Summary
The COVID-19 pandemic revealed an urgent need for rapid profiling of neutralizing antibody responses and development of antibody therapeutics. The current Food and Drug Administration-approved serological tests do not measure antibody-mediated viral neutralization, and there is a need for standardized quantitative neutralization assays. We report a high-throughput two-step profiling approach for identifying neutralizing convalescent plasma. Screening and downselection for serum antibody binding to the receptor-binding domain are followed by quantitative neutralization testing using a chimeric vesicular stomatitis virus expressing spike protein of SARS-CoV-2 in a real-time cell analysis assay. This approach enables a predictive screening process for identifying plasma units that neutralize SARS-CoV-2. To calibrate antibody neutralizing activity in serum from convalescent plasma donors, we introduce a neutralizing antibody standard reagent composed of two human antibodies that neutralize SARS-CoV strains, including SARS-CoV-2 variants of concern. Our results provide a framework for establishing a standardized assessment of antibody-based interventions against COVID-19.
Subject areas: Immunology, Virology
Graphical abstract

Highlights
-
•
A high-throughput approach enabling antibody activity testing in COVID-19 plasma
-
•
SARS-CoV-2 IgG binding screen coupled with quantitative neutralization testing
-
•
Neutralization testing is necessary for profiling COVID-19 convalescent plasma
-
•
Two broad monoclonal antibodies identified as a neutralization testing standard
Immunology; Virology
Introduction
Since the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as a major threat to global public health, many studies have focused efforts on identifying and evaluating antibody-based therapeutic interventions (Baum et al., 2020; Chen et al., 2021b; Jones et al., 2021; Liu et al., 2020; Robbiani et al., 2020; Rogers et al., 2020; Taylor et al., 2021; Zost et al., 2020a). Neutralizing antibodies to the spike (S) protein of SARS-CoV-2 elicited during naturally occurring infection, or vaccination, are considered a serological correlate of protection (Corbett et al., 2021; Krammer, 2021). Among the most low-cost and easily accessible sources of human antiviral antibodies are plasma from SARS-CoV-2 convalescent donors to treat infection in high-risk patients. Accordingly, numerous large-scale randomized, controlled trials are ongoing to determine if COVID-19 convalescent plasma is a safe and effective treatment for COVID-19, and the Food and Drug Administration (FDA) issued an Emergency Use Authorization (EUA) in August of 2020 for passive transfer of COVID-19 convalescent plasma for treatment of COVID-19 (FDA, 2020). Establishing antibody binding and neutralizing titers for convalescent plasma correlated with protection would be significant for public health considerations and patient management. However, the current FDA-approved serological tests for screening activity in serum samples from convalescent plasma donors do not specifically measure antibody-mediated viral neutralization (FDA, 2021b), and relatively few studies have attempted a systematic analysis to correlate quantitative antibody binding results with neutralization results in serum or plasma. In addition, the lack of standardized laboratory neutralization assays and potency standards (Gundlapalli et al., 2021; Khoury et al., 2020) hinders the evaluation and use of convalescent plasma both within and across these large-scale studies.
This study describes a systematic evaluation of antibody reactivity and neutralization in a large set of human convalescent plasma units designated for transfusion in the Passive Immunity Trial for Our Nation (PassITON) (Self et al., 2021) using currently available clinical-use and newly developed laboratory methodologies. To harmonize the results from different studies and calibrate neutralizing activity in convalescent plasma, we propose a GMP-grade immunoglobulin standard composed of two recombinant human mAbs that broadly neutralize SARS-CoV-2 strains, including variants of concern. Our data provide a framework for standardized assessment of neutralizing activity in convalescent plasma and the other antibody-based interventions against COVID-19.
Results
Development of a quantitative high-throughput assay for neutralizing antibody evaluation in serum
For benchmarking purposes, we analyzed matched serum samples collected from convalescent individuals of COVID-19 who donated their convalescent plasma for transfusion. To determine neutralizing antibody activity in serum, we explored the utility of a high-throughput real-time cell analysis (RTCA)-based methodology we previously developed for monoclonal antibody studies. RTCA has demonstrated high performance with representative strains from several virus families and offers potential advantages over conventional laboratory plaque or focus reduction neutralization tests (PRNT or FRNT) (Gilchuk et al., 2020a, 2020b; Zost et al., 2020b). RTCA provides a continuous automated measurement of cellular impedance in a 6 × 96-well plate format using an xCELLigence RTCA MP Analyzer (Agilent Technologies) instrument to quantify virus-induced cytopathic effect (CPE) in virus-infected cell culture monolayers. We recently deployed RTCA-based assays using Vero cells in a BSL3 environment (using authentic SARS-CoV-2 live virus suspensions) or in a BSL2 environment (using chimeric vesicular stomatitis virus [VSV] expressing SARS-CoV-2 S protein) for large-scale discovery of human monoclonal antibodies (mAbs) that neutralize SARS-CoV-2 (Suryadevara et al., 2021; Zost et al., 2020b). We also described the application of RTCA to measure antibody-mediated neutralizing activity in serum from an individual after vaccination with mRNA-encoded spike protein (Chen et al., 2021a). Overall, these prior studies suggested this automated CPE measurement approach for quantitative and high-throughput antibody screening might be of utility for serological screening.
We assessed the performance, precision, and reproducibility of RTCA for neutralizing antibody testing in serum, which serves as a proxy for antibody levels and neutralizing activity in contemporaneously collected plasma. For our workflow, we used a replication-competent chimeric VSV expressing the S protein from SARS-CoV-2 (WA1/2020 strain) (Case et al., 2020b). The ability of this fully infectious virus to induce rapid CPE allows for high-throughput and quantitative neutralization assessment in a BSL2 laboratory.
Serum is a complex biological matrix that may differentially impact measurements of antibody neutralization by different assays. To assess if the RTCA assay could reliably measure neutralizing activity of an antibody in the context of human serum, we used the previously developed potently neutralizing mAb COV2-2196 (Zost et al., 2020a) or a combination of two potently neutralizing human mAbs COV2-2130 and COV2-2381 (Zost et al., 2020a) that form a basis of investigational drug product against COVID-19-designated ADM03820 (ClinicalTrials.gov Identifier: NCT04592549). Purified neutralizing mAbs carried in normal human serum or cell culture growth medium showed equivalent performance in measuring antibody neutralization in the RTCA assay (Figure 1A).
Figure 1.
Analysis of accuracy and reproducibility of high-throughput automated RTCA neutralization assay measurements
(A) Dose-response neutralization curves for potently neutralizing human monoclonal antibodies that were diluted in incubation medium or medium containing normal human serum at 1:50 dilution for the assay.. Antibodies tested included mAb COV2-2196 and a combination of two mAbs COV2-2130 and COV2-2381 designated as ADM03826. Neutralization was assessed against VSV-SARS-CoV-2 WA1/2020 virus. Data show mean ± SD of quadruplicates.
(B) Representative dose-response RTCA neutralization curves for 76 convalescent serum samples as in (A). Each curve represents one sample. Data show mean of duplicates for one of two independent experiments.
(C) Orthogonal (Deming) regression analysis of area under the curve mean values from two independent experiments as in (B). Duplicate measurements were performed for each independent experiment. Each dot represents one sample, the regression line is indicated with solid line, and line of identity is dotted. p value is indicated. LOD is 2.7 log10AUC.
(D) Bland–Altman plot of AUC values from two independent experiments as in (C). Duplicate measurements were performed for each independent experiment. The 95% limits of agreement are shown as two dotted lines.
(E) Orthogonal (Deming) regression analysis of NT50 values from two independent experiments as in (B). Duplicate measurements were performed for each independent experiment. Only 17 of 76 samples that revealed NT50 ≥ 50 are shown. The regression line is indicated with solid line, and dotted line indicates line of equal X and Y coordinates for every point. p value is indicated.
(F) Bland–Altman plot of NT50 values from two independent experiments as in (E). Duplicate measurements were performed for each independent experiment. The 95% limits of agreement are shown as two dotted lines.
To define quantitative measurements of antibody neutralizing activity in serum, we assembled a training set of 76 SARS-CoV-2 serum samples from individuals who had recovered from COVID-19. This panel represented individuals with a broad spectrum of clinical severity from severe disease (n = 6) to mild to moderate disease (n = 63), a wide range of ages (20–70 years), and a balance of sexes (54.3% female/44.3% male/1.4% unknown) (Table S1). Serially diluted serum samples neutralized VSV-SARS-CoV-2 in a dose-dependent manner. This analysis identified 6 samples with high (>95% virus neutralization at the lowest tested [1:50] serum dilution), 11 samples with medium (50%–90%), and 59 samples with low (<50%) neutralizing activity (Figure 1B). Determination of half-maximal neutralization titer (NT50) or neutralization titer that reduced virus infectivity by 80% (NT80) provided a quantitative assessment of samples exhibiting high or moderate neutralizing activity. Area under the curve (AUC) assessment allowed for potency ranking across the entire sample set, including serum with low neutralizing activity.
To assess assay precision and reproducibility, we analyzed the full set of 76 samples from two independent experiments, using the mean of duplicate measurements for each experiment. Orthogonal regression showed excellent agreement (1.000∗X + 0.0736; P < 0.0001) (Figure 1C). The Bland–Altman plot showed little difference between repeated measurements, although the variability of AUC measurements was higher for samples exhibiting low neutralizing activity (log10AUC<3.5 with the limit of detection [LOD] = 2.7) (Figure 1D). Serum samples that exhibited NT50 values ≥ 50 showed strong agreement (0.9364∗X + 0.4147; P < 0.0001) (Figure 1E) and low variability (bias = 0.1990; 95% limits of agreement from −2.57% to 2.97% difference) (Figure 1F) between the two independent experiments. This highlights the high assay accuracy and reproducibility in quantifying high to moderate levels of neutralizing activity in potentially clinically relevant samples.
In summary, our results demonstrated high performance of RTCA methodology in a BSL2 laboratory setting for quantitative neutralizing antibody assessment in human serum.
Antibody neutralization and reactivity relationship in serum
We next defined a workflow for evaluating anti-SARS-CoV-2 antibody response by performing a multifactorial assessment of antibody features in serum samples. For benchmarking purposes, we used 76 SARS-CoV-2 serum samples from individuals who had recovered from COVID-19 infections (Figure 1B; Table S1). We carried out three quantitative or semi-quantitative assays to understand the relationship between these assay measurements in convalescent plasma evaluation: 1) antibody binding to SARS-CoV-2 S protein, 2) inhibition of SARS-CoV-2 spike RBD (SRBD) binding to human angiotensin-converting enzyme 2 (ACE2) receptor protein, or 3) virus neutralization.
