ABSTRACT
The causative agents of recurrent Escherichia coli bacteremia can be genetically identical or discordant, but the differences between them remain unclear. This study aimed to explore these differences, with regard to their clinical and microbiological features. Patients were recruited from a Japanese tertiary teaching hospital based on blood culture data and the incidence of recurrent E. coli bacteremia. We compared the patients' clinical and microbiological characteristics between the two groups (those with identical or discordant E. coli bacteremia) divided by the result of enterobacterial repetitive intergenic consensus PCR. Among 70 pairs of recurrent E. coli bacteremia strains, 49 pairs (70%) were genetically identical. Patients with genetically identical or discordant E. coli bacteremia were more likely to have renal failure or neoplasms, respectively. The virulence factor (VF) scores of genetically identical E. coli strains were significantly higher than those of genetically discordant strains, with the prevalence of eight VF genes being significantly higher in genetically identical E. coli strains. No significant differences were found between the two groups regarding antimicrobial susceptibility and biofilm formation potential. This study showed that genetically identical E. coli bacteremia strains have more VF genes than genetically discordant strains in recurrent E. coli bacteremia.
IMPORTANCE Escherichia coli causes bloodstream infection, although not all strains are pathogenic to humans. In some cases, this infection reoccurs, and several reports have described the clinical characteristics and/or molecular microbiology of recurrent Escherichia coli bacteremia. However, these studies focused on patients with specific characteristics, and they included cases caused by microorganisms other than Escherichia coli. Hence, little is known about the pathogenicity of Escherichia coli isolated from the recurrent one. The significance of our study is in evaluating the largest cohorts to date, as no cohort studies have been conducted on this topic.
KEYWORDS: recurrent Escherichia coli bacteremia, enterobacterial repetitive intergenic consensus polymerase chain reaction, genetic identity, virulence factor
INTRODUCTION
Escherichia coli is the most frequent pathogen causing bloodstream infections in many situations (1–4). However, not all E. coli strains are pathogenic to humans, and commensal E. coli represents a colonizer of the human gastrointestinal microbiota (5). The majority of pathogenic E. coli strains belong to the phylogenetic group B2 (6). Conversely, groups A and B1 account for a larger proportion of commensal strains (7).
E. coli has various virulence factors (VFs), including toxins, adhesins, siderophores, and polysaccharide capsules (8, 9). Numerous VFs are related to the pathogenicity of E. coli, with a wide range of pathogenic activities (10). In addition to VFs, antimicrobial resistance and biofilm formation also make it difficult to eliminate microorganisms from the human body (11, 12).
Two or more E. coli bacteremia episodes occur in 3–28% of all cases (13–20), although the prevalence of recurrent E. coli bacteremia depends on the situation and its definition. Several reports have described the clinical characteristics and/or molecular microbiology of recurrent E. coli bacteremia (13, 15, 18–21). However, these studies are case series, studies focused on patients with specific characteristics, such as hematological malignancy, or studies of bacteremia caused by microorganisms other than E. coli. Moreover, little is known about the pathogenicity of E. coli isolated from recurrent E. coli bacteremia.
The aim of this study was to identify the differences in clinical and microbiological characteristics of E. coli bacteremia depending on genetic identity. To our knowledge, this study is the largest cohort ever conducted solely on patients with recurrent E. coli bacteremia.
RESULTS
Inclusion of the study participants and differentiation by using ERIC-PCR.
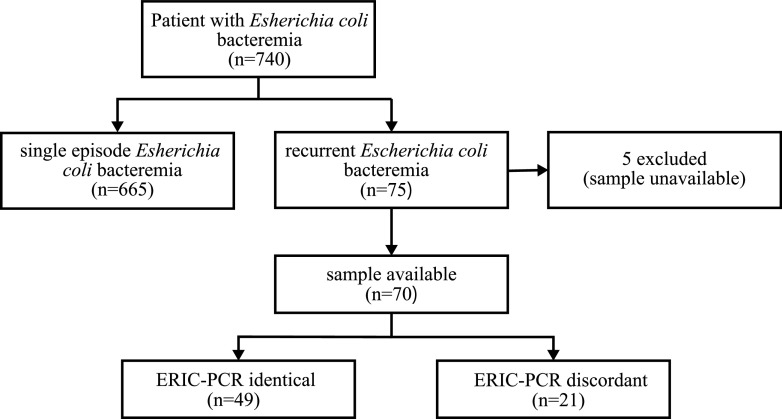
In total, 740 patients with bloodstream infections caused by E. coli were identified between April 2013 and March 2019. Among them, 75 patients (10.1%) had a second episode of E. coli bacteremia. Five patients were excluded because their culture samples were unavailable. ERIC-PCR was performed by using 70 pairs of E. coli isolates from the remaining 70 patients. Among the 70 pairs of E. coli isolates, 49 pairs (70%) were genetically identical (Fig. 1).
FIG 1.
Flow chart of the classification of study participants according to the ERIC-PCR results. ERIC-PCR, enterobacterial repetitive intergenic consensus PCR.
Comparison of patients with genetically identical and genetically discordant E. coli bacteremia.
The demographic and clinical characteristics of patients in the two groups are listed in Table 1. Most of the clinical characteristics were not significantly different between the two groups. Compared with patients with genetically discordant E. coli bacteremia, those with genetically identical E. coli bacteremia were significantly more likely to have renal failure (P = 0.0079). In contrast, patients with genetically discordant E. coli bacteremia were more likely to have neoplasms (P = 0.038).
TABLE 1.
