Abstract
Haploid yeast cells initiate pheromone signaling upon the binding of pheromone to its receptor and activation of the coupled G protein. A regulatory process termed receptor inhibition blocks pheromone signaling when the a-factor receptor is inappropriately expressed in MATa cells. Receptor inhibition blocks signaling by inhibiting the activity of the G protein β subunit, Ste4p. To investigate how Ste4p activity is inhibited, its subcellular location was examined. In wild-type cells, α-factor treatment resulted in localization of Ste4p to the plasma membrane of mating projections. In cells expressing the a-factor receptor, α-factor treatment resulted in localization of Ste4p away from the plasma membrane to an internal compartment. An altered version of Ste4p that is largely insensitive to receptor inhibition retained its association with the membrane in cells expressing the a-factor receptor. The inhibitory function of the a-factor receptor required ASG7, an a-specific gene of previously unknown function. ASG7 RNA was induced by pheromone, consistent with increased inhibition as the pheromone response progresses. The a-factor receptor inhibited signaling in its liganded state, demonstrating that the receptor can block the signal that it initiates. ASG7 was required for the altered localization of Ste4p that occurs during receptor inhibition, and the subcellular location of Asg7p was consistent with its having a direct effect on Ste4p localization. These results demonstrate that Asg7p mediates a regulatory process that blocks signaling from a G protein β subunit and causes its relocalization within the cell.
The pheromone response of the yeast Saccharomyces cerevisiae is initiated by the binding of a peptide pheromone to its specific receptor on a responding cell. Haploid yeast cells containing the MATa allele secrete a-factor and express receptors for α-factor; cells containing the MATα allele secrete α-factor and express receptors for a-factor (19, 33, 36). The a- and α-factor receptors are G protein-coupled receptors, and they activate a heterotrimeric G protein that is common to both cell types. Activation of the G protein by occupied receptors results in guanine nucleotide exchange on the G protein α subunit, which causes the β and γ subunits to be released from the α subunit. The free βγ complex interacts with downstream components of the pathway, resulting in activation of a mitogen-activated protein (MAP) kinase cascade. Signaling through this pathway produces a number of changes in cellular physiology, including arrest in the G1 phase of the cell cycle, induction of gene expression, redistribution of cell surface proteins, and formation of cellular projections.
In addition to the classical G protein activation process described above, the pheromone response pathway is subject to a process that inhibits G protein signaling. This process, termed receptor inhibition, was uncovered by a mutation that causes the a-factor receptor to be inappropriately expressed in MATa cells. The STE3DAF mutation was isolated in a screen for dominant mutations that conferred resistance to pheromone-induced cell cycle arrest (4). STE3DAF, which is an allele of the a-factor receptor gene STE3, contains a rearranged 5′ regulatory region that causes it to be expressed in MATa cells (13). Expression of STE3 in MATa cells confers resistance to pheromone-induced cell cycle arrest by blocking signaling through the pheromone response pathway. Although MATa haploid cells do not normally express the α-specific gene STE3, a- and α-specific gene products do come into contact with each other immediately after the fusion of a MATa cell and a MATα cell during the process of mating. Therefore, a potential physiological function of receptor inhibition is to inhibit signaling in mating cells that have recently undergone cell fusion. This process may function to promote recovery from mating and allow cell cycle progression to resume.
Cells that contain a STE3DAF allele display a characteristic pattern of signaling. In STE3DAF cells treated with pheromone, activation of the MAP kinase is normal early in the response but is gradually inhibited at later time points (3). This pattern suggests that prior activation of the signaling pathway must occur before the inhibitory process can function. Another characteristic of receptor inhibition is that it is independent of the G protein α subunit (4, 13). In the pheromone response pathway, the α subunit plays a negative role by sequestering the βγ complex and keeping it inactive. Deletion of GPA1, the G protein α subunit gene, causes constitutive signaling due to release of the βγ complex. The constitutive signaling phenotype of cells with a GPA1 deletion is suppressed by the STE3DAF mutation, indicating that receptor inhibition of signaling can occur in the absence of Gpa1p. This result demonstrates that the target of receptor inhibition is a signaling component that is downstream of the G protein α subunit.
Two types of evidence suggest that the G protein β subunit, Ste4p, is the signaling component that is targeted by receptor inhibition. Initial results supporting this idea involve the phenotypes of double mutants that carry both STE3DAF and a constitutive signaling mutation. The STE3DAF allele blocks signaling in cells that contain constitutive or overexpression alleles of STE4 (3, 13). However, STE3DAF does not block signaling in cells that overexpress STE20 or that contain a constitutive allele of STE5 (3). STE20 encodes a kinase that activates the MAP kinase cascade, and STE5 encodes a scaffolding protein for the MAP kinase cascade, so both of these signaling components act downstream of Ste4p. These studies are therefore consistent with the idea that receptor inhibition acts at the level of Ste4p, the G protein β subunit. This conclusion was further supported by the isolation of altered versions of Ste4p that can signal normally but that are insensitive to receptor inhibition (17). The altered residues in Ste4p are not in the regions that contact the α subunit, suggesting that they constitute part of a binding site for another protein that interacts with Ste4p. Such a protein could be a negative regulator that prevents Ste4p from interacting with its downstream targets. One potential explanation for the phenotype of the STE3DAF allele is that a negative regulator of Ste4p is expressed only in MATa cells.
Receptor inhibition appears to require an a-specific component based on the following observation. Deletion of GPA1 causes constitutive signaling in both MATa and MATα cells due to release of the βγ complex. Expression of STE3 inhibits the constitutive signaling conferred by a GPA1 deletion in MATa cells but has no effect in MATα cells (17). One important difference between MATa and MATα cells is that Ste2p, the α-factor receptor, is expressed only in MATa cells. However, receptor inhibition does not require the presence of Ste2p because the ability of STE3DAF to block signaling in MATa gpa1Δ cells is unaffected by a ste2Δ mutation (17). These findings indicate that an a-specific component other than Ste2p is required for inhibition of β-subunit activity by the a-factor receptor.
Here we investigate further the process by which Ste4p signaling is blocked by receptor inhibition. Examination of the subcellular location of Ste4p indicates that it undergoes altered localization during receptor inhibition. In addition, we show that altered localization of Ste4p and inhibition of signaling require an a-specific gene called ASG7.
MATERIALS AND METHODS
Plasmid construction.
Plasmid YCpΔ36 was constructed by replacing the 392-bp HpaI-XhoI fragment in plasmid YCpSTE4 (17) with a 284-bp HpaI-XhoI fragment from a plasmid containing the STE4 gene in which the 108 bp encoding amino acids 310 to 346 of Ste4p had been deleted by site-directed mutagenesis.
