Abstract
The inappropriate expression of the a-factor pheromone receptor (Ste3p) in the MATa cell leads to a striking inhibition of the yeast pheromone response, the result of a functional interaction between Ste3p and some MATa-specific protein. The present work identifies this protein as Asg7p. Normally, expression of Ste3p and Asg7p is limited to distinct haploid mating types, Ste3p to MATα cells and Asg7p to MATa cells. Artificial coexpression of the two in the same cell, either a or α, leads to dramatic inhibition of the pheromone response. Ste3p-Asg7p coexpression also perturbs the membrane trafficking of Ste3p: Ste3p turnover is slowed, a result of an Asg7p-mediated retardation of the secretory delivery of the newly synthesized receptor to the plasma membrane. However, in the absence of ectopic Ste3p expression, the asg7Δ mutation is without consequence either for pheromone signaling or overall mating efficiency of a cells. Indeed, the sole phenotype that can be assigned to MATa asg7Δ cells is observed following zygotic fusion to its α mating partner. Though formed at wild-type efficiency, zygotes from these pairings are morphologically abnormal. The pattern of growth is deranged: emergence of the first mitotic bud is delayed, and, in its place, growth is apparently diverted into a novel structure superficially resembling the polarized mating projection characteristic of haploid cells responding to pheromone. Together these results suggest a mechanism in which, following the zygotic fusion event, Ste3p and Asg7p gain access to one another and together act to repress the pheromone response, promoting the transition of the new diploid cell to vegetative growth.
Sexual identity in the yeast Saccharomyces cerevisiae is controlled by the MAT locus. Transcriptional activators and repressors encoded by MATa or MATα alleles control the expression of a limited number of cell type-specific gene products that distinguish the two haploid mating types, the a cell and the α cell (24, 31). Chief among the cell type-specific products expressed is a receptor-ligand system which is used to direct the mating of the a and the α cells to form the a/α diploid. The a cell secretes the farnesylated peptide a-factor and expresses at its surface the α-factor receptor (Ste2p), a G protein-coupled receptor which confers detection of the α-factor peptide specifically produced and secreted by the α cell. Likewise, the α cell expresses a distinct G protein-coupled receptor, the a-factor receptor (Ste3p), which detects the a-factor secreted by the a cell.
This system of pheromones and receptors enables the communication of mating (for reviews, see references 18 and 19). Detection of pheromone alerts the cell to the proximity of a potential mating partner and prepares the cell for conjugation both through transcriptional induction of mating genes and through arrest of the cell cycle in G1 at Start. Through polarized growth of the cell body, the two mating partners extend mating projections towards one another. The tips of the mating projections meet, cells adhere to one another, and, with the joining of the cell walls, the prezygote is formed. Finally, with dissolution of the intervening cell walls and subsequent fusion of cytoplasms and nuclei, the diploid zygote is formed: a cell with a characteristic dumbbell morphology, having two terminal bulbs derived from the cell bodies of the two haploid mating partners connected via a conjugation bridge derived from the tip-to-tip fusion of the two mating projections.
The mating process culminates with the transition of the new diploid cell to vegetative growth: the cells transition from G1 to S, DNA replication is initiated, and a first mitotic bud begins to emerge from the midpoint of the zygotic conjugation bridge. Prior to zygote formation, during mating, pheromone signaling activates the cell cycle kinase inhibitor protein Far1p, which binds to and inhibits the Cdc28/Cln cell cycle kinase, and thus causes G1 arrest. The reinitiation of the mitotic cycle for the new diploid cell requires that this inhibition be relieved.
The STE3DAF mutation is a STE3 promoter mutation which confers cell type-independent expression of the a-factor receptor (Ste3p) (13). With this mutant it was found that the inappropriate expression of Ste3p in the a cell context leads to a striking inhibition of the pheromone signal transduction pathway, apparently exerted at the level of the Gβγ component of the heterotrimeric G protein, i.e., the first postreceptor signaling step (6, 13, 17). Inhibition depends on Ste3p and at least one MATa-specific gene product that is neither a-factor nor Ste2p (13). As Ste3p and the hypothetical a-specific interactor normally reside in different cell types, the biological relevance of this striking interaction has remained a mystery. Kim et al. (17) have suggested that Ste3p and its a cell interactor may gain access to one another in the zygote following fusion of the two mating partners and that the resulting inhibition of the pheromone response could serve to promote the transition of the zygote into a vegetative mode of growth. The present work identifies the a-specific Ste3p interactor as Asg7p and provides evidence which suggests that the inhibition of the pheromone response provided by Ste3p-Asg7p may indeed play a role in promoting the transition of the zygote to vegetative growth.
MATERIALS AND METHODS
Plasmids.
The LEU2 CEN ARS pRS315-derived plasmid pND541 expresses an N-terminally hemagglutinin (HA)-tagged Ura3p from the GAL1 promoter. Construction of pND541 first required fusion of an 820-bp GAL1,10 promoter fragment to the PstI site just upstream of the URA3 open reading frame (ORF). A restriction site for XhoI was then introduced by oligonucleotide-directed mutagenesis between codons 3 and 4 of the URA3 ORF (the introduced sequence translates to the dipeptide Leu-Glu). Finally, a DNA duplex encoding a single copy of the HA epitope, creating by annealing the two oligonucleotides 5′-pTCGAGTACCCATACGATGTTCCAGATTACGCTG-3′ and 5′-pTCGACAGCGTAATCTGGAACATCGTATGGGTAC-3′ was introduced into the XhoI site.
The wild-type ASG7 gene was isolated from a YCp50-based yeast genomic library (25) through complementation of the asg7Δ::G418R allele of GAL1-STE3 GAL1-STE4 MATa strain NDY1050, isolating transformants capable of growth on galactose plates. The isolated library plasmid pND978 carries a 12-kb insert of genomic DNA that includes the ASG7 locus. To construct pND997, a 950-bp ADH1 promoter fragment isolated from pLexA (Clontech Laboratories, Inc., Palo Alto, Calif.) was inserted upstream of a 3.2-kb ASG7 ORF-containing fragment from pND978 carried on URA3 CEN ARS vector pRS316 (30). ADH1 and ASG7 sequences were fused through ligation at BamHI restriction sites introduced by oligonucleotide mutagenesis into the two sequences at positions 35 bp upstream of the initiator ATG codons for both ORFs. Plasmid p2664 carries the coding sequence for a green fluorescent protein (GFP)-Ras2 fusion construct (34) on the 2 μm LEU2 vector plasmid YEplac181 (12).
Strains.
The strains used are listed in Table 1. The asg7Δ::G418R disruption allele, which replaces the entire ASG7 ORF with the G418R marker from plasmid pFA-kanMX, was generated by PCR as described previously (33). The fus1Δ::URA3 allele of plasmid construct pSL671 and the fus2Δ::URA3 allele of p268 (complete descriptions of these plasmids are available from C. Boone upon request) were used to chromosomally transplace wild-type FUS1 and FUS2 alleles, respectively.
TABLE 1.
