ABSTRACT
Salmonella enterica serotype Typhimurium, a nontyphoidal Salmonella (NTS), results in a range of enteric diseases, representing a major disease burden worldwide. There is still a significant portion of Salmonella genes whose mechanistic basis to overcome host innate defense mechanisms largely remains unknown. Here, we have applied transposon insertion sequencing (Tn-seq) method to unveil the genetic factors required for the growth or survival of S. Typhimurium under various host stressors simulated in vitro. A highly saturating Tn5 library of S. Typhimurium 14028s was subjected to selection during growth in the presence of short-chain fatty acid (100 mM propionate), osmotic stress (3% NaCl), or oxidative stress (1 mM H2O2) or survival in extreme acidic pH (30 min in pH 3) or starvation (12 days in 1× phosphate-buffered saline [PBS]). We have identified a total of 339 conditionally essential genes (CEGs) required to overcome at least one of these conditions mimicking host insults. Interestingly, all eight genes encoding FoF1-ATP synthase subunit proteins were required for fitness in all five stresses. Intriguingly, a total of 88 genes in Salmonella pathogenicity islands (SPI), including SPI-1, SPI-2, SPI-3, SPI-5, SPI-6, and SPI-11, are also required for fitness under the in vitro conditions. Additionally, by comparative analysis of the genes identified in this study and the genes previously shown to be required for in vivo fitness, we identified novel genes (marBCT, envF, barA, hscA, rfaQ, rfbI, and the genes encoding putative proteins STM14_1138, STM14_3334, STM14_4825, and STM_5184) that have compelling potential for the development of vaccines and antibacterial drugs to curb Salmonella infection.
IMPORTANCE Salmonella enterica serotype Typhimurium is a major human bacterial pathogen that enters the food chain through meat animals asymptomatically carrying this pathogen. Despite the rich genome sequence data, a significant portion of Salmonella genes remain to be characterized for their potential contributions to virulence. In this study, we used transposon insertion sequencing (Tn-seq) to elucidate the genetic factors required for growth or survival under various host stressors, including short-chain fatty acids, osmotic stress, oxidative stress, extreme acid, and starvation. Among the total of 339 conditionally essential genes (CEGs) that are required under at least one of these five stress conditions were 221 previously known virulence genes required for in vivo fitness during infection in at least one of four animal species, including mice, chickens, pigs, and cattle. This comprehensive map of virulence phenotype-genotype in S. Typhimurium provides a roadmap for further interrogation of the biological functions encoded by the genome of this important human pathogen to survive in hostile host environments.
KEYWORDS: Salmonella, Tn-seq, conditionally essential genes, in vivo fitness, host stressors, virulence genes
INTRODUCTION
Nontyphoidal Salmonella (NTS), a Gram-negative bacterial pathogen, causes 93 million enteric infections, 155,000 diarrheal deaths, and 3.4 million bloodstream infections worldwide annually (1, 2). Salmonella enterica serotype Typhimurium (S. Typhimurium) is one of the leading causes of NTS (3, 4). Despite that Salmonella infection is an enormous global burden on disease worldwide and the first complete genome sequence of S. Typhimurium LT2 became available nearly 2 decades (2002) ago followed by additional complete genomes of >348,000 Salmonella strains (www.ncbi.nlm.nih.gov/pathogens), the mechanistic basis for S. Typhimurium genes required for in vivo survival is still unknown for a large portion of the genes (5, 6). Researchers have tried to delve into the pathogenesis of S. Typhimurium using different variations of high-throughput screening of transposon mutants based on negative selection (7–9). Chan et al. discovered 157 and 264 genes required by S. Typhimurium strain SL1344 for acute infection in mice (A-Mice) and survival inside macrophages (MΦ), respectively, using a microarray-based tracking method (9). Lawley et al. used the same method to identify 118 genes of S. Typhimurium SL1344 required for long-term persistent infection in mice (P-Mice) using spleen samples collected 28 days postinfection (8). Additionally, Chaudhuri et al. comprehensively assigned a core set of 611 genes of S. Typhimurium strain ST4/74 required for effective gut colonization in calves, pigs, and chickens (10). Recently, Silva-Valenzuela et al. identified 224 mutants of S. Typhimurium 14028s that were negatively selected using two pools of single-gene deletion mutants recovered from spleen and liver at 2 days postinfection in mice (Sp-Liv) (11). Previously, our laboratory conducted transposon insertion sequencing (Tn-seq) screening to identify an overlapping set of 105 coding genes of S. Typhimurium 14028s required for in vitro growth in diluted Luria-Bertani (dLB) medium, LB medium plus bile acid, and LB medium at 42°C (12). However, there is still a gap in the above approaches to correlate the genes required for growth or survival by S. Typhimurium between in vitro and in vivo conditions, which will help us delve into the biochemical and/or molecular basis of virulence and potentially pave a roadmap toward the development of novel vaccines, antibiotics, and/or control strategies.