The fusion of STE4 with the gene encoding green fluorescent protein (GFP) was constructed as follows. STE4 was amplified by PCR using oligodeoxynucleotides 5′-CCACTAGTGCATGCATGGCAGCACATCAGATG-3′ and 5′-AGGAGCTCCTACCCGGGTTGATAACCTGGAGAC-3′ (the newly created SpeI and SacI sites are underlined; the newly created SphI and SmaI sites are in boldface) as primers and pL38 (18) as the template and cloned into the SpeI and SacI sites of pRS316-GAL (20). The GFPS65T mutant (11) was amplified by PCR using oligodeoxynucleotides 5′-CGGGATCCGCTAGCATGAGTAAAGGAGAAGAAC-3′ and 5′-GCTCTAGATTAGCATGCACTAGTTTTGTATAGT-3′ (the newly created BamHI and XbaI sites are underlined; the newly created NheI and SphI sites are in boldface) as primers and pRSET-B-GFPS65T (obtained from R. Tsien) as the template and cloned into the XbaI and BamHI sites of pRS316-GAL to yield plasmid pBTL32. Plasmid pBTL34 carrying STE4 fused to the carboxyl terminus of GFPS65T was created by cloning the SphI-to-SacI fragment of pBTL29 into pBTL32. The promoter region of STE4 was amplified by PCR using oligodeoxynucleotides 5′-CGGAATTCAATGTTTCAGGAAGAGAT-3′ and 5′-GCGGATCCCGTAATGTGTACCTGATT-3′ (the newly created EcoRI and BamHI sites are underlined) as primers and pL38 as the template and cloned into the EcoRI and BamHI sites of pRS313, creating plasmid pBTL42. Plasmid pBTL60 carrying a fusion of STE4 with the carboxyl terminus of GFPS65T under the control of the STE4 promoter was then constructed by cloning the BamHI-to-SacI fragment from plasmid pBTL34 into plasmid pBTL42, and pBTL49 was constructed by transferring the promoter-GFP fusion construct contained within the PvuI fragment to plasmid pRS316. Plasmid YCpGFP-SD10 was constructed by replacing the 2.1-kb BamHI-SacI fragment in pBTL49, which contains STE4, with a 2.1-kb BamHI-SacI fragment that contains the STE4SD10 allele (17).
The fragment used for construction of asg7::URA3 null alleles was synthesized by two-step PCR (35). In the first step, oligonucleotides W1517 (5′-CCGCATTAGTGGGCTATCAGTAGCAC) and W1519 (5′-TATCAGTTATTACCCTATGCGGTGTGCCAAGGGTTCTCATCGTTCTCGAGGGCGC) were used as the primers and yeast genomic DNA was used as the template to generate a 427-bp fragment in which the first 401-bp region is homologous to the 5′ untranslated region of ASG7 and the last 26-bp region is homologous to the 5′ region of the URA3 gene. Oligonucleotides W1520 (5′-CCTTCTGTTCGGAGATTACCGAATCAGTAGATCTAAAGACAGAAAATGATATCAGCC) and W1518 (5′-CGACTGAGGTCCACTGGCAGCGACTG) were used as the primers to generate a 353-bp fragment in which the first 26 bp is homologous to the 3′ region of the URA3 gene and the last 327 bp is homologous to the 3′ untranslated region of the ASG7 gene. In the second step, the 427- and 353-bp fragments from the first PCRs served as the primers and plasmid pRS426, which contains the URA3 gene, was used as the template in the PCR. The product of this reaction was used to transform yeast strains.
A plasmid containing the wild-type ASG7 gene was isolated by complementation of the asg7::URA3 mutation. DNA from yeast genomic library 2J351 (8) was transformed into a MATa STE3DAF asg7::URA3 strain (H67-6C.a7), and transformants were screened for their ability to grow in the presence of α-factor. Plasmid pASG7-351.1, which contains the ASG7 gene, conferred resistance to α-factor-induced cell cycle arrest. Plasmid pASG7-351.2 was constructed by subcloning the 1.6-kb HindIII-BglII fragment from pASG7-351.1 into the HindIII-BamHI sites of vector YEp351. The ASG7-GFP fusion gene was constructed using a 1.8-kb HindIII fragment of genomic DNA from pASG7-351.1 that contains ASG7. A 0.7-kb fragment containing GFP flanked by NotI sites was subcloned into a NotI site that was inserted immediately before the stop codon in ASG7 by site-directed mutagenesis. The 2.5-kb HindIII fragment containing the ASG7-GFP fusion gene was subcloned into the HindIII site of YCplac111 (10) to create plasmid pASG7-111.GFP.
Strains and media.
Strains used in this study are listed in Table 1. The asg7::URA3 null allele was made by transformation of strains with a 1.8-kb fragment generated by two-step PCR, as described above. The gpa1::TRP1 allele was created by transformation of a strain containing the gpa1::URA3 allele (7) with a 3.2-kb EcoRI-XhoI fragment from plasmid pTU10 (5). The far1::LEU2 allele was created by transformation of a strain containing the far1::URA3 allele (3) with a 4.6-kb SmaI fragment from plasmid pUL9 (5). The asg7::HIS3 allele was created by transformation of a strain containing the asg7::URA3 allele with a 3.6-kb XbaI fragment from plasmid pUH7 (5).
TABLE 1.
Yeast strains used in this study
| Strain | Genotypea | Reference or source |
|---|---|---|
| W3031A | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 | R. Rothstein |
| W3031B | MATα | R. Rothstein |
| W3031A.Ba | MATa sst1::hisG | This study |
| H67-9D.Ba | MATa mfa1-Δ3::HIS3 mfa2-Δ2::HIS3 sst1::hisG | 3 |
| H67-9D.a7 | MATa mfa1-Δ3::HIS3 mfa2-Δ2::HIS3 sst1::hisG asg7::URA3 | This study |
| H67-6C.Ba | MATa mfa1-Δ3::HIS3 mfa2-Δ2::HIS3 STE3DAF2.5 sst1::hisG | 3 |
| H67-6C.a7 | MATa mfa1-Δ3::HIS3 mfa2-Δ2::HIS3 STE3DAF2.5 sst1::hisG asg7::URA3 | This study |
| AC17-7B | MATa mfa1-Δ3::HIS3 mfa2-Δ2::HIS3 sst1::hisG ste4::HIS3 | 17 |
| AC17-2B | MATa mfa1-Δ3::HIS3 mfa2-Δ2::HIS3 STE3DAF2.5 sst1::hisG ste4::HIS3 | 17 |
| AC17-2B.aL | MATa mfa1-Δ3::HIS3 mfa2-Δ2::HIS3 STE3DAF2.5 sst1::hisG ste4::HIS3 asg7::LEU2 | This study |
| H125-7D | MATa gpa1::TRP1 STE3DAF2.5 | This study |
| H125-7D.aH | MATa gpa1::TRP1 STE3DAF2.5 asg7::HIS3 | This study |
All strains other than W3031A are isogenic to W3031A.
Strains were grown on yeast extract-peptone-dextrose (2% glucose) or yeast extract-peptone–Gal (3% galactose), and strains under selection were grown on synthetic dropout media, as described previously (32).
Yeast methods.
Yeast transformations were performed by the lithium acetate method (15) modified as described previously (13). Yeast RNA was extracted from cells as described previously (6).
Halo assays were performed by plating a lawn of cells to be tested and placing a filter paper disk containing 5 μl of 1 mM α-factor onto the plate. Plates were then incubated at 30°C for 1 to 2 days.
Northern blots.
Cells were treated with 0.1 μM α-factor (Sigma), 60 nM a-factor, or 300 nM a-factor (generously provided by Fred Naider) for various periods of time, and RNA was isolated. RNA was transferred to a nitrocellulose membrane after formaldehyde-agarose gel electrophoresis as described previously (31). The membranes were UV cross-linked using a Stratalinker UV box. Prehybridization and hybridization were done at 65°C in a buffer containing 0.9 M NaCl, 0.09 M sodium citrate, 0.1% Ficoll, 0.1% polyvinylpyrrolidone, 0.1% bovine serum albumin, 33 mM sodium pyrophosphate, and 50 mM sodium phosphate monobasic. The probes used were gel-purified DNA restriction fragments 32P labeled by random primer labeling using a Prime-It kit (Stratagene). The fragments used were FUS1, a 1.4-kb EcoRI-HindIII fragment from plasmid pSL589 (25), phosphoglycerate kinase gene PGK1, a 0.5-kb BamHI-XbaI fragment from pPGK1, and ASG7, a 0.75-kb HpaI-SacI fragment from pASG7-351.2.
Immunoblots.
