Abstract
COVID‐19 disease is the manifestation of syndrome coronavirus 2 (SARS‐CoV‐2) infection, which is causing a worldwide pandemic. This disease can lead to multiple and different symptoms, being lymphopenia associated with severity one of the most persistent. Natural killer cells (NK cells) are part of the innate immune system, being fighting against virus‐infected cells one of their key roles. In this study, we determined the phenotype of NK cells after COVID‐19 and the main characteristic of SARS‐CoV‐2‐specific‐like NK population in the blood of convalescent donors. CD57+ NKG2C+ phenotype in SARS‐CoV‐2 convalescent donors indicates the presence of ‘memory’/activated NK cells as it has been shown for cytomegalovirus infections. Although the existence of this population is donor dependent, its expression may be crucial for the specific response against SARS‐CoV‐2, so that, it gives us a tool for selecting the best donors to produce off‐the‐shelf living drug for cell therapy to treat COVID‐19 patients under the RELEASE clinical trial (NCT04578210).
Keywords: cell therapy, COVID‐19, HLA, KIR, NK cells
SARS‐CoV‐2 infection induce a pro‐inflammatory environment that could lead to NK cells secreting IFN‐γ, increasing HLA‐E expression and being activated through NKG2C‐HLA‐E recognition. The NK cells become adaptive or “memory” cells that can recognize SARS‐CoV‐2 soluble peptides via NKG2C, triggering a specific inflammatory response to fight this virus.
INTRODUCTION
Coronavirus disease 2019 (COVID‐19) reached pandemic state in a few weeks since in December 2019, severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) emerged in Wuhan, China, and spread worldwide. Until October 2021, 223 different countries were affected by COVID‐19, reaching more than 238 000 000 confirmed cases and more than 4 800 000 confirmed deaths [1]. Patients with COVID‐19 may develop various symptoms, such as fever and cough, pneumonia and dyspnoea, and occasionally may progress to multiorgan failure [2]. Another common symptom is lymphopenia correlated with severity of disease, which makes patients more prompt to co‐infections [3].
Up to now, the treatments used for COVID‐19 are mainly supportive, as there is not a specific treatment that cures SARS‐CoV‐2 infection. Antiviral drug lopinavir/ritonavir has been used mainly for treating COVID‐19 patients with less severe symptoms and in the early stages of the disease [4], although its use in more severe cases did not lead to significant clinical improvements [5]. Remdesivir is currently used in moderate and severe COVID‐19. Study results show that remdesivir may have therapeutic advantages in patients with severe COVID‐19 [6]. There are other alternative therapies, cell therapies against COVID‐19 such as the use of mesenchymal stem cells which have shown promising results [7] and T cells from convalescent COVID‐19 patients as a ‘living drug’ with promising results as well [8]. However, there is still room for new cell therapy approaches in order to treat this disease.
Natural killer (NK) cells are innate lymphoid cells (ILC) phenotypically described as CD3‐/CD56+ cells within the lymphocyte population [9]. ILCs act early in the immune response by reacting promptly to signals or inducer cytokines, expressed by tissue‐resident cells. ILCs are divided into groups 1, 2 and 3; NK cells, along with ILC1, form the ILC group 1 due to their similar function, especially in terms of cytokine output. They can be distinguished from the other ILC subsets by Tbet expression. NK cells and ILC1 differ mainly in their function; while ILC1 is weakly cytotoxic, NK cells are dedicated cytotoxic cells [10]. Their function is to defend the organism from viral infections [11], such as Epstein–Barr virus [12], varicellA−zoster virus [13], cytomegalovirus (CMV) [14] or herpesvirus‐6B [15] as well as fighting against malignant cells [16]; they have been reported to get to a functional exhaustion level along with disease progression, characterized by elevated expression of regulatory receptors such as PD1 and CD244 [17], lower levels of CD16 marker [18] and/or higher levels of NKG2A marker [19]. Peripheral blood NK cells can be divided in two different subsets regarding the expression of CD56: CD56bright and CD56dim cells [20]. These two subsets have different functions, as CD56bright cells are in charge of cytokine production that may modulate the immune responses [21], while CD56dim/CD16+ cells have cytotoxic activity through CD16‐activating receptor, which mediates antibody‐dependent cellular cytotoxicity (ADCC) [22, 23]. Moreover, there are many different NK cell subsets regarding the presence or absence of other markers such as NCR family (NKp46, NKp44 and NKp30) [24], CD94/NKG2A, KIR, [25] CD57 or NKG2C [26]. More than 95% of peripheral blood and 85% of spleen NK cells are CD56dimCD16+ [27]. Furthermore, NK cells are key pieces of the puzzle that integrates the innate and adaptive immune responses [28]. Although historically immunological memory was found in the adaptive part of the immune system, recent studies show that NK cells also own several characteristics of the immunological memory, as they have the capacity to respond at a higher magnitude to the exposure of certain stimuli [29]. In addition, NK cells share several features with B and T cells, such as a common lymphoid progenitor stem cell, undergoing several processes that are indications of adaptive immunity including developmental education clonal‐like expansion, memory cell generation and recall responses [30]. In fact, it has been proved that previously activated NK cells can produce long‐lived progeny able to mediate robust secondary responses in vitro and in vivo [31]. The particular subset of NK cells that express NKG2C+ is rising much interest for their unique cytotoxic potential against viral infections, such as CMV, hantavirus [32], chikungunya virus [33] and tumour cells, in comparison with their counterparts NKG2A+ NK cells [34].
The study of NK cell response to CMV shows an expansion of NKG2C+ NK cells [35], suggesting the implication of this receptor in the response to CMV infection [36]. The subset that expanded in response to CMV also expresses CD57 marker, leading to a unique population of CD57+/NKG2C NK cells [37]. This subset expands early after CMV infection and is very specific to the virus [38]. This subset expands in the post‐transplantation setting after CMV reactivation, especially when effective T lymphocytes are still not differentiated from the CD34+ progenitors’ cells, described to as memory‐like NKG2C+ NK cells [39]. NK cell rapidly expands and persists in response to hantavirus infection, along with the upregulation of NKG2C marker mediated by hantavirus‐infected endothelial cells [32]. NK cell exposure to varicellA−zoster virus (VZV) also leads to an increase in CD57 marker, providing NK cell maturity [13]. These patterns raise the possibility of an NK cell subset selectively responding against a specific pathogen and accruing memory [40].
In this study, we aimed at characterizing NK cells from convalescent donors that overcame COVID‐19 less than 6 months prior to their donation, separating them by severity of the disease, as well as the study of NK cell‐specific populations that respond to the presence of SARS‐CoV‐2 peptides. These NK cells could help to fight the infection, so that they could represent an off‐the‐shelf living drug for cell therapy to treat COVID‐19 patients under the RELEASE clinical trial (NCT04578210).
MATERIALS AND METHODS
Donors’ characteristics
This preclinical study based on the interferon gamma (IFN‐γ) release study included 6 COVID‐19 convalescent donors and 4 healthy controls (Table 1), and the immunophenotype study included 40 convalescent donors and 6 healthy controls (Table 2). All the COVID‐19 convalescent donors were tested for SARS‐CoV‐2. The median age of donors for IFN‐γ study was 49·6 years (range 30–60), all of them were men. Convalescent donors for the immunophenotype study were divided into disease severities, asymptomatic, mild (no hospitalization required), moderate (hospitalization required) and severe (ICU hospitalization required). There were 11 asymptomatic donors, with a median age of 42·7 years, and IgG median of 3·88 (range 1·09–10·6); 22 mild donors, with a median age of 42·5 years, and IgG median of 4·23 (range 0·17–8·74); 6 moderate donors, with a median age of 53·3 years, and IgG median of 6·05 (range 0·49–8·96); and one severe donor, age 56 and IgG 7·7. Healthy donors enrolled in these studies had not been exposed to SARS‐CoV‐2. All samples were collected through the Basque Biobank (http://www.biobancovasco.org) under an institutional review board‐approved protocol by the Basque Committee of Ethics and Clinical Research (PI2020063). The methods were carried out in accordance with the approved guidelines. The Basque Biobank complies with the quality management, traceability and biosecurity, set out in the Spanish Law 14/2007 of Biomedical Research and in the Royal Decree 1716/2011. All study subjects were provided written informed consent.
TABLE 1.
