Abstract
Spike (S) protein cleavage is a crucial step in coronavirus infection. In this review, this process is discussed, with particular focus on the novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). Compared with influenza virus and paramyxovirus membrane fusion proteins, the cleavage activation mechanism of coronavirus S protein is much more complex. The S protein has two cleavage sites (S1/S2 and S2′), and the cleavage motif for furin protease at the S1/S2 site that results from a unique four‐amino acid insertion is one of the distinguishing features of SARS‐CoV‐2. The viral particle incorporates the S protein, which has already undergone S1/S2 cleavage by furin, and then undergoes further cleavage at the S2′ site, mediated by the type II transmembrane serine protease transmembrane protease serine 2 (TMPRSS2), after binding to the receptor angiotensin‐converting enzyme 2 (ACE2) to facilitate membrane fusion at the plasma membrane. In addition, SARS‐CoV‐2 can enter the cell by endocytosis and be proteolytically activated by cathepsin L, although this is not a major mode of SARS‐CoV‐2 infection. SARS‐CoV‐2 variants with enhanced infectivity have been emerging throughout the ongoing pandemic, and there is a close relationship between enhanced infectivity and changes in S protein cleavability. All four variants of concern carry the D614G mutation, which indirectly enhances S1/S2 cleavability by furin. The P681R mutation of the delta variant directly increases S1/S2 cleavability, enhancing membrane fusion and SARS‐CoV‐2 virulence. Changes in S protein cleavability can significantly impact viral infectivity, tissue tropism, and virulence. Understanding these mechanisms is critical to counteracting the coronavirus pandemic.
Keywords: cleavage, furin, SARS‐CoV‐2, spike protein, TMPRSS2
Abbreviations
- ACE2
angiotensin‐converting enzyme 2
- COVID‐19
coronavirus disease 2019
- FCM
furin cleavage motif
- FP
fusion peptide
- HA
hemagglutinin
- HAE‐ALI culture
human airway epithelia cultured at an air–liquid interface
- hAO
human airway organoid
- HCoV
human coronavirus
- HR
heptad repeat
- MERS
Middle East respiratory syndrome
- NTD
N‐terminal domain
- RBD
receptor‐binding domain
- S
spike
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- TMD
transmembrane domain
- TMPRSS2
transmembrane protease serine 2
- TTSP
type II transmembrane serine protease
- VOCs
variants of concern
INTRODUCTION
Many viruses infect host cells using surface glycoproteins on viral particles to bind host receptors and fuse with cell membranes. In coronaviruses, the viral protein responsible for these functions is the spike (S) protein. Similar to many other viral proteins that facilitate membrane fusion, the S protein must undergo cleavage by protease(s) at the appropriate position(s) in the S protein to exert its membrane fusion function. The proteases responsible for this cleavage are not encoded in the viral genome, and the virus uses host proteases to cleave (proteolytically activate) the S protein so that membrane fusion can occur. Therefore, understanding the proteolytic activation mechanism of the S protein is critical for understanding the infectivity of coronaviruses and the pathogenesis of coronavirus infections. The activation mechanism of coronaviruses is more complex than that of, for example, influenza viruses or paramyxoviruses, and many aspects of activation remain to be elucidated. In this review, the current knowledge regarding the cleavage activation mechanisms of coronavirus S proteins is described with a focus on the newly emerged coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), which causes the pandemic disease, coronavirus disease 2019 (COVID‐19).
STRUCTURE AND DOMAINS OF THE SARS‐CoV‐2 S PROTEIN
The S proteins of coronaviruses are type I transmembrane proteins and are composed of the S1 subunit responsible for receptor binding and the S2 subunit responsible for membrane fusion (Figure 1). Within the S1 subunit, there is an N‐terminal domain (NTD) and a receptor‐binding domain (RBD). 3 The RBD, as its name suggests, is responsible for binding to the receptor, and in the case of SARS‐CoV‐2, it binds to angiotensin‐converting enzyme 2 (ACE2) and heparan sulfate. 4 , 5 However, the NTD in certain coronaviruses and possibly in SARS‐CoV‐2 can also bind receptors, 6 including proteinaceous receptors (CEACAM1 for mouse hepatitis virus) as well as glycans. 3 Presumably in many coronaviruses, both domains bind to different molecules, and together they are involved in the binding of the S protein to the cell. The S protein has two major cleavage sites: the S1/S2 site that divides the S1 subunit from the S2 subunit, and the S2′ site within the S2 subunit 7 , 8 (Figure 1). In addition to the N‐terminal signal peptide and the transmembrane region, the S protein has a hydrophobic fusion peptide (FP), located immediately downstream of the S2′ cleavage site in the S2 subunit. This FP shares structural features with FPs of membrane fusion proteins of many other viruses. The presence of this FP at the amino terminus of the S2 subunit is thought to be critical for membrane fusion because S2′ cleavage, but not S1/S2 cleavage, is essential for the S protein to exhibit membrane fusion activity. One of the major features of the SARS‐CoV‐2 S protein is presence of a suboptimal furin cleavage motif (FCM) (682‐RRAR↓S‐686; the arrow indicates the cleavage site) at the S1/S2 site by insertion of unique four amino acids (681‐PRRA‐684; Figure 2). This feature is absent in SARS‐CoV, which caused an outbreak in 2002–2003, and the closest relative of SARS‐CoV‐2, bat RaTG13 strain, or any other related bat coronaviruses. 9 This distinguishing feature is one of the grounds for the nonscientific argument that SARS‐CoV‐2 may be artificially generated. However, this type of insertion can occur during the natural evolution of coronaviruses, 10 , 11 although the cause of this insertion in SARS‐CoV‐2 is not yet known. Regardless, this FCM is undoubtedly responsible for some of the properties of SARS‐CoV‐2. Acquisition of an FCM greatly increases virulence for avian influenza A viruses and is a characteristic of highly pathogenic avian influenza viruses. However, this feature is not necessarily correlated with high virulence in coronaviruses. This is because seasonal human coronaviruses (HCoV‐HKU1 and HCoV‐OC43), which are clearly less virulent than SARS‐CoV or SARS‐CoV‐2, also have an FCM at the S1/S2 site. 12 , 13 However, it is possible that this amino acid insertion is involved in host range expansion and adaptation to humans by SARS‐CoV‐2. 5 , 9 , 14
Figure 1.