For immunoblots, cells were treated with 0.1 μM α-factor (Sigma) for various periods of time, and 10-ml aliquots of cells were pelleted and washed once with 10 mM Tris-HCl (pH 7.8)–1 mM EDTA. The washed cells were resuspended in 350 μl of lysis buffer (50 mM Tris-HCl [pH 8.0], 1% sodium dodecyl sulfate [SDS], 1 mM phenylmethylsulfonyl fluoride, and 1 μg each of leupeptin, aprotinin, chymostatin, and pepstatin/ml). The cell suspension was lysed by adding approximately 0.25 ml of acid-washed glass beads (0.5 mm; Biospec Products) and vortexing at high speed for 10 min at 4°C. The lysate was cleared by centrifuging for 2 min at 4°C. The protein concentrations of the samples were determined using a bicinchoninic protein assay kit (Pierce), and equal amounts of protein were loaded onto SDS-polyacrylamide gels (10% acrylamide). Separated proteins were transferred to nitrocellulose, and the blot was probed with anti-Ste4p rabbit antiserum (14) at a dilution of 1:1,000. Donkey anti-rabbit immunoglobulin conjugated to horseradish peroxidase (Amersham) was used at a dilution of 1:10,000, and immune complexes were detected with an enhanced chemiluminescence kit (Amersham).
Microscopy.
For fluorescence microscopy, cells containing GFP-Ste4p or Asg7p-GFP fusion proteins were observed on a Zeiss Axioskop microscope with a ×100 (1.3-numerical-aperture) objective and a fluorescein isothiocyanate (FITC) filter set (Chroma Technology). Digital images were captured with a Photometrics SenSys 1400-C1.cCCD camera using IPLab Spectrum image acquisition software (Scanalytics).
RESULTS
Expression of the a-factor receptor in MATa cells inhibits signal transduction through the pheromone response pathway by a process called receptor inhibition. In cells undergoing receptor inhibition, the signaling pathway is blocked at the level of Ste4p, the G protein β subunit (3, 17). To investigate the mechanism responsible for the signaling block, the effect of receptor inhibition on properties of Ste4p that are required for signaling was examined.
Expression of the a-factor receptor in a cells affects Ste4p localization.
Previous studies have shown that association of the βγ complex with the plasma membrane is required for activation of the signaling pathway (29). Therefore, one mechanism to account for signaling inhibition in the presence of the a-factor receptor is that the Ste4p β subunit is not properly localized to the plasma membrane. To investigate this possibility, the localization pattern of a GFP-Ste4p fusion protein was observed in wild-type MATa cells and STE3DAF MATa cells that were treated with pheromone for various amounts of time. STE3 encodes the a-factor receptor, and the STE3DAF allele causes expression of STE3 in MATa cells due to an insertion in its promoter region (13). To eliminate any effects of autocrine signaling through the a-factor receptor, both the wild-type and STE3DAF strains used in these experiments have deletions of the genes encoding a-factor (13).
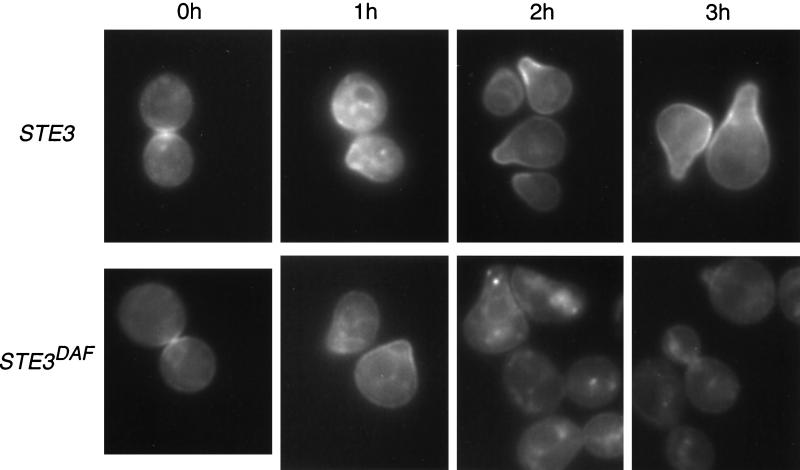
Both wild-type and STE3DAF untreated cells displayed a signal that appeared to be partially localized at the membrane and partially localized to an internal compartment (Fig. 1). This result is in agreement with cell fractionation studies done by others, which have shown that Ste4p partitions 40% with the plasma membrane, 30% with internal membranes, and 30% with nonmembrane fractions (14). After 1 h of α-factor treatment, GFP-Ste4p was concentrated at the sites of incipient mating projection formation in both strains. At this time point, the similar localization patterns of GFP-Ste4p in the two strains are expected, given the previous finding that STE3DAF cells undergo a detectable level of signaling after 1 h of pheromone treatment (3). After 2 h of α-factor treatment of wild-type cells, GFP-Ste4p was localized predominantly at the membrane in regions where mating projections had formed (Fig. 1, 2 h). In contrast, STE3DAF cells treated with α-factor for 2 h showed a dramatic reduction in the amount of GFP-Ste4p that was membrane associated and an increase in the amount that localized in an internal particulate pattern. This effect was observed even in cells that still contained mating projections. After 3 h of α-factor treatment, GFP-Ste4p remained at the sites of mating projections in wild-type cells but was predominantly localized to an internal compartment in STE3DAF cells. At this time point, the STE3DAF cells had recovered from cell cycle arrest and displayed a high percentage of budded cells. Significant inhibition of signaling occurs in STE3DAF cells at about 2 h after pheromone treatment (3). Thus, localization of GFP-Ste4p away from the membrane correlates with the period of signaling inhibition.
FIG. 1.
Localization of GFP-Ste4p in wild-type and STE3DAF cells. The following strains were treated with α-factor (0.1 μM) for the indicated periods of time: AC17-7B, a MATa STE3 ste4::HIS3 strain (STE3), and AC17-2B, a MATa STE3DAF ste4::HIS3 strain (STE3DAF). Both strains contained the low-copy-number GFP-STE4 plasmid BTL49. The live cells were viewed by fluorescence microscopy using an FITC filter set.
Expression of the a-factor receptor in a cells does not affect Ste4p abundance.
In cells undergoing receptor inhibition, the localization pattern of Ste4p resembled that of an endocytic compartment (Fig. 1, STE3DAF, 2 h). If receptor inhibition causes Ste4p to undergo endocytosis, it is possible that the level of Ste4p would decrease under these conditions. To determine the effect of receptor inhibition on Ste4p abundance, the levels of Ste4p in wild-type MATa cells and STE3DAF MATa cells were investigated.
