Abstract
We present the results of classical and QM/MM simulations for the inhibition of SARS‐CoV‐2 3CL protease by a hydroxymethylketone inhibitor, PF‐00835231. In the noncovalent complex the carbonyl oxygen atom of the warhead is placed in the oxyanion hole formed by residues 143 to 145, while P1–P3 groups are accommodated in the active site with interactions similar to those observed for the peptide substrate. According to alchemical free energy calculations, the P1′ hydroxymethyl group also contributes to the binding free energy. Covalent inhibition of the enzyme is triggered by the proton transfer from Cys145 to His41. This step is followed by the nucleophilic attack of the Sγ atom on the carbonyl carbon atom of the inhibitor and a proton transfer from His41 to the carbonyl oxygen atom mediated by the P1′ hydroxyl group. Computational simulations show that the addition of a chloromethyl substituent to the P1′ group may lower the activation free energy for covalent inhibition
Keywords: 3CL protease, inhibitors, molecular modeling, PF-00835231, SARS-CoV-2
Multiscale simulations unveil the binding and reaction mechanism of the SARS‐CoV‐2 main protease inhibitor, PF‐00835231 inhibitor. This compound contains a hydroxymethyl group that plays a relevant role in the formation of the noncovalent and covalent complexes. In silico modifications show a possible strategy for the design of new inhibitors.
Introduction
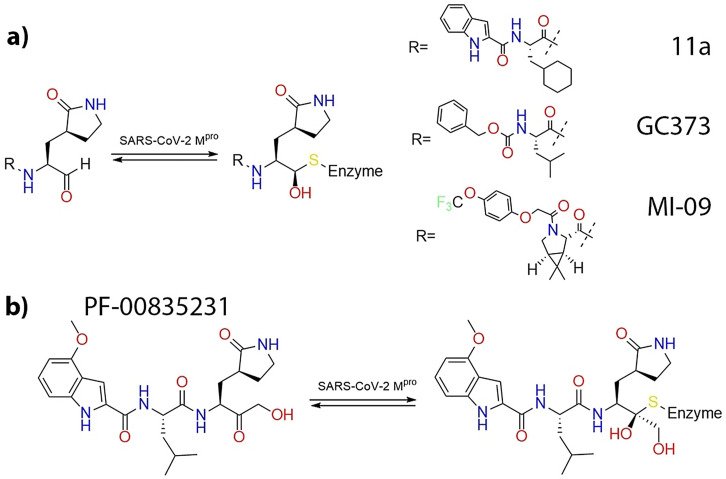
Inhibition of the activity of the SARS‐CoV‐2 3CL protease (or main protease) is one of the therapeutic strategies to treat the COVID‐19 pandemic. This enzyme is essential in the vital cycle of this and other related coronaviruses, being in charge of the cleavage of the long polyproteins resulting from the translation of the viral genome, in order to produce the non‐structural proteins needed for virus replication. [1] The 3CL or main protease of SARS‐CoV‐2 is a cysteine protease that uses a Cys‐His catalytic dyad to hydrolyze peptide bonds at specific positions of the polyprotein chain. The proteolysis mechanism of the 3CL protease involves two basic steps: i) the formation of a thiohemiacetal intermediate resulting from the attack of the Sγ atom of the catalytic cysteine at the carbonyl carbon atom of the peptide bond, which results in the release of the N‐fragment and ii) the hydrolysis of the acyl‐enzyme intermediate to release the C‐terminal fragment and to recover the resting state of the active site (see Figure 1 a).[ 2 , 3 , 4 , 5 ] The 3CL protease exclusively cleaves the polyprotein after a glutamine residue, a sequence specificity not shown by any known human protease, [5] which may facilitate the development of antiviral drugs with reduced side effects.
Figure 1.
a) Proposed proteolysis mechanism of SARS‐CoV‐2 3CL protease. [3] b) Schematic representation of the interactions in the active site of the proteases according to the Schechter and Berger nomenclature.
The reactive cysteine present in 3CL protease is an attractive target for the development of covalent inhibitors of this enzyme, using as warheads functional groups able to form a chemical bond with the Sγ atom. [6] Examples of warheads already tested in inhibitors of the SARS‐CoV‐2 3CL protease are Michael acceptors, [7] α‐ketoamides, [8] aldehydes,[ 9 , 10 , 11 ] ketones [12] and others. [13] In these inhibitors warheads are flanked by different groups that try to mimic the interactions established by the fragments of the peptide substrate placed before (Pi) and after (Pi′) the cleaved peptide bond with the Si and Si′ sites of the protease (see Figure 1 b).
Covalent inhibitors first bind into the active site of the protease forming a complex (EI) governed by noncovalent interactions. After binding, the noncovalent EI complex reacts with the thiol group of the catalytic cysteine to yield the E‐I covalent complex.

