Summary
Bicuspid aortic valve (BAV) with ∼1%–2% prevalence is the most common congenital heart defect (CHD). It frequently results in valve disease and aorta dilation and is a major cause of adult cardiac surgery. BAV is genetically linked to rare left-heart obstructions (left ventricular outflow tract obstructions [LVOTOs]), including hypoplastic left heart syndrome (HLHS) and coarctation of the aorta (CoA). Mouse and human studies indicate LVOTO is genetically heterogeneous with a complex genetic etiology. Homozygous mutation in the Pcdha protocadherin gene cluster in mice can cause BAV, and also HLHS and other LVOTO phenotypes when accompanied by a second mutation. Here we show two common deletion copy number variants (delCNVs) within the PCDHA gene cluster are associated with LVOTO. Analysis of 1,218 white individuals with LVOTO versus 463 disease-free local control individuals yielded odds ratios (ORs) at 1.47 (95% confidence interval [CI], 1.13–1.92; p = 4.2 × 10−3) for LVOTO, 1.47 (95% CI, 1.10–1.97; p = 0.01) for BAV, 6.13 (95% CI, 2.75–13.7; p = 9.7 × 10−6) for CoA, and 1.49 (95% CI, 1.07–2.08; p = 0.019) for HLHS. Increased OR was observed for all LVOTO phenotypes in homozygous or compound heterozygous PCDHA delCNV genotype comparison versus wild type. Analysis of an independent white cohort (381 affected individuals, 1,352 control individuals) replicated the PCDHA delCNV association with LVOTO. Generalizability of these findings is suggested by similar observations in Black and Chinese individuals with LVOTO. Analysis of Pcdha mutant mice showed reduced PCDHA expression at regions of cell-cell contact in aortic smooth muscle and cushion mesenchyme, suggesting potential mechanisms for BAV pathogenesis and aortopathy. Together, these findings indicate common variants causing PCDHA deficiency play a significant role in the genetic etiology of common and rare LVOTO-CHD.
Keywords: bicuspid aortic valve, coarctaction, left ventricular outflow obstruction, genetics, protocadherin, copy number variants, common variants
Bicuspid aortic valve is the most common congenital heart defect, but its genetic etiology is not well understood. We showed common deletion copy number variants in the PCDHA gene cluster play a significant role in the genetic etiology of BAV and also in other rare left-sided congenital heart defects.
Introduction
Bicuspid aortic valve (BAV) is the most common congenital heart defect (CHD), exhibiting 1%–2% prevalence in the general population.1 It is a structural heart defect resulting from maldevelopment of the aortic valve such that only two functional valve leaflets are formed instead of the normal tricuspid aortic valve (TAV).2 BAV in isolation is largely benign during early life and may remain undiagnosed until clinical presentation with aortic valve dysfunction or aorta dilation in later adult life. Given its prevalence, BAV is a major cause for adult cardiac surgery, accounting for more than 50% of valve replacement surgery.3 Approximately 25% of individuals with BAV will require aorta surgery, and 50%–75% will require aortic valve replacement.4,5 The surgical management of BAV is challenging, as the pathogenic mechanism for BAV-associated aortopathy remains unknown.
Familial studies have identified a genetic etiology for BAV.6, 7, 8, 9 These studies have also shown BAV is closely associated with less common CHDs involving other defects on the left side of the heart, such as coarctation of the aorta (CoA; prevalence of 0.04%) and hypoplastic left heart syndrome (HLHS; prevalence of 0.02%).10 Collectively, these malformations are referred to as left ventricular outflow tract obstructions (LVOTOs). Thus, family members of probands with either BAV or HLHS may have isolated BAV, CoA, or other LVOTO lesions. Together, these findings suggest a broad spectrum of LVOTO is genetically related. LVOTO also has been shown to be genetically heterogenous and characterized by reduced penetrance, such that subjects carrying a known pathogenic mutation may not have disease, and variable expressivity in which different disease phenotypes may be associated with the same mutation.11 These findings suggest LVOTO is mediated by complex genetics, with familial studies providing data supporting an oligogenic/digenic model of disease.9,12, 13, 14
Currently, only a few genes have been suggested to cause BAV. They include rare pathogenic variants in genes such as NOTCH1, SMAD6, or ROBO4, all recovered from familial studies with evidence of incomplete penetrance and variable expressivity.15, 16, 17, 18, 19 A recent study leveraging the close ancestry of subjects in Iceland also identified a rare pathogenic MYH6 coding variant linked to CoA and likely BAV.20 Analysis of 8 mutant mouse lines exhibiting HLHS and other LVOTO phenotypes recovered from a large-scale mouse mutagenesis screen has confirmed the genetic heterogeneity of LVOTO.21,22 The finding of non-Mendelian transmission in these HLHS mouse models also supports complex genetics.22 Analysis of one such mouse line, Ohia, identified a mutation in Pcdha9, a protocadherin gene mediating cell-cell adhesion, to cause BAV.21 When accompanied by a second mutation in the chromatin modifying protein Sap130, HLHS and other LVOTO lesions are observed.21
The finding of a role for Pcdha9 in HLHS/LVOTO was notable, given a previous report of a common 16.8 kb deletion copy number variant (delCNV) spanning PCDHA8-10 in the human genome.23 The prevalence of this delCNV varied by race (Minor Allele Frequency (MAF), 0.11–0.033), being most common in white individuals, less common in Black individuals, and least common in Asian individuals. This similarity to the racial prevalence of BAV suggested a possible genetic link between the PCDHA delCNV and BAV and perhaps other closely associated LVOTOs.24,25 This was investigated in the present study with the recruitment of a large discovery cohort of white subjects with LVOTO. Replication analysis was conducted in independent cohorts of white, Black, and Chinese LVOTO subjects. Using the Pcdha9 mutant mice, we further explored potential developmental mechanisms of heart malformation.
These studies differ from traditional approaches for investigating the genetic causes of CHD, which typically focus on rare variants in genes causing CHD. Alternatively, common variants are investigated and used to track and identify genomic regions linked to disease in genome-wide association studies. In the latter instance, the common variants typically found in noncoding intergenic regions serve merely as markers for tracking genomic regions linked to disease. In contrast, our study investigated common variants in the PCDHA gene cluster as possible disease alleles that can cause BAV/LVOTO. These studies are motivated by findings from the Ohia mouse model showing Pcdha9 deficiency can cause BAV/LVOTO and supported by the observation that a common PCDHA delCNV has a racial prevalence that mirrors the racial prevalence of BAV.
Material and methods
Human study participants
This study was performed in accordance with the Declaration of Helsinki for the ethical conduct of research involving human subjects. Ethics approval was obtained from the institutional ethics committee of each participating institution, with written informed consent obtained from all participants or from the parent of minor participants. Adult and pediatric cases for the white discovery cohort were recruited from several institutions (Table S1), including the University of Pittsburgh Medical Center (UPMC) for Thoracic Aortic Disease, Children’s Hospital of Pittsburgh (CHP), Hospital for Sick Children (Sickkids), Nationwide Children’s Hospital (NCH), Cincinnati Children’s Hospital Medical Center (CCHMC), and Children’s Healthcare of Atlanta. The Chinese cohort was recruited from the Chinese University of Hong Kong and Beijing Anzhen Hospital.
The pediatric LVOTO cases included isolated or complex BAV, CoA, HLHS, or other LVOTO as defined in Tables S2 and S3. Adult individuals with isolated BAV were either from clinic-based recruitment (UPMC Center for Thoracic Aortic Disease) or family-based recruitment from screening relatives of pediatric LVOTO probands (only one subject per family) (Table S1). In total, 1,322 individuals with LVOTO were recruited, comprising 1,218 white and 104 Chinese (self-reported) subjects. We also recruited 976 local control subjects comprising 463 white adult subjects from UPMC Heart and Vascular Institute and 513 Chinese adult subjects from the Chinese University of Hong Kong (Table S1). All control subjects had echocardiographic or magnetic resonance imaging (MRI) examination confirming normal TAV with no CHD. For replication analysis, whole-exome sequencing (WES) data were obtained for white LVOTO subjects from the Pediatric Cardiac Genomics Consortium (PCGC).26,27 This analysis was conducted using WES data from the Framingham Heart Study (FHS) as an independent control population.28 While BAV was not curated in the FHS, valve and/or aorta surgeries were recorded, allowing removal of these individuals to minimize the inclusion of BAV subjects.
PCDHA delCNV genotype determination
Multiplexed PCR genotyping
The PCDHA delCNV genotypes were determined using a multiplexed PCR assay. For the wild-type and Δ16.8 allele, PCR was performed according to a previously published protocol that yielded a wild-type PCR product of 469 bp and a 16.8 kb delCNV (nsv4655880) PCR product of 554 bp.23 Similarly, multiplexed PCR was performed to detect the wild-type and 13.6 kb PCDHA del CNV (nsv4684081). Primers for these PCR reactions are shown in Table S4, and PCR reactions were conducted using 60°C annealing temperature. The wild-type PCR product generated is 1,440 bp, and the 13.6 kb del CNV PCR product is approximately 2,500–3,000 bp. The PCR genotyping protocols were validated with demonstration that the delCNV genotype calls obtained from the WES data available for 166 CHP and 89 Sickkids individuals with LVOTO matched those obtained by PCR analysis performed by independent laboratories in CHP and Sickkids.
PCDHA delCNV genotyping using WES and whole-genome sequencing (WGS) data
WES data were analyzed from 166 CHP subjects, 89 Sickkids subjects, Chinese pediatric and PCGC LVOTO subjects (database for Genotype and Phenotype [dbGAP]: phs001194.v2.p2), and FHS subjects (FHS, dbGaP: phs000007.v30.p11). The alignment BAM files was generated by mapping the sequencing reads to the reference genome (GRCh38) with BWA-MEM v.0.7.17, with further processing using the GATK Best Practices workflows (GATK v.4.0.8.1).29, 30, 31 Plink v.1.932 was used to identify and remove cryptic relatedness in Framingham and PCGC cohorts using methods previously described.33 Briefly, second-degree or closer relatives with shared ancestry with PI_HAT > 0.25 were removed, such that only unrelated individuals were used for downstream analyses.