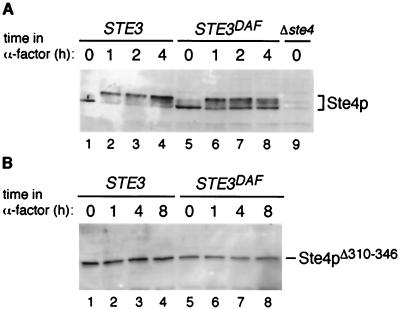
Cell extracts were prepared from wild-type and STE3DAF strains that had been treated with α-factor for various amounts of time, and immunoblots prepared from these extracts were probed with a polyclonal anti-Ste4p antibody (14). In wild-type cells treated with α-factor for 1 to 4 h, Ste4p displayed a mobility shift characteristic of the phosphorylated form (Fig. 2A, lanes 1 to 4), as described previously (2). In STE3DAF cells, Ste4p was present in both the unmodified and phosphorylated forms after 1 h of exposure to α-factor and the unmodified form increased in abundance at later time points (Fig. 2A, lanes 5 to 8). These results are consistent with previous observations showing that STE3DAF cells undergo an initial response to pheromone that is gradually inhibited at later times after pheromone treatment (3). There did not appear to be a significant difference between the abundance of Ste4p in wild-type cells and that in STE3DAF cells at the 2- and 4-h time points, when signaling is inhibited in STE3DAF cells. However, the presence of multiple forms of Ste4p made it difficult to compare its abundances in the two different strains. To eliminate this complication, the effect of the STE3DAF allele on Ste4p abundance was investigated in a strain containing a form of Ste4p that does not undergo phosphorylation in response to pheromone (2). This form of Ste4p, which lacks residues 310 to 346, is fully capable of signal transmission because phosphorylation is not required for the Ste4p signaling function (22). Moreover, the signaling activity of Ste4pΔ310–346 is capable of being inhibited by expression of STE3 in MATa cells, indicating that phosphorylation does not play a role in receptor inhibition (J. Kim and J. P. Hirsch, unpublished data). Immunoblots of samples from wild-type and STE3DAF strains were probed with anti-Ste4p antibody to detect the level of Ste4pΔ310–346 at various times after pheromone treatment. At all time points, wild-type cells contained slightly higher levels of Ste4pΔ310–346 than STE3DAF cells (Fig. 2B, lanes 1 to 8). However, the level of Ste4pΔ310–346 did not change significantly in STE3DAF cells after treatment with pheromone. These results demonstrate that receptor inhibition does not cause a major change in the steady-state level of the Ste4p β subunit.
FIG. 2.
Abundances of Ste4p and Ste4pΔ310–346 in wild-type and STE3DAF cells. (A) The following strains were treated with α-factor (0.1 μM) for the indicated periods of time: H67-9D.Ba, a MATa STE3 strain (lanes 1 to 4), H67-6C.Ba, a MATa STE3DAF strain (lanes 5 to 8), and AC17-7B, a MATa STE3 ste4::HIS3 strain (lane 9). Cell extracts were prepared, and immunoblots containing these extracts were probed with anti-Ste4p polyclonal antibody. (B) The following strains were treated with α-factor (0.1 μM) for the indicated periods of time: AC17-7B, a MATa STE3 ste4::HIS3 strain carrying plasmid YCpΔ36, which contains the STE4Δ310–346 allele (lanes 1 to 4), and AC17-2B, a MATa STE3DAF ste4::HIS3 strain carrying plasmid YCpΔ36 (lanes 5 to 8). Cell extracts were prepared, and immunoblots containing these extracts were probed with an anti-Ste4p polyclonal antibody.
An altered version of Ste4p localizes normally in cells expressing the a-factor receptor.
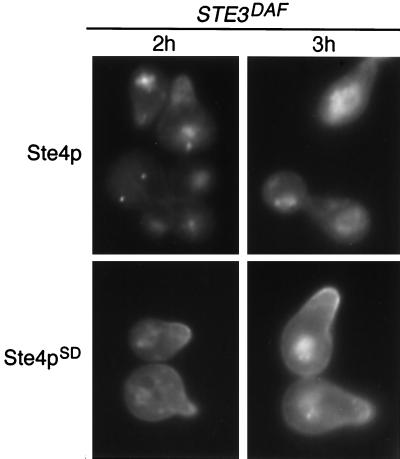
STE4SD alleles are mutated versions of the STE4 gene that produce Ste4p variants that are largely insensitive to inhibition by the a-factor receptor (17). In wild-type MATa cells, the Ste4pSD10 variant signals in a manner indistinguishable from that of wild-type Ste4p. In STE3DAF MATa cells, Ste4pSD10 produces a much greater signal than wild-type Ste4p at late time points after α-factor treatment. To test whether signaling correlates with localization of Ste4p at the plasma membrane, the localization pattern of Ste4pSD10 was investigated in MATa cells expressing the a-factor receptor. In STE3DAF cells treated with α-factor for 2 or 3 h, GFP-Ste4p was localized in an internal particulate pattern (Fig. 3, Ste4p). In contrast, a large proportion of the GFP-Ste4pSD10 signal was localized at the membrane at regions of mating projections (Fig. 3, Ste4pSD). Therefore, the increased signaling conferred by the Ste4pSD10 variant correlates with increased membrane localization. These results suggest that inhibition of signaling by the a-factor receptor is effected by localization of Ste4p away from the plasma membrane.
FIG. 3.
Localization of GFP-Ste4p and GFP-Ste4pSD10 in STE3DAF cells. Strain AC17-2B (MATa STE3DAF ste4::HIS3) containing a GFP-STE4 plasmid (pBTL49) or a GFP-STE4SD10 plasmid (YCpGFP-SD10) was treated with α-factor (0.1 μM) for the indicated periods of time. The live cells were viewed by fluorescence microscopy using an FITC filter set.
ASG7 is required for inhibition of signaling by the a-factor receptor.
Previous results indicate that receptor inhibition requires a component that is only present in MATa cells. However, it has been shown that inhibition of signaling does not require any of the known a-specific genes that might be expected to be involved in this process, including STE2 (a-factor receptor gene), MFA1 and MFA2 (a-factor genes), SST1 (α-factor protease gene), and STE6 (a-factor transporter gene) (13; A. Couve and J. P. Hirsch, unpublished data). A comprehensive approach to identify a-specific genes was recently performed by screening the Saccharomyces Genome Database for Matα2p-Mcm1p binding sites in the 5′ flanking regions of open reading frames (38). The Matα2p-Mcm1p complex functions to repress transcription in MATα cells, and thus these binding sites identify genes that are only expressed in MATa cells. A previously uncharacterized a-specific gene, ASG7, was identified by the screen. The requirement for ASG7 in receptor inhibition was tested by deleting the ASG7 gene in MATa cells containing a STE3DAF allele.
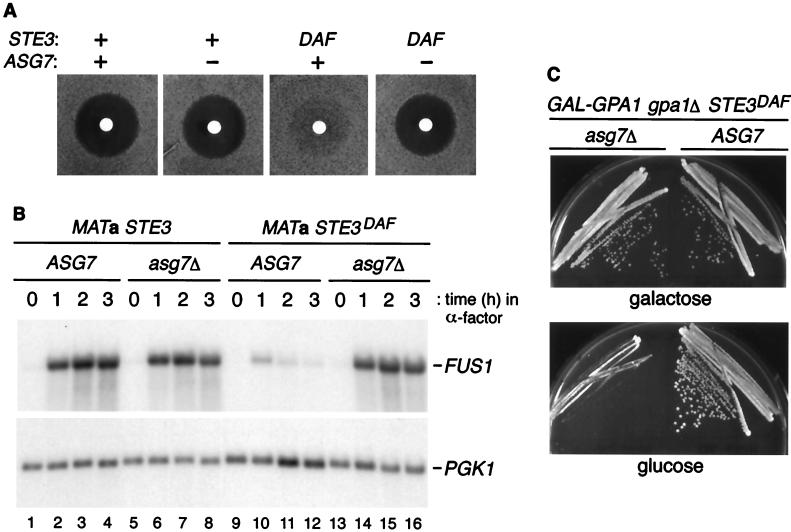
As described previously, MATa cells containing a STE3DAF allele did not undergo cell cycle arrest in response to pheromone as measured by a halo assay (Fig. 4A). However, deletion of ASG7 in STE3DAF cells completely eliminated the inhibition of cell cycle arrest seen in the STE3DAF strain. Deletion of ASG7 had no effect on pheromone-induced cell cycle arrest in wild-type MATa cells. These results demonstrate that a functional ASG7 gene is required for inhibition of the cell cycle arrest response by STE3DAF.
FIG. 4.
