In this review, Phan et al. discuss the different models that have been proposed to explain how centrosome dysfunction impairs cortical development, and review the evidence supporting a unified model in which centrosome defects reduce cell proliferation in the developing cortex by prolonging mitosis and activating a mitotic surveillance pathway. Last, they also extend their discussion to centrosome-independent microcephaly mutations, such as those involved in DNA replication and repair
Keywords: brain development, centriole, centrosome, cilia, microcephaly
Abstract
Primary microcephaly is a brain growth disorder characterized by a severe reduction of brain size and thinning of the cerebral cortex. Many primary microcephaly mutations occur in genes that encode centrosome proteins, highlighting an important role for centrosomes in cortical development. Centrosomes are microtubule organizing centers that participate in several processes, including controlling polarity, catalyzing spindle assembly in mitosis, and building primary cilia. Understanding which of these processes are altered and how these disruptions contribute to microcephaly pathogenesis is a central unresolved question. In this review, we revisit the different models that have been proposed to explain how centrosome dysfunction impairs cortical development. We review the evidence supporting a unified model in which centrosome defects reduce cell proliferation in the developing cortex by prolonging mitosis and activating a mitotic surveillance pathway. Finally, we also extend our discussion to centrosome-independent microcephaly mutations, such as those involved in DNA replication and repair.
The cerebral cortex is the primary site of neural integration in the brain and serves as the ultimate control and information-processing center of the central nervous system. Thus, generating the correct number of cortical neurons is essential for proper brain development and the performance of higher-order brain functions. Microcephaly is a brain disorder where defects in corticogenesis lead to the formation of fewer cells, resulting in a thinner cerebral cortex and reduced brain size. Curiously, many of the genes mutated in patients with primary microcephaly encode proteins functioning at the centrosome, illustrating that cortical neurogenesis is highly sensitive to disruptions in centrosome function (Faheem et al. 2015; Jayaraman et al. 2018; Naveed et al. 2018; Marthiens and Basto 2020).
Centrosomes function as microtubule organizing centers (MTOCs). In proliferating animal cells, centrosomes organize the interphase microtubule network that provides cell shape and polarity and helps traffic material within the cell (Nigg and Raff 2009; Bornens 2012; Conduit et al. 2015; Fu et al. 2015). Centrosomes also catalyze the formation of the bipolar spindle in mitosis that directs the movement of chromosomes into the two daughter cells. In early G1 of cycling cells and in most quiescent cells, the centrosome migrates to the cell surface to initiate the formation of a primary cilium that serves as a signaling antenna to sense extracellular cues (Fırat-Karalar and Stearns 2014; Sánchez and Dynlacht 2016). Mutations in centrosome proteins can lead to disruption in any of these cellular processes, but it remains to be determined which of these pathways are central to microcephaly pathogenesis.
In this review, we provide an overview of cerebral cortical development and the centrosome genes that are mutated in microcephaly. We then discuss which cellular processes are impacted by centrosome defects and highlight which microcephaly models show evidence of delayed spindle assembly and prolonged mitosis. We follow this with a summary of recent advances in our understanding of how cells sense and respond to time in mitosis. We also review the evidence supporting the hypothesis that prolonged mitosis is the central cause of impaired cortical development in microcephaly caused by mutations in centrosome and spindle genes. Finally, we extend our discussion to additional microcephaly mutations that are not thought to impact centrosome function, such as those involved in DNA replication and repair.
Primary microcephaly
Microcephaly, derived from the Greek word for “small head,” is a clinical term used to describe a cranium that is significantly smaller than the average head size of the population (three or more standard deviations below the mean). Primary microcephaly (also known as congenital microcephaly) refers to cases in which problems in prenatal developmental result in a reduced head circumference and small brain at birth (Woods 2004; Woods et al. 2005). This is distinct from secondary microcephaly, which is associated with progressive neurodegeneration and has postnatal onset.
Primary microcephaly can have both genetic and nongenetic origins. Some examples of environmental factors that can affect prenatal brain development and manifest in reduced brain growth include Zika viral infection, congenital infection with toxoplasma, and alcohol overconsumption during pregnancy (Popova et al. 2016; Devakumar et al. 2018; Antoniou et al. 2020). To distinguish primary microcephaly caused by genetic defects from environmentally induced congenital microcephaly, a subclass was established called microcephaly primary hereditary (MCPH), also known as autosomal recessive microcephaly (Woods et al. 2005). The most common clinical feature of MCPH is a reduction in size of the cerebral cortex without significant defects in cortical layering architecture, although simplified cortical folding patterns are observed in some patients. Depending on the severity of cerebral growth impairment, MCPH patients can display mild to severe mental retardation. In some cases, symptoms also include frequent spasticity or seizure.
In addition to MCPH, an additional group of disorders referred to as microcephalic primordial dwarfism (MPD) has been described in which autosomal recessive microcephaly is accompanied by pre- and postnatal body growth retardation. Syndromes classified under MPD include Seckel syndrome, Meier-Gorlin syndrome, and microcephalic osteodysplastic primordial dwarfism (MOPD) (Khetarpal et al. 2016). Strikingly, many MCPH- and MPD-linked microcephaly genes encode centrosome proteins (Table 1). Moreover, mutations in some centrosome genes can cause either MCPH or MPD, further strengthening the connection between these disorders and arguing for a common cellular origin for the associated brain development defects. Given the phenotypic and mechanistic overlap, we discuss how mutations in MCPH- or MPD-linked microcephaly genes (referred to here as “microcephaly genes”) lead to defects in the brain, and more specifically in cerebral cortical growth.
Table 1.
MCPH- and MPD-linked genes
Cortical development and microcephaly pathogenesis
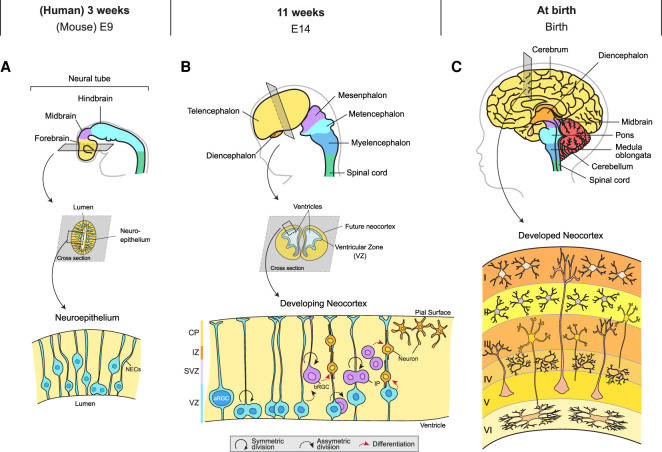
Cerebral cortical development starts with a rudimental tubule with a single layer of highly proliferative neuroepithelial cells known as the neural tube (Fig. 1A; Sauer 1935). During the early stages of neurogenesis, neuroepithelial cells undergo rapid divisions to expand the stem cell pool and thicken the wall of the neural tube (Fujita 1964; Rakic 1995). At the midstage of neurogenesis, the neural progenitor cells (NPCs) within the neural tube can be subdivided into apical and basal progenitors (Florio and Huttner 2014). Apical progenitor cells lie adjacent to the ventricle surface in a region known as the ventricular zone (VZ). The subventricular zone (SVZ) lies adjacent to the VZ and contains basal progenitor cells (Fig. 1B).
Figure 1.
Cortical development at different stages in humans and the equivalent time line in mice. (A) As the neural tube develops, it enlarges into three vesicles corresponding to the forebrain, midbrain, and hindbrain regions of the adult brain. At this stage, the neural tube wall has a single layer of neuroepithelial cells (NECs). These cells migrate to the lumen of the neural tube to divide, a phenomenon also known as interkinetic nuclear migration. (B) At the midstage of neurogenesis, apical radial glial cells (aRGCs) that derived from NECs can undergo either symmetric division to expand the progenitor pool size or asymmetric division, giving rise to basal RGCs (bRGCs) or intermediate progenitors (IPs). Note that bRGCs are almost absent in rodents. bRGCs, IPs, and aRGCs will eventually generate postmitotic neurons that migrate toward the pial surface. (VZ) Ventricular zone, (SVZ) subventricular zone, (IZ) intermediate zone, (CP) cortical plate. (C) The fully formed adult cerebral cortex consists of six layers of neurons.
The VZ contains several types of progenitors, including apical radial glial cells (aRGCs), apical intermediate progenitors, and subapical progenitors (Schultze and Korr 1981; Malatesta et al. 2003; Götz and Huttner 2005; Gal et al. 2006; Pilz et al. 2013; Tyler and Haydar 2013). Among these, aRGC is the predominant class of progenitor within the VZ (Noctor et al. 2002). These cells are named after their distinct morphology, where long radial processes connect the cell body to the outer and inner border of the neural tube wall. During each cell cycle, the nuclei of aRGCs translocate toward the lumen of the neural tube, a phenomenon known as interkinetic nuclear migration, where mitosis takes place (Sauer and Walker 1959; Taverna and Huttner 2010). Once created, the two daughter nuclei migrate away from the ventricular surface. aRGCs initially undergo several rounds of symmetric, or proliferative, divisions to generate two identical daughters, further expanding the number of progenitors within the developing cortex. As neurogenesis progresses, aRGCs undergo asymmetric divisions, giving rise to one stem cell and one fate-restricted daughter cell such as a basal progenitor or postmitotic neuron (Götz and Huttner 2005; Taverna et al. 2014). Cell divisions that give rise to neurons are considered neurogenic or nonproliferative divisions.
In rodents, basal progenitors mainly consist of intermediate progenitors (IPs) (Miyata et al. 2004; Noctor et al. 2004; Kowalczyk et al. 2009; Franco and Müller 2013). IPs can undergo either one to two additional round of division to generate more IPs or commit to terminal division, generating two daughter neurons (Haubensak et al. 2004; Wu et al. 2005). In primates such as humans, basal progenitors are comprised of IPs and a second cell type termed the basal RGCs (bRGCs, also known as outer RGCs) (Fietz et al. 2010; Hansen et al. 2010; Reillo et al. 2011). bRGCs arise from divisions of aRGCs and retain their basal processes that connect them to the pial surface. bRGCs are highly neurogenic and are implicated in the development of cortical folds or gyri (Penisson et al. 2019). The low abundance of bRGCs is thought to contribute to the lack of cortical folds in rodents, resulting in lissencephalic (smooth) brains (Betizeau et al. 2013).
Once a cell has committed to a neuronal fate, it migrates radially outward along the basal processes that attach RGCs to the outer pial wall (Rakic 1971). Neurons that are generated earlier in development are destined for the deeper layers of the cortex, while later-forming neurons migrate past pre-established sections to successively add the superficial layers (McConnell and Kaznowski 1991). At the time of birth, the newborn neocortex has a six-layer neuronal structure (Fig. 1C). At this stage, most of the NPC population has been exhausted, and all the neurons that contribute to cortical architecture and function have been produced. It is worth mentioning that toward the end of neurogenesis, RGCs also participate in generating cells of the glial lineage such as astrocytes and microglial, a process termed gliogenesis (Qian et al. 2000). Glial cells are essential for proper neuronal function, as they are involved in both synaptic formation and synaptic pruning to ensure proper neuronal connections are generated (Jäkel and Dimou 2017).
The formation of a smaller brain and a thinner cerebral cortex in microcephaly patients originates from defects that arise during NPC development. Specifically, the pool of NPCs is significantly depleted through either premature commitment to neurogenic division or death of progenitors and their subsequent progeny. This eventually leads to the generation of fewer cells in the cerebral cortex. Thus, understanding what triggers NPCs to undergo these fate changes is fundamental to deciphering how cortical development is impaired in microcephaly.
The molecular genetics of primary microcephaly
Studies in the past two decades have identified many genetic mutations that are responsible for causing MCPH and MPD. These discoveries have revealed a strikingly consistent theme: Cortical development is highly sensitive to the disruption of mitotic structures such as the centrosome or the mitotic spindle (Jayaraman et al. 2018; Naveed et al. 2018; Degrassi et al. 2020; Marthiens and Basto 2020). Indeed, some of the earliest MCPH genes identified included WDR62, CDK5RAP2, ASPM, CPAP (CENPJ/SAS4), STIL, CEP135, and CEP152, all of which encode proteins that localize and function at the centrosome (Roberts et al. 1999; Moynihan et al. 2000; Pattison et al. 2000; Leal et al. 2003; Kumar et al. 2009; Guernsey et al. 2010; Hussain et al. 2012). In this section, we highlight the microcephaly mutations that fall into two broad functional categories: mutations in genes required for centrosome function and those that operate within the mitotic spindle. We also discuss how microcephaly mutations in these genes compromise protein function, and how this in turn influences centrosome integrity, mitotic progression, and the outcome of cell division.
Centrioles, centrosomes, and cilia
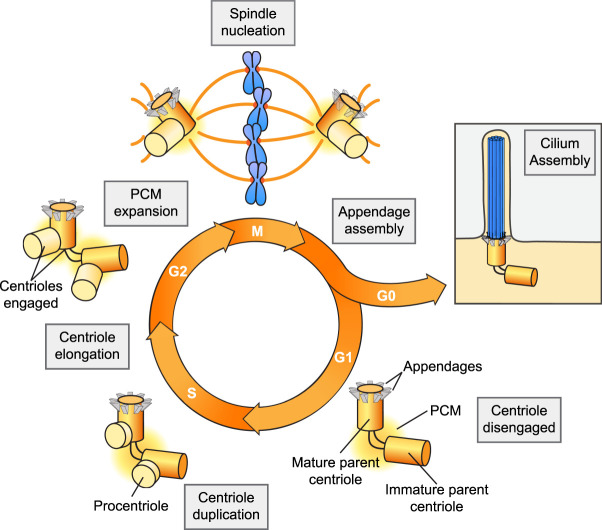
A centrosome is comprised of two main components: centrioles and the surrounding pericentriolar material (PCM) (Fig. 2). Centrioles are microtubule-based structures with an evolutionarily conserved ninefold rotational symmetry that form the core of centrosomes and recruit the proteins of the surrounding PCM. The PCM is a layered assembly of proteins that recruits factors responsible for nucleating and anchoring microtubules (Woodruff et al. 2014). Centrioles, on the other hand, are the replicating unit of the centrosome, and their duplication controls centrosome copy number.
Figure 2.
Centrosome biogenesis cycle. G1 cells contain a single centrosome comprising a pair of centrioles surrounded by the pericentriolar material (PCM). The mature parent centriole is decorated with distal and subdistal appendages, while the immature centriole lacks these structures. In S phase, a single procentriole forms on both parent centrioles. Throughout S and G2 phases, the procentrioles elongate, and in late G2, the two centrosomes separate and undergo PCM expansion. During mitosis, the two centrosomes act to catalyze the assembly of the bipolar microtubule spindle apparatus on which chromosomes are segregated. The mature parent centriole can also dock at the cell membrane and initiate the assembly of a primary cilium in early G1 in the case of dividing cells, or during G0 phase in quiescent cells.
Centrioles have dual lives: In addition to forming the core of centrosomes, centrioles are also required for the formation of cilia and flagella (Breslow and Holland 2019). Cilia are hair-like projections that protrude from the cell surface and are classified into either motile cilia that drive fluid movement or nonmotile primary cilia that function in signaling. In quiescent cells, centrioles migrate to the plasma membrane where the parent centriole docks to act as a basal body that directs the assembly of a cilium. Primary cilia are found in most cell types and act to sense extracellular cues and regulate intracellular signaling pathways, including Hedgehog and Wnt signaling. In cycling cells, cilia are formed in early G1 and disassembled prior to mitosis so that the centrioles can be released to form centrosomes that participate in organizing the mitotic spindle.
Mutations in centriole genes are frequent in MCPH and MPD patients. However, because centrioles play important roles in the formation of both centrosomes and cilia, it can be challenging to define to what extent microcephaly is caused by centrosome or cilium dysfunction. Primary cilia are present in most cells of the brain and play important roles in early cortical patterning, expansion of NPCs, and the specification of adult neural stem cells (Hasenpusch-Theil and Theil 2021). Consequently, patients with primary cilium dysfunction, or ciliopathies, often display a broad collection of brain phenotypes that includes microcephaly (Andreu-Cervera et al. 2021). However, while microcephaly can occur in isolation in MCPH patients, microcephaly that is linked to ciliopathies occurs in the context of syndromes with wide-ranging phenotypes that manifest across multiple organ systems. Furthermore, mutations in genes that are only required to build cilia and not centrosomes have never been identified in MCPH patients, suggesting that cilium defects are unlikely to be the only feature driving the pathology in these patients.
Mutations in genes required for centriole assembly
Centriole copy number is tightly regulated in proliferating cells. At the beginning of the cell cycle, each cell has a single centrosome containing a pair of centrioles (Fig. 2). In G1, the two parent centrioles differ in age and structure, with the older parent centriole possessing distal and subdistal appendages at the distal end. The distal appendages are required for the docking of the centriole to the plasma membrane and building a cilium, while the subdistal appendages are involved in microtubule anchoring. In cycling cells, the two parent centrioles duplicate once in S phase to produce two procentrioles. Work in multiple model systems has revealed that centriole assembly is controlled by a conserved subset of proteins that includes PLK4, SASS6, STIL, CPAP (CENPJ), CEP135, CEP152, and RTTN (Nigg and Holland 2018). Centriole duplication begins in S phase, when PLK4, the master regulator of centriole biogenesis, is recruited to the wall of the two parent centrioles by its receptor proteins, CEP152 and CEP192 (Cizmecioglu et al. 2010; Hatch et al. 2010; Kim et al. 2013; Sonnen et al. 2013; Park et al. 2014). Through mechanisms that are still not understood, PLK4 coalesces into a single site on each parent centriole to mark the site for future procentriole assembly. Local PLK4 kinase activity helps recruit STIL, followed by additional proteins required to assemble the procentriole such as SASS6, CEP135, CPAP, and RTTN (Dzhindzhev et al. 2014; Ohta et al. 2014; Arquint et al. 2015; Kratz et al. 2015; Moyer et al. 2015; Sharma et al. 2016). This process ensures that by the time the cell reaches mitosis, there are two centrosomes, each of which contains a pair of centrioles. At the end of mitosis, the two centrosomes are divided equally so that each daughter cell inherits a single copy. Thus, a single cycle of duplication followed by the equal partitioning of the centrioles during cell division ensures that centriole and centrosome copy number is strictly maintained in dividing cells.