Characteristics of convalescent donors used for IFN‐γ assay in response to SARS‐CoV‐2 peptides
| Donor | Age | Sex | IgG | Blood type | Time after COVID−19 | Severity |
|---|---|---|---|---|---|---|
| D1 | 50 | M | 5·9 | 0− | – | |
| D2 | 58 | M | 5·21 | 0+ | 4 months | |
| D2 bis | 58 | M | 0+ | 9 months | ||
| D3 | 30 | M | 5·24 | A+ | 3 months | Moderate |
| D4 | 41 | H | 7·62 | A− | – | |
| D5 | 59 | H | 6·7 | A+ | 5 months | |
| D6 | 60 | H | 7·1 | A+ | – |
TABLE 2.
Characteristic of convalescent donors for immunophenotyping
| Severity | Donor ID | Age | Sex | IgG | Blood type |
|---|---|---|---|---|---|
| Asymptomatic | A1 | 45 | M | 3·02 | 0+ |
| A2 | 52 | M | 1·09 | A+ | |
| A3 | 34 | M | 10·6 | 0+ | |
| A4 | 51 | M | 1·68 | B‐ | |
| A5 | 32 | F | 5·6 | 0+ | |
| A6 | 64 | F | 5·52 | A− | |
| A7 | 59 | M | 1·61 | A+ | |
| A8 | 22 | F | 2·33 | A+ | |
| A9 | 42 | M | 2·07 | A+ | |
| A10 | 18 | M | 6·64 | 0+ | |
| A11 | 51 | M | 2·56 | A+ | |
| MILD | M1 | 29 | M | 4·76 | 0+ |
| M2 | 48 | M | 7·38 | 0+ | |
| M3 | 31 | F | 8·74 | A− | |
| M4 | 48 | M | 7·65 | 0+ | |
| M5 | 31 | M | 5·26 | A+ | |
| M6 | 49 | M | 3·79 | 0+ | |
| M7 | 46 | M | 1·74 | A+ | |
| M8 | 43 | M | 1·38 | 0+ | |
| M9 | 54 | F | 4·18 | A+ | |
| M10 | 59 | M | 3·03 | 0+ | |
| M11 | 26 | F | 1·92 | A+ | |
| M12 | 41 | M | 1·27 | A+ | |
| M13 | 48 | M | 0·17 | 0− | |
| M14 | 57 | M | 7·45 | 0+ | |
| M15 | 58 | M | 2·14 | A+ | |
| M16 | 46 | F | 3·16 | A− | |
| M17 | 33 | F | 5·52 | A+ | |
| M18 | 45 | M | 6·83 | A+ | |
| M19 | 53 | M | 1·81 | A+ | |
| M20 | 19 | M | 0·76 | A− | |
| M21 | 30 | M | 1·9 | A+ | |
| M22 | 41 | M | 5·62 | A− | |
| Moderate | Mo1 | 57 | M | 6·78 | 0− |
| Mo2 | 59 | M | 7·42 | 0+ | |
| Mo3 | 47 | M | 6·44 | 0− | |
| Mo4 | 56 | M | 8·96 | A+ | |
| Mo5 | 57 | M | 6·22 | 0+ | |
| Mo6 | 44 | M | 0·49 | 0+ | |
| Severe | S1 | 56 | M | 7·7 | A+ |
The acronyms correspond as following: A – asymptomatic; M – mild; Mo – moderate; S – severe.
Cell processing and immunophenotype of convalescent donors’ NK cells determined by flow cytometry assay
Peripheral blood mononuclear cells (PBMCs) from COVID‐19 convalescent and healthy donors were obtained from blood samples (buffy coat and/or EDTA 9‐ml tubes) by density gradient using Ficoll™ Paque Plus (GE Healthcare 17‐1440‐02). Cells were frozen in FBS (Gibco) +10% of DMSO using slow freeze method; after 48 h, cells were stored in liquid nitrogen. For immunophenotype studies, PBMCs were thawed and rested for 1 h at 37°C. Next, cells were stained for 30 min at 4°C using the antibodies listed in Table S1. Cell acquisition was performed using a BD FACSCanto™ II, acquiring an average of 200,000 cells. The analysis was performed using Flow Jo 10·5.3 (Flow Jo LLC).
NK cell response to SARS‐CoV‐2 peptides by IFN‐γ detection by flow cytometry
PBMCs from convalescent and healthy donors were thawed and rested for 1 h at 37°C. Next, NK cells were purified using NK cell isolation kit human (Miltenyi 130‐092‐657). NK cells were plated at a density of 1*106 cells per millilitre and rested overnight (o/n) at 37°C in RPMI (Gibco 72400‐021), 10% AB serum (Innovative Research, Inc. ISERABHI100ml‐30238), 1% penicillin/streptomycin (Gibco 15140‐122) and 1% GlutaMAX (Gibco 35050‐061). The following day, cells were harvested and plated in a 96‐well plate at 10*106/ml, plating 100 µl per well. For positive control, 1*106 of the cells were stimulated by adding 500 U/ml of IL‐2 (Miltenyi 130‐097‐745) and 20 ng/ml of IL‐15 (Miltenyi 130‐095‐762) to the culture medium. NK cells were stimulated with individual and pooled SARS‐CoV‐2 peptides (M, N, S) (Miltenyi) [41] at a final concentration of 0·6 nmol/ml. NK cells with no cytokines or peptides were used as a negative control. After 5 h of stimulation at 37°C, cells were labelled with anti‐CD3 FITC (clone OKT3; BD Bioscience), anti‐CD56 PE‐Cy7 (clone MEM‐188; BioLegend) and anti‐CD16 BV421 (clone 3G8; BD Bioscience) for detecting NK cell subset; anti‐C57 APC (clone NK‐1; BD Bioscience) and anti‐NKG2C BV510 (clone 134591; BD Bioscience) for detecting ‘activated/memory’ NK cells; anti‐CD62L and anti‐NKG2A for detecting naive NK cells and 7AAD PerCP‐Cy5·5 (BD Bioscience) viability marker. The peptides cover the immunodominant sequence domains of the surface glycoprotein S, the complete sequence of the nucleocapsid phosphoprotein N and the membrane glycoprotein M (GenBank MN908947·3, Protein QHD43416·1, Protein QHD43423·2, Protein QHD43419·1; Miltenyi Biotec, Germany) [8]. Then, NK cells were permeabilized and labelled with anti‐IFN‐γ PE (Miltenyi Biotec) clone antibody. Cells were analysed by flow cytometry.
Freezing and thawing effect on purified NK cells by degranulation assay
PBMCs from healthy donor buffy coats were obtained by density gradient using Ficoll™ Paque Plus. Then, NK cells were purified using NK cell isolation kit human. 2*106 of the NK cells were plate and rested o/n at 37°C in RPMI, 10% AB serum, 1% penicillin/streptomycin, 1% GlutaMAX, 500 U/ml of IL‐2 and 10 ng/ml IL‐15. The following day, a degranulation assay was performed with these NK cells. Briefly, NK cells were cocultured with K562 target cells at a ratio of 1:1 in a 24‐well plate for 4 h at 37°C. At the beginning of the assay, anti‐CD107a BV421 (clone H4A3; BD Biosciences) was added in order to detect the degranulation activity of the effector cells against the target cells. Golgi Stop™ (BD Biosciences) (monensin) was added following the manufacturer's protocol. After the incubation, cells were collected, washed and labelled with anti‐CD3‐PerCP/Cy5·5 and anti‐CD56‐APC. Degranulating NK cells (CD107a+) were determined in the CD56+/CD3− cells. Moreover, NK cells were labelled with previously mentioned, ‘memory/activated NK cells panel’. The rest of the NK cells were frozen using several freezing media: freezing medium 1 (FM1) (human plasma +5% dimethyl sulfoxide (DMSO)), freezing medium 2 (FM2) (human plasma +10% DMSO) or freezing medium 3 (FM3) (50% plasmalyte +40% AB serum +10% DMSO). 2*106 of NK cells were frozen per cryovial. 5 days later, NK cells from the 3 different conditions were thaw and performed the same assays.