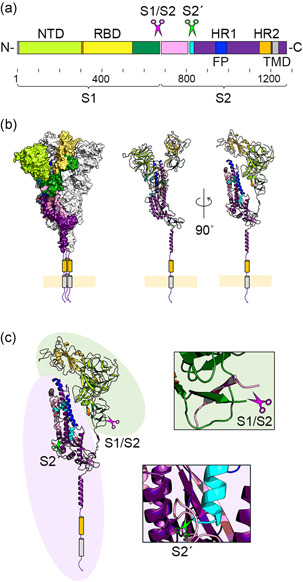
Structural domains of SARS‐CoV‐2 S protein and cleavage sites. (a) The SARS‐CoV‐2 S protein is a type I transmembrane protein, consisting of S1 and S2 domains, with a total length of 1273 amino acids. NTD (14–303), RBD (319–541), FP (816–854), HR1 (942–990), HR2 (1163–1202), and TMD (1214–1234) are shown in light green, yellow, cyan, blue, orange, and gray, respectively. 1 , 2 The S1/S2 (685/686) and S2′ (815/816) sites are shown by magenta and green scissors, respectively. (b) The homotrimeric structure is shown in the surface presentation model. One protomer is shown in the cartoon model. (c) A magnified view of the two cleavage sites (S1/S2 and S2′) is shown. The light green area shows the S2 domain and the light purple area shows the S2 domain. (b, c) Data for PDB 6XR8 were used to create these figures
Figure 2.

Amino acid sequences of S1/S2 cleavage sites of SARS‐CoV‐2 and closely related coronaviruses. Magenta scissors indicate the S1/S2 cleavage site. NTD, RBD, FP, HR1, HR2, and TMD are shown in light green, yellow, cyan, blue, orange, and gray, respectively. Basic arginine and lysine residues are shown in blue boxes. Amino acids are shown as single characters, and hyphens indicate gaps in the sequence
CLEAVAGE AND CONFORMATIONAL CHANGES OF SARS‐CoV‐2 S PROTEIN
The S protein undergoes a dynamic structural change in a sequential process from receptor binding to membrane fusion, including the process of S protein cleavage 7 , 15 , 16 , 17 , 18 , 19 , 20 (Figure 3). The timing of the conformational change must be highly regulated because this alteration during the fusion process is irreversible. Once the S protein undergoes the final conformational change, it completely loses its membrane fusion function. Furthermore, the timing of cleavage must be properly controlled because cleavage may affect the structural stability of the S protein, especially in intracellular organelles where the pH environment is different, such as transport vesicles, endosomes, and lysosomes, and therefore precise control is required to prevent structural changes at inappropriate times. Furin is a proprotein convertase that is ubiquitously expressed in the protein synthesis and transport pathways of cells. Thus, the S1/S2 site of SARS‐CoV‐2 S protein undergoes cleavage by furin (priming cleavage) during biosynthesis of the S protein and virus particle formation 5 , 21 (Figure 4a). The degree of cleavage (the percentage of S proteins that are cleaved) is greatly affected by the amino acid sequence of the cleavage site, and the cleavage is often partial. 13 Although furin is the primary protease responsible for this priming cleavage, a certain level of S1/S2 cleavage occurs even when furin is completely absent, presumably due to the action of other proprotein convertases. 22 , 23
Figure 3.
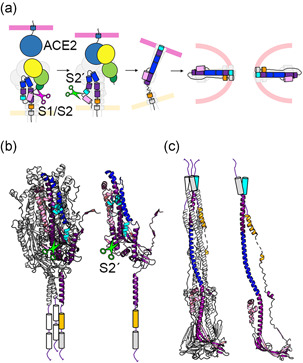
Structural changes in S protein during the membrane fusion process. (a) This illustration shows the sequence of structural changes from the prefusion form before receptor binding, binding to the receptor ACE2, cleavage of the S2′ site, and the final postfusion form. NTD, RBD, FP, HR1, HR2, and TMD are shown in light green, yellow, cyan, blue, orange, and gray, respectively. Magenta and green scissors indicate the S1/S2 and S2′ cleavage sites, respectively. (b) S2 domain in a prefusion form. The trimeric structure is shown on the left, and one of the protomers is shown on the right, both as cartoon models. Green scissors indicate the S2′ cleavage site. (c) S2 domain in a postfusion form. The trimeric structure is shown on the left, and one of the protomers is shown on the right, both as cartoon models. (b, c) Data for 6XR8 (prefusion form) and 6XRA (postfusion form) were used to create these figures
Figure 4.
