Abstract
Background
Nosocomial respiratory virus outbreaks represent serious public hgealth challenges. Rapid and precise identification of cases and tracing of transmission chains is critical to end outbreaks and to inform prevention measures.
Methods
We combined conventional surveillance with influenza A virus (IAV) genome sequencing to identify and contain a large IAV outbreak in a metropolitan healthcare system. A total of 381 individuals, including 91 inpatients and 290 healthcare workers (HCWs), were included in the investigation.
Results
During a 12-day period in early 2019, infection preventionists identified 89 HCWs and 18 inpatients as cases of influenza-like illness (ILI), using an amended definition without the requirement for fever. Sequencing of IAV genomes from available nasopharyngeal specimens identified 66 individuals infected with a nearly identical strain of influenza A H1N1pdm09 (43 HCWs, 17 inpatients, and 6 with unspecified affiliation). All HCWs infected with the outbreak strain had received the seasonal influenza virus vaccination. Characterization of 5 representative outbreak viral isolates did not show antigenic drift. In conjunction with IAV genome sequencing, mining of electronic records pinpointed the origin of the outbreak as a single patient and a few interactions in the emergency department that occurred 1 day prior to the index ILI cluster.
Conclusions
We used precision surveillance to delineate a large nosocomial IAV outbreak, mapping the source of the outbreak to a single patient rather than HCWs as initially assumed based on conventional epidemiology. These findings have important ramifications for more-effective prevention strategies to curb nosocomial respiratory virus outbreaks.
Keywords: precision surveillance, respiratory viruses, influenza A virus, nosocomial outbreak, next-generation pathogen sequencing
Nosocomial respiratory virus outbreaks represent serious public health challenges We used precision surveillance to delineate a large nosocomial Influenza A virus outbreak, mapping the source of the outbreak to a single patient rather than Health care workers as initially assumed based on conventional epidemiology.
Nosocomial outbreaks represent major challenges for healthcare providers. It is critical for hospitals and health systems to not only quickly identify infected cases but also determine the source of the outbreak in order to mitigate the threat to patients and healthcare workers (HCWs). Nosocomial influenza virus outbreaks have been described worldwide [1–3]; children, the elderly, and institutionalized and immunocompromised patients are particularly vulnerable. In some instances, nosocomial outbreaks have been caused by HCWs who work while ill [4].
Influenza A virus (IAV) is a single-stranded, negative-sense, segmented RNA virus that causes an acute infection of the upper respiratory tract. Influenza A virus is further divided into subtypes based on the hemagglutinin (H) and neuraminidase (N) surface proteins. Both IAV subtypes H1N1 and H3N2 circulate in humans and were prevalent in the winter/spring of 2018–2019 in New York City.
Viral genome sequencing has been used previously to complement traditional epidemiological investigations to trace nosocomial influenza outbreaks [2, 5, 6]. However, co-circulation and ongoing transmission of multiple virus subtypes in the surrounding community can complicate the determination of whether there is a clonal outbreak and make identifying the source of the outbreak challenging. In order to develop effective intervention strategies, approaches that enable rapid detection of the origin and spread of a clonal influenza virus outbreak in the context of seasonally circulating strains are needed. The precision surveillance approach we implemented in our healthcare system, therefore, incorporates both traditional outbreak investigation and routine surveillance of influenza viruses causing disease in our community.
Here we report precision surveillance integrating conventional infection prevention measures, sequencing of IAV genomes from clinical biospecimens, and data mining of electronic medical records to successfully manage a large nosocomial IAV outbreak among inpatients and HCWs.
METHODS
Ethics Statement
Study protocols for the collection and viral genome sequencing of discarded clinical specimens by the Pathogen Surveillance Program (protocol HS#13–00981) and chart reviews of outbreak investigation cases (protocol HS#19–00686) were reviewed and approved by the Mount Sinai Hospital Institutional Review Board.
Summary of Infection-Prevention Measures and Investigation
When we detected symptoms suggestive of influenza-like illness (ILI) in inpatients in early 2019, infection-prevention measures, including extensive screening of inpatients and HCWs for ILI and collection of nasopharyngeal (NP) swab specimens, were implemented. Once the outbreak was identified, an amended ILI case definition was developed within 3 days. This specific case definition included any HCWs or patients with cough, rhinorrhoea, sore throat, body aches, with or without fever, and positive diagnostic test for influenza virus by molecular polymerase chain reaction (PCR) testing (Xpert Xpress Flu/RSV test; Cepheid) of an NP swab specimen in universal transport medium (NP-UTM).
