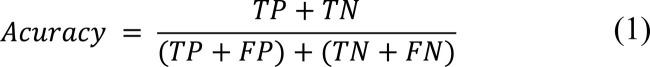Abstract
Objective
Automatic prediction of COVID-19 using deep convolution neural networks based pre-trained transfer models and Chest X-ray images.
Methods
This research employs the advantages of computer vision and medical image analysis to develop an automated model that has the clinical potential for early detection of the disease. Using Deep Learning models, the research aims at evaluating the effectiveness and accuracy of different convolutional neural networks models in the automatic diagnosis of COVID-19 from X-ray images as compared to diagnosis performed by experts in the medical community.
Results
Due to the fact that the dataset available for COVID-19 is still limited, the best model to use is the InceptionNetV3. Performance results show that the InceptionNetV3 model yielded the highest accuracy of 98.63% (with data augmentation) and 98.90% (without data augmentation) among the three models designed. However, as the dataset gets bigger, the Inception ResNetV2 and NASNetlarge will do a better job of classification. All the performed networks tend to over-fit when data augmentation is not used, this is due to the small amount of data used for training and validation.
Conclusion
A deep transfer learning is proposed to detecting the COVID-19 automatically from chest X-ray by training it with X-ray images gotten from both COVID-19 patients and people with normal chest X-rays. The study is aimed at helping doctors in making decisions in their clinical practice due its high performance and effectiveness, the study also gives an insight to how transfer learning was used to automatically detect the COVID-19.
Keywords: Deep transfer learning, coronavirus, X-ray, CNN, inceptioNetV3, inceptionResNetV2
1. INTRODUCTION
Coronavirus disease 2019 (COVID-19), caused by a novel coronavirus has spread to over 214 countries and areas in the world. The first infected novel coronavirus case was found in Hubei, China towards the end of 2019. COVID-19 is easily spread in human-human transmissions. The World Health Organization (WHO) recognized COVID-19 as a global pandemic in March 2020 [1]. WHO and US Centers for Disease Control and Prevention (CDC) have released technical guidelines to support the response to this pandemic [2]. COVID-19 can be fatal with a mortality rate of 2%. Symptoms can include alveolar damage and progressive respiratory failure, which in the most serious cases can lead to death.
Governments are making efforts to address the pandemic e.g. applying regional lockdown to limit the spread of infection, ensuring healthcare facilities can accommodate patient numbers, quarantining for suspected infected individuals without clinical symptoms, and taking steps to alleviate impacts on the economy [3].
Reverse Transcription-Polymerase Chain Reaction (RT-PCR) test is a standard for confirming COVID-19 infection. But, RT-PCR is costly, time consuming, and insufficiently sensitivity. The RT-PCR method is also not suitable to meet the rapid detection rate required during this COVID-19 pandemic [4].
Covid-19 diagnoses and health status monitoring of patients has increased the number of X-ray images and Computerized Tomography (CT) scans. Chest X-ray and CT scans are widely used in the diagnosis of COVID-19 [5, 6]. These medical images can be considered as a valuable dataset for researchers. This dataset has the potential to assist in the identification, tracking and prediction of COVID-19 infection.
Machine Learning and Deep Learning are parts of Artificial Intelligence that have contributed actively in the classification of medical images. The application of machine learning and deep learning has become a focus for research, analysis, and pattern recognition. Recent improvements in these areas, as well as the growth in medical images and radiography datasets, provide new opportunities to augment medical decision-making systems [7]. In the case of COVID-19, X-ray images and CT scans can be used as the input of Deep Learning models for medical image analysis [6].
Deep learning models have shown superior performance over classical machine learning models in many studies. Mayr et al. [8] showed superior performance of Feed-Forward Neural Networks over other models such as SVM and Random Forest in drug target prediction. Bui et al. [9] showed similar results where deep learning neural networks out-performed MLP-NN, SVM and Random Forest in landslide susceptibility assessment. Korotcov et al. [10] ranked deep neural networks higher than SVM in diverse drug discovery. All the studies mentioned above belongs to different fields, yet all studies have shown a similar outcome with deep learning models outperformed classical machine learning models. Other superior qualities of deep learning over classical machine learning models are that they are capable of multi-task learning [11, 12] and can extract complex features automatically [12]. This shows that using deep learning for the crucial problem of COVID-19 detection could be very beneficial.