Yeast strains
| Strain | Genotype | Reference or source |
|---|---|---|
| IH1792 | MATα lys1 | Herskowitz laboratory |
| IH1793 | MATa lys1 cry1 | Herskowitz laboratory |
| Sc252 | MATa ura3-52 leu2-3,112 ade1 | 35 |
| SY1817a | MATα ste3Δ mfa1Δ mfa2Δ HIS3::FUS1-HIS3 | 7 |
| SY2150a | MATα GAL1-STE3 mfa1Δ mfa2Δ HIS3::FUS1-HIS3 | 7 |
| SY2555a | MATa GAL1-STE3 mfa1Δ mfa2Δ HIS33::FUS1-HIS3 | This work |
| SY2560a | MATα GAL1-STE3 rad16::GAL1-STE4 mfa1Δ mfa2Δ HIS3::FUS1-HIS3 | This work |
| SY2561a | MATa GAL1-STE3 rad16::GAL1-STE4 mfa1Δ mfa2Δ HIS3::FUS1-HIS3 | This work |
| NDY1050a | MATa asg7Δ::G418RGAL1-STE3 rad16::GAL1-STE4 mfa1Δ mfa2Δ HIS3::FUS1-HIS3 | This work |
| NDY1077a | MATa ste3Δ::LEU2 rad16::GAL1-STE4 mfa1Δ mfa2Δ HIS3::FUS1-HIS3 | This work |
| NDY1078a | MATa asg7Δ::G418Rste3Δ::LEU2 rad16::GAL1-STE4 mfa1Δ mfa2Δ HIS3::FUS1-HIS3 | This work |
| NDY1086a | MATa GAL1-STE3 asg7Δ::G418Rmfa1Δ mfa2Δ HIS3::FUS1-HIS3 | This work |
| NDY1123a | MATa GAL1-STE3 lys2::FUS1-LacZ bar1Δ mfa1Δ mfa2Δ HIS3::FUS1-HIS3 | This work |
| NDY1124a | MATa GAL1-STE3 asg7Δ::G418Rlys2::FUS1-LacZ bar1Δ mfa1Δ mfa2Δ HIS3::FUS1-HIS3 | This work |
| NDY1125a | MATa GAL1-STE3Δ365 asg7Δ::G418Rbar1Δ mfa1Δ mfa2Δ HIS3::FUS1-HIS3 | This work |
| NDY1131a | MATa GAL1-STE3 ASG7::ADH1-ASG7 lys2::FUS1-LacZ bar1Δ mfa1Δ mfa2Δ HIS3::FUS1-HIS3 | This work |
| NDY1132a | MATα GAL1-STE3 | This work |
| NDY1140a | MATa GAL1-STE3 | This work |
| NDY1141a | MATa GAL1-STE3Δ365 ASG7::ADH1-ASG7 bar1Δ mfa1Δ mfa2Δ HIS3::FUS1-HIS3 | This work |
| NDY1172a | MATa ste3Δ::LEU2 lys2::FUS1-LacZ bar1Δ mfa1Δ mfa2Δ HIS3::FUS1-HIS3 | This work |
| NDY1173a | MATa ste3Δ::LEU2 asg7Δ::G418Rlys2::FUS1-LacZ bar1Δ mfa1Δ mfa2Δ HIS3::FUS1-HIS3 | This work |
| NDY1174a | MATa ste3Δ::LEU2 ASG7::ADH1-ASG7 lys2::FUS1-LacZ bar1Δ mfa1Δ mfa2Δ HIS3::FUS1-HIS3 | This work |
| W303-1A | MATa ura3-1 leu2-3 his3-11,-15 trp1-1 ade2-1 can1-100 | Rothstein laboratory |
| W303-1Bb | MATα | Rothstein laboratory |
| NDY1089b | MATa asg7Δ::G418R | This work |
| NDY1171b | MATa ASG7::ADH1-ASG7 | This work |
| NDY1179b | MATα ASG7::ADH1-ASG7 | This work |
| NDY1200b | MATα GAL1-STE3Δ365 | This work |
| NDY1204b | MATα GAL1-STE3Δ365 ASG7::ADH1-ASG7 | This work |
| NDY1225b | MATα GAL1-STE3Δ365 ASG7::ADH1-ASG7 ste4Δ::LEU2 | This work |
| Y405b | MATa fus2Δ::URA3 | This work |
| Y411b | MATα fus2Δ::URA3 HIS3::FUS1-HIS3 mfa2Δ::FUS1-LacZ | This work |
| Y427b | MATa fus1Δ::URA3 HIS3::FUS1-HIS3 mfa2Δ::FUS1-LacZ | This work |
| Y428b | MATα fus1Δ::URA3 TRP1 | This work |
| Y2615b | MATa asg7Δ::G418Rfus2Δ::URA3 | This work |
| Y2617b | MATa asg7Δ::G418Rfus1Δ::URA3 | This work |
| NDY343 | MATα ste3Δ::LEU2 ura3 leu2 his4 bar1-1 | 28 |
| NDY349c | MATα GAL1-STE3Δ365 | This work |
| NDY1176 | MATa/α GAL1-STE3Δ365/STE3 ASG7+/ASG7+ | This work |
| NDY1178d | MATa/α GAL1-STE3Δ365/STE3 ASG7::ADH1-ASG7/ASG7 | This work |
| NDY1185d | MATα/α GAL1-STE3Δ365/STE3 ASG7+/ASG7+ | This work |
| NDY1186d | MATα/α GAL1-STE3Δ365/STE3 ASG7::ADH1-ASG7/ASG7 | This work |
Isogenic to Sc252.
Isogenic to W303-1A.
Isogenic to NDY343.
Isogenic to NDY1176.
Most of the other strains constructed for this work utilized the two-step gene replacement strategy (29) as previously described (27). Many of these integrating plasmid constructs used for these replacements have been described previously, including replacement of MFA2 by mfa2Δ::FUS1-LacZ (14), replacement of RAD16 by rad16::GAL1-STE4 (14), replacement of BAR1 by bar1Δ (14), and finally replacement of STE3 by either ste3Δ::LEU2 (27), GAL1-STE3 (26), or GAL1-STE3Δ365 (10).
Several additional gene replacement plasmids were constructed for the present work. The integrating plasmid for ste4Δ::LEU2, pND1103, derives from pRS306 (30), having a functional LEU2 fragment replacing an internal, 605-bp HindIII fragment of STE4. Chromosomal integration was directed by cleavage at a unique XhoI site within the remaining C-terminal portion of the STE4 ORF sequence. For construction of the lys2::FUS1-LacZ integrating plasmid, a 6.7-kb PstI-to-SalI FUS1-LacZ fragment from pSL553 (14) having the PstI end modified by an added KpnI linker was inserted into a 4.8-kb EcoRI-HindIII LYS2 fragment between a KpnI site and an XhoI site, 1.14 and 2.87 kb downstream of the LYS2 initiator ATG. Chromosomal integration utilized cleavage at a unique BglII site within sequences upstream of LYS2. The ASG7::ADH1-ASG7 gene replacement construct, carried on URA3 integrating plasmid pRS306 (30), is derived from pND997 through introduction of a 3.6-kb fragment of ASG7 upstream flanking sequence from pND978. In the resulting construct the 950-bp ADH1 promoter fragment replaces the putative ASG7 promoter element (removing sequences 420 to 35 bp upstream of the ASG7 initiator codon). Chromosomal integration of this construct into asg7Δ::G418R strains was directed by cleavage at a unique XhoI site located just downstream of the ASG7 ORF.
NDY1176 and NDY1178, are the a/α diploid products of matings of MATα GAL1-STE3Δ365 strain NDY349 with wild-type MATa strain W303-1A or with MATa ADH1-ASG7 strain NDY1171, respectively. The α/α diploid strains NDY1185 and NDY1186 were derived from NDY1176 and NDY1178, respectively, via HO-mediated mating type switching.
Ste3p turnover.
Ste3p turnover was monitored via a nonradioactive pulse-chase protocol as previously described (26). Briefly, a pulse of Ste3p synthesis was induced from GAL1-STE3 strains with the addition of galactose (2%) to cultures growing in yeast extract-peptone (YP)-raffinose (2%) medium. The chase period was initiated with the addition of glucose (3%), and, at various times thereafter, culture aliquots were removed for protein extract preparation (26). Extracts were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then Western analysis with affinity-purified rabbit polyclonal Ste3p-specific antibodies (28) or with the HA.11 monoclonal antibody (Berkeley Antibody Co., Berkeley, Calif.).
β-Galactosidase assays.
FUS1-LacZ bar1Δ MATa cells were cultured in YP-galactose (2%) medium and then treated with different concentrations of α-factor for 1 h. Culture aliquots were collected by centrifugation, cells were permeabilized, and β-galactosidase activity was determined as described previously (15).
Protease digestion of intact cells.
The treatment of whole cells with pronase (Calbiochem-Novabiochem Corp., La Jolla, Calif.) and subsequent extract preparation were as described previously (7).
Matings.
For quantitation of mating, aliquots containing approximately 106 cells taken from log-phase cultures of MATa and MATα strains were mixed together and then applied, under gentle vacuum pressure, as an approximately 1-cm-diameter spot onto a 2.5-cm-diameter, 0.45-μm-pore-size nitrocellulose filter (Millipore, Inc.). The filter was then transferred to a 30°C yeast extract-peptone-dextrose (YPD) plate and incubated for 3 h. Cells were then eluted from filters with 1 ml of synthetic dextrose medium, subjected to a 3-s sonication at the lowest power setting of a Branson Sonifier 450, and then immediately diluted and plated onto medium selective for the diploid cells. For the microscopic inspection of zygotic and prezygotic morphology, matings were performed as described above except that 107 cells of the two mating partners were applied to filters, filters were incubated on YPD plates at 30°C for various times, and then cells were eluted with 1 ml of 0.15 M NaCl–3.7% formaldehyde and stored at 4°C prior to analysis.
RESULTS
Impaired Ste3p turnover in MATa cells.
The yeast a-factor receptor (Ste3p) is rapidly turned over via an endocytic mechanism that transports the receptor from the cell surface to the vacuole for degradation by the resident proteases (7). In its normal MATα cell context, the Ste3p turnover half-life is about 15 min whether the protein is expressed from its natural α-cell-specific promoter (7) or from the GAL1 promoter (Fig. 1) (26). We find a different result for Ste3p ectopically expressed in the MATa context (Fig. 1). Quantification of the rate of this Ste3p loss indicates that turnover is indeed slowed in the a cell context relative to that in α cells (data not shown), indicating either that some α-specific protein(s) acts to augment Ste3p turnover or, alternatively, that an a-specific gene product somehow interferes with the Ste3p turnover mechanism. In the a/α diploid cell context, we find rapid Ste3p turnover equivalent to that seen in α cells (data not shown), suggesting that the slowed turnover of a cells likely is the result of interference by some a-specific gene product.
