SUMMARY
For precise motor control, distinct subpopulations of corticospinal neurons (CSN) must extend axons to distinct spinal segments, from proximal targets in the brainstem and cervical cord to distal targets in thoracic and lumbar spinal segments. We find that developing CSN subpopulations exhibit striking axon targeting specificity in spinal white matter, which establishes the foundation for durable specificity of adult corticospinal circuitry. Employing developmental retrograde and anterograde labeling, and their distinct neocortical locations, we purified developing CSN subpopulations using fluorescence-activated cell sorting to identify genes differentially expressed between bulbar-cervical and thoracolumbar-projecting CSN subpopulations at critical developmental times. These segmentally distinct CSN subpopulations are molecularly distinct from the earliest stages of axon extension, enabling prospective identification even before eventual axon targeting decisions are evident in the spinal cord. This molecular delineation extends beyond simple spatial separation of these subpopulations in the cortex. Together, these results identify candidate molecular controls over segmentally specific corticospinal axon projection targeting.
Graphical Abstract
In brief
Sahni et al. show that corticospinal neuron (CSN) axons exhibit targeting specificity in spinal white matter in development, well before encountering spinal neurons or making connections with spinal circuits. CSNs projecting to distinct spinal segments can now be prospectively identified molecularly and investigated even before their axons reach the cord.
INTRODUCTION
The corticospinal system is essential for skilled movements (Martin, 2005); fine motor control relies on corticospinal neurons (CSN) projecting axons to appropriate and specific segmental targets. CSN in multiple cortical areas, beyond the primary motor cortex (M1), form corticospinal circuitry with distinct functional outputs, including spinal motor neuron excitation and inhibition by direct and indirect pathways, descending control of ascending inputs, and autonomic control (Lemon, 2008; Lemon and Griffiths, 2005; Sahni et al., 2020; Welniarz et al., 2017). Such diverse functional output, especially for precise motor control, requires distinct CSN subpopulations to project to segmentally specific targets in the pons, medulla, and cervical, thoracic, and lumbar cord for head, forelimb, trunk, and hindlimb motor control.
Cortical organization of segmentally distinct CSN subpopulations, their spinal projections, and their role in motor control has been intensely researched. Penfield’s homunculus in the primary motor cortex (M1) in humans (Penfield and Boldrey, 1937; Penfield and Rasmussen, 1950) and Woolsey’s primate simiusculus (Woolsey et al., 1952) demonstrate that cortical regions are organized to control movement of distinct body parts. Many anatomical (Akintunde and Buxton, 1992; Li et al., 1990; Miller, 1987; Tennant et al., 2011; Ullan and Artieda, 1981; Wise et al., 1979) and physiological (Donoghue and Wise, 1982; Neafsey et al., 1986; Tennant et al., 2011) studies have also revealed somatotopic mapping in rodents, extending beyond M1.
In mice, intracortical microstimulation identified cortical regions controlling forelimb movement, including the rostral and caudal forelimb areas (RFA and CFA, respectively). The RFA is surrounded by representations of the whiskers, jaw, and other proximal structures (Tennant et al., 2011). CSN axons from RFA versus CFA have distinct spinal projections and control distinct elements of forelimb reaching (Wang et al., 2017). There appears to be only one ‘‘hindlimb’’ area, with lumbar projections (Kamiyama et al., 2015; Wang et al., 2018). Understanding how CSN axons target specific spinal segments is a major goal toward identifying the developmental basis for the organization of segmentally specific function, including motor, sensory, and autonomic control. Further, in distinct variants of amyotrophic lateral sclerosis (ALS), distinct subsets of CSN are preferentially vulnerable such as bulbar-projecting CSN in bulbar ALS and lumbar-projecting CSN in hereditary spastic paraplegia (HSP).
Previous work investigating transcriptional regulators controlling differentiation of projection neuron subtypes, such as CSN versus corticothalamic projection neurons (CThPN) versus callosal projection neurons (CPN) (reviewed in Fame et al., 2011; Franco and Müller, 2013; Greig et al., 2013; Leone et al., 2008; Lodato et al., 2015; Molyneaux et al., 2007), has identified that CSN development is regulated first by controls over broad corticofugal neuron differentiation, then by controls over all sub-cerebral projection neurons (SCPN), and then by CSN-specific controls (Arlotta et al., 2005; Cederquist et al., 2013; Chen et al., 2005a, 2005b; Galazo et al., 2016; Greig et al., 2016; Han et al., 2011; Joshi et al., 2008; Kwan et al., 2008; Lai et al., 2008; Lodato et al., 2014; McKenna et al., 2011; Molyneaux et al., 2005; Ozdinler and Macklis, 2006; Shim et al., 2012; Tomassy et al., 2010; Woodworth et al., 2012, 2016). CSN-specific controls identified to date do not distinguish between hodologically distinct CSN subpopulations, and the molecular basis for the segmentally specific CSN diversity described above remains entirely unknown.
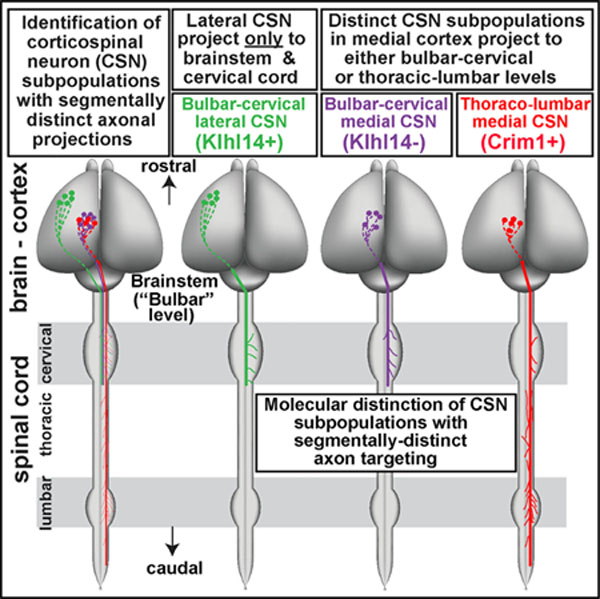
To provide a foundational framework for this eventually specific circuit establishment, we aimed in experiments presented here and in the accompanying functional investigation (Sahni et al., 2021) to identify in mice distinct, developmentally specified, segmentally specific subpopulations and investigated molecular control over CSN segmental target specificity. Using retrograde and anterograde physical and viral labeling, we find that CSN axons exhibit remarkable specificity of targeting in spinal white matter well before encountering any spinal neurons in the gray matter. CSN in the lateral sensorimotor cortex extend axons exclusively to targets in the brainstem and cervical cord (bulbar-cervical) and do not extend even transient axons past these targets. In contrast, CSN in the medial sensorimotor cortex are diverse, with a thoracolumbar-projecting subpopulation interspersed with a bulbar-cervical-projecting subpopulation. Further, these developmental axon-extension decisions in spinal white matter establish the durable foundation for later stages of segmentally distinct axon collateralization in spinal gray matter. Anatomical and molecular analyses define three segmentally distinct CSN subpopulations through development into maturity. This indicates that segmentally distinct CSN subpopulations are molecularly substantially predetermined for their segmentally specific axon targeting. In the accompanying study (Sahni et al., 2021), we functionally investigate a subset of these controls in regulating segmentally specific CSN axon targeting. These results provide reagents with potential for molecular- and circuit-level investigation of CSN organization and functional output.
RESULTS
Differential spinal segmental axon targeting in white matter by distinct CSN subpopulations during development
In contrast to prior methods like spinal gray matter injections (Kamiyama et al., 2015) and rabies tracing (Ueno et al., 2018), which can only label CSN already collateralized into the spinal gray matter, we investigated whether CSN axon targeting specificity exists from the earliest stages of axon extension to the cord before branching or innervation. We retrogradely labeled CSN from spinal white matter as axons traverse the dorsal funiculus (DF) as defined levels.
We investigated neocortical locations of CSN that project axons to only bulbar-cervical segments (CSNBC) versus CSN that project axons past T2 to thoracic and lumbar segments (CSNTL) during development. These subpopulations were selected because (1) transition from the cervical to thoracic cord reflects a functional distinction of motor control, with cervical segments executing more skilled forelimb movements; (2) studies suggest that cervical-projecting CSN represent a distinct subpopulation with seemingly more specific axon targeting (Akintunde and Buxton, 1992); (3) molecular programs downstream of Hox genes delimit cervical from thoracic circuits (Dasen et al., 2003; Jung et al., 2010), which suggests that developing CSN axons encounter distinct signals as they navigate between these segments; and (4) the transition is reliably and reproducibly identifiable.
We first differentially labeled CSNBC and CSNTL by retrograde labeling from the spinal cord of postnatal mice at thoracic T2 (to specifically label CSNTL) at P4, followed by retrograde with a distinct label at cervical C1 (to label all CSN, both CSNBC and CSNTL) at P6 (Figures 1A and A’). Mice were perfused at P8. We included CSNBC and CSNTL regardless of the nominal functional area in which they are located, as CSN reside in multiple cortical locations, including outside M1 (Donoghue and Wise, 1982; Lemon, 2008; Tennant et al., 2011).
Figure 1. Distinct CSN subpopulations in the developing sensorimotor cortex exhibit axon extension specificity.

(A) Dual retrograde labeling to investigate developmental CSN axon extension specificity. Retrograde label CTB-647 injection into cervical C1 (green micropipet) labels all CSN (green oval in the cortex), while retrograde label CTB-555 injection into thoracic T2 (red micropipet) labels only CSNTL, which are doubly fluorescent (yellow oval in the cortex; CSNTL are a subset of all CSN).
(A’) P8 mouse CNS showing injection sites of CTB-555 (red) into T2 at P4 followed by CTB-647 (green) into C1 at P6.
(A’’) Same brain showing green fluorescence in the cortex occupies a broader area, while yellow (green + red) fluorescence (CSNTL) occupies a smaller area within the green area.
(B–M’’) Coronal sections of the same brain at four rostrocaudal levels (dotted lines in A’’). CSNTL reside medially; most reside at midcaudal (H’ and J–J’’) and caudal (K’ and M–M’’) levels. CSNTL are excluded from the lateral cortex at all levels (C’, F’, I’, and L’) where CSN are almost exclusively CSNBC (green only; C, C’, F, F’, I, I’, L, and L’). CSNBC are present both laterally and medially; at caudal levels, a subset of CSNBC (CSNBC-med) are intermingled with CSNTL in the medial cortex.
(N) Quantification of CSN in the medial versus lateral cortex retrogradely labeled by C1 versus T2 label. n = 5 mice. Graphs show average counts ± SEM.
Scale bars, 1 mm in A’ and A’’ and 500 μm in B–K.
Whole-mount analysis shows that CSNTL occupy a more restricted area of the sensorimotor cortex compared to CSNBC (Figure 1A’’). (Approximately 25% of CSN projecting to thoracolumbar levels at P4 are later pruned by P28 [below]; we refer to CSN projecting to thoracolumbar segments at P4 as CSNTL.) We quantified the number of CSNBC versus CSNTL in the medial versus lateral cortex at four distinct rostrocaudal levels (Figures 1B–1M’’ and 1N). Similar distributions were observed when retrograde injections were performed in separate mice (data not shown). Retrograde analyses confirm that, at P4, all CSNTL are located medially (Figures 1B, 1E, 1H, and 1K) and excluded from the lateral cortex (Figures 1C’, 1F’, 1I’, and 1L’). While a minority of CSNTL are located in the rostral medial cortex (Figures 1D, 1D’’, and 1N), most reside in the caudomedial cortex in an area including the eventual representations of the hindlimb and part of the trunk (Tennant et al., 2011) (Figures 1H’, 1H’’, 1J, 1J’’, 1K’, 1K’’, 1M, 1M’’, and 1N). In contrast, CSNBC occupy a broader cortical area extending medially and laterally (Figures 1B–K’’ and 1N). While most CSN in the caudomedial cortex are CSNTL, they are intermingled with CSNBC, and almost all CSN in the lateral cortex are CSNBC (Figure 1N). Therefore, CSN subpopulations exhibit axon targeting specificity between cervical and thoracolumbar segments early in development.
Early axon targeting specificity by CSN in the lateral cortex is durably maintained
To confirm these retrograde labeling results, we performed anterograde labeling from distinct cortical areas with adeno-associated virus (AAV) particles engineered to express either EGFP (AAV-EGFP) or tdTomato (AAV-tdTomato). AAV-EGFP was injected into the rostrolateral cortex and AAV-tdTomato was injected into the caudomedial cortex at P0 (Figures 2A–2J). We analyzed labeled axons from these distinct CSN subpopulations at specific spinal levels at P4 while corticospinal tract (CST) axons are still extending toward distal targets (Bareyre et al., 2005) and at P7 when they have reached the lumbar cord (Figures 2E–2J). As expected, CSN axons from both cortical locations are present in DF at C1 (Figures 2E and 2F); essentially all EGFP-labeled CSN axons from the rostrolateral cortex terminate within the cervical cord, with only a few axons present at T1-T2 (Figure 2G) and none extending to L1-L2 (Figure 2I). In contrast, tdTomato-labeled CSN axons from the caudomedial cortex extend to thoracic and lumbar levels (Figures 2H and 2J). We also investigated this differential axon targeting at P28 with equivalent results (Figures 2K–2M, 2O, and 2O’). Interestingly, while axons from both the lateral and medial cortex collateralize extensively in the cervical gray matter, EGFP+ axon collaterals from CSNBC in the rostrolateral cortex occupy a more ventral location compared to tdTomato+ collaterals from the caudomedial cortex (Figures 2N–2N’’). Together, these anterograde and retrograde data define a somatotopic distribution of CSNBC versus CSNTL in the developing cortex, which persists with maturity. CSNBC in the lateral sensorimotor cortex appear relatively homogeneous, with projections exclusively to targets in the brainstem and cervical cord (bulbar-cervical), referred to here as ‘‘CSNBC-lat.’’ In contrast, CSN in the medial sensorimotor cortex (CSNmedial) project to both bulbar-cervical and thoracolumbar targets.
Figure 2. Early projection targeting specificity by CSN in the lateral cortex is durably maintained.
