Abstract
The main protease (3CLp) of the SARS‐CoV‐2, the causative agent for the COVID‐19 pandemic, is one of the main targets for drug development. To be active, 3CLp relies on a complex interplay between dimerization, active site flexibility, and allosteric regulation. The deciphering of these mechanisms is a crucial step to enable the search for inhibitors. In this context, using NMR spectroscopy, we studied the conformation of dimeric 3CLp from the SARS‐CoV‐2 and monitored ligand binding, based on NMR signal assignments. We performed a fragment‐based screening that led to the identification of 38 fragment hits. Their binding sites showed three hotspots on 3CLp, two in the substrate binding pocket and one at the dimer interface. F01 is a non‐covalent inhibitor of the 3CLp and has antiviral activity in SARS‐CoV‐2 infected cells. This study sheds light on the complex structure‐function relationships of 3CLp and constitutes a strong basis to assist in developing potent 3CLp inhibitors.
Keywords: drug discovery, fragment screening, NMR spectroscopy, protein structure, viruses
We report the liquid‐sate NMR spectroscopy analysis of the dimeric SARS‐CoV‐2 main protease (3CLp), including its backbone assignments, to study its complex conformational regulation. Using fragment‐based NMR screening, we highlighted three hotspots on the protein, two in the substrate binding pocket and one at the dimer interface, and we identified a non‐covalent reversible inhibitor of 3CLp that has antiviral activity in infected cells.
Introduction
Since the end of 2019, the world faces the global COVID‐19 pandemic that is a major health burden worldwide with strong societal and economic impacts. The etiological agent is the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) with a case fatality rate of ca. 2 %. [1] This virus represents the seventh coronavirus that infects humans and causes the third β‐coronavirus outbreak that emerged in the 21st century. Even though, both vaccines[ 2 , 3 , 4 , 5 ] and neutralizing antibodies[ 6 , 7 , 8 ] are now available to fight against SARS‐CoV‐2, specific and efficient antivirals against β‐coronaviruses are urgently needed to overcome the limited vaccine coverage, variant escapes from antibodies and the future outbreaks.
The RNA genome of SARS‐CoV‐2 encodes for up to 27 different proteins:[ 9 , 10 ] the structural proteins, the nonstructural proteins (Nsp) and finally several accessory proteins. The Nsp, corresponding to the replicase‐transcriptase, are first translated in two polyproteins, pp1a and pp1ab, which are then cleaved by two viral proteases, the main protease (Mpro or 3CLp) and papain‐like protease to release 16 functional proteins. 3CLp cleaves at 11 sites (Nsp4‐Nsp16), including its own release. Native 3CLp (306 aa) is composed of three domains. [11] Domains I and II are chymotrypsin‐like domains with a β‐barrel fold and domain III is a 5 α‐helices globular domain that is involved in the regulation of 3CLp dimerization. A long linker (L3) [12] connects the domains II and III whereas the N‐ter and C‐ter (N‐terminal and C‐terminal) ends are located at the interface between the protomers (Figure S1). The functional and active SARS‐CoV‐2 3CLp corresponds to a homodimeric [13] cysteine protease with an unusual catalytic dyad (Cys145, His41). These are buried in a cleft between the domains I and II that is highly conserved among coronaviruses. The recognition sequence, (L,F)Q↓(S,A,G), [14] for the proteolytic cleavage (↓) requires a Gln at position P1 that is a hallmark feature shared by 3CLp of others coronaviruses,[ 15 , 16 ] and which in contrast is not present in human proteases. [17] The substrate binding site is made by 4 pockets named S1′, S1, S2 and S4[ 11 , 18 ] formed by residues from domains I and II and also by residues from the linker (L3). The active conformation of the active site is further stabilized by Ser1 from the second protomer, which stresses the functional importance of 3CLp dimerization.
The function, conservation, substrate specificity, and the absence of human homologue all contribute to make 3CLp an attractive drug target. Structural biology here plays a tremendous role as it helps to select the ligands, to get the molecular details of their interactions, and to find the proper ways to improve their potency. So far, the 3CLp inhibitors correspond either to compounds that covalently react with the catalytic Cys145, or to non‐covalent molecules that bind either in the active site or at several allosteric sites. Among the first category, there are boceprevir,[ 19 , 20 ] GC376, [21] inhibitors 11a [22] and 13b; [11] and more recently PF‐00835231.[ 23 , 24 ] In the second, molecules that bind either in its active site (MUT056399, 23R) or elsewhere on its molecular surface, including two allosteric sites (Pelitinib, AT7519), have been identified.[ 25 , 26 ] Moreover, fragment screening has also been performed to identify fragments [27] that can be grown, linked or merged to develop potent 3CLp inhibitors. [28] Numerous structural biology methods, including crystallography (X‐ray[ 11 , 18 , 22 , 25 ] or neutrons [29] ), mass spectrometry,[ 13 , 19 , 27 ] computational analyses,[ 30 , 31 , 32 ] have been used to get a better understanding of the complex structure‐function relationships in 3CLp, including the conformational flexibility of its active site,[ 11 , 12 , 18 , 19 , 25 , 30 , 33 , 34 , 35 , 36 ] and then to find or conceive inhibitors.
In this work, we used NMR spectroscopy to study the dimeric SARS‐CoV‐2 3CLp. We obtained its NMR chemical shift backbone assignment and used these data in a fragment‐based screening that led to the identification of 38 fragment hits. The deciphering of their binding sites and the conformational consequences they induced in 3CLp led to the identification of 3 protein hotspots, two located in the active site of the protease, with two different NMR signatures, and one at the dimerization interface. We further show that the fragment lead F01 binds in the active site and is, without optimization, a reversible 3CLp inhibitor with antiviral activity in SARS‐CoV‐2 infected Vero‐81 cells. The crystal structure of F01‐bound 3CLp that we have solved will help its optimization. These NMR data should help to get a better understanding of the complex interplays between the active site plasticity, the dimerization and the enzymatic activity of 3CLp. This also constitutes a new tool to assist the development of potent 3CLp inhibitors for the present or future outbreaks.
