Abstract
As the pathogen of COVID‐19, SARS‐CoV‐2 encodes two essential cysteine proteases that process the pathogen's two large polypeptide products pp1a and pp1ab in the human cell host to form 15 functionally important, mature nonstructural proteins. One of the two enzymes is papain‐like protease or PLPro. It possesses deubiquitination and deISGylation activities that suppress host innate immune responses toward SARS‐CoV‐2 infection. To repurpose drugs for PLPro, we experimentally screened libraries of 33 deubiquitinase and 37 cysteine protease inhibitors on their inhibition of PLPro. Our results showed that 15 deubiquitinase and 1 cysteine protease inhibitors exhibit strong inhibition of PLPro at 200 μM. More comprehensive characterizations revealed seven inhibitors GRL0617, SJB2‐043, TCID, DUB‐IN‐1, DUB‐IN‐3, PR‐619, and S130 with an IC50 value below 40 μM and four inhibitors GRL0617, SJB2‐043, TCID, and PR‐619 with an IC50 value below 10 μM. Among four inhibitors with an IC50 value below 10 μM, SJB2‐043 is the most unique in that it does not fully inhibit PLPro but has a noteworthy IC50 value of 0.56 μM. SJB2‐043 likely binds to an allosteric site of PLPro to convene its inhibition effect, which needs to be further investigated. As a pilot study, the current work indicates that COVID‐19 drug repurposing by targeting PLPro holds promise, but in‐depth analysis of repurposed drugs is necessary to avoid omitting critical allosteric inhibitors.
Keywords: COVID-19, SARS-CoV-2, papain-like protease, deubiquitinase, cysteine protease
Papain‐like protease (PLpro) is a cysteine protease with deubiquitination and deISGylation activities, and is essential for SARS‐CoV‐2 replication and pathogenesis. By screening 33 deubiquitinase inhibitors and 37 cysteine protease inhibitors, we discovered that a number of molecules are potent PLpro inhibitors such as TCID, PR‐619, and SJB2‐043. Among these inhibitors, SJB2‐043 has an outstanding IC50 value of 0.56 μM, but only shows partial inhibition of papain‐like protease. It binds likely to an allosteric site of PLpro to convene its inhibition effect.
Introduction
The current prevailing COVID‐19 pandemic has caused global health and economic consequences that are often compared to that of the 1918 influenza pandemic. [1] As of June 1st, 2021, the total number of confirmed global COVID‐19 cases has exceeded 170 million, of which more than 3.5 million have succumbed to death (WHO data). Encouragingly, three COVID‐19 vaccines developed by Pfizer/BioNtech, Moderna, and Johnson & Johnson have been approved by FDA for human immunization in the United States. Although vaccines are promising in containing the pandemic, their availability does not diminish the urgent need for other effective antiviral drugs. Existing COVID‐19 vaccines are targeting the membrane Spike protein of SARS‐CoV‐2, the pathogen of COVID‐19. Spike is highly mutable. [2] New viral strains with critical mutations in Spike such as UK and South Africa SARS‐CoV‐2 strains have emerged. Effectiveness of vaccines against these new strains will be a concern. [2] Vaccines are also preventative making them not an option for the treatment of COVID‐19 patients. For quick access to effective antivirals for COVID‐19, repurposing of existing drugs has been broadly conducted. This effort has led to the identifications of a number of potential antivirals for SARS‐CoV‐2. [3] So far, remdesivir is the only antiviral that has been approved for the treatment of COVID‐19. Remdesivir is a nucleoside analog that inhibits RNA‐dependent RNA polymerase (RdRp) of SARS‐CoV‐2. It shows high potency in inhibiting SARS‐CoV‐2 in vitro but appears to be only modestly effective in treating COVID‐19 patients. [4] In view of the ongoing pandemic in its colossal scale and apocalyptic impact, there is a dire need of more effective antivirals with novel mechanisms of action to save lives of COVID‐19 patients.
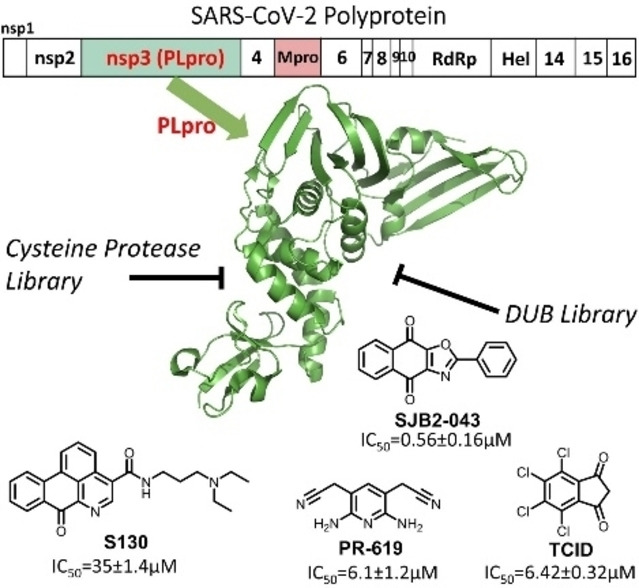
As the pathogen of COVID‐19, SARS‐CoV‐2 has a positive‐sensed genomic RNA. [5] It encodes 10 open reading frames (ORFs). As the largest ORF, ORF1ab comprises more than two thirds of the whole genome. In an infected host cell, ORF1ab is translated to two large polypeptide products, pp1a (∼500 kDa) and pp1ab (∼800 kDa), [6] by the host protein translation system. The pp1ab polypeptide is formed due to a frameshift during protein translation. Both pp1a and pp1ab need to undergo proteolytic cleavage to form 15 mature proteins. These mature proteins are nonstructural proteins (nsps) essential for the virus in its reproduction and pathogenesis. The proteolytic cleavage of pp1a and pp1ab is an autocatalytic process. Two internal polypeptide regions, nsp3 and nsp5, possess cysteine protease activities that cleave themselves and all other nsps in pp1a and pp1ab. Nsp3 is commonly referred to as papain‐like protease (PLPro) and nsp5 as main protease (MPro). [7] Both PLPro and MPro are essential to SARS‐CoV‐2. Of the two enzymes, PLPro recognizes the tetrapeptidyl LXGG motif juxtaposed between viral proteins nsp1 and nsp2, nsp2 and nsp3, and nsp3 and nsp4. Its cleavage after the LXGG motif leads to the release of nsp1, nsp2, and nsp3 which are essential for host modulation and viral replication. [8] In addition, recent studies have shown that PLPro can proteolytically remove K48‐crosslinked ubiquitin (Ub) and interferon‐stimulated gene 15 product (ISG15) that play important roles in the regulation of host innate immune responses to viral infection. [9] PLPro is also conserved across various coronaviruses. [10] For these reasons, PLPro is considered an attractive COVID‐19 drug target for the development of antivirals that may be potentially used as broad‐spectrum inhibitors for other coronaviruses as well. [11]
In view of the urgent need of antivirals for effective COVID‐19 treatments, repurposing approved drugs and late‐stage clinical drug candidates against SARS‐CoV‐2 is an efficient strategy. Significant progress has been made in drug repurposing for MPro. The first orally administered SARS‐CoV‐2 MPro inhibitor PF‐07321332 that was developed by Pfizer is currently undergoing evaluation in a Phase 1b multi‐dose study in hospitalized COVID‐19 participants. In contrast, inhibitors for PLPro remain relatively less explored because PLPro is comparably a more challenging drug target. It has a flatter active site pocket than MPro. Since PLpro is both a cysteine protease and a deubiquitinase, we reasoned that screening of cysteine protease inhibitors and deubiquitinase inhibitors would provide fast identification of PLPro inhibitors as initial hits for further optimization. In this study, we report our progress in identifying PLPro inhibitors by screening 33 deubiquitinase and 37 cysteine protease inhibitors. Among these molecules, we identified seven candidates that can potently inhibit PLPro with an IC50 value below 40 μM. Four molecules have an IC50 value below 10 μM. One molecule SBJ2‐043 can only partially inhibit PLPro but has a remarkable low IC50 value of 0.56±0.16 μM. More in‐depth investigation of these inhibitors in their mechanisms of action will likely reveal unique inhibition mechanisms for PLPro for advanced development of potent PLpro inhibitors.
