Abstract
Reliable methods to detect the presence of SARS‐CoV‐2 at venues where people gather are essential for epidemiological surveillance to guide public policy. Communal screening of air in a highly crowded space has the potential to provide early warning on the presence and potential transmission of SARS‐CoV‐2 as suggested by studies early in the epidemic. As hospitals and public facilities apply varying degrees of restrictions and regulations, it is important to provide multiple methodological options to enable environmental SARS‐CoV‐2 surveillance under different conditions. This study assessed the feasibility of using high‐flowrate air samplers combined with RNA extraction kit designed for environmental sample to perform airborne SARS‐CoV‐2 surveillance in hospital setting, tested by RT‐qPCR. The success rate of the air samples in detecting SARS‐CoV‐2 was then compared with surface swab samples collected in the same proximity. Additionally, positive RT‐qPCR samples underwent viral culture to assess the viability of the sampled SARS‐CoV‐2. The study was performed in inpatient ward environments of a quaternary care university teaching hospital in Singapore housing active COVID‐19 patients within the period of February to May 2020. Two types of wards were tested, naturally ventilated open‐cohort ward and mechanically ventilated isolation ward. Distances between the site of air sampling and the patient cluster in the investigated wards were also recorded. No successful detection of airborne SARS‐CoV‐2 was recorded when 50 L/min air samplers were used. Upon increasing the sampling flowrate to 150 L/min, our results showed a high success rate in detecting the presence of SARS‐CoV‐2 from the air samples (72%) compared to the surface swab samples (9.6%). The positive detection rate of the air samples along with the corresponding viral load could be associated with the distance between sampling site and patient. The furthest distance from patient with PCR‐positive air samples was 5.5 m. The airborne SARS‐CoV‐2 detection was comparable between the two types of wards with 60%–87.5% success rate. High prevalence of the virus was found in toilet areas, both on surfaces and in air. Finally, no successful culture attempt was recorded from the environmental air or surface samples.
Keywords: communal testing, COVID‐19, environmental surveillance, high‐flowrate air sampling
Practical Implications.
Deploying high‐flowrate air samplers was found to improve the success rate of airborne SARS‐CoV‐2 detection.
An opposing association was observed between airborne SARS‐CoV‐2 detection and the distance of air sampling site to the patients.
Our air surveillance approach produced a higher success rate of environmental SARS‐CoV‐2 detection than the surface sampling.
Both air and surface sampling highlighted the high prevalence of the virus in toilet areas of the hospital wards
None of the RT‐qPCR‐positive air or surface samples was successfully cultured.
1. INTRODUCTION
Countries, regions, and cities have individually and collectively gone under varying degrees of physical distancing in a bid to control the spread of the COVID‐19 pandemic. International travel has been significantly reduced, and numerous social and educational activities have been severely restricted. These measures have had an extraordinary impact on a global scale. A robust surveillance method for the presence of SARS‐CoV‐2 in the environment is essential to allow safe resumption of normal activities, given that it could require multiple years to achieve reasonable vaccination rate globally. 1 There is also no assurance that vaccines will eliminate the need for continued physical distancing. 2
A large‐scale and accurate test regime that could detect the virus will aid in designing practical screening strategies. Currently, SARS‐CoV‐2 testing is performed on an individual level, which has proven to be laborious and costly. 3 To overcome these limitations, community testing using air, surface, and sewage samples are being explored. In practice, wastewater testing is currently the most advanced 4 ; however, by its very nature, wastewater sampling is retrospective, and cannot give early information about the presence or absence of SARS‐CoV‐2 in an occupied environment. More timely measures for early detection may be reached by SARS‐CoV‐2 environmental surveillance through air and surface monitoring. In this regard, several studies have demonstrated mixed results in validating the presence of SARS‐CoV‐2 RNA in air and on surfaces in hospital environments, based on the reverse transcriptase qPCR (RT‐qPCR) analysis, as approved for nasopharyngeal testing. 5 , 6 , 7 , 8 , 9 , 10 Further, successful environmental surveillance through air is especially relevant as infection via SARS‐CoV‐2 containing aerosols is increasingly being considered as alternative route of transmission. 11 While the definition of “aerosol” varies across different practitioners, 12 there has been growing evidence that supports airborne transmission route of the virus. 13 , 14 , 15 Though limited in number, some studies on airborne SARS‐CoV‐2 have demonstrated success in culturing the virus from aerosol samples. 16 , 17 Recently, human challenge studies have also been approved in the UK that seek to test airborne transmission in healthy human individuals. 18
To contribute to the existing efforts of assessing the potential of air sampling as a tool to detect the presence of SARS‐CoV‐2 in the environment, we undertook air and surface sampling in naturally ventilated open‐cohort ward and mechanically ventilated isolation ward within a large hospital. As regulations and restrictions between hospitals and other facilities where COVID‐19 patients, either symptomatic or asymptomatic, may congregate differ worldwide, it is important to provide multiple methodological options to enable environmental SARS‐CoV‐2 surveillance in a variety of settings. The aims of this study are threefold. First, this study assesses the feasibility of deploying high‐flowrate air sampler in combination with environmental sample RNA extraction kit for the purpose of SARS‐CoV‐2 surveillance in hospital setting. Second, this study aims to investigate the spatial prevalence of the virus in various areas of the hospital when it was fully utilized to care for COVID‐19 patients (ie, in toilets, in different wards, at hallways or at a certain distance from the patient clusters). Finally, the success rate of the tested air surveillance approach is compared to the surface swab sampling and analysis pipeline, which has been more established in our hospital. The two surveillance approaches were tested for direct detection of SARS‐CoV‐2 in the environment and in terms of their potential to conduct culture‐based assessment.
