Abstract
This article highlights recent pioneering work by Günther et al. towards the discovery of potential repurposed antiviral compounds (peptidomimetic and non‐peptidic) against the SARS‐CoV‐2 main protease (Mpro). The antiviral activity of the most potent drugs is discussed along with their binding mode to Mpro as observed through X‐ray crystallographic screening.
Keywords: allosteric inhibitors, antiviral drugs, drug screening, SARS-CoV-2, X-ray crystallographic screening
The highly contagious novel coronavirus disease or COVID‐19 has turned out to be a major pandemic of the 21st century. With nearly 200 million cases and 4 million deaths worldwide to date, it has created havoc in society and immense human suffering. In a race against time to stop the spread of the disease, concurrent efforts of the scientific community worldwide have brought forth several potential small‐molecule inhibitors of SARS‐CoV‐2 infection within a very limited span of time. [1]
Reports have shown that the viral entry and replication within the host cells involve multiple molecular factors from both the host and the virus.[ 2 , 3 ] One of those important molecular factors is the main protease (Mpro), also referred to as the 3C‐like protease (3CLpro). Mpro is essential for the cleavage of the viral polyprotein pp1ab at 11 discrete sites, Leu‐Gln‐↓‐Ser/Gly/Ala being the cleavage motif. After cleavage, the released non‐structural proteins form a replicase complex, which in turn is responsible for the viral replication. [2] Inhibition of this proteolytic cleavage can prevent SARS‐CoV‐2 replication inside host cells. Hence, Mpro is a prime target for antiviral drug discovery against SARS‐CoV‐2.
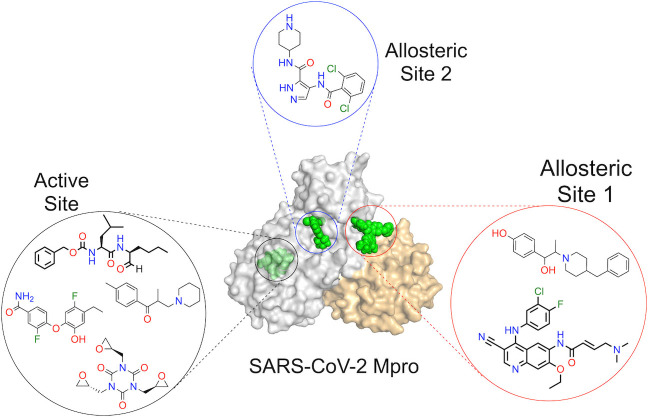
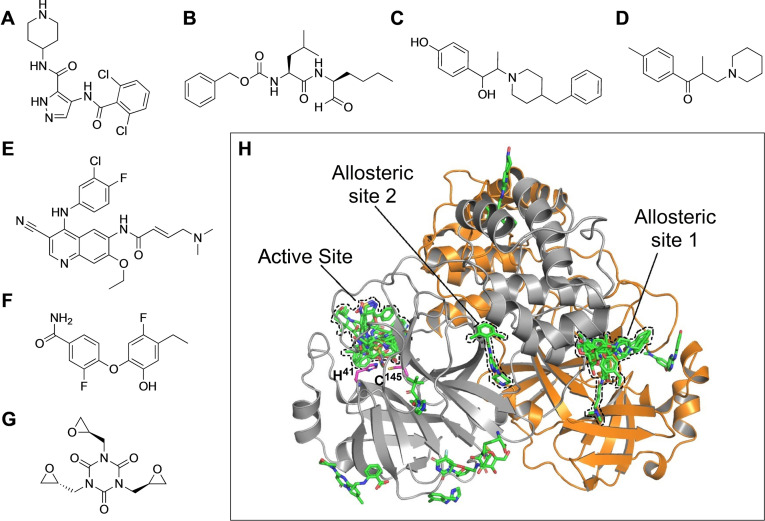
Within a year of the first reported case of COVID‐19, several candidate drug molecules targeting Mpro appeared in the literature, which include both ab initio designed[ 2 , 4 ] as well as repurposed drugs.[ 3 , 5 ] Particularly notable is the report by Douangamath et al. that demonstrated electrophilic as well as noncovalent fragment screening against Mpro using a combination of mass spectrometry and crystallographic techniques with hits found both in the active site as well as in the dimerization interface. [6] Recently, using a very similar yet elegant drug‐repurposing crystallographic screen, Günther et al. [7] identified several highly potent small‐molecule inhibitors of SARS‐CoV‐2 Mpro. The all‐inclusive nature of the work by Günther et al. is vindicated by the discovery of both active‐site as well as allosteric inhibitors of Mpro from the library of molecules which are either already approved drugs or are in the clinical trial. They had shown 37 compounds that bind to SARS‐CoV‐2 Mpro, among which seven exhibited exceptional antiviral properties at nontoxic concentrations in cell‐based viral reduction assays. These seven compounds consist of six nonpeptidic and one peptidomimetic small‐molecule binders (Figure 1 A–G).