Results and Discussion
PLPro is a cysteine protease with a classical Cys‐His‐Asp catalytic triad (Cys111, His272 and Asp286). It is responsible for processing three cleavage sites in the viral polypeptides pp1a and pp1ab to release nsp1, nsp2, and nsp3. Besides this function, PLPro has also deubiquitinase and deISGylase activities to help SARS‐CoV‐2 evade host immune responses.[ 9 , 12 ] Due to its both general protease and deubiquitinase nature, we screened existing deubiquitinase and cysteine protease inhibitors on their inhibition of PLPro.
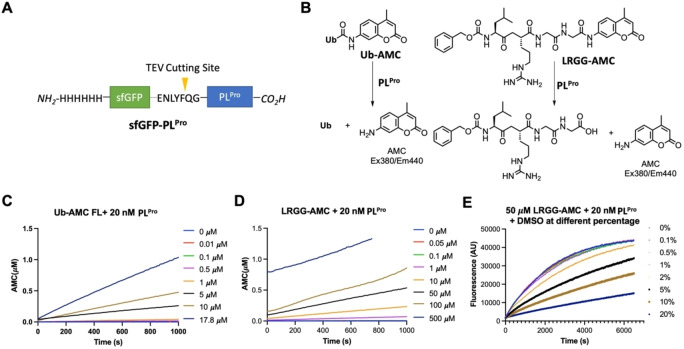
To express PLPro for experimental characterization of existing deubiquitinase and cysteine protease inhibitors, we constructed a PLPro vector for expression in E. coli (Figure 1A). In this construct, PLPro was fused to the C terminus of superfolder green fluorescent protein (sfGFP) that is known to stabilize a fused partner. [13] A 6×His tag was added to the N terminus of sfGFP for affinity purification using Ni‐NTA resins. A TEV cleavage site was inserted between sfGFP and PLPro for the proteolytic removal of PLPro from sfGFP by TEV protease. During the purification and treatment of the sfGFP‐PLPro fusion, we noticed that the cleaved PLPro quickly aggregates. Therefore, for long term storage of PLPro, we have chosen to save and use sfGFP‐PLPro instead of PLPro for all our assays. In order to test catalytic activities of PLPro, we synthesized a fluorogenic protein substrate Ub‐AMC (AMC: 7‐amino‐4‐methylcoumarin) using our recently developed activated cysteine‐directed protein ligation technique and purchased a fluorogenic peptide substrate Z‐LRGG‐AMC (Figure 1B). [14] The hydrolysis of Ub‐AMC and Z‐LRGG‐AMC releases AMC whose strong blue fluorescence can be detected by a fluorometer or a fluorescent plate reader. To obtain an optimal assay condition for inhibitor characterizations, both Ub‐AMC and Z‐LRGG‐AMC are titrated at 20 nM PLPro (Figure 1C and 1D). Based on the product formation curves, we deemed that 5 μM Ub‐AMC and 50 μM Z‐LRGG‐AMC provide easily detectable linear production formation within 1000 s and therefore used these two conditions in our inhibitor characterization assays. We have also characterized the influence of DMSO to the PLPro catalysis since most commercial small molecules are provided as 10 mM DMSO stocks. The stability of PLPro in the presence of DMSO restricts the highest drug concentration we can test. We titrated DMSO from 0.1 % to 20 % (Figure 1E). It showed that PLPro had slightly reduced activity at 2 % DMSO and this inhibition trend by DMSO increases with the DMSO concentration. Considering that most of commercial small molecules are provided as 10 mM DMSO stocks, their concentrations will be 200 μM when they are diluted by 50‐fold leading to 2 % DMSO in the final assay conditions. At 2 % DMSO, PLPro remains almost 100 % active and is good for quantifying drug inhibition effects.
Figure 1.
(A) The sfGFP‐PLPro fusion protein whose expression in E. coli has been tested. (B) Two PLPro substrates and their catalytic release of AMC. (C) The catalytic release of AMC at various concentrations of Ub‐AMC by 20 nM PLPro. (D) The catalytic release of AMC at various concentrations of Z‐LRGG‐AMC by 20 nM PLPro. (E) The catalytic release of AMC at 50 μM LRGG‐AMC by 20 nM PLPro in the presence of various concentrations of DMSO.