2. MATERIALS AND METHODS
2.1. Hospital wards and condition of patients
Air and surface samples were collected from one isolation ward and two open‐cohort wards housing laboratory‐confirmed COVID‐19 patients at the National University Hospital, an academic medical center, in Singapore. The isolation ward, defined as ward 62 in this study, consists of negative pressure single rooms, each with a dedicated toilet, as previously described. 19 It is equipped with a mechanical ventilation with a design Air Change per Hour (ACH) of 14. The room temperature (T) and relative humidity (RH) were strictly maintained at 23°C and 60%, respectively, during operation. The two open‐cohort wards, defined as ward 42 and ward 43, are located on the same floor and have similar indoor settings. At the beginning of the pandemic in Singapore, the wards were re‐purposed to exclusively care for COVID‐19 patients. The wards were segregated into staff areas, designated as “clean” zones, and patient care areas, designated as “contaminated” zones. The contaminated zones consisted of five open cubicles, each housing six patients, and two smaller rooms, each housing one patient. Each room or cubicle has a dedicated toilet, and these rooms and cubicles share a common corridor. The main difference between the two wards is the distance between the designated clean and contaminated zones. As ward 42 had a slightly smaller total ward area, the distance between the two zones was shorter in ward 42 (4.0–5.5 m), as compared to ward 43 (9.8 m). Both open‐cohort wards adopt natural ventilations with open‐window and ceiling fans. The ACH of such naturally ventilated hospital ward is estimated to range between 20 and 50. The T and RH in the wards fluctuated according to the outdoor air with a range of 26.0–28.0°C and 65% to 75%.
The study was conducted within the period of February to May 2020. This period was within the peak of the first wave of COVID‐19 cases in Singapore. Due to the large number of cases, all three wards were fully occupied by COVID‐19 patients at the time of sampling (confirmed by nasopharyngeal swab (N/P) test). We were unable to obtain the exact Ct values of the N/P test for the involved patients due to logistic issues and data access restrictions from a number of clinical laboratories across Singapore. All patients involved during the study, however, could be considered as mildly symptomatic without significant hypoxia or need for oxygen, as such patients would have been admitted to the intensive care ward instead. All patients were masked with regular surgical mask whenever possible. There was no distinction between the patients housed in ward 62, 42, or 43 as they were admitted to the next available slot as they arrive in the hospital.
2.2. Air sampling and RNA extraction
All air sampling was conducted with filter‐based SASS 3100 air samplers (Research International). The sampler collects total suspended particle (TSP) with no particle size cutoff. The filter media were the default 44 mm diameter SASS bioaerosol filter (polyester material, no electrostatic charge, Research International) with two different pore‐sizes. As per the manufacturer's specification, the small pore‐size filter has an efficiency of 90% for particles <0.5 µm with 50 L/min sampling flowrate, whereas the large pore‐size filter has an efficiency of 50% for particles <0.5 µm with 150 L/min sampling flowrate. The air sampler, along with the selected filter media, has been well studied for their use in sampling and analyzing environmental biological 20 , 21 , 22 and chemical aerosols. 23
All air samples were duplicated for environmental SARS‐CoV‐2 test and for culturing (only for positive samples). Three sampling campaigns were conducted in ward 62 (isolation ward). For the first sampling campaign, three sets of air samplers were placed in the toilet, windowsill (closed), and cardiac table next to the patient. This arrangement corresponds to 0.9–3 m of distance to the patient (details in Table 1). The three sets of samplers were running simultaneously using the small pore‐size filter (90% efficiency, 50 L/min flowrate) for 8 h duration (24 m 3 of air sampled) starting at 12:00 pm. The sampling was repeated three times on February 17, February 18, and March 4, 2020, for replication purpose (n = 9 samples). The second sampling campaign for ward 62 was conducted on 19 and 30 March 2020 with the same sampling start time and duration. An additional set of samplers were added near the air exhaust of the ward, while the other three had identical placements. For flowrate comparison, this second campaign used the large pore‐size filters (50% efficiency, 150 L/min for 8 h, and 72 m3 of air sampled) (n = 7 samples, one toilet sample was not collected on 19 March). Noting the higher frequency of positive detection near air exhaust, an additional third sampling campaign was conducted on May 6, 2020. This time, with the large pore‐size filter (50% efficiency, 150 L/min for 8 h), samplers were only placed near the air exhaust of the ward (n = 1 sample).
TABLE 1.