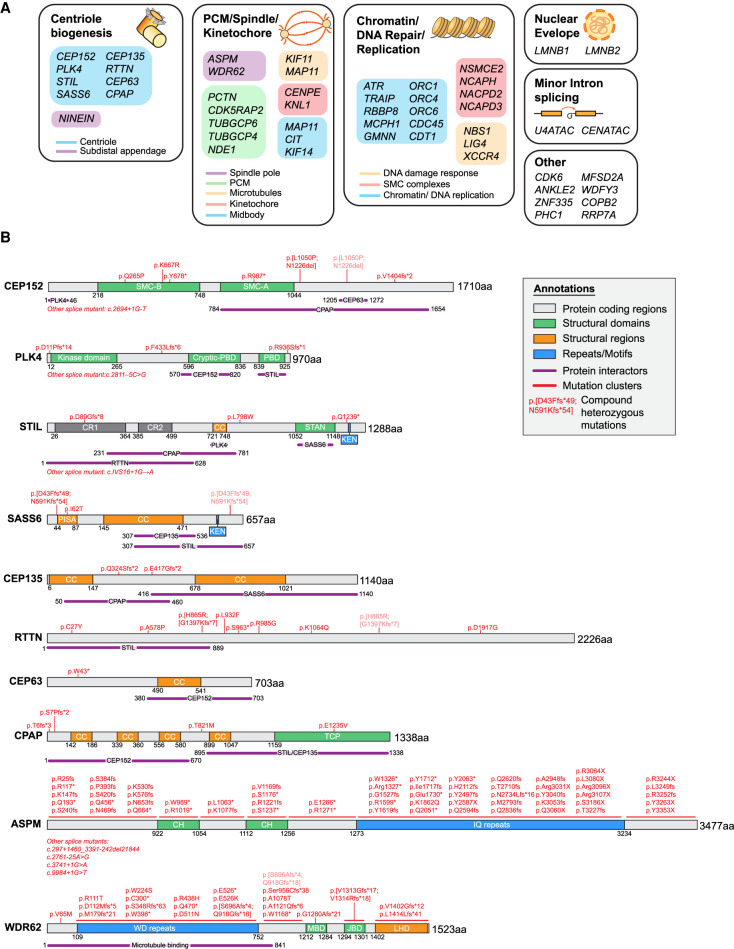
Strikingly, many of the central regulators of centriole assembly have been found to be mutated in microcephaly patients (Fig. 3A). In all cases where it has been tested, complete loss of these genes leads to embryonic or perinatal lethality in mice, and consequently patient mutations in the core centriole assembly factors are likely to be hypomorphic (Izraeli et al. 1999; Hudson et al. 2001; Smart 2002; Ko et al. 2005; Basto et al. 2006; Pfaff et al. 2007; Kia et al. 2012; Bazzi and Anderson 2014; González-Martínez et al. 2021). Many microcephaly mutants of centriole assembly genes are nonsense or frameshift mutations that introduce a premature termination codon and produce truncated proteins that lack key functional domains (Fig. 3B). In some cases, microcephaly mutations reduce protein abundance or generate point mutations within interaction domains required for centriole biogenesis (Bond et al. 2005; Papari et al. 2013; Chen et al. 2017; Khan et al. 2017). For instance, an MCPH-linked E1235V mutation in CPAP significantly reduces its binding to STIL and consequently disrupts procentriole assembly (Tang et al. 2011; Cottee et al. 2013; Hatzopoulos et al. 2013; Zheng et al. 2014). Microcephaly mutations can also alter RNA splicing, through either deletion of known splice sites or the creation of new splice acceptors (Farooq et al. 2016; Dinçer et al. 2017). Thus, disruption of centriole assembly and a subsequent reduction in centriole number appear to be common consequences of microcephaly mutations.
Figure 3.
The molecular genetics of primary microcephaly. (A) Genes implicated in MCPH and MPD classified by functional group and subcellular localization. (B) Schematic of representative centrosome and spindle proteins mutated in microcephaly. Protein domains and regions of interactions are depicted based on studies by Gillingham and Munro (2000), Kohlmaier (2009), Carvalho-Santos et al. (2010), Hatch et al. (2010), Holland et al. (2010), Van Breugel et al. (2011), Issa et al. (2013), Kim et al. (2013), Lin et al. (2013), Sonnen et al. (2013), Arquint et al. (2015), Mori et al. (2015), Chen et al. (2017), and Patwardhan et al. (2018). (SMC-A/B) Structural maintenance of chromosomes (SMC)-like domain A/B, (PBD) polo-box domain, (CR1/2) conserved region 1/2, (CC) coiled-coil region, (STAN) Stil/Ana2 domain, (PISA) present in SAS-6, (TCP) T complex protein 10 domain, (CH) calponin homology domain, (MBD) MKK7β1 binding domain, (JBD) JNK binding domain, (LHD) loop helix domain.
Nevertheless, microcephaly mutations in centriolar genes do not always lead to centriole loss. Two different nonsense mutations in STIL have been found to increase protein stability and cause centriole amplification (Arquint and Nigg 2014). Furthermore, driving centriole amplification in the brains of mice can also lead to NPC depletion and microcephaly (Marthiens et al. 2013). In addition, some microcephaly mutations that have been experientially tested do not have a clear impact on centriole number, although alterations in centriole structure were not examined (Chen et al. 2017). Importantly, there are few examples where centriole number has been examined in cells derived from patients with microcephaly-linked mutations. This underscores the importance of functionally characterizing more microcephaly mutations to obtain insight on how they cause disease in human patients.
Microcephaly mutations in PCM and spindle proteins
In addition to mutations in genes directly involved in centriole assembly, a second class of MCPH mutations is found in genes involved in mitotic spindle organization and microtubule dynamics. This includes genes encoding PCM proteins, proteins that regulate mitotic spindle assembly or microtubule dynamics, and those that localize to kinetochores. Despite having diverse functional roles, all these proteins contribute to timely and faithful progression through mitosis.
Following centriole duplication in S phase, centrosomes undergo PCM expansion in G2 in preparation for mitotic spindle formation (Fig. 2). This is marked by both the increased recruitment of the PCM scaffolding proteins PCNT, CDK5RAP2, and CEP192 and the increased docking of microtubule-nucleating γ-Tubulin complexes (Woodruff et al. 2014). In mitosis, the two centrosomes catalyze the formation of a bipolar spindle on which chromosomes are segregated. Mutations in PCM scaffolding proteins have been shown to cause microcephaly. For example, mutations in CDK5RAP2 are linked to MCPH and Seckel syndrome, while MOPD type II is caused by mutations in PCNT (Fig. 3B; Bond et al. 2005; Hassan et al. 2007; Rauch et al. 2008; Pagnamenta et al. 2012; Pachajoa et al. 2014; Li et al. 2015; Yigit et al. 2015; Dehghan Tezerjani et al. 2020). Intriguingly, the depletion of the PCM scaffolding proteins in cultured cells showed that CEP192, but not PCNT and CDK5RAP2, is essential for bipolar spindle assembly (Watanabe et al. 2020; Chinen et al. 2021), providing an explanation for why no microcephaly mutations in CEP192 have been identified to date. In addition to alterations in PCNT and CDK5RAP2, mutations in components of the γ-Tubulin complex have also been identified in MPD patients (Puffenberger et al. 2012; Scheidecker et al. 2015; Martin et al. 2016). Meanwhile, mutations in NINEIN, a protein that recruits the γ-Tubulin complex to the subdistal appendages of the mature parent centriole, are linked to Seckel syndrome (Dauber et al. 2012). Although the effect of these microcephaly mutations on PCM function have yet to be evaluated, it is plausible that they function to reduce the microtubule-nucleating activity of centrosomes while still allowing spindle assembly and chromosome segregation to take place.
Some proteins mutated in MCPH and Seckel patients, such as WDR62 and ASPM, localize to the spindle poles but are not part of the PCM. ASPMor WDR62knockout animals are viable (Pulvers et al. 2010; Jayaraman et al. 2016; Johnson et al. 2018), which may explain why mutations in these two genes are prevalent in human microcephaly patients (Bond et al. 2002; Kumar et al. 2004; Shen et al. 2005; Saadi et al. 2009; Bilgüvar et al. 2010; Nicholas et al. 2010; Yu et al. 2010; Bhat et al. 2011; Murdock et al. 2011). Indeed, many ASPMor WDR62MCPH mutations are frameshifting or nonsense mutations distributed across the entire length of both genes (Fig. 3B). Considering that both genes are nonessential, it is plausible that at least some of these mutations create true null alleles. In cells, loss of ASPM or WDR62 leads to multiple mitotic abnormalities, including defective spindle orientation (Fish et al. 2006; Higgins et al. 2010; Bogoyevitch et al. 2012; Gai et al. 2016; Miyamoto et al. 2017), delayed spindle assembly and mitotic progression (Bogoyevitch et al. 2012; Chen et al. 2014b), and an increased frequency of lagging chromosomes (Guerreiro et al. 2021).
Other MCPH and MPD genes with clear mitotic functions include genes encoding the kinetochore proteins KNL1 (or CASC5) and CENPE (Jamieson et al. 1999; Genin et al. 2012; Mirzaa et al. 2014b; Saadi et al. 2016; Zarate et al. 2016), the microtubule motor proteins KIF11 and KIF14 (Ostergaard et al. 2012; Jones et al. 2014; Robitaille et al. 2014; Li et al. 2016b; Moawia et al. 2017; Makrythanasis et al. 2018), citron kinase (CIT), and the microtubule-associated protein MAP11 (Di Cunto et al. 2000; Basit et al. 2016; Gai et al. 2016; Harding et al. 2016; Li et al. 2016a; Shaheen et al. 2016; Perez et al. 2019). Kinetochore proteins such as KNL1 and CENPE function to ensure the correct attachment of chromosomes to microtubules of the mitotic spindle, and knockout of these genes increases the frequency of chromosome missegregation and aneuploidy. Widespread aneuploidy is not tolerated in vivo, explaining why knockout of KNL1 or CENPE is embryonic lethal in mice (Weaver et al. 2003; Shi et al. 2019). Studies of several microcephaly mutations in KNL1 have shown significantly reduced protein levels (Saadi et al. 2016; Zarate et al. 2016), while similar analyses suggest that some microcephaly mutations in CENPE have impaired kinetochore localization (Mirzaa et al. 2014b). KIF11 (or EG5) is an essential kinesin required for the formation of the mitotic spindle (Blangy et al. 1995). Inhibition of KIF11 prevents bipolar spindle assembly and arrests cells in prometaphase, while mutations in its motor domain slow down progression through mitosis (Kwok et al. 2004; Skoufias et al. 2006). MAP11, KIF14, and CIT have all been described to function at the midbody where they play important roles in cytokinesis (Gruneberg et al. 2006; Basit et al. 2016; Moawia et al. 2017; Perez et al. 2019). Consistently, microcephaly-linked mutations in KIF14 and CIT are reported to increase the frequency of cytokinesis failure and the generation of polyploid progeny (Bianchi et al. 2017; Moawia et al. 2017). Polyploidy can be tolerated in certain tissues such as the liver but has yet to be reported in patients or animal models of microcephaly (Sladky et al. 2021).
As the list of causative mutations in MCPH and MPD patients continues to grow, more effort will be required to characterize these mutations and how they impact protein abundance and function. Much of the current research has modeled microcephaly mutations with the knockout of gene function in cell culture systems. The ability to generate precise edits at the endogenous locus now opens the exciting possibility of modeling microcephaly mutations in vivo to investigate their impact in the context of cortical development. Together with the emergence of new model systems such as ferrets and brain organoids that more closely mimic human brain development, we can look forward to a better understanding of how mutations in primary microcephaly genes link to disease phenotypes.
Centrosome and spindle dysfunction in microcephaly
The abundance of microcephaly mutations in centrosome genes strongly suggests that brain development is especially susceptible to disruptions in this organelle. In the following section, we explore the ways in which centrosomes contribute to normal cortical development and how mutations in centrosome genes can perturb these processes (Jayaraman et al. 2018; Naveed et al. 2018; O'Neill et al. 2018; Marthiens and Basto 2020; Yang et al. 2021). We also outline the various models for how defects in centrosome proteins lead to premature differentiation or cell death in NPCs. Considering that centrosomes are present in many cell types, we discuss how alterations in ubiquitously expressed centrosome proteins can, in some cases, cause selective defects in brain development.
Spindle orientation, centrosome positioning, and asymmetric centrosome inheritance
In the developing mammalian cortex, aRGCs initially divide symmetrically to produce identical daughter cells that can continue to proliferate through many rounds of division. As cortical development progress, these cells start switching to asymmetric divisions, with only one of the two daughter cells maintaining the original aRGC identity (Götz and Huttner 2005; Taverna et al. 2014). The timing of this commitment to asymmetric division has a direct impact on how long RCGs continue to self-renew, and subsequently how many neurons are generated.
Previous studies of cortical development have proposed that cells that divide with their spindle oriented parallel to the ventricular surface undergo symmetric division, while cells with spindles that deviate from this angle divide asymmetrically (Chenn and McConnell 1995; Kosodo et al. 2004; Sanada and Tsai 2005). Since centrosome positioning dictates the orientation of the mitotic spindle, it seems reasonable to assume that mutations in centrosome genes could deplete NPCs by disrupting spindle alignment and affecting the onset and/or frequency of asymmetric divisions. Nevertheless, this model has been challenged by several studies in mice where disruption of the epithelial architecture changes spindle orientation but does not lead to microcephaly (Imai et al. 2006; Rašin et al. 2007). Moreover, knockout of LGN, a protein that links astral microtubules to the cell cortex, randomizes spindle orientation in aRGCs without affecting the rate of neuron production (Konno et al. 2008). Finally, several mouse models that lack microcephaly genes exhibit microcephaly without showing changes in spindle orientation or the ratio of symmetric to asymmetric cell divisions (Table 2). This argues that spindle misorientation is unlikely to be a common cause of neuronal reduction and cortical thinning in microcephaly.
Table 2.
In vivo models of microcephaly caused by defects in centrosome or spindle proteins
In addition to the cleavage plane orientation, spindle size asymmetry has also been proposed to influence the outcome of NPC divisions. Drosophila neuroblasts generate an asymmetric mitotic spindle that leads to the production of a highly proliferative neuroblast and a smaller transit-amplifying ganglion mother cell that is committed to generating neurons or glial cells (Kaltschmidt et al. 1999). This led to the hypothesis that spindle asymmetry in neuroblasts helps determine the asymmetric fate choice of the two daughter cells. A similar model has been proposed for NPC division in mice, though the asymmetry in spindle size in NPCs is much less pronounced (Delaunay et al. 2014). Thus, whether asymmetric spindle size strongly correlates with cell fate in mouse cortical development remains unclear.
An alternative proposal for how centrioles can influence the outcome of aRGC division is through the asymmetric inheritance of the two parent centrioles. In G1-phase cells, only the older mature parent centriole is decorated with distal appendage proteins required for cilium assembly. Each aRGC possesses a single primary cilium that connects its apical process to the ventricular surface. When aRGCs undergo cell division, their primary cilia disassemble, and the parent centriole undocks from the membrane through endocytosis and remains associated with ciliary membrane remnants (Paridaen et al. 2013). The centriole-associated ciliary membrane retains receptors for Sonic Hedgehog signaling that persist through mitosis and are inherited by the daughter cell that acquires the older mature parent centriole. In the following cell cycle, the ciliary remnants are thought to allow the rapid reassembly of a primary cilium. During the peak of neurogenesis, the older parent centriole is preferentially inherited by the aRGC, while the younger parent centriole is selectively passed on to the differentiating progeny (Wang et al. 2009). This led to the proposal that preferential inheritance of the older centriole promotes the early reassembly of a primary cilium that is used to maintain aRGC identity (Paridaen et al. 2013). Despite being an attractive model for asymmetric cell fate determination, this model does not readily explain how aRGCs undergo symmetric divisions to generate daughter cells with the same fate, since centriole inheritance is always asymmetric. This suggests that additional mechanisms must exist to regulate the fate of aRGC progeny.
A recent study reported that mice lacking CEP83, a distal appendage protein required for docking of the centriole at the plasma membrane, overproduce NPCs, leading to megacephaly or brain overgrowth (Shao et al. 2020). In this case, the loss of centrosome anchorage at the ventricle surface increases the stiffness of the apical membrane, which in turn activates mechanically sensitive HIPPO signaling to induce excessive NPC proliferation. This suggests that centrosomes can regulate the mechanical properties of NPCs to influence the size of the developing cortex.
Cell cycle regulation
Is has long been appreciated that the cell cycle length of NPCs increases with the progression of cortical development, and this coincides with an increase in neurogenic divisions (Schultze and Korr 1981; Dehay et al. 1993; Takahashi et al. 1995; Kornack and Rakic 1998). In mice, this increase in total cell cycle length is predominantly due to the lengthening of G1 phase. This has led to the “cell cycle length hypothesis” (Calegari et al. 2005; Lange et al. 2009; Pilaz et al. 2009), which postulates that differences in G1 duration determine whether NPCs continue to proliferate or commit to differentiation. In support of this theory, extending time in G1 through down-regulation of CDK activity was sufficient to induce neuronal differentiation in whole-embryo mouse cultures (Calegari and Huttner 2003).
This raises the question of whether G1 lengthening could be responsible for the onset of precocious neurogenic divisions in microcephaly patients. Studies in patient-derived fibroblasts carrying a CPAP truncating mutation revealed defects in cilium disassembly in G1/G0, which in turn delayed cell cycle progression (Gabriel et al. 2016). Brain organoids generated from these fibroblasts showed evidence of premature neuronal differentiation, supporting a model in which the lengthening of G1 through delayed cilium resorption could contribute to reduced neuronal production. Similar defects in cilium disassembly and cell cycle re-entry have also been reported from studies in mice lacking WDR62 (Sgourdou et al. 2017). An additional component of the cilium disassembly complex NDE1 has also been implicated in microcephaly accompanied by lissencephaly but whether microcephaly-linked mutations in NDE1 alter cell cycle length is unclear (Alkuraya et al. 2011).
It is important to note that a correlation between changes in the cell cycle and neuronal fate is not just limited to variation in the length of G1. Studies in the developing mouse cortex have revealed that S phase becomes shorter as progenitor cells commit to neurogenic divisions (Arai et al. 2011). In addition, studies using ferret models have revealed that shortening of S phase, rather than an extension of G1, is the main source of cell cycle variation during neocortical development (Turrero García et al. 2016), while similar work performed in macaques show a combination of shorter S phase and longer G1 (Betizeau et al. 2013). This indicates that there may be differences in how cell cycle regulation dictates cell fate in lissencephalic mouse brains versus gyrencephalic animals such as ferrets and primates. Furthermore, premature neuronal differentiation because of cell cycle lengthening cannot account for the cell death commonly observed in microcephaly models (Table 2), suggesting that additional mechanisms are likely involved.
Spindle assembly and delay in mitosis
Centrosomes serve as a major site of microtubule nucleation in most dividing cells and were originally thought to be critical for spindle assembly. However, it is now clear that although centrosomes are a dominant source of microtubule nucleation in mitosis, mitotic spindle assembly and cell division can still occur in the absence of centrosomes. In other words, cells have additional microtubule nucleation pathways that can also contribute to mitotic spindle formation (Petry 2016). One centrosome-independent source of microtubule nucleation emanates from chromosomes and relies on the generation of Ran-GTP from chromatin that leads to the release of spindle assembly factors in the vicinity of chromosomes (McKim and Hawley 1995; Heald et al. 1996). New microtubules can also be generated from existing microtubules within the mitotic spindle. This amplification pathway depends on the Augmin complex, which binds to the microtubule wall and nucleates a new microtubule parallel to the template fiber (Goshima et al. 2008; Kamasaki et al. 2013). How the centrosome, chromatin, and Augmin-mediated microtubule nucleation pathways are coordinated to generate a functional spindle is an area of active research.