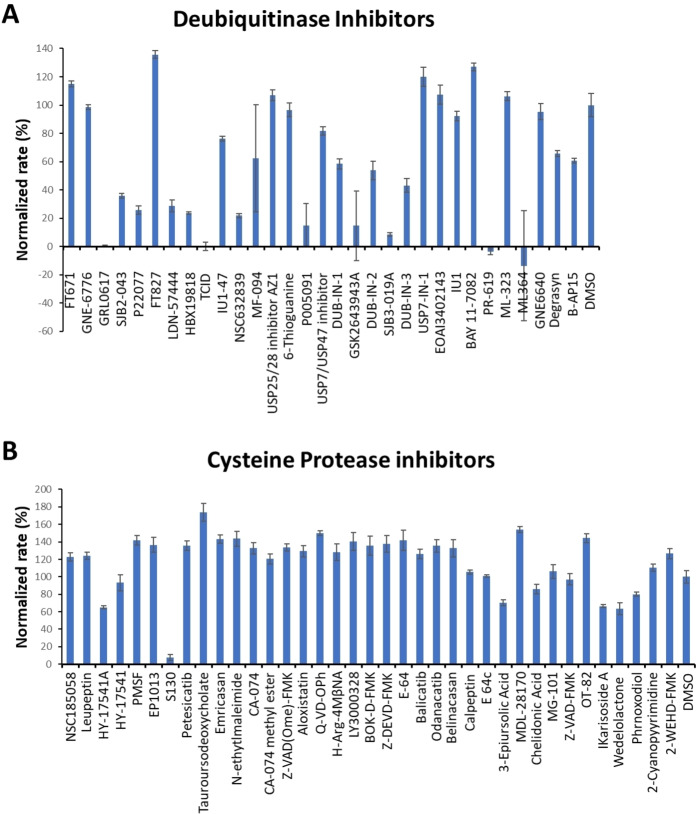
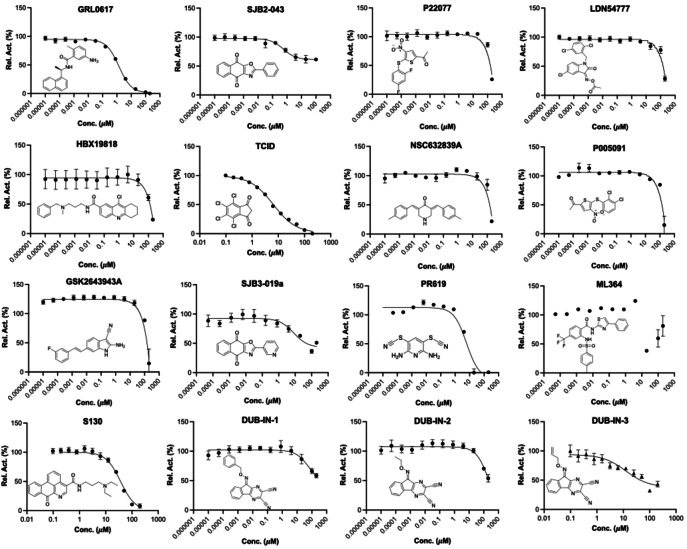
We initiated our search of PLPro inhibitors by testing 31 deubiquitinase inhibitors (Figure 2) and 37 cysteine protease inhibitors (Figure 3) that we purchased from MedChemExpress. These molecules were provided as 10 mM stocks in DMSO without further purification. Two additional deubiquitinase inhibitors C527 and VLX1570 were acquired from Cayman Scientific (Figure 2). In an initial screening assay, all acquired small molecules were analyzed at a 200 μM final concentration. In this assay, an inhibitor (400 μM) was incubated with 40 nM PLPro, 4 mM DTT, and 4 % DMSO in a reaction buffer containing 20 mM Tris and 300 mM NaCl at pH 7.5 for 30 min. 50 μL of this incubation solution was then mixed with an equal volume of a substrate solution containing 100 μM Z‐LRGG‐AMC in the reaction buffer to initiate the catalytic release of AMC. The AMC fluorescence (Ex380/Em440) was recorded in a plate reader and the linear slope within the first 10 min was calculated as the initial product formation rate. Among the deubiquitinase inhibitors at 200 μM, GRL0617 and TCID inhibited PLPro activity almost completely, PR‐619 and ML364 had significant quenching effects leading to negative signals detected for the two compounds, and SJB2‐043, P22077, LDN‐57444, HBX19818, NSC632839, P005091, GSK2643943 A and SJB3‐019 A inhibited PLPro partially but at significant levels (Figure 4A). In contrast, among 37 cysteine protease inhibitors, only S130 displayed strong inhibition of PLPro at the 200 μM level (Figure 4B and Table 2). The weak PLPro inhibition by most cysteine protease inhibitors is likely due to a much shallower active site in PLPro compared to other cysteine proteases.
Figure 2.
Structures of 33 selected deubiquitinase inhibitors.
Figure 3.
Structures of 37 selected cysteine protease inhibitors.
Figure 4.
(A) Initial screening of PLPro inhibition by 33 deubiquitinase inhibitors. (B) Initial screening of PLPro inhibition by 37 cysteine protease inhibitors. Reaction conditions: 20 nM PLPro, 50 μM Z‐LRGG‐AMC, 200 μM inhibitor, 20 mM Tris‐HCl, 300 mM NaCl, pH 7.5.
Table 2.
IC50 values of identified cysteine protease inhibitors against PLPro
|
Name |
IC50 (μM)[a] |
|---|---|
|
NSC18058 |
n.d. |
|
Leupeptin |
n.d. |
|
HY‐17541 A |
n.d. |
|
HY‐17541 |
n.d. |
|
PMSF |
n.d. |
|
EP1013 |
n.d. |
|
S130 |
35.0±1.4 |
|
Petesicatib |
n.d. |
|
Tauroursodeoxycholate |
n.d. |
|
Emricasan |
n.d. |
|
N‐Ethylmaleimide |
n.d. |
|
CA‐074 |
n.d. |
|
CA‐074 methyl ester |
n.d. |
|
Z‐VAD(OMe)‐FMK |
n.d. |
|
Aloxistatin |
n.d. |
|
Q‐VD‐OPh |
n.d. |
|
H‐Arg‐4 MβNA |
n.d. |
|
LY 3000328 |
n.d. |
|
BOC‐D‐FMK |
n.d. |
|
Z‐DEVD‐FMK |
n.d. |
|
E‐64 |
n.d. |
|
Balicatib |
n.d. |
|
Odanacatib |
n.d. |
|
Belnacasan |
n.d. |
|
Calpetin |
n.d. |
|
E 64c |
n.d. |
|
3‐Epiursolic Acid |
n.d. |
|
MDL‐28170 |
n.d. |
|
Chelidonic acid |
n.d. |
|
MG‐101 |
n.d. |
|
Z‐VAD‐FMK |
n.d. |
|
OT‐82 |
n.d. |
|
IKarisoside A |
n.d. |
|
Wedelolactone |
n.d. |
|
Phenoxodiol |
n.d. |
|
2‐Cyanopyrimidine |
n.d. |
|
Z‐WEHD‐FMK |
n.d. |
[a] n.d.: not determined.
