Abstract
In the present work we examined nonhomologous integration of plasmid DNA in a yku70 mutant. Ten of 14 plasmids integrated as composite elements, including Ty sequences probably originating from erroneous strand-switching and/or priming events. Three additional plasmids integrated via Ty integrase without cointegrating Ty sequences, as inferred from 5-bp target site duplication and integration site preferences. Ty integrase-mediated integration of non-Ty DNA has never been observed in wild-type cells, although purified integrase is capable of using non-Ty DNA as a substrate in vitro. Hence our data implicate yKu70 as the cellular function preventing integrase from accepting non-Ty DNA as a substrate.
Random integration of DNA fragments into chromosomes is potentially mutagenic and can result in inactivation of genes located at the integration site. In addition, by uptake and integration of open reading frames (ORFs) of foreign DNA, the recipient cells may acquire new phenotypes. Recent work suggests that horizontal gene transfer resulting from the uptake and integration of foreign DNA occurs readily. For example, after mice were fed with M13 DNA, M13 sequences could be recovered from different tissues, and at least in some cases the foreign DNA appeared to have integrated in the recipient cell's genome (30, 31). Examples of horizontal gene transfer are mainly from prokaryotic systems, in which transfer and integration apparently involved mobile DNA elements (4). In other examples the mechanism of DNA integration is less clear. If the foreign DNA shares homology with the recipient genome, integration may result from homologous recombination. Integration of DNA fragments in the absence of homology is called nonhomologous integration (NHI). Although NHI seems to be very efficient in mammalian cells, the molecular mechanisms involved are largely unclear.
Recent work has uncovered some of the features of NHI in Saccharomyces cerevisiae. By transformation of linear URA3 fragments or plasmids containing the URA3 gene into yeast cells that lacked the URA3 gene, and subsequent selection for a Ura+ phenotype, different classes of Ura+ transformants were obtained (27–29, 41). In most transformants, the plasmid DNA had integrated into chromosomal targets exhibiting 1 to 4 bp of homology to one or both ends of the transforming DNA (microhomology-mediated integration). In the second class of events, no homology was observed between target site DNA and the ends of the transforming DNA. A fraction of such events were mediated by topoisomerase I. In the third class, by joining of the plasmid DNA and pieces of mitochondrial DNA, extrachromosomal circles were formed that were stable, presumably due to autonomously replicating sequences on the mitochondrial DNA. In general, loss of base pairs from the transforming DNA occurred very infrequently, and exclusively at the single-stranded overhangs of the restriction fragments. Target sites in general were found to be unaltered or to contain small deletions or insertions due to fill-in reactions; in a few cases, larger deletions or rearrangements were observed. Apart from microhomologies and/or topoisomerase I cleavage sites, chromosomally integrated plasmids did not exhibit any obvious target site preference.
The frequency of NHI of linearized plasmids can be enhanced considerably by cotransformation with restriction enzymes. In this case most of the transformants had integrated the plasmid DNA into chromosomal cleavage sites of the respective restriction enzyme (22, 28, 29), suggesting that the open double-strand break (DSB) attracted the integrating plasmids. Integration of DNA fragments at sites of DSBs may be mediated by a nonhomologous end-joining (NHEJ) mechanism of DSB repair, which requires little or no homology at the ends to be joined. Indeed, inactivation of RAD50, which is involved in certain aspects of NHEJ, reduces the frequency of spontaneous NHI events 20- to 100-fold (29).
Among the proteins known to be involved in NHEJ, the heterodimeric Ku protein has been studied most intensely. In yeast, its components are encoded by the YKU70 (also called HDF1) and YKU80 (also called HDF2) genes, which exhibit significant homology to the mammalian Ku70 and Ku80 genes, respectively (10, 23). Both in yeast and in mammalian cells, the Ku heterodimer binds to double-stranded DNA ends and some other irregularities, such as hairpins (7). Depending on the structure of the DNA ends, Ku-mediated joining results in ligation of the ends without nucleotide loss or in a microhomology-mediated joining accompanied by the loss of a few nucleotides (9). The Ku-dependent process has the potential of joining promiscuous ends, as is seen from the fact that radiation-induced exchange-type aberrations in yeast depend largely on the presence of functional Ku (11).
Ty elements are yeast retrotransposons that are structurally related to vertebrate retroviruses (reviewed by Boeke and Chapman [1]). Five classes of Ty elements, Ty1 through Ty5, are found in the yeast genome, with Ty1 being most abundant. Ty elements contain long terminal repeats (LTRs) on both ends that in Ty1 and Ty2 are called delta elements. The LTRs are subdivided into domains U3, R, and U5, and they play an important role in Ty transcription and in cDNA formation and integration. Transcription starts in the 5′ LTR and terminates in the 3′ LTR in such a way that the RNA molecule is terminally redundant. The redundant sequences define the R domain of the LTR, whereas U3 and U5 represent sequences unique to the RNA 3′ end and 5′ end, respectively. Synthesis of the cDNA minus strand is primed by a specific tRNA bound to the complementary primer binding site (pbs) that is located adjacent to the 5′ LTR. Minus-strand DNA is synthesized until the 5′ end of the genomic RNA is reached, thus generating minus-strand strong-stop DNA. A template switch is necessary to allow elongation of the minus strand to proceed. After RNase H-mediated degradation of the RNA hybridized with freshly synthesized DNA, the minus-strand DNA can, because of the redundant R domain sequences, anneal to the 3′ end of Ty genomic RNA molecules. The minus strand is then extended until the RNA 5′ end is reached. Plus-strand synthesis is primed by short RNA sequences that are created by specific RNase H cleavage at so-called polypurine tract (PPT) sequences. PPT1 lies adjacent to the 3′ LTR, and a second PPT site is found within the Ty coding region. Synthesis of the plus strand proceeds until the 3′ end of the minus strand is reached, thus creating plus-strand strong-stop DNA. A second template switch is required to allow completion of plus-strand synthesis and thus generation of full-length Ty cDNA. In vivo Ty1 transposition requires that the cDNA terminate in the sequence 5′-TG…CA-3′ (32), but it is not clear how Ty integrase differentiates between Ty cDNA and other extrachromosomal DNA fragments.
Here we report that the absence of yKu70 results in a novel mode of integration of linearized plasmid DNA, not observed in wild-type cells, that is most likely mediated by Ty integrase.
MATERIALS AND METHODS
Strains and media.