Collection of NP-UTM for Influenza A Subtyping and Sequencing
A total of 486 IAV-positive NP-UTM specimens, including 104 samples from the outbreak investigation, 150 background influenza surveillance samples at Hospital A, and 231 background influenza surveillance samples at Hospital B, were collected and IAV-subtyped and sequenced. The time frame from which surveillance and investigation samples were included covered a total of 27 days, starting from 6 days before to 7 days after the 12-day outbreak investigation.
Influenza Virus Sequencing
RNA from the NP samples and viral isolates was used for multisegment reverse transcription (RT)–PCR genome amplification of IAV with Opti1 primers [7], followed by next-generation sequencing (NGS).
Identification of Clonal Outbreak Isolates
To detect clusters of highly related outbreak isolates and reconstruct the early outbreak timeline, we used the open-source PathoSPOT (Pathogen Sequencing Phylogenomic Outbreak Toolkit) software (https://pathospot.org), setting a threshold of 3 or less genome-wide single-nucleotide variants (SNVs) to identify transmission events. The admission/transfer/discharge histories for outbreak cases were combined with patient–HCW interaction data to reconstruct a network of all known contacts in Cytoscape [8].
Additional Material and Methods Information
See the Supplementary Methods for descriptions and experimental details not included in the main manuscript [10–14].
RESULTS
Epidemiology of the Nosocomial Influenza Outbreak
In early 2019, symptoms suggestive of ILI were observed in several HCWs as well as in inpatients receiving critical care at Hospital A. The hospital’s incident command system was activated, and an extensive outbreak investigation was started that included mandatory staff symptom checks and testing of all inpatients with any respiratory symptoms, regardless of fever status. Terminal cleaning of patient care areas and clinical staff workspaces was also performed.
Over the course of the hospital-wide outbreak investigation, a total of 381 individuals (91 inpatients and 290 HCWs) were screened by regular temperature checks, symptom surveys, and molecular diagnostic testing for IAV, influenza B virus, and respiratory syncytial virus. A total of 18 inpatients (19.8%) and 89 HCWs (29.7%) included in the epidemiological investigation tested positive for IAV (Figure 1A).
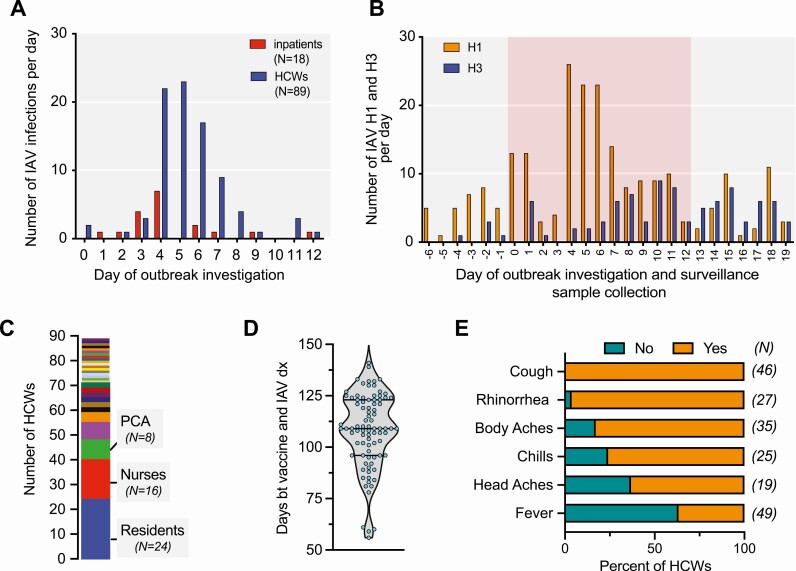
Figure 1.
Epidemiology of the nosocomial IAV outbreak. A, Timeline of the nosocomial IAV outbreak at a metropolitan hospital. Day 0: initiation of the infection-prevention investigation. B, Distribution of IAV subtypes detected in individuals identified in the outbreak investigation and patients seeking care at the hospital and testing positive for IAV between 6 days before (day −6) and 19 days after (day 19) the initiation (day 0) of the infection-prevention investigation. The red background highlights the period of the outbreak investigation. C, The distribution of professions (29 job categories) of the 89 HCWs who tested positive for IAV. D, The distribution of days between receiving seasonal influenza virus vaccination and testing positive for IAV among HCWs. Of note, 87 of the 89 HCWs who tested positive for IAV were vaccinated. E, Clinical signs and symptoms reported by HCWs who tested positive for IAV. The data available for each symptom differ with respect to the number of employees (“N” listed in parentheses provides the absolute numbers for each). Note that 63% of HCWs were afebrile. Abbreviations: bt, between; dx, IAV diagnosis; HCW, healthcare worker; IAV, influenza A virus; PCA, patient-care assistant.