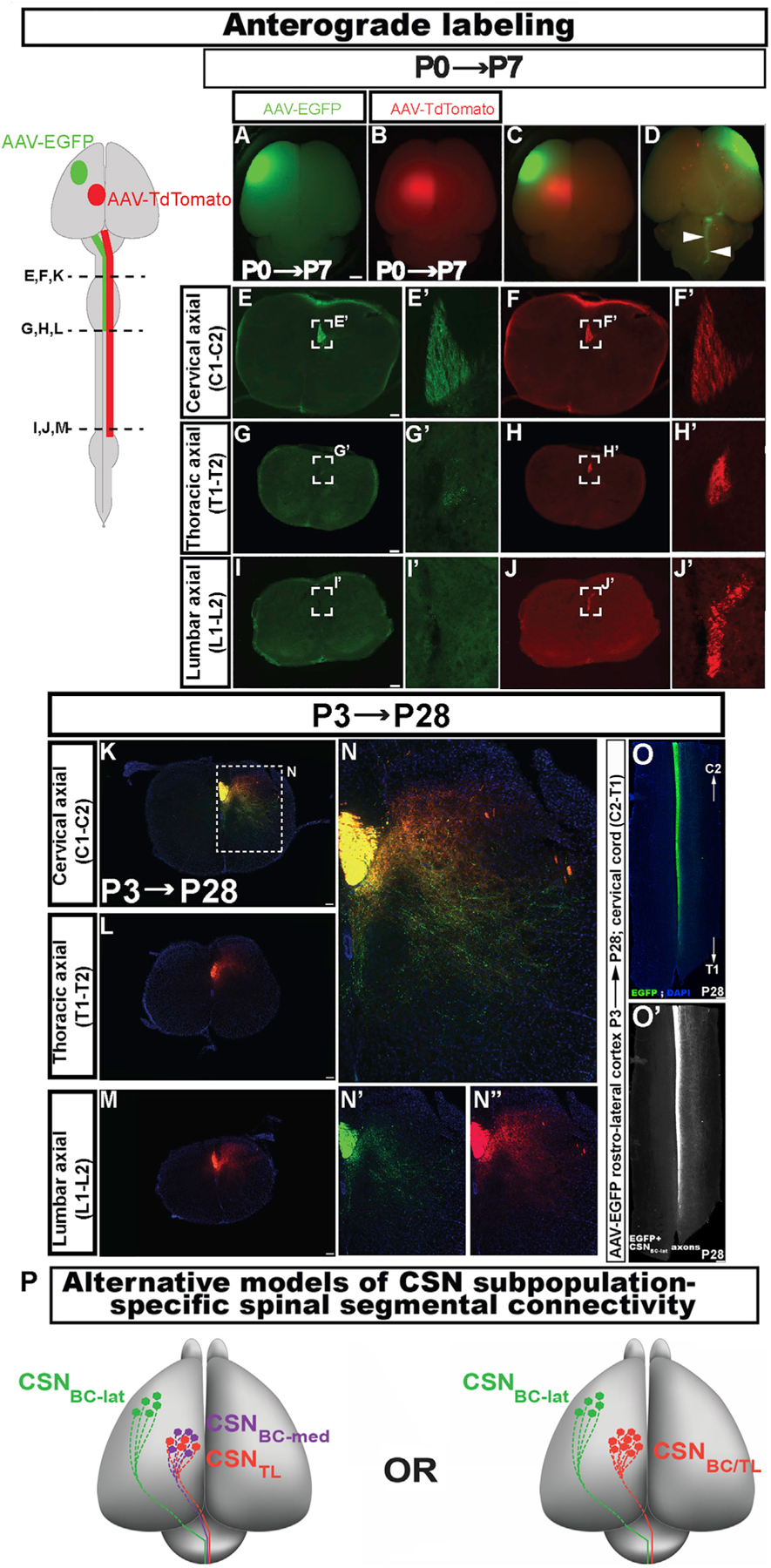
(A–D) Whole-mount view of a P7 brain injected at P0 with AAV-EGFP rostrolaterally (green, A) and AAV-tdTomato caudomedially (red, B; merged dorsal view showing both sites in C) to anterogradely label the CST (arrowheads, ventral view in D).
(E–J) Axial spinal sections from the same mouse at cervical (E and F), thoracic (G, H), and lumbar (I and J) segments. Only tdTomato+ axons extend to the lumbar cord. (E’–J’) Magnified views of the dorsal funiculus (boxed regions in EJ).
(K–M) Axial spinal sections from a P28 mouse that was similarly injected with the two AAVs at P3, at cervical (K), thoracic (L), and lumbar (M) levels. Only tdTomato+ axons extend to the lumbar cord.
(N–N’’) Magnified view of the boxed region in K. At P28, EGFP+ CSNBC-lat axons (green) extend collaterals into more ventral laminae in cervical gray matter compared to tdTomato+ CSNmedial axons (red) (individual channels in N’ and N’’).
(O and O’) Horizontal section of a cervical cord from the same mouse as in K–N (O). CSNBC-lat axons traverse and extend collaterals throughout the rostrocaudal extent of the cervical cord (O’).
(P) Schematic representation of two distinct possibilities that might theoretically give rise to segmentally distinct CSNmedial axon projections. In both, CSN in the lateral cortex (CSNBC-lat; green) project exclusively to the brainstem and cervical cord. (Left) CSNBC-med (purple) and CSNTL (red) are developmentally distinct, and this specification persists into maturity. (Right) CSNBC and CSNTL in the medial cortex are developmentally equivalent early, but later give rise to segmentally specific projections (CSNBC/TL in red).
Scale bars, 1 mm in A–D and 100 μm in E–O.
CSNBC-lat do not significantly extend even transient projections past thoracic T2
We next investigated two alternative possibilities underlying this differential targeting: (1) that it arises from different rates of axon extension by these CSN subpopulations (e.g., CSNBC-lat axons might ‘‘lag behind’’ CSNmedial axons during early development, resulting in differential targeting later [at P4, P7, or P28]); or (2) CSNBC-lat axons might theoretically initially extend into the thoracic cord, with subsequent pruning between P1 and P4. We therefore investigated the time course of axon extension past T2 by CSNBC-lat and CSNmedial by retrograde labeling from C8 at P1, T2-T3 at P2, and T3-T4 at P4 (Figures S1A, S1F, and S1K). We find that both CSNBC-lat and CSNmedial are labeled from C8 at P1, but neither population is labeled from T2, consistent with previous results (Bareyre et al., 2005). Labeling from T2 at P2 very specifically labels a small subset of CSN in the medial cortex (Figures S1F–S1J’’), with a larger subset labeled from T3 to T4 at P4 (Figures S1K–S1O’’); no CSN in the lateral cortex are labeled at any time investigated (Figures S1L’’–S1O’’ and S1G’’–S1J’’).
These experiments reveal that although axons from both subpopulations arrive at C8 by P1, they then exhibit strikingly different behaviors. CSNmedial (of which CSNTL are a subset) extend axons past T2, while CSNBC-lat do not. Further, once CSNBC-lat axons execute this decision, they maintain this specificity from P1 into maturity at P28 (Figures 2K–2M).
Developmental axon extension specificity in the medial sensorimotor cortex is durably maintained
Since there is known promiscuity of CSN axon projections during development (Kuang and Kalil, 1994), it could be possible that some of the early CSNmedial axonal projections past T2 are promiscuous and subsequently pruned between P4 and P28. In this scenario, CSNBC and CSNTL in the medial cortex could be molecularly equivalent at P4, and therefore not yet specified with regard to their later segmental axon targeting (Figure 2P, schematic on the right with both populations shown in red). Alternatively, this early axon targeting specificity could reflect early developmental specification of CSN segmental projection. In this scenario, CSN projecting past T2 at P4 are already committed, and thus likely molecularly specified, toward thoracolumbar axon targeting at maturity. The medial sensorimotor cortex would therefore contain CSNTL interspersed with medial CSNBC, with the two populations already expressing distinct genes that determine their segmental target specificity (Figure 2P, schematic on the left in which CSNBC are shown in purple and CSNTL are shown in red).
To directly address this question, we used an intersectional viral strategy to permanently label segmentally distinct CSN subpopulations during development to then investigate their axon projections into maturity. We injected AAV-FLEX-tdTomato into the medial sensorimotor cortex, combined with AAV-Cre into either C1 or T2 DF at P4 (Figures 3A and 3E). AAV-Cre injection at C1 labels all CSNmedial, regardless of their segmental targets at P4 (both CSNBC and CSNTL). In contrast, AAV-Cre injected at T2 only labels CSN projecting to thoracolumbar segments (CSNTL). This approach specifically and permanently labels segmentally distinct CSN subpopulations at P4, enabling investigation of their cortical location, and eventual segmental axon targeting at P28.
Figure 3. CSN axon extension specificity during development is maintained with maturation, preceding specificity of axon collateralization at thoracolumbar versus cervical segments.

(A and E schematics) At P3, Cre-dependent AAV-FLEX-tdTomato was injected into the medial cortex (red micropipet), followed by AAV-Cre injection at P4 (green micropipet) into DF at cervical C1 (CSNmedial; A–D, I, K, and M) or thoracic T2 (CSNTL; E–H, J, L, and N).
(A, E) P28 CNS whole mount showing Cre injection (via EGFP, green) at C1 (A) or T2 (E), with tdTomato (red) in the cortex.
(B–D) Axial spinal sections from the same mice at C1-C2 (B and F), T1-T2 (C and G), and L1-L2 (D and H).
(B’–D’ and F’–H’) High-magnification single plane confocal images of DF (areas boxed in B–D and F–H). In B–D, the majority of tdTomato+ CSN axons at C1-C2 do not extend to thoracolumbar segments.
(F–H) Approximately 75% of tdTomato+ CSN axons at C1-C2 extend to thoracolumbar segments.
(I–N) Flattened two-dimensional projections of digitally reconstructed P28 spinal cords from the same mice in B–H (red, CSN axons; blue, DAPI). Binned monochrome images in Figure S2; three-dimensional reconstructions in Videos S1–S6. Projections of cervical C2–C8 (I and I’), thoracic T2–T13 (K and K’), and lumbar L2–L6 (M and M’) segments after P4 injection with either C1 AAV-Cre (same mouse as A–D) or T2 AAV-Cre (same mouse as E–H).
(I’–N’) tdTomato+ CSN axons without DAPI.
(I’’–N’’) Monochrome magnified views of respective areas boxed in I’–N’.
(O) Percentage of tdTomato+ axons in DF at C1-C2 that extend to T1-T2 and L1-L2 in P28 mice injected with either C1 or T2 AAV-Cre at P4. Graphs show average percentages ± SEM. *p < 0.05 by Student’s t test. The dotted line indicates one outlier in the C1 AAV-Cre-injected group (mouse 2 in Figure S2; details in Figure S2 legend).
(P) Percentage of tdTomato+ axon collateral area in cervical, thoracic, and lumbar segments in P28 mice injected with either C1 or T2 AAV-Cre at P4. Graphs show average percentages ± SEM.
(Q) Quantification of finer, binned segmental distribution of tdTomato+ axon collateral area within the cervical, thoracic, and lumbar cord in P28 mice injected with C1 versus T2 AAV-Cre at P4 (7 bins for ~C2–C8, 12 bins for ~T2–T13, and 5 bins for ~L2–L6). Graphs show average percentages ± SEM. The peak collateral distribution in mice injected with C1 AAV-Cre is over bins approximately overlying cervical enlargement (C3–C7), while peak collateral distribution in mice injected with T2 AAV-Cre is over bins approximately overlying lumbar enlargement (L2–L5).
Scale bars, 1 mm in A and E and 100 μm in B–D and F–N.
All tdTomato+ CSNmedial at P28 labeled by either C1 or T2 AAV-Cre at P4 were in layer V, validating labeling specificity (Figures S2A’–S2O’). tdTomato+ CSN labeled by C1 AAV-Cre reside throughout the rostrocaudal medial cortex (Figures S2A–S2J), while most CSN labeled by T2 AAV-Cre reside more caudally in the medial cortex (Figures S2K–S2O). These P28 results are consistent with retrograde labeling at P4.
Next, we investigated whether differential axon extension by CSNmedial at P4 is durably maintained by analyzing axon projections at P28 from CSN labeled at P4 with AAV-Cre injected at C1 (CSNmedial; both CSNBC and CSNTL) versus T2 (only CSNTL). We first quantified the number of C1-labeled tdTomato+ CSN axons present at cervical, thoracic, and lumbar levels. Of total labeled CSN axons in DF at C1, only roughly half (48% ± 13%) extend to the thoracic DF and only roughly a quarter (22% ± 7%) extend to the lumbar DF (Figures 3B–3D and 3O), indicating that only approximately half of CSNmedial extend axons to the thoracolumbar cord at P28. In one outlier, all labeled CSN were CSNBC (see Figure S2 for details; ‘‘outlier’’ in Figure 3O).
We next analyzed axon extension at P28 by CSNmedial labeled with AAV-Cre from T2 at P4 (CSNTL). In striking contrast to CSN labeled from C1, nearly 75% (74% ± 3%) of CSN labeled from T2 extend axons to the thoracic DF and more than half (54% ± 5%) extend to lumbar DF (Figures 3F–3H and 3O). These results indicate that CSNTL within the medial cortex are substantially predetermined by P4 toward thoracolumbar axon projection and that this early specificity is largely and durably maintained from P4 to P28.
Together, P4 developmental retrograde analyses combined with intersectional viral labeling enable further delineation of spatially interdigitated CSN subpopulations in the medial cortex based on differential axon targeting in DF at thoracic T2: (1) CSN that extend axons to cervical cord and not beyond T2 (referred to here as CSNBC-med); and (2) CSN that extend axons past T2 to thoracolumbar segments (CSNTL) at P4, approximately 25% of which are later pruned. Further, these data indicate that axon targeting specificity by CSNBC-med versus CSNTL at P4 is largely maintained into maturity and therefore represents a first step in establishing eventual segmentally specific axon targeting.
CSN axon targeting specificity establishes an early foundation for later segmentally distinct axon collateralization
We next investigated the extent to which specificity of axon extension in DF between CSNBC-med versus CSNTL dictates specificity of axon collateralization in the cervical versus thoracolumbar cord. We investigated at P28 the distribution of tdTomato+ axon collaterals across the entire cord by CSNmedial intersectionally labeled by AAV-Cre at P4 as described above (Figure 3; Videos S1–S6). We quantified, for each mouse, the percentage of total tdTomato+ CSN axon collateral area in the cervical versus thoracic versus lumbar cord.
We find that the specificity of axon extension between CSNBC-med versus CSNTL correlates with the specificity of axon collateralization in the cervical versus thoracolumbar cord. Nearly 70% (70% ± 11%) of the total CSNmedial axon collateral area is in cervical cord (Figures 3I and 3P), with only approximately 30% (30% ± 11%) in the thoracolumbar cord (Figures 3K, 3M, and 3P; Video S7). In striking contrast, ~70% (72% ± 7%) of the CSNTL axon collateral area is present in the thoracolumbar cord (Figures 3L, 3N, and 3P; Video S7), with only 30% (28% ± 7%) in the cervical cord (Figures 3J and 3P; Video S7). This links axon targeting specificity in spinal white matter with the specificity of axon collateralization in spinal gray matter. This finding is consistent with prior electrophysiology results in cats showing that CSN innervating caudal to thoracic T3 also collateralize at cervical levels (Shinoda et al., 1986), but it also reveals that this is a relative minority.
While CSNmedial axons are ~2.3 times more likely to branch in the cervical versus thoracolumbar cord, CSNTL are four times more likely to collateralize in the lumbar cord than the overall CSNmedial population (axon collateral area in lumbar cord: CSNTL = 32% ± 5% and CSNmedial = 9% ± 4%). At P4, a CSNmedial axon extending past T2 is four times more likely to collateralize in the lumbar cord at P28.
We also investigated approximate segmental distribution of tdTomato+ CSN axon collaterals and find that the peak of CSNmedial axon collateral density occurs in spinal gray matter bins spanning C3–C7, approximately overlying the cervical enlargement (Figure 3Q). In contrast, the peak of CSNTL axon collateral density occurs in spinal gray matter bins spanning L2–L5 approximately overlying the lumbar enlargement (Figure 3Q). Together, these results indicate that CSNTL axons preferentially extend collaterals into thoracolumbar segments, and that early axon extension specificity at P4 at T2 establishes a durable foundation for later stages of segmentally specific axon collateralization.
Together, our retrograde and viral labeling experiments indicate that axon targeting specificity in the white matter identifies segmentally distinct subpopulations during development (CSNBC-lat, CSNBC-med, and CSNTL). CSNBC-lat reside outside the ‘‘classic’’ motor cortex in the lateral cortex and project only to bulbar-cervical segments. In contrast, in the medial sensorimotor cortex, some CSN project only to bulbar-cervical segments and not beyond (CSNBC-med), and others (CSNTL) extend axons past T2 to thoracolumbar segments. These subpopulations remain distinct into maturity, and they continue to exhibit distinct axon projection and gray matter collateralization. Further, early axon extension specificity between CSNBC-med and CSNTL appears to reflect molecularly specified diversity, with distinct interspersed subsets of medially located CSN molecularly controlled either to extend axons past T2 or to limit axon extension rostral to T2.