Figure 1.
The compounds which showed antiviral effects in cell‐based assays. A) AT7519. B) Calpeptin (peptidomimetic drug). C) Ifenprodil. D) Tolperisone. E) Pelitinib. F) MUT056399. G) Triglycidyl isocyanurate. H) A schematic of the Mpro dimer structure (the two protomers are shown in grey and orange). The active site and allosteric binding sites 1 and 2 along with 29 unambiguous binders bound to them are shown in green. Catalytic residues His41 and Cys145 in one of the protomers are also highlighted. [7]
The Mpro enzyme consists of a C‐terminal helical domain and a catalytic duo consisting of Cys145 and His41 in its active site. [2] Most of the binders use the active site for binding to Mpro, but, very interestingly, some of the binders employ novel allosteric binding sites, providing interesting insights into drug development against SARS‐CoV‐2 (Figure 1 H). Günther et al. selected 5953 compounds from two repurposing libraries, The Fraunhofer IME Repurposing Collection and Safe‐in‐man library from Dompé Farmaceutici S.p.A, [8] for co‐crystallization and then crystallographically screened them against Mpro. After X‐ray diffraction data collection followed by structure refinement, cluster analysis, [9] and pan dataset density analysis, [10] 29 binders were unambiguously distinguished. Sixteen of them bound to the active site of the enzyme, whereas the remaining 13 bound to various other sites. The binding mode also varied for the 16 active‐site binders. Six of them bound covalently via thioether linkage to Cys145, one bound covalently as a thiohemiacetal to Cys145, one was zinc‐coordinated, and the remaining eight bound noncovalently. Hence, even though the binding mode was different for the different compounds at the same active site, most of them produced almost similar antiviral effects. This underscores time and again the beauty of the underlying chemistry involved.
The advantage of utilizing compounds from drug‐repurposing libraries is that these molecules have already proven cell permeability and bioactivity. [11] Günther et al. harnessed this advantage very effectively to target Mpro. From the hits obtained from the X‐ray screen, they examined several compounds for antiviral activities in SARS‐CoV‐2 cell‐based assays. Surprisingly, nine compounds were able to reduce viral RNA replication by a factor of 100. Two of them (AT7519 and Ifenprodil) showed reduction in viral RNA replication to a slightly lesser extent than that but had different binding sites outside the active site, making them worth further investigation. These 11 compounds were then further examined to evaluate the effective concentration at which 50 % of the SARS‐CoV‐2 infectious particles were reduced (EC50). This evaluation finally resulted seven compounds (AT7519, Calpeptin, Ifenprodil, Tolperisone, Pelitinib, MUT056 399, Triglycidyl isocyanurate; Figure 1 A–G) that were antivirally active with nearly 100‐fold reduction of infectious particles and a selectivity index (CC50/EC50) greater than 5, representing no cytotoxicity in tested concentration range. Here CC50 stands for 50 % cytotoxic concentration.
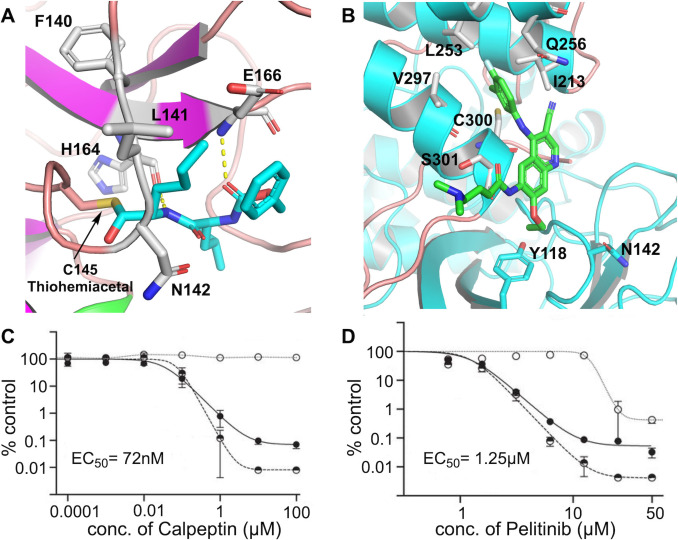
Among these seven antiviral candidates, the two most potent ones are Calpeptin (EC50=72 nM, CC50>100 μM) and Pelitinib (EC50=1.25 μM, CC50=13.96 μM), which are suitable for preclinical testing and deserve more discussion in this context. Calpeptin is a peptidomimetic inhibitor that binds to the active‐site residue Cys145, forming a thiohemiacetal through its aldehyde warhead. The backbone is involved in hydrogen bonding with His164 and Glu166, whereas the norleucine side chain is engaged in van der Waals interaction with Phe140, Leu141, and Asn142 (Figure 2 A). The inhibitory effect of this molecule on SARS‐CoV‐2 replication in Vero E6 cells is shown (Figure 2 C). Calpeptin also inhibits Cathepsin L [12] and thus this dual targeting of Mpro and Cathepsin L can provide a new avenue to drug discovery. Pelitinib, an anticancer drug [13] with the second highest anti SARS‐CoV‐2 activity, on the other hand, is unique because it does not bind to the active site of Mpro. It rather makes use of the hydrophobic pocket in the C‐terminal dimerization domain which forms the first allosteric binding site. Pelitinib inserts its halogenated aromatic moiety into the hydrophobic groove formed by Ile213, Leu253, Gln256, Val297, and Cys300. Although Pelitinib is a Michael acceptor, no covalent linkage with the active‐site Cys145 was observed in the electron density maps, as it binds far away from the active site. The function of Mpro depends on the structure of the active site and the correct orientation of the subdomains. The ethyl ether substituent of Pelitinib pushes the Tyr118 and Asn142 residues of the opposing protomer within the dimer and most likely hampers the dimerization process as well as the correct structural orientation of the pocket necessary for Mpro function (Figure 2 B). Hence, by binding to the first allosteric site it is capable of stopping viral replication. The inhibitory effect of Pelitinib on SARS‐CoV‐2 replication in Vero E6 cells is shown in Figure 2 D.