In this study, we conducted Tn-seq analysis of S. Typhimurium 14028s under the five in vitro conditions mimicking host stressors found during enteric and systemic infection. Tn-seq is a powerful tool for functional analysis of bacterial genomes based on the use of random transposon mutagenesis and next-generation sequencing technology (7, 13, 14). We have applied a highly efficient method for Tn-seq library preparation that requires only a small amount of DNA without the need for enzymatic digestion or physical shearing of genomic DNA (15–18). To cause enteric infection, S. Typhimurium has to overcome host insults, such as low acidic pH in the stomach, osmotic pressure, and short-chain fatty acid (SCFAs) in the intestinal tract (19–22). Eventually, for systemic infection, S. Typhimurium has to vanquish macrophage stresses, such as oxidative stress, starvation, and hyperosmotic conditions (23–25). We hypothesized that a comparative analysis of the comprehensive sets of the in vitro fitness genes (for resistance against host stressors [from this and previous studies]) and in vivo fitness genes (required for enteric and systemic infection in different hosts [from previous studies]) will allow a better understanding of the mechanistic basis of the genetic determinants of S. Typhimurium required for host infection and provide enhanced resolution to link genotype to phenotype. Thus, we performed Tn-seq screenings under the five different host stressors simulated in vitro, which was then followed by a comparative analysis between the in vivo and in vitro fitness genes identified from previous studies and the current study.
RESULTS AND DISCUSSION
Stress conditions used for the Tn-seq screenings.
Among various stressors in the host environment, we selected five conditions for Tn-seq screenings in this study. They include NaCl, propionate (PA), H2O2, pH 3, and starvation, representing high osmolarity (intestinal tract), SCFAs (intestinal tract and macrophages), oxidative stress (macrophages), extreme acid (stomach and macrophages), and limited nutrients (macrophages), respectively, which S. Typhimurium encounters in different tissues during the course of infection in the host (26). For NaCl, propionate, and H2O2, we used growth-based selection, as these stressors at the range of concentration found in the host tissues are bacteriostatic. On the contrary, pH 3 and starvation operate as the bactericidal stressors S. Typhimurium encounters under conditions where the pathogen does not multiply, such as acidic stomach and acidified vacuole in macrophages (26). Therefore, survival-based selection was used for pH 3 and starvation stressors. To determine the concentrations for the growth-based selections, we performed growth assays in LB medium containing each stressor at the concentrations commonly used to mimic host stressors in the literature. The growth curves of S. Typhimurium 14028s wild-type strain in the presence of each of the three stressors at the respective concentrations used in the Tn-seq screenings in this study are shown in Fig. S2 in the supplemental material.
Overall evaluation of resulting Tn-seq profiles.
