Abstract
Coronavirus disease 2019 (COVID‐19) manifests with high clinical variability and warrants sensitive and specific assays to analyze immune responses in infected and vaccinated individuals. Using Single Molecule Arrays (Simoa), we developed an assay to assess antibody neutralization with high sensitivity and multiplexing capabilities based on antibody‐mediated blockage of the ACE2‐spike interaction. The assay does not require live viruses or cells and can be performed in a biosafety level 2 laboratory within two hours. We used this assay to assess neutralization and antibody levels in patients who died of COVID‐19 and patients hospitalized for a short period of time and show that neutralization and antibody levels increase over time. We also adapted the assay for SARS‐CoV‐2 variants and measured neutralization capacity in pre‐pandemic healthy, COVID‐19 infected, and vaccinated individuals. This assay is highly adaptable for clinical applications, such as vaccine development and epidemiological studies.
Keywords: COVID-19, multiplexing, neutralization assays, single molecule arrays
Using Single Molecule Arrays (Simoa), we developed a simple neutralization assay based on antibody‐mediated blockage of the ACE2‐spike interaction. This assay can assess antibody neutralization with high sensitivity and multiplexing capabilities. We used this assay to profile neutralization capacity in patients infected with SARS‐CoV‐2 and vaccinated individuals and show that this assay is useful for various clinical applications.
Introduction
The coronavirus disease 2019 (COVID‐19), which is caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), has led to high morbidity and mortality globally. Serological assays for detecting SARS‐CoV‐2 antibodies are of critical importance for several reasons. First, serological assays provide a better understanding of the virus‐induced host immune response.[ 1 , 2 ] Second, screening convalescent plasma for immunoglobulin content can improve the efficacy of antibody therapy. [3] Third, there is an urgent need to evaluate novel vaccines by detecting the levels and types of immunoglobulins produced in response to different vaccines, doses, and schedules. [4] Finally, serological assays can identify asymptomatic individuals with previous exposure to SARS‐CoV‐2.
There are currently two main serological assay formats. The first format is based on the enzyme‐linked immunosorbent assay (ELISA). ELISA can provide relative quantification of antibody levels and can be implemented at the point‐of care; however, it cannot distinguish between neutralizing and non‐neutralizing antibodies, limiting its utility. The second format is the live‐virus neutralization assay.[ 5 , 6 , 7 ] This assay detects neutralizing antibodies in a biological sample, such as blood, and is currently the gold standard method for determining antibody efficacy. However, it requires use of infectious SARS‐CoV‐2 virus particles, which pose safety considerations and require handling in a biosafety level (BSL) 3 lab. As a result, there have been efforts to develop simpler antibody neutralization assays. [8] For example, the pseudovirus neutralization assay overcomes the need for live virus and can be conducted in a BSL‐2 lab.[ 9 , 10 ] However, the pseudovirus neutralization assay still has several challenges. [9] First, generating pseudoviruses is a complex process and requires many steps including packaging, purifying, and titrating the recombinant virus. Second, the assay requires use of live cells, which adds additional complexity and reduces assay reproducibility. Finally, despite being a BSL‐2 assay, using a live cell that has been engineered to be sensitive to infection has biosafety implications. The presence of a single replication‐competent viral particle in the sample can cause amplification of the virus. Having a purely protein‐based assay circumvents these challenges.
In this work, we developed a SARS‐CoV‐2 neutralization assay based on antibody‐mediated blockage of the ACE2‐spike interaction without the need for live cells or viruses using Single Molecule Arrays (Simoa). Previously, we have developed competitive Simoa assays, which are ≈50 fold more sensitive than a conventional competitive ELISA. [11] Here, we show that a competitive Simoa assay can be adapted and used to assess antibody neutralization in blood. This assay has high sensitivity and multiplexing capabilities. We used this assay for two applications. First, we measured the neutralization capacity and antibody levels in hospitalized COVID‐19 patients. We show that there is an overall increase in neutralization capacity and antibody levels over time. Second, we developed a multiplex Simoa neutralization assay against the SARS‐CoV‐2 receptor binding domain and three variants, and measured the neutralization capacity in pre‐pandemic healthy, COVID‐19 positive, and vaccinated individuals. The Simoa‐based SARS‐CoV‐2 neutralization assay presented here is a cell‐free, quick, and highly sensitive alternative to live‐virus neutralization assays and can be used for a variety of clinical applications. The assay is versatile and can be easily adapted towards future detection of antibody neutralization for other infectious diseases.