NHI experiments were performed with S. cerevisiae strain RSY12 (MATa leu2-3,112 his3-11,15 ura3Δ::HIS3), which contains a replacement of the entire ORF of URA3 with the HIS3 gene (28). Strain RSY12 yku70 was generated as follows. Plasmid pBRWS-HDF1, which contains the ORF of YKU70, was a gift from W. Siede, Emory University, Atlanta, Ga. By restriction with HindIII a 415-bp fragment of the YKU70 sequence was eliminated, and a 1.6-kb HindIII fragment containing LEU2 was inserted to create plasmid phdf-LEU. For transformation the phdf-LEU plasmid was restricted with XmnI to liberate the disrupted YKU70 gene. To generate strains RSY12 spt3 and RSY12 yku70 spt3, a spt3Δ::kanMX deletion was introduced by PCR-based gene disruption (37), using primers spt3-Kan-N (5′-ATG ATG GAC AAG CAT AAG TAT CGT GTG GAG ATT CAA CAG ATG GAT GGC GGC GTT AGT ATC-3′) and spt3-Kan-C (5′-TTA CAT GAT AAT TGG TTT AGA ACT GAG TCT ACC ACC TTT AAA GTT GGG TCA CCC GGC CAG CG-3′). Correct gene replacement was verified by PCR analysis. Standard techniques (33) were used for cell cultivation and medium preparation.
Plasmids.
Plasmid pM151 (22) is a pUC18 derivative containing the S. cerevisiae URA3 gene. Plasmid YEplac195 contains the URA3 gene and the 2μm origin of replication (14). Plasmids were maintained and amplified in Escherichia coli strain DH5α. Large-scale plasmid preparations were done using the Qiagen (Valencia, Calif.) Maxi Kit.
Yeast transformation.
Yeast cells were transformed using the lithium acetate–single-stranded (ss) DNA–polyethylene glycol transformation method (13). For NHI experiments, plasmid pM151 was digested to completion with BglII, extracted with phenol, precipitated with ethanol, and dissolved in sterile water. Seven micrograms of BglII-digested pM151 was used per transformation reaction. For parallel control transformations (using the same batch of cells), 10 ng of circular YEplac195 was used. Transformants were selected by growth on synthetic dextrose-Ura plates for 4 days. Relative transformation frequencies were determined by normalizing the rate of linearized pM151 transformation (per microgram of DNA) relative to the rate of transformation with YEplac195.
Plasmid rescue.
To recover the integrated plasmids and adjacent chromosomal sequences, genomic yeast DNA was isolated as described previously (22) and digested with ClaI, which does not cut within pM151. After ethanol precipitation, about 5 μg of DNA was allowed to self-ligate in a 100-μl ligation mixture containing 1,000 U of T4 DNA ligase (New England Biolabs, Beverly, Mass.). The ligated DNA was precipitated with ethanol and resuspended in sterile water. Ten micrograms was used to transform E. coli DH5α by electroporation, using a high-efficiency protocol (21). Transformants containing pM151-derived sequences were selected by growth on an ampicillin-containing medium before plasmid preparation. Plasmids with flanking chromosomal DNA, as identified by migration behavior in agarose gels, were sequenced by Genome Therapeutics (Waltham, Mass.) using sequencing primers P1 (5′-AGC GGA TAA CAA TTT CAC ACA GGA-3′) and P2 (5′-CGG AGA TTA CCG AAT CAA-3′). Sequencing data were analyzed by a BLASTN homology search on the Saccharomyces Genome Database (SGD; available at http://genome-www2.stanford.edu/cgi-bin/SGD/).
Analysis of integration sites.
To determine the identity of chromosomes containing integrated plasmid sequences, yeast chromosomal DNA was prepared, separated by pulsed-field gel electrophoresis (PFGE), Southern blotted, and hybridized as described previously (11), using a 32P-labeled URA3 PCR fragment. PCR analysis of regions of suspected plasmid integration was done using the Expand Long template PCR kit (Roche, Mannheim, Germany), with elongation times in the range of 8 to 15 min. PCR primers used were MK18fw-VI-263742 (5′-ACA CCA TAT TCA CGC TCG-3′), MK18rev-VI-264195 (5′-TCA TTG TAG TGA ATA TTT TGG-3′), MK20fw-IV-1256721 (5′-TCC TTT AAC AAT TCA GAC C-3′), MK20rev-IV-1257671 (5′-CAA GTC ATT CCA AAT GGC-3′), MK26fw-VII-205343 (5′-CGC TCA CTT GAG CAG TTC-3′), MK26rev-VII-205674 (5′-GTA GCT TGA TCA CAG AAC C-3′), MK34fw-XIII-420387 (5′-ACT ATG TAC TGC ATT TAG G-3′), and MK34rev-XIII-421203 (5′-AAG TCT AGA GAT GTA ATG G-3′). The PCR products obtained for sample MK26 were sequenced using primer MK26-seq2 (5′-CCA CCC GAT TTC ACA CC-3′) in order to determine the second junction between chromosomal and integrated DNA.
RESULTS
The frequency of spontaneous NHI events is not reduced in yku70 mutant cells.
To determine the frequency of NHI events, strain RSY12 and its derivative, RSY12 yku70, were transformed with the BglII-linearized plasmid pM151, which contains the S. cerevisiae URA3 gene and lacks any homology to the genome of strain RSY12. Transformants were selected by plating on SD-Ura medium, and the transformation rate per microgram of transforming DNA was determined and normalized relative to the transformation rate obtained in parallel experiments using circular YEplac195. Surprisingly, inactivation of the YKU70 gene did not reduce the transformation frequency. The relative transformation frequencies (transformants per microgram of DNA per 104 YEplac195 transformants) determined in three independent experiments were 1.06 ± 0.45 for strain RSY12 yku70 and 0.86 ± 0.24 for strain RSY12. In additional experiments, which will be presented in detail elsewhere, we observed that introduction of chromosomal breaks by γ-irradiation strongly induced NHI in wild-type cells, whereas no increase was seen in the yku70 mutant strain. These data suggest that YKU70-mediated NHEJ is essential for integration events taking place at open break sites. There are two possible explanations for the fact that spontaneous transformation events are not reduced in Ku-deficient mutants. First, the end-joining mechanism may not be involved in spontaneous NHI. Alternatively, the absence of a functional Ku protein may favor a type of spontaneous integration event that is not observed in wild-type cells.
