Abstract
The direct visualization of vaccine fate is important to investigate its immunoactivation process to elucidate the detailed molecular reaction process at single‐molecular level. Yet, visualization of the spatiotemporal trafficking of vaccines remains poorly explored. Here, we show that quantum dot (QD) nanomaterials allow for monitoring vaccine dynamics and for amplified immune response. Synthetic QDs enable efficient conjugation of antigen and adjuvants to target tissues and cells, and non‐invasive imaging the trafficking dynamics to lymph nodes and cellular compartments. The nanoparticle vaccine elicits potent immune responses and anti‐tumor efficacy alone or in combination with programmed cell death protein 1 blockade. The synthetic QDs showed high fluorescence quantum yield and superior photostability, and the reliable and long‐term spatiotemporal tracking of vaccine dynamics was realized for the first time by using the synthetic QDs, providing a powerful strategy for studying immune response and evaluating vaccine efficacy.
Keywords: imaging, immune response, nanotechnology, nanovaccines, quantum dots
Visualization of spatiotemporal trafficking of vaccines is achieved with synthetic QDs for potent immune activation.
Introduction
Vaccines aim to prevent infectious diseases or treat established diseases such as cancer through manipulation of immune response.[ 1 , 2 ] Over traditional treatments, vaccines can orchestrate the body's own immune system to eradicate cancers and pathogens for durable control and reduced recurrence.[ 3 , 4 ] In vaccine development, besides the rational design of vaccine to boost immune responses,[ 5 , 6 ] the monitoring of vaccine dynamics is a very significant issue and indispensable step for immune responses considering that vaccines’ immunoactivation only proceeded after the efficient delivery of these vaccines to target tissues and cells. [7] Immune response is related to the vaccine formulation, the kinetics and biodistribution of vaccine exposure, crucial to potent immunoactivation.[ 7 , 8 ] For example, the physiological trafficking to different tissues, cells and intracellular compartments, the cellular uptake and processing pathways, will influence the antigen‐presenting cell (APC) activation and T cell priming, and will enhance or shape immune response in a different way. [9] Therefore, tracking vaccine dynamics and events are critical to improve mechanistic understanding, as well as assess and optimize vaccine design[ 10 , 11 ] or efficacy of immunotherapy.[ 12 , 13 , 14 ]
Nanotechnology is proved valuable for directing immune response in vaccine development, and benefits modern vaccine design including the COVID‐19 vaccine development,[ 1 , 15 ] as nanoparticles and pathogens operate at similar length scale, and nanomaterials mimic the key structural features of microbes that are sensed by immune system, ideal for co‐delivery of antigen and/or adjuvant, invoking signaling and immune response, and improving drug bioavailability.[ 16 , 17 , 18 ] The nanoparticle vaccine (nanovaccine) has additional unique advantages by targeting possible tissues or cells, enhancing intracellular delivery and antigen cross‐presentation in APCs.[ 19 , 20 , 21 ] More importantly, by exploiting the inherent physicochemical properties of nanoparticles or by exploring their facile conjugation capability to label diverse molecules of interest,[ 22 , 23 ] synthetic materials would encode striking features for tracking of vaccines and predicting of immune responses, [24] offering an innovative approach to visualization of vaccine dynamics. As such, well‐designed nanomaterials are anticipated to enable real‐time tracking vaccine dynamics in vitro and in vivo for monitoring immune responses and enhanced immunotherapy; however, remain elusive.
Here, we report synthetic quantum dots (QDs) nanomaterials that enable visualization of the spatiotemporal trafficking of vaccines for immunotherapy (Figure 1 A,B). Owing to excellent brightness, superior resistance to photo‐bleaching, small particle size and versatile surface modifications,[ 25 , 26 ] QDs are ideal labels for spatiotemporal tracking of important biological events.[ 27 , 28 ] To demonstrate the non‐invasive real‐time visualization of dynamic immune response, we functionalize different types of core–shell semiconductor QDs that are in the red wavelength range and near‐infrared (NIR) window as trackers. Our synthetic QD materials facilely conjugate immune components, antigen (ovalbumin, OVA) and adjuvant (unmethylated cytosine‐phosphate‐guanine, CpG), acting as labels as well as nanocarriers. Such nanovaccines enable optimal physical sites targeting by trafficking the nanoparticles to the draining lymph nodes and subcellular locations of lysosomes in APCs, inducing strong APC activation and robust T cell priming. These trafficking dynamics are simultaneously visualized by the QDs‐integrated nanovaccine for its high fluorescence quantum yield and superior photostability through in vitro and in vivo imaging. With this approach, we achieve long‐term spatiotemporal tracking the in situ transmembrane procedure between nanovaccine and cellular membrane in a live cell and the intracellular transfer of vaccine into lysosome process, facilitating the study of immunoactivation at the single‐particle‐level as well as the detailed molecular mechanism of Toll‐like receptor 9 (TLR9) activation. Besides, the synthetic QDs endow monitoring long‐term spatiotemporal trafficking of the vaccine in vivo with potent immunogenicity and antitumor efficacy, providing a good guidance for appropriate vaccination interval. A further combination immunotherapy with anti‐programmed cell death protein 1 (anti‐PD‐1) suppresses tumors growth substantially. As far as we know, it is the first time to use synthetic QDs as trackers to visualize vaccine dynamics.
Figure 1.