Results and Discussion
Development of an Antibody Neutralization Assay Using Single Molecule Arrays (Simoa)
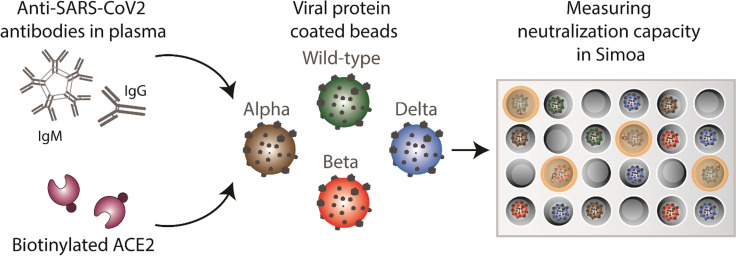
We developed a competitive bead‐based antibody neutralization assay using Single Molecule Arrays (Simoa). [12] In this assay format (Figure 1), SARS‐CoV‐2 spike protein is first conjugated to micron‐sized beads, creating a spike‐decorated surface that resembles the virus. Then, biotinylated ACE2 and a sample containing antibodies against SARS‐CoV‐2 spike are incubated with the beads, followed by addition of streptavidin coated beta‐galactosidase, which can bind to the biotinylated ACE2. The beads are then resuspended in a fluorogenic substrate and loaded onto an array of femtoliter sized wells in which each well can fit only one bead. The wells are sealed with oil and active wells are counted using the units of average enzyme per bead (AEB). [13] In the absence of anti‐spike antibodies, the biotinylated ACE2 binds to the spike‐coated beads, leading to high signal. In the presence of anti‐spike antibodies, the biotinylated ACE2 competes with the antibodies for the binding sites on the bead and thus, as the concentration of anti‐spike antibodies in the sample increases, the signal in the assay decreases. It is important to note that the timing of incubations must be kept constant across different samples in this competitive immunoassay since the amount of antibody as well as antibody avidity varies between the plasma samples.[ 11 , 14 ] To determine the antibody neutralization capacity (NT50), we serially diluted each patient plasma sample and then determined the dilution factor that corresponded to 50 % loss in assay signal.
Figure 1.
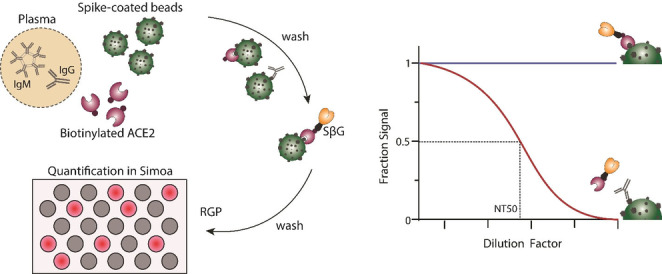
Simoa neutralization assay. The assay is based on antibody‐mediated blockage of the ACE2‐spike interaction. Spike‐coated beads, biotinylated ACE2, and patient plasma are mixed. After several washing steps, streptavidin conjugated beta‐galactosidase (SβG) is added. The beads are then washed again, resuspended in resorufin β‐D‐galactopyranoside (RGP), and loaded into a microwell array for imaging. As the concentration of anti‐spike antibodies in the sample increases, the signal in the assay decreases. To determine neutralization capacity (NT50), the plasma sample is diluted and the signal at each dilution factor is normalized to the AEB of the highest dilution factor.
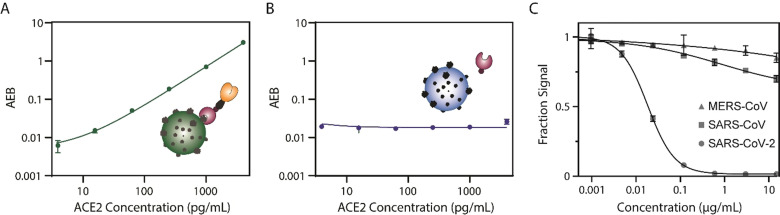
To develop the Simoa assay, we first prepared beads coated with the SARS‐CoV‐2 spike protein (Methods section and Figure S1). We then added increasing concentrations of ACE2 and showed that the signal also increased, confirming that ACE2 interacts with the spike‐coated beads (Figure 2 A). We also tested beads coated with the SARS‐CoV‐2 nucleocapsid protein (Figure S1) as a control, and observed that as the concentration of ACE2 increases, the signal of the assay does not change, as expected (Figure 2 B). As another validation, we tested the assay using a neutralizing anti‐spike monoclonal antibody (40592‐R001) and observed that as the concentration of the neutralizing antibody increases, the assay signal decreases to background level, as expected (Figure 2 C). To assess the potential cross‐reactivity of the assay, we tested antibodies against other coronaviruses, including SARS‐CoV and MERS‐CoV (Figure 2 C). Even at a high concentration (15 μg mL−1) of anti SARS‐CoV (40069‐R723) and anti MERS‐CoV (CR‐3022) antibodies, there is minimal neutralization capacity and the assay signal decreases by only 15 % and 30 %, respectively. These results suggest that this assay could be used for various applications, such as comparing and quantifying the neutralizing capacity of different therapeutic antibodies.