Figure 2.
Compounds bound to Mpro and their effect on SARS‐CoV‐2 replication in Vero E6 cells. A) Calpeptin (shown in cyan) bound to the Mpro active site (PDB 7AKU). The hydrogen bonds are shown as dashed lines and the thiohemiacetal covalent bond with Cys145 is also highlighted. B) The binding mode of Pelitinib (shown in green) to the first allosteric binding site of Mpro (PDB 7AXM). The viral RNA (v‐RNA) yield (filled circles), viral titers (half‐filled circles), and cell viability (empty circles) for Calpeptin (C) and Pelitinib (D); for each, the EC50 for viral titer reduction is given. [7] Portions of this figure are reprinted with permission from ref. [7], Copyright 2021, American Association for the Advancement of Science.
The pioneering work by Günther et al. provides us valuable insights on functional antiviral drugs against the deadly SARS‐CoV‐2 obtained from high‐throughput crystallographic screening, several of which are suitable for preclinical studies. The two allosteric sites identified during the screening could be exploited further as additional targets to accelerate drug discovery efforts. The potential of peptidomimetic drugs has also been excellently demonstrated along with non‐peptidic drugs, highlighting the fact that screening against such repurposing drug libraries has great potential and can be used to target other regions of the coronavirus like the spike receptor binding domain and/or N‐terminal domain. The covalent binding strategy would be an added advantage for the same. Also, the beautiful example of allosteric inhibition as well as dual inhibition provides a new vista to the science of drug discovery. The study by Günther et al. is a promising start to the production of even more potent drug candidates against this deadly coronavirus and may have opened the door to a deeper perception in drug discovery against any virus that might have the power to create future pandemics.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
The authors acknowledge the intramural funds at TIFR Hyderabad from the Department of Atomic Energy (DAE) to K.M.
A. Sarkar, K. Mandal, Angew. Chem. Int. Ed. 2021, 60, 23492.
References
- 1. Scudellari M., Nature 2020, 581, 252–255. [DOI] [PubMed] [Google Scholar]
- 2. Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R., Science 2020, 368, 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Guddat L. W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H., Nature 2020, 582, 289–293. [DOI] [PubMed] [Google Scholar]
- 4.
- 4a. Dai W., Zhang B., Jiang X., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L., Xu Y., Yang H., Liu H., Science 2020, 368, 1331–1335; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4b. Rathnayake A. D., Zheng J., Kim Y., Perera K. D., Mackin S., Meyerholz D. K., Kashipathy M. M., Battaile K. P., Lovell S., Perlman S., Groutas W. C., Chang K., Sci. Transl. Med. 2020, 12, eabc5332; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4c. Kreutzer A. G., Krumberger M., Diessner E. M., Parrocha C. M. T., Morris M. A., Guaglianone G., Butts C. T., Nowick J. S., Eur. J. Med. Chem. 2021, 221, 113530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.