Antibody binding was assessed by two commercial automated clinical-use tests that carry an EUA from the FDA: (1) Ortho VITROS Anti-SARS-CoV-2 IgG (S protein binding, semi-quantitative assay) (FDA, 2021d) and (2) Abbott AdviseDx II SARS-CoV-2 IgG (SRBD protein binding; quantitative assay) (FDA, 2021a) (see STAR Methods). We also tested other antibody-binding assays, including an automated research-use assay, Leinco Trace IgG micro-ELISA (Leinco Technologies, semi-quantitative assay), and a recently reported high-throughput fluorescence-based quantitative binding assay using MagPlex microspheres (Bennett et al., 2021), both of which measure IgG binding to the SRBD. Additionally, we evaluated a bead-based ACE2-inhibition semi-quantitative assay (Leinco Technologies) that measures the ability of an antibody-containing sample to inhibit the interaction of recombinant SRBD with the recombinant extracellular domain of human ACE2 and therefore is considered a surrogate measurement of antibody neutralization.
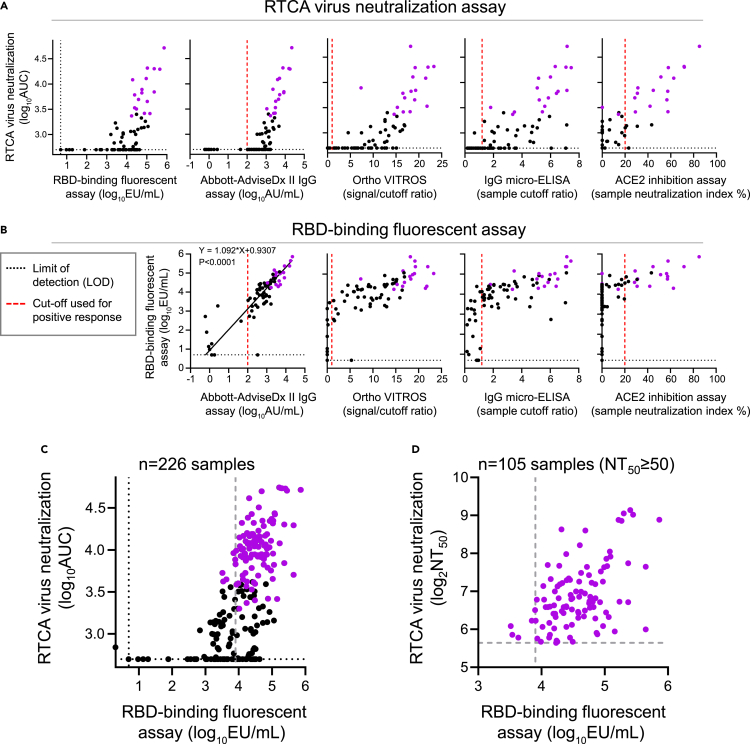
We first evaluated the relationship between antibody binding and neutralization. Measurements from each binding assay for individual serum samples were compared with their respective AUC measurements estimated from the dose-response RTCA assay neutralization curves (Figure 1A). The analysis revealed a relationship between antibody binding and neutralization suggestive of a binding threshold below which samples did not exhibit detectable neutralizing activity based on half-maximal neutralization titer measurement (i.e., samples with NT50≥ 50 indicated with violet dots in Figure 2). Moreover, samples with relatively high binding activity measured by each assay varied from high to low neutralizing activity. Better agreement was observed between RTCA assay neutralizing activity and ACE2 inhibition assay measurements, consistent with previous studies that demonstrated a significant fraction of neutralizing antibodies in serum or isolated mAbs inhibit ACE2 binding to the SRBD(Liu et al., 2020; Robbiani et al., 2020; Zost et al., 2020b). We conclude that high antibody-binding titers to SARS-CoV-2 antigens are required for virus neutralization but do not fully predict the level of neutralizing activity in serum.
Figure 2.
Relationship of the RTCA neutralization and binding measurements for convalescent human serum
(A) Comparison of measurements from the high-throughput RTCA chimeric VSV-SARS-CoV-2 virus neutralization assay with those from the indicated binding assays using 76 serum samples. Seventeen samples from Figure 1D with NT50 values ≥50 are indicated using violet dots. Red dotted line indicates designated cutoff for commercial tests, and black dotted line depicts the assay limit of detection.
(B) Comparison of measurements from high-throughput RBD-binding fluorescent assay with those from the indicated binding assays using 76 serum samples as in (A). Seventeen samples from Figure 1E with NT50 values ≥50 are shown using violet dots. Orthogonal (Deming) regression analysis was performed to compare RBD-binding fluorescent assay measurements to those from Abbot AdviseDx II IgG assay, and p value is indicated.
(C) Relationship of RTCA and RBD-binding fluorescent assay measurements using a larger collection of 226 serum samples, which included 76 samples as in (A and B). Violet dots indicate 105 samples with NT50 values ≥50. Samples with an antibody-binding level above 8,000 EU/mL and NT50 values ≥50 were selected for transfusion in the trial. Gray dashed line is empirically defined cutoff for positive binding response as detailed in the STAR Methods section.
(D) Relationship of RTCA and RBD-binding fluorescent assay measurements using 105 serum samples from (C) with NT50 values ≥50 are shown. Gray dashed lines indicate empirically defined cutoffs for positive binding and neutralizing responses as detailed in the STAR Methods section.
As the key measure of antibody-antigen binding in this study, we implemented a fluorescent SARS-CoV-2 RBD-coupled MagPlex microsphere binding assay (Bennett et al., 2021) (defined as RBD-binding fluorescent assay). This microsphere fluorescence-based assay has advantages for convalescent plasma testing in a research laboratory setting owing to high throughput, wide dynamic range, and high accuracy compared to ELISA binding assays. In addition, this assay measures antibody binding to SRBD, the principal target for anti-SARS-CoV-2 neutralizing antibodies. A comparison of the RBD-binding fluorescent assay with the other binding assays measurements using the same set of 76 samples identified the majority of 17 top neutralizing samples (NT50≥ 50; Figure 1D) within the category of moderate-to-high binding activity for each assay (Figure 2B). These results demonstrated better performance by the RBD-binding fluorescent assay relative to the semi-quantitative commercial SARS-CoV-2 antigen-binding assays in its ability to eliminate a large fraction of samples with low neutralizing activity and narrow the focus to samples that are most likely to have medically relevant levels of neutralizing antibodies. Importantly, RBD-binding fluorescent assay measurements revealed excellent agreement with the quantitative FDA-approved Abbott AdviseDx II assay (Y = 1.092∗X+0.9307; P < 0.001), which measures binding to SRBD. Both assays reliably identified 17 samples with RTCA NT50≥ 50 within the category of high binders (Figure 2B).
Plasma transfusions immediately followed serum testing and transfusion unit downselection, and therefore, serum testing was performed in small batches and based on plasma availability collected from donors. For these reasons, the thresholds for RBD binding (≥8,000 EU/mL) and neutralizing (NT50 ≥ 50) activities in our two-step screening approach were selected empirically from the limited data available to allow a sufficient fraction of convalescent plasma samples to be used for transfusions (17 of 76 or ∼22% of the original units would have met this criterion).
A large-scale comparison of RTCA and RBD-binding fluorescent assays carried out using 226 serum samples (Table S1), further supported our findings that beyond an empirically defined threshold for antibody-antigen binding (8,000 EU/mL), above which most neutralizing samples were found (NT50 ≥ 50), no further discrimination of antibody neutralization potency could be inferred from binding data (Figures 2C and 2D). Samples with an antibody binding level above 8,000 EU/mL in the RBD-binding fluorescent assay and half-maximal neutralization titer ≥50 were selected for transfusion in the PassITON trial (Self et al., 2021). The sensitivity and specificity to detect neutralizing activity above pre-specified binding threshold was 64% and 93%, respectively, as detailed in the STAR Methods.
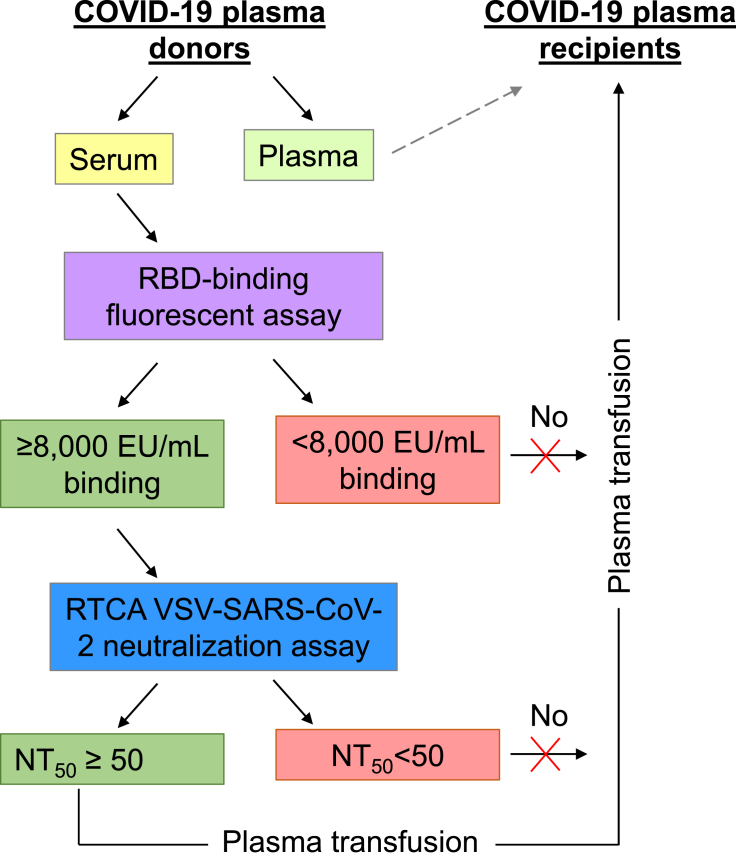
In summary, these results revealed a discrepancy between binding activity and neutralization activity that highlights the importance of directly assessing neutralizing potency of antibody-containing samples. Our results suggested a threshold level of RBD-binding activity in the RBD-binding fluorescent assay (8,000 EU/mL of serum) correlated with reproducible detection of neutralizing activity (RTCA NT50≥ 50; Figure 2D). Together, these studies offered a streamlined workflow for accurate profiling of SARS-CoV-2 binding and neutralizing activity of human serum under BSL2 conditions via a sequential algorithm consisting of screening by RBD-binding fluorescent assay followed by potency quantification using RTCA assay (Figure 3).