Figure 2.
Development of the Simoa neutralization assay for SARS‐CoV‐2 spike and nucleocapsid. A) As the concentration of ACE2 increases using spike‐coated beads, so does the signal. B) As the concentration of ACE2 increases using nucleocapsid‐coated beads, the signal does not increase. C) As the concentration of a neutralizing SARS‐CoV‐2 antibody increases, the assay signal decreases. As the concentration of SARS‐CoV and MERS‐CoV antibodies increases, the assay signal decreases slightly. All measurements were obtained in duplicate.
Simoa Assay Validation in Serological Samples
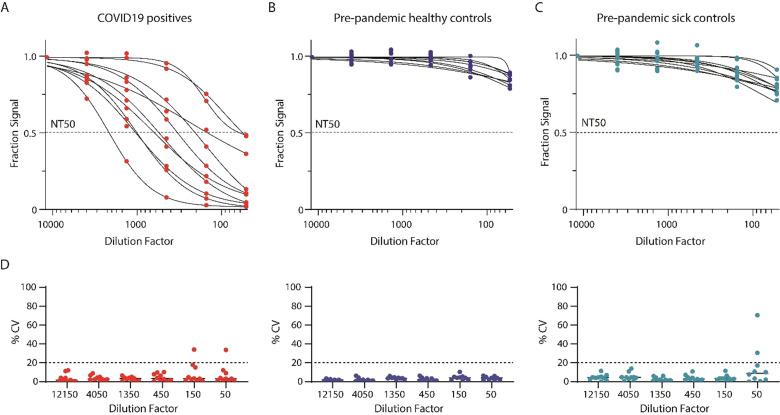
To assess whether the assay can be used in clinical samples, we tested plasma from COVID‐19 patients (Figure 3 A) and pre‐pandemic patients, including patients with various respiratory diseases, (Figure 3 B and C) using the neutralization assay. All ten of the COVID‐19 patients had antibodies with neutralizing capacity in their plasma (Figure 3 A and Table S1). For the ten pre‐pandemic controls, we could not calculate NT50 values, indicating that anti‐spike antibodies are not present in the plasma (Figure 3 B). We also could not calculate NT50 values in samples from patients with other documented respiratory infections prior to October 1st, 2019 (Figure 3 C), indicating that cross‐reactivity is minimal. We note that the patients infected with other respiratory diseases have lower overall assay signals, corresponding to higher neutralization, compared to the pre‐pandemic healthy controls. Patients with different respiratory diseases likely have antibodies that minimally cross‐react with antibodies against SARS‐CoV‐2. To evaluate the precision of our assay, we measured each sample in duplicate and calculated the coefficient of variation (CV) for each dilution factor (Figure 3 D). We found high agreement between technical replicates, with most CVs falling below 20 %. To assess run‐to‐run variability, we measured eight samples, including COVID‐19 positive samples and pre‐pandemic controls, in two consecutive runs (Figure S3). We observed high agreement between the NT50 values for the two measurements with low CVs (<20 %). Overall, these results show that the Simoa neutralization assay can be used to assess antibody neutralization capacity in the plasma of COVID‐19 patients with good precision.
Figure 3.
Simoa neutralization assay validation using patient plasma samples. A) Plasma samples from ten COVID‐19 patients were serially diluted and measured using the Simoa neutralization assay. B) Plasma samples from ten pre‐pandemic controls were serially diluted and measured using the Simoa neutralization assay. C) Plasma samples from ten pre‐pandemic controls with respiratory illnesses were serially diluted and measured using the Simoa neutralization assay. All measurements were performed in duplicate. D) CVs of duplicate measurements for each of the sample dilutions presented in (A), (B), and (C).