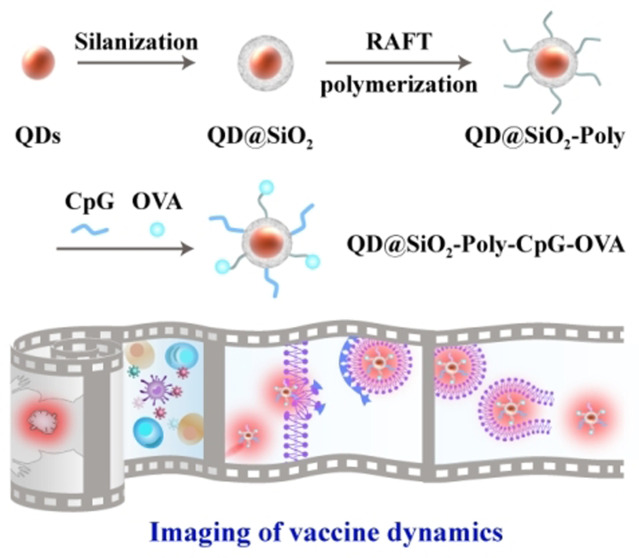
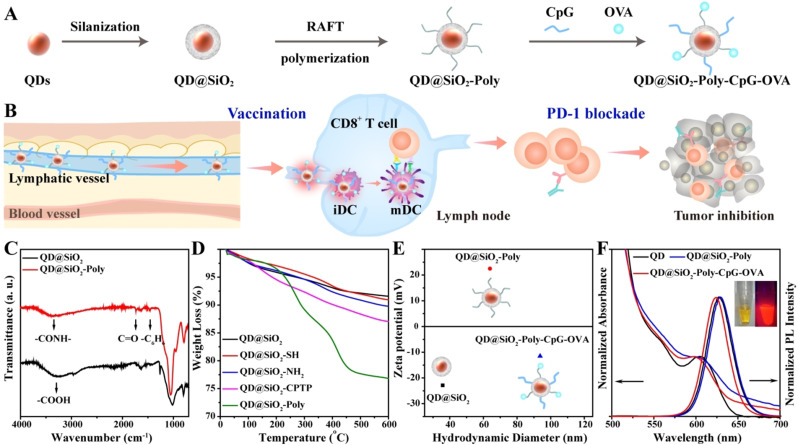
Visualization of the spatiotemporal trafficking of vaccines with amplified immune response and characterization of QD nanomaterials. A) Functionalization of core–shell QDs to construct nanovaccines. B) Synthetic QDs enable imaging trafficking dynamics of QD@SiO2‐Poly‐CpG‐OVA to lymph nodes and immune cells, allow real‐time monitoring intracellular events and kinetics, and induce potent immune responses for cancer immunotherapy by vaccination alone or combination immunotherapy with anti‐PD‐1. C) FTIR spectra of QD@SiO2 and QD@SiO2‐Poly. D) TGA curves of the indicated materials. E) Mean hydrodynamic sizes and zeta potential of the indicated materials (n=3, mean ± SD). F) Visible light absorption and photoluminescence (PL) spectra of the pristine QDs, QD@SiO2‐Poly and QD@SiO2‐Poly‐CpG‐OVA. Inset: photographs of QD@SiO2‐Poly‐CpG‐OVA under daylight (left) and UV lamp (right).
Results and Discussion
QD modification involves three steps including silica coating (Experimental procedures and Figure S1), surface polymerization (detailed synthetic process showing in Figure S2, S3), and vaccine components conjugation (Figure 1 A). Fourier transform infrared (FTIR) spectroscopy showed a strong absorption peak of QD@SiO2 appeared at 3296 cm−1, which corresponded to the stretching vibration of ‐COOH relating to the surface ligands of oleic acid on QDs (Figure 1 C). After cationic polymerization on the surface of QD@SiO2, apparent absorption peaks of poly (2‐(dimethylamino) ethyl methacrylate) (PDMAEMA)‐modified QDs (QD@SiO2‐Poly) appeared at 3386 cm−1, 1725 cm−1 and 1463 cm−1, attributed to the stretching vibration of ‐CONH‐, C=O and ‐C6H6, respectively. These data indicated the successful cationic PDMAEMA conjugation. In addition, comparisons via thermogravimetric analysis (TGA) showed an obviously weight loss with pyrolysis or combustion for QD@SiO2‐Poly, certifying the polymerization on the surface of QDs (Figure 1 D). Dynamic light scattering (DLS) analysis showed that the hydrodynamic diameter of QD@SiO2 was 35.8 nm and zeta potential was −22.9 mV, while the hydrodynamic diameter of QD@SiO2‐Poly increased to 63.9 nm and zeta potential reversed to +22.5 mV (Figure 1 E). All these results confirmed the successful fabrication of QD@SiO2‐Poly. Finally, negatively charged OVA (isoelectric point, pI 4.7) and CpG were conjugated with positively charged QD@SiO2‐Poly via electrostatic interaction in HEPES buffer (10 mM, pH 7.4), and various feed molar ratios of QD@SiO2‐Poly, OVA and CpG were investigated for optimization (Figure S4). To ensure both loading amount and modulate particle size for lymphatic drainage (20–100 nm),[ 19 , 29 ] the feed molar ratio of QD@SiO2‐Poly, OVA and CpG of 1/100/100 was chosen for the following experiments. After OVA and CpG conjugation, the hydrodynamic diameter of nanovaccines further elevated to 93.7 nm and zeta potential shifted to −11.5 mV (Figure 1 E). The amounts of OVA and CpG modified onto QD@SiO2‐Poly were quantified with Bradford method and fluorescence spectra of Quasar labeled‐CpG, respectively (Figure S5).
We next investigated fluorescence performance and stability of the materials. The QD@SiO2‐Poly exhibited high quantum yield (QY, 20.1 %, Table S1) and stable photoluminescence in a wide pH range from 5 to 10 (Figure S6). After RAFT polymerization on QD surface or antigen/adjuvant conjugation, QDs kept the broad absorption bands and strong and narrow emission bands (Figure 1 F, and Figure S7 for NIR QD). Over 5 days, the cumulative released vaccine components from the nanovaccines were less than 14 % (Figure S8). Methyl thiazolyl tetrazolium (MTT) and hemolysis assays showed that the synthetic QDs were nontoxic to living cells and exhibited good blood compatibility (Figure S9). These results demonstrated the synthetic QDs showed high brightness, well photostability and biocompatibility, crucial for in vitro and in vivo applications.