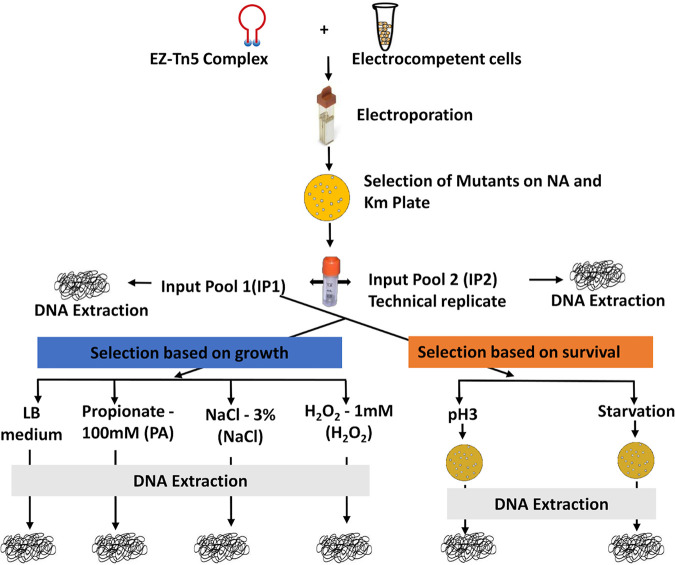
We have constructed a highly saturated transposon mutant library of S. Typhimurium 14028s with approximately 350,000 transposon mutants created via transformation of the EZ-Tn5 transposome complex to electrocompetent cells. The complex Tn5 library, Input pool 1 (IP1), was then subjected to negative selection under the in vitro stress conditions encountered by S. Typhimurium during enteric and systemic infection as described in Materials and Methods. Input pool 2 (IP2) was the technical replicate of IP1 to evaluate the reproducibility of our Tn-seq method (Fig. 1). The Tn-seq amplicon library for Illumina sequencing was prepared for each of the input and output pools (Fig. S1A and S1B). This efficient Tn-seq protocol was developed in our laboratory and offers distinct advantages over other Tn-seq library preparation methods, including a small amount (∼100 ng) of DNA required and no need for physical shearing or restriction digestion (7, 15–17).
FIG 1.
Schematic overview of the experimental design. A highly saturating Tn5 library was constructed through electroporation of EZ-Tn5 transposome complex to S. Typhimurium 14028s. Approximately 350,000 Tn5 mutants were collected on LB (Km + NA) plates. Complex Tn5 mutant library (IP1) was selected based on growth (LB medium [LB], 100 mM propionate in LB medium [PA], 3% NaCl in LB medium [NaCl], and 1 mM hydrogen peroxide in LB medium [H2O2]) and survival (exposed to pH 3 for 30 min [pH 3] and incubated for 12 days in 1× PBS [starvation]). Input pool 2 (IP2) was a technical replicate of input pool 1 (IP1).
Illumina sequencing using a HiSeq 3000 produced 163,943,475 reads from a single flow cell lane. The raw reads were demultiplexed, allowing a perfect match for the sample barcodes used (Table S1) with the exception of up to two mismatches within the Tn5 mosaic end (ME) using a custom Perl script. H2O2 (19,250,956) had the highest number of reads followed by IP1 (10,842,764), starvation (9,518,226), IP2 (6,345,173), LB (5,004,934), pH 3 (3,841,401), PA (2,113,033), and NaCl (1,970,072) (Fig. S3A).
After demultiplexing, Illumina reads were trimmed of barcode and transposon sequences. The Tn5 junction sequences of 20 bp were extracted and mapped to the complete genome of S. Typhimurium 14028s (NC_016856.1) using Bowtie. The overall alignment rate throughout all Tn5 libraries was 85.19% (standard error [SE] ± 1.79). Additionally, we looked for unique insertion sites in the genome in each library. IP1 had the highest number of unique insertions (186,621) followed by LB (157,915), H2O2 (149,752), IP2 (149,740), PA (127,722), NaCl (125,918), starvation (118,607), and pH 3 (92,008) (Fig. S3A). Similarly, H2O2 had the highest average read per unique insertion site in the genome (96.007 ± 1.11) with 40 median reads, whereas NaCl had the lowest (13.53 ± 0.99) with 5 median reads (Fig. S3A).