Locations of air sampling, distance from most proximate SARS‐CoV‐2–infected patient and corresponding RT‐qPCR results
| Location | qPCR result | |||
|---|---|---|---|---|
| Distance from patient (m) | Positive samples | Ct value E‐gene | Ct value N‐gene | |
| Open‐Cohort Ward (150 L/min) | ||||
| Patient Care Area ("Contaminated" zone) | ||||
| Patient cubicle | ||||
| Patient cubicle, ward 43 | 3.5 | 0/1 | ND | ND |
| Patient cubicle, ward 42 | 3.5 | 2/2 | 35.049, 32.732 | ND, 37.343 |
| Toilet | ||||
| Toilet, ward 42 | 2.5 | 1/1 | 29.555 | 34.307 |
| Adjacent to patient area | ||||
| Corridor outside patient cubicles, ward 42 | 5.5 | 1/1 | 35.763 | ND |
| Doffing area, ward 42 | 4 | 1/1 | 36.011 | ND |
| Doffing area, ward 43 | 9.8 | 0/1 | ND | ND |
| Staff Area ("Clean" zone) | ||||
| Donning area, ward 42 | 5 | 1/2 | 28.168 | 35,217 |
| Donning area, ward 43 | 13.3 | 0/1 | ND | ND |
| Negative pressure isolation rooms (50 L/min) | ||||
| Patient Care Area | ||||
| Patient room | ||||
| Cardiac table | 0.9 | 0/3 | ND | ND |
| Windowsill | 3 | 0/3 | ND | ND |
| Toilet | ||||
| Toilet | 0.9 | 0/3 | ND | ND |
| Negative pressure isolation rooms (150 L/min) | ||||
| Patient Care Area | ||||
| Patient room | ||||
| Cardiac table | 0.9 | 2/2 | 36.367, 36.704 | ND |
| Windowsill | 3 | 1/2 | 37.221 | 38.954 |
| Air exhaust | 0.9 | 3/3 | 33.482, 34.882, 37.141 | 37.103, ND, ND |
| Toilet | ||||
| Toilet | 0.9 | 1/1 | 32.149 | 36.107 |
Two sampling campaigns were conducted at the open‐cohort wards on April 15 and May 6, 2020. The large pore‐size filters (50% efficiency, 150 L/min for 8 h starting 12:00 pm) were used for both campaigns. The first campaign sampled the air simultaneously in both ward 42 and ward 43 with the samplers placed at personal protective equipment (PPE) donning area (clean zone), doffing area and patient care area (contaminated zone). An extra set of samplers were available and placed in the corridor area of ward 42. In total, 7 air samples were collected in this first campaign (n = 7 samples). This sampling arrangement covered a range of 2.5 m to 13 m distance between sampling sites and patients (details in Table 1). The second campaign only interrogated ward 42 as ward 43 was not available for sampling. Three sets of samplers with large pore‐size filters were placed in donning area (clean zone), patient care area and toilet (contaminated zone). In total, three air samples were collected in this second campaign (n = 3 samples). Overall, 27 air samples were collected from the three investigated hospital wards. Schematic diagram showing the locations of the air samplers in the three wards can be found in the supplementary material (Figure S1a‐c).
Air samples from the same sampling site were placed in a sterile container and soaked in 1 ml viral Universal Transport Medium (UTM) (COPAN diagnostic) to be transported to the laboratory for storage at −80°C until analysis. RNA was extracted from samples not received in UTM using the RNeasy® PowerWater® Kit (QIAGEN) as per the manufacturer's instructions with the following modification: Each air filter was put in 1 ml of the provided lysis buffer (solution PM1) and incubated at 55°C for 15 min prior to continuing with the default extraction steps. The RNA is then subjected to RT‐qPCR to test for presence of SARS‐CoV‐2 in the sample. In case of positive detection, the UTM inoculated with air filter would then be processed for culturing. The UTM was first filtered with 0.4 µm membrane filter. RNA was then extracted from 600 µl of the UTM with QIAamp Viral RNA Mini Kit (QIAGEN) and subjected to RT‐qPCR. Any qPCR‐positive samples were then cultured to assess viral infectivity using the remaining 400 µl of the UTM.
2.3. Surface sampling and RNA extraction.
Surface samples were obtained using sterile FLOQ swab (COPAN diagnostic) moistened with the aforementioned viral UTM. For each site swabbed, the swabbing motion was repeated twice in two different directions. Swab samples were collected once during the last campaign of air sampling in the same wards. The chosen swab sites were not cleaned for at least 8 h prior to swabbing.
Surfaces from the patient care, staff, and toilet areas of the two ward types were swabbed. Patient care and staff area surfaces included the floor, table, bed handrail, nurse call button, cell phone, the sink, and door handles. The toilet area surfaces were door handle, the sink, toilet ledge, toilet bowl, and the flush button. A portion of some swab sites, for example, the bedside table in the patient care areas and ledge in the toilet, were covered with a sieve one day before swabbing to avoid direct contact from the patient. During swabbing, the sieves were first removed and different sections of the area with and without the sieves were swabbed with separate swab sticks to distinguish the possible difference. A total of seventy‐three surface samples were collected. After sampling, all swab samples were stored in 1 ml of the viral UTM and stored at −80°C until further processing. For extraction, the UTM inoculated with swab was filtered once with a 0.4 µm filter membrane. RNA was then extracted from 600 µl of the samples using the QIAamp® Viral RNA Mini Kit (QIAGEN) as per manufacturer's instructions. RT‐qPCR was then performed to test for presence of SARS‐CoV‐2 in the samples. Any qPCR‐positive samples were then processed for culturing with the remaining 400 µl of the UTM.