Donor selection, human leukocyte antigen typing, killer immunoglobulin‐like receptors typing and large‐scale NK cell purification
The criteria for selecting convalescent donors for leukapheresis were as follows: (1) specific immunophenotype which leads to IFN‐γ secretion in response to SARS‐CoV‐2‐specific peptides (M, N, S) and (2) time passed since COVID‐19 infection. In addition, HLA and KIR genotype of the convalescent donors was performed at the Basque Center for Blood Transfusion and Human Tissues (Basque Country, Spain) by NGS and Luminex, respectively. The first chosen donor genotype was the following HLA: A*23:01P, A*30:02P/B*35:01P,B*49:01P/C*04:01P,C*07:01P/DRB1*13:02P,DRB1*15:01P/DQB1*06:02P,DQB1*06:09/DPB1*04:01P,DPB1*04:01P. KIR: KIR2DL1, KIR2DL3, KIR2DL4, KIR2DL5, KIR2DP1, KIR2DS1, KIR2DS2, KIR2DS4, KIR2DS5, KIR3DL1, KIR3DL2, KIR3DL3, KIR3DP1, KIR3DS1; haplotype AB/AB. The second chosen donor genotype was the following HLA: A*01:01P, A*01:01P/B*44:05P,B*55:01P/C*01:02P,C*02:02P/DRB1*07:01P,DRB1*13:01P/DQB1*03:03P,DQB1*06:03/DPB1*04:01P,DPB1*14:01P. KIR: KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL4, KIR2DP1, KIR2DS2, KIR2DS4, KIR3DL1, KIR3DL2, KIR3DL3, KIR3DP1; haplotype AB/AA.
Non‐mobilized apheresis was performed at the Basque Center for Blood Transfusion and Human Tissues (Galdakao, Spain), and the NK cell product enrichment was performed at the Bone Marrow Transplantation and Cell Therapy Unit of University Hospital La Paz (Madrid, Spain) using a CliniMACS Plus cell separation system (Miltenyi Biotec). The donors provided written informed consent, and the study was conducted according to the Declaration of Helsinki protocol and the guidelines of the local ethics committee (Basque Committee of Ethics and Clinical Research (PI2020063) and IRB number 5579). Both units, Basque Center for Blood Transfusion and Human Tissues and Bone Marrow Transplantation and Cell Therapy Unit of University Hospital La Paz, were responsible for complying with the requirements regarding the quality and safety of the donation, obtaining, storage, distribution and cryopreservation of human blood cells and tissues under the Spanish‐specific regulation. Following apheresis, CD56+ cells were purified following a two‐step protocol: first, CD3 population was depleted, and next, CD56 population was enriched by immunomagnetic separation using CliniMACS CD3/CD56 Complete Kit and the CliniMACS Plus system (both from Miltenyi Biotec), following the manufacturer's instructions. CD3−/CD56+ cells were frozen using the following freezing medium: 50% plasmalyte, 40% autologous plasma and 10% DMSO and stored. We were able to cryopreserve 5 aliquots at 1*106 cells/kg body weight for a 100 kg patient. The purity of the CD3‐/CD56+ was analysed by flow cytometry.
Statistical analysis
The quantitative variables are expressed as mean ± standard error of the mean (SEM), and qualitative variables are expressed as percentages (%). An unpaired t‐test was used for comparison using GraphPad Prim 7 (GraphPad Software, San Diego, CA) A p‐value < 0·05 was considered statistical significant.
RESULTS
Immunophenotype of COVID‐19 convalescent NK cells
After analysing different NK markers previously described in “materials and methods” (see Table S1), including FMO for each antibody, from COVID‐19 convalescent donors with different severities within six months post‐infection, we observed several differences between them. NKG2C marker is downregulated in asymptomatic and moderate donors, although there are not significant differences. CD57 expression is very similar severity groups, except in the severe case, which is higher than healthy controls and all the conditions. CD16 expression is significantly downregulated in asymptomatic, mild and moderate donors, comparing to healthy donors; it is also significantly lower in mild donors comparing to moderate donors (Figure 1a). Next, NKp30 levels are very similar in all the severity groups, while NKG2D is downregulated in asymptomatic and moderate donors, with no significant differences. On the other hand, NKp46 is highly significantly downregulated in all convalescent groups, being inexistent expression in severe condition. Its expression is also significantly lower in asymptomatic donors in comparison with mild donors (Figure 1b). The expression of CD158a, CD158b and CD158e is very similar in all severity groups. Meanwhile, NKG2A is highly upregulated in the severe donor (Figure 1c). Finally, we barely got any expression of the CTLA4, TIGIT and TIM3 markers. PD1 marker is significantly downregulated in asymptomatic and moderate donors in comparison with mild donors and healthy donors (Figure 1d). Gating strategy was done based on lymphocyte population, live cells, single cells, CD3− cells and CD56+ cells (Figure S1a). No antibody overlaid as FMO showed fluorescence overlapping (Figure S1b).
FIGURE 1.
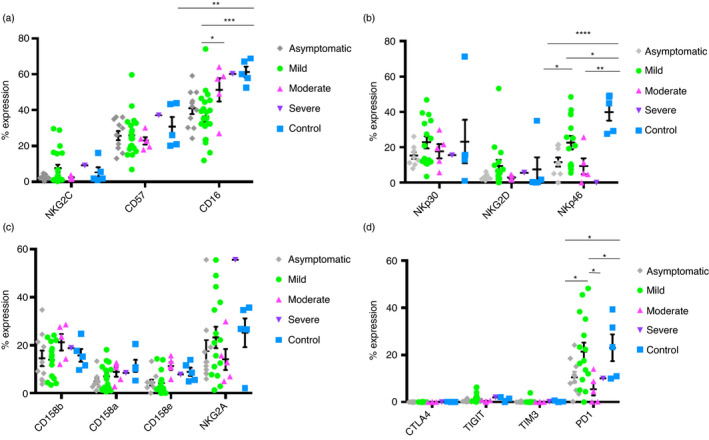
Comparison of the expression levels of different surface markers in NK cells of COVID‐19 convalescent donors of different severities. (a) Expression of CD57, NKG2C and CD16 markers in asymptomatic, mild, moderate, severe convalescent donors and healthy donors. (b) Expression of NKp30, NKG2D and NKp46 markers in asymptomatic, mild, moderate, severe convalescent donors and healthy donors. (c) Expression of CD158b, CD158a, CD158e and NKG2A markers in asymptomatic, mild, moderate, severe convalescent donors and healthy donors. (d) Expression of CTLA4, TIGIT, TIM3 and PD1 markers in asymptomatic, mild, moderate, severe convalescent donors and healthy donors. Asymptomatic (n = 11), mild (=22), moderate (n = 6), severe (n = 1) convalescent donors and healthy (n = 6) donors. Each dot corresponds to an individual, and the mean with the standard error of the mean (SEM) is shown. Student's t‐test was used to analyse the data. p‐value: *p < 0·05, **p < 0·005, ***p < 0·001. ****p < 0·0001
Characterization of IFN‐γ and memory naive COVID‐19 convalescent NK cells
We recovered less percentage of NK cells from all COVID‐19 convalescent donor samples (4·34 ± 2·1) than from healthy donor samples (5·6 ± 2) (Figure 2a). We analysed the specific response of convalescent donors NK cells in the presence of SARS‐CoV‐2 peptides (Figure 2b). We observed no response in negative control, as well as in healthy donors (Figure S2). Not all the peptides lead to the same response in the NK cell population, secreting different levels of IFN‐γ within the same convalescent donor (Figure 2e). Regarding cytolytic (CD56+/CD16−) and cytotoxic (CD56+/CD16+) NK cells, we observed in the best responder (D2) that the cytolytic population are releasing the IFN‐γ for the most part (Figure 2b). We also observed a general decrease in the CD16+ population, except for two donors (D3 and D5); when it is gating within the CD57+/NKG2C+ population (Figure 2c) in the responder donors, it is even lower, except for D5. Moreover, when analysing the CD57+/NKG2C+ population within the IFN‐γ secreting NK cells, most of the CD16+ cells are not releasing IFN‐γ, except for D5 (Figure 2d). We obtained samples from D2 at different times during COVID‐19 recovery, at 4 months (D2) and 9 months (D2 bis), and we observed a decrease in the specificity and amount of the response. In addition, the specific response to SARS‐CoV‐2 peptides is not developed in all the convalescent donors, being a donor‐dependent feature (Figure 2e). This response is related to the presence of a ‘activated/memory’ NK cell population, in which the presence of a robust CD57+/NKG2C+ population leads to a better response against the SARS‐CoV‐2 peptides (Figure 3a). We also observed a decrease in this population in the D2 samples as times went by. However, in despite of this population being a good indicator, it seems to not be the only way of action, D1 has a good response, but lack this ‘activated/memory’ NK cell population. Moreover, convalescent donors with lower response to SARS‐CoV‐2 peptides have a higher percentage of naive NK cell markers such as CD62L and NKG2A, regardless COVID‐19 severity (Figure 3b).