There are sophisticated endocytosis pathways of nanoparticles into cells, and different uptake pathways not only affect uptake efficiency of nanovaccines, but also alter intracellular trafficking of nanovaccines in APCs, which may further affect immunoactivation of APCs.[ 30 , 31 ] Therefore, we studied the endocytosis mechanism and intracellular trafficking dynamics to APCs by real‐time imaging. When incubated with the nanovaccines, rapid cellular uptake was observed within 2 h, and the fluorescence signal increased with the incubation time elongation, showing 8.17‐fold changs at 6 h compared with that of 2 h (Figure S10). To verify the co‐delivery capacity of CpG and OVA, nanovaccines were estimated by confocal fluorescence microscopy using Quasar‐labeled CpG (Figure 2 A). The nanovaccines and the Quasar‐CpG were incubated with RAW264.7 cells. Little fluorescence signal was detected in cells incubated with Quasar‐CpG alone, which was consistent with the facts that naked oligonucleotides are difficult to pass through the cytoplasmic membrane and prone to degradation by nucleases. The co‐localization fluorescence signals of QDs and Quasar‐CpG indicated that QD@SiO2‐Poly acted as efficient nanocarriers for delivering CpG into RAW264.7 cells. Additionally, confocal images also indicated the co‐localization of Quasar‐CpG and FITC‐OVA in RAW264.7 cells (Figure S11), suggesting that QD@SiO2‐Poly could act efficient nanocarriers for co‐delivery of CpG and OVA.
Figure 2.
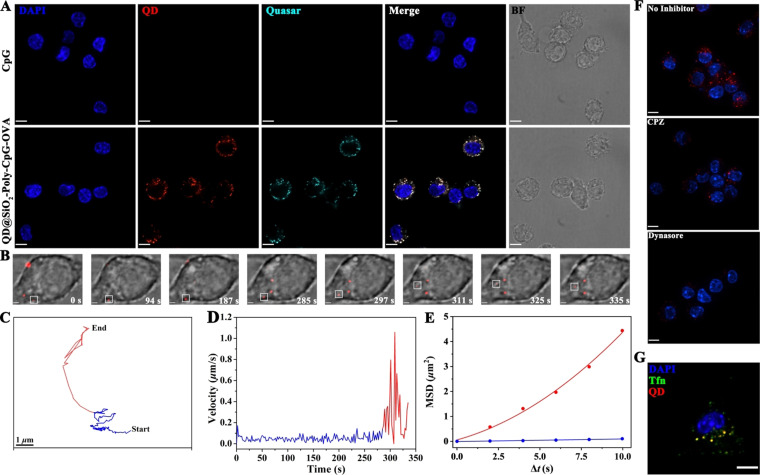
Visualization of nanovaccine's multiple‐staged slow‐to‐fast transmembrane process and its clathrin‐mediated endocytosis. A) Efficient intracellular delivery of nanovaccines revealed by confocal images. RAW264.7 cells were incubated with Quasar‐CpG (50 nM) and QD@SiO2‐Poly‐CpG‐OVA (50 μg mL−1) for 8 h at 37 °C. Scale bar is 10 μm. B) Spatiotemporal trafficking dynamics of vaccine to APC demonstrated by the snapshots of transmembrane process of the nanovaccine shown in Movie S4 (highlighted in white squares). Scale bar is 2 μm. C and D) Trajectory (C) and velocity curve (D) of the nanovaccine diffusion shown in (B). E) Dependence of mean square displacement (MSD) against Δt during diffusion of the nanovaccine. The blue and red lines represent the fits to corresponding equations of restricted and directed diffusion, respectively. F and G) Clathrin‐mediated endocytosis of the nanovaccines confirmed by inhibitor assays (F) and transferrin endocytosis (G). Scale bar is 7 μm. For images in (G), RAW264.7 cells were incubated with transferrin (Tfn, labeled with Alexa Fluor 488) and nanovaccines for 15 min at 37 °C. Scale bar is 10 μm.
The photostability of the synthetic QDs was evaluated by long‐term imaging (Figure S12, Movies S1–S3), crucial factor for single particle tracking (SPT).[ 27 , 32 ] The fluorescence intensity of QDs was stable for acquiring time‐series images within 5 min. In comparison, the fluorescence decay of small molecules (Cy5 or FITC) was obviously observed with approximately 40 % decrease. These results demonstrated the synthetic QDs showed superior photostability for long‐term tracking the vaccine dynamics in live cells compared with fluorescent small molecules. With the SPT technology, the nanovaccine was dynamically tracked its real‐time transmembrane procedure in a live cell to elucidate the mechanism of endocytosis process of nanovaccine (n=15). RAW264.7 cells were incubated with the prechilled nanovaccines at 4 °C for 10 min, and then shifted to 37 °C for real‐time observation (the negative and positive controls, Figure S13, S14). The dynamic transmembrane procedure of nanovaccine was tracked in a RAW264.7 cell (Figure 2 B). The whole trajectory was displayed in Figure 2 C and Movie S4. Surprisingly, the time‐dependent instantaneous speed showed that the nanovaccine entered into a RAW264.7 cell through a multiple‐staged slow‐to‐fast process (Figure 2 D). The nanovaccine first confined to the cellular membrane and then rapidly transported into the cytosol. The movement modes of nanovaccine in two phases were also analyzed based on the mean square displacement (MSD, Figure 2 E). The nanovaccine firstly experienced a slow and confined (diffusion coefficient, ca. 0.0052 μm2 s−1) motion mode at the edge of cellular membranes (blue line). Subsequently, the nanovaccine moved rapidly towards the cytoplasm with a mean speed of ca. 0.31 μm s−1 (red line) and a diffusion coefficient of ca. 0.092 μm2 s−1. Furthermore, whether the clathrin‐mediated entry [33] was mainly involved in endocytosis of nanovaccines into RAW264.7 cells was next investigated. Cells were pretreated with chlorpromazine (CPZ) and dynasore, which specifically inhibit clathrin‐mediated endocytosis at noncytotoxic dosages (Figure S15). Confocal images and flow cytometry analysis indicated that cellular internalization of nanovaccines was drastically reduced in cells treated with both inhibitors (Figure 2 F, S16). Additionally, transferrin, a classical marker for clathrin‐mediated endocytosis, was imaged for its involvement in the nanovaccines entry into RAW264.7 cells (Figure 2 G), suggesting that 79.8 % (calculated by Pearson's correlation) of the nanovaccines were co‐localized with Tfn spots in the cytoplasm. These results illustrated that the nanovaccines entered into RAW264.7 cells through a multiple‐staged slow‐to‐fast transmembrane process via clathrin‐mediated endocytosis.