For analysis of the delCNV genotypes, depth information directly extracted from BAM files was used to determine the 16.8/13.6 kb delCNV genotype by visually reviewing the depth distribution across the region encompassing the overlapping 16.8 kb and13.6 kb deletion intervals in each subject (Figures S1A–S1E). The CHP and Sickkids samples were further analyzed by multiplexed PCR, which yielded consistent results, confirming accuracy of the WES-determined genotypes. For analysis of whole-genome sequencing data from the 1000 Genomes Project (1KG), depth information of positions spanning 16.8 kb and 13.6 kb deletion intervals were directly extracted from joint-calling VCF files downloaded from the 1KG website.34 The allele frequency observed for the top CNVs in the PCDHA cluster is shown in Figure S2C. Similar visual inspection of coverage depth in the whole-genome sequencing data was used to determine the 16.8/13.6 kb delCNV genotypes in subjects from the IKG (Figures S1F–S1J).
Ancestral identification using principal component analysis
Germline single-nucleotide variants (SNVs) and small indels (InDels) from WES data of subjects in the Pittsburgh, PCGC, and FHS cohorts were jointly called from BAM files using GATK HaplotypeCaller, and variants that pass filtering based on GATK4 best practices, have a cohort MAF > 0.05, and have MAF > 0.1 across non-Finnish European samples in gnomAD exome v.2.1 were considered high-quality common variants.26, 27, 28, 29,31,35 To find white samples with ancestry similar to Pittsburgh subjects in the PCGC and FHS cohorts, genotypes of these common variants were extracted, and principal component analysis (PCA) was performed with Smartpca in EIGENSOFT v.6.1.4.36,37 White subjects were extracted with PC1 < −0.003 and PC2 < 0.018 and ggplot2 via a centroid delineated ellipse encompassing most of the white subjects in the Pittsburgh cohort in the PCA plot of PC1 versus PC3 (Figure S3). PCGC and FHS subjects within this ellipse are considered white subjects with ancestry similar to the Pittsburgh cohort (Figure S3) and were used for further association analyses. Black samples from Pittsburgh and PCGC were directly extracted with PC1 > 0.015 and were used for downstream analyses.
PCDHA9 pathogenic variant detection
To obtain potentially pathogenic variants in PCDHA9, variants including SNVs and InDels were extracted from joint-calling variant files generated by above analyses and annotated by Annovar.38 High-quality variants were recovered that: (1) passed GATK Variant Score Quality Recalibration (VSQR); (2) have minimum 5 supported reads; (3) have genotype quality ≥20 or 60 for SNVs or InDels, respectively; (4) SNVs or InDels not within 10 bp or 5 bp, respectively, of an indel were removed to eliminate false positives from mismatched reads from repetitive genomic sequences. This latter filter had no impact on the variants recovered.
We noted no PCDHA9 high-quality variants were recovered with MAF < 0.005 across non-Finnish European samples in gnomAD exome v.2.1.1.35 These were further filtered to identify potentially pathogenic variants, defined as loss-of-function (LoF) mutations (nonsense, canonical splice-site, frameshift indels, and start loss), inframeshift indels, and predicted deleterious missense mutation (D-Mis), that were called damaging by at least 6 of 9 prediction algorithms (SIFT, PolyPhen2_HDIV, LRT, MutationTaster, MutationAssessor, FATHMM, PROVEAN, MetaSVM, M_CAP annotated using dbNSFP v.35a), with CADD score > 20 and GERP++ > 2.39
Pcdha9 mutant mouse breeding and BAV phenotyping
Mouse studies were conducted under an animal study protocol approved by the University of Pittsburgh Institutional Animal Care and Use Committee. The Pcdha9 (c.2389_2399del; [p.Asp796Phefs∗]) mutant mice generated by CRISPR gene editing were maintained in the C57BL/6J background.21 Pcdha9 mutant mouse embryos at embryonic day 14.5 (E14.5) or term were analyzed for BAV using histological analysis with episcopic confocal microscopy.21 For immunoconfocal microscopy, embryos were cryoembedded (see below). BAV diagnosis in adult mice was determined by cardiac MRI (CMR). In vivo cardiac MRI was carried out on a Bruker Biospec 7T/30 system (Bruker Biospin MRI, Billerica, MA, USA) with the 35-mm quadrature coil for both transmission and reception. Free-breathing no-gating cine MRI with retrospective navigators was acquired with the Bruker Intragate module. Multi-planar short-axis, 2-chamber and 4-chamber long-axis cine MRI covering the whole heart volume was acquired with the following parameters: field of view (FOV) = 2.5 cm × 2.5 cm, slice thickness = 1 mm, in-plane resolution = 0.97 μm, flip angle (FA) = 30 degrees, echo time (TE) = 1.872 ms, repetition time (TR) = 38.293 ms; FOV = 1.8 cm × 1.8 cm, slice thickness = 0.8 mm, in-plane resolution = 0.070 mm, FA = 30 degrees, TE = 1.872 ms, TR = 38.293 ms, 50 cardiac phases with equivalent temporal resolution around 6 ms per time frame.
Analysis of protein expression with immunostaining and confocal imaging
Wild-type C57BL/6J and homozygous Pcdha9 mutant embryos were obtained at E14.5. The embryos were fixed in 4% paraformaldehyde and processed for cryoembedding. Frozen sections were collected for immunostaining. Cryosections of mouse embryonic heart tissue were immunostained with rabbit anti-alpha-protocadherin antibody (190003, Synaptic Systems, 1:500), goat anti-PECAM-1 antibody (sc-1506, Santa Cruz Biotechnology, 1:500), and mouse anti-alpha smooth muscle actin antibody (1A4) (ab7817, Abcam, 1:500). Human adult aortic tissues were stained with the alpha-protocadherin antibody and also imaged for elastin autofluorescence. Secondary antibodies included goat anti-rabbit IgG (Invitrogen, #A21429, 1:1,000) and goat anti-mouse IgM (Invitrogen, #A21042, 1:1,000). DAPI was used to label nuclei (D1306, Invitrogen). Sections were imaged with confocal microscopy and quantitatively analyzed with ImageJ.40
Human aortic tissue transcript analysis
RNA was isolated from the medial layer of human aortic tissues from individuals with BAV and TAV who underwent aortic valve replacement surgery at the UPMC Center for Thoracic Aorta Disease. For human gene expression analysis, after cDNA was prepared, real-time PCR was performed for transcripts from PCDHA7–11 using Power SYBR Green PCR Master Mix (Applied Biosystems) on an ABI 7900HT Fast Real Time PCR system. Gapdh was used to normalize the expression data. Primers used for this analysis are shown in Table S4.
Pcdha9 siRNA knockdown analysis
Mouse mammary gland cell line (NMuMg) was cultured in 12-well plates for gene knockdown analysis. Pcdha9 small interfering RNA (siRNA) (Santa Cruz, #sc-106386) and negative control siRNA (Santa Cruz, #sc-37007) were delivered into NMuMg cells by transfection (FuGeneHD, Promega). 24 h after siRNA treatment, cells were treated with recombinant TGF-β1 (4 ng/mL, CellSignaling, #8915) and harvested at day 3. Three independent experiments were performed. For mRNA analysis, total RNA isolated using Trizol (Invitrogen) was reverse transcribed. Quantitative real-time PCR was used to analyze transcripts for Notch1, Postn, Rac1, Snai1, Tgfb1, and Tgfb2. Gapdh was used as internal control. Primers used for this analysis are shown in Table S4.
Bioinformatic analysis of RNA sequencing data from human fetal heart
Single-cell RNA-sequencing (RNA-seq) data of human embryos spanning 5 to 20 weeks of gestation from a previously published study were used to examine expression of genes in the PCDHA gene cluster.41 The gene-cell UMI (unique molecular identifier) count sparse matrix of single-cell RNA-seq data was directly downloaded from GEO database under GEO: GSE106118. Cell type annotation was obtained from the original publication.41 The sum of UMI counts for each cell was normalized to 10,000 and then log-transformed. A total of 2,500 cardiac cells were used for these analyses.
Statistics
For the CNV allelic and genotypic association tests, we conducted 2 × 2 Pearson’s chi-square test (or Fisher’s exact test if the cell frequency was <5), and odds ratio (OR) with 95% confidence interval (CI) was calculated for each association test. As both the 13.6 and 16.8 kb PCDHA delCNV eliminated PCDHA9, primary analyses considered both delCNVs. Secondary analyses considered each delCNV separately. Sex-adjusted allelic association tests were performed using logistic regression (plink2-glm sex-ci 0.95).32 Sex-adjusted genotypic association was conducted using MASS R package (glm[PHENOTYPE-1∼GENO+SEX,data,family = binomial]).42 While our primary analysis focused on the entirety of the LVOTO cohort compared to control subjects, we also conducted additional subanalyses to evaluate whether specific LVOTO phenotypes were driving the association with the PCHDA delCNVs. The animal experiments were not randomized; however, the investigators were blinded to the identity of the samples. In the mouse studies, comparison was conducted with unpaired Student’s t test. p values for t tests are reported in the respective figures, and p < 0.05 was considered statistically significant. All values are shown either as mean ± SEM (standard error of the mean) or as mean ± SD (standard deviation). Statistical analysis was performed using SPSS, SAS, or R package.
Results
Individuals in the discovery cohort and PCDHA deletion CNV analysis
We recruited 1,218 white subjects in a discovery cohort of individuals with LVOTO from Pittsburgh, Columbus, Cincinnati, Atlanta, and Toronto. This included pediatric individuals with BAV, CoA, HLHS, and other LVOTO phenotypes (n = 927), and also adult individuals with isolated BAV (n = 291) (Figures 1C–1F) (Table S5). In addition, 463 local control subjects were recruited from Pittsburgh, comprising white adult subjects all clinically confirmed to have normal cardiac anatomy with the same rigorous phenotype characterization as the case group with echocardiography or cardiac MRI.