Performance of the Simoa Assay Compared to the Orthogonal Pseudovirus Neutralization Assay and ELISA
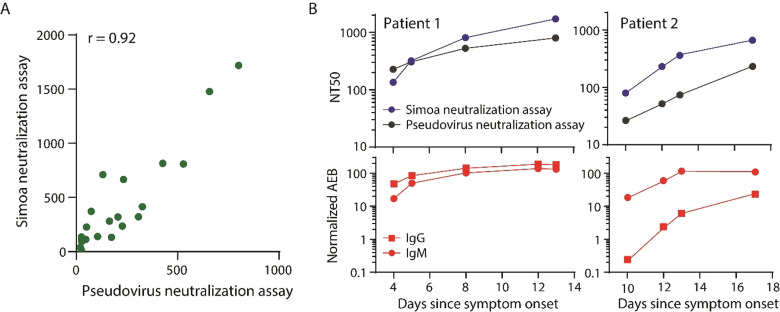
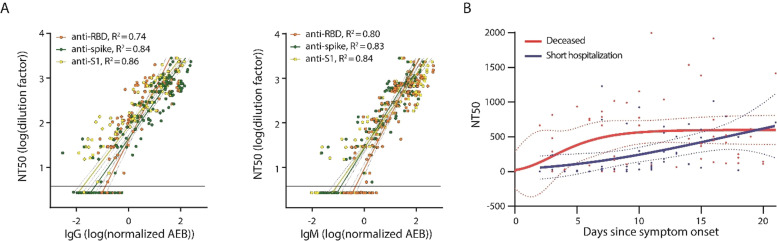
We then sought to validate our Simoa assay using an orthogonal neutralization assay. We obtained 28 plasma samples from patients with COVID‐19 and measured the NT50 values using both Simoa and pseudovirus neutralization assays. We show that overall, there is good correlation between our method and the pseudovirus neutralization assay across the different samples, with a Pearson correlation coefficient of 0.92 (Figure 4 A). We note that there may be additional mechanisms of neutralization that are not dependent on blocking the spike‐ACE2 interaction.[ 15 , 16 ] For example, antibodies can still bind to the spike protein and neutralize the virus by halting membrane fusion without disrupting the spike‐ACE2 interaction. However, due to the high correlation between the assays, these other mechanisms are unlikely to be the dominant neutralization factors. We then obtained serial samples from two different COVID‐19 patients and measured antibody neutralization over time using both Simoa and pseudovirus neutralization assays and observed good agreement between the two methods (Figure 4 B). We also measured anti‐spike IgG and IgM levels in these samples and show similar trends between antibody levels and antibody neutralization over time (Figure 4 B).
Figure 4.
Validation of the Simoa assay using the orthogonal pseudovirus neutralization assay. A) 28 samples from patients with COVID‐19 were used to correlate the Simoa and the pseudovirus neutralization assays. The Pearson correlation coefficient was 0.92. B) Patient samples over time were measured using both the Simoa and the pseudovirus neutralization assays and show good agreement (top). Anti‐spike IgM and anti‐spike IgG levels were measured using Simoa and correlated with neutralization capacity (bottom). All Simoa measurements were performed in duplicate.
We then evaluated the sensitivity of the Simoa neutralization assay by comparing the assay performance to two commercially available ELISA kits. We first measured SARS‐CoV‐2 neutralizing and SARS‐CoV non‐neutralizing antibodies using these assays (Figure 5 A). All three assays show good agreement, with expected signal decrease for the neutralizing antibody and minimal signal decrease for the non‐neutralizing antibody. We then measured NT50 values for six COVID‐19 patients with low neutralization capacity (Figure 5 B). We show that the Simoa assay is substantially more sensitive than ELISA and enables quantification of neutralization capacity. This improved sensitivity is important for assessing the immune response in patients with COVID‐19.
Figure 5.
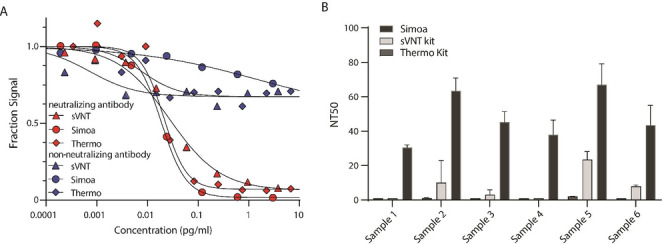
Simoa has higher sensitivity than ELISA. A) Neutralizing SARS‐CoV‐2 and non‐neutralizing SARS‐CoV antibodies were serially diluted and measured using Simoa and two ELISA neutralization assays. B) NT50 for six patient samples were measured using Simoa and two ELISA neutralization assays. Error bars represent the standard deviation of two measurements.
Assessing Antibody Levels and Neutralization Capacity in Patients Hospitalized with COVID‐19
To assess the utility of the Simoa assay in clinical settings, we compared the neutralization capacity with antibody levels in 130 samples from patients who were admitted to the hospital with COVID‐19. Our sample cohort consisted of patients hospitalized with COVID‐19 for less than ten days who did not require treatment in the intensive care unit (ICU) and hospitalized patients who died of COVID‐19. We measured the neutralization capacity and antibody levels of IgG and IgM isotypes against the receptor binding domain (RBD), S1, spike, and nucleocapsid, for a total of eight antibody measurements, as previously described. [17] We then compared the NT50 values to the antibody levels and observed that as the antibody levels against RBD, S1, and spike increased, so do the NT50 values (Figure 6 A). On the other hand, antibodies against nucleocapsid show poor correlation with NT50 (Figure S4). The neutralization capacity varied by approximately three orders of magnitude, with an NT50 range of ≈5 to ≈3000. The capability of this assay to measure a wide range of sample dilution factors, along with the high assay sensitivity, allows us to quantify neutralization capacities throughout disease progression.