Plasmid integration in yku70 mutants is associated with retrotransposon sequences.
To characterize the integration events in the yku70 mutant, the integrated pM151 plus adjacent chromosomal sequences were recovered by plasmid rescue. The sequences of junction sites and adjacent chromosomal DNA were determined and analyzed by a homology search with the SGD. Seven plasmids that had integrated after transformation of unirradiated cells were recovered. In addition, we analyzed seven integration events from cells that were irradiated shortly after transformation. For 12 of those 14 samples, sequencing from both primers yielded yeast-specific sequences. The remaining two samples (data not shown in detail) yielded yeast-specific (Ty-related) sequences with one primer and pUC-related sequences with the other primer. In 10 of the 12 samples for which data are shown, we detected sequences derived from Ty retrotransposon elements at or close to one or both of the junction sites. Such an association between NHI and Ty sequences has not been observed in any of 66 events from wild-type cells and top1 or rad52 mutants (27–29, 41). The structure of the recovered sequences appeared not to be affected by the irradiation treatment of the cells, and irradiated yku70 mutants exhibited the same relative integration frequency as unirradiated cells. Therefore, we pooled the results obtained from unirradiated and irradiated cells.
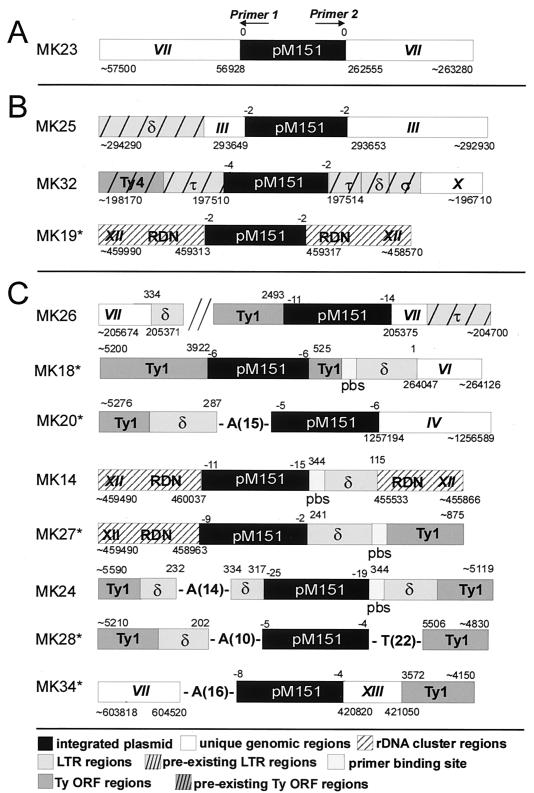
DNA elements identified in the regions surrounding the integrated plasmid are schematically shown in Fig. 1, while Fig. 2 shows the sequences at the plasmid junction sites. In only one of the samples, MK23, the integration event displayed phenotypic similarity to events detected in wild-type cells. In this sample, both ends of the plasmid were intact and joined to different regions of chromosome VII. At the P1 junction site (i.e., the site sequenced by using primer P1), a topoisomerase I consensus target site (5′-CTT-3′) and a potential 3-bp microhomology were found. Results from electrophoretic karyotyping (11), using PFGE and subsequent hybridization with a URA3 fragment, were consistent with integration of the plasmid into chromosome VII. The affected chromosome was, however, found to be about 200 kbp longer than that in the parental strain, while the remaining chromosomal bands appeared normal (data not shown). This observation, and the fact that the recovered chromosome VII-derived sequences at both plasmid ends were in antiparallel orientation, may hint at a gross intrachromosomal rearrangement, possibly a large inverted duplication. Rearrangements associated with an integration event have also been observed previously in wild-type cells (27, 41).
FIG. 1.
Sequence elements identified adjacent to pM151 sequences after analysis of rescued plasmids. Indicated are the areas sequenced by using primer P1 (on the left side of the plasmid) and primer P2 (on the right side). For better comparison, different sequence element types (unique, rDNA, LTR, tRNA pbs, and Ty ORF) are differentiated by shading types (see the key); elements were not drawn to scale. Numbers below the elements refer to SGD genomic coordinates, while coordinates given above the Ty elements refer to those Ty1 element sequences which gave the best fit in BLASTN analysis (see Table 1). The numbers of nucleotides missing from the 5′ protruding single-stranded plasmid ends are given just above the plasmid junctions. Asterisks indicate samples obtained after irradiation with 50 Gy. (A) Integration event probably mediated by topoisomerase I. (B) Three plasmid integrations without cointegrating Ty sequences. These events were most likely mediated by Ty integrase, since they created a 5-bp target site duplication. (C) Apparent cointegration events of pM151 and Ty sequences. For MK26 the junction between chromosomal DNA and cointegrating Ty sequences is also shown. RDN, rDNA.
FIG. 2.
Nucleotide sequences of junctions between plasmid and chromosomal or Ty1-derived DNA. Sequences determined after plasmid rescue (middle row) are depicted in 5′-to-3′ orientation and aligned with the plasmid sequence (top row) and chromosomal or Ty1-derived sequences (bottom row). Sequence coordinates are given as described for Fig. 1. For MK26, the junction between chromosome VII DNA and Ty1-derived sequences is also given. Underlined sequences indicate confirmed 5-bp duplications at chromosomal target sites. Shading in the MK23 sequence indicates a putative topoisomerase I target site.
Except for MK23, a 5-bp duplication of the target site was detected in all clones in which both integration junctions were identified.
In three samples, MK25, MK32, and MK19, the plasmid integrated into a contiguous chromosomal sequence. In MK25, the plasmid integrated into chromosome III. Sequence analysis revealed the presence of a delta LTR element 77 bp distant from the P1 junction. At the same chromosomal position, a delta element is found in the yeast genome database, suggesting that the LTR sequence existed at this place before the integration event. In MK32, the plasmid had inserted into a tau element, i.e., a Ty4 LTR. Adjacent to the tau element at the P1 side, a Ty4 coding region was found. At the P2 side, the tau element was followed by sequences derived from delta and sigma elements (i.e., Ty3 LTR), and unique sequences derived from chromosome X. This arrangement of Ty4 ORF, tau, delta, sigma, and chromosome X sequences is also present in the yeast genome database, suggesting that plasmid pM151 had integrated into a preexisting tau element. In MK19, the plasmid integrated into the ribosomal DNA (rDNA) locus, about 300 bp upstream of the 5S rDNA promoter. In all three events, a 5-bp duplication of the target site had apparently occurred (Fig. 2), although the P1 junction in MK25 could not be determined unambiguously because of a 1-bp sequence overlap. Five-base-pair duplication of target sites is a typical feature of Ty integrase-mediated integration (8, 12) that has, to our knowledge, never been detected in integrase-independent reactions.