Clinical characteristics of patients with recurrent E. coli bacteremiaa
| Characteristics | ERIC-PCR identical strains (n = 49) | ERIC-PCR discordant strains (n = 21) | P-value |
|---|---|---|---|
| Demographics | |||
| Median age (IQR) | 72 (63–78.5) | 75 (68–80) | 0.175 |
| Male-no. (%) | 29 (59.2) | 11 (52.3) | 0.61 |
| Acquisition-no. (%) | 0.387 | ||
| Hospital-acquired | 16 (32.7) | 4 (19.0) | |
| Community-acquired | 33 (67.3) | 17 (81.0) | |
| Underlying disease/condition-no. (%) | |||
| Diabetes mellitus | 21 (42.9) | 14 (66.7) | 0.117 |
| Neoplasm | 21 (42.9) | 15 (71.4) | 0.038 |
| Immunosuppressant use | 20 (40.8) | 4 (19.1) | 0.103 |
| Transplantation | 5 (10.2) | 1 (4.8) | 0.661 |
| Renal failure | 17 (34.7) | 1 (4.8) | 0.0079 |
| Surgery within 30 days | 2 (4.1) | 0 (0.0) | 1 |
| Foreign body | 14 (28.6) | 5 (23.8) | 0.776 |
| Chemotherapy | 10 (20.4) | 6 (28.6) | 0.538 |
| Source of bacteremia-no. (%) | 0.072 | ||
| Biliary tract | 13 (26.5) | 12 (57.1) | |
| Urinary | 19 (38.8) | 3 (14.3) | |
| Others | 16 (32.7) | 7 (33.3) | |
| Pitt bacteremia score ≧4-no. (%) | 2 (4.1) | 0 (0.0) | 1 |
| Relapse within 60 days-no. (%) | 20 (40.8) | 7 (33.3) | 0.603 |
| Polymicrobial bacteremia-no. (%) | 4 (8.2) | 3 (14.3) | 0.421 |
IQR, interquartile range; ERIC-PCR, enterobacterial repetitive intergenic consensus PCR.
Distribution of phylogenetic groups and sequence types among recurrent E. coli bacteremia isolates.
As shown in Tables 2 and 3, there were significant differences between genetically identical strains and discordant strains in the distribution of phylogenetic groups (P = 0.0001) and sequence types (STs) (P = 0.0063). In both groups, that with the largest number of strains was categorized as the phylogenetic group B2. The percentages of phylogenetic groups B2, ST 131, ST 73, and ST 1193 were higher in genetically identical strains than in genetically discordant strains.
TABLE 2.
Distribution of phylogenetic groups among recurrent E. coli bacteremia isolates
| Phylogenetic group | ERIC-PCR identical strains (n = 49) (no. [%])a | ERIC-PCR discordant strains (n = 21) (no. [%]) | P-value |
|---|---|---|---|
| A | 0 (0.0) | 1 (4.8) | 0.0001 |
| B1 | 0 (0.0)* | 6 (28.6)* | |
| B2 | 43 (87.8)* | 10 (47.6)* | |
| C | 0 (0.0) | 0 (0.0) | |
| D | 0 (0.0) | 0 (0.0) | |
| E | 4 (8.2) | 2 (9.5) | |
| F | 2 (4.1) | 2 (9.5) |
ERIC-PCR, enterobacterial repetitive intergenic consensus PCR.
*Significant, P = 0.002 (Fisher's exact test with the Bonferroni correction).
TABLE 3.
Distribution of STs among recurrent E. coli bacteremia isolates
| Sequence type | ERIC-PCR identical strains (n = 49) (no. [%])a | ERIC-PCR discordant strains (n = 21) (no. [%]) | P-value |
|---|---|---|---|
| ST131 | 18 (36.7) | 3 (14.2) | 0.0063 |
| ST95 | 9 (18.4) | 4 (19.1) | |
| ST73 | 7 (14.3) | 0 (0.0) | |
| ST1193 | 4 (8.2) | 0 (0.0) | |
| ST357 | 1 (2.0) | 1 (4.8) | |
| Others | 10 (20.4) | 13 (61.9) |
ERIC-PCR, enterobacterial repetitive intergenic consensus PCR.
In post hoc analyses performed to determine which groups differed, the distribution of STs was not significantly different for all combinations. The phylogenetic group B2 was significantly more common than phylogenetic group B1 in the genetically identical strains (Bonferroni-corrected P = 0.002).
Distribution of VFs.
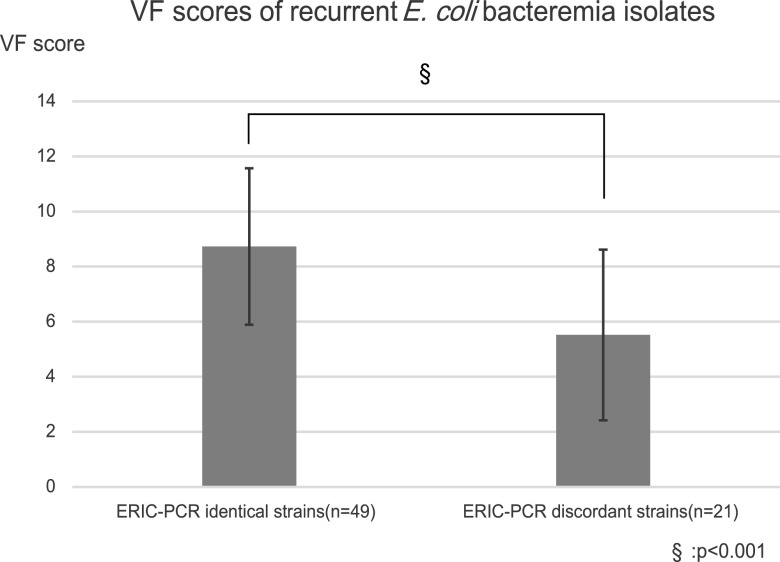
The difference in the distribution of 20 VFs between genetically identical and discordant strains is provided in Table 4. Of the 20 VFs evaluated, the prevalence rates of eight VF genes (sfaD/E [P = 0.049], iha [P = 0.0095], fyuA [P = 0.0019], cnf1 [P = 0.027], sat [P = 0.017], hlyA [P = 0.027], KpsMT2 [P = 0.041], and usp [P = 0.002]) were significantly higher in genetically identical E. coli strains than in the discordant strains. In addition, VF scores of genetically identical E. coli strains were significantly higher than those of genetically discordant strains (P = 0.0003) (Fig. 2).
TABLE 4.