Subtyping of IAV from the NP samples collected during the epidemiological investigation (N = 104) and the routine influenza surveillance at Hospital A (N = 150) and Hospital B (N = 231) revealed a stark increase in IAV/H1 at days 4, 5, and 6 of the investigation (Figure 1B). Of note, all the samples from inpatients and HCWs included in the investigation that we successfully subtyped harbored IAV/H1N1pdm09, suggesting a single transmission chain.
The 89 positive HCWs were distributed across 29 different work-assignment categories (Figure 1C), including 24 resident physicians (residents, fellows, or interns), 16 registered nurses, 8 patient-care assistants, and 6 attending physicians. Eighty-seven of these 89 HCWs (>90%) had been vaccinated with the quadrivalent seasonal influenza virus vaccine 2 to 5 months (average: 108 days) prior to testing positive for IAV (Figure 1D). Importantly, these infected HCWs mostly had minor symptoms and usually would not have been classified as having ILI given the lack of fever (Figure 1E). Recognition of the altered clinical presentation in vaccinated HCWs prompted removal of the requirement for fever from our case definition early in the investigation.
Genomics of the Nosocomial Influenza Virus Outbreak
In order to determine whether there was transmission of a single IAV strain or several independent introductions into the hospital system, we performed NGS of IAV from the NP specimens that were banked following the initial diagnostic testing. As part of our institution’s Pathogen Surveillance Program, we routinely sequence influenza viruses from a subset of the patients seeking care at our hospitals (termed “surveillance”) to identify circulating community strains. Thus, we were able to compare IAV sequences from cases identified in the outbreak investigation and sequences from surveillance samples in our analysis.
Complete genomic sequences were obtained from 214 IAV isolates (Figure 2A), including 126 from Hospital A (investigation and surveillance) and 88 from Hospital B (surveillance only). Pairwise comparison of these viral genomes showed a large cluster of 66 viral isolates that differed by no more than 3 SNVs, indicating that a single virus clone was responsible for a large portion of the nosocomial outbreak (Figure 2B). Additionally, our analyses indicated that other independent introductions of IAV H1N1pdm09 strains, with limited forward transmissions, had caused smaller clusters of ILI at both Hospital A and Hospital B. We also noted 2 small independent clusters due to transmission of IAV H3N2 viruses.
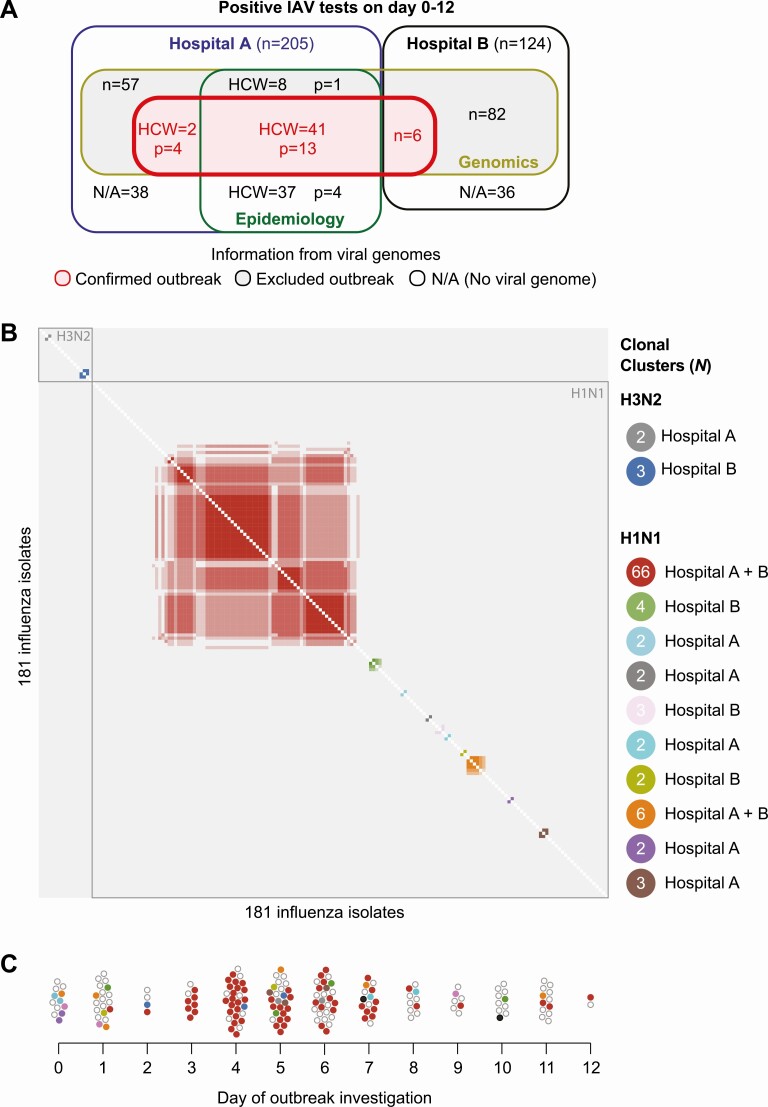
Figure 2.
