinucleotide PAM for In Vivo Genome Editing. Mol. Cel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M., Mikami M., Toki S. (2016). Biallelic Gene Targeting in rice. Plant Physiol. 170 (2), 667–677. 10.1104/pp.15.01663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt K. M., Mali P., Braff J. L., Moosburner M., Yaung S. J., Church G. M. (2013). Orthogonal Cas9 Proteins for RNA-Guided Gene Regulation and Editing. Nat. Methods 10 (11), 1116–1121. 10.1038/nmeth.2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Mao Y., Xu N., Zhang B., Wei P., Yang D.-L., et al. (2014). Multigeneration Analysis Reveals the Inheritance, Specificity, and Patterns of CRISPR/Cas-induced Gene Modifications in Arabidopsis. Proc. Natl. Acad. Sci. 111 (12), 4632–4637. 10.1073/pnas.1400822111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay G. M., Boyle E. A., Hause R. J., Klein J. C., Shendure J. (2014). Saturation Editing of Genomic Regions by Multiplex Homology-Directed Repair. Nature 513 (7516), 120–123. 10.1038/nature13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers G. P., Timberlake A. T., Mclean K. C., Monaghan J. R., Crews C. M. (2014). Highly Efficient Targeted Mutagenesis in Axolotl Using Cas9 RNA-Guided Nuclease. Development 141 (10), 2165–2171. 10.1242/dev.105072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I., Le Rhun A., Chylinski K., Makarova K. S., Lécrivain A.-L., Bzdrenga J., et al. (2013). Phylogeny of Cas9 Determines Functional Exchangeability of Dual-RNA and Cas9 Among Orthologous Type II CRISPR-Cas Systems. Nucleic Acids Res. 42 (4), 2577–2590. 10.1093/nar/gkt1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E., Baral R., Singhal P., Loveluck K., Shen S., Sanchez M., et al. (2015). Characterization of Staphylococcus aureus Cas9: a Smaller Cas9 for All-In-One Adeno-Associated Virus Delivery and Paired Nickase Applications. Genome Biol. 16 (1), 257. 10.1186/s13059-015-0817-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiácovo M. P., Church G. M., Calarco J. A. (2013). Heritable Genome Editing in C. elegans via a CRISPR-Cas9 System. Nat. Methods 10 (8), 741–743. 10.1038/nmeth.2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Foden J. A., Khayter C., Maeder M. L., Reyon D., Joung J. K., et al. (2013). High-frequency Off-Target Mutagenesis Induced by CRISPR-Cas Nucleases in Human Cells. Nat. Biotechnol. 31 (9), 822–826. 10.1038/nbt.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Sander J. D., Reyon D., Cascio V. M., Joung J. K. (2014). Improving CRISPR-Cas Nuclease Specificity Using Truncated Guide RNAs. Nat. Biotechnol. 32 (3), 279–284. 10.1038/nbt.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T., Fujii H. (2013). Efficient Isolation of Specific Genomic Regions and Identification of Associated Proteins by Engineered DNA-Binding Molecule-Mediated Chromatin Immunoprecipitation (enChIP) Using CRISPR. Biochem. biophysical Res. Commun. 439 (1), 132–136. 10.1016/j.bbrc.2013.08.013 [DOI] [PubMed] [Google Scholar]
- Fujita T., Fujii H. (2014). Identification of Proteins Associated with an IFNγ-Responsive Promoter by a Retroviral Expression System for enChIP Using CRISPR. PloS one 9 (7), e103084. 10.1371/journal.pone.0103084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller K. K., Chen S., Loros J. J., Dunlap J. C. (2015). Development of the CRISPR/Cas9 System for Targeted Gene Disruption in Aspergillus fumigatus. Eukaryot. Cel 14 (11), 1073–1080. 10.1128/ec.00107-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz V. M., Jasinskiene N., Tatarenkova O., Fazekas A., Macias V. M., Bier E., et al. (2015). Highly Efficient Cas9-Mediated Gene Drive for Population Modification of the Malaria Vector Mosquito Anopheles stephensi . Proc. Natl. Acad. Sci. USA 112 (49), E6736–E6743. 10.1073/pnas.1521077112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Zhao Y. (2014). Self‐processing of Ribozyme‐flanked RNAs into Guide RNAs In Vitro and In Vivo for CRISPR‐mediated Genome Editing. J. Integr. Plant Biol. 56 (4), 343–349. 10.1111/jipb.12152 [DOI] [PubMed] [Google Scholar]
- Garneau J. E., Dupuis M.-È., Villion M., Romero D. A., Barrangou R., Boyaval P., et al. (2010). The CRISPR/Cas Bacterial Immune System Cleaves Bacteriophage and Plasmid DNA. Nature 468 (7320), 67–71. 10.1038/nature09523 [DOI] [PubMed] [Google Scholar]
- Garrett R., Shah S., Erdmann S., Liu G., Mousaei M., León-Sobrino C., et al. (2015). CRISPR-cas Adaptive Immune Systems of the Sulfolobales: Unravelling Their Complexity and Diversity. Life 5 (1), 783–817. 10.3390/life5010783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G., Barrangou R., Horvath P., Siksnys V. (2012). Cas9-crRNA Ribonucleoprotein Complex Mediates Specific DNA Cleavage for Adaptive Immunity in Bacteria. Proc. Natl. Acad. Sci. 109 (39), E2579–E2586. 10.1073/pnas.1208507109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger J. M., Turan S., Hernandez S., Spector L. P., Calos M. P. (2016). In Vivoblunt-End Cloning through CRISPR/Cas9-facilitated Non-homologous End-Joining. Nucleic Acids Res. 44 (8), e76. 10.1093/nar/gkv1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L. A., Horlbeck M. A., Adamson B., Villalta J. E., Chen Y., Whitehead E. H., et al. (2014). Genome-scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159 (3), 647–661. 10.1016/j.cell.2014.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L. A., Larson M. H., Morsut L., Liu Z., Brar G. A., Torres S. E., et al. (2013). CRISPR-mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 154 (2), 442–451. 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González F., Zhu Z., Shi Z.-D., Lelli K., Verma N., Li Q. V., et al. (2014). An iCRISPR Platform for Rapid, Multiplexable, and Inducible Genome Editing in Human Pluripotent Stem Cells. Cell stem cell 15 (2), 215–226. 10.1016/j.stem.2014.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gori J. L., Hsu P. D., Maeder M. L., Shen S., Welstead G. G., Bumcrot D. (2015). Delivery and Specificity of CRISPR/Cas9 Genome Editing Technologies for Human Gene Therapy. Hum. Gene Ther. 26 (7), 443–451. 10.1089/hum.2015.074 [DOI] [PubMed] [Google Scholar]
- Govind C. K., Yoon S., Qiu H., Govind S., Hinnebusch A. G. (2005). Simultaneous Recruitment of Coactivators by Gcn4p Stimulates Multiple Steps of Transcription In Vivo . Mol. Cel Biol 25 (13), 5626–5638. 10.1128/mcb.25.13.5626-5638.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., Cummings A. M., et al. (2014). Highly Specific and Efficient CRISPR/Cas9-catalyzed Homology-Directed Repair in Drosophila. Genetics 196, 961–971. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., Harrison M. M., et al. (2013). Genome Engineering of Drosophila with the CRISPR RNA-Guided Cas9 Nuclease. Genetics 194 (4), 1029–1035. 10.1534/genetics.113.152710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greely H. T. (2019). CRISPR'd Babies: Human Germline Genome Editing in the 'He Jiankui Affair'*. J. L. Biosciences 6 (1), 111–183. 10.1093/jlb/lsz010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger J. P., Thompson D. B., Liu D. R. (2014). Fusion of Catalytically Inactive Cas9 to FokI Nuclease Improves the Specificity of Genome Modification. Nat. Biotechnol. 32 (6), 577–582. 10.1038/nbt.2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft D. H., Selengut J., Mongodin E. F., Nelson K. E. (2005). A Guild of 45 CRISPR-Associated (Cas) Protein Families and Multiple CRISPR/Cas Subtypes Exist in Prokaryotic Genomes. Plos Comput. Biol. 1 (6), e60. 10.1371/journal.pcbi.0010060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C. R., Cocozaki A., Li H., Terns R. M., Terns M. P. (2014). Target RNA Capture and Cleavage by the Cmr Type III-B CRISPR-Cas Effector Complex. Genes Dev. 28 (21), 2432–2443. 10.1101/gad.250712.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C. R., Majumdar S., Elmore J., Pfister N., Compton M., Olson S., et al. (2012). Essential Features and Rational Design of CRISPR RNAs that Function with the Cas RAMP Module Complex to Cleave RNAs. Mol. Cel. 45 (3), 292–302. 10.1016/j.molcel.2011.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C. R., Zhao P., Olson S., Duff M. O., Graveley B. R., Wells L., et al. (2009). RNA-guided RNA Cleavage by a CRISPR RNA-Cas Protein Complex. Cell 139 (5), 945–956. 10.1016/j.cell.2009.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T., Chandrashekhar M., Aregger M., Steinhart Z., Brown K. R., MacLeod G., et al. (2015). High-resolution CRISPR Screens Reveal Fitness Genes and Genotype-specific Cancer Liabilities. Cell 163 (6), 1515–1526. 10.1016/j.cell.2015.11.015 [DOI] [PubMed] [Google Scholar]