Covalent inhibitors can be reversible or irreversible depending on the relative stability of the E‐I complex.[ 14 , 15 ]
One of the most promising families of inhibitors is constituted by aldehyde and ketone derivatives. At least three aldehydes presenting large inhibitory capacities have been already successfully tested in animals.[ 9 , 10 , 11 ] These compounds, see Figure 2 a, present a γ‐lactam ring at the P1 position, taking advantage of the selectivity of this enzyme for a glutamine residue before the bond to be cleaved. Another common characteristic is the presence of a hydrophobic group at P2 position, reproducing the preference of the enzyme for a leucine residue in the natural substrate. Combination of kinetic and structural studies demonstrated that aldehydes react with the enzyme forming a hemithioacetal complex, where the electrophilic carbon atom of the aldehyde group is bonded to the Sγ atom of the cysteine.[ 9 , 10 , 11 ] We recently performed computational simulations that showed that a water molecule is recruited in the reaction mechanism to participate in the proton transfer from the catalytic histidine to the aldehyde oxygen atom. [16] A very promising inhibitor, already under clinical trial, is the ketone‐based inhibitor developed by Pfizer and known as PF‐00835231 (see Figure 2 b) that inhibits the 3CL protease forming also a hemithioacetal complex. [12] PF‐00835231 is a hydroxymethyl ketone obtained from the metabolization of the phosphate prodrug PF‐07304814 that shows potent SARS‐CoV‐2 inhibition, good solubility and stability in antiviral assays, converting it to an excellent candidate for therapeutic treatment of COVID‐19. [12] The hydroxymethyl group of this ketone inhibitor could potentially mimic the interactions established by a serine residue at the P1′ position of the peptide substrate of the protease, interactions that are obviously absent in aldehyde inhibitors. In addition, the hydroxymethyl group could also actively participate in the reaction mechanism, playing the role of the recruited water molecule in 3CL inhibition by aldehyde derivatives. [16]
Figure 2.
a) Aldehyde inhibitors of the SARS‐CoV‐2 3CL protease. b) PF‐00835231 ketone‐based inhibitor.
In this work we present the results of classical and hybrid QM/MM molecular dynamics (MD) simulations of the ketone‐based inhibitor PF‐00835231 in the active site of the SARS‐CoV‐2 3CL protease. We have first carried out classical MD simulations of the noncovalent complex (EI) identifying the most relevant hydrogen‐bond interactions established between the inhibitor and the active site residues. Second, we used hybrid QM/MM methods to explore the reaction mechanism for the covalent inactivation of the enzyme. As reported previously,[ 3 , 4 , 16 ] the reaction process is initiated with the activation of the catalytic dyad: a proton transfer from Cys145 to His41 that results in the catalytic dyad ion pair (IP). After this proton transfer the reaction proceeds with the nucleophilic attack of the activated cysteine on the carbonyl carbon atom and the proton transfer from the catalytic histidine to the carbonyl oxygen atom. In this last step the hydroxyl group of the inhibitor acts as a proton rely, accepting the proton from His41 and giving a proton to the carbonyl oxygen atom. The structure obtained for the reaction product agrees with the X‐ray structure of the 3CL protease inhibited by PF‐00835231 (6XHM). [12] From the knowledge of the interactions established in the noncovalent complex and the mechanistic details of the covalent inhibition reaction we propose a chemical modification of the PF‐00835231 inhibitor that could present improved kinetic properties. Our design is computationally tested by means of alchemical free energy calculations and QM/MM analysis of the reaction process. The simulations presented in this work could be used to improve, by rational design, future generations of antivirals.
Results and Discussion
Binding of the PF‐00835231 Inhibitor