Distribution of virulence factor (VF) genes among recurrent E. coli bacteremia isolates
| VF genes | ERIC-PCR identical strains (n = 49) (no. [%])a | ERIC-PCR discordant strains (n = 21) (no. [%]) | P-value |
|---|---|---|---|
| afaB/C | 3 (6.1) | 1 (4.8) | 1 |
| fimH | 49 (100.0) | 20 (95.2) | 0.3 |
| sfaD/E | 9 (18.4) | 0 (0.0) | 0.049 |
| papC | 21 (42.9) | 4 (19.0) | 0.064 |
| papG2 | 14 (28.6) | 3 (14.3) | 0.24 |
| iha | 26 (53.1) | 4 (19.0) | 0.0095 |
| iucD | 29 (59.2) | 8 (38.1) | 0.124 |
| iutA | 29 (59.2) | 8 (38.1) | 0.124 |
| fyuA | 46 (93.9) | 13 (61.9) | 0.0019 |
| iroN | 13 (26.5) | 4 (19.0) | 0.561 |
| cnf1 | 11 (22.4) | 0 (0.0) | 0.027 |
| sat | 25 (51.0) | 4 (19.0) | 0.017 |
| hlyA | 11 (22.4) | 0 (0.0) | 0.027 |
| KpsMT2 | 39 (79.6) | 11 (52.4) | 0.041 |
| cvaC | 3 (6.1) | 4 (19.0) | 0.186 |
| ibeA | 10 (20.4) | 3 (14.3) | 0.741 |
| ompT | 3 (6.1) | 4 (19.0) | 0.186 |
| TcpC | 8 (16.3) | 2 (9.5) | 0.712 |
| usp | 42 (85.7) | 10 (47.6) | 0.002 |
| traT | 38 (77.6) | 13 (61.9) | 0.242 |
ERIC-PCR, enterobacterial repetitive intergenic consensus PCR.
FIG 2.
Differences in the proportion of VF genes between ERIC-PCR identical strains and genetically discordant strains. ERIC-PCR identical strains have significantly more VF genes than genetically discordant strains. ERIC-PCR, enterobacterial repetitive intergenic consensus PCR; VF, virulence factor.
Possession of ESBL genes/antimicrobial susceptibilities.
Among 70 E. coli bacteremia isolates that caused the first bacteremia episode, 13 strains (18.6%) carried ESBL genes. Of those 13 ESBL-producing E. coli strains, 10 belonged to CTX M-9 group and three to CTX M-1 group. All isolates that were positive in the ESBL phenotypic test carried ESBL genes.
As shown in Table 5, no significant differences in antimicrobial susceptibilities including ESBL production were found between the two groups.
TABLE 5.
Antimicrobial susceptibilities among recurrent E. coli bacteremia isolatesa
| Antimicrobial susceptibilities | ERIC-PCR identical strains (n = 49) (no. [%]) | ERIC-PCR discordant strains (n = 21) (no. [%]) | P-value |
|---|---|---|---|
| ESBL production | 11 (22.4) | 2 (9.5) | 0.317 |
| Quinolone resistance | 22 (44.9) | 4 (19.1) | 0.059 |
| Ampicillin resistance | 28 (57.1) | 8 (38.1) | 0.194 |
ERIC-PCR, enterobacterial repetitive intergenic consensus PCR; ESBL, extended-spectrum β-lactamase.
Biofilm formation ability.
Compared with genetically discordant strains, there were no significant differences in the biofilm formation abilities of genetically identical strains (Table 6).
TABLE 6.
Biofilm formation ability among recurrent E. coli bacteremia isolatesa
| Biofilm formation | ERIC-PCR identical strains (n = 49) (no. [%]) | ERIC-PCR discordant strains (n = 21) (no. [%]) | P-value |
|---|---|---|---|
| Biofilm formation (LB Lennox broth) | 21 (42.9) | 6 (28.6) | 0.296 |
| Biofilm formation (BHI broth) | 17 (34.7) | 8 (38.1) | 0.792 |
ERIC-PCR, enterobacterial repetitive intergenic consensus PCR; BHI, brain heart infusion.
DISCUSSION
To our knowledge, among comparative studies limited to recurrent E. coli bacteremia classified by genetic homology, this study evaluated the largest cohorts to date. The prevalence of recurrent E. coli bacteremia in this study was 10.1%, which is within the range reported in previous studies (13–20). However, the prevalence of recurrent E. coli bacteremia varies across the studies (13–20). This variation is presumably influenced by the definition of recurrence, study population, and study design.
In previous studies that categorized recurrent E. coli bacteremia by genetic identity, E. coli strains were genetically characterized using pulsed-field gel electrophoresis (PFGE) (15, 20), repetitive extragenic palindromic sequence PCR (13), and ribotyping (15, 21). Compared with other PCR-based typing techniques, PFGE is a powerful and accurate technique for strain typing, but it is laborious because of its low throughput (22). In a large sample size study, Casarez et al. reported that ERIC-PCR was similarly effective to PFGE in the differentiation of E. coli strains (23). With the advent of next-generation genome sequencing techniques, whole-genome sequencing (WGS) provides higher discrimination power for typing of pathogens including E coli compared to PCR-based typing techniques (24, 25). To the best of our knowledge, there are no studies that directly compare the performance of WGS to that of ERIC-PCR, so the quantitative difference in resolution between these two methods is unknown. A study of recurrent Staphylococcus aureus bacteremia showed that the amount of SNP variation by WGS is consistent with the PFGE, and authors concluded that no novel insights were provided by WGS in the study (26). Although the result may not be applicable to our study because of the difference in the species of bacteria studied, as for repeated bacteremia in a single patient, the result of WGS may be consistent with that of PCR-based typing techniques, which has lower discrimination power compared to WGS. This consistency may well be the case in repeated infections due to the colonization of the same bacteria. WGS technique has changed the landscape of genomic science; however, its high costs and the need for bioinformatics analysis can contribute to delays of its replacement of PCR-based typing techniques (27). Considering the above, we used ERIC-PCR to classify the genetic identity of recurrent E. coli bacteremia in this study, and we found that 70% of the recurrent E. coli bacteremia cases were caused by genetically identical strains. This percentage was similar to that reported for ESBL-producing E. coli and Klebsiella pneumoniae (67.8%) (20) but was higher than that reported in a previous study on E. coli (47.7%) (13).
The comparison between genetically identical strains and genetically discordant strains showed neoplasms as a factor that correlated with genetic discordance. In contrast, patients with genetically identical E. coli bacteremia were significantly more likely to have renal failure. In a previous study of recurrent bacteremia caused by ESBL-producing E. coli and Klebsiella pneumoniae, there was no significant difference in comorbidity, including neoplasms and renal failure, between the two groups based on genetic identity (20). Although it is difficult to interpret the results of our study because there are no similar studies referring to these comorbidities on E. coli alone, genetically identical E. coli may have not been eliminated from the patients during the treatment of the prior bacteremia episodes. Zerr et al. indicated that Enterobacteriales strains of the same STs and resistance genotypes can persistently colonize the intestinal tract even after treatment with effective antimicrobial agents, which may lead to recurrent infections (28).