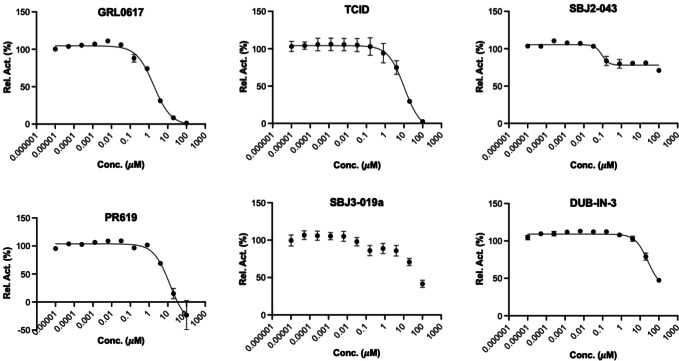
For 16 small molecules that exhibited significant inhibition of PLPro at the 200 μM level (PR‐619 and ML364 were selected as well), we further characterized their IC50 values. For IC50 characterization, AMC product formation rates at 20 nM PLPro, 50 μM Z‐LRGG‐AMC and varied concentrations of a small molecule were measured in triplicates. Data of the AMC product formation rate vs the small molecule concentration were fitted nonlinearly to a sigmodal inhibition equation to determine IC50 values for all compounds in GraphPad Prism 9. All data are presented in Figure 5. For PR‐619 and ML364, we adjusted the initial rate by subtracting autofluorescence and adding photobleaching from a provided inhibitor. After this adjustment, we noticed that ML364 did not show strong concentration‐dependent inhibition of PLPro but PR‐619 strongly inhibited PLPro at concentrations above 1 μM. Among all tested compounds, seven has a determined IC50 value below 40 μM. These include SJB2‐043 (0.56±0.16 μM), GRL0617 (1.37±0.10 μM), PR‐619 (6.1±1.2 μM), TCID (6.42±0.32 μM), SJB3‐019 A (8.15±3.59 μM), DUB‐IN‐3 (12.5±3.9 μM), and S130 (35.0±1.4 μM); see Table 1. GRL0617, TCID, PR‐619 and S130 fully inhibited PLPro at 200 μM. SJB2‐043, SJB3‐019a and DUB‐IN‐3 achieved only partial inhibition of PLPro at 200 μM. GRL0617 is a naphthalene‐based compound that was previously shown as a non‐covalent inhibitor of PLPro from SARS‐CoV with no significant inhibition of host proteases. [15] PLPro proteins from SARS‐CoV and SARS‐CoV‐2 share 83 % sequence identity and have a very high level of sequence and structural similarity in their substrate binding pockets.[ 9b , 11e , 16 ] Recent studies have showed that RL0617 can effectively inhibit PLPro of SARS‐CoV‐2.[ 9a , 16a , 17 ] In line with these recent studies, our results show that GRL0617 is effective at inhibiting PLPro from SARS‐CoV‐2 (IC50: 1.4 μM). Given its strong potency and relatively small size, structure‐activity relationship studies of GRL0617 will likely lead to more potent PLPro inhibitors with high antiviral activities. TCID is an inhibitor for ubiquitin C‐terminal hydrolase L3 with a reported IC50 of 0.6 μM. [18] TCID has two ketones that are conjugated to a highly electron withdrawing tetrachlorobenzene. Both ketones are highly prone to hydration and reaction with nucleophilic cysteine residue in PLPro. It is likely that it interacts with PLPro covalently. A structural investigation of its complex with PLPro will serve as a starting point for the development of more potent inhibitors. PR619 is a reversible and cell‐permeable inhibitor that broadly inhibits deubiquitinases. [19] The propensity of exchanging the cyano group with a protein thiolate makes PR619 likely to target cysteine proteases broadly. How exactly PR619 inactivates PLPro needs to be investigated further. S130 is an inhibitor that was originally discovered as an ATG4B inhibitor. [20] ATG4B functions similar to a deubiquitinase and hydrolyzes the preprotein of ATG8. Since mutating the catalytic cysteine in ATG4B does not significantly affect the binding of S130 to ATG4B, S130 likely interacts with ATG4B noncovalently. A similar mechanism to inhibit PLPro is also expected. S130 has a large aromatic moiety in which four rings are conjugated. It will be interesting to see how this extended large aromatic moiety interacts with PLPro to achieve strong binding. SJB2‐043, SJB3‐019a and DUB‐IN‐3 are three molecules that inhibit PLPro only partially at 200 μM. SJB2‐043 and SJB3‐019a are two structurally similar compounds that inhibit USP1. [21] Both have two quinone oxygens that potentially interact with nucleophilic residues in PLPro. Since both molecules did not inhibit PLPro completely but displayed clearly measurable IC50 values, they likely bind to an allosteric site of PLPro. Further investigations of these two molecules in their mechanisms of inhibiting PLPro will likely reveal novel targeting sites for the development of PLPro inhibitors. SJB2‐043 has the lowest IC50 value among all tested small molecules, indicating very strong binding to PLPro. A potential application of this strong binding is to use it to develop a proteasome targeting chimera for PLPro. This is a direction that we are actively exploring. DUB‐IN‐3 is a USP8 inhibitor that has two nitrile groups conjugated to a diazine. [22] The electron deficient nature of the diazine makes the two nitriles reactive toward nucleophilic residues in PLPro. It is possible that one of these two nitriles will hit on the catalytic cysteine in PLpro to form a reversible covalent complex. The low Hill coefficient of the DUB‐IN‐3 inhibition curve also indicates a complicated inhibition mechanism.
Figure 5.
IC50 characterization for 16 small molecules on their inhibition of PLPro using 50 μM LRGG‐AMC as a substrate. Experiments at different conditions were performed in triplicates.
Table 1.
