Here, Li et al. systematically analyzed Argonaute-CLASH (cross-linking, ligation, and sequencing of miRNA–target RNA hybrids) data and identified numerous candidate target RNA-directed miRNA degradation (TDMD) triggers, focusing on their ability to induce nontemplated nucleotide addition at the miRNA 3′ end. They uncovered widespread TDMD triggers in target RNAs and demonstrated that they could functionally cooperate with the encoded proteins.
Keywords: microRNA, TDMD, AGO-CLASH, BCL2L11, apoptosis
Abstract
Binding of microRNAs (miRNAs) to mRNAs normally results in post-transcriptional repression of gene expression. However, extensive base-pairing between miRNAs and target RNAs can trigger miRNA degradation, a phenomenon called target RNA-directed miRNA degradation (TDMD). Here, we systematically analyzed Argonaute-CLASH (cross-linking, ligation, and sequencing of miRNA–target RNA hybrids) data and identified numerous candidate TDMD triggers, focusing on their ability to induce nontemplated nucleotide addition at the miRNA 3′ end. When exogenously expressed in various cell lines, eight triggers induce degradation of corresponding miRNAs. Both the TDMD base-pairing and surrounding sequences are essential for TDMD. CRISPR knockout of endogenous trigger or ZSWIM8, a ubiquitin ligase essential for TDMD, reduced miRNA degradation. Furthermore, degradation of miR-221 and miR-222 by a trigger in BCL2L11, which encodes a proapoptotic protein, enhances apoptosis. Therefore, we uncovered widespread TDMD triggers in target RNAs and demonstrated an example that could functionally cooperate with the encoded protein.
MicroRNAs (miRNAs) are ∼22-nt noncoding RNAs that play a critical role in post-transcriptional regulation of gene expression in animals and plants. Typically, miRNA–Argonaute (AGO) complexes are recruited to mRNAs via Watson-Crick base-pairing between target sites in the 3′ untranslated region (UTR) and the seed region (nucleotides 2–7) of the miRNA, allowing miRNAs to influence translation or stability of target mRNA molecules (Bartel 2018). As a group, miRNAs are relatively stable with long half-lives (Kingston and Bartel 2019; Reichholf et al. 2019). Surprisingly, extensive base-pairing between target RNA and miRNA could lead to miRNA degradation, a phenomenon termed “target RNA-directed miRNA degradation” (TDMD) (Wightman et al. 2018).
The initial discovery of TDMD came from synthetic and viral transcripts (Ameres et al. 2010; Cazalla et al. 2010). In Drosophila and human cells, synthetic target RNAs that form extensive base-pairing with the miRNA induce 3′ end adenosine (A) and/or uridine (U) tailing, trimming, and degradation of the miRNA (Ameres et al. 2010). Here, we refer to the ∼20-nt sequence that forms extensive base-pairing with the miRNA and can induce miRNA degradation as the “TDMD trigger.” Additional studies using synthetic targets in a variety of experimental systems confirmed the importance of base-pairing in both the miRNA seed and the 3′ region in inducing tailing and trimming (Xie et al. 2012; de la Mata et al. 2015; Haas et al. 2016; Park et al. 2017). Tailing and trimming occur when miRNA is bound by AGO, while the terminal uridylyl transferases (TUT4/TUT7) and the 3′ exonuclease DIS3L2 are implicated in this process (Haas et al. 2016; Yang et al. 2020). Together, these data and structural studies suggest a model in which extensively base-paired miRNA and target RNA induce a conformational change of AGO to expose miRNA for 3′ end tailing and trimming, marking the miRNA for subsequent degradation (Sheu-Gruttadauria et al. 2019).
As noted above, several virus-encoded RNAs can degrade host miRNAs through TDMD (Cazalla et al. 2010; Libri et al. 2012; Marcinowski et al. 2012; Lee et al. 2013). The founding example of a naturally occurring TDMD exists in Herpesvirus saimiri (HVS) transformed marmoset T cells, in which the viral small nuclear RNA HSUR1 can base-pair with and degrade miR-27 (Cazalla et al. 2010). Similarly, miR-27 degradation is induced by viral RNA m169 in murine cytomegalovirus (CMV)-infected mouse cells (Libri et al. 2012; Marcinowski et al. 2012). As a result, miR-27's target genes are up-regulated in infected cells, highlighting a strategy by which viruses manipulate host gene expression through miRNA degradation (Guo et al. 2014). An additional example of viral TDMD trigger lies in the UL/b′ region of the human cytomegalovirus genome, which mediates the degradation of miR-17 and miR-20a (Lee et al. 2013).
TDMD is now recognized as an endogenous gene regulation mechanism, as several cellular transcripts were found to induce miRNA degradation (Bitetti et al. 2018; Ghini et al. 2018; Kleaveland et al. 2018). First, mammalian NREP mRNA, or its noncoding homolog libra in fish, is almost exclusively expressed in the brain to limit miR-29b in cerebellar granule cells (Bitetti et al. 2018). Mutation within the NREP trigger leads to impaired motor coordination and motor learning deficits in mice. Second, the long noncoding (lnc)RNA CYRANO also induces miR-7 degradation in the brain, although disruption of this trigger did not yield a clear phenotype other than up-regulation of miR-7 and repression of miR-7 targets (Kleaveland et al. 2018). Third, a trigger in the Serpine1 3′ UTR suppresses miR-30b/c, which affect cell cycle re-entry and apoptosis (Ghini et al. 2018). In all cases, the base-pairing pattern between the endogenous trigger and the miRNA resembles the synthetic and viral TDMD pairs. However, tailing and trimming appear to be dispensable for TDMD, as knockdown of cytoplasmic poly(A) polymerase PAPD4 (a tailing enzyme) affected tailing but not the levels of miR-7 (Kleaveland et al. 2018). Consistent with these observations, recent studies demonstrated that TDMD base-pairing induces a conformational change in AGO protein, which recruits ZSWIM8 E3 ubiquitin ligase to ubiquitinate and mark AGO for degradation (Han et al. 2020; Shi et al. 2020). Losing protection from AGO, miRNAs are then quickly degraded by ribonucleases. Importantly, TDMD only causes degradation of the miRNAs but not the target RNAs (Bitetti et al. 2018; Kleaveland et al. 2018). Given the long half-lives of miRNAs, TDMD is apparently a mechanism that could swiftly clear a specific miRNA and influence its corresponding gene regulatory network in response to environmental cues. There appears to be abundant endogenous TDMD triggers in the transcriptome, because >30 miRNAs increased in a variety of ZSWIM8 knockout (KO) cell lines and animals (Han et al. 2020; Shi et al. 2020). To better understand how TDMD molds the gene regulatory network, it is essential to identify endogenous TDMD triggers.
Results
A wide range of transcripts contain potential TDMD triggers
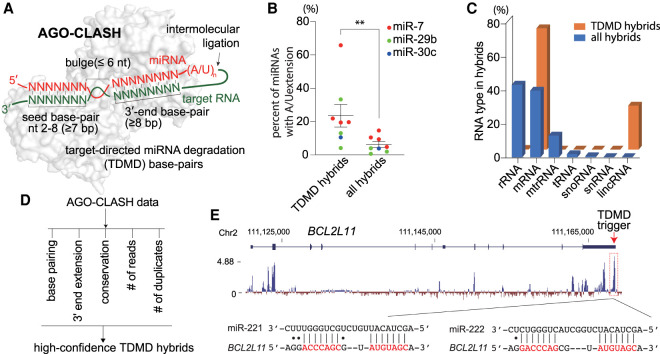
To identify TDMD pairs in mammals, we analyzed target RNA/miRNA hybrids in our AGO-CLASH (cross-linking, ligation, and sequencing of hybrids) data obtained from HCT116 colorectal cancer cells (Fields et al. 2021), as well as five publicly available AGO-CLASH data sets, which include three data sets from human cells and two data sets from murine samples (Supplemental Fig. S1A; Helwak et al. 2013; Moore et al. 2015; Gay et al. 2018; Bullard et al. 2019). In AGO-CLASH experiments, target RNA/miRNA pairs are captured by UV cross-linking and intermolecular ligation, with resulting target RNA/miRNA hybrid reads obtained from Illumina sequencing (Fig. 1A). Based on known TDMD pairs, the trigger interacts with both the 5′ seed region and the 3′ portion of the miRNA with more than seven and eight consecutive base pairs, respectively, separated by a bulge no longer than 6 nt (Fig. 1A; Wightman et al. 2018). According to this base-pairing pattern, we have broadly identified 671 candidate TDMD hybrids in all AGO-CLASH data sets (Supplemental Table S1). When analyzed for gene ontology enrichment, candidate TDMD transcripts are involved in essential pathways such as regulation of RNA polymerase II transcription, peptidyl-threonine phosphorylation, mitotic cell cycle, and brain development (Supplemental Fig. S1B).
Figure 1.
A wide range of transcripts contain potential TDMD triggers. (A) Schematic of AGO-CLASH and target-directed miRNA degradation (TDMD) base-pairing pattern. AGO2 structure (PDB: 6MDZ) is used in the illustration. A detailed description is given in the text. (B) The percentage of miRNA with extension in the hybrids of CYRANO/miR-7, NREP/miR-29b, SERPINE1/miR-30c, or all RNA/miRNA. Each dot represents a data point from one AGO-CLASH data set. (**) P < 0.01, ratio paired t test. (C) The composition of different types of RNA in the AGO-CLASH miRNA-containing hybrids obtained from HCT116 cells, comparing all hybrids and TDMD hybrids. (D) High-confidence TDMD hybrid screening based on five criteria. See the Materials and Methods for details. (E) Organization and conservation of the human BCL2L11 locus. Shown below the gene model ([blue boxes] exons, [blue line] introns) is a conservation plot, which is based on a 100-vertebrate basewise conservation by PhyloP. Diagramed below is the pairing of miR-221 and miR-222 to its potential TDMD trigger, located within the most conserved region of BCL2L11.
miRNA degradation by TDMD is usually accompanied by the addition of A and/or U residues to the 3′ end of the miRNA. Indeed, when we examined hybrids that contain known TDMD triggers (CYRANO, NREP, and SERPINE1), the A/U extension rate of their corresponding miRNAs (miR-7, miR-29b, and miR-30c) is significantly higher than when these miRNAs are paired with all RNAs, most of which do not induce TDMD (Fig. 1B; Supplemental Fig. S1C). Interestingly, while rRNAs (43.8%) and mRNAs (40.3%) make up the majority of the miRNA hybrids in AGO-CLASH from HCT116 cells, candidate TDMD hybrids are enriched in mRNAs (73.1%) and lncRNAs (26.5%), consistent with the distribution of known triggers (Fig. 1C). The candidate TDMD hybrids’ composition across all six AGO-CLASH data sets remains the same (Supplemental Fig. S1D).