Fluorescence-activated cell sorting purification and transcriptional analysis of developing CSNBC-lat versus CSNmedial reveals binary, subpopulation-specific molecular differences
We hypothesized that CSN subpopulations extending axons rostral versus caudal to T2 might express distinct regulators over differential axon targeting. We aimed to identify distinct molecular controls for each subpopulation (CSNBC-lat, CSNBC-med, and CSNTL). We took advantage of the spatial separation between CSNmedial and CSNBC-lat to selectively purify each subpopulation by retrogradely labeling CSN from C1 and micro-dissecting the rostrolateral versus caudomedial sensorimotor cortex followed by FACS purification (details and justification in the Star Methods Method details). This allowed us to prospectively isolate CSNTL, as part of the broader CSNmedial subpopulation, even before their differential axon targeting occurred (P1) and at later stages of differential targeting (P4, P7) for differential gene expression analysis (Figures 4A–4E’).
Figure 4. Genes previously identified as CSN specific additionally exhibit differential expression between CSNCBC-lat and CSNmedial.

(A–E’) Experimental design to isolate CSNBC-lat and CSNmedial before their axons reach their ultimate segmental targets. (A) Retrogradely labeled CSN (green) in the P4 cortex. (B) Custom-made four-blade instrument to microdissect 700-μm-thick tissue blocks of sensorimotor cortex. Caudomedial (CSNmedial; arrowhead in C) and rostrolateral (CSNBC-lat; arrowhead in D) tissue blocks are collected separately. (E) Cortical tissue collected for FACS; labeled CSN seen as a bright band (E’).
(F) FACS profiles showing purification of retrogradely labeled CSNmedial at P4. Purified CSNmedial comprised 2.67% of total cells from the caudomedial cortex. PE-, phycoerythrin.
(G, J, M, P, S, and V) Microarray expression profiles at postnatal ages P1, P4, and P7. CSNBC-lat profiles in blue, and CSNmedial profiles in red. The y-axis represents normalized fluorescence intensity. Data are presented as mean ± SEM.
(H, I, K, L, N, O, Q, R, T, U, W, and X) In situ hybridization on coronal brain sections at ages indicated.
(H’, I’, K’, L’, N’, O’, Q’, R’, T’, U’, W’, and X’) Magnification of boxed regions showing the rostrolateral versus caudomedial cortex. All genes are specific to layer V. CSNBC-lat-specific genes are expressed in the rostrolateral and excluded from the caudomedial layer V, while the converse is true for CSNmedial-specific genes. sp, subplate; II/III– VI, neocortical layers II/III–VI. Scale bars, 1 mm in A–D and 500 μm in H, I, K, L, N, O, Q, R, T, U, W, and X.
We confirmed the reproducibility and consistency of data (Figure S3A) and used multiple statistical tools (Star Methods Method details) to identify differentially expressed genes specifically and mutually exclusively expressed by CSNBC-lat or CSNmedial either at all three times or at specific developmental stages (Figure S3B; Table S1).
In situ hybridization analyses confirm binary transcriptional differences between developing CSNBC-lat versus CSNmedial and highlight further diversity within CSNmedial
We confirmed subtype-specific expression of CSNBC-lat and CSNmedial candidate genes by in situ hybridization. We first examined the expression of candidate genes previously identified as predominantly CSN specific (versus CPN; Arlotta et al., 2005; Molyneaux et al., 2009). As expected, these genes are expressed in layer V, and consistent with our hypothesis, CSNBC-lat-specific genes Klhl14 (Figures 4G–4I’), Ermin (Figures 4J–4L’), Cartpt (Figures 4M–4O’), and Afap1l2 (Figures S3C–S3E’) are expressed only in lateral layer V and excluded from medial layer V. Conversely, CSNmedial-specific genes Crim1 (Figures 4P–4R’), Cry-mu (Figures 4S–4U’), St6galnac5 (Figures 4V–4X’), and Chst8 (Figures S3O–S3Q’) are specifically expressed only in medial, but not lateral, layer V. These results identify that subsets of CSN-specific genes exhibit additional CSNBC-lat versus CSNmedial expression.
Because differentiation of CSNBC-lat versus CSNmedial might also be regulated by area-specific genes expressed across multiple projection neuron subtypes (Greig et al., 2013), we investigated genes that are not predominantly CSN specific. For example, CSNBC-lat-specific Pappa2 (Figures S3F–S3H’) and Alcam (Figures S3I–S3K’) are also expressed by CThPN (Table S3; Molyneaux et al., 2015), while Frzb (Figures S3L–S3N’) is also expressed by CPN. All of these genes are expressed in lateral, but not medial, layer V. Reciprocally, Igsf4a (Figures S3R–S3T’), Zbtb16 (Figures S3U–S3W’), and Wnt4 (Figures S3X–S3Z’) are selectively expressed by CSNmedial versus CSNBC-lat but are also expressed by other projection neuron subtypes in the medial cortex. Some genes were further restricted to subpopulations of CSNmedial located at different rostrocaudal levels and might potentially represent additional segmental (thoracic versus lumbar) diversity, given that retrograde tracing from T13/L1 only labels a subset of CSNTL (lumbar-projecting ‘‘CSNL’’) in the caudomedial sensorimotor cortex (Figures S4B–S4E). For instance, St6galnac5 is expressed throughout the medial cortex (Figures S4F–S4F’’); Chst8 has graded expression from low rostral to high caudal (Figures S4G–S4G’’), while Igsf4a and Wnt4 are mostly restricted to the caudomedial sensorimotor cortex (Figures S4H–S4I’’). Overall, these data indicate that spatially distinct CSN subpopulations express complements of genes giving rise to considerable early molecular diversity, some of which might control their distinct segmental axon targeting and connectivity.
Klhl14 and Crim1 expression molecularly distinguish and parcellate CSNBC-lat versus CSNTL during early development
We selected Crim1 in CSNmedial and Klhl14 in CSNBC-lat for more detailed analysis using a priori criteria (see STAR Methods and Tables S2 and S3). In situ hybridization confirms that Crim1 and Klhl14 exhibit complementary expression in layer V (Figures 5A–5C); Klhl14 is expressed in the lateral cortex, where CSNBC-lat reside, while Crim1 is expressed in the medial cortex. Importantly, expression of both genes is absent in the Fezf2 null cortex, which completely lacks CSN (Chen et al., 2005a, 2005b; Lodato et al., 2014; Molyneaux et al., 2005), confirming their CSN specificity (Figures 5D–5G). We combined in situ hybridization with retrograde labeling, and we confirmed that the domain of Klhl14+ CSN in the lateral cortex does not overlap with the region occupied by CSNTL in the medial cortex (Figures 5H–5J). Using real-time PCR following laser-capture microdissection, we confirmed that Crim1 is enriched in CSNTL, while Klhl14 is enriched in CSNBC-lat (Figures S5A–S5C).
Figure 5. Klhl14 and Crim1 expression molecularly distinguishes and parcellates CSNBC-lat versus CSNTL during early development.

(A and B) In situ hybridization on serial coronal sections of a P4 brain showing Klhl14 (A) and Crim1 (B) expression.
(C) Overlay of inverted and pseudo-colored images from A and B. Klhl14 and Crim1 exhibit complementary expression.
(D–G’) There is no Klhl14 (D, E, and E’) or Crim1 (F, G, and G’) expression in the Fezf2 null cortex (E and E’), which lacks CSN, confirming that Klhl14+ and Crim1+ neurons are CSN. (D’, E’, F’, and G’) Magnified views of regions boxed in (D), (E), (F), and (G) respectively.
(H–J, L, and N) CSNTL were retrogradely labeled (red in I, L, and N). (H) In situ hybridization showing that Klhl14 is excluded from CSNTL (merged in J). (H’–J’) Caudal view of the same brain in (H)–(J) showing that Klhl14 is excluded from from CSNTL even in caudal sensorimotor cortex.
(K–L’’) Crim1 (cyan in K) is expressed by almost all CSNTL (magnified merged view in L, white; individual channels in L’ and L’’).
(M–N’’) Cry-mu (cyan in M), another CSNmedial-specific gene, is not expressed by all CSNTL (magnified merged view in N, white; individual channels in N’ and N’’; arrowheads indicate labeled CSNTL that do not express Cry-mu).
Scale bars, 500 μm.
We next investigated temporal expression of both Klhl14 and Crim1 (Figure S6). Neither gene is detected in the cortical progenitors that give rise to CSN at E13.5 (data not shown). Consistent with transcriptomic analysis, Klhl14 expression peaks early, with the highest expression at E18.5 and P1 (Figures S6A and S6B). Klhl14 levels decline in the first postnatal week, once CSNBC-lat axons begin to collateralize in cervical gray matter (Figures S6C and S6D), with few Klhl14+ cells with low-level expression by P10 (Figure S6E). At P14, there is no detectable expression in the cortex (data not shown). Crim1 expression begins by E18.5 and peaks at P4 (Figures S6F–S6H). Crim1 expression declines from P7 to P14 (Figures S6I and S6J), with low-level Crim1 expression by P28 (Figure S6K). Crim1 expression remains restricted to medial layer V, i.e., where CSNTL reside (Figures S6L and S6M), at all times. Thus, Klhl14 and Crim1 are expressed in nonoverlapping cortical domains through development.
We quantified colocalization of retrogradely labeled CSNTL with Crim1 using single molecule fluorescence in situ hybridization (smFISH, via RNAscope) or Cry-mu, another CSNmedial-specific gene (Figure 4S). Nearly all retrogradely labeled CSNTL (95% ± 2%) express Crim1 (Figures 5K and 5L). In contrast, Cry-mu does not exhibit the same extent of colocalization with CSNTL; only 63% ± 5% of CSNTL express Cry-mu (Figures 5M and 5N), suggesting that, unlike Crim1, it is also expressed by CSNBC-med.
We combined CTIP2 immunocytochemistry, CSNTL retrograde labeling, and Crim1 expression analysis (Figures 6 A–6C’’’) to quantify the percentage of high CTIP2+ CSNmedial that are either Crim1 positive or Crim1 negative. We identified that 66% ± 1% of CSNmedial are Crim1 positive; they reside interdigitated with a high CTIP2+, Crim1-negative (34% ± 1%) subpopulation (Figures 6D and 6E). Strikingly, CSNBC-med comprise a very similar percentage of CSNmedial (36% ± 7%), while CSNTL comprise the remaining 64% ± 7% of CSNmedial (Figure 6H; retrograde label data from Figure 1N are shown again in this integrated panel). These results indicate that Crim1 delineates more than spatial separation between CSNBC-lat versus CSNmedial; Crim1 is expressed by an exceptionally high percentage of developing CSNTL and is mostly not expressed by developing CSNBC-med.
Figure 6. The CSNmedial subpopulation is composed of both CSNTL, which are largely Crim1 positive, and CSNBC-med, which are largely Crim1 negative.
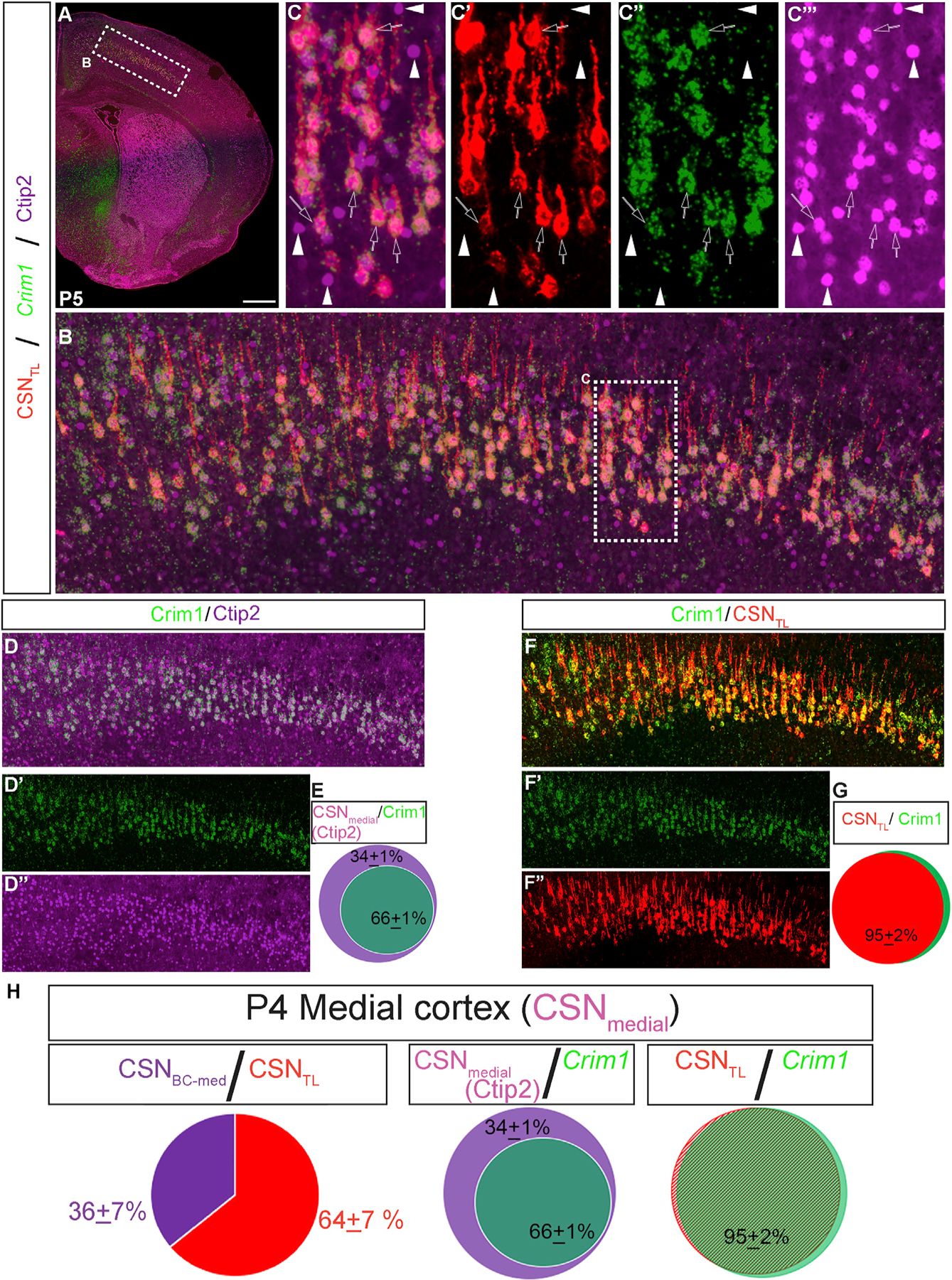
(A) Coronal hemisection of a P5 mouse brain injected with CTB-555 into T2 at P4, showing retrogradely labeled CSNTL (red), Crim1 via smFISH (green), and CTIP2 immunocytochemistry (purple).
(B) Magnified view of medial layer V (boxed in A).
(C–C’’’) Further magnified view of region boxed in (B) showing all CSNmedial labeled by high-level CTIP2. All CSNmedial that are retrogradely labeled (CSNTL) are also Crim1 positive (green; arrows). A subset of high CTIP2+ CSNmedial are Crim1 negative and are not retrogradely labeled (CSNBC-med; white arrowheads).
(D–D’’’) Same image as (B), showing Crim1 and CTIP2 expression.
(E) Approximately 66% ± 1% of all CTIP2+ CSNmedial are Crim1 positive, while ~34% ± 1% are Crim1 negative (counts represent average counts ± s.e.m).
(F–F’’’) Same image as (B), showing Crim1 expression and retrogradely labeled CSNTL.
(G) Approximately 95% of all CSNTL are Crim1 positive.
(H) Summary of quantification for the P4 medial cortex. CSNBC-med constitute ~36% of overall CSNmedial. An equivalent number of CSNmedial identified by high CTIP2 are Crim1 negative, while nearly all CSNTL (~95%) are Crim1 positive.
Scale bars, 500 μm.
Crim1 expression prospectively delineates a subpopulation highly enriched in CSNTL: Crim1+ CSN largely extend axons past thoracic T2 at maturity
We next investigated whether Crim1 expression at P4 might identify CSNTL prospectively—i.e., whether Crim1-expressing CSN during early development (1) maintain axon projections to thoracolumbar segments and (2) if so, whether they also collateralize more extensively in thoracolumbar gray matter.