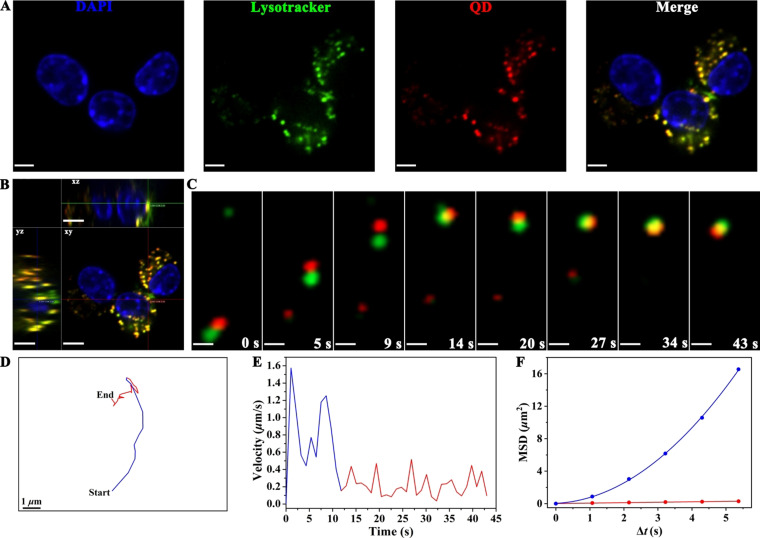
Owing to the residence of TLR9 in the lysosome, [34] clathrin‐mediated endocytic pathway of nanovaccines would be beneficial to potentiate the activity of CpG, thereby enhancing the immunoactivation of RAW264.7 cells. The interaction between nanovaccines and lysosome directly affects the efficacy of TLR9 activation.[ 34 , 35 ] Therefore, the co‐localization of nanovaccines with lysosome and its transfer process into lysosome in RAW264.7 cells was next investigated. Confocal images revealed that 93.0 % (calculated by Pearson's correlation) of the internalized nanovaccines co‐localized with lysosome in RAW264.7 cells (Figure 3 A,B), ensuring sufficient binding of CpG with TLR9 for immunoactivation. Furthermore, the nanovaccine was dynamically tracked its transfer process into lysosome with the whole trajectory in real‐time via SPT (n=15, Figure 3 C,D and Movie S5). Time‐dependent speed suggested that the nanovaccine entered into lysosome through a multiple‐staged fast‐to‐slow process (Figure 3 E). The nanovaccine and lysosome firstly experienced rapid and directed movement towards the same direction with a mean speed of ca. 0.78 μm s−1 and a diffusion coefficient of ca. 0.052 μm2 s−1 (blue line in Figure 3 E,F). Then, they collided with each other in a slow and confined motion mode with a diffusion coefficient of ca. 0.014 μm2 s−1 (red line in Figure 3 E,F). These results demonstrated that the nanovaccine transfered into lysosome through a multiple‐staged fast‐to‐slow process after clathrin‐mediated endocytosis for the TLR9 activation. Our rational designed nanovaccine could provide a powerful platform for long‐term spatiotemporal tracking the in situ transmembrane procedure and the intracellular transfer of vaccine into lysosome process, thus facilitating the study of immunoactivation at the single‐nanoparticle‐level as well as the detailed molecular mechanism of TLR9 activation.[ 34 , 36 , 37 ]
Figure 3.
Tracking the multiple‐staged fast‐to‐slow transfer process of nanovaccine into lysosome. A) Co‐localization of nanovaccines with lysosome in RAW264.7 cells by confocal images. RAW264.7 cells were incubated with nanovaccines (50 μg mL−1) for 4 h at 37 °C. Scale bar is 10 μm. B) Orthogonal image of RAW264.7 cells. Scale bar is 10 μm. C) Snapshots of the intracellular nanovaccine transferring into lysosome in a RAW264.7 cell. Scale bar is 1 μm. D and E) Trajectory (D) and velocity curve (E) of the nanovaccine diffusion shown in (C). F) Dependence of MSD against Δt during diffusion of the nanovaccine. The blue and red lines represent the fits to corresponding equations of directed and restricted diffusion, respectively.
Considering the efficient intracellular endocytosis and well co‐localization of the materials with lysosome, the immunoactivation of nanovaccines in RAW264.7 cells was explored, including the costimulatory molecules (CD40) expression, nitric oxide (NO) production, proinflammatory cytokine secretion, phagocytic activity and morphology of the cells. The nanovaccines induced 7.30‐fold higher expression of costimulatory molecules CD40 than that of soluble CpG and OVA (Figure S17), indicating the activation of the RAW264.7 cell. NO is an important signaling molecule in immunomodulation, [38] and the NO production in macrophages is essential for the defense mechanisms against tumor cells. Compared with CpG + OVA, the nanovaccines induced 3.85‐fold higher level of NO production (Figure S18A), owing to efficient intracellular co‐delivery of the antigen and adjuvant, and well co‐localization with the lysosome for immunomodulation. Also, secreting proinflammatory cytokines such as tumor necrosis factor (TNF‐α) and interleukin (IL)‐6 are an important mechanism to directly kill tumor cells.[ 39 , 40 ] Enzyme‐linked immunosorbent assay (ELISA) analysis revealed that the nanovaccines dramatically increased secretion of TNF‐α (3.72‐fold) and IL‐6 (20.2‐fold) than that of CpG + OVA (Figure S18B,C). Phagocytosis of pathogens by macrophages initiates the innate immune response, which in turn orchestrates the adaptive response. Consistently, neutral red uptake test indicated that cells treated with the nanovaccines exhibited enhanced phagocytic activity over other groups (Figure S18D). Additionally, morphology change of RAW264.7 cells was used as a potential indication of cell function and investigated by confocal images. For inactivated RAW264.7 cells, cells tended to be uniform and round. By contrast, once activated by the nanovaccines, cells turned to be rougher and larger (Figure S18E). Lysosome escape of the antigen would contribute to priming CD8+ T cells for potent antitumor response. [41] The lysosome escape capacity of nanovaccines was mimicked in vitro by the red blood cell (RBC) lysis assay, exhibiting evident pH‐dependent membrane disruptive properties (Figure S19A). The long‐term incubation of nanovaccines with RAW264.7 cells showed intense cytosolic fluorescence signal, indicating the lysosome escape of the nanovaccines (Figure S19B). Taken together, these results demonstrated a potent immunoactivation of nanovaccines for its efficient intracellular delivery and lysosome escape capacity, promising for strong immunoactivation in vivo.