Results and Discussion
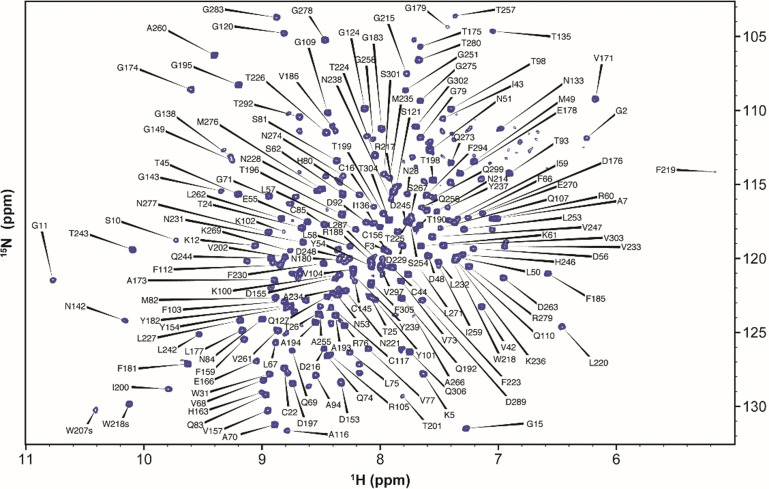
We produced SARS‐CoV‐2 3CLp samples with different isotopic labeling schemes to study by liquid‐state NMR spectroscopy. The purified protease (306 aa, 67.6 kDa) has both native N‐ and C‐terminal ends (SI and Figure S1), which is crucial for both its enzymatic activity and its proper dimerization. We obtained good quality 1H,15N‐TROSY HSQC spectrum, with ca. 280 resonances (Figure 1) and then recorded 3D 1H,15N,13C‐TROSY‐ HNCACB, ‐HN(CO)CACB, ‐HNCO, ‐HN(CA)CO, ‐HN(CO)CA spectra. Due to unfavorable magnetic relaxation properties some 13C signals were not observed and thus we had to record additional data on other samples, including 3CLp bound to boceprevir, and a monomeric 3CLp R298A mutant, to reduce the protein dynamics and the molecular weight, respectively. To perform the NMR backbone assignments of SARS‐CoV‐2 3CLp, we used a combined and integrated strategy that includes classical sequential assignment, analyses of chemical shift perturbations (CSPs) upon boceprevir binding, CS predictions and previous NMR assignments for the isolated N‐ter and C‐ter domains of SARS‐CoV 3CLp [37] (see SI). We assigned 183 proton amide correlations (183/293, 63 %) and further obtained 239, 207 and 234 chemical shifts for Cα, Cβ and C′, respectively (Figure 1, SI, BMRB entry 50780).
Figure 1.
2D 1H,15N‐TROSY‐HSQC NMR spectrum of SARS‐CoV‐2 3CLp dimer. The assignments are annotated with black labels.
Most of the unassigned proton amides lie in the first two β‐barrel domains or at the dimerization interface (Figure S2). Whereas previous attempts to record multidimensional NMR data on SARS‐CoV [38] and SARS‐CoV‐2 [39] 3CLp have failed, these new NMR data open the field to a large range of future studies of the dimeric 3CLp in solution and at temperature close to physiological, an important parameter when considering dynamics. To assess the potential of our experimental system, we analyzed the 3CLp spectral perturbations upon binding of either boceprevir or GC376 (Figures S3,S4). In both cases, the perturbations induced are highest in the active site but also propagate further in the two catalytic domains, and even toward its C‐terminal end with GC376. NMR perturbations may arise from ligand binding but also from the subsequent conformational changes. GC376 indeed induces perturbations both at the active site and at the dimerization interface, the two regions of the protease that are targeted to develop inhibitors.[ 13 , 25 , 27 , 40 ] Moreover, in the presence of GC376, a few 3CLp NMR resonances split into two new ones (Figure S5), probably highlighting the two conformations of the P3 moiety of the bound inhibitor. [20] The split resonances notably match with Val42, Asn142, Gln192 and Gly2. The later one showing that we can detect inter‐protomer conformational consequences. Interestingly, when using a R298A 3CLp monomeric mutant, we observed ca. 115 additional resonances in the 2D 1H,15N NMR spectrum that is ca. 100 more than expected. This could be due to the two orientations of the domain III that have been described for SARS‐CoV 3CLp R298A. [41] These data highlight the potential for in‐solution studies of the 3CLp. Based on the NMR assignments we are able to not only detect ligand binding and map the binding site(s), but also to analyze the conformational rearrangement(s) throughout the dimer, providing essential molecular detail for medicinal chemistry.
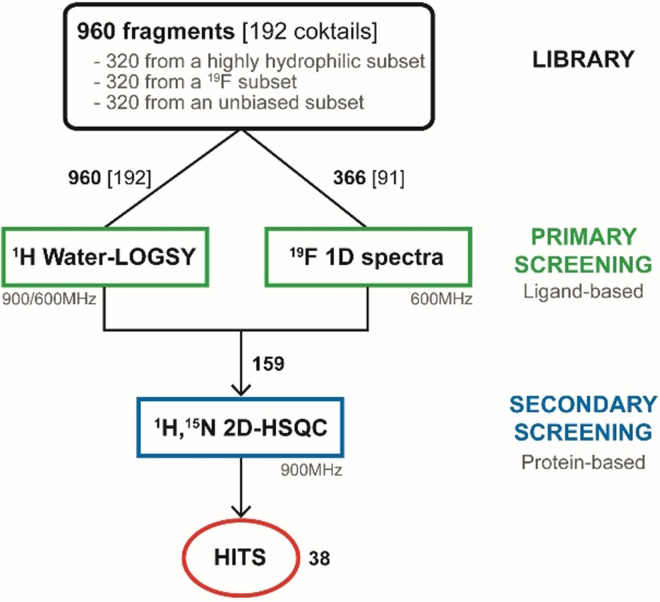
Fragment screening is widely used in drug discovery as it allows to efficiently probe the chemical space while keeping reasonable the numbers of molecule that have to be assessed. [28] The fragment hits identified (low MW) that bind to the target are then optimized to give lead compounds. We used a library of 960 commercially available fragments with physio‐chemical properties that mostly fulfill the “rule of three” criteria [42] (Figure S7a–d). We designed a strategy with a primary and a secondary screening using ligand‐ and protein‐observed NMR methods, respectively (Figure 2). The screening steps were performed in the presence of DTT, a nucleophile and reducing agent, to minimize the selection of highly electrophilic and nonspecific compounds that would covalently bind to the protease.