We first bred Crim1CreERT2 (Crim1GCE) mice (Harding et al., 2011; see STAR Methods) with Thy1-STOP-YFP reporter mice (Bareyre et al., 2005; Buffelli et al., 2003) (schematized in Figure 7A). Recombination was induced in Crim1+ neurons at P0, when CST axons first reach the cord, and P3.5, when CSNTL axons are extending to thoracolumbar segments. We confirmed Cre expression by YFP+ CSN at P5 (CTIP2+, SATB2-; Figures S7A–S7C’’, presumptive CSNTL) and that the molecular identity and axon trajectories of CSN labeled with tamoxifen at P0 and P3.5 do not differ.
Figure 7. Crim1 expression in early development prospectively identifies a CSN subpopulation highly enriched in CSNTL.
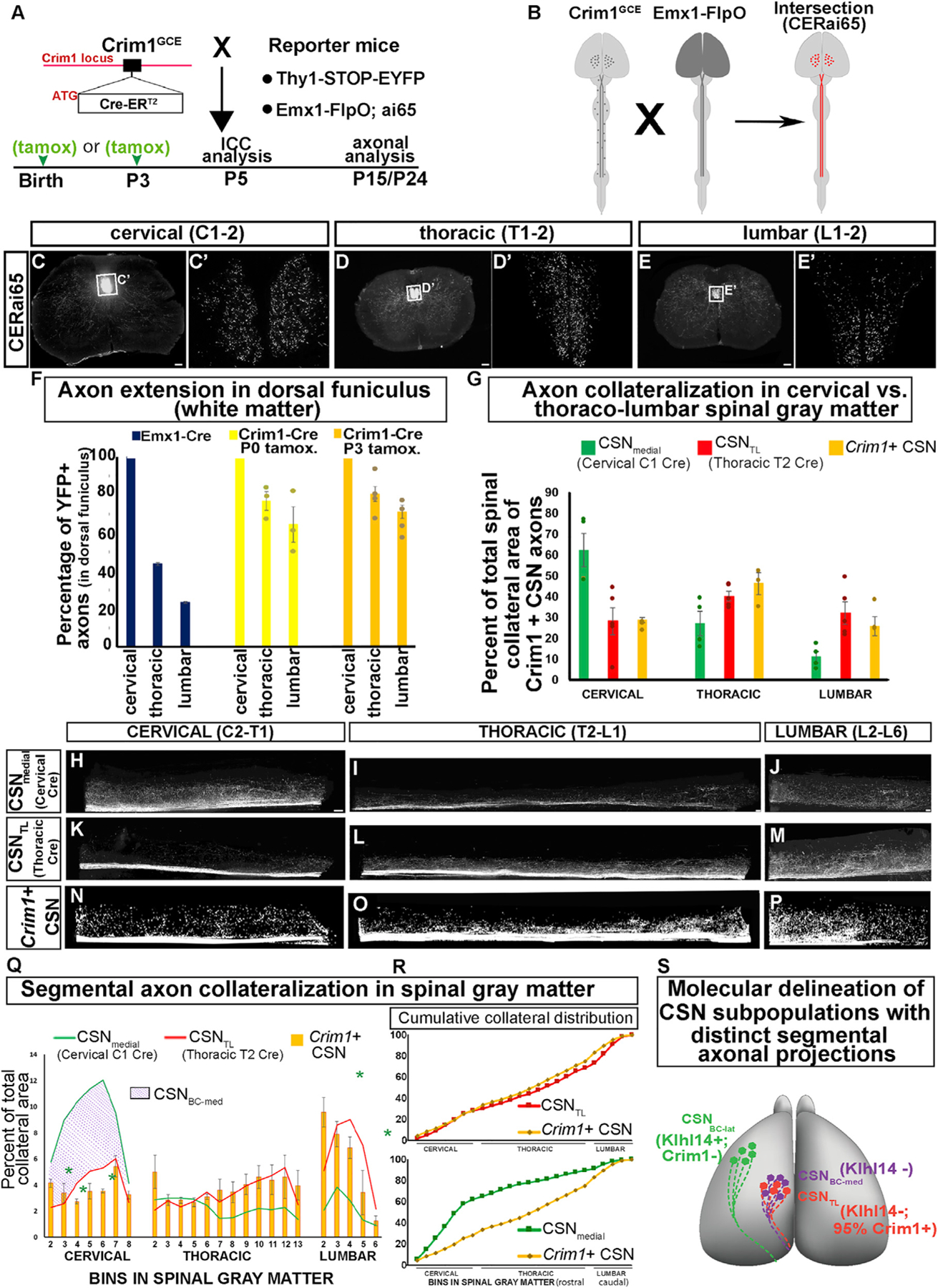
(A) Crim1GCE were bred with Thy1-STOP-YFP mice and pulsed with tamoxifen, at P0 or at P3.5. A YFP reporter is used to investigate molecular identity (at P5) and axon projections (at P15) of Crim1+ neurons.
(B) Crim1GCE; Emx1-FlpO; ai65 (CERai65) intersectional reporter mice. In Emx1-FlpO mice, FlpO is expressed by all cortical projection neurons (dark gray cortex), which labels all CSN axons (dark gray line in cord). Crim1GCE mice express CreERT2 in Crim1+ CSN (dark gray ovals in the cortex) and in Crim1+ spinal neurons (dark gray ovals in cord). In CERai65 mice, only Crim1+ CSN axons are labeled by tdTomato in the cord (red line in cord in CERai65).
(C–E) Axial spinal sections at C1-C2 (C), T1-T2 (D), and L1-L2 (E) from a P24 CERai65 mouse pulsed with tamoxifen at P3.5.
(C’–E’) Single plane confocal images of regions boxed in C, D, and E. tdTomato-labeled Crim1+ CSN axons extend to lumbar cord.
(F) Percentage of axons at C1-C2 that reach T1-T2 and L1-L2 in CST-YFP mice (data from axon counts in Bareyre et al., 2005), compared with Crim1GCE; Thy1-STOP-YFP mice pulsed with tamoxifen at P0 (n = 3 mice) or at P3.5 (n = 4 mice).
(G) Total Crim1+ CSN spinal axon collateral area in CERai65 mice in the cervical, thoracic, and lumbar cord identifies preferential branching in thoracolumbar segments. Collateral area distribution of CSNmedial and CSNTL axons defined by intersectional viral labeling (C1 or T2 AAV-Cre) is shown for comparison (quantification of data from Figure 4G).
(H–M) Flattened 2D projections of digitally reconstructed cervical (H and K), thoracic (I and L), and lumbar (J and M) spinal hemisections from mice injected at P4 with either C1 AAV-Cre (i.e., CSNmedial; H–J; hemisections from images in Figures 3I’, K’, and M’; binning data in Figures 3Q and 7Q) or T2 AAV-Cre (i.e., CSNTL; K–M; hemisections from images in Figures 3J’, L’, and N’; binning data in Figures 3Q and 7Q).
(N–P) Flattened two-dimensional projections of digitally reconstructed cervical (N), thoracic (O), and lumbar (P) spinal hemisections from a CERai65 intersectional mouse pulsed with tamoxifen at P3.5 (same mouse shown in C–E; images of entire spinal cord from this mouse are in Figures S7R–S7T’).
(Q) Binned segmental distribution of Crim1+ CSN axon collaterals in cervical (7 bins; ~C2–C8), thoracic (12 bins; ~T2–T13), and lumbar (5 bins; ~L2–L6) gray matter in CERai65 mice pulsed with tamoxifen at P3.5. For comparison, similarly binned distributions for CSNmedial (green; C1 AAV-Cre) and CSNTL (red; T2 AAV-Cre) axons are shown (line plots are drawn using dataset used for bar histograms in Figure 3Q). Strikingly, orange binned segmental collateral area distributions of Crim1+ CSN axons are nearly identical to CSNTL (red line plots) and are significantly different from CSNmedial axons (green line plots). The peak collateral distribution of CSNmedial axons is in cervical enlargement (this peak likely represents the included CSNBC-med subpopulation, indicated with purple hatching). This peak is the distinguishing feature from Crim1+ CSN. Green asterisks indicate significant difference between Crim1+ CSN axon collateral area at specific segmental bins and CSNmedial (p < 0.05 by Student’s t test). There is no significant difference at any segmental bin between Crim1+ CSN axons and CSNTL. The peak of collateral distribution in CERai65 mice occurs over L2–L5 bins approximately overlying the lumbar enlargement similar to CSNTL.
(R) Cumulative collateral area compared using two-sample Kolmogorov-Smirnov test between Crim1+ axons (CERai65) and CSNTL axons (T2 AAV-Cre at P4). There is no significant difference between the groups. In contrast, there is significant difference between Crim1+ axons and CSNmedial axons (C1 AAV-Cre at P4) (p < 0.005).
(S) Schematic integrating expression, axon extension, and collateralization analyses, showing molecular delineation during development of CSN subpopulations with persistent, distinct segmental axon projection targeting before their final axonal connectivity is established. Klhl14 expression delineates Klhl14-positive CSNBC-lat (green) from Klhl14-negative CSNBC-med (purple); all CSNTL (red) are Klhl14 negative, and 95% ± 2% of CSNTL express Crim1 in early development.
Scale bars, 100 μm.
We then investigated their axon projections at later stages. Crim1+ CSN axons in the internal capsule traverse medially (Figures S7F and S7G), where anatomically identified CSNmedial axons traverse (Figures S7D–S7E’), extending to all spinal levels (Figures S7H–S7L). We compared segmental targets of Crim1+ CSN axons, labeled by tamoxifen at P3.5, with published results examining all CSN projections to the spinal cord in CST-YFP mice (Bareyre et al., 2005). The results showed that 44% ± 1% of total CSN axons at C1-C2 reach T1-T2, and less than a quarter (24% ± 0.5%) extend to L1 (calculated from Bareyre et al., 2005; Figure 7F). In striking contrast, following tamoxifen administration at P3.5, 80% ± 4.0% of Crim1+ CSN axons in DF at C1-C2 reach T1-T2, and 71% ± 3.0% reach L1-L2 (Figure 7F). Interestingly, these percentages are similar after tamoxifen administration at P0 (76% ± 5.0% of axons at C1-C2 reach T1-T2, and 64% ± 9.0% reach lumbar L1-L2 (Figure 7F). This is striking since CSN axons have not reached the caudal cervical cord at P0, but Crim1+ CSN appear to be already partially specified to extend thoracolumbar projections. Together, these data identify that compared with the total CSN population, Crim1+ CSN largely extend axons past cervical to thoracolumbar segments.
Crim1 expression prospectively delineates a subpopulation highly enriched in CSNTL: Crim1+ CSN axons preferentially collateralize in thoracolumbar gray matter
Because Crim1 is also expressed by spinal neurons (Kolle et al., 2000), we were not able to investigate axon branching by Crim1+ CSN using Crim1GCE; Thy1-STOP-YFP mice as described above, since their spinal gray matter is also invaded by YFP+ collaterals from Crim1+ spinal neurons (Figures S7P–S7P’’’). To circumvent this limitation, we employed a mouse line, Emx1-IRES-FlpO (Greig, 2015), which drives FlpO recombinase expression in neocortical projection neurons, with no spinal expression (Gorski et al., 2002). We used intersectional reporter Ai65 (RCFL-tdT) mice, in which tdTomato expression occurs only upon both Cre and Flp expression (Madisen et al., 2015). We generated Crim1GCE; Emx1-IRES-FlpO; Ai65(RCFL-tdT) (referred to here as ‘‘CERai65’’) intersectional genetic reporter mice and administered tamoxifen at P3.5 (schematized in Figure 7B). We confirmed that CERai65 mice eliminate all noncortical reporter expression (observed in Crim1GCE; Thy1-STOP-YFP mice). As expected, we detect YFP+ neurons in the striatum and medial septum of Crim1GCE; Thy1-STOP-YFP mice (Figures S7N–S7N’’’), and this labeling is eliminated in CERai65 mice. Similarly, spinal neuronal labeling in Crim1GCE; Thy1-STOP-YFP mice (Figures S7P–S7P’’’) is also eliminated in CERai65 mice (Figures S7Q–S7Q’’). tdTomato expression in the medial cortex, where Crim1+ CSN reside (Figures S7O–S7O’’’), is maintained in CERai65 mice and tdTomato+ axons extend to all spinal levels (Figures 7C–7E).
We therefore utilized CERai65 mice to investigate whether segmental axonal branching by Crim1+ CSN is similar to CSNTL. We compared the Crim1+ CSN axon collateral area with CSNTL (labeled by T2 AAV-Cre at P4) and the overall CSNmedial subpopulation (labeled by C1 AAV-Cre at P4). We find that 76% ± 1% of the total Crim1+ CSN axon collateral area is distributed over thoracolumbar segments, with 24% ± 1% branched into the cervical cord (Figure 7G). Therefore, Crim1+ CSN axon collateral distribution is nearly identical to CSNTL (Figures 3P and 7G) and distinct from overall CSNmedial (Figure 7G).
We next investigated whether segmental collateral distribution of Crim1+ CSN axons matches the distribution of CSNTL (Figure 3Q). As with CSNTL, the highest peaks of the Crim1+ axon collateral area occur in the L2–L5 bins of the lumbar enlargement (Figures 7H–7Q). We statistically compared the overall cumulative segmental axon collateral distribution between Crim1+ CSN and CSNTL as well as CSNmedial using the two-sample Kolmogorov-Smirnov test (Kirkman, 1996). We find that the overall cumulative segmental axonal collateral distribution of Crim1+CSN is not significantly different from that of CSNTL (Figure 7R) but differs significantly from that of CSNmedial (p < 0.005; Figure 7R). Moreover, Pearson pairwise correlation analyses of segmental axon collateral distribution reveal a significant correlation between Crim1+ CSN and CSNTL (Pearson correlation; r = 0.578, p < 0.003; Figure S7U), but no correlation between Crim1+ CSN and CSNmedial (Figure S7V) or between CSNTL and CSNmedial (Figure S7W). These results indicate that Crim1+ CSN axons exhibit significantly different segmental collateral distribution when compared to all other CSNmedial axons. Together, the combined results of matched segmental specificity of axon targeting, matched axon collateralization within lumbar enlargement, identical cumulative segmental axon collateral distribution, and correlation of segmental collateralization strongly indicate that Crim1 expression during development is a reliable indicator for CSNTL or a subpopulation highly enriched in CSNTL.
These results validate the hypothesis that molecular delineation of defined CSN subpopulations during development can prospectively identify their segmentally distinct spinal axon connectivity into maturity. Together, these analyses enable delineation during development of CSN subpopulations with persistent distinct segmental axon targeting, well before final axon connectivity is established: (1) Klhl14 expression delineates Klhl14-positive CSNBC-lat from Klhl14-negative CSNBC-med, (2) all CSNTL are Klhl14 negative, and (3) ~95% of CSNTL express Crim1 (schematized in Figure 7S).
DISCUSSION
Segmentally specific CSN connectivity is critical for precise execution of skilled movement. Further, individual human neurodegenerative diseases predominantly affect segmentally specific CSN (e.g., bulbar ALS or HSPs). In this report, we identify (1) that segmentally specific corticospinal connectivity is initially established by axon targeting specificity and (2) that distinct CSN subpopulations are molecularly specified during development to target distinct spinal segments at maturity. We identify molecular controls that distinguish distinct CSN subpopulations during development and prospectively identify them before differential axon targeting in the cord. Thus, despite CSN sharing a broad developmental program, additional controls expressed by distinct CSN subpopulations likely control their distinct connectivity, physiology, and function.