Among the E. coli strains included in this study, phylogenetic group B2 strains accounted for the largest percentage in both groups. However, the proportion of phylogenetic group B2 strains was significantly higher in the genetically identical group. Given that phylogenetic group B2 strains are more likely to cause infections than other strains (6), this result is logical. As for STs, the percentages of ST 131, ST 73, and ST 1193, which belong to phylogenetic group B2, were higher in the genetically identical group. However, the proportion of the ST 95 strain, which also belongs to phylogenetic group B2, was almost the same between the two groups. Thus, not all phylogenetic B2 strains may be associated with genetic identity. The results of the post hoc analysis did not reveal any differences in the combinations of STs between the two groups. The lack of significant differences in the STs between groups may be partly due to limited statistical power due to the small sample size. ST 131 is one of the main ESBL-producing E. coli clones isolated worldwide (29); however, there were no significant differences in antimicrobial susceptibility between the two groups, including ESBL production.
Among the eight VF genes that showed significant differences, cnf1, hlyA, and sat encode toxins; sfaD/E and iha regulate adhesins; fyuA regulates siderophores; KpsMT2 and usp encode capsules and uropathogenic-specific proteins, respectively. Previous studies that analyzed recurrent urinary tract infection (UTI) in women have showed the presence of numerous VF genes that regulate various factors, including toxins, adhesins, iron-acquisition systems, capsules, fimbriae, and other factors in relation to recurrent E. coli UTI (30–33). In our study, genetically identical strains possessed significantly more VFs than genetically discordant strains. There was no statistically significant difference, but there was a trend toward more UTIs in the genetically identical group. According to a previous report, E. coli bacteremia strains originating from urinary tract infective foci harbored more VF genes than those from non-urinary tract infective foci (34). Furthermore, the percentages of ST 131, ST 73, and ST 1193 were higher in the genetically identical group, as aforementioned. There are some reports indicating that these strains carry prototypic VF genes (35, 36). Thus, the high prevalence of UTIs and several STs in the genetically identical group may be one of the reasons for the higher VF score. However, making comparisons of these studies is difficult because their selection of VF genes differs from each other. Moreover, only a few reports of recurrent E. coli bacteremia have described the distribution of VF genes; therefore, further investigation is required to clarify the distribution of VF genes in recurrent E. coli bacteremia and how VFs contribute to recurrence. The roles of VFs are diverse, suggesting the involvement of multiple VFs in the establishment of recurrent infections.
Several VFs have been reported to be related to biofilm formation in ESBL E. coli strains (37). Biofilms are a layer of bacteria attached to biological tissues or artificial device surfaces, and biofilms have been reported to have several advantages for the survival of bacteria: increased capacity of bacterial conjugation (38), increased interspecific metabolic cooperation (39), and increased needs for higher concentrations of antibiotics (40). A study focused on E. coli associated with recurrent cystitis showed that recurrent infection isolates had better biofilm formation capability than single infection isolates (41), although this difference was not significant.
This study has several limitations. First, our study was a hospital-based, retrospective, single-center study. The description of E. coli strains identified in this study might not be entirely generalizable to E. coli strains collected in different situations. Second, we analyzed only in-hospital data, and the sample size was too small to perform multivariate analyses. For post hoc multiple comparison analyses, the small sample size limited the statistical power to detect significant differences. Third, relapse and reinfection were not distinguished in this study, both of which can be included in genetically identical strains group. To exclude cases of relapse to the extent possible, we selected cases in which the interval between the first and second positive blood cultures was more than 4 weeks. Fourth, WGS was not performed as mentioned above, which leads to the reduction of discrimination power regarding genetic identity. Despite these limitations, this study can help identify the clinical and microbiological characteristics that predispose patients to recurrent E. coli bacteremia.
In conclusion, among recurrent E. coli bacteremia strains, genetically identical strains have significantly more VF genes than genetically discordant strains. We have also shown the clinical characteristics of patients with recurrent E. coli bacteremia. Further research is required to explore how VFs may contribute to E. coli bacteremia recurrence.
MATERIALS AND METHODS
Study design and patients.
This retrospective study was conducted at the University of Tokyo Hospital, a 1,217-bed tertiary teaching hospital in Japan. Patients with E. coli bacteremia were selected from a database of blood culture results between April 2013 and March 2019. Patients with recurrent E. coli bacteremia were selected from these patients. Information on demographics, underlying diseases/conditions, site of acquisition, source of bacteremia, Pitt bacteremia score (42) calculated on the day of positive blood culture, time between first and second episode, and presence or absence of polymicrobial infection was extracted from the medical records. Recurrent E. coli bacteremia strains were categorized into two groups based on genetic identity. As for microbiological characteristics, we analyzed E. coli strains that caused the first bacteremia episode. The study protocol was approved by the ethics committee of the Graduate School of Medicine and Faculty of Medicine, University of Tokyo (approval number 10799). The requirement for written informed consent was waived because of the observational, retrospective design of the study.
Classification by genetic identity using ERIC PCR.
Enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) analysis was performed using primers ERIC-1 and ERIC-2R, as described previously (43). PCR was performed using EmeraldAmp MAX PCR Master Mix (TaKaRa Bio Inc., Shiga, Japan). The results of ERIC-PCR were compared using GelJ v2.0, a software tool for analyzing DNA fingerprint gel images (44). Isolates with more than 90% identical profiles were defined as genetically identical pairs.
Definitions.
Recurrent E. coli bacteremia was defined as a second episode of E. coli bacteremia occurring at least 4 weeks from the date of the positive blood culture of the first E. coli bacteremia episode. All patients' first episodes of bacteremia were appropriately treated prior to the second episode. E. coli bacteremia was categorized as community-acquired or nosocomial. Nosocomial E. coli bacteremia was defined as that which occurred at 48 h or more after hospital admission, and community-acquired E. coli bacteremia was defined as anything other than nosocomial E. coli bacteremia.
The diagnosis of UTI was based on the detection of E. coli that has grown in the quantity of 10^5 CFU per milliliter in the urine culture, with clinical symptoms, such as fever (≥37.5°C), pain during urination, or costovertebral angle tenderness.