IC50 values of identified deubiquitinase inhibitors against PLPro
|
Name |
IC50 (μM)[a] |
IC50 (μM)[b] |
|---|---|---|
|
FT671 |
n.d. |
|
|
GNE‐6776 |
n.d. |
|
|
GRL0617 |
1.37±0.10 |
1.80±0.21 |
|
SJB2‐043 |
0.56±0.16 |
0.091±0.024 |
|
>100 |
|
|
|
FT827 |
n.d. |
|
|
LDN‐57444 |
>100 |
|
|
HBX19818 |
>100 |
|
|
TCID |
6.42±0.32 |
10.5±2.7 |
|
IU1‐47 |
n.d. |
|
|
NSC63283 |
>100 |
|
|
MF‐094 |
n.d. |
|
|
USP25/28 inhibitor AZ1 |
n.d. |
|
|
6‐Thioguanine |
n.d. |
|
|
P005091 |
>100 |
|
|
USP7/USP47 inhibitor |
n.d. |
|
|
DUB‐IN‐1 |
>100 |
|
|
GSK2643943 A |
>100 |
|
|
DUB‐IN‐2 |
>100 |
|
|
SJB3‐019 A |
8.15±3.59 |
n.d. |
|
DUB‐IN‐3 |
12.5±3.9 |
>10 |
|
USP7‐IN‐1 |
n.d. |
|
|
EOAI3402143 |
n.d. |
|
|
IU1 |
n.d. |
|
|
BAY 11–7082 |
n.d. |
|
|
PR‐619 |
6.1±1.2 |
12.9±2.4 |
|
ML‐323 |
n.d. |
|
|
ML364 |
n.d. |
|
|
GNE‐6640 |
n.d. |
|
|
Degrasyn |
n.d. |
|
|
b‐AP15 |
n.d. |
|
|
VLX1570 |
>100 |
|
|
C527 |
n.d. |
[a] Determined using Z‐LRGG‐AMC as a PLPro substrate (n.d.: not determined). [b] Determined using Ub‐AMC as a PLPro substrate.
For six deubiquitinase inhibitors that showed high potency in inhibiting PLPro‐catalyzed AMC release from Z‐LRGG‐AMC, we have also characterized their inhibition of PLPro in its hydrolysis of Ub‐AMC. Ub‐AMC is much larger than Z‐LRGG‐AMC and involves a much bigger interface than Z‐LRGG‐AMC to interact with PLPro. For inhibitors that do not directly target the active site, they may display different inhibition characteristics when Ub‐AMC instead of Z‐LRGG‐AMC is used as a substrate. We tested all six inhibitors in the presence of 20 nM PLPro and 5 μM Ub‐AMC (Figure 6). When 5 μM Ub‐AMC was used as a substrate, IC50 values of GRL0617, TCID and PR619 were determined as 1.80±0.21, 10.5±2.7 and 12.9±2.4 μM, respectively. These IC50 values are higher than but comparable to IC50 values determined when 50 μM Z‐LRGG‐AMC was used as a substrate, indicating that all three inhibitors are likely involved in a similar mechanism in inhibiting PLPro. GRL0617 is known to bind the PLPro active site. It is highly likely that TCID and PR619 bind to the PLPro active site as well. When Ub‐AMC was used as a substrate, SJB2‐043 had a determined IC50 value of 0.091±0.024 μM. This IC50 value is much lower than the determined IC50 value when Z‐LRGG‐AMC was used as a substrate. Similar to the observed pattern when Z‐LRGG‐AMC was used as a substrate, SJB2‐043 did not inhibit PLpro completely. It is obvious that SJB2‐043 behaves very differently from GRL0617, TCID and PR619. It binds likely to an allosteric site of PLPro. This allosteric binding influences apparently the catalytic hydrolysis of Ub‐AMC more than Z‐LRGG‐AMC. This observation is possibly attributed to the much larger binding interface of Ub‐AMC that responds more sensitively to the PLPro structural perturbation. When Ub‐AMC was used as a substrate, SBJ3‐019a displayed an inhibition curve that is more complicated than that from SBJ2‐043. It showed two inhibition stages with the first leading to partial inhibition and the second continuously inhibiting PLPro without reaching its inhibition plateau at the highest concentration we tested. Since it could not be fitted to a simple inhibition mechanism, we did not calculate its IC50 value. When Ub‐AMC was used as a substrate, DUB‐IN‐3 displayed a simple inhibition curve that did not reach its plateau at the highest inhibitor concentration. Its estimated IC50 value was above 10 μM and similar to the IC50 value determined when Z‐LRGG‐AMC was used as a substrate. This similarity indicates that DUB‐IN‐3 likely binds to the PLPro active site to achieve its inhibition of PLPro.
Figure 6.
IC50 assays for 6 deubiquitinase inhibitors on their inhibition of 20 nM PLPro using 5 μM Ub‐AMC as substrate. Experiments at different conditions were performed in triplicates.
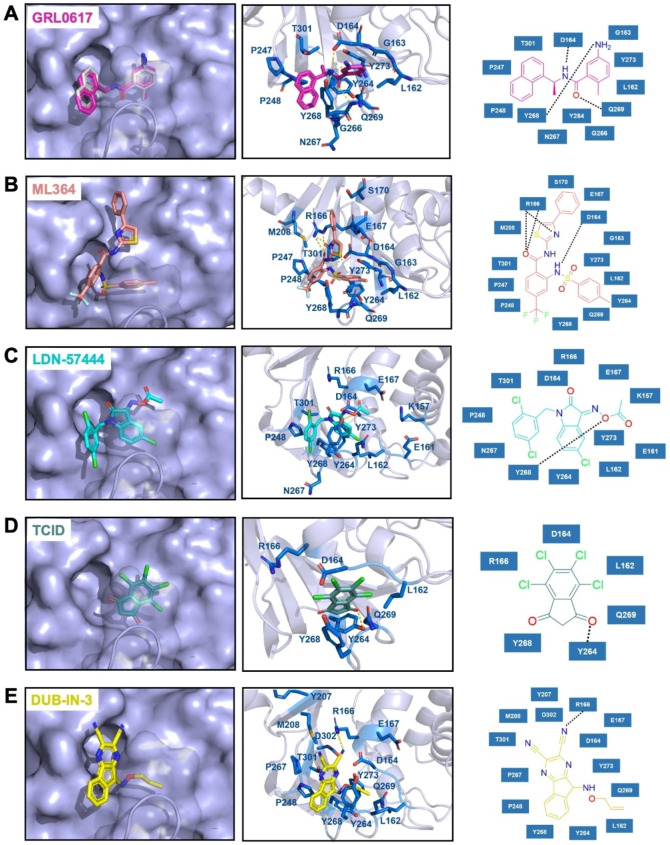
To understand how some inhibitors interact with PLPro, we conducted molecular docking using AutoDock Vina. As a starting point for docking, we used the crystal structure of PLPro bound with GRL0617 (PDB ID: 7CMD).[ 16a , 17 ] GRL0617 and water molecules were removed from the active site to prepare PLPro for the docking analysis. To simplify our analysis, we limited docking around the active site. A binding pocket was defined based on the known residues of the S3/S4 binding pocket site of PLPro. 3D Conformers of selected compounds were generated using OpenBabel. We validated the docking protocol by conducting re‐docking of GRL0617. Our docking results suggested a very similar binding mode of GRL0617 with PLPro as shown in the co‐crystal structure (Figure 7A). GRL0617 formed two hydrogen bonds between its amides and PLPro residues Asp164 and Tyr268 and one hydrogen bond between its oxygen atom and Gln269. In addition, the naphthalene group of GRL0617 forms hydrophobic interactions with aromatic residues Tyr264 and Tyr268. Other inhibitors ML364, LDN‐57444, TCID and DUB‐IN‐3 were also docked against PLPro. The protein‐ligand interactions and their detailed binding modes were illustrated in Figure 7. In the docked model, ML364 forms a hydrogen bond with Asp164 and three hydrogen bonds with Arg166 (Figure 7B). LDN‐57444, TCID, and DUB‐IN‐3 make hydrogen bonds with Tyr268, Tyr264, and Arg166, respectively (Figure 7C−E). There is no strong correlation between calculated binding energies and determined IC50 values. This discrepancy may be explained by limited factors involved in the calculation that would lead to missing some of the potentially important interactions. Other contributing factors include potential covalent interactions with the enzyme. Further crystallographic investigation will provide critical insights on how these molecules inhibit PLpro.