Topographic organization of CSN subpopulations in the sensorimotor cortex from development into maturity
Functional maps have identified CSN connectivity from distinct cortical regions, several outside M1 (Cisek et al., 2003; Donoghue and Wise, 1982; Kwan et al., 1978; Neafsey et al., 1986; Tennant et al., 2011), consistent with our findings that a substantial CSNBC subset resides outside M1. Recent work has additionally highlighted that, even in rodents, CSN residing outside M1 have distinct axon projections in the spinal gray matter, have distinct synaptic connectivity, and control distinct functional outputs in comparison with M1 projections (Liu et al., 2018; Ueno et al., 2018; Wang et al., 2017). Our results are broadly consistent with the overall topography of established functional maps in adult rodents: CSNBC are enriched in areas that will eventually be the adult forelimb cortex, and the majority of CSNTL reside in what will eventually become the hindlimb cortex (Tennant et al., 2011). Collectively, our data, combined with prior analyses in adults, indicate that the overall topography of segmentally specific CSN projections is established relatively early in development.
Our data also indicate that molecular and connectivity analyses are complementary to microstimulation experiments in more fully identifying CSN connectivity and diversity. Many cervical-projecting CSNBC-lat reside in the rostrolateral sensorimotor cortex, extending into the ‘‘jaw representation’’ (Tennant et al., 2011). From our anterograde analyses, we identify that these CSN send projections throughout the rostrocaudal extent of the cervical cord (Figure 2). It is likely that interspersed CSN populations with jaw versus forelimb connectivity reside in the same cortical domain that would have been identified by microstimulation as the ‘‘jaw area.’’ There are likely other subsets of interspersed projections with connectivity distinct from previously functionally identified cortical domains. This indicates that while microstimulation can identify the presence of CSN connectivity, the absence of movement by microstimulation does not mean an absence of projection from a cortical area to a specific spinal level.
CSNBC-lat cervical collaterals extend more ventrally than cervical collaterals from CSNmedial, which is reminiscent of CSN collaterals from the rostral versus caudal forelimb cortex in cats (Martin, 1996). CSNBC-lat projections might play distinct function(s) compared to CSNmedial projections; for example, digit movement in rats is almost entirely evoked by stimulating RFA and not CFA (Kleim et al., 1998). Future investigations of now molecularly distinct CSN subpopulations can elucidate their contributions to distinct aspects of motor control and nonmotor functions.
CSNBC and CSNTL exhibit striking axon targeting specificity during development
Previous studies using diffusible dyes suggested some promiscuity of axon extension by which some CSN axons from the forelimb motor cortex extended to the lumbar cord (Kuang and Kalil, 1994). Our approach of using intersectional viral labeling during development directly addresses this question of promiscuity versus specificity of axon extension during development. We find that there is some promiscuity of axon targeting, with ~25% of CSNmedial projecting to T2 at P4, pruning this projection by P28. However, ~75% of these P4 projections are still present at P28, indicating that axon extension specificity from precircuit development is largely maintained. This early axon segmental targeting specificity of CSNTL also appears to establish the specificity of axon collateralization in the spinal gray matter at thoracolumbar versus cervical segments, with 70% versus 30% of total CSNTL spinal collateral area within these segmental regions, respectively.
We find that CSNBC-lat versus CSNTL differential targeting is not due to either differences in axon extension rates, or promiscuous growth with later pruning, but likely reflects tight molecular control over axon extension by molecularly distinct subpopulations (investigated in Sahni et al., 2021). This might reflect additional levels of control over connectivity between CSN subpopulations, since ‘‘promiscuous’’ CSN reside in M1, and we identify them as distinct from CSNBC-lat, anatomically and molecularly. We speculate that CSNBC-lat targeting specificity reflects tighter control for specialized function.
Identification of CSNBC-med as a distinct subpopulation in the medial sensorimotor cortex
Consistent with previous investigations (Kamiyama et al., 2015), we find CSN subpopulations that innervate both cervical and lumbar segments. These previous investigations injected tracers into spinal gray matter at cervical C7 or lumbar L4, thereby focusing on CSN that had already collateralized into those gray matter segments. Our results indicate that these dual, cervical- and lumbar-projecting CSNare a subset of CSNTL. However, we injected retrograde tracers and AAV vectors directly into DF (i.e., white matter), which identified all axons projecting to and beyond that spinal level; tracer injection at T2 labels all CSN projecting past the cervical cord to thoracolumbar segments, not just projections at T2. This key difference enabled us to screen entire subpopulations, rather than sampling CSN innervation at just two levels. Therefore, while prior studies (Kamiyama et al., 2015) established that some CSNarborize at both C7 and L4, they could not identify the presence of CSNBC-med, which we now identify as an anatomically and molecularly distinct CSN subpopulation in the medial cortex.
Segmentally distinct CSN subpopulations are molecularly distinct, including differential expression of genes known to direct CST pathfinding
We find that some genes differentially expressed by CSNBC-lat versus CSNmedial can function in modulating downstream signaling known to regulate CSN axon growth, such as insulin-like growth factor-1 (IGF-I) (Ozdinler and Macklis, 2006) and Wnt-Ryk (Liu et al., 2005). CSNBC-lat express genes known to negatively regulate these mechanisms. For example, CSNBC-lat express pappalysin 2 (Pappa2), which cleaves IGFBP-5 (Overgaard et al., 2001). IGFBP5 normally augments IGF signaling in the brain (Pera et al., 2001; Salih et al., 2004); thus, its cleavage is expected to negatively regulate IGF signaling. CSNBC-lat also express Frizzled related protein (Frzb), a known Wnt inhibitor in the CNS (Jang et al., 2013; Lodewyckx et al., 2012) (Figure S3). Conversely, CSNmedial-specific genes include positive regulators of pathways that promote CSN axon growth: (1) Wnt ligands Wnt4 and Wnt5a (Figure S3; Tables S1 and S2) and (2) genes positively regulating the thyroid hormone (TH) pathway, which is required for CST extension to the lumbar cord (Hsu et al., 2008), including Mu-crystallin (Cry-mu) (Mori et al., 2002) and Slc16a2 (Dumitrescu et al., 2006; Trajkovic et al., 2007) (Figure 4; Table S1).
Finally, more subtly graded expression differences over space and time are likely to be important in the fine control of targeting and final connectivity. Molecular differences at later developmental times likely control collateral branching, synaptic connectivity, and pruning. CSN collateralization occurs at distinct times between cervical and lumbar segments (Kamiyama et al., 2015; Schreyer and Jones, 1988), which is possibly regulated by such distinct molecular controls. Further, in addition to genes exhibiting sharp boundaries, we also identified CSN-specific genes exhibiting gradients of expression. While these are more highly expressed by one CSN subpopulation versus another, this suggests that gene dosage might affect differential connectivity.
Crim1 expression prospectively identifies a segmentally distinct CSN subpopulation
We previously identified Crim1 as CSN specific (Arlotta et al., 2005; Molyneaux et al., 2005) and Crim1+ cortical neurons extend subcortical axons (Leighton et al., 2001). Here, we find that Crim1 expression by CSN during development predicts largely thoracolumbar projection at maturity. Crim1 expression is regulated by area-specific controls, such as Ctip1 (Greig et al., 2016). However, Crim1 specificity goes beyond CSN spatial locations, with interspersed Crim1-positive and Crim1-negative subpopulations in the medial cortex, indicating that Crim1 expression is not only controlled by areal patterning mechanisms. Acquisition of cortical projection neuron subtype identity involves specification of SCPN versus CThPN and CPN. Delineation of CSN subpopulations via Crim1 expression likely reflects more specific refinement of subtype identity acquisition. Fezf2, a critical determinant of SCPN, and other known molecular controls required for establishing SCPN identity (e.g., Ctip2) are not differentially expressed between CSNBC-lat and CSNmedial. Therefore, transcriptional control over Crim1 expression is likely mediated by additional interconnected mechanisms, yet to be identified.
Subpopulation-specific molecular controls have relevance for motor neuron disease
CSN degeneration in motor neuron diseases, such as ALS (Bruijn et al., 2004), HSP (Salinas et al., 2008), and primary lateral sclerosis (PLS) (Singer et al., 2007; Strong and Gordon, 2005), causes spasticity, dyscoordination, and paralysis. ALS and related CSN diseases exhibit well-known heterogeneity; for example, brainstem-projecting CSN preferentially degenerate in bulbar ALS, and primarily lumbar-projecting CSN degenerate in HSP. Early developmental controls, when dysregulated and/or when they have unique function in a specialized subtype, might predispose CSN subpopulations for preferential vulnerability to later degeneration in maturity. This might be due to increased complexity, an inherent increase in vulnerability with specific circuitry maturation, imperfect circuitry formation, and/or other causes.
It is now clear that multiple mechanisms cause ALS (Ravits et al., 2013) and that subtle interactions of genetics with other factors might lead to subtype-specific vulnerability. There is emerging evidence that molecular controls over CSN development are important candidate genes linked to CSN disease. Resequencing of CSN developmental genes (Arlotta et al., 2005) has identified novel candidate ALS-disease genes, including Crim1 and Cry-mu investigated here (Daoud et al., 2011).
Implications for organization, plasticity, and evolution of cortical function in motor control
The results presented here might also shed light on CST plasticity after spinal cord injury (SCI), in which changes in segmental innervation have been reported. Such plasticity has been extensively studied in both animal models (Fouad et al., 2001; Raineteau and Schwab, 2001) and humans (Oudega and Perez, 2012). For example, after thoracic SCI, CSN in the hindlimb cortex increase collateralization into the cervical cord (Fouad et al., 2001). This has been presumed to reflect increased sprouting by lumbar-projecting CSN (likely CSNTL) axons into the cervical cord. Our data that CSNBC-med and CSNTL are spatially interdigitated suggest that, rather than solely cervical collateralization by CSNTL axons in such cases, there might be increased collateralization by axons from CSNBC-med. Future experiments using Crim1-labeled CSNTL axons (e.g., using CERai65 mice) might distinguish between these possibilities.
M1 in primates can be divided into an ‘‘older’’ and ‘‘newer’’ M1 based on the absence or presence of CM cells (cortico-motoneuronal cells, i.e., CSN with monosynaptic connections on spinal motor neurons) that are critical for fine dexterous movement (Heffner and Masterton, 1975; Rathelot and Strick, 2009). Skilled movements are now known to be phylogenetically older than primates, present in rodents, especially in the forelimb (Whishaw, 2003). This correlates with an increase in cortical representation of the forelimb versus hindlimb, even in mice (Tennant et al., 2011). We find that CSNBC-lat largely reside in the evolutionarily newer cortex (Krubitzer, 2007) and that CSNBC-lat-specific Klhl14 is a relatively recent evolutionary addition; Klhl14 orthologs are present in vertebrates, but not invertebrates (no orthologs in Drosophila melanogaster, Caenorhabditis elegans, or Ciona intestinalis; data from https://ensembl.org). Conversely, a significant number of CSNmedial-specific genes, including Crim1, are also expressed in the cingulate cortex, a phylogenetically older region.
Klhl14 expression is conserved in the developing human neocortex (Miller et al., 2014). At 21 weeks postconception (pc), Klhl14 is expressed in the cortical plate and excluded from germinal zones. Further, Klhl14 is highly expressed at 21 weeks pc, and its levels sharply decline after 24 weeks pc (Figure S6N), coincident with CSN axons reaching cervical C8 (Eyre et al., 2000). Hence, similar to mice, Klhl14 is expressed by human postmitotic neurons in development; its levels decline once CSN axons reach the caudal cervical cord.
Our identification of molecular controls that distinguish segmentally specific CSN subpopulations offers insight (1) into the precision of circuit development for motor control and (2) into potential abnormalities of their development that might underlie later disease vulnerability. These results suggest that early regulators play critical roles in establishing specificity of corticospinal connectivity. In the accompanying study (Sahni et al., 2021), we functionally investigate a subset of these genes identifying their role(s) in controlling segmentally specific CSN axon targeting. Future investigations will likely identify additional nested subpopulations with more molecular diversity, highlighting potentially even greater early differentiation and specialization of CSN during early development that underlie the establishment of this complex motor control circuitry.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Jeffrey D. Macklis (jeffrey_macklis@harvard.edu).
Materials availability
We plan to deposit Emx1-IRES-FlpO mice to the Jackson Laboratory or MMRC. All unique/stable reagents generated in this study are available with a materials transfer agreement from the lead contact for academic, non-commercial use; negotiation and completion of a materials transfer agreement with Harvard University is required if there is potential for commercial application.
Data and code availability
The microarray data profiling lateral versus medial CSN subpopulations at P1, P4, and P7 have been deposited at GEO and are publicly available. Accession number is listed in the key resources table. Gene intensities using Rosetta Resolver for all genes have been deposited on Mendeley and are publicly available. The DOI is listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-CTIP2 | Abcam | Cat# ab28448; RRID:AB_1140055 |
| Mouse anti-SATB2 | Abcam | Cat# ab51502; RRID:AB_882455 |
| Rabbit anti-GFP | Molecular Probes | Cat# A-1112; RRID:AB_221569 |
| Chicken anti-GFP | Thermo Fisher Scientific | Cat# A10262; RRID:AB_2534023 |
| Rabbit anti-RFP | Rockland | Cat# 600–401-379; RRID:AB_2209751 |
|
| ||
| Bacterial and virus strains | ||
| AAV-8 hsyn-GFP-Cre | UNC Vector core | N/A |
| AAV-2/1 CAG-FleX-TdTomato | From University of Pennsylvania vector core (now at Addgene) | N/A |
| AAV 2/1 CAG-EGFP | vector core at Massachusetts General Hospital, Boston, MA | Maguire et al. (2013) |
| AAV 2/1 CAG-tdTomato | This paper | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
| Green fluorescent microspheres | LumaFluor | Green Retrobeads IX |
| Cholera Toxin B subunit, Alexa 555 conjugate | ThermoFisher | C34776 |
| Cholera Toxin B subunit, Alexa 647 conjugate | ThermoFisher | C34778 |
|
| ||
| Critical commercial assays | ||
| Mouse Genome Affymetrix 430.2 arrays | Affymetrix | N/A |
| Thermoscript Reverse transcriptase kit | Fisher scientific | 12236 |
| RNeasy FFPE | QIAGEN | 73504 |
| SYBR® Green PCR Master Mix | ThermoFisher | 4309155 |
| RNAscope 2.5 HD Assay-RED | ACD Bio | Made to order-Crim1 (550751) |
|
| ||
| Deposited data | ||
| Gene expression profiling of CSN subpopulations by microarray | This paper | GEO: GSE77311; Mendeley: https://doi.org/10.17632/9jj6jm2bdp.1 |
|
| ||
| Experimental models: Organisms/strains | ||
| Crim1GCE mice | The Jackson Laboratory | GUDMAP database Harding et al. (2011) |
| Fezf2 null | N/A | Hirata et al. (2004) and Molyneaux et al. (2005) |
| Thy1-STOP-EYFP | N/A | Buffelli et al. (2003) and Bareyre et al. (2005) |
| Emx1-IRES-FlpO | This Paper | N/A |
| Ai65(RCFL-tdT)-D or Ai65D | The Jackson Laboratory | Stock number: 021875 |
|
| ||
| Oligonucleotides | ||
| See Table S4 for oligonucleotide information | N/A | |
|
| ||
| Recombinant DNA | ||
| pCAG-tdTomato-t2A-STOP | This paper | N/A |
|
| ||
| Software and algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Rosetta Resolver software | Rosetta (now Microsoft) | N/A |
| Bioconductor | Bioconductor.org | Bioconductor.org |
| GraphPad Prism 8.0 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
No original code was generated as part of this study.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice used in this study
Wild-type mice on a pure C57BL/6 and CD1 background were obtained from Charles River Laboratories (Wilmington, MA). The day of vaginal plug detection was designated as E0.5. The day of birth was designated as P0. All mouse studies were approved by the Harvard University IACUC, and were performed in accordance with institutional and federal guidelines. Thy1-STOP-YFP mice were generously provided by Dr. Joshua Sanes at Harvard University. Fezf2 –/– mice were generated previously (Hirata et al., 2004) and have been previously described (Molyneaux et al., 2005).