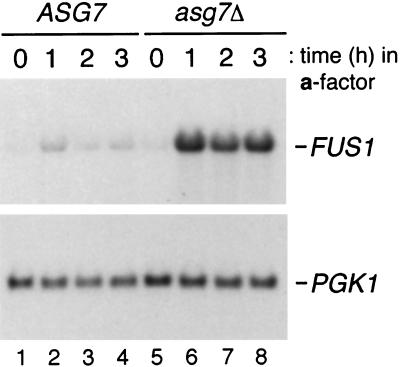
Effect of ASG7 on signaling in STE3DAF cells. (A) Halo assays were performed with 5 μl of 1 mM α-factor using the following strains (from left to right): H67-9D.Ba, a MATa STE3 ASG7 strain, H67.9D.a7, a MATa STE3 asg7::URA3 strain, H67-6C.Ba, a MATa STE3DAF ASG7 strain, and H67-6C.a7, a MATa STE3DAF asg7::URA3 strain. (B) The following strains were treated with α-factor (0.1 μM) for the indicated periods of time: H67-9D.Ba, a MATa STE3 ASG7 strain (lanes 1 to 4), H67-9D.a7, a MATa STE3 asg7::URA3 strain (lanes 5 to 8), H67-6C.Ba, a MATa STE3DAF ASG7 strain (lanes 9 to 12), and H67-6C.a7, a MATa STE3DAF asg7::URA3 strain (lanes 13 to 16). RNA was isolated, transferred to nitrocellulose, hybridized with a FUS1 probe, and rehybridized with a PGK1 probe. (C) MATa STE3DAF gpa1::TRP1 ASG7 (H125-7D) and MATa STE3DAF gpa1::TRP1 asg7::HIS3 (H125-7D.aH) strains containing a plasmid with GPA1 under the control of the GAL promoter were streaked onto galactose- or glucose-containing plates and grown at 30°C for 2 to 3 days.
The STE3DAF allele causes inhibition of pheromone-inducible transcription at late times after pheromone treatment (3). To determine the effect of ASG7 on transcriptional induction, a time course of FUS1 RNA expression was performed in cells containing wild-type or null alleles of ASG7. Deletion of ASG7 had no effect on the level of FUS1 RNA induced in wild-type cells (Fig. 4B, lanes 1 to 8). As described previously, STE3DAF cells displayed a decrease in FUS1 RNA induction that was most pronounced at the 2- and 3-h time points (Fig. 4B, lanes 9 to 12). In STE3DAF cells containing an asg7Δ mutation, the levels of FUS1 RNA were the same as those seen in wild-type cells (Fig. 4B, lanes 1 to 4 and 13 to 16). Therefore, the asg7Δ mutation completely eliminated the inhibitory effect of expressing STE3 in MATa cells.
One of the characteristics of receptor inhibition is that it blocks the constitutive signaling conferred by deletion of GPA1, the G protein α-subunit gene, in an a-specific manner (4, 13, 17). In a wild-type background, a gpa1Δ mutation causes permanent cell cycle arrest due to constitutive activation of the pheromone response pathway by the βγ complex. In MATa cells, the STE3DAF allele suppresses the cell cycle arrest phenotype of a gpa1Δ strain. If ASG7 is required for receptor inhibition, then deletion of the ASG7 gene would be expected to result in cell cycle arrest in STE3DAF MATa cells that contain a gpa1Δ mutation. The effect of deleting ASG7 in MATa STE3DAF gpa1Δ cells was tested using a strain that contains GPA1 under the control of the GAL promoter. When this strain was grown in galactose, expression of GPA1 complemented the gpa1Δ mutation and both the ASG7 and asg7Δ strains formed colonies (Fig. 4C). In glucose, the cell cycle arrest phenotype was suppressed by the STE3DAF allele in the ASG7 strain. In the STE3DAF strain that contains an asg7Δ mutation, cell cycle arrest occurred in the absence of GPA1 expression. These findings demonstrate that ASG7 is required for the inhibitory effect of the STE3DAF allele that occurs in cells lacking the G protein α subunit.
All of the results presented above show that ASG7 is required for the inhibition of signaling conferred by expression of STE3 in MATa cells. Although these results do not prove that ASG7 is the only a-specific gene that mediates receptor inhibition, there is at present no evidence to indicate that other a-specific genes are required for this process.
Expression of ASG7 is a specific and pheromone inducible.
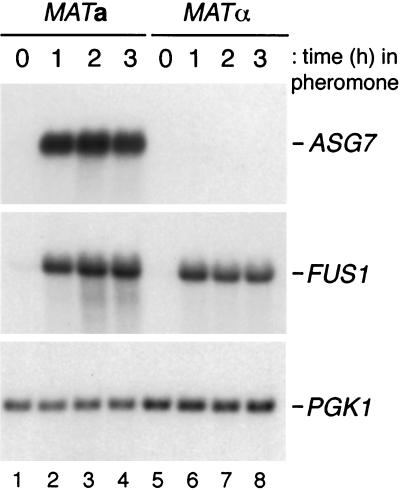
Previous results have shown that expression of ASG7 RNA is a specific (38). To investigate further the regulation of ASG7, a time course of RNA expression was performed in both MATa cells treated with α-factor and MATα cells treated with a-factor. In these experiments, ASG7 RNA was not detectable in untreated MATa or MATα cells (Fig. 5, lanes 1 and 5). Treatment of MATa cells with α-factor for 1 h caused a large increase in the abundance of ASG7 RNA, and this increase was maintained for 3 h (Fig. 5, lanes 2 to 4). Treatment of MATα cells with a-factor had no effect on the expression of ASG7 RNA, although FUS1 RNA was induced normally (Fig. 5, lanes 6 to 8). These results demonstrate that ASG7 RNA expression is completely a specific and that it is pheromone inducible to a large degree.
FIG. 5.
Abundance of ASG7 RNA in MATa and MATα cells treated with pheromone. Strain W3031A.Ba was treated with α-factor (0.1 μM) for the indicated periods of time (lanes 1 to 4); strain W3031B was treated with a-factor (60 nM) for the indicated periods of time (lanes 5 to 8). RNA was isolated, transferred to nitrocellulose, hybridized with an ASG7 or FUS1 probe, and rehybridized with a PGK1 probe.
The a-factor receptor inhibits signaling in its liganded state.
In the experiments described above, the a-factor receptor is not occupied by a ligand because the strains used have deletions of the genes encoding a-factor. Therefore, it was not known whether the a-factor receptor could function in receptor inhibition in its liganded state. To test this idea, the effect of the ASG7 gene on signaling that originates from the liganded a-factor receptor was investigated. Thus, this experiment tests whether the a-factor receptor can carry out its two independent functions, signal transduction and receptor inhibition, in the same cell.
To determine the effect of ASG7 on signaling through the a-factor receptor, a time course of FUS1 RNA expression was performed in MATa STE3DAF cells containing wild-type or null alleles of ASG7. The cells were treated with a-factor at a concentration of 300 nM, which is approximately 10,000-fold higher than the lowest concentration required to induce a transcriptional response in MATα cells (24). Because STE3 is expressed at comparable levels in MATa STE3DAF cells and wild-type MATα cells (13), it is expected that the majority of cell surface receptors on STE3DAF cells would be occupied by a-factor at this concentration. In the strain containing wild-type ASG7, there was only a slight induction of FUS1 RNA in response to treatment with a-factor (Fig. 6, lanes 1 to 4). In the strain containing an asg7Δ mutation, there was a large increase in the induction of FUS1 RNA in response to treatment with a-factor (Fig. 6, lanes 5 to 8). The simplest interpretation of these results is that the a-factor receptor can block the signal that it initiates. In the strain that contains wild-type ASG7, the low level of FUS1 RNA induction was due to the presence of both the a-factor receptor and Asg7p in the same cell. These results imply that the a-factor receptor can function in the process of receptor inhibition while it is bound to the ligand.