Inception ResNet V2, InceptionNet V3 and NASNetLarge are proven state-of-the-art models and have been used previously using less data and without the contribution of auxiliary classifiers at the end of classification. InceptionNet V3 helps improve the accuracy at the end and does help in improving the overall accuracy of the model. InceptionNet V3 also contains features of V1 and V2 and helps working with scaled images [9]. Inception ResNet V2 has been used for Classification of Breast Cancer Histology Images with 90% accuracy, and has also shown better performance with the use of data augmentation [10]. Other research has shown that pretrained image-net models perform better [11]. ResNet has been used in X-ray images for the detection of COVID-19. This model distinguished between COVID-19 suspect or non-COVID-19. The result showed sensitivity of 96.0% along with AUC of 0.952 [12]. Hyper-tuning of the deep learning models can be achieved using transfer learning. Transfer learning can tune the parameters of the deep layer especially for the image classification model for COVID-19 infected patients [13]. To handle a small dataset data augmentation is available. Data augmentation can improve the training process efficiency in a Deep Learning image classification problem [14]. Hence, we are using Inception ResNet V2, InceptionNet V3 and NASNetLarge and applying data augmentation to get better results.
Our main purpose is to help the research community perform chest X-ray image analysis to detect different stages of COVID-19, using extraction and selection models that are completely automated. We also develop a state-of-the-art solution of chest X-ray image and Pneumonia analysis by exploring the latest deep learning and computer vision methods and verifying the performance over special collected dataset. This result will help doctors make decisions in their clinical practice due its high performance and effectiveness. The study also gives an insight into how transfer learning was used to automatically detect COVID-19.
This work is organized as follows. Section II presents the literature review of existing techniques related to the detection of COVID-19 from medical images. The proposed technique is explained in detail in Section III, followed by experimental evaluation and results with the discussion in Section IV. Section V concludes the paper and states possible future directions.
2. RELATED WORK
2.1. Overview of Deep Learning in Image Analysis
There have been some Machine Learning and Deep Learning research works available for effective detection of chest diseases in general rather than being performed specifically for COVID-19 detection. Arimura et al. [15] employed a template matching technique to identify chest-related diseases. Lehmann et al. [16] exploited traditional K-nearest neighbor technique to work with hernia identification. Kao et al. [17] utilized a two features-based methodology which can exploit body symmetry and background information for the classification of radiographic images. Jian-xin Yang et al. [18] computed tomography detected 154 lung atelectasis consolidation in 324 lung regions in 81 patients with 80% accuracy.
Pietka et al. [19] proposed a method which can identify view-based radiographic images for the detection of chest related infections. Boone, et al. [20] used projection profiles features and applied a neural network approach to classify the radiographic images. Similarly, Kao et al. [21] provided a technique to provide a view invariant chest infection identification methodology. Luo et al. [22] employed Bayes’s decision theory to identify Pneumonia. The features they used included the existence, shape, and spatial relation of medial axes of anatomic structures, as well as the average intensity of region of interest.
In Deep Learning, Dong et al. [23] used lung images that were divided into ten different classes using Vgg16 and ResNet-101 CNN with 90% accuracy. Gao et al. [24] used a 3D block based residual deep learning network to predict levels of tuberculosis using CT pulmonary images.
2.2. State-of-the-art of COVID-19 Issues
There are state-of-the-art studies on Deep Learning and Machine Learning models for COVID-19. A study by Apostolopoulos et al. [9] advantages of convolutional neural networks (CNN) for the automatic diagnosis of COVID-19 from chest X-ray images. Transfer learning was used to solve the small dataset problem. The dataset of COVID-19 consists of 224 sample images. Despite the limitation of dataset size, the results showed effective automatic detection of COVID-19 related diseases.
Abbas et al. [25] used a CNN based DeTraC framework and also used transfer learning to achieve best performance.
This model achieved 95.12% accuracy and 97.91% sensitivity. Chen et al. [26] provided prediction of COVID-19 infected patients or non-COVID-19 using a UNet++ based segmentation model. Narin et al. [8] classified Chest X-Rays using a RestNet50 model and got the highest classification performance with 98% accuracy using only 50 COVID-19 and 50 Normal samples for the dataset. Li et al. using 468 COVID-19 data, 1551 CAP and 1445 Non-Pneumonia using ResNet-50 achieved 90.0% sensitivity. Using deep learning approaches to extract and transform features has proved its ability in COVID-19 diagnosis [11].
2.3. Transfer Learning with CNNs
Deep learning requires a large amount of data for training, otherwise the model will over-fit. Over-fitting occurs when a model captures noise from the data and as a result the model fits the data too well due to the data being small. The convolution neutral network is used to process new sets of images of a different nature and extract features from these images; this is done by referring to previous training because transfer learning enables us to transfer information. Pre-trained convolutional neural networks are used in two ways, first, they are used for feature extraction which means after the data is analyzed and scanned, knowledge of important features is kept and used by another model that is designed for classification. This method is mainly used because it is much cheaper to set up as compared to building a very deep network from scratch, or to retain useful features extractors learned in the initial stage. The second strategy is a more sophisticated procedure where specific modifications are made to pre-trained models to achieve optimal results, these modifications may include architecture adjustment and parameter tuning. Transfer learning is used to tune the initial parameter of the deep layers [13]. Transfer learning is good in convolutional neural network especially for using DCNN in the medical domain that has a limited availability of experts, cost pressures and ethical concerns, and avoids DCNN from random weights [27].