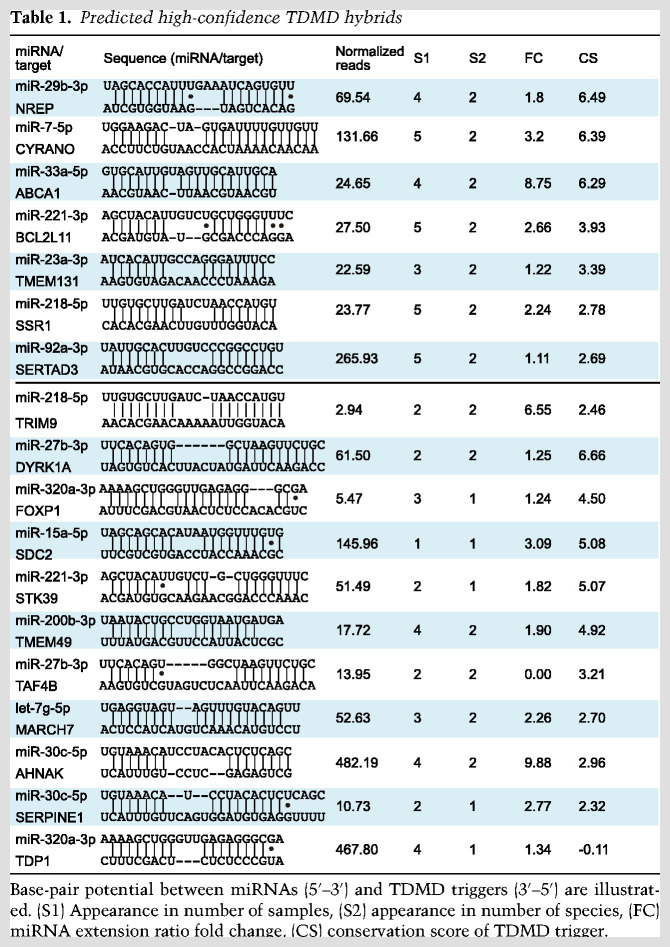
To narrow down probable TDMD triggers, we applied five criteria to filter high-confidence TDMD hybrids: (1) a TDMD-like base-pairing pattern with more stringent base-pairing at the miRNA 3′ end, (2) a higher rate of miRNA 3′ end A/U extension when compared with all hybrids, (3) a conservation score of target sequence >2, (4) a normalized hybrid read abundance >10, and (5) that candidate hybrids exist in both human and mouse data sets (Fig. 1D, see the Materials and Methods for details). Only seven hybrids satisfy all five criteria, while 11 hybrids can satisfy four of the five criteria (Table 1). We refer to the target RNA sequences in these 18 hybrids as high-confidence TDMD triggers. It was reassuring to find that all three known triggers in CYRANO, NREP, and SERPINE1 are among the list of 18 high-confidence triggers, with conservation scores of 6.39, 6.49, and 2.32, respectively (Table 1). Many candidates on our list exhibit similar conservation scores, suggesting the existence of additional bona fide TDMD triggers. Except for CYRANO, which is a lncRNA, all other triggers reside in mRNAs. Among these, nine are located in the 3′ UTR of the gene and eight are within the coding region (Fig. 1E; Supplemental Fig. S2). Taken together, we have potentially identified multiple endogenous TDMD triggers, consistent with recent findings that suggest TDMD is more widespread than previously appreciated (Han et al. 2020; Shi et al. 2020).
Table 1.
Predicted high-confidence TDMD hybrids
Exogenous expression of candidate TDMD triggers in cells decreases corresponding miRNAs
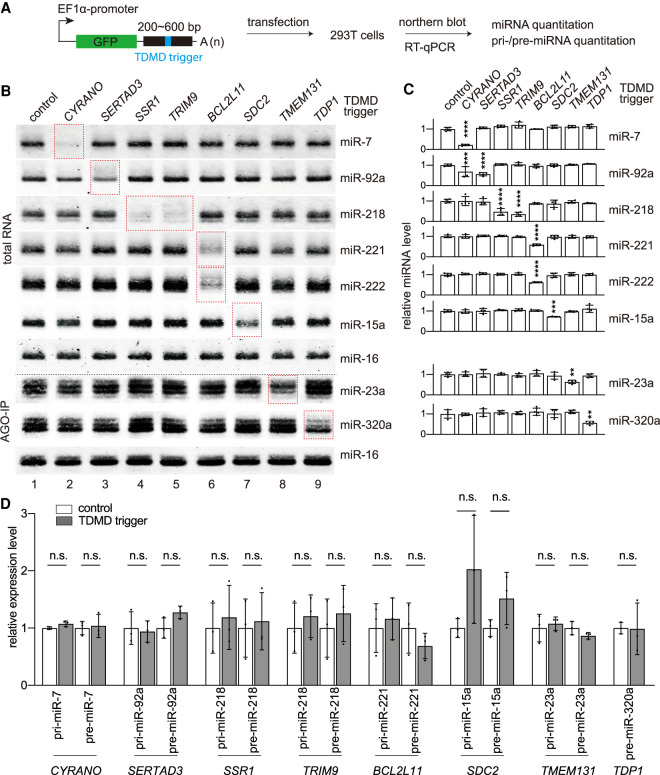
We next tested whether high-confidence TDMD triggers can degrade their corresponding miRNAs. To this end, we cloned ∼200- to ∼600-bp conserved sequences of the 11 high-confidence triggers downstream from a GFP reporter and transfected these reporters into human embryonic kidney (HEK) 293T cells (Fig. 2A). Near-infrared Northern blot analyses for miRNAs were performed on total RNAs extracted from 293T cells transfected with the reporter plasmids for 48 h (Miller et al. 2018). As a positive control, CYRANO triggers reduced miR-7 levels to ∼20% (Fig. 2B, lane 2), consistent with a previous report (Kleaveland et al. 2018). Seven new high-confidence triggers from SERTAD3, SSR1, TRIM9, BCL2L11, SDC2, TMEM131, and TDP1 were able to specifically and significantly lower the levels of their corresponding miRNAs in 293T cells by 30%–70% (Fig. 2B [lanes 3–9], C). In these experiments, low-abundance miR-23a and miR-320a signals are obscured by high background in Northern blots using total RNA (data not shown), we therefore enriched AGO-bound miRNAs by immunoprecipitation (IP) and confirmed that these two miRNAs are reduced by TMEM131 and TDP1 triggers, respectively (Fig. 2B, bottom panels, lanes 8,9). Additionally, AGO-IP experiments confirmed that functional miRNAs associating with AGO decreased in response to the expression of triggers in SERTAD3, SSR1, TRIM9, BCL2L11, and SDC2 (Supplemental Fig. S3A). We also tested two other “low-confidence” triggers that are nonetheless highly conserved (MTF1 and RARS1) (Supplemental Fig. S3B). miR-106 levels decreased nearly 40% upon MTF1 trigger expression, while miR-16 levels were not decreased in response to the RARS1 trigger (Supplemental Fig. S3C).
Figure 2.
Exogenous expression of candidate TDMD triggers in cells decreases corresponding miRNAs. (A) Schematic of high-confidence TDMD trigger validation in HEK293T cells. Two-hundred base pairs to 600 bp of the TDMD regions were inserted into a GFP reporter. The plasmids were transfected in HEK293T cells for 2 d and cells were collected to detect miRNA abundance by Northern blot or pri-miRNA abundance by RT-qPCR. The TDMD trigger, flanking region, and GFP are represented by blue, black, and green rectangles, respectively. (B) Representative Northern blots measuring miR-7, miR-92a, miR-218, miR-221, miR-222, miR-15a, miR-23a, miR-320a, and miR-16 levels in HEK293T cells transfected with the TDMD reporters illustrated in A. RNA samples were either total RNA or AGO-associating RNA. Red dotted rectangles highlight the lanes with corresponding miRNA and the TDMD trigger. (C) Quantification of miRNA levels in Northern blots as represented in B. miRNA abundance was normalized to miR-16. Statistically significant changes compared with the GFP control are indicated. (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001, ordinary ANOVA with Dunnett's test. n = 4 biological replicates. (D) The influence of TDMD transcripts on pri-miRNA and pre-miRNA expression in control plasmid transfected cells and TDMD reporter transfected cells, as determined by RT-qPCR, normalized to the geometric mean of the values for ACTIN housekeeping gene. Error bars indicate SD. (n.s.) P > 0.05, unpaired Welch's t-test. n = 3 biological replicates.
To examine whether TDMD would occur without overexpression of the TDMD triggers by plasmid transfection, we transduced 293T cells or U87MG glioblastoma cells with lentiviruses carrying CYRANO, TRIM9, or mouse Bcl2l11 triggers, which induced the most robust miRNA degradation in transfection assays (Fig. 2A–C). Lentiviral transduced CYRANO trigger level is ∼8.5-fold higher than the endogenous trigger level in 293T cells, which express much lower CYRANO than the nervous system (Supplemental Fig. S3D; Kleaveland et al. 2018). Therefore, trigger expression levels in the lentivirus transduced cells are within the physiological range. Mouse Bcl2l11 trigger is almost identical to its human counterpart except for one nucleotide, and maintains base-pairing with miR-221 and miR-222 (miR-221/222), two polycistronic miRNAs encoded in the same primary transcript (see detail below). Like the transfection experiments, stable expression of triggers in transduced cells led to a 30%–60% reduction of their corresponding miRNAs (Supplemental Fig. S3E).