The starting point for our simulations is the X‐ray structure with PDB code 6XHM. This structure corresponds to the dimeric enzyme with the two active sites inhibited by PF‐00835231. In both active sites (corresponding to chains A and B) the bond between the Sγ atom of Cys145 and the carbonyl carbon atom is formed, with distances of 1.86 and 1.80 Å in chains A and B, respectively. In both cases the pose of the inhibitor is very similar (see Figure S1a) and the only significant difference appears in the rotameric state of the catalytic histidine (His41). In both active sites the Nϵ atom of His41 is close to the Sγ atom of Cys145, displaying the same distance, 3.71 Å. However, in chain B the Nϵ atom is significantly closer to the inhibitor than in chain A, suggesting that this conformation would be more adequate for a subsequent proton transfer to the inhibitor. The distance from this atom to the hydroxyl oxygen atom of the inhibitor is only 2.65 Å in chain B, while in chain A the distance is increased to 3.80 Å. In the active site of this chain the Nδ atom of His41 is only slightly closer, 3.67 Å. These two rotameric states, hereafter denoted as ϵ‐rotamer (chain B) and δ‐rotamer (chain A), are connected by means of a 180° rotation around the Cβ‐Cγ bond of His41. We thus started our simulations of the noncovalent EI complex studying the preferred rotameric state of the catalytic His41 (see Figure S1b). To this aim (see SI), we traced the free energy profiles from the ϵ‐rotamer to the δ‐rotamer and backwards. The PMF displayed in Figure S1c correspond to the average of the two profiles. The ϵ‐rotamer is the preferred conformation, being more stable than the δ‐rotamer by 6.1 kcal mol−1. This finding agrees with the simulations performed for the noncovalent complex formed between the 3CL protease and the 11a aldehyde inhibitor shown in Figure 2. Also in this case the ϵ‐rotamer was found to be more stable than the δ‐rotamer, by 3.2 kcal mol−1. [16] It must be also noticed that the larger stability observed for the ϵ‐rotamer agrees with the observation that this conformer appears more frequently in the X‐ray structures of the hemithioacetal complexes formed between the SARS‐CoV‐2 3CL protease and aldehyde or ketone inhibitors. [16]
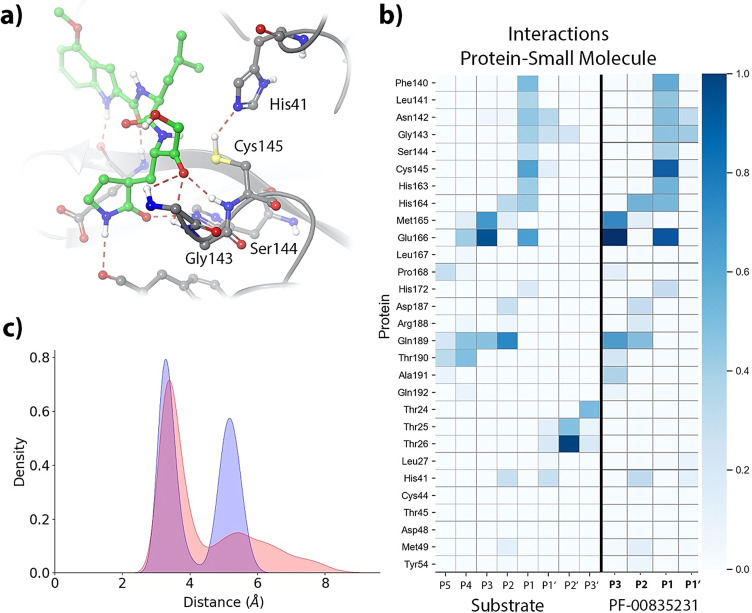
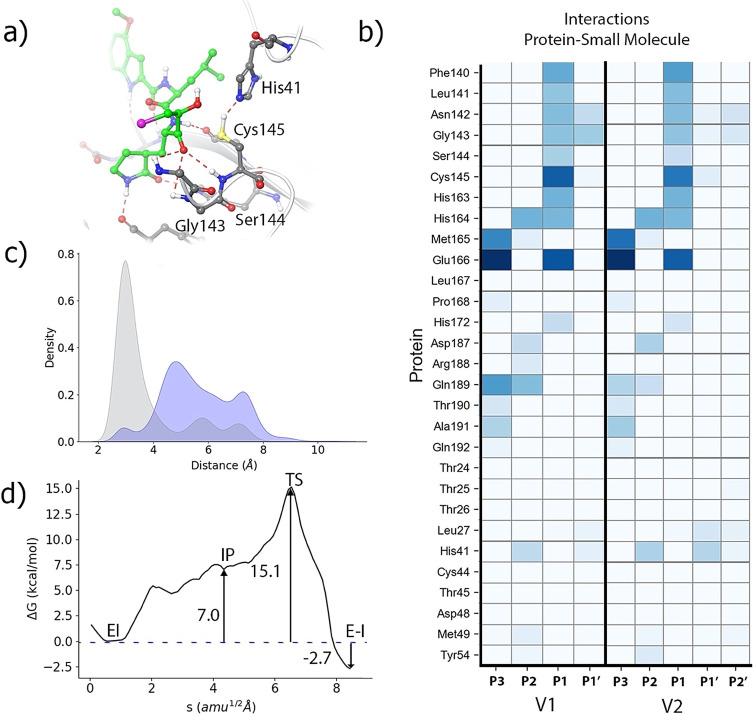
Once the preferred conformation for the catalytic histidine has been determined, we analyze the binding pose of the inhibitor and the interactions established with the enzyme in the noncovalent EI complex by means of MD simulations (5 replicas of 1 μs each). These MD simulations were stable in all cases (see RMSD time evolutions in Figure S2), showing a binding pose consistent with the X‐ray structure of the hemithioacetal complex (see Figure 3 a). P1, P2 and P3 sites of the inhibitor present an interaction pattern similar to that of a peptide substrate with sequence ‐Val‐Leu‐Gln|Ser‐ (where the vertical line indicates the scissile bond) [3] and also similar to those of other peptidyl inhibitors, such as Michael acceptors [17] and aldehyde derivatives. [16] Figure 3 b compares the fraction of hydrogen bonds established between the peptide substrate or the inhibitor with enzymatic residues as obtained from MD simulations. The P1 group establishes hydrogen bonds with His163, Glu166 and Phe140. The isobutyl hydrocarbon group at the P2 position stacks with the His41 imidazole ring, interacting also with other nearby residues, such as His164, Met165 and Gln189. Finally, the P3 group of the PF‐00835231 inhibitor is exposed to the solvent and stabilized by hydrogen bond interactions with main chain atoms of Met165, Glu166 and Glu189. The position of the carbonyl oxygen atom of the inhibitor is stabilized by means of hydrogen bond interactions with the main chain NH groups of Cys145 (2.3±0.3 Å), Ser144 (2.8±0.4 Å) and Gly143 (2.5±0.3 Å), as seen in Figure 3 a. These interactions are also observed in the X‐ray structure of the hemithioacetal product.
Figure 3.
Noncovalent complex formed between PF‐00835231 and the 3CL protease of SARS‐CoV‐2. a) Binding pose of the inhibitor in the active site of the protease, showing the location of the catalytic dyad and the oxyanion hole. Note that the carbonyl oxygen is accommodated in the oxyanion hole. b) Fraction of hydrogen bond contacts between residues of PF‐00835231 and a peptide substrate [3] and those of the protease. A hydrogen bond contact is counted when the donor–acceptor distance is <3.8 Å and the hydrogen bond angle is >120°. c) Pair distribution functions between the Cys145‐Sγ atom and the carbonyl carbon atom of the substrate (in blue) and to the Nϵ atom of His41 (in red).