Transfer learning is used by Abbasian et al. [28] to optimize the CNN to manage COVID-19 in routine clinical practice using CT images. Pathak et al. [13] also built a deep transfer learning-based classification model for COVID-19 disease. They added a top-2 smooth loss function with cost-sensitive attributes to handle a noisy and imbalanced COVID-19 dataset. The classification result achieved 96.2% training accuracy and 93.01% testing accuracy.
2.4. Data Augmentation
Data augmentation is the technique for increasing the amount of data by scaling existing images i.e. stretching, rotation, reflection, scaling and shearing. Data augmentation also uses geometric distortions to increase the number of samples for training the deep neural models, to balance the size of datasets [14]. To solve the lack of training data, recent methods have used data augmentation techniques to enlarge the dataset. Data augmentation reduces overfitting [29, 30].
3. PROPOSED METHODS
This research is aimed at evaluating the effectiveness and accuracy of different convolutional neural networks models. The automatic diagnosis of COVID-19 from Chest X-rays was compared to diagnosis performed by experts in the medical community. The effectiveness of state-of-the-art methods was demonstrated using a collection of 1765 X-ray radiographs from a well-known dataset by Joseph et al in GitHub [31, 32] and another dataset obtained from the Kaggle repository named “COVID-19 chest X-ray” [33]. These datasets were processed and used to train and validate convolutional neural networks models. Specifically, 850 X-ray scans of confirmed COVID-19 cases and 915 images of normal X-rays that have no disease were used.
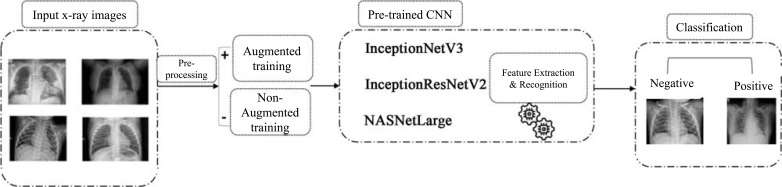
Transfer learning is the preferred strategy to train deep convolutional neural networks [34]. The reason behind this is that other convolutional neural networks learning strategies require large-scale datasets to perform accurate feature extraction and classification [35]. The retention of knowledge extracted from one task is the key to perform an alternative task. For this problem, data augmentation has been applied to overcome over-fitting while using less data. As shown in Fig. (1), the input test images are entered in a preprocessing process [36]. The input data is processed through 2 paths, one that performed data augmentation and the other that did not perform data augmentation. Deep convolutional neural networks designed using InceptionNetV3, InceptionResNetV2 and NASNetLarge models were trained using these datasets. ImageNet is used as a last step for transfer learning to tune the parameters and produce high accuracy.
Fig. (1).
Architecture diagram for proposed virus recognition phase. Different Deep conventional neural network architectures implemented to figure out the COVID-19. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
To build a COVID-Net method, we selected 3 models: Inception ResNet V2, InceptionNet V3 and NASNetLarge.
3.1. The InceptionNetV3
The InceptionNetV3 model has 48 deep layers and is pre-trained using an ImageNet dataset. The head of this model is also replaced with a modified head that will be used for two classes classification. In order to reduce the number of trainable parameters, the convolutional layers were set to ‘non-trainable’. By doing this, the number of trainable parameters was reduced from 22,982,626 to 1,179,842. The architecture of InceptionNetV3 is purposely designed to have a deep structure even though it has significantly less training parameters as compared to other neural networks. For example, VGG16 has about 90 million parameters but when compared to InceptionNetV3 it is found to have much less depth, and it is this depth that sharpens the accuracy of a model by allowing the model to capture finer details.
3.2. InceptionResNetV2
The Inception ResnetV2 is an advanced model as compared to InceptionNetV3 because it has 162 deep network layers, but for this research, we modified the architecture of the model in a similar way to the InceptionNetV3 by replacing the head with the required dense and classification layers. The Inception ResnetV2 combines the properties of both Inception and ResNet. The main property of ResNet is that the residual variants always show instability and the last layer results to zero whenever a large number of filters are used. This problem still continued even when the learning rate was changed or when we applied batch normalization to the layer. The best way to overcome this problem is to scale down the residuals before adding them to the previous activation layer.