Figure 3.
Two-step testing of antibody activity in serum to select COVID-19 convalescent plasma for transfusion trial
A schematic of the sequential approach consisting of donor’s serum screening by RBD-binding fluorescent assay followed by antiviral potency quantification using the RTCA VSV-SARS-CoV-2 neutralization assay is shown. For transfusion trial, plasma units were selected based on serum screening results and included units with an estimated ≥8,000 EU/mL binding and NT50 ≥ 50 neutralizing activities.
Relationship between RTCA and live SARS-CoV-2 virus neutralization assays for neutralizing antibody assessment in serum
Numerous in vitro assays assessing the neutralizing activity of anti-SARS-CoV-2 antibodies have been reported (Khoury et al., 2020). The plaque reduction neutralization test (PRNT) using live SARS-CoV-2 in BSL3 containment was widely employed for monoclonal and polyclonal antibody neutralization assessment, including profiling of neutralizing antibody responses in serum of SARS-CoV-2 mRNA vaccines in clinical trials (Anderson et al., 2020; Jackson et al., 2020). Several factors, including the density of spike protein on the virion, geometry of viral particle, replication kinetics, and differences in parental virus (VSV) proteins, can potentially affect the mechanics of cell entry and the ability of antibodies to neutralize chimeric infectious VSV-SARS-CoV-2 relative to native SARS-CoV2 (Case et al., 2020b). In addition, there are inherent differences between PRNT and CPE-based RTCA assays in how these assays measure the level of infection at the endpoint. Therefore, we next determined the relationship between quantitative measurements from the VSV-SARS-CoV-2 RTCA and authentic SARS-CoV-2 PRNT assays by comparing neutralizing antibody titers in serum.
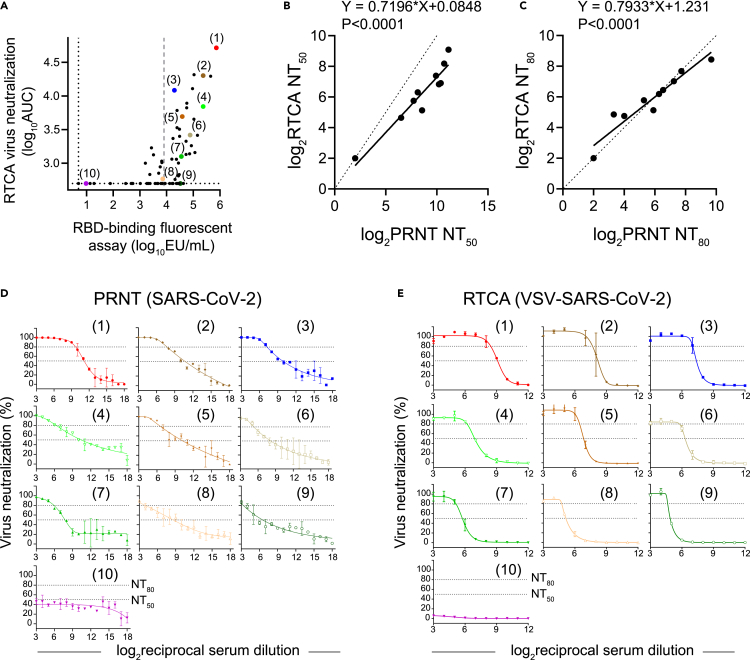
From the 76-sample training set described in Figure 2A, ten samples representing a broad range of neutralizing activity by RTCA were selected (Figure 4A). Nine of these ten samples had RBD-binding activity above the suggested RBD-binding fluorescent assay threshold level (≥8,000 EU/mL of serum) but varying neutralization levels from high to below the limit of detection (1:50 was the lowest tested serum dilution). One sample had low binding and low neutralizing activities (sample 10) and was used as a control. Of note, this panel of 10 included samples that showed similar and high binding to RBD but varying neutralization levels (e.g., samples 3, 5, 7, and 9).
Figure 4.
Relationship between VSV-SARS-CoV-2 RTCA and authentic SARS-CoV-2 PRNT neutralization assay measurements
(A) Ten samples from a panel of 76 selected for parallel testing using RTCA and PRNT neutralization assays are shown with color dots and numbers. Black dotted line indicates assay LOD, and gray dashed line indicates suggested cutoff for positive response as in Figure 1A.
(B) Orthogonal (Deming) regression analysis of NT50 values from PRNT and RTCA neutralization assays. Each dot represents one sample, the regression line is indicated with solid line, and the line of identity is dotted. p value is indicated.
(C) Orthogonal (Deming) regression analysis of NT80 values from PRNT and RTCA neutralization assays. Each dot represents one sample, the regression line is indicated with solid line, and the line of identity is dotted. p value is indicated.
(D) Dose-response neutralization curves obtained from PRNT assay using authentic SARS-CoV-2 WA1/2020 virus. Data show mean ± SD of two independent experiments. Numbers in parentheses indicate designated identifiers for ten samples as in (A).
(E) Dose-response neutralization curves obtained from RTCA assay using chimeric VSV-SARS-CoV-2 WA1/2020 virus. Numbers in parentheses indicate designated identifiers for ten samples as in (A). Data show mean ± SD of duplicates from one experiment (n = 2).
We performed parallel RTCA and PRNT assays with these samples using a broader range of serum dilutions. Estimated NT50 and NT80 values from the dose-response neutralization curves demonstrated strong agreement between RTCA and PRNT assays. Additionally, all ten samples ranked similarly by neutralization results in both assays (Figures 4B–4E). A noticeable difference was observed between the slopes of the curves, with RTCA assay curve slopes being considerably “steeper” than those observed by PRNT (Figures 4D and 4E). We speculate that this result might be explained by inherent differences between the endpoint measurements in these two assays; For example, PRNT reflects the fractional reduction in the number of plaque-forming units in the initial inoculum, while CPE quantification relies on endpoint measurement after multiple cycles of virus release and entry. Nevertheless, a high agreement was observed for the NT80 values determined from the PRNT and RTCA dose-response curves for all ten tested samples. Given that PRNT-estimated NT80 values were used in previous studies as a measure for serum-neutralizing activity in clinical trials that tested the efficacy of COVID-19 mRNA vaccines (Anderson et al., 2020; Jackson et al., 2020), this finding supports RTCA NT80 estimation as a predictor of antibody-neutralizing potency against authentic SARS-CoV-2. Together, these results demonstrate that the VSV-SARS-CoV-2 RTCA assay performed similarly to authentic SARS-CoV-2 PRNT assay for quantitative assessment and potency ranking of convalescent serum neutralization.
Standardization of convalescent serum testing by introducing an internal reference reagent composed of two human mAbs that broadly neutralize SARS CoV-2 strains, including variants of concern
Quantitative standardization of COVID-19 convalescent plasma or antibody-based interventions testing is critical for comparing the results across different studies (Gundlapalli et al., 2021). However, standardization methods for neutralization testing, especially CPE-based assays, have limitations. These limitations include variation in operator performance, differences in implementation across laboratories, and the potential for lack of consistency over time as new virus stocks and/or cell culture stocks and/or cell lines are used. Using a biological standard antibody solution with defined neutralizing activity would allow for calibration and harmonization of the data from different studies and laboratories, as has been done in the past for the other viruses (Mattiuzz et al., 2019). We developed such an approach to calibrate serum antibody binding activity in two commercial automated clinical-use tests that carry an EUA from the FDA.
For neutralization testing assays, we initially implemented the first WHO International Standard for anti-SARS-CoV-2 immunoglobulin (defined as “WHO Standard” for simplicity). This standard is composed of pooled plasma obtained from eleven individuals recovered from SARS-CoV-2 infection. This WHO Standard was developed and evaluated to calibrate and harmonize serological assays detecting anti-SARS-CoV-2 neutralizing antibodies by WHO International Collaborative study (Mattiuzzo et al., 2020). The assigned potency of the WHO Standard for SARS-CoV-2 is 1,000 International Units (IU)/mL for neutralizing antibody activity against the WA1/2020 strain of SARS-CoV-2. Several factors, however, limit the utility of the WHO Standard for worldwide applications in the current pandemic scenario: (1) the limited supply of WHO Standard preparations, (2) the challenging requirement for repeated plasma collection from the same or other individuals to match the assigned biological potency and replenish current stocks of the standard, and (3) considerable variation of neutralizing activity against circulating SARS-CoV-2 variants of concern in human plasma collected from different individuals (Chen et al., 2021c; Wang et al., 2021). These findings suggest that testing of convalescent plasma using a standard plasma reagent calibrated only against historical SARS-CoV-2 strains is no longer advisable.