In most NHI events, retrotransposon sequences appear to cointegrate with the plasmid.
In most of the remaining clones, determination of the sequence recovered by plasmid rescue allowed us to identify unique sequences only from one side of the plasmid, or not at all, thus making an unequivocal determination of the integration site impossible. Where possible, we therefore attempted to verify suspected integration sites by electrophoretic karyotyping and PCR analysis.
In MK26, the P2 end of the plasmid was found joined to a unique sequence derived from chromosome VII, and results from electrophoretic karyotyping were consistent with an integration into chromosome VII. Sequence analysis revealed the presence of a tau element 34 bp distant from the P2 junction; a tau element is also found at this position on chromosome VII in the yeast genome database. On the P1 side, the plasmid was joined to a sequence derived from the TyB ORF. In the database, no Ty1 element is found close to the suspected integration site. To test whether strain RSY12 yku70 differs in this region from the published strain, PCR primers that bind on both sides of the suspected integration site were chosen on the basis of the information in the database. With the parental strain these primers gave rise to an amplification product of the expected size (about 330 bp). With clone MK26, however, a PCR product of about 10 kb was obtained by using the same primers (data not shown), in agreement with a large de novo insertion. To characterize the second junction between chromosome VII sequences and integrated sequences, part of the PCR product was sequenced. This revealed that the unique chromosome VII sequence was joined to the 3′ end of a 3′ delta element, suggesting that a composite element that consisted of Ty1-derived sequences and pM151 sequences and that terminated at one end in an LTR element had integrated into chromosome VII. The chromosomal target sequence again exhibited the 5-bp duplication expected for Ty integrase-mediated integration events (Fig. 2).
In MK18, on both sides of pM151, Ty1-derived sequences were found. On one side (P1), part of a TyB ORF was joined to the plasmid sequence. On the P2 side, after a region containing part of a TyA ORF, a pbs, and a 5′ LTR element, unique sequences derived from chromosome VI could be recovered, and results from PFGE analysis were consistent with an integration of pM151 into chromosome VI. The published yeast genomic sequence does not contain Ty1-derived sequences at or close to the suspected integration site of pM151. Based on the published sequence of chromosome VI, we chose PCR primers that should bind to both sides of the suspected integration site. With these primers, the expected 460-bp PCR amplification product was obtained for the parental strain RSY12 yku70. For MK18, the same primers gave rise to a fragment of about 7 kb (data not shown). Again, these data are in agreement with an integration of a composite plasmid-Ty element.
Integration of a composite pM151-Ty1 element appears also to have occurred in sample MK20: one plasmid end (P2) was joined to a unique sequence derived from chromosome IV, while the other end was connected, via a poly(A) stretch, to the 3′ end of the R segment of a 3′ LTR element. Again, PCR analysis of the parental strain gave the product expected in the absence of insertional sequences from the suspected integration area. Unfortunately, PCR analysis of MK20 was not possible because we could not recover this strain.
A common feature of samples MK26, MK18, and MK20 is that the plasmid DNA was joined, on one or both ends, to Ty1-derived sequences that appear not to have existed at the integration site prior to the integration event. Junctions of plasmid DNA to Ty1-derived sequences were also observed in other samples, where the absence of unique sequence information precluded PCR analysis of the suspected integration region. In MK14, plasmid end P1 was joined to an rDNA sequence about 300 bp downstream of the 5S rDNA promoter. The P2 side of the plasmid was joined to the 3′ end of a pbs, which was followed by an incomplete LTR and rDNA sequences. The rDNA sequences found on both sides did not form the contiguous stretch of typical rDNA units. Because of the repetitive nature of rDNA units, it was not possible to determine whether the targeted unit was already incomplete before integration took place, or whether the Ty1-derived sequences preexisted at this site.
Junction of the integrated plasmid with rDNA sequences was also observed in MK27. The P1 side of the plasmid was joined to a region about 600 bp upstream of the 5S rDNA promoter, while the P2 side was joined to the R segment of a 5′ LTR element. In total, integrations into the rDNA locus were observed in three of the events, and in each case, integration of at least one end of the integrated element occurred into the nontranscribed regions flanking the 5S rDNA gene.
In two samples, MK24 and MK28, the pM151 sequence was surrounded by noncontiguous sequences derived from Ty1 elements, and no unique sequences could be recovered by plasmid rescue. In MK28, the plasmid sequence was bracketed by a poly(A) and a poly(T) stretch, while in MK24 a poly(A) stretch was found close to the P1 junction, at the border between two incomplete delta elements which were in antiparallel orientation. PFGE analysis showed that MK24 and MK28 had integrated into chromosomes VII/XV and XIII/XVI, respectively (data not shown). The actual chromosomes could not be determined because these specific chromosomes are not resolved by PFGE.
An apparently complex event took place in sample MK34. The P1 side of the plasmid was connected, via a poly(A) stretch, to unique sequences derived from chromosome VII. The P2 side, however, was joined to unique sequences derived from chromosome XIII. In PFGE gels, the normal bands corresponding to chromosomes VII and XIII (about 1,100 and 1,000 kb long, respectively) were missing for this clone, and two new bands of about 1,600 and 500 kb had appeared (data not shown), suggesting a reciprocal translocation. Hybridization revealed that the plasmid is present on the 1,600-kb chromosome. Interestingly, about 240 bp distant from the P2 integration site, sequences derived from the TyB ORF were detected; as inferred from a database search and PCR analysis of the parental strain, these sequences were not present at this position prior to the integration event.
In a total of 10 junctions, the plasmid ends were connected directly (7 cases) or via A or T stretches (3 cases) to Ty-derived sequences that either were shown to have newly integrated (MK26, MK18, and MK20) or whose origin could not be determined. The identities of the chromosomal Ty elements that showed the best match with these recovered Ty sequences in a BLASTN search are given in Table 1. Interestingly, for 9 of the 10 sequences analyzed, the best matches were obtained with a subgroup of Ty1 elements, called Ty1/2, that were proposed to have arisen from replicative strand-switching events between Ty1 and Ty2 elements (17, 18). For the remaining sequence, equally high scores were obtained with elements from the Ty1 group and the Ty1/2 evolutionary subgroup. The biological significance of this preferential involvement of Ty1/2 is unclear at present.