3.3. NASNetLarge
Neural Architecture Search (NAS) is employed when designing the NASNetLarge. Inception cells are used to construct the layer of the model as in the case of InceptionNetV3. The two types of cells used for designing NASNet are normal cell and reduction cell and they are both presented pictorially below.
4. RESULTS AND DISCUSSION
In this research, we propose a COVID-Net method to predict and diagnose COVID-19 cases. It should be noted that COVID-19 is a new disease and there is a limited amount of research published to date. In the light of this, key contributions are expected as regards to image structure-based feature extraction and image structure analysis to predict positive COVID-19 infected patient.
To build COVID-Net, InceptionNetV3, InceptionResNetV2 and NASNetLarge models are designed and tasked with classifying X-ray images to predict COVID-19 positive and negative patients. Transfer learning is used due to the scarcity of data. Each convolutional neural network has an input shape of 512x512x3. In the experiments, all three models are trained both with and without data augmentation to observe the effects on each of the models. In data augmentation we have only used the rotation parameter which generates data by rotating the images in a range of 15 degrees angle. Data was distributed as 80% for training and 20% for test.
The hyperparameters of the models are initial learning rate of 0.001, batch size of 16, 10 epochs. Activation functions used are Softmax and ReLU.
For the results, Table 1 shows the comparison of all the training and validation accuracies of the different convolution neutral network architectures used in our experiments. Performance was measured using a well-known metric in the deep learning area called Accuracy which is presented in equation (1):
Table 1.
A comparison of training and validation accuracies of InceptionNetV3, InceptionResNetV2 and NASNetLarge.