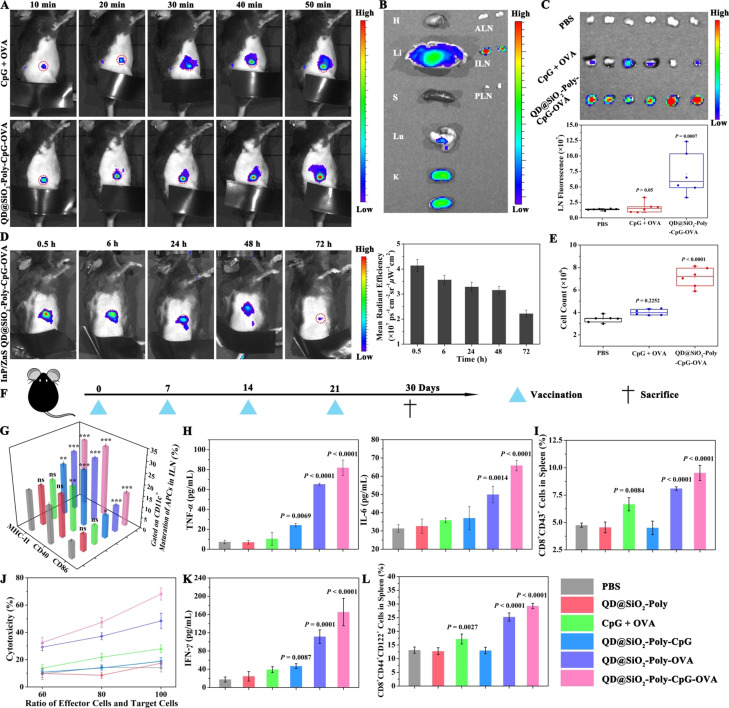
Effective lymph node accumulation and durable retention of nanovaccines are vital to activate potent immunoactivation. Correspondingly, in vivo lymphatic drainage of nanovaccines was investigated. Firstly, C57BL/6j mice were subcutaneously (s.c.) injected with different materials at the tail base for in vivo fluorescence imaging. The nanovaccines showed rapid lymphatic drainage within 10 min, and exhibited stronger fluorescence intensity than the CpG and OVA mixture, indicating an enhanced accumulation in inguinal lymph node (ILN, Figure 4 A). After 24 h post injection, major organs and lymph nodes from mice injected with nanovaccines were harvested for ex vivo imaging. Except for some accumulation in liver and kidney due to the metabolic clearance, the mean radiant efficiency showed 45.0 % of nanovaccines accumulation in ILNs (Figure 4 B). Consistently, ex vivo images of ILNs treated with nanovaccines showed 4.24‐fold higher fluorescence signal than that of CpG + OVA (Figure 4 C). The intravital spatiotemporal tracking of lymphatic drainage with NIR InP/ZnS QDs‐based nanovaccines indicated that there was a durable retention in ILNs from 0.5 h to 72 h (Figure 4 D). These results showed a long‐termed activation of immune response in vivo, which could guide 3 days as an interval for further therapeutic antitumor study. Furthermore, cell number quantification of the resected ILNs showed increased cell number by the nanovaccines over PBS with 2.09‐fold, indicating recruitment of immune cells for immunomodulation by nanovaccines (Figure 4 E). These data on spatiotemporal tracking of lymphatic drainage demonstrated that the nanovaccines exhibited rapid accumulation and durable retention in the lymph nodes for a long‐term immune stimulus.
Figure 4.
In vivo monitoring the nanovaccines trafficking to draining lymph nodes and the enhanced immunoactivation. A) C57BL/6j mice were s.c. injected with the CpG and OVA mixture or nanovaccines at the tail base for in vivo fluorescence imaging. The red circle referred to the ILN. CpG is labeled with Cy5. B) Ex vivo fluorescence imaging of major organs of mice injected with nanovaccines after 24 h. H, Li, S, Lu, K, ALN, ILN and PLN represent heart, liver, spleen, lung, kidney, axillary lymph node, inguinal lymph node and popliteal lymph node, respectively. C) The ILNs of three different groups were harvested at 24 h post injection for ex vivo imaging and quantification of mean fluorescence signals. n=6, mean ± SD. D) C57BL/6j mice were s.c. injected near‐infrared InP/ZnS QD‐based nanovaccines at the tail base for in vivo imaging from 30 min to 72 h (left) and for calculating mean radiant efficiency of ILN (right). The red circle referred to the ILN. n=3, mean ± SD. E) Total cell number quantification of the resected ILNs as indicated in (C). F) C57BL/6j mice were s.c. injected at the tail base with different formulations of QD@SiO2‐Poly (4 mg kg−1, 50 μL), or equivalent, on day 0, 7, 14 and 21, and sacrificed for immunogenicity study on day 30. G) Flow cytometry analysis of APCs (CD40, CD86 and MHC‐II expression on CD11c+ cells) in ILNs. H) ELISA analysis of TNF‐α (left) and IL‐6 (right). I) Percentage of CD8+CD43+ T cells in spleen. J) Cytotoxic lymphocytes (CTL)‐mediated immune responses determined by incubating B16‐OVA cells with various effector/target ratios of re‐stimulated splenocytes. K) ELISA analysis of IFN‐γ in serum. L) Percentage of CD8+ central memory T cells (CD8+CD44+CD122+ cells) in spleen. Data in (G)–(L) were expressed as n=4, mean ± SD. Statistical significance was calculated by one‐way ANOVA with the Bonferroni post hoc test. ***P<0.001, **P<0.01, *P<0.05, ns, not significant.