Figure 2.
NMR fragment screening.
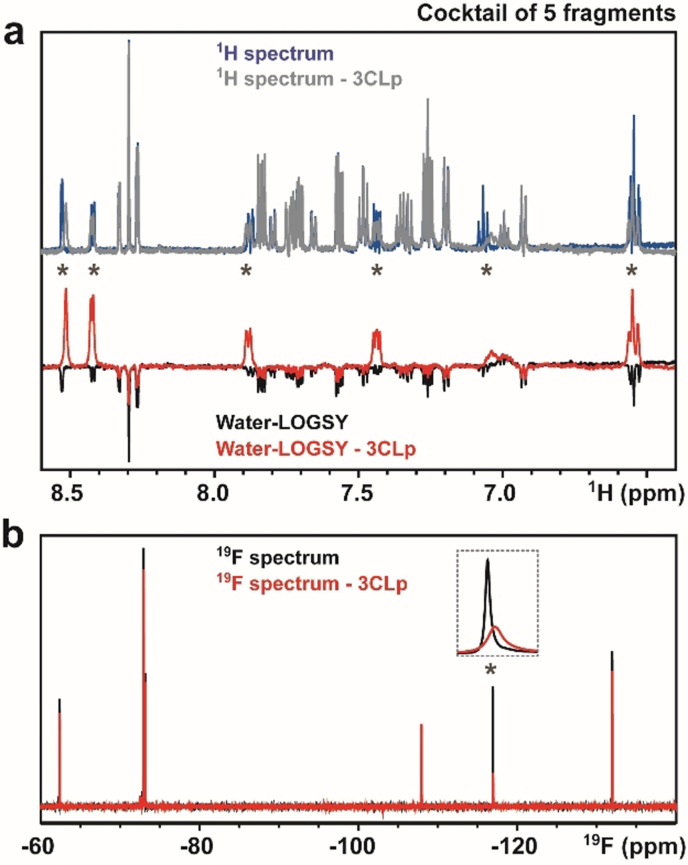
The 960 fragments were split into 192 cocktails of 5 fragments, as this strategy already proved efficient. [43] All the cocktails have been analyzed with 1H Water‐LOGSY [44] and additionally with 19F spectroscopy for 91 of them (Figure 2), as our library contains 427 fluorine fragments in total. With Water‐LOGSY, the detection of the hits is straightforward since their signals have opposite phase (Figure 3 a). When using 19F spectroscopy, the spectra only contain one NMR signal for each 19F‐fragment present in the cocktail and we monitored both CSPs and signal broadening (Figure 3 b). The primary screening led to the identification of 159 binders (Scheme S1), corresponding to a 16.6 % hit rate (Figure 2).
Figure 3.
Ligand‐based NMR primary screening. Analyses of a 5‐fragment cocktail in the absence and in the presence of unlabeled 3CLp. a) 1D 1H and 1H Water‐LOGSY spectra. b) 1D 19F‐NMR of the same cocktail. Signals, annotated with an asterisk, correspond to the F04 fragment that is a direct binder. See Scheme S1 for other cocktails.
We performed the secondary screening using 2D 1H,15N TROSY‐HSQC spectra that have been acquired on SARS‐CoV‐2 3CLp in the presence of each of the 159 binders identified in the primary screening. Using both CSPs and signal broadening (Figure 4), we confirmed 38 fragments as direct binders of 3CLp, corresponding to an overall ca. 4 % hit rate (Figures 2 and 4 and Scheme S2, Tables S1,S2). This value can be compared with the ca. 6 % obtained in a combined MS and X‐ray approach. [27] The ratio of 19F‐containing fragments in the hits (ca. 40 %) is close to the ratio in the library used. In contrast, both the average MW and lipophilicity of the fragment hits are higher than those in the entire library (Figure S7a–d).
Figure 4.
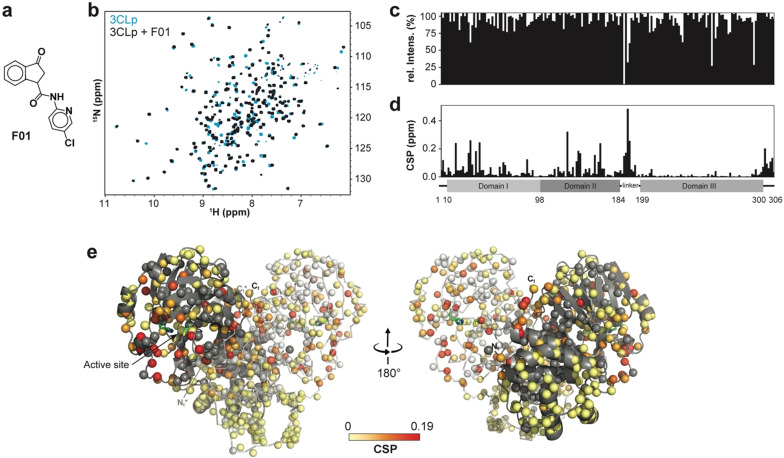
Protein‐based NMR secondary screening. a) Fragment F01 structure. b) Overlay of two 2D 1H,15N‐TROSY‐HSQC spectra acquired on 3CLp in the absence (in light blue) and in the presence (in black) of fragment F01. The broadening of the resonances and 1H and 15N‐combined CSPs induced upon fragment binding are shown along the 3CLp sequence in (c) and (d), respectively. e) Structure of the 3CLp dimer (PDB: 7k3t), with protomers A and B shown in grey and white, respectively. Each small ball represents a proton amide and thus should correspond to a resonance in the 1H,15N 2D spectrum. The CSPs, shown in (d), have been color coded (from light yellow to red) and are displayed on these balls. Unassigned residues were kept in the original color of the protomer. Catalytic His41 and Cys145 are shown in green. See Scheme S2 and Table S2 for other hits. See the SI for a color‐blind‐friendly version of this Figure.