FIGURE 2.
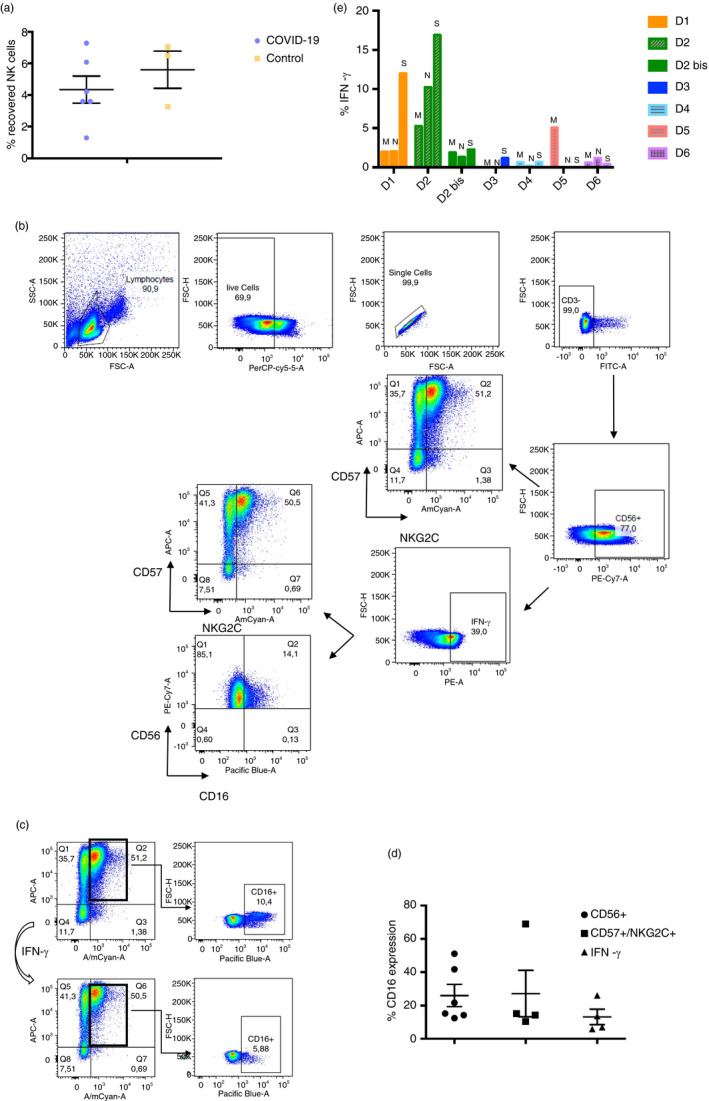
Response of convalescent donor NK cells to SARS‐CoV‐2‐specific peptides measured by IFN‐γ production. (a) Percentage of NK cells recovered from PBMCs of blood samples, in purple from convalescent donors (n = 6), in yellow from healthy donors (n = 3). Each dot corresponds to an individual, and the mean with the standard error of the mean (SEM) is shown. (b) Representative figure of gating strategy of the different cell subsets within de CD3‐ CD56+ population (NK cells). On the right below, figure expression of IFN‐γ production by healthy donors’ NK cells. (c) Representative figure of CD16+ population analysis within the CD57+/NKG2C+ population (top), and the CD57+/NKG2C+/IFN‐γ+ population (bottom). (d) Percentage of CD16+ population expression within CD56+ subset, CD57+/NKG2C+ subset and IFN‐γ+ subset of convalescent donors. Each dot corresponds to an individual, and the mean with the standard error of the mean (SEM) is shown. (e) Percentage of IFN‐γ production done by convalescent donors’ NK cells in response to the presence of three different SARS‐CoV‐2 peptides (M, N and S)
FIGURE 3.
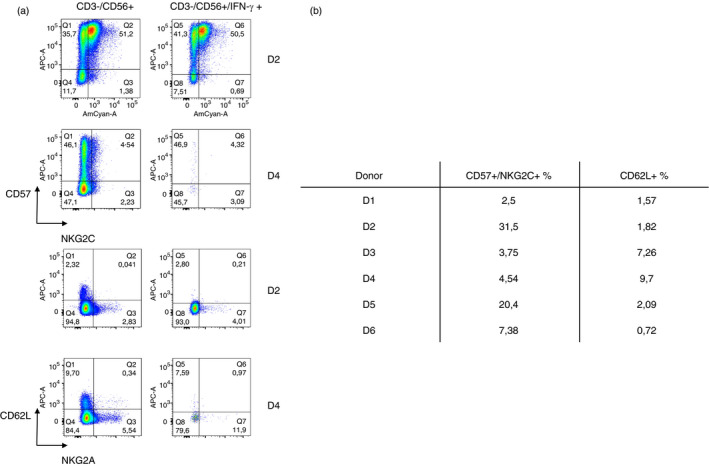
Expression of the activated population CD57 + NKG2C+ and the naive NK cells population CD62L+ NKG2A+ in the convalescent donors. (a) Representative figure of the expression of CD57 + NKG2C+ and the naive NK cell population CD62L+ NKG2A+ of the best responder (D2) and the worst responder (D4). On the left column, these populations are contained within the CD3‐ CD56+ population, and on the right column, these populations are contained within the CD3‐ CD56+ IFN‐γ + population. (b) Table summarizing the percentage of CD57+ NKG2C+ and CD62L+ in each convalescent donor of IFN‐γ production assay
CliniMACS depletion and enrichment
Apheresis product contained 41·72% of CD3+/CD56− population, 0·84% of CD3+/CD56+ population and 6·98% of CD3−/CD56+ population (NK cells). After depletion of the CD3+ cells (see Materials and Methods section), 1·07% of the cells were CD3+. NK cell percentage was 12·05. Next, after CD56+ enrichment, we obtain a purified NK cell population of 93%.
Immunophenotype of COVID‐19 convalescent NK cells after CliniMACS
A blood sample was taken from the donor at day of the leukapheresis. The final cell product had a 96·62 ± 4·1% of NK cell purified population (CD56+/CD3−). Similar data as we previously reported [42]. The remaining cells from the final product of the NK cell purification were frozen in freezing medium 3 (FM3) in cryovials containing 2*106 cells. Flow cytometry analysis was performed of these two samples. We observed a CD57+/NKG2C+ population of 8·76% before the CliniMACS purification, and a 29·5% population after purification (Figure 4a). Moreover, cytotoxic NK cell population (CD56+/CD16+) was higher after the NK cell purification procedure (57%), while before was 23·7%. (Figure 4b). There was an increase on rest of the markers studied in the phenotype section after CliniMACS purification of the NK cells, except for TIM3 (Figure 4c).
FIGURE 4.
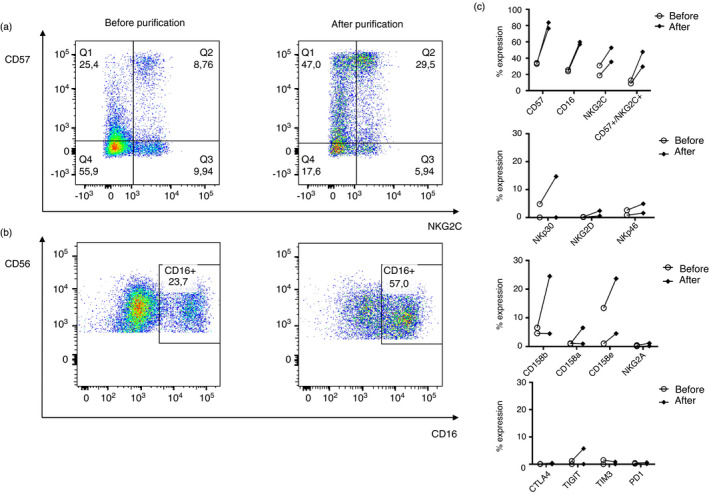
Flow cytometry analysis of NK cells from leukapheresis donors before and after the purification of NK cells. (a) Representative figure of CD57+ NKG2C+ activated population expression on NK cells. On the left, before the purification, and on the right, after purification. (b) Representative figure of CD16+ NK cell maturation marker expression. On the left, before the purification, and on the right, after purification. (c) Comparison of the expression levels of different surface markers in NK cells before and after CliniMACS NK cell purification. (n = 2). Each dot corresponds to an individual, and the mean with the standard error of the mean (SEM) is shown
HLA and KIR genotype of COVID‐19 convalescent donors
We obtained a high‐resolution HLA profile from the convalescent donors (Table S2). We observe a higher presence in DRB1:15:01 allele as well as in DQB1:06:02 allele, sometimes being co‐expressed in the same individual due to linkage disequilibrium and proximity of genes. We also observe a more numerous appearance of HLA*C16 and DPB1:04:01.