| - | Training Accuracy | Validation Accuracy | Data Augmentation |
|---|---|---|---|
| InceptionResNetV2 | 98.64% | 97.87% | Yes |
| 99.47% | 87.23% | No | |
| InceptionNetV3 | 98.63% | 97.87% | Yes |
| 98.90% | 87.23% | No | |
| NASNetLarge | 99.54% | 92% | Yes |
| 98.93% | 61.70% | No |
Where:
TP (True Positives): the number of correctly labeled instances of the class under observation.
FP (False Positives): the number of miss-classified labeled of rest of the classes.
TN (True Negatives): the number of correctly labeled instances of rest of the classes.
FN (False Negatives): the number of miss-classified labeled of the class under observation.
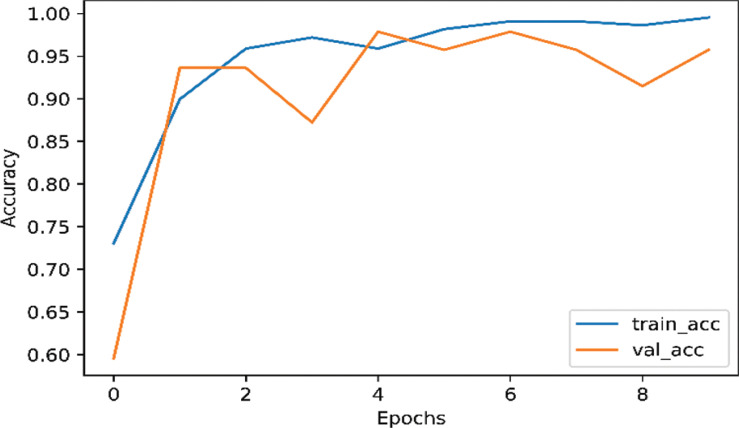
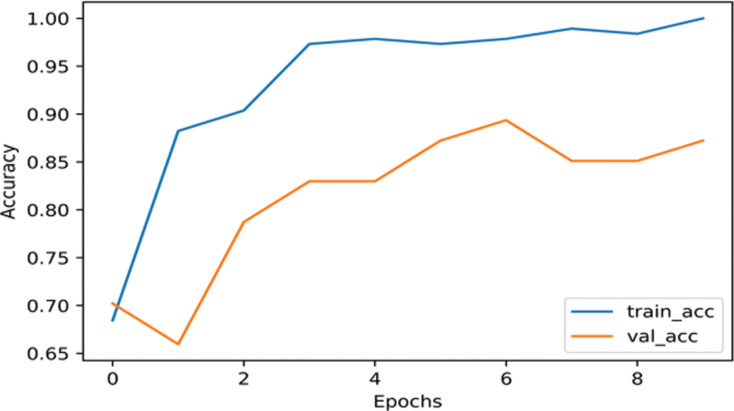
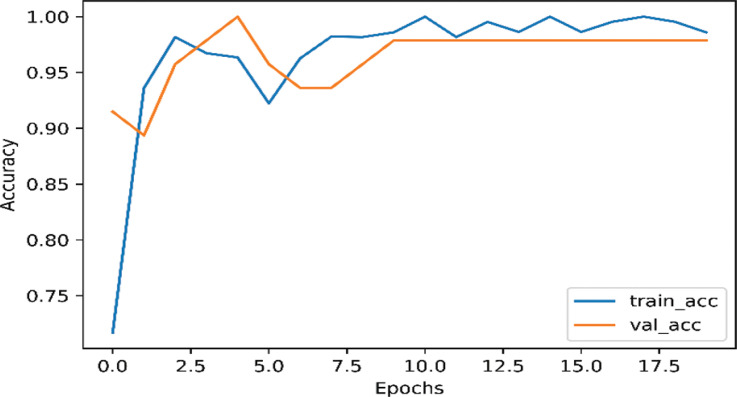
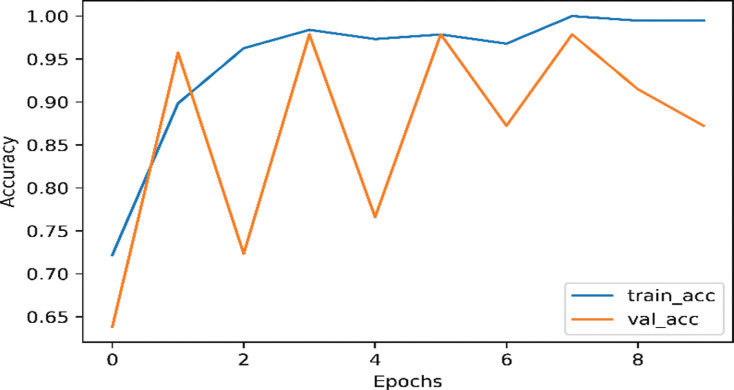
Each of our models was divided into two part, using data augmentation and non-augmentation. As shown in Fig. (2), the InceptionNetV3 model training accuracy is 98.63% and the validation accuracy 97.87% on the augmented data. As shown in Fig. (3), the InceptionNetv3 model training accuracy is 98.90% and the validation accuracy is 87.23% in non-augmented data.
Fig. (2).
The accuracy of InceptionNetV3 for nCOVID-19 when data augmentation is performed. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Fig. (3).
The accuracy of InceptionNetV3 on nCOVID-19 dataset when data augmentation is not performed. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
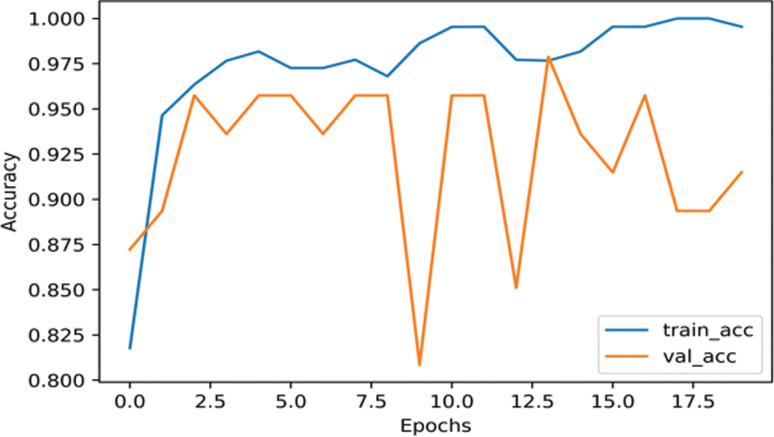
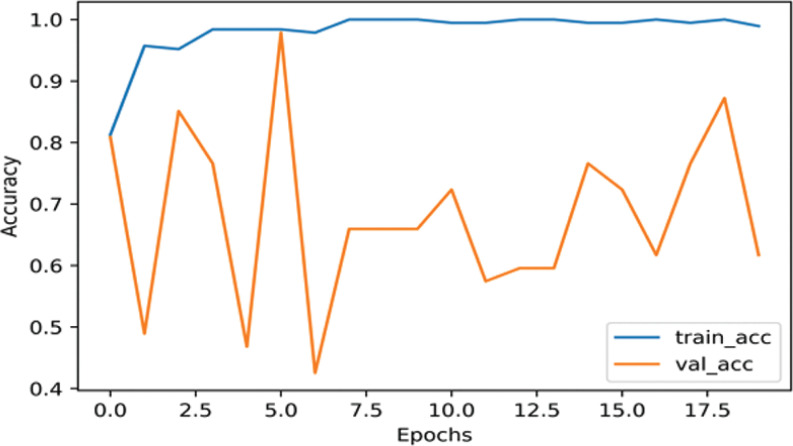
InceptionResNetV2 had 98.64% training accuracy and 97.87% validation accuracy using augmented COVID-19 dataset as shown in Fig. (4). As shown in Fig. (5), the training accuracy is 99.47% and validation accuracy is 87.23% using un-augmented data, which is 0.57% higher than that of the InceptionNetV3 model.