Biliary tract infection was defined based on the Tokyo guidelines 2018 (45). A definite diagnosis of cholangitis was made when all the following were positive: i) systemic inflammation signs, such as fever and/or shaking chills, elevation of inflammation sign on blood test; ii) signs of cholestasis, such as jaundice, abnormal liver function, and biliary function tests; and ii) biliary dilatation or evidence of the etiology on imaging. Definite cholecystitis was defined as the patient meeting all the following criteria: i) localized clinical signs, such as Murphy’s sign and pain or tenderness in the right upper quadrant; ii) systemic inflammation signs, such as fever, elevation of C-reactive protein level or white blood cell count; and iii) imaging findings and characteristic of acute cholecystitis.
Renal failure was defined as having a serum creatinine level ≥1.5 mg/dl or requirement of dialysis therapy.
Identification of E. coli and antimicrobial susceptibility testing.
Blood samples were cultured using the BacT/Alert 3D Microbial Detection System (bioMérieux, Inc., Durham, NC).
All E. coli isolates were identified using the MicroScan WalkAway system (Beckman Coulter, Tokyo, Japan) or matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI Biotyper; Bruker Daltonics, Germany). Antimicrobial susceptibility testing was performed using a MicroScan WalkAway system (Beckman Coulter K.K., Tokyo, Japan). Antimicrobial susceptibility was defined according to the Clinical and Laboratory Standards Institute M100, 28th ed (46).
Detection of extended-spectrumβ-lactamase genes, phylogenetic classification, and multilocus sequence typing.
The phenotypic detection of extended-spectrum β-lactamase (ESBL) production was performed according to the Clinical and Laboratory Standards Institute M100, 28th ed (46). For confirmation of ESBL genotype, PCR analysis was performed, as described previously (47).
Affiliation of E. coli isolates to phylogenetic groups, such as A, B1, B2, C, D, E, and F, was determined by the PCR method, as described previously (6, 48–51). For the rapid identification of E. coli STs 69, 73, 95, and 131, a multilocus sequence typing PCR method (52) was used. Other STs were determined according to the Achtman's seven-locus multilocus sequence typing method. Fragments of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) distributed around the E. coli chromosome were amplified by using primers, as described previously (53). PCR was performed using TaKaRa Ex Taq (TaKaRa Bio Inc., Shiga, Japan). The amplified PCR products were sequenced bidirectionally using the Sanger DNA sequencing method at Eurofins Genomics (Ebersberg, Germany), and the sequences were assembled into single contigs using ChromasPro software (version 2.1.8; Technelysium Pty. Ltd, South Brisbane, Australia). The allelic profile for each E. coli isolate was determined, and the STs were assigned using the Enterobase genome database (http://enterobase.warwick.ac.uk/mlst/dbs/Ecoli).
Virulence factor determination.
E.coli isolates were screened for carriage of the following 20 VF genes: afaB/C (afimbrial adhesin), fimH (type 1 fimbriae), sfaD/E (S fimbriae), papC, papG2 (P fimbriae), iha (IrgA homologue adhesin), iucD (aerobactin), iutA (aerobactin), fyuA (yersiniabactin), iroN (catecholate siderophore), cnf1 (cytotoxic necrotizing factor 1), hlyA (alpha hemolysin), sat (secreted autotransporter toxin), KpsMT2 (protectin), usp (uropathogenic specific protein), ibeA (invasion of brain endothelium), traT (serum/complement resistance), cvaC (colicin V), ompT (outer membrane protease T), and TcpC (Toll/interleukin-1 receptor domain-containing protein C). PCR assays were used to reveal the prevalence of these virulence genes using specific primers, as described previously (54–66). PCR was performed using EmeraldAmp MAX PCR Master Mix (TaKaRa Bio Inc., Shiga, Japan). The primers used in this study are listed in Table S1.
VF scores were defined as the number of VFs detected for each isolate.
Biofilm formation assay.
The biofilm formation of E. coli strains was investigated in microtiter plates as described previously (67), with minor modifications. Briefly, each E. coli strain was cultured overnight in 10 ml LB Lennox broth or brain heart infusion (BHI) broth at 37°C, and then the bacterial culture was diluted into fresh LB Lennox broth or BHI broth (Becton, Dickinson, and Company, Sparks, MD) and adjusted to the turbidity of a 0.5 McFarland standard (McFarland Densitometer DEN-1B, Wakenbtech Co., Ltd., Kyoto, Japan). For LB Lennox broth, 1 g tryptone, 0.5 g yeast extract, and 0.5 g NaCl were suspended in 100 ml of distilled water. The dilutions were added to a polystyrene 96-well dish (100 μl/well) with six replicate wells for each strain, and the microtiter plate was incubated for 24 h at 37°C. After incubation, each well was washed three times with sterile distilled water and 0.1% crystal violet solution (Wako Pure Chemical Industries, Ltd, Osaka, Japan), and incubated at room temperature for 10 min. The microtiter plate was rinsed three times by submerging in sterile distilled water and drying overnight. Next, the biofilm stain was dissolved using 125 μl of 30% acetic acid (Wako Pure Chemical Industries, Ltd, Osaka, Japan), and the absorbance was quantified using a plate reader (MultiSkan FC, Thermo Fisher Scientific K.K., Tokyo, Japan) at 550 nm. The tested strains were classified according to their ability to form biofilms, as previously suggested (68). The optical density (OD) cutoff value was defined as three standard deviations above the average OD of the negative control. If the average OD of an E. coli strain was larger than the OD cutoff value, the E. coli strain was defined as a biofilm former. The averages are the results of at least three trials.
Statistical analysis.
Continuous variables were compared using Student's t test. Categorical variables were expressed as numbers and percentages and were compared using Fisher’s two-tailed exact test. For multiple comparison tests, post hoc analyses were performed with the Bonferroni correction. Statistical significance was set at P < 0.05. All statistical analyses were performed using JMP Pro version 14 software (SAS Institute Inc., Cary, NC).
ACKNOWLEDGMENTS
We thank Editage (www.editage.com) for English language editing. This manuscript was presented at the World Microbe Forum, June 20–24, 2021.
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Shu Okugawa, Email: okugawa-tky@g.ecc.u-tokyo.ac.jp.