FIG. 6.
Effect of ASG7 on signaling in STE3DAF cells treated with a-factor. The following strains were treated with a-factor (300 nM) for the indicated periods of time: H67-6C.Ba, a MATa STE3DAF ASG7 strain (lanes 1 to 4), and H67-6C.a7, a MATa STE3DAF asg7::URA3 strain (lanes 5 to 8). RNA was isolated, transferred to nitrocellulose, hybridized with a FUS1 probe, and rehybridized with a PGK1 probe.
ASG7 affects Ste4p localization.
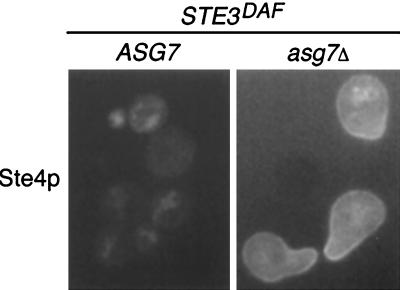
The results described above show that receptor inhibition results in altered localization of the Ste4p β subunit. In addition, they show that ASG7 is required for inhibition of signaling by the a-factor receptor. It was therefore of interest to determine whether deletion of the ASG7 gene would have an effect on the subcellular location of Ste4p. To investigate this possibility, the localization pattern of a GFP-Ste4p fusion protein in MATa STE3DAF cells that contained the wild-type ASG7 gene or an asg7Δ mutation was observed. The cells were treated with α-factor for 2 h to maximize the difference between signaling and nonsignaling cells (Fig. 2). GFP-Ste4p was localized in an internal particulate pattern in STE3DAF cells that contained the wild-type ASG7 gene (Fig. 7). In contrast, GFP-Ste4p was localized predominantly at the cell membrane in STE3DAF cells that contained the asg7Δ mutation. These results demonstrate that, in addition to its effect on signaling, ASG7 has an effect on Ste4p localization in cells that express the a-factor receptor. Moreover, localization of Ste4p to the cell membrane correlated with activation of the signaling pathway in all experiments, as would be expected for a protein that transmits a signal to membrane-associated target proteins.
FIG. 7.
Effect of ASG7 on localization of GFP-Ste4p in STE3DAF cells. The following strains were treated with α-factor (0.1 μM) for 2 h: AC17-2B, a MATa STE3DAF ste4::HIS3 ASG7 strain (ASG7), and AC17-2B.aL, a MATa STE3DAF ste4::HIS3 asg7::LEU2 strain (asg7Δ). Both strains contained the low-copy-number STE4-GFP plasmid pBTL49. The live cells were viewed by fluorescence microscopy using an FITC filter set.
Asg7p localization is consistent with a direct effect of Asg7p on Ste4p.
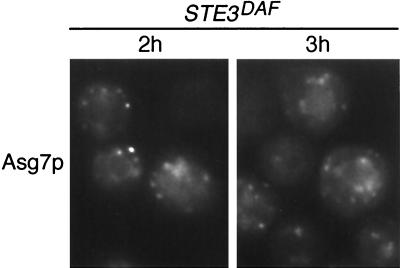
The effect of Asg7p on Ste4p localization could occur through an indirect mechanism, such as transcriptional activation of other genes, or through a more direct mechanism, such as binding of Ste4p to a complex containing Asg7p. To determine whether the location of Asg7p in the cell is consistent with a direct effect on Ste4p, a fully functional ASG7-GFP fusion construct was expressed in STE3DAF cells containing an asg7Δ mutation. In cells treated with α-factor for 2 or 3 h, Asg7p-GFP was localized in an internal particulate pattern (Fig. 8). Asg7p-GFP was observed in this pattern as soon as the signal became visible, at about 1 h after α-factor treatment, and the pattern was the same in both STE3DAF and wild-type cells (E. Bortz and J. P. Hirsch, unpublished data). The subcellular location of Asg7p is similar to that of Ste4p during receptor inhibition. Therefore, these results are consistent with Asg7p having a direct effect on Ste4p localization.
FIG. 8.
Localization of Asg7p in STE3DAF cells. Strain H67-6C.a7 (MATa STE3DAF asg7::URA3) containing an ASG7-GFP plasmid (pASG7-111.GFP) was treated with α-factor (0.1 μM) for the indicated periods of time. The live cells were viewed by fluorescence microscopy using an FITC filter set.
DISCUSSION
Localization and signaling function of Ste4p.
Expression of the a-factor receptor in MATa cells inhibits signaling through the pheromone response pathway at the level of Ste4p, the G protein β subunit (3, 17). To investigate the way in which signaling is inhibited, the normal localization pattern of Ste4p during pheromone signaling was examined. Biochemical fractionation of cell membranes had indicated that the fraction of Ste4p that is associated with the plasma membrane does not change after pheromone stimulation (14). We used microscopic examination of live cells to show that treatment of cells with pheromone causes a redistribution of Ste4p to regions of mating projection formation. These studies were therefore able to detect a change in the location of Ste4p that had not been seen previously. Other signaling components of the pheromone response pathway, such as the pheromone receptors, the kinase Ste20p, and the scaffolding protein Ste5p, have also been shown to localize to the membranes of mating projections (16, 20, 23, 26, 29). These results are consistent with a model in which Ste5p is recruited to the plasma membrane by the released Ste4p β subunit upon activation of the G protein (29). Recruitment of Ste5p to the membrane would bring Ste11p, which is bound to Ste5p, into close proximity with the membrane-associated kinase Ste20p. Phosphorylation of the MAP kinase kinase kinase Ste11p by Ste20p would then activate the MAP kinase cascade. This model is also consistent with our observation that localization of Ste4p away from the cell membrane to an internal compartment correlates with inhibition of signaling. In this case, Ste4p would not be able to recruit Ste5p to the plasma membrane and phosphorylation of Ste11p would not occur. Therefore, sequestration of the G protein β subunit away from the plasma membrane provides an efficient posttranslational mechanism for inhibition of signal transduction.
In cells undergoing receptor inhibition, Ste4p was localized to a region of the cell that resembles an endocytic compartment. However, it should be noted that the yeast βγ complex does not undergo endocytosis during pheromone signaling in wild-type cells (14) and that the steady-state level of Ste4p does not change under conditions of receptor inhibition. Therefore, although Ste4p appears to be associated with vesicles or other subcellular structures, this association probably does not result in its degradation in the lysosome.
Gβ subunit binding partners and regulation of localization.
ASG7 was identified as an a-specific gene that is required for the inhibitory effect of the a-factor receptor on pheromone signaling. In addition, it was shown that the subcellular location of Asg7p and Ste4p is consistent with Asg7p having a direct effect on localization of the Ste4p β subunit. Several proteins that bind directly to βγ subunits and inhibit their signaling activity have been identified. One such protein is phosducin, which undergoes regulated phosphorylation and which binds to βγ in its unphosphorylated form. The crystal structure of the phosducin-βγ complex reveals that βγ would not be able to bind to a G protein α subunit when it is bound to phosducin (9). The binding of phosducin to βγ is also predicted to disrupt the orientation of βγ relative to the membrane (9). This idea is in agreement with experimental evidence demonstrating that the binding of phosducin to βγ causes it to shift from a membrane subcellular fraction to a soluble fraction (21, 37). The ability of phosducin to cause the translocation of βγ to a different subcellular compartment and to inhibit its signaling activity appears analogous to the action of Asg7p on the yeast βγ complex during receptor inhibition. However, unlike the phosducin-βγ complex, the yeast βγ complex remains in the pellet fraction after it has translocated away from the plasma membrane (J. Kim and J. P. Hirsch, unpublished data). Therefore, the process of receptor inhibition does not cause complete solubilization of the βγ complex.