Genomics of the nosocomial outbreak. A, Venn diagram illustrating the number of sequenced outbreak-confirmed H1N1 strains (N = 66) and outbreak-excluded IAV, including unrelated H1N1 strains (N = 113) and H3N2 strains (N = 36), identified in Hospital A (investigation and surveillance) and Hospital B (surveillance). Epidemiology describes the cases identified by infection prevention. B, Pairwise comparison of the complete viral genomes. Note the tight cluster of the outbreak H1N1 strains (N = 66, red cluster) at the center and the 2 small H3N2 clusters at the top left of the pairwise comparison. C, Dynamics of the case numbers and IAV strains during the investigation period. All color coding used in this panel are the same as those used in the panel B. The outbreak H1N1 strain (shown in red) was first detected on the day after the initiation of the investigation (day 1) in a hospitalized patient. The first 2 employees who tested positive for IAV on the day of the initiation of the investigation (day 0) harbored unrelated H1N1 strains. Abbreviations: HCW, healthcare worker; IAV, influenza A virus; N/A, no viral genome available; p, patient.
Correlating virus genomes with the timing and the source of these isolates showed that all of the virus isolates obtained on day 0 and most of the virus isolates collected on day 1 of the infection-prevention investigation were distinct from the strain that caused the large outbreak. Although 2 HCWs tested positive for IAV on day 0, their viruses were genetically different from the strain that caused the outbreak and not associated with any nosocomial transmission. Some patients and all HCWs infected with the outbreak virus had received the seasonal influenza virus vaccine. The first isolate that clustered with the outbreak virus strain was obtained from a patient identified on day 1 of the investigation (Figure 2C).
Altogether, the genomic analyses of available clinical influenza isolates showed that cases identified by the conventional epidemiological investigation encompassed patients and HCWs who together harbored 12 different IAV strains, but that only 1 strain was associated with widespread nosocomial transmission.
Phylogenetic and Functional Properties of the Influenza A Virus Outbreak Strains
Phylogenetic analysis of the sequenced IAV genomes showed that the outbreak strain tightly clustered within a specific H1N1pdm09 6b1A subclade. Other IAV H1N1 isolates obtained in the infection-prevention investigation and routine surveillance mapped throughout the H1N1pdm09 6b1A clade (Figure 3A, compare green to red dots) and likely reflected the predominance of seasonally spreading strains in the community. A detailed analysis of genomic sequences of all outbreak IAV strains showed that they were highly conserved; most of the nucleotide variations occurred in the hemagglutinin (HA) and the NA segments encoding the viral surface proteins and the majority of predicted amino acid changes occurred only in the neuraminidase (NA) segment (Figure 3B). We selected 5 outbreak virus strains with representative variants to be propagated in cell culture for functional characterization (Figure 3C). Hemagglutination inhibition assays performed with these strains confirmed that none had drifted when compared with the H1N1pdm09 vaccine strain used in that season (Figure 3D).
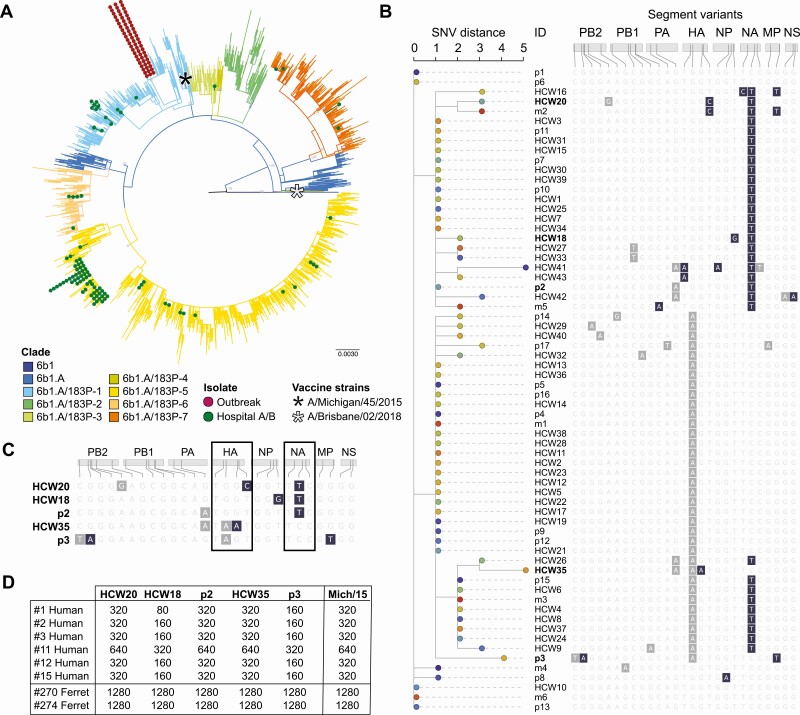
Figure 3.