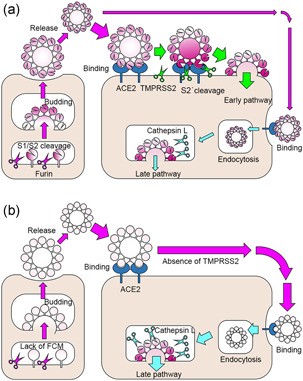
Entry pathways of SARS‐CoV‐2 and timing of S protein cleavage activation. (a) The S1/S2 site of the S protein is cleaved during virus particle formation. After binding to the receptor, ACE2, the S2′ site is cleaved by TMPRSS2 and enters the cell (early pathway); if TMPRSS2 is not available, the virus particles are endocytosed and the S protein is cleaved and activated by lysosomal cathepsin L (late pathway). (b) If SARS‐CoV‐2 is cultured in cells that do not express TMPRSS2, the FCM at the S1/S2 site is lost due to mutation, and the virus infects the cells using only the late pathway
As mentioned above, S2′ cleavage is essential for inducing membrane fusion, and thus S1/S2 cleavage alone does not trigger membrane fusion. However, S1/S2 cleavage destabilizes the S protein, and likely promotes its binding to ACE2 17 , 24 (Figure 5). This is because, through a hinge‐like movement, the RBD of the S protein changes its position in the S protein trimer from a folded (“down” or “closed”) to an exposed (“up” or “open”) conformation to efficiently access and bind to its receptor, ACE2 5 , 25 (Figure 6). S protein cleaved by furin at the S1/S2 site may more easily achieve the “up” conformation. 24 , 26 Binding of one protomer to ACE2 promotes sequential conversion of other protomers to the open conformation ideal for binding to the receptor. 17 SARS‐CoV‐2 with the D614G mutation, which emerged early in the epidemic, has better infectivity and proliferative potential and has subsequently become the predominant strain in the pandemic worldwide. 27 , 28 The 614th amino acid on the S1 subunit is involved in the interaction of the S1 subunit with the S2 subunit in another protomer 17 , 28 (Figure 7). The D614, but not G614, acts as a “latch” that secures the two protomers together 28 (Figure 7). The S protein with the D614G mutation, which therefore affects the interprotomer interactions between the S1 and S2 subunits, tends to adopt the up/open conformation 17 , 28 , 29 (Figure 5). Besides, the 614th amino acid is located in the vicinity of the S1/S2 cleavage site (Figure 7), and the D614G mutation increases S1/S2 cleavability by furin 26 (Figure 5). Thus, S protein that contains the D614G mutation may be more likely to adopt the up/open conformation 28 , 29 (Figure 5). In fact, the D614G mutation is of great significance when the S protein contains an FCM. 28
Figure 5.
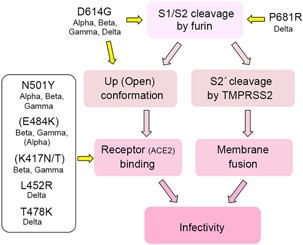
Chart notation of enhanced infectivity by furin cleavage and significance of major mutations in VOC
Figure 6.
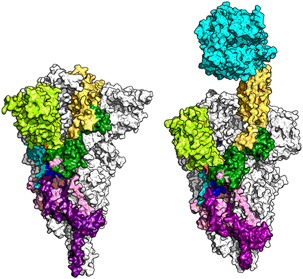
S protein in down conformation and in up conformation binding to the receptor, ACE2.The homotrimeric structure is shown in the surface presentation model. One protomer is shown in color, the other two in white. NTD, RBD, FP, HR1, HR2, and TMD are shown in light green, yellow, cyan, blue, orange, and gray, respectively. ACE2 is shown in cyan. The S protein in down conformation is shown on the left, and in up conformation binding to ACE2 on the right. Data for PDB 6XR8 (down conformation) and 7LNB (up conformation) were used to create these figures
Figure 7.
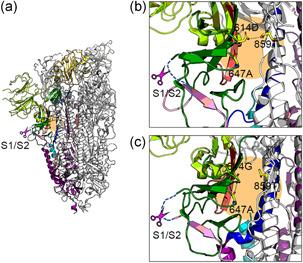
Alteration of interprotomer interaction by D614G mutation. (a–c) The homotrimeric structure is shown in the cartoon model. One protomer is shown in color, the other two in white. NTD, RBD, FP, HR1, HR2, and TMD are shown in light green, yellow, cyan, blue, orange, and gray, respectively. Magenta scissors indicate the S1/S2 cleavage site. The blue dashed lines indicate the loop containing the FCM. The orange area shows the surrounding area of the 614th amino acid. (a, b) The S protein trimer with 614D. (c) The S protein trimer with 614G. (b, c) Magnified views of the area around the 614th amino acid showing the hydrogen bonds formed with the 614th amino acid (yellow dotted lines). (a–c) Data for PDB 6VSB (614D) and 6XS6 (614G) were used to create these figures
As discussed above, cleavage of the S2′ site is essential for coronavirus infection, and cleavage of this site occurs after the S protein binds to the receptor, not during the biosynthesis or intracellular transport of S protein (Figures 3a and 4a). This is because the conformational change of S protein induced by receptor binding is necessary for the S2′ site to be accessed and cleaved by specific proteases 15 , 17 , 20 (Figure 3a). For SARS‐CoV‐2, transmembrane protease serine 2 (TMPRSS2) is responsible for this S2′ cleavage 30 , 31 (Figures 3a and 4a). TMPRSS2 is a type II transmembrane serine protease (TTSP) that is most highly expressed in the prostate and in a variety of epithelial tissues, including the airway epithelium. This enzyme is expressed in intracellular transport vesicles, as well as on the plasma membrane. TMPRSS2 gene‐knockout mice exhibit perfectly healthy growth and fertility, 32 , 33 but its physiological functions remain unknown. While cleavage of the S1/S2 sites is not necessarily critical for the infectivity of coronaviruses, for SARS‐CoV‐2 to become fully infectious, S1/S2 cleavage by furin is essential. 21 During the receptor‐binding process, the N‐terminal region of the S2 protein generated by furin‐mediated S1/S2 cleavage (in some studies, this N‐terminal region is also referred to as FP) may be targeted to the plasma membrane, and after receptor binding, S2 is further cleaved at the S2′ site by TMPRSS2 on the plasma membrane, placing the FP at the new N terminus of the S2 subunit (in some papers, this region is referred to as internal FP). 12 , 34 This S2′ cleavage finally triggers an extensive and irreversible conformational change to cause membrane fusion 34 (Figure 3). The S1/S2 cleavage may also have additional significance. It has been shown that neuropilin‐1 binds to the RRAR sequence at the carboxyl terminus of the S1 subunit generated by furin‐mediated S1/S2 cleavage, and this binding enhances ACE2‐mediated infection by SARS‐CoV‐2. 35 , 36