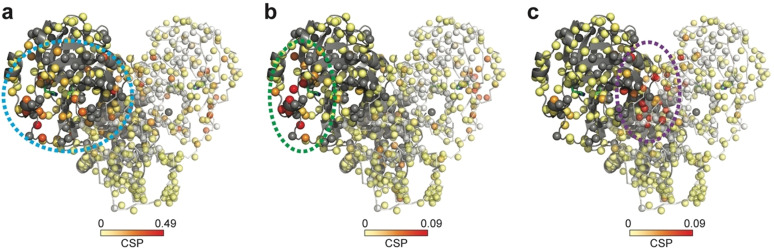
Using the backbone assignments, the analysis of the CSPs induced by the 38 hits shows that they can be grouped into three classes corresponding to three 3CLp hotspots (Figure 5; Figure S8). In Class I (24 hits), CSPs are observed for resonances assigned to residues distributed in the active site cleft, in the loop L3, and in the C‐ter end, whereas residues from the N‐ter end are only moderately affected. Class II is made by 8 hits that induce CSPs for only a restricted set of residues, in the substrate binding site, that belong exclusively to either the domain I or the tip of the loop L3 and that corresponds to the S2 and S3 binding sites. Class III (5 hits) is defined by CSPs for residues located at the dimerization interface of 3CLp (N‐ter and C‐ter ends). As to the fragment F27, it induces a strong reduction in the signal intensity all along the 3CLp sequence (Figure S8 and Table S2), and may correspond to a false positive.
Figure 5.
The 38 hits identified in the NMR screening can be grouped into three classes according to the CSPs they induced on the 2D NMR spectrum of 2H,15N‐3CLp upon binding. The representation is similar to that in Figure 4 e. a) Class I (F01): The CSPs are distributed in all the active site cleft, including the S1‐S4 substrate pockets, and extend toward the dimerization interface of the protease. b) Class II (F30): The CSPs induced correspond to a binding of the fragments in the S2 and S3 pockets, with the highest perturbations observed for residues located in a short α‐helix (Ser46‐Leu50). c) Class III (F15): Upon binding these fragments induce CSPs at the dimerization interface of 3CLp. See Figures S8‐S9. See the SI for a color‐blind‐friendly version of this Figure.
The CSPs pattern in Class I, illustrated by fragment F01, is similar to the ones observed in 3CLp upon binding of either boceprevir or GC376 (Figure S8), two potent inhibitors. The NMR CSPs induced upon binding of F01 (see Figure 4) correspond to residues distributed all along the 3CLp active site cleft (S1–S4 pockets) and indeed match with the residues involved in the binding of GC376 (Figure S9a). Moreover, the CSPs propagate toward the 3CLp dimerization interface, as with GC376 (Figure S4).
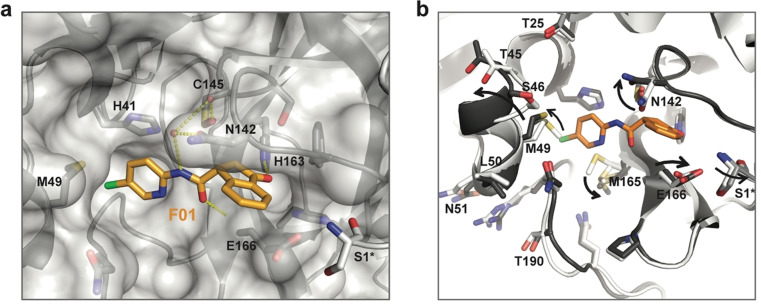
These NMR data are fully supported by the crystal structure of fragment F01‐bound 3CLp that we solved (Figure 6 and Figure S10 and Table S3; PDB: 7p51). The 3‐oxo‐2,3‐dihydro‐indene ring and 5‐chloro‐2‐pyridyl group of F01 occupy the S1 and S2 pockets of 3CLp, respectively. Three hydrogen bonds are formed between F01 and 3CLp. One of them involves the ketone in the indene ring of F01 that is electrophilic and could covalently react with the catalytic Cys145. This group, located in a key position of the active site, rather behaves as a H‐bond acceptor and interacts with His163 (see SI). The binding of F01 induces conformational changes in all the active site of 3CLp (see SI, Figure 6 and Figure S10b). It induces the displacement of: the α‐helix (Ser46–Leu50) around the S2 pocket, the loop L3 and of Asn142 and Glu166 residues around the S1 pocket. This last movement propagates to the 3CLp dimeric interface with Ser1 of protomer B being slightly displaced. It has been shown that in the 3CLp dimer, Ser1 from protomer B interacts with Glu166 of protomer A and stabilizes the active conformation of the S1 pocket.[ 11 , 18 ] Thus, the CSPs observed in 3CLp spectrum upon F01 binding both match with its binding site and the induced conformational changes through allosteric pathways (Figure S10c).
Figure 6.
Crystal structure of the fragment F01‐bound 3CLp. a) Close‐up view of the F01 binding in the active site. Protomer A is shown in grey and with surface representation, whereas protomer B is displayed in white and in cartoon representation. Three hydrogen bonds between F01 and 3CLp are displayed as yellow dashes. Residue from protomer A are labeled in black and residue S1 from protomer B is marked with an asterisk. b) Conformational changes in the F01‐bound 3CLp structure (PDB: 7p51) compared to the apo 3CLp structure (PDB: 7nts). See Figure S10.
Our data show that conformational plasticity[ 29 , 36 ] and allosteric regulations[ 13 , 25 , 35 ] within 3CLp can be studied using NMR spectroscopy, especially the tight interplay between substrate binding, active site conformation and dimerization.
The hits from Class II, such as F30, induced CSPs that would correspond to their binding into the S2 and S3 pockets located in the domain I‐side of the 3CLp substrate binding site, as SEN1269 [25] (Figure S9b). This molecule binds to S2 and induced the displacement of the short α‐helix (Ser46‐Leu50), for which we observed the highest CSPs upon binding of Class II hits (Figure S8).
The NMR CSPs induced upon binding of the Class III hits, which includes F15, are localized at the 3CLp dimeric interface and could be predicted to resemble the binding of x1086 and x1187[ 13 , 27 ] in the hydrophobic pocket made by residues both in the N‐ter (Met6, Phe8) and C‐ter (Arg298, Gln299, Val303) ends (Figure S9c). With F15, we also observed a high CSP for the resonance corresponding to Gln127, which is at the dimeric interface, and that has been shown to make a hydrogen bond with x1086.