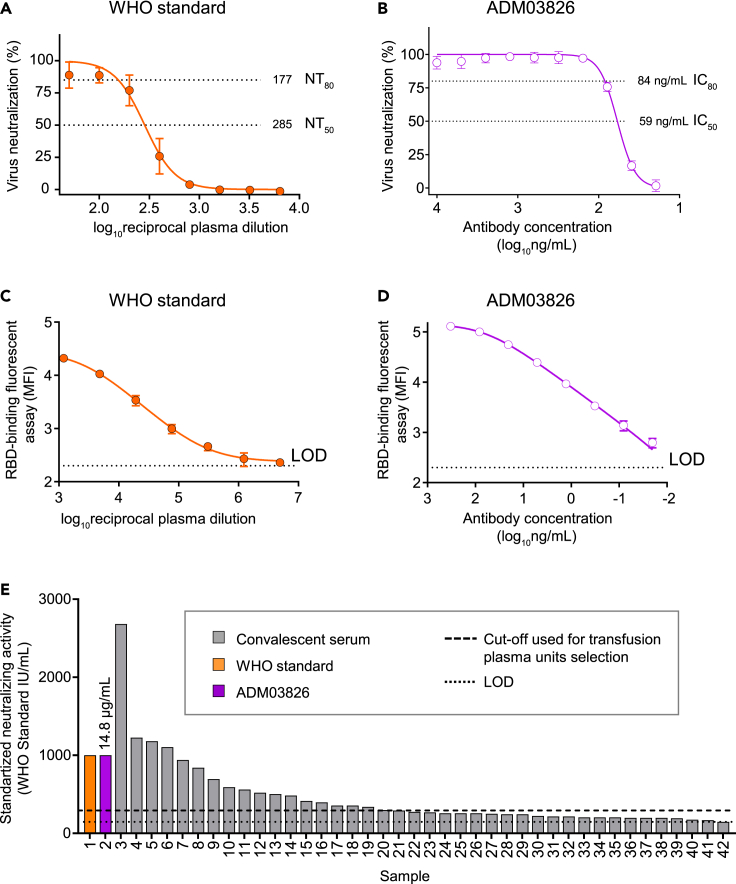
Two potently neutralizing human mAbs COV2-2130 and COV2-2381, targeting non-overlapping epitopes on the RBD of the SARS-CoV-2 spike protein (Zost et al., 2020a), form the basis of the investigational drug product ADM03820 against COVID-19 (ClinicalTrials.gov Identifier: NCT04592549). Given a virtually unlimited supply of these antibodies and their high potency in SARS-CoV-2 neutralization, we used a GMP-grade formulation of ADM03820 as the immunoglobulin standard for calibration of neutralizing antibody activity in convalescent serum. First, we assessed neutralization by WHO Standard or ADM03820 formulations across a range of plasma dilutions or mAb concentrations (Figures 5A and 5B). The activity of ADM03826 was expressed as IU/mg of IgG protein, and it was calculated using NT80 values estimated from dose-response curves for the WHO Standard and ADM03826. The potency observed using 1,000 IU/mL of the WHO Standard reagent corresponded to that afforded by a 14.8 μg/mL IgG concentration of ADM03826. Because our convalescent serum evaluation workflow involves sequential RBD binding + RTCA neutralization testing, we next demonstrated the utility of ADM03826 for calibration of antibody RBD binding against the WHO Standard using the RBD-binding fluorescent assay (Figures 5C and 5D).
Figure 5.
Standardization of RTCA neutralization assay measurements using internal reference anti-SARS-CoV-2 human immunoglobulin reagents
(A) Dose-response RTCA neutralization curve for WHO international anti-SARS-CoV-2 human immunoglobulin standard using VSV-SARS-CoV-2 WA1/2020. Data show mean ± SD of quadruplicates. One representative of two independent experiments is shown.
(B) Dose-response RTCA neutralization curve for ADM03826 that formulated with two potent human monoclonal antibodies using VSV-SARS-CoV-2 WA1/2020. Data show mean ± SD of quadruplicates. One representative of two independent experiments is shown.
(C and D) Dose-response RBD-binding curve for WHO international anti-SARS-CoV-2 human immunoglobulin standard (C) and ADM03826 (D) using RBD-binding fluorescent assay. Data show mean ± SD of triplicates. MFI – median fluorescence intensity. One representative of two independent experiments is shown.
(E) Forty convalescent serum samples (gray bars) are rank-ordered based on their neutralizing activity against VSV-SARS-CoV-2 WA1/202, which was determined using international units (IU) of neutralizing activity. ADM03826 was used as an internal reference to calibrate the activity in serum samples, and the activity of ADM03826 was pre-determined using WHO international anti-SARS-CoV-2 human immunoglobulin standard as in (A and B). The amount of ADM03826 monoclonal antibody with estimated activity of 1,000 IU (14.8 μg/mL) is shown for a comparative purpose (violet bar). Serum samples were analyzed in duplicates from serial two-fold dilutions starting from 1:25, and ADM03826 was analyzed in quadruplicates from serial two-fold dilutions starting from 1 μg/mL. Only 40 samples that revealed equal or greater than 80% virus neutralization at the lowest tested dilution (1:25) from total 76 tested samples are shown.
Next, we quantified neutralizing antibody activity in 76 serum samples (described in Figures 1 and 2) by performing a separate RTCA assay in which ADM03826 was used as an internal reference. Serum samples were tested across a broad range of dilutions along with ADM03826. Only 40 of 76 samples exhibited neutralizing potency that allowed for NT80 estimation (1:25 was the lowest tested serum dilution). These results demonstrated that the activity in these neutralizing sera ranged from 150 to 2,680 IU/mL (Figure 5E). In addition, a large fraction of these 40 samples exhibited considerably lower activity relative to WHO Standard, highlighting that most of the samples in the panel are unlikely to be clinically useful owing to the low neutralizing antibody titers observed.
Finally, in parallel, we performed focus reduction neutralization test (FRNT) assays in a BSL3 laboratory with WHO Standard and ADM03826 using parental strain WA1/2020 SARS-CoV-2, SARS-CoV-2 WA1/2020 bearing a D614G mutation, authentic B.1.1.7 virus, authentic B.1.617.2 virus, as well as, chimeric Wash-B.1.351 and Wash-B.1.1.28 viruses, which contain an S gene from B.1.351 or B.1.1.28, respectively, in the backbone of WA1/2020 (Chen et al., 2021c). The WHO Standard neutralizing titer was relatively low, with full neutralization of tested viruses achieved only at the 1:50 to 1:100 dilutions of the Standard plasma reagent (Figure 6A). In contrast, ADM03826 fully neutralized the viruses at relatively low antibody concentrations (greater than 120 ng/mL), demonstrating a much higher neutralizing potency for the ADM03826 monoclonal antibodies compared with polyclonal antibodies in the WHO Standard plasma reagent (Figure 6B). WHO Standard revealed 2.2- to 3.1-fold reduced potency against B.1.1.7, Wash-B.1.351, and B.1.617.2 viruses compared to the result with WA1/2020 as estimated from the NT80values. ADM03826 neutralized all tested viruses with similar high potency (IC80 ranged from 37 to 58 ng/mL) except for even greater potency against Wash-B.1.1.28 (NT80 = 16) (Figure 6C).
Figure 6.
Neutralizing activity against SARS-CoV-2 variants of concern by the reagents used for standardization of RTCA neutralization assay
(A and B) WHO Standard reagent (A) and ADM03826 (B) were assessed for neutralizing activity against authentic SARS-CoV-2 WA1/2020 bearing a D614G mutation (blue), a B.1.1.7 isolate (red), chimeric Wash-B.1.351 (green), chimeric Wash-B.1.1.28 (black), and authentic B.1.617.2 (gray) viruses by FRNT assay. Authentic SARS-CoV-2 WA1/2020 virus was used as a control for the parental virus (violet).
(C) NT80 values were estimated from dose-response neutralization curves in (A and B). Serum samples were analyzed in duplicate from serial two-fold dilutions starting from 1:50, and ADM03826 was analyzed in duplicate from serial three-fold dilutions starting from 10 μg/mL. Data show mean ± SD of duplicates of two independent experiments (n = 4).
Together, these studies showed broad neutralizing activity mediated by ADM03826 against SARS-CoV-2 variants of concern and demonstrated the utility of ADM03826 for the standardization and calibration of serological antibody neutralization assays.
Discussion
This study describes an optimized approach and methodology for selecting convalescent plasma for clinical use based on neutralizing antibody functionality. We demonstrated that antiviral activity could not be assessed adequately with antibody-binding assays alone owing to a complex relationship between antibody binding and virus neutralization. The studies reveal that antibody-mediated antiviral activity is best evaluated through a combination of both antigen binding and virus neutralization assays. In addition, we developed a universal neutralizing antibody standard that can be manufactured reproducibly, and that is aligned to the WHO anti-SARS-CoV-2 immunoglobulin standard for use as a calibrator to ensure reproducibility of serological antibody neutralization assays.
Quantifying neutralizing antibody titers against SARS-CoV-2 can inform patient management and guide clinical trials with convalescent plasma or monoclonal antibody-based interventions. Although many consider the development of highly potent neutralizing antibodies to be a principal correlate of protective immunity (Addetia et al., 2020; Baden et al., 2021; Chen et al., 2021b; Corbett et al., 2021; Jackson et al., 2020; McMahan et al., 2021; Polack et al., 2020), current FDA-approved serological tests do not measure antibody-mediated viral neutralization. Disparate assays and methodologies are being used to evaluate the level of SARS-CoV-2 antibodies in convalescent plasma in clinical trials, including many qualitative or semi-quantitative assessments of SARS-CoV-2 antibody presence or absence (Gundlapalli et al., 2021; Krammer and Simon, 2020). More rigorous quantitative assessments of convalescent plasma antibody titers are primarily based on detecting antibodies binding to the spike protein or RBD. For example, Abbott AdviseDx II SARS-CoV-2 IgG is an approved screening test under the EUA from the FDA issued on February 2021 (FDA, 2021a). On June 2, 2021, the US FDA amended the COVID-19 convalescent plasma EUA to include the DiaSorin LIASON SARS-CoV-2 TrimericS IgG test (FDA, 2021c) as an acceptable test for use in the manufacture of high-titer COVID-19 convalescent plasma. Notably, while many SARS-CoV-2 elicited antibodies bind the spike protein, only a subset of binding antibodies confers antiviral neutralization (Liu et al., 2020; Robbiani et al., 2020; Rogers et al., 2020; Zost et al., 2020b), which is highly predictive of immune protection (Khoury et al., 2021). Another EUA methodology is based on inhibition of RBD binding to the human ACE2 receptor, and this test has been suggested to be a surrogate of neutralization testing (Laboratory Corporation of America, 2020). However, the application of this assay for inferring SARS-CoV-2 neutralization by plasma antibodies is limited. Recent studies revealed that prevalent neutralizing plasma antibody responses in some individuals are directed against the N-terminal domain (NTD) of the SARS-CoV-2 spike protein, which lies outside of the RBD (Voss et al., 2021); however, these neutralizing antibodies do not inhibit binding to ACE2 (Chi et al., 2020; McCallum et al., 2021; Suryadevara et al., 2021). In addition, several RBD-specific antibodies have been described that neutralize SARS-CoV-2 but do not block ACE2 binding, including S309, an antibody in clinical use (Pinto et al., 2020). The initial RBD binding screening in the two-step plasma profiling approach allows pre-selection of samples most likely to exhibit high neutralizing activity based on our observation of a threshold effect, where samples with NT50≥ 50 all had high RBD binding. Notably, preliminary analyses with either 76 or 226 samples not pre-selected based on binding supported the utility of pre-screening based on RBD binding, as we were unable to observe neutralization in samples with RBD binding below the observed threshold. The second step then involves a neutralization screen that measures overall activity by all classes of neutralizing antibodies, unlike an ACE2 inhibition assay where performance is limited to detecting neutralizing activity against RBD.