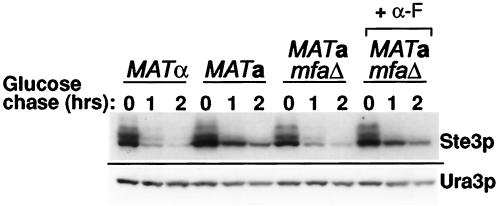
FIG. 1.
Slowed Ste3p turnover in MATa cells responding to pheromone. Turnover of Ste3p was monitored in three isogenic yeast strains carrying a GAL1-STE3 construct in place of the wild-type STE3 locus: the wild-type MATα strain NDY1132, the wild-type MATa strain NDY1140, and the mfa1Δ mfa2Δ MATa strain SY2555. In addition, strains were transformed by pND541, a plasmid which expresses an HA-tagged Ura3p from the GAL1 promoter. A 3-h period of Ste3p and HA-Ura3p expression was induced from cultures growing in raffinose medium with the addition of galactose and subsequent termination by glucose addition. One of two SY2555 cultures was treated with two doses of α-factor: a dose of 6 × 10−6 M administered 30 min prior to glucose addition and a dose of 15 × 10−5 M administered 30 min after the glucose addition. Protein extracts were prepared from culture aliquots, removed at the time of glucose addition (0-h time point) and at indicated times thereafter, and subjected to Western analysis both with Ste3p-specific antibodies (top) and with the HA.11 monoclonal antibody (bottom). The HA-Ura3p control provides the experiment with an internal standard of a protein not subject to rapid turnover.
Two obvious candidates for the responsible a-specific gene product are the α-factor receptor (Ste2p) and the a-factor pheromone. While deletion of the α-factor receptor structural gene (STE2) is without consequence for the slowed Ste3p turnover seen in MATa cells (data not shown), deletion of the two structural genes for a-factor, MFA1 and MFA2, in fact restores rapid Ste3p turnover to these MATa cells (Fig. 1). This finding could indicate that the liganded receptor turns over more slowly. Alternatively, slowed turnover could be a secondary consequence of pheromone response pathway activation; forced expression of Ste3p in MATa cells initiates an autocrine signaling loop (1). We were interested to see, therefore, if added α-factor, acting through the α-factor receptor (Ste2p) of these MATa GAL1-STE3 mfa1Δ mfa2Δ cells, might compensate for the a-factor deficiency. With the addition of α-factor, Ste3p turnover is again slowed (Fig. 1), indicating that the slow turnover, rather than being a direct consequence of a-factor binding, is instead likely a secondary consequence of pheromone response pathway activation.
While the cell type-specific effect on Ste3p turnover could indicate that endocytic mechanisms are deranged in cells responding to pheromone, we thought it also might fit within the constellation of phenotypes described for the STE3DAF allele (13). The inhibition of the pheromone signaling pathway induced through the inappropriate expression of Ste3p in MATa cells depends on the presence of at least one a-specific gene product that is neither a-factor nor Ste2p (13). If the impaired Ste3p turnover seen in MATa cells also reflects an interaction with this hypothetical a-specific gene product, then our results (Fig. 1) would indicate that this gene also is likely to be pheromone inducible.
ASG7.
Genome-wide analyses have identified ASG7 as an a-specific gene that is strongly induced by α-factor (24, 36). ASG7 encodes a protein of 214 residues with two potential transmembrane domains (Saccharomyces Genome Database). Asg7p shows no significant homology to other yeast or mammalian proteins. Below, we test if ASG7 is the a-specific gene responsible for both the STE3DAF phenotypes and slowed Ste3p turnover in MATa cells.
MATa STE3DAF cells overcome the terminal G1 arrest instigated by high doses of α-factor, a mutational disabling of the Gα subunit (a gpa1ts mutation), or overproduction of the Gβ subunit Ste4p (13). Haploid cells carrying a GAL1-STE4 construct fail to plate on galactose medium (Fig. 2): Gβ overproduction is thought to allow escape of the Gβγ component from Gα inhibition, thus activating the signaling pathway (5, 21, 35). The Daf phenotype is apparent with introduction of GAL1-STE3 into the GAL1-STE4 MATa cells, i.e., robust growth is restored (Fig. 2). This suppression of G1 arrest is a cell specific: MATα GAL1-STE4 GAL1-STE3 cells grow quite poorly on galactose (Fig. 2).
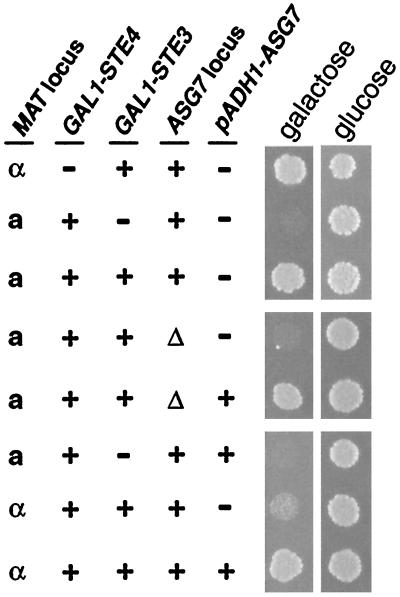
FIG. 2.
ASG7 is the a-specific gene responsible for the Daf phenotype. Approximately 300 cells were spotted onto rich YP plates containing either 2% glucose or 2% galactose and allowed to grow for 2 days at 30°C. Cells from eight isogenic strains SY2150, SY2560, SY2561, NDY1050, and NDY1077 (see Table 1 for genotypes), as well as SY2560, NDY1050, and NDY1078, transformed by pND997 (pADH1-ASG7), a plasmid construct which constitutively expresses ASG7 from the ADH1 promoter, were tested. The relevant features of the strain genotypes are indicated to the left of growth spots.
To test if the suppression of G1 arrest mediated by Ste3p in a cells requires ASG7, a asg7Δ::G418R version of the MATa GAL1-STE4 GAL1-STE3 strain was constructed. With ASG7 disrupted, the strain failed to plate to galactose medium (Fig. 2). The failed growth is due to failed GAL1-STE4 repression and not to a requirement of ASG7 for growth: in the absence of GAL1-STE4, the asg7Δ mutation results in no growth defect in a or α cells growing on either galactose or glucose medium (data not shown). We conclude that ASG7 is indeed the a-specific gene that collaborates with Ste3p to suppress activation of the pheromone response pathway.
As ASG7 expression is limited to a cells (24, 36), we have constructed an ADH1-ASG7 allele to examine the effects of ASG7 expression in the α-cell context. Constitutive ASG7 expression, we find, is not deleterious to cell growth in a or α cells (data not shown). The ADH1-ASG7 allele complements asg7Δ: introduction of the ADH1-ASG7 plasmid into MATa GAL1-STE4 GAL1-STE3 asg7Δ cells restores growth on galactose (Fig. 2). This suppression of GAL1-STE4 remains dependent on Ste3p coexpression: the ADH1-ASG7 plasmid fails to restore growth on galactose to MATa GAL1-STE4 asg7Δ cells (Fig. 2). In the MATα context, forced ASG7 expression restores robust growth on galactose medium to MATα GAL1-STE4 GAL1-STE3 cells (Fig. 2), indicating that ASG7 indeed is the only a-specific gene required for suppression of GAL1-STE4. Again, as in a cells, suppression in α cells also depends on Ste3p coexpression: the MATα GAL1-STE4 ste3Δ ADH1-ASG7 strain fails to grow on galactose (data not shown).
Coexpression of Asg7p and Ste3p blocks pheromone-induced transcription.
We have examined the effects of Asg7p and Ste3p coexpression on the α-factor-induced transcription of the pheromone-inducible gene FUS1 by assessing β-galactosidase levels induced from a FUS1-LacZ reporter construct. To eliminate the potential for autocrine signaling through Ste3p, mfa1Δ mfa2Δ MATa cells were used. Responses were assessed over a 10,000-fold α-factor concentration range. Consistent with the previous description of the Daf phenotype (13), ectopic expression of Ste3p in MATa strongly impairs the α-factor response (Fig. 3; compare GAL1-STE3 ASG7+ to ste3Δ ASG7+). Wild-type responsiveness is restored to MATa GAL1-STE3 cells through ASG7 disruption (Fig. 3; see GAL1-STE3 asg7Δ), indicating again that Ste3p and Asg7p together are required for the inhibition of pheromone signaling. We have also investigated the effects of constitutive expression of ASG7 from the ADH1-ASG7 construct. In terms of FUS1-LacZ induction, a much more pronounced inhibition was apparent: MATa GAL1-STE3 ADH1-ASG7 cells failed to show a detectable response to α-factor (Fig. 3). Again, this inhibition wholly depended on Ste3p coexpression (Fig. 3; see ste3Δ ADH1-ASG7). The difference between the responsiveness of MATa GAL1-STE3 ADH1-ASG7 cells and that of MATa GAL1-STE3 ASG7wt cells may be a reflection of the initial levels of Asg7p present in the two types of cells at the time of α-factor addition. ASG7 is strongly inducible, and expression is virtually undetectable in MATa cells unstimulated by pheromone (16, 24, 36). Inhibition of signaling in the GAL1-STE3 ASG7wt cells may await the new Asg7p that is synthesized following the pheromone challenge. Indeed, a previous kinetic analysis of the α-factor response of MATa STE3DAF cells showed that inhibition of signaling occurred only following an initial lag period: at early times following α-factor addition, the response was found to be nearly wild type (17). With constitutive Asg7p expression from the ADH1 promoter, both Ste3p and Asg7p would be present in the cell at the time of the α-factor challenge, possibly eliminating this lag in the institution of the Asg7p-Ste3p repression.