Prealigned reads of the Tn5 library in default SAM mapping file format were fed to the “analysis of high-resolution transposon-insertion sequences technique” (ARTIST) pipeline (27). Tn5 insertions were mapped into 100-bp genome-wide windows. We observed the highest Spearman correlation coefficients (a commonly used numerical measure to describe a statistical relationship between two variables) between IP1 and IP2 and IP1 and LB (0.98, P < 0.0001). However, there was a lower Spearman correlation of IP1 with NaCl (0.97, P < 0.0001), PA (0.96, P < 0.0001), and H2O2 (0.93, P < 0.0001). We observed the lowest correlation of IP1 with pH 3 and starvation (0.84 and 0.91, respectively, P < 0.0001) (Fig. S3B). These relationships corroborate well with the Tn5 library selection strategies used, with a higher correlation for the selections based on growth fitness (NaCl, PA, and H2O2) and a lower correlation for the selections based on survival (pH 3 and starvation).
Additionally, we looked for the occurrence of any hot spots of Tn5 insertions in the sample libraries. We found an even distribution of Tn5 insertion reads across the libraries throughout the genome. Some of the genomic regions lacking insertions have white stripes that are clearly visible (Fig. S4) across all the samples that represent essential loci in the S. Typhimurium 14028s genome.
Identification of conditionally essential genes.
In this study, we used two strategies to identify conditionally essential genes (CEGs) of S. Typhimurium to overcome host stressors. The first strategy was a negative selection of the complex Tn5 mutant library based on growth fitness under mild stressors (3% NaCl, 100 mM propionate, 1 mM H2O2), and the second strategy was based on survival under harsher stressors (12 days of starvation and pH 3), as shown in Fig. 1.
The ARTIST pipeline can identify if genes are entirely essential or domain essential under a given condition. In our study, only a few of the genes were identified as domain essential, and the majority of them were entirely essential. For simplicity, we assigned both categories of the genes entirely essential and domain essential into one category, CEGs. We deliberately compared each of the output pool PA, NaCl, and H2O2 with both IP1 and LB separately. As expected, most of the CEGs were overlapped with these two comparisons. For the conditions PA, NaCl, and H2O2, we considered the common set of identified CEGs via the comparison of output libraries with both IP1 and LB as CEGs for each condition. However, the output libraries for pH 3 and starvation were compared only with IP1 because the selection of the Tn5 library was based on survived mutants, and the mutant cells did not multiply during selection in liquid medium.
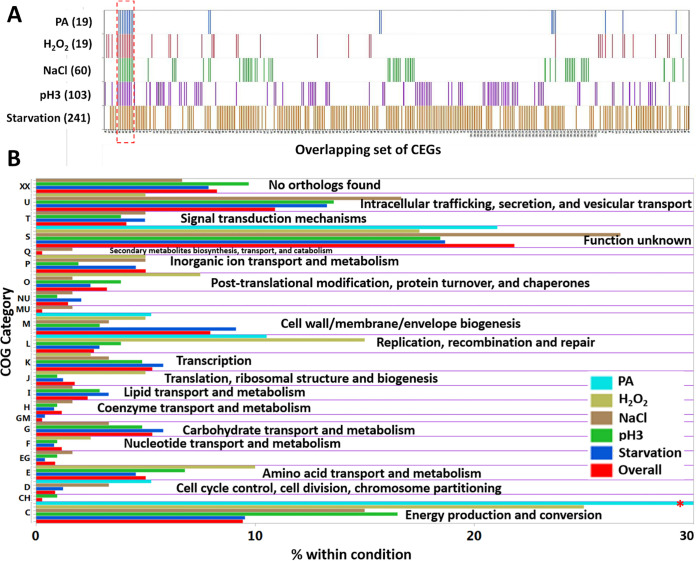
We identified a total of 339 CEGs that are required for the fitness of S. Typhimurium 14028s under at least one of the five conditions (Fig. 2A). Starvation identified the highest number of CEGs (241), followed by pH 3 (103), NaCl (60), H2O2 (40), and PA (19), as shown in Tables S2 and S3. This might likely reflect that starvation is a severe stressor involving diverse genetic pathways for survival, while PA is a mild stressor for the fitness of S. Typhimurium. More than one-half of CEGs were on the lagging strand (56.63%), which is somewhat contrary to the responsive genes in Escherichia coli and Streptococcus pneumoniae (28, 29). We assigned a functional role to 96 CEGs that were putative proteins and 21 CEGs belonging to hypothetical proteins. The stress-tolerant proteins commonly identified in at least two of the in vitro stressors included ATP synthase, a transcriptional regulator, 3-dehydoroquinate synthase, site-specific tyrosine recombinase xerC, flavin mononucleotide phosphatase, ribulose-phosphate 3-epimerase, and DNA-dependent helicase II among others (Tables S2 and S3).