Swaine L. Chen, The National University of Singapore and the Genome Institute of Singapore
REFERENCES
- 1.Madsen KM, Schønheyder HC, Kristensen B, Sørensen HT. 1999. Secular trends in incidence and mortality of bacteraemia in a Danish county 1981–1994. APMIS 107:346–352. doi: 10.1111/j.1699-0463.1999.tb01563.x. [DOI] [PubMed] [Google Scholar]
- 2.Uslan DZ, Crane SJ, Steckelberg JM, Cockerill FR, 3rd, St Sauver JL, Wilson WR, Baddour LM. 2007. Age- and sex-associated trends in bloodstream infection: a population-based study in Olmsted County, Minnesota. Arch Intern Med 167:834–839. doi: 10.1001/archinte.167.8.834. [DOI] [PubMed] [Google Scholar]
- 3.Skogberg K, Lyytikäinen O, Ruutu P, Ollgren J, Nuorti JP. 2008. Increase in bloodstream infections in Finland, 1995–2002. Epidemiol Infect 136:108–114. doi: 10.1017/S0950268807008138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson J, Elgohari S, Livermore DM, Cookson B, Johnson A, Lamagni T, Chronias A, Sheridan E. 2011. Trends among pathogens reported as causing bacteraemia in England, 2004–2008. Clin Microbiol Infect 17:451–458. doi: 10.1111/j.1469-0691.2010.03262.x. [DOI] [PubMed] [Google Scholar]
- 5.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 6.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun 67:546–553. doi: 10.1128/IAI.67.2.546-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Köhler CD, Dobrindt U. 2011. What defines extraintestinal pathogenic Escherichia coli? Int J Med Microbiol 301:642–647. doi: 10.1016/j.ijmm.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Tivendale KA, Logue CM, Kariyawasam S, Jordan D, Hussein A, Li G, Wannemuehler Y, Nolan LK. 2010. Avian-pathogenic Escherichia coli strains are similar to neonatal meningitis E. coli strains and are able to cause meningitis in the rat model of human disease. Infect Immun 78:3412–3419. doi: 10.1128/IAI.00347-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croxen MA, Finlay BB. 2010. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol 8:26–38. doi: 10.1038/nrmicro2265. [DOI] [PubMed] [Google Scholar]
- 11.Sharma G, Sharma S, Sharma P, Chandola D, Dang S, Gupta S, Gabrani R. 2016. Escherichia coli biofilm: development and therapeutic strategies. J Appl Microbiol 121:309–319. doi: 10.1111/jam.13078. [DOI] [PubMed] [Google Scholar]
- 12.Poirel L, Madec JY, Lupo A, Schink AK, Kieffer N, Nordmann P, Schwarz S. 2018. Antimicrobial Resistance in Escherichia coli. Microbiol Spectr 6. doi: 10.1128/microbiolspec.ARBA-0026-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanz-García M, Fernández-Cruz A, Rodríguez-Créixems M, Cercenado E, Marin M, Muñoz P, Bouza E. 2009. Recurrent Escherichia coli bloodstream infections: epidemiology and risk factors. Medicine (Baltimore, MD) 88:77–82. doi: 10.1097/MD.0b013e31819dd0cf. [DOI] [PubMed] [Google Scholar]
- 14.Giannella M, Pascale R, Toschi A, Ferraro G, Graziano E, Furii F, Bartoletti M, Tedeschi S, Ambretti S, Lewis RE, Viale P. 2018. Treatment duration for Escherichia coli bloodstream infection and outcomes: retrospective single-centre study. Clin Microbiol Infect 24:1077–1083. doi: 10.1016/j.cmi.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Maslow JN, Mulligan ME, Arbeit RD. 1994. Recurrent Escherichia coli bacteremia. J Clin Microbiol 32:710–714. doi: 10.1128/jcm.32.3.710-714.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wendt C, Messer SA, Hollis RJ, Pfaller MA, Wenzel RP, Herwaldt LA. 1999. Recurrent gram-negative bacteremia: incidence and clinical patterns. Clin Infect Dis 28:611–617. doi: 10.1086/515152. [DOI] [PubMed] [Google Scholar]
- 17.Marschall J, Doherty J, Warren DK. 2010. The epidemiology of recurrent Gram-negative bacteremia in a tertiary-care hospital. Diagn Microbiol Infect Dis 66:456–459. doi: 10.1016/j.diagmicrobio.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Samet A, Sledzińska A, Krawczyk B, Hellmann A, Nowicki S, Kur J, Nowicki B. 2013. Leukemia and risk of recurrent Escherichia coli bacteremia: genotyping implicates E. coli translocation from the colon to the bloodstream. Eur J Clin Microbiol Infect Dis 32:1393–1400. doi: 10.1007/s10096-013-1886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cattaneo C, Antoniazzi F, Tumbarello M, Skert C, Borlenghi E, Schieppati F, Cerqui E, Pagani C, Petullà M, Re A, Rossi G. 2014. Relapsing bloodstream infections during treatment of acute leukemia. Ann Hematol 93:785–790. doi: 10.1007/s00277-013-1965-0. [DOI] [PubMed] [Google Scholar]
- 20.Lee CH, Su LH, Chen FJ, Tang YF, Chien CC, Liu JW. 2015. Clinical and microbiologic characteristics of adult patients with recurrent bacteraemia caused by extended-spectrum beta-lactamase-producing Escherichia coli or Klebsiella pneumoniae. Clin Microbiol Infect 21:1105.e1-8–1105.e8. doi: 10.1016/j.cmi.2015.07.025. [DOI] [PubMed] [Google Scholar]
- 21.Olesen B, Kolmos HJ, Orskov F, Orskov I. 1998. Escherichia coli bacteraemia in patients with and without haematological malignancies: a study of strain characters and recurrent episodes. J Infect 36:93–100. doi: 10.1016/s0163-4453(98)93378-3. [DOI] [PubMed] [Google Scholar]
- 22.Sampaio JL, Viana-Niero C, de Freitas D, Höfling-Lima AL, Leão SC. 2006. Enterobacterial repetitive intergenic consensus PCR is a useful tool for typing Mycobacterium chelonae and Mycobacterium abscessus isolates. Diagn Microbiol Infect Dis 55:107–118. doi: 10.1016/j.diagmicrobio.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 23.Casarez EA, Pillai SD, Mott JB, Vargas M, Dean KE, Di Giovanni GD. 2007. Direct comparison of four bacterial source tracking methods and use of composite data sets. J Appl Microbiol 103:350–364. doi: 10.1111/j.1365-2672.2006.03246.x. [DOI] [PubMed] [Google Scholar]