Fig. (4).
The accuracy of InceptionResNetV2 on nCOVID-19 dataset with data augmentation. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Fig. (5).
The accuracy of InceptionResNetV2 on nCOVID-19 dataset without performing data augmentation. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
NASNetLarge had 99.54% training accuracy and 92% validation accuracy using augmented dataset as shown in Fig. (6), which is higher than that of the COVID-19 un-augmented dataset. As shown in Fig. (1), the NASNetLarge model without data augmentation has result of 98.90% training accuracy and 61.70% validation accuracy. This validation accuracy of NASNetLarge model is lower than the other models.
Fig. (6).
The accuracy of NASNetLarge on nCOVID-19 dataset using data augmentation. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Fig. (7).
The accuracy of NASNetLarge on nCOVID-19 dataset when data augmentation is not performed. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Comparing with other models in Table 1, the NASNetLarge consistently gets the higher training and validation accuracy when using data augmentation. But comparing with the other validation accuracy, InceptionNetV3 and InceptionResNetV2 has the same result. As presented in Figs. (2-7), the models tend to over-fit when data augmentation is not performed. This over-fitting is happening as a result of limited data available for the research. By performing data augmentation, we introduce the sense of generalization in models and as a result, they yield better and more effective results and are free of over-fitting. It can be seen that NASNetLarge only has 61.70% accuracy when data augmentation is not performed, and this may be because the models is overfitting.
The highest training accuracy is obtained with the NASNetLarge model when data augmentation is performed. The training accuracy of NASNetLarge is also better than all of the other architectures used in this experiment. However, NASNetLarge tends to overfit even when the data augmentation is not used. For validation accuracy, the highest accuracy is obtained with both of InceptionNetV3 and InceptionResNetV2 when data augmentation is performed.
By looking at Table 2 it can be said that our proposed approach gives a comparable accuracy to other state of the art models with a small amount of data. This shows that performance improvement can be achieved using data augmentation.
Table 2.
A comparison of different studies and models for detection of COVID-19.
| Reference | Model | Accuracy |
|---|---|---|
| Narin et al. [13] | Inception-V3 ResNet50 Inception ResNet V2 |
100% 100% 95% |
| Wang et al. [16] | M-Inception | 82.9% |
| Abbas et al. [30] | DeTrac | 95.12% |
| Pathak et al. [18] | GCNN (proposed) | 3.01% |
| Ardakani et al. [33] | AlexNet VGG-16 VGG-19 SqueezNet GoogleNet MobileNet-V2 ResNet-18 ResNet-50 ResNet-101 Xception |
78.92% 83.33% 85.29% 82.84% 85.29% 92.16% 91.67% 94.12% 99.51% 99.02% |
For all three architectures used in this research; the networks tend to overfit when data augmentation is not used especially in validation accuracy. Data augmentation improves both training and validation accuracy in all cases except for validation using NASNetLarge. NASNetLarge had the worst validation results when compared to the other models.
Therefore, our results show that InceptionNetV3 and InceptionResNetV2 are the optimal choices for this classification task using data augmentation. Both of InceptionNetV3 and InceptioResNetV2 have 97.87% validation accuracy. They also have a small gap between the training and the validation result. If more data is made available, there is a higher possibility that the deeper networks, i.e., NASNetLarge will perform better than both of them.
CONCLUSION
COVID-19 is a disease that has spread all over the world. The early and effective prediction of COVID-19 in suspected patients is vital to prevention, detection and tracking of the spread. In this research, we propose a COVID-Net that consists of deep learning architecture with transfer learning models and data augmentation for detecting COVID-19 automatically from chest X-ray of COVID-19 patients and people with normal chest X-rays. The research demonstrates that InceptionNetV3 and InceptionResNetV2 models yielded the highest accuracy compared with the other models. Both have 97.87% validation accuracy using data augmentation.
Therefore, the proposed method can be used as an alternative to predict COVID-19 in suspected patients. Despite the limited data availability, COVID-19 research is still ongoing with possibilities biomarkers in X-ray images being discovered which could be used for detection and diagnosis with high performance in image classification.
For future work, image classification with high accuracy can be increased using multi-classification techniques. Huge datasets will be required for effective, efficient, and accurate detection of COVID-19. Moreover, more information such as: patient age, gender, status, symptoms, etc. can be analyzed beside the x-ray images to assist radiologists in differential diagnosis. Thus, more information can be added to the x-ray scan as metadata and this can be used to feed the model with more details to reach its decision with greater certainty.