FIG 2.
Conditionally essential genes (CEGs) of S. Typhimurium 14028s and cluster of orthologous groups (COG). (A) Distribution of the 339 CEGs required for fitness under at least one of the five stress conditions. Numbers inside the bracket indicate the number of CEGs identified. The red dashed box indicates the CEGs (ATP synthase genes) common to all five conditions. (B) Functional assignments of CEGs into COG category. Overall is the COG assigned to all the 339 CEGs. The red asterisk indicates that the abundance of COG C in PA was 57.89%.
Intriguingly, we found that many genes in the Salmonella pathogenicity islands (SPI) were required for fitness in the presence of the in vitro stressors used in this study. Numerous genes in SPI-1, SPI-2, SPI-3, SPI-5, SPI-6, and SPI-11 were required for resistance against starvation (n = 68), NaCl (n = 28), and pH 3 (n = 27) (Table S4). However, no SPI genes were required for fitness in PA and H2O2. SPI-5 and SPI-11 genes were only conditionally essential in pH 3 (n = 4 and 6, respectively), while SPI-3 genes were only conditionally essential in NaCl (n = 7) and SPI-6 genes in starvation (n = 7). Tn-seq profiles for the SPI-1 region are shown in Fig. S5A as an example.
For broader insight into pathways involved in stress resistance, we assigned each CEG to the cluster of orthologous groups (COG) using the evolutionary genealogy of genes: nonsupervised orthologous groups (eggNOG) database (30). The CEGs having a top hit for the COG in S. Typhimurium LT2 were kept, and CEGs with no orthologous group were allotted to group XX (Fig. 2B; Table S3). Overall, 21.83% of CEGs belonged to the category “function unknown”, followed by “intracellular trafficking, secretion, and vesicular transport” (10.91%), “energy production and conversion” (9.44%), and “no orthologs found” (8.26%) among others. A substantial portion of CEGs (30.6%) falling into either “function unknown” or “no orthologs found” shows that our data set is rich in novel genotype-phenotype relationships.
Additionally, we were interested to see if any CEGs identified in our study fell into the essential genomes of S. Typhimurium in other strain backgrounds. Essential genomes of S. Typhimurium strain SL3261 (selected on LB agar) (31) and S. Typhimurium strain LT2 (selected on rich medium) (32, 33) were compared with the CEGs of S. Typhimurium 14028s identified in this study. Genes in different strain backgrounds were examined for the corresponding orthologous genes in the S. Typhimurium 14028s background. Interestingly, 10 and 15 CEGs in this study were shared with the essential genes of S. Typhimurium SL3261 and LT2, respectively (Table S5 and Fig. S6). This indicates that these genes that are essential in other strain backgrounds are dispensable in the S. Typhimurium 14028s strain background.
Phenotypic basis for the requirement of CEGs in S. Typhimurium.