It is worth asking: If cells can use several pathways to build a mitotic spindle, what advantage does the centrosome provide? One advantage of centrosome-mediated spindle assembly is that centrosomes increase the fidelity of chromosome transmission during cell division by ensuring the formation of exactly two spindle poles. In addition, centrosomes promote efficient microtubule nucleation through PCM activity and thereby act to speed up progression through mitosis. As a result, cells lacking centrosomes have delayed spindle assembly and take, on average, two to three times longer to complete mitosis (Fong et al. 2016; Lambrus et al. 2016; Meitinger et al. 2016). In NPCs, spindle assembly occurs rapidly and mitosis generally completes in ≤30 min (Pilaz et al. 2016; Phan et al. 2021). Therefore, mutations in centrosome proteins or other factors that participate in mitotic spindle assembly are expected to delay NPC progression through cell division and possibly increase the frequency of cell division errors. In the following section, we outline the evidence that suggests a mitotic delay may be a common mechanism by which microcephaly mutations lead to the depletion of NPCs in the developing cerebral cortex.
The mitotic surveillance pathway
Mitosis is a highly dynamic phase of the cell cycle when cells are susceptible to genomic alterations through the breakage or missegregation of chromosomes (Levine and Holland 2018). Cells reduce the risk of these defects by carefully monitoring DNA integrity and kinetochore–microtubule attachments with quality control checkpoints (Barnum and O'Connell 2014). A well-established example of this is the spindle assembly checkpoint (SAC), which functions to delay the onset of anaphase until all chromosomes are correctly attached to the mitotic spindle. Recently, an additional mitotic fail-safe was identified, known as the mitotic surveillance pathway (MSP), which prevents the growth of cells that had undergone a prolonged mitosis (Uetake and Sluder 2010; Fong et al. 2016; Lambrus et al. 2016; Meitinger et al. 2016). Specifically, nontransformed cells in culture that arise from a delayed mitosis (∼120 min or longer instead of the average ∼30 min) undergo a G1 arrest even if cell division errors are avoided. This suggests that there exists an optimal window of time for the completion of mitosis: At their quickest, cells need to be in mitosis long enough to satisfy the SAC and accurately segregate their chromosomes; however, there is an upper threshold for how long cells can stay in mitosis before the MSP is activated, and further proliferation is suppressed.
A central question is why a pathway would evolve to curb the growth of cells that experience prolonged mitosis, even in the absence of cell division errors. An attractive possibility is that cells use mitotic length as a surrogate for cell health and/or mitotic fidelity (Lambrus and Holland 2017). Although mitotic errors that produce complex karyotypes with altered chromosomal structures trigger a P53-dependent cell cycle arrest, modest levels of whole-chromosome segregation errors often fail to activate this response (Santaguida et al. 2017; Soto et al. 2017). Thus, the MSP may serve as an additional fail-safe to prevent the growth of cells with an increased risk of accumulating mitotic errors. This fits with the observation that the frequency of cell division errors typically increases with mitotic duration.
Components of the mitotic surveillance pathway
Through several genome-wide screens, three central players in the MSP have been identified: 53BP1, USP28, and P53. 53BP1 directly interacts with both USP28 and P53 through distinct interfaces in 53BP1's C-terminal BRCT repeats. Knockout of 53BP1 or USP28 prevents the stabilization of P53 in cells that experience prolonged mitosis, indicating that 53BP1 and USP28 lie upstream of P53 in this signaling axis (Fong et al. 2016; Lambrus et al. 2016; Meitinger et al. 2016). While it remains unclear how cells monitor mitotic length and activate the MSP, a mitotic timekeeper that relies on post-translational modifications of 53BP1 and/or USP28 remains an attractive hypothesis (Gliech and Holland 2020).
Although 53BP1 and P53 play established roles in DNA damage signaling, several lines of evidence from work in cultured cells argue that their role in the MSP is independent of DNA damage signaling. First, while cells that undergo extreme increases in mitotic length (≥6 h) accumulate extensive DNA damage as a result of telomere deprotection (Hayashi et al. 2012), short mitotic delays (<1.5 h) do not induce detectable levels of DNA breaks, despite triggering robust activation of the MSP (Uetake and Sluder 2010). Second, the loss of other components involved in DNA damage signaling does not prevent the cell cycle arrest following a mitotic delay, arguing that inactivation of the MSP and ablation of DNA damage signaling are genetically separable (Lambrus et al. 2016). Third, preventing the recruitment of 53BP1 to sites of DNA damage does not affect MSP signaling (Fong et al. 2016; Lambrus et al. 2016). Finally, the BRCT domain of 53BP1 is dispensable for DNA double-strand break repair (Ward et al. 2006), but is required for MSP function (Fong et al. 2016). Taken together, the available evidence argues that DNA damage is not the trigger for the activation of the MSP in cells that delay in mitosis.
The function of the mitotic surveillance pathway in vivo
An interesting question is under what circumstances cells in vivo would experience mitotic delays that then activate the MSP. Mitosis normally occurs rapidly, and perturbation of any process that alters the levels or activity of mitotic proteins will often result in a mitotic delay. Exposure to environmental toxins or spontaneous genetic alterations are two possible sources of MSP activation. Moreover, heritable mutations in proteins that are required for efficient mitotic progression can also slow down mitosis. A good example of this is primary microcephaly, where mutations in genes that encode proteins functioning at the centrosome or the spindle apparatus result in an increased mitotic length in NPCs (Chavali et al. 2014). NPCs harboring these mutations often complete mitosis normally, but their progeny differentiate prematurely or undergo apoptosis (Phan et al. 2021).
In mice, 53BP1 and USP28 are expressed in the early stages of embryogenesis (E5.5), but functional signaling through the MSP is thought to be established around gastrulation (E7.5) (Xiao et al. 2021). The timing of MSP activation coincides with the centrosome taking over as a dominant source of microtubule-nucleating activity, suggesting that MSP signaling is closely linked to centrosomal MTOC function. 53BP1 plays an important role outside of the MSP in DNA damage signaling, and mice lacking 53BP1 are radiation-sensitive, exhibit immune deficiencies, and are cancer-prone. In contrast, USP28 knockout mice do not display an obvious phenotype (Diefenbacher et al. 2014), though there is accumulating evidence to suggest that USP28 could act as a tumor suppressor in humans (Richter et al. 2018). Importantly, in the context of brain development, knockout of USP28 or 53BP1 does not lead to an overproduction of neurons, arguing that the MSP does not globally suppress NPC proliferation.
Role of the mitotic surveillance pathway in microcephaly
Prolonged mitosis has been observed across many different animal models of microcephaly, including most models where genes encoding centriole assembly proteins, PCM proteins, or spindle pole proteins are knocked out (Table 2). In addition, several studies using brain organoids generated from patient-derived cells as models for microcephaly have also reported evidence of extended mitotic duration (Bershteyn et al. 2017; Zhang et al. 2019a). A delay in mitosis has often been considered a secondary consequence of microcephaly mutations. However, pioneering work from the Silver group (Pilaz et al. 2016; Mitchell-Dick et al. 2020; Sheehan et al. 2020) revealed that delaying NPC progression through mitosis, either genetically or pharmacologically, is sufficient to induce premature neurogenic divisions, while exceedingly long mitoses result in the production of apoptotic progeny. These data suggest that an increased mitotic duration could be a cause of the NPC depletion observed in microcephaly.
The demonstration that the fate of NPCs is tightly correlated with the duration of mitosis raised the question of whether a causal link exists between activation of the MSP and the altered neurogenesis observed in microcephaly. Microcephaly mutations in mitotic genes are frequently observed to trigger P53 activation in NPCs, and knockout of P53 is sufficient to suppress cell death and restore cortical size in mouse models (Insolera et al. 2014; Marjanović et al. 2015). However, P53 activation is a common outcome of signaling pathways that respond to genomic instability, and thus the nature of the upstream trigger for P53 remained unclear. Recently, it was shown that loss of USP28 or 53BP1, two upstream components of MSP, can rescue NPC proliferation in microcephaly models with centrosome defects (Phan et al. 2021). Furthermore, knockout of USP28 prevented P53 up-regulation, consistent with the notion that P53 activation in these microcephaly models occurs through MSP signaling. Deletion of the MSP genes restored NPC proliferation and cortical size without correcting the upstream centrosome defects or the extended mitosis. This suggests that mutations in centrosome genes, and possibly other genes that function in mitosis, trigger pathological activation of the MSP in NPCs, leading to microcephaly.
One may expect that inactivation of the MSP will rescue NPC loss and restore cortical size in all microcephaly models that exhibit a mitotic delay. However, while the loss of some mitotic regulators leads to catastrophic cell division errors, the removal of centrosomes does not dramatically impact the fidelity of chromosome segregation in NPCs (Phan et al. 2021). As a result, inactivation of the MSP in NPCs with centrosome defects enables the continued proliferation of the euploid daughter cells generated following a prolonged mitosis. In contrast, ablating the MSP in NPCs that exhibit an extended mitosis accompanied by significant mitotic errors suppresses cell death but results in the accumulation of aneuploid daughter cells. Cells with aneuploidy exhibit defects in cell cycle progression, proteotoxic stress, and genome instability (Chunduri and Storchová 2019) and are often outcompeted or eliminated in vivo (Bolton et al. 2016; Pfau et al. 2016; Singla et al. 2020). This likely explains why the loss of P53 in microcephaly models with significant aneuploidy fails to fully restore brain growth (Marthiens et al. 2013; Shi et al. 2019; Viais et al. 2021).
An interesting contrast between NPCs and cultured RPE1 cells is their different responses to MSP activation. In RPE1 cells, daughter cells that have activated the MSP arrest in G1 and undergo senescence in a P21-dependent manner (Lambrus et al. 2016). Transcriptomic analysis of NPCs following an acute mitotic delay suggests that in addition to P21, there is up-regulation of other proapoptotic P53 target genes (Mitchell-Dick et al. 2020). The same study also reported an up-regulation of transcripts associated with neuronal differentiation, but this occurs prior to an increase in P53 expression. Future work analyzing changes to the proteome and/or post-translational modifications in NPCs that undergo mitotic delays may provide additional insight into the molecular players linking time in mitosis to MSP activation.
Why is the brain preferentially impacted by mutations in centrosome genes?
In contrast to MCPH, where a reduction in brain size usually occurs without a substantial alteration in stature, in Seckel syndrome, microcephaly occurs in combination with severe body growth retardation. Several of the genes linked to Seckel syndrome function in centriole duplication such as CPAP, CEP152, and CEP63 (Al-Dosari et al. 2010; Kalay et al. 2011; Sir et al. 2011). In addition, some centriole proteins are classified as both MCPH and Seckel syndrome genes (Table 1), further arguing that these disorders exist within a phenotypic spectrum where microcephaly can occur with varying degrees of body size reduction. This raises the question of why mutations in centrosome proteins have a variable impact on prenatal growth. A simple explanation is that the proliferation of NPCs in the cortex is more sensitive to defects in centrosome function than dividing cells in other tissues. As a result, weaker hypomorphic mutations in centrosome genes would selectively affect brain size, while more complete loss-of-function alleles would impact the growth of the brain and other tissues.
Mice lacking CEP63 offer a useful model to study Seckel syndrome since they exhibit both microcephaly and primordial dwarfism. Importantly, while inactivation of the MSP rescued cortical growth in CEP63-deficient mice, loss of this pathway failed to restore body size (Marjanović et al. 2015; Phan et al. 2021). This indicates that the microcephaly and dwarfism phenotypes are caused by activation of different cellular pathways: Microcephaly can be caused by activation MSP, while dwarfism may be a result of delayed progression through the cell cycle, leading to a general defect in cell proliferation. This reinforces the view that the brain may be uniquely sensitive to centrosome defects because only a modest mitotic delay is required to activate the MSP. Indeed, work in mouse NPCs has shown that an average increase of ∼30 min in mitotic duration can promote a dramatic shift in the fate of the progeny, leading to cell death or premature differentiation (Pilaz et al. 2016).
An alternative, nonmutually exclusive hypothesis is that the variability in response to a mitotic delay arises from differences in tissue-specific sensitivity to P53 activation. A study using a series of mouse models to generate different magnitudes of P53 activation during development found that neuronal lineages are highly sensitive to P53-driven apoptosis (Bowen et al. 2019). Therefore, the unique susceptibility of the brain to centrosome abnormalities may arise from NPCs’ hypersensitivity to even modest P53 activation. Finally, to generate the ∼16 billion neurons of the human neocortex, NPCs must undergo massive proliferation in a restricted period during embryonic development (Azevedo et al. 2009). With few cell divisions occurring later in life, there is little opportunity to rescue cortical size if too few cells are created embryonically. Thus, the narrow developmental window for neurogenesis offers an additional explanation for the disproportionate effect of mutations that reduce cell proliferation on cortex size.
Microcephaly caused by mutations in genes that do not function at the centrosome or mitotic spindle
Besides mutations in genes that function at the centrosome or mitotic spindle, there are several other broad pathways into which MCPH and MPD genes can be organized (Fig. 3A). Below we discuss these pathways and highlight what is currently known about how defects in these processes contribute to microcephaly.
Replication defects
Along with mutations in centrosome proteins, Seckel syndrome can also be caused by mutations in DNA damage signaling proteins, including ATR and two proteins required for its activation, TRAIP and RBBP8 (O'Driscoll et al. 2003; Qvist et al. 2011; Ogi et al. 2012). ATR is an essential gene that promotes progression through S phase by facilitating recovery from replication fork stalling. Mutations in DONSON, which functions to stabilize stalled replication forks and activate ATR-dependent signaling in response to replication stress, also cause MPD (Evrony et al. 2017; Reynolds et al. 2017). This suggests that chronic replication stress is a driver of MPD. A pathogenic role of DNA replication defects in MPD is further implicated in Meier-Gorlin syndrome, a subclass of MPD caused by mutations in the prereplication complex proteins ORC1, ORC4, ORC6, CDC45, CDT1, and GMNN (Bicknell et al. 2011a,b; Guernsey et al. 2011; Burrage et al. 2015; Fenwick et al. 2016). Taken together, the available data suggest that mutations that reduce the efficiency of DNA replication in MPD patients cause a general impairment of cell proliferation that leads to reduced cell number and global growth failure.
Mutations in SMC complex protein
The structural maintenance of chromosomes (SMC) protein complexes play critical roles in maintaining chromosome structure and have been strongly linked to microcephaly pathogenesis. There are three SMC complexes: cohesin, condensin, and SMC5/6. Cohesin is critical for sister chromatid cohesion, while condensin functions to promote chromosome compaction. The third SMC complex, SMC5/6, is less well understood but has been proposed to function in DNA repair and promoting fork restart after replication stress. Mutations in the SMC5/6 complex subunit NSMCE2 cause MPD in humans (Payne et al. 2014). Patient cells with NSMCE2 mutations showed increased chromatin bridges, elevated micronucleation, and delayed recovery following replication stress. Mutations in genes that encode subunits of the condensin complex were recently shown to cause MCPH (Martin et al. 2016). Condensins have essential roles in cell division, and thus the condensin mutations observed in MCPH patients are all functionally hypomorphic. Patient-derived primary fibroblasts carrying MCPH-linked condensin mutations showed defective decatenation of the replicated sister chromatids and an increase in lagging anaphase chromosomes and micronucleation. Moreover, a hypomorphic condensin II mutant mouse exhibited reduced brain size and an increased frequency of chromatin bridges in NPCs. Intriguingly, MCPH1 was the first characterized MCPH gene and encodes a protein that binds and inhibits condensin II (Trimborn et al. 2006; Yamashita et al. 2011). Premature chromosome condensation is one of the phenotypes observed in cells carrying an MCPH1 mutation (Pfau et al. 2013), suggesting that defects in this process due to misregulated condensin activity may also contribute to the reduced brain size in these patients. Notably, knockout of MCPH1 leads to an increased mitotic index in murine NPC (Gruber et al. 2011). In the future it would be interesting to determine the effect of condensin mutations on the duration of mitosis in NPCs.
DNA damage signaling
NPCs are especially sensitive to DNA damage, perhaps because of their low threshold for initiating apoptosis in response to P53 activation (McKinnon 2013). Mutations in several DNA damage repair genes produce syndromes that feature developmental microcephaly, often in conjunction with increased cancer predisposition and immunodeficiency (Carney et al. 1998; Matsuura et al. 1998; Varon et al. 1998; Seemanová and Jarolím 1999; O'Driscoll et al. 2001, 2003; Ben-Omran et al. 2005; Buck et al. 2006a,b; Matsumoto et al. 2011). Work in mouse models has shown that DNA lesions activate P53 through the ATM–CHK2 and/or ATR–CHK1 pathway to induce cell death in the developing cortex (Frappart et al. 2005; Foster et al. 2012; Li et al. 2012). This suggests that increased rates of DNA damage or defective DNA repair can contribute to NPC attrition in the developing cortex. In addition to controlling chromatin compaction through regulation of condensin activity, MCPH1 has also been implicated in DNA damage signaling and DNA repair (Jackson et al. 2002; Lin et al. 2010). However, the relative contribution of the DNA damage response and other functions of MCPH1 to the development of microcephaly remains to be established.
Defects in the minor spliceosome
MOPD type I is caused by mutations in the U4atac snRNA component of the minor spliceosome that is responsible for recognizing and excising a small subset of introns (He et al. 2011; Farach et al. 2018; Hallermayr et al. 2018; Wang et al. 2018). Additionally, mutations in the minor spliceosome subunit CENATATC/CCDC84 have also been identified in patients with mosaic variegated aneuploidy, a syndrome characterized by microcephaly, high rates of aneuploidy, and childhood cancers (Wolf et al. 2021). In these cases, deficiencies in the splicing of minor introns likely results in defective RNA processing of multiple mitotic regulators, leading to an increased frequency of chromosome segregation errors. Interestingly, brain-specific knockout of the minor spliceosome component Rnu11 in mice leads to misregulated splicing of several cell cycle genes, an extended mitosis, and accompanying microcephaly in mice (Baumgartner et al. 2018). This illustrates a crucial link between minor intron splicing, mitotic progression, chromosome segregation fidelity, and brain development.
Nuclear envelope defects
A final pathway implicated in MCPH pathogenesis are defects in the integrity of the nuclear lamina. Mutations in the nuclear lamina proteins LMNB1 and LMNB2 cause a form of autosomal dominant primary microcephaly (Cristofoli et al. 2020; Parry et al. 2021). Knockout of either LMNB1 or LMNB2 in the forebrains of mice leads to reduced brain growth and accompanying defects in neuronal migration and cortical layering (Coffinier et al. 2010; Kim et al. 2011). Defects in B-type lamins impair nuclear envelope integrity and may interfere with the reassembly of the nuclear envelope following mitosis. B-type lamins are also associated with the mitotic spindle, raising the possibility that mutations in these genes could lead to spindle defects that contribute to the microcephaly phenotype (Tsai et al. 2006).
Additional MCPH genes that have been identified include CDK6, ANKLE2, ZNF335, PHC1, MFSD2A, WDFY3 (ALFY), COPB2, and RRP7A (Table 1). As yet, there are no common pathways that functionally link these genes, and it remains unclear how mutations in these genes cause microcephaly.