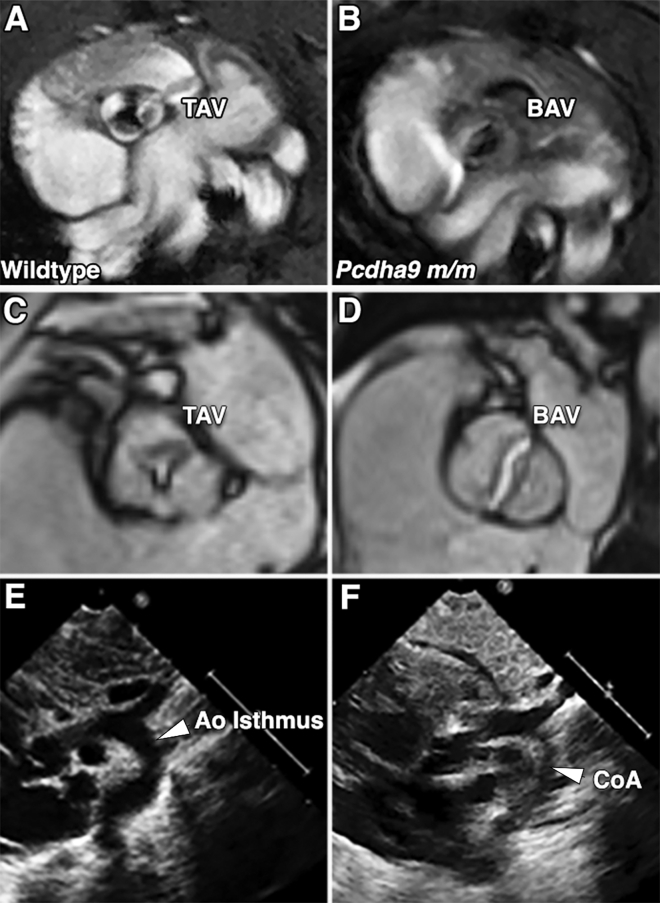
Figure 1.
Human and mouse LVOTO phenotypes
(A–D) Cardiac MRI showed TAV (A and C) and BAV (B and D) in homozygous Pcdha9 mutant mice (A and B) and in adult human subjects (C and D). TAV is characterized by three distinct cusps with three distinct Y-shaped closure lines (A and C), whereas BAV has the two cusps forming a single valve coaptation line with thickened leaflets (B and D).
(E and F) Two-dimensional (2D) transthoracic echocardiogram in suprasternal view shows subject with normal arch (E) and a CHD individual with severe CoA (F).
TAV, tricuspid aortic valve; BAV, bicuspid aortic valve; Ao isthmus, aortic isthmus; CoA, coarctation of aorta.
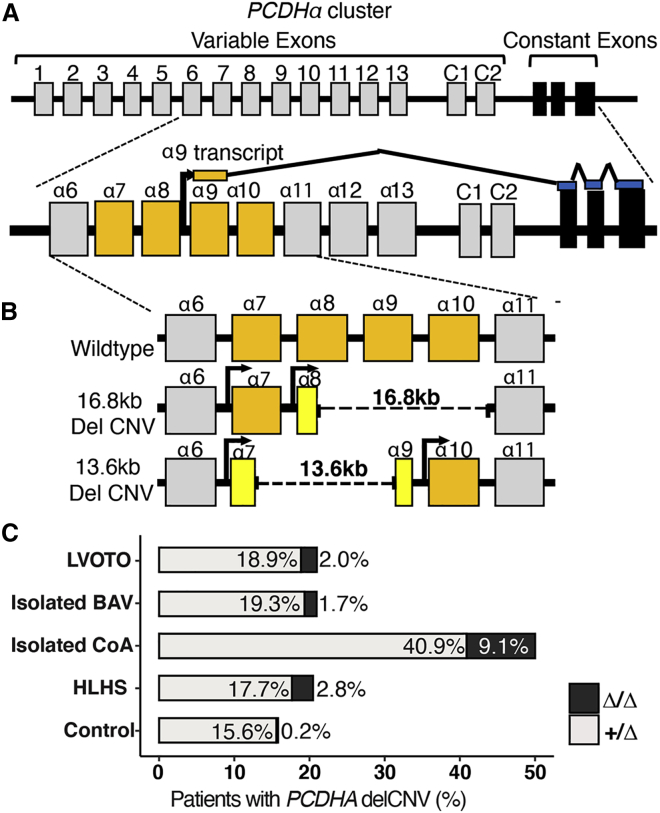
PCR genotyping was conducted to identify those carrying the known common 16.8 kb PCDHA delCNV (nsv4655880) and an additional less-common overlapping 13.6 kb delCNV (nsv4684081) discovered during the course of this study (Figures 2A and 2B).23 The protocadherin gene cluster comprises 15 variable exons, each transcribed from its own promoter and spliced to the same three terminal exons (Figure 2A).43 The 16.8 kb delCNV deletes variable exons spanning PCDHA8–10, while the 13.6 kb delCNV removes PCDHA7– 9 (Figure 2B). Hence, the two delCNVs share the common loss of PCDHA8 and PCDHA9. As WES was available on 166 of the individuals with LVOTO from Pittsburgh, we were able to confirm fidelity of the genotyping protocol in detection of both the 16.8 and 13.6 kb PCDHA delCNVs.
Figure 2.
Association of PCDHA delCNV with LVOTO
(A) Schematic of the organization of the PCDHA gene cluster. The human PCDHA gene cluster contains 15 variable exons (gray) and 3 constant exons (black). Each gene defined by a variable exon is transcribed from its own promotor and is spliced to 3 constant exons. The variable exon encodes the extracellular domain, and the constant exons encode the common distal intracellular domain.
(B) Schematic representation of the 16.8 kb delCNV (16.8 kb dashed line) spanning α8 to α10, and the ~13.6 kb delCNV (13.6 kb dashed line) spanning α7 to α9. Exons that are partially deleted are represented in yellow.
(C) Bar graph showing prevalence of the 13.6 kb and 16.8 kb PCDHA delCNV in individuals of the white discovery cohort with different LVOTO phenotypes.
Significant association of common PCDHA deletion CNV with BAV and other LVOTOs
A case-control analysis for the prevalence of the two PCDHA delCNVs (16.8/13.6 kb) in the discovery LVOTO cohort versus local control subjects showed apparent enrichment of the delCNV in the individuals with LVOTO. Thus, presence of the 16.8/13.6 kb delCNV (Δ/+ or Δ/Δ) was observed in 21% of individuals with LVOTO versus 15.8% of control subjects (Figure 2C). Most striking was the finding that 50% of individuals with CoA had the delCNV (Figure 2C). Also interesting to note is the much higher prevalence of the Δ/Δ genotype. While this was seen in only 0.2% of control subjects, it was observed in 2.1% of individuals with LVOTO, including 2.8% of those with HLHS and 9.1% with CoA (Figure 2C).
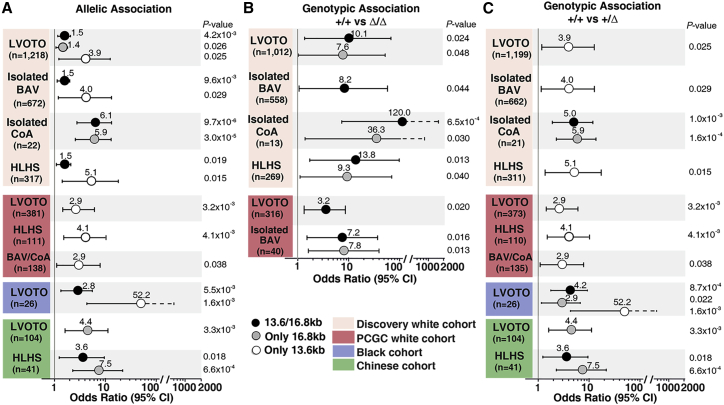
To investigate the role of the PCDHA delCNVs in LVOTO, we conducted an allelic association analysis of the 13.6 kb and 16.8 kb PCDHA delCNV. Adjustment for sex was carried out to account for the greater proportion of males observed among individuals with LVOTO.44 In the control cohort, the 16.8 kb delCNV has MAF of 0.077, while the 13.6 kb delCNV is less frequent, with MAF of 0.003 (Table S5). The 13.6/16.8 kb delCNV alleles combined showed significant association, with OR of 1.47 (95% CI, 1.13–1.92) (p = 4.2 × 10−3) for all LVOTOs combined, 1.47 (95% CI, 1.10–1.97) (p = 0.01) for isolated BAV, 6.13 (95% CI, 2.75–13.69) (p = 9.7 × 10−6) for isolated CoA, and 1.49 (95% CI, 1.07–2.08) (p = 0.019) for HLHS (Table S5A; Figure 2C). Allelic association analysis conducted separately for the 16.8 kb or 13.6 kb delCNV showed each CNV was significantly associated with LVOTO. Higher OR (3.9) was observed for the 13.6 kb delCNV as compared to the 16.8 kb (OR, 1.36) or 13.6/16.8 kb delCNV (OR, 1.47) combined (Figure 3A; Table S5). This likely reflects the much lower prevalence of the 13.6 kb delCNV. Among the LVOTO phenotypes, the 13.6 kb delCNV showed significant association with isolated BAV and HLHS, and the 16.8 kb delCNV showed significant association with isolated CoA (Figure 3A; Table S5).
Figure 3.
Association of the PCDHA delCNV with LVOTO in different cohorts
(A) Significant allelic association is observed for the PCDHA delCNV with LVOTO in different cohorts. The odds ratio (OR) and 95% confidence interval (CI) are shown. The total number of affected individuals analyzed is indicated.
(B and C) Genotype comparisons show significant association of homozygous/compound heterozygous (Δ/Δ) (B) or heterozygous (Δ/+) (C) genotypes with LVOTO phenotypes. The OR and 95% CI are shown. The total number of subjects analyzed with the different phenotypes is indicated.
As Pcdha9 mutant mice with either Pcdha9 missense or frameshift mutation (generated by CRISPR gene editing) yielded similar LVOTO phenotypes (Figures 1A and 1B), we used WES data available from 166 subjects with LVOTO from CHP to interrogate for possible coding mutations in PCDHA9. Analysis of the WES data showed 37 (22%) of these individuals had PCDHA delCNV, consistent with the PCR genotype. In three individuals, none with the PCDHA delCNV, three rare damaging variants were found in PCDHA9 (Table S6). This consisted of one stop gain mutation (MAF = 6.7 × 10−5) and two missense variants predicted to be damaging by six prediction algorithms (Table S6). These findings suggest coding sequence variation in the PCDHA gene cluster should be further investigated for a possible role in LVOTO pathogenesis.