2.4. RT‐qPCR Assays
The presence of SARS‐CoV‐2 was detected by RT‐qPCR of the E‐gene and N‐gene as described by Corman et al 24 (PMID: 31992387) using the SuperScript™ III One‐Step RT‐PCR System with Platinum™ Taq DNA Polymerase Kit (Invitrogen) as per the manufacturer's instructions. The positive controls of the RT‐qPCR assay were two serial dilutions of RNA extracted from pure culture of SARS‐CoV‐2 with concentrations equivalent to 1 and 0.1 infectious viral particle (PFU) per PCR reaction. In all assays, our positive control Ct values ranged between 30.0 and 30.8 for the 1 PFU‐positive control and 33.8–35.1 for the 0.1 PFU‐positive control. The negative controls were reagent blanks without any sample and were all negative. The reported limit of detection (LOD) of the RT‐qPCR assay is 3.9 copies per reaction. 24 Accounting for the abovementioned sampling efficiency and assuming complete extraction of viral particles trapped on the filter media, the estimated LOD of our air sampling and processing pipeline is 2.2 copies/m3 of air for the small pore‐size filter and 1.3 copies/m3 of air for the large pore‐size filter. For swab samples, we could not define a fixed surface area for all samples due to the large variety of surfaces to be swabbed. Overall, with the same assumption as the air samples, each swab sample has an LOD of 78.2 copies/swabbed surface. For example, if a 5 cm × 5 cm surface is swabbed for a particular sample, the estimated LOD of the sample will be 3.1 copies/cm2 of surface.
2.5. SARS‐CoV‐2 Culturing
Culture was performed using African green monkey kidney cells (Vero E6; ATCC CRL‐1586™) grown in Dulbecco's Modified Eagle's Medium (DMEM) (Sigma‐Aldrich) supplemented with 2% heat‐inactivated fetal calf serum (FCS) and buffered with 2 g sodium hydrogen carbonate. 400 µl of each qPCR‐positive UTM filtrate was added to Vero E6 cells and incubated at 37°C with 5% CO2. After 4–7 days incubation, cytopathic effect (CPE) was monitored using light microscopy (Olympus). The supernatant was passaged twice more following a four to seven‐day incubation period at 37°C with 5% CO2. RNA was then extracted from these cultures after the third passage using the QIAamp® Viral RNA Mini Kit (QIAGEN) and qPCR performed as described above.
2.6. Limitations
As the study was conducted at the early stage of the pandemic, the chosen timing of sample collection and the subsequent analysis were always subjected to the availability of the trained medical staff, consent of patients, and the capacity of the BSL‐3 processing laboratory. RNA extractions, qPCR assays, and cultures for SARS‐CoV‐2 were all performed in a BSL‐3 laboratory. We, therefore, must strictly adhere to protocols that were, at the time, pre‐approved specifically for each type of sample by the BSL‐3 Biosafety Committee and Institutional Biosafety Committee of the National University of Singapore as well as the Institutional Review Board of the National University of Singapore. Thus, we use Ct values to express viral loads in this study as we were unable to include RT‐qPCR standard curve which could provide estimations of viral RNA copies . As noted above, the positive control of our qPCR assays was SARS‐CoV‐2 RNA expressed in equivalent infectious viral particle (PFU). This study was also exempt from hospital Institutional Board Review according to protocols of the National Healthcare Group domain‐specific institutional review board that governs clinical research at the National University Hospital.
3. RESULTS AND DISCUSSIONS
3.1. High‐flowrate air sampling and distance from patient
The first air sampling campaign of the study was conducted in the isolation ward 62 with a flowrate of 50 L/min. In all three repeats of the first campaign, no successful detection of airborne SARS‐CoV‐2 was recorded. Air sampling flowrate was subsequently increased to 150 L/min for the remainder of our study in both the isolation and open‐cohort wards. As a result, the presence of airborne SARS‐CoV‐2 was detected with a high success rate of 60%–87.5% (Figure 1a,1c). This result highlights the importance of using high‐flowrate air samplers (eg, 150 L/min) to improve the success rate of airborne SARS‐CoV‐2 surveillance, particularly when sampling at a certain distance from the patients and/or in highly ventilated areas such as hospitals (14–50 ACH). Previous studies have also supported this finding. Guo et al. 9 reported up to 44% successful detection of airborne SARS‐CoV‐2 using a 300 L/min air sampler. In a similar case, a study by Ding et al. 10 found positive SARS‐CoV‐2 detection from a single air sample only when a 500 L/min sampler was deployed in a hospital hallway collecting 10 m3 of air. Direct quantitative comparison between studies is challenging due to differences in methodologies and the studied hospital settings. Despite our results, however, we would like to also highlight that lower flowrate air sampler has been successfully deployed in past air surveillance studies. For instance, Chia et al. 7 detected airborne SARS‐CoV‐2 RNA in hospital wards using 5–9 L/min air sampler, while Santarpia et al. 17 detected the virus using a mobile personal sampler with 4 L/min air flowrate. It must be noted that airborne virus concentrations reported by both studies were very high. Their reported concentrations were in thousands of copies per m3 of air, with a peak of 48,000 copies per m3 of air during nasal canula procedure. These concentrations are significantly higher than other studies 6 , 9 , 10 who reported successful detections with high‐flowrate samplers (in tens to hundreds of copies/m3 of air). The elevated viral loads in the environments could explain the higher success rate of the studies using low flowrate air samplers.
FIGURE 1.