Interestingly, no NMR perturbations observed in our screening match with fragment binding into the allosteric sites 1 and 2 that have been identified by Günther et al. [25] It could be that the binding in these two sites requires bigger and more complex molecule structures, or simply that the fragment library used did not allow to probe all the possible binding sites.
Looking at the chemical properties of the fragment hits on the basis of their Class I, II or III belonging, we found that in average Class II hits are smaller than Class I hits (avg. 233.3 Da vs. 245.7 Da), and that Class III hits are even smaller (avg. 206.85 Da) and are also more hydrophobic (80 % with 2<AlogP<3) (Figure S7).
Among the 38 hits identified in this work, F01 induced the highest CSPs in the NMR spectrum of SARS‐CoV‐2 3CLp (Table S2 and Scheme S2).
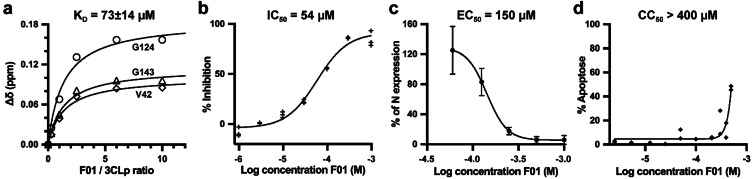
We further characterized F01, the main hit of our screening. First, using NMR titration experiments, we determined a dissociation constant K D=73±14 μM for the interaction between F01 and 3CLp (Figure 7 a and Figure S11). This affinity is higher than expected, as initial hits from fragment‐based screening usually bind to their target with a low affinity, in the 1–10 mM range. [45] Second, using an in vitro enzymatic assay, we showed that F01 is an inhibitor of 3CLp with a moderate potency (IC50=54 μM) (Figure 7 b). Third, using jump dilution assay, we showed that F01 is a reversible inhibitor of the protease (Figure S12), which agrees with the crystal structure (see Figure 6). Finally, Vero‐81 cells were infected with SARS‐CoV‐2 in the presence of increasing concentrations of F01 and then both the viral N protein cellular content was assayed and the number of infectious viral particles was determined in the cell supernatants. The results showed that F01 has antiviral activity (EC50=150 μM) against SARS‐CoV‐2 (Figure 7 c and Figure S13) while displaying a low cytotoxicity (CC50 >400 μM) (Figure 7 d).
Figure 7.
F01 is an inhibitor of 3CLp and is active against SARS‐CoV‐2 in Vero‐81 cells. a) Affinity of the Interaction between F01 and 3CLp. NMR titration curves where the 1H,15N‐combined CSPs (Δδ, ppm) were plotted as a function of the F01/3CLp ratios. The K D value (μM) corresponds to the mean (±SD) calculated over 18 3CLp resonances (Figure S11). b) F01 inhibits the in vitro enzymatic activity of 3CLp. The half‐maximal inhibitory concentration (IC50) has been calculated using the initial velocities of the reactions. c) The antiviral activity of F01 against SARS‐CoV‐2 has been tested on Vero‐81 infected cells. After infection in the presence of increasing F01 concentrations, the cells were lysed (t=16 h) and the viral N‐protein content was quantified and was used to determine the half‐maximal effective concentration (EC50). Viral titers were also measured in the cell supernatants (Figure S13). d) The 50 % cytotoxic concentration (CC50) of F01 has been assayed on Vero‐81 cells (t=20 h).
Usually, the initial fragment hits have neither in vitro nor biological activity, as they often are too small and bind to their target with very low affinity. In this work, we identified the fragment F01 that even without optimization has antiviral activity against SARS‐CoV‐2. Very recently, Bajusz et al. have reported a fragment, SX013, that blocks the SARS‐CoV‐2 replication in Vero E6 cells with an EC50 of 304 μM, [46] which is double of that for F01 in Vero‐81 cells. The ligand efficiency of F01 is 0.29–0.30 kcal mol−1 heavy atom−1 showing that F01 is a good fragment lead and deserved to be optimized in order to increase its potency and other drug related properties.[ 28 , 45 ]
Conclusion
Whereas structural biology plays a central role in drug discovery and drug development, up to date, NMR spectroscopy has not successfully been pushed forward to study the 3CLp from coronaviruses.[ 37 , 38 , 39 ] In this work, we used solution‐state NMR spectroscopy to study the dimeric 3CLp protease of the SARS‐CoV‐2, which is one of the main targets to develop efficient antivirals to fight against the COVID‐19 pandemic. Considering the high sequence conservation between the 3CLps,[ 20 , 40 ] our data will also be valuable for others β‐coronaviruses, such as MERS‐CoV and SARS‐CoV (67 % and 98 % sequence similarity, respectively), and possibly for future emerging β‐coronaviruses. Even being incomplete, the 3CLp backbone chemical shift assignment, obtained at pH and temperature close to physiological ones, has proved to be highly valuable in a medicinal chemistry project as these new NMR data allowed the study of both the structure and conformation of the dimeric protease. As a complement to the molecular dynamics,[ 12 , 30 , 35 , 47 ] these data also provide, for future studies, an experimental mean to assess the 3CLp dynamics in solution, an important point to consider in drug development.
Since mid‐2020, the world faces the apparition of SARS‐CoV‐2 variants that may, at least partially, escape to currents vaccines. This stresses that there is a need for direct acting antiviral(s) and also that there is a high risk for emergence of resistance mutations in 3CLp if targeted. To help resolve this common issue in drug development, a promising strategy consists in the combination of both orthosteric and allosteric drugs.[ 48 , 49 ] In this way, our NMR data could be valuable to identify both the allosteric sites of SARS‐CoV‐2 3CLp and the molecules that bind into, and to identify the allosteric pathways along which resistance mutations may also occur.