ACKNOWLEDGEMENTS
Declared none.
Funding Statement
This study was funded by the Qassim University, represented by the Deanship of Scientific Research, on the financial support for this research under the number (Coc–2020- 1-1-L- 9950) during the academic year 1441 AH / 2020 AD”.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
This study was funded by the Qassim University, represented by the Deanship of Scientific Research, on the financial support for this research under the number (Coc–2020- 1-1-L- 9950) during the academic year 1441 AH / 2020 AD”.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.World Health Organization (WHO) announces COVID-19 outbreak a pandemic. 2020. Available at: https://www.euro.who.int/en/health-topics/health-emergencies/coronavirus- covid-19/news/news/2020/3/who-announces-covid-19-outbreak-a- pandemic
- 2.Information for Healthcare Professionals about Coronavirus (COVID-19). 2020. Available at: https://www.cdc.gov/coronavirus/2019-ncov/hcp/index.html
- 3.Pham Q.V., Nguyen D.C., Hwang W.J., Pathirana P.N. Artificial intelligence (AI) and big data for coronavirus (COVID-19) pandemic: A survey on the state-of-the-arts. IEEE Access. 2020;99:1–1. doi: 10.1109/ACCESS.2020.3031614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L., Wong A. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-Ray images. arXiv preprint. 2003. [DOI] [PMC free article] [PubMed]
- 6.Albahli S. A deep neural network to distinguish COVID-19 from other chest diseases using X-ray images. Cur Med Imaging. 2021;17(1):109–19. doi: 10.2174/1573405616666200604163954. [DOI] [PubMed] [Google Scholar]
- 7.Greenspan H., Van Ginneken B., Summers R.M. Guest editorial deep learning in medical imaging: Overview and future promise of an exciting new technique. IEEE Trans. Med. Imaging. 2016;35(5):1153–1159. doi: 10.1109/TMI.2016.2553401. [DOI] [Google Scholar]
- 8.Mayr A., Klambauer G., Unterthiner T., Steijaert M., Wegner J.K., Ceulemans H., Clevert D.A., Hochreiter S. Large-scale comparison of machine learning methods for drug target prediction on ChEMBL. Chem. Sci. (Camb.) 2018;9(24):5441–5451. doi: 10.1039/C8SC00148K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bui D.T., Tsangaratos P., Nguyen V.T., Van Liem N., Trinh P.T. Comparing the prediction performance of a Deep Learning Neural Network model with conventional machine learning models in landslide susceptibility assessment. Catena. 2020;188:104426. doi: 10.1016/j.catena.2019.104426. [DOI] [Google Scholar]
- 10.Korotcov A., Tkachenko V., Russo D.P., Ekins S. Comparison of deep learning with multiple machine learning methods and metrics using diverse drug discovery data sets. Mol. Pharm. 2017;14(12):4462–4475. doi: 10.1021/acs.molpharmaceut.7b00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caruana R. Multitask learning. Mach. Learn. 1997;28(1):41–75. doi: 10.1023/A:1007379606734. [DOI] [Google Scholar]
- 12.Bengio Y., Courville A., Vincent P. Representation learning: a review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 2013;35(8):1798–1828. doi: 10.1109/TPAMI.2013.50. [DOI] [PubMed] [Google Scholar]
- 13.Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. arXiv preprint. 2003. [DOI] [PMC free article] [PubMed]
- 14.Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phy. Eng. Sci. Med. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kermany D.S., Goldbaum M., Cai W., Valentim C.C.S., Liang H., Baxter S.L., McKeown A., Yang G., Wu X., Yan F., Dong J., Prasadha M.K., Pei J., Ting M.Y.L., Zhu J., Li C., Hewett S., Dong J., Ziyar I., Shi A., Zhang R., Zheng L., Hou R., Shi W., Fu X., Duan Y., Huu V.A.N., Wen C., Zhang E.D., Zhang C.L., Li O., Wang X., Singer M.A., Sun X., Xu J., Tafreshi A., Lewis M.A., Xia H., Zhang K. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. e9. [DOI] [PubMed] [Google Scholar]