Figure 6.
Antibody neutralization capacity in COVID‐19 patients. A) Correlation between NT50 values and IgG levels (left) and IgM levels (right). B) NT50 levels over time in the two patient groups. All measurements were performed in duplicate. Dotted lines represent the 95 % confidence interval. Horizontal line represents the detection limit.
We assessed antibody levels over time in the two patient groups and observed that, overall, the group of patients who died of COVID‐19 had higher antibody levels compared to the group of patients with a shorter period of hospitalization (Figure S5). When we evaluated antibody neutralization over time in the two patient groups (Figure 6 B), we observed that the antibody levels and neutralization capacities increase over the first 20 days post symptom onset, which is consistent with previous reports. [18] Our results demonstrate the utility of Simoa assays to comprehensively characterize the immune response by profiling both antibody levels and neutralization capacity for various clinical applications.
Multiplex Simoa Neutralization Assay to SARS‐CoV‐2 RBD and Its Variants
In response to the on‐going emergence of SARS‐CoV‐2 variants, we adapted the Simoa assay to a multiplex assay format to assess antibody neutralization capacities against three SARS‐CoV‐2 variants of concern (VOCs), as classified by the US Department of Health and Human Services.[ 19 , 20 ] The first VOC has an N501Y mutation (B.1.1.7, Alpha variant), the second VOC has K417N, E484K, and N501Y mutations (B.1.351, Beta variant), and the third VOC has T478K and L452R mutations (B.1.617.2, Delta variant) on the receptor binding domain (RBD) epitope. A major advantage of Simoa compared to traditional ELISA is the ability to multiplex and thereby measure multiple target analytes in one sample. In addition to interrogating more than one target, multiplexing is advantageous as it increases throughput and saves precious patient sample volume.
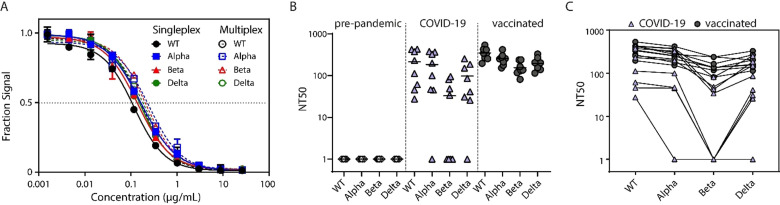
We first developed four singleplex Simoa neutralization assays: one assay against the wild‐type (WT) RBD, and three against the variants (Figure S6). We used these assays to quantify the neutralization capacity of a SARS‐CoV‐2 monoclonal neutralizing antibody (40591‐MM43) against each of the four RBD epitopes. As the concentration of antibody increased, the assay signal decreased for each variant (Figure 7 A), indicating that this antibody had neutralization potential against SARS‐CoV‐2 VOCs. We then developed a four‐plex Simoa assay, which can measure all four of these targets simultaneously, and measured the neutralization capacity of the antibody. The assay signals for the singleplex and multiplex assays showed excellent agreement for all four targets (Figure 7 A). We also tested the singleplex and multiplex assays using a second monoclonal neutralizing antibody (40592‐R001) against each of the four variants (Figure S7). The neutralizing antibody had a similar neutralization capacity for the Alpha and Delta variants and the WT RBD.[ 21 , 22 , 23 ] The neutralizing antibody had a substantially lower neutralization capacity (≈9 fold) against the Beta variant, which could be due to the E484K mutation that has been shown to escape serum antibody responses.[ 24 , 25 ]
Figure 7.
Multiplex Simoa neutralization assay for the SARS‐CoV‐2 wild‐type, Alpha, Beta, and Delta variants. A) Comparison of singleplex and multiplex Simoa assays using increasing concentrations of a SARS‐CoV‐2 neutralizing antibody (40591‐MM43). B) NT50 for the SARS‐CoV‐2 wild‐type, Alpha, Beta, and Delta variants in pre‐pandemic controls, COVID‐19 patients and vaccinated subjects. C) NT50 for each subject in the COVID‐19 and vaccinated groups.