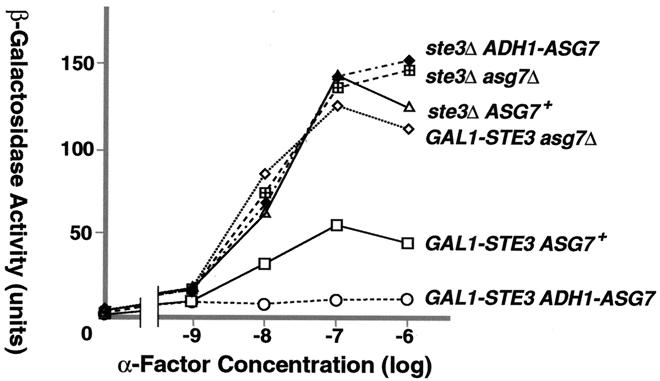
FIG. 3.
Ste3p and Asg7p coexpression impairs the response to α-factor. β-Galactosidase activity from a FUS1-LacZ reporter construct was used as a measure of the α-factor-induced transcriptional response in six isogenic FUS1-LacZ bar1Δ STE2+ MATa strains: NDY1123, NDY1124, NDY1131, NDY1172, NDY1173, and NDY1174 (see Table 1 for strain genotypes). Cultures growing in YP-galactose (2%) medium were treated for 1 h with 10−9, 10−8, 10−7, 10−6, or 10−5 M α-factor or were mock treated in parallel with no pheromone. Dose-response curves for induced β-galactosidase activity versus α-factor concentration are shown. As indicated to the right of each of the plots, the six strains differ both at the STE3 locus, which is ste3Δ or GAL1-STE3, and at the ASG7 locus, which is wild-type ASG7, asg7Δ, or ADH1-ASG7.
We conclude that Ste3p and Asg7p coexpression leads to a striking inhibition of the pheromone response. However, in the absence of Ste3p coexpression (the usual situation in a cells), we can discern no effect of either ASG7 disruption or overproduction on the cell's response to pheromone. Inhibition requires the presence of both proteins. By itself, Asg7p has no capacity for modulating the pheromone response.
Effects of Asg7p on Ste3p turnover and localization.
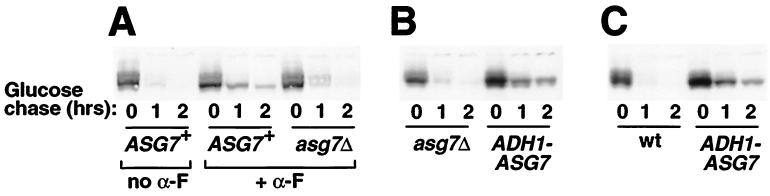
Next, we examined if Asg7p is responsible for the slowed Ste3p turnover observed in a cells (Fig. 1). MATa mfa1Δ mfa2Δ GAL1-STE3 cells that were either ASG7+ or asg7Δ were treated with α-factor to induce ASG7 expression, and Ste3p turnover was monitored (Fig. 4A). Disruption of ASG7 restores rapid turnover to these a cells, indicating that ASG7 is required for the slowed turnover previously observed in a cells (Fig. 1). Furthermore, with expression of ASG7 from the ADH1 promoter, slowed Ste3p turnover is apparent in both the a- and α-cell contexts in the absence of added pheromone (Fig. 4B and C), indicating that for the slowed Ste3p turnover phenotype, pheromone is required simply for establishing elevated levels of Asg7p within the cell and not for activating Asg7p or Ste3p in some other way.
FIG. 4.
Asg7p is responsible for the slowed Ste3p turnover. Ste3p turnover was assessed by Western blotting as in Fig. 1; the loss of Ste3 antigen following the glucose repression of GAL1-STE3 constructs was monitored. (A) ASG7 encodes the pheromone-inducible factor responsible for slowed Ste3p turnover. The isogenic ASG7+ and asg7Δ MATa GAL1-STE3 mfa1Δ mfa2Δ strains SY2555 and NDY1086 were cultured and treated with α-factor or were mock treated as described above for Fig. 1. (B) Constitutively expressed ASG7 suffices to retard Ste3p turnover in a cells. The isogenic asg7Δ, and ADH1-ASG7 MATa GAL1-STE3 mfa1Δ mfa2Δ strains NDY1124 and NDY1131 were cultured as described above for Fig. 1, except α-factor treatment was omitted. (C) Ectopic ASG7 expression in α cells retards Ste3p turnover. The GAL1-STE3 MATα SY2150 cells transformed by the ADH1-ASG7 plasmid pND997 or by the empty-vector plasmid (wt) were cultured as described for Fig. 1, except α-factor treatment was omitted.
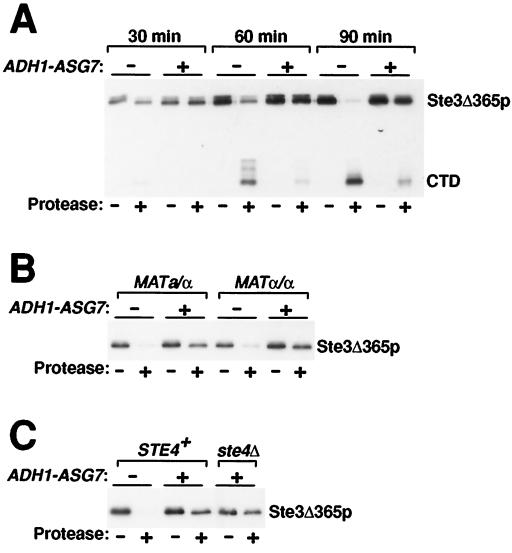
Impaired turnover could reflect impaired Ste3p endocytosis. Alternatively, Asg7p could act at a prior step. For instance, impaired delivery of newly synthesized Ste3p to the cell surface also would have the effect of slowing overall turnover. To test if Asg7p might act to impair the secretory delivery of Ste3p to the cell surface, we compared the rate at which the newly synthesized Ste3p truncation mutant Ste3Δ365p arrives at the cell surface in MATa GAL1-STE3Δ365 cells that were asg7Δ with that in cells that were ADH1-ASG7. The Δ365 mutation removes the signal for constitutive endocytosis; consequently, Ste3Δ365p does not turn over and instead stably accumulates at the plasma membrane. Synthesis of Ste3Δ365p was induced with galactose addition, and the rate of appearance at the cell surface was assessed using our standard protease-shaving protocol (7, 27). For this, intact cells are subjected to digestion by added, extracellular proteases; receptors that localize to the plasma membrane are susceptible to digestion, while receptors that localize intracellularly are resistant. Besides causing the loss of plasma membrane-localized receptor proteins, protease shaving results in the appearance of a characteristic 15-kDa digestion product that corresponds to the protected cytoplasmic tail domain plus the most C-terminal of the seven receptor transmembrane domains (7). At the initial time point, 30 min following the induction of synthesis, the bulk of the Δ365 receptor resisted digestion in both the asg7Δ and ADH1-ASG7 cells (Fig. 5A); presumably the bulk of the newly synthesized receptor had yet to arrive at the plasma membrane. In the asg7Δ background, with increasing time, an increasing fraction of the total receptor population became available at the cell surface for digestion (Fig. 5A). This is apparent at the 60-min time point and is especially clear following a 30-min chase period in which continued receptor synthesis was shut down through glucose-mediated repression of the GAL1 promoter (Fig. 5A; the 90-min time point): >90% of the receptor protein in wild-type cells now resided at the plasma membrane. Quite a different outcome is observed with the ADH1-ASG7 cells. Even following the 30-min glucose chase period, only a small fraction of the receptor population arrived at the cell surface (Fig. 5A). We conclude that Asg7p severely disrupts Ste3Δ365p delivery to the cell surface. Thus, the impaired turnover seen for Ste3p with Asg7p coexpression (Fig. 1 and 4) is likely secondary to its impaired surface delivery; for endocytosis to commence, the receptor must first reach the plasma membrane.
FIG. 5.