Next, the immunostimulatory effects of nanovaccines were investigated in naive mice. C57BL/6j mice were s.c. administrated at the tail base with indicated materials and sacrificed for immunogenicity study (Figure 4 F). Flow cytometry analysis of ILNs indicated that QD@SiO2‐Poly‐CpG‐OVA exhibited higher maturation effect of APCs (1.74, 1.52 and 1.63‐fold increase of CD40, CD86 and MHC‐II expression on CD11c+ cells, respectively) in ILNs than that of CpG + OVA (Figure 4 G), which was attributed to efficient lymphatic drainage and durable retention in the lymph nodes. Consistently, nanovaccines significantly enhanced secretion of pro‐inflammatory cytokines (7.84 and 1.84‐fold increased secretion of TNF‐α and IL‐6, respectively) than that of CpG + OVA (Figure 4 H). These results demonstrated that combination of CpG and OVA in the nanoparticles induced efficient co‐delivery of vaccine components and significant APC maturation in vivo. APC maturation facilitates the T cell priming, leading to eliminating tumor cells. Thus, T cell response is important to evaluate the vaccine‐induced immune response. QD@SiO2‐Poly‐CpG‐OVA elicited significantly more CD8+ T cells with 1.43 and 2.11‐fold increase than that of the CpG + OVA or QD@SiO2‐Poly‐CpG, respectively, indicating the potent immunigenicity of QD@SiO2‐Poly‐CpG‐OVA (Figure 4 I, S20A). The mice immunized with QD@SiO2‐Poly‐CpG‐OVA showed more proliferation of CD8+ T cells in comparison with QD@SiO2‐Poly‐OVA, owing to adjuvant effect of CpG. The antigen‐specific cytotoxicity was estimated by stimulating splenocytes from immunized mice with OVA for 60 h and co‐incubating with B16‐OVA cells. Splenocytes obtained from QD@SiO2‐Poly‐CpG‐OVA treated mice exhibited higher cytotoxicity to tumor cells than other groups under the same effector/target ratio (Figure 4 J). Furthermore, with the increased ratio of effector cells and target cells, the nanovaccines showed gradually enhanced cytotoxicity of CTLs. Similarly, secretion of interferon gamma (IFN‐γ) was prominently enhanced with 4.23‐fold after treatment by QD@SiO2‐Poly‐CpG‐OVA over the CpG + OVA mixture (Figure 4 K). Additionally, on days 30, the mice immunized with nanovaccines showed larger fractions of central memory CD8+ T cells (CD8+CD44+CD122+ cells) compared with other groups (Figure 4 L, S20B), indicating durable control of T cell immune response. Biosafety of the nanovaccines was studied by in vivo clearance detection, hematoxylin‐eosin (H&E) staining and body weight evaluation (Figure S21–S23), showing rapid metabolic clearance of nanovaccines in 72 h with well biocompatibility. Collectively, these results demonstrated that the QDs performed as spatiotemporal trackers for monitoring rapid and durable lymphatic drainage in vivo, and mediated synergistic enhancement of immunoactivation such as robust APC maturation and T cell responses in vivo.
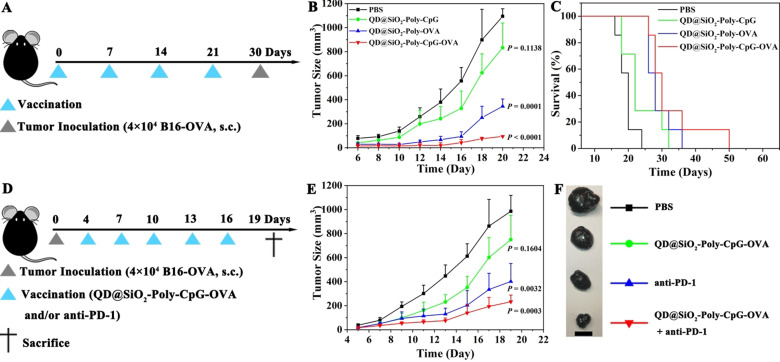
To estimate whether the enhanced adaptive immune response of nanovaccines could eliminate tumor cells, in vivo prophylactic study for cancer immunotherapy was carried out (Figure 5 A, and average body weights of mice see Figure S24). Compared with PBS, the mice vaccinated with QD@SiO2‐Poly‐OVA exhibited modest inhibition of tumor growth, extending medium survival from 20 days to 28 days. QD@SiO2‐Poly‐CpG‐OVA significantly suppressed tumor growth and extended medium survival to 30 days (Figure 5 B,C, and S25). Taken together, nanovaccines exerted potent prophylactic efficiency for tumor inhibition in vivo and significantly extended medium survival of the vaccinated mice. To further amplify the therapeutic responses and advance its practical potential, we develop a combination immunotherapy with vaccines and anti‐PD‐1 (Figure 5 D, and average body weights of mice see Figure S26). The combination treatment exerted enhanced control of tumor growth over other groups (Figure 5 E,F and S27). H&E staining of tumor slices showed more necrotic/apoptotic tumor cells under the combination treatment than other groups (Figure S28A). The immunohistochemical (IHC) analysis indicated that compared with other groups, combination treatment exhibited higher expression CD4+ T and CD8+ T cells in tumor (Figure S28B), contributing to enhanced immune response to eliminate tumor cells. Conclusively, these results illustrated that nanovaccines exerted potent antitumor prophylactic and therapeutic efficacy, and could act synergistically with other immunotherapies, such as immune checkpoint blockade, activating different aspects of immune response for better elimination of tumor cells.
Figure 5.
In vivo prophylactic and therapeutic studies of nanovaccines alone or in combination with anti‐PD‐1 for tumor suppression. A) Experiment setup for prophylactic treatment. C57BL/6j mice were s.c. injected at the tail base with different formulations of QD@SiO2‐Poly (4 mg kg−1, 50 μL), or equivalent, on day 0, 7, 14 and 21. On day 30, the mice were s.c. inoculated with 4×104 B16‐OVA cells on the right flank. B and C) Average tumor sizes (B) and survival ratio (C) for prophylactic study (n=7, mean ± SD). D) Experiment setup for therapeutic treatment. C57BL/6j mice were s.c. inoculated with 4×104 B16‐OVA cells on day 0. Then the mice were s.c. injected at the tail base with QD@SiO2‐Poly‐CpG‐OVA (4 mg kg−1, 50 μL) and on day 4, 7, 10, 13 and 16. Anti‐PD‐1 (10 μg) was intravenously (i.v.) administered on the same day. E and F) Average tumor sizes (E; n=7, mean ± SD) and representative tumor tissues (F) under these indicated treatments for therapeutic study. Scale bar is 1 cm. Statistical significance was calculated by one‐way ANOVA with the Bonferroni post hoc test.