- Hauswirth W. W., Aleman T. S., Kaushal S., Cideciyan A. V., Schwartz S. B., Wang L., et al. (2008). Treatment of Leber Congenital Amaurosis Due toRPE65Mutations by Ocular Subretinal Injection of Adeno-Associated Virus Gene Vector: Short-Term Results of a Phase I Trial. Hum. Gene Ther. 19 (10), 979–990. 10.1089/hum.2008.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havlicek S., Shen Y., Alpagu Y., Bruntraeger M. B., Zufir N. B. M., Phuah Z. Y., et al. (2017). Re-engineered RNA-Guided FokI-Nucleases for Improved Genome Editing in Human Cells. Mol. Ther. 25 (2), 342–355. 10.1016/j.ymthe.2016.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Weintraub S. J. (1998). Stepwise Recruitment of Components of the Preinitiation Complex by Upstream Activators In Vivo . Mol. Cel Biol 18 (5), 2876–2883. 10.1128/mcb.18.5.2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckl D., Kowalczyk M. S., Yudovich D., Belizaire R., Puram R. V., McConkey M. E., et al. (2014). Generation of Mouse Models of Myeloid Malignancy with Combinatorial Genetic Lesions Using CRISPR-Cas9 Genome Editing. Nat. Biotechnol. 32 (9), 941–946. 10.1038/nbt.2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel A., Bak R. O., Clark J. T., Kennedy A. B., Ryan D. E., Roy S., et al. (2015). Chemically Modified Guide RNAs Enhance CRISPR-Cas Genome Editing in Human Primary Cells. Nat. Biotechnol. 33 (9), 985–989. 10.1038/nbt.3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton I. B., D'ippolito A. M., Vockley C. M., Thakore P. I., Crawford G. E., Reddy T. E., et al. (2015). Epigenome Editing by a CRISPR-Cas9-Based Acetyltransferase Activates Genes from Promoters and Enhancers. Nat. Biotechnol. 33 (5), 510–517. 10.1038/nbt.3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz J. M., Laughery M. F., Wyrick J. J. (2015). Nucleosomes Inhibit Cas9 Endonuclease Activity In Vitro . Biochemistry 54 (48), 7063–7066. 10.1021/acs.biochem.5b01108 [DOI] [PubMed] [Google Scholar]
- Hotta A., Ellis J. (2008). Retroviral Vector Silencing during iPS Cell Induction: an Epigenetic beacon that Signals Distinct Pluripotent States. J. Cel. Biochem. 105 (4), 940–948. 10.1002/jcb.21912 [DOI] [PubMed] [Google Scholar]
- Hou Z., Zhang Y., Propson N. E., Howden S. E., Chu L.-F., Sontheimer E. J., et al. (2013). Efficient Genome Engineering in Human Pluripotent Stem Cells Using Cas9 from Neisseria Meningitidis. Proc. Natl. Acad. Sci. 110 (39), 15644–15649. 10.1073/pnas.1313587110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes R., Schofield C. (2015). Genome Engineering Using Adeno-Associated Virus (AAV). Methods Mol. Biol. 1239, 75–103. 10.1007/978-1-4939-1862-1_5 [DOI] [PubMed] [Google Scholar]
- Hsu P. D., Scott D. A., Weinstein J. A., Ran F. A., Konermann S., Agarwala V., et al. (2013). DNA Targeting Specificity of RNA-Guided Cas9 Nucleases. Nat. Biotechnol. 31 (9), 827–832. 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. H., Miller S. M., Geurts M. H., Tang W., Chen L., Sun N., et al. (2018). Evolved Cas9 Variants with Broad PAM Compatibility and High DNA Specificity. Nature 556 (7699), 57–63. 10.1038/nature26155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Kaminski R., Yang F., Zhang Y., Cosentino L., Li F., et al. (2014). RNA-directed Gene Editing Specifically Eradicates Latent and Prevents New HIV-1 Infection. Proc. Natl. Acad. Sci. 111 (31), 11461–11466. 10.1073/pnas.1405186111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Chang N., Wang X., Zhou F., Zhou X., Zhu X., et al. (2013). Heritable Gene-Targeting with gRNA/Cas9 in Rats. Cel Res 23 (11), 1322–1325. 10.1038/cr.2013.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert J. L. (2008). Four-stranded Nucleic Acids: Structure, Function and Targeting of G-Quadruplexes. Chem. Soc. Rev. 37 (7), 1375–1384. 10.1039/b702491f [DOI] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., Sander J. D., et al. (2013). Efficient Genome Editing in Zebrafish Using a CRISPR-Cas System. Nat. Biotechnol. 31 (3), 227–229. 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y., Kim J., Cho S. W., Choi Y., Kim J.-S., Coupland G. (2015). Site-directed Mutagenesis in Arabidopsis thaliana Using Dividing Tissue-Targeted RGEN of the CRISPR/Cas System to Generate Heritable Null Alleles. Planta 241 (1), 271–284. 10.1007/s00425-014-2180-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y., Shinagawa H., Makino K., Amemura M., Nakata A. (1987). Nucleotide Sequence of the Iap Gene, Responsible for Alkaline Phosphatase Isozyme Conversion in Escherichia coli, and Identification of the Gene Product. J. Bacteriol. 169 (12), 5429–5433. 10.1128/jb.169.12.5429-5433.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. L., Bartz S. R., Schelter J., Kobayashi S. V., Burchard J., Mao M., et al. (2003). Expression Profiling Reveals Off-Target Gene Regulation by RNAi. Nat. Biotechnol. 21 (6), 635–637. 10.1038/nbt831 [DOI] [PubMed] [Google Scholar]
- Jacobs T. B., LaFayette P. R., Schmitz R. J., Parrott W. A. (2015). Targeted Genome Modifications in Soybean with CRISPR/Cas9. BMC Biotechnol. 15 (1), 16. 10.1186/s12896-015-0131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R., Embden J. D. A. v., Gaastra W., Schouls L. M. (2002). Identification of Genes that Are Associated with DNA Repeats in Prokaryotes. Mol. Microbiol. 43 (6), 1565–1575. 10.1046/j.1365-2958.2002.02839.x [DOI] [PubMed] [Google Scholar]
- Javaid N., Choi S. (2017). Acetylation- and Methylation-Related Epigenetic Proteins in the Context of Their Targets. Genes 8 (8), 196. 10.3390/genes8080196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Bikard D., Cox D., Zhang F., Marraffini L. A. (2013a). RNA-guided Editing of Bacterial Genomes Using CRISPR-Cas Systems. Nat. Biotechnol. 31 (3), 233–239. 10.1038/nbt.2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Marraffini L. A. (2015). CRISPR-cas: New Tools for Genetic Manipulations from Bacterial Immunity Systems. Annu. Rev. Microbiol. 69, 209–228. 10.1146/annurev-micro-091014-104441 [DOI] [PubMed] [Google Scholar]
- Jiang W., Yang B., Weeks D. P. (2014). Efficient CRISPR/Cas9-mediated Gene Editing in Arabidopsis thaliana and Inheritance of Modified Genes in the T2 and T3 Generations. PloS one 9 (6), e99225. 10.1371/journal.pone.0099225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Zhou H., Bi H., Fromm M., Yang B., Weeks D. P. (2013b). Demonstration of CRISPR/Cas9/sgRNA-mediated Targeted Gene Modification in Arabidopsis, Tobacco, Sorghum and rice. Nucleic Acids Res. 41, 188. 10.1093/nar/gkt780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E. (2012a). A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337, 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E. (2012b). A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. science 337 (6096), 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabadi A. M., Ousterout D. G., Hilton I. B., Gersbach C. A. (2014). Multiplex CRISPR/Cas9-based Genome Engineering from a Single Lentiviral Vector. Nucleic Acids Res. 42 (19), e147. 10.1093/nar/gku749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallimasioti-Pazi E. M., Thelakkad Chathoth K., Taylor G. C., Meynert A., Ballinger T., Kelder M. J. E., et al. (2018). Heterochromatin Delays CRISPR-Cas9 Mutagenesis but Does Not Influence the Outcome of Mutagenic DNA Repair. Plos Biol. 16 (12), e2005595. 10.1371/journal.pbio.2005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K. B., Büning H., Galy A., Schambach A., Grez M. (2013). Gene Therapy on the Move. EMBO Mol. Med. 5 (11), 1642–1661. 10.1002/emmm.201202287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns N. A., Pham H., Tabak B., Genga R. M., Silverstein N. J., Garber M., et al. (2015). Functional Annotation of Native Enhancers with a Cas9-Histone Demethylase Fusion. Nat. Methods 12 (5), 401–403. 10.1038/nmeth.3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiani S., Chavez A., Tuttle M., Hall R. N., Chari R., Ter-Ovanesyan D., et al. (2015). Cas9 gRNA Engineering for Genome Editing, Activation and Repression. Nat. Methods 12 (11), 1051–1054. 10.1038/nmeth.3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Bae S., Park J., Kim E., Kim S., Yu H. R., et al. (2015). Digenome-seq: Genome-wide Profiling of CRISPR-Cas9 Off-Target Effects in Human Cells. Nat. Methods 12 (3), 237–243. 10.1038/nmeth.3284 [DOI] [PubMed] [Google Scholar]