Next, we assessed the neutralization capacity in plasma from eight healthy pre‐pandemic controls, eight COVID‐19 patients who had seroconverted, and eight vaccinated individuals (mRNA‐1273 vaccine, 56 days post 1st dose) (Figure 7 B and Figure 7 C). We measured these samples using both singleplex and multiplex Simoa assays to ensure agreement between the two assay formats (Figures S8 and S9). As expected, no neutralization was found in the healthy pre‐pandemic controls. The COVID‐19 patients had overall higher neutralization capacity against the WT, Alpha, and Delta variants compared to the Beta variant. No neutralization against the Beta variants was observed for some of the patients. On the other hand, the neutralization capacity for all the variants was high in the vaccinated individuals, suggesting that this vaccine can lead to production of neutralizing antibodies against the variants at the time at which the samples were collected. [26] We demonstrate that this multiplex Simoa assay can be rapidly adapted to detect the neutralization capacity against novel SARS‐CoV‐2 variants. Particularly, it has been recently shown that the levels of neutralizing antibodies are related to vaccine efficacy.[ 27 , 28 ] We anticipate similar adaptability of the assay for future infectious disease outbreaks.
Conclusion
In this study, we developed a simple assay for determining antibody neutralization using Single Molecule Arrays (Simoa). This assay shows high sensitivity and multiplexing capabilities, enabling high resolution profiling of neutralization capacity for several targets. Additionally, the assay can complement traditional serology assays that measure antibody levels by quantifying the antibodies that inhibit the viral antigen‐ACE2 interaction. More specifically, antibodies that have higher off‐rates, or lower affinities, to spike than ACE2 will be displaced. As a result, the information obtained from this assay closely resembles the biological immune response, in which antibodies with lower dissociation constants for spike, or higher affinities than ACE2, demonstrate substantial neutralization.
We demonstrated the utility of this assay for two applications. First, we used both the neutralization assay and the Simoa serology assay to characterize the immune response in hospitalized COVID‐19 patients. We observed an overall increase in neutralization capacity over time in our sample cohort. We found that patients who died of COVID‐19 had higher levels of antibodies than patients who were hospitalized for a shorter period. These results are in agreement with previous reports.[ 29 , 30 , 31 , 32 ] However, the relationship between disease severity and the production of neutralizing antibodies is presently unclear. Future studies can use this assay format to determine the antibody potency by measuring the neutralization capacity and levels of antibody isotypes for predicting disease outcome. [31] Second, we developed a multiplex Simoa assay to assess the neutralization capacity against SARS‐CoV‐2 variants. We measured the neutralization capacity in pre‐pandemic healthy, COVID‐19 positive, and vaccinated individuals against WT RBD and three variants. The ability to quantify neutralization capacity against the different variants can be used to guide vaccine development and epidemiological studies. The Simoa neutralization assay we developed here has high reproducibility and specificity, with minimal cross‐reactivity. The assay is highly versatile and is broadly applicable to future detection of antibody neutralization for SARS‐CoV‐2 and other infectious diseases.
Conflict of interest
David Walt has a financial interest in Quanterix Corporation, a company that develops an ultra‐sensitive digital immunoassay platform. He is an inventor of the Simoa technology, a founder of the company and also serves on its Board of Directors. Dr. Walt's interests were reviewed and are managed by BWH and Partners Healthcare in accordance with their conflict of interest policies.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
The authors would like to thank Kathleen Bellon for her assistance with the experiments. Funding for this work came from a generous donation from Barbara and Amos Hostetter and the Chleck Foundation. This work was also largely funded through a grant from the Massachusetts Consortium for Pathogen Readiness. Funding was also provided by the Synthetic Biology Platform at the Wyss Institute for Biologically Inspired Engineering.
T. Gilboa, L. Cohen, C.-A. Cheng, R. Lazarovits, A. Uwamanzu-Nna, I. Han, K. Griswold, N. Barry, D. B. Thompson, R. E. Kohman, A. E. Woolley, E. W. Karlson, D. R. Walt, Angew. Chem. Int. Ed. 2021, 60, 25966.