- 24.Salipante SJ, SenGupta DJ, Cummings LA, Land TA, Hoogestraat DR, Cookson BT. 2015. Application of whole-genome sequencing for bacterial strain typing in molecular epidemiology. J Clin Microbiol 53:1072–1079. doi: 10.1128/JCM.03385-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rumore J, Tschetter L, Kearney A, Kandar R, McCormick R, Walker M, Peterson CL, Reimer A, Nadon C. 2018. Evaluation of whole-genome sequencing for outbreak detection of Verotoxigenic Escherichia coli O157:H7 from the Canadian perspective. BMC Genomics 19:870. doi: 10.1186/s12864-018-5243-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi SH, Dagher M, Ruffin F, Park LP, Sharma-Kuinkel BK, Souli M, Morse AM, Eichenberger EM, Hale L, Kohler C, Warren B, Hansen B, Medie FM, McIntyre LM, Fowler VG. 2021. Risk factors for recurrent staphylococcus aureus bacteremia. Clin Infect Dis 72:1891–1899. doi: 10.1093/cid/ciaa801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fricke WF, Rasko DA. 2014. Bacterial genome sequencing in the clinic: bioinformatic challenges and solutions. Nat Rev Genet 15:49–55. doi: 10.1038/nrg3624. [DOI] [PubMed] [Google Scholar]
- 28.Zerr DM, Qin X, Oron AP, Adler AL, Wolter DJ, Berry JE, Hoffman L, Weissman SJ. 2014. Pediatric infection and intestinal carriage due to extended-spectrum-cephalosporin-resistant Enterobacteriaceae. Antimicrob Agents Chemother 58:3997–4004. doi: 10.1128/AAC.02558-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother 66:1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 30.Foxman B, Zhang L, Tallman P, Palin K, Rode C, Bloch C, Gillespie B, Marrs CF. 1995. Virulence characteristics of Escherichia coli causing first urinary tract infection predict risk of second infection. J Infect Dis 172:1536–1541. doi: 10.1093/infdis/172.6.1536. [DOI] [PubMed] [Google Scholar]
- 31.Johnson JR, O’Bryan TT, Delavari P, Kuskowski M, Stapleton A, Carlino U, Russo TA. 2001. Clonal relationships and extended virulence genotypes among Escherichia coli isolates from women with a first or recurrent episode of cystitis. J Infect Dis 183:1508–1517. doi: 10.1086/320198. [DOI] [PubMed] [Google Scholar]
- 32.Soto SM, Smithson A, Horcajada JP, Martinez JA, Mensa JP, Vila J. 2006. Implication of biofilm formation in the persistence of urinary tract infection caused by uropathogenic Escherichia coli. Clin Microbiol Infect 12:1034–1036. doi: 10.1111/j.1469-0691.2006.01543.x. [DOI] [PubMed] [Google Scholar]
- 33.Stapleton A, Moseley S, Stamm WE. 1991. Urovirulence determinants in Escherichia coli isolates causing first-episode and recurrent cystitis in women. J Infect Dis 163:773–779. doi: 10.1093/infdis/163.4.773. [DOI] [PubMed] [Google Scholar]
- 34.Dale AP, Pandey AK, Hesp RJ, Belogiannis K, Laver JR, Shone CC, Read RC. 2018. Genomes of Escherichia coli bacteraemia isolates originating from urinary tract foci contain more virulence-associated genes than those from non-urinary foci and neutropaenic hosts. J Infect 77:534–543. doi: 10.1016/j.jinf.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riley LW. 2014. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin Microbiol Infect 20:380–390. doi: 10.1111/1469-0691.12646. [DOI] [PubMed] [Google Scholar]
- 36.Duan Y, Gao H, Zheng L, Liu S, Cao Y, Zhu S, Wu Z, Ren H, Mao D, Luo Y. 2020. Antibiotic resistance and virulence of extraintestinal pathogenic Escherichia coli (ExPEC) vary according to molecular types. Front Microbiol 11:598305. doi: 10.3389/fmicb.2020.598305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Surgers L, Boyd A, Girard PM, Arlet G, Decré D. 2019. Biofilm formation by ESBL-producing strains of Escherichia coli and Klebsiella pneumoniae. Int J Med Microbiol 309:13–18. doi: 10.1016/j.ijmm.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 38.Hennequin C, Aumeran C, Robin F, Traore O, Forestier C. 2012. Antibiotic resistance and plasmid transfer capacity in biofilm formed with a CTX-M-15-producing Klebsiella pneumoniae isolate. J Antimicrob Chemother 67:2123–2130. doi: 10.1093/jac/dks169. [DOI] [PubMed] [Google Scholar]
- 39.Shapiro JA. 1998. Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol 52:81–104. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- 40.Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. 1999. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol 37:1771–1776. doi: 10.1128/JCM.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agarwal J, Mishra B, Srivastava S, Srivastava R, Pandey A. 2014. Virulence determinants in Escherichia coli associated with recurrent cystitis in sexually active women. Microb Pathog 74:38–41. doi: 10.1016/j.micpath.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 42.Hilf M, Yu VL, Sharp J, Zuravleff JJ, Korvick JA, Muder RR. 1989. Antibiotic therapy for Pseudomonas aeruginosa bacteremia: outcome correlations in a prospective study of 200 patients. Am J Med 87:540–546. doi: 10.1016/S0002-9343(89)80611-4. [DOI] [PubMed] [Google Scholar]
- 43.Versalovic J, Koeuth T, Lupski JR. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed]
- 44.Heras J, Domínguez C, Mata E, Pascual V, Lozano C, Torres C, Zarazaga M. 2015. GelJ–a tool for analyzing DNA fingerprint gel images. BMC Bioinformatics 16:270. doi: 10.1186/s12859-015-0703-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayumi T, Okamoto K, Takada T, Strasberg SM, Solomkin JS, Schlossberg D, Pitt HA, Yoshida M, Gomi H, Miura F, Garden OJ, Kiriyama S, Yokoe M, Endo I, Asbun HJ, Iwashita Y, Hibi T, Umezawa A, Suzuki K, Itoi T, Hata J, Han HS, Hwang TL, Dervenis C, Asai K, Mori Y, Huang WS, Belli G, Mukai S, Jagannath P, Cherqui D, Kozaka K, Baron TH, de Santibañes E, Higuchi R, Wada K, Gouma DJ, Deziel DJ, Liau KH, Wakabayashi G, Padbury R, Jonas E, Supe AN, Singh H, Gabata T, Chan ACW, Lau WY, Fan ST, Chen MF, Ker CG, et al. 2018. Tokyo guidelines 2018: management bundles for acute cholangitis and cholecystitis. J Hepatobiliary Pancreat Sci 25:96–100. doi: 10.1002/jhbp.519. [DOI] [PubMed] [Google Scholar]