Perspective
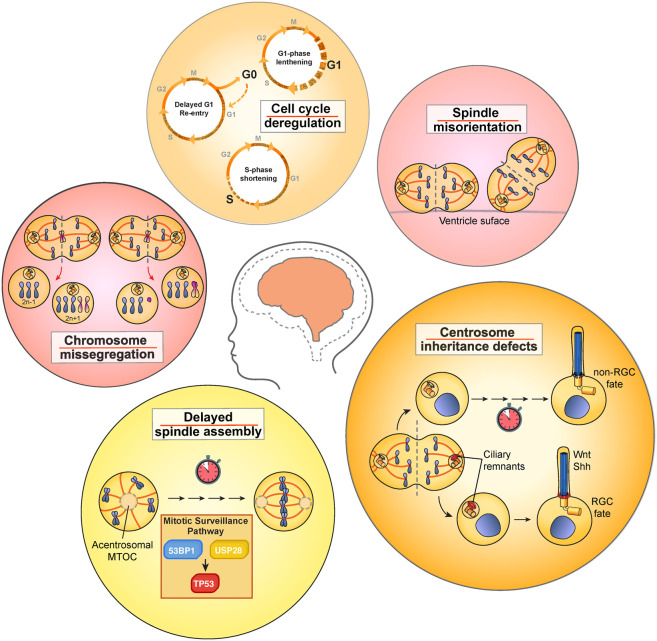
In the past two decades, our understanding of primary microcephaly has greatly expanded thanks to the identification of many new causative mutations. The striking frequency of microcephaly mutations in genes that encode centrosome proteins has led to several proposals for how centrosome dysfunction can impair brain development. These models include mitotic spindle misorientation, deregulation of centrosome inheritance, cell cycle delays, and defects in mitotic progression and fidelity (Fig. 4). While these models are in keeping with the known roles of centrosomes in cortical development, recent studies have indicated that some of these defects are correlated with, but may not be causative of, microcephaly.
Figure 4.
Cellular processes disrupted in neural progenitor cells by microcephaly mutations in centrosomal proteins. Mutations in genes that encode centrosome proteins have been proposed to cause microcephaly through several mechanisms. This includes deregulating of cell cycle progression, altering spindle orientation, increasing the frequency of chromosome missegregation, disrupting asymmetric centrosome inheritance, and delaying mitotic spindle assembly.
In this review, we highlight evidence in support of an emerging model for how mutations in centrosome and mitotic spindle-associated proteins can lead to microcephaly (Table 2). This model postulates that mutations in these proteins slow down progression through mitosis and trigger pathological activation of the MSP to induce cell death and differentiation of NPC progeny. So far, this model has only been tested in two mouse microcephaly models, and whether this holds true for other models of microcephaly remains to be seen. In addition, many primary microcephaly mutations do not lie in genes encoding proteins that function at the centrosome or mitotic spindle. Therefore, activation of the MSP is only one of several possible pathways that can result in reduced cortical size. In fact, it was shown that microcephaly caused by loss of SMC5 in the brains of mice is independent of the MSP and thus cannot be rescued by knocking out the MSP signaling components 53BP1 and USP28 (Atkins et al. 2020; Phan et al. 2021). However, this same microcephaly model can be rescued by knocking out CHK2 or P53, indicating that DNA damage signaling is predominantly responsible for the impaired cortical growth in these animals (Atkins et al. 2020). Identifying which microcephaly mutations activate the MSP and which trigger other pathogenic pathways is an important area of future research. These studies will not only aid our understanding of primary microcephaly and its etiology but also cast light on fundamental mechanisms that are important for normal brain development.
Acknowledgments
We thank members of our laboratory for the helpful discussions and input on this review and apologize to colleagues whose work could not be cited due to space limitations. Work in the Holland laboratory was supported by the National Institutes of Health grants R01GM114119 and R01GM133897, and American Cancer Society Mission Boost grant MBG-19-173-01-MBG.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.348866.121.
Competing interest statement
The authors declare no competing interests.
References
- Abdel-Hamid MS, Ismail MF, Darwish HA, Effat LK, Zaki MS, Abdel-Salam GMH. 2016. Molecular and phenotypic spectrum of ASPM-related primary microcephaly: identification of eight novel mutations. Am J Med Genet Part A 170: 2133–2140. 10.1002/ajmg.a.37724 [DOI] [PubMed] [Google Scholar]
- Abdel-Salam GMH, Abdel-Hamid MS, Issa M, Magdy A, El-Kotoury A, Amr K. 2012. Expanding the phenotypic and mutational spectrum in microcephalic osteodysplastic primordial dwarfism type I. Am J Med Genet Part A 158A: 1455–1461. 10.1002/ajmg.a.35356 [DOI] [PubMed] [Google Scholar]
- Alakbarzade V, Hameed A, Quek DQY, Chioza BA, Baple EL, Cazenave-Gassiot A, Nguyen LN, Wenk MR, Ahmad AQ, Sreekantan-Nair A, et al. 2015. A partially inactivating mutation in the sodium-dependent lysophosphatidylcholine transporter MFSD2A causes a non-lethal microcephaly syndrome. Nat Genet 47: 814–817. 10.1038/ng.3313 [DOI] [PubMed] [Google Scholar]
- Al-Dosari MS, Shaheen R, Colak D, Alkuraya FS. 2010. Novel CENPJ mutation causes Seckel syndrome. J Med Genet 47: 411–414. 10.1136/jmg.2009.076646 [DOI] [PubMed] [Google Scholar]
- Alkuraya FS, Cai X, Emery C, Mochida GH, Al-Dosari MS, Felie JM, Hill RS, Barry BJ, Partlow JN, Gascon GG, et al. 2011. Human mutations in NDE1 cause extreme microcephaly with lissencephaly. Am J Hum Genet 88: 536–547. 10.1016/j.ajhg.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu-Cervera A, Catala M, Schneider-Maunoury S. 2021. Cilia, ciliopathies and hedgehog-related forebrain developmental disorders. Neurobiol Dis 150: 105236. 10.1016/j.nbd.2020.105236 [DOI] [PubMed] [Google Scholar]
- Antoniou E, Orovou E, Sarella A, Iliadou M, Rigas N, Palaska E, Iatrakis G, Dagla M. 2020. Zika virus and the risk of developing microcephaly in infants: a systematic review. Int J Environ Res Public Health 17: 3806. 10.3390/ijerph17113806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai Y, Pulvers JN, Haffner C, Schilling B, Nüsslein I, Calegari F, Huttner WB. 2011. Neural stem and progenitor cells shorten S-phase on commitment to neuron production. Nat Commun 2: 154. 10.1038/ncomms1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arquint C, Nigg EA. 2014. STIL microcephaly mutations interfere with APC/C-mediated degradation and cause centriole amplification. Curr Biol 24: 351–360. 10.1016/j.cub.2013.12.016 [DOI] [PubMed] [Google Scholar]
- Arquint C, Gabryjonczyk AM, Imseng S, Böhm R, Sauer E, Hiller S, Nigg EA, Maier T. 2015. STIL binding to Polo-box 3 of PLK4 regulates centriole duplication. Elife 4: e07888. 10.7554/eLife.07888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins A, Xu MJ, Li M, Rogers NP, Pryzhkova MV, Jordan PW. 2020. Smc5/6 is required for replication fork stability and faithful chromosome segregation during neurogenesis. Elife 9: e61171. 10.7554/eLife.61171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad S, Al-Dosari MS, Al-Yacoub N, Colak D, Salih MA, Alkuraya FS, Poizat C. 2013. Mutation in PHC1 implicates chromatin remodeling in primary microcephaly pathogenesis. Hum Mol Genet 22: 2200–2213. 10.1093/hmg/ddt072 [DOI] [PubMed] [Google Scholar]
- Azevedo FAC, Carvalho LRB, Grinberg LT, Farfel JM, Ferretti REL, Leite REP, Filho WJ, Lent R, Herculano-Houzel S. 2009. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J Comp Neurol 513: 532–541. 10.1002/cne.21974 [DOI] [PubMed] [Google Scholar]
- Barnum KJ, O'Connell MJ. 2014. Cell cycle regulation by checkpoints. Methods Mol Biol 1170: 29–40. 10.1007/978-1-4939-0888-2_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera JA, Kao LR, Hammer RE, Seemann J, Fuchs JL, Megraw TL. 2010. CDK5RAP2 regulates centriole engagement and cohesion in mice. Dev Cell 18: 913–926. 10.1016/j.devcel.2010.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basit S, Al-Harbi KM, Alhijji SAM, Albalawi AM, Alharby E, Eldardear A, Samman MI. 2016. CIT, a gene involved in neurogenic cytokinesis, is mutated in human primary microcephaly. Hum Genet 135: 1199–1207. 10.1007/s00439-016-1724-0 [DOI] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. 2006. Flies without Centrioles. Cell 125: 1375–1386. 10.1016/j.cell.2006.05.025 [DOI] [PubMed] [Google Scholar]
- Baumgartner M, Olthof AM, Aquino GS, Hyatt KC, Lemoine C, Drake K, Sturrock N, Nguyen N, al Seesi S, Kanadia RN. 2018. Minor spliceosome inactivation causes microcephaly owing to cell cycle defects and death of self-amplifying radial glial cells. Development 145: dev166322. 10.1242/dev.166322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi H, Anderson KV. 2014. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proc Natl Acad Sci 111: E1491–E1500. 10.1073/pnas.1400568111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Omran TI, Cerosaletti K, Concannon P, Weitzman S, Nezarati MM. 2005. A patient with mutations in DNA ligase IV: clinical features and overlap with Nijmegen breakage syndrome. Am J Med Genet 137A: 283–287. 10.1002/ajmg.a.30869 [DOI] [PubMed] [Google Scholar]
- Bershteyn M, Nowakowski TJ, Pollen AA, Di Lullo E, Nene A, Wynshaw-Boris A, Kriegstein AR. 2017. Human iPSC-derived cerebral organoids model cellular features of lissencephaly and reveal prolonged mitosis of outer radial glia. Cell Stem Cell 20: 435–449.e4. 10.1016/j.stem.2016.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betizeau M, Cortay V, Patti D, Pfister S, Gautier E, Bellemin-Ménard A, Afanassieff M, Huissoud C, Douglas RJ, Kennedy H, et al. 2013. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron 80: 442–457. 10.1016/j.neuron.2013.09.032 [DOI] [PubMed] [Google Scholar]
- Bhat V, Girimaji S, Mohan G, Arvinda H, Singhmar P, Duvvari M, Kumar A. 2011. Mutations in WDR62, encoding a centrosomal and nuclear protein, in Indian primary microcephaly families with cortical malformations. Clin Genet 80: 532–540. 10.1111/j.1399-0004.2011.01686.x [DOI] [PubMed] [Google Scholar]
- Bianchi FT, Tocco C, Pallavicini G, Liu Y, Vernì F, Merigliano C, Bonaccorsi S, El-Assawy N, Priano L, Gai M, et al. 2017. Citron kinase deficiency leads to chromosomal instability and TP53-sensitive microcephaly. Cell Rep 18: 1674–1686. 10.1016/j.celrep.2017.01.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell LS, Bongers EMHF, Leitch A, Brown S, Schoots J, Harley ME, Aftimos S, Al-Aama JY, Bober M, Brown PAJ, et al. 2011a. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat Genet 43: 356–359. 10.1038/ng.775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell LS, Walker S, Klingseisen A, Stiff T, Leitch A, Kerzendorfer C, Martin CA, Yeyati P, Al Sanna N, Bober M, et al. 2011b. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat Genet 43: 350–355. 10.1038/ng.776 [DOI] [PubMed] [Google Scholar]
- Bilgüvar K, Öztürk AK, Louvi A, Kwan KY, Choi M, Tatlı B, Yalnizoǧlu D, Tüysüz B, Çaǧlayan AO, Gökben S, et al. 2010. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature 467: 207–210. 10.1038/nature09327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A, Lane HA, d'Hérin P, Harper M, Kress M, Nigg EA. 1995. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83: 1159–1169. 10.1016/0092-8674(95)90142-6 [DOI] [PubMed] [Google Scholar]
- Bogoyevitch MA, Yeap YYC, Qu Z, Ngoei KR, Yip YY, Zhao TT, Heng JI, Ng DCH. 2012. WD40-repeat protein 62 is a JNK-phosphorylated spindle pole protein required for spindle maintenance and timely mitotic progression. J Cell Sci 125: 5096–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton H, Graham SJL, Van Der Aa N, Kumar P, Theunis K, Fernandez Gallardo E, Voet T, Zernicka-Goetz M. 2016. Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential. Nat Commun 7: 11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Roberts E, Mochida GH, Hampshire DJ, Scott S, Askham JM, Springell K, Mahadevan M, Crow YJ, Markham AF, et al. 2002. ASPM is a major determinant of cerebral cortical size. Nat Genet 32: 316–320. 10.1038/ng995 [DOI] [PubMed] [Google Scholar]
- Bond J, Scott S, Hampshire DJ, Springell K, Corry P, Abramowicz MJ, Mochida GH, Hennekam RCM, Maher ER, Fryns JP, et al. 2003. Protein-truncating mutations in ASPM cause variable reduction in brain size. Am J Hum Genet 73: 1170–1177. 10.1086/379085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Roberts E, Springell K, Lizarraga S, Scott S, Higgins J, Hampshire DJ, Morrison EE, Leal GF, Silva EO, et al. 2005. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet 37: 353–355. 10.1038/ng1539 [DOI] [PubMed] [Google Scholar]
- Bornens M. 2012. The centrosome in cells and organisms. Science 335: 422–426. 10.1126/science.1209037 [DOI] [PubMed] [Google Scholar]
- Bowen ME, McClendon J, Long HK, Sorayya A, Van Nostrand JL, Wysocka J, Attardi LD. 2019. The spatiotemporal pattern and intensity of p53 activation dictates phenotypic diversity in p53-driven developmental syndromes. Dev Cell 50: 212–228.e6. 10.1016/j.devcel.2019.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Holland AJ. 2019. Mechanism and regulation of centriole and cilium biogenesis. Annu Rev Biochem 88: 691–724. 10.1146/annurev-biochem-013118-111153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck D, Malivert L, De Chasseval R, Barraud A, Fondanèche MC, Sanal O, Plebani A, Stéphan JL, Hufnagel M, Le Deist F, et al. 2006a. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124: 287–299. 10.1016/j.cell.2005.12.030 [DOI] [PubMed] [Google Scholar]
- Buck D, Moshous D, de Chasseval R, Ma Y, le Deist F, Cavazzana-Calvo M, Fischer A, Casanova JL, Lieber MR, de Villartay JP. 2006b. Severe combined immunodeficiency and microcephaly in siblings with hypomorphic mutations in DNA ligase IV. Eur J Immunol 36: 224–235. 10.1002/eji.200535401 [DOI] [PubMed] [Google Scholar]
- Burrage LC, Charng WL, Eldomery MK, Willer JR, Davis EE, Lugtenberg D, Zhu W, Leduc MS, Akdemir ZC, Azamian M, et al. 2015. De novo GMNN mutations cause autosomal-dominant primordial dwarfism associated with Meier-Gorlin syndrome. Am J Hum Genet 97: 904–913. 10.1016/j.ajhg.2015.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabernard C, Doe CQ. 2009. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in drosophila. Dev Cell 17: 134–141. 10.1016/j.devcel.2009.06.009 [DOI] [PubMed] [Google Scholar]
- Calegari F, Huttner WB. 2003. An inhibition of cyclin-dependent kinases that lengthens, but does not arrest, neuroepithelial cell cycle induces premature neurogenesis. J Cell Sci 116: 4947–4955. 10.1242/jcs.00825 [DOI] [PubMed] [Google Scholar]
- Calegari F, Haubensak W, Haffner C, Huttner WB. 2005. Selective lengthening of the cell cycle in the neurogenic subpopulation of neural progenitor cells during mouse brain development. J Neurosci 25: 6533–6538. 10.1523/JNEUROSCI.0778-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, Hays L, Morgan WF, Petrini JHJ. 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93: 477–486. 10.1016/S0092-8674(00)81175-7 [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Machado P, Branco P, Tavares-Cadete F, Rodrigues-Martins A, Pereira-Leal JB, Bettencourt-Dias M. 2010. Stepwise evolution of the centriole-assembly pathway. J Cell Sci 123: 1414–1426. 10.1242/jcs.064931 [DOI] [PubMed] [Google Scholar]
- Chavali PL, Pütz M, Gergely F. 2014. Small organelle, big responsibility: the role of centrosomes in development and disease. Philos Trans R Soc B Biol Sci 369: 20130468. 10.1098/rstb.2013.0468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CT, Hehnly H, Yu Q, Farkas D, Zheng G, Redick SD, Hung HF, Samtani R, Jurczyk A, Akbarian S, et al. 2014a. A unique set of centrosome proteins requires pericentrin for spindle–pole localization and spindle orientation. Curr Biol 24: 2327–2334. 10.1016/j.cub.2014.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Zhang Y, Wilde J, Hansen KC, Lai F, Niswander L. 2014b. Microcephaly disease gene Wdr62 regulates mitotic progression of embryonic neural stem cells and brain size. Nat Commun 5: 3885. 10.1038/ncomms4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H-Y, Wu C-T, Tang C-JC, Lin Y-N, Wang W-J, Tang TK. 2017. Human microcephaly protein RTTN interacts with STIL and is required to build full-length centrioles. Nat Commun 8: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A, McConnell SK. 1995. Cleavage orientation and the asymmetric inheritance of Notchl immunoreactivity in mammalian neurogenesis. Cell 82: 631–641. 10.1016/0092-8674(95)90035-7 [DOI] [PubMed] [Google Scholar]
- Chinen T, Yamazaki K, Hashimoto K, Fujii K, Watanabe K, Takeda Y, Yamamoto S, Nozaki Y, Tsuchiya Y, Takao D, et al. 2021. Centriole and PCM cooperatively recruit CEP192 to spindle poles to promote bipolar spindle assembly. J Cell Biol 220: e202006085. 10.1083/jcb.202006085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chunduri NK, Storchová Z. 2019. The diverse consequences of aneuploidy. Nat Cell Biol 21: 54–62. [DOI] [PubMed] [Google Scholar]
- Cizmecioglu O, Arnold M, Bahtz R, Settele F, Ehret L, Haselmann-Weiß U, Antony C, Hoffmann I. 2010. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J Cell Biol 191: 731–739. 10.1083/jcb.201007107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffinier C, Chang SY, Nobumori C, Tu Y, Farber EA, Toth JI, Fong LG, Young SG. 2010. Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc Natl Acad Sci 107: 5076–5081. 10.1073/pnas.0908790107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit PT, Wainman A, Raff JW. 2015. Centrosome function and assembly in animal cells. Nat Rev Mol Cell Biol 16: 611–624. 10.1038/nrm4062 [DOI] [PubMed] [Google Scholar]
- Cottee MA, Muschalik N, Wong YL, Johnson CM, Johnson S, Andreeva A, Oegema K, Lea SM, Raff JW, van Breugel M. 2013. Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly. Elife 2: e01071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofoli F, Moss T, Moore HW, Devriendt K, Flanagan-Steet H, May M, Jones J, Roelens F, Fons C, Fernandez A, et al. 2020. De novo variants in LMNB1 cause pronounced syndromic microcephaly and disruption of nuclear envelope integrity. Am J Hum Genet 107: 753–762. 10.1016/j.ajhg.2020.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvish H, Esmaeeli-Nieh S, Monajemi GB, Mohseni M, Ghasemi-Firouzabadi S, Abedini SS, Bahman I, Jamali P, Azimi S, Mojahedi F, et al. 2010. A clinical and molecular genetic study of 112 Iranian families with primary microcephaly. J Med Genet 47: 823–828. 10.1136/jmg.2009.076398 [DOI] [PubMed] [Google Scholar]