The formation of a covalent bond between the enzyme and the inhibitor requires the activation of the Sγ atom of Cys145 by means of a proton transfer from Cys145 to His41 and the subsequent nucleophilic attack of this atom on the electron deficient carbonyl carbon atom of the inhibitor.[ 3 , 17 ] We monitored the distances of the Cys145 Sγ atom with the His41 Nϵ atom and with the carbonyl carbon atom of the inhibitor. The pair distribution functions are shown in Figure 3 c. Both of them display bimodal distributions that can be attributed to the presence of trans and gauche conformers of the Cys145 side chain. [18] The Sγ‐C and Sγ‐Nϵ distributions are peaked at 3.4/5.4 and 3.3/5.2 Å, respectively; showing that a significant fraction of the noncovalent EI complex conformations observed during the MD simulations are ready to proceed to the formation of the hemithioacetal product.
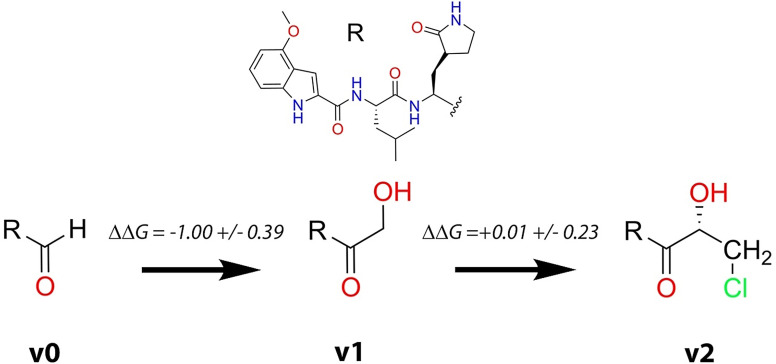
One of the main novelties of the PF‐00835231 inhibitor is the inclusion of a hydroxymethyl group at the P1′ position. This group resembles a serine residue, which is one of the preferences of 3CL proteases at this position. In fact, comparison of the hydrogen bond interactions established by Ser‐P1′ in the peptide substrate and the hydroxymethyl group in the PF‐00835231 inhibitor shows that this last is able to recover a fraction of the interactions established by the peptide. The hydroxyl group of the inhibitor partially mimics the interactions made by the serine side chain, in particular with the catalytic dyad, His41 and Cys145. The main difference is caused by the larger rotational freedom of the P1′ group in the inhibitor than in the peptide substrate (compare the P1′ interactions for the peptide and the inhibitor in Figure 3 b). In the case of the peptide substrate, the presence of P2′ and subsequent groups reduces the conformational flexibility of the Ser‐P1′ side chain, favoring the formation of more stable interactions with the catalytic dyad. Instead, in the case of the inhibitor, the P1′ group can rotate and then establish more frequent interactions with other residues of the active site, mainly with Gly143 and Asn142. We have evaluated the contribution of the hydroxymethyl group to the binding free energy of the inhibitor by means of the alchemical transformation from the corresponding aldehyde, where the hydroxymethyl group is substituted by a hydrogen atom (see v0 → v1 transformation in Figure 4). The binding free energy difference between the PF‐00835231 inhibitor and the aldehyde was computed by means of Thermodynamic Integration to be −1.00±0.39 kcal mol−1 (Table S1 provides the values corresponding to the five independent alchemical transformations in solution and in the protein environment, see SI for details). This means that the P1′ group moderately contributes to increase the affinity of the inhibitor by the protein, a contribution that can be rationalized in terms of the protein‐inhibitor interactions observed in the noncovalent complex.
Figure 4.
Alchemical transformations considered to evaluate the impact of chemical substitutions on the binding free energy. The original PF‐00835231 inhibitor is denoted as v1, the aldehyde derivative as v0 and the inhibitor containing a chloromethyl substituent as v2. The free energy changes provided here correspond to the average of five independent simulations in solution and in the protein environment. Details of free energy simulations are given in Table S1.
Formation of the Covalent Hemithioacetal Product
In order to explore the reaction mechanism for the formation of the covalent E‐I complex from the noncovalent one, we traced the corresponding MFEP using the string method at the B3LYPD3/6–31+G*/MM level (see SI for details). According to previous studies on the 3CL protease activity with peptide substrates[ 3 , 4 ] and other inhibitors,[ 16 , 17 , 19 , 20 ] formation of a covalent bond with the Sγ atom of Cys145 requires its activation by means of a proton transfer to His41. The reaction must be completed with a proton transfer from His41 to the inhibitor. In our previous study with aldehyde derivatives, we found that this proton transfer takes place mediated by a water molecule that occupies the same position as the hydroxyl group in the PF‐00835231 inhibitor, suggesting the participation of this group as a proton rely during the proton transfer from His41 to the carbonyl oxygen atom of the inhibitor. [16]