Phylogenetic and functional properties of the IAV outbreak strains. A, Analysis of IAV H1N1 genomic sequence diversity. Note that the outbreak strains form a tight cluster that maps to 1 subclade of the H1N1 6b1A clade, whereas those not responsible for the outbreak are diverse and map to other various subclades. IAV H1N1 vaccine strains A/Michigan/2015 (included in the 2017–2018 and 2018–2019 vaccines) and A/Brisbane/2018 (included in the 2019–2020 vaccine) are included in the analysis for reference. B, Phylogenetic relationships based on SNV distance among the outbreak virus strains. All 8 viral segments are shown, with gray indicating synonymous changes and dark blue indicating nonsynonymous substitutions. Predicted amino acid changes are listed at the bottom. C, Genotype of the 5 representative outbreak strains that were grown in cell culture. Gray, synonymous mutations; dark blue, nonsynonymous changes. The HA and NA are indicated by boxes. D, Hemagglutination inhibition titers of 5 representative outbreak strains with the sera of 6 recently vaccinated individuals. Sera from ferrets infected with the vaccine strain A/Michigan/45/2015 served as a positive control for antisera and the A/Michigan/45/2015 virus was the antigen positive control. Abbreviations: HA, hemagglutinin; HCW, healthcare worker; IAV, influenza A virus; NA, neuraminidase; NP, nucleoprotein; NS, nonstructural protein 1; PB1 and PB2, RNA-directed RNA polymerase catalytic subunit 1 and 2; p, patient; SNV, single-nucleotide variant.
Reconstruction of the Transmission Chain in the Early Days of the Outbreak
In order to understand the origin of the outbreak and the factors that facilitated its rapid spread, we focused on the earliest events. We further analyzed IAV genomes from 10 of the 12 cases identified by the conventional investigation as occurring during days 0–3 of the outbreak (83%) and from 34 of the 46 IAV-positive surveillance samples obtained during the corresponding period at Hospitals A and B (74%). Of these, IAV from 8 cases and 1 surveillance sample matched the outbreak strain. We used the PathoSPOT framework (https://pathospot.org) to query various electronic hospital records in order to create a timeline comprising these 9 cases (Figure 4A). These data revealed that 4 of the 9 earliest nosocomial IAV cases were treated (patient [p] 1, p2, and p3) or worked (HCW1) in the emergency department (ED) on the same day (day −1) during overlapping time periods. The 3 patients were admitted from the ED to different inpatient units and had no other shared interactions with HCWs, indicating that the common exposure most likely occurred in the ED. Similarly, because one of the patients who acquired nosocomial IAV did not have direct contact with HCW1 and had already been transferred from the ED prior to the time HCW1 was present in the ED, the evidence indicates that HCW1 was exposed during that specific work shift in the ED rather than being the primary case.