Using a two‐step fragment screening, we identified 38 hits, including the promising fragment F01, and three binding sites, or hotspots, located in the active site and at the dimerization interface of 3CLp. It has been shown that 3CLp can indeed be efficiently targeted at its active site, at its dimerization interface and even at different allosteric sites.[ 11 , 13 , 14 , 18 , 22 , 24 , 25 , 27 , 29 , 50 ] We showed that F01 binds to 3CLp active site with a rather good affinity (K D=73 μM), is a non‐covalent reversible inhibitor of the protease (IC50=54 μM) and demonstrates antiviral activity against SARS‐CoV‐2 (EC50=150 μM), despite no optimization. Our results indicates that F01 is a promising fragment lead that deserved to be optimized to give more potent compounds.[ 28 , 51 ] Structure‐activity relationship studies, guided by the crystal structure, will help this process and two approaches could be considered: first, F01 (Class I) could be linked or merged to Class II hits, and second, F01 could be studied in combination with fragments from Class III that bind at the dimerization interface.
This work and our NMR results will benefit to the better understanding of the complex structure‐function relationships in the dimer of 3CLp and assist the rational design of potent 3CLp inhibitors, that may both block its active site and interfere with its dimerization, to tackle current, or even future, coronaviruses pandemics. [52]
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Supporting Information
Acknowledgements
The NMR facilities were funded by the Nord Region Council, CNRS, Institut Pasteur de Lille, European Union (FEDER), French Research Ministry and University of Lille. Financial support from the IR‐RMN‐THC (FR 3050 CNRS) for the infrastructure is gratefully acknowledged. This study was supported by the I‐site ULNE (project 3CLPRO‐SCREEN‐NMR), The CPER CTRL (Transdisciplinary Research Center on Longevity) program, and the Institut Pasteur de Lille. Prof. B. Luy (Karlsruhe Institute of Technology) and Dr. D. Sinnaeve are thanked for advice about the 19F BURBOP pulses. We would like to thank T. Isabet, S. Sirigu and W. Shepard for their valuable support during data collection at beamlines PX1 and PX2A at the SOLEIL synchrotron facility (Paris, France). We thank Dr V. Villeret and Dr E. Dupre for their advice on crystallogenesis and data processing.
F.-X. Cantrelle, E. Boll, L. Brier, D. Moschidi, S. Belouzard, V. Landry, F. Leroux, F. Dewitte, I. Landrieu, J. Dubuisson, B. Deprez, J. Charton, X. Hanoulle, Angew. Chem. Int. Ed. 2021, 60, 25428.
Contributor Information
Prof. Benoit Deprez, Email: benoit.deprez@univ-lille.fr.
Dr. Xavier Hanoulle, Email: xavier.hanoulle@univ-lille.fr.
References
- 1.World Health Organization, “WHO Coronavirus (COVID-19) Dashboard,” can be found under https://covid19.who.int, 2021.
- 2. Mullard A., Lancet 2020, 395, 1751–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.F. P. Polack, S. J. Thomas, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, J. L. Perez, G. Pérez Marc, E. D. Moreira, C. Zerbini, R. Bailey, K. A. Swanson, S. Roychoudhury, K. Koury, P. Li, W. V. Kalina, D. Cooper, R. W. Frenck, L. L. Hammitt, Ö. Türeci, H. Nell, A. Schaefer, S. Ünal, D. B. Tresnan, S. Mather, P. R. Dormitzer, U. Şahin, K. U. Jansen, W. C. Gruber, C4591001 Clinical Trial Group, N. Engl. J. Med. 2020, 383, 2603–2615.
- 4.L. R. Baden, H. M. El Sahly, B. Essink, K. Kotloff, S. Frey, R. Novak, D. Diemert, S. A. Spector, N. Rouphael, C. B. Creech, J. McGettigan, S. Khetan, N. Segall, J. Solis, A. Brosz, C. Fierro, H. Schwartz, K. Neuzil, L. Corey, P. Gilbert, H. Janes, D. Follmann, M. Marovich, J. Mascola, L. Polakowski, J. Ledgerwood, B. S. Graham, H. Bennett, R. Pajon, C. Knightly, B. Leav, W. Deng, H. Zhou, S. Han, M. Ivarsson, J. Miller, T. Zaks, COVE Study Group, N. Engl. J. Med. 2021, 384, 403–416.
- 5. Voysey M., Clemens S. A. C., Madhi S. A., Weckx L. Y., Folegatti P. M., Aley P. K., Angus B., Baillie V. L., Barnabas S. L., Bhorat Q. E., Bibi S., Briner C., Cicconi P., Collins A. M., Colin-Jones R., Cutland C. L., Darton T. C., Dheda K., Duncan C. J. A., Emary K. R. W., Ewer K. J., Fairlie L., Faust S. N., Feng S., Ferreira D. M., Finn A., Goodman A. L., Green C. M., Green C. A., Heath P. T., Hill C., Hill H., Hirsch I., Hodgson S. H. C., Izu A., Jackson S., Jenkin D., Joe C. C. D., Kerridge S., Koen A., Kwatra G., Lazarus R., Lawrie A. M., Lelliott A., Libri V., Lillie P. J., Mallory R., Mendes A. V. A., Milan E. P., Minassian A. M., McGregor A., Morrison H., Mujadidi Y. F., Nana A., O'Reilly P. J., Padayachee S. D., Pittella A., Plested E., Pollock K. M., Ramasamy M. N., Rhead S., Schwarzbold A. V., Singh N., Smith A., Song R., Snape M. D., Sprinz E., Sutherland R. K., Tarrant R., Thomson E. C., Török M. E., Toshner M., Turner D. P. J., Vekemans J., Villafana T. L., Watson M. E. E., Williams C. J., Douglas A. D., Hill A. V. S., Lambe T., Gilbert S. C., Pollard A. J., Oxford COVID Vaccine Trial Group, Lancet 2021, 397, 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang C., Li W., Drabek D., Okba N. M. A., van Haperen R., Osterhaus A. D. M. E., van Kuppeveld F. J. M., Haagmans B. L., Grosveld F., Bosch B.-J., Nat. Commun. 