Regarding KIR gene expression, it is quite significant the amount of donors that do no express KIR2DS3. We did not find a pattern in KIR haplotype regarding the severity of the disease (Table S3).
Comparative study to choose the best freezing medium for COVID‐19 convalescent NK cell therapy
We tried three different freezing media (FM) for preserving properly the purified NK cells. Although viability dropped with all three different media, FM3 preserved better the percentage of living cells after thawing (Figure 5a). We also checked the maintenance of the functionality and the ‘active/memory’ population in all samples thawed. As we can observe in Figure 5b and c, in spite of the decreasing of these two features, FM3 is still the best freezing medium for preserving these qualities. Finally, we checked the percentage of cytotoxic NK cells (CD56 dim/CD16+), barely noticing any significant drop (Figure 5d).
FIGURE 5.
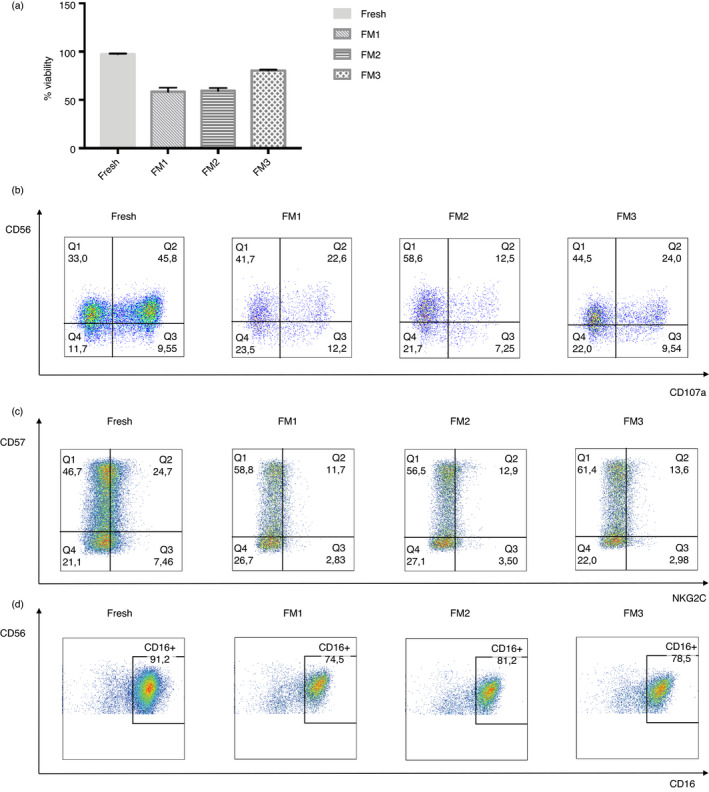
Comparison of the condition of NK cells without freezing and after thawing with different freezing media (FM). (a) Viability of the NK cells in the fresh, FM1, FM2 and FM3 condition (n = 2). (b) Representative figure of the functionality measured by degranulation assay against K562 target cells in the four different conditions. (c) Representative figure of CD57+ NKG2C+ activated population expression on NK cells in the four different conditions. (d) Representative figure of CD16+ marker expression in the four different conditions. The bars represent the mean and error bars represent SEM
DISCUSSION
Due to their specific features, NK cells are a very interesting source for immunotherapy. In fact, there are several experimental trials in which NK cells are used in order to boost the immune system to fight against different diseases. Autologous or allogenic peripheral blood NK cells can be purified and stimulate ex vivo to treat cancer patients in risk of receiving chemotherapy or relapsed [43]. NK cells are also infused after human leukocyte antigen (HLA)‐haploidentical hematopoietic cell transplantation, and they seem to decrease post‐transplantation progression of acute leukaemia [44]. Recently, umbilical cord NK cells have been used along with CAR‐based therapy, as CAR‐NK, cells to treat B‐cell malignancies [45]. Although most of the applications are directed to treat neoplastic malignancies, NK cells also play a key role fighting viral infections [46], so that, these cells could benefit patients affected with some viruses, such as SARS‐CoV‐2. In fact, there are several clinical trials using NK cell‐based products and NK cell stimulants to fight COVID‐19 disease [47].
As we observe in this work, the specific NK cell response to SARS‐CoV‐2 is donor dependent, being crucial to find key characteristics that will allow to predict a good response against this virus. CD16 marker is described to be downregulated in COVID‐19 patients, being more pronounced in severe patients [48]. This marker is still in significantly lower percentage in asymptomatic, mild and moderate convalescent donors; that is, they did not recover yet the basal levels of CD16. NKG2D, NKp30 and NKp46 receptors work as direct activators, targeting virus‐infected cells [24, 49]. Elevated IL‐6 in SARS‐CoV‐2‐infected patients reduces the NKG2D expression, diminishing the capacity of NK cells of killing viral‐infected cells [50]. In our study, asymptomatic and moderate convalescent donors have not recover NKG2D basal expression yet. We found that NKp46 receptor is also significantly downregulated in all the conditions of the convalescent donors. Even though NKG2A marker is known to be overexpressed in COVID‐19 patients [51], several months after passing the disease, we observed that convalescent donors recover the basal levels of this marker, except for the severe convalescent donor, whose NKG2A expression is still upregulated. In vivo, NK cell anti‐tumour function is suppressed by the presence of immune check point molecules such as PD1 [52]. We found that in asymptomatic, moderate and severe donors, this marker is downregulated, while in mild donors, its expression has already been restored. The decrease in expression of this marker could facilitate the activation of NK cells during viral infection.
CD57+NKG2C+ NK cell population is detected in higher frequencies in COVID‐19 patients, and it has been reported to increase along with severity of the disease [53]. The co‐expression of CD57 and NKG2C surface markers is a phenotypic signature coinciding with that observed in adaptive NK cells [54], characterizing mature NK cells and appearing in different viral infections such as CMV [55]. Frequency of CD57+ NK cell population is also increased when culture them with VZV, suggesting a maturation when facing viral infections [40]. Upregulation of NKG2C marker has been observed in chronic hepatitis B virus (HBV) and HCV infections. The activation of this receptor could occur due to a viral imprinting on NK cells [56]. In our study, we observed that convalescent donor NK cells seem to present a more pro‐inflammatory phenotype, as CD16 marker is expressed in lower rates than expected [57]. This pro‐inflammatory phenotype, characterize by the downregulation of CD16 marker, might represent an important mechanism for regulating NK cell function [58], as NK cells rapidly enter a refractory period where CD16 molecules are shed from the surface of the cells after NK cell activation through CD16 receptor [59]. As other studies show, this pro‐inflammatory environment usually leads to an enhancement of the HLA‐E expression (Figure 6a) [60]. We also noticed a correlation between the presences of double CD57/NKG2C‐positive cells with specific immunoresponse against SARS‐CoV‐2 peptides. Previous studies in mouse models of NK activating receptor Ly49H and m157 murine CMV (MCMV) glycoprotein have shown the importance of activating receptor‐induced activation to maintain the NK cell population during the prolonged MCMV infection [61]. It is suggested that in humans, Ly49H receptors correlate with NKG2C expression, in reactive memory cells [62]. HLA‐E interaction with NKG2C marker leads to an activation of the NK cell (Figure 6b) [63], which could influence an adaptive status of the NK cell, also expressing CD57 maturation marker. Taking these data into account, we suggest that adaptive/‘memory’ NK cells are capable of recognizing soluble‐specific peptides through NKG2C receptor instead of using the activation of NKG2C via HLA‐E (Figure 6c) [64], and this could be the mechanism for which some of the convalescent donors respond to the presence of SARS‐CoV‐2 peptides, by secreting IFN‐γ (Figure 6d). Moreover, the higher the expression of the double population, the better response we obtain. However, D1 responded satisfactorily to the presence of the peptides without having this ‘adaptive’ NK cell population. This could lead to think that this is not the only parameter that will dictate a response, but it is one of the more reliable ones, as the cells that express these markers respond to the presence of the virus. However, most of the phenotypically analysed convalescent donors do not present this CD57+/NKG2C+ population showing that not all of them acquire an adaptive/‘memory’ function within their NK cells, being more similar to healthy donors’ levels.