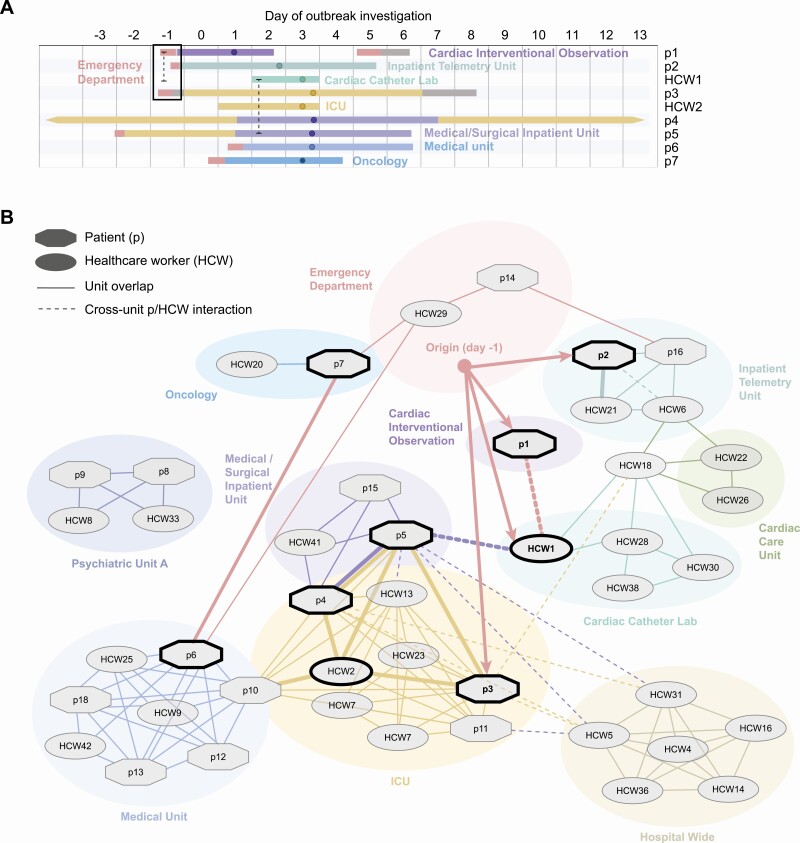
Figure 4.
Reconstruction of the transmission chain in the early days of the outbreak. A, Timeline and locations inside the hospital of the first 9 cases harboring the IAV/H1N1 outbreak strain. Day 0 is the day the investigation started. Dots on the line point to the day on which patient/HCW tested positive for IAV. The dashed lines indicate presumed contacts between infected individuals. The black box highlights the 3 patients who received care in the emergency department 1 day prior to the start of the infection-control investigation (day −1). B, Interaction network of HCWs and patients (p) who tested positive for the IAV H1N1 outbreak strain. The cases identified by thick black outlines were critical in seeding the transmission within the hospital. Abbreviations: HCW, healthcare worker; IAV, influenza A virus; ICU, intensive care unit.
In contrast, p3, the putative primary case for this outbreak, was brought to the ED in the morning of day −1, several hours before p1 and p2, and interacted with HCW1 before being admitted to a medical unit the same day (Figure 4A) with fever, shaking chills, dyspnea, and abdominal pain. Patient 3 subsequently developed systemic inflammatory response syndrome and was transferred to the intensive care unit (ICU) where the patient was intubated, a procedure that can generate significant aerosols [9]. Because blood cultures of p3 grew gram-negative bacteria, the diagnosis of IAV was delayed. However, the patient remained febrile despite antibiotics, which prompted diagnostic testing for influenza virus on day 3.
The next 3 early cases (HCW2, p4, and p5) most likely acquired IAV in the ICU from p3, although p4 and p5 had further overlapping stays following transfer to the same medical/surgical inpatient unit from the ICU. Our data suggest that cases p6 and p7, who were admitted through the ED several days after the start of the outbreak, acquired IAV from ED HCWs who had been exposed to p3 and then tested positive for IAV in the days thereafter.
The interaction network based on available contact records (Figure 4B) indicates that almost all later cases can be traced back in some way to the initial 9 cases shown in the timeline (Figure 4A), with transfers of patients acting as the major vectors for spread to other hospital units. A direct transmission link could not be documented for 2 patients hospitalized on a closed psychiatric unit and 2 HCWs assigned to the same closed unit, suggesting that indirect interactions may have occurred elsewhere in the hospital.
DISCUSSION
In this report, we describe application of a precision surveillance approach integrating conventional infection-prevention measures, genomic analysis, and data mining to delineate and manage a large nosocomial IAV outbreak. Widespread molecular testing and genomic analysis enabled differentiation of IAV outbreak cases from community-acquired IAV and the correct identification of the likely primary case and consequently highlighted transmission risks and opportunities for mitigation in addition to those identified by the conventional investigation.
The most important step in bringing the outbreak under control was early identification of the potential nosocomial spread, which was followed by rapid implementation of control measures including mandatory masking and limiting admissions to certain wards. Additionally, recognition of IAV in specimens from vaccinated HCWs and modification of our case definition were essential to identify and suppress transmission from mildly symptomatic/asymptomatic employees with influenza but without fever.
Almost one-third of the 290 HCWs included in the conventional epidemiological investigation tested positive for IAV (Figure 1); the vast majority had received the seasonal influenza virus vaccination. Among the infected hospital employees for whom viral influenza genotypes were available (49/89), 41 harbored the outbreak strain, whereas 8 were infected with unrelated H1N1pdm09 viruses (Figure 2). In the absence of our infection-prevention intervention, many of these cases would have gone undiagnosed, pointing to the fact that influenza virus infections in vaccinated HCWs remain largely underdiagnosed due to the milder disease presentation. Thus, diagnostic testing of vaccinated HCWs with mild symptoms should be considered, especially when the seasonal influenza vaccine is well matched to circulating viruses.