References
- 1. Borremans B., Gamble A., Prager K., Helman S. K., McClain A. M., Cox C., Savage V., Lloyd-Smith J. O., eLife 2020, 9, e60122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Krammer F., Simon V., Science 2020, 368, 1060–1061. [DOI] [PubMed] [Google Scholar]
- 3. Zost S. J., Gilchuk P., Case J. B., Binshtein E., Chen R. E., Nkolola J. P., Schäfer A., Reidy J. X., Trivette A., Nargi R. S., Sutton R. E., Suryadevara N., Martinez D. R., Williamson L. E., Chen E. C., Jones T., Day S., Myers L., Hassan A. O., Kafai N. M., Winkler E. S., Fox J. M., Shrihari S., Mueller B. K., Meiler J., Chandrashekar A., Mercado N. B., Steinhardt J. J., Ren K., Loo Y.-M., Kallewaard N. L., McCune B. T., Keeler S. P., Holtzman M. J., Barouch D. H., Gralinski L. E., Baric R. S., Thackray L. B., Diamond M. S., Carnahan R. H., Crowe J. E., Nature 2020, 584, 443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.A. M. Sholukh, A. Fiore-Gartland, E. S. Ford, Y. J. Hou, V. Tse, H. Kaiser, A. Park, F. A. Lempp, R. Saint Germain, E. Bossard, J. J. Kee, K. Diem, A. B. Stuart, P. B. Rupert, C. Brock, M. Buerger, M. K. Doll, A. K. Randhawa, L. Stamatatos, R. K. Strong, C. McLaughlin, K. R. Jerome, R. S. Baric, D. Montefiori, L. Corey, medRxiv 2020, 10.1101/2020.12.07.20245431. [DOI]
- 5. Perera R. A. P. M., Mok C. K. P., Tsang O. T. Y., Lv H., Ko R. L. W., Wu N. C., Yuan M., Leung W. S., Chan J. M. C., Chik T. S. H., Choi C. Y. C., Leung K., Chan K. H., Chan K. C. K., Li K.-C., Wu J. T., Wilson I. A., Monto A. S., Poon L. L. M., Peiris M., Euro Surveill. 2020, 25, 2000421. [Google Scholar]
- 6. Muruato A. E., Fontes-Garfias C. R., Ren P., Garcia-Blanco M. A., Menachery V. D., Xie X., Shi P.-Y., Nat. Commun. 2020, 11, 4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Okba N. M. A., Müller M. A., Li W., Wang C., Geurts van Kessel C. H., Corman V. M., Lamers M. M., Sikkema R. S., de Bruin E., Chandler F. D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C. B. E. M., Bosch B.-J., Drosten C., Koopmans M. P. G., Haagmans B. L., Emerging Infect. Dis. 2020, 26, 1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tan C. W., Chia W. N., Qin X., Liu P., Chen M. I.-C., Tiu C., Hu Z., Chen V. C.-W., Young B. E., Sia W. R., Tan Y.-J., Foo R., Yi Y., Lye D. C., Anderson D. E., Wang L.-F., Nat. Biotechnol. 2020, 38, 1073–1078. [DOI] [PubMed] [Google Scholar]
- 9. Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Fan C., Huang W., Xu M., Wang Y., Emerging Microbes Infect. 2020, 9, 680–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang R., Huang B., R., Li W., Wang W., Deng Y., Tan W., Biosaf. Heal. 2020, 2, 226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang X., Cohen L., Wang J., Walt D. R., J. Am. Chem. Soc. 2018, 140, 18132—18139. [DOI] [PubMed] [Google Scholar]
- 12. Cohen L., Walt D. R., Annu. Rev. Anal. Chem. 2017, 10, 345–363. [DOI] [PubMed] [Google Scholar]
- 13. Rissin D. M., Kan C. W., Campbell T. G., Howes S. C., Fournier D. R., Song L., Piech T., Patel P. P., Chang L., Rivnak A. J., Ferrell E. P., Randall J. D., Provuncher G. K., Walt D. R., Duffy D. C., Nat. Biotechnol. 2010, 28, 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davies C. in The Immunoassay Handbook (Eds.: Wild D.), Elsevier, Oxford, 2013, pp. 29–59. [Google Scholar]
- 15.M. Sikora, S. von Bülow, F. E. C. Blanc, M. Gecht, R. Covino, G. Hummer, bioRxiv 2020, 10.1101/2020.07.03.186825. [DOI]
- 16. Ling R., Dai Y., Huang B., Huang W., Yu J., Lu X., Jiang Y., Peptides 2020, 130, 170328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Norman M., Gilboa T., Ogata A. F., Maley A. M., Cohen L., Busch E. L., Lazarovits R., Mao C.-P., Cai Y., Zhang J., Feldman J. E., Hauser B. M., Caradonna T. M., Chen B., Schmidt A. G., Alter G., Charles R. C., Ryan E. T., Walt D. R., Nat. Biomed. Eng. 2020, 4, 1180—1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang K., Long Q.-X., Deng H.-J., Hu J., Gao Q.-Z., Zhang G.-J., He C.-L., Huang L.-Y., Hu J.-L., Chen J., Tang N., Huang A.-L., Clin. Infect. Dis. 2021, 73, e531—e539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fontanet A., Autran B., Lina B., Kieny M. P., Karim S. S. A., Sridhar D., Lancet 2021, 397, 952–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Happi A. N., Ugwu C. A., Happi C. T., Nat. Med. 2021, 27, 372–373. [DOI] [PubMed] [Google Scholar]