2020, 11, 2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen X., Li R., Pan Z., Qian C., Yang Y., You R., Zhao J., Liu P., Gao L., Li Z., Huang Q., Xu L., Tang J., Tian Q., Yao W., Hu L., Yan X., Zhou X., Wu Y., Deng K., Zhang Z., Qian Z., Chen Y., Ye L., Cell. Mol. Immunol. 2020, 17, 647–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., Chai X., He R., Li X., Lv Q., Zhu H., Deng W., Xu Y., Wang Y., Qiao L., Tan Y., Song L., Wang G., Du X., Gao N., Liu J., Xiao J., Su X.-D., Du Z., Feng Y., Qin C., Qin C., Jin R., Xie X. S., Cell 2020, 182, 73–84.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E. C., Zhang Y.-Z., Nature 2020, 579, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen Y., Liu Q., Guo D., J. Med. Virol. 2020, 92, 418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R., Science 2020, 368, 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Verma N., Henderson J. A., Shen J., J. Am. Chem. Soc. 2020, 142, 21883–21890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. El-Baba T. J., Lutomski C. A., Kantsadi A. L., Malla T. R., John T., Mikhailov V., Bolla J. R., Schofield C. J., Zitzmann N., Vakonakis I., Robinson C. V., Angew. Chem. Int. Ed. 2020, 59, 23544–23548; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 23750–23754. [Google Scholar]
- 14. Rut W., Groborz K., Zhang L., Sun X., Zmudzinski M., Pawlik B., Wang X., Jochmans D., Neyts J., Młynarski W., Hilgenfeld R., Drag M., Nat. Chem. Biol. 2021, 17, 222–228. [DOI] [PubMed] [Google Scholar]
- 15. Fan K., Wei P., Feng Q., Chen S., Huang C., Ma L., Lai B., Pei J., Liu Y., Chen J., Lai L., J. Biol. Chem. 2004, 279, 1637–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chuck C.-P., Chow H.-F., Wan D. C.-C., Wong K.-B., PLoS One 2011, 6, e27228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang L., Lin D., Kusov Y., Nian Y., Ma Q., Wang J., von Brunn A., Leyssen P., Lanko K., Neyts J., de Wilde A., Snijder E. J., Liu H., Hilgenfeld R., J. Med. Chem. 2020, 63, 4562–4578. [DOI] [PubMed] [Google Scholar]
- 18. Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L. W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H., Nature 2020, 582, 289–293. [DOI] [PubMed] [Google Scholar]
- 19. Ma C., Sacco M. D., Hurst B., Townsend J. A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M. T., Chen Y., Wang J., Cell Res. 2020, 30, 678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., Zhao C., Sun H., Huang B., Niu P., Song H., Shi Y., Li X., Tan W., Qi J., Gao G. F., Nat. Commun. 2020, 11, 4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vuong W., Khan M. B., Fischer C., Arutyunova E., Lamer T., Shields J., Saffran H. A., McKay R. T., van Belkum M. J., Joyce M. A., Young H. S., Tyrrell D. L., Vederas J. C., Lemieux M. J., Nat. Commun. 2020, 11, 4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dai W., Zhang B., Jiang X.-M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.-K., Xu Y., Yang H., Liu H., Science 2020, 368, 1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hoffman R. L., Kania R. S., Brothers M. A., Davies J. F., Ferre R. A., Gajiwala K. S., He M., Hogan R. J., Kozminski K., Li L. Y., Lockner J. W., Lou J., Marra M. T., Mitchell L. J., Murray B. W., Nieman J. A., Noell S., Planken S. P., Rowe T., Ryan K., Smith G. J., Solowiej J. E., Steppan C. M., Taggart B., J. Med. Chem. 2020, 63, 12725—12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boras B., Jones R. M., Anson B. J., Arenson D., Bakowski M. A., Beutler N., Binder J., Chen E., Eng H., Hammond H., Hammond J., Haupt R. E., Hoffman R., Kadar E. P., Kania R., Kimoto E., Kirkpatrick M. G., Lanyon L., Lendy E. K., Lillis J. R., Logue J., Luthra S. A., Ma C., Mason S. W., McGrath M. E., Noell S., Obach R. S., Brien M. N. O., O'Connor R., Ogilvie K., Owen D., Pettersson M., Reese M. R., Rogers T. F., Rosales R., Rossulek M. I., Sathish J. G., Shirai N., Steppan C., Ticehurst M., Updyke L. W., Weston S., Zhu Y., White K. M., García-Sastre A., Wang J., Chatterjee A. K., Mesecar A. D., Frieman M. B., Anderson A. S., Allerton C., Nat. Commun. 2021, 12, 6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Günther S., Reinke P. Y. A., Fernández-García Y., Lieske J., Lane T. J., Ginn H. M., Koua F. H. M., Ehrt C., Ewert W., Oberthuer D., Yefanov O., Meier S., Lorenzen K., Krichel B., Kopicki J.-D., Gelisio L., Brehm W., Dunkel I., Seychell B., Gieseler H., Norton-Baker B., Escudero-Pérez B., Domaracky M., Saouane S., Tolstikova A., White T. A., Hänle A., Groessler M., Fleckenstein H., Trost F., Galchenkova M., Gevorkov Y., Li C., Awel S., Peck A., Barthelmess M., Schluenzen F., Xavier P. L., Werner N., Andaleeb H., Ullah N., Falke S., Srinivasan V., França B. A., Schwinzer M., Brognaro H., Rogers C., Melo D., Zaitseva-Doyle J. J., Knoska J., Peña-Murillo G. E., Mashhour A. R., Hennicke V., Fischer P., Hakanpää J., Meyer J., Gribbon P., Ellinger B., Kuzikov M., Wolf M., Beccari A. R., Bourenkov G., von Stetten D., Pompidor G., Bento I., Panneerselvam S., Karpics I., Schneider T. R., Garcia-Alai M. M., Niebling S., Günther C., Schmidt C., Schubert R., Han H., Boger J., Monteiro D. C. F., Zhang L., Sun X., Pletzer-Zelgert J., Wollenhaupt J., Feiler C. G., Weiss M. S., Schulz E.-C., Mehrabi P., Karničar K., Usenik A., Loboda J., Tidow H., Chari A., Hilgenfeld R., Uetrecht C., Cox R., Zaliani A., Beck T., Rarey M., Günther S., Turk D., Hinrichs W., Chapman H. N., Pearson A. R., Betzel C., Meents A., Science 2021, 372, 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kitamura N., Sacco M. D., Ma C., Hu Y., Townsend J. A., Meng X., Zhang F., Zhang X., Ba M., Szeto T., Kukuljac A., Marty M. T., Schultz D., Cherry S., Xiang Y., Chen Y., Wang J., J. Med. Chem. 2021, 10.1021/acs.jmedchem.1c00509. [DOI] [Google Scholar]