TABLE 1.
Chromosomal Ty1 elements yielding the best match in a BLASTN search with Ty-derived sequences that apparently had cointegrated with pM151a
| Recovered Ty sequence | Chromosomal Ty1 element(s) yielding the best match | Evolutionary group(s) |
|---|---|---|
| MK26/P1 (TyB) | YARCTy1-1, YPRCTy1-2, YDRWTy1-4, YGRWTy1-1 | Ty1 and Ty1/2 |
| MK18/P1 (TyB) | YPRCTy1-2 | Ty1/2 |
| MK18/P2 (LTR + TyA) | YPRCTy1-2 | Ty1/2 |
| MK20/P1 (TyB + LTR) | YARCTy1-1, YERCTy1-1, YMLWTy1-2, YPLWTy1-1, YLRCTy1-1 | Ty1/2 |
| MK14/P2 (LTR + pbs) | YMLWTy1-2 | Ty1/2 |
| MK27/P2 (LTR + TyA) | YERCTy1-1, YPLWTy1-1, YLRCTy1-1 | Ty1/2 |
| MK24/P1 (TyB + LTR) | YLRWTy1-3, YDRCTy1-2, YORWTy2-2 | Ty1/2 |
| MK24/P2 (TyB + LTR) | YNLWTy1-2 | Ty1/2 |
| MK28/P1 (TyB + LTR) | YJRWTy1-1/YJRWdelta12 | Ty1/2 |
| MK28/P2 (TyB) | YARCTy1-1 | Ty1/2 |
Plasmid sequences directly joined to chromosomal DNA preferentially end in a 3′-terminal A residue.
A typical feature of nonhomologous plasmid integration in wild-type cells is that, apart from rare resections of protruding single-stranded ends, little processing of the plasmid ends occurs. This is also seen in sample MK23, in which the plasmid sequence can be considered complete if one assumes that integration at the P1 side was initiated by a topoisomerase I-mediated break. In the remaining 11 samples, 2 to 25 bp were lost from the plasmid ends (Fig. 1). Of the 22 plasmid ends, 11 were joined directly to unique chromosomal sequences, rDNA sequences, or retrotransposon sequences that appeared to have preexisted at the integration site. In 7 of these 11 junctions (P1 ends of MK32, MK19, and MK14; P2 ends of MK25, MK32, MK19, and MK26), the end of the plasmid sequence could be unequivocally determined, and in each case the 3′ end of the plasmid terminated in 5′-GA-3′ or 5′-CA-3′. In the remaining four junctions, microhomologies between the plasmid and the target sequence precluded an exact determination of the plasmid ends. In one of these (the P1 end of MK25 [MK25/P1]), the plasmid DNA also ended in 5′-GA-3′, if one assumes a 5-bp duplication of the chromosomal target site.
Seven plasmid ends were joined directly to Ty1-derived sequences that had apparently cointegrated or whose origin could not be determined. Among these, only at one junction, MK24/P1, was no potential sequence overlap with the Ty sequence found, and the plasmid sequence ended in 5′-CG-3′. At the junctions MK26/P1, MK18/P1, and MK18/P2, each of which included sequences derived from the Ty1 ORF, extensive (8- to 15-bp), albeit in part imperfect, microhomologies were found. A 2-bp microhomology was found at the plasmid-delta junction MK27/P2. In events MK14/P2 and MK24/P2, where the plasmid DNA was joined to the 3′ end of the pbs, a 2-bp microhomology is present if one assumes an interaction of the plasmid with Ty1 plus-strand molecules terminating in a 12-bp tRNA-complementary sequence (see Discussion).
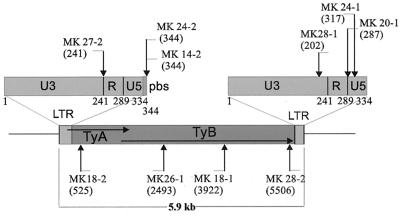
In Fig. 3, the locations of junctions between plasmid DNA and those Ty1-derived sequences which could not be shown to have preexisted at the target sites are indicated relative to a standard Ty1 element sequence. Six of the 10 junctions involved the LTR regions, although these represent only about 1/10 of the total Ty1 DNA.
FIG. 3.
Locations of junctions between pM151 and those Ty1 sequences that could not be demonstrated to have preexisted at the integration site, relative to a standard Ty1 element.
Most NHI events in yku70 mutants require Ty1 transcription.
Our data suggested that Ty1 metabolism is involved in most of the NHI events taking place in Ku-deficient cells. To test whether Ty1 transcription is necessary for these integration events, we deleted the SPT3 gene in strains RSY12 and RSY12 yku70. SPT3 codes for a general transcription factor that is required for transcription of natural Ty1 and Ty2 elements (39). In a Ku-proficient background, inactivation of SPT3 did not reduce the frequency of NHI events (1.51 events per μg of DNA per 104 control transformants). In a Ku-deficient background, however, inactivation of SPT3 reduced the relative NHI frequency by a factor of about 10 (from 1.06 ± 0.45 to 0.11 ± 0.09 per μg of DNA per 104 control transformants). This indicates that most of the events that occur in the absence of a functional Ku protein require Ty1 mRNA and probably Ty1-encoded proteins and/or cDNA for integration.
DISCUSSION
In the present work, we characterized spontaneous integration of a linearized plasmid, which lacked homology to the genome of the recipient cell, into chromosomal DNA of yeast cells deficient in the Ku70 subunit of the DNA end-binding factor Ku. Of 14 integration events, only 1 (MK23) exhibited phenotypic similarity with events observed previously in wild-type cells. In this case, integration probably was mediated by topoisomerase I, as is the case in about 40% of spontaneous NHI events in wild-type cells (41).