- Kim D., Kim J., Hur J. K., Been K. W., Yoon S.-h., Kim J.-S. (2016). Genome-wide Analysis Reveals Specificities of Cpf1 Endonucleases in Human Cells. Nat. Biotechnol. 34 (8), 863–868. 10.1038/nbt.3609 [DOI] [PubMed] [Google Scholar]
- Kim H., Um E., Cho S.-R., Jung C., Kim H., Kim J.-S. (2011). Surrogate Reporters for Enrichment of Cells with Nuclease-Induced Mutations. Nat. Methods 8 (11), 941–943. 10.1038/nmeth.1733 [DOI] [PubMed] [Google Scholar]
- Kim S., Kim D., Cho S. W., Kim J., Kim J.-S. (2014). Highly Efficient RNA-Guided Genome Editing in Human Cells via Delivery of Purified Cas9 Ribonucleoproteins. Genome Res. 24 (6), 1012–1019. 10.1101/gr.171322.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B. P., Pattanayak V., Prew M. S., Tsai S. Q., Nguyen N. T., Zheng Z., et al. (2016). High-fidelity CRISPR-Cas9 Nucleases with No Detectable Genome-wide Off-Target Effects. Nature 529 (7587), 490–495. 10.1038/nature16526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B. P., Prew M. S., Tsai S. Q., Nguyen N. T., Topkar V. V., Zheng Z., et al. (2015a). Broadening the Targeting Range of Staphylococcus aureus CRISPR-Cas9 by Modifying PAM Recognition. Nat. Biotechnol. 33 (12), 1293–1298. 10.1038/nbt.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B. P., Prew M. S., Tsai S. Q., Topkar V. V., Nguyen N. T., Zheng Z., et al. (2015b). Engineered CRISPR-Cas9 Nucleases with Altered PAM Specificities. Nature 523 (7561), 481–485. 10.1038/nature14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S. C., Xie L., Deng W., Guglielmi B., Witkowsky L. B., Bosanac L., et al. (2015). Dynamics of CRISPR-Cas9 Genome Interrogation in Living Cells. Science 350 (6262), 823–826. 10.1126/science.aac6572 [DOI] [PubMed] [Google Scholar]
- Koike-Yusa H., Li Y., Tan E.-P., Velasco-Herrera M. D. C., Yusa K. (2014). Genome-wide Recessive Genetic Screening in Mammalian Cells with a Lentiviral CRISPR-Guide RNA Library. Nat. Biotechnol. 32 (3), 267–273. 10.1038/nbt.2800 [DOI] [PubMed] [Google Scholar]
- Konermann S., Brigham M. D., Trevino A. E., Hsu P. D., Heidenreich M., Le Cong L., et al. (2013). Optical Control of Mammalian Endogenous Transcription and Epigenetic States. nature 500 (7463), 472–476. 10.1038/nature12466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S., Brigham M. D., Trevino A. E., Joung J., Abudayyeh O. O., Barcena C., et al. (2015). Genome-scale Transcriptional Activation by an Engineered CRISPR-Cas9 Complex. Nature 517 (7536), 583–588. 10.1038/nature14136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotterman M. A., Schaffer D. V. (2014). Engineering Adeno-Associated Viruses for Clinical Gene Therapy. Nat. Rev. Genet. 15 (7), 445–451. 10.1038/nrg3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotterman M. A., Yin L., Strazzeri J. M., Flannery J. G., Merigan W. H., Schaffer D. V. (2015). Antibody Neutralization Poses a Barrier to Intravitreal Adeno-Associated Viral Vector Gene Delivery to Non-human Primates. Gene Ther. 22 (2), 116–126. 10.1038/gt.2014.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuhn M., Adams F. F., Ng M., Knoess S., Schambach A., Charpentier E. M., et al. (2017). Refined sgRNA Efficacy Prediction Improves Large- and Small-Scale CRISPR-Cas9 Applications. Nucleic Acids Res. 46 (3), 1375–1385. 10.1093/nar/gkx1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctôt C., Cheutin T., Cremer M., Cavalli G., Cremer T. (2007). Dynamic Genome Architecture in the Nuclear Space: Regulation of Gene Expression in Three Dimensions. Nat. Rev. Genet. 8 (2), 104–115. 10.1038/nrg2041 [DOI] [PubMed] [Google Scholar]
- Larcher T., Lafoux A., Tesson L., Remy S., Thepenier V., François V., et al. (2014). Characterization of Dystrophin Deficient Rats: a New Model for Duchenne Muscular Dystrophy. PloS one 9 (10), e110371. 10.1371/journal.pone.0110371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., Kwon H.-K., Shin H.-J., Nam G.-H., Kim J.-H., Choi S. (2017). Quasi-Stem Cells Derived from Human Somatic Cells by Chemically Modified Carbon Nanotubes. ACS Appl. Mater. Inter. 10 (10), 8417–8425. 10.1021/acsami.7b12914 [DOI] [PubMed] [Google Scholar]
- Lee J. K., Jeong E., Lee J., Jung M., Shin E., Kim Y. H., et al. (2018). Directed Evolution of CRISPR-Cas9 to Increase its Specificity. Nat. Commun. 9 (1), 1–10. 10.1038/s41467-018-05477-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.-F., Norville J. E., Aach J., McCormack M., Zhang D., Bush J., et al. (2013). Multiplex and Homologous Recombination-Mediated Genome Editing in Arabidopsis and Nicotiana Benthamiana Using Guide RNA and Cas9. Nat. Biotechnol. 31 (8), 688–691. 10.1038/nbt.2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Li X., Zhou Z., Wu P., Fang M., Pan X., et al. (2016). Reassessment of the Four Yield-Related Genes Gn1a, DEP1, GS3, and IPA1 in rice Using a CRISPR/Cas9 System. Front. Plant Sci. 7, 377. 10.3389/fpls.2016.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Kleinstiver B. P., Leon M. Y., Prew M. S., Navarro-Gomez D., Greenwald S. H., et al. (2018). Allele-specific CRISPR-Cas9 Genome Editing of the Single-Base P23H Mutation for Rhodopsin-Associated Dominant Retinitis Pigmentosa. CRISPR J. 1 (1), 55–64. 10.1089/crispr.2017.0009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-Q., Du D. (2014). Variation, Evolution, and Correlation Analysis of C+G Content and Genome or Chromosome Size in Different Kingdoms and Phyla. PLoS One 9 (2), e88339. 10.1371/journal.pone.0088339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Pan S., Zhang Y., Ren M., Feng M., Peng N., et al. (2015a). Harnessing Type I and Type III CRISPR-Cas Systems for Genome Editing. Nucleic Acids Res. 44 (4), e34. 10.1093/nar/gkv1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang Y., Lin J., Pan S., Han W., Peng N., et al. (2017). Cmr1 Enables Efficient RNA and DNA Interference of a III-B CRISPR-Cas System by Binding to Target RNA and crRNA. Nucleic Acids Res. 45 (19), 11305–11314. 10.1093/nar/gkx791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Liu Z.-B., Xing A., Moon B. P., Koellhoffer J. P., Huang L., et al. (2015b). Cas9-guide RNA Directed Genome Editing in Soybean. Plant Physiol. 169 (2), 960–970. 10.1104/pp.15.00783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Zhang H., Lou D., Yu D. (2016). Selection of Highly Efficient sgRNAs for CRISPR/Cas9-based Plant Genome Editing. Sci. Rep. 6, 21451. 10.1038/srep21451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P., Xu Y., Zhang X., Ding C., Huang R., Zhang Z., et al. (2015). CRISPR/Cas9-mediated Gene Editing in Human Tripronuclear Zygotes. Protein Cell 6 (5), 363–372. 10.1007/s13238-015-0153-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z., Chen K., Li T., Zhang Y., Wang Y., Zhao Q., et al. (2017). Efficient DNA-free Genome Editing of Bread Wheat Using CRISPR/Cas9 Ribonucleoprotein Complexes. Nat. Commun. 8, 14261. 10.1038/ncomms14261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. I., Paik E., Mishra B., Burkhardt D., Kernytsky A., Pettiglio M., et al. (2017). CRISPR/Cas9 Genome Editing to Treat Sickle Cell Disease and B-Thalassemia: Re-creating Genetic Variants to Upregulate Fetal Hemoglobin Appear Well-Tolerated, Effective and Durable. Am. Soc. Hematol. 130, 238. 10.1182/blood.V130.Suppl_1.284.284 [DOI] [Google Scholar]