FIGURE 6.
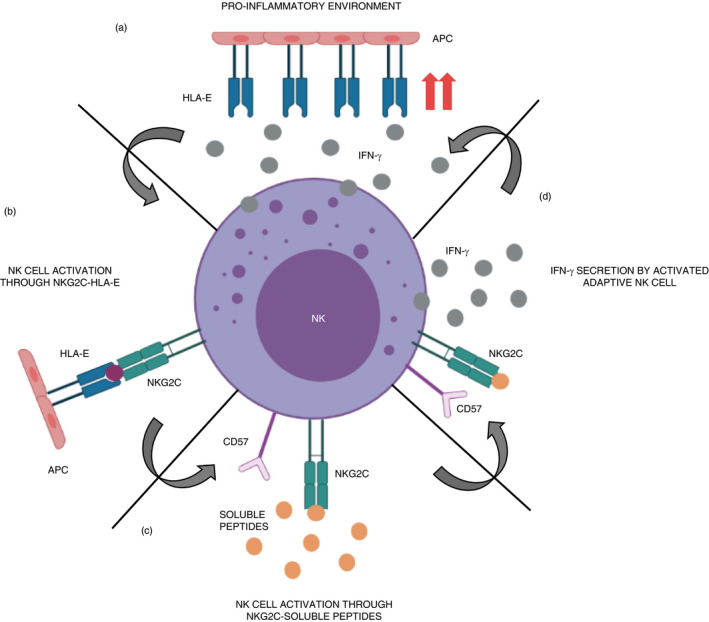
Potential mechanism of adaptive/‘memory’ NK cells with CD57+/NKG2C+ phenotype activation in SARS‐CoV‐2 infection. (a) Pro‐inflammatory environment with NK cells secretes IFN‐γ that leads to an increase in HLA−E expression. (b) NK cells expressing NKG2C are activated through this receptor by HLA−E presentation. This could lead to an adaptive NK cell. (c) Adaptive NK cells could be activated by the recognition of SARS‐CoV‐2‐specific soluble peptides via NKG2C, which could enter a pro‐inflammatory status secreting IFN‐γ. (d) Activated adaptive/‘memory’ NK cells secrete IFN‐γ in response to SARS‐CoV‐2 soluble peptide‐specific activation
Time passed after COVID‐19 recovery might be crucial in order to choose an optimal donor. As other studies suggest, SARS‐CoV‐2‐specific memory is persistent 1–3 months after infection [65]; another one shows that specific IgG starts decreasing at month 3 being lower at Month 6 after infection [66]. Actually, taking into account the different time points of D2, a strong decrease in the specific response is detected, suggesting that NK cells lose the specificity over time.
All these data suggest that the presence of a NK cell population co‐expressing CD57 and NKG2C, and the time passed from the infection are key points to select a convalescent donor for NK cell infusion to patients for our clinical trial: NCT04578210.
Regarding the status of the NK cells before and after the purification, it seems that the CliniMACS procedure activates the NK cells, making the CD57+ NKG2C+ population more noticeable, as well as increasing the cytotoxic CD16+ population. This could offer an advantage when transfusing this product to COVID‐19 patients.
HLA sequencing showed a high prevalence of DPB1:04:01 allele among convalescent donors. This allele has already been reported to be the most common one between Spaniard COVID‐19 patients [67]. Moreover, DRB1:15:01 and DQB1:06:02 alleles strongly present in Italian COVID‐19 population [68] have also a representative expression in our donors, sometimes being present at the same time, which is expectable due to linkage disequilibrium and proximity of genes. B39 and C16 alleles were found in higher rate in COVID‐19 patients than in healthy subjects [69], being present in some of our donors too. Although there has not been described a correlation between KIR gene expression and SARS‐CoV‐2 infection, there are some studies concerning other viruses. KIR3DS1 in homozygosis is more frequent in HIV‐negative population, suggesting a protective role [70]. However, half of the donors are positive for this gene, which does not imply a key role in the infection. KIR2DL3 expression in decreased in chronic hepatitis B patients [71], while in our donors is mainly present. There is a remarkable absence of expression of KIR2DS3, which has an activating role, among our donors. This could be a key prognosis for susceptibility to SARS‐CoV‐2 infection.
An optimal cryopreservation of purified NK cells is crucial for the ideal performance of the therapy. We tried three different freezing media, and plasmalyte with 40% AB serum and 10% DMSO resulted the best in order to preserve the viability and functionality. Other groups already used this medium with great results in conserving the NK cells [72]. Nevertheless, in most cases, fresh NK cells are infused, so that there are few studies regarding NK cell freezing media. Finding an optimum cryoprotectant for NK cells would help to drive more ‘off‐the‐shelf’ NK treatments.
Despite the effectiveness of different vaccines against SARS‐CoV‐2, there are still numerous COVID‐19 patients in need to be treated. With no gold standard in treatment, several clinical trials have emerged to cure this disease. Cell therapy has shown therapeutic efficacy against other viral respiratory infections, becoming an attractive approach for COVID‐19 [73]. Mesenchymal stem cells (MSCs) have been used due to their intense immunomodulatory effects, being a safe and effective treatment [7]. Cells from the immune system such as NK cells, T cells and macrophage are involved in several clinical trials for treating COVID [74, 75].
Taking all of these data into account, these allogenic activated NK cells could be very useful for the treatment of COVID‐19, providing the tools from trained immune system cells to attack virus‐infected cells, as it is already been proved with CD45RA− T memory cells [8, 76]. In fact, the study of this memory capacity of NK cells against certain viral infections might open a door for targeted cell therapy, giving patients a more personalized treatment against new emerging virus.
CONFLICT OF INTERESTS
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
L.H. collected and/or assembled the data, in vitro experiments, data analysis and interpretation, and manuscript writing and final approval of manuscript. M.M‐I., M‐I‐F., A.A. and M.P‐V. collected and/or assembled the data. S.S. involved in HLA and KIR typing. M.A.V. involved in final approval of manuscript and financial support. C.E. conceptualized and designed, collected and/or assembled the data, analysed and interpreted the data, wrote the manuscript and involved in final approval of manuscript. All authors revised the manuscript, participated in the interpretation of the data and the approval and submission of the manuscript.
Supporting information
Figure S1
Figure S2
Supplementary Material
ACKNOWLEDGEMENT
We apologize to our colleagues whose work was not cited due to space limitations.
Herrera L, Martin‐Inaraja M, Santos S, Inglés‐Ferrándiz M, Azkarate A, Perez‐Vaquero MA, et al. Identifying SARS‐CoV‐2 ‘memory’ NK cells from COVID‐19 convalescent donors for adoptive cell therapy. Immunology. 2022;165:234–249. 10.1111/imm.13432
Funding information
This work was supported by the Health Department of the Basque Government (Grant 2020111058 and 2020333032), Economic Development and Infrastructures Department of the Basque Government (KK‐2020/00068), Project ‘PI18/01299’ and ‘PI21/01187’, funded by Instituto de Salud Carlos III and co‐funded by European Union (ERDF) ‘A way to make Europe’, ‘ICI21/00095’ funded by Instituto de Salud Carlos III and co‐funded by European Union (NextGenerationEU), Inocente Inocente Foundation (FII18‐003‐CPS) and EITB Maratoia (BIO21/COV/030). LH was supported by the Jesus Gangoiti Barrera Foundation and the Asociación Española contra el Cáncer (AECC), the Fundación Mutua Madrileña (AP176182020).