Figure 7.
The top binding modes of the selected compounds along with their corresponding interactions within the active site of PLPro.
Conclusion
Since 2003, there have been three coronavirus disease outbreaks. Many researchers have predicted that additional coronavirus diseases will emerge with higher frequency. For both combating the current pandemic and preparing to contain future coronavirus disease outbreaks, it is imperative to develop small molecule antivirals that may be applied generally to inhibit coronaviruses. Due to the highly conserved sequences among coronaviruses, PLPro is an attractive drug target for developing broad‐spectrum antivirals. In this study, we experimentally characterized 33 deubiquitinase inhibitors and 37 cysteine protease inhibitors on their inhibition of PLPro from SARS‐CoV‐2. From this study, we identified seven molecules that potently inhibit PLPro with an IC50 value below 40 μM when Z‐LRGG‐AMC was used a substrate. Five inhibitors GRL0617, SJB2‐043, TCID, SJB3‐019 A, and PR‐619 have an IC50 value below 10 μM. Interestingly, SBJ2‐043 only partially inhibits PLPro but has an outstanding IC50 value of 0.56 μM. When Ub‐AMC was used as a substrate, an even lower IC50 value of 0.091 μM was determined. SJB2‐043 binds likely to an allosteric site of PLPro to exert its inhibition effect. As a pilot study, the current work indicates that drug repurposing for COVID‐19 by targeting PLPro holds promises but in‐depth investigation of these inhibitors in their mechanisms of action is necessary for the development of more potent PLPro inhibitors based on them.
Materials and Methods
Chemicals
The deubiquitinase inhibitor library and cysteine protease library were purchased from MedChemExpress. Two other deubiquitinase inhibitors, C527 and VLX1579, were purchased from Cayman Chemicals. The PLPro substrate, Z‐LRGG‐AMC, was purchased from Cayman Chemicals.
In‐silico molecular docking study
Protein preparation: The crystal structure of the SARS‐CoV‐2 PLPro in complex with inhibitor GRL0617 (PDB ID : 7CMD) was obtained from the RCSB Protein Data Bank (https://www.rcsb.org/). Only chain A of the homo‐tetrameric structure was used in our docking study. The cognate ligand was removed from the structure that was applied in the following redocking test. In AutoDockTools‐1.5.7, water molecules were deleted and polar hydrogens were added to the structure. Finally, the prepared protein structure was converted into a PDBQT file for further docking studies.
Ligand preparation: Compounds in SDF format were converted into PDB files using OpenBabel‐3.1.1. These PDB files were further converted into PDBQT files via AutoDockTools‐1.5.7 with default options.
Docking parameters and method: In addition to receptor and ligand preparations, AutoDockTools‐1.5.7 was also used for grid parameter setting. The cognate ligand of crystal structure 7CMD suggested the inhibitor binding site. A grid box with dimensions 30×30×30 centered at the coordinates X=−30.0, Y=−12.0, and Z=−30.0 was used to represent the search space. Then we applied AutoDock Vina docking protocol with options of 20 CPUs to use and maximum 100 binding modes to generate. Only modes with the highest binding affinities were shown in this paper.
Docking validation: We redocked the cognate ligand GRL0617 into its cognate receptor in order to verify the correctness of our docking method and parameters. The top row of Figure 7A showed the docked conformation in magenta and the crystal conformation in grey. The superposition of two conformations demonstrated the feasibility of our molecular docking method.
Expression and purification of PLPro
PLPro gene fragment was purchased from Integrated DNA Technologies (IDT), amplified with the primer pair that has XbaI and XhoI cleavage sites. The amplified gene was inserted into pBAD‐sfGFP vector. In the final construct (pBAD‐sfGFP‐ PLPro), a hexahistidine tag is located at the N‐terminus of sfGFP. A TEV cutting site is placed after the C‐terminus of sfGFP which is followed by PLPro. Chemically competent Top10 E. coli cells were transformed with the pBAD‐sfGFP‐ PLPro plasmid. Then a single colony was picked, inoculated into 10 mL of 2YT with ampicillin and grew overnight at 37 °C. The overnight culture was inoculated to 1 L of 2YT with 100‐fold dilution and grew until OD 600 (optical density at 600 nm) reached 0.6. Then, the medium was cooled down to 20 °C and induced with 0.2 % L‐arabinose for 24 h at 20 °C. The cells were collected by centrifuging at 6000 rpm for 30 mins and resuspended in lysis buffer (20 mM Tris, 250 mM NaCl, 5 % glycerol, 0.2 % TritonX‐100 and 1 mg/mL lysozyme at pH 7.8). Collected cells were lysed by sonication at 60 % amplitude with 1 sec on 4 sec off for a total of 5 min. The cell debris was removed by centrifuging at 16000 rpm for 30 min and the supernatant was loaded onto 3 mL of Ni‐NTA resins (Genscript). A gravity flow Ni‐NTA chromatography was performed. The Ni‐NTA resin was washed with 10 times resin bed volume of wash buffer (20 mM Tris, 250 mM NaCl and 10 mM imidazole at pH 7.8) and eluted with 20 mL elution buffer (20 mM Tris, 30 mM NaCl and 300 mM imidazole at pH 7.8). The buffer was changed to 20 mM Tris and 30 mM NaCl with 20 % glycerol by using HiPrep 26/10 desalting column. Finally, the protein was aliquoted, flash frozen with liquid nitrogen and stored in –80 °C freezer.