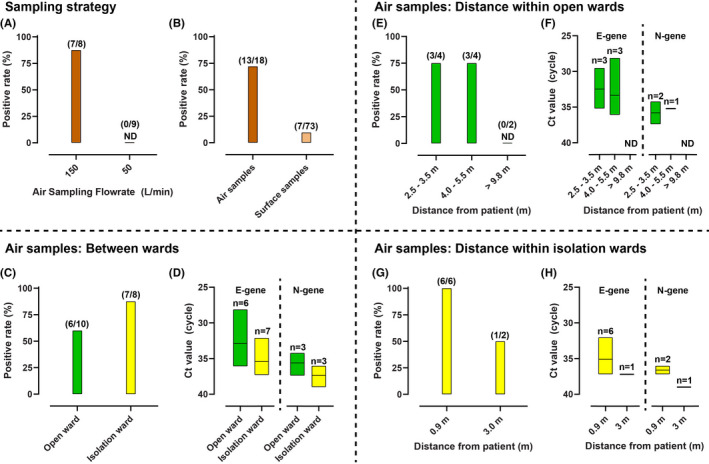
Positive detection rate and Ct values representing viral load of environmental SARS‐CoV‐2 samples collected in hospital wards. a. positive detection rate comparison between air samples collected with high (150 L/min) and low (50 L/min) flowrate in isolation ward. b. Positive detection rate comparing air samples collected with high‐flowrate (150 L/min) and surface swab samples. c. Positive detection rate comparison of air samples collected between wards and d. their corresponding Ct values. e. Positive detection rate comparison between air samples collected in open‐cohort wards and f. the Ct values. g. Positive detection rate comparison between air samples collected in isolation ward and h. the resulted Ct values. The y‐axes for Ct values in d, f, and h were reversed to illustrate the lower viral load when higher Ct values are recorded. The box plot upper and lower boundary represent the maximum and minimum values among positive samples. The line represents the mean of positive samples
Our study also demonstrates an opposite association between the distance of air sampling site from infected patients and the positive detection rate of airborne SARS‐CoV‐2 as well as the corresponding viral load (Ct values) (Figure 1e–h). All six air samples collected with patient present within 0.9 meters in the negative pressure isolation rooms were PCR positive for SARS‐CoV‐2 RNA. One air sample which was negative for SARS‐CoV‐2 RNA was from a sampler placed on a windowsill three meters from the patient's bed. In the open wards, three of four air samples in the patient care areas, with patients within four meters, and two of three air samples in areas adjacent to patient care areas, were PCR positive. In contrast, all three of the air samples from the clean zones in the open wards (>9.8 m distance) were found to be PCR negative, with the exception of one of two samples from a clean area (PPE donning zone) directly adjacent to (~5 m distance) the contaminated area (Table 1). The presence of blowers at the donning area should provide enough positive pressure to prevent air from the contaminated area to enter the clean zone. The positive detection in one of these areas indicates that the airflow direction could be counter‐acted by the use of high‐flowrate air samplers, which provided higher sensitivity in detecting environmental SARS‐CoV‐2 in air when conducting surveillance in such indoor setting. The higher detection success rate with closer distance to patient is consistent with the finding reported by Guo et al. 9
3.2. Comparison to surfaces and prevalence of the virus in certain areas
Of the seventy‐three surface samples obtained, only seven samples were PCR positive. Surface samples in both isolation and open wards were PCR negative, except for samples taken in the toilets, and one sample from the patient bedside table in the isolation wards (Table 2). The other side of the table which was covered with sieve was PCR negative, implying that touch frequency is the more likely source of the virus on the table surface. The lower detection rate of surface samples compared to air (Figure 1b) agrees with a recent study conducted by Zhou et al. 25 The equally high success rate between air and surface samples reported by Chia et al. 7 and Santarpia et al. 17 could be explained by the above‐discussed high viral load at the sites at the time of sampling. It suggests that, in clean hospital settings, air sampling is a more sensitive surveillance tool as compared to surface swabs. Possible reasons include the fact that air is a more well‐mixed medium as opposed to surface where viral load depends on touch frequency. In addition, hospital surfaces are frequently cleaned with hydrogen peroxide or phenolic disinfectant, which prevents long‐term accumulation of the virus. It is, therefore, worth noting that environmental surveillance in personal quarantine spaces outside hospital, such as private hotel rooms or home environments could yield higher success rate for surface swab samples due to substantially less stringent cleaning regime.
TABLE 2.