TWO MAJOR CELL ENTRY PATHWAYS OF SARS‐CoV‐2 AND INDIVIDUAL ROLES OF HOST PROTEASES (FURIN, TMPRSS2, AND CATHEPSIN L)
It has been established that the S1/S2 site is cleaved by furin and the S2′ site is cleaved by TMPRSS2. 14 , 30 , 37 , 38 Notably, cleavage by both furin and TMPRSS2 is required for efficient SARS‐CoV‐2 infection of lung epithelial cells (Calu‐3 cells). 14 , 37 , 39 Each enzyme plays a different role because one enzyme cannot compensate for the other. For SARS‐CoV‐2 to be efficiently activated by TMPRSS2, the FCM is required. 31 , 39 , 40 These results indicate that furin and TMPRSS2 (S1/S2 and S2′ cleavage) work in concert. However, the following complicates our understanding of coronavirus cleavage activation. In many studies, VeroE6 cells, which do not express TMPRSS2, have been used to isolate and grow SARS‐CoV‐2. These findings indicate that TMPRSS2 may not necessarily be required for SARS‐CoV‐2 infection. As has been shown with other coronaviruses, 41 SARS‐CoV‐2 can enter the cell by the endocytic pathway without the support of furin or TMPRSS2 and can be activated by lysosomal cathepsin L 21 , 42 , 43 (Figure 4a). This entry route of infection is called “the late pathway,” while the route from the plasma membrane using TMPRSS2 is called “the early pathway” (Figure 4a). Cathepsin L is thought to cleave near the S1/S2 site, 34 , 44 , 45 but it may also cleave near the S2′ site. 18 Analyses of lung epithelial Calu‐3 cells, intestinal epithelial Caco‐2 cells, and primary human airway epithelia cultured at an air–liquid interface (HAE‐ALI culture) have shown that the cathepsin L pathway is inefficiently used and that the early pathway using furin and TMPRSS2 is the major route of SARS‐CoV‐2 infection in these cells. 37 , 46 , 47 In fact, the expression level of cathepsin L is very low in Calu‐3 cells and HAE‐ALI culture, whereas it is very high in VeroE6 cells. 18 , 48 In the tissues of the nose and lungs, both TMPRSS2 and cathepsin L are detected clearly, although the expression levels are not so high. 48 Therefore, the late pathway may also play a role in infecting the respiratory tract in vivo. However, the dependence of SARS‐CoV‐2 on TMPRSS2 in the early pathway is more evident than that of SARS‐CoV 49 because SARS‐CoV‐2, but not SARS‐CoV, has an FCM. 40 The fact that the major mode of SARS‐CoV‐2 infection in vivo is the early pathway using TMPRSS2 is explained in more detail in the following section.
SIGNIFICANCE OF THE FCM VARIES BY CELL TYPE
When SARS‐CoV‐2 is propagated in VeroE6 (or Vero) cells, the virus is clearly under a strong selection pressure that causes mutations in the FCM or deletion in the loop containing the FCM. 45 , 47 , 50 , 51 , 52 , 53 , 54 The length of the loop is also important for cleavage by furin because it affects the accessibility of the protease. 48 Viruses whose S protein is not cleaved by furin (SARS‐CoV‐2 ∆FCM) are more infectious to VeroE6 cells and replicate faster in these cells than wild‐type SARS‐CoV‐2. 5 , 48 , 54 , 55 Therefore, it is clear that when propagating in VeroE6 cells, S1/S2 cleavage by furin is not necessary 39 and may even be detrimental for SARS‐CoV‐2 propagation (Figure 4b). The disadvantage of having an FCM is not observed in VeroE6 cells expressing TMPRSS2 (VeroE6/TMPRSS2). 30 , 55 When infecting VeroE6/TMPRSS2 cells, SARS‐CoV‐2 can use both the TMPRSS2‐mediated early and cathepsin‐mediated late pathways; however, the TMPRSS2‐mediated early pathway is preferentially used 47 , 49 (Figure 4a). Nevertheless, this does not mean that presence of the FCM is advantageous for SARS‐CoV‐2. Why, then, does SARS‐CoV‐2 have an FCM? Some answers can be obtained from the following observations. SARS‐CoV‐2 ∆FCM has reduced infectivity and proliferative potential in respiratory epithelial Calu‐3 cells 14 , 48 , 49 , 55 , 56 and in human airway organoids (hAOs). 49 Therefore, in most cases, the FCM is maintained in HAE‐ALI cultures 57 and hAOs. 58 However, in one case of HAE‐ALI culture, deletion mutations in the FCM increased as observed in VeroE6 cells. 57 These observations may be due to interexperimental differences in HAE‐ALI cultures. Presumably, in cases where FCM mutations accumulate, the expression level of TMPRSS2 may be expected to be very low. The expression level of hepatocyte growth activator inhibitor‐2, a physiological inhibitor of TMPRSS2, may also have influenced these results 59 , 60 , 61 (Figure 4a). Leastwise, the presence of an FCM is a distinctive feature of SARS‐CoV‐2, and its conservation has been confirmed by genetic analysis of a vast number of clinical strains. Therefore, the mode of entry via the late pathway in VeroE6 cells that affects (deletes or destroys) the FCM (Figure 4b) would not be the main mode of entry of SARS‐CoV‐2 in patients.