Crim1GCE mice (Harding et al., 2011) were obtained from Jackson Laboratories; they contain an EGFP coding sequence followed by a tamoxifen-inducible Cre recombinase coding sequence placed 3′ to the ATG of the first codon. The original depositing investigator observed no EGFP expression, and we confirm no EGFP expression at any age. Cre was induced using tamoxifen (Sigma) dissolved in corn oil (Sigma). A 3.5 mg/ml stock solution was prepared, and each pup received 350 μg (100 μL of stock / pup). For investigating and quantification of axon segmental targeting by Crim1+ CSN axons, we used Crim1GCE; Thy1-STOP-EYFP double heterozygous mice. These mice were pulsed with tamoxifen either at P0 (Figure 7F) or at P3.5 (Figures 7F, S7A–S7C, S7F–S7L, S7N, and S7P).
Emx1-IRES-FlpO mice were generated and validated (Greig, 2015) following the same strategy employed by Jones and colleagues(Gorski et al., 2002) to introduce an IRES-FlpO cassette into the 3′ untranslated region of the Emx1 gene.
Ai65 (RCFL-tdT) mice were obtained from Jackson Laboratories (stock number 21875), and genotyped using their recommended protocol.
For establishing CERai65 triple transgenic mice, we bred generated Crim1GCE/+, Emx1-IRES-FlpO/+, and ai65 (RCFL-tdT)/+ mice to obtain Crim1GCE/+; Emx1-IRES-FlpO homozygous; ai65 (RCFL-tdT) homozygous mice. These mice were then pulsed with tamoxifen (as described above) and used for downstream analyses of axon projection targeting and collateralization.
The genders of early postnatal mice were not determined. Mice were used at the following ages:
Crim1GCE mice: were used at P5; both males and females were used at P15, and at P24.
Thy1-STOP-EYFP mice: were used at P5; both males and females were used at P15.
Emx1-IRES-FlpO mice: both males and females were used at P24.
Ai65 (RCFL-tdT) mice: both males and females were used at P24.
METHOD DETAILS
Anterograde and retrograde labeling
CSN were retrogradely labeled bilaterally from specific spinal levels by injecting the retrograde label Cholera Toxin B subunit (CTB; Thermo Scientific) into each side of the midline using ultrasound backscatter microscopy (Vevo 770; VisualSonics, Toronto, Canada) via a pulled glass micropipette with a nanojector (Nanoject II, Drummond Scientific, Broomall, PA). For cervical and thoracic cord labeling (P1, P2, and P4), we counted the vertebral segments from C1 to C8. An identical approach was used to inject AAV-Cre at P4 into cervical C1 versus thoracic T2 for intersectional viral labeling experiments. The landmark for labeling CSN whose axons reach thoracic T12/13 -lumbar L1 was determined by examining the relative position of the vertebral column as it approaches the dorsal surface of the body (moving from caudal thoracic levels rostrally, where the column is located more deeply and closer to the viscera, to lumbar levels caudally, where the column is situated closer to the dorsal surface). The central landmarks for all intraspinal injections are the midline, vertebral bodies, dorsal aspect of the spinal cord, and echo density in the dorsal funiculus. For these neonatal injections, pups were anesthetized under ice for 4 minutes. The pups were placed on a heating pad for recovery.
For AAV-mediated anterograde labeling, AAV2/1 particles expressing fluorescent protein were injected at P0 into specific cortical sub-regions under guidance by ultrasound backscatter microscopy, using techniques similar to those described above. The central landmarks for the intracranial injections that provide both accuracy and precision are the midline, dorsal and lateral aspects of the lateral ventricle, anterior aspect of the hippocampus, posterolateral aspect of the striatum, corpus callosum, and its genu. All virus work was approved by the Harvard Committee on Microbiological Safety, and conducted according to institutional guidelines.
Retrograde labeling and FACS purification
Green fluorescent microspheres (LumaFluor, Naples, FL) were injected under guidance by high-resolution ultrasound backscatter microscopy into the cerebral peduncle at the midbrain-hindbrain boundary at P0, or into the CST at cervical segments C1/C2 at P3 and P5.5. Twenty-four hours (P0 and P3 injection) or 36 hr (P5.5 injection) later, neocortical tissue was microdissected for dissociation. The meninges were removed from fluorescently-labeled cortices. A custom-designed multi-blade microtome was used to precisely and distinctly microdissect the rostrolateral and caudomedial regions of motor cortex, using a fluorescence dissecting microscope to precisely visualize the labeled cortical regions during dissection (shown stepwise in Figure 4). Retrogradely labeled neurons from each sub-region were purified by fluorescence activated cell sorting (FACS) using established methods (Arlotta et al., 2005; Catapano et al., 2001; Molyneaux et al., 2009).
Experimental approach for purifying CSN subpopulations for transcriptional analysis
We aimed to identify candidate molecular controls distinct for each subpopulation– CSNBC-lat, CSNBC-med, and CSNTL– that controlled their differential axon targeting. One possible approach might have been to differentially label CSNBC-med and CSNTL by using distinct retrograde labels from cervical versus thoracic cord (as in Figure 1) once this axon targeting had been established, then transcriptionally analyze these differentially labeled subpopulations. However, this approach would not detect earlier molecular differences and controls over CSN axon targeting that might be downregulated by the time distinct axonal targeting is actually achieved. We reasoned from much prior knowledge in the field of cortical projection neuron development, identifying a number of early-expressed transcriptional controls that are absent or substantially downregulated by P0-P4, that it would be likely that relevant controls over spinal segmental targeting might be similarly substantially downregulated once their early axon targeting functions had been served (and the earliest possible stage to label from thoracolumbar segments would have been P4).
We used an alternative approach to discover earlier molecular controls. Since CSNBC-lat are relatively homogeneous, with only bulbar-cervical projections, we compared CSNBC-lat gene expression with the more heterogeneous CSNmedial subpopulation (which includes CSNTL). We performed transcriptional analysis at developmentally informative stages to identify candidate gene expression that might be specifically required for CSNTL axon projection past cervical cord. For example, at P4, CSNTL are the only CSN in medial cortex that project axons past the cervical cord toward thoracolumbar segments, so potential early molecular controls over axon extension to distal spinal segments would be predicted to be expressed only by the CSNTL subset of CSNmedial (and not at all by CSNBC-lat).
We isolated CSNmedial and CSNBC-lat at developmentally critical stages of P1, P4, and P7. At P1, CSNmedial and CSNBC-lat axons have simultaneously reached only the cervical cord–CSNBC-lat axons have halted their white matter extension, while CSNmedial axons will continue white matter extension. At P4, CSNBC-lat axons are innervating the cervical cord, while a subset of CSNmedial (CSNTL) are extending axons through thoracic cord. At P7, CSNTL are innervating thoracic and lumbar target segments.
We purified CSNBC-lat and CSNmedial from rostrolateral versus caudomedial cortex respectively, via fluorescence-activated cell sorting (FACS) following retrograde labeling with green fluorescent microspheres under high-resolution ultrasound backscatter microscopic guidance. We performed retrograde labeling from cervical C1 at P0, P3, or P5.5 for purification of CSN subpopulations at P1, P4, and P7, respectively
We microdissected the non-overlapping rostrolateral versus caudomedial regions of developing sensorimotor cortex (Figures 4A–4E’) using a custom-made four-blade instrument to microdissect 700 μm tissue blocks. We then dissociated isolated neuron suspensions (Figure 4F) using our previously validated approaches (Arlotta et al., 2005; Catapano et al., 2001; Molyneaux et al., 2009; Ozdinler and Macklis, 2006) and performed differential gene expression analysis.
Affymetrix microarrays
RNA samples for microarrays were collected from independent FACS purifications from each age to rigorously validate reproducibility and significance (n = 2 for P4 and P7; n = 3 for P1). Samples were hybridized to Mouse Genome Affymetrix 430.2 arrays (Affymetrix, Santa Clara, CA) arrays per Affymetrix GeneChip Expression Analysis protocol, stained using an Affymetrix GeneChip Fluidics Station 450, and scanned with a GeneChip Scanner 3000. All microarray data have been deposited in the Gene Expression Omnibus database at NCBI (Accession GSE77311).
Data files from microarrays were uploaded via Rosetta Resolver software (version 5.0; Rosetta Biosoftware, Seattle, WA) for normalization and intensity ratio calculations, or with Bioconductor (www.bioconductor.org) for normalization with three independent methods: (1) Microarray Suite 5.0 (MAS5), in which arrays are scaled to have the same mean value; (2) Robust Multi-Array Analysis (RMA), which normalizes the data at the probe level, and across all microarrays; (3) GC-RMA, a modification of RMA, which accounts for GC content of the individual probes. Statistical analysis was performed by pairwise comparisons at each age with Rosetta Resolver, or with Statistical Analysis of Microarrays (SAM; performed on RMA, GC-RMA and MAS5 normalized data). The temporal expression profile plots are intensities obtained from the Rosetta Resolver software (Figures 4 and S3). Error bars represent standard error of mean (SEM).
We applied a cut-off of a ≥ 2-fold difference in expression with a p value ≤ 0.005 based on Rosetta Resolver parameters, to select genes significantly differentially expressed between the two CSN subpopulations. In addition, we performed cross-correlation with other statistical approaches such as Statistical Analysis of Microarrays (SAM) (applied to data normalized with RMA, GCRMA or MAS5). Although different statistical methods can include some differences in highly significant genes, genes that are repeatedly top ranking are highly likely to be true positives.
Validating reproducibility of Microarray data
We confirmed the reproducibility and consistency of data by comparing gene expression between independent biological replicates (3 replicates for P1, and 2 replicates each for P4, and P7). Correlation coefficients for biological replicates range from 0.96 to 0.99, indicating high reliability of the data. Anterograde and retrograde analyses (presented in Figures 1 and 2) identify that CSNBC-lat represents a relatively more homogeneous subpopulation as compared to CSNmedial. Consistent with this, we find that CSNBC-lat biological replicates are more highly correlated than CSNmedial biological replicates (Figure S3A) likely reflecting the heterogeneity in CSNmedial (which includes both CSNBC-med + CSNTL).
Statistical criteria used for selecting candidate genes from microarray data for further analysis
Genes differentially expressed in lateral sensorimotor cortex are expressed specifically by CSNBC-lat. However, since CSNBC-med and CSNTL are interdigitated in medial sensorimotor cortex, genes differentially expressed by CSNmedial might be expressed either by CSNBC-med or CSNTL or both subpopulations. Our combined anterograde and retrograde analyses identified P4 as the peak time of CSNTL axon extension toward distal spinal segments. Therefore, to molecularly delineate CSNTL, as a distinct subset within CSNmedial, we specifically investigated genes that exhibited peak differential expression by CSNmedial at P4. Our hypothesis was that such genes might control axon extension to distal spinal segments and would therefore be expressed only by CSNTL and not CSNBC-med, and not at all by CSNBC-lat. This would therefore enable molecular delineation of CSNTL as a molecularly distinct subpopulation within CSNmedial. In addition, CSNBC-lat axons target the cervical cord at P1 (Figures S1A–S1O). We therefore investigated genes that are differentially expressed by CSNBC-lat at P1 to identify potential molecular controls that potentially limit CSNBC-lat axon extension to the cervical cord. We next detail the criteria utilized for identifying such candidate genes for further analyses.
We prioritized candidate genes for additional expression analysis and functional investigation based on three central criteria: (1) we examined genes found to be most statistically significantly differentially expressed by either subpopulation; (2) we examined temporal expression, and prioritized genes exhibiting peak differential expression at P1 in CSNBC-lat (when they target cervical cord) and P4 in CSNmedial (when CSNTL target thoracic and lumbar cord); and (3) we cross-referenced these potential top candidates with published datasets of genes with CSN-specific expression (using our previously published analyses comparing CSN with CPN, CThPN, and corticotectal projection neurons (Arlotta et al., 2005; Galazo et al., 2016; Molyneaux et al., 2009), and the DeCoN database (Molyneaux et al., 2015)). We prioritized genes that exhibit CSN-specific expression (in comparison with CPN and CThPN). Of the top ranked CSNmedial genes that are differentially expressed at P4, Crim1 (cysteine rich transmembrane BMP regulator 1) is the highest ranked gene that exhibits CSN specificity (genes highlighted in red font in Table S2). While other genes are ranked higher than Crim1 based solely on level of differential expression and significance (e.g., Gpr88, Hmgb1), these genes either exhibit no enrichment by CSN (e.g., Hmgb1) or exhibit significantly higher expression levels in other projection neuron populations (e.g., Cav1, Gpr88 are highly expressed by CPN). Further, Crim1 differential expression is highly statistically significant only at P4, and not at the other two times examined - P1 or P7 (Figure 4P, Table S2), indicating that Crim1 exhibits peak differential expression just when CSNTL axons target thoracolumbar cord. Kelch-like 14 (Klhl14) is the highest ranked differentially expressed gene for CSNBC-lat (Table S3). Using these a priori criteria, the highest ranked CSN-specific genes are Crim1 in CSNmedial and Klhl14 in CSNBC-lat, each showing peak differential expression at the appropriate times for each CSN subpopulation (Crim1 by two separate probes, overall SAM RMA ranks 5, 45; Klhl14 by two separate probes, overall SAM RMA ranks 1, 3; see Tables S2 and S3).
Interestingly, microarray analysis finds ~1.7-fold enrichment of Crim1 in CSNmedial at P1 (compared to ~5-fold enrichment at P4). This suggests that, even though CSNTL exhibit relatively selective Crim1 expression at P0, differential analysis comparing overall CSNmedial at P1 was unable to stringently detect this selective expression. This is likely because this modest enrichment is diluted by heterogeneity of CSNmedial.
Generation of AAV particles
AAV2/1 particles were generated at the Massachusetts General Hospital Virus Core using established protocols (Maguire et al., 2013). EGFP (for AAV-EGFP), and tdTomato (for AAV-tdTomato) coding sequences were cloned into a shuttle plasmid that contains the following elements flanked by AAV2 ITRs: a CMV/b-actin promoter to drive the expression of the gene of interest, followed by the woodchuck hepatitis virus post-transcriptional regulatory element (WPRE), a bovine GH pA signal, and an SV40 pA signal.
AAV8 hsyn-GFP-Cre was obtained from the vector core at the University of North Carolina at Chapel Hill (UNC Vector Core). This virus is retrogradely trafficked at a lower efficiency, which enabled accurate and reproducible visualization of individually labeled CSN axons in the cord. AAV2/1 CAG-FLEX-tdTomato (originally generated by the Allen Institute) was obtained from the vector core at the University of Pennsylvania.
Immunocytochemistry and in situ hybridization
Brains were fixed and stained using standard methods (Galazo et al., 2016; Greig et al., 2016; Woodworth et al., 2016). Primary antibodies and dilutions used: rat anti-CTIP2, 1:500 (Abcam); chicken anti-GFP, 1:500 (Invitrogen); rabbit anti-GFP, 1:500 (Invitrogen); rabbit anti-RFP 1:500 (Rockland Immunochemicals), mouse anti-SATB2, 1:500 (Abcam). In situ hybridization was performed as previously described(Galazo et al., 2016; Greig et al., 2016; Woodworth et al., 2016). cDNA clones for riboprobes are listed in Table S4. Fluorescence in situ hybridization (FISH) for Cry-mu using DIG-labeled probes was performed as previously described(Molyneaux et al., 2009).
Single molecule Crim1 in situ hybridization
Single molecule in situ hybridization was performed using the RNAscope 2.5 HD RED kit according to the manufacturer’s instructions (Advanced Cell Diagnostics). In brief, 16 μm thick brain cryosections were mounted onto glass slides in phosphate buffer (pH 7.2), airdried, and baked at 60°C for 30 min, followed by pretreatment with hydrogen peroxide for 10 min, target retrieval, and ethanol dehydration. After the pretreatment with Protease Plus (1:10 diluted in PBS) at 40°C for 30 min, brain sections were incubated with Crim1 probe (550751, Entrez Gene: NM_015800.3) at 40°C for 2 h, and the standard RNAscope protocol was followed. Incubation time of amplification step 5 and color reaction were optimized at 12 min and 5 min, respectively.