- 27. Douangamath A., Fearon D., Gehrtz P., Krojer T., Lukacik P., Owen C. D., Resnick E., Strain-Damerell C., Aimon A., Ábrányi-Balogh P., Brandão-Neto J., Carbery A., Davison G., Dias A., Downes T. D., Dunnett L., Fairhead M., Firth J. D., Jones S. P., Keeley A., Keserü G. M., Klein H. F., Martin M. P., Noble M. E. M., O'Brien P., Powell A., Reddi R. N., Skyner R., Snee M., Waring M. J., Wild C., London N., von Delft F., Walsh M. A., Nat. Commun. 2020, 11, 5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Erlanson D. A., Fesik S. W., Hubbard R. E., Jahnke W., Jhoti H., Nat. Rev. Drug Discovery 2016, 15, 605–619. [DOI] [PubMed] [Google Scholar]
- 29. Kneller D. W., Phillips G., Weiss K. L., Pant S., Zhang Q., O'Neill H. M., Coates L., Kovalevsky A., J. Biol. Chem. 2020, 295, 17365–17373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mody V., Ho J., Wills S., Mawri A., Lawson L., Ebert M. C. C. J. C., Fortin G. M., Rayalam S., Taval S., Commun. Biol. 2021, 4, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sencanski M., Perovic V., Pajovic S. B., Adzic M., Paessler S., Glisic S., Molecules 2020, 25, 3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Breidenbach J., Lemke C., Pillaiyar T., Schäkel L., Hamwi G. A., Diett M., Gedschold R., Geiger N., Lopez V., Mirza S., Namasivayam V., Schiedel A. C., Sylvester K., Thimm D., Vielmuth C., Vu L. P., Zyulina M., Bodem J., Gütschow M., Müller C. E., Angew. Chem. Int. Ed. 2021, 60, 10423–10429; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 10515–10521. [Google Scholar]
- 33. Walsh M. A., Grimes J. M., Stuart D. I., Biochem. Biophys. Res. Commun. 2021, 538, 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qiao J., Li Y.-S., Zeng R., Liu F.-L., Luo R.-H., Huang C., Wang Y.-F., Zhang J., Quan B., Shen C., Mao X., Liu X., Sun W., Yang W., Ni X., Wang K., Xu L., Duan Z.-L., Zou Q.-C., Zhang H.-L., Qu W., Long Y.-H.-P., Li M.-H., Yang R.-C., Liu X., You J., Zhou Y., Yao R., Li W.-P., Liu J.-M., Chen P., Liu Y., Lin G.-F., Yang X., Zou J., Li L., Hu Y., Lu G.-W., Li W.-M., Wei Y.-Q., Zheng Y.-T., Lei J., Yang S., Science 2021, 371, 1374–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suárez D., Díaz N., J. Chem. Inf. Model. 2020, 60, 5815–5831. [DOI] [PubMed] [Google Scholar]
- 36. Kneller D. W., Phillips G., O'Neill H. M., Jedrzejczak R., Stols L., Langan P., Joachimiak A., Coates L., Kovalevsky A., Nat. Commun. 2020, 11, 3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang S., Zhong N., Ren X., Jin C., Xia B., Biomol. NMR Assignments 2011, 5, 143–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shi J., Song J., FEBS J. 2006, 273, 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kantsadi A. L., Cattermole E., Matsoukas M.-T., Spyroulias G. A., Vakonakis I., J. Biomol. NMR 2021, 75, 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Goyal B., Goyal D., ACS Comb. Sci. 2020, 22, 297–305. [DOI] [PubMed] [Google Scholar]
- 41. Shi J., Sivaraman J., Song J., J. Virol. 2008, 82, 4620–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Congreve M., Carr R., Murray C., Jhoti H., Drug Discovery Today 2003, 8, 876–877. [DOI] [PubMed] [Google Scholar]
- 43. Valenti D., Neves J. F., Cantrelle F.-X., Hristeva S., Santo D. L., Obšil T., Hanoulle X., Levy L. M., Tzalis D., Landrieu I., Ottmann C., MedChemComm 2019, 10, 1796–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dalvit C., Fogliatto G., Stewart A., Veronesi M., Stockman B., J. Biomol. NMR 2001, 21, 349–359. [DOI] [PubMed] [Google Scholar]
- 45. Murray C. W., Rees D. C., Nat. Chem. 2009, 1, 187–192. [DOI] [PubMed] [Google Scholar]
- 46. Bajusz D., Wade W. S., Satała G., Bojarski A. J., Ilaš J., Ebner J., Grebien F., Papp H., Jakab F., Douangamath A., Fearon D., von Delft F., Schuller M., Ahel I., Wakefield A., Vajda S., Gerencsér J., Pallai P., Keserű G. M., Nat. Commun. 2021, 12, 3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Macchiagodena M., Pagliai M., Procacci P., Chem. Phys. Lett. 2020, 750, 137489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ni D., Li Y., Qiu Y., Pu J., Lu S., Zhang J., Trends Pharmacol. Sci. 2020, 41, 336–348. [DOI] [PubMed] [Google Scholar]
- 49. Lu S., Qiu Y., Ni D., He X., Pu J., Zhang J., Drug Discovery Today 2020, 25, 177–184. [DOI] [PubMed] [Google Scholar]
- 50. Liu C., Boland S., Scholle M. D., Bardiot D., Marchand A., Chaltin P., Blatt L. M., Beigelman L., Symons J. A., Raboisson P., Gurard-Levin Z. A., Vandyck K., Deval J., Antiviral Res. 2021, 187, 105020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hopkins A. L., Keserü G. M., Leeson P. D., Rees D. C., Reynolds C. H., Nat. Rev. Drug Discovery 2014, 13, 105–121. [DOI] [PubMed] [Google Scholar]
- 52.Accession codes: Backbone NMR assignments of SARS-CoV-2 3CLp have been deposited in the Biological Magnetic Resonance Data Bank (Entry 50780). Crystal structures of apo 3CLp and 3CLp in complex with fragment F01 have been deposited in the Protein Data Bank as entries 7NTS and 7P51, respectively.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Supporting Information