The reduction of mature miRNA via TDMD should not be a result of decreased transcription or processing of primary or precursor miRNA (pri/pre-miRNA) (Bitetti et al. 2018; Ghini et al. 2018; Kleaveland et al. 2018). To confirm, we performed real-time polymerase chain reactions after reverse transcription (RT-qPCR) to measure pri/pre-miRNA levels of the corresponding miRNAs. Our data show that none of these pri/pre-miRNAs reduced in cells after transfection or lentiviral transduction of the trigger reporters, suggesting that the mature miRNA reduction is caused by TDMD (Fig. 2D; Supplemental Fig. S3F).
In total, we tested 13 TDMD triggers with the GFP reporter. Other than the eight positive triggers and the one negative trigger, four could not be assessed accurately, since their corresponding miRNAs (ABCA1/miR-33a, TMEM49/miR-200b, NNAT/miR-708, and SEPT8/miR-151) are not detectable by Northern blot, even after AGO-IP enrichment (data not shown). Nonetheless, our success in confirming multiple TDMD triggers’ efficacy in inducing miRNA degradation suggests that AGO-CLASH is a reliable experimental tool to search for triggers.
TDMD triggers promote miRNA degradation and miRNA 3′ end extension, and influence miRNA function
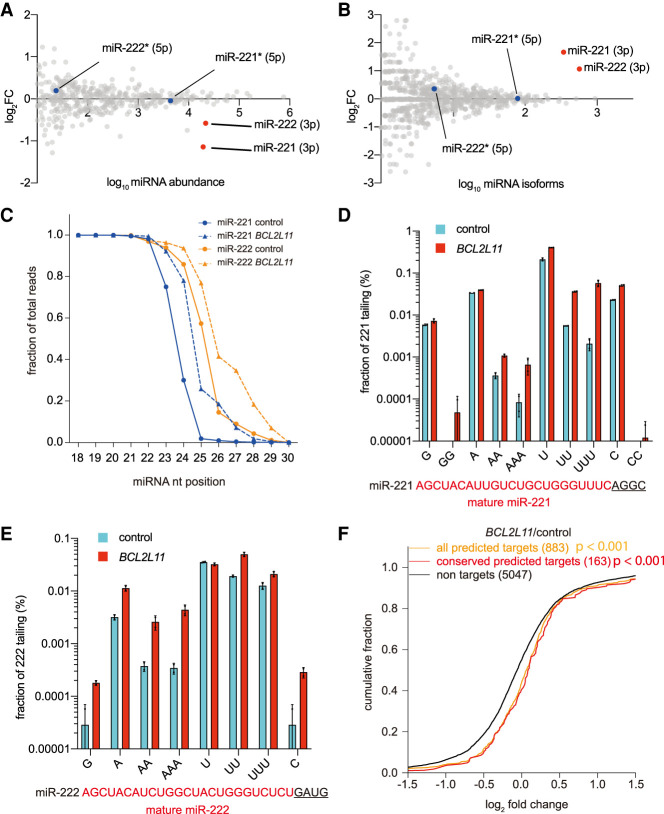
To globally determine whether TDMD triggers affected only the corresponding miRNA or affected a wider network of miRNAs, we performed small RNA sequencing in 293T cells expressing the BCL2L11 or TRIM9 trigger (Fig. 2B). From two biological replicates of the small RNA sequencing data, we calculated the total abundance of miRNA between 21 and 26 nt. Consistent with the results of Northern blots, levels of targeted miRNAs (miR-221/222 or miR-218) in trigger reporter (BCL2L11 or TRIM9) transfected cells were reduced by 30%–55% (Fig. 3A; Supplemental Fig. S4A, red dots). Meanwhile, the global miRNA population, including the passenger strand of the targeted miRNA, remained largely unchanged, again supporting the notion that TDMD is specific to mature miRNAs (Fig. 3A; Supplemental Fig. S4A, gray and blue dots).
Figure 3.
TDMD triggers promote miRNA degradation and miRNA 3′ end extension, and influence miRNA function. The influence of BCL2L11 trigger on miRNA levels (A) and miRNA isoforms (B) in 293T cells. Relative miRNA levels (A) and isoforms (B) were measured by small RNA-seq after transfection of BCL2L11 trigger reporter and compared with the control reporter. n = 2 biological replicates. miR-221 and miR-222 are indicated by red dots, and the blue dots represent their passenger strands. (C) The fraction of sRNA-seq reads with coverage of 18–30 nt for miR-221 and miR-222. For each miRNA, solid lines delineate the control samples, and dashed lines delineate the BCL2L11 trigger-expressing samples. n = 2 biological replicates. Frequency of tailing for miR-221 (D) and miR-222 (E) in BCL2L11 trigger reporter transfected 293T cells. Shown are the percent fractions of the tailed miR-221/222 reads relative to the total miR-221/222 reads. miR-221 and miR-222 sequences (red capital letters), together with the genomic sequence downstream (black underlined letters), are given below the plot. (F) Repression of miR-221/222 targets in BCL2L11 transfected 293T cells. Plotted are cumulative fractions of mRNA fold changes observed after degradation of miR-221/222 in BCL2L11 trigger reporter transfected 293T cells, comparing the impact on all predicted miR-221/222 targets (orange line) and conserved predicted miR-221/222 targets (red line) with that of nontargets (black).
TDMD triggers are also known to induce miRNA 3′ end tailing and trimming. We therefore calculated the number of isoforms for each miRNA from 18 to 30 nt. As expected, in BCL2L11 and TRIM9 trigger-expressing cells, miR-221/222 and miR-218 have the largest numbers of miRNA isoforms (Fig. 3B; Supplemental Fig. S4B, red dots). These isoforms mainly result from 3′ end tailing (Fig. 3C; Supplemental Fig. S4C). Upon closer examination, single or multiple nucleotide A/U tailing accounts for most of the tailing events on the miR-221/222 and miR-218 that are targeted by TDMD (Fig. 3D,E; Supplemental Fig. S4D), consistent with previous observations (Ameres et al. 2010; Ghini et al. 2018; Kleaveland et al. 2018).
To examine the functional consequences of TDMD, we simultaneously analyzed the trigger-expressing 293T cells by poly(A) RNA sequencing. We compared fold changes of TargetScan (version 7.2) predicted and conserved miR-221/222 or miR-218 target mRNAs with fold changes of nontarget mRNAs. After filtering out low-abundance mRNAs, more predicted and conserved miR-221/222 target mRNAs increased compared with nontarget mRNAs in BCL2L11 trigger-expressing cells (Fig. 3F). Similarly, conserved miR-218 targets showed higher fold changes than nontargets in TRIM9 trigger-expressing cells, although predicted miR-218 targets did not significantly differ from nontargets (Supplemental Fig. S4E). Collectively, these data confirmed that TDMD triggers in BCL2L11 and TRIM9 induce degradation and 3′ end A/U extension of miR-221/222 and miR-218, respectively, which in turn results in up-regulation of the mRNAs targeted by the corresponding miRNAs.
Sequence requirement for TDMD triggers
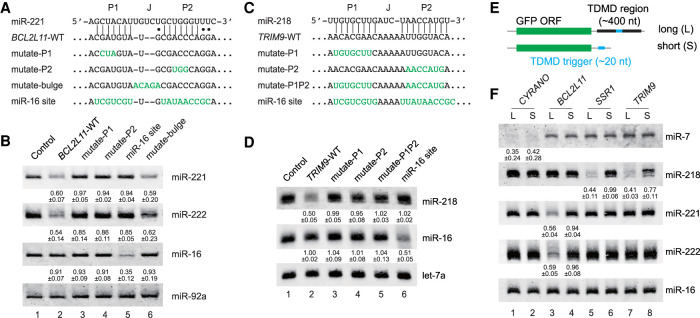
We next carried out a series of mutational analyses to probe sequence features essential for our newly identified TDMD triggers. Previous studies suggested that the base-pairing in the miRNA seed region (P1 duplex) and miRNA 3′ end region (P2 duplex), as well as the central bulge (J region), are all important to TDMD both in vivo and in vitro (Fig. 4A,C; Xie et al. 2012; Bitetti et al. 2018; Ghini et al. 2018; Kleaveland et al. 2018; Sheu-Gruttadauria et al. 2019). For example, let-7a was most strongly attenuated by the fully complementary target sequence, compared with other let-7 family members that contain mismatch(es) in the nonseed region (Xie et al. 2012). In addition, a TDMD trigger against one miRNA can be mutated to base-pair with a different miRNA, leading to its degradation (Cazalla et al. 2010; Libri et al. 2012; Kleaveland et al. 2018). When we introduced mutations as short as 3 nt in BCL2L11 and TRIM9 triggers to disrupt the P1 and P2 duplexes, the ability to degrade their corresponding miRNAs was abolished when these mutation constructs were tested in 293T cells (Fig. 4A,B [lanes 3,4], C,D [lanes 3–5]). Furthermore, mutating both the BCL2L11 and TRIM9 triggers to pair them with miR-16 causes degradation of miR-16 (Fig. 4B, lane 5; Fig. 4D, lane 6). Finally, when the J region in the BCL2L11 trigger was converted to the perfect complementary sequence of the corresponding miRNA, the mutant was ∼10% less efficient in miRNA decay (Fig. 4B, lane 6). This is possibly because the mismatch in the J region inhibits target RNA cleavage by AGO2, allowing the target to induce multiple rounds of miRNA degradation (Sheu-Gruttadauria et al. 2019).
Figure 4.