- Lin S., Staahl B. T., Alla R. K., Doudna J. A. (2014a). Enhanced Homology-Directed Human Genome Engineering by Controlled Timing of CRISPR/Cas9 Delivery. elife 3, e04766. 10.7554/eLife.04766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Cradick T. J., Brown M. T., Deshmukh H., Ranjan P., Sarode N., et al. (2014b). CRISPR/Cas9 Systems Have Off-Target Activity with Insertions or Deletions between Target DNA and Guide RNA Sequences. Nucleic Acids Res. 42, 7473–7485. 10.1093/nar/gku402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Cradick T. J., Brown M. T., Deshmukh H., Ranjan P., Sarode N., et al. (2014c). CRISPR/Cas9 Systems Have Off-Target Activity with Insertions or Deletions between Target DNA and Guide RNA Sequences. Nucleic Acids Res. 42 (11), 7473–7485. 10.1093/nar/gku402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Hu R., Palla K. J., Tuskan G. A., Yang X. (2016a). Advances and Perspectives on the Use of CRISPR/Cas9 Systems in Plant Genomics Research. Curr. Opin. Plant Biol. 30, 70–77. 10.1016/j.pbi.2016.01.007 [DOI] [PubMed] [Google Scholar]
- Liu K. I., Ramli M. N. B., Woo C. W. A., Wang Y., Zhao T., Zhang X., et al. (2016b). A Chemical-Inducible CRISPR-Cas9 System for Rapid Control of Genome Editing. Nat. Chem. Biol. 10.1038/nchembio.2179 [DOI] [PubMed] [Google Scholar]
- Liu X., Homma A., Sayadi J., Yang S., Ohashi J., Takumi T. (2016c). Sequence Features Associated with the Cleavage Efficiency of CRISPR/Cas9 System. Sci. Rep. 6, 19675. 10.1038/srep19675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C., McAnally J. R., Shelton J. M., Mireault A. A., Bassel-Duby R., Olson E. N. (2014). Prevention of Muscular Dystrophy in Mice by CRISPR/Cas9-mediated Editing of Germline DNA. Science 345 (6201), 1184–1188. 10.1126/science.1254445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Naseri A., Reyes-Gutierrez P., Wolfe S. A., Zhang S., Pederson T. (2015a). Multicolor CRISPR Labeling of Chromosomal Loci in Human Cells. Cell Biology, 112, 3002–3007. 10.1073/pnas.1420024112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M., Ye A. Y., Zheng W., Kong L. (20132013). A Guide RNA Sequence Design Platform for the CRISPR/Cas9 System for Model Organism Genomes. Biomed. Research International. 10.1155/2013/270805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Zhang Q., Zhu Q., Liu W., Chen Y., Qiu R., et al. (2015b). A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 8 (8), 1274–1284. 10.1016/j.molp.2015.04.007 [DOI] [PubMed] [Google Scholar]
- MacLaren R. E., Groppe M., Barnard A. R., Cottriall C. L., Tolmachova T., Seymour L., et al. (2014). Retinal Gene Therapy in Patients with Choroideremia: Initial Findings from a Phase 1/2 Clinical Trial. The Lancet 383 (9923), 1129–1137. 10.1016/s0140-6736(13)62117-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddalo D., Manchado E., Concepcion C. P., Bonetti C., Vidigal J. A., Han Y.-C., et al. (2014). In Vivo engineering of Oncogenic Chromosomal Rearrangements with the CRISPR/Cas9 System. Nature 516 (7531), 423–427. 10.1038/nature13902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder M. L., Linder S. J., Cascio V. M., Fu Y., Ho Q. H., Joung J. K. (2013). CRISPR RNA-Guided Activation of Endogenous Human Genes. Nat. Methods 10 (10), 977–979. 10.1038/nmeth.2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder M. L., Stefanidakis M., Wilson C. J., Baral R., Barrera L. A., Bounoutas G. S., et al. (2019). Development of a Gene-Editing Approach to Restore Vision Loss in Leber Congenital Amaurosis Type 10. Nat. Med. 25 (2), 229–233. 10.1038/s41591-018-0327-9 [DOI] [PubMed] [Google Scholar]
- Majowicz A., Maczuga P., Kwikkers K. L., Marel S., Logtenstein R., Petry H., et al. (2013). Mir‐142‐3p Target Sequences Reduce Transgene‐directed Immunogenicity Following Intramuscular Adeno‐associated Virus 1 Vector‐mediated Gene Delivery. J. Gene Med. 15 (6-7), 219–232. 10.1002/jgm.2712 [DOI] [PubMed] [Google Scholar]
- Makarova K. S., Grishin N. V., Shabalina S. A., Wolf Y. I., Koonin E. V. (2006). A Putative RNA-Interference-Based Immune System in Prokaryotes: Computational Analysis of the Predicted Enzymatic Machinery, Functional Analogies with Eukaryotic RNAi, and Hypothetical Mechanisms of Action. Biol. Direct 1 (1), 7. 10.1186/1745-6150-1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K. S., Haft D. H., Barrangou R., Brouns S. J. J., Charpentier E., Horvath P., et al. (2011). Evolution and Classification of the CRISPR-Cas Systems. Nat. Rev. Microbiol. 9 (6), 467–477. 10.1038/nrmicro2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K. S., Koonin E. V. (2015). Annotation and Classification of CRISPR-Cas Systems. Methods Mol. Biol. 1311, 47–75. 10.1007/978-1-4939-2687-9_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K. S., Wolf Y. I., Alkhnbashi O. S., Costa F., Shah S. A., Saunders S. J., et al. (2015). An Updated Evolutionary Classification of CRISPR-Cas Systems. Nat. Rev. Microbiol. 13 (11), 722–736. 10.1038/nrmicro3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Aach J., Stranges P. B., Esvelt K. M., Moosburner M., Kosuri S., et al. (2013a). CAS9 Transcriptional Activators for Target Specificity Screening and Paired Nickases for Cooperative Genome Engineering. Nat. Biotechnol. 31 (9), 833–838. 10.1038/nbt.2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., et al. (2013b). RNA-guided Human Genome Engineering via Cas9. Science 339 (6121), 823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malina A., Cameron C. J. F., Robert F., Blanchette M., Dostie J., Pelletier J. (2015). PAM Multiplicity marks Genomic Target Sites as Inhibitory to CRISPR-Cas9 Editing. Nat. Commun. 6, 10124. 10.1038/ncomms10124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandegar M. A., Huebsch N., Frolov E. B., Shin E., Truong A., Olvera M. P., et al. (2016). CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell 18 (4), 541–553. 10.1016/j.stem.2016.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Zhang H., Xu N., Zhang B., Gou F., Zhu J.-K. (2013). Application of the CRISPR-Cas System for Efficient Genome Engineering in Plants. Mol. Plant 6 (6), 2008–2011. 10.1093/mp/sst121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Zhang Z., Feng Z., Wei P., Zhang H., Botella J. R., et al. (2016). Development of Germ-line-specific CRISPR-Cas9 Systems to Improve the Production of Heritable Gene Modifications inArabidopsis. Plant Biotechnol. J. 14 (2), 519–532. 10.1111/pbi.12468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini L. A., Sontheimer E. J. (2008). CRISPR Interference Limits Horizontal Gene Transfer in Staphylococci by Targeting DNA. science 322 (5909), 1843–1845. 10.1126/science.1165771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini L. A., Sontheimer E. J. (2010). Self versus Non-self Discrimination during CRISPR RNA-Directed Immunity. Nature 463 (7280), 568–571. 10.1038/nature08703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., Dougan S. K., Truttmann M. C., Bilate A. M., Ingram J. R., Ploegh H. L. (2015). Increasing the Efficiency of Precise Genome Editing with CRISPR-Cas9 by Inhibition of Nonhomologous End Joining. Nat. Biotechnol. 33 (5), 538–542. 10.1038/nbt.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H., Fujimoto N., Sasakawa N., Ohinata Y., Shima M., Yamanaka S., et al. (2014). Delivery of Full-Length Factor VIII Using a piggyBac Transposon Vector to Correct a Mouse Model of Hemophilia A. PloS one 9 (8), e104957. 10.1371/journal.pone.0104957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger L., Iliakis G. (1991). Kinetics of DNA Double-Strand Break Repair throughout the Cell Cycle as Assayed by Pulsed Field Gel Electrophoresis in CHO Cells. Int. J. Radiat. Biol. 59 (6), 1325–1339. 10.1080/09553009114551201 [DOI] [PubMed] [Google Scholar]
- Mikami M., Toki S., Endo M. (2015). Parameters Affecting Frequency of CRISPR/Cas9 Mediated Targeted Mutagenesis in rice. Plant Cel Rep 34 (10), 1807–1815. 10.1007/s00299-015-1826-5 [DOI] [PubMed] [Google Scholar]