- 21. Muik A., Wallisch A.-K., Sänger B., Swanson K. A., Mühl J., Chen W., Cai H., Maurus D., Sarkar R., Türeci Ö., Dormitzer P. R., Şahin U., Science 2021, 371, 1152–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rees-Spear C., Muir L., Griffith S. A., Heaney J., Aldon Y., Snitselaar J. L., Thomas P., Graham C., Seow J., Lee N., Rosa A., Roustan C., Houlihan C. F., Sanders R. W., Gupta R. K., Cherepanov P., Stauss H. J., Nastouli E., Doores K. J., van Gils M. J., McCoy L. E., Cell Rep. 2021, 34, 108890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.X. Shen, H. Tang, C. McDanal, K. Wagh, W. Fischer, J. Theiler, H. Yoon, D. Li, B. F. Haynes, K. O. Sanders, S. Gnanakaran, N. Hengartner, R. Pajon, G. Smith, F. Dubovsky, G. M. Glenn, B. Korber, D. C. Montefiori, bioRxiv 2021, 10.1101/2021.01.27.428516. [DOI]
- 24. Baum A., Fulton B. O., Wloga E., Copin R., Pascal K. E., Russo V., Giordano S., Lanza K., Negron N., Ni M., Wei Y., Atwal G. S., Murphy A. J., Stahl N., Yancopoulos G. D., Kyratsous C. A., Science 2020, 369, 1014–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Greaney A. J., Loes A. N., Crawford K. H. D., Starr T. N., Malone K. D., Chu H. Y., Bloom J. D., Cell Host Microbe 2021, 29, 463–476.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shen X., Tang H., Pajon R., Smith G., Glenn G. M., Shi W., Korber B., Montefiori D. C., N. Engl. J. Med. 2021, 384, 2352–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M. M., Planchais C., Porrot F., Robillard N., Puech J., Prot M., Gallais F., Gantner P., Velay A., Le Guen J., Kassis-Chikhani N., Edriss D., Belec L., Seve A., Courtellemont L., Péré H., Hocqueloux L., Fafi-Kremer S., Prazuck T., Mouquet H., Bruel T., Simon-Lorière E., Rey F. A., Schwartz O., Nature 2021, 596, 276—280. [DOI] [PubMed] [Google Scholar]
- 28. Bergwerk M., Gonen T., Lustig Y., Amit S., Lipsitch M., Cohen C., Mandelboim M., Gal Levin E., Rubin C., Indenbaum V., Tal I., Zavitan M., Zuckerman N., Bar-Chaim A., Kreiss Y., Regev-Yochay G., N. Engl. J. Med. 2021, 10.1056/NEJMoa2109072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shrock E., Fujimura E., Kula T., Timms R. T., Lee I.-H., Leng Y., Robinson M. L., Sie B. M., Li M. Z., Chen Y., Logue J., Zuiani A., McCulloch D., Lelis F. J. N., Henson S., Monaco D. R., Travers M., Habibi S., Clarke W. A., Caturegli P., Laeyendecker O., Piechocka-Trocha A., Li J. Z., Khatri A., Chu H. Y., Villani A.-C., Kays K., Goldberg M. B., Hacohen N., Filbin M. R., Yu X. G., Walker B. D., Wesemann D. R., Larman H. B., Lederer J. A., Elledge S. J., Science 2020, 370, eabd4250.32994364 [Google Scholar]
- 30. Secchi M., Bazzigaluppi E., Brigatti C., Marzinotto I., Tresoldi C., Rovere-Querini P., Poli A., Castagna A., Scarlatti G., Zangrillo A., Ciceri F., Piemonti L., Lampasona V., J. Clin. Invest. 2020, 130, 6366–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garcia-Beltran W. F., Lam E. C., Astudillo M. G., Yang D., Miller T. E., Feldman J., Hauser B. M., Caradonna T. M., Clayton K. L., Nitido A. D., Murali M. R., Alter G., Charles R. C., Dighe A., Branda J. A., Lennerz J. K., Lingwood D., Schmidt A. G., Iafrate A. J., Balazs A. B., Cell 2021, 184, 476–488.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lau E. H. Y., Tsang O. T. Y., Hui D. S. C., Kwan M. Y. W., Chan W., Chiu S. S., Ko R. L. W., Chan K. H., Cheng S. M. S., Perera R. A. P. M., Cowling B. J., Poon L. L. M., Peiris M., Nat. Commun. 2021, 12, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information