Asg7p delays Ste3p delivery to the cell surface. A 60-min period of receptor expression from GAL1-STE3Δ365 was induced with galactose addition and terminated with the subsequent addition of glucose. Cell growth was continued for an additional 30 min, affording the newly synthesized receptor the opportunity to reach the cell surface. Culture aliquots at various time points were either treated with proteases (+) or were mock treated in parallel (−) (see Materials and Methods). Protein extracts prepared from these cells were subjected to Western analysis using Ste3p-specific antibodies. (A) Constitutive ASG7 expression delays the delivery of Ste3p to the cell surface in MATa cells. MATa GAL1-STE3Δ365 mfa1Δ mfa2Δ cells, either asg7Δ (NDY1125) or ADH1-ASG7 (NDY1141), were cultured as described above. At 30 and 60 min following initiation of the galactose-induced pulse and 30 min after the subsequent addition of glucose (the 90-min time point), culture aliquots were removed for protease shaving. Indicated at right are the positions both of the undigested Ste3Δ365p receptor and of a receptor digestion product corresponding to the protected cytoplasmic tail domain (CTD) plus the most C-terminal of the seven receptor transmembrane domains. (B) The Asg7p-mediated delay in Ste3p surface delivery occurs in a/α diploid cells which do not express the subunits of the heterotrimeric G protein. Localization of the Ste3Δ365p expressed in MATa/α and MATα/α cells cultured as described above was assessed at the 90-min time point (following the 30-min glucose chase period) via the protease-shaving protocol. The MATa/α strains used, NDY1176 and NDY1178, and the MATα/α strains used, NDY1185 and NDY1186, were ASG7+/ASG7+ and ADH1-ASG7/ASG7+, respectively. (Wild-type ASG7 is expected to be transcriptionally silent in both a/α and α/α contexts [24]). For brevity, the portion of the gel displaying the lower-molecular-weight CTD fragment is not shown. (C) The Asg7p-mediated delay in Ste3p surface delivery occurs in cells with the Gβ subunit-encoding gene STE4 deleted. MATα GAL1-STE3Δ365 cells (NDY1200) as well as MATα GAL1-STE3Δ365 ADH1-ASG7 (NDY1204) and MATα GAL1-STE3Δ365 ADH1-ASG7 ste4Δ::LEU2 cells (NDY1225) were cultured and treated with the protease-shaving protocol as described for panel B.
Previous studies have indicated a central role for Gβγ in the Daf phenotype (6, 17). We were interested to investigate, therefore, G protein involvement in Asg7p inhibition of Ste3p transport to the cell surface. For this, we investigated the ability of Asg7p to block the surface delivery of Ste3Δ365p in the MATa/α diploid cell context; in a/α cells, the three G protein subunits are transcriptionally repressed (31). As a control, we constructed isogenic α/α pseudodiploids; like MATa or MATα haploid cells, MATα/α cells are expected to express the G protein subunits. In the α/α cells, Asg7p coexpression was found to retard the delivery of Ste3Δ365p to the cell surface (Fig. 5B) just as it did in the MATa cell context (Fig. 5A). Identical effects are seen in the a/α context (Fig. 5B): Asg7p still retards Ste3Δ365p surface delivery, indicating a lack of G protein requirement for this Asg7p action. Likewise, the introduction of a ste4Δ mutation into MATa ADH1-ASG7 GAL1-STE3Δ365 cells also is without consequence for Asg7p-mediated impaired delivery of Ste3p to the surface (Fig. 5C). For the other Asg7p-Ste3p phenotype, i.e., inhibition of the pheromone response, a similar assessment of G protein involvement is not possible since the G proteins play a critical role in the experimental test (e.g., either for FUS1-LacZ induction or for suppression of GAL1-STE4). Nonetheless, the G protein independence demonstrated in the present experiment (Fig. 5B and C) suggests the possibility of direct interaction between Asg7p and Ste3p.
ASG7 and mating.
The striking phenotypes noted above for ASG7 disruption or overproduction all involve the artificial coexpression of Asg7p and Ste3p in the same cell. In MATa cells not expressing STE3, both ASG7 deletion and overexpression were without discernible effect on the FUS1 transcriptional response (Fig. 3). Furthermore, we also find that asg7Δ and ADH1-ASG7 MATa cells show a morphogenetic response similar to that of wild-type MATa cells following 3 h of treatment with 10−5 M α-factor: normal mating projections were observed (data not shown).
Table 2 shows mating efficiencies from matings between wild-type α cells and either wild-type a cells or the equivalent isogenic asg7Δ or ADH1-ASG7 mutant. We discern no effect of either ASG7 disruption or overproduction on overall mating fitness. The only clear effect of ASG7 status on mating occurred with ASG7 expressed inappropriately in α cells (MATα ADH1-ASG7); introduction of the ADH1-ASG7 allele into the MATα context blocked mating (Table 2). This cross, of course, differs from the other matings in that one of the cells, the α cell, coexpresses both Ste3p and Asg7p. In such ADH1-ASG7 MATα cells, two negative actions of Asg7p on mating are anticipated: transport of Ste3p to the cell surface is expected to be impaired, and, for receptors that are delivered to the surface, signaling is expected to be blocked through the Asg7p-Ste3p repression mechanism.
TABLE 2.
Mating efficiency
| Genotype of strain used for matinga
|
Mating efficiencyb | |
|---|---|---|
| MATa | MATα | |
| wt | wt | 0.13 ± 0.01 |
| asg7Δ | wt | 0.10 ± 0.01 |
| ADH1-ASG7 | wt | 0.11 ± 0.01 |
| wt | wt | 0.07 ± 0.01 |
| wt | ADH1-ASG7 | <10−5 |
Matings were performed as described in Materials and Methods. For the top three matings, the three isogenic MATa strains W303-1A, NDY1089, and NDY1171 were mated to MATα strain IH1792. For the bottom two matings, two isogenic MATα strains W303-1B and NDY1179 were mated to MATa strain IH1793. wt, wild type.
Mating efficiency is the number of diploid cells formed divided by the total number of MATa cells (top three matings) or MATα cells (bottom two matings) input into the mating reaction. Mating efficiencies averaged from three separate experiments ± standard deviations are reported.
In addition to the matings reported in Table 2, we have also examined the effects of the asg7Δ mutation in a variety of different mating contexts that are known to reveal more subtle mating defects. Defects in a variety of late-acting mating functions affecting the chemotropic response or zygotic cell fusion are often poorly revealed in matings to wild-type mating partners but can be accentuated in matings to impaired mating partners or when the mutant genes are tested in synthetic combination with mutations in other genes affecting these same processes (4, 8, 9). Such tests reveal striking mating defects for mutations in a large number of functions, including SPA2, AXL1, RVS161, FUS1, FUS2, BNI1, SST2, and FAR1 (3, 8, 9, 11). As a first test, we introduced the asg7Δ allele into sst2Δ or fus1Δ MATa cells; no added, synthetic defect was conferred by the asg7 mutation (data not shown). We also tested the mating of asg7Δ MATa cells to impaired partners, either fus1Δ, sst2Δ or far1 MATα cells. While this approach has proved effective in uncovering subtle mating defects (4), again by this approach no defect was apparent for asg7Δ a cells (data not shown). Thus, by these varied measures of mating, we can discern no significant contribution of Asg7p to overall mating fitness.
Deranged zygotic morphology in asg7 matings.
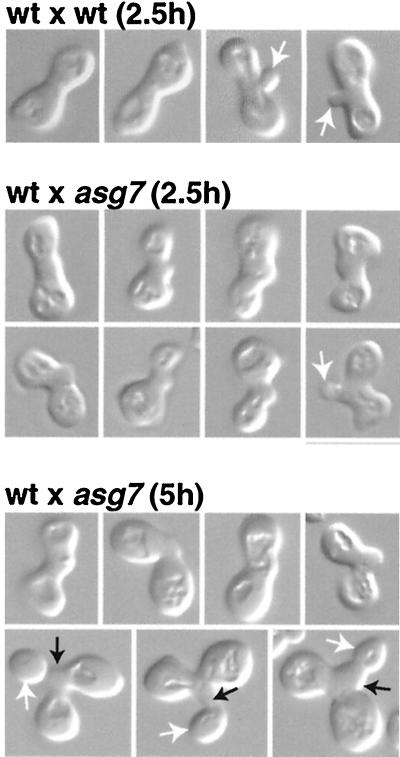
We have performed microscopic analysis of zygotes formed from matings between wild-type MATα cells and asg7Δ MATa cells (Fig. 6). Here, we note several striking effects of the asg7Δ mutation. While zygotes are formed with wild-type efficiency in the mating of wild-type α cells to asg7Δ a cells (Table 3), these new diploid cells show a variety of morphologic abnormalities, often showing an unusual protuberant structure at the midregion of the conjugation bridge that connects the two cell bodies (Fig. 6). While approximately situated where the first diploid mitotic bud normally emerges, this protrusion does not show the neck-like constriction typical of emerging buds. These novel structures can be detected at very early mating time points (Fig. 6, 2.5-h time point) and often appear to be enlarged at later time points (Fig. 6, 5-h time point), suggesting that these may represent sites of aberrant polarized growth. Indeed, consistent with this being a locus of cell growth, mitotic buds are often found to emerge from the tips of these protuberances at late time points, (Fig. 6, 5-h time point). Zygotes with these protrusions generally manifest an overall bent morphology: instead of the linear dumbbell structure typical of wild-type zygotes, the two mating partners often have the aspect of having connected on an angle (Fig. 6). Finally, unlike the zygotes derived from wild-type matings, very few of the zygotes derived from early times of mating between wild-type MATα and the asg7Δ MATa cells are found with discernible emerging mitotic buds.