SIGNIFICANCE OF THE FCM IN THE PATHOGENICITY AND TRANSMISSION OF SARS‐CoV‐2
Demonstrating the importance of the FCM in SARS‐CoV‐2 pathogenicity, mutant strains that have lost furin‐mediated cleavability (SARS‐CoV‐2 ∆FCM) are attenuated in animal models (hamsters, transgenic mice). 51 , 54 , 55 Pathological changes and inflammation in the lungs are also significantly reduced in SARS‐CoV‐2 ∆FCM‐infected animals. 54 , 55 The SARS‐CoV‐2 ∆FCM mutant strains retain proliferative ability in the nasal cavity, but the virus titers in the lungs are markedly reduced in animal models. 54 Changes in the cell types infected have also been observed in animals infected with SARS‐CoV‐2 ∆FCM mutants. In animals infected with the wild‐type SARS‐CoV‐2 strain, infection can be observed in both epithelial cells and pneumocytes, but in animals infected with SARS‐CoV‐2 ∆FCM mutants, the infectivity of epithelial cells is reduced while infection of pneumocytes is mainly observed. 54 This change in cell types due to differential sensitivity to proteases (furin) is very interesting. The importance of the FCM in transmission of SARS‐CoV‐2 is also shown with a ferret model. 46 For SARS‐CoV and Middle East respiratory syndrome‐related (MERS‐) CoV, TMPRSS2 has been already demonstrated to play an important role in the in vivo growth. 62 In TMPRSS2 gene‐knockout mice, the infectivity of SARS‐CoV in bronchial epithelium (the primary infection site in this animal model) is severely affected, while the infectivity in pneumocytes is relatively stable, although the viral titer in the lungs is significantly decreased. 62 Similar analysis is required to show the role of TMPRSS2 in in vivo proliferation of SARS‐CoV‐2.
VARIANTS OF CONCERN AND CHANGES IN PROTEASE SENSITIVITY
The emergence of variants with increased infectivity or reduced vaccine efficacy has become a public health concern, and there has been much discussion about mutations within the S protein in particular. As mentioned earlier, SARS‐CoV‐2 with the D614G mutation, which emerged early in the epidemic and has become a major cause of the global pandemic, 27 , 28 , 63 has gained increased infectivity because of the D614G mutation. This mutation in the S protein increases the efficiency of S1/S2 cleavage by furin 26 and thus facilitates the adoption of an “up” conformation that favors binding to ACE2 28 , 29 (Figure 5). Currently, SARS‐CoV‐2 strains with increased infectivity and transmissibility or altered antigenicity are designated as variants of concern (VOCs), and as of August 8, 2021, four variants labeled alpha, beta, gamma, and delta are considered VOCs. All of these VOCs have the D614G mutation (Figure 5). The alpha‐variant emerged in the United Kingdom in November 2020 64 and has an N501Y mutation that directly affects the interaction with ACE2, making it more contagious than previous strains 65 (Figure 5). The alpha variant also has an amino acid mutation, P681H, in the S1/S2 site (681‐HRRAR↓S‐686; the mutated amino acid is underlined; Figure 2). A study showed that this mutation increases the cleavability of the S protein but has no significant effect on viral infectivity or membrane fusion ability. 66 The beta and gamma variants emerged in South Africa and Brazil, respectively, 67 , 68 and in addition to N501Y, they have mutations, K417N/T and E484K, which may directly affect the interaction with ACE2, resulting in enhanced ACE2 binding 69 , 70 (Figure 5). However, there are conflicting observations that E484K and K417N/T are mutations acquired to escape from neutralizing antibodies and rather reduce binding to ACE2. 71 The delta variant has spread rapidly in India since the end of 2020. It does not have mutations found in the aforesaid three VOCs, such as N501Y, E484K, and K417N/T. Instead, it has L452R and T478K mutations. Both of these mutations are located in positions that can affect the interaction with ACE2 and are likely to increase ACE2 binding (Figure 5). Similar to the alpha variant, the S protein of the delta variant has the P681R mutation in addition to L452R and T478K (Figure 5). Notably, this mutation adds a strong basic amino acid, arginine, to the S1/S2 cleavage site (681‐RRRAR↓S‐686; the mutated amino acid is underlined; Figure 2). The P681R mutation promotes furin‐mediated S1/S2 cleavage and confers the strong membrane fusion activity characteristic of the delta variant, which has been shown to have increased virulence 72 (Figure 5). The same mutation (P681R) has been found in another variant that is not a VOC, and a study with this variant has also demonstrated that the P681R mutation increases the cleavability of the S protein. 73 However, in that study, the P681R mutation alone was not sufficient to increase the infectivity of the original Wuhan SARS‐CoV‐2 strain. 73 Nevertheless, given the effect of the D614G mutation and the characteristics of the delta variant, it is likely that cleavability by furin is deeply involved in the infectivity and transmissibility of SARS‐CoV‐2.
ROLES OF OTHER TTSPs
Analysis using knockout mice has demonstrated that TMPRSS2 plays a major role in the in vivo replication and pathogenicity of SARS‐CoV, MERS‐CoV, and influenza A viruses. 32 , 62 , 74 , 75 TMPRSS2 is a member of the TTSP family, which contains about 20 different TTSPs. 76 For influenza viruses, TMPRSS4 and matriptase (suppression of tumorgenicity 14) have also been shown to have hemagglutinin (HA) cleavage activity, but it is unclear whether these TTSPs are involved in HA cleavage in vivo. 77 , 78 , 79 For SARS‐CoV‐2, TMPRSS13 (mosaic serine protease large‐form) and TMPRSS11D (human airway trypsin‐like protease) also activate S protein, although they may be less efficient than TMPRSS2. 48 , 60 , 80 , 81 , 82 TMPRSS4, TMPRSS11E (differentially expressed in squamous cell carcinoma gene 1), and TMPRSS11F may also have the ability to activate the S protein of SARS‐CoV‐2, but the results have varied greatly among published studies. 48 , 60 , 80 , 81 , 83 Thus, the importance of these TTSPs in vivo in the pathogenicity of SARS‐CoV‐2 requires further analysis.