The remaining 13 integration events exhibited unexpected features that have not been observed before in any of 66 events from wild-type cells and top1 or rad52 mutants (27–29, 41). These features include a close association between plasmid DNA and retrotransposon-derived sequences and/or hallmarks of Ty integrase-mediated reactions at plasmid-chromosome junctions. In 12 of those 13 cases, retrotransposon-derived sequences were found at or close to one or both plasmid ends. From 10 of these 12 events, both junctions contained yeast-specific sequences. In 6 of the 10 cases, the presence of unique sequences within the sequenced region allowed further characterization of the suspected target site by sequence comparison to the yeast genomic database or by PCR analysis. We found that two different types of events can lead to an association between plasmid and retrotransposon DNA. (i) In MK32, MK25/P1, and MK26/P2, the plasmid integrated into or close to retrotransposon sequences which were found at the same chromosomal position in the yeast genomic database. Therefore, they appear to have been present prior to the plasmid integration event. (ii) In the cases of MK18, MK26/P1, MK20/P1, and MK34/P2, the Ty-derived sequence in question is not found in the database and PCR analysis of the parental strain indicated that it did not preexist in strain RSY12 yku70 in the respective region. We conclude that the Ty-derived sequences have cointegrated with the plasmid DNA.
In the remaining four Ty-associated integrations, the nature of the Ty sequences could not be determined, since no unique sequence was found. Since these Ty sequences were, in general, truncated, we assume that at least some of them also originate from cointegration events.
Models for generation of composite plasmid-Ty elements.
How can composite plasmid-Ty elements arise? One possibility would be that plasmid and extrachromosomal Ty cDNA are joined by an end-joining mechanism. We consider this possibility unlikely, given that the Ku protein is a major player in NHEJ. We favor a model in which composite plasmid-Ty elements originate from Ty replication-associated events that involve template switching and/or erroneous priming mechanisms. Ty virus-like particles (VLPs), like retrovirus particles, contain two genomic RNA molecules that serve as templates for reverse transcription. During cDNA formation, two so-called strand transfer events that may occur either intra- or intermolecularly are necessary, and there is evidence for frequent additional strand-switching events during minus-strand synthesis (38). If the two template RNAs are different, intermolecular strand-switching events can give rise to recombinant cDNA molecules. For example, sequence analysis of all Ty1 and Ty2 elements in the SGD revealed that 14 of 32 Ty1 elements actually contain Ty2-derived sequence stretches that appear to have been incorporated by means of intermolecular strand-switching events (17, 18). Furthermore, several examples of junctions between retrovirus- or retrotransposon- and gene-derived sequences are known which were proposed to have arisen from template-switching events between virus or transposon RNA and cellular RNA that had accidentally been copackaged in the virus particle or the VLP (5, 15, 35). We cannot exclude the possibility that occasionally, by read-through transcription of the URA3 gene, nearly full-length transcripts of pM151 are formed. If these were involved in strand-switching events during Ty reverse transcription, one would expect the P2 end of the integrated pM151 sequences to lack the region between the BglII site and the URA3 transcription start, i.e., about 180 bp. However, we observed deletions of only 19 bp at most, and therefore we propose that plasmid DNA rather than RNA interacted with Ty replication intermediates. After recessing of plasmid DNA ends, single-stranded 3′ overhangs could allow for base pairing with nascent Ty1 cDNA strands, thus facilitating a jump of the reverse transcriptase onto the plasmid DNA. In addition, these overhangs could serve as primers for the synthesis of a second, complementary cDNA strand. In such a scenario, the junctions between plasmid and Ty sequences would be formed by polymerases (reverse transcriptase and, possibly, cellular DNA polymerase) rather than by an end-joining mechanism.
Potential role of replication intermediates in formation of the plasmid-Ty composites.
If aberrant strand-switching events are responsible for the generation of plasmid-Ty junctions, one should expect that those Ty replication intermediates (which frequently serve as substrates for natural strand-switching processes) should also be frequently involved in the aberrant processes. Indeed, we observed several plasmid-Ty1 junctions that appear to have involved pausing Ty1 replication intermediates. (For a recent overview of replication intermediates, see Mules et al. [26].) Such a potential intermediate is nearly full-length minus-strand cDNA. If it was generated after an intermolecular transfer of the minus-strand strong-stop DNA to a second Ty1 RNA molecule, it is predicted to terminate at the R/U3 border. An interaction of its 3′ end with plasmid DNA should result in the junction structure seen in MK27 (Fig. 1). Another potential pausing intermediate is plus-strand strong-stop DNA. If, during plus-strand synthesis, the tRNA primer molecule is still covalently linked to the 5′ end of the minus-strand template, the plus-strand sequence is predicted to extend to the first modified base of the tRNA 3′ end, i.e., 2 bp beyond the 10-bp region of pbs complementarity. Current models for Ty1 replication propose that plus-strand molecules terminating in a 12-bp tRNA complementary sequence are dead-end products that may accumulate in the cell (20). The P2 junction of sample MK24 is consistent with a model in which plus-strand molecules terminating in a 12-bp tRNA complementary sequence (5′-TGG TAG CGC CGC-3′) anneal, via a 2-bp microhomology, with a 3′ protruding plasmid P2 end. The 3′ plasmid end would then have primed synthesis of the minus strand. The P2 junction of MK14 may be explained by a similar mechanism.
Protruding plasmid DNA ends may interact not only with cDNA single strands but also with Ty1 RNA molecules. For example, the Ty1-derived sequences found at the P1 junction of sample MK20 correspond to the 3′ end of polyadenylated Ty1 RNA. We can imagine a model in which, starting in the poly(A) tract, the plasmid 3′ end would have primed synthesis of the corresponding minus-strand cDNA. Such a reaction may be facilitated by previous reverse transcriptase-mediated addition of untemplated T residues onto the plasmid DNA. The respective plus strand would then have been primed by a plus-strand strong-stop DNA provided in trans. Similar mechanisms may have occurred in event MK28.
It is not clear where the proposed strand-switching and erroneous priming events take place. According to present knowledge, cytoplasmic VLPs are the most likely place for the interactions of transformed DNA with Ty replication intermediates. However, the possibility that these interactions occur in the nucleus cannot be excluded. The junctions between plasmid and Ty sequences described here share certain similarities with junctions between chromosomal DNA and Ty sequences captured at sites of HO endonuclease-mediated chromosome breaks (24, 36, 40): in both cases, the Ty-derived sequences appear to originate from Ty replication intermediates. Furthermore, Yu and Gabriel (40) report that about half of the Ty-derived sequences captured in their experiments were apparently derived from the endogenous elements YLRWTy1-2 and YMLWTy1-2. These elements belong to the Ty1/2 evolutionary group (17, 18).