Laser-capture microdissection and quantitative PCR
35 μm sections of labeled brains were stained for EGFP, and mounted on membrane PEN slides (Carl Zeiss). Labeled neurons were visualized, and captured into adhesive cap tubes (Carl Zeiss) using a PALM laser microdissection system (Carl Zeiss) using a uv laser to cut and flip tissue samples directly into inverted adhesive caps placed above slides. RNA was extracted using an RNeasy FFPE kit (QIAGEN). Reverse transcription was performed using random hexamers and Thermoscript reverse transcriptase (Fisher Scientific). Real time PCR was performed using SYBR® Green PCR Master Mix (Thermo Fisher) on a Realplex4 cycler (Eppendorf). We confirmed the specificity and size of the amplicons by running the PCR product on agarose gels, and by melting curve analysis. For Klhl14-overexpression experiments, data were analyzed using an unpaired two-sided t test. The PCR primers are listed in Key Resources Table.
Imaging and quantification
For analysis of retrogradely labeled CSN from cervical versus thoracic levels, 50 μm coronal brain sections of retrogradely labeled brains were imaged on an Axioscan Z1. We set a priori anatomical criteria for distinguishing medial versus lateral locations of CSN in sensorimotor cortex. For counting CSN distributed in medial versus lateral locations in cortex, images of coronal brain sections at specific rostro-caudal levels (shown in Figure 1) were binned into 5 medio-lateral bins spanning the width of each cortical hemisphere, and medial versus lateral distinction was made by combining the 3 medial bins for medial CSN counts and the 2 lateral bins for lateral CSN counts. We confirmed reproducibility of this binning by also evaluating the separation of medial versus lateral cortical locations with respect to histological landmarks in each section. At the mid-rostral level, the lateral edge of the medial bin aligns with lateral aspect of the striatum. At the mid-caudal and caudal levels, the barrels in primary somatosensory cortex (S1) were completely included in the two lateral bins. At the caudal level, the hippocampus was completely included in medial cortex. CSNmedial include all CSN in agranular cortex, including both primary and secondary motor cortex. Therefore, primary motor cortex, including the transition zone between agranular motor cortex and granular sensory cortex, is completely included in medial cortex. This medial versus lateral division aligned with and informed the 700 μm tissue blocks that were made on retrogradely labeled brains following microdissection for FACS purification of spatially distinct CSN subpopulations. Both cortical hemispheres were analyzed for each mouse, and every labeled neuron was counted in each section using the Cell Counter function in ImageJ.
For analysis of collateral distribution, CSN axonal collaterals in intersectional viral labeling experiments were visualized by native tdTomato fluorescence from AAV-FLEX-tdTomato. Crim1+ CSN axonal collaterals in CERai65 mice were first amplified using anti-RFP antibody (Rockland Immunochemicals) before imaging and analysis. All serial horizontal 70 μm spinal cord sections were imaged on an Axioscan Z1 (Carl Zeiss Microscopy). Each section was imaged in its entirety, from rostral to caudal and in the Z axis. These Z stacks were collapsed using the ‘‘Extended Depth of Focus’’ function on ZEN blue 2.6 image processing software (Carl Zeiss Microscopy). Collapsed images of each section were then exported as a single TIF image and imported into Adobe Photoshop. Each individual section (from dorsal to ventral) was manually aligned with its nearest neighbor using its rostral and caudal ends and its midline as landmarks. For quantification of collateral area, we first identified the boundary between gray and white matter in the spinal cord using Hoechst counter stain. The white matter was then cropped out from all the sections that contained it including the labeled, tdTomato+ corticospinal tract. TdTomato+ pixels in the spinal gray matter (representing CSN axonal collaterals) were then identified by thresholding in NIH ImageJ. The total area of thresholded pixels was measured using the ‘‘Analyze Particles’’ function, and the percentage of total area present in the entire cervical, thoracic, and lumbar cord was calculated. For the binned segmental analysis, the thresholded images of each section were first divided into the appropriate number of bins (7 in cervical, 12 in thoracic, 5 in lumbar) in Adobe Photoshop using the ‘‘Divide Slice’’ tool, and each slice was imported and analyzed in NIH ImageJ as above. For 3D rendering, aligned horizontal sections of the spinal cord were imported as an image stack in NIH ImageJ, and the rendered using the 3D project function.
For all quantification of axon counts (Figures 3I and 7F) quantification on axial sections, 60X confocal Z stacks of the entire CST in the dorsal funiculus were obtained on either a BioRad Radiance 2000 or Zeiss LSM 780 confocal microscope. Cervical, thoracic, and lumbar cord axial sections were imaged using identical parameters. Axons were counted manually in each section using the Cell Counter function in ImageJ.
QUANTIFICATION AND STATISTICAL ANALYSIS
The details of imaging quantification methodologies have been described in Method details. All n values, as well as p values obtained are also listed in the figure legends. GraphPad Prism version 8 was used to perform statistical tests in this study.
Data distributions were assumed to be normal, but this was not formally tested. No statistical methods were used to pre-determine sample sizes. For axon counts comparing intersectional viral labeling with Crim1+ axons in Crim1GCE mice, we used a one-way ANOVA followed by Tukey’s posthoc test. For comparing the total collateral area and binned collateral area distribution between cervical AAV-Cre versus thoracic AAV-Cre injected mice, we used a two-sided t test. For, comparing segmentally binned axonal collateral distribution between Crim1CreERT2-labeled CSN axons and CSN axons labeled using intersectional viral labeling with either cervical versus thoracic AAV-Cre, we used a two-sample Kolmogorov-Smirnov test as well as Pearson correlation in GraphPad Prism 8.0 (GraphPad Software, Inc., San Diego, CA).
The reason for employing distinct statistical tests to investigate axon extension versus axon collateralization is because analysis metric of axon collateralization is quite different from the metric of white matter axon extension. For axon extension analyses, axon numbers are counted at specific sites along the thoracic cord. Because this is not a cumulative distribution, we employed the statistically appropriate ANOVA. For the axon collateralization analysis in the spinal gray matter, we measured the total axonal collateral area throughout the entire spinal gray matter, then analyzed the percentage of this total collateral area at each spinal segmental bin from cervical C2 to lumbar L6. Since this is a cumulative distribution (total percentage across all bins equals 100%), we used the two sample Kolmogorov-Smirnov test, in which the null hypothesis states that there is no difference between the cumulative distributions being compared.
Supplementary Material
Highlights.
Developing corticospinal axons specifically target spinal segments before connectivity
Corticospinal neurons (CSN) targeting distinct segments are molecularly distinct
CSNBC-med is an anatomically and molecularly distinct subpopulation
CSN subpopulations can be prospectively identified through development into maturity
ACKNOWLEDGMENTS
We thank Erica Gornstein, Ryan Richardson, Gustav Cederquest, Lincoln Pasquina, Ted Yamamoto, Aaron Wheeler, Eva Gillis-Buck, Thomas Addison, and Bonnie Wall for technical assistance; members of the Macklis laboratory for input; and the Harvard Center for Biological Imaging for infrastructure support. This work was supported by the NIH (grants R01NS045523 and R01NS075672 and additional infrastructure grants NS049553, NS104055, and DP1NS1066 65), the ALS Association, the Packard Center for ALS Research at Johns Hopkins, the Travis Roy Foundation, and the Massachusetts Department of Public Health (to J.D.M.). V.S. was partially supported by the NIH (NTRAIN/Eunice Kennedy Shriver National Institute of Child Health and Human Development [NICHD] grant K12HD093427) and the DEARS Foundation. S.J.S. was partially supported by the NIH (predoctoral Ruth L. Kirschstein National Research Service Award [NRSA] F31NS063516). Y.I. had partial support from the Uehara Memorial Foundation, the Kanae Foundation, the Murata Overseas Scholarship Foundation, and the DEARS Foundation. L.C.G. was partially supported by the Harvard Medical Scientist Training Program, the NIH (predoctoral NRSA F31 NS080343), and the DEARS Foundation. J.D.M. is an Allen Distinguished Investigator of the Paul G. Allen Frontiers Group.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109843.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Akintunde A, and Buxton DF (1992). Differential sites of origin and collateralization of corticospinal neurons in the rat: a multiple fluorescent retrograde tracer study. Brain Res 575, 86–92. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, and Macklis JD (2005). Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 45, 207–221. [DOI] [PubMed] [Google Scholar]
- Bareyre FM, Kerschensteiner M, Misgeld T, and Sanes JR (2005). Transgenic labeling of the corticospinal tract for monitoring axonal responses to spinal cord injury. Nat. Med 11, 1355–1360. [DOI] [PubMed] [Google Scholar]
- Bruijn LI, Miller TM, and Cleveland DW (2004). Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu. Rev. Neurosci 27, 723–749. [DOI] [PubMed] [Google Scholar]
- Buffelli M, Burgess RW, Feng G, Lobe CG, Lichtman JW, and Sanes JR (2003). Genetic evidence that relative synaptic efficacy biases the outcome of synaptic competition. Nature 424, 430–434. [DOI] [PubMed] [Google Scholar]
- Catapano LA, Arnold MW, Perez FA, and Macklis JD (2001). Specific neurotrophic factors support the survival of cortical projection neurons at distinct stages of development. J. Neurosci 21, 8863–8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederquist GY, Azim E, Shnider SJ, Padmanabhan H, and Macklis JD (2013). Lmo4 establishes rostral motor cortex projection neuron subtype diversity. J. Neurosci 33, 6321–6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Schaevitz LR, and McConnell SK (2005a). Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc. Natl. Acad. Sci. USA 102, 17184–17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Rasin MR, Kwan KY, and Sestan N (2005b). Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc. Natl. Acad. Sci. USA 102, 17792–17797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisek P, Crammond DJ, and Kalaska JF (2003). Neural activity in primary motor and dorsal premotor cortex in reaching tasks with the contralateral versus ipsilateral arm. J. Neurophysiol 89, 922–942. [DOI] [PubMed] [Google Scholar]
- Daoud H, Valdmanis PN, Gros-Louis F, Belzil V, Spiegelman D, Henrion E, Diallo O, Desjarlais A, Gauthier J, Camu W, et al. (2011). Resequencing of 29 candidate genes in patients with familial and sporadic amyotrophic lateral sclerosis. Arch. Neurol 68, 587–593. [DOI] [PubMed] [Google Scholar]
- Dasen JS, Liu J-PP, and Jessell TM (2003). Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature 425, 926–933. [DOI] [PubMed] [Google Scholar]
- Donoghue JP, and Wise SP (1982). The motor cortex of the rat: cytoarchitecture and microstimulation mapping. J. Comp. Neurol 212, 76–88. [DOI] [PubMed] [Google Scholar]
- Dumitrescu AM, Liao XH, Weiss RE, Millen K, and Refetoff S (2006). Tissue-specific thyroid hormone deprivation and excess in monocarboxylate transporter (mct) 8-deficient mice. Endocrinology 147, 4036–4043. [DOI] [PubMed] [Google Scholar]
- Eyre JA, Miller S, Clowry GJ, Conway EA, and Watts C (2000). Functional corticospinal projections are established prenatally in the human foetus permitting involvement in the development of spinal motor centres. Brain 123, 51–64. [DOI] [PubMed] [Google Scholar]
- Fame RM, MacDonald JL, and Macklis JD (2011). Development, specification, and diversity of callosal projection neurons. Trends Neurosci 34, 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouad K, Pedersen V, Schwab ME, and Brösamle C (2001). Cervical sprouting of corticospinal fibers after thoracic spinal cord injury accompanies shifts in evoked motor responses. Curr. Biol 11, 1766–1770. [DOI] [PubMed] [Google Scholar]
- Franco SJ, and Müller U (2013). Shaping our minds: stem and progenitor cell diversity in the mammalian neocortex. Neuron 77, 19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galazo MJ, Emsley JG, and Macklis JD (2016). Corticothalamic projection neuron development beyond subtype specification: Fog2 and intersectional controls regulate intraclass neuronal diversity. Neuron 91, 90–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, and Jones KR (2002). Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci 22, 6309–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig LC (2015). Transcriptional controls over specification of neocortical projection neuron subtype and area diversity (Harvard University), PhD dissertation
- Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, and Macklis JD (2013). Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci 14, 755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Greppi C, and Macklis JD (2016). Ctip1 controls acquisition of sensory area identity and establishment of sensory input fields in the developing neocortex. Neuron 90, 261–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Kwan KY, Shim S, Lam MM, Shin Y, Xu X, Zhu Y, Li M, and Sestan N (2011). TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc. Natl. Acad. Sci. USA 108, 3041–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding SD, Armit C, Armstrong J, Brennan J, Cheng Y, Haggarty B, Houghton D, Lloyd-MacGilp S, Pi X, Roochun Y, et al. (2011). The GUD-MAP database–an online resource for genitourinary research. Development 138, 2845–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffner R, and Masterton B (1975). Variation in form of the pyramidal tract and its relationship to digital dexterity. Brain Behav. Evol 12, 161–200. [DOI] [PubMed] [Google Scholar]
- Hirata T, Suda Y, Nakao K, Narimatsu M, Hirano T, and Hibi M (2004). Zinc finger gene fez-like functions in the formation of subplate neurons and thalamocortical axons. Dev. Dyn 230, 546–556. [DOI] [PubMed] [Google Scholar]
- Hsu JY, Stein SA, and Xu XM (2008). Abnormal growth of the corticospinal axons into the lumbar spinal cord of the hyt/hyt mouse with congenital hypothyroidism. J. Neurosci. Res 86, 3126–3139. [DOI] [PubMed] [Google Scholar]
- Jang M-HH, Bonaguidi MA, Kitabatake Y, Sun J, Song J, Kang E, Jun H, Zhong C, Su Y, Guo JU, et al. (2013). Secreted frizzled-related protein 3 regulates activity-dependent adult hippocampal neurogenesis. Cell Stem Cell 12, 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi PS, Molyneaux BJ, Feng L, Xie X, Macklis JD, and Gan L (2008). Bhlhb5 regulates the postmitotic acquisition of area identities in layers II-V of the developing neocortex. Neuron 60, 258–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Lacombe J, Mazzoni EO, Liem KF Jr., Grinstein J, Mahony S, Mukhopadhyay D, Gifford DK, Young RA, Anderson KV, et al. (2010). Global control of motor neuron topography mediated by the repressive actions of a single hox gene. Neuron 67, 781–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiyama T, Kameda H, Murabe N, Fukuda S, Yoshioka N, Mizukami H, Ozawa K, and Sakurai M (2015). Corticospinal tract development and spinal cord innervation differ between cervical and lumbar targets. J. Neurosci 35, 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkman T (1996). Statistics to use http://www.physics.csbsju.edu/stats/.