- Miller J. C., Tan S., Qiao G., Barlow K. A., Wang J., Xia D. F., et al. (2010). A TALE Nuclease Architecture for Efficient Genome Editing. Nat. Biotechnol. 29 (2), 143–148. 10.1038/nbt.1755 [DOI] [PubMed] [Google Scholar]
- Mojica F. J. M., Díez-Villaseíor C. s., García-Martínez J. s., Soria E. (2005). Intervening Sequences of Regularly Spaced Prokaryotic Repeats Derive from Foreign Genetic Elements. J. Mol. Evol. 60 (2), 174–182. 10.1007/s00239-004-0046-3 [DOI] [PubMed] [Google Scholar]
- Mojica F. J. M., Diez-Villasenor C., Soria E., Juez G. (2000). Biological Significance of a Family of Regularly Spaced Repeats in the Genomes of Archaea, Bacteria and Mitochondria. Mol. Microbiol. 36 (1), 244–246. 10.1046/j.1365-2958.2000.01838.x [DOI] [PubMed] [Google Scholar]
- Moreno-Mateos M. A., Vejnar C. E., Beaudoin J.-D., Fernandez J. P., Mis E. K., Khokha M. K., et al. (2015). CRISPRscan: Designing Highly Efficient sgRNAs for CRISPR-Cas9 Targeting In Vivo . Nat. Methods 12 (10), 982–988. 10.1038/nmeth.3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou M. J., Bogdanove A. J. (2009). A Simple Cipher Governs DNA Recognition by TAL Effectors. Science 326 (5959), 1501. 10.1126/science.1178817 [DOI] [PubMed] [Google Scholar]
- Nakayama T., Fish M. B., Fisher M., Oomen-Hajagos J., Thomsen G. H., Grainger R. M. (2013). Simple and Efficient CRISPR/Cas9-mediated Targeted Mutagenesis inXenopus Tropicalis. genesis 51 (12), 835–843. 10.1002/dvg.22720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathwani A. C., Tuddenham E. G. D., Rangarajan S., Rosales C., McIntosh J., Linch D. C., et al. (2011). Adenovirus-Associated Virus Vector-Mediated Gene Transfer in Hemophilia B. N. Engl. J. Med. 365 (25), 2357–2365. 10.1056/nejmoa1108046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihongaki Y., Kawano F., Nakajima T., Sato M. (2015a). Photoactivatable CRISPR-Cas9 for Optogenetic Genome Editing. Nat. Biotechnol. 33 (7), 755–760. 10.1038/nbt.3245 [DOI] [PubMed] [Google Scholar]
- Nihongaki Y., Yamamoto S., Kawano F., Suzuki H., Sato M. (2015b). CRISPR-Cas9-based Photoactivatable Transcription System. Chem. Biol. 22 (2), 169–174. 10.1016/j.chembiol.2014.12.011 [DOI] [PubMed] [Google Scholar]
- Nishimasu H., Cong L., Yan W. X., Ran F. A., Zetsche B., Li Y., et al. (2015). Crystal Structure of Staphylococcus aureus Cas9. Cell 162 (5), 1113–1126. 10.1016/j.cell.2015.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H., Ran F. A., Hsu P. D., Konermann S., Shehata S. I., Dohmae N., et al. (2014). Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell 156 (5), 935–949. 10.1016/j.cell.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H., Shi X., Ishiguro S., Gao L., Hirano S., Okazaki S., et al. (2018). Engineered CRISPR-Cas9 Nuclease with Expanded Targeting Space. Science 361 (6408), 1259–1262. 10.1126/science.aas9129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim L., Perli S. D., Fridkin A., Perez-Pinera P., Lu T. K. (2014). Multiplexed and Programmable Regulation of Gene Networks with an Integrated RNA and CRISPR/Cas Toolkit in Human Cells. Mol. Cel. 54 (4), 698–710. 10.1016/j.molcel.2014.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y., Shen B., Cui Y., Chen Y., Wang J., Wang L., et al. (2014). Generation of Gene-Modified Cynomolgus Monkey via Cas9/RNA-Mediated Gene Targeting in One-Cell Embryos. Cell 156 (4), 836–843. 10.1016/j.cell.2014.01.027 [DOI] [PubMed] [Google Scholar]
- O'Geen H., Henry I. M., Bhakta M. S., Meckler J. F., Segal D. J. (2015). A Genome-wide Analysis of Cas9 Binding Specificity Using ChIP-Seq and Targeted Sequence Capture. Nucleic Acids Res. 43 (6), 3389–3404. 10.1093/nar/gkv137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orioli A., Pascali C., Pagano A., Teichmann M., Dieci G. (2012). RNA Polymerase III Transcription Control Elements: Themes and Variations. Gene 493 (2), 185–194. 10.1016/j.gene.2011.06.015 [DOI] [PubMed] [Google Scholar]
- Osakabe Y., Watanabe T., Sugano S. S., Ueta R., Ishihara R., Shinozaki K., et al. (2016). Optimization of CRISPR/Cas9 Genome Editing to Modify Abiotic Stress Responses in Plants. Sci. Rep. 6, 26685. 10.1038/srep26685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino D., Sharon I., Morovic W., Stahl B., Thomas B. C., Barrangou R., et al. (2015). CRISPR Immunity Drives Rapid Phage Genome Evolution in Streptococcus Thermophilus. MBio 6 (2), e00262–00215. 10.1128/mBio.00262-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Folkmann A., Rasoloson D., Seydoux G. (2015). High Efficiency, Homology-Directed Genome Editing in Caenorhabditis elegans Using CRISPR-Cas9 Ribonucleoprotein Complexes. Genetics 201, 47–54. 10.1534/genetics.115.179382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C., Ye L., Qin L., Liu X., He Y., Wang J., et al. (2016a). CRISPR/Cas9-mediated Efficient and Heritable Targeted Mutagenesis in Tomato Plants in the First and Later Generations. Sci. Rep. 6, 24765. 10.1038/srep24765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C., Ye L., Qin L., Liu X., He Y., Wang J., et al. (2016b). CRISPR/Cas9-mediated Efficient and Heritable Targeted Mutagenesis in Tomato Plants in the First and Later Generations. Sci. Rep. 6, 24765. 10.1038/srep24765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S., Boulton S. J. (2014). Double-strand Break Repair: 53BP1 Comes into Focus. Nat. Rev. Mol. Cel Biol 15 (1), 7–18. 10.1038/nrm3719 [DOI] [PubMed] [Google Scholar]
- Pattanayak V., Lin S., Guilinger J. P., Ma E., Doudna J. A., Liu D. R. (2013). High-throughput Profiling of Off-Target DNA Cleavage Reveals RNA-Programmed Cas9 Nuclease Specificity. Nat. Biotechnol. 31 (9), 839–843. 10.1038/nbt.2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson A. G., Chang J. T., Taylor C., Fineran P. C. (2015). Regulation of the Type I-F CRISPR-Cas System by CRP-cAMP and GalM Controls Spacer Acquisition and Interference. Nucleic Acids Res. 43 (12), 6038–6048. 10.1093/nar/gkv517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P., Kocak D. D., Vockley C. M., Adler A. F., Kabadi A. M., Polstein L. R., et al. (2013). RNA-guided Gene Activation by CRISPR-Cas9-Based Transcription Factors. Nat. Methods 10 (10), 973–976. 10.1038/nmeth.2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D., Meuleman W., Pagie L., Bruggeman S. W. M., Solovei I., Brugman W., et al. (2010). Molecular Maps of the Reorganization of Genome-Nuclear Lamina Interactions during Differentiation. Mol. Cel. 38 (4), 603–613. 10.1016/j.molcel.2010.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt R. J., Chen S., Zhou Y., Yim M. J., Swiech L., Kempton H. R., et al. (2014). CRISPR-Cas9 Knockin Mice for Genome Editing and Cancer Modeling. Cell 159 (2), 440–455. 10.1016/j.cell.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein L. R., Gersbach C. A. (2015). A Light-Inducible CRISPR-Cas9 System for Control of Endogenous Gene Activation. Nat. Chem. Biol. 11 (3), 198–200. 10.1038/nchembio.1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel C., Salvignol G., Vergnaud G. (2005). CRISPR Elements in Yersinia pestis Acquire New Repeats by Preferential Uptake of Bacteriophage DNA, and Provide Additional Tools for Evolutionary Studies. Microbiology 151 (3), 653–663. 10.1099/mic.0.27437-0 [DOI] [PubMed] [Google Scholar]