Another example of a protein that binds to βγ subunits is the mammalian β-adrenergic receptor kinase (βARK). In this case, the βγ complex targets βARK to the membrane and facilitates phosphorylation of the receptor by the kinase (28). The direct binding of βγ subunits to a protein involved in down-regulating the response suggests a parallel with the process of receptor inhibition in yeast. However, the complex of βγ and βARK localizes to a membrane where βγ is thought to be active, unlike the altered subcellular location of βγ under conditions of receptor inhibition.
βγ subunits also bind to the mammalian protein KSR-1, a kinase that was originally identified as a regulator of the Ras signaling pathway (1). The binding of KSR-1 to βγ inhibits the ability of βγ to activate the MAP kinase ERK1. In contrast to the change in subcellular location that is associated with inhibition of the yeast βγ, the inactive complex of KSR-1 and βγ remains associated with the plasma membrane. Thus, although receptor inhibition has features in common with other systems in which βγ forms complexes with known proteins, it is not strictly analogous to any previously described process.
Models for Asg7p function.
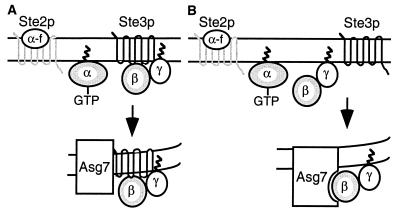
The a-specific gene ASG7 was shown to be required for receptor inhibition by demonstrating that deletion of ASG7 eliminated the inhibitory effects of STE3 expression on signal transduction. Moreover, MATa cells expressing STE3 but lacking ASG7 displayed normal localization of Ste4p at the cell membrane. These results are consistent with several different models for the way in which Asg7p functions with the a-factor receptor to inhibit signaling by Ste4p. One potential model for receptor inhibition involves a direct interaction between Asg7p and the a-factor receptor that takes place within an intracellular membrane (Fig. 9A). The Asg7p protein is predicted to contain two hydrophobic regions that could function as transmembrane domains. In this model, the alpha-helical bundle of the receptor transmembrane domains would interact with the transmembrane domains of Asg7p. This interaction is not expected to take place in the plasma membrane because Asg7p is not observed on the cell surface. However, the pheromone receptors are associated with several different intracellular membranes during their life cycle. This model assumes that the cytoplasmic domains of the a-factor receptor directly associate with the βγ subunits. The interaction of the receptor with Asg7p would produce a high-affinity binding site for βγ, which would result in the association of βγ with a specific internal compartment. This model is consistent with several studies of mammalian systems, which have indicated that βγ subunits may bind directly to their associated receptors in the absence of an α subunit (12, 27, 34). Moreover, Roth and colleagues have also identified the role of ASG7 in receptor inhibition using a genomics approach and have shown that Asg7p inhibits delivery of the Ste3p receptor to the cell surface by a process that is independent of βγ (30). This finding provides support for the idea that Asg7p and the a-factor receptor directly interact with each other.
FIG. 9.
Models for ASG7 function. See text for details.
The other potential model for receptor inhibition involves a direct interaction between Asg7p and the Ste4p β subunit (Fig. 9B). In this model, the presence of the a-factor receptor would promote loading of Asg7p into a complex that contains Ste4p. Targeting signals on Asg7p would then localize the complex to an internal compartment. This model is consistent with the observation that Ste4p and Asg7p display similar subcellular localization patterns in cells undergoing receptor inhibition. Activation of a downstream component by an unliganded G protein-coupled receptor would represent a novel function for this class of receptors.
Altered versions of Ste4p that are resistant to receptor inhibition and thus are capable of signaling in MATa cells in which the a-factor receptor is expressed have been identified (17). The effect of these changes in Ste4p can be interpreted in different ways when considering the two models presented above. If the first model is correct (Fig. 9A), then these changes in Ste4p are expected to affect its ability to bind to the cytoplasmic domains of the a-factor receptor. If the second model is correct (Fig. 9B), then these changes in Ste4p are expected to affect its ability to bind to a protein interaction domain of Asg7p. It is also possible that additional components that have not yet been identified could transmit a signal between the receptor, Asg7p, and Ste4p.
Analysis of the kinetics of signaling in MATa cells expressing the a-factor receptor has shown that normal initiation of signaling is followed by a gradual inhibition of the response (3). The finding that ASG7 is a pheromone-inducible gene provides an explanation for this pattern of signaling. When cells are first exposed to pheromone, the ASG7 gene product is present at a low level that does not affect activation of the signal transduction pathway. As the response proceeds, the accumulation of Asg7p results in gradual inhibition of signaling activity. Induction of ASG7 RNA by pheromone in a wild-type MATa cell, where Asg7p does not affect signaling, could function to prepare the cell for signaling inhibition after the fusion of two mating partners, as described below.
Physiological role of receptor inhibition.
The results presented here document inhibition of signaling by the βγ complex when Asg7p and the a-factor receptor are present in the same cell. Although this situation was generated by a mutant allele in these experiments, it occurs naturally during several transient phases of the yeast life cycle. For example, homothallic strains of yeast undergo mating type switching during haploid growth. During the transition from one mating type to another, a single cell could produce both a particular pheromone receptor and the pheromone that binds to that receptor. This situation could activate the pheromone response pathway and induce the expression of ASG7, resulting in the presence of Asg7p and the a-factor receptor in the same cell. Activation of the process of receptor inhibition would then turn off the pheromone response pathway and allow the cell to resume cycling. Another example of such a situation occurs immediately after fusion of two haploid cells during the mating process. The long-term mechanism for eliminating pheromone signaling in diploids is the establishment of transcriptional inhibition of genes encoding components of the pheromone response pathway. However, it is also possible that a short-term mechanism acts to inhibit signaling in zygotes. The fusion of two mating partners could bring Asg7p from the MATa mating partner into contact with a-factor receptor from the MATα mating partner. In the newly fused zygote, both pheromone receptors are expected to be occupied. The finding that the occupied a-factor receptor can participate in receptor inhibition indicates that signaling could be blocked under these conditions. The function of receptor inhibition in this case would be to prevent multiple rounds of mating or to allow the zygote to recover rapidly from cell cycle arrest. In support of this idea, Roth and colleagues have demonstrated that there is a delay in the emergence of the first mitotic bud from zygotes in which the MATa mating partner has a deletion of ASG7 (30). Identification of ASG7 as an a-specific component required for receptor inhibition provides an opportunity to test multiple physiological conditions for their ability to be affected by this unique process.
ACKNOWLEDGMENTS
This project was supported by Research Project Grant RPG-96-119-03/4-MBC from the American Cancer Society (to J.P.H.) and grant GM49265 from the National Institutes of Health (to A.K.V.).
We thank J. Kurjan and I. Karpichev for providing plasmids used in this work, J. Hirschman and D. Jenness for providing anti-Ste4p antibody, R. Tsien for providing the GFP plasmid, and F. Naider for providing synthetic a-factor. We also thank N. Davis for communicating results prior to publication.