Next, we delved into the phenotypic mechanisms related to the CEGs identified in our study. For convenience, we split the section into specific CEGs required for fitness in only one stressor and common CEGs shared in at least two stressors out of five host stressors.
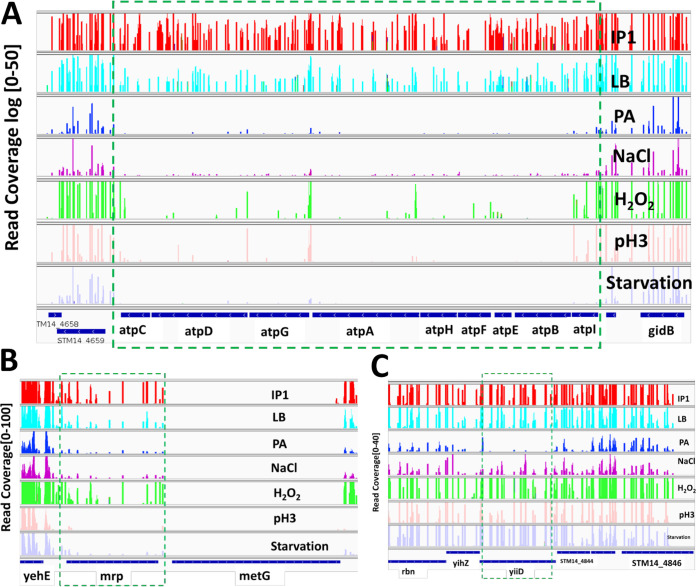
(i) CEGs specifically required for propionate (100 mM PA) stress resistance. CEGs specific for the fitness of S. Typhimurium in propionate were yiiD and sdhAD (Fig. S7). YiiD is a putative acetyltransferase protein (read coverage shown in Fig. 3C). Acetylation, a posttranslation protein modification, was previously shown to enable prokaryotes to increase stress resistance (34). Additionally, succinate dehydrogenase flavoprotein (sdhA) and cytochrome b566 (sdhD) subunit proteins were upregulated by intestinal SCFA in S. Typhimurium (35). Chowdhury and Shimizu reported that sdhA in the tricarboxylic acid cycle (TCA) was highly induced during temperature upshift in E. coli (36).
FIG 3.
Tn-seq profiles for selected genes across 7 conditions. Along the y axis, the numbers in the bracket indicate the raw read coverage. (A) ATP synthase genes conditionally essential under all five conditions (PA, NaCl, H2O2, pH 3, and starvation). (B) Gene mrp essential in pH 3 and starvation. (C) Gene yiiD essential in PA only.
(ii) CEGs specifically required for osmotic (3% NaCl) stress resistance. Twenty-six resistance genes of S. Typhimurium were required for fitness in osmotic stress (3% NaCl) alone (Fig. S7). Protein-protein network analysis using STRING database version 11.5 (http://string-db.org) against S. enterica LT2 showed three distinct clusterings of genes, SPI-3 (mgtBC, misL, cigR, slsA, fidL, and marT), two-component system (TC; dcuBRS), and sodium ion transport (yihPO) along with other nodes (https://version-11-5.string-db.org/cgi/network?networkId=bLmjZQ9kZpsE). SPI-3 genes are important for intracellular replication inside the phagosome, where Salmonella experiences hyperosmotic stress (37). The virulence genes mgtC and mgtB, Mg2+ were expressed 5-fold higher when S. Typhimurium was exposed to 0.3 M NaCl (38). MisL, an autotransporter protein is an intestinal colonization factor (activated by marT, a transcriptional regulator) that binds to extracellular matrix fibronectin in an animal host and is also involved in adhesion to plant tissue (39). Deletion of cigR in Salmonella enterica serotype Pullorum resulted in significantly decreased biofilm formation and increased virulence (40). Additionally, Figureueira et al. showed that the ΔcigR strain of S. Typhimurium had attenuated replication in mouse bone marrow-derived macrophages (41).