At present, we cannot judge whether the proposed strand-switching and/or erroneous priming events involving plasmid DNA occur more often in Ku-deficient cells than in wild-type cells. Ku might, by its association with VLPs (6) or by binding to the ends of DNA fragments, prevent extranuclear DNA from entering VLPs. Alternatively, by binding to the ends of DNA fragments, Ku might prevent interactions of plasmid ends with Ty RNA, cDNA, or reverse transcriptase. Yet another possibility is that erroneous priming events occur equally often in the presence and in the absence of Ku, but that integration of the resulting composite elements is blocked when Ku is present.
Ku70 is involved in conferring substrate specificity on Ty integrase.
Several observations suggest that the absence of functional Ku affects the substrate specificity of Ty integrase. In four cases (MK25, MK32, MK19, and MK26), we were able to retrieve both chromosomal junction sites, and in all these cases a 5-bp duplication of the chromosomal target site occurred. Target site duplications, a typical feature of integration events mediated by transposases or integrases, occur because the integration-mediating enzymes produce a staggered cut of the target site, which, after joining of the mobile element to the protruding ends, leaves single-stranded gaps that have to be filled in. The extent of the duplicated region is specific to the integration-mediating enzyme; 5-bp duplications occur in Ty integrase-mediated events (8, 12). In three of the cases where we were able to demonstrate target site duplication, only plasmid DNA sequences, but no newly integrated Ty sequences, were found at the integration site. To our knowledge, it has so far never been reported that the integration of foreign DNA that lacks identifiable similarity to known mobile DNA elements was, in vivo, accompanied by a 5-bp target site duplication. Previously, with the same system for integration, 1- to 4-bp duplications, but never 5-bp target site duplications, have been found among 66 NHI target sequence junctions from wild-type cells and top1 and rad52 mutants (27–29, 41). These events always resulted when the endpoints of homology of the junctions were 1 to 4 bp apart.
In vitro studies showed that the efficiency of Ty1 integrase is highest when the substrate DNA ends with a 3′-terminal A residue (2, 25). Furthermore, the integrase can utilize staggered substrate ends, provided that the double-stranded region terminates in a 3′ A (25). In 7 of 11 junctions between plasmid and chromosomal DNA, we could unambiguously determine the end of the plasmid sequence. In all those cases, the plasmid ended in a 3′-terminal A, which is consistent with the substrate end preference of Ty1 integrase.
If these integrations of plasmid or composite plasmid-Ty1 elements were mediated by Ty1 integrase, one would expect these events to show additional characteristics of Ty1 integrase-mediated integration, such as target site preference. Integration of Ty1, Ty2, Ty3, and Ty4 elements does not exhibit target site sequence specificity, but it has been observed that these elements integrate preferentially upstream of RNA polymerase III promoter regions (19). An analysis of Ty1 transposition events on chromosome III showed that the majority of integrations occurred within a distance of several hundred base pairs upstream of tRNA genes or within or near preexisting Ty elements or LTRs (16). The integrations in MK25, MK32, and MK26 are located 0 to 77 bp from preexisting LTRs and 154 to 557 bp upstream of tRNA genes. In addition, we found three integrations into rDNA. Four of the five junctions between rDNA sequences and integrated sequences recovered were located in the nontranscribed spacer regions, which are known as efficient targets for Ty1 integration (3, 34). Whether the integration sites in MK18 and MK20 relate to preferential sites of insertion is less clear. While the SGD database contains a tRNA gene close (126 bp) to the integration site of MK18, we did not find any tRNA gene when sequencing this region. In MK20, integration occurred into a hypothetical ORF, and according to the database, there is no tRNA gene in the vicinity of this region.
Taken together, our data strongly suggest that most of the plasmid integration events detected in the yku70-deficient mutant were mediated by Ty integrase. This has never been found in wild-type cells. In vitro studies showed that Ty1 integrase from purified VLPs can mediate integration-like events of plasmid-derived sequences (2), suggesting that a cellular factor prevents such integrations. Our data suggest that Ku is this factor and that the absence of functional Ku proteins alleviates the specificity of the integration reaction for Ty cDNA sequences. In those events where the plasmid DNA was found to be connected to chromosomal DNA at one end and to new Ty-derived sequences at the other end, it is possible that one-sided integration events involving one normal LTR end took place. The presence of three samples where an apparently integrase-mediated integration of plasmid DNA in the absence of Ty sequences occurred clearly indicates, however, that LTR ends are not always needed in the ku70 background. Hence, Ku may have a role in preventing integrase proteins from using extrachromosomal non-Ty DNA fragments as substrates. Given that an integration of pieces of foreign DNA may threaten a cell's genomic integrity, this proposed function would represent a novel mechanism by which the yeast Ku proteins ensure genomic integrity, in addition to their function in DSB repair and telomere stabilization.
ACKNOWLEDGMENTS
We thank David Garfinkel for useful discussions and comments on the manuscript. We also thank Dan Gietz and Wolfram Siede for plasmids. Sequencing was generously provided by Genome Therapeutics.
This research was supported by grant CN-83 from the American Cancer Society and Research Career Development Award ES00299 from the National Institute of Environmental Health Sciences to R.H.S., by grant FI4P-CT95-0010 from the European Commission to F.E.-S., and by travel grants from the Neuherberger Forschungsfoerderung to M.K.
M.K. and A.A.F. contributed equally to this work.