Locations of environmental surfaces swabbed, and corresponding RT‐qPCR results
| Location | qPCR result | ||
|---|---|---|---|
| Positive samples | Ct value E‐gene | Ct value N‐gene | |
| Open‐Cohort Ward (Ward 43) | |||
| Patient Care Area ("Contaminated" zone) | |||
| Patient cubicle | |||
| Floor, 1 m from patient | 0/1 | ND | ND |
| Floor, 2 m from patient | 0/1 | ND | ND |
| Floor, 3 m from patient | 0/1 | ND | ND |
| Patient bedside table—covered with sieve | 0/1 | ND | ND |
| Patient bedside table—not covered with sieve | 0/1 | ND | ND |
| Bed hand rail | 0/1 | ND | ND |
| Nurse call button | 0/1 | ND | ND |
| Intravenous pole | 0/1 | ND | ND |
| Cell phone | 0/1 | ND | ND |
| Room sink, rim | 0/1 | ND | ND |
| Room sink, drain | 0/1 | ND | ND |
| Toilet Door handle, ward side | 0/1 | ND | ND |
| Toilet | |||
| Door handle, toilet side | 0/1 | ND | ND |
| Sink, handle | 1/1 | 39.633 | ND |
| Sink, rim* | 1/1 | 32.811 | 33.418 |
| Sink, drain* | 1/1 | 35.955 | 36.357 |
| Toilet bowl* | 0/1 | ND | ND |
| Toilet flush button | 0/1 | ND | ND |
| Staff area ("Clean" zone) | |||
| Door knob (Staff rest area) | 0/1 | ND | ND |
| Cardiac table | 0/1 | ND | ND |
| Computer On Wheels Keyboard | 0/1 | ND | ND |
| Patient Service Associate table | 0/1 | ND | ND |
| *denotes positive CPE from first passage of viral culture | |||
| Negative pressure isolation rooms | |||
| Patient care area | |||
| Patient room | |||
| Floor, 1 m from patient | 0/1 | ND | ND |
| Floor, 2 m from patient | 0/1 | ND | ND |
| Floor, 3 m from patient | 0/1 | ND | ND |
| Patient bedside table—covered with sieve | 0/5 | ND | ND |
| Patient bedside table | 1/9 | 39.328 | ND |
| Bed hand rail | 0/1 | ND | ND |
| Nurse call button | 0/1 | ND | ND |
| Cell phone | 0/1 | ND | ND |
| Room sink, rim | 0/1 | ND | ND |
| Room sink, drain | 0/1 | ND | ND |
| Door knob, patient‐side | 0/1 | ND | ND |
| Area under bed | 0/1 | ND | ND |
| Toilet Door handle, ward side | 0/1 | ND | ND |
| Gloves in contact with patient | 0/3 | ND | ND |
| Gloves in contact with patient surroundings | 0/4 | ND | ND |
| Toilet | |||
| Door handle, toilet side | 0/1 | ND | ND |
| Sink, handle | 1/1 | 39.261 | ND |
| Sink, rim | 0/1 | ND | ND |
| Sink, drain | 0/1 | ND | ND |
| Toilet bowl | 1/1 | 35.016 | 36.155 |
| Toilet flush button | 0/1 | ND | ND |
| Toilet ledge—covered with sieve | 1/4 | 37.424 | ND |
| Toilet ledge | 0/8 | ND | ND |
| Staff area | |||
| Door knob outside room | 0/1 | ND | ND |
Comparing the airborne SARS‐CoV‐2 surveillance between the two types of wards, the positive detection rates of the virus were comparable. The rate in the isolation ward was 27.5% higher than the open ward (Figure 1c). However, if only the contaminated area samples were considered, this difference was reduced to 12.5%. Interestingly, despite the higher positive rate of the isolation ward air samples, the open ward air samples had an averagely higher viral load (Figure 1d). In addition to the abovementioned distance factor, differences in ventilation scheme between the two wards (mechanical vs natural ventilation) as well as variabilities associated with the patients that were present at the time of sampling are the likely confounding factors for the observed dynamics between the investigated wards. For instance, while the open wards housed a larger number of occupants, the wards are much bigger in size compared to the isolation ward. The ACH of the open ward is also expected to be higher than the isolation ward, counter‐acting the high number of patients. It is important to note, however, that there is a possibility of airborne cross‐contamination between different patient care areas within the open ward.
Finally, we would like to highlight the high prevalence of environmental SARS‐CoV‐2 in our hospital toilet areas. PCR‐positive swab samples were only found in toilet surfaces. In this regard, air sampling conducted in the toilet also yielded higher SARS‐CoV‐2 detection (Ct: 29.6 in open‐cohort ward and 32.2 in isolation ward) relative to other areas. The higher viral load in toilet areas is supported by the finding reported by Ding et al., 10 which showed that toilet areas dominate the environmental detection of SARS‐CoV‐2 in a hospital in China. The high viral shedding of infected individuals in the toilet also supports the concept of wastewater screening as an adjunctive, “early warning” surveillance for SARS‐CoV‐2 infection in the community, as has beena component of the “endgame” for polio eradication. 26
3.3. RNA Extraction, RT‐qPCR Assay, and SARS‐CoV‐2 Culturing from Environmental Samples
While it is tempting to directly compare viral loads between different studies, it is not recommended due to differences in sampling and analytical approaches. In addition, hospitals are operated under different ventilation regimes and airflow patterns. Comprehensive review studies for airborne SARS‐CoV‐2 surveillance 5 , 27 have mainly focused on sampling methodologies. However, an equally important factor is the RNA extraction protocol. Several airborne SARS‐CoV‐2 studies have reported success with RNA extraction kits designed for pure viral culture or human clinical samples (eg, sputum or saliva) such as King Fisher Viral Total NA Kit 9 or QIAamp Viral RNA Mini Kit. 7 Our study provides an alternative for environmental SARS‐CoV‐2 sample processing by confirming the possibility of using an extraction kit specifically designed for environmental samples (RNeasy Power Water Kit). An important difference for this kit is the inclusion of an inhibitor removal step. PCR inhibition has been reported for air filter samples that amassed large quantities of particulate matter (PM). 28 The use of extraction kits designed for environmental samples is therefore potentially more relevant when conducting air surveillance outside of clean hospital environments.
In all positive environmental samples (air and surface), it was noted that qPCR assay targeting the E‐gene is more sensitive than the N‐gene both in terms of positive detection rate and Ct values (Figure 1d,1f,and 1h). While this finding is consistent with Corman et al., 24 their study tested the two RT‐qPCR assays on clinical samples. Our study highlighted that this finding is consistent with SARS‐CoV‐2 detected from environmental air and surface samples. Interestingly, we also found that the differences in Ct values were higher in the air samples (3–5 Ct differences, Table 1) as compared to surface swab samples (1–2 Ct differences, Table 2). The E‐gene is the envelope protein gene, whereas the N‐gene is the nucleocapsid protein gene. A future study involving a more controlled laboratory‐based experiment is necessary to pinpoint the exact cause. However, we suspect that these are caused by differences in assay efficiencies. It is also possible that it is an artifact of the two sampling approaches and the subsequent processing pipelines. Drawing conclusions from quantitative comparison of multiple studies must therefore be mindful of not only differences in sampling details but also in the sample processing steps and the RT‐qPCR assays.