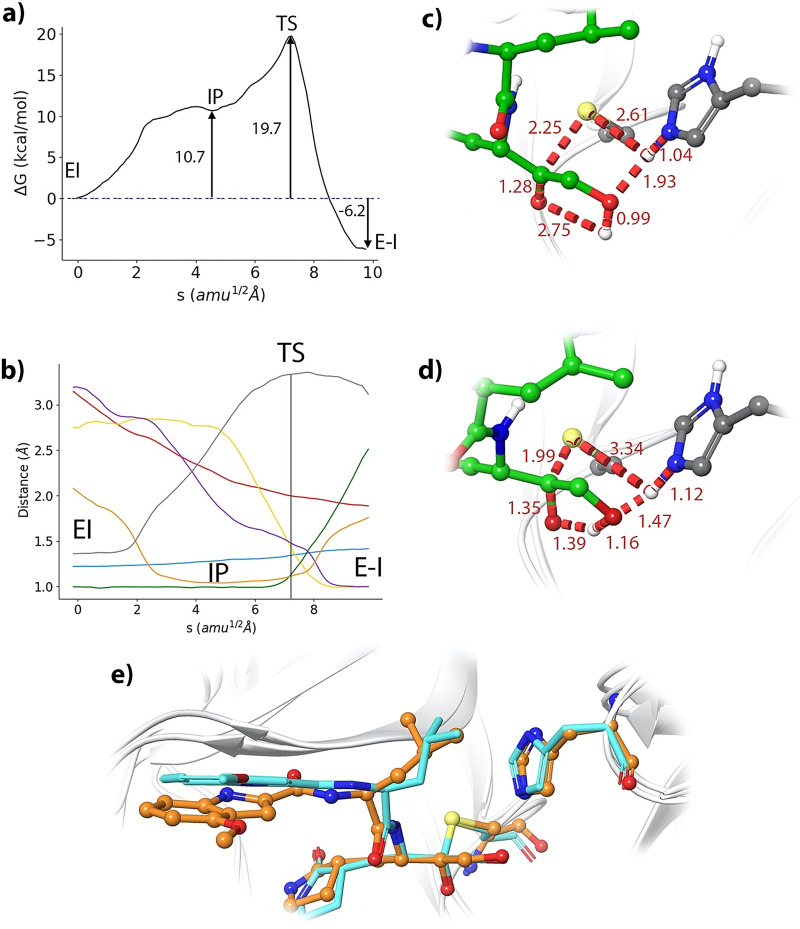
Our string simulations converged to a two‐steps reaction mechanism, as shown in Figures 5 a and b (and video S1). In the first step a metastable ion pair (IP) catalytic dyad is formed after the proton transfer from Cys145 to His41. The IP is found to be 10.7 kcal mol−1 above the noncovalent EI complex (see Figure 5 a). We confirmed this value by means of umbrella sampling simulations along the antisymmetric proton transfer coordinate (d(SγH)‐d(NϵH)), which gave a free energy difference of 10.5 kcal mol−1 (see Figure S3). This free energy difference between the neutral and IP forms of the dyad is very close to the values reported for other inhibitors. In particular, formation of the IP in presence of the 11a aldehyde [16] requires 9.3 kcal mol−1 and 10.3 in presence of a Michael acceptor. [17] Instead, formation of the IP has a significantly smaller free energy cost (4.8 kcal mol−1) when a peptide with a serine residues at the P1′ position is present in the active site [3] because in that case the hydroxyl group is better positioned to make favorable contacts with the catalytic dyad. The IP, which is represented in Figure 5 c, is found in a free energy plateau, with a very low free energy barrier for its reversion back to the neutral dyad. At the IP, the proton transfer from Cys145 to His45 has been completed (the Nϵ‐H and Sγ‐H distances are 1.04 and 2.61 Å, respectively) and His41 is now oriented towards the hydroxyl group of the inhibitor, forming a hydrogen bond with a NϵH‐O distance of 1.93 Å. This first proton transfer from Cys145 to His41 is accompanied by the approach of the activated Sγ atom to the carbonyl carbon: from 3.09 to 2.25 Å, when going from the noncovalent EI complex to the IP. From this state, the reaction proceeds completing the nucleophilic attack of the Sγ atom of Cys145 to the carbonyl carbon atom of the inhibitor and the proton transfer from His41 to the hydroxyl group of the inhibitor and from this to the carbonyl oxygen atom of the ketone group (see evolution of the CVs in Figure 5 b). The rate‐limiting transition state (TS) for the formation of the hemithioacetal is presented in Figure 5 d. At the TS the enzyme‐inhibitor covalent bond (Sγ‐C) is almost completely formed, displaying a distance of 1.99 Å and the carbonyl double bond of the inhibitor has been elongated from 1.22 (EI complex) to 1.35 Å. The two proton transfers events are not very advanced at the TS. The Nϵ‐H distance has been slightly lengthened up to 1.12 Å, while the distance of this proton to the hydroxyl oxygen atom of the inhibitor is 1.47 Å. The proton transfer from the hydroxyl group to the carbonyl oxygen atom is found at a slightly more advanced stage, being the distances of the proton to the donor and acceptor atoms of 1.16 and 1.39 Å, respectively. From this TS the reaction is completed to yield the hemithioacetal product (see Figure 5 e) where the two proton transfers are now completed. In our simulations, the covalent Sγ−C bond presents a distance of 1.88 Å in the product state, in agreement with the X‐ray observations (1.80/1.86 Å). In general, the configuration obtained for the covalent E‐I complex agrees with the X‐ray 6XHM structure (see Figure 5 e), with an average RMSD calculated for the main chain non‐hydrogen atoms, including those of the inhibitor, of 1.54 Å.
Figure 5.
Formation of the (S)‐hemithioacetal product. a) B3LYPD3/6–31+G*/MM free energy profile along the path‐CV for the formation of the covalent E‐I complex from the EI complex. b) Evolution of the selected CVs along the MFEP: Sγ‐C (red), Sγ‐H (grey), Nϵ‐H (orange), H‐Ohid (purple), Ohid‐Hhid (green), Hhid‐O (yellow), C‐O (pale blue); see Scheme S1. c) Representation of the IP state. The values of the distances (in Å) correspond to the coordinates of the MFEP where the IP is located. d) Representation of the rate‐limiting TS. e) Overlap of the product structure (balls and sticks with carbon atoms in orange) with the X‐ray structure 6XHM containing a hydroxymethylketone inhibitor PF‐00835231 (rods with carbon atoms in light blue).