CONCLUSION
Focusing on SARS‐CoV‐2, we have sought to discuss the proteolytic activation of coronavirus S protein. Unlike coronaviruses, the mechanism of cleavage activation of paramyxoviruses and influenza viruses is simple because there is only one cleavage site for the membrane fusion protein, and this cleavage occurs during protein synthesis, transport, and the formation of viral particles. 84 , 85 , 86 Although cleavage is essential for paramyxoviruses and influenza viruses, the cleavage itself is not directly involved in the triggering of membrane fusion. In Newcastle disease virus (paramyxovirus) and avian influenza viruses, there is a clear difference in pathogenicity closely related to whether the membrane fusion protein has an FCM or not. 84 , 86 , 87 In comparison, cleavage of the coronavirus S protein is much more complex. However, we are yet to fully understand this mechanism. As discussed in this review, presence of an FCM at the S1/S2 site is one of the distinguishing features of SARS‐CoV‐2, and some of the characteristics of the mutant viruses seem to be explained by differences in the cleavability of the S protein. Changes in cleavage properties and the proteases available for cleavage can cause significant changes in viral proliferation, tissue tropism, and virulence. 84 , 85 , 86 , 87 Coronaviruses, including SARS‐CoV‐2, seem to be no exception to this. Therefore, elucidating the mechanism of S protein cleavage activation, which is closely related to S protein‐mediated membrane fusion activity, is essential for understanding the infection mechanism of coronaviruses and the diseases by which coronavirus infections may again cause future pandemics. Such understanding will contribute to the risk assessment, early detection, and prevention of further pandemics and the development of new therapeutic agents against coronaviruses.
DISCLOSURE
The authors declare that there are no conflict of interests.
ACKNOWLEDGMENTS
I am very grateful to Dr Shutoku Matsuyama for his comments on this manuscript. This work was supported by a Grant‐in‐Aid from the Japanese Ministry of Health Labour Sciences Research (20HA2007).
Takeda M. Proteolytic activation of SARS‐CoV‐2 spike protein. Microbiol and Immunol. 2022;66:15–23. 10.1111/1348-0421.12945
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study. Data sharing not applicable to this article.
REFERENCES
- 1. Khelashvili G, Plante A, Doktorova M, Weinstein H Ca2+‐dependent mechanism of membrane insertion and destabilization by the SARS‐CoV‐2 fusion peptide. Biophys J. 2021;120: (6):1105–1119. 10.1016/j.bpj.2021.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xia X Domains and Functions of Spike Protein in SARS‐Cov‐2 in the Context of Vaccine Design. Viruses. 2021;13:(1):109. 10.3390/v13010109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clausen TM, Sandoval DR, Spliid CB, et al. SARS‐CoV‐2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1043–1057.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell. 2020;181:281–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang S, Qiu Z, Hou Y, et al. AXL is a candidate receptor for SARS‐CoV‐2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31:126–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci U S A. 2009;106:5871–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Madu IG, Roth SL, Belouzard S, Whittaker GR. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J Virol. 2009;83:7411–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS‐CoV‐2. Nat Med. 2020;26:450–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu Y, Zhao S. Furin cleavage sites naturally occur in coronaviruses. Stem Cell Res. 2020;50:102115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou H, Chen X, Hu T, et al. A novel bat coronavirus closely related to SARS‐CoV‐2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr Biol. 2020;30:2196–2203.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019‐nCoV contains a furin‐like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Millet JK, Whittaker GR. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoffmann M, Kleine‐Weber H, Pohlmann S. A multibasic cleavage site in the spike protein of SARS‐CoV‐2 is essential for infection of human lung cells. Mol Cell. 2020;78:779–784.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Song W, Gui M, Wang X, Xiang Y. Cryo‐EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14:e1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cai Y, Zhang J, Xiao T, et al. Distinct conformational states of SARS‐CoV‐2 spike protein. Science. 2020;369:1586–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Benton DJ, Wrobel AG, Xu P, et al. Receptor binding and priming of the spike protein of SARS‐CoV‐2 for membrane fusion. Nature. 2020;588:327–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park JE, Li K, Barlan A, et al. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc Natl Acad Sci U S A. 2016;113:12262–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Millet JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two‐step, furin‐mediated activation of the spike protein. Proc Natl Acad Sci U S A. 2014;111:15214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matsuyama S, Taguchi F. Two‐step conformational changes in a coronavirus envelope glycoprotein mediated by receptor binding and proteolysis. J Virol. 2009;83:11133–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shang J, Wan Y, Luo C, et al. Cell entry mechanisms of SARS‐CoV‐2. Proc Natl Acad Sci U S A. 2020;117:11727–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Papa G, Mallery DL, Albecka A, et al. Furin cleavage of SARS‐CoV‐2 Spike promotes but is not essential for infection and cell‐cell fusion. PLoS Pathog. 2021;17:e1009246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jaimes JA, Millet JK, Whittaker GR. Proteolytic cleavage of the SARS‐CoV‐2 spike protein and the role of the novel S1/S2 site. iScience. 2020;23:101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wrobel AG, Benton DJ, Xu P, et al. SARS‐CoV‐2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin‐cleavage effects. Nat Struct Mol Biol. 2020;27:763–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wrapp D, Wang N, Corbett KS, et al. Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science. 