Impact of PCDHA delCNV genotype
The enrichment observed for homozygous/compound heterozygous delCNV genotypes in individuals with LVOTO suggested gene dosage may impact penetrance of LVOTO phenotypes (Figure 2C). This is supported by findings from genotype comparison of homozygous/compound heterozygous 13.6/16.8 kb delCNV (Δ/Δ) versus wild-type genotypes showing significant association of the Δ/Δ genotypes with LVOTO, CoA, BAV, and HLHS (Figure 3B; Table S5). Compared to allelic association analysis, the ORs were higher at 8.2 (95% CI, 1.06–68.9) (p = 0.044) for BAV, 119.98 (95% CI, 7.66–1878) (p = 6.5 × 10−4) for isolated CoA, 13.83 (95% CI, 1.74–109.91) (p = 0.013) for HLHS, and 10.12 (95% CI, 1.37–74.89) (p = 0.024) for all LVOTO combined (Figure 3B; Table S5). The increased CI likely reflects the small number of Δ/Δ genotypes observed (Figure 3B; Table S5). These values are all substantially higher than when modeling an additive allelic effect. Further analysis of the 13.6 versus 16.8 kb delCNV individually showed significant Δ/Δ genotype association only with the 16.8 kb delCNV. This was observed for LVOTO, CoA, and HLHS (Figure 3B). Similar comparison of heterozygous versus wild-type genotypes showed the combined 13.6/16.8 kb delCNVs or the 16.8 kb delCNV alone showed significant association only for isolated CoA (Figure 3C; Table S5). In contrast, the 13.6 kb Δ/+ genotype, which is less frequent, showed significant association in BAV, HLHS, and all LVOTO combined (Figure 3C; Table S5). Overall, these findings indicate gene dosage effects related to the delCNV genotype may significantly impact LVOTO disease penetrance.
To further investigate whether the delCNV genotype also may impact penetrance of adult-onset aortopathy, we investigated the PCDHA delCNV genotype distribution among adult isolated BAV subjects from clinic-based recruitment at the UPMC Center for Thoracic Aortic Disease versus those from family-based recruitment with the screening of relatives of pediatric LVOTO proband (only 1 per family). Homozygous 16.8 kb delCNV genotypes were observed in 5 of the clinic-based individuals, 3 with aortic aneurysm, but no homozygous genotype was found among subjects from the family-based recruitment (Table 1). The genotype distribution in the clinic-based cohort deviated significantly from Hardy-Weinberg equilibrium due to the enrichment of the homozygous delCNV (p = 0.019). In contrast, such deviation was not observed in the family-based cohort (Table 1). These observations suggest homozygous deficiency may have more severe clinical impact. While Hardy-Weinberg deviation could reflect genotyping error, this seems unlikely, since both cohorts were genotyped in parallel, and reliability of the delCNV genotyping protocol was independently confirmed by WES analysis.
Table 1.
Hardy-Weinberg equilibrium for PCDHA delCNV in BAV subjects
| Genotypea | Observed | Expected | Deviation |
|---|---|---|---|
| Clinic-based BAV recruitment | |||
| +/+ | 173 | 170 | 0.05 |
| +/Δ | 31 | 36.98 | 0.97 |
| Δ/Δ | 5 | 2.01 | 4.44 |
| Total | 209 | 209 | 5.46 |
| Hardy-Weinberg equilibrium p | 0.019 | ||
| Family-based BAV recruitment | |||
| +/+ | 56 | 57.09 | 0.02 |
| +/Δ | 18 | 15.81 | 0.30 |
| Δ/Δ | 0 | 1.09 | 1.09 |
| Total | 74 | 74 | 1.42 |
| Hardy-Weinberg equilibrium p | 0.234 | ||
Δ: 16.8 or 13.6 kb PCDHA delCNV.
Replication analysis in ancestry-matched white LVOTO cohort
To confirm findings in the discovery cohort, we conducted similar analysis using WES data from dbGAP for white LVOTO subjects from the PCGC and white subjects from the FHS as an independent control. Related individuals were removed using Plink32 (see Material and methods), and PCA was used to ancestry match the PCGC and FHS white subjects with the Pittsburgh LVOTO subjects. This resulted in the recovery of 381 PCGC LVOTO subjects and 1,352 FHS control subjects (Figure S3). Case-control analysis for the 13.6/16.8 kb delCNV, either together or individually, showed significant association of the 13.6 kb delCNV with LVOTO, with OR at 2.94 (95% CI, 1.44–6.04) (p = 0.0032), HLHS with OR at 4.05 (95% CI, 1.56–10.50) (p = 0.004), and BAV/CoA with OR 2.94 (95% CI, 1.06–8.12) (p = 0.038) (Figure 3A; Table S7). Further analysis with genotype comparisons showed significant association of the 13.6/16.8 kb Δ/Δ delCNV genotype with LVOTO with OR at 3.20 (95% CI, 1.20–8.50) (p = 0.020) and BAV with OR at 7.18 (95% CI,1.45–35.56) (p = 0.016) and the 16.8 kb Δ/Δ delCNV genotype with isolated BAV with OR at 7.84 (95% CI, 1.56–39.49) (p = 0.013) (Figure 3C; Table S7). Analysis for Δ/+ genotype showed significant association only for the 13.6 kb delCNV. This was observed for LVOTO with OR at 2.94 (95% CI, 1.44–6.04) (p = 0.0032), HLHS with OR at 4.05 (95% CI, 1.56–10.50) (p = 0.0041), and BAV/CoA with OR at 2.94 (95% CI, 1.06–8.12) (p = 0.038) (Figure 3C; Table S7). Overall, these findings support the association of the PCDHA delCNV with LVOTO seen in the white discovery cohort.
PCDHA delCNV in Black LVOTO cohort
PCA analysis yielded 24 Black subjects from PCGC and 2 from the Pittsburgh cohort (Figure S2). PCDHA delCNVs in these 26 Black LVOTO subjects were examined using 661 African individuals from the 1KG database as control subjects, given the FHS control subjects had few Black individuals. Consistent with prior study, the PCDHA delCNVs in the 1KG Black subjects were at much lower prevalence, with MAF of 0.06623 (Table S8). Without cardiac imaging to exclude BAV, this MAF is likely to be inflated. Among the 1KG control subjects, 12.3% had the 13.6/16.8 kb PCDHA delCNV, while the delCNVs were seen in 34.6% (9) of the Black LVOTO subjects (Table S9). This indicated significant allelic association, with OR at 2.81 (95% CI, 1.35–5.81) (p = 0.0055) (Figure 3A; Table S9). Heterozygous genotype association with LVOTO was also observed, with the 13.6/16.8 delCNVs yielding OR at 4.22 (95% CI, 1.81–9.84) (p = 8.7 × 10−4) and the 16.8 kb delCNV with OR at 2.88 (95% CI, 1.17–7.10) (p = 0.022). These findings suggest generalizability of the association of PCDHA delCNVs with LVOTO in the Black as well as white populations.
PCDHA delCNV association in Chinese LVOTO cohort
To further assess the broader role of the PCDHA delCNVs on LVOTO, we investigated their association with LVOTO in the Asian population. For this analysis, we recruited 104 Chinese individuals with LVOTO, including 88 pediatric and 16 adult individuals with BAV, and genotyped them for the two PCDHA delCNVs (Table S5). For comparison, 513 Chinese control subjects confirmed to have normal heart structure were recruited and genotyped for the two PCDHA delCNVs (Table S5). We found the PCDHA delCNVs at much lower prevalence in the Chinese subjects. We observed a combined MAF of only 0.019 for the two PCDHA delCNVs in the Chinese control subjects. This was elevated to 0.038 in the LVOTO subjects (Table S5). Significant association of the 13.6 kb PCDHA delCNV with LVOTO was observed, with OR at 4.43 (95% CI, 1.64–12.0) (p = 0.0033) and with HLHS at OR 7.45 (95% CI, 2.34–23.67) (p = 0.015) (Figure 3C; Table S9). For HLHS, significant allelic association was also observed for the 13.6/16.8 kb delCNV combined with OR at 3.55 (95% CI, 1.25–10.1) (p = 0.018) (Figure 3A; Table S9). Genotype association analysis showed similar findings for heterozygous genotype associations (Figure 3C; Table S9). Overall, these results confirm the PCDHA delCNVs are also significantly associated with LVOTO in the Chinese population.