- 46.CLSI. 2018. Performance standards for antimicrobial susceptibility testing. 28th ed. CLSI supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 47.Shibata N, Kurokawa H, Doi Y, Yagi T, Yamane K, Wachino J-i, Suzuki S, Kimura K, Ishikawa S, Kato H, Ozawa Y, Shibayama K, Kai K, Konda T, Arakawa Y. 2006. PCR classification of CTX-M-type beta-lactamase genes identified in clinically isolated gram-negative bacilli in Japan. Antimicrob Agents Chemother 50:791–795. doi: 10.1128/AAC.50.2.791-795.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 49.Clermont O, Bonacorsi S, Bingen E. 2004. Characterization of an anonymous molecular marker strongly linked to Escherichia coli strains causing neonatal meningitis. J Clin Microbiol 2:1770–1772. doi: 10.1128/JCM.42.4.1770-1772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lescat M, Clermont O, Woerther PL, Glodt J, Dion S, Skurnik D, Djossou F, Dupont C, Perroz G, Picard B, Catzeflis F, Andremont A, Denamur E. 2013. Commensal Escherichia coli strains in Guiana reveal a high genetic diversity with host-dependant population structure. Environ Microbiol Rep 5:49–57. doi: 10.1111/j.1758-2229.2012.00374.x. [DOI] [PubMed] [Google Scholar]
- 51.Clermont O, Lescat M, O’Brien CL, Gordon DM, Tenaillon O, Denamur E. 2008. Evidence for a human-specific Escherichia coli clone. Environ Microbiol 10:1000–1006. doi: 10.1111/j.1462-2920.2007.01520.x. [DOI] [PubMed] [Google Scholar]
- 52.Doumith M, Day M, Ciesielczuk H, Hope R, Underwood A, Reynolds R, Wain J, Livermore DM, Woodford N. 2015. Rapid identification of major Escherichia coli sequence types causing urinary tract and bloodstream infections. J ClinMicrobiol 53:160–166. doi: 10.1128/JCM.02562-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson JR, Brown JJ. 1996. A novel multiply primed polymerase chain reaction assay for identification of variant papG genes encoding the Gal (alpha 1-4)Gal-binding PapG adhesins of Escherichia coli. J Infect Dis 173:920–926. doi: 10.1093/infdis/173.4.920. [DOI] [PubMed] [Google Scholar]
- 55.Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 56.Nakano M, Yamamoto S, Terai A, Ogawa O, Makino SI, Hayashi H, Nair GB, Kurazono H. 2001. Structural and sequence diversity of the pathogenicity island of uropathogenic Escherichia coli which encodes the USP protein. FEMS Microbiol Lett 205:71–76. doi: 10.1111/j.1574-6968.2001.tb10927.x. [DOI] [PubMed] [Google Scholar]
- 57.Ananias M, Yano T. 2008. Serogroups and virulence genotypes of Escherichia coli isolated from patients with sepsis. Braz J Med Biol Res 41:877–883. doi: 10.1590/S0100-879X2008001000008. [DOI] [PubMed] [Google Scholar]
- 58.Cirl C, Wieser A, Yadav M, Duerr S, Schubert S, Fischer H, Stappert D, Wantia N, Rodriguez N, Wagner H, Svanborg C, Miethke T. 2008. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat Med 14:399–406. doi: 10.1038/nm1734. [DOI] [PubMed] [Google Scholar]
- 59.Le Bouguenec C, Archambaud M, Labigne A. 1992. Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J Clin Microbiol 30:1189–1193. doi: 10.1128/jcm.30.5.1189-1193.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson JR, Russo TA, Tarr PI, Carlino U, Bilge SS, Vary JC, Jr, Stell AL. 2000. Molecular epidemiological and phylogenetic associations of two novel putative virulence genes, iha and iroN(E. coli), among Escherichia coli isolates from patients with urosepsis. Infect Immun 68:3040–3047. doi: 10.1128/IAI.68.5.3040-3047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamamoto S, Terai A, Yuri K, Kurazono H, Takeda Y, Yoshida O. 1995. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol 12:85–90. doi: 10.1111/j.1574-695X.1995.tb00179.x. [DOI] [PubMed] [Google Scholar]
- 62.Schubert S, Rakin A, Karch H, Carniel E, Heesemann J. 1998. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect Immun 66:480–485. doi: 10.1128/IAI.66.2.480-485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodriguez-Siek KE, Giddings CW, Doetkott C, Johnson TJ, Fakhr MK, Nolan LK. 2005. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology (Reading) 151:2097–2110. doi: 10.1099/mic.0.27499-0. [DOI] [PubMed] [Google Scholar]
- 64.Huang SH, Wass C, Fu Q, Prasadarao NV, Stins M, Kim KS. 1995. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect Immun 63:4470–4475. doi: 10.1128/iai.63.11.4470-4475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson TJ, Wannemuehler Y, Doetkott C, Johnson SJ, Rosenberger SC, Nolan LK. 2008. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J Clin Microbiol 46:3987–3996. doi: 10.1128/JCM.00816-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson JR, Stapleton AE, Russo TA, Scheutz F, Brown JJ, Maslow JN. 1997. Characteristics and prevalence within serogroup O4 of a J96-like clonal group of uropathogenic Escherichia coli O4:H5 containing the class I and class III alleles of papG. Infect Immun 65:2153–2159. doi: 10.1128/iai.65.6.2153-2159.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O’Toole GA. 2011. Microtiter dish biofilm formation assay. J Vis Exp 47:2437. doi: 10.3791/2437.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M. 2000. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods 40:175–179. doi: 10.1016/s0167-7012(00)00122-6.. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM01399-21_Supp_1_seq7.pdf, PDF file, 0.4 MB (437.6KB, pdf)