- Kleim JA, Barbay S, and Nudo RJ (1998). Functional reorganization of the rat motor cortex following motor skill learning. J. Neurophysiol 80, 3321–3325. [DOI] [PubMed] [Google Scholar]
- Kolle G, Georgas K, Holmes GP, Little MH, and Yamada T (2000). CRIM1, a novel gene encoding a cysteine-rich repeat protein, is developmentally regulated and implicated in vertebrate CNS development and organogenesis. Mech. Dev 90, 181–193. [DOI] [PubMed] [Google Scholar]
- Krubitzer L (2007). The magnificent compromise: cortical field evolution in mammals. Neuron 56, 201–208. [DOI] [PubMed] [Google Scholar]
- Kuang RZ, and Kalil K (1994). Development of specificity in corticospinal connections by axon collaterals branching selectively into appropriate spinal targets. J. Comp. Neurol 344, 270–282. [DOI] [PubMed] [Google Scholar]
- Kwan HC, Mackay WA, Murphy JT, and Wong YC (1978). An intracortical microstimulation study of output organization in precentral cortex of awake primates. J. Physiol. (Paris) 74, 231–233. [PubMed] [Google Scholar]
- Kwan KY, Lam MM, Krsnik Z, Kawasawa YI, Lefebvre V, and Sestan N (2008). SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc. Natl. Acad. Sci. USA 105, 16021–16026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T, Jabaudon D, Molyneaux BJ, Azim E, Arlotta P, Menezes JR, and Macklis JD (2008). SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron 57, 232–247. [DOI] [PubMed] [Google Scholar]
- Leighton PA, Mitchell KJ, Goodrich LV, Lu X, Pinson K, Scherz P, Skarnes WC, and Tessier-Lavigne M (2001). Defining brain wiring patterns and mechanisms through gene trapping in mice. Nature 410, 174–179. [DOI] [PubMed] [Google Scholar]
- Lemon RN (2008). Descending pathways in motor control. Annu. Rev. Neurosci 31, 195–218. [DOI] [PubMed] [Google Scholar]
- Lemon RN, and Griffiths J (2005). Comparing the function of the corticospinal system in different species: organizational differences for motor specialization? Muscle Nerve 32, 261–279. [DOI] [PubMed] [Google Scholar]
- Leone DP, Srinivasan K, Chen B, Alcamo E, and McConnell SK (2008). The determination of projection neuron identity in the developing cerebral cortex. Curr. Opin. Neurobiol 18, 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XG, Florence SL, and Kaas JH (1990). Areal distributions of cortical neurons projecting to different levels of the caudal brain stem and spinal cord in rats. Somatosens. Mot. Res 7, 315–335. [DOI] [PubMed] [Google Scholar]
- Liu Y, Shi J, Lu CC, Wang ZB, Lyuksyutova AI, Song XJ, and Zou Y (2005). Ryk-mediated Wnt repulsion regulates posterior-directed growth of corticospinal tract. Nat. Neurosci 8, 1151–1159. [DOI] [PubMed] [Google Scholar]
- Liu Y, Latremoliere A, Li X, Zhang Z, Chen M, Wang X, Fang C, Zhu J, Alexandre C, Gao Z, et al. (2018). Touch and tactile neuropathic pain sensitivity are set by corticospinal projections. Nature 561, 547–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato S, Molyneaux BJ, Zuccaro E, Goff LA, Chen HH, Yuan W, Meleski A, Takahashi E, Mahony S, Rinn JL, et al. (2014). Gene co-regulation by Fezf2 selects neurotransmitter identity and connectivity of corticospinal neurons. Nat. Neurosci 17, 1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato S, Shetty AS, and Arlotta P (2015). Cerebral cortex assembly: generating and reprogramming projection neuron diversity. Trends Neurosci 38, 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodewyckx L, Cailotto F, Thysen S, Luyten FP, and Lories RJ (2012). Tight regulation of wingless-type signaling in the articular cartilage - subchondral bone biomechanical unit: transcriptomics in Frzb-knockout mice. Arthritis Res. Ther 14, R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Garner AR, Shimaoka D, Chuong AS, Klapoetke NC, Li L, van der Bourg A, Niino Y, Egolf L, Monetti C, et al. (2015). Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire CA, Bovenberg MS, Crommentuijn MH, Niers JM, Kerami M, Teng J, Sena-Esteves M, Badr CE, and Tannous BA (2013). Triple bioluminescence imaging for in vivo monitoring of cellular processes. Mol. Ther. Nucleic Acids 2, e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JH (1996). Differential spinal projections from the forelimb areas of the rostral and caudal subregions of primary motor cortex in the cat. Exp. Brain Res 108, 191–205. [DOI] [PubMed] [Google Scholar]
- Martin JH (2005). The corticospinal system: from development to motor control. Neuroscientist 11, 161–173. [DOI] [PubMed] [Google Scholar]
- McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, Rubenstein JL, and Chen B (2011). Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J. Neurosci 31, 549–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MW (1987). The origin of corticospinal projection neurons in rat. Exp. Brain Res 67, 339–351. [DOI] [PubMed] [Google Scholar]
- Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. (2014). Transcriptional landscape of the prenatal human brain. Nature 508, 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, and Macklis JD (2005). Fezl is required for the birth and specification of corticospinal motor neurons. Neuron 47, 817–831. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, and Macklis JD (2007). Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci 8, 427–437. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Fame RM, MacDonald JL, MacQuarrie KL, and Macklis JD (2009). Novel subtype-specific genes identify distinct subpopulations of callosal projection neurons. J. Neurosci 29, 12343–12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Goff LA, Brettler AC, Chen HH, Hrvatin S, Rinn JL, and Arlotta P (2015). DeCoN: genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron 85, 275–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori J, Suzuki S, Kobayashi M, Inagaki T, Komatsu A, Takeda T, Miyamoto T, Ichikawa K, and Hashizume K (2002). Nicotinamide adenine dinucleotide phosphate-dependent cytosolic T(3) binding protein as a regulator for T(3)-mediated transactivation. Endocrinology 143, 1538–1544. [DOI] [PubMed] [Google Scholar]
- Neafsey EJ, Bold EL, Haas G, Hurley-Gius KM, Quirk G, Sievert CF, and Terreberry RR (1986). The organization of the rat motor cortex: a microstimulation mapping study. Brain Res 396, 77–96. [DOI] [PubMed] [Google Scholar]
- Oudega M, and Perez MA (2012). Corticospinal reorganization after spinal cord injury. J. Physiol 590, 3647–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard MT, Boldt HB, Laursen LS, Sottrup-Jensen L, Conover CA, and Oxvig C (2001). Pregnancy-associated plasma protein-A2 (PAPP-A2), a novel insulin-like growth factor-binding protein-5 proteinase. J. Biol. Chem 276, 21849–21853. [DOI] [PubMed] [Google Scholar]
- Ozdinler PH, and Macklis JD (2006). IGF-I specifically enhances axon outgrowth of corticospinal motor neurons. Nat. Neurosci 9, 1371–1381. [DOI] [PubMed] [Google Scholar]
- Penfield W, and Boldrey E (1937). Somatic motor and sensory representation in the cerebral cortex of man as studied by electrical stimulation. Brain 60, 389–443. [Google Scholar]
- Penfield W, and Rasmussen T (1950). The Cerebral Cortex of Man: A Clinical Study of Localization of Function (Macmillan)
- Pera EM, Wessely O, Li SY, and De Robertis EM (2001). Neural and head induction by insulin-like growth factor signals. Dev. Cell 1, 655–665. [DOI] [PubMed] [Google Scholar]
- Raineteau O, and Schwab ME (2001). Plasticity of motor systems after incomplete spinal cord injury. Nat. Rev. Neurosci 2, 263–273. [DOI] [PubMed] [Google Scholar]
- Rathelot JA, and Strick PL (2009). Subdivisions of primary motor cortex based on cortico-motoneuronal cells. Proc. Natl. Acad. Sci. USA 106, 918–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravits J, Appel S, Baloh RH, Barohn R, Brooks BR, Elman L, Floeter MK, Henderson C, Lomen-Hoerth C, Macklis JD, et al. (2013). Deciphering amyotrophic lateral sclerosis: what phenotype, neuropathology and genetics are telling us about pathogenesis. Amyotroph. Lateral Scler. Frontotemporal Degener 14 (Suppl 1), 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni V, Engmann A, Ozkan A, and Macklis JD (2020). Motor cortex connections. In Neural Circuit and Cognitive Development, Second Edition, Rubenstein J, Rakic P, Chen B, and Kwan KY, eds. (Academic Press; ), pp. 167–199. [Google Scholar]
- Sahni V, Itoh Y, Shnider SJ, and Macklis JD (2021). Crim1 and Kelch-like 14 exert dual-directional, complementary developmental control over segmentally specific corticospinal axon projection targeting. Cell Rep 37, 109842–1–109842–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salih DA, Tripathi G, Holding C, Szestak TA, Gonzalez MI, Carter EJ, Cobb LJ, Eisemann JE, and Pell JM (2004). Insulin-like growth factor-binding protein 5 (Igfbp5) compromises survival, growth, muscle development, and fertility in mice. Proc. Natl. Acad. Sci. USA 101, 4314–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas S, Proukakis C, Crosby A, and Warner TT (2008). Hereditary spastic paraplegia: clinical features and pathogenetic mechanisms. Lancet Neurol 7, 1127–1138. [DOI] [PubMed] [Google Scholar]
- Schreyer DJ, and Jones EH (1988). Topographic sequence of outgrowth of corticospinal axons in the rat: a study using retrograde axonal labeling with Fast blue. Brain Res 466, 89–101. [DOI] [PubMed] [Google Scholar]
- Shim S, Kwan KY, Li M, Lefebvre V, and Sestan N (2012). Cis-regulatory control of corticospinal system development and evolution. Nature 486, 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda Y, Yamaguchi T, and Futami T (1986). Multiple axon collaterals of single corticospinal axons in the cat spinal cord. J. Neurophysiol 55, 425–448. [DOI] [PubMed] [Google Scholar]
- Singer MA, Statland JM, Wolfe GI, and Barohn RJ (2007). Primary lateral sclerosis. Muscle Nerve 35, 291–302. [DOI] [PubMed] [Google Scholar]
- Strong MJ, and Gordon PH (2005). Primary lateral sclerosis, hereditary spastic paraplegia and amyotrophic lateral sclerosis: discrete entities or spectrum? Amyotroph. Lateral Scler. Other Motor Neuron Disord 6, 8–16. [DOI] [PubMed] [Google Scholar]
- Tennant KA, Adkins DL, Donlan NA, Asay AL, Thomas N, Kleim JA, and Jones TA (2011). The organization of the forelimb representation of the C57BL/6 mouse motor cortex as defined by intracortical microstimulation and cytoarchitecture. Cereb. Cortex 21, 865–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomassy GS, De Leonibus E, Jabaudon D, Lodato S, Alfano C, Mele A, Macklis JD, and Studer M (2010). Area-specific temporal control of corticospinal motor neuron differentiation by COUP-TFI. Proc. Natl. Acad. Sci. USA 107, 3576–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic M, Visser TJ, Mittag J, Horn S, Lukas J, Darras VM, Raivich G, Bauer K, and Heuer H (2007). Abnormal thyroid hormone metabolism in mice lacking the monocarboxylate transporter 8. J. Clin. Invest 117, 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno M, Nakamura Y, Li J, Gu Z, Niehaus J, Maezawa M, Crone SA, Goulding M, Baccei ML, and Yoshida Y (2018). Corticospinal circuits from the sensory and motor cortices differentially regulate skilled movements through distinct spinal interneurons. Cell Rep 23, 1286–1300.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullan J, and Artieda J (1981). Somatotopy of the corticospinal neurons in the rat. Neurosci. Lett 21, 13–18. [DOI] [PubMed] [Google Scholar]
- Wang X, Liu Y, Li X, Zhang Z, Yang H, Zhang Y, Williams PR, Alwahab NSA, Kapur K, Yu B, et al. (2017). Deconstruction of corticospinal circuits for goal-directed motor skills. Cell 171, 440–455.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Maunze B, Wang Y, Tsoulfas P, and Blackmore MG (2018). Global connectivity and function of descending spinal input revealed by 3D microscopy and retrograde transduction. J. Neurosci 38, 10566–10581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welniarz Q, Dusart I, and Roze E (2017). The corticospinal tract: Evolution, development, and human disorders. Dev. Neurobiol 77, 810–829. [DOI] [PubMed] [Google Scholar]
- Whishaw IQ (2003). Did a change in sensory control of skilled movements stimulate the evolution of the primate frontal cortex? Behav. Brain Res 146, 31–41. [DOI] [PubMed] [Google Scholar]
- Wise SP, Murray EA, and Coulter JD (1979). Somatotopic organization of corticospinal and corticotrigeminal neurons in the rat. Neuroscience 4, 65–78. [DOI] [PubMed] [Google Scholar]
- Woodworth MB, Greig LC, Kriegstein AR, and Macklis JD (2012). SnapShot: cortical development. Cell 151, 918.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth MB, Greig LC, Liu KX, Ippolito GC, Tucker HO, and Macklis JD (2016). Ctip1 regulates the balance between specification of distinct projection neuron subtypes in deep cortical layers. Cell Rep 15, 999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolsey CN, Settlage PH, Meyer DR, Sencer W, Pinto Hamuy T, and Travis AM (1952). Patterns of localization in precentral and ‘‘supplementary’’ motor areas and their relation to the concept of a premotor area. Res. Publ. Assoc. Res. Nerv. Ment. Dis 30, 238–264. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data profiling lateral versus medial CSN subpopulations at P1, P4, and P7 have been deposited at GEO and are publicly available. Accession number is listed in the key resources table. Gene intensities using Rosetta Resolver for all genes have been deposited on Mendeley and are publicly available. The DOI is listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-CTIP2 | Abcam | Cat# ab28448; RRID:AB_1140055 |
| Mouse anti-SATB2 | Abcam | Cat# ab51502; RRID:AB_882455 |
| Rabbit anti-GFP | Molecular Probes | Cat# A-1112; RRID:AB_221569 |
| Chicken anti-GFP | Thermo Fisher Scientific | Cat# A10262; RRID:AB_2534023 |
| Rabbit anti-RFP | Rockland | Cat# 600–401-379; RRID:AB_2209751 |
|
| ||
| Bacterial and virus strains | ||
| AAV-8 hsyn-GFP-Cre | UNC Vector core | N/A |
| AAV-2/1 CAG-FleX-TdTomato | From University of Pennsylvania vector core (now at Addgene) | N/A |
| AAV 2/1 CAG-EGFP | vector core at Massachusetts General Hospital, Boston, MA | Maguire et al. (2013) |
| AAV 2/1 CAG-tdTomato | This paper | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
| Green fluorescent microspheres | LumaFluor | Green Retrobeads IX |
| Cholera Toxin B subunit, Alexa 555 conjugate | ThermoFisher | C34776 |
| Cholera Toxin B subunit, Alexa 647 conjugate | ThermoFisher | C34778 |
|
| ||
| Critical commercial assays | ||
| Mouse Genome Affymetrix 430.2 arrays | Affymetrix | N/A |
| Thermoscript Reverse transcriptase kit | Fisher scientific | 12236 |
| RNeasy FFPE | QIAGEN | 73504 |
| SYBR® Green PCR Master Mix | ThermoFisher | 4309155 |
| RNAscope 2.5 HD Assay-RED | ACD Bio | Made to order-Crim1 (550751) |
|
| ||
| Deposited data | ||
| Gene expression profiling of CSN subpopulations by microarray | This paper | GEO: GSE77311; Mendeley: https://doi.org/10.17632/9jj6jm2bdp.1 |
|
| ||
| Experimental models: Organisms/strains | ||
| Crim1GCE mice | The Jackson Laboratory | GUDMAP database Harding et al. (2011) |
| Fezf2 null | N/A | Hirata et al. (2004) and Molyneaux et al. (2005) |
| Thy1-STOP-EYFP | N/A | Buffelli et al. (2003) and Bareyre et al. (2005) |
| Emx1-IRES-FlpO | This Paper | N/A |
| Ai65(RCFL-tdT)-D or Ai65D | The Jackson Laboratory | Stock number: 021875 |
|
| ||
| Oligonucleotides | ||
| See Table S4 for oligonucleotide information | N/A | |
|
| ||
| Recombinant DNA | ||
| pCAG-tdTomato-t2A-STOP | This paper | N/A |
|
| ||
| Software and algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Rosetta Resolver software | Rosetta (now Microsoft) | N/A |
| Bioconductor | Bioconductor.org | Bioconductor.org |
| GraphPad Prism 8.0 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
No original code was generated as part of this study.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