Several studies have demonstrated the utility of IgG-binding assays for predicting SARS-CoV-2 neutralizing activity in COVID-19 plasma (Boonyaratanakornkit et al., 2021; Legros et al., 2021). However, several lines of evidence highlight the necessity of direct virus neutralization testing for assessment of the protective capacity of convalescent plasma: (1) a complex relationship between binding and neutralization titers against SARS-CoV-2 in convalescent plasma that varies depending on the clinical assay used and the cohort studied ((Chia et al., 2021; Gniadek et al., 2021; Grzelak et al., 2020; Muecksch et al., 2021; Rathe et al., 2021); this study)); (2) limited suitability of commercial serological assays for inferring neutralizing activity against SARS-CoV-2 in convalescent plasma ((Tang et al., 2020; Valdivia et al., 2021); this study)); (3) transfusion of convalescent plasma with low neutralizing titers predictably did not increase recipient plasma neutralizing activity (Bradfute et al., 2020); and (4) several studies have reported accelerated viral evolution and the emergence of viruses with mutations in persistently infected immunocompromised individuals treated with convalescent plasma (Choi et al., 2020; Kemp et al., 2021; McCarthy et al., 2021), prompting consideration that transfusion of convalescent plasma with a high titer of binding antibodies but low or sub-optimal neutralizing activity may have adverse consequences.
Results of some trials suggested that COVID-19 convalescent plasma with high-binding antibody titer may be beneficial (Joyner et al., 2021; Libster et al., 2021; Salazar et al., 2021). In contrast, a meta-analysis of the existing data from ten randomized clinical trials showed that treatment with convalescent plasma was not significantly associated with any benefit for clinical outcome compared to placebo or standard-of-care (Janiaud et al., 2021). Together, the current evidence from the literature and our findings suggest a more complex, nonlinear relationship between binding and neutralization in plasma than was initially expected and indicate that high binding is not equivalent to potent neutralization. Our work suggests that quantitative antibody neutralization screening is necessary to definitively evaluate the efficacy of convalescent plasma in clinical trials.
Numerous laboratory assays measure neutralization of the infection of cells with either the authentic SARS-CoV-2, SARS-CoV-2 containing luciferase reporter genes, replicating VSV-SARS-CoV-2 chimeric viruses, or VSV- or lentivirus-based non-replicative pseudoviruses incorporating SARS-CoV-2 spike and reporter genes. Infection is measured by quantifying either the number of infected cells, signal from the reporter gene, the production of viral RNA or infectious virus, or virus-induced CPE (Khoury et al., 2020). Trials that have included assessing neutralizing titers in convalescent plasma have primarily used BSL3 assays with authentic SARS-CoV-2 or BSL2 pseudovirus assays (Anderson et al., 2020; Jackson et al., 2020). Given the limited availability of BSL3 laboratory space and low throughput, the use of authentic virus screening is not scalable across the many sites seeking to conduct convalescent plasma studies. The RTCA-based testing methodology we describe in this study offers several advantages over traditional neutralization assays. These include (1) achieving a high level of accuracy and reproducibility in CPE quantification by implementing an objective and quantitative measurement of cellular impedance in infected cells, (2) allowing for automation and kinetic measurements, (3) reducing hands-on time while providing capacity for medium-to-high throughput testing that includes hundreds of samples per instrument per testing, and (4) testing in BSL2 laboratory settings using chimeric VSV-SARS-CoV-2. The assay could also be adapted for BSL3 testing with authentic viruses, including variants of concern. Therefore, plasma neutralization screening can be conducted immediately in a BSL3 laboratory after rapid isolation of authentic circulating viruses. This would precede the availability of validated recombinant antigens or chimeric viruses. To increase the efficiency of convalescent plasma screening further, we also developed a sequential RBD binding + neutralization-testing two-step workflow, which has been implemented to select transfusion units in the PassITON convalescent plasma trial (Self et al., 2021).
A significant problem for all SARS-CoV-2 antibody screening methodologies is the lack of a reliable and inexhaustible standard for anti-SARS-CoV-2 neutralizing antibody activity to allow rigorous comparison and aggregation of neutralization results across numerous trials. The WHO recently developed an International Standard for neutralizing antibodies to facilitate the standardization of these SARS-CoV-2 serological methods and compare and harmonize convalescent plasma datasets across studies (Mattiuzzo et al., 2020). The WHO Standard comprises a plasma pool from SARS-CoV-2 convalescent individuals and is a critical first step toward standardization across vaccine and convalescent plasma studies and clinical trials. However, this reagent's limited and non-renewable nature necessitates the development of a secondary standard of high quality and unlimited renewability that is traceable to the WHO standard and allows wide-scale utilization. Ultimately, the use of assays aligned to antiviral neutralization tests and the broad implementation of a high-quality antibody standard will help determine the neutralizing antibody levels required for efficacious vaccines and therapeutics.
This study addressed these issues by implementing a potent broadly neutralizing recombinant monoclonal antibody cocktail as a universal standard for neutralizing antibody assessment in our convalescent plasma testing workflow. We chose to incorporate antibodies in the standard with demonstrated protective capacity in animal models. These antibodies are also currently in Phase II efficacy trials for prevention or therapy in humans. Unlike the WHO polyclonal antibody plasma standard collected from humans, this recombinant monoclonal antibody cocktail shows equivalent performance across historical SARS-CoV-2 strains and current variants of concern. It can also be produced and distributed on a massive scale for assay standardization across the globe.
A recent report defined minimal SARS-CoV-2 neutralizing titer in recipient plasma sufficient for protection in prophylaxis or therapy studies with passively transferred polyclonal antibodies in a non-human primate challenge model (McMahan et al., 2021). Defining a protective titer against COVID-19 in humans is an important next step for designing antibody and vaccine interventions, and quantifying functional antibody levels will require the implementation of calibrated neutralization assays. As SARS-CoV-2 variants of concern arise that may have reduced capacity for neutralization by serum or plasma antibodies from previously infected or vaccinated individuals, rapid and standardized neutralization assays will be essential to assessing the impact of SARS-CoV-2 variants on neutralization titers of convalescent plasma.
In summary, our findings have broad implications for standardized testing and the design of antibody-based interventions against COVID-19.
Limitations of the study
Additional studies are needed to compare the performance of RTCA to widely used methods such as pseudovirus neutralization assays. The introduction of the ADM03826 standard sample is the first step of a complex multistep assay harmonization process. Further studies are needed to ensure that the reported result and interpretation are the same regardless of assay methods and conditions. Because both mAbs in the proposed standard (ADM03826) are RBD-targeting, this performance of the standard will not reflect SARS-CoV-2 neutralization via the S2 domain. While the RTCA array is unlikely to be widely deployable in laboratory settings owing to high instrument cost and the availability of alternative established neutralization assays, this assay is suited to a clinical environment for situations in which standardization of neutralization testing and reduction of assay setup steps are desirable.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| COV2-2196 (recombinant CHO-produced) | Zost et al. (2020a, 2020b) | GenBank: MT763531, MT763532 |
| CR3022 (recombinant CHO-produced) | Bennett et al. (2021) | GenBank: DQ168569, DQ168570 |
| ADM03826 drug product | Ology Bioservices | NA |
| WHO international standard for anti-SARS-CoV-2 human immunoglobulin | NIBSC | Cat# 20/136 |
| R-Phycoerythrin AffiniPure F(ab')₂ fragment goat anti-human IgG, Fcγ fragment specific | Jackson ImmunoResearch | Cat # 109-116-170; RRID: AB_2337681 |
| Bacterial and virus strains | ||
| SARS-CoV-2 WA1/2020 | CDC | NA |
| SARS-CoV-2 D614G | Chen et al. (2021c) | NA |
| Wash-B.1.351 | Chen et al. (2021c) | NA |
| Wash-B.1.1.28 (or Wash-BR-B.1.1.248) | Chen et al. (2021c) | NA |
| VSV-SARS-CoV-2 | Case et al. (2020b) | NA |
| B.1.1.7 | Chen et al. (2021c) | NA |
| B.1.617.2 | Human isolate | NA |
| Biological samples | ||
| Human serum | This study | NA |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 RBD | Zost et al. (2020b) | NA |
| Critical commercial assays | ||
| Ortho VITROS anti-SARS-CoV-2 IgG | Ortho Clinical Diagnostics | NA |
| Abbott AdviseDx II SARS-CoV-2 IgG | Abbott | NA |
| COVID-19 trace IgG MICRO-ELISA kit | Leinco | S1500 |
| COVID-19 ImmunoRank neutralization MICRO-ELISA kit | Leinco | S2000 |
| Experimental models: Cell lines | ||
| Monkey: Vero | ATCC | ATCC: CCL-81 RRID:CVCL_0059 |
| Monkey: Vero E6 | ATCC | NA |
| Monkey: Vero-TMPRSS2 | Zang et al. (2020) | NA |
| Monkey: MA104 | ATCC | CRL-2378.1 RRID: CVCL_3845 |
| Software and algorithms | ||
| GraphPad prism 8.4.3 | GraphPad Software, Inc. | GraphPad Prism, RRID:SCR_002798 |
| RTCA version 2.1.0 | Agilent Technologies | RTCA Software, RRID:SCR_014821 |
| Xponent version 4.3 | Luminex | Luminex software |
| Other | ||
| MagPix system | Luminex | NA |
| xCELLigence RTCA MP analyzer | Agilent Technologies | NA |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Robert H. Carnahan (robert.carnahan@vumc.org).
Materials availability
Materials described in this paper are available for distribution under the Uniform Biological Material Transfer Agreement, a master agreement that was developed by the NIH to simplify transfers of biological research materials.