PCDHA expression in the developing mouse aorta and aortic cushion
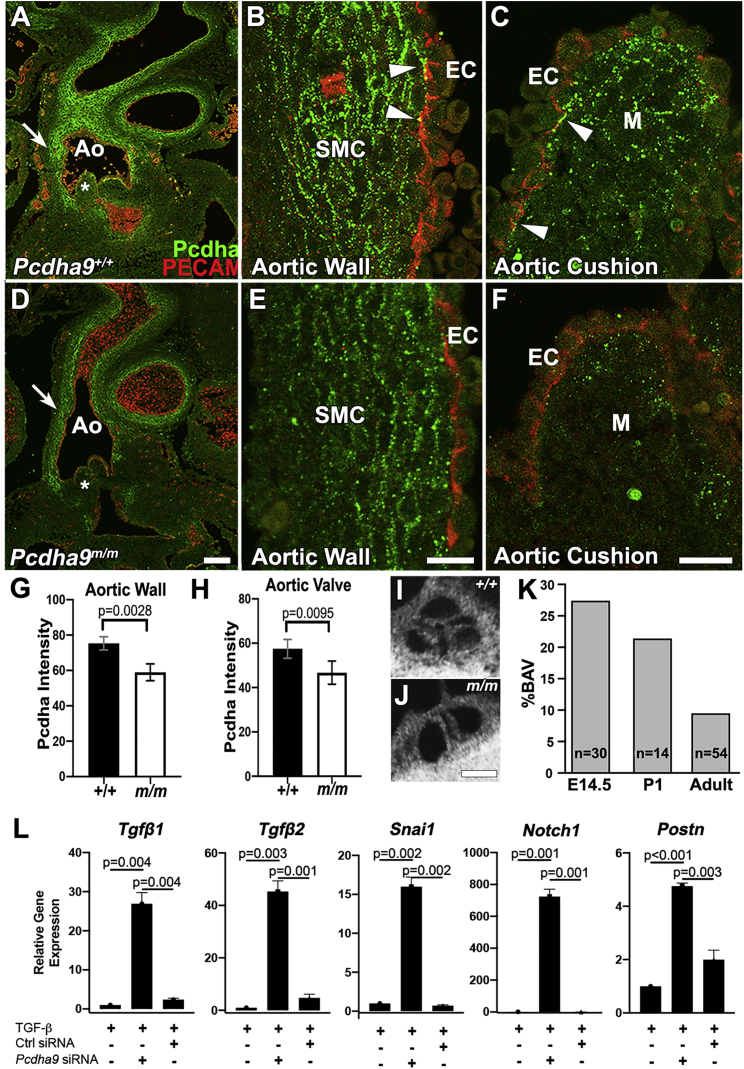
The role of protocadherins in heart development has not been examined previously, although their role in brain development is well described.45, 46, 47 Using an antibody to the constant region of the Pcdha cluster that detects protein expressed by all genes in the cluster (Figure 2A), we conducted immunostaining and confocal microscopy on sections of E14.5 wild-type mouse embryos, examining expression in the developing aorta and aortic cushions (Figure 4A).48 As expected, strong immunostaining was observed at regions of cell-cell contact. This was prominently seen along the intimal side of the aortic media (Figures 4A and 4B). Immunostaining was also observed in mesenchymal cells in the aortic cushions (Figure 3C). In contrast, little or no immunostaining was observed between endothelial cells, although occasional punctate staining was observed between aortic endothelial cells and adjacent vascular smooth muscle cells (see arrowheads in Figure 4B) or between endothelial cells of the cardiac cushions and underlying cushion mesenchyme (see arrowheads in Figure 4C).
Figure 4.
PCDHA protein expression in Pcdha9 mutant mice and knockdown of mouse Pcdha9
(A–C) Immunostaining and confocal imaging show PCDHA protein expression in the wild-type E14.5 mouse embryo aorta (A), with magnified views showing expression in the smooth muscle cells (SMC) on the intimal side (B), and also in mesenchyme (M) of the aortic cushion (C). Occasional staining also can be seen at the interface between the endothelial (PECAM positive) and underlying smooth muscle cells. Scale bar, 25 μm.
(D–F) Immunostaining and confocal imaging of PCDHA protein expression in E14.5 CRISPR targeted Pcdha9m/m mutant embryo showed reduced immunostaining, indicating reduced PCDHA protein expression in the aorta (D). Magnified views show reduced immunostaining both in the aorta wall (E) and the aortic cushion (F). Scale bar, 25 μm.
(G and H) Quantification of staining intensity showed reduced PCDHA protein expression in both the aorta wall (G) and the aortic cushion (H) of Pcdha9m/m embryos. The values shown are mean ± SD with statistical analysis conducted using t test. (I,J) Episcopic confocal microscopy images of a tricuspid aortic valve at P1 (I) and a bicuspid aortic valve at P1 (J). (K) Percent of Pcdha9 mutant animalsfound to have bicupsid aortic valve phenotype at E14.5, P1, and adult.
(L) Real-time PCR analysis shows upregulation of downstream EMT gene expression following mouse Pcdha9 siRNA treatment and TGFβ induction in mouse mammary gland cell line (NMuMg). Control scrambled siRNA treatment and blank controls show little or no expression of EMT genes.
Ao, aorta; EC, endothelial cells; +/+, wild-type; m/m, Pcdha9.
To determine the impact of the Pcdha9 mutation on PCDHA protein expression, similar confocal imaging analysis was conducted on aortic sections from homozygous Pcdha9 mutant embryos carrying a functional null Pcdha9 frameshift mutation.21 These mutant embryos showed an identical pattern of PCDHA protein localization as in the wild-type embryos, but staining intensity was significantly reduced in both the aortic media and the aortic cushion mesenchyme (Figures 4D and 4E). Immunostaining between the endothelial cells and underlying smooth muscle cells or cushion mesenchyme were markedly reduced or absent (Figures 4D and 4E). These findings suggest PCDHA deficiency in the aorta and aortic cushion can contribute to BAV and LVOTO phenotypes in the Ohia mutant mice.
Reduced penetrance of BAV in Pcdha9 mutant mice
We investigated penetrance of the BAV phenotype in the Pcdha9m/m mice, as familial studies have shown reduced penetrance for BAV and other LVOTO phenotypes. Analysis of E14.5 embryos showed over 25% of the Pcdha9m/m mutant mice have BAV, but this declined to approximately 20% in mutant mice found at postnatal day 1 (PN1) and was further reduced to 10% in adult Pcdha9m/m mice (Figures 3I–3K). This suggests mutant mice with BAV die neonatally/postnatally; whether this is due to complication of aortic valve disease or aortopathy or both is unknown. Given that abnormal epithelial-mesenchymal transition (EMT) has been observed in the aortic intima and media of individuals with BAV, and defects in EMT can cause cardiac valve abnormalities, we investigated whether Pcdha9 deficiency may promote EMT.49, 50, 51, 52 For this analysis, we conducted Pcdha9 siRNA-mediated gene knockdown in a well-characterized in vitro model of TGFβ-induced EMT in the mouse mammary epithelial cell line NMuMG.53 Pcdha9 gene knockdown in NMuMG cells caused marked induction of multiple EMT-associated genes, such as Tgfb1, Tgfb2, Snail1, Notch1, and Postn (Figure 3L). These findings suggest protocadherin deficiency can perturb EMT and thus potentially contribute to defects in aortic valve development and/or in the adult onset of aortopathies.
PCDHA expression in human aorta
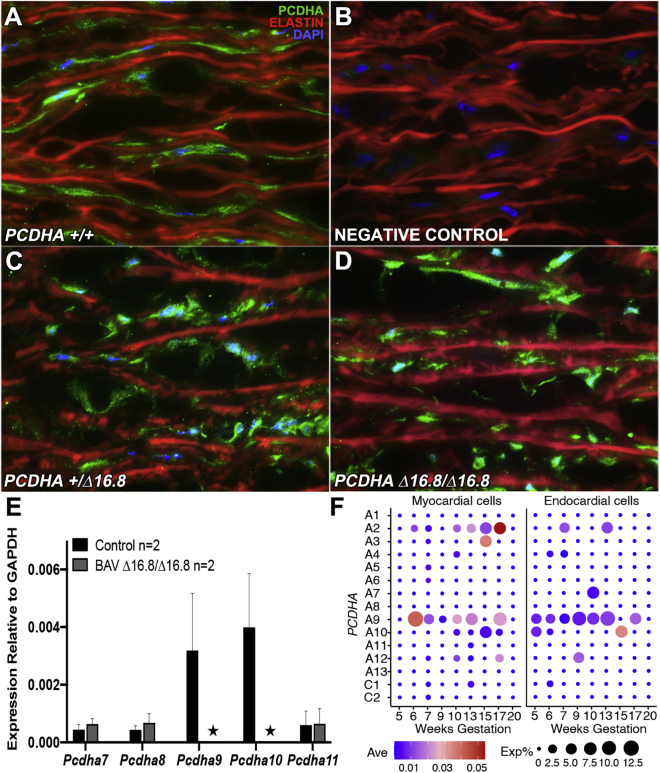
We further examined PCDHA expression in human aortic tissue with confocal microscopy using the same PCDHA constant region antibody. This analysis included tissue from individuals that are wild type and individuals with BAV who are heterozygous or homozygous for the 16.8 kb PCDHA delCNV. Staining was observed in smooth muscle cells in the aortic media, but the level of immunostaining was highly variable, with no consistent correlation with genotypes (Figures 5A–5D). Given the antibody cannot discriminate between different genes within the PCDHA gene cluster, we also conducted real-time PCR analysis with RNA extracted from aorta tissue to assess PCDHA transcript expression. In wild-type individuals without the delCNV, transcript expression was observed for PCDHA9 and 10, but not for the flanking genes PCDHA7, 8, and 11 (Figure 5E). In contrast, in individuals with BAV homozygous for the 16.8 kb delCNV (Figure 5E), few or no transcripts were observed for any of these genes, including the three within the delCNV—PCDHA 8, 9, and 10 (Figure 5E). Using publicly available human single-cell RNaseq data, we examined PCDHA expression in human embryonic heart tissue spanning 5–20 weeks gestation. This analysis showed PCDHA9 as the predominant gene expressed from the PCDHA gene cluster (Figure 4F). This was observed both in cardiomyocytes and in endocardial cells of the developing human embryonic heart (Figure 5F).41 While comparable scRNaseq data are not available for the human aorta or aortic valves (3′ sequencing used for most scRNaseq precludes PCDHA gene identification), it is significant to note that upregulated expression of PCDHA9 has been documented in the human fetal aortic valve by bulk RNA sequencing analysis.54 Together, these findings suggest PCDHA9, the predominant PCDHA gene expressed in the embryonic human aorta, is also expressed in the adult aorta.
Figure 5.
PCDHA transcript and protein expression in human aortic tissue
(A–D) Immunostaining and confocal imaging using an antibody to detect the constant region of the PCDHA gene cluster showed evidence of PCDHA protein expression in smooth muscle cells of the human aorta of BAV subjects with wild-type (+/+), heterozygous (+/Δ16.8), and homozygous PCDHA 16.8 kb deletion (Δ16.8/Δ16.8). No consistent differences were observed in the intensity or pattern of immunostaining with genotype. Elastin, red; PCDHA, green; DAPI, blue.
(E) Real time PCR analysis was carried out to examine PCDHA7-11 expression in aortic tissue from 2 control subjects without the delCNV and 2 BAV subjects homozygous for the 16.8 kb PCDHA delCNV (Δ/Δ). In the control subjects, transcripts were detected for PCDHA9 and 10, but in BAV subjects homozygous for the 16.8 kb delCNV, neither PCDHA9 nor 10 transcripts were detected.The values shown are mean ± SD.