Finally, our study could not show conclusive evidence for successful viral culture from both air and swab samples. Positive CPEs with Ct values greater than 36 were only detected from the first passage viral cultures from three of the positive surface swab samples (obtained from toilet sink rim, drain, and toilet bowl, Table 2). Subsequent passages did not find any viable virus. None of the PCR‐positive air samples were successfully cultured. While we recommend sampling with higher flowrate to improve detection with molecular analysis, prolonged high‐flowrate air sampling on dry filter media has been shown to damage and desiccate the viral particles during the sampling process, thus significantly reducing the success rate of culturing the virus. This finding is consistent with previous reports. 29
4. CONCLUSION
In summary, our findings continue to support the suitability of using air sampling as a tool for environmental surveillance of airborne SARS‐CoV‐2 in hospital environment. This study further demonstrated that opting for higher air sampling flowrate improves the chance of successful airborne SARS‐CoV‐2 surveillance especially in sites that are highly ventilated or in situations where air samplers could only be placed at a certain distance from the patient (3–5 m), albeit with a much lower chance of successful culture‐based assessment due to harsh sampling condition. In hospitals with a high daily census of COVID‐19 patients, employing a routine air surveillance program with a high success rate could prove beneficial in detecting the presence of the virus early in certain unsuspecting spaces/rooms for better protection of the involved healthcare workers. Future air surveillance studies will need to be tested in locations outside of hospital environments where mass gatherings occur for rapid and sensitive high throughput communal testing at the population level. One potential complication of such large‐scale expansion for airborne SARS‐CoV‐2 surveillance outside hospital is the high background concentration of other biological agents or non‐biotic particulate matter (PM) which could interfere with analysis. 30
CONFLICT OF INTEREST
The authors declare no conflict of interests.
AUTHOR CONTRIBUTIONS
Paul A Tambyah, David M Allen, Stephan C Schuster, and Daniela I Drautz‐Moses involved in conceptualization of study and study design. Alicia XY Ang, Sai Meng Tham, and David M Allen involved in data collection. Bintou A Ahidjo, Justin JH Chu, and Chee Keng Mok involved in laboratory testing. Alicia XY Ang, Irvan Luhung, Bintou A Ahidjo, Paul A Tambyah, and David M Allen involved in data analysis. Alicia XY Ang, Bintou A Ahidjo, Paul A Tambyah, David M Allen, Daniela I Drautz‐Moses, Irvan Luhung, and Stephan C Schuster involved in drafting of the manuscript. Alicia XY Ang, Paul A Tambyah, David M Allen, Stephan C Schuster, Kenny JX Lau, Irvan Luhung, and Daniela I Drautz‐Moses involved in manuscript revision. Stephan C Schuster, David M Allen, and Paul A Tambyah involved in acquisition of funds. Stephan C Schuster, Daniela I Drautz‐Moses, Irvan Luhung, and Kenny JX Lau involved in development of air sampling and sample extraction protocol.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/ina.12930.
Supporting information
Fig S1
ACKNOWLEDGEMENT
An unrestricted educational grant from Freepoint Commodities, Pvt Ltd, Singapore supported the laboratory work, biosafety level 3 procedures and consumables and equipment, manpower, sample collection, and NMRC grant MOH‐000411 supported the air samplers, associated consumables, protocol development, and RNA extraction work (PI Stephan Christoph Schuster). Bintou A Ahidjo was supported by NMRC/CG/M009/2017‐NUH/NUHS. Chee Keng Mok is supported by NMRC/CG/M009/2017‐NUH/NUHS.
Ang AXY, Luhung I, Ahidjo BA, et al. Airborne SARS‐CoV‐2 surveillance in hospital environment using high‐flowrate air samplers and its comparison to surface sampling. Indoor Air. 2022;32:e12930. 10.1111/ina.12930
Alicia XY Ang, Irvan Luhung and Bintou A. Ahidjo contributed equally.
David M. Allen and Stephan C. Schuster are Co‐corresponding authors.
Contributor Information
Alicia XY Ang, Email: alicia_ang@nuhs.edu.sg.
Irvan Luhung, Email: lirvan@ntu.edu.sg.
David M. Allen, Email: mdcdma@nus.edu.sg.