REFERENCES
- 1.Boeke J D, Chapman K B. Retrotransposition mechanisms. Curr Opin Cell Biol. 1991;3:502–507. doi: 10.1016/0955-0674(91)90079-e. [DOI] [PubMed] [Google Scholar]
- 2.Braiterman L T, Boeke J D. In vitro integration of retrotransposon Ty1: a direct physical assay. Mol Cell Biol. 1994;14:5719–5730. doi: 10.1128/mcb.14.9.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryk M, Banerjee M, Murphy M, Knudsen K E, Garfinkel D J, Curcio M J. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- 4.Davison J. Genetic exchange between bacteria in the environment. Plasmid. 1999;42:73–91. doi: 10.1006/plas.1999.1421. [DOI] [PubMed] [Google Scholar]
- 5.Derr L K, Strathern J N, Garfinkel D J. RNA-mediated recombination in S. cerevisiae. Cell. 1991;67:355–364. doi: 10.1016/0092-8674(91)90187-4. [DOI] [PubMed] [Google Scholar]
- 6.Downs J A, Jackson S P. Involvement of DNA end-binding protein Ku in Ty element retrotransposition. Mol Cell Biol. 1999;19:6260–6268. doi: 10.1128/mcb.19.9.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dynan W S, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farabaugh P J, Fink G R. Insertion of the eukaryotic transposable element Ty1 creates a 5-base pair duplication. Nature. 1980;286:352–356. doi: 10.1038/286352a0. [DOI] [PubMed] [Google Scholar]
- 9.Featherstone C, Jackson S P. Ku, a DNA repair protein with multiple cellular functions? Mutat Res. 1999;434:3–15. doi: 10.1016/s0921-8777(99)00006-3. [DOI] [PubMed] [Google Scholar]
- 10.Feldmann H, Winnacker E L. A putative homologue of the human autoantigen Ku from Saccharomyces cerevisiae. J Biol Chem. 1993;268:12895–12900. [PubMed] [Google Scholar]
- 11.Friedl A A, Kiechle M, Fellerhoff B, Eckardt-Schupp F. Radiation-induced chromosome aberrations in Saccharomyces cerevisiae: influence of DNA repair pathways. Genetics. 1998;148:975–988. doi: 10.1093/genetics/148.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gafner J, Philippsen P. The yeast transposon Ty1 generates duplications of target DNA on insertion. Nature. 1980;286:414–418. doi: 10.1038/286414a0. [DOI] [PubMed] [Google Scholar]
- 13.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 15.Hajjar A M, Linial M L. A model system for nonhomologous recombination between retroviral and cellular RNA. J Virol. 1993;67:3845–3853. doi: 10.1128/jvi.67.7.3845-3853.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji H, Moore D P, Blomberg M A, Braiterman L T, Voytas D F, Natsoulis G, Boeke J D. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell. 1993;73:1007–1018. doi: 10.1016/0092-8674(93)90278-x. [DOI] [PubMed] [Google Scholar]
- 17.Jordan I K, McDonald J F. Evidence for the role of recombination in the regulatory evolution of Saccharomyces cerevisiae Ty elements. J Mol Evol. 1998;47:14–20. doi: 10.1007/pl00006358. [DOI] [PubMed] [Google Scholar]
- 18.Jordan I K, McDonald J F. The role of interelement selection in Saccharomyces cerevisiae Ty element evolution. J Mol Evol. 1999;49:352–357. doi: 10.1007/pl00006558. [DOI] [PubMed] [Google Scholar]
- 19.Kim J M, Vanguri S, Boeke J D, Gabriel A, Voytas D F. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 20.Lauermann V, Boeke J D. Plus-strand strong-stop DNA transfer in yeast Ty retrotransposons. EMBO J. 1997;16:6603–6612. doi: 10.1093/emboj/16.21.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manivasakam P, Schiestl R H. High efficiency transformation of Saccharomyces cerevisiae by electroporation. Nucleic Acids Res. 1993;21:4414–4415. doi: 10.1093/nar/21.18.4414. . (Erratum, 21:4856.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manivasakam P, Schiestl R H. Nonhomologous end joining during restriction enzyme-mediated DNA integration in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:1736–1745. doi: 10.1128/mcb.18.3.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milne G T, Jin S, Shannon K B, Weaver D T. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4189–4198. doi: 10.1128/mcb.16.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore J K, Haber J E. Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature. 1996;383:644–646. doi: 10.1038/383644a0. [DOI] [PubMed] [Google Scholar]
- 25.Moore S P, Powers M, Garfinkel D J. Substrate specificity of Ty1 integrase. J Virol. 1995;69:4683–4692. doi: 10.1128/jvi.69.8.4683-4692.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mules E H, Uzun O, Gabriel A. In vivo Ty1 reverse transcription can generate replication intermediates with untidy ends. J Virol. 1998;72:6490–6503. doi: 10.1128/jvi.72.8.6490-6503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schiestl R H, Dominska M, Petes T D. Transformation of Saccharomyces cerevisiae with nonhomologous DNA: illegitimate integration of transforming DNA into yeast chromosomes and in vivo ligation of transforming DNA to mitochondrial DNA sequences. Mol Cell Biol. 1993;13:2697–2705. doi: 10.1128/mcb.13.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schiestl R H, Petes T D. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:7585–7589. doi: 10.1073/pnas.88.17.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schiestl R H, Zhu J, Petes T D. Effect of mutations in genes affecting homologous recombination on restriction enzyme-mediated and illegitimate recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:4493–4500. doi: 10.1128/mcb.14.7.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schubbert R, Hohlweg U, Renz D, Doerfler W. On the fate of orally ingested foreign DNA in mice: chromosomal association and placental transmission to the fetus. Mol Gen Genet. 1998;259:569–576. doi: 10.1007/s004380050850. [DOI] [PubMed] [Google Scholar]
- 31.Schubbert R, Renz D, Schmitz B, Doerfler W. Foreign (M13) DNA ingested by mice reaches peripheral leukocytes, spleen, and liver via the intestinal wall mucosa and can be covalently linked to mouse DNA. Proc Natl Acad Sci USA. 1997;94:961–966. doi: 10.1073/pnas.94.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharon G, Burkett T J, Garfinkel D J. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol Cell Biol. 1994;14:6540–6551. doi: 10.1128/mcb.14.10.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 34.Smith J S, Boeke J D. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 35.Temin H M. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci USA. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teng S C, Kim B, Gabriel A. Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature. 1996;383:641–644. doi: 10.1038/383641a0. [DOI] [PubMed] [Google Scholar]
- 37.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 38.Wilhelm M, Boutabout M, Heyman T, Wilhelm F X. Reverse transcription of the yeast Ty1 retrotransposon: the mode of first strand transfer is either intermolecular or intramolecular. J Mol Biol. 1999;288:505–510. doi: 10.1006/jmbi.1999.2723. [DOI] [PubMed] [Google Scholar]
- 39.Winston F, Durbin K J, Fink G R. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell. 1984;39:675–682. doi: 10.1016/0092-8674(84)90474-4. [DOI] [PubMed] [Google Scholar]
- 40.Yu X, Gabriel A. Patching broken chromosomes with extranuclear cellular DNA. Mol Cell. 1999;4:873–881. doi: 10.1016/s1097-2765(00)80397-4. [DOI] [PubMed] [Google Scholar]
- 41.Zhu J, Schiestl R H. Topoisomerase I involvement in illegitimate recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1805–1812. doi: 10.1128/mcb.16.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]