FIG. 6.
Zygotes from asg7 matings are morphologically deranged. Wild-type W303-1B MATα cells were mated to either wild-type W303-1A MATa cells or the isogenic asg7Δ strain NDY1089 for 2.5 or 5 h. Selected zygotes visualized by Nomarski optics are shown. White arrows, new mitotic buds; black arrows, stalk-like structures from the mitotic bud often seen to emerge for asg7 zygotes.
TABLE 3.
Zygotic morphology and budding
| Matinga | Time (h) | % Zygotesb | % of zygotesc that were:
|
|
|---|---|---|---|---|
| Abnormal | Budded | |||
| MATa (wt) × MATα (wt) | 1 | 0 | ndd | nd |
| 1.5 | 1 ± 1 | nd | nd | |
| 2 | 4 ± 1 | 5 ± 3 | 21 ± 3 | |
| 2.5 | 4 ± 1 | 8 ± 1 | 39 ± 2 | |
| 3 | 5 ± 1 | 7 ± 2 | 52 ± 2 | |
| MATa (asg7Δ) × MATα (wt) | 1 | 0 | nd | nd |
| 1.5 | 1 ± 1 | nd | nd | |
| 2 | 3 ± 1 | 42 ± 7 | 2 ± 1 | |
| 2.5 | 4 ± 1 | 57 ± 2 | 9 ± 3 | |
| 3 | 5 ± 1 | 63 ± 3 | 31 ± 1 | |
Wild-type (wt) MATα cells (W303-1B) were mated on filters to either wild-type MATa cells (W303-1A) or isogenic asg7Δ MATa cells (NDY1089).
The mean percentages of the total cells within the mating mixture showing typical zygotic (dumbbell-like) morphology ± standard deviations are reported. Five hundred cells were counted from three experiments. Prezygotes are not distinguished from zygotes by this analysis and are thus included within the percent zygotes.
For each mating time point, 200 cells identified as zygotes (or prezygotes) were subclassed as having either normal or abnormal morphology and also subclassed as to whether or not they displayed a bud. Zygotes were considered morphologically abnormal if they either showed a bent morphology or displayed a bulge-like protrusion emerging from the conjugation bridge (see Fig. 6 for examples). Buds were distinguished (from bulges) by the neck-like constriction present at the emergence site. Both the percentages of total zygotes (plus prezygotes) that were considered to be morphologically abnormal and the percentages that displayed obvious buds were determined for three experiments with the means ± the standard deviations being reported. As some zygotes with small buds certainly escaped detection, the percentages of zygotes with buds reported are likely to be underestimates.
nd, not determined.
In Table 3, we have quantified both the morphologic defect and budding defect for asg7 zygotes, comparing zygotes from a fully wild-type mating to zygotes from matings between wild-type MATα cells and asg7Δ MATa cells. Zygotes were classed as being normal or morphologically deranged. Only zygotes with clear morphological derangements, displaying a midregion protrusion and/or an overall bent morphology, were scored as abnormal. From the asg7 matings a consistently high fraction of the zygotes were classed as abnormal at each of the time points: at the 2-, 2.5-, and 3-h mating time points, 42, 57, and 63% of the zygotes, respectively, were classed as abnormal. While similar derangements can be found among zygotes from fully wild-type mating mixtures, these occur at far lower frequencies (Table 3). Zygotes from matings of wild-type α cells to a cells constitutively overexpressing ASG7 from the ADH1 promoter were found to be morphologically wild type (data not shown).
In addition to subclassing zygotes by morphology, we classed the same zygotes as to whether a mitotic bud was displayed (Table 3). As this analysis focuses on early mating time points (2 to 3 h), the observed buds are expected to be the first buds to emerge from the new zygotes. For wild-type zygotes, bud initiation is expected to begin soon after cell fusion coincident with the G1-to-S transition. For the wild-type pairing at the early 2-h time point, discernible buds were identifiable on 21% of the new zygotes (Table 3). This fraction increased with time (Table 3, 2.5- and 3-h time points), reflecting both new bud initiation and also continued growth of preexisting buds (larger buds are more likely to be detected by our microscopic analysis). For zygotes derived from the asg7 pairing, the emergence of this first mitotic bud was quite significantly delayed (Table 3; compare the fraction of zygotes with buds at the 2-, 2.5-, and 3-h time points for the wild-type and asg7 matings). The delay in first bud emergence for the asg7 zygotes could be secondary to the deranged morphology; for instance, derangement of the budding site could delay bud emergence. Alternatively, delayed budding could reflect a cell cycle delay, with asg7 zygotes being slow to transition from G1 to S.
Despite abnormal morphology and delayed budding, other aspects of the zygotic developmental pathway appeared to proceed normally. Consistent with the wild-type mating efficiency measured for asg7 matings (Table 2), zygotes arose from wild-type and asg7 mating mixtures at roughly equivalent frequencies (Table 3). In addition, the kinetics of the zygotic cell fusion event was monitored for the two matings. Zygotes were distinguished from prezygotes by monitoring the redistribution of a plasma membrane-localized GFP-Ras2p fusion protein present initially in just the α mating partner (8). Prezygotes, i.e., intermediates in which the two mating partners have attached but in which the intervening cell wall has not yet been dissolved, localize the GFP fluorescence to just one of the two partners (the α cell). With fusion, the fluorescence signal rapidly distributes from the donor α cell throughout the entire cell surface of the zygote. Following 2.5 h of mating, similar ratios of prezygotes to zygotes from both wild-type and asg7 matings were found (see Table 5). Thus, the asg7Δ a cells are not fusion defective. In addition, we have used the fluorescent DNA stain DAPI (4′,6′-diamidino-2-phenylindole) to visualize nuclei and to monitor the congress of the two haploid nuclei into the single diploid nucleus, i.e., karyogamy. No obvious defect in karyogamy was observed for the asg7 zygotes; in general, the vast majority of both the wild-type and asg7 zygotes examined at early mating time points showed a single locus of DNA staining localizing to the midsection of the conjugation bridge (data not shown).
TABLE 5.
Prezygote morphologies from fusion-competent matings
| Mating (α × a)a | % Prezygotes + zygotesb | % Prezygotesc | % Abnormal zygotesd | % Abnormal prezygotese |
|---|---|---|---|---|
| wt × wt | 3 ± 1 | 19 ± 2 | 3 ± 2 | 2 ± 2 |
| wt × asg7 | 4 ± 2 | 17 ± 5 | 53 ± 3 | 2 ± 4 |
Yeast strains were identical to those used in Table 4 (see footnote a), except that the MATα mating partner W303-1B was transformed by GFP-RAS2 plasmid p2664. Cells were mated for 2.5 h. wt, wild type.
The percentages of the total cells within the mating mixture showing a zygotic morphology (both prezygotes and zygotes) were estimated as described for Table 3 (footnote b).
A total of 200 cells having both zygotic morphology (either zygotes or prezygotes) as well as GFP fluorescence were identified and then classified as being either zygotes or prezygotes. Zygotes show the GFP fluorescence, contributed from the MATα mating partner, distributed throughout the zygotic plasma membrane. For prezygotes, fluorescence remained limited to just one of the two partners (the α cell). The percentages of the total zygote plus prezygote population that are prezygotes are reported.
Zygotes were classified by their morphological presentation, using the criteria of Table 3 (footnote c). The percentages of the total zygote population showing abnormal morphology are reported.
Prezygotes also were classified by their morphological presentation, using the criteria of Table 4 (footnote c). The percentages of the total prezygote population showing abnormal morphology are reported.
Epistasis of asg7 with fusion-defective mutations.
The linear dumbbell presentation of the wild-type zygote results from tip-to-tip fusion of the two mating partners. The bent or angled morphology seen for many of the asg7-derived zygotes (Fig. 6) suggested the possibility that the asg7Δ a cells might be defective for this prezygotic connection to their α partners. Indeed, an angled connection between mating partners might also account for the midpoint protuberance seen for the asg7 zygotes, with the protuberance being derived from the angled fusion of the two mating projections. We were interested therefore to examine the morphology of prezygotes derived from asg7 pairings to see if the abnormalities seen for the zygotes are also present at this early step.
Matings in which both partners are defective either for FUS1 or for FUS2 are blocked at the prezygote stage (20, 32). Mating partners stably connect through a process that involves remodeling and fusion of exterior cell walls, but the intervening cell wall between the two partners fails to be dissolved and, consequently, cell and nuclear fusion fails to occur (3, 11). For matings between MATα fus1Δ cells and MATa fus1Δ cells or the equivalent fus2Δ bilateral pairing, prezygotes accumulated with kinetics similar to that seen for zygote accumulation in wild-type matings (Table 4). In gross outline, the prezygotes from either of these pairings generally resembled the linear dumbbell structure typical of wild-type zygotes (not shown). To examine the effects of the asg7Δ mutation on prezygote morphology, double-mutant MATa strains, either fus1Δ asg7Δ or fus2Δ asg7Δ, were constructed and then tested in bilateral matings to the appropriate MATα fus mutant. The morphologies of the prezygotes derived from these matings were compared to those derived from the fusion-defective matings involving ASG7+ MATa cells (Table 4). In contrast to the severe effects of the asg7Δ mutation on zygotic morphology (Fig. 6), we were not able to discern any effect on prezygote morphology: no enhanced derangement was seen with the introduction of asg7Δ into either the fus1 or fus2 bilateral matings (Table 4). We conclude that the fus1 and fus2 mutations are epistatic to asg7Δ, suggesting that asg7-mediated derangements are initiated at steps downstream of the zygotic fusion event.