Experimental model and subject details
Research participants
We studied 168 participants with an exposure history to SARS-CoV-2 infection that donated their plasma in the United States (Table S1). The studies were approved by the Vanderbilt University Medical Center Institutional Review Board, and samples were collected after written informed consent was obtained by research personnel. Participants were included in the study if they were between 18 – 80 years old and met US FDA requirements for donation of blood products. Specific to COVID-19 convalescent plasma donation, participants must have had a reported history of positive diagnostic SARS-CoV-2 PCR test and free of symptoms for 14 days with a negative PCR test for COVID-19 or 28 days without repeat testing. Participants must have satisfied requirements per the apheresis device (e.g., Alyx). Persons with blood type AB were allowed to participate without a positive SARS-CoV-2 PCR; however, plasma was only collected if antibody testing was positive. There were no additional exclusion criteria. As the study progressed, participants were asked to rate their COVID-19 symptoms subjectively on a scale of 1 to 10 (1 being mild, 10 being severe), and only participants with a symptom score of 3 or greater were invited to participate in plasma collection. Participants were allowed to donate plasma every 7 days for 4 donations without laboratory monitoring. After the fourth donation, participants required a laboratory assessment of total protein and albumin prior to additional donations to assure safety of the procedure.
Cell lines
Vero E6 (ATCC, CRL-1586) and Vero-TMPRSS2 (Zang et al., 2020) (a gift of S. Ding, Washington University) cells were cultured at 37°C in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES (pH 7.3), 1 mM sodium pyruvate, 1× nonessential amino acids, and 100 U/mL of penicillin and 100 U/mL streptomycin. For PRNTs, Vero E6 cell monolayers were cultured at 37°C in DMEM supplemented with 10% FBS, 100 U/mL of penicillin and 100 U/mL streptomycin, and 0.25 μg/mL amphotericin B (Corning). Vero-TMPRSS2 cell cultures were supplemented with 5 μg/mL of blasticidin. Vero (ATCC, CCL-81) cells were maintained at 37°C in 5% CO2 in DMEM containing 10% FBS, 10 mM HEPES (pH 7.3), 1 mM sodium pyruvate, and 100 U/mL of penicillin and 100 U/mL streptomycin.
Viruses
For PRNTs and FRNTs, we used SARS-CoV-2 strain 2019 n-CoV/USA_WA1/2020 obtained from the Centers for Disease Control and Prevention (a gift from N. Thornburg). The D614G virus was produced by introducing the mutation into an infectious clone of WA1/2020, and the B.1.351 and B.1.1.28 spike genes were cloned into the WA1/2020 infectious clone to produce Wash-B.1.351 and Wash-B.1.1.28 chimeric viruses, as described previously (Chen et al., 2021c). B.1.1.7 and B.1.617.2 were isolated from infected individuals. Viruses were propagated in Vero E6, Vero CCL81 or Vero-TMPRSS2 cells and titrated by plaque assay on Vero E6 or Vero-TMPRSS2 cell culture monolayers as previously described (Case et al., 2020a; Chen et al., 2021c; Jackson et al., 2020). The generation of a replication-competent vesicular stomatitis virus (VSV) expressing SARS-CoV-2 S protein with a 21 aminoacid C-terminal deletion that replaces the VSV G protein (VSV-SARS-CoV-2) was described previously (Case et al., 2020b). The S protein-expressing VSV virus was propagated in MA104 cell culture monolayers (African green monkey, ATCC CRL-2378.1) as described previously (Case et al., 2020a). All work with infectious SARS-CoV-2 was performed in Institutional Biosafety Committee approved BSL3 facilities at Vanderbilt University Medical Center and Washington University School of Medicine using appropriate positive pressure air respirators and protective equipment.
Monoclonal antibodies, plasma, and serum
Human recombinant mAbs COV2-2196 and CR3022 were described previously (Zost et al., 2020a). ADM03826 Drug Product was formulated with two human mAbs that were produced based on antibody variable region sequences of two previously described neutralizing human mAbs, COV2-2381 and COV2-2130 (Zost et al., 2020a). The ADM03826 Drug Product was kindly provided by Ron Cobb (Ology Bioservices, Inc). WHO International Standard for anti-SARS-CoV-2 human immunoglobulin was purchased from NIBSC. Plasma and serum samples were collected from 168 research participants in this study.
Method details
RBD antibody binding quantification fluorescent assay
Quantification of binding IgG against the receptor-binding domain (RBD) of SARS-CoV-2 was performed on donor sera using a liquid bead-array assay as previously described (Bennett et al., 2021). Briefly, recombinant RBD protein was produced and purified as described previously (Zost et al., 2020b). Purified RBD protein was conjugated to MagPlex microspheres (Luminex Corp.) and incubated in 96- well plates with serially diluted serum samples and a SARS-CoV spike-reactive monoclonal antibody CR3022 as a standard. Serum IgG bound to SARS-CoV-2 RBD was detected by R-Phycoerythrin conjugated F(ab')2 fragment goat anti-human IgG Fc gamma conjugate (Jackson ImmunoResearch). Beads were acquired from 96-well plates using a Luminex MagPix Instrument at 100 beads per well, with Xponent software version 4.3.
Commercial anti-SARS-CoV-2 antibody serological tests
Serum samples were tested using two automated clinical-use methodologies: [1] Ortho VITROS Anti-SARS-CoV-2 IgG, a chemiluminescent assay measuring reactivity to S protein (FDA, 2021d); and [2] Abbott AdviseDx II SARS-CoV-2 IgG, a quantitative microparticle chemiluminescent assay measuring reactivity to SARS-CoV-2 RBD (FDA, 2021a). Two automated research-use methodologies were also utilized: [3] Leinco COVID-19 Trace IgG MICRO-ELISA Kit, a chemiluminescent assay measuring reactivity to immobilized RBD domain of S protein; and [4] Leinco COVID-19 ImmunoRank Neutralization MICRO-ELISA Kit, a quantitative chemiluminescent assay measuring Ig antibodies that bind S protein RBD domain and are capable of blocking the binding of the RBD to ACE2 (Leinco Technologies). The Ortho and Abbot assays currently carry EUA from the FDA, although a pre-commercial-release lot of [2] was utilized for the samples here, obtained under research agreement. Testing was performed per manufacturer instruction on the 5600 platform (for [1]) and Architect i2000 platform (for [2]). All samples were characterized both according to categorical results (Positive/Negative, per fixed thresholds) and numeric signal output (S/CO for [1], not part of the clinically reportable result; AU/mL for [2], an EUA reportable result).
Plaque-reduction neutralization test
Neutralizing activity of convalescent donor serum against authentic SARS-CoV-2 was determined using PRNT as previously described (Jackson et al., 2020). Briefly, serum samples were heat-inactivated at 56°C, serially diluted two-fold in gelatin saline (0.3% [wt/vol] gelatin [Sigma-Aldrich] in Dulbecco's phosphate-buffered saline supplemented with CaCl2 and MgCl2 [Gibco]), and combined with an equal volume of SARS-CoV-2 clinical isolate, SARS-CoV-2/human/USA/USA-WA1/2020 (GenBank: MN985325.1), in gelatin saline. Final serum dilutions ranged from 1:8 to 1:262,144 of the original sample and contained an average of 960 plaque-forming units (PFU) of virus per mL. After incubation for 1 h at 37°C, 0.1 mL of virus-serum mixtures were applied in duplicate on Vero E6 cell monolayers, overlaid with 1% agar in culture medium following virus adsorption, and incubated for 3 days. Plaques were counted by direct visualization without monolayer staining. Average number of plaques in virus/serum (duplicate) and virus-only (quadruplicate) wells was used to calculate percent virus neutralization at each serum dilution according to the following formula: 1 - ([mean number of plaques in the presence of serum]/[mean number of plaques in the absence of serum]). Each sample was tested in two independent assays performed at different times. Fractional neutralization from duplicate testing was plotted as a function of log2 serum dilution, and the dose-response relationship was estimated using five-parameter logistic regression analysis using Prism software (version 9.1.2; GraphPad). PRNT50 and PRNT80 titer values, expressed as the reciprocal of the highest serum dilution reducing virus infectivity by 50% or 80%, respectively, were interpolated from regression curves. If a serum titration failed to generate 50% inhibition within the range of dilutions tested a titer value of one-half of the lowest serum dilution (highest serum concentration) tested was assigned to it.
Real-time cell analysis neutralization assay
To determine neutralizing activity of purified recombinant IgG or human serum, we used a real-time cell analysis (RTCA) assay on an xCELLigence RTCA MP Analyzer (Agilent Technologies) that measures virus-induced cytopathic effect (CPE) (Gilchuk et al., 2020a; Suryadevara et al., 2021; Zost et al., 2020b). Briefly, 50 μL of cell culture medium (DMEM supplemented with 2% FBS, 10 mM HEPES (pH 7.3), 1 mM sodium pyruvate, and 100 U/mL of penicillin and streptomycin) was added to each well of a 96-well E-plate using a ViaFlo384 liquid handler (Integra Biosciences) to obtain background reading. A suspension of 18,000 Vero cells in 50 μL of cell culture medium was seeded in each well, and the plate was placed on the analyzer. Measurements were taken automatically every 15 min, and the sensograms were visualized using RTCA software version 2.1.0 (Agilent Technologies). VSV-SARS-CoV-2 (0.01 MOI, ∼120 PFU per well) was mixed 1:1 with a respective dilution of human serum sample or monoclonal antibody in a total volume of 100 μL using cell culture medium as a diluent and incubated for 1 h at 37°C in 5% CO2. At 16 h after seeding the cells, the virus-sample mixtures were added in replicates to the cells in 96-well E-plates. Triplicate wells containing virus only (maximal CPE in the absence of human serum) and wells containing only Vero cells in medium (no-CPE wells) were included as controls. Plates were measured continuously (every 15 min) for 48 h to assess virus neutralization. Normalized cellular index (CI) values at the endpoint (48 h after incubation with the virus) were determined using the RTCA software version 2.1.0 (Agilent Technologies). Results are expressed as percent neutralization in a presence of respective mAb relative to control wells with no CPE minus CI values from control wells with maximum CPE. Curves for virus neutralization were fitted using a four-parameter log-logistic (4PL) analysis by Prism software (version 9.1.2; GraphPad). Serum titer was expressed as the reciprocal of serum dilution. Half-maximal neutralization titer (NT50) and neutralization titer that reduced virus infectivity by 80% (NT80) were interpolated from regression curves. If a serum titration failed to generate 50% inhibition within the range of dilutions tested a titer value of one-half of the lowest serum dilution (highest serum concentration) tested was assigned to it.