(F) Dot plot showing transcript expression of the PCDHA gene cluster in heart tissue of human fetuses 5 to 20 weeks gestation from single-cell RNA-seq data of Cui et al.41 The size of the circle corresponds to the percentage of cells expressing the indicated gene (Exp%) and the color represents the average expression values (Ave).
Discussion
We found two common delCNVs spanning the PCDHA gene cluster that are significantly associated with LVOTO, including BAV, CoA, and HLHS. This was observed in a large white cohort. Smaller Black and Chinese cohorts also showed trends for similar enrichment. These delCNVs were most prevalent in white individuals, less common in Black individuals and least common in Chinese individuals, similar to the racial prevalence for BAV. PCDHA was found to be highly expressed in the aortic media and cushion mesenchyme of mouse embryos. This expression was reduced in the Pcdha mutant mice, suggesting PCDHA deficiency or haploinsufficiency can contribute to maldevelopment of the aorta/aortic valve. Incomplete penetrance was observed in both the Pcdha mutant mice and human subjects with the PCDHA delCNV. Together, these findings demonstrate common variants can contribute to the reduced penetrance and variable expressivity that characterize the complex genetics of rare LVOTO CHD.
In the discovery white cohort, the 13.6/16.8 kb delCNV combined, and the 13.6 kb or 16.8 kb delCNV independently, showed significant association with LVOTO. For the PCGC white cohort, only the rarer 13.6 kb delCNV showed significant association with LVOTO, while for the Chinese cohort, only the more common 16.8 kb delCNV showed significance. The significant association of the 13.6 kb delCNV in the smaller PCGC cohort likely reflects the increased statistical power associated with analysis of the rarer 13.6 kb delCNV. We note as the Framingham cohort used as control subjects in the PCGC analysis were not screened to exclude subjects with BAV, the background prevalence of BAV in these control subjects also may have reduced power.
Generalizability of PCDHA delCNV association with LVOTO
In the Chinese cohort, the very low prevalence of the PCDHA delCNVs overall may account for the significant association observed only for the more common 16.8 kb delCNV. In the Black cohort, the prevalence of the delCNV is intermediate between that of the white and Chinese cohorts, and only the combined 13.6/16.8 kb delCNV showed significant allelic association with LVOTO. However, the small sample size of the Black cohort is a major limitation and points to the need for more diversification in subject recruitment. Nevertheless, these findings support generalizability of the association of PCDHA delCNV with LVOTO.
Role of the 13.6 kb and 16.8 kb PCDHA delCNV in LVOTO and aortopathy
Our findings indicate the 13.6 and 16.8 kb PCDHA delCNVs both contribute to LVOTO and may have equivalent roles in LVOTO pathogenesis. These two delCNVs share the common deletion of coding exons for PCDHA8 and 9, but only PCDHA9 is expressed in the human embryonic heart at the time of aorta/aortic valve formation.55 Hence, it may be PCDHA9 deficiency that contributes to LVOTO. However, in the adult human aorta, expression of both PCDHA9 and 10 are observed. While both genes are deleted in the 16.8 kb delCNV, PCDHA10 is unaffected by the 13.6 kb delCNV. This suggests only the 16.8 kb delCNV may contribute to aortopathy risk. Interestingly, in the clinic-based recruitment, all 5 individuals with the Δ/Δ delCNV genotype were homozygous for the 16.8 kb delCNV, three of whom had aneurysm.
Role of PCDHA delCNV in common and rare LVOTOs
We observed the PCDHA delCNVs in 21% of BAV and 20.5% of HLHS subjects in the white discovery cohort, showing these delCNVs are significant genetic factors contributing to one of the most common (BAV) and also one of the rarer (HLHS) CHDs. This somewhat puzzling finding can be explained by a multigenic model of disease suggested by observations in the Ohia mutant mice and in human clinical findings.9,21 In Ohia mutants, the Pcdha9 mutation alone can cause BAV, while in conjunction with a second mutation HLHS can arise.21 Hence, the common PCDHA delCNV can account for the high prevalence of BAV, while the requirement for a second hit may explain HLHS being rarer. Together, these findings suggest the genetic architecture of LVOTO may involve a role for the PCDHA delCNVs in mediating the common and rarer LVOTO phenotypes.
PCDHA delCNV association with coarctation
The PCDHA delCNV showed the highest OR for association with isolated CoA, with 50% of 22 individuals with CoA in the discovery cohort harboring the delCNV. However, this finding was not replicated in the PCGC cohort, indicating further studies are needed with larger number of CoA subjects. We note a recent Icelandic population study reported significant association of a rare missense variant in MYH6 with isolated CoA. This missense variant was observed in 20% of 39 individuals with CoA, but penetrance was very low, with only one CoA individual noted per 123 carriers.20 While the PCDHA delCNVs also show incomplete penetrance for LVOTO, penetrance is likely much higher than the MYH6 variant if we assume 1% incidence of BAV and 10% prevalence for the delCNV.
Protocadherin and the developmental etiology of BAV and aortopathy
Our analysis in mouse embryos showed abundant PCDHA expression in the aortic smooth muscle cells of neural crest origin, a cell lineage derived from the neural tube with abundant protocadherin expression.56 PCDHA expression was also observed in the aortic endothelial cells and cushion mesenchyme. In the Pcdha9 mutant mouse embryos, significant reduction of the PCDHA protein expression was observed in the aortic cushion mesenchyme and smooth muscle cells in the aortic media. As endocardial EMT generates the cushion mesenchyme that forms the cardiac valve leaflets, dysregulated EMT could impact formation of the aortic valve leaflets. Supporting a role for PCDHA regulation of EMT, we showed abnormal activation of EMT-related marker gene expression upon Pcdha9 gene knockdown in a well-described in vitro model of EMT. These findings suggest enhanced EMT from PCDHA deficiency can contribute to abnormal valvular morphogenesis and the emergence of bicuspid aortic valves.
Additionally, such dysregulated EMT in the aortic media could contribute to aortopathy with aorta dilation and risk of aortic aneurysm. Thus, several studies have reported the aortic intima and media of individuals with BAV have a maturation defect associated with an abnormal mesenchymal state.49,50,57 Also notable is the report that pediatric individuals with BAV can exhibit aorta dilation early in life, even during fetal development, supporting a developmental etiology for BAV-associated aortopathy.58,59 Consistent with this, various studies have shown aortopathy can occur independent of hemodynamic disturbances (Dumitrascu-Biris et al., British Congenital Cardiac Association Annual Meeting, 2019).58,60,61 Nevertheless, a study of pediatric individuals with BAV showed aortic stenosis and aortic insufficiency are independently associated with aorta dilation, suggesting the involvement of hemodynamic factors.58 Based on our findings, we propose PCDHA deficiency may contribute to aortopathy risk via the disruption of cell-cell adhesion and enhanced EMT, processes that could be exacerbated by altered hemodynamics.
Epigenetic regulation of the PCDHA gene cluster
The cause for the late age onset of aortopathy is not well understood, and without mechanistic insight, it has not been possible to predict which individuals with BAV will require valve replacement or aorta surgery. In this regard, it is interesting to note that the PCDHA gene cluster is well described to be epigenetically regulated by DNA methylation. As methylation of the PCDHA gene cluster is observed to increase with advancing age, this could contribute to the late age onset of aortopathy.62, 63, 64 A role for epigenetics in LVOTO is supported by observations in Williams-Beuren syndrome (WBS [MIM: 609757]), a genetic disorder characterized by ∼75% incidence of CHD that includes BAV and aortic stenosis.65,66 WBS subjects show epigenetic disturbance with increased DNA methylation, including hypermethylation of PCDHA9, the predominant gene expressed in the aorta and during human heart development.65 Thus, epigenetic modulation of the PCDHA locus also might contribute to the reduced penetrance of LVOTO and impact the risk of aortopathy.
Common variants as disease-causing genes
Our pursuit of the possible role of common variants in the genetic etiology of CHD is informed by our mouse models showing Pcdha deficiency can cause BAV/LVOTO. Unlike common variants identified in GWAS, the common variants recovered in our study are likely pathogenic as a result of reduced protein expression, as seen in the Pcdha mutant mice. It should be noted that our study was not designed as a genome-wide study and does not surpass traditional genome-wide thresholds for significance. Rather, our study demonstrates the value of leveraging animal models to inform genetic studies in the clinical setting, where the larger sample sizes required for genome-wide analysis are often difficult or virtually impossible to acquire.
Interrogating the possible role of common variants in human disease is also inherently challenging. As such variants are not likely to be under strong selection, it is difficult to establish proper filtering criteria to distinguish signal from noise. Indeed, the loss-of-function intolerance index often used to assess the functional importance of gene loss predicted PCDHA gene loss would have little or no functional impact.67,68 However, this index has been shown to be uninformative for genes whose essential function does not impact reproductive fitness, as is the case for BAV, the most common LVOTO.68 Also, possibly confounding analysis of common variants is the potential impact of ancestry on variant prevalence. We addressed this by using regional control subjects in the discovery cohort analysis and using WES data available for a small subset of the white discovery cohort to further ancestry match the white replication LVOTO (PCGC) and control (FHS) cohorts.
Conclusions
The combined mouse and human studies provide strong evidence that common variants in a disease-causing gene can contribute to the genetic etiology of LVOTO. These studies also demonstrate a role for the PCDHA gene cluster in BAV and LVOTO. Our studies in mice suggest BAV and the risk of aortopathy associated with PCDHA deficiency may arise from the disturbance of cell-cell adhesion and enhanced EMT. The possible additional involvement of PCDHA methylation silencing may warrant further investigation.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
We are indebted to all family members for their research participation and contribution to this project. We acknowledge Tara Paton from the Centre for Applied Genomics at the Hospital for Sick Children for assistance with PCDHA9 genotyping of Canadian samples. This work was supported by NIH grants HL132024, HL142788 (C.W.L), HL109132 (T.G.G.), F30HD097967 (G.C.G.), and R01 HL109759-A1 and Nationwide Children’s Hospital Foundation (K.L.M.), Ted Rogers Centre for Heart Research (S.M.), and Heart and Stroke Foundation of Canada/Robert M. Freedom Chair (S.M.). Additional support was provided by start-up funds to P.T. from the Chinese University of Hong Kong.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2021.100037.