Sequence requirement for TDMD triggers. Schematic of the WT BCL2L11 trigger (A) and TRIM9 trigger (C) constructs with a mutated miR-221/222 or miR-218 site. The miR-221/222 site in BCL2L11 or the miR-218 site in TRIM9 was mutated to either disrupt or expand pairing to the corresponding miRNA or introduce an extensively complementary site to miR-16. The importance of the complementary site for the BCL2L11 trigger (B) or TRIM9 trigger (D) function. Representative Northern blot measuring miR-221, miR-222, miR-218, miR-16, miR-92a, and let-7a levels in HEK293T cells transfected with the constructs described in A and C. The normalized miRNA abundance and standard deviation are shown below each miRNA band. miRNA abundance in B and D was normalized to miR-92a and let-7a, respectively. The miRNA abundance in control samples was normalized to 1. n = 3 biological replicates. (E) Schematic of long and short TDMD trigger constructs. The long TDMD trigger includes the trigger sequence as well as the conserved flanking regions, while the short TDMD trigger contains only the TDMD trigger sequence. (F) The importance of the TDMD-flanking region on miRNA degradation. Representative Northern blot measuring miR-7, miR-218, miR-221, miR-222, and miR-16 levels in HEK293T cells transfected with either the long or short TDMD constructs described in E. The normalized miRNA abundance and standard deviation are shown below each miRNA band. miRNA abundance was normalized to miR-16. n = 3 biological replicates.
One interesting case for the influence of sequence variation occurs naturally in the BCL2L11 trigger. Human BCL2L11 and mouse Bcl2l11 triggers differ in only one nucleotide in the P2 region, but both maintain TDMD-like base-pairing with miR-221 (Supplemental Fig. S5A). However, this G-to-A variation prevents an extensive P2 duplex between Bcl2l11 and miR-222 (Supplemental Fig. S5A). We thus predicted that BCL2L11 and Bcl2l11 triggers would induce miR-222 degradation to different extents. To test this hypothesis, we created reporters containing BCL2L11 or Bcl2l11 triggers and transfected them into 293T cells. While miR-221 was degraded equally at ∼40% by the two triggers, miR-222 was, as predicted, ∼20% more degraded by BCL2L11 (Supplemental Fig. S5B,C). Importantly, in the reporter transfection assays, pri-miR-221/222 levels were unchanged (Supplemental Fig. S5D). These results support the notion that more extensive base-pairing in the P2 region leads to more efficient TDMD. In addition, the Watson-Crick A-U pair and G-U wobble pair appear to be interchangeable toward the edge of the P2 duplex, as seen in BCL2L11/miR-221 and Bcl2l11/miR-221 TDMD pairs (Supplemental Fig. S5A).
Requirement of the trigger-flanking sequences for TDMD
The structural flexibility of the transcript surrounding the TDMD trigger is another crucial factor in mediating miRNA degradation. In HSUR1, the flanking sequences form secondary structures to present an open and flexible trigger for base-pairing with miR-27 (Pawlica et al. 2016). Similarly, an evolutionarily conserved secondary structure spanning the trigger in CYRANO is believed to facilitate the interaction with miR-7 (Jones et al. 2020). We therefore tested whether the flanking sequences in our newly identified TDMD triggers are also required for miRNA degradation. To this end, we created reporters containing only the trigger sequence downstream from GFP (Fig. 4E). Compared with the “long” reporters, including hundreds of nucleotides spanning the entire conserved TDMD region, the “short” reporters containing only the ∼20-nt TDMD trigger from BCL2L11, SSR1, or TRIM9 barely degrade the corresponding miR-221/222 or miR-218 (Fig. 4F, cf. “S” and “L” lanes). Interestingly, the control CYRANO trigger could induce miR-7 degradation when present in both “long” and “short” reporters. We analyzed potential secondary structures of 100-nt sequences centered around the triggers using the RNAFold program (Gruber et al. 2008). The CYRANO trigger is situated in a cloverleaf structure in the “long” transcript, as reported (Supplemental Fig. S6A, left; Jones et al. 2020). Surprisingly, the CYRANO trigger was completely exposed with 100% unpaired nucleotides in the “short” transcript (Supplemental Fig. S6A, right), even more so than the “long” transcript that has 31% unpaired nucleotides, correlating with its potency in inducing miR-7 degradation. In contrast, other TDMD triggers in “short” transcripts contain fewer unpaired nucleotides than “long” transcripts (Supplemental Fig. S6B [SSR1 20% vs. 35%],C [BCL2L11 33% vs. 39%], D [TRIM9 32% vs. 55%]), limiting their abilities to base-pair with and induce degradation of the corresponding miRNAs (Fig. 4F). Therefore, the TDMD triggers are necessary but not sufficient for TDMD, and an optimal secondary structure offered by the conserved surrounding sequences is also required.
Involvement of ZSWIM8 in TDMD
Recently, the Bartel and Mendell groups (Han et al. 2020; Shi et al. 2020) reported that the ZSWIM8 cullin-RING E3 ubiquitin ligase is a key component in the TDMD pathway. ZSWIM8 ubiquitinates AGO proteins that bear TDMD pairs inside and signals the proteasome to degrade such AGO proteins (Han et al. 2020; Shi et al. 2020). Knockout of ZSWIM8 not only increases miRNAs subject to TDMD regulation, it also accentuates their 3′ end tailing and trimming (Shi et al. 2020). While the CYRANO-regulated miR-7 was broadly up-regulated in ZSWIM8 KO cell lines, other miRNAs are sensitive to ZSWIM8 loss in specific cell lines (Han et al. 2020; Shi et al. 2020). To test whether miR-221/222 and miR-218 degradation induced by BCL2L11 and TRIM9 is also ZSWIM8-dependent, we transfected the trigger reporters in 293T cells with or without ZSWIM8 (Han et al. 2020). ZSWIM8 KO in 293T cells increased baseline levels for miR-7 (because of TDMD induced by endogenous CYRANO), but not for miR-221 or miR-218 (Supplemental Fig. S7A). After transfection of the corresponding TDMD trigger reporters, miR-7, miR-221, and miR-218 all exhibit more extensive tailing in the absence of ZSWIM8 (Supplemental Fig. S7A, cf. lanes 5–7 and 1–3). Although we did not observe a clear increase in the abundance of miR-221 and miR-218 in ZSWIM8 KO 293T cells, the increase in miRNA tailing suggests the involvement of ZSWIM8 in the TDMD induced by BCL2L11 and TRIM9.
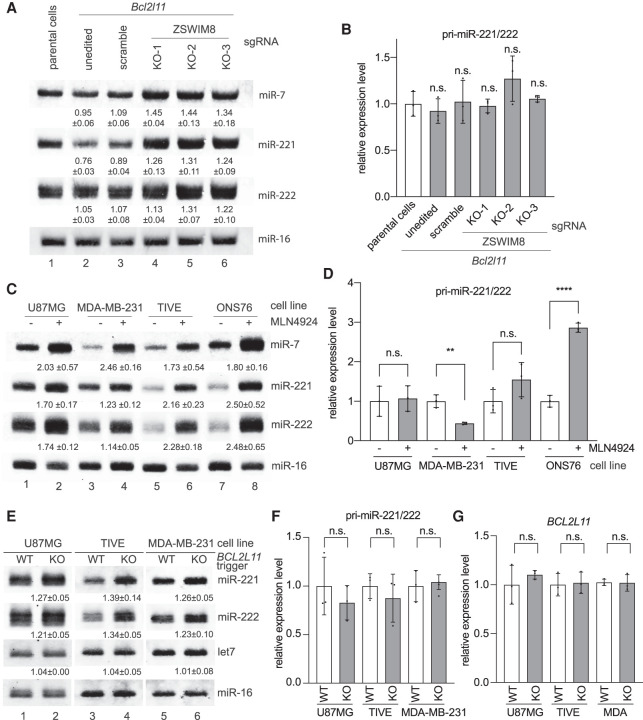
To further verify that the TDMD of miR-221/222 is related to ZSWIM8, we used the U87MG cell line stably expressing the Bcl2l11 trigger described in Supplemental Figure S3E. Stable expression of the Bcl2l11 trigger leads to significant degradation of miR-221 rather than miR-222 (Fig. 5A, cf. lanes 2 and 1), consistent with the transfection experiments (Supplemental Fig. S5B). Using lentivirus-delivered CRISPR–Cas9 and three previously designed sgRNAs (Han et al. 2020; Shi et al. 2020), we disrupted the ZSWIM8 gene in these U87MG cells. Levels of miR-7 and miR-221/222 increased by 20%–40% in ZSWIM8 KO compared with control cells, suggesting that TDMD of these miRNAs is ZSWIM8-dependent (Fig. 5A, cf. lanes 3 and 4–6). The lack of a tailing and trimming pattern on both miR-7 and miR-221/222 in ZSWIM8 KO cells may result from low terminal transferase activity in U87MG cells. RT-qPCR verified that pri-miR-221/222 levels were unchanged in ZSWIM8 KO cell lines (Fig. 5B). A similar set of experiments was conducted in 293T cells. However, we did not observe decreased miR-221/222 levels upon Bcl2l11 trigger expression (Supplemental Fig. S7B, cf. lanes 1 and 2). Consistently, ZSWIM8 KO did not increase miR-221/222, but increased miR-7 by ∼60% (Supplemental Fig. S7B, cf. lanes 3 and 4–6). Therefore, miR-221/222 TDMD appears to be cell type-specific. Collectively, these data suggest that, like miR-7, TDMD of miR-221/222 involves ZSWIM8. Since ZSWIM8 KO increased miR-221/222 levels by 20%–30% compared with wild-type (WT) cells (Fig. 5A, cf. lanes 1 and 4–6), there may be endogenous TDMD triggers, including BCL2L11, contributing to miR-221/222 degradation in U87MG cells.
Figure 5.