Conclusion
We have demonstrated visualization of spatiotemporal trafficking of vaccine dynamics to target physiological sites with the synthetic QDs for enhanced immunity, including the efficient delivery into dendritic cells, well localization with lysosome, and targeted lymph node accumulation. The synthetic QDs showed high fluorescence quantum yield and superior photostability, enabling reliable and long‐term spatiotemporal tracking the kinetics and biodistribution of vaccine, such as the multiple‐staged slow‐to‐fast transmembrane process in live cells via clathrin‐mediated endocytosis and the fast‐to‐slow transfer of vaccine into lysosome process. The trafficking dynamics of nanovaccines also revealed durable retention in lymph nodes for sustained immune cell activation, robust APC maturation, and potent T cell responses. Correspondingly, vaccination with the nanovaccines strongly amplifies immunization in vivo, and the combination of nanovaccines and PD‐1 blockade induces stronger immune response to combat cancer. Taken together, this work describes the spatiotemporal dynamics of nanovaccines using QD materials to help evaluate and boost immune response. Such a nanoparticle platform suggests great potentials in vaccine development and immunotherapy. Considering that more bright, stable and compatible QDs with diverse functionalities have been emerged,[ 42 , 43 , 44 ] these QD materials indicate additional merits for multimodal tracking of vaccines, and for a better understanding of immune response in clinical immunotherapy. [45] In addition, the QDs could conjugate with different types of therapeutic components, such as therapeutic nucleic acids, neoantigens, and more,[ 46 , 47 ] thus offering a versatile platform to develop more advanced RNA vaccines for personalized therapy and combinational therapy.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Acknowledgements
The research was supported by the National Natural Science Foundation of China (21874103, 22074112, 91859123), and Fundamental Research Funds for the Central Universities (No. 2042018kf1006). We thank Dr. Bo Tang for his help.
J. Sun, F. Liu, W. Yu, D. Fu, Q. Jiang, F. Mo, X. Wang, T. Shi, F. Wang, D.-W. Pang, X. Liu, Angew. Chem. Int. Ed. 2021, 60, 24275.
Contributor Information
Prof. Fuan Wang, Email: fuanwang@whu.edu.cn.
Prof. Dai‐Wen Pang, Email: dwpang@whu.edu.cn.
Prof. Xiaoqing Liu, Email: xiaoqingliu@whu.edu.cn.
References
- 1. Shin M. D., Shukla S., Chung Y. H., Beiss V., Chan S. K., Ortega-Rivera O. A., Wirth D. M., Chen A., Sack M., Pokorski J. K., Steinmetz N. F., Nat. Nanotechnol. 2020, 15, 646–655. [DOI] [PubMed] [Google Scholar]
- 2. Wang C., Ye Y., Hu Q., Bellotti A., Gu Z., Adv. Mater. 2017, 29, 1606036. [DOI] [PubMed] [Google Scholar]
- 3. Sharma P., Hu-Lieskovan S., Wargo J. A., Ribas A., Cell 2017, 168, 707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Banchereau J., Palucka K., Nat. Rev. Clin. Oncol. 2018, 15, 9–10. [DOI] [PubMed] [Google Scholar]
- 5. Yu W., Sun J., Liu F., Yu S., Hu J., Zhao Y., Wang X., Liu X., ACS Appl. Mater. Interfaces 2020, 12, 40002–40012. [DOI] [PubMed] [Google Scholar]
- 6. Cai H., Shukla S., Wang C., Masarapu H., Steinmetz N. F., J. Am. Chem. Soc. 2019, 141, 6509–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pittet M. J., Garris C. S., Arlauckas S. P., Weissleder R., Sci. Immunol. 2018, 3, eaaq0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lindsay K. E., Bhosle S. M., Zurla C., Beyersdorf J., Rogers K. A., Vanover D., Xiao P., Araínga M., Shirreff L. M., Pitard B., Baumhof P., Villinger F., Santangelo P. J., Nat. Biomed. Eng. 2019, 3, 371–380. [DOI] [PubMed] [Google Scholar]
- 9. Schudel A., Francis D. M., Thomas S. N., Nat. Rev. Mater. 2019, 4, 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Luo M., Wang H., Wang Z., Cai H., Lu Z., Li Y., Du M., Huang G., Wang C., Chen X., Porembka M. R., Lea J., Frankel A. E., Fu Y. X., Chen Z. J., Gao J., Nat. Nanotechnol. 2017, 12, 648–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alam I. S., Mayer A. T., Sagiv-Barfi I., Wang K., Vermesh O., Czerwinski D. K., Johnson E. M., James M. L., Levy R., Gambhir S. S., J. Clin. Invest. 2018, 128, 2569–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Woodham A. W., Zeigler S. H., Zeyang E. L., Kolifrath S. C., Cheloha R. W., Rashidian M., Chaparro R. J., Seidel R. D., Garforth S. J., Dearling J. L., Mesyngier M., Duddempudi P. K., Packard A. B., Almo S. C., Ploegh H. L., Nat. Methods 2020, 17, 1025–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhong Y., Ma Z., Wang F., Wang X., Yang Y., Liu Y., Zhao X., Li J., Du H., Zhang M., Cui Q., Zhu S., Sun Q., Wan H., Tian Y., Liu Q., Wang W., Garcia K. C., Dai H., Nat. Biotechnol. 2019, 37, 1322–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ramesh A., Kumar S., Brouillard A., Nandi D., Kulkarni A., Adv. Mater. 2020, 32, 2000648. [DOI] [PubMed] [Google Scholar]
- 15. Liu Y., Chen C., Adv. Drug Delivery Rev. 2016, 103, 76–89. [DOI] [PubMed] [Google Scholar]