REFERENCES
- 1. WHO . Coronavirus disease (COVID‐19) outbreak situation reports (available at https://www.who.int/emergencies/diseases/novel‐coronavirus‐2019
- 2. Hui DS, Azhar EI, Madani TA, Ntoumi F, Kock R, Dar O, et al. The continuing 2019‐nCoV epidemic threat of novel coronaviruses to global health – the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhao Q, Meng M, Kumar R, Wu Y, Huang J, Deng Y, et al. Lymphopenia is associated with severe coronavirus disease 2019 (COVID‐19) infections: a systemic review and metA−analysis. Int J Infect Dis. 2020;96:131–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stasi C, Fallani S, Voller F, Silvestri C. Treatment for COVID‐19: an overview. Eur J Pharmacol. 2020;889:173644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, et al. A trial of lopinavir‐ritonavir in adults hospitalized with severe covid‐19. N Engl J Med. 2020;382:1787–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goldman JD, Lye DCB, Hui DS, Marks KM, Bruno R, Montejano R, et al. G.‐U.‐540−5773 investigators, remdesivir for 5 or 10 days in patients with severe Covid‐19. N Engl J Med. 2020;383:1827–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leng Z, Zhu R, Hou W, Feng Y, Yang Y, Han Q, et al. Transplantation of ACE2(‐) mesenchymal stem cells improves the outcome of patients with COVID‐19 pneumonia. Aging Dis. 2020;11:216–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ferreras C, Pascual‐Miguel B, Mestre‐Durán C, Navarro‐Zapata A, Clares‐Villa L, Martín‐Cortázar C, et al. SARS‐CoV‐2‐specific memory T lymphocytes from COVID‐19 convalescent donors: identification, biobanking, and large‐scale production for adoptive cell therapy. Front Cell Dev Biol. 2021;9:620730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Van Acker HH, Capsomidis A, Smits EL, Van Tendeloo VF. CD56 in the immune system: more than a marker for cytotoxicity? Front Immunol. 2017;8:892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells: 10 years on. Cell. 2018;174:1054–66. [DOI] [PubMed] [Google Scholar]
- 11. Zwirner NW, Domaica CI, Fuertes MB. Regulatory functions of NK cells during infections and cancer. J Leukoc Biol. 2021;109:185–94. [DOI] [PubMed] [Google Scholar]
- 12. George LC, Rowe M, Fox CP. Epstein‐Barr virus and the pathogenesis of T and NK lymphoma: a mystery unsolved. Curr Hematol Malig Rep. 2012;7:276–84. [DOI] [PubMed] [Google Scholar]
- 13. Campbell TM, McSharry BP, Steain M, Ashhurst TM, Slobedman B, Abendroth A. Varicella zoster virus productively infects human natural killer cells and manipulates phenotype. PLoS Pathog. 2018;14:e1006999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Truitt LL, Yang D, Espinoza DA, Fan X, Ram DR, Moström MJ, et al. Impact of CMV infection on natural killer cell clonal repertoire in CMV‐Naïve Rhesus Macaques. Front Immunol. 2019;10:2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gasior M, Ferreras C, de Paz R, Bueno D, Mozo Y, Sisinni L, et al. The role of early natural killer cell adoptive infusion before engraftment in protecting against human herpesvirus‐6B encephalitis after naïve T‐cell‐depleted allogeneic stem cell transplantation. Transfusion. 2021;61:1505–17. [DOI] [PubMed] [Google Scholar]
- 16. Nicholson SE, Keating N, Belz GT. Natural killer cells and anti‐tumor immunity. Mol Immunol. 2019;110:40–7. [DOI] [PubMed] [Google Scholar]
- 17. Li M, Guo W, Dong Y, Wang X, Dai D, Liu X, et al. Elevated exhaustion levels of NK and CD8(+) T cells as indicators for progression and prognosis of COVID‐19 disease. Front Immunol. 2020;11:580237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Srpan K, Ambrose A, Karampatzakis A, Saeed M, Cartwright ANR, Guldevall K, et al. Shedding of CD16 disassembles the NK cell immune synapse and boosts serial engagement of target cells. J Cell Biol. 2018;217:3267–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, et al. Functional exhaustion of antiviral lymphocytes in COVID‐19 patients. Cell Mol Immunol. 2020;17:533–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Montaldo E, Del Zotto G, Della Chiesa M, Mingari MC, Moretta A, De Maria A, et al. Human NK cell receptors/markers: a tool to analyze NK cell development, subsets and function. Cytom Part A. 2013;83A:702–13. [DOI] [PubMed] [Google Scholar]
- 21. Keating SE, Zaiatz‐Bittencourt V, Loftus RM, Keane C, Brennan K, Finlay DK, et al. Metabolic reprogramming supports IFN‐γ production by CD56bright NK cells. J Immunol. 2016;196:2552–60. [DOI] [PubMed] [Google Scholar]
- 22. Zhang C, Hu Y, Shi C. Targeting natural killer cells for tumor immunotherapy. Front Immunol. 2020;11:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Romee R, Foley B, Lenvik T, Wang Y, Zhang B, Ankarlo D, et al. NK cell CD16 surface expression and function is regulated by a disintegrin and metalloprotease‐17 (ADAM17). Blood. 2013;121:3599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barrow AD, Martin CJ, Colonna M. The natural cytotoxicity receptors in health and disease. Front Immunol. 2019;10:909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marcenaro E, Notarangelo LD, Orange JS, Vivier E. Editorial: NK cell subsets in health and disease: new developments. Front Immunol. 2017;8:1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dogra P, Rancan C, Ma W, Toth M, Senda T, Carpenter DJ, et al. Tissue determinants of human NK cell development, function, and residence. Cell. 2020;180:749–763 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ferlazzo G, Thomas D, Lin S‐L, Goodman K, Morandi B, Muller WA, et al. The abundant NK cells in human secondary lymphoid tissues require activation to express killer cell Ig‐like receptors and become cytolytic. J Immunol. 2004;172:1455–62. [DOI] [PubMed] [Google Scholar]
- 28. Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or adaptive immunity? Example of natural killer cells. Science. 2011;331:44–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cerwenka A, Lanier LL. Natural killer cell memory in infection, inflammation and cancer. Nat Rev Immunol. 2016;16:112–23. [DOI] [PubMed] [Google Scholar]
- 30. Sun JC, Lopez‐Verges S, Kim CC, DeRisi JL, Lanier LL. NK cells and immune “memory”. J Immunol. 2011;186:1891–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sun JC, Lanier LL. Versatility in NK cell memory. Immunol Cell Biol. 2011;89:327–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Björkström NK, Lindgren T, Stoltz M, Fauriat C, Braun M, Evander M, et al. Rapid expansion and long‐term persistence of elevated NK cell numbers in humans infected with hantavirus. J Exp Med. 2010;208:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Petitdemange C, Becquart P, Wauquier N, Béziat V, Debré P, Leroy EM, et al. Unconventional repertoire profile is imprinted during acute chikungunya infection for natural killer cells polarization toward cytotoxicity. PLoS Pathog. 2011;7:e1002268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zuo W, Zhao X. Natural killer cells play an important role in virus infection control: Antiviral mechanism, subset expansion and clinical application. Clin Immunol. 2021;227:108727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gumá M, Budt M, Sáez A, Brckalo T, Hengel H, Angulo A, et al. Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus‐infected fibroblasts. Blood. 2006;107:3624–31. [DOI] [PubMed] [Google Scholar]
- 36. Djaoud Z, Riou R, Gavlovsky P‐J, Mehlal S, Bressollette C, Gérard N, et al. Cytomegalovirus‐infected primary endothelial cells trigger NKG2C+ natural killer cells. J Innate Immun. 2016;8:374–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lopez‐Vergès S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA, et al. Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci USA. 2011;108:14725–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Paust S, Blish CA, Reeves RK. Redefining memory: building the case for adaptive NK cells. J Virol. 2017;91:e00169–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bigley AB, Rezvani K, Shah N, Sekine T, Balneger N, Pistillo M, et al. Latent cytomegalovirus infection enhances anti‐tumour cytotoxicity through accumulation of NKG2C+ NK cells in healthy humans. Clin Exp Immunol. 2016;185:239–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Newhook N, Fudge N, Grant M. NK cells generate memory‐type responses to human cytomegalovirus‐infected fibroblasts. Eur J Immunol. 2017;47:1032–9. [DOI] [PubMed] [Google Scholar]