By sequencing influenza virus genomes from the infected patients and HCWs, we could focus the investigation into the source of the outbreak on only those cases who actually were infected with the identified outbreak virus strain. Integrating data from various hospital electronic records with molecular confirmation of which patients and HCWs were infected with the outbreak strain enabled reconstruction of the dynamics of the outbreak and identification of the likely primary case, and therefore ensured reassessment of transmission risk and heightened remediation for areas where transmission likely occurred.
Of note, the 2 HCWs who were the first to be diagnosed with ILI and incorrectly identified by the traditional epidemiologic investigation as the likely source(s) of the outbreak were found to be infected with viruses distinct from the outbreak strain (Figure 2). It is also important to note that the time of diagnosis or date of a positive test may not be a good indicator for the actual chronology of an outbreak, particularly in vaccinated HCWs who may ignore or downplay symptoms. Indeed, we observed that almost 3 days transpired between the onset of symptoms and a positive IAV test in HCWs compared to the average of one-half day for inpatients.
Since we routinely sequence influenza virus isolates from patients receiving care at our health system as part of our Pathogen Surveillance Program, we could compare the strains from the outbreak investigation conducted at Hospital A with the strains found in the surveillance of Hospital A as well as Hospital B (Figure 3). These additional data allowed us not only to identify previously unrecognized smaller transmission events (4 inpatients and 2 HCWs at Hospital A and 6 patients at Hospital B) but also to ascertain that there was a large (in number) but limited (in time) outbreak of H1N1pdm09 in our health system (Figures 2 and 3). Importantly, this specific H1N1pdm09 virus outbreak strain did not spread further in the community.
A limitation of our study is that we did not have access to biospecimens from 22 HCWs whose tests were performed at laboratories outside our health system. Additionally, only partial viral genomes could be retrieved from 2 of the available biospecimens linked to the epidemiological outbreak investigation. However, we were able to obtain viral genomes for all the patients identified during the first 3 days of the outbreak, providing a solid foundation for the reconstruction of the transmission chain. Our data suggest that the outbreak began in the ED most likely through introduction of the virus by a single patient, who had received aggressive resuscitative care and was subsequently transferred to the ICU of Hospital A (Figure 4). Enhanced screening and isolation of patients coming into the ED with any respiratory symptoms, even when an alternative diagnosis seems to be the predominant complaint, is an important step to mitigate the risk of respiratory virus transmission. Additionally, recognition by hospital leadership of the potential for transmission even from HCWs with mild ILI resulted in administrative support for intensified education of staff to avoid working while ill, extended sick leave when needed, and a move away from the HCW culture of “presenteeism,” which can contribute to nosocomial transmission of influenza [4] and other respiratory viruses. Potential influenza virus transmission from vaccinated HCWs with mild symptoms can be limited by masking, especially in high-risk areas of the hospital. The requirement for universal masking implemented in most hospitals during the ongoing coronavirus disease 2019 (COVID-19) pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) will provide data on the impact of employee masking on the frequency of nosocomial influenza outbreaks.
It is critical for patient care that any healthcare organization quickly detects the occurrence of hospital-acquired infections and limits their spread through swift identification of their origins. Conventional infection-prevention approaches, however, are challenged if a hospital outbreak occurs in the context of widespread community-acquired infections (eg, during the peak of the influenza season as in this study). Our findings are applicable to a wide range of highly transmissible respiratory viral pathogens, including SARS-CoV-2. Indeed, emerging evidence suggests that a high percentage of HCWs became infected with SARS-CoV-2 before screening for acute infection became more available. Implementation of precision surveillance measures as outlined here will be of critical importance for the identification and mitigation of the risks of nosocomial transmission for patients and HCWs alike.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. W. J., J. E., B. K., B. B., M. M., and I. H. were involved in the outbreak investigation. W. J., J. E., M. R. G., and E. M. S. provided clinical evaluations. W. J., J. E., M. R. G., and E. M. S. collected and verified clinical data. E. H., J. T., T. L., F. C., L. P., M. M. H., M. G., E. M. S., V. S., and H. v. B. performed clinical sample accessioning. E. H., M. M. H., and H. A. performed RNA extraction and viral subtyping. J. M. C. and V. S. selected and grew viral isolates. R. A. A. and V. S. provided banked serum samples. J. M. C. performed hemagglutination inhibition assays. A. S. G.-R., Z. K., and A. M. performed NGS experiments. A. S. G.-R., R. S., and M. S. provided NGS services. A. S. G.-R., D. K., T. R. P., and H. v. B. performed genome assembly and/or comparative genome analyses. J. E., T. R. P., E. M. S., and H. v. B. performed mining of electronic medical records. W. J., J. E., A. S. G.-R., M. M. H., F. K., E. M. S., V. S., and H. v. B. analyzed, interpreted, and/or discussed data. W. J., A. S. G.-R., E. M. S., V. S., and H. v. B. wrote the manuscript. E. M. S., V. S., and H. v. B. conceived and supervised the study and raised financial support.