- 16. Lynn G. M., Sedlik C., Baharom F., Zhu Y., Ramirez-Valdez R. A., Coble V. L., Tobin K., Nichols S. R., Itzkowitz Y., Zaidi N., Gammon J. M., Blobel N. J., Denizeau J., de la Rochere P., Francica B. J., Decker B., Maciejewski M., Cheung J., Yamane H., Smelkinson M. G., Francica J. R., Laga R., Bernstock J. D., Seymour L. W., Drake C. G., Jewell C. M., Lantz O., Piaggio E., Ishizuka A. S., Seder R. A., Nat. Biotechnol. 2020, 38, 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Callmann C. E., Cole L. E., Kusmierz C. D., Huang Z. Y., Horiuchi D., Mirkin C. A., Proc. Natl. Acad. Sci. USA 2020, 117, 17543–17550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun J., Pan M., Liu F., Yu W., Wang W., Mo F., Yu S., Zhou Y., Liu X., ACS Mater. Lett. 2020, 2, 1606–1614. [Google Scholar]
- 19. Luo M., Samandi L. Z., Wang Z., Chen Z. J., Gao J., J. Controlled Release 2017, 263, 200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang H., Mooney D. J., Nat. Mater. 2018, 17, 761–772. [DOI] [PubMed] [Google Scholar]
- 21. Zhu G., Zhang F., Ni Q., Niu G., Chen X., ACS Nano 2017, 11, 2387–2392. [DOI] [PubMed] [Google Scholar]
- 22. Pan J., Wang Y., Zhang C., Wang X., Wang H., Wang J., Yuan Y., Wang X., Zhang X., Yu C., Sun S. K., Yan X. P., Adv. Mater. 2018, 30, 1704408. [Google Scholar]
- 23. Van Herck S., Deswarte K., Nuhn L., Zhong Z., Portela Catani J. P., Li Y., Sanders N. N., Lienenklaus S., De Koker S., Lambrecht B. N., David S. A., De Geest B. G., J. Am. Chem. Soc. 2018, 140, 14300–14307. [DOI] [PubMed] [Google Scholar]
- 24. Zhang Y., He S., Chen W., Liu Y., Zhang X., Miao Q., Pu K., Angew. Chem. Int. Ed. 2021, 60, 5921–5927; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 5986–5992. [Google Scholar]
- 25. Liu F., Sun J., Yu W., Jiang Q., Pan M., Xu Z., Mo F., Liu X., Biomaterials 2020, 242, 119928. [DOI] [PubMed] [Google Scholar]
- 26. Sun J., Liu F., Yu W., Jiang Q., Hu J., Liu Y., Wang F., Liu X., Nanoscale 2019, 11, 5014–5020. [DOI] [PubMed] [Google Scholar]
- 27. Liu S. L., Wang Z. G., Xie H. Y., Liu A. A., Lamb D. C., Pang D. W., Chem. Rev. 2020, 120, 1936–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo Y., Sakonsinsiri C., Nehlmeier I., Fascione M. A., Zhang H., Wang W., Pöhlmann S., Turnbull W. B., Zhou D., Angew. Chem. Int. Ed. 2016, 55, 4738–4742; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 4816–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Irvine D. J., Swartz M. A., Szeto G. L., Nat. Mater. 2013, 12, 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dixit S., Sahu R., Verma R., Duncan S., Giambartolomei G. H., Singh S. R., Dennis V. A., Biomaterials 2018, 159, 130–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Druzd D., Matveeva O., Ince L., Harrison U., He W., Schmal C., Herzel H., Tsang A. H., Kawakami N., Leliavski A., Uhl O., Yao L., Sander L. E., Chen C. S., Kraus K., de Juan A., Hergenhan S. M., Ehlers M., Koletzko B., Haas R., Solbach W., Oster H., Scheiermann C., Immunity 2017, 46, 120–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shen H., Tauzin L. J., Baiyasi R., Wang W., Moringo N., Shuang B., Landes C. F., Chem. Rev. 2017, 117, 7331–7376. [DOI] [PubMed] [Google Scholar]
- 33. Li Q., Li W., Yin W., Guo J., Zhang Z. P., Zeng D., Zhang X., Wu Y., Zhang X. E., Cui Z., ACS Nano 2017, 11, 3890–3903. [DOI] [PubMed] [Google Scholar]
- 34. Latz E., Schoenemeyer A., Visintin A., Fitzgerald K. A., Monks B. G., Knetter C. F., Lien E., Nilsen N. J., Espevik T., Golenbock D. T., Nat. Immunol. 2004, 5, 190–198. [DOI] [PubMed] [Google Scholar]
- 35. Yu W., Sun J., Liu F., Yu S., Xu Z., Wang F., Liu X., ACS Appl. Mater. Interfaces 2020, 12, 17167–17176. [DOI] [PubMed] [Google Scholar]
- 36. Shukoor M. I., Natalio F., Tahir M. N., Wiens M., Tarantola M., Therese H. A., Barz M., Weber S., Terekhov M., Schroder H. C., Müller W. E. G., Janshoff A., Theato P., Zentel R., Schreiber L. M., Tremel W., Adv. Funct. Mater. 2009, 19, 3717–3725. [Google Scholar]
- 37. Combes A., Camosseto V., N'Guessan P., Arguello R. J., Mussard J., Caux C., Bendriss-Vermare N., Pierre P., Gatti E., Nat. Commun. 2017, 8, 913–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rhule A., Navarro S., Smith J. R., Shepherd D. M., J. Ethnopharmacol. 2006, 106, 121–128. [DOI] [PubMed] [Google Scholar]
- 39. Dayer J. M., Best Pract. Res. Clin. Rheumatol. 2004, 18, 31–45. [DOI] [PubMed] [Google Scholar]
- 40. Diehl S., Rincon M., Mol. Immunol. 2002, 39, 531–536. [DOI] [PubMed] [Google Scholar]
- 41. Neefjes J. J., Stollorz V., Peters P. J., Geuze H. J., Ploegh H. L., Cell 1990, 61, 171–183. [DOI] [PubMed] [Google Scholar]
- 42. Lu H., Carroll G. M., Neale N. R., Beard M. C., ACS Nano 2019, 13, 939–953. [DOI] [PubMed] [Google Scholar]
- 43. Shu Y., Lin X., Qin H., Hu Z., Jin Y., Peng X., Angew. Chem. Int. Ed. 2020, 59, 22312–22323; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 22496–22507. [Google Scholar]
- 44. Shashkov E. V., Everts M., Galanzha E. I., Zharov V. P., Nano Lett. 2008, 8, 3953–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cazaux M., Grandjean C. L., Lemaitre F., Garcia Z., Beck R. J., Milo I., Postat J., Beltman J. B., Cheadle E. J., Bousso P., J. Exp. Med. 2019, 216, 1038–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Riel D., de Wit E., Nat. Mater. 2020, 19, 810–812. [DOI] [PubMed] [Google Scholar]
- 47. Liu S., Jiang Q., Zhao X., Zhao R., Wang Y., Wang Y., Liu J., Shang Y., Zhao S., Wu T., Zhang Y., Nie G., Ding B., Nat. Mater. 2021, 20, 421–430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information