- Pozzoli U., Menozzi G., Fumagalli M., Cereda M., Comi G. P., Cagliani R., et al. (2008). Both selective and Neutral Processes Drive GC Content Evolution in the Human Genome. BMC Evol. Biol. 8 (1), 99. 10.1186/1471-2148-8-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., Arkin A. P., et al. (2013). Repurposing CRISPR as an RNA-Guided Platform for Sequence-specific Control of Gene Expression. Cell 152 (5), 1173–1183. 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahdar M., McMahon M. A., Prakash T. P., Swayze E. E., Bennett C. F., Cleveland D. W. (2015). Synthetic CRISPR RNA-Cas9-Guided Genome Editing in Human Cells. Proc. Natl. Acad. Sci. USA 112 (51), E7110–E7117. 10.1073/pnas.1520883112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna S., Kwaku Dad A.-B., Beloor J., Gopalappa R., Lee S.-K., Kim H. (2014). Gene Disruption by Cell-Penetrating Peptide-Mediated Delivery of Cas9 Protein and Guide RNA. Genome Res. 24 (6), 1020–1027. 10.1101/gr.171264.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Cong L., Yan W. X., Scott D. A., Gootenberg J. S., Kriz A. J., et al. (2015). In Vivo genome Editing Using Staphylococcus aureus Cas9. Nature 520 (7546), 186–191. 10.1038/nature14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Lin C.-Y., Gootenberg J. S., Konermann S., Trevino A. E., et al. (2013a). Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell 154 (6), 1380–1389. 10.1016/j.cell.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., Zhang F. (2013b). Genome Engineering Using the CRISPR-Cas9 System. Nat. Protoc. 8 (11), 2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan V., Wahlin K., Maruotti J., Zack D. J. (2014). Expansion of the CRISPR-Cas9 Genome Targeting Space through the Use of H1 Promoter-Expressed Guide RNAs. Nat. Commun. 5, 4516. 10.1038/ncomms5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath D., Amlinger L., Hoekzema M., Devulapally P. R., Lundgren M. (2014). Efficient Programmable Gene Silencing by Cascade. Nucleic Acids Res. 43 (1), 237–246. 10.1093/nar/gku1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner H. K., Sampson T. R., Weiss D. S. (2015). I Can See CRISPR Now, Even when Phage Are Gone. Curr. Opin. Infect. Dis. 28 (3), 267–274. 10.1097/qco.0000000000000154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud J.-B., Boix C., Charpentier M., De Cian A., Cochennec J., Duvernois-Berthet E., et al. (2016). Improved Genome Editing Efficiency and Flexibility Using Modified Oligonucleotides with TALEN and CRISPR-Cas9 Nucleases. Cel Rep. 14 (9), 2263–2272. 10.1016/j.celrep.2016.02.018 [DOI] [PubMed] [Google Scholar]
- Richardson C. D., Ray G. J., DeWitt M. A., Curie G. L., Corn J. E. (2016). Enhancing Homology-Directed Genome Editing by Catalytically Active and Inactive CRISPR-Cas9 Using Asymmetric Donor DNA. Nat. Biotechnol. 10.1038/nbt.3481 [DOI] [PubMed] [Google Scholar]
- Rodriguez E. (2016). Ethical Issues in Genome Editing Using Crispr/Cas9 System. J. Clin. Res. Bioeth. 07. 10.4172/2155-9627.1000266 [DOI] [Google Scholar]
- Rodriguez E., Keiser M., McLoughlin H., Zhang F., Davidson B. L. (2016). MOLECULAR THERAPY: NATURE PUBLISHING GROUP 75 VARICK ST, 9TH FLR. NEW YORK, NY 10013-1917 USA, S94. 10.1016/S1525-0016(16)35260-1()"AAV-CRISPR: a New Therapeutic Approach to Nucleotide Repeat Diseases [DOI] [Google Scholar]
- Rudin N., Sugarman E., Haber J. E. (1989). Genetic and Physical Analysis of Double-Strand Break Repair and Recombination in Saccharomyces cerevisiae . Genetics 122 (3), 519–534. 10.1093/genetics/122.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T., Nakade S., Sakane Y., Suzuki K.-I. T., Yamamoto T. (2016). MMEJ-assisted Gene Knock-In Using TALENs and CRISPR-Cas9 with the PITCh Systems. Nat. Protoc. 11 (1), 118–133. 10.1038/nprot.2015.140 [DOI] [PubMed] [Google Scholar]
- Sakuma T., Nishikawa A., Kume S., Chayama K., Yamamoto T. (2014). Multiplex Genome Engineering in Human Cells Using All-In-One CRISPR/Cas9 Vector System. Sci. Rep. 4, 5400. 10.1038/srep05400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rivera F. J., Papagiannakopoulos T., Romero R., Tammela T., Bauer M. R., Bhutkar A., et al. (2014). Rapid Modelling of Cooperating Genetic Events in Cancer through Somatic Genome Editing. Nature 516 (7531), 428–431. 10.1038/nature13906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J. D., Joung J. K. (2014). CRISPR-cas Systems for Editing, Regulating and Targeting Genomes. Nat. Biotechnol. 32 (4), 347–355. 10.1038/nbt.2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapranauskas R., Gasiunas G., Fremaux C., Barrangou R., Horvath P., Siksnys V. (2011). The Streptococcus Thermophilus CRISPR/Cas System Provides Immunity in Escherichia coli . Nucleic Acids Res. 39 (21), 9275–9282. 10.1093/nar/gkr606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer N. J., Narváez-Vásquez J., Mozoruk J., Miller R. B., Warburg Z. J., Woodward M. J., et al. (2016). Oligonucleotide-mediated Genome Editing Provides Precision and Function to Engineered Nucleases and Antibiotics in Plants. Plant Physiol. 170 (4), 1917–1928. 10.1104/pp.15.01696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk J. L., Gao L., Li D., Gardner Z., Strecker J., Lash B., et al. (2020). Highly Parallel Profiling of Cas9 Variant Specificity. Mol. Cel. 78 (4), 794e798–800. 10.1016/j.molcel.2020.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R., Grosschedl R. (2007). Dynamics and Interplay of Nuclear Architecture, Genome Organization, and Gene Expression. Genes Development 21 (23), 3027–3043. 10.1101/gad.1604607 [DOI] [PubMed] [Google Scholar]
- Schwank G., Koo B.-K., Sasselli V., Dekkers J. F., Heo I., Demircan T., et al. (2013). Functional Repair of CFTR by CRISPR/Cas9 in Intestinal Stem Cell Organoids of Cystic Fibrosis Patients. Cell stem cell 13 (6), 653–658. 10.1016/j.stem.2013.11.002 [DOI] [PubMed] [Google Scholar]
- Schwartz C. M., Hussain M. S., Blenner M., Wheeldon I. (2016). Synthetic RNA Polymerase III Promoters Facilitate High-Efficiency CRISPR-Cas9-Mediated Genome Editing in Yarrowia Lipolytica. ACS Synth. Biol. 5 (4), 356–359. 10.1021/acssynbio.5b00162 [DOI] [PubMed] [Google Scholar]
- Senís E., Fatouros C., Große S., Wiedtke E., Niopek D., Mueller A.-K., et al. (2014). CRISPR/Cas9-mediated Genome Engineering: An Adeno-Associated Viral (AAV) Vector Toolbox. Biotechnol. J. 9 (11), 1402–1412. 10.1002/biot.201400046 [DOI] [PubMed] [Google Scholar]
- Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., Mikkelsen T. S., et al. (2014). Genome-scale CRISPR-Cas9 Knockout Screening in Human Cells. Science 343 (6166), 84–87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J., Wang M., Yu G., Zhu S., Yu Y., Heng B. C., et al. (2018). Synthetic Far-Red Light-Mediated CRISPR-dCas9 Device for Inducing Functional Neuronal Differentiation. Proc. Natl. Acad. Sci. USA 115 (29), E6722–E6730. 10.1073/pnas.1802448115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechner D. M., Hacisuleyman E., Younger S. T., Rinn J. L. (2015). Multiplexable, Locus-specific Targeting of Long RNAs with CRISPR-Display. Nat. Methods 12 (7), 664–670. 10.1038/nmeth.3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Zhang W., Zhang J., Zhou J., Wang J., Chen L., et al. (2014). Efficient Genome Modification by CRISPR-Cas9 Nickase with Minimal Off-Target Effects. Nat. Methods 11 (4), 399–402. 10.1038/nmeth.2857 [DOI] [PubMed] [Google Scholar]
- Shin S. E., Lim J. M., Koh H. G., Kim E. K., Kang N. K., Jeon S., et al. (2016). CRISPR/Cas9-induced Knockout and Knock-In Mutations in Chlamydomonas Reinhardtii. Sci. Rep. 6, 27810. 10.1038/srep27810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S., Abudayyeh O. O., Makarova K. S., Wolf Y. I., Gootenberg J. S., Semenova E., et al. (2015). Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol. Cel. 60 (3), 385–397. 10.1016/j.molcel.2015.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigoillot F. D., Lyman S., Huckins J. F., Adamson B., Chung E., Quattrochi B., et al. (2012). A Bioinformatics Method Identifies Prominent Off-Targeted Transcripts in RNAi Screens. Nat. Methods 9 (4), 363–366. 10.1038/nmeth.1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi M., Hasnain S. E., Rosenthal B., Phan T., Luciani F., Shaw M.-A., et al. (2020). SARS-CoV-2 and COVID-19: A Genetic, Epidemiological, and Evolutionary Perspective. Infect. Genet. Evol. 84, 104384. 10.1016/j.meegid.2020.104384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker I. M., Gao L., Zetsche B., Scott D. A., Yan W. X., Zhang F. (2016). Rationally Engineered Cas9 Nucleases with Improved Specificity. Science 351 (6268), 84–88. 10.1126/science.aad5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J., Grizot S., Arnould S., Duclert A., Epinat J.-C., Chames P., et al. (2006). A Combinatorial Approach to Create Artificial Homing Endonucleases Cleaving Chosen Sequences. Nucleic Acids Res. 34 (22), e149. 10.1093/nar/gkl720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Yang D., Xu J., Zhu T., Chen Y. E., Zhang J. (2016). RS-1 Enhances CRISPR/Cas9- and TALEN-Mediated Knock-In Efficiency. Nat. Commun. 7, 10548. 10.1038/ncomms10548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg S. H., Doudna J. A. (2015). Expanding the Biologist's Toolkit with CRISPR-Cas9. Mol. Cel. 58 (4), 568–574. 10.1016/j.molcel.2015.02.032 [DOI] [PubMed] [Google Scholar]