- 5a. Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F. G., Horby P. W., Zhang D., Wang C., N. Engl. J. Med. 2020, 382, 1787–1799; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5b. Ma C., Sacco M. D., Hurst B., Townsend J. A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M. T., Chen Y., Wang J., Cell Res. 2020, 30, 678–692; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5c. Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., De Jesus P. D., Teriete P., Hull M. V., Chang M. W., Chan J. F., Cao J., Poon V. K., Herbert K. M., Cheng K., Nguyen T. H., Rubanov A., Pu Y., Nguyen C., Chol A., Rathnasinghe R., Schotsaert M., Miorin L., Dejosez M., Zwaka T. P., Sit K., Martinez-Sobrido L., Liu W., White K. M., Chapman M. E., Lendy E. K., Glynne R. J., Albrecht R., Ruppin E., Mesecar A. D., Johnson J. R., Benner C., Sun R., Schultz P. G., Su A. J., Garcia-Sastre A., Chatterjee A. K., Yuen K., Chanda S. K., Nature 2020, 586, 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Douangamath A., Fearon D., Gehrtz P., Krojer T., Lukacik P., Owen C. D., Resnick E., Strain-Damerell C., Aimon A., Ábrányi-Balogh P., Brandão-Neto J., Carbery A., Davison G., Dias A., Downes T. D., Dunnett L., Fairhead M., Firth J. D., Jones S. P., Keeley A., Keserii G. M., Klein H. F., Martin M. P., Noble M. E. M., O'Brien P., Powell A., Reddi R. N., Skyner R., Snee M., Waring M. J., Wild C., London N., von Delft F., Walsh M. A., Nat. Commun. 2020, 11, 5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Günther S., Reinke P. Y. A., Fernández-Garcia Y., Lieske J., Lane T. J., Ginn H. M., Koua F. H. M., Ehrt C., Ewert W., Oberthuer D., Yefanov O., Meier S., Lorenzen K., Krichel B., Kopicki J., Gelisio L., Brehm W., Dunkel I., Seychell B., Gieseler H., Norton-Baker B., Escudero-Pérez B., Domaracky M., Saouane S., Tolstikova A., White T. A., Hänle A., Groessler M., Fleckenstein H., Trost F., Galchenkova M., Gevorkov Y., Li C., Awel S., Peck A., Barthelmess M., Schlünzen F., Xavier P. L., Werner N., Andaleeb H., Ullah N., Falke S., Srinivasan V., Franca B., Schwinzer M., Brognaro H., Rogers C., Melo D., Zaitseva-Doyle J. J., Knoska J., Peña-Murillo G. E., Mashhour A. R., Hennicke V., Fischer P., Hakanpää J., Meyer J., Gribbon P., Ellinger B., Kurzikov M., Wolf M., Beccari A. R., Bourenkov G., Stetten D. V., Pompidor G., Bento I., Panneerselvam S., Karpics I., Schneider T. R., Garcia-Alai M. M., Niebling S., Günther C., Schmidt C., Schubert R., Han H., Boger J., Monteiro D. C. F., Zhang L., Sun X., Pletzer-zelgert J., Wollenhaupt J., Feiler C. G., Weiss M. S., Schulz E., Mehrabi P., Karničar K., Usenik A., Loboda J., Tidow H., Chari A., Hilgenfeld R., Uetrecht C., Cox R., Zaliani A., Beck T., Rarey M., Günther S., Turk D., Hinrichs W., Chapman H. N., Pearson A. R., Betzel C., Meents A., Science 2021, 372, 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuzikov M., Constanzi E., reinshagen J., Esposito F., Vangeel L., Wolf M., Ellinger B., Claussen C., Geisslinger G., Corona A., Taconis D., Talarico C., Manelfi C., Cannalire R., Rosselti G., Gossen J., Albani S., Musiani F., Herzog K., Ye Y., Giabbai B., Demitri N., Jochmans D., DeJonghe S., Rymenants J., Summa V., Tramontano E., Beccari A. R., Leyssen P., Historians P., Neyts J., Gribbon P., Zaliani A., ACS Pharmacol. Transl. Sci. 2021, 4, 1096–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ginn H. M., Acta Crystallogr. Sect. D 2020, 76, 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pearce N. M., Krojer T., Bradley A. R., Collins P., Nowak R. P., Talon R., Marsden B. D., Kelm S., Shi J., Deane C. M., von Delft F., Nat. Commun. 2017, 8, 15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pushpakom S., Iorio F., Eyers P. A., Escott K. J., Hopper S., Wells A., Doig A., Guilliams T., Latimer J., McNamee C., Norris A., Sanseau P., Cavalla D., Pirmohamed M., Nat. Rev. Drug Discovery 2019, 18, 41–58. [DOI] [PubMed] [Google Scholar]
- 12. Sasaki T., Kishi M., Saito M., Tanaka T., Higuchi N., Kominami E., Katunuma N., Murachi T., J. Enzyme Inhib. 1990, 3, 195–201. [DOI] [PubMed] [Google Scholar]
- 13. Erlichman C., Hidalgo M., Boni J. P., Martins P., Quinn S. E., Zacharchuk C., Amorusi P., Adjei A. A., Rowinsky E. K., J. Clin. Oncol. 2006, 24, 2252–2260. [DOI] [PubMed] [Google Scholar]