Data and code availability
Sequencing coverage for PCDHA9 in the Pittsburgh cohort and in the region of the PCDHA delCNV for the Anzhen Hospital cohort are available under SRA accession numbers BioProject: PRJNA632119 and GSA: PRJCA002663, respectively. WES data from the PCGC and the FHS are accessible in NCBI dbGAP database under accession numbers dbGAP: phs001194.v2.p2 and phs000007.v30.p11, respectively. Single-cell RNA-seq data of human embryonic hearts are available from GEO database under accession number GEO: GSE106118.
Web resources
BioProject, https://www.ncbi.nlm.nih.gov/bioproject/
OMIM, https://www.omim.org
1000 genome project, ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp//data_collections/1000G_2504_high_coverage/working
Supplemental information
References
- 1.Siu S.C., Silversides C.K. Bicuspid aortic valve disease. J. Am. Coll. Cardiol. 2010;55:2789–2800. doi: 10.1016/j.jacc.2009.12.068. [DOI] [PubMed] [Google Scholar]
- 2.Wu B., Wang Y., Xiao F., Butcher J.T., Yutzey K.E., Zhou B. Developmental Mechanisms of Aortic Valve Malformation and Disease. Annu. Rev. Physiol. 2017;79:21–41. doi: 10.1146/annurev-physiol-022516-034001. [DOI] [PubMed] [Google Scholar]
- 3.Borger M.A., Fedak P.W.M., Stephens E.H., Gleason T.G., Girdauskas E., Ikonomidis J.S., Khoynezhad A., Siu S.C., Verma S., Hope M.D., et al. The American Association for Thoracic Surgery consensus guidelines on bicuspid aortic valve-related aortopathy: Full online-only version. J. Thorac. Cardiovasc. Surg. 2018;156:e41–e74. doi: 10.1016/j.jtcvs.2018.02.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michelena H.I., Khanna A.D., Mahoney D., Margaryan E., Topilsky Y., Suri R.M., Eidem B., Edwards W.D., Sundt T.M., 3rd, Enriquez-Sarano M. Incidence of aortic complications in patients with bicuspid aortic valves. JAMA. 2011;306:1104–1112. doi: 10.1001/jama.2011.1286. [DOI] [PubMed] [Google Scholar]
- 5.Yassine N.M., Shahram J.T., Body S.C. Pathogenic mechanisms of bicuspid aortic valve aortopathy. Front. Physiol. 2017;8:687. doi: 10.3389/fphys.2017.00687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duran A.C., Frescura C., Sans-Coma V., Angelini A., Basso C., Thiene G. Bicuspid aortic valves in hearts with other congenital heart disease. J. Heart Valve Dis. 1995;4:581–590. [PubMed] [Google Scholar]
- 7.Lewin M.B., McBride K.L., Pignatelli R., Fernbach S., Combes A., Menesses A., Lam W., Bezold L.I., Kaplan N., Towbin J.A., Belmont J.W. Echocardiographic evaluation of asymptomatic parental and sibling cardiovascular anomalies associated with congenital left ventricular outflow tract lesions. Pediatrics. 2004;114:691–696. doi: 10.1542/peds.2003-0782-L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cripe L., Andelfinger G., Martin L.J., Shooner K., Benson D.W. Bicuspid aortic valve is heritable. J. Am. Coll. Cardiol. 2004;44:138–143. doi: 10.1016/j.jacc.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 9.McBride K.L., Pignatelli R., Lewin M., Ho T., Fernbach S., Menesses A., Lam W., Leal S.M., Kaplan N., Schliekelman P., et al. Inheritance analysis of congenital left ventricular outflow tract obstruction malformations: Segregation, multiplex relative risk, and heritability. Am. J. Med. Genet. A. 2005;134A:180–186. doi: 10.1002/ajmg.a.30602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y., Chen S., Zühlke L., Black G.C., Choy M.-K., Li N., Keavney B.D. Global birth prevalence of congenital heart defects 1970-2017: updated systematic review and meta-analysis of 260 studies. Int. J. Epidemiol. 2019;48:455–463. doi: 10.1093/ije/dyz009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benson D.W., Martin L.J., Lo C.W. Genetics of hypoplastic left heart syndrome. J. Pediatr. 2016;173:25–31. doi: 10.1016/j.jpeds.2016.02.052. [DOI] [PubMed] [Google Scholar]
- 12.Hinton R.B., Martin L.J., Rame-Gowda S., Tabangin M.E., Cripe L.H., Benson D.W. Hypoplastic left heart syndrome links to chromosomes 10q and 6q and is genetically related to bicuspid aortic valve. J. Am. Coll. Cardiol. 2009;53:1065–1071. doi: 10.1016/j.jacc.2008.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin L.J., Ramachandran V., Cripe L.H., Hinton R.B., Andelfinger G., Tabangin M., Shooner K., Keddache M., Benson D.W. Evidence in favor of linkage to human chromosomal regions 18q, 5q and 13q for bicuspid aortic valve and associated cardiovascular malformations. Hum. Genet. 2007;121:275–284. doi: 10.1007/s00439-006-0316-9. [DOI] [PubMed] [Google Scholar]
- 14.McBride K.L., Zender G.A., Fitzgerald-Butt S.M., Koehler D., Menesses-Diaz A., Fernbach S., Lee K., Towbin J.A., Leal S., Belmont J.W. Linkage analysis of left ventricular outflow tract malformations (aortic valve stenosis, coarctation of the aorta, and hypoplastic left heart syndrome) Eur. J. Hum. Genet. 2009;17:811–819. doi: 10.1038/ejhg.2008.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garg V., Muth A.N., Ransom J.F., Schluterman M.K., Barnes R., King I.N., Grossfeld P.D., Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 16.Bonachea E.M., Chang S.-W., Zender G., LaHaye S., Fitzgerald-Butt S., McBride K.L., Garg V. Rare GATA5 sequence variants identified in individuals with bicuspid aortic valve. Pediatr. Res. 2014;76:211–216. doi: 10.1038/pr.2014.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gould R.A., Aziz H., Woods C.E., Seman-Senderos M.A., Sparks E., Preuss C., Wünnemann F., Bedja D., Moats C.R., McClymont S.A., et al. Baylor-Hopkins Center for Mendelian Genomics. MIBAVA Leducq Consortium ROBO4 variants predispose individuals to bicuspid aortic valve and thoracic aortic aneurysm. Nat. Genet. 2019;51:42–50. doi: 10.1038/s41588-018-0265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKellar S.H., Tester D.J., Yagubyan M., Majumdar R., Ackerman M.J., Sundt T.M., 3rd Novel NOTCH1 mutations in patients with bicuspid aortic valve disease and thoracic aortic aneurysms. J. Thorac. Cardiovasc. Surg. 2007;134:290–296. doi: 10.1016/j.jtcvs.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 19.Tan H.L., Glen E., Töpf A., Hall D., O’Sullivan J.J., Sneddon L., Wren C., Avery P., Lewis R.J., ten Dijke P., et al. Nonsynonymous variants in the SMAD6 gene predispose to congenital cardiovascular malformation. Hum. Mutat. 2012;33:720–727. doi: 10.1002/humu.22030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bjornsson T., Thorolfsdottir R.B., Sveinbjornsson G., Sulem P., Norddahl G.L., Helgadottir A., Gretarsdottir S., Magnusdottir A., Danielsen R., Sigurdsson E.L., et al. A rare missense mutation in MYH6 associates with non-syndromic coarctation of the aorta. Eur. Heart J. 2018;39:3243–3249. doi: 10.1093/eurheartj/ehy142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X., Yagi H., Saeed S., Bais A.S., Gabriel G.C., Chen Z., Peterson K.A., Li Y., Schwartz M.C., Reynolds W.T., et al. The complex genetics of hypoplastic left heart syndrome. Nat. Genet. 2017;49:1152–1159. doi: 10.1038/ng.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yagi H., Liu X., Gabriel G.C., Wu Y., Peterson K., Murray S.A., Aronow B.J., Martin L.J., Benson D.W., Lo C.W. The Genetic Landscape of Hypoplastic Left Heart Syndrome. Pediatr. Cardiol. 2018;39:1069–1081. doi: 10.1007/s00246-018-1861-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noonan J.P., Li J., Nguyen L., Caoile C., Dickson M., Grimwood J., Schmutz J., Feldman M.W., Myers R.M. Extensive linkage disequilibrium, a common 16.7-kilobase deletion, and evidence of balancing selection in the human protocadherin α cluster. Am. J. Hum. Genet. 2003;72:621–635. doi: 10.1086/368060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandra S., Lang R.M., Nicolarsen J., Gayat E., Spencer K.T., Mor-Avi V., Hofmann Bowman M.A. Bicuspid aortic valve: inter-racial difference in frequency and aortic dimensions. JACC Cardiovasc. Imaging. 2012;5:981–989. doi: 10.1016/j.jcmg.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Wei X., Zhao Z., Liao Y., He J., Xiong T., Xu Y., Lv W., Ou Y., Tang H., et al. Prevalence and complications of bicuspid aortic valve in Chinese according to echocardiographic database. Am. J. Cardiol. 2017;120:287–291. doi: 10.1016/j.amjcard.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 26.Gelb B., Brueckner M., Chung W., Goldmuntz E., Kaltman J., Kaski J.P., Kim R., Kline J., Mercer-Rosa L., Porter G., et al. Pediatric Cardiac Genomics Consortium The Congenital Heart Disease Genetic Network Study: rationale, design, and early results. Circ. Res. 2013;112:698–706. doi: 10.1161/CIRCRESAHA.111.300297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Homsy J., Zaidi S., Shen Y., Ware J.S., Samocha K.E., Karczewski K.J., DePalma S.R., McKean D., Wakimoto H., Gorham J., et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsao C.W., Vasan R.S. Cohort Profile: The Framingham Heart Study (FHS): overview of milestones in cardiovascular epidemiology. Int. J. Epidemiol. 2015;44:1800–1813. doi: 10.1093/ije/dyv337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poplin R., Ruano-Rubio V., DePristo M.A., Fennell T.J., Carneiro M.O., Van der Auwera G.A., Kling D.E., Gauthier L.D., Levy-Moonshine A., Roazen D., et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv. 2017 doi: 10.1101/201178. [DOI] [Google Scholar]