2020;367:1260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gobeil SM, Janowska K, McDowell S, et al. D614G mutation alters SARS‐CoV‐2 spike conformation and enhances protease cleavage at the S1/S2 junction. Cell Rep. 2021;34:108630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Korber B, Fischer WM, Gnanakaran S, et al. Tracking changes in SARS‐CoV‐2 spike: evidence that D614G increases infectivity of the COVID‐19 virus. Cell. 2020;182:812–827.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yurkovetskiy L, Wang X, Pascal KE, et al. Structural and functional analysis of the D614G SARS‐CoV‐2 spike protein variant. Cell. 2020;183:739–751.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weissman D, Alameh MG, de Silva T, et al. D614G spike mutation increases SARS CoV‐2 susceptibility to neutralization. Cell Host Microbe. 2021;29:23–31.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matsuyama S, Nao N, Shirato K, et al. Enhanced isolation of SARS‐CoV‐2 by TMPRSS2‐expressing cells. Proc Natl Acad Sci U S A. 2020;117:7001–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sakai K, Ami Y, Tahara M, et al. The host protease TMPRSS2 plays a major role in in vivo replication of emerging H7N9 and seasonal influenza viruses. J Virol. 2014;88:5608–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim TS, Heinlein C, Hackman RC, Nelson PS. Phenotypic analysis of mice lacking the Tmprss2‐encoded protease. Mol Cell Biol. 2006;26:965–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu G, Wang Q, Gao GF. Bat‐to‐human: spike features determining ‘host jump’ of coronaviruses SARS‐CoV, MERS‐CoV, and beyond. Trends Microbiol. 2015;23:468–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cantuti‐Castelvetri L, Ojha R, Pedro LD, et al. Neuropilin‐1 facilitates SARS‐CoV‐2 cell entry and infectivity. Science. 2020;370:856–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Daly JL, Simonetti B, Klein K, et al. Neuropilin‐1 is a host factor for SARS‐CoV‐2 infection. Science. 2020;370:861–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bestle D, Heindl MR, Limburg H, et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS‐CoV‐2 in human airway cells. Life Sci Alliance. 2020;3:e202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hornich BF, Grosskopf AK, Schlagowski S, et al. SARS‐CoV‐2 and SARS‐CoV spike‐mediated cell‐cell fusion differ in their requirements for receptor expression and proteolytic activation. J Virol. 2021;95:e00002‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tang T, Jaimes JA, Bidon MK, Straus MR, Daniel S, Whittaker GR. Proteolytic activation of SARS‐CoV‐2 spike at the S1/S2 boundary: potential role of proteases beyond furin. ACS Infect Dis. 2021;7:264–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ou T, Mou H, Zhang L, Ojha A, Choe H, Farzan M. Hydroxychloroquine‐mediated inhibition of SARS‐CoV‐2 entry is attenuated by TMPRSS2. PLoS Pathog. 2021;17:e1009212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci U S A. 2005;102:11876–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS‐CoV‐2 on virus entry and its immune cross‐reactivity with SARS‐CoV. Nat Commun. 2020;11:1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Daniloski Z, Jordan TX, Wessels HH, et al. Identification of required host factors for SARS‐CoV‐2 infection in human cells. Cell. 2021;184:92–105.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bosch BJ, Bartelink W, Rottier PJ. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J Virol. 2008;82:8887–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu Z, Zheng H, Lin H, et al. Identification of common deletions in the spike protein of severe acute respiratory syndrome coronavirus 2. J Virol. 2020;94:e00790‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peacock TP, Goldhill DH, Zhou J, et al. The furin cleavage site in the SARS‐CoV‐2 spike protein is required for transmission in ferrets. Nat Microbiol. 2021;6:899–909. [DOI] [PubMed] [Google Scholar]
- 47. Sasaki M, Uemura K, Sato A, et al. SARS‐CoV‐2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2‐deficient cells. PLoS Pathog. 2021;17:e1009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Laporte M, Raeymaekers V, Van Berwaer R, et al. The SARS‐CoV‐2 and other human coronavirus spike proteins are fine‐tuned towards temperature and proteases of the human airways. PLoS Pathog. 2021;17:e1009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mykytyn AZ, Breugem TI, Riesebosch S, et al. SARS‐CoV‐2 entry into human airway organoids is serine protease‐mediated and facilitated by the multibasic cleavage site. eLife. 2021;10:e64508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ogando NS, Dalebout TJ, Zevenhoven‐Dobbe JC, et al. SARS‐coronavirus‐2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J Gen Virol. 2020;101:925–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lau SY, Wang P, Mok BW, et al. Attenuated SARS‐CoV‐2 variants with deletions at the S1/S2 junction. Emerg Microbes Infect. 2020;9:837–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Klimstra WB, Tilston‐Lunel NL, Nambulli S, et al. SARS‐CoV‐2 growth, furin‐cleavage‐site adaptation and neutralization using serum from acutely infected hospitalized COVID‐19 patients. J Gen Virol. 2020;101:1156–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Davidson AD, Williamson MK, Lewis S, et al. Characterisation of the transcriptome and proteome of SARS‐CoV‐2 reveals a cell passage induced in‐frame deletion of the furin‐like cleavage site from the spike glycoprotein. Genome Med. 2020;12:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang P, Lau SY, Deng S, et al. Characterization of an attenuated SARS‐CoV‐2 variant with a deletion at the S1/S2 junction of the spike protein. Nat Commun. 2021;12:2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Johnson BA, Xie X, Bailey AL, et al. Loss of furin cleavage site attenuates SARS‐CoV‐2 pathogenesis. Nature. 2021;591:293–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chu H, Hu B, Huang X, et al. Host and viral determinants for efficient SARS‐CoV‐2 infection of the human lung. Nat Commun. 2021;12:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zou W, Xiong M, Hao S, et al. The SARS‐CoV‐2 transcriptome and the dynamics of the S gene furin cleavage site in primary human airway epithelia. mBio. 2021;12:e01006‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lamers MM, Mykytyn AZ, Breugem TI, et al. Human airway cells prevent SARS‐CoV‐2 multibasic cleavage site cell culture adaptation. eLife. 2021;10:e66815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tomita Y, Matsuyama S, Fukuhara H, et al. The physiological TMPRSS2 inhibitor HAI‐2 alleviates SARS‐CoV‐2 infection. J Virol. 2021;95:e00434‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fuentes‐Prior P. Priming of SARS‐CoV‐2 S protein by several membrane‐bound serine proteinases could explain enhanced viral infectivity and systemic COVID‐19 infection. J Biol Chem. 2020;296:100135. 10.1074/jbc.REV120.015980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kataoka H, Kawaguchi M, Fukushima T, Shimomura T. Hepatocyte growth factor activator inhibitors (HAI‐1 and HAI‐2): emerging key players in epithelial integrity and cancer. Pathol Int. 2018;68:145–58. [DOI] [PubMed] [Google Scholar]