TABLE 4.
Prezygote morphologies from fusion-defective matings
| Matinga | % Prezygotesb | % Abnormal prezygotesc |
|---|---|---|
| a fus1Δ × α fus1Δ | 9 ± 1 | 17 ± 2d |
| a fus1Δ asg7Δ × α fus1Δ | 10 ± 1 | 16 ± 5 |
| a fus2Δ × α fus2Δ | 12 ± 2 | 2 ± 1 |
| a fus2Δ asg7Δ × α fus2Δ | 9 ± 1 | 2 ± 1 |
Matings were for 2.5 h. Bilateral fus1 matings were between Y428 and either Y427 or Y2617. Bilateral fus2 matings were between Y411 and either Y405 or Y2615.
The percentages of total cells within the mating mixtures showing prezygotic morphology (dumbbells) were determined as for the percentages of zygotes for Table 3.
A total of 200 cells showing prezygotic morphology were identified and classified as being either morphologically normal or abnormal. Prezygotes were classified as abnormal if the two mating partners were seen to connect at an angle. The fractions of the 200 counted prezygotes found to be morphologically normal were determined for three experiments, and the means ± standard deviations are reported.
A somewhat higher percentage of the prezygotes from the bilateral fus1 matings were scored as being morphologically abnormal (relative to those from bilateral fus2 matings). These abnormal prezygotes appear to result from the fus1 partners connecting on an angle rather than via the typical tip-to-tip connection.
We have also examined the prezygotes which may be found as intermediate structures in fusion-competent matings at early mating time points (Table 5). Following 2.5 h of mating, similar ratios of prezygotes to zygotes were found in both the wild-type and asg7 matings (Table 5). While the zygotes from the asg7 mating showed the usual array of deranged morphologies, prezygotes examined from the same mating mixtures were morphologically normal, indeed indistinguishable from the prezygotes found in the wild-type mating mixtures (Table 5). Again, these results indicate that the deranged morphology of the asg7 zygotes is a consequence of events that occur subsequent to the fusion of the two haploid cells.
DISCUSSION
Asg7p-Ste3p coexpression phenotypes.
We find that ASG7 is the a-specific gene responsible for the phenotype of the STE3DAF allele. Though defined in a cells (13), the Daf phenotype, we also find, may be reproduced in α cells with the forced, inappropriate expression of Asg7p in this context. Thus, Asg7p is the only a-specific gene product and Ste3p is the only α-specific gene product required for the inhibition. The key requirement for inhibition is coexpression of Asg7p and Ste3p in the same cell. We also report effects of ASG7 on Ste3p turnover and localization. Asg7p slows Ste3p turnover (Fig. 4). However, instead of being a direct result of Ste3p endocytosis, slowed turnover appears to be a secondary consequence of the Asg7p-mediated inhibition of the secretory delivery of Ste3p to the cell surface (Fig. 5).
The striking phenotypes described above depend on coexpression of Ste3p and Asg7p in the same cell. While the strength and specificity of these phenotypes argue for a functional linkage between these two proteins in the mating process, the two proteins reside in distinct cell types and thus are not normally available to one another. A possible explanation for this paradox, offered by Hirsch and colleagues, is that the response inhibition seen with STE3DAF MATa cells recapitulates regulation that normally functions in the zygote (17). The zygotic fusion event affords Ste3p the opportunity to interact with its a-specific coinhibitor (now known to be Asg7p). The inhibition provided by this interaction could be used to shut down the pheromone response in the newly formed zygote, promoting its transition to a vegetative mode of growth. As discussed below, our results are nicely consistent with this model.
Asg7p-Ste3p inhibition in the zygote.
MATa asg7Δ cells respond to pheromone normally and mate with wild-type efficiency. However, while normal numbers of zygotes are formed, the zygotes are morphologically deranged and show a delay to the emergence of the first diploid mitotic bud. Since prezygotes examined from these same matings are morphologically normal, we concluded that perturbed zygotic morphology results from postfusion events and not from an initially misoriented connection of the two mating partners.
The two asg7 zygotic phenotypes, namely, deranged morphology and delayed budding, may be explained in terms of aberrant growth patterns in the mutant zygote. In the wild-type zygotic developmental progression, following cell and nuclear fusion, the new zygote transitions out of G1 to S, DNA replication is initiated, and a new mitotic bud begins to emerge from a central position in the connecting conjugation bridge. For the asg7 zygote, delayed bud emergence and the aberrant morphology may be coupled. With delayed bud initiation, ongoing growth of the cell must be channeled elsewhere, perhaps into the bulge-like structure which emanates from the approximate site where the first bud normally emerges (the conjugation bridge). In overall aspect, the bulge somewhat resembles a growing mating projection. A possibility, therefore, is that the asg7 zygote is slow at making the transition from the pheromone-stimulated growth pattern of haploid cells (i.e., mating projection formation) to the vegetative, budding pattern of growth characteristic of the new diploid cell. While slow, asg7 zygotes make this transition; the wild-type mating efficiency of asg7Δ MATa cells (Table 2) indicates that the resulting zygotes give rise to vegetatively growing colonies of diploid cells. Indeed, this transition is also apparent at the microscopic level; though the budding index of asg7 zygotes initially lags behind that of wild-type zygotes, the asg7 budding index increases sharply at later time points (Table 3). Thus, bud initiation is delayed but not blocked for the asg7 zygotes.
In haploid cells responding to pheromone, maintenance of both the G1 cell cycle arrest and the polarized pattern of cell growth depends on continued pheromone signaling. Following successful fusion of the two mating partners, the reinitiation of the budding cycle requires that pheromone signaling be turned off. How is this accomplished? Part of the answer may involve the known transcriptional changes that accompany the formation of the new a/α diploid genome. Many of the proteins that mediate pheromone signaling are haploid specific and are transcriptionally repressed in the a/α cell. Included in this set are the pheromones, the pheromone receptors, the subunits of the heterotrimeric G protein, and several of the components of the downstream signaling cascade (31). With the loss of these proteins from the new diploid cell, the capacity for signaling is blocked and the cell may then exit G1 and initiate budding. The rapidity of this transition obviously should depend on the rate at which these signaling proteins are removed, depending both on the rate at which the transcriptional repression is imposed and the rate at which preexisting proteins and mRNA turn over. At the time of the zygotic fusion event, all these proteins remain present and pheromone signaling is expected to be intense. Layered onto the regulation provided by the a1/α2 transcriptional repression mechanism is a second, potentially more rapid and effective means of shutting down signaling provided by Asg7p and Ste3p, which gain access to one another through the zygotic fusion event. The formation of an inhibitor from preexisting components uniquely contributed from the two cell types provides a simple and direct mechanism for shutting down signaling and terminating the mating process once successful fusion has occurred.
Support for the hypothesis that Asg7p and Ste3p function together to shut down signaling in the zygote is presently based in large part both on the bud emergence delay observed for asg7 zygotes and on our interpretation of the novel structure that emerges in these zygotes as being a mating projection (and not a morphologically deranged mitotic bud). Proof will await further experiments. Microarray analyses should reveal if the pattern of gene expression in the newly formed asg7 zygote is consistent with a failure to shut down pheromone-induced gene expression. In addition, the expectation that the G1-S transition is delayed for asg7 zygotes will be directly tested by analyzing DNA content in new zygotes by flow cytometry.
Ste3p-Asg7p negative regulation of the pheromone response pathway may function in other situations as well. In the mating type switching of HO+ yeast cells, the newly switched cell is expected to transiently express receptors and pheromones derived from both mating types. Asg7p-Ste3p might serve to shut down this unnecessary and potentially deleterious autocrine signaling. Along the same lines, Asg7p-Ste3p inhibition likely provides at least part of the explanation for the long-appreciated but poorly understood phenotype of matα2 mutant cells. matα2 cells, which are defective for repression of a-specific genes, constitutively express both a- and α-cell-specific gene products (thus, both Ste3p and Asg7p). In light of the present understanding of Asg7p-Ste3p inhibition, it is not surprising that matα2 mutants are severely impaired in their response to pheromone (1).
Mechanism of Asg7p-Ste3p inhibition.
Little is understood regarding the mechanism of Asg7p-Ste3p inhibition. From the present analysis, it is clear that Ste3p is the only α-specific protein required and Asg7p is the only a-specific protein required. It is not clear if these two proteins physically interact. Furthermore, it is not clear to what extent other proteins, present in both cell types, participate.