- Dauber A, LaFranchi SH, Maliga Z, Lui JC, Moon JE, McDeed C, Henke K, Zonana J, Kingman GA, Pers TH, et al. 2012. Novel microcephalic primordial dwarfism disorder associated with variants in the centrosomal protein ninein. J Clin Endocrinol Metab 97: E2140–E2151. 10.1210/jc.2012-2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrassi F, Damizia M, Lavia P. 2020. The mitotic apparatus and kinetochores in microcephaly and neurodevelopmental diseases. Cells 9: 49. 10.3390/cells9010049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay C, Giroud P, Berland M, Smart I, Kennedy H. 1993. Modulation of the cell cycle contributes to the parcellation of the primate visual cortex. Nature 366: 464–466. 10.1038/366464a0 [DOI] [PubMed] [Google Scholar]
- Dehghan Tezerjani M, Vahidi Mehrjardi MY, Hozhabri H, Rahmanian M. 2020. A novel PCNT frame shift variant (c.7511delA) causing osteodysplastic primordial dwarfism of majewski type 2 (MOPD II). Front Pediatr 8: 340. 10.3389/fped.2020.00340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay D, Cortay V, Patti D, Knoblauch K, Dehay C. 2014. Mitotic spindle asymmetry: a Wnt/PCP-regulated mechanism generating asymmetrical division in cortical precursors. Cell Rep 6: 400–414. 10.1016/j.celrep.2013.12.026 [DOI] [PubMed] [Google Scholar]
- De Munnik SA, Bicknell LS, Aftimos S, Al-Aama JY, Van Bever Y, Bober MB, Clayton-Smith J, Edrees AY, Feingold M, Fryer A, et al. 2012. Meier-Gorlin syndrome genotype-phenotype studies: 35 individuals with pre-replication complex gene mutations and 10 without molecular diagnosis. Eur J Hum Genet 20: 598–606. 10.1038/ejhg.2011.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desir J, Cassart M, David P, Van Bogaert P, Abramowicz M. 2008. Primary microcephaly with ASPM mutation shows simplified cortical gyration with antero–posterior gradient pre- and post-natally. Am J Med Genet Part A 146A: 1439–1443. 10.1002/ajmg.a.32312 [DOI] [PubMed] [Google Scholar]
- Devakumar D, Bamford A, Ferreira MU, Broad J, Rosch RE, Groce N, Breuer J, Cardoso MA, Copp AJ, Alexandre P, et al. 2018. Infectious causes of microcephaly: epidemiology, pathogenesis, diagnosis, and management. Lancet Infect Dis 18: e1–e13. 10.1016/S1473-3099(17)30398-5 [DOI] [PubMed] [Google Scholar]
- Di Cunto F, Imarisio S, Hirsch E, Broccoli V, Bulfone A, Migheli A, Atzori C, Turco E, Triolo R, Dotto GP, et al. 2000. Defective neurogenesis in citron kinase knockout mice by altered cytokinesis and massive apoptosis. Neuron 28: 115–127. 10.1016/S0896-6273(00)00090-8 [DOI] [PubMed] [Google Scholar]
- Diefenbacher ME, Popov N, Blake SM, Schülein-Völk C, Nye E, Spencer-Dene B, Jaenicke LA, Eilers M, Behrens A. 2014. The deubiquitinase USP28 controls intestinal homeostasis and promotes colorectal cancer. J Clin Invest 124: 3407–3418. 10.1172/JCI73733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinçer T, Yorgancıoğlu-Budak G, Ölmez A, Er İ, Dodurga Y, Özdemir ÖMA, Toraman B, Yıldırım A, Sabir N, Akarsu NA, et al. 2017. Analysis of centrosome and DNA damage response in PLK4 associated Seckel syndrome. Eur J Hum Genet 25: 1118–1125. 10.1038/ejhg.2017.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W, Wu Q, Sun L, Pan NC, Wang X. 2019. Cenpj regulates cilia disassembly and neurogenesis in the developing mouse cortex. J Neurosci 39: 1994–2010. 10.1523/JNEUROSCI.1849-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiStasio A, Driver A, Sund K, Donlin M, Muraleedharan RM, Pooya S, Kline-Fath B, Kaufman KM, Prows CA, Schorry E, et al. 2017. Copb2 is essential for embryogenesis and hypomorphic mutations cause human microcephaly. Hum Mol Genet 26: 4836–4848. 10.1093/hmg/ddx362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev NS, Tzolovsky G, Lipinszki Z, Schneider S, Lattao R, Fu J, Debski J, Dadlez M, Glover DM. 2014. Plk4 phosphorylates ana2 to trigger SAS6 recruitment and procentriole formation. Curr Biol 24: 2526–2532. 10.1016/j.cub.2014.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery P, Marcaillou C, Sahbatou M, Labalme A, Chastang J, Touraine R, Tubacher E, Senni F, Bober MB, Nampoothiri S, et al. 2011. Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science 332: 240–243. 10.1126/science.1202205 [DOI] [PubMed] [Google Scholar]
- Evrony GD, Cordero DR, Shen J, Partlow JN, Yu TW, Rodin RE, Hill RS, Coulter ME, Lam ATN, Jayaraman D, et al. 2017. Integrated genome and transcriptome sequencing identifies a noncoding mutation in the genome replication factor DONSON as the cause of microcephaly–micromelia syndrome. Genome Res 27: 1323–1335. 10.1101/gr.219899.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faheem M, Naseer MI, Rasool M, Chaudhary AG, Kumosani TA, Ilyas AM, Pushparaj PN, Ahmed F, Algahtani HA, Al-Qahtani MH, et al. 2015. Molecular genetics of human primary microcephaly: an overview. BMC Med Genomics 8: S4. 10.1186/1755-8794-8-S1-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farach LS, Little ME, Duker AL, Logan CV, Jackson A, Hecht JT, Bober M. 2018. The expanding phenotype of RNU4ATAC pathogenic variants to Lowry Wood syndrome. Am J Med Genet Part A 176: 465–469. 10.1002/ajmg.a.38581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq M, Fatima A, Mang Y, Hansen L, Kjaer KW, Baig SM, Larsen LA, Tommerup N. 2016. A novel splice site mutation in CEP135 is associated with primary microcephaly in a Pakistani family. J Hum Genet 61: 271–273. 10.1038/jhg.2015.138 [DOI] [PubMed] [Google Scholar]
- Farooq M, Lindbæk L, Krogh N, Doganli C, Keller C, Mönnich M, Gonçalves AB, Sakthivel S, Mang Y, Fatima A, et al. 2020. RRP7A links primary microcephaly to dysfunction of ribosome biogenesis, resorption of primary cilia, and neurogenesis. Nat Commun 11: 5816. 10.1038/s41467-020-19658-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Walsh CA. 2004. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron 44: 279–293. 10.1016/j.neuron.2004.09.023 [DOI] [PubMed] [Google Scholar]
- Fenwick AL, Kliszczak M, Cooper F, Murray J, Sanchez-Pulido L, Twigg SRF, Goriely A, McGowan SJ, Miller KA, Taylor IB, et al. 2016. Mutations in CDC45, encoding an essential component of the pre-initiation complex, cause Meier-Gorlin syndrome and craniosynostosis. Am J Hum Genet 99: 125–138. 10.1016/j.ajhg.2016.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fietz SA, Kelava I, Vogt J, Wilsch-Bräuninger M, Stenzel D, Fish JL, Corbeil D, Riehn A, Distler W, Nitsch R, et al. 2010. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat Neurosci 13: 690–699. 10.1038/nn.2553 [DOI] [PubMed] [Google Scholar]
- Fırat-Karalar EN, Stearns T. 2014. The centriole duplication cycle. Philos Trans R Soc B Biol Sci 369: 20130460. 10.1098/rstb.2013.0460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JL, Kosodo Y, Enard W, Pääbo S, Huttner WB. 2006. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci 103: 10438–10443. 10.1073/pnas.0604066103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio M, Huttner WB. 2014. Neural progenitors, neurogenesis and the evolution of the neocortex. Development 141: 2182–2194. 10.1242/dev.090571 [DOI] [PubMed] [Google Scholar]
- Fong CS, Mazo G, Das T, Goodman J, Kim M, O'Rourke BP, Izquierdo D, Tsou MFB. 2016. 53BP1 and USP28 mediate p53-dependent cell cycle arrest in response to centrosome loss and prolonged mitosis. Elife 5: e16270. 10.7554/eLife.16270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SS, De S, Johnson LK, Petrini JHJ, Stracker TH. 2012. Cell cycle- and DNA repair pathway-specific effects of apoptosis on tumor suppression. Proc Natl Acad Sci 109: 9953–9958. 10.1073/pnas.1120476109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco SJ, Müller U. 2013. Shaping our minds: stem and progenitor cell diversity in the mammalian neocortex. Neuron 77: 19–34. 10.1016/j.neuron.2012.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappart PO, Tong WM, Demuth I, Radovanovic I, Herceg Z, Aguzzi A, Digweed M, Wang ZQ. 2005. An essential function for NBS1 in the prevention of ataxia and cerebellar defects. Nat Med 11: 538–544. 10.1038/nm1228 [DOI] [PubMed] [Google Scholar]
- Fu J, Hagan IM, Glover DM. 2015. The centrosome and its duplication cycle. Cold Spring Harb Perspect Med 7: a015800. 10.1101/cshperspect.a015800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura K, Setsu T, Tanigaki K, Abe T, Kiyonari H, Terashima T, Sakisaka T. 2013. Kif14 mutation causes severe brain malformation and hypomyelination. PLoS One 8: e53490. 10.1371/journal.pone.0053490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S. 1964. Analysis of neuron differentiation in the central nervous system by tritiated thymidine autoradiography. J Comp Neurol 122: 311–327. 10.1002/cne.901220303 [DOI] [PubMed] [Google Scholar]
- Gabriel E, Wason A, Ramani A, Gooi LM, Keller P, Pozniakovsky A, Poser I, Noack F, Telugu NS, Calegari F, et al. 2016. CPAP promotes timely cilium disassembly to maintain neural progenitor pool. EMBO J 35: 803–819. 10.15252/embj.201593679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai M, Bianchi FT, Vagnoni C, Vernì F, Bonaccorsi S, Pasquero S, Berto GE, Sgrò F, Chiotto AM, Annaratone L, et al. 2016. ASPM and CITK regulate spindle orientation by affecting the dynamics of astral microtubules. EMBO Rep 17: 1396–1409. 10.15252/embr.201541823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal JS, Morozov YM, Ayoub AE, Chatterjee M, Rakic P, Haydar TF. 2006. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J Neurosci 26: 1045–1056. 10.1523/JNEUROSCI.4499-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garshasbi M, Motazacker MM, Kahrizi K, Behjati F, Abedini SS, Nieh SE, Firouzabadi SG, Becker C, Rüschendorf F, Nürnberg P, et al. 2006. SNP array-based homozygosity mapping reveals MCPH1 deletion in family with autosomal recessive mental retardation and mild microcephaly. Hum Genet 118: 708–715. 10.1007/s00439-005-0104-y [DOI] [PubMed] [Google Scholar]
- Genin A, Desir J, Lambert N, Biervliet M, Van der Aa N, Pierquin G, Killian A, Tosi M, Urbina M, Lefort A, et al. 2012. Kinetochore KMN network gene CASC5 mutated in primary microcephaly. Hum Mol Genet 21: 5306–5317. 10.1093/hmg/dds386 [DOI] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. 2000. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep 1: 524–529. 10.1093/embo-reports/kvd105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gliech CR, Holland AJ. 2020. Keeping track of time: the fundamentals of cellular clocks. J Cell Biol 219: e202005136. 10.1083/jcb.202005136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Martínez J, Cwetsch AW, Martínez-Alonso D, López-Sainz LR, Almagro J, Melati A, Gómez J, Pérez-Martínez M, Megías D, Boskovic J, et al. 2021. Deficient adaptation to centrosome duplication defects in neural progenitors causes microcephaly and subcortical heterotopias. JCI Insight 6: e146364. 10.1172/jci.insight.146364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. 2008. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol 181: 421–429. 10.1083/jcb.200711053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz M, Huttner WB. 2005. The cell biology of neurogenesis. Nat Rev Mol Cell Biol 6: 777–788. 10.1038/nrm1739 [DOI] [PubMed] [Google Scholar]
- Griffith E, Walker S, Martin CA, Vagnarelli P, Stiff T, Vernay B, Sanna N Al, Saggar A, Hamel B, Earnshaw WC, et al. 2008. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat Genet 40: 232–236. 10.1038/ng.2007.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber R, Zhou Z, Sukchev M, Joerss T, Frappart PO, Wang ZQ. 2011. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1–Cdc25 pathway. Nat Cell Biol 13: 1325–1334. 10.1038/ncb2342 [DOI] [PubMed] [Google Scholar]
- Gruneberg U, Neef R, Li X, Chan EHY, Chalamalasetty RB, Nigg EA, Barr FA. 2006. KIF14 and citron kinase act together to promote efficient cytokinesis. J Cell Biol 172: 363–372. 10.1083/jcb.200511061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guemez-Gamboa A, Nguyen LN, Yang H, Zaki MS, Kara M, Ben-Omran T, Akizu N, Rosti RO, Rosti B, Scott E, et al. 2015. Inactivating mutations in MFSD2A, required for omega-3 fatty acid transport in brain, cause a lethal microcephaly syndrome. Nat Genet 47: 809–813. 10.1038/ng.3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guernsey DL, Jiang H, Hussin J, Arnold M, Bouyakdan K, Perry S, Babineau-Sturk T, Beis J, Dumas N, Evans SC, et al. 2010. Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4. Am J Hum Genet 87: 40–51. 10.1016/j.ajhg.2010.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guernsey DL, Matsuoka M, Jiang H, Evans S, MacGillivray C, Nightingale M, Perry S, Ferguson M, Leblanc M, Paquette J, et al. 2011. Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nat Genet 43: 360–364. 10.1038/ng.777 [DOI] [PubMed] [Google Scholar]
- Guerreiro A, De Sousa F, Liaudet N, Ivanova D, Eskat A, Meraldi P. 2021. WDR62 localizes katanin at spindle poles to ensure synchronous chromosome segregation. J Cell Biol 220: e202007171. 10.1083/jcb.202007171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gul A, Hassan MJ, Mahmood S, Chen W, Rahmani S, Naseer MI, Dellefave L, Muhammad N, Rafiq MA, Ansar M, et al. 2006. Genetic studies of autosomal recessive primary microcephaly in 33 Pakistani families: novel sequence variants in ASPM gene. Neurogenetics 7: 105–110. 10.1007/s10048-006-0042-4 [DOI] [PubMed] [Google Scholar]
- Guven A, Gunduz A, Bozoglu TM, Yalcinkaya C, Tolun A. 2012. Novel NDE1 homozygous mutation resulting in microhydranencephaly and not microlyssencephaly. Neurogenetics 13: 189–194. 10.1007/s10048-012-0326-9 [DOI] [PubMed] [Google Scholar]
- Hallermayr A, Graf J, Koehler U, Laner A, Schönfeld B, Benet-Pagès A, Holinski-Feder E. 2018. Extending the critical regions for mutations in the non-coding gene RNU4ATAC in another patient with Roifman syndrome. Clin Case Reports 6: 2224–2228. 10.1002/ccr3.1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PRL, Kriegstein AR. 2010. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464: 554–561. 10.1038/nature08845 [DOI] [PubMed] [Google Scholar]
- Harding BN, Moccia A, Drunat S, Soukarieh O, Tubeuf H, Chitty LS, Verloes A, Gressens P, El Ghouzzi V, Joriot S, et al. 2016. Mutations in citron kinase cause recessive microlissencephaly with multinucleated neurons. Am J Hum Genet 99: 511–520. 10.1016/j.ajhg.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel T, Quek DQY, Wong BH, Cazenave-Gassiot A, Wenk MR, Fan H, Berger I, Shmueli D, Shaag A, Silver DL, et al. 2018. Homozygous mutation in MFSD2A, encoding a lysolipid transporter for docosahexanoic acid, is associated with microcephaly and hypomyelination. Neurogenetics 19: 227–235. 10.1007/s10048-018-0556-6 [DOI] [PubMed] [Google Scholar]
- Hasenpusch-Theil K, Theil T. 2021. The multifaceted roles of primary cilia in the development of the cerebral cortex. Front Cell Dev Biol 9: 630161. 10.3389/fcell.2021.630161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MJ, Khurshid M, Azeem Z, John P, Ali G, Chishti MS, Ahmad W. 2007. Previously described sequence variant in CDK5RAP2 gene in a Pakistani family with autosomal recessive primary microcephaly. BMC Med Genet 8: 58. 10.1186/1471-2350-8-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. 2010. Cep152 interacts with Plk4 and is required for centriole duplication. J Cell Biol 191: 721–729. 10.1083/jcb.201006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzopoulos GN, Erat MC, Cutts E, Rogala KB, Slater LM, Stansfeld PJ, Vakonakis I. 2013. Structural analysis of the G-box domain of the microcephaly protein CPAP suggests a role in centriole architecture. Structure 21: 2069–2077. 10.1016/j.str.2013.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB. 2004. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci 101: 3196–3201. 10.1073/pnas.0308600100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi MT, Cesare AJ, Fitzpatrick JAJ, Lazzerini-Denchi E, Karlseder J. 2012. A telomere-dependent DNA damage checkpoint induced by prolonged mitotic arrest. Nat Struct Mol Biol 19: 387–394. 10.1038/nsmb.2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Liyanarachchi S, Akagi K, Nagy R, Li J, Dietrich RC, Li W, Sebastian N, Wen B, Xin B, et al. 2011. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD i. Science 332: 238–240. 10.1126/science.1200587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. 1996. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382: 420–425. 10.1038/382420a0 [DOI] [PubMed] [Google Scholar]
- Higgins J, Midgley C, Bergh AM, Bell SM, Askham JM, Roberts E, Binns RK, Sharif SM, Bennett C, Glover DM, et al. 2010. Human ASPM participates in spindle organisation, spindle orientation and cytokinesis. BMC Cell Biol 11: 85. 10.1186/1471-2121-11-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. 2010. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J Cell Biol 188: 191–198. 10.1083/jcb.200911102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlihan SL, Feng Y. 2014. The scaffold protein Nde1 safeguards the brain genome during S phase of early neural progenitor differentiation. Elife 3: e03297. 10.7554/eLife.03297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Xiao X, Li S, Jia X, Guo X, Zhang Q. 2016. KIF11 mutations are a common cause of autosomal dominant familial exudative vitreoretinopathy. Br J Ophthalmol 100: 278–283. 10.1136/bjophthalmol-2015-306878 [DOI] [PubMed] [Google Scholar]