ZSWIM8 and the endogenous BCL2L11 TDMD trigger control miR-221/222 abundance. (A) The influence of ZSWIM8 on miR-221/222 accumulation in U87MG cells. Representative Northern blot measuring miR-7 (positive control), miR-221, miR-222, and miR-16 levels in U87MG cells stably expressing control or the Bcl2l11 trigger. In Bcl2l11 trigger cell lines, a scramble sgRNA, or one of the three sgRNAs (KO-1, KO-2, and KO-3) targeting ZSWIM8, was used for CRISPR-mediated KO. The normalized miRNA abundance and standard deviation are shown below each miRNA band. miRNA abundance was normalized to miR-16. The miRNA abundance in parental cells was normalized to 1. n = 3 biological replicates. (B) Primary transcripts for miR-221/222 measured by RT-qPCR in U87MG cell lines related to A, normalized to the levels of the ACTIN housekeeping gene. Error bars indicate SD. (n.s.) P > 0.05, unpaired Welch's t-test. n = 3 biological replicates. (C) The influence of neddylation inhibitor MLN4924 on miR-221/222 accumulation. Northern blot analysis of four cells (U87MG, MDA-MB-231, TIVE, and ONS76) following 48 h of 1 μM MLN4924 treatment. The normalized miRNA abundance and standard deviation are shown below each miRNA band. miRNA abundance was normalized to miR-16. The miRNA abundance in untreated cells was normalized to 1. n = 3 biological replicates. (D) Primary transcripts for the miR-221/222 measured by RT-qPCR in four cells (U87MG, MDA-MB-231, TIVE, and ONS76) related to C, normalized to the levels of the ACTIN housekeeping gene. Error bars indicate SD. (n.s.) P > 0.05, (**) P < 0.01, (****) P < 0.0001, unpaired Welch's t-test. n = 3 biological replicates. (E) The influence of the BCL2L11 TDMD trigger on miR-221/222 accumulation. Shown is a representative Northern blot measuring miR-221, miR-222, let-7, and miR-16 levels in three independently derived U87MG, TIVE, and MDA-MB-231 cell populations with or without BCL2L11 TDMD trigger KO. The normalized miRNA abundance and standard deviation are shown below each miRNA band. miRNA abundance was normalized to miR-16. The miRNA abundance in wild-type (WT) cells was normalized to 1. n = 3 biological replicates. (F,G) Primary transcripts for the miR-221/222 (F) and BCL2L11 (G) transcripts measured by RT-qPCR in three cell populations (U87MG, MDA-MB-231, and TIVE) related to E, normalized to the levels of the ACTIN housekeeping gene. Error bars indicate SD. (n.s.) P > 0.05, unpaired Welch's t-test. n = 3 biological replicates.
Degradation of miR-221/222 by the endogenous BCL2L11 TDMD trigger
Given that miR-221/222 degradation involves ZSWIM8, we next used various proteasome inhibitors to survey different cell lines whose miR-221/222 may be regulated by endogenous TDMD triggers. MLN4924 is a neddylation inhibitor that blocks NEDD8-cullin conjugates, and MG-132 directly occupies the active site of 20S proteasome; therefore, both inhibitors abolish ZSWIM8-dependent TDMD (Merlet et al. 2009; Han et al. 2020; Shi et al. 2020). We treated 12 different cell lines with either MLN4924 or MG-132. Unlike miR-7, which increased in most inhibitor-treated cell lines, miR-221/222 increased in only four cell lines, including U87MG, MDA-MB-231 breast cancer cells, telomerase immortalized vein endothelial cells (TIVE), and ONS76 medulloblastoma cells (Fig. 5C; Supplemental Fig. S7C). With the exception of ONS76 cells, pri-miR-221/222 levels were not significantly up-regulated by the MLN4924 treatment (Fig. 5D). miR-221/222 in 293T cells did not respond to MLN4924 treatment (Supplemental Fig. S7C), consistent with the ZSWIM8 KO experiment (Supplemental Fig. S7B).
We next examined whether the endogenous BCL2L11 trigger is responsible for degrading miR-221/222. To this end, we used CRISPR–-Cas9 with a single sgRNA to knock out the BCL2L11 trigger in U87MG, TIVE, and MDA-MB-231 cells. The Cas9 cleavage site within the BCL2L11 trigger allows mutations that can disrupt the base-pairings with miR-221/222 (Supplemental Fig. S8A). TIDE (tracking of indels by decomposition) analyses showed that ∼90% of the CRISPR–Cas9-modified cells had mutations in the BCL2L11 trigger (Supplemental Fig. S8B; Brinkman et al. 2014). A T7 endonuclease I mismatch cleavage assay on the PCR-amplified DNAs from the BCL2L11 trigger region in KO cells confirmed high mutation efficiency (Supplemental Fig. S8C). Note that many of the mutations are single-nucleotide insertions or deletions that may not affect TDMD. In all three KO cell lines, we detected a 20%–40% miR-221/222 increase compared with miR-16 or let-7 (Fig. 5E, cf. KO and WT lanes), without changes of pri-miR-221/222 or BCL2L11 mRNA (Fig. 5F,G). We conclude that the endogenous BCL2L11 trigger induces miR-221/222 degradation in specific cell lines.
The BCL2L11 TDMD trigger enhances BIM-induced apoptosis
BCL2L11 encodes BIM (BCL-2-interacting mediator of cell death), which belongs to the BCL-2 (B-cell lymphoma 2) family of proteins that can induce apoptosis. Specifically, BIM can directly interact with many antiapoptotic BCL-2 family members, leading to activation of apoptosis in both normal and cancer cells (Harada and Grant 2012; Sionov et al. 2015). On the other hand, miR-221/222 antagonizes apoptosis by inhibiting proapoptotic proteins (Zhang et al. 2010a; Zhao et al. 2015). Thus, it is intriguing that BCL2L11 mRNA contains an evolutionarily conserved TDMD trigger for miR-221/222 degradation.
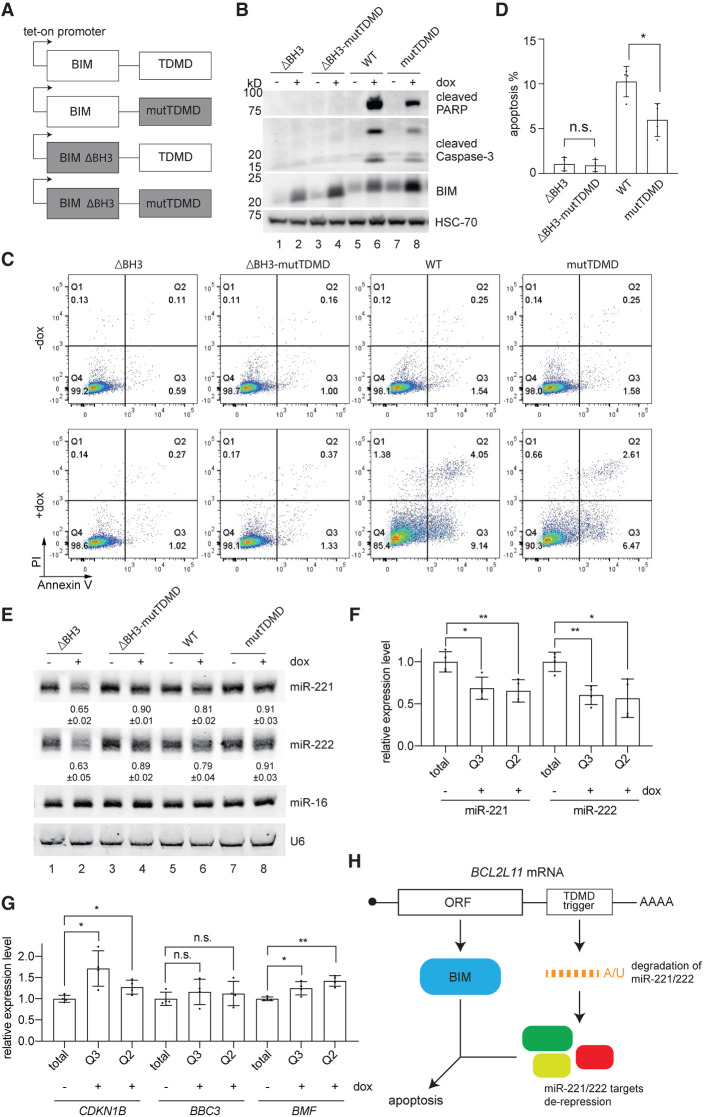
To test whether BIM and the TDMD trigger could function on apoptosis cooperatively, we engineered a set of four different BCL2L11-expressing constructs in a piggybac transposon vector driven by a doxycycline-inducible promoter (Fig. 6A). The WT construct had the intact BIM coding sequence and, in the 3′ UTR, a TDMD trigger capable of inducing miR-221/222 degradation. The mutTDMD construct contained a mutated trigger, which would not base-pair with the seed region of miR-221/222, and was incapable of inducing miR-221/222 degradation (Fig. 4A, mutate-P1). The ΔBH3 construct contained a deletion of nine amino acids in the BH-3 domain of BIM. This mutation was reported to render the resulting mutant BIM protein incompetent in interacting with BCL-2 family proteins and therefore incapable of inducing apoptosis (O'Connor et al. 1998). The double mutant (ΔBH3-mutTDMD) combined the mutations of mutTDMD and ΔBH3 into a single construct.
Figure 6.
The BCL2L11 TDMD trigger enhances BIM-induced apoptosis. (A) Schematic of WT or mutation form of the BCL2L11 TDMD region fused to either the WT or BH3 domain-deleted (ΔBH3) BIM coding sequence. The tet-on promoter-containing constructs were induced by 500 ng/mL doxycycline for 24 h in B–G. (B) Expression of cleaved PARP, cleaved caspase3, and BIM in four HCT116 lines containing different doxycycline-inducible BIM constructs as illustrated in A, with or without induction. HSC-70 protein was used as a loading reference. (C) Flow cytometry analyses of Annexin V- and propidium iodide-stained HCT116 cells as in A and B; the percentage of cells in each quadrant is labeled. (D) Quantification of apoptotic cells (Annexin-V+-stained; induced − uninduced) in four independently derived HCT116 lines in C. The asterisk marks significant values. (*) P < 0.05, ratio paired t test. n = 4 biological replicates. (E) Representative Northern blot measuring miR-221, miR-222, and miR-16 levels in four HCT116 lines containing different doxycycline-inducible BIM constructs as illustrated in A, with or without induction. The normalized miRNA abundance and standard deviation are shown below each miRNA band. miRNA abundance was normalized to miR-16. The miRNA abundance in uninduced samples was normalized to 1. n = 3 biological replicates. The influence of the BCL2L11 TDMD trigger on miR-221/222 (F) and three targets’ (G) abundance in sorted apoptotic cells expressing the TDMD construct compared with cells expressing the mutTDMD construct, normalized to the levels of miR-26a for miRNA or ACTIN housekeeping gene for targets. Error bars indicate SD. (n.s.) P > 0.05, (*) P < 0.05, (**) P < 0.01, unpaired Welch's t-test. n = 4 biological replicates. (H) Model of cooperative apoptosis induction by the BCL2L11 mRNA.