- Stovicek V., Borodina I., Forster J. (2015). CRISPR-cas System Enables Fast and Simple Genome Editing of Industrial Saccharomyces cerevisiae Strains. Metab. Eng. Commun. 2, 13–22. 10.1016/j.meteno.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Zhang X., Wu C., He Y., Ma Y., Hou H., et al. (2016). Engineering Herbicide-Resistant rice Plants through CRISPR/Cas9-mediated Homologous Recombination of Acetolactate Synthase. Mol. Plant 9 (4), 628–631. 10.1016/j.molp.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Sunagawa G. A., Sumiyama K., Ukai-Tadenuma M., Perrin D., Fujishima H., Ukai H., et al. (2016). Mammalian Reverse Genetics without Crossing Reveals Nr3a as a Short-Sleeper Gene. Cel Rep. 14 (3), 662–677. 10.1016/j.celrep.2015.12.052 [DOI] [PubMed] [Google Scholar]
- Sung Y. H., Kim J. M., Kim H.-T., Lee J., Jeon J., Jin Y., et al. (2014). Highly Efficient Gene Knockout in Mice and Zebrafish with RNA-Guided Endonucleases. Genome Res. 24 (1), 125–131. 10.1101/gr.163394.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitashev S., Schwartz C., Lenderts B., Young J. K., Mark Cigan A. (2016). Genome Editing in maize Directed by CRISPR-Cas9 Ribonucleoprotein Complexes. Nat. Commun. 7, 13274. 10.1038/ncomms13274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitashev S., Young J. K., Schwartz C., Gao H., Falco S. C., Cigan A. M. (2015). Targeted Mutagenesis, Precise Gene Editing, and Site-specific Gene Insertion in maize Using Cas9 and Guide RNA. Plant Physiol. 169 (2), 931–945. 10.1104/pp.15.00793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiat M. A., Dashko S., den Ridder M., Wijsman M., van der Oost J., Daran J. M., et al. (2017). FnCpf1: a Novel and Efficient Genome Editing Tool for Saccharomyces cerevisiae . Nucleic Acids Res. 45 (21), 12585–12598. 10.1093/nar/gkx1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiech L., Heidenreich M., Banerjee A., Habib N., Li Y., Trombetta J., et al. (2015). In Vivo interrogation of Gene Function in the Mammalian Brain Using CRISPR-Cas9. Nat. Biotechnol. 33 (1), 102–106. 10.1038/nbt.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabebordbar M., Zhu K., Cheng J. K. W., Chew W. L., Widrick J. J., Yan W. X., et al. (2016). In Vivo gene Editing in Dystrophic Mouse Muscle and Muscle Stem Cells. Science 351 (6271), 407–411. 10.1126/science.aad5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taleei R., Nikjoo H. (2013). Biochemical DSB-Repair Model for Mammalian Cells in G1 and Early S Phases of the Cell Cycle. Mutat. Res. 756 (1), 206–212. 10.1016/j.mrgentox.2013.06.004 [DOI] [PubMed] [Google Scholar]
- Tanenbaum M. E., Gilbert L. A., Qi L. S., Weissman J. S., Vale R. D. (2014). A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging. Cell 159 (3), 635–646. 10.1016/j.cell.2014.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T.-H., Bachellerie J.-P., Rozhdestvensky T., Bortolin M.-L., Huber H., Drungowski M., et al. (2002). Identification of 86 Candidates for Small Non-messenger RNAs from the Archaeon Archaeoglobus Fulgidus. Proc. Natl. Acad. Sci. 99 (11), 7536–7541. 10.1073/pnas.112047299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. Q., Wyvekens N., Khayter C., Foden J. A., Thapar V., Reyon D., et al. (2014). Dimeric CRISPR RNA-Guided FokI Nucleases for Highly Specific Genome Editing. Nat. Biotechnol. 32 (6), 569–576. 10.1038/nbt.2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. Q., Zheng Z., Nguyen N. T., Liebers M., Topkar V. V., Thapar V., et al. (2015). GUIDE-seq Enables Genome-wide Profiling of Off-Target Cleavage by CRISPR-Cas Nucleases. Nat. Biotechnol. 33 (2), 187–197. 10.1038/nbt.3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov F. D., Miller J. C., Lee Y.-L., Beausejour C. M., Rock J. M., Augustus S., et al. (2005). Highly Efficient Endogenous Human Gene Correction Using Designed Zinc-finger Nucleases. Nature 435 (7042), 646–651. 10.1038/nature03556 [DOI] [PubMed] [Google Scholar]
- Vakulskas C. A., Dever D. P., Rettig G. R., Turk R., Jacobi A. M., Collingwood M. A., et al. (2018). A High-Fidelity Cas9 Mutant Delivered as a Ribonucleoprotein Complex Enables Efficient Gene Editing in Human Hematopoietic Stem and Progenitor Cells. Nat. Med. 24 (8), 1216–1224. 10.1038/s41591-018-0137-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkuijl S. A., Rots M. G. (2019). The Influence of Eukaryotic Chromatin State on CRISPR-Cas9 Editing Efficiencies. Curr. Opin. Biotechnol. 55, 68–73. 10.1016/j.copbio.2018.07.005 [DOI] [PubMed] [Google Scholar]
- Vouillot L., Thélie A., Pollet N. (2015). Comparison of T7E1 and Surveyor Mismatch Cleavage Assays to Detect Mutations Triggered by Engineered Nucleases. G3: Genes Genome| Gen.cs 5 (3), 407–415. 10.1534/g3.114.015834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton R. T., Christie K. A., Whittaker M. N., Kleinstiver B. P. (2020). Unconstrained Genome Targeting with Near-PAMless Engineered CRISPR-Cas9 Variants. Science 368 (6488), 290–296. 10.1126/science.aba8853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Wang C., Liu P., Lei C., Hao W., Gao Y., et al. (2016). Enhanced rice Blast Resistance by CRISPR/Cas9-targeted Mutagenesis of the ERF Transcription Factor Gene OsERF922. PloS one 11 (4), e0154027. 10.1371/journal.pone.0154027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng A. W., Zhang F., et al. (2013). One-step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-mediated Genome Engineering. cell 153 (4), 910–918. 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Zhang S., Wang W., Xiong X., Meng F., Cui X. (2015a). Efficient Targeted Mutagenesis in Potato by the CRISPR/Cas9 System. Plant Cel Rep 34 (9), 1473–1476. 10.1007/s00299-015-1816-7 [DOI] [PubMed] [Google Scholar]
- Wang T., Wei J. J., Sabatini D. M., Lander E. S. (2014). Genetic Screens in Human Cells Using the CRISPR-Cas9 System. Science 343 (6166), 80–84. 10.1126/science.1246981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.-P., Xing H.-L., Dong L., Zhang H.-Y., Han C.-Y., Wang X.-C., et al. (2015b). Egg Cell-specific Promoter-Controlled CRISPR/Cas9 Efficiently Generates Homozygous Mutants for Multiple Target Genes in Arabidopsis in a Single Generation. Genome Biol. 16 (1), 144. 10.1186/s13059-015-0715-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra E. R., Buckling A., Fineran P. C. (2014). CRISPR-cas Systems: beyond Adaptive Immunity. Nat. Rev. Microbiol. 12 (5), 317–326. 10.1038/nrmicro3241 [DOI] [PubMed] [Google Scholar]
- Whitworth K. M., Lee K., Benne J. A., Beaton B. P., Spate L. D., Murphy S. L., et al. (2014). Use of the CRISPR/Cas9 System to Produce Genetically Engineered Pigs from In Vitro-derived Oocytes and Embryos. Biol. Reprod. 91 (378), 78–13. 10.1095/biolreprod.114.121723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong N., Liu W., Wang X. (2015). Wu-CRISPR: Characteristics of Functional Guide RNAs for the CRISPR/Cas9 System. Genome Biol. 16 (1), 218. 10.1186/s13059-015-0784-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J. W., Kim J., Kwon S. I., Corvalán C., Cho S. W., Kim H., et al. (2015). DNA-free Genome Editing in Plants with Preassembled CRISPR-Cas9 Ribonucleoproteins. Nat. Biotechnol. 33 (11), 1162–1164. 10.1038/nbt.3389 [DOI] [PubMed] [Google Scholar]
- Wu X., Scott D. A., Kriz A. J., Chiu A. C., Hsu P. D., Dadon D. B., et al. (2014). Genome-wide Binding of the CRISPR Endonuclease Cas9 in Mammalian Cells. Nat. Biotechnol. 32 (7), 670–676. 10.1038/nbt.2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Liang D., Wang Y., Bai M., Tang W., Bao S., et al. (2013). Correction of a Genetic Disease in Mouse via Use of CRISPR-Cas9. Cell stem cell 13 (6), 659–662. 10.1016/j.stem.2013.10.016 [DOI] [PubMed] [Google Scholar]