- Hudson JW, Kozarova A, Cheung P, Macmillan JC, Swallow CJ, Cross JC, Dennis JW. 2001. Late mitotic failure in mice lacking Sak, a polo-like kinase. Curr Biol 11: 441–446. 10.1016/S0960-9822(01)00117-8 [DOI] [PubMed] [Google Scholar]
- Hussain MS, Baig SM, Neumann S, Nürnberg G, Farooq M, Ahmad I, Alef T, Hennies HC, Technau M, Altmüller J, et al. 2012. A truncating mutation of CEP135 causes primary microcephaly and disturbed centrosomal function. Am J Hum Genet 90: 871–878. 10.1016/j.ajhg.2012.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain MS, Baig SM, Neumann S, Peche VS, Szczepanski S, Nürnberg G, Tariq M, Jameel M, Khan TN, Fatima A, et al. 2013. CDK6 associates with the centrosome during mitosis and is mutated in a large Pakistani family with primary microcephaly. Hum Mol Genet 22: 5199–5214. 10.1093/hmg/ddt374 [DOI] [PubMed] [Google Scholar]
- Imai F, Hirai SI, Akimoto K, Koyama H, Miyata T, Ogawa M, Noguchi S, Sasaoka T, Noda T, Ohno S. 2006. Inactivation of aPKCλ results in the loss of adherens junctions in neuroepithelial cells without affecting neurogenesis in mouse neocortex. Development 133: 1735–1744. 10.1242/dev.02330 [DOI] [PubMed] [Google Scholar]
- Insolera R, Bazzi H, Shao W, Anderson KV, Shi SH. 2014. Cortical neurogenesis in the absence of centrioles. Nat Neurosci 17: 1528–1535. 10.1038/nn.3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa L, Mueller K, Seufert K, Kraemer N, Rosenkotter H, Ninnemann O, Buob M, Kaindl AM, Morris-Rosendahl DJ. 2013. Clinical and cellular features in patients with primary autosomal recessive microcephaly and a novel CDK5RAP2 mutation. Orphanet J Rare Dis 8: 59. 10.1186/1750-1172-8-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izraeli S, Lowe LA, Bertness VL, Good DJ, Dorward DW, Kirsch IR, Kuehn MR. 1999. The SIL gene is required for mouse embryonic axial development and left–right specification. Nature 399: 691–694. 10.1038/21429 [DOI] [PubMed] [Google Scholar]
- Jackson AP, Eastwood H, Bell SM, Adu J, Toomes C, Carr IM, Roberts E, Hampshire DJ, Crow YJ, Mighell AJ, et al. 2002. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am J Hum Genet 71: 136–142. 10.1086/341283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäkel S, Dimou L. 2017. Glial cells and their function in the adult brain: a journey through the history of their ablation. Front Cell Neurosci 11: 24. 10.3389/fncel.2017.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson CR, Govaerts C, Abramowicz MJ. 1999. Primary autosomal recessive microcephaly: homozygosity mapping of MCPH4 to chromosome 15. Am J Hum Genet 65: 1465–1469. 10.1086/302640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson CR, Fryns JP, Jacobs J, Matthijs G, Abramowicz MJ. 2000. Primary autosomal recessive microcephaly: MCPH5 maps to 1q25-q32. Am J Hum Genet 67: 1575–1577. 10.1086/316909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman D, Kodani A, Gonzalez DM, Mancias JD, Mochida GH, Vagnoni C, Johnson J, Krogan N, Harper JW, Reiter JF, et al. 2016. Microcephaly proteins Wdr62 and Aspm define a mother centriole complex regulating centriole biogenesis, apical complex, and cell fate. Neuron 92: 813–828. 10.1016/j.neuron.2016.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman D, Bae B Il, Walsh CA. 2018. The genetics of primary microcephaly. Annu Rev Genomics Hum Genet 19: 177–200. 10.1146/annurev-genom-083117-021441 [DOI] [PubMed] [Google Scholar]
- Johnson MB, Sun X, Kodani A, Borges-Monroy R, Girskis KM, Ryu SC, Wang PP, Patel K, Gonzalez DM, Woo YM, et al. 2018. Aspm knockout ferret reveals an evolutionary mechanism governing cerebral cortical size. Nat 556: 370–375. 10.1038/s41586-018-0035-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones GE, Ostergaard P, Moore AT, Connell FC, Williams D, Quarrell O, Brady AF, Spier I, Hazan F, Moldovan O, et al. 2014. Microcephaly with or without chorioretinopathy, lymphoedema, or mental retardation (MCLMR): review of phenotype associated with KIF11 mutations. Eur J Hum Genet 22: 881–887. 10.1038/ejhg.2013.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadir R, Harel T, Markus B, Perez Y, Bakhrat A, Cohen I, Volodarsky M, Feintsein-Linial M, Chervinski E, Zlotogora J, et al. 2016. ALFY-controlled DVL3 autophagy regulates Wnt signaling, determining human brain size. PLoS Genet 12: e1005919. 10.1371/journal.pgen.1005919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakar N, Ahmad J, Morris-Rosendahl DJ, Altmüller J, Friedrich K, Barbi G, Nürnberg P, Kubisch C, Dobyns WB, Borck G. 2015. STIL mutation causes autosomal recessive microcephalic lobar holoprosencephaly. Hum Genet 134: 45–51. 10.1007/s00439-014-1487-4 [DOI] [PubMed] [Google Scholar]
- Kalay E, Yigit G, Aslan Y, Brown KE, Pohl E, Bicknell LS, Kayserili H, Li Y, Tüysüz B, Nürnberg G, et al. 2011. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat Genet 43: 23–26. 10.1038/ng.725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltschmidt JA, Davidson CM, Brown NH, Brand AH. 1999. Rotation and asymmetry of the mitotic spindle direct asymmetric cell division in the developing central nervous system. Nat Cell Biol 2: 7–12. [DOI] [PubMed] [Google Scholar]
- Kamasaki T, O'Toole E, Kita S, Osumi M, Usukura J, McIntosh RR, Goshima G. 2013. Augmin-dependent microtubule nucleation at microtubule walls in the spindle. J Cell Biol 202: 25–33. 10.1083/jcb.201304031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantaputra P, Tanpaiboon P, Porntaveetus T, Ohazama A, Sharpe P, Rauch A, Hussadaloy A, Thiel CT. 2011. The smallest teeth in the world are caused by mutations in the PCNT gene. Am J Med Genet Part A 155: 1398–1403. 10.1002/ajmg.a.33984 [DOI] [PubMed] [Google Scholar]
- Kavaslar GN, Önengüt S, Derman O, Kaya A, Tolun A. 2000. The novel genetic disorder microhydranencephaly maps to chromosome 16p13.34-12.1. Am J Hum Genet 66: 1705–1709. 10.1086/302898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Rupp VM, Orpinell M, Hussain MS, Altmüller J, Steinmetz MO, Enzinger C, Thiele H, Höhne W, Nürnberg G, et al. 2014. A missense mutation in the PISA domain of HsSAS-6 causes autosomal recessive primary microcephaly in a large consanguineous Pakistani family. Hum Mol Genet 23: 5940–5949. 10.1093/hmg/ddu318 [DOI] [PubMed] [Google Scholar]
- Khan MA, Windpassinger C, Ali MZ, Zubair M, Gul H, Abbas S, Khan S, Badar M, Mohammad RM, Nawaz Z. 2017. Molecular genetic analysis of consanguineous families with primary microcephaly identified pathogenic variants in the ASPM gene. J Genet 96: 383–387. 10.1007/s12041-017-0759-x [DOI] [PubMed] [Google Scholar]
- Khetarpal P, Das S, Panigrahi I, Munshi A. 2016. Primordial dwarfism: overview of clinical and genetic aspects. Mol Genet Genomics 291: 1–15. 10.1007/s00438-015-1110-y [DOI] [PubMed] [Google Scholar]
- Kia SK, Verbeek E, Engelen E, Schot R, Poot RA, De coo IFM, Lequin MH, Poulton CJ, Pourfarzad F, Grosveld FG, et al. 2012. RTTN mutations link primary cilia function to organization of the human cerebral cortex. Am J Hum Genet 91: 533–540. 10.1016/j.ajhg.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Sharov AA, McDole K, Cheng M, Hao H, Fan CM, Gaiano N, Ko MSH, Zheng Y. 2011. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science 334: 1706–1710. 10.1126/science.1211222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TS, Park JE, Shukla A, Choi S, Murugan RN, Lee JH, Ahn M, Rhee K, Bang JK, Kim BY, et al. 2013. Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proc Natl Acad Sci 110: E4849–E4857. 10.1073/pnas.1319656110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko MA, Rosario CO, Hudson JW, Kulkarni S, Pollett A, Dennis JW, Swallow CJ. 2005. Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat Genet 37: 883–888. 10.1038/ng1605 [DOI] [PubMed] [Google Scholar]
- Kohlmaier G. 2009. “Functional characterization of the SAS-4-related protein CPAP in centrosome biology of human cells.” PhD thesis, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland.. 10.5075/epfl-thesis-4491 [DOI] [Google Scholar]
- Konno D, Shioi G, Shitamukai A, Mori A, Kiyonari H, Miyata T, Matsuzaki F. 2008. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat Cell Biol 10: 93–101. 10.1038/ncb1673 [DOI] [PubMed] [Google Scholar]
- Kornack DR, Rakic P. 1998. Changes in cell-cycle kinetics during the development and evolution of primate neocortex. Proc Natl Acad Sci 95: 1242–1246. 10.1073/pnas.95.3.1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosodo Y, Röper K, Haubensak W, Marzesco AM, Corbeil D, Huttner WB. 2004. Asymmetric distribution of the apical plasma membrane during neurogenic divisions of mammalian neuroepithelial cells. EMBO J 23: 2314–2324. 10.1038/sj.emboj.7600223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk T, Pontious A, Englund C, Daza RAM, Bedogni F, Hodge R, Attardo A, Bell C, Huttner WB, Hevner RF. 2009. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb Cortex 19: 2439–2450. 10.1093/cercor/bhn260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz AS, Bärenz F, Richter KT, Hoffmann I. 2015. Plk4-dependent phosphorylation of STIL is required for centriole duplication. Biol Open 4: 370–377. 10.1242/bio.201411023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Blanton SH, Babu M, Markandaya M, Girimaji SC. 2004. Genetic analysis of primary microcephaly in Indian families: novel ASPM mutations. Clin Genet 66: 341–348. 10.1111/j.1399-0004.2004.00304.x [DOI] [PubMed] [Google Scholar]
- Kumar A, Girimaji SC, Duvvari MR, Blanton SH. 2009. Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am J Hum Genet 84: 286–290. 10.1016/j.ajhg.2009.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok BH, Yang JG, Kapoor TM. 2004. The rate of bipolar spindle assembly depends on the microtubule-gliding velocity of the mitotic kinesin Eg5. Curr Biol 14: 1783–1788. 10.1016/j.cub.2004.09.052 [DOI] [PubMed] [Google Scholar]
- Lambrus BG, Holland AJ. 2017. A new mode of mitotic surveillance. Trends Cell Biol 27: 314–321. 10.1016/j.tcb.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrus BG, Daggubati V, Uetake Y, Scott PM, Clutario KM, Sluder G, Holland AJ. 2016. A USP28-53BP1–p53–p21 signaling axis arrests growth after centrosome loss or prolonged mitosis. J Cell Biol 214: 143–153. 10.1083/jcb.201604054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. 2013. Cerebral organoids model human brain development and microcephaly. Nature 501: 373–379. 10.1038/nature12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, Huttner WB, Calegari F. 2009. Cdk4/CyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell 5: 320–331. 10.1016/j.stem.2009.05.026 [DOI] [PubMed] [Google Scholar]
- Leal GF, Roberts E, Silva EO, Costa SMR, Hampshire DJ, Woods CG. 2003. A novel locus for autosomal recessive primary microcephaly (MCPH6) maps to 13q12.2. J Med Genet 40: 540–542. 10.1136/jmg.40.7.540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerit DA, Rusan NM. 2013. PLP inhibits the activity of interphase centrosomes to ensure their proper segregation in stem cells. J Cell Biol 202: 1013–1022. 10.1083/jcb.201303141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MS, Holland AJ. 2018. The impact of mitotic errors on cell proliferation and tumorigenesis. Genes Dev 32: 620–638. 10.1101/gad.314351.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yang YG, Gao Y, Wang ZQ, Tong WM. 2012. A distinct response to endogenous DNA damage in the development of Nbs1-deficient cortical neurons. Cell Res 22: 859–872. 10.1038/cr.2012.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FF, Wang XD, Zhu MW, Lou ZH, Zhang Q, Zhu CY, Feng HL, Lin ZG, Liu SL. 2015. Identification of two novel critical mutations in PCNT gene resulting in microcephalic osteodysplastic primordial dwarfism type II associated with multiple intracranial aneurysms. Metab Brain Dis 30: 1387–1394. 10.1007/s11011-015-9712-y [DOI] [PubMed] [Google Scholar]
- Li H, Bielas SL, Zaki MS, Ismail S, Farfara D, Um K, Rosti RO, Scott EC, Tu S, Chi NC, et al. 2016a. Biallelic mutations in citron kinase link mitotic cytokinesis to human primary microcephaly. Am J Hum Genet 99: 501–510. 10.1016/j.ajhg.2016.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JK, Fei P, Li Y, Huang QJ, Zhang Q, Zhang X, Rao YQ, Li J, Zhao P. 2016b. Identification of novel KIF11 mutations in patients with familial exudative vitreoretinopathy and a phenotypic analysis. Sci Rep 6: 26564. 10.1038/srep26564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Sun L, Fang A, Li P, Wu Q, Wang X. 2017. Recapitulating cortical development with organoid culture in vitro and modeling abnormal spindle-like (ASPM related primary) microcephaly disease. Protein Cell 8: 823–833. 10.1007/s13238-017-0479-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Li H, Xu X, Wang L, Liu B, Zheng W, Lian L, Song Y, Xia X, Hou L, et al. 2020. Haploinsufficiency of GCP4 induces autophagy and leads to photoreceptor degeneration due to defective spindle assembly in retina. Cell Death Differ 27: 556–572. 10.1038/s41418-019-0371-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Liang Y, Li K. 2010. Multiple roles of BRIT1/MCPH1 in DNA damage response, DNA repair, and cancer suppression. Yonsei Med J 51: 295–301. 10.3349/ymj.2010.51.3.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Chang CW, Hsu W Bin, Tang CJC, Lin YN, Chou EJ, Wu CT, Tang TK. 2013. Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. EMBO J 32: 1141–1154. 10.1038/emboj.2013.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YN, Lee YS, Li SK, Tang TK. 2020. Loss of CPAP in developing mouse brain and its functional implication in human primary microcephaly. J Cell Sci 133: jcs243592. 10.1242/jcs.243592 [DOI] [PubMed] [Google Scholar]
- Lizarraga SB, Margossian SP, Harris MH, Campagna DR, Han AP, Blevins S, Mudbhary R, Barker JE, Walsh CA, Fleming MD. 2010. Cdk5rap2 regulates centrosome function and chromosome segregation in neuronal progenitors. Development 137: 1907–1917. 10.1242/dev.040410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrythanasis P, Maroofian R, Stray-Pedersen A, Musaev D, Zaki MS, Mahmoud IG, Selim L, Elbadawy A, Jhangiani SN, Coban Akdemir ZH, et al. 2018. Biallelic variants in KIF14 cause intellectual disability with microcephaly. Eur J Hum Genet 26: 330–339. 10.1038/s41431-017-0088-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malatesta P, Hack MA, Hartfuss E, Kettenmann H, Klinkert W, Kirchhoff F, Götz M. 2003. Neuronal or glial progeny: regional differences in radial glia fate. Neuron 37: 751–764. 10.1016/S0896-6273(03)00116-8 [DOI] [PubMed] [Google Scholar]
- Marjanović M, Sánchez-Huertas C, Terré B, Gómez R, Scheel JF, Pacheco S, Knobel PA, Martínez-Marchal A, Aivio S, Palenzuela L, et al. 2015. CEP63 deficiency promotes p53-dependent microcephaly and reveals a role for the centrosome in meiotic recombination. Nat Commun 6: 7676. 10.1038/ncomms8676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthiens V, Basto R. 2020. Centrosomes: the good and the bad for brain development. Biol Cell 112: 153–172. 10.1111/boc.201900090 [DOI] [PubMed] [Google Scholar]
- Marthiens V, Rujano MA, Pennetier C, Tessier S, Paul-Gilloteaux P, Basto R. 2013. Centrosome amplification causes microcephaly. Nat Cell Biol 15: 731–740. 10.1038/ncb2746 [DOI] [PubMed] [Google Scholar]
- Martin CA, Ahmad I, Klingseisen A, Hussain MS, Bicknell LS, Leitch A, Nürnberg G, Toliat MR, Murray JE, Hunt D, et al. 2014. Mutations in PLK4, encoding a master regulator of centriole biogenesis, cause microcephaly, growth failure and retinopathy. Nat Genet 46: 1283–1292. 10.1038/ng.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CA, Murray JE, Carroll P, Leitch A, Mackenzie KJ, Halachev M, Fetit AE, Keith C, Bicknell LS, Fluteau A, et al. 2016. Mutations in genes encoding condensin complex proteins cause microcephaly through decatenation failure at mitosis. Genes Dev 30: 2158–2172. 10.1101/gad.286351.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Campos M, Basto R, Baker J, Kernan M, Raff JW. 2004. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J Cell Biol 165: 673–683. 10.1083/jcb.200402130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Miyamoto T, Sakamoto H, Izumi H, Nakazawa Y, Ogi T, Tahara H, Oku S, Hiramoto A, Shiiki T, et al. 2011. Two unrelated patients with MRE11A mutations and Nijmegen breakage syndrome-like severe microcephaly. DNA Repair 10: 314–321. 10.1016/j.dnarep.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Matsuura S, Tauchi H, Nakamura A, Kondo N, Sakamoto S, Endo S, Smeets D, Solder B, Belohradsky BH, Der Kaloustian VM, et al. 1998. Positional cloning of the gene for Nijmegen breakage syndrome. Nat Genet 19: 179–181. 10.1038/549 [DOI] [PubMed] [Google Scholar]
- McConnell SK, Kaznowski CE. 1991. Cell cycle dependence of laminar determination in developing neocortex. Science 254: 282–285. 10.1126/science.254.5029.282 [DOI] [PubMed] [Google Scholar]
- McKim KS, Hawley RS. 1995. Chromosomal control of meiotic cell division. Science 270: 1595–1601. 10.1126/science.270.5242.1595 [DOI] [PubMed] [Google Scholar]