Acknowledgments. The authors thank the numerous team members of the Clinical Microbiology laboratories at the Mount Sinai Health System and the Department of Microbiology for expert support and their willingness to go the extra mile. The authors thank Ms M. C. Bermudez and the team of the Personalized Virology Initiative for expert processing of clinical samples. They also extend their gratitude to Dr A. Garcia-Sastre and Dr P. Palese for guidance and many thought-provoking discussions. They are indebted to Dr C. Cordon-Cardo for continued support of the Pathogen Surveillance Program. They also thank the staff; the Nursing leadership, including Christine Mahoney and Maria Latrace; the Residency Program Leadership, including Daniel Steinberg; the Hospital Leadership, including Dr J. Boal at Mount Sinai Beth Israel; and the New York State Department of Health, especially Dr K. Southwick and Rafael Fernandez, for their extraordinary help and support during the outbreak. They gratefully acknowledge the authors and the originating and submitting laboratories of sequences from GISAID’s EpiFlu (www.gisaid.org) that were used as background for the phylogenetic inferences. The list of authors and submitting laboratories is shown in Supplementary Table 1. An early version of the manuscript was edited by Life Science Editors.
Financial support. This work was supported, in part, by the National Institute of Allergy and Infectious Disease (NIAID) Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract HHSN272201400008C, the Office of Research Infrastructure of the National Institutes of Health (NIH) (grant numbers S10OD018522 and S10OD026880), and institutional seed funds. T. R. P. reports NIH/NIAID grant F30AI122673.
Potential conflicts of interest. R. S. serves as Vice President of Technology Development at Sema4, a Mount Sinai venture, outside the submitted work. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Blackburn RM, Frampton D, Smith CM, et al. ; ICONIC Group . Nosocomial transmission of influenza: a retrospective cross-sectional study using next generation sequencing at a hospital in England (2012-2014). Influenza Other Respir Viruses 2019; 13:556–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Godoy P, Torner N, Soldevila N, et al. ; Working Group on the Surveillance of Severe Influenza Hospitalized Cases in Catalonia . Hospital-acquired influenza infections detected by a surveillance system over six seasons, from 2010/2011 to 2015/2016. BMC Infect Dis 2020; 20:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huzly D, Kurz S, Ebner W, Dettenkofer M, Panning M. Characterisation of nosocomial and community-acquired influenza in a large university hospital during two consecutive influenza seasons. J Clin Virol 2015; 73:47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wilson KE, Wood SM, Schaecher KE, et al. Nosocomial outbreak of influenza A H3N2 in an inpatient oncology unit related to health care workers presenting to work while ill. Am J Infect Control 2019; 47:683–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cummings MJ, Tokarz R, Bakamutumaho B, et al. Precision surveillance for viral respiratory pathogens: virome capture sequencing for the detection and genomic characterization of severe acute respiratory infection in Uganda. Clin Infect Dis 2019; 68:1118–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Houlihan CF, Frampton D, Ferns RB, et al. Use of whole-genome sequencing in the investigation of a nosocomial influenza virus outbreak. J Infect Dis 2018; 218:1485–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mena I, Nelson MI, Quezada-Monroy F, et al. Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. Elife 2016; 5:e16777. doi: 10.7554/eLife.16777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tran K, Cimon K, Severn M, Pessoa-Silva CL, Conly J. Aerosol generating procedures and risk of transmission of acute respiratory infections to healthcare workers: a systematic review. PLoS One 2012; 7:e35797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 2014; 15:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu STH, Behzadi MA, Sun W, et al. Antigenic sites in influenza H1 hemagglutinin display species-specific immunodominance. J Clin Invest 2018; 128:4992–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tan GS, Krammer F, Eggink D, Kongchanagul A, Moran TM, Palese P. A pan-H1 anti-hemagglutinin monoclonal antibody with potent broad-spectrum efficacy in vivo. J Virol 2012; 86:6179–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nachbagauer R, Wohlbold TJ, Hirsh A, et al. Induction of broadly reactive anti-hemagglutinin stalk antibodies by an H5N1 vaccine in humans. J Virol 2014; 88:13260–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.