Expression and purification of Ub‐G76 C‐6H
The Ub‐G76 C‐6H was expressed according to the protocol described before. [14] Briefly, an overnight starting culture was inoculated in 2YT medium using a 1 : 100 dilution ratio. The cells were grown at 37 °C with 250 rpm. The protein was induced with 1 mM of IPTG overnight at 18 °C when OD 600 reached 0.7–0.9. The next morning, cells were harvested by centrifugation at 6000 rpm for 30 min and lysed by sonication in lysis buffer (50 mM NaH2PO4, pH 7.8, 500 mM NaCl, 5 mM imidazole, 1 mM TCEP). The whole cell lysate was then clarified by centrifugation (16000 rpm, 30 min, 4 °C). The supernatant was collected and 6 M HCl was added dropwise until it reached pH 2. The acidic supernatant was stirred for 5–10 min to allow impurities to precipitate. The impurities were removed by centrifugation (10000 rpm, 30 min, 4 °C) and the supernatant was neutralized to pH 7.8 using 6 M NaOH solution. Subsequently, the neutralized supernatant was loaded onto Ni‐NTA resins to perform a gravity flow chromatography. The resin was washed with wash buffer (50 mM NaH2PO4, 500 mM NaCl, 30 mM imidazole, 1 mM TCEP at pH 7.8). Then the target protein was eluted with elution buffer (washing buffer substituting 20 mM imidazole with 300 mM imidazole). The elution was desalted to 50 mM ammonium bicarbonate (ABC) buffer using a HiPrep Desalting column and lyophilized into 100 nmol per aliquot for further use.
Synthesis of Ub‐AMC
Ub‐AMC was synthesized using the Activated Cysteine‐directed Protein Ligation (ACPL) technique described before. [14] 100 nmol of lyophilized Ub‐G76 C‐6H was dissolved in 194.5 μL of 50 mM borate buffer (pH 9) supplied with 0.5 μL of 500 mM TCEP stock solution. It was then mixed with 300 μL of 40 mM Gly‐AMC solution in DMSO to achieve a reaction mixture containing 24 mM Gly‐AMC, 20 mM borate and 60 % DMSO. The ACPL reaction was initiated by adding 5 μL of 500 mM NTCB stock solution in DMSO and the pH value was carefully adjusted to 9 using 6 M NaOH solution. The reaction was performed at 37 °C for 16 h and terminated by desalting to 50 mM ABC buffer using a HiTrap Desalting column. The desalted sample was further purified using Ni‐NTA purification by collecting the flow‐through. The purified Ub‐AMC was analyzed by Orbitrap ESI‐MS, then flash frozen and stored in −80 °C freezer.
Screening assay
To have a preliminary test of inhibition ability of each drug, a screening assay was performed. 50 μL reaction solutions with 40 nM of PLPro and 400 μM of each drug (deubiquitinase inhibitors and cysteine protease inhibitors) were preincubated in PLPro assay buffer (20 mM Tris, 300 mM NaCl, pH 7.5) with 4 mM DTT in 37 °C for 30 min. It was then mixed with 50 μL solutions with 100 μM of Z‐LRGG‐AMC. The final assay solution had 200 μM of each compound, 20 nM PLPro, 2 mM DTT and 50 μM Z‐LRGG‐AMC. The fluorescence of AMC that was generated as a result of PLPro enzymatic activity was recorded in a plate reader using 380 nm as the excitation wavelength and 440 nm as the emission wavelength. The initial slope of the fluorescence‐vs‐time graph for each drug was analyzed by calculating the slope of the 0–10 min curve. The calculation was done by using linear regression analysis in GraphPad 9.0.
Inhibition analysis
The inhibition assay was performed in the same way as the screening assay except a series of inhibition concentrations were used. All the reactions were performed in triplicates. Reaction rates that were normalized against the rate in the absence of an inhibitor were used to determine IC50 values using GraphPad 9.0.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported in part by Welch Foundation (grant A‐1715), Texas A&M Presidential Impact Fellowship Fund and Texas A&M X‐Grant. The development of ACPL and its use to generate different ubiquitin related products was supported by National Institutes of Health (Grant R01 GM127575).
C.-C. Cho, S. G. Li, T. J. Lalonde, K. S. Yang, G. Yu, Y. Qiao, S. Xu, W. Ray Liu, ChemMedChem 2022, 17, e202100455.
Contributor Information
Shiqing Xu, Email: shiqing.xu@tamu.edu.
Wenshe Ray Liu, Email: wsliu2007@tamu.edu.
References
- 1.
- 1a. Gates B., N. Engl. J. Med. 2020, 382, 1677–1679; [DOI] [PubMed] [Google Scholar]
- 1b. Morens D. M., Daszak P., Taubenberger J. K., N. Engl. J. Med. 2020, 382, 1293–1295. [DOI] [PubMed] [Google Scholar]
- 2. Morse J. S., Lalonde T., Xu S., Liu W. R., ChemBioChem 2020, 21, 730–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vatansever E. C., Yang K. S., Drelich A. K., Kratch K. C., Cho C. C., Kempaiah K. R., Hsu J. C., Mellott D. M., Xu S., Tseng C. K., Liu W. R., Proc. Natl. Acad. Sci. USA 2021, 118, e2012201118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y., Luo G., Wang K., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yang J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y., Cao L., Guo T., Wan Y., Qin H., Jiang Y., Jaki T., Hayden F. G., Horby P. W., Cao B., Wang C., Lancet 2020, 395, 1569–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.