The free energy profile for the covalent inactivation of the 3CL protease with PF‐00835231 (see Figure 5 a) can be compared with experimental and theoretical results obtained for the inhibition with aldehyde derivatives 11a and GC‐373. Recent analyses indicate that the PF‐00835231 inhibitor has similar or higher potency against SARS‐CoV‐2 in human A549 cells than GC‐376 (the prodrug of GC‐373). [21] Our calculations predict that the formation of the hemithioacetal product from the noncovalent complex is exergonic, with a reaction free energy of −6.2 kcal mol−1. This value is larger, in absolute value, than the reaction free energy obtained for SARS‐CoV‐2 3CL inhibition with aldehyde 11a using the same computational Scheme, −2.8 kcal mol−1; [16] but smaller than the value predicted for a Michael acceptor, −15.0 kcal mol−1. [17] These two inhibitors, the 11a aldehyde and the N3 Michael acceptor, are examples of covalent reversible [10] and irreversible [7] inhibitors, respectively. Thus, our calculations predict that the ketone‐based PF‐00835231 inhibitor would display an intermediate behavior, with more irreversible character than other aldehyde inhibitors. The main difference found between the hemithioacetal products obtained with the aldehyde and the ketone‐based inhibitors that can explain the larger stabilization of the latter is the formation of a hydrogen bond between the hydroxyl group of the inhibitor and the Nϵ atom of His41. The average O‐Nϵ distance observed in our simulations of the hemithioacetal product of the PF‐00835231 inhibitor was 2.77±0.11 Å, close to the observed X‐ray value (2.65 Å, see Figure S1).
The predicted activation free energy for the process is 19.7 kcal mol−1. Unfortunately, there are not experimental determinations of the first order inactivation rate constant for this inhibitor, but we can again compare our results with the values obtained for the inhibition of the SARS‐CoV‐2 3CL protease with aldehyde derivatives. The rate constant for the inhibition of the protease with the inhibitor GC373 has been measured to be 2.45×10−3 s−1 at 30 °C, [22] a value that, according to Transition State Theory, can be translated into an activation free energy of 21.1 kcal mol−1. For 11a our simulations predicted an activation free energy of 18.5 kcal mol−1, [16] the PF‐00835231 inhibitor would present similar kinetic properties for the covalent inhibition of the SARS‐CoV‐2 3CL enzyme as the aforementioned aldehyde‐based inhibitors.
Improving the Kinetic Properties of the Inhibitor
After the analysis of the characteristics of the binding and covalent inactivation processes of the PF‐00835231 inhibitor, our next goal was to try to find a strategy to design an inhibitor with improved properties. This in silico design can also be considered as a further test of the conclusions reached with our simulations. As discussed in the previous section, our simulations indicate that a significant contribution to the inactivation free energy barrier comes from the formation of the IP and that this free energy cost significantly depends on the nature of the substrate/inhibitor and the interactions established in the active site. Our initial hypothesis was that the substitution of one of the hydrogen atoms of the P1′ site of the inhibitor by a bulkier group could reduce its rotational freedom, favoring the interaction of the hydroxyl moiety with the catalytic dyad. This substitution could then lower the energetic cost due to IP formation, at the cost of reducing the binding entropy. A comparison between the binding pose of the PF‐00835231 inhibitor at the non‐covalent EI complex with that of a peptide substrate (see Figure S3) shows that such a substitution may be carried out on the pro‐R hydrogen atom of the hydroxymethyl group. We selected a chloromethyl group (see variant v2 in Figure 4) as an appropriate candidate to be placed at the pro‐R position for two reasons: i) it is a relatively small substituent and ii) this group polarizes the ketone group of the inhibitor, increasing the positive charge on the electrophilic carbon atom.
The binding pose of this new inhibitor (2 MD replicas of 1.5 μs) is showed in Figure 6 a. The interaction mode of the chloromethyl containing variant in the active site of the 3CL protease is very similar to that observed for the original inhibitor (compare Figures 3 a and 6 a). The new group is accommodated in the active site by means of hydrogen bond contacts with the side chain of Asn142 and the main chain NH group of Gly143 (see Figure 6 b). As expected, the attachment of the methylchoride group reduces the rotation freedom of the P1′ fragment of the inhibitor in the active site. This restriction is reflected in the distribution function of the distance between the oxygen atom of the hydroxyl group and the Nϵ atom of His41 (see Figure 6 c) as compared to that of PF‐00835231 However, these changes found in the interaction pattern are not reflected in an improved binding free energy. As explained in the methodological section, we used Thermodynamic Integration to evaluate the impact of the alchemical transformation of the original inhibitor into the chloromethyl variant (v1 → v2 in Figure 4). According to our simulations, addition of the chloromethyl group has an almost neutral impact on the binding free energy, 0.01±0.23 kcal mol−1 (see Table S1 for details). The entropic effect associated to the rotational restriction of the P1′ fragment seems to compensate the enthalpic gain obtained from the new interactions established between the inhibitor and the protein.
Figure 6.