- 30.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 https://arxiv.org/abs/1303.3997 arXiv:1303.3997. [Google Scholar]
- 31.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., et al. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo Y., de Lange K.M., Jostins L., Moutsianas L., Randall J., Kennedy N.A., Lamb C.A., McCarthy S., Ahmad T., Edwards C., et al. Exploring the genetic architecture of inflammatory bowel disease by whole-genome sequencing identifies association at ADCY7. Nat. Genet. 2017;49:186–192. doi: 10.1038/ng.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 37.Patterson N., Price A.L., Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X., Jian X., Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat. 2013;34:E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cui Y., Zheng Y., Liu X., Yan L., Fan X., Yong J., Hu Y., Dong J., Li Q., Wu X., et al. Single-Cell Transcriptome Analysis Maps the Developmental Track of the Human Heart. Cell Rep. 2019;26:1934–1950. doi: 10.1016/j.celrep.2019.01.079. e5. [DOI] [PubMed] [Google Scholar]
- 42.Ripley B.D. Springer; 2002. Modern applied statistics with S. [Google Scholar]
- 43.Wu Q., Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97:779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- 44.Miller-Hance W.C., Tacy T.A. Gender differences in pediatric cardiac surgery: the cardiologist’s perspective. J. Thorac. Cardiovasc. Surg. 2004;128:7–10. doi: 10.1016/j.jtcvs.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 45.Canzio D., Maniatis T. The generation of a protocadherin cell-surface recognition code for neural circuit assembly. Curr. Opin. Neurobiol. 2019;59:213–220. doi: 10.1016/j.conb.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Esumi S., Kakazu N., Taguchi Y., Hirayama T., Sasaki A., Hirabayashi T., Koide T., Kitsukawa T., Hamada S., Yagi T. Monoallelic yet combinatorial expression of variable exons of the protocadherin-α gene cluster in single neurons. Nat. Genet. 2005;37:171–176. doi: 10.1038/ng1500. [DOI] [PubMed] [Google Scholar]
- 47.Chen W.V., Maniatis T. Clustered protocadherins. Development. 2013;140:3297–3302. doi: 10.1242/dev.090621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ing-Esteves S., Kostadinov D., Marocha J., Sing A.D., Joseph K.S., Laboulaye M.A., Sanes J.R., Lefebvre J.L. Combinatorial effects of alpha-and gamma-protocadherins on neuronal survival and dendritic self-avoidance. J. Neurosci. 2018;38:2713–2729. doi: 10.1523/JNEUROSCI.3035-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maleki S., Kjellqvist S., Paloschi V., Magné J., Branca R.M., Du L., Hultenby K., Petrini J., Fuxe J., Lehtiö J., et al. MIBAVA Leducq Consortium Mesenchymal state of intimal cells may explain higher propensity to ascending aortic aneurysm in bicuspid aortic valves. Sci. Rep. 2016;6:35712. doi: 10.1038/srep35712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maleki S., Poujade F.A., Bergman O., Gådin J.R., Simon N., Lång K., Franco-Cereceda A., Body S.C., Björck H.M., Eriksson P. Endothelial/Epithelial Mesenchymal Transition in Ascending Aortas of Patients With Bicuspid Aortic Valve. Front. Cardiovasc. Med. 2019;6:182. doi: 10.3389/fcvm.2019.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fischer A., Steidl C., Wagner T.U., Lang E., Jakob P.M., Friedl P., Knobeloch K.-P., Gessler M. Combined loss of Hey1 and HeyL causes congenital heart defects because of impaired epithelial to mesenchymal transition. Circ. Res. 2007;100:856–863. doi: 10.1161/01.RES.0000260913.95642.3b. [DOI] [PubMed] [Google Scholar]
- 52.Kostina A.S., Uspensky V.Е., Irtyuga O.B., Ignatieva E.V., Freylikhman O., Gavriliuk N.D., Moiseeva O.M., Zhuk S., Tomilin A., Kostareva А.А., Malashicheva A.B. Notch-dependent EMT is attenuated in patients with aortic aneurysm and bicuspid aortic valve. Biochim. Biophys. Acta. 2016;1862:733–740. doi: 10.1016/j.bbadis.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J., Tian X.-J., Zhang H., Teng Y., Li R., Bai F., Elankumaran S., Xing J. TGF-β-induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Sci. Signal. 2014;7:ra91. doi: 10.1126/scisignal.2005304. [DOI] [PubMed] [Google Scholar]
- 54.Gottlieb Sen D., Halu A., Razzaque A., Gorham J.M., Hartnett J., Seidman J.G., Aikawa E., Seidman C.E. The Transcriptional Signature of Growth in Human Fetal Aortic Valve Development. Ann. Thorac. Surg. 2018;106:1834–1840. doi: 10.1016/j.athoracsur.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 55.Dhanantwari P., Lee E., Krishnan A., Samtani R., Yamada S., Anderson S., Lockett E., Donofrio M., Shiota K., Leatherbury L., Lo C.W. Human cardiac development in the first trimester: a high-resolution magnetic resonance imaging and episcopic fluorescence image capture atlas. Circulation. 2009;120:343–351. doi: 10.1161/CIRCULATIONAHA.108.796698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sawada H., Rateri D.L., Moorleghen J.J., Majesky M.W., Daugherty A. Smooth muscle cells derived from second heart field and cardiac neural crest reside in spatially distinct domains in the media of the ascending aorta—brief report. Arterioscler. Thromb. Vasc. Biol. 2017;37:1722–1726. doi: 10.1161/ATVBAHA.117.309599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grewal N., Gittenberger-de Groot A.C., Poelmann R.E., Klautz R.J., Lindeman J.H., Goumans M.-J., Palmen M., Mohamed S.A., Sievers H.-H., Bogers A.J., DeRuiter M.C. Ascending aorta dilation in association with bicuspid aortic valve: a maturation defect of the aortic wall. J. Thorac. Cardiovasc. Surg. 2014;148:1583–1590. doi: 10.1016/j.jtcvs.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 58.Grattan M., Prince A., Rumman R.K., Morgan C., Petrovic M., Hauck A., Young L., Franco-Cereceda A., Loeys B., Mohamed S.A., et al. Predictors of Bicuspid Aortic Valve-Associated Aortopathy in Childhood: A Report From the MIBAVA Consortium. Circ. Cardiovasc. Imaging. 2020;13:e009717. doi: 10.1161/CIRCIMAGING.119.009717. [DOI] [PubMed] [Google Scholar]
- 59.Simpson J.M., Pushparajah K. Dilatation of the Aorta in Bicuspid Aortic Valve Disease. Circ. Cardiovasc. Imaging. 2020;13:e010448. doi: 10.1161/CIRCIMAGING.120.010448. [DOI] [PubMed] [Google Scholar]
- 60.Yasuda H., Nakatani S., Stugaard M., Tsujita-Kuroda Y., Bando K., Kobayashi J., Yamagishi M., Kitakaze M., Kitamura S., Miyatake K. Failure to prevent progressive dilation of ascending aorta by aortic valve replacement in patients with bicuspid aortic valve: comparison with tricuspid aortic valve. Circulation. 2003;108(Suppl 1):II291–II294. doi: 10.1161/01.cir.0000087449.03964.fb. [DOI] [PubMed] [Google Scholar]
- 61.Grewal N., Girdauskas E., DeRuiter M., Goumans M.J., Poelmann R.E., Klautz R.J.M., Gittenberger-de Groot A.C. The role of hemodynamics in bicuspid aortopathy: a histopathologic study. Cardiovasc. Pathol. 2019;41:29–37. doi: 10.1016/j.carpath.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 62.Toyoda S., Kawaguchi M., Kobayashi T., Tarusawa E., Toyama T., Okano M., Oda M., Nakauchi H., Yoshimura Y., Sanbo M., et al. Developmental epigenetic modification regulates stochastic expression of clustered protocadherin genes, generating single neuron diversity. Neuron. 2014;82:94–108. doi: 10.1016/j.neuron.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 63.Lomvardas S., Maniatis T. Histone and DNA Modifications as Regulators of Neuronal Development and Function. Cold Spring Harb. Perspect. Biol. 2016;8:a024208. doi: 10.1101/cshperspect.a024208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim S., Wyckoff J., Morris A.-T., Succop A., Avery A., Duncan G.E., Jazwinski S.M. DNA methylation associated with healthy aging of elderly twins. Geroscience. 2018;40:469–484. doi: 10.1007/s11357-018-0040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strong E., Butcher D.T., Singhania R., Mervis C.B., Morris C.A., De Carvalho D., Weksberg R., Osborne L.R. Symmetrical dose-dependent DNA-methylation profiles in children with deletion or duplication of 7q11.23. Am. J. Hum. Genet. 2015;97:216–227. doi: 10.1016/j.ajhg.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Collins R.T., 2nd Cardiovascular disease in Williams syndrome. Circulation. 2013;127:2125–2134. doi: 10.1161/CIRCULATIONAHA.112.000064. [DOI] [PubMed] [Google Scholar]
- 67.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ziegler A., Colin E., Goudenège D., Bonneau D. A snapshot of some pLI score pitfalls. Hum. Mutat. 2019;40:839–841. doi: 10.1002/humu.23763. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing coverage for PCDHA9 in the Pittsburgh cohort and in the region of the PCDHA delCNV for the Anzhen Hospital cohort are available under SRA accession numbers BioProject: PRJNA632119 and GSA: PRJCA002663, respectively. WES data from the PCGC and the FHS are accessible in NCBI dbGAP database under accession numbers dbGAP: phs001194.v2.p2 and phs000007.v30.p11, respectively. Single-cell RNA-seq data of human embryonic hearts are available from GEO database under accession number GEO: GSE106118.