For standardization of RTCA neutralization assay measurements in human serum, we used WHO international anti-SARS-CoV-2 human immunoglobulin standard (NIBSC; code:20/136) with arbitrary assigned neutralizing activity of 1,000 IU/mL. The activity of ADM03826 (IU/mg of IgG protein) was calculated using WHO international anti-SARS-CoV-2 human immunoglobulin standard after determining interpolated NT80 values from neutralization dose-response curves obtained for WHO Standard reagent and ADM03826. Then the ADM03826 was used as an internal reference in each RTCA study to calibrate the activity in human serum samples in IU/mL by determining NT80 values as detailed above.
Selection of COVID-19 convalescent plasma units for transfusion
Serum testing was performed in small batches and based on plasma availability collected from donors, and the thresholds for RBD binding (≥8,000 EU/mL) and neutralizing (NT50 ≥ 50) activities in our two-step screening approach were selected empirically from the limited data available. We chose NT50 ≥ 50 as a cut-off for the neutralization threshold. This is two-fold over the limit of the detection of RTCA assay (1:25 was the first serum dilution tested) and the threshold allowed a sufficient fraction of convalescent plasma samples to be used for transfusions (17 of 76 or ∼22% of the original units would have met this criterion). The binding threshold was chosen to incorporate all neutralizers with NT50 ≥ 50 in the initial studies with 76 samples. Sensitivity was estimated as the proportion of samples above binding threshold (≥8,000 EU/mL) that are positive for neutralizing activity (NT50 ≥ 50), and specificity was estimated as the proportion of samples below binding threshold (<8,000 EU/mL) that are negative for neutralizing activity (NT50< 50).
Focus reduction neutralization test
The assay was performed as described previously (Chen et al., 2021c). Briefly, serial dilutions of mAbs or serum were incubated with 102 focus-forming units of different strains or variants of SARS-CoV-2 for 1 h at 37°C. Antibody–virus complexes were added to Vero-TMPRSS2 cell monolayers in 96-well plates and incubated at 37°C for 1 h. Subsequently, cells were overlaid with 1% (w/v) methylcellulose in MEM supplemented with 2% FBS. Plates were collected 30 h later by removing overlays and fixed with 4% paraformaldehyde in PBS for 20 min at room temperature. Plates were washed and sequentially incubated with an oligoclonal pool of SARS2-2, SARS2-11, SARS2-16, SARS2-31, SARS2-38, SARS2-57 and SARS2-71 mouse anti-S antibodies (Liu et al., 2021; VanBlargan et al., 2021) and HRP-conjugated goat anti-mouse IgG (Sigma-Aldrich) in PBS supplemented with 0.1% saponin and 0.1% bovine serum albumin. SARS-CoV-2-infected cell foci were visualized using TrueBlue peroxidase substrate (KPL) and quantitated on an ImmunoSpot microanalyzer (Cellular Technologies).
Quantification and statistical analysis
Mean ± SD were determined for continuous variables as noted. Technical and biological replicates are described in the figure legends. An agreement between two methods was assessed using an orthogonal (Deming) regression analysis, and variability between two methods measurements was visually assessed using Bland-Altman plotting in Prism (version 9.1.2; GraphPad).
Acknowledgments
We thank Agilent Technologies for their assistance with RTCA methods development and technical support, Leinco Technologies for performing Leinco Trace IgG Micro-ELISA and ImmunoRank Neutralization assays, Xiaotao Lu (VUMC) and Tia Hughes (VUMC) for technical support of SARS-CoV-2 neutralization testing, and Thomas Stewart, Wang Li, and Chris Lindsell for their advice for statistical analysis. VSV-SARS-CoV-2 was a kind gift from Sean Whelan (Washington University School of Medicine). This work was supported by the National Institute of Health (NIH) National Center for Advancing Translational Sciences (NCATS) grant 3UL1TR002243-04S4, Vanderbilt Institute for Infection, Immunology, and Inflammation (VI4), the Hays Foundation COVID-19 Research Fund (to I.T.), Defense Advanced Research Projects Agency (DARPA) grant HR0011-18-2-0001, U.S. N.I.H. contract 75N93019C00074, the Dolly Parton COVID-19 Research Fund at Vanderbilt, and a grant from Fast Grants, Mercatus Center, George Mason University. Reagents for the AdviseDx II IgG assay were provided under research agreement between Abbott Laboratories and VUMC. J.E.C. is a recipient of the 2019 Future Insight Prize from Merck KGaA, which supported this work with a grant. The content is solely the responsibility of the authors and does not represent the official views of the U.S. government or other sponsors. We thank the anonymous donors of the plasma samples for their consent, which has allowed WHO International standard for anti-SARS-CoV-2 human immunoglobulin to be prepared. We express our gratitude to those who have coordinated the collection of the convalescent plasma: Malcom Semple (University of Liverpool, UK), Lance Turtle (University of Liverpool, UK), Peter Openshaw (Imperial College London, UK), and Kenneth Baillie (University of Edinburgh) on behalf of the ISARIC4C Investigators; Heli Harvala Simmonds and David Roberts (National Health Service Blood and Transplant, UK). We also thank NIBSC Standards Production and Development staff for the formulation and distribution of materials.
Author contributions
P.G., I.T., J.D.C., M.R.D., J.S., J.M.P., M.S.D., G.R.B., A.P.W., J.E.C., and R.H.C. planned the studies. J.P.R. and A.P.W. recruited the participants and coordinated plasma and serum samples collection. P.G., S.Y., E.B., L.J.S., R.E.S., R.E.C., L.A.V., and N.S. conducted experiments. P.G., I.T., S.Y., J.D.C., L.J.S., R.E.C., and S.Z., analyzed data, P.G., I.T., J.D.C., J.S., J.M.P., M.S.D., G.R.B., W.H.S., and T.W.R interpreted the studies. P.G., I.T., J.E.C., and R.H.C wrote the first draft of the paper. I.T., M.S.D., G.R.B., and J.E.C. obtained funding. All authors reviewed, edited, and approved the paper.
Declaration of interests
I.T. reports grants from NIH/NIAID, during the conduct of the study, and has served as a consultant for Nashville Biosciences and Horizon Therapeutics. J. D. C., L.J.S., and M.R.D. report grants from NIH/NCATS, during the conduct of the study. T.W.R. reports grants from NIH/NCATS, during the conduct of the study; personal fees from Cumberland Pharmaceuticals, Inc, personal fees from Sanofi Pharma, and personal fees from Cytovale, outside the submitted work. T.G.S. reports grants from NIH, during the conduct of the study. W.H.S. reports grants from NCATS of the NIH, during the conduct of the study. M.S.D. is a consultant for Inbios, Vir Biotechnology, Fortress Biotech, and Carnival Corporation and on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received funding support in sponsored research agreements from Moderna, Vir Biotechnology, and Emergent BioSolutions. J.E.C. has served as a consultant for Luna Biologics, is a member of the Scientific Advisory Board of Meissa Vaccines and is Founder of IDBiologics. The Crowe laboratory at Vanderbilt University Medical Center has received sponsored research agreements from Takeda Vaccines, IDBiologics, and AstraZeneca and grants from NIH, and DARPA during the conduct of the study. Vanderbilt University has applied for patents related to antibodies described in this paper. All other authors declare no competing interests.
Published: January 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103602.
Supplemental information
Data and code availability
-
•
Data - All data needed to evaluate the conclusions in the paper are present in the paper or the supplemental information; source data for each of the display items is provided in key resources table.
-
•
Code – No custom computer code or algorithms to report.
-
•
Other – No “All other items” to report.