- McKinnon PJ. 2013. Maintaining genome stability in the nervous system. Nat Neurosci 16: 1523–1529. 10.1038/nn.3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Anzola JV, Kaulich M, Richardson A, Stender JD, Benner C, Glass CK, Dowdy SF, Desai A, Shiau AK, et al. 2016. 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J Cell Biol 214: 155–166. 10.1083/jcb.201604081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaa GM, Enyedi L, Parsons G, Collins S, Medne L, Adams C, Ward T, Davitt B, Bicknese A, Zackai E, et al. 2014a. Congenital microcephaly and chorioretinopathy due to de novo heterozygous KIF11 mutations: five novel mutations and review of the literature. Am J Med Genet Part A 164: 2879–2886. 10.1002/ajmg.a.36707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaa GM, Vitre B, Carpenter G, Abramowicz I, Gleeson JG, Paciorkowski AR, Cleveland DW, Dobyns WB, O'Driscoll M. 2014b. Mutations in CENPE define a novel kinetochore-centromeric mechanism for microcephalic primordial dwarfism. Hum Genet 133: 1023–1039. 10.1007/s00439-014-1443-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Dick A, Chalem A, Pilaz L-J, Silver DL. 2020. Acute lengthening of progenitor mitosis influences progeny fate during cortical development in vivo. Dev Neurosci 41: 300–317. 10.1159/000507113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T, Akutsu SN, Fukumitsu A, Morino H, Masatsuna Y, Hosoba K, Kawakami H, Yamamoto T, Shimizu K, Ohashi H, et al. 2017. PLK1-mediated phosphorylation of WDR62/MCPH2 ensures proper mitotic spindle orientation. Hum Mol Genet 26: 4429–4440. 10.1093/hmg/ddx330 [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. 2004. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development 131: 3133–3145. 10.1242/dev.01173 [DOI] [PubMed] [Google Scholar]
- Moawia A, Shaheen R, Rasool S, Waseem SS, Ewida N, Budde B, Kawalia A, Motameny S, Khan K, Fatima A, et al. 2017. Mutations of KIF14 cause primary microcephaly by impairing cytokinesis. Ann Neurol 82: 562–577. 10.1002/ana.25044 [DOI] [PubMed] [Google Scholar]
- Mori Y, Inoue Y, Tanaka S, Doda S, Yamanaka S, Fukuchi H, Terada Y. 2015. Cep169, a novel microtubule plus-end-tracking centrosomal protein, binds to CDK5RAP2 and regulates microtubule stability. PLoS One 10: e0140968. 10.1371/journal.pone.0140968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer TC, Clutario KM, Lambrus BG, Daggubati V, Holland AJ. 2015. Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J Cell Biol 209: 863–878. 10.1083/jcb.201502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan L, Jackson AP, Roberts E, Karbani G, Lewis I, Corry P, Turner G, Mueller RF, Lench NJ, Woods CG. 2000. A third novel locus for primary autosomal recessive microcephaly maps to chromosome 9q34. Am J Hum Genet 66: 724–727. 10.1086/302777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad F, Baig SM, Hansen L, Hussain MS, Inayat IA, Aslam M, Qureshi JA, Toilat M, Kirst E, Wajid M, et al. 2009. Compound heterozygous ASPM mutations in Pakistani MCPH families. Am J Med Genet Part A 149A: 926–930. 10.1002/ajmg.a.32749 [DOI] [PubMed] [Google Scholar]
- Murdock DR, Clark GD, Bainbridge MN, Newsham I, Wu YQ, Muzny DM, Cheung SW, Gibbs RA, Ramocki MB. 2011. Whole-exome sequencing identifies compound heterozygous mutations in WDR62 in siblings with recurrent polymicrogyria. Am J Med Genet Part A 155: 2071–2077. 10.1002/ajmg.a.34165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim V, Imarisio S, Di Cunto F, Gatti M, Bonaccorsi S. 2004. Drosophila citron kinase is required for the final steps of cytokinesis. Mol Biol Cell 15: 5053–5063. 10.1091/mbc.e04-06-0536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveed M, Kazmi SK, Amin M, Asif Z, Islam U, Shahid K, Tehreem S. 2018. Comprehensive review on the molecular genetics of autosomal recessive primary microcephaly (MCPH). Genet Res 100: e7. 10.1017/S0016672318000046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neitzel H, Neumann LM, Schindler D, Wirges A, Tönnies H, Trimborn M, Krebsova A, Richter R, Sperling K. 2002. Premature chromosome condensation in humans associated with microcephaly and mental retardation: a novel autosomal recessive condition. Am J Hum Genet 70: 1015–1022. 10.1086/339518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas AK, Swanson EA, Cox JJ, Karbani G, Malik S, Springell K, Hampshire D, Ahmed M, Bond J, Di Benedetto D, et al. 2009. The molecular landscape of ASPM mutations in primary microcephaly. J Med Genet 46: 249–253. 10.1136/jmg.2008.062380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas AK, Khurshid M, Désir J, Carvalho OP, Cox JJ, Thornton G, Kausar R, Ansar M, Ahmad W, Verloes A, et al. 2010. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat Genet 42: 1010–1014. 10.1038/ng.682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Holland AJ. 2018. Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat Rev Mol Cell Biol 19: 297–312. 10.1038/nrm.2017.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell 139: 663–678. 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Wong WS, Clinton BK, Kriegstein AR. 2002. Dividing precursor cells of the embryonic cortical ventricular zone have morphological and molecular characteristics of radial glia. J Neurosci 22: 3161–3173. 10.1523/JNEUROSCI.22-08-03161.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR. 2004. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci 7: 136–144. 10.1038/nn1172 [DOI] [PubMed] [Google Scholar]
- Novorol C, Burkhardt J, Wood KJ, Iqbal A, Roque C, Coutts N, Almeida AD, He J, Wilkinson CJ, Harris WA. 2013. Microcephaly models in the developing zebrafish retinal neuroepithelium point to an underlying defect in metaphase progression. Open Biol 3: 130065. 10.1098/rsob.130065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Driscoll M, Cerosaletti KM, Girard PM, Dai Y, Stumm M, Kysela B, Hirsch B, Gennery A, Palmer SE, Seidel J, et al. 2001. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol Cell 8: 1175–1185. 10.1016/S1097-2765(01)00408-7 [DOI] [PubMed] [Google Scholar]
- O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. 2003. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet 33: 497–501. 10.1038/ng1129 [DOI] [PubMed] [Google Scholar]
- Ogi T, Walker S, Stiff T, Hobson E, Limsirichaikul S, Carpenter G, Prescott K, Suri M, Byrd PJ, Matsuse M, et al. 2012. Identification of the first ATRIP-deficient patient and novel mutations in ATR define a clinical spectrum for ATR-ATRIP Seckel syndrome. PLoS Genet 8: e1002945. 10.1371/journal.pgen.1002945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Ashikawa T, Nozaki Y, Kozuka-Hata H, Goto H, Inagaki M, Oyama M, Kitagawa D. 2014. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat Commun 5: 5267. 10.1038/ncomms6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill RS, Schoborg TA, Rusan NM. 2018. Same but different: pleiotropy in centrosome-related microcephaly. Mol Biol Cell 29: 241–246. 10.1091/mbc.E17-03-0192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard P, Simpson MA, Mendola A, Vasudevan P, Connell FC, Van Impel A, Moore AT, Loeys BL, Ghalamkarpour A, Onoufriadis A, et al. 2012. Mutations in KIF11 cause autosomal-dominant microcephaly variably associated with congenital lymphedema and chorioretinopathy. Am J Hum Genet 90: 356–362. 10.1016/j.ajhg.2011.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachajoa H, Ruiz-Botero F, Isaza C. 2014. A new mutation of the PCNT gene in a Colombian patient with microcephalic osteodysplastic primordial dwarfism type II: a case report. J Med Case Rep 8: 191. 10.1186/1752-1947-8-191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnamenta AT, Murray JE, Yoon G, Akha ES, Harrison V, Bicknell LS, Ajilogba K, Stewart H, Kini U, Taylor JC, et al. 2012. A novel nonsense CDK5RAP2 mutation in a Somali child with primary microcephaly and sensorineural hearing loss. Am J Med Genet Part A 158A: 2577–2582. 10.1002/ajmg.a.35558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papari E, Bastami M, Farhadi A, Abedini S, Hosseini M, Bahman I, Mohseni M, Garshasbi M, Moheb LA, Behjati F, et al. 2013. Investigation of primary microcephaly in Bushehr province of Iran: novel STIL and ASPM mutations. Clin Genet 83: 488–490. 10.1111/j.1399-0004.2012.01949.x [DOI] [PubMed] [Google Scholar]
- Paridaen JTML, Wilsch-Bräuninger M, Huttner WB. 2013. Asymmetric inheritance of centrosome-associated primary cilium membrane directs ciliogenesis after cell division. Cell 155: 333–344. 10.1016/j.cell.2013.08.060 [DOI] [PubMed] [Google Scholar]
- Park SY, Park JE, Kim TS, Kim JH, Kwak MJ, Ku B, Tian L, Murugan RN, Ahn M, Komiya S, et al. 2014. Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis. Nat Struct Mol Biol 21: 696–703. 10.1038/nsmb.2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry DA, Martin CA, Greene P, Marsh JA, Ambrose JC, Arumugam P, Baple EL, Bleda M, Boardman-Pretty F, Boissiere JM, et al. 2021. Heterozygous lamin B1 and lamin B2 variants cause primary microcephaly and define a novel laminopathy. Genet Med 23: 408–414. 10.1038/s41436-020-00980-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passemard S, Titomanlio L, Elmaleh M, Afenjar A, Alessandri JL, Andria G, De Villemeur TB, Boespflug-Tanguy O, Burglen L, Del Giudice E, et al. 2009. Expanding the clinical and neuroradiologic phenotype of primary microcephaly due to ASPM mutations. Neurology 73: 962–969. 10.1212/WNL.0b013e3181b8799a [DOI] [PubMed] [Google Scholar]
- Pattison L, Crow YJ, Deeble VJ, Jackson AP, Jafri H, Rashid Y, Roberts E, Woods CG. 2000. A fifth locus for primary autosomal recessive microcephaly maps to chromosome 1q31. Am J Hum Genet 67: 1578–1580. 10.1086/316910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan D, Mani S, Passemard S, Gressens P, El Ghouzzi V. 2018. STIL balancing primary microcephaly and cancer. Cell Death Dis 9: 65. 10.1038/s41419-017-0101-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlisz AS, Mutch C, Wynshaw-Boris A, Chenn A, Walsh CA, Feng Y. 2008. Lis1-Nde1-dependent neuronal fate control determines cerebral cortical size and lamination. Hum Mol Genet 17: 2441–2455. 10.1093/hmg/ddn144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne F, Colnaghi R, Rocha N, Seth A, Harris J, Carpenter G, Bottomley WE, Wheeler E, Wong S, Saudek V, et al. 2014. Hypomorphism in human NSMCE2 linked to primordial dwarfism and insulin resistance. J Clin Invest 124: 4028–4038. 10.1172/JCI73264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penisson M, Ladewig J, Belvindrah R, Francis F. 2019. Genes and mechanisms involved in the generation and amplification of basal radial glial cells. Front Cell Neurosci 13: 381. 10.3389/fncel.2019.00381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Y, Bar-Yaacov R, Kadir R, Wormser O, Shelef I, Birk OS, Flusser H, Birnbaum RY. 2019. Mutations in the microtubule-associated protein MAP11 (C7orf43) cause microcephaly in humans and zebrafish. Brain 142: 574–585. 10.1093/brain/awz004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S. 2016. Mechanisms of mitotic spindle assembly. Annu Rev Biochem 85: 659–683. 10.1146/annurev-biochem-060815-014528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff KL, Straub CT, Chiang K, Bear DM, Zhou Y, Zon LI. 2007. The zebrafish cassiopeia mutant reveals that SIL is required for mitotic spindle organization. Mol Cell Biol 27: 5887–5897. 10.1128/MCB.00175-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfau RB, Thrush DL, Hamelberg E, Bartholomew D, Botes S, Pastore M, Tan C, del Gaudio D, Gastier-Foster JM, Astbury C. 2013. MCPH1 deletion in a newborn with severe microcephaly and premature chromosome condensation. Eur J Med Genet 56: 609–613. 10.1016/j.ejmg.2013.09.007 [DOI] [PubMed] [Google Scholar]
- Pfau SJ, Silberman RE, Knouse KA, Amon A. 2016. Aneuploidy impairs hematopoietic stem cell fitness and is selected against in regenerating tissues in vivo. Genes Dev 30: 1395–1408. 10.1101/gad.278820.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan TP, Maryniak AL, Boatwright CA, Lee J, Atkins A, Tijhuis A, Spierings DC, Bazzi H, Foijer F, Jordan PW, et al. 2021. Centrosome defects cause microcephaly by activating the 53BP1–USP28–TP53 mitotic surveillance pathway. EMBO J 40: e106118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon B, Vankerckhove S, Bourrouillou G, Duprez L, Abramowicz MJ. 2004. A translocation breakpoint disrupts the ASPM gene in a patient with primary microcephaly. Eur J Hum Genet 12: 419–421. 10.1038/sj.ejhg.5201169 [DOI] [PubMed] [Google Scholar]
- Pilaz LJ, Patti D, Marcy G, Ollier E, Pfister S, Douglas RJ, Betizeau M, Gautier E, Cortay V, Doerflinger N, et al. 2009. Forced G1-phase reduction alters mode of division, neuron number, and laminar phenotype in the cerebral cortex. Proc Natl Acad Sci 106: 21924–21929. 10.1073/pnas.0909894106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilaz LJ, McMahon JJ, Miller EE, Lennox AL, Suzuki A, Salmon E, Silver DL. 2016. Prolonged mitosis of neural progenitors alters cell fate in the developing brain. Neuron 89: 83–99. 10.1016/j.neuron.2015.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilz GA, Shitamukai A, Reillo I, Pacary E, Schwausch J, Stahl R, Ninkovic J, Snippert HJ, Clevers H, Godinho L, et al. 2013. Amplification of progenitors in the mammalian telencephalon includes a new radial glial cell type. Nat Commun 4: 2125. 10.1038/ncomms3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova S, Lange S, Shield K, Mihic A, Chudley AE, Mukherjee RAS, Bekmuradov D, Rehm J. 2016. Comorbidity of fetal alcohol spectrum disorder: a systematic review and meta-analysis. Lancet 387: 978–987. 10.1016/S0140-6736(15)01345-8 [DOI] [PubMed] [Google Scholar]
- Puffenberger EG, Jinks RN, Sougnez C, Cibulskis K, Willert RA, Achilly NP, Cassidy RP, Fiorentini CJ, Heiken KF, Lawrence JJ, et al. 2012. Genetic mapping and exome sequencing identify variants associated with five novel diseases. PLoS One 7: e28936. 10.1371/journal.pone.0028936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulvers JN, Bryk J, Fish JL, Wilsch-Bräuninger M, Arai Y, Schreier D, Naumann R, Helppi J, Habermann B, Vogt J, et al. 2010. Mutations in mouse Aspm (abnormal spindle-like microcephaly associated) cause not only microcephaly but also major defects in the germline. Proc Natl Acad Sci 107: 16595–16600. 10.1073/pnas.1010494107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Shen Q, Goderie SK, He W, Capela A, Davis AA, Temple S. 2000. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron 28: 69–80. 10.1016/S0896-6273(00)00086-6 [DOI] [PubMed] [Google Scholar]
- Qvist P, Huertas P, Jimeno S, Nyegaard M, Hassan MJ, Jackson SP, Børglum AD. 2011. CtIP mutations cause Seckel and Jawad syndromes. PLoS Genet 7: e1002310. 10.1371/journal.pgen.1002310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. 1971. Guidance of neurons migrating to the fetal monkey neocortex. Brain Res 33: 471–476. 10.1016/0006-8993(71)90119-3 [DOI] [PubMed] [Google Scholar]
- Rakic P. 1995. A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci 18: 383–388. 10.1016/0166-2236(95)93934-P [DOI] [PubMed] [Google Scholar]
- Ramdas Nair A, Singh P, Salvador Garcia D, Rodriguez-Crespo D, Egger B, Cabernard C. 2016. The microcephaly-associated protein Wdr62/CG7337 is required to maintain centrosome asymmetry in drosophila neuroblasts. Cell Rep 14: 1100–1113. 10.1016/j.celrep.2015.12.097 [DOI] [PubMed] [Google Scholar]
- Rao FQ, Cai XB, Cheng FF, Cheng W, Fang XL, Li N, Huang XF, Li LH, Jin ZB. 2017. Mutations in LRP5, FZD4, TSPAN12, NDP, ZNF408, or KIF11 genes account for 38.7% of Chinese patients with familial exudative vitreoretinopathy. Investig Ophthalmol Vis Sci 58: 2623–2629. 10.1167/iovs.16-21324 [DOI] [PubMed] [Google Scholar]
- Rašin MR, Gazula VR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li HS, Jan LY, Jan YN, Rakic P, et al. 2007. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci 10: 819–827. 10.1038/nn1924 [DOI] [PubMed] [Google Scholar]
- Rauch A, Thiel CT, Schindler D, Wick U, Crow YJ, Ekici AB, Van Essen AJ, Goecke TO, Al-Gazali L, Chrzanowska KH, et al. 2008. Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science 319: 816–819. 10.1126/science.1151174 [DOI] [PubMed] [Google Scholar]
- Reillo I, De Juan Romero C, García-Cabezas MÁ, Borrell V. 2011. A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb Cortex 21: 1674–1694. 10.1093/cercor/bhq238 [DOI] [PubMed] [Google Scholar]
- Reuter MS, Tawamie H, Buchert R, Gebril OH, Froukh T, Thiel C, Uebe S, Ekici AB, Krumbiegel M, Zweier C, et al. 2017. Diagnostic yield and novel candidate genes by exome sequencing in 152 consanguineous families with neurodevelopmental disorders. JAMA Psychiatry 74: 293–299. 10.1001/jamapsychiatry.2016.3798 [DOI] [PubMed] [Google Scholar]
- Reynolds JJ, Bicknell LS, Carroll P, Higgs MR, Shaheen R, Murray JE, Papadopoulos DK, Leitch A, Murina O, Tarnauskaite Z, et al. 2017. Mutations in DONSON disrupt replication fork stability and cause microcephalic dwarfism. Nat Genet 49: 537–549. 10.1038/ng.3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K, Paakkola T, Mennerich D, Kubaichuk K, Konzack A, Kippari HA, Kozlova N, Koivunen P, Haapasaari KM, Jukkola-Vuorinen A, et al. 2018. USP28 deficiency promotes breast and liver carcinogenesis as well as tumor angiogenesis in a HIF-independent manner. Mol Cancer Res 16: 1000–1012. 10.1158/1541-7786.MCR-17-0452 [DOI] [PubMed] [Google Scholar]
- Roberts E, Jackson AP, Carradice AC, Deeble VJ, Mannan J, Rashid Y, Jafri H, McHale DP, Markham AF, Lench NJ, et al. 1999. The second locus for autosomal recessive primary microcephaly (MCPH2) maps to chromosome 19q13.1-13.2. Eur J Hum Genet 7: 815–820. 10.1038/sj.ejhg.5200385 [DOI] [PubMed] [Google Scholar]