Four HCT116 colorectal cell lines were engineered with the inducible BIM constructs. HCT116 cells were chosen for this assay due to the lack of endogenous BCL2L11 TDMD activity (Supplemental Fig. S7C). BIM protein expression was induced in response to doxycycline in all four cells, as shown by immunoblot (Fig. 6B). In cells expressing intact BCL2L11, ∼10% underwent apoptosis. In contrast, cells expressing BCL2L11 without the TDMD trigger underwent less apoptosis (∼5%), as measured by Annexn V staining (Fig. 6C,D, cf. WT and mutTDMD). Additionally, cleaved PARP and cleaved caspase-3 were less evident in response to induction of the TDMD trigger mutant construct (Fig. 6B, cf. lanes 6 and 8). Deletion of the BH3 domain failed to induce apoptosis as expected (Fig. 6C,D). Only constructs with an intact trigger induced miR-221/222 degradation (Fig. 6E, lanes 2,6), and this correlated with increased apoptosis in the intact BCL2L11 construct. The presence of the TDMD trigger and degradation of miR-221/222 did not rescue the inability of ΔBH3 BIM to induce apoptosis (Fig. 6C,D, cf. ΔBH3 and ΔBH3-mutTDMD). We note that there was a modest ∼20% reduction of miR-221/222 in the WT construct (Fig. 6E). This is probably an underestimation, as there were more dead cells expressing the WT construct and the RNAs in dead cells may have leaked out. Therefore, we measured the levels of miR-221/222 in early (Q3) and late (Q2) apoptotic cells after cell sorting (Fig. 6C). In both early and late apoptotic cells, there was an ∼40% reduction of miR-221/222 in WT cells compared with mutTDMD cells (Fig. 6F). This is consistent with the reduction observed in cells expressing ΔBH3 BIM (Fig. 6E, lane 2). Reduction of miR-221/222 in WT cells resulted in a significant increase of previously identified miR-221/222 targets that are related to apoptosis, including CDKN1B and BMF (Fig. 6G; Fornari et al. 2008; Gramantieri et al. 2009). Another apoptosis-related miR-221/222 target, BBC3, was slightly increased, although the increase was not significant (Zhang et al. 2010b).
In addition, we prepared four BIM constructs as described above under the control of a constitutive CMV promoter and transfected them in U87MG cells. Similar results were obtained. Apoptosis induced by BIM decreased from 50% to 30% (Supplemental Fig. S9A–C) when the TDMD trigger was absent from the construct. ΔBH3 BIM failed to induce apoptosis regardless of the presence of an intact trigger (Supplemental Fig. S9A–C). Northern blots confirmed the reduction of miR-221/222 in the presence of the BCL2L11 trigger (Supplemental Fig. S9D, lanes 2,4). Similar to the experiments performed in HCT116 cells with inducible BIM constructs (Fig. 6E), a decrease of miR-221/222 was less apparent in cells with WT BIM at ∼15% (Supplemental Fig. S9D, lane 2), while miR-221/222 decreased ∼40% in ΔBH3 BIM-expressing cells. Taken together, our data lend support to the model that both BIM and the TDMD trigger cooperate in inducing apoptosis (Fig. 6H).
Discussion
How many endogenous TDMD triggers exist? The recent discovery of the central role of ZSWIM8 in TDMD has revealed >30 ZSWIM8-sensitive miRNAs in various cell lines and model organisms (Han et al. 2020; Shi et al. 2020). Presumably, each of the ZSWIM8-sensitive miRNA has one or more triggers to induce their degradation. Previously, the Nicassio group (Ghini et al. 2018) used bioinformatic methods to predict the existence of hundreds of potential TDMD triggers. However, only the Serpine1 trigger was validated to degrade miR-30b/c during cell cycle re-entry in mouse 3T9 cells. Here, we based our TDMD trigger discovery on experimental data from six different sets of AGO-CLASH (Helwak et al. 2013; Moore et al. 2015; Gay et al. 2018; Bullard et al. 2019; Fields et al. 2021). In AGO-CLASH, intermolecular ligation between miRNA and target RNA physically connects 3′-tailed miRNA with the triggers that induce such modifications (Fig. 1). Even though miRNA 3′ tailing is not a prerequisite for TDMD, this feature allowed highly effective identification of TDMD triggers (Table 1).
Three previously known triggers, CYRANO, NREP, and SERPINE1, were all identified in this search. Furthermore, seven out of the 15 new high-confidence candidate triggers and one additional conserved trigger tested positive for degrading their corresponding miRNAs in various cell lines (Figs. 2, 3). Therefore, AGO-CLASH represents a preferred method for TDMD trigger identification owing to its foundation built on experimental data.
Consistent with previously identified TDMD examples, the new TDMD triggers require extensive base-pairing with the miRNA, an optimal flanking region, and ZSWIM8 to induce miRNA degradation (Figs. 4 and 5). It is worth noting that we modeled the trigger search based on knowledge learned from known TDMD examples (Fig. 1A). It is possible that different trigger–miRNA base-pairing patterns exist to recruit ZSWIM8. Such new patterns may be identified in AGO-CLASH hybrids by emphasizing both the existence of 3′ tailing on miRNAs and the increase of corresponding miRNAs in ZSWIM8 knockout or knockdown cells. On the other hand, some TDMD pairs may not trigger miRNA 3′ tailing under certain conditions. For example, based on the Northern blots, we did not observe appreciable miR-7 or miR-221 tailing in U87MG cells with ZSWIM8 KO (Fig. 5A). Because the adenylation of miRNAs appears to be the predominant 3′ tailing during certain TDMD events (de la Mata et al. 2015; Ghini et al. 2018; Kleaveland et al. 2018), future TDMD hybrid searches in AGO-CLASH data may be geared toward finding enrichment of 3′ adenylation.
miR-7 degradation by CYRANO is highly efficient compared with other known TDMD events (Kleaveland et al. 2018). In our reporter transfection assays, the CYRANO trigger induced the most efficient degradation of its corresponding miR-7 (Fig. 2A–C). The different degrees of miRNA degradation could be influenced by multiple factors, one of which is the length of the P2 duplex. We note that the CYRANO/miR-7 pair contains the most extensive P2 duplex compared with other TDMD pairs, with 14 continuous Watson-Crick base pairs stretching to the very 3′ end of the miRNA (Kleaveland et al. 2018). Such an extensive P2 duplex can destabilize miRNA independently of the seed region base-pairing (P1 duplex) in an in vitro TDMD assay (Park et al. 2017). Our assays comparing the BCL2L11 and Bcl2l11 triggers confirmed that shortening the P2 duplex significantly reduces the TDMD of miR-222 (Supplemental Fig. S5).
Another factor contributing to TDMD efficiency lies within conserved sequences flanking triggers. We found that conserved sequences flanking the BCL2L11, TRIM9, and SSR1 triggers are critical for TDMD (Fig. 4E,F). Secondary structure predictions indicated that the flanking regions modulate the accessibility of the triggers (Supplemental Fig. S6). Although more detailed in vivo structural probing will be necessary to investigate the contribution of flanking regions, our data suggest that trigger sequence alone is not sufficient for TDMD. This is another reason that predicting TDMD triggers by bioinformatic methods may result in many false positives. However, in the future, bioinformatic searches may be improved by taking into account common features that may exist in the flanking regions.
miR-7 is almost universally sensitive to ZSWIM8 KO in all of the systems examined (Han et al. 2020; Shi et al. 2020), probably due to the high efficiency of TDMD by CYRANO and its ubiquitous expression. In contrast, other TDMD-regulated miRNAs, including miR-29b, miR-30b/c, miR-221/222, and miR-218 levels, were not significantly elevated in several ZSWIM8 KO cell lines, including 293T, K562, and mouse embryonic fibroblasts cells (Han et al. 2020; Shi et al. 2020). In our study, the levels of miR-221/222 increased in MLN4924-treated or ZSWIM8 KO U87MG cells, but not in 293T cells (Fig. 5A–D; Supplemental Fig. S7B,C). These results suggest cell or tissue specificity for most miRNAs under TDMD regulation. For example, miR-30b/c are more strongly degraded by Serpine1 in mouse fibroblasts during cell cycle re-entry than in growing conditions. Knocking out the endogenous Serpine1 trigger resulted in a mild miR-30b/c increase in growing cells (Ghini et al. 2018). Similarly, in several cell lines, we found comparable miR-221/222 up-regulation by MLN4924 treatment or BCL2L11 trigger KO (Fig. 5). Additionally, the TDMD of miR-221/222 significantly impacts apoptosis, specifically when BIM protein (encoded by BCL2L11) is expressed (Fig. 6). One explanation for cell-specific TDMD is the expression levels of TDMD trigger transcripts. In addition, high-efficiency and high-abundance TDMD pairs, such as CYRANO/miR-7, may be more effective in attracting ZSWIM8, especially in cells where ZSWIM8 is limiting, rendering lower-efficiency and lower-abundance TDMD triggers ineffective. Only in cells and tissues that are free from competition of high-efficiency TDMD pairs can low-efficiency TDMD triggers be functional.