Improved design of a hydroxymethylketone inhibitor. a) Binding pose of the chloromethyl‐containing inhibitor (v2 in Figure 4) in the active site of the SARS‐CoV‐2 3CL protease (chlorine atom shown in purple). b) Fraction of hydrogen bond contacts between residues of PF‐00835231 inhibitor (v1) and the chloromethyl variant (v2) with the protease. c) Pair distribution functions from the oxygen atom of the inhibitor's hydroxyl group to the Nϵ atom of His41; the PF‐00835231 inhibitor in blue and the chloromethyl variant in grey. d) B3LYPD3/6‐31+G*/MM free energy profile along the path‐CV for the formation of the covalent E‐I complex from the EI complex for the chloromethyl inhibitor.
We also evaluated the effect of the inclusion of the chloromethyl group on the formation of the covalent complex, tracing the corresponding free energy profile with the string method. These calculations were carried out using the same QM/MM description used for the PF‐00835231 inhibitor, but including now the chloromethyl group into the QM region. The new profile, showed in Figure 6 d, displays an activation free energy of 15.1 kcal mol−1, significantly smaller than the value predicted for the original inhibitor (19.7 kcal mol−1, see Figure 5 a). This reduction is essentially due to the lower free energy cost associated to the formation of the IP, 7.0 kcal mol−1 in the presence of the new inhibitor versus 10.7 kcal mol−1 obtained with the PF‐00835231 inhibitor (we compare similar configurations of the IPs). The larger stabilization of the IP observed for the chloromethyl variant with respect to the original inhibitor is due to the enhanced interactions observed between the hydroxyl group of the inhibitor and the catalytic dyad as a consequence of the restricted mobility of the P1′ group. The hydroxyl group has now the ability to stabilize the two charged fragments of the ionic pair catalytic dyad, acting simultaneously as proton donor and proton acceptor with Cys145 and His41, respectively. As expected, the reaction mechanism, described by the evolution of the collective variables and the geometry of the TS, remains essentially unchanged with respect to the original PF‐00835231 inhibitor (see Figure S5 for details). The observed reduction in the activation free energy barrier in the new inhibitor can be translated into an increase of the covalent inactivation rate constant [k 3 in Eq. (1)] of 3 orders of magnitude at 300 K. This result shows that the kinetic properties of hydroxymethylketone inhibitors could probably be improved attaching new substituents to the hydroxymethyl group. These substituents play the role of P′ fragments in peptidic substrates. Regarding the reaction free energy, that determines the irreversibility of the process, the process is now exergonic by only −2.7 kcal mol−1, smaller in absolute value than for the original inhibitor (−6.2 kcal mol−1).
Conclusion
We have used a combination of classical and hybrid QM/MM molecular dynamics simulations to explore the covalent inhibition mechanism of the SARS‐CoV‐2 3CL protease by a ketone‐based inhibitor that is currently under clinical trial, PF‐00835231. We first explored the binding mode and interactions established by the inhibitor starting from the X‐ray structure of the hemithioacetal complex. We determined the preferred rotameric state of the catalytic His41, since the X‐ray structure shows a different conformation for this residue in each of the active sites of the dimer. The conformation determined as the most stable is that presenting the Nϵ atom of His41 close to the inhibitor warhead.
Our simulations emphasize the role played by the hydroxymethyl group at the P1′ position of this inhibitor. Alchemical transformations from the corresponding aldehyde derivative show that the hydroxymethyl group contributes to increase the binding free energy of the inhibitor. Furthermore, this group plays also an active role during the formation of the hemithioacetal complex, mediating the proton transfer from the catalytic histidine to the carbonyl oxygen atom. The formation of the covalent complex is initiated after a proton transfer from Cys145 to His41 to form an ion pair. Once the catalytic dyad is activated, the process continues with the nucleophilic attack of the cysteine sulfur atom on the carbonyl carbon atom of the inhibitor and the proton transfer from His41 to the carbonyl oxygen atom. The rate limiting TS shows a short carbon‐sulfur bond distance, while the proton transfers from the catalytic histidine to the hydroxyl group of the inhibitor and from this group to the carbonyl oxygen atom are found at an earlier stage. The activation free energy associated to this TS is similar to that found for aldehyde inhibitors, while formation of the hemithioacetal complex is more exergonic for the ketone‐based inhibitor.
Finally, we have explored the possibility to improve the kinetic properties of the inhibitor adding a substituent on the pro‐R position of the P1′ hydroxymethyl group. We tested the effect of the addition of a chloromethyl group which was introduced to favor the positioning of the P1′ hydroxyl group in the vicinity of the catalytic dyad and to increase the positive charge on the carbonyl carbon atom. Our simulations show that the modification had a neutral effect on the binding free energy but increased the inactivation rate constant, due to the stabilization of the ion pair formed by the catalytic dyad. This new inhibitor should not be considered as end point in the design process, but rather as an indication of a possible route to further improve the design of hydroxymethylketone inhibitors. Our simulations indicate that substitutions at the pro‐R hydrogen atom of the hydroxymethyl group of the PF‐ PF‐00835231 inhibitor offer the possibility to improve the kinetic properties of this inhibitor of the SARS‐CoV‐2 3CL protease.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Supporting Information
Supporting Information
Acknowledgements
The authors acknowledge financial support from Consellería de Innovación, Universidades, Ciencia y Sociedad Digital, Generalitat Valenciana (GVCOV19/Decreto180/2020) and from Feder funds and the Ministerio de Ciencia, Innovación y Universidades (project PGC2018‐094852‐B‐C22). We want to acknowledge Barcelona Supercomputing Center (BSC) for providing us access to MareNostrum and the staff from BSC for the technical support (Project QSB‐2021‐1‐0006). We also acknowledge the use of the Tirant supercomputer at the Universitat de València (financed by the FEDER funds for Scientific Infrastructures; IDIFEDER‐2018‐063) and the support of Alejandro Soriano from Servei d'Informàtica from the Universitat de València.