- 62. Iwata‐Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol. 2019;93:e01815‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Volz E, Hill V, McCrone JT, et al. Evaluating the effects of SARS‐CoV‐2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;18:64–75.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Leung K, Shum MH, Leung GM, Lam TT, Wu JT. Early transmissibility assessment of the N501Y mutant strains of SARS‐CoV‐2 in the United Kingdom, October to November 2020. Euro Surveill. 2021;26:2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Davies NG, Abbott S, Barnard RC, et al. Estimated transmissibility and impact of SARS‐CoV‐2 lineage B.1.1.7 in England. Science. 2021;372:eabg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lubinski B, Tang T, Daniel S, Jaimes JA, Whittaker GR. Functional evaluation of proteolytic activation for the SARS‐CoV‐2 variant B.1.1.7: role of the P681H mutation. bioRxiv. 2021. 10.1101/2021.04.06.438731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tegally H, Wilkinson E, Giovanetti M, et al. Emergence and rapid spread of a new severe acute respiratory syndrome‐related coronavirus 2 (SARS‐CoV‐2) lineage with multiple spike mutations in South Africa. medRxiv. 2020. [Google Scholar]
- 68. Mahase E. Covid‐19: what new variants are emerging and how are they being investigated? BMJ. 2021;372:n158. [DOI] [PubMed] [Google Scholar]
- 69. Khan A, Zia T, Suleman M, et al. Higher infectivity of the SARS‐CoV‐2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J Cell Physiol. 2021;236:7045–57. 10.1002/jcp.30367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ramanathan M, Ferguson ID, Miao W, Khavari PA. SARS‐CoV‐2 B.1.1.7 and B.1.351 spike variants bind human ACE2 with increased affinity. Lancet Infect Dis. 2021;21:1070. 10.1016/S1473-3099(21)00262-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cai Y, Zhang J, Xiao T, et al. Structural basis for enhanced infectivity and immune evasion of SARS‐CoV‐2 variants. Science. 2021;373:642–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Saito A, Irie T, Suzuki R, et al. SARS‐CoV‐2 spike P681R mutation, a hollmark of the delta variant, enhances viral fusogenicity and pathogenicity. bioRxiv. 2021. 10.1101/2021.06.17.448820 [DOI] [Google Scholar]
- 73. Lubinski B, Frazier LE, Phan MVT, et al. Spike protein cleavage‐activation mediated by the SARS‐CoV‐2 P681R mutation: a case‐study from its first appearance in variant of interest (VOI) A.23.1 identified in Uganda. bioRxiv. 2021. 10.1101/2021.06.30.450632 [DOI] [Google Scholar]
- 74. Tarnow C, Engels G, Arendt A, et al. TMPRSS2 is a host factor that is essential for pneumotropism and pathogenicity of H7N9 influenza A virus in mice. J Virol. 2014;88:4744–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hatesuer B, Bertram S, Mehnert N, et al. Tmprss2 is essential for influenza H1N1 virus pathogenesis in mice. PLoS Pathog. 2013;9:e1003774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bugge TH, Antalis TM, Wu Q. Type II transmembrane serine proteases. J Biol Chem. 2009;284:23177–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bertram S, Glowacka I, Blazejewska P, et al. TMPRSS2 and TMPRSS4 facilitate trypsin‐independent spread of influenza virus in Caco‐2 cells. J Virol. 2010;84:10016–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Baron J, Tarnow C, Mayoli‐Nussle D, et al. Matriptase, HAT, and TMPRSS2 activate the hemagglutinin of H9N2 influenza A viruses. J Virol. 2013;87:1811–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Beaulieu A, Gravel E, Cloutier A, et al. Matriptase proteolytically activates influenza virus and promotes multicycle replication in the human airway epithelium. J Virol. 2013;87:4237–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hoffmann M, Hofmann‐Winkler H, Smith JC, et al. Camostat mesylate inhibits SARS‐CoV‐2 activation by TMPRSS2‐related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine. 2021;65:103255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kishimoto M, Uemura K, Sanaki T, et al. TMPRSS11D and TMPRSS13 Activate the SARS‐CoV‐2 Spike Protein. Viruses. 2021;13:384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Xia S, Lan Q, Su S, et al. The role of furin cleavage site in SARS‐CoV‐2 spike protein‐mediated membrane fusion in the presence or absence of trypsin. Signal Transduct Target Ther. 2020;5:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zang R, Gomez Castro MF, McCune BT, et al. TMPRSS2 and TMPRSS4 promote SARS‐CoV‐2 infection of human small intestinal enterocytes. Sci Immunol. 2020;5:eabc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Galloway SE, Liang B, Steinhauer DA. Activation of the hemagglutinin of influenza viruses. In: Bottcher‐Friebertshauser E, Garten W, Klenk DH, editors. Activation of viruses by host proteases. Cham, Switzerland: Springer; 2018. p. 3–26. [Google Scholar]
- 85. Smith EC, Dutch RE. Proteolytic activation of paramyxoviruses and pneumoviruses. In: Bottcher‐Friebertshauser E, Garten W, Klenk DH (ed) Activation of viruses by host proteases. Cham, Switzerland: Springer; 2018. p. 27–45. [Google Scholar]
- 86. Bertram S, Glowacka I, Steffen I, Kuhl A, Pohlmann S. Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Rev Med Virol. 2010;20:298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Morrison TG Structure and function of a paramyxovirus fusion protein. Biochimica et Biophysica Acta (BBA) ‐ Biomembranes. 2003;1614: (1):73–84. 10.1016/s0005-2736(03)00164-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study. Data sharing not applicable to this article.