To the best of our knowledge, BCL2L11 presents the first example in which both the coding region and the TDMD trigger contribute to the same pathway. The mRNA encodes the BIM protein, which triggers apoptosis through mitochondrial membrane permeabilization (Sionov et al. 2015). The noncoding 3′ UTR causes the degradation of antiapoptotic miR-221/222, which in turn shapes a cellular environment to enhance apoptosis induced by BIM (Garofalo et al. 2009; Zhang et al. 2010a,b; Kneitz et al. 2014; Wang et al. 2015; Zhao et al. 2015). Therefore, TDMD offers a second layer of gene regulatory potential for protein-coding transcripts, which may be involved in a variety of essential processes (Supplemental Fig. S1B). To date, less than a handful of cellular RNAs have been discovered to degrade miRNA (Bitetti et al. 2018; Ghini et al. 2018; Kleaveland et al. 2018), yet TDMD is potentially a general mechanism for miRNA turnover that has been largely unexplored (Han et al. 2020; Shi et al. 2020). Based on six AGO-CLASH data sets, we identified many potential TDMD triggers, significantly expanding the gene regulatory network influenced by TDMD.
This study focused on the most conserved and abundant TDMD pairs occurring across the AGO-CLASH data sets. In the future, it will be important to search for cell- or developmental stage-specific TDMD events, as different triggers might coordinate temporal and spatial degradation of the miRNAs. With the discovery of ZSWIM8 E3 ubiquitin ligase as a key component in TDMD and the advances in biochemical techniques such as AGO-CLASH, it would be possible to discover more endogenous triggers and deepen our understanding of TDMD, an emerging mechanism that may have a significant effect on gene expression.
Materials and methods
Identification of TDMD triggers
Six AGO-CLASH data sets (GSE: 46039, 164634, 101978, 73059, 73058, and 124687) from different cell types and two species were used to identify target RNA/miRNA hybrids that can potentially induce TDMD. Briefly, adapter sequences of raw reads were trimmed with Trimmomatic (Bolger et al. 2014) or Cutadapt software (Martin 2011), and reads <18 nt were removed. Paired FastQ files were merged by Pear software (Zhang et al. 2014) and collapsed by fastx_collapser (http://hannonlab.cshl.edu/fastx_toolkit) based on the random index sequences at the 5′ end of the reads to remove PCR duplications. Adapter sequences are listed in Supplemental Table S2. Processed reads then underwent mapping, hybrid calling, base-pairing prediction, and annotation by the Hyb software with default settings (Travis et al. 2014). Note that in the default settings, the target RNA reads were extended 25 nt on the 3′ end to compensate for possible trimming of the sequences that can base-pair with the miRNA.
Custom Python scripts (“TDMD identification” in https://github.com/UF-Xie-Lab/TDMD) were written to screen candidate TDMD hybrids that satisfied three criteria based on reported TDMD pairs (Wightman et al. 2018): First, the seed region (nucleotides 2–8) of the miRNA must base-pair with the target RNA (nucleotide 8 of the seed could be engaged in a G-U pair). Second, the 3′ end of the miRNA must contain more than eight consecutive base pairs with the target RNA. Third, the central bulge of the target RNA/miRNA hybrids should be <7 nt, but >0 nt. TDMD hybrids that met these criteria are summarized in Supplemental Table S1.
Furthermore, we applied five stringent standards to screen for high-confidence TDMD triggers using custom Python scripts (“high-confidence screen” and “miRNA_extension_in_hybrids” in https://github.com/UF-Xie-Lab/TDMD). To qualify as a high-confidence TDMD pair, it had to satisfy at least four of the five criteria described in the Supplemental Material.
Plasmid construction
For expression of the candidate TDMD transcripts, we cloned TDMD triggers with or without the ∼200- to 600-nt flanking region using specific oligonucleotides (Supplemental Table S3) from the human genomic DNA into the 3′ UTR of GFP in the AVA2590 plasmid (Addgene 85442) via the MluI restriction site. For generation of doxycycline-inducible BCL2L11 TDMD trigger cell lines, BCL2L11 sequences (with ΔBH3 or TDMD trigger mutations) from the plasmids used for transfection were PCR-amplified and inserted into pENTR_D TOPO (Thermo Fisher) via NotI and AscI sites. Subsequently, the inserts in pENTR plasmids were cloned into pB-TAG-ERP2 (Addgene 80479) with the LR clonase II (Invitrogen 11791020).
Cell culture
HEK293T, TIVE, MDA-MB-231, ONS76, and U87MG cells were cultured in Dulbecco's modified Eagle medium (DMEM; Sigma-Aldrich) with 5% CO2 at 37°C. HCT116 cells were cultured in McCoy's 5A medium (Hyclone). All media contained 1% penicillin and streptomycin (Gibco) and 10% fetal bovine serum (FBS; Gibco). HEK293T cells were transfected with polyethylenimine (PEI) MAX 40K (Polysciences) or Lipofectamine 3000 (Invitrogen) according to the manufacturers’ protocols. Briefly, 5×105 cells/well were seeded in six-well plates for 24 h. Doxycycline-inducible HCT116 cells expressing the BCL2L11 TDMD trigger were seeded at 2.5 × 105 cells/mL in tetracycline-depleted McCoy's 5A media (Atlanta Biologicals S10350). Twenty-four hours later, the medium was supplemented with doxycycline at 500 ng/mL. Doxycycline (250 ng/mL) was replenished after 24 h. Cells were harvested for analysis 72 h after seeding.
For protease inhibitor treatment, 12 different cell lines (HEK293T, MCF7, MDA-MB-231, TIVE, HCT116, MED8A, DAOY, ONS76, MOLT4, RPMI-8402, K562, and U87MG) were cultured to 50% confluency and treated with 1 µM MLN4924 in the medium for 48 h, or 10 µM MG-132 for 24 h, and then the total RNA was collected for Northern blot and RT-qPCR.
RNA quantification
Northern blots using near-infrared probes and EDC [N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide] (Thermo Scientific) cross-linking were performed as described (Pall and Hamilton 2008; Miller et al. 2018) and the results were analyzed with ImageStudio (v5.0) or ImageQuant TL (v7.0). miRNA cDNA synthesis and RT-qPCR were performed using TaqMan Advanced miRNA assays kit (Applied Biosystems) according to the manufacturer's instructions. RT-qPCR for long RNAs was performed and quantified using the ΔΔCT method as described (Stribling et al. 2021). Northern blot probes and RT-qPCR primers are in Supplemental Table S3. The quantification graphs and statistical analyses of RT-qPCR and Northern blot results were generated by GraphPad Prism 7. Unpaired two-tailed t-tests were used for comparisons between two different groups. A minimum of three biological replicates was used to calculate the standard deviation. A P-value < 0.05 was considered statistically significant.
Flow cytometry analysis
The dox-induced HCT116 cells or the transfected U87MG cells, which express the BCL2L11 TDMD trigger, were suspended in 200 µL of 0.25% trypsin per well in a six-well plate. All cells were collected by centrifugation at 300g for 5 min and washed once with 1× PBS. The cell pellets were resuspended in 100 µL of Annexin V binding buffer. Five microliters of APC-Annexin V and 10 µL of propidium iodide solution were added, and the cells were vortexed gently and incubated in the dark for 15 min at room temperature (25°C). The cells were mixed with 400 µL of Annexin V binding buffer and analyzed using the BD LSRFortessa flow cytometer. Furthermore, the apoptotic cells from gates Q2 and Q3 were sorted by Aria-II flow cytometer.
Supplementary Material
Acknowledgments
We thank Dr. Xin Chen, Dr. Tongjun Gu, Dr. Jianping Huang, Dr. Xi Huang, Dr. Lingtao Jin, Dr. Joshua T. Mendell, Dr. Lien Nguyen, Dr. Zhijian Qian, Dr. Laura Ranum, Dr. Rolf Renne, Dr. Fang Yu, and Dr. Eric Wang for kindly providing reagents, protocols, and technical support; Dr. J. Bert Flanegan and Dr. Demian Cazalla for stimulating discussions; Savannah Weeks and Augustine Vinson for assistance in TDMD trigger plasmid construction; and Tracy Ouncap for proofreading the manuscript. This work was supported by grants from the National Institutes of Health (R35GM128753 to M.X., T32CA257923 to N.M.H., and R01CA195732 to J.D.L.), the Florida Department of Health (Live Like Bella Pediatric Cancer initiative 21L03 to M.X.), the Elsa U. Pardee Foundation (to M.X.), the Harry T. Mangurian Jr. Foundation (to J.D.L), and the Leukemia and Lymphoma Society (Special Fellow grant 3399-20 to J. L.).
Authors contributions: L.L., P.S., and M.X. conceived the project and were involved in all experiments. T.L. validated RNA expression by RT-qPCR in 293T cell lines. C.J.F. conducted cumulative fraction curve analysis of predicted targets of miR-221/222 and miR-218. N.M.H. generated the doxycycline-inducible BIM HCT116 cell lines and performed inhibitor treatment in medulloblastoma cells. Y.W. performed validation Northern blots to measure miR-221/222 and miR-218 reduction upon plasmid transfection. J.L. supported apoptosis assays and flow cytometry analysis. C.M.G. extracted total RNA from TDMD trigger expressing 293T cells. L.L., J.D.L., and M.X. wrote the paper.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.348874.121.
Competing interest statement
The authors declare no competing interests.