C. A. Ramos-Guzmán, J. J. Ruiz-Pernía, I. Tuñón, Angew. Chem. Int. Ed. 2021, 60, 25933.
A previous version of this manuscript has been deposited on a preprint server (https://doi.org/10.33774/chemrxiv‐2021‐tndbs).
Contributor Information
Dr. J. Javier Ruiz‐Pernía, Email: j.javier.ruiz@uv.es.
Prof. Iñaki Tuñón, Email: ignacio.tunon@uv.es.
References
- 1. Bangham C. R. M., J. Gen. Virol. 2003, 84, 3177–3189. [DOI] [PubMed] [Google Scholar]
- 2. Solowiej J., Thomson J. A., Ryan K., Luo C., He M., Lou J., Murray B. W., Biochemistry 2008, 47, 2617–2630. [DOI] [PubMed] [Google Scholar]
- 3. Ramos-Guzmán C. A., Ruiz-Pernía J. J., Tuñón I., ACS Catal. 2020, 10, 12544–12554. [DOI] [PubMed] [Google Scholar]
- 4. Świderek K., Moliner V., Chem. Sci. 2020, 11, 10626–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hilgenfeld R., FEBS J. 2014, 281, 4085–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Steuten K., Kim H., Widen J. C., Babin B. M., Onguka O., Lovell S., Bolgi O., Cerikan B., Neufeldt C. J., Cortese M., Muir R. K., Bennett J. M., Geiss-Friedlander R., Peters C., Bartenschlager R., Bogyo M., ACS Infect. Dis. 2021, 7, 1457—1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L. W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H., Nature 2020, 582, 289–293. [DOI] [PubMed] [Google Scholar]
- 8. Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R., Science 2020, 368, 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dai W., Zhang B., Jiang X.-M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.-K., Xu Y., Yang H., Liu H., Science 2020, 368, 1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vuong W., Khan M. B., Fischer C., Arutyunova E., Lamer T., Shields J., Saffran H. A., McKay R. T., van Belkum M. J., Joyce M. A., Young H. S., Tyrrell D. L., Vederas J. C., Lemieux M. J., Nat. Commun. 2020, 11, 4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Qiao J., Li Y.-S., Zeng R., Liu F.-L., Luo R.-H., Huang C., Wang Y.-F., Zhang J., Quan B., Shen C., Mao X., Liu X., Sun W., Yang W., Ni X., Wang K., Xu L., Duan Z.-L., Zou Q.-C., Zhang H.-L., Qu W., Long Y.-H.-P., Li M.-H., Yang R.-C., Liu X., You J., Zhou Y., Yao R., Li W.-P., Liu J.-M., Chen P., Liu Y., Lin G.-F., Yang X., Zou J., Li L., Hu Y., Lu G.-W., Li W.-M., Wei Y.-Q., Zheng Y.-T., Lei J., Yang S., Science 2021, 371, 1374–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoffman R. L., Kania R. S., Brothers M. A., Davies J. F., Ferre R. A., Gajiwala K. S., He M., Hogan R. J., Kozminski K., Li L. Y., Lockner J. W., Lou J., Marra M. T., Mitchell L. J., Murray B. W., Nieman J. A., Noell S., Planken S. P., Rowe T., Ryan K., Smith G. J., Solowiej J. E., Steppan C. M., Taggart B., J. Med. Chem. 2020, 63, 12725–12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Menéndez C. A., Byléhn F., Perez-Lemus G. R., Alvarado W., de Pablo J. J., Sci. Adv. 2020, 6, eabd0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Powers J. C., Asgian J. L., Ekici Ö. D., James K. E., Chem. Rev. 2002, 102, 4639–4750. [DOI] [PubMed] [Google Scholar]
- 15. Drag M., Salvesen G. S., Nat. Rev. Drug Discovery 2010, 9, 690–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ramos-Guzmán C. A., Ruiz-Pernía J. J., Tuñón I., ACS Catal. 2021, 11, 4157–4168. [DOI] [PubMed] [Google Scholar]
- 17. Ramos-Guzmán C. A., Ruiz-Pernía J. J., Tuñón I., Chem. Sci. 2021, 12, 3489–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G. F., Anand K., Bartlam M., Hilgenfeld R., Rao Z., Proc. Natl. Acad. Sci. USA 2003, 100, 13190–13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mondal D., Warshel A., Biochemistry 2020, 59, 4601–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arafet K., Serrano-Aparicio N., Lodola A., Mulholland A. J., González F. V., Świderek K., Moliner V., Chem. Sci. 2021, 12, 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Vries M., Mohamed A. S., Prescott R. A., Valero-Jimenez A. M., Desvignes L., O'Connor R., Steppan C., Devlin J. C., Ivanova E., Herrera A., Schinlever A., Loose P., Ruggles K., Koralov S. B., Anderson A. S., Binder J., Dittmann M., J. Virol. 2021, 95, e01819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma C., Sacco M. D., Hurst B., Townsend J. A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M. T., Chen Y., Wang J., Cell Res. 2020, 30, 678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Supporting Information
Supporting Information