REFERENCES
- 1.Bell B, Xing H, Yan K, Gautam N, Muslin A J. KSR-1 binds to G-protein βγ subunits and inhibits βγ-induced mitogen-activated protein kinase activation. J Biol Chem. 1999;274:7982–7986. doi: 10.1074/jbc.274.12.7982. [DOI] [PubMed] [Google Scholar]
- 2.Cole G M, Reed S I. Pheromone-induced phosphorylation of a G protein β subunit in S. cerevisiae is associated with an adaptive response to mating pheromone. Cell. 1991;64:703–716. doi: 10.1016/0092-8674(91)90500-x. [DOI] [PubMed] [Google Scholar]
- 3.Couve A, Hirsch J P. Loss of sustained Fus3p kinase activity and the G1 arrest response in cells expressing an inappropriate pheromone receptor. Mol Cell Biol. 1996;16:4478–4485. doi: 10.1128/mcb.16.8.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cross F R. The DAF2–2 mutation, a dominant inhibitor of the STE4 step in the α-factor signalling pathway of Saccharomyces cerevisiae MATa cells. Genetics. 1990;126:301–308. doi: 10.1093/genetics/126.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cross F R. ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast. 1997;13:647–653. doi: 10.1002/(SICI)1097-0061(19970615)13:7<647::AID-YEA115>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 6.Cross F R, Tinkelenberg A H. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- 7.Dietzel C, Kurjan J. The yeast SCG1 gene: a Gα-like protein implicated in the a- and α-factor response pathway. Cell. 1987;50:1001–1010. doi: 10.1016/0092-8674(87)90166-8. [DOI] [PubMed] [Google Scholar]
- 8.Engebrecht J, Hirsch J, Roeder G S. Meiotic gene conversion and crossing over: their relationship to each other and to chromosome synapsis and segregation. Cell. 1990;62:927–937. doi: 10.1016/0092-8674(90)90267-i. [DOI] [PubMed] [Google Scholar]
- 9.Gaudet R, Bohm A, Sigler P B. Crystal structure at 2.4 Å resolution of the complex of transducin βγ and its regulator, phosducin. Cell. 1996;87:577–588. doi: 10.1016/s0092-8674(00)81376-8. [DOI] [PubMed] [Google Scholar]
- 10.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 11.Heim R, Tsien R Y. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 12.Heithier H, Fröhlich M, Dees C, Baumann M, Häring M, Gierschik P, Schiltz E, Vaz W L C, Hekman M, Helmreich E J M. Subunit interactions of GTP-binding proteins. Eur J Biochem. 1992;204:1169–1181. doi: 10.1111/j.1432-1033.1992.tb16744.x. [DOI] [PubMed] [Google Scholar]
- 13.Hirsch J P, Cross F R. The pheromone receptors inhibit the pheromone response pathway in Saccharomyces cerevisiae by a process that is independent of their associated Gα protein. Genetics. 1993;135:943–953. doi: 10.1093/genetics/135.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirschman J E, De Zutter G S, Simonds W F, Jenness D D. The Gβγ complex of the yeast pheromone response pathway: subcellular fractionation and protein-protein interactions. J Biol Chem. 1997;272:240–248. doi: 10.1074/jbc.272.1.240. [DOI] [PubMed] [Google Scholar]
- 15.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson C L, Konopka J B, Hartwell L H. S. cerevisiae α pheromone receptors activate a novel signal transduction pathway for mating partner discrimination. Cell. 1991;67:389–402. doi: 10.1016/0092-8674(91)90190-a. [DOI] [PubMed] [Google Scholar]
- 17.Kim J, Couve A, Hirsch J P. Receptor inhibition of pheromone signaling is mediated by the Ste4p Gβ subunit. Mol Cell Biol. 1999;19:441–449. doi: 10.1128/mcb.19.1.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leberer E, Dignard D, Hougan L, Thomas D Y, Whiteway M. Dominant-negative mutants of a yeast G-protein β subunit identify two functional regions involved in pheromone signalling. EMBO J. 1992;11:4805–4813. doi: 10.1002/j.1460-2075.1992.tb05586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leberer E, Thomas D Y, Whiteway M. Pheromone signalling and polarized morphogenesis in yeast. Curr Opin Genet Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- 20.Leberer E, Wu C, Leeuw T, Fourest-Lieuvin A, Segall J E, Thomas D Y. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 1997;16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee R H, Whelan J P, Lolley R N, McGinnis J F. The photoreceptor-specific 33 kDa phosphoprotein of mammalian retina: generation of monospecific antibodies and localization by immunocytochemistry. Exp Eye Res. 1988;46:829–840. doi: 10.1016/s0014-4835(88)80035-6. [DOI] [PubMed] [Google Scholar]
- 22.Li E, Cismowski M J, Stone D E. Phosphorylation of the pheromone-responsive Gβ protein of Saccharomyces cerevisiae does not affect its mating-specific signaling function. Mol Gen Genet. 1998;258:608–618. doi: 10.1007/s004380050774. [DOI] [PubMed] [Google Scholar]
- 23.Mahanty S K, Wang Y, Farley F W, Elion E A. Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell. 1999;98:501–512. doi: 10.1016/s0092-8674(00)81978-9. [DOI] [PubMed] [Google Scholar]
- 24.Marcus S, Caldwell G A, Miller D, Xue C-B, Naider R, Becker J M. Significance of C-terminal cysteine modifications to the biological activity of the Saccharomyces cerevisiaea-factor mating pheromone. Mol Cell Biol. 1991;11:3603–3612. doi: 10.1128/mcb.11.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCaffrey G, Clay F J, Kelsay K, Sprague G F., Jr Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2680–2690. doi: 10.1128/mcb.7.8.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peter M, Neiman A M, Park H-O, van Lohuizen M, Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- 27.Phillips W J, Cerione R A. Rhodopsin/transducin interactions. I. Characterization of the binding of the transducin-βγ subunit complex to rhodopsin using fluorescence spectroscopy. J Biol Chem. 1992;267:17032–17039. [PubMed] [Google Scholar]
- 28.Pitcher J A, Inglese J, Higgins J B, Arriza J L, Casey P J, Kim C, Benovic J L, Kwatra M M, Caron M G, Lefkowitz R J. Role of βγ subunits of G proteins in targeting the β-adrenergic receptor kinase to membrane-bound receptors. Science. 1992;257:1264–1267. doi: 10.1126/science.1325672. [DOI] [PubMed] [Google Scholar]
- 29.Pryciak P M, Huntress F A. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roth A, Nelson B, Boone C, Davis N G. Asg7p-Ste3p repression of pheromone signaling: regulation of the zygotic transition to vegetative growth. Mol Cell Biol. 2000;20:8815–8825. doi: 10.1128/mcb.20.23.8815-8825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Sherman F, Fink G R, Hicks J B. Laboratory course manual for methods in yeast genetics. Plainview, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Sprague G F, Jr, Thorner J W. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces: gene expression. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 657–744. [Google Scholar]
- 34.Taylor J M, Jacob-Mosier G G, Lawton R G, VanDort M, Neubig R R. Receptor and membrane interaction sites on Gβ. J Biol Chem. 1996;271:3336–3339. doi: 10.1074/jbc.271.7.3336. [DOI] [PubMed] [Google Scholar]
- 35.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 36.Wittenberg C, Reed S I. Plugging it in: signaling circuits and the yeast cell cycle. Curr Opin Cell Biol. 1996;8:223–230. doi: 10.1016/s0955-0674(96)80069-x. [DOI] [PubMed] [Google Scholar]
- 37.Yoshida T, Willardson B M, Wilkins J F, Jensen G J, Thornton B D, Bitensky M W. The phosphorylation state of phosducin determines its ability to block transducin subunit interactions and inhibit transducin binding to activated rhodopsin. J Biol Chem. 1994;269:24050–24057. [PubMed] [Google Scholar]
- 38.Zhong H, McCord R, Vershon A K. Identification of target sites of the α2-Mcm1 repressor complex in the yeast genome. Genome Res. 1999;9:1040–1047. doi: 10.1101/gr.9.11.1040. [DOI] [PubMed] [Google Scholar]