Stephan C. Schuster, Email: scschuster@ntu.edu.sg, Email: stephan.c.schuster@gmail.com.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Xing Y, Wong GW, Ni W, Xing Q. Rapid response to an outbreak in qingdao, China. N Engl J Med. 2020;383(23):e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aschwanden C. Five reasons why COVID herd immunity is probably impossible. Nature. 2021;591(7851):520‐522. 10.1038/d41586-021-00728-2 [DOI] [PubMed] [Google Scholar]
- 3. Sahajpal NS, Mondal AK, Njau A, et al. Proposal of reverse transcription‐PCR–based mass population screening for SARS‐CoV‐2 (COVID‐19). J Mol Diagn. 2020;22(10):1294‐1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ahmed W, Bertsch PM, Angel N, et al. Detection of SARS‐CoV‐2 RNA in commercial passenger aircraft and cruise ship wastewater: a surveillance tool for assessing the presence of COVID‐19 infected travellers. J Travel Med. 2020;27(5):taaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Birgand G, Peiffer‐Smadja N, Fournier S, et al. Assessment of air contamination by SARS‐CoV‐2 in hospital settings. JAMA Netw Open. 2020;3(12):e2033232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu Y, Ning Z, Chen Y, et al. Aerodynamic analysis of SARS‐CoV‐2 in two Wuhan hospitals. Nature. 2020;582:557‐560. [DOI] [PubMed] [Google Scholar]
- 7. Chia PY, Coleman KK, Tan YK, et al. Detection of air and surface contamination by SARS‐CoV‐2 in hospital rooms of infected patients. Nat Commun. 2020;11:e2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li YH, Fan YZ, Jiang L, Wang HB. Aerosol and environmental surface monitoring for SARS‐CoV‐2 RNA in a designated hospital for severe COVID‐19 patients. Epidemiol Infect. 2020;148:e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo ZD, Wang ZY, Zhang SF, et al. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, Wuhan, China, 2020. Emerg Infect Dis. 2020;26(7):1583‐1591. 10.3201/eid2607.200885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ding Zhen, Qian Hua, Xu Bin, et al. Toilets dominate environmental detection of severe acute respiratory syndrome coronavirus 2 in a hospital. Sci Total Environ. 2021;753:141710. 10.1016/j.scitotenv.2020.141710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coronavirus is in the air—there’s too much focus on surfaces. Nature. 2021;590:7. 10.1038/d41586-021-00277-8 [DOI] [PubMed] [Google Scholar]
- 12. Tang JW, Bahnfleth WP, Bluyssen PM, et al. Dismantling myths on the airborne transmission of severe acute respiratory syndrome coronavirus (SARS‐CoV‐2). J Hosp Infect. 2021;110:89‐96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Richard M, Kok A, de Meulder D, et al. SARS‐CoV‐2 is transmitted via contact and via the air between ferrets. Nat Commun. 2020;11:e3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khanh N, Thai P, Quach H, et al. Transmission of SARS‐CoV 2 During Long‐Haul Flight. Emerg Infect Dis. 2020;26(11):2617‐2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Murphy N, Boland M, Bambury N, et al. A large national outbreak of COVID‐19 linked to air travel, Ireland, summer 2020. Euro Surveill. 2020;25(42):2001624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lednicky JA, Lauzard M, Fan ZH, et al. Viable SARS‐CoV‐2 in the air of a hospital room with COVID‐19 patients. Int J Infect Dis. 2020;100:476‐482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Santarpia JL, Rivera DN, Herrera VL, et al. Aerosol and surface contamination of SARS‐CoV‐2 observed in quarantine and isolation care. Sci Rep. 2020;10(1):1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kirby T. COVID‐19 human challenge studies in the UK. Lancet Respir Med. 2020;8(12):e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ong SW, Coleman KK, Chia PY, et al. Transmission modes of severe acute respiratory syndrome coronavirus 2 and implications on infection control: a review. Singapore Med J. 2021;1:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mbareche H, Veillette M, Bilodeau GJ, Duchaine C. Bioaerosol sampler choice should consider efficiency and ability of samplers to cover microbial diversity. Appl Environ Microbiol. 2018;84(23):e01589‐e1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dybwad M, Skogan G, Blatny JM. Comparative testing and evaluation of nine different air samplers: end‐to‐end sampling efficiencies as specific performance measurements for bioaerosol applications. Aerosol Sci Technol. 2014;48(3):282‐295. [Google Scholar]
- 22. Luhung I, Uchida A, Lim SB, et al. Experimental parameters defining ultra‐low biomass bioaerosol analysis. NPJ biofilms and microbiomes. 2021;7(1):1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Daniel D, Lin M, Luhung I, et al. Effective design of barrier enclosure to contain aerosol emissions from COVID‐19 patients. Indoor Air. 2021;31(5):1639‐1644. 10.1111/ina.12828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Eurosurveillance. 2020;25(3):2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou L, Yao M, Zhang X, et al. Breath‐, air‐and surface‐borne SARS‐CoV‐2 in hospitals. J Aerosol Sci. 2021;152:105693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. World Health Organization . Guidelines for Environmental Surveillance Of Poliovirus Circulation. No. WHO/V&B/03.03. Switzerland: World Health Organization, 2003;1‐19. https://apps.who.int/iris/handle/10665/67854 [Google Scholar]
- 27. Morawska L, Tang JW, Bahnfleth W, et al. How can airborne transmission of COVID‐19 indoors be minimised? Environ Int. 2020;142:105832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luhung I, Wu Y, Xu S, Yamamoto N, Wei‐Chung Chang V, Nazaroff WW. Exploring temporal patterns of bacterial and fungal DNA accumulation on a ventilation system filter for a Singapore university library. PLoS One. 2018;13(7):e0200820. 10.1371/journal.pone.0200820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pan M, Lednicky JA, Wu CY. Collection, particle sizing and detection of airborne viruses. J Appl Microbiol. 2019;127(6):1596‐1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lednicky JA, Shankar SN, Elbadry MA, et al. Collection of SARS‐CoV‐2 virus from the air of a clinic within a university student health care center and analyses of the viral genomic sequence. Aerosol and air quality research. 2020;20(6):1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