- 41. Sattler A, Angermair S, Stockmann H, Heim KM, Khadzhynov D, Treskatsch S, et al. SARS‐CoV‐2‐specific T cell responses and correlations with COVID‐19 patient predisposition. J Clin Invest. 2020;130:6477–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fernández L, Leivas A, Valentín J, Escudero A, Corral D, de Paz R, et al. How do we manufacture clinical‐grade interleukin‐15–stimulated natural killer cell products for cancer treatment? Transfusion. 2018;58:1340–7. [DOI] [PubMed] [Google Scholar]
- 43. Chabannon C, Mfarrej B, Guia S, Ugolini S, Devillier R, Blaise D, et al. Manufacturing natural killer cells as medicinal products. Front Immunol. 2016;7:504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chamorro‐Viña C, Valentín J, Fernández L, González‐Vicent M, Pérez‐Ruiz M, Lucía A, et al. Influence of a moderate‐intensity exercise program on early NK cell immune recovery in pediatric patients after reduced‐intensity hematopoietic stem cell transplantation. Integr Cancer Ther. 2017;16:464–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu E, Marin D, Banerjee P, Macapinlac HA, Thompson P, Basar R, et al. Use of CAR‐transduced natural killer cells in CD19‐positive lymphoid tumors. N Engl J Med. 2020;382:545–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Björkström NK, Strunz B, Ljunggren H‐G. Natural killer cells in antiviral immunity. Nat Rev Immunol. 2021. 10.1038/s41577-021-00558-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Market M, Angka L, Martel AB, Bastin D, Olanubi O, Tennakoon G, et al. Flattening the COVID‐19 curve with natural killer cell based immunotherapies. Front Immunol. 2020;11:1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu Y, Huang X, Sun J, Xie T, Lei Y, Muhammad J, et al. Clinical Characteristics and Immune Injury Mechanisms in 71 Patients with COVID‐19. mSphere 2020;5:e00362–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Brandstadter JD, Yang Y. Natural killer cell responses to viral infection. J Innate Immun. 2011;3:274–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Osman MS, van Eeden C, Cohen Tervaert JW. Fatal COVID‐19 infections: Is NK cell dysfunction a link with autoimmune HLH? Autoimmun Rev. 2020;19:102561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yaqinuddin A, Kashir J. Innate immunity in COVID‐19 patients mediated by NKG2A receptors, and potential treatment using Monalizumab, Chloroquine, and antiviral agents. Med Hypotheses. 2020;140:109777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Del Zotto G, Marcenaro E, Vacca P, Sivori S, Pende D, Della Chiesa M, et al. Markers and function of human NK cells in normal and pathological conditions. Cytom Part B Clin Cytom. 2017;92:100–14. [DOI] [PubMed] [Google Scholar]
- 53. Maucourant C, Filipovic I, Ponzetta A, Aleman S, Cornillet M, Hertwig L, et al. Natural killer cell immunotypes related to COVID‐19 disease severity. Sci. Immunol. 2020;5:eabd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Béziat V, Liu LL, Malmberg J‐A, Ivarsson MA, Sohlberg E, Björklund AT, et al. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood. 2013;121:2678–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kared H, Martelli S, Tan SW, Simoni Y, Chong ML, Yap SH, et al. Adaptive NKG2C(+)CD57(+) natural killer cell and Tim‐3 expression during viral infections. Front Immunol. 2018;9:686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vidal SM, Khakoo SI, Biron CA. Natural killer cell responses during viral infections: flexibility and conditioning of innate immunity by experience. Curr Opin Virol. 2011;1:497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Poli A, Michel T, Thérésine M, Andrès E, Hentges F, Zimmer J. CD56bright natural killer (NK) cells: an important NK cell subset. Immunology. 2009;126:458–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Goodier MR, Lusa C, Sherratt S, Rodriguez‐Galan A, Behrens R, Riley EM. Sustained immune complex‐mediated reduction in CD16 expression after vaccination regulates NK cell function. Front Immunol. 2016;7:384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Grzywacz B, Kataria N, Verneris MR. CD56dimCD16+ NK cells downregulate CD16 following target cell induced activation of matrix metalloproteinases. Leukemia. 2007;21:356–9. [DOI] [PubMed] [Google Scholar]
- 60. Duygu B, Olieslagers TI, Groeneweg M, Voorter CEM, Wieten L. HLA class I molecules as immune checkpoints for NK cell alloreactivity and anti‐viral immunity in kidney transplantation. Front Immunol. 2021;12:680480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee S‐H, Biron CA. Here today–not gone tomorrow: roles for activating receptors in sustaining NK cells during viral infections. Eur J Immunol. 2010;40:923–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wight A, Parsons BD, Rahim MMA, Makrigiannis AP. A central role for Ly49 receptors in NK cell memory. J Immunol. 2020;204:2867–75. [DOI] [PubMed] [Google Scholar]
- 63. Lauterbach N, Wieten L, Popeijus HE, Voorter CEM, Tilanus MGJ. HLA−E regulates NKG2C+ natural killer cell function through presentation of a restricted peptide repertoire. Hum Immunol. 2015;76:578–86. [DOI] [PubMed] [Google Scholar]
- 64. Hammer Q, Rückert T, Borst EM, Dunst J, Haubner A, Durek P, et al. Peptide‐specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat Immunol. 2018;19:453–63. [DOI] [PubMed] [Google Scholar]
- 65. Rodda LB, Netland J, Shehata L, Pruner KB, Morawski PA, Thouvenel CD, et al. Functional SARS‐CoV‐2‐specific immune memory persists after mild COVID‐19. Cell. 2021;184:169–183.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jagannathan P, Wang TT. Immunity after SARS‐CoV‐2 infections. Nat Immunol. 2021;22:539–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tomita Y, Ikeda T, Sato R, Sakagami T. Association between HLA gene polymorphisms and mortality of COVID‐19: An in silico analysis. Immunity Inflamm Dis. 2020;8:684–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Novelli A, Andreani M, Biancolella M, Liberatoscioli L, Passarelli C, Colona VL, et al. HLA allele frequencies and susceptibility to COVID‐19 in a group of 99 Italian patients. HLA. 2020;96:610–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lorente L, Martín MM, Franco A, Barrios Y, Cáceres JJ, Solé‐Violán J, et al. HLA genetic polymorphisms and prognosis of patients with COVID‐19 TT – Polimorfismos genéticos de los HLA y pronóstico de pacientes con COVID‐19. Med. Intensiva. 2021;45:96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bashirova AA, Thomas R, Carrington M. HLA/KIR restraint of HIV: surviving the fittest. Annu Rev Immunol. 2011;29:295–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Di Bona D, Aiello A, Colomba C, Bilancia M, Accardi G, Rubino R, et al. KIR2DL3 and the KIR ligand groups HLA−A−Bw4 and HLA−C2 predict the outcome of hepatitis B virus infection. J Viral Hepat. 2017;24:768–75. [DOI] [PubMed] [Google Scholar]
- 72. Khatua S, Cooper LJN, Sandberg DI, Ketonen L, Johnson JM, Rytting ME, et al. Phase I study of intraventricular infusions of autologous ex vivo expanded NK cells in children with recurrent medulloblastoma and ependymoma. Neuro. Oncol. 2020;22:1214–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Esmaeilzadeh A, Elahi R. Immunobiology and immunotherapy of COVID‐19: a clinically updated overview. J Cell Physiol. 2021;236:2519–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lega S, Naviglio S, Volpi S, Tommasini A. Recent insight into SARS‐CoV2 immunopathology and rationale for potential treatment and preventive strategies in COVID‐19. Vaccines. 2020;8:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tu Y‐F, Chien C‐S, Yarmishyn AA, Lin Y‐Y, Luo Y‐H, Lin Y‐T, et al. A review of SARS‐CoV‐2 and the ongoing clinical trials. Int J Mol Sci. 2020;21:2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Perez‐Martinez A, Ferreras C, MorA−Rillo M, Guerra P, Pascual‐Miguel B, Mestre‐Durán C, et al. A phase I/II dose‐escalation single center study to evaluate the safety of infusion of memory t cells as adoptive therapy in coronavirus pneumonia and /or lymphopenia (release). Cytotherapy. 2021;23:S29. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Supplementary Material