- 16.Wang S, Kang B, Ma J, et al. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19). MedRxiv. 2020 doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Z Jianpeng, X Yutong. Viral pneumonia screening on chest X-ray images using confidence-aware anomaly detection. MedRxiv. 2020 doi: 10.1109/TMI.2020.3040950. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pathak Y., Shukla P.K., Tiwari A., Stalin S., Singh S., Shukla P.K. Deep transfer learning based classification model for COVID-19 disease. Online ahead of print. Ing Rech Biomed. 2020 doi: 10.1016/j.irbm.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mikołajczyk A, Grochowski M. In 2018 international interdisciplinary PhD workshop (IIPhDW) IEEE; 2018. Data augmentation for improving deep learning in image classification problem. pp. 117 –122 . [DOI] [Google Scholar]
- 20.Arimura H., Katsuragawa S., Li Q., Ishida T., Doi K. Development of a computerized method for identifying the posteroanterior and lateral views of chest radiographs by use of a template matching technique. Med. Phys. 2002;29(7):1556–1561. doi: 10.1118/1.1487426. [DOI] [PubMed] [Google Scholar]
- 21.Lehmann T.M., Güld O., Keysers D., Schubert H., Kohnen M., Wein B.B. Determining the view of chest radiographs. J. Digit. Imaging. 2003;16(3):280–291. doi: 10.1007/s10278-003-1655-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao E.F., Lee C., Jaw T.S., Hsu J.S., Liu G.C. Projection profile analysis for identifying different views of chest radiographs. Acad. Radiol. 2006;13(4):518–525. doi: 10.1016/j.acra.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 23.Yang J.X., Zhang M., Liu Z.H., Ba L., Gan J.X., Xu S.W. Detection of lung atelectasis/consolidation by ultrasound in multiple trauma patients with mechanical ventilation. Crit. Ultrasound J. 2009;1(1):13–16. doi: 10.1007/s13089-009-0003-x. [DOI] [Google Scholar]
- 24.Pietka E., Huang H.K. Orientation correction for chest images. J. Digit. Imaging. 1992;5(3):185–189. doi: 10.1007/BF03167768. [DOI] [PubMed] [Google Scholar]
- 25.Boone J.M., Seshagiri S., Steiner R.M. Recognition of chest radiograph orientation for picture archiving and communications systems display using neural networks. J. Digit. Imaging. 1992;5(3):190–193. doi: 10.1007/BF03167769. [DOI] [PubMed] [Google Scholar]
- 26.Kao E.F., Lin W.C., Hsu J.S., Chou M.C., Jaw T.S., Liu G.C. A computerized method for automated identification of erect posteroanterior and supine anteroposterior chest radiographs. Phys. Med. Biol. 2011;56(24):7737–7753. doi: 10.1088/0031-9155/56/24/004. [DOI] [PubMed] [Google Scholar]
- 27.Luo H., Hao W., Foos D.H., Cornelius C.W. Automatic image hanging protocol for chest radiographs in PACS. IEEE Trans. Inf. Technol. Biomed. 2006;10(2):302–311. doi: 10.1109/TITB.2005.859872. [DOI] [PubMed] [Google Scholar]
- 28.Dong Y., Pan Y., Zhang J., Xu W. 2017 IEEE/ACM International Conference on Connected Health: Applications, Systems and Engineering Technologies (CHASE) IEEE.; 2017. Learning to read chest X-ray images from 16000+ examples using CNN. pp. 51–57. [DOI] [Google Scholar]
- 29.Gao X.W., James-Reynolds C., Currie E. Analysis of tuberculosis severity levels from ct pulmonary images based on enhanced residual deep learning architecture. Neurocomputing. 2020;392:233–244. doi: 10.1016/j.neucom.2018.12.086. [DOI] [Google Scholar]
- 30.Abbas A., Abdelsamea M.M., Gaber M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. arXiv preprint . 2003. [DOI] [PMC free article] [PubMed]
- 31.Chen J, Wu L, Zhang J, et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study. . MedRxiv. . 2020 doi: 10.1038/s41598-020-76282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharif Razavian A., Azizpour H., Sullivan J., Carlsson S. CNN features off-the-shelf: an astounding baseline for recognition; InProceedings of the IEEE conference on computer vision and pattern recognition workshops; 2014. pp. 806–813. [Google Scholar]
- 33.Ardakani A.A., Kanafi A.R., Acharya U.R., Khadem N., Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput. Biol. Med. 2020;121:103795. doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin H.C., Lee K.I., Lee C.E. 2020 IEEE International Conference on Big Data and Smart Computing (Big Comp) IEEE.; 2020. Data Augmentation Method of Object Detection for Deep Learning in Maritime Image. pp. 463–466. [DOI] [Google Scholar]
- 35.Cohen J.P., Morrison P., Dao L. COVID-19 image data collection. arXiv:2003. 2020. p. 11597. https://github.com/ieee8023/covid-chestxray-dataset .
- 36.Kaggle. COVID-19 chest x-ray. 2020. https://www.kaggle.com/bachrr/covid-chest-xray .
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.