- 5a. Lam T. T., Jia N., Zhang Y. W., Shum M. H., Jiang J. F., Zhu H. C., Tong Y. G., Shi Y. X., Ni X. B., Liao Y. S., Li W. J., Jiang B. G., Wei W., Yuan T. T., Zheng K., Cui X. M., Li J., Pei G. Q., Qiang X., Cheung W. Y., Li L. F., Sun F. F., Qin S., Huang J. C., Leung G. M., Holmes E. C., Hu Y. L., Guan Y., Cao W. C., Nature 2020, 583, 282–285; [DOI] [PubMed] [Google Scholar]
- 5b.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, Nat. Microbiol. 2020, 5, 536–544;
- 5c. Zhou P., Yang X. L., Wang X. G., Hu B., Zhang L., Zhang W., Si H. R., Zhu Y., Li B., Huang C. L., Chen H. D., Chen J., Luo Y., Guo H., Jiang R. D., Liu M. Q., Chen Y., Shen X. R., Wang X., Zheng X. S., Zhao K., Chen Q. J., Deng F., Liu L. L., Yan B., Zhan F. X., Wang Y. Y., Xiao G. F., Shi Z. L., Nature 2020, 579, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Phan T., Infect. Genet. Evol. 2020, 81, 104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen Y. W., Yiu C. B., Wong K. Y., F1000Research 2020, 9, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Naqvi A. A. T., Fatima K., Mohammad T., Fatima U., Singh I. K., Singh A., Atif S. M., Hariprasad G., Hasan G. M., Hassan M. I., Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.
- 9a. Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., Schulz L., Widera M., Mehdipour A. R., Tascher G., Geurink P. P., Wilhelm A., van der Heden van Noort G. J., Ovaa H., Muller S., Knobeloch K. P., Rajalingam K., Schulman B. A., Cinatl J., Hummer G., Ciesek S., Dikic I., Nature 2020, 587, 657–662; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9b. Klemm T., Ebert G., Calleja D. J., Allison C. C., Richardson L. W., Bernardini J. P., Lu B. G., Kuchel N. W., Grohmann C., Shibata Y., Gan Z. Y., Cooney J. P., Doerflinger M., Au A. E., Blackmore T. R., van der Heden van Noort G. J., Geurink P. P., Ovaa H., Newman J., Riboldi-Tunnicliffe A., Czabotar P. E., Mitchell J. P., Feltham R., Lechtenberg B. C., Lowes K. N., Dewson G., Pellegrini M., Lessene G., Komander D., EMBO J. 2020, 39, e106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baez-Santos Y. M., St John S. E., Mesecar A. D., Antiviral Res. 2015, 115, 21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.
- 11a. McClain C. B., Vabret N., Signal Transduct. Target Ther. 2020, 5, 223; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11b. Totura A. L., Bavari S., Expert Opin. Drug Discovery 2019, 14, 397–412; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11c. Liu C., Zhou Q., Li Y., Garner L. V., Watkins S. P., Carter L. J., Smoot J., Gregg A. C., Daniels A. D., Jervey S., Albaiu D., ACS Cent. Sci. 2020, 6, 315–331; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11d. Petushkova A. I., A. A. Zamyatnin, Jr. , Pharmaceuticals (Basel) 2020, 13, 277; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11e. Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S. J., El Oualid F., Huang T. T., Bekes M., Drag M., Olsen S. K., Sci. Adv. 2020, 6, eabd4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.
- 12a. Lei J., Kusov Y., Hilgenfeld R., Antiviral Res. 2018, 149, 58–74; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12b. Lindner H. A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Menard R., J. Virol. 2005, 79, 15199–15208; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12c. Ratia K., Saikatendu K. S., Santarsiero B. D., Barretto N., Baker S. C., Stevens R. C., Mesecar A. D., Proc. Natl. Acad. Sci. USA 2006, 103, 5717–5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu M., Wang B., Wang F., Yang Z., Gao D., Zhang C., Ma L., Yu X., Appl. Microbiol. Biotechnol. 2019, 103, 6071–6079. [DOI] [PubMed] [Google Scholar]
- 14. Qiao Y., Yu G., Kratch K. C., Wang X. A., Wang W. W., Leeuwon S. Z., Xu S., Morse J. S., Liu W. R., J. Am. Chem. Soc. 2020, 142, 7047–7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.
- 15a. Ratia K., Pegan S., Takayama J., Sleeman K., Coughlin M., Baliji S., Chaudhuri R., Fu W., Prabhakar B. S., Johnson M. E., Baker S. C., Ghosh A. K., Mesecar A. D., Proc. Natl. Acad. Sci. USA 2008, 105, 16119–16124; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15b. Baez-Santos Y. M., Barraza S. J., Wilson M. W., Agius M. P., Mielech A. M., Davis N. M., Baker S. C., Larsen S. D., Mesecar A. D., J. Med. Chem. 2014, 57, 2393–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.
- 16a. Fu Z., Huang B., Tang J., Liu S., Liu M., Ye Y., Liu Z., Xiong Y., Zhu W., Cao D., Li J., Niu X., Zhou H., Zhao Y. J., Zhang G., Huang H., Nat. Commun. 2021, 12, 488; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16b. Ratia K., Pegan S., Takayama J., Sleeman K., Coughlin M., Baliji S., Chaudhuri R., Fu W., Prabhakar B. S., Johnson M. E., Baker S. C., Ghosh A. K., Mesecar A. D., Proc. Natl. Acad. Sci. USA 2008, 105, 16119–16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gao X., Qin B., Chen P., Zhu K., Hou P., Wojdyla J. A., Wang M., Cui S., Acta Pharm. Sin. B 2021, 11, 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu Y., Lashuel H. A., Choi S., Xing X., Case A., Ni J., Yeh L. A., Cuny G. D., Stein R. L., P. T. Lansbury, Jr. , Chem. Biol. 2003, 10, 837–846. [DOI] [PubMed] [Google Scholar]
- 19. Altun M., Kramer H. B., Willems L. I., McDermott J. L., Leach C. A., Goldenberg S. J., Kumar K. G., Konietzny R., Fischer R., Kogan E., Mackeen M. M., McGouran J., Khoronenkova S. V., Parsons J. L., Dianov G. L., Nicholson B., Kessler B. M., Chem. Biol. 2011, 18, 1401–1412. [DOI] [PubMed] [Google Scholar]
- 20. Fu Y., Hong L., Xu J., Zhong G., Gu Q., Gu Q., Guan Y., Zheng X., Dai Q., Luo X., Liu C., Huang Z., Yin X. M., Liu P., Li M., Autophagy 2019, 15, 295–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mistry H., Hsieh G., Buhrlage S. J., Huang M., Park E., Cuny G. D., Galinsky I., Stone R. M., Gray N. S., D′Andrea A. D., Parmar K., Mol. Cancer Ther. 2013, 12, 2651–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Colombo M., Vallese S., Peretto I., Jacq X., Rain J. C., Colland F., Guedat P., ChemMedChem 2010, 5, 552–558. [DOI] [PubMed] [Google Scholar]