- Roberts E, Hampshire DJ, Pattison L, Springell K, Jafri H, Corry P, Mannon J, Rashid Y, Crow Y, Bond J, et al. 2002. Autosomal recessive primary microcephaly: an analysis of locus heterogeneity and phenotypic variation. J Med Genet 39: 718–721. 10.1136/jmg.39.10.718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robitaille JM, Gillett RM, LeBlanc MA, Gaston D, Nightingale M, Mackley MP, Parkash S, Hathaway J, Thomas A, Ells A, et al. 2014. Phenotypic overlap between familial exudative vitreoretinopathy and microcephaly, lymphedema, and chorioretinal dysplasia caused by KIF11 mutations. JAMA Ophthalmol 132: 1393–1399. 10.1001/jamaophthalmol.2014.2814 [DOI] [PubMed] [Google Scholar]
- Rujano MA, Sanchez-Pulido L, Pennetier C, Le Dez G, Basto R. 2013. The microcephaly protein Asp regulates neuroepithelium morphogenesis by controlling the spatial distribution of myosin II. Nat Cell Biol 15: 1294–1306. 10.1038/ncb2858 [DOI] [PubMed] [Google Scholar]
- Saadi A, Borck G, Boddaert N, Chekkour MC, Imessaoudene B, Munnich A, Colleaux L, Chaouch M. 2009. Compound heterozygous ASPM mutations associated with microcephaly and simplified cortical gyration in a consanguineous Algerian family. Eur J Med Genet 52: 180–184. 10.1016/j.ejmg.2009.03.013 [DOI] [PubMed] [Google Scholar]
- Saadi A, Verny F, Siquier-Pernet K, Bole-Feysot C, Nitschke P, Munnich A, Abada-Dendib M, Chaouch M, Abramowicz M, Colleaux L. 2016. Refining the phenotype associated with CASC5 mutation. Neurogenetics 17: 71–78. 10.1007/s10048-015-0468-7 [DOI] [PubMed] [Google Scholar]
- Sajid Hussain M, Marriam Bakhtiar S, Farooq M, Anjum I, Janzen E, Reza Toliat M, Eiberg H, Kjaer KW, Tommerup N, Noegel AA, et al. 2013. Genetic heterogeneity in Pakistani microcephaly families. Clin Genet 83: 446–451. 10.1111/j.1399-0004.2012.01932.x [DOI] [PubMed] [Google Scholar]
- Sanada K, Tsai LH. 2005. G protein βγ subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell 122: 119–131. 10.1016/j.cell.2005.05.009 [DOI] [PubMed] [Google Scholar]
- Sánchez I, Dynlacht BD. 2016. Cilium assembly and disassembly. Nat Cell Biol 18: 711–717. 10.1038/ncb3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Richardson A, Iyer DR, Saad OM, Zasadil L, Knouse KA, Wong YL, Rhind N, Desai A, Amon A. 2017. Chromosome mis-segregation generates cell cycle-arrested cells with complex karyotypes that are eliminated by the immune system. Dev Cell 41: 638–651. 10.1016/j.devcel.2017.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato R, Takanashi JI, Tsuyusaki Y, Kato M, Saitsu H, Matsumoto N, Takahashi T. 2016. Association between invisible basal ganglia and ZNF335 mutations: a case report. Pediatrics 138: e20160897. 10.1542/peds.2016-0897 [DOI] [PubMed] [Google Scholar]
- Sauer FC. 1935. Mitosis in the neural tube. J Comp Neurol 62: 377–405. 10.1002/cne.900620207 [DOI] [Google Scholar]
- Sauer ME, Walker BE. 1959. Radioautographic study of interkinetic nuclear migration in the neural tube. Proc Soc Exp Biol Med 101: 557–560. 10.3181/00379727-101-25014 [DOI] [PubMed] [Google Scholar]
- Scheidecker S, Etard C, Haren L, Stoetzel C, Hull S, Arno G, Plagnol V, Drunat S, Passemard S, Toutain A, et al. 2015. Mutations in TUBGCP4 alter microtubule organization via the γ-tubulin ring complex in autosomal-recessive microcephaly with chorioretinopathy. Am J Hum Genet 96: 666–674. 10.1016/j.ajhg.2015.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze B, Korr H. 1981. Cell kinetic studies of different cell types in the developing and adult brain of the rat and the mouse: a review. Cell Tissue Kinet 14: 309–325. [DOI] [PubMed] [Google Scholar]
- Seemanová E, Jarolím P. 1999. Nijmegen breakage syndrome (NBS). Ces Pediatr 54: 97–101. [Google Scholar]
- Sgourdou P, Mishra-Gorur K, Saotome I, Henagariu O, Tuysuz B, Campos C, Ishigame K, Giannikou K, Quon JL, Sestan N, et al. 2017. Disruptions in asymmetric centrosome inheritance and WDR62-Aurora kinase B interactions in primary microcephaly. Sci Rep 7: 43708. 10.1038/srep43708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen R, Al Tala S, Almoisheer A, Alkuraya FS. 2014. Mutation in PLK4, encoding a master regulator of centriole formation, defines a novel locus for primordial dwarfism. J Med Genet 51: 814–816. 10.1136/jmedgenet-2014-102790 [DOI] [PubMed] [Google Scholar]
- Shaheen R, Hashem A, Abdel-Salam GMH, Al-Fadhli F, Ewida N, Alkuraya FS. 2016. Mutations in CIT, encoding citron rho-interacting serine/threonine kinase, cause severe primary microcephaly in humans. Hum Genet 135: 1191–1197. 10.1007/s00439-016-1722-2 [DOI] [PubMed] [Google Scholar]
- Shaheen R, Maddirevula S, Ewida N, Alsahli S, Abdel-Salam GMH, Zaki MS, Tala S Al, Alhashem A, Softah A, Al-Owain M, et al. 2019. Genomic and phenotypic delineation of congenital microcephaly. Genet Med 21: 545–552. 10.1038/s41436-018-0140-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalev SA, Khayat M, Etty DS, Elpeleg O. 2015. Further insight into the phenotype associated with a mutation in the ORC6 gene, causing Meier-Gorlin syndrome 3. Am J Med Genet Part A 167: 607–611. 10.1002/ajmg.a.36906 [DOI] [PubMed] [Google Scholar]
- Shao W, Yang J, He M, Yu XY, Lee CH, Yang Z, Joyner AL, Anderson KV, Zhang J, Tsou MFB, et al. 2020. Centrosome anchoring regulates progenitor properties and cortical formation. Nature 580: 106–112. 10.1038/s41586-020-2139-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Aher A, Dynes NJ, Frey D, Katrukha EA, Jaussi R, Grigoriev I, Croisier M, Kammerer RA, Akhmanova A, et al. 2016. Centriolar CPAP/SAS-4 imparts slow processive microtubule growth. Dev Cell 37: 362–376. 10.1016/j.devcel.2016.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan CJ, McMahon JJ, Serdar LD, Silver DL. 2020. Dosage-dependent requirements of Magoh for cortical interneuron generation and survival. Dev 147: dev182295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Eyaid W, Mochida GH, Al-Moayyad F, Bodell A, Woods CG, Walsh CA. 2005. ASPM mutations identified in patients with primary microcephaly and seizures. J Med Genet 42: 725–729. 10.1136/jmg.2004.027706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Qalieh A, Lam MM, Keil JM, Kwan KY. 2019. Robust elimination of genome-damaged cells safeguards against brain somatic aneuploidy following Knl1 deletion. Nat Commun 10: 2588. 10.1038/s41467-019-10411-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla S, Iwamoto-Stohl LK, Zhu M, Zernicka-Goetz M. 2020. Autophagy-mediated apoptosis eliminates aneuploid cells in a mouse model of chromosome mosaicism. Nat Commun 11: 2958. 10.1038/s41467-020-16796-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir J-H, Barr AR, Nicholas AK, Carvalho OP, Khurshid M, Sossick A, Reichelt S, D'Santos C, Woods CG, Gergely F. 2011. A primary microcephaly protein complex forms a ring around parental centrioles. Nat Genet 43: 1147–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias DA, DeBonis S, Saoudi Y, Lebeau L, Crevel I, Cross R, Wade RH, Hackney D, Kozielski F. 2006. S-trityl-L-cysteine is a reversible, tight binding inhibitor of the human kinesin Eg5 that specifically blocks mitotic progression. J Biol Chem 281: 17559–17569. 10.1074/jbc.M511735200 [DOI] [PubMed] [Google Scholar]
- Sladky VC, Eichin F, Reiberger T, Villunger A. 2021. Polyploidy control in hepatic health and disease. J Hepatol 75: 1177–1191. 10.1016/j.jhep.2021.06.030 [DOI] [PubMed] [Google Scholar]
- Smart IHM. 2002. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Smart, Iain H M 12: 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen KF, Gabryjonczyk AM, Anselm E, Nigg EA, Stierhof YD. 2013. Human cep192 and cep152 cooperate in plk4 recruitment and centriole duplication. J Cell Sci 126: 3223–3233. [DOI] [PubMed] [Google Scholar]
- Soto M, Raaijmakers JA, Bakker B, Spierings DCJ, Lansdorp PM, Foijer F, Medema RH. 2017. p53 prohibits propagation of chromosome segregation errors that produce structural aneuploidies. Cell Rep 19: 2423–2431. 10.1016/j.celrep.2017.05.055 [DOI] [PubMed] [Google Scholar]
- Stouffs K, Stergachis AB, Vanderhasselt T, Dica A, Janssens S, Vandervore L, Gheldof A, Bodamer O, Keymolen K, Seneca S, et al. 2018. Expanding the clinical spectrum of biallelic ZNF335 variants. Clin Genet 94: 246–251. 10.1111/cge.13260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Nowakowski RS, Caviness VSJ. 1995. The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J Neurosci 15: 6046–6057. 10.1523/JNEUROSCI.15-09-06046.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CA, Topper S, Ward Melver C, Stein J, Reeder A, Arndt K, Das S. 2014. The first case of CDK5RAP2-related primary microcephaly in a non-consanguineous patient identified by next generation sequencing. Brain Dev 36: 351–355. 10.1016/j.braindev.2013.05.001 [DOI] [PubMed] [Google Scholar]
- Tang CJC, Lin SY, Hsu W Bin, Lin YN, Wu CT, Lin YC, Chang CW, Wu KS, Tang TK. 2011. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J 30: 4790–4804. 10.1038/emboj.2011.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna E, Huttner WB. 2010. Neural progenitor nuclei IN motion. Neuron 67: 906–914. 10.1016/j.neuron.2010.08.027 [DOI] [PubMed] [Google Scholar]
- Taverna E, Götz M, Huttner WB. 2014. The cell biology of meurogenesis: toward an understanding of the development and evolution of the neocortex. 30: 465–502. [DOI] [PubMed] [Google Scholar]
- Trimborn M, Bell SM, Felix C, Rashid Y, Jafri H, Griffiths PD, Neumann LM, Krebs A, Reis A, Sperling K, et al. 2004. Mutations in microcephalin cause aberrant regulation of chromosome condensation. Am J Hum Genet 75: 261–266. 10.1086/422855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimborn M, Schindler D, Neitzel H, Hirano T. 2006. Misregulated chromosome condensation in MCPH1 primary microcephaly is mediated by condensin II. Cell Cycle 5: 322–326. 10.4161/cc.5.3.2412 [DOI] [PubMed] [Google Scholar]
- Tsai MY, Wang S, Heidinger JM, Shumaker DK, Adam SA, Goldman RD, Zheng Y. 2006. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science 311: 1887–1893. 10.1126/science.1122771 [DOI] [PubMed] [Google Scholar]
- Turrero García M, Chang Y, Arai Y, Huttner WB. 2016. S-phase duration is the main target of cell cycle regulation in neural progenitors of developing ferret neocortex. J Comp Neurol 524: 456–470. 10.1002/cne.23801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler WA, Haydar TF. 2013. Multiplex genetic fate mapping reveals a novel route of neocortical neurogenesis, which is altered in the Ts65Dn mouse model of down syndrome. Ann Intern Med 158: 5106–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. 2010. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr Biol 20: 1666–1671. 10.1016/j.cub.2010.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Breugel M, Hirono M, Andreeva A, Yanagisawa HA, Yamaguchi S, Nakazawa Y, Morgner N, Petrovich M, Ebong IO, Robinson C V, et al. 2011. Structures of SAS-6 suggest its organization in centrioles. Science 331: 1196–1199. 10.1126/science.1199325 [DOI] [PubMed] [Google Scholar]
- Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanová E, Cooper PR, Nowak NJ, et al. 1998. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 93: 467–476. 10.1016/S0092-8674(00)81174-5 [DOI] [PubMed] [Google Scholar]
- Viais R, Fariña-Mosquera M, Villamor-Payà M, Watanabe S, Palenzuela L, Lacasa C, Lüders J. 2021. Augmin deficiency in neural stem cells causes p53-dependent apoptosis and aborts brain development. Elife 10: e67989. 10.7554/eLife.67989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield JG, Bonaccorsi S, Gatti M. 2001. The Drosophila protein Asp is involved in microtubule organization during spindle formation and cytokinesis. J Cell Biol 153: 637–648. 10.1083/jcb.153.4.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB, Shi SH. 2009. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature 461: 947–955. 10.1038/nature08435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wu X, Du L, Zheng J, Deng S, Bi X, Chen Q, Xie H, Férec C, Cooper DN, et al. 2018. Identification of compound heterozygous variants in the noncoding RNU4ATAC gene in a Chinese family with two successive foetuses with severe microcephaly. Hum Genomics 12: 3. 10.1186/s40246-018-0135-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I, Kim JE, Minn K, Chini CC, Mer G, Chen J. 2006. The tandem BRCT domain of 53BP1 is not required for its repair function. J Biol Chem 281: 38472–38477. 10.1074/jbc.M607577200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Meitinger F, Shiau AK, Oegema K, Desai A. 2020. Centriole-independent mitotic spindle assembly relies on the PCNT-CDK5RAP2 pericentriolar matrix. J Cell Biol 219: e202006010. 10.1083/jcb.202006010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BAA, Bonday ZQ, Putkey FR, Kops GJPL, Silk AD, Cleveland DW. 2003. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol 162: 551–563. 10.1083/jcb.200303167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss K, Ekhilevitch N, Cohen L, Bratman-Morag S, Bello R, Martinez AF, Hadid Y, Shlush LI, Kurolap A, Paperna T, et al. 2020. Identification of a novel PCNT founder pathogenic variant in the Israeli Druze population. Eur J Med Genet 63: 103643. 10.1016/j.ejmg.2019.03.007 [DOI] [PubMed] [Google Scholar]
- Willems M, Geneviève D, Borck G, Baumann C, Baujat G, Bieth E, Edery P, Farra C, Gerard M, Héron D, et al. 2010. Molecular analysis of pericentrin gene (PCNT) in a series of 24 Seckel/microcephalic osteodysplastic primordial dwarfism type II (MOPD II) families. J Med Genet 47: 797–802. 10.1136/jmg.2009.067298 [DOI] [PubMed] [Google Scholar]
- Wolf B, Oghabian A, Akinyi MV, Hanks S, Tromer EC, Hooff JJE, Voorthuijsen L, Rooijen LE, Verbeeren J, Uijttewaal ECH, et al. 2021. Chromosomal instability by mutations in the novel minor spliceosome component CENATAC. EMBO J 40: e106536. 10.15252/embj.2020106536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB, Wueseke O, Hyman AA. 2014. Pericentriolar material structure and dynamics. Philos Trans R Soc B Biol Sci 369: 20130459. 10.1098/rstb.2013.0459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods CG. 2004. Human microcephaly. Curr Opin Neurobiol 14: 112–117. 10.1016/j.conb.2004.01.003 [DOI] [PubMed] [Google Scholar]
- Woods CG, Bond J, Enard W. 2005. Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. Am J Hum Genet 76: 717–728. 10.1086/429930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SX, Goebbels S, Nakamura K, Nakamura K, Kometani K, Minato N, Kaneko T, Nave KA, Tamamaki N. 2005. Pyramidal neurons of upper cortical layers generated by NEX-positive progenitor cells in the subventricular zone. Proc Natl Acad Sci 102: 17172–17177. 10.1073/pnas.0508560102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Grzonka M, Meyer-Gerards C, Mack M, Figge R, Bazzi H. 2021. Gradual centriole maturation associates with the mitotic surveillance pathway in mouse development. EMBO Rep 22: e51127. 10.15252/embr.202051127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Jaiswal M, Charng WL, Gambin T, Karaca E, Mirzaa G, Wiszniewski W, Sandoval H, Haelterman NA, Xiong B, et al. 2014. A Drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell 159: 200–214. 10.1016/j.cell.2014.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita D, Shintomi K, Ono T, Gavvovidis I, Schindler D, Neitzel H, Trimborn M, Hirano T. 2011. MCPH1 regulates chromosome condensation and shaping as a composite modulator of condensin II. J Cell Biol 194: 841–854. 10.1083/jcb.201106141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YJ, Baltus AE, Mathew RS, Murphy EA, Evrony GD, Gonzalez DM, Wang EP, Marshall-Walker CA, Barry BJ, Murn J, et al. 2012. Microcephaly gene links trithorax and REST/NRSF to control neural stem cell proliferation and differentiation. Cell 151: 1097–1112. 10.1016/j.cell.2012.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Hu X, Ma J, Shi S-H. 2021. Centrosome regulation and function in mammalian cortical neurogenesis. Curr Opin Neurobiol 69: 256–266. 10.1016/j.conb.2021.06.003 [DOI] [PubMed] [Google Scholar]
- Yigit G, Brown KE, Kayserili H, Pohl E, Caliebe A, Zahnleiter D, Rosser E, Bögershausen N, Uyguner ZO, Altunoglu U, et al. 2015. Mutations in cdk5rap2 cause Seckel syndrome. Mol Genet Genomic Med 3: 467–480. 10.1002/mgg3.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TW, Mochida GH, Tischfield DJ, Sgaier SK, Flores-Sarnat L, Sergi CM, Topçu M, McDonald MT, Barry BJ, Felie JM, et al. 2010. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat Genet 42: 1015–1020. 10.1038/ng.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarate YA, Kaylor JA, Bosanko K, Lau S, Vargas J, Gao H. 2016. First clinical report of an infant with microcephaly and CASC5 mutations. Am J Med Genet Part A 170: 2215–2218. 10.1002/ajmg.a.37726 [DOI] [PubMed] [Google Scholar]
- Zhang W, Yang SL, Yang M, Herrlinger S, Shao Q, Collar JL, Fierro E, Shi Y, Liu A, Lu H, et al. 2019a. Modeling microcephaly with cerebral organoids reveals a WDR62–CEP170–KIF2A pathway promoting cilium disassembly in neural progenitors. Nat Commun 10: 2612. 10.1038/s41467-019-10497-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li H, Pang J, Peng Y, Shu L, Wang H. 2019b. Novel SASS6 compound heterozygous mutations in a Chinese family with primary autosomal recessive microcephaly. Clin Chim Acta 491: 15–18. 10.1016/j.cca.2019.01.007 [DOI] [PubMed] [Google Scholar]
- Zheng X, Gooi LM, Wason A, Gabriel E, Mehrjardi NZ, Yang Q, Zhang X, Debec A, Basiri ML, Avidor-Reiss T, et al. 2014. Conserved TCP domain of Sas-4/CPAP is essential for pericentriolar material tethering during centrosome biogenesis. Proc Natl Acad Sci 111: E354–E363. 10.1073/pnas.1317535111 [DOI] [PMC free article] [PubMed] [Google Scholar]