- Wu Y., Zhou H., Fan X., Zhang Y., Zhang M., Wang Y., et al. (2015). Correction of a Genetic Disease by CRISPR-Cas9-Mediated Gene Editing in Mouse Spermatogonial Stem Cells. Cel Res 25 (1), 67–79. 10.1038/cr.2014.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyvekens N., Topkar V. V., Khayter C., Joung J. K., Tsai S. Q. (2015). Dimeric CRISPR RNA-Guided FokI-dCas9 Nucleases Directed by Truncated gRNAs for Highly Specific Genome Editing. Hum. Gene Ther. 26 (7), 425–431. 10.1089/hum.2015.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K., Minkenberg B., Yang Y. (2015). Boosting CRISPR/Cas9 Multiplex Editing Capability with the Endogenous tRNA-Processing System. Proc. Natl. Acad. Sci. USA 112 (11), 3570–3575. 10.1073/pnas.1420294112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Xiao T., Chen C.-H., Li W., Meyer C. A., Wu Q., et al. (2015a). Sequence Determinants of Improved CRISPR sgRNA Design. Genome Res. 25 (8), 1147–1157. 10.1101/gr.191452.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R.-F., Li H., Qin R.-Y., Li J., Qiu C.-H., Yang Y.-C., et al. (2015b). Generation of Inheritable and "transgene Clean" Targeted Genome-Modified rice in Later Generations Using the CRISPR/Cas9 System. Sci. Rep. 5, 11491. 10.1038/srep11491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Chen S., Yin H., Tammela T., Papagiannakopoulos T., Joshi N. S., et al. (2014a). CRISPR-mediated Direct Mutation of Cancer Genes in the Mouse Liver. Nature 514 (7522), 380–384. 10.1038/nature13589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z., Wu M., Wen K., Ren M., Long L., Zhang X., et al. (2014b). CRISPR/Cas9 Mediates Efficient Conditional Mutagenesis in Drosophila. G3: Genes, Genomes, Genet. 4 (11), 2167–2173. 10.1534/g3.114.014159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Wei S., Wu Y., Hu R., Li H., Yang W., et al. (2015). High-efficiency Genome Editing in Arabidopsis Using YAO Promoter-Driven CRISPR/Cas9 System. Mol. Plant 8 (12), 1820–1823. 10.1016/j.molp.2015.10.004 [DOI] [PubMed] [Google Scholar]
- Yang H., Wang H., Jaenisch R. (2014). Generating Genetically Modified Mice Using CRISPR/Cas-mediated Genome Engineering. Nat. Protoc. 9 (8), 1956–1968. 10.1038/nprot.2014.134 [DOI] [PubMed] [Google Scholar]
- Yang H., Wang H., Shivalila C. S., Cheng A. W., Shi L., Jaenisch R. (2013). One-step Generation of Mice Carrying Reporter and Conditional Alleles by CRISPR/Cas-mediated Genome Engineering. Cell 154 (6), 1370–1379. 10.1016/j.cell.2013.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Wang L., Bell P., McMenamin D., He Z., White J., et al. (2016). A Dual AAV System Enables the Cas9-Mediated Correction of a Metabolic Liver Disease in Newborn Mice. Nat. Biotechnol. 34 (3), 334–338. 10.1038/nbt.3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Steentoft C., Hauge C., Hansen L., Thomsen A. L., Niola F., et al. (2015). Fast and Sensitive Detection of Indels Induced by Precise Gene Targeting. Nucleic Acids Res. 43 (9), e59. 10.1093/nar/gkv126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesudhas D., Anwar M. A., Choi S. (2019). Structural Mechanism of DNA-Mediated Nanog-Sox2 Cooperative Interaction. RSC Adv. 9 (14), 8121–8130. 10.1039/c8ra10085c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesudhas D., Anwar M. A., Panneerselvam S., Kim H.-K., Choi S. (2017a). Evaluation of Sox2 Binding Affinities for Distinct DNA Patterns Using Steered Molecular Dynamics Simulation. FEBS open bio 7 (11), 1750–1767. 10.1002/2211-5463.12316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesudhas D., Batool M., Anwar M., Panneerselvam S., Choi S. (2017b). Proteins Recognizing DNA: Structural Uniqueness and Versatility of DNA-Binding Domains in Stem Cell Transcription Factors. Genes 8 (8), 192. 10.3390/genes8080192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Xue W., Chen S., Bogorad R. L., Benedetti E., Grompe M., et al. (2014). Genome Editing with Cas9 in Adult Mice Corrects a Disease Mutation and Phenotype. Nat. Biotechnol. 32 (6), 551–553. 10.1038/nbt.2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Liu M., Liu Y., Wu J., Gan T., Zhang W., et al. (2019). Optimizing Genome Editing Strategy by Primer-Extension-Mediated Sequencing. Cell Discov 5 (1), 18–11. 10.1038/s41421-019-0088-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka S., Fujii W., Ogawa T., Sugiura K., Naito K. (2015). Development of a Mono-Promoter-Driven CRISPR/Cas9 System in Mammalian Cells. Sci. Rep. 5, 18341. 10.1038/srep18341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Liu Y., Ma T., Liu K., Xu S., Zhang Y., et al. (2015). Small Molecules Enhance CRISPR Genome Editing in Pluripotent Stem Cells. Cell stem cell 16 (2), 142–147. 10.1016/j.stem.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K. (2013). Seamless Genome Editing in Human Pluripotent Stem Cells Using Custom Endonuclease-Based Gene Targeting and the piggyBac Transposon. Nat. Protoc. 8 (10), 2061–2078. 10.1038/nprot.2013.126 [DOI] [PubMed] [Google Scholar]
- Zalatan J. G., Lee M. E., Almeida R., Gilbert L. A., Whitehead E. H., La Russa M., et al. (2015). Engineering Complex Synthetic Transcriptional Programs with CRISPR RNA Scaffolds. Cell 160 (1-2), 339–350. 10.1016/j.cell.2014.11.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B., Gootenberg J. S., Abudayyeh O. O., Slaymaker I. M., Makarova K. S., Essletzbichler P., et al. (2015a). Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 163 (3), 759–771. 10.1016/j.cell.2015.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B., Volz S. E., Zhang F. (2015b). A Split-Cas9 Architecture for Inducible Genome Editing and Transcription Modulation. Nat. Biotechnol. 33 (2), 139–142. 10.1038/nbt.3149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Meng X., Wei X., Lu L. (2016a). Highly Efficient CRISPR Mutagenesis by Microhomology-Mediated End Joining in Aspergillus fumigatus. Fungal Genet. Biol. 86, 47–57. 10.1016/j.fgb.2015.12.007 [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhang J., Wei P., Zhang B., Gou F., Feng Z., et al. (2014). The CRISPR/Cas9 System Produces Specific and Homozygous Targeted Gene Editing in rice in One Generation. Plant Biotechnol. J. 12 (6), 797–807. 10.1111/pbi.12200 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Long C., Li H., McAnally J. R., Baskin K. K., Shelton J. M., et al. (2017). CRISPR-Cpf1 Correction of Muscular Dystrophy Mutations in Human Cardiomyocytes and Mice. Sci. Adv. 3 (4), e1602814. 10.1126/sciadv.1602814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liang Z., Zong Y., Wang Y., Liu J., Chen K., et al. (2016b). Efficient and Transgene-free Genome Editing in Wheat through Transient Expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617. 10.1038/ncomms12617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhang C., Liu W., Gao W., Liu C., Song G., et al. (2016). An Alternative Strategy for Targeted Gene Replacement in Plants Using a Dual-sgRNA/Cas9 Design. Sci. Rep. 6, 23890. 10.1038/srep23890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhu S., Cai C., Yuan P., Li C., Huang Y., et al. (2014). High-throughput Screening of a CRISPR/Cas9 Library for Functional Genomics in Human Cells. Nature 509 (7501), 487–491. 10.1038/nature13166 [DOI] [PubMed] [Google Scholar]
- Zhu Z., Verma N., González F., Shi Z.-D., Huangfu D. (2015). A CRISPR/Cas-mediated Selection-free Knockin Strategy in Human Embryonic Stem Cells. Stem Cel. Rep. 4 (6), 1103–1111. 10.1016/j.stemcr.2015.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuris J. A., Thompson D. B., Shu Y., Guilinger J. P., Bessen J. L., Hu J. H., et al. (2015). Cationic Lipid-Mediated Delivery of Proteins Enables Efficient Protein-Based Genome Editing In Vitro and In Vivo . Nat. Biotechnol. 33 (1), 73–80. 10.1038/nbt.3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.