References
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. 2010. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328: 1534–1539. 10.1126/science.1187058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2018. Metazoan microRNAs. Cell 173: 20–51. 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitetti A, Mallory AC, Golini E, Carrieri C, Carreño Gutiérrez H, Perlas E, Pérez-Rico YA, Tocchini-Valentini GP, Enright AJ, Norton WHJ, et al. 2018. MicroRNA degradation by a conserved target RNA regulates animal behavior. Nat Struct Mol Biol 25: 244–251. 10.1038/s41594-018-0032-x [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman EK, Chen T, Amendola M, van Steensel B. 2014. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 42: e168. 10.1093/nar/gku936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard WL, Kara M, Gay LA, Sethuraman S, Wang YP, Nirmalan S, Esemenli A, Feswick A, Hoffman BA, Renne R, et al. 2019. Identification of murine γherpesvirus 68 miRNA-mRNA hybrids reveals miRNA target conservation among γherpesviruses including host translation and protein modification machinery. PLoS Pathog 15: e1007843. 10.1371/journal.ppat.1007843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Yario T, Steitz JA. 2010. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 328: 1563–1566. 10.1126/science.1187197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Gaidatzis D, Vitanescu M, Stadler MB, Wentzel C, Scheiffele P, Filipowicz W, Großhans H. 2015. Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO Rep 16: 500–511. 10.15252/embr.201540078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields CJ, Li L, Hiers NM, Li T, Sheng P, Huda T, Shan J, Gay L, Gu T, Bian J, et al. 2021. Sequencing of Argonaute-bound microRNA/mRNA hybrids reveals regulation of the unfolded protein response by microRNA-320a. PLoS Genet (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Giovannini C, Croce CM, Bolondi L, et al. 2008. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 27: 5651–5661. 10.1038/onc.2008.178 [DOI] [PubMed] [Google Scholar]
- Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, et al. 2009. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 16: 498–509. 10.1016/j.ccr.2009.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gay LA, Sethuraman S, Thomas M, Turner PC, Renne R. 2018. Modified cross-linking, ligation, and sequencing of hybrids (qCLASH) identifies Kaposi's sarcoma-associated herpesvirus microRNA targets in endothelial cells. J Virol 92: e02138–e02117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghini F, Rubolino C, Climent M, Simeone I, Marzi MJ, Nicassio F. 2018. Endogenous transcripts control miRNA levels and activity in mammalian cells by target-directed miRNA degradation. Nat Commun 9: 3119. 10.1038/s41467-018-05182-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramantieri L, Fornari F, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Croce CM, Bolondi L, Negrini M. 2009. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin Cancer Res 15: 5073–5081. 10.1158/1078-0432.CCR-09-0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. 2008. The Vienna RNA websuite. Nucleic Acids Res 36: W70–W74. 10.1093/nar/gkn188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YE, Riley KJ, Iwasaki A, Steitz JA. 2014. Alternative capture of noncoding RNAs or protein-coding genes by herpesviruses to alter host T cell function. Mol Cell 54: 67–79. 10.1016/j.molcel.2014.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas G, Cetin S, Messmer M, Chane-Woon-Ming B, Terenzi O, Chicher J, Kuhn L, Hammann P, Pfeffer S. 2016. Identification of factors involved in target RNA-directed microRNA degradation. Nucleic Acids Res 44: 2873–2887. 10.1093/nar/gkw040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, LaVigne CA, Jones BT, Zhang H, Gillett F, Mendell JT. 2020. A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming. Science 370: eabc9546. 10.1126/science.abc9546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Grant S. 2012. Targeting the regulatory machinery of BIM for cancer therapy. Crit Rev Eukaryot Gene Expr 22: 117–129. 10.1615/CritRevEukarGeneExpr.v22.i2.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwak A, Kudla G, Dudnakova T, Tollervey D. 2013. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153: 654–665. 10.1016/j.cell.2013.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AN, Pisignano G, Pavelitz T, White J, Kinisu M, Forino N, Albin D, Varani G. 2020. An evolutionarily conserved RNA structure in the functional core of the lincRNA Cyrano. RNA 26: 1234–1246. 10.1261/rna.076117.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston ER, Bartel DP. 2019. Global analyses of the dynamics of mammalian microRNA metabolism. Genome Res 29: 1777–1790. 10.1101/gr.251421.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleaveland B, Shi CY, Stefano J, Bartel DP. 2018. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell 174: 350–362.e17. 10.1016/j.cell.2018.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneitz B, Krebs M, Kalogirou C, Schubert M, Joniau S, van Poppel H, Lerut E, Kneitz S, Scholz CJ, Ströbel P, et al. 2014. Survival in patients with high-risk prostate cancer is predicted by miR-221, which regulates proliferation, apoptosis, and invasion of prostate cancer cells by inhibiting IRF2 and SOCS3. Cancer Res 74: 2591–2603. 10.1158/0008-5472.CAN-13-1606 [DOI] [PubMed] [Google Scholar]
- Lee S, Song J, Kim S, Kim J, Hong Y, Kim Y, Kim D, Baek D, Ahn K. 2013. Selective degradation of host MicroRNAs by an intergenic HCMV noncoding RNA accelerates virus production. Cell Host Microbe 13: 678–690. 10.1016/j.chom.2013.05.007 [DOI] [PubMed] [Google Scholar]
- Libri V, Helwak A, Miesen P, Santhakumar D, Borger JG, Kudla G, Grey F, Tollervey D, Buck AH. 2012. Murine cytomegalovirus encodes a miR-27 inhibitor disguised as a target. Proc Natl Acad Sci 109: 279–284. 10.1073/pnas.1114204109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcinowski L, Tanguy M, Krmpotic A, Rädle B, Lisnić VJ, Tuddenham L, Chane-Woon-Ming B, Ruzsics Z, Erhard F, Benkartek C, et al. 2012. Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog 8: e1002510. 10.1371/journal.ppat.1002510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Merlet J, Burger J, Gomes JE, Pintard L. 2009. Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell Mol Life Sci 66: 1924–1938. 10.1007/s00018-009-8712-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BR, Wei T, Fields CJ, Sheng P, Xie M. 2018. Near-infrared fluorescent northern blot. RNA 24: 1871–1877. 10.1261/rna.068213.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Scheel TKH, Luna JM, Park CY, Fak JJ, Nishiuchi E, Rice CM, Darnell RB. 2015. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat Commun 6: 8864. 10.1038/ncomms9864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor L, Strasser A, O'Reilly LA, Hausmann G, Adams JM, Cory S, Huang DC. 1998. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J 17: 384–395. 10.1093/emboj/17.2.384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall GS, Hamilton AJ. 2008. Improved northern blot method for enhanced detection of small RNA. Nat Protoc 3: 1077–1084. 10.1038/nprot.2008.67 [DOI] [PubMed] [Google Scholar]
- Park JH, Shin SY, Shin C. 2017. Non-canonical targets destabilize microRNAs in human Argonautes. Nucleic Acids Res 45: 1569–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlica P, Moss WN, Steitz JA. 2016. Host miRNA degradation by Herpesvirus saimiri small nuclear RNA requires an unstructured interacting region. RNA 22: 1181–1189. 10.1261/rna.054817.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichholf B, Herzog VA, Fasching N, Manzenreither RA, Sowemimo I, Ameres SL. 2019. Time-resolved small RNA sequencing unravels the molecular principles of microRNA homeostasis. Mol Cell 75: 756–768.e7. 10.1016/j.molcel.2019.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu-Gruttadauria J, Pawlica P, Klum SM, Wang S, Yario TA, Schirle Oakdale NT, Steitz JA, MacRae IJ. 2019. Structural basis for target-directed microRNA degradation. Mol Cell 75: 1243–1255.e7. 10.1016/j.molcel.2019.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CY, Kingston ER, Kleaveland B, Lin DH, Stubna MW, Bartel DP. 2020. The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation. Science 370: eabc9359. 10.1126/science.abc9359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sionov RV, Vlahopoulos SA, Granot Z. 2015. Regulation of Bim in health and disease. Oncotarget 6: 23058–23134. 10.18632/oncotarget.5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stribling D, Lei Y, Guardia CM, Li L, Fields CJ, Nowialis P, Opavsky R, Renne R, Xie M. 2021. A noncanonical microRNA derived from the snaR-A noncoding RNA targets a metastasis inhibitor. RNA 27: 694–709. 10.1261/rna.078694.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis AJ, Moody J, Helwak A, Tollervey D, Kudla G. 2014. Hyb: a bioinformatics pipeline for the analysis of CLASH (crosslinking, ligation and sequencing of hybrids) data. Methods 65: 263–273. 10.1016/j.ymeth.2013.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu C, Li C, Xue J, Zhao S, Zhan P, Lin Y, Zhang P, Jiang A, Chen W. 2015. Effects of microRNA-221/222 on cell proliferation and apoptosis in prostate cancer cells. Gene 572: 252–258. 10.1016/j.gene.2015.07.017 [DOI] [PubMed] [Google Scholar]
- Wightman FF, Giono LE, Fededa JP, de la Mata M. 2018. Target RNAs strike back on MicroRNAs. Front Genet 9: 435. 10.3389/fgene.2018.00435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Ameres SL, Friedline R, Hung J-H, Zhang Y, Xie Q, Zhong L, Su Q, He R, Li M, et al. 2012. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat Methods 9: 403–409. 10.1038/nmeth.1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Shao TJ, Bofill-De Ros X, Lian C, Villanueva P, Dai L, Gu S. 2020. AGO-bound mature miRNAs are oligouridylated by TUTs and subsequently degraded by DIS3L2. Nat Commun 11: 2765. 10.1038/s41467-020-16533-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han L, Jia ZF, Yang WD, Wang GX, Jiang T, You YP, et al. 2010a. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol Cancer 9: 229. 10.1186/1476-4598-9-229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Zhang JX, Zhang AL, Wang YY, Han L, You YP, Pu PY, Kang CS. 2010b. PUMA is a novel target of miR-221/222 in human epithelial cancers. Int J Oncol 37: 1621–1626. 10.3892/ijo_00000816 [DOI] [PubMed] [Google Scholar]
- Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate illumina paired-End reAd mergeR. Bioinformatics 30: 614–620. 10.1093/bioinformatics/btt593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JJ, Chu ZB, Hu Y, Lin J, Wang Z, Jiang M, Chen M, Wang X, Kang Y, Zhou Y, et al. 2015. Targeting the miR-221-222/PUMA/BAK/BAX pathway abrogates dexamethasone resistance in multiple myeloma. Cancer Res 75: 4384–4397. 10.1158/0008-5472.CAN-15-0457 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.