References
- Addetia A., Crawford K.H.D., Dingens A., Zhu H., Roychoudhury P., Huang M.L., Jerome K.R., Bloom J.D., Greninger A.L. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with a high attack rate. J. Clin. Microbiol. 2020;58:e02107–e02120. doi: 10.1128/JCM.02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N.Engl. J. Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N.Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M., Yoder S., Brady E., Pulley J.M., Rhoads J.P., Stewart T.G., Bernard G.R., Creech C.B., Wheeler A.P., Thomsen I. A high-throughput liquid bead array assay confirms strong correlation between SARS-CoV-2 antibody level and COVID-19 severity. iScience. 2021;24:102052. doi: 10.1016/j.isci.2021.102052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonyaratanakornkit J., Morishima C., Selke S., Zamora D., McGuffin S., Shapiro A.E., Campbell V.L., McClurkan C.L., Jing L., Gross R., et al. Clinical, laboratory, and temporal predictors of neutralizing antibodies against SARS-CoV-2 among COVID-19 convalescent plasma donor candidates. J. Clin. Invest. 2021;131:e144930. doi: 10.1172/JCI144930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradfute S.B., Hurwitz I., Yingling A.V., Ye C., Cheng Q., Noonan T.P., Raval J.S., Sosa N.R., Mertz G.J., Perkins D.J., et al. Severe acute respiratory syndrome coronavirus 2 neutralizing antibody titers in convalescent plasma and recipients in New Mexico: an open treatment study in patients with coronavirus disease 2019. J. Infect. Dis. 2020;222:1620–1628. doi: 10.1093/infdis/jiaa505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Bailey A.L., Kim A.S., Chen R.E., Diamond M.S. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology. 2020;548:39–48. doi: 10.1016/j.virol.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Rothlauf P.W., Chen R.E., Liu Z., Zhao H., Kim A.S., Bloyet L.M., Zeng Q., Tahan S., Droit L., et al. Neutralizing antibody and soluble ACE2 Inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe. 2020;28:475–485.e5. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.C., Gilchuk P., Zost S.J., Suryadevara N., Winkler E.S., Cabel C.R., Binshtein E., Sutton R.E., Rodriguez J., Day S., et al. Convergent antibody responses to the SARS-CoV-2 spike protein in convalescent and vaccinated individuals. bioRxiv. 2021 doi: 10.1101/2021.05.02.442326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Winkler E.S., Case J.B., Aziati I.D., Bricker T.L., Joshi A., Darling T.L., Ying B., Errico J.M., Shrihari S., et al. In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains. Nature. 2021 doi: 10.1038/s41586-021-03720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia W.N., Zhu F., Ong S.W.X., Young B.E., Fong S.W., Le Bert N., Tan C.W., Tiu C., Zhang J., Tan S.Y., et al. Dynamics of SARS-CoV-2 neutralising antibody responses and duration of immunity: a longitudinal study. Lancet Microbe. 2021;2:e240–e249. doi: 10.1016/S2666-5247(21)00025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B., Cernadas M., Li J.Z. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N.Engl. J. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Nason M.C., Flach B., Gagne M., S O.C., Johnston T.S., Shah S.N., Edara V.V., Floyd K., Lai L., et al. Immune correlates of protection by mRNA-1273 immunization against SARS-CoV-2 infection in nonhuman primates. bioRxiv. 2021 doi: 10.1101/2021.04.20.440647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA FDA issues emergency use authorization for convalescent plasma as potential promising COVID–19 treatment, another achievement in administration’s fight against pandemic. 2020. https://www.fda.gov/news-events/press-announcements/fda-issues-emergency-use-authorization-convalescent-plasma-potential-promising-covid-19-treatment
- FDA AdviseDx SARS-CoV-2 IgG II - instructions for use (ARCHITECT) 2021. https://www.fda.gov/media/146371/download
- FDA EUA authorized serology test performance. 2021. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/eua-authorized-serology-test-performance
- FDA LIAISON® SARS-CoV-2 TrimericS IgG. 2021. https://www.fda.gov/media/149059/download
- FDA VITROS immunodiagnostic products anti-SARS-CoV-2 IgG reagent pack - IFU. 2021. https://www.fda.gov/media/137363/download
- Gilchuk P., Bombardi R.G., Erasmus J.H., Tan Q., Nargi R., Soto C., Abbink P., Suscovich T.J., Durnell L.A., Khandhar A., et al. Integrated pipeline for the accelerated discovery of antiviral antibody therapeutics. Nat. Biomed. Eng. 2020;4:1030–1043. doi: 10.1038/s41551-020-0594-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchuk P., Murin C.D., Milligan J.C., Cross R.W., Mire C.E., Ilinykh P.A., Huang K., Kuzmina N., Altman P.X., Hui S., et al. Analysis of a therapeutic antibody cocktail reveals determinants for cooperative and broad ebolavirus neutralization. Immunity. 2020;52:388–403.e12. doi: 10.1016/j.immuni.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gniadek T.J., Thiede J.M., Matchett W.E., Gress A.R., Pape K.A., Fiege J.K., Jenkins M.K., Menachery V.D., Langlois R.A., Bold T.D. SARS-CoV-2 neutralization and serology testing of COVID-19 convalescent plasma from donors with nonsevere disease. Transfusion. 2021;61:17–23. doi: 10.1111/trf.16101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzelak L., Temmam S., Planchais C., Demeret C., Tondeur L., Huon C., Guivel-Benhassine F., Staropoli I., Chazal M., Dufloo J., et al. A comparison of four serological assays for detecting anti-SARS-CoV-2 antibodies in human serum samples from different populations. Sci. Transl. Med. 2020;12:eabc3103. doi: 10.1126/scitranslmed.abc3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundlapalli A.V., Salerno R.M., Brooks J.T., Averhoff F., Petersen L.R., McDonald L.C., Iademarco M.F., Response C.C.-. SARS-CoV-2 serologic assay needs for the next phase of the US COVID-19 pandemic response. Open Forum Infect. Dis. 2021;8:ofaa555. doi: 10.1093/ofid/ofaa555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. An mRNA Vaccine against SARS-CoV-2 - preliminary report. N.Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiaud P., Axfors C., Schmitt A.M., Gloy V., Ebrahimi F., Hepprich M., Smith E.R., Haber N.A., Khanna N., Moher D., et al. Association of convalescent plasma treatment with clinical outcomes in patients with COVID-19: a systematic review and meta-analysis. JAMA. 2021;325:1185–1195. doi: 10.1001/jama.2021.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021;13:eabf1906. doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner M.J., Carter R.E., Senefeld J.W., Klassen S.A., Mills J.R., Johnson P.W., Theel E.S., Wiggins C.C., Bruno K.A., Klompas A.M., et al. Convalescent plasma antibody levels and the risk of death from Covid-19. N.Engl. J. Med. 2021;384:1015–1027. doi: 10.1056/NEJMoa2031893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp S.A., Collier D.A., Datir R.P., Ferreira I.A.T.M., Gayed S., Jahun A., Hosmillo M., Rees-Spear C., Mlcochova P., Lumb I.U., et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury D.S., Wheatley A.K., Ramuta M.D., Reynaldi A., Cromer D., Subbarao K., O'Connor D.H., Kent S.J., Davenport M.P. Measuring immunity to SARS-CoV-2 infection: comparing assays and animal models. Nat. Rev. Immunol. 2020;20:727–738. doi: 10.1038/s41577-020-00471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- Krammer F. Correlates of protection from SARS-CoV-2 infection. Lancet. 2021;397:1421–1423. doi: 10.1016/S0140-6736(21)00782-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F., Simon V. Serology assays to manage COVID-19. Science. 2020;368:1060–1061. doi: 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- Laboratory Corporation of America LabCorp launches new neutralizing antibody test: neutralizing antibody assay could be used to assist in development of COVID-19 vaccines and plasma-based treatments. 2020. https://www.labcorp.com/coronavirus-disease-covid-19/news/labcorp-launches-new-neutralizing-antibody-test
- Legros V., Denolly S., Vogrig M., Boson B., Siret E., Rigaill J., Pillet S., Grattard F., Gonzalo S., Verhoeven P., et al. A longitudinal study of SARS-CoV-2-infected patients reveals a high correlation between neutralizing antibodies and COVID-19 severity. Cell. Mol. Immunol. 2021;18:318–327. doi: 10.1038/s41423-020-00588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libster R., Perez Marc G., Wappner D., Coviello S., Bianchi A., Braem V., Esteban I., Caballero M.T., Wood C., Berrueta M., et al. Early high-titer plasma therapy to prevent severe Covid-19 in older adults. N.Engl. J. Med. 2021;384:610–618. doi: 10.1056/NEJMoa2033700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Liu Z.M., VanBlargan L.A., Bloyet L.M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H.Y., Errico J.M., Theel E.S., Liebeskind M.J., et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiuzz G., Knezevic I., Hassall M., Ashall J., Myhill S., Faulkner V., Hockley J., Rigsby P., Wilkinson D.E., Page M., et al. Harmonization of Zika neutralization assays by using the WHO International Standard for anti-Zika virus antibody. NPJ Vaccin. 2019;4 doi: 10.1038/s41541-019-0135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiuzzo G., Bentley E.M., Hassall M., Routley S., Richardson S., Bernasconi V., Kristiansen P., Harvala H., Roberts D. WHO Expert Committee on Biological Standardization WHO/BS/2020.2403; 2020. Establishment of the WHO International Standard and Reference Panel for Anti-SARS-CoV-2 Antibody. [Google Scholar]
- McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy K.R., Rennick L.J., Nambulli S., Robinson-McCarthy L.R., Bain W.G., Haidar G., Duprex W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. 2021;371:1139–1142. doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan K., Yu J.U., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J.Y., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590 doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muecksch F., Wise H., Batchelor B., Squires M., Semple E., Richardson C., McGuire J., Clearly S., Furrie E., Greig N., et al. Longitudinal serological analysis and neutralizing antibody levels in coronavirus disease 2019 convalescent patients. J. Infect. Dis. 2021;223:389–398. doi: 10.1093/infdis/jiaa659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N.Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathe J.A., Hemann E.A., Eggenberger J., Li Z.Q., Knoll M.L., Stokes C., Hsiang T.Y., Netland J., Takehara K.K., Pepper M., et al. SARS-CoV-2 serologic assays in control and unknown populations demonstrate the necessity of virus neutralization testing. J. Infect. Dis. 2021;223:1120–1131. doi: 10.1093/infdis/jiaa797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar E., Christensen P.A., Graviss E.A., Nguyen D.T., Castillo B., Chen J., Lopez B.V., Eagar T.N., Yi X., Zhao P.C., et al. Significantly decreased mortality in a large cohort of coronavirus disease 2019 (COVID-19) patients transfused early with convalescent plasma containing high-titer anti-severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein IgG. Am. J. Pathol. 2021;191:90–107. doi: 10.1016/j.ajpath.2020.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self W.H., Stewart T.G., Wheeler A.P., El Atrouni W., Bistran-Hall A.J., Casey J.D., Cataldo V.D., Chappell J.D., Cohn C.S., Collins J.B., et al. Passive Immunity Trial for Our Nation (PassITON): study protocol for a randomized placebo-control clinical trial evaluating COVID-19 convalescent plasma in hospitalized adults. Trials. 2021;22:221. doi: 10.1186/s13063-021-05171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryadevara N., Shrihari S., Gilchuk P., VanBlargan L.A., Binshtein E., Zost S.J., Nargi R.S., Sutton R.E., Winkler E.S., Chen E.C., et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell. 2021;184:2316–2331.e5. doi: 10.1016/j.cell.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M.S., Case J.B., Franks C.E., Chen R.E., Anderson N.W., Henderson J.P., Diamond M.S., Gronowski A.M., Farnsworth C.W. Association between SARS-CoV-2 neutralizing antibodies and commercial serological assays. Clin. Chem. 2020;66:1538–1547. doi: 10.1093/clinchem/hvaa211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P.C., Adams A.C., Hufford M.M., de la Torre I., Winthrop K., Gottlieb R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021;21:382–393. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia A., Torres I., Latorre V., Frances-Gomez C., Albert E., Gozalbo-Rovira R., Alcaraz M.J., Buesa J., Rodriguez-Diaz J., Geller R., et al. Inference of SARS-CoV-2 spike-binding neutralizing antibody titers in sera from hospitalized COVID-19 patients by using commercial enzyme and chemiluminescent immunoassays. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:485–494. doi: 10.1007/s10096-020-04128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBlargan L., Adams L., Liu Z., Chen R.E., Gilchuk P., Raju S., Smith B., Zhao H., Case J.B., Winkler E.S., et al. A potently neutralizing anti-SARS-CoV-2 antibody inhibits variants of concern by binding a highly conserved epitope. bioRxiv. 2021 doi: 10.1101/2021.04.26.441501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss W.N., Hou Y.J., Johnson N.V., Delidakis G., Kim J.E., Javanmardi K., Horton A.P., Bartzoka F., Paresi C.J., Tanno Y., et al. Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes. Science. 2021;372:1108–1112. doi: 10.1126/science.abg5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schafer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data - All data needed to evaluate the conclusions in the paper are present in the paper or the supplemental information; source data for each of the display items is provided in key resources table.
-
•
Code – No custom computer code or algorithms to report.
-
•
Other – No “All other items” to report.