While we have demonstrated that Ste3p delivery to the cell surface is impaired in cells that express Asg7p, this mislocalization may not be part of the natural zygotic regulatory mechanism. Both proteins are membrane proteins and are expected to be initially inserted into the endoplasmic reticulum (ER) following synthesis. With the artificial coexpression of these two proteins in the same cell, an interaction between newly synthesized Asg7p and Ste3p in the ER could be recognized as inappropriate, with the consequence being Ste3p retention (2). In the newly formed zygote, the interactions that mediate the repressive effects on signaling would likely be between preexisting, not newly synthesized, Asg7p and Ste3p. Nonetheless, an aspect of the Asg7p-mediated inhibition of Ste3p surface delivery that may be of wider significance in terms of the zygotic repression mechanism is the G protein independence of this phenotype (Fig. 5B). While the Gβγ component likely is a central player for Asg7p-Ste3p response inhibition (see the discussion below), the lack of a G protein requirement for the Ste3p trafficking phenotype demonstrates that Asg7p and Ste3p are capable of functionally interacting in the absence of the Gβγ subunit.
Previous work has indicated that the repression associated with the STE3DAF allele (i.e., Asg7p-Ste3p repression) is likely exerted on the signaling pathway at the level of the Gβγ component of the G protein (6, 13, 17). Consistent with this, Kim et al. (16) have demonstrated that a key part of the Asg7p-Ste3p repression mechanism is the mislocalization of Ste4p away from the plasma membrane (its normal site of action) to an intracellular locale. Significantly, Kim et al. (16) also found a similar intracellular localization for a functional Asg7p-GFP fusion, suggesting the possibility that Asg7p and Ste4p may localize to the same intracellular compartment and that Asg7p may act directly in the mislocalization of Ste4p. Indeed, our own preliminary analyses of a functional HA epitope-tagged Asg7p expressed in resting MATa cells from the GAL1 promoter reveal a clear localization both to the vacuolar membrane and to perivacuolar compartments (data not shown). While Ste3p localization has not been monitored in this context, Ste3p is known to traverse similarly located endosomal compartments on its endocytic route to the vacuole (7, 22). One possibility, therefore, is that Asg7p increases the affinity of Ste3p for Gβγ. Tight binding may cause Gβγ to be internalized together with the endocytosed receptor. Gβγ depletion from the plasma membrane should block pheromone signal transduction (23). Future studies will focus on the Asg7p-Ste3p repression mechanism. Do Asg7p and Ste3p directly interact? Does Asg7p alter the interaction of Ste3p with Gβγ?
ACKNOWLEDGMENTS
We thank our colleagues Linyi Chen and Ying Feng for input and support throughout the course of this work, and we thank Jeff Loeb for the use of his microscope facility.
This work was supported by grants to C.B. from the National Science and Engineering Council of Canada and from the National Cancer Institute of Canada and by a grant to N.G.D. from the National Science Foundation (MCB 99-06839).
REFERENCES
- 1.Bender A, Sprague G F., Jr Pheromones and pheromone receptors are the primary determinants of mating specificity in the yeast Saccharomyces cerevisiae. Genetics. 1989;121:463–476. doi: 10.1093/genetics/121.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonifacino J S, Weissman A M. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu Rev Cell Dev Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brizzio V, Gammie A E, Rose M D. Rvs161p interacts with Fus2p to promote cell fusion in Saccharomyces cerevisiae. J Cell Biol. 1998;141:567–584. doi: 10.1083/jcb.141.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chenevert J, Valtz N, Herskowitz I. Identification of genes required for normal pheromone-induced cell polarization in Saccharomyces cerevisiae. Genetics. 1994;136:1287–1296. doi: 10.1093/genetics/136.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole G M, Stone D E, Reed S I. Stoichiometry of G protein subunits affects the Saccharomyces cerevisiae mating pheromone signal transduction pathway. Mol Cell Biol. 1990;10:510–517. doi: 10.1128/mcb.10.2.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Couve A, Hirsch J P. Loss of sustained Fus3p kinase activity and the G1 arrest response in cells expressing an inappropriate pheromone receptor. Mol Cell Biol. 1996;16:4478–4485. doi: 10.1128/mcb.16.8.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis N G, Horecka J L, Sprague G F., Jr Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J Cell Biol. 1993;122:53–65. doi: 10.1083/jcb.122.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorer R, Boone C, Kimbrough T, Kim J, Hartwell L H. Genetic analysis of default mating behavior in Saccharomyces cerevisiae. Genetics. 1997;146:39–55. doi: 10.1093/genetics/146.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorer R, Pryciak P M, Hartwell L H. Saccharomyces cerevisiae cells execute a default pathway to select a mate in the absence of pheromone gradients. J Cell Biol. 1995;131:845–861. doi: 10.1083/jcb.131.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng Y, Davis N G. Akr1p and the type I casein kinases act prior to the ubiquitination step of yeast endocytosis: Akr1p is required for kinase localization to the plasma membrane. Mol Cell Biol. 2000;20:5350–5359. doi: 10.1128/mcb.20.14.5350-5359.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gammie A E, Brizzio V, Rose M D. Distinct morphological phenotypes of cell fusion mutants. Mol Biol Cell. 1998;9:1395–1410. doi: 10.1091/mbc.9.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 13.Hirsch J P, Cross F R. The pheromone receptors inhibit the pheromone response pathway in Saccharomyces cerevisiae by a process that is independent of their associated G α protein. Genetics. 1993;135:943–953. doi: 10.1093/genetics/135.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horecka J, Sprague G F., Jr Identification and characterization of FAR3, a gene required for pheromone-mediated G1 arrest in Saccharomyces cerevisiae. Genetics. 1996;144:905–921. doi: 10.1093/genetics/144.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarvis E E, Hagen D C, Sprague G F., Jr Identification of a DNA segment that is necessary and sufficient for α-specific gene control in Saccharomyces cerevisiae: implications for regulation of α-specific and a-specific genes. Mol Cell Biol. 1988;8:309–320. doi: 10.1128/mcb.8.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J, Bortz E, Zhong H, Leeuw T, Leberer E, Vershon A K, Hirsch J P. Localization and signaling of Gβ subunit Ste4p are controlled by a-factor receptor and the a-specific protein Asg7p. Mol Cell Biol. 2000;20:8826–8835. doi: 10.1128/mcb.20.23.8826-8835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, Couve A, Hirsch J P. Receptor inhibition of pheromone signaling is mediated by the Ste4p Gβ subunit. Mol Cell Biol. 1999;19:441–449. doi: 10.1128/mcb.19.1.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leberer E, Thomas D Y, Whiteway M. Pheromone signalling and polarized morphogenesis in yeast. Curr Opin Genet Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- 19.Marsh L, Rose M D. The pathway of cell and nuclear fusion during mating in S. cerevisiae. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces cerevisiae: cell cycle and cell biology. Vol. 3. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 827–888. [Google Scholar]
- 20.McCaffrey G, Clay F J, Kelsay K, Sprague G F., Jr Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2680–2690. doi: 10.1128/mcb.7.8.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nomoto S, Nakayama N, Arai K, Matsumoto K. Regulation of the yeast pheromone response pathway by G protein subunits. EMBO J. 1990;9:691–696. doi: 10.1002/j.1460-2075.1990.tb08161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piper R C, Cooper A A, Yang H, Stevens T H. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pryciak P M, Huntress F A. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts C J, Nelson B, Marton M J, Stoughton R, Meyer M R, Bennett H A, He Y D, Dai H, Walker W L, Hughes T R, Tyers M, Boone C, Friend S H. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 25.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 26.Roth A F, Davis N G. Ubiquitination of the PEST-like endocytosis signal of the yeast a-factor receptor. J Biol Chem. 2000;275:8143–8153. doi: 10.1074/jbc.275.11.8143. [DOI] [PubMed] [Google Scholar]
- 27.Roth A F, Davis N G. Ubiquitination of the yeast a-factor receptor. J Cell Biol. 1996;134:661–674. doi: 10.1083/jcb.134.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roth A F, Sullivan D M, Davis N G. A large PEST-like sequence directs the ubiquitination, endocytosis, and vacuolar degradation of the yeast a-factor receptor. J Cell Biol. 1998;142:949–961. doi: 10.1083/jcb.142.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 30.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sprague G F, Jr, Thorner J W. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces cerevisiae: gene expression. Vol. 2. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 657–744. [Google Scholar]
- 32.Trueheart J, Boeke J D, Fink G R. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987;7:2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 34.Whistler J L, Rine J. Ras2 and Ras1 protein phosphorylation in Saccharomyces cerevisiae. J Biol Chem. 1997;272:18790–18800. doi: 10.1074/jbc.272.30.18790. [DOI] [PubMed] [Google Scholar]
- 35.Whiteway M, Hougan L, Thomas D Y. Overexpression of the STE4 gene leads to mating response in haploid Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:217–222. doi: 10.1128/mcb.10.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong H, McCord R, Vershon A K. Identification of target sites of the α2-Mcm1 repressor complex in the yeast genome. Genome Res. 1999;9:1040–1047. doi: 10.1101/gr.9.11.1040. [DOI] [PubMed] [Google Scholar]