yihPO genes are essential for capsule assembly that is required by Salmonella for environmental stress persistence, such as desiccation (42). The absence of ompL (ortholog of yshA) leads to solvent hypersensitivity, as it helps in the stabilization of cell wall integrity protecting from solvent penetrance as a physical barrier (43). In E. coli, genes under the control of dcuS-dcuR, a two-component system, were not affected following hyperosmotic shock (44). However, dcuBRS was conditionally essential in S. Typhimurium for fitness during osmotic stress. Putative cytoplasmic protein (STM14_4542, STM14_4828, and STM14_5175), putative inner membrane protein (STM14_4824 and STM14_5184), and putative hydrolase (STM14_4823) were also required for osmotic stress tolerance.
(iii) CEGs specifically required for oxidative (1 mM H2O2) stress resistance. We identified 16 specific resistance genes required for the fitness of S. Typhimurium in the presence of 1 mM H2O2 (Fig. S7), and the functional protein association network analysis among the genes was constructed using STRING against S. enterica LT2 (https://version-11-5.string-db.org/cgi/network?networkId=b5BBHR89UGpJ). Major resistance genes were those involved in the two-component system (glnD, rpoN, arcA [STM4598], and arcB [STM3328]), DNA recombination (recJ and xerD), and metal ion transport (corA and trkA).
Hydrogen peroxide kills E. coli cells with two distinct modes: mode 1 killing occurs at a lower concentration of H2O2 due to DNA damage, and mode 2 killing occurs at a higher concentration of H2O2 due to damage of other structures such as proteins and lipids (45). Nucleic acid metabolic process genes involved in oxidative stress resistance were recJ, xerD, sun, and rpoN. RecJ protein, a single-stranded DNA (ssDNA)-specific 5′ to 3′ exonuclease/deoxribophophodiesterase, plays a role in homologous recombination, mismatch repair, and base excision repair (46). In E. coli, xerD-knockout mutants are hypersensitive to tightly bound DNA-protein complexes (TBCs) that block replication forks in vivo (47). rpoN, the alternative sigma factor 54 (σ54), is an important regulator of stress resistance and virulence genes in many bacterial species (48). σ54 is involved in carbon/nitrogen limitation, nucleic acid damage, the cell envelope, and nitric oxide stress (49). However, Hwang et al. found that an rpoN mutant in Campylobacter jejuni was more resistant to 1 mM H2O2 (50).
Additionally, cellular component genes crucial for fitness in H2O2 stress were dsbC, glmS, trkA, and corA, including sun and xerD. DsbC, a protein essential for disulfide bond isomerization in the periplasm, has a new role in E. coli in protection against oxidative stress (51). In E. coli, GlmS plays an important role in cell wall synthesis, thus protecting cell envelope stress response (52). HscB, a chaperone-encoding gene is upregulated after exposure to oxidative stress in Burkholderia pseudomallei (53). YbgF, an outer membrane vesicle protein, increases the survival of bacteria during exposure to stress or from toxic unfolded proteins by releasing the unwanted periplasmic component (54).
(iv) CEGs specifically required for higher acidic (pH 3) stress resistance. We found 49 specific stress resistance genes required only for the survival of S. Typhimurium under extreme acidic conditions (pH 3) among other stressors (Fig. S7). Formate dehydrogenase (fdoHI and fdhDE), curli proteins (csgBDEFG), virulence and envelope proteins (SPI-2 [orf245, orf408, ssaB], SPI-5 [pipBC, sopB], and SPI-11 [envEF, pagCD, msgA, and STM14_1486 where ssaB, pipB, and sopB are effector proteins]), and biopolymer transport protein (exbD and exbB) were clustered in functional protein association network analysis using STRING (https://version-11-5.string-db.org/cgi/network?networkId=bS96Nh4Mgvxr).
Formate dehydrogenase catalyzes the oxidation of formate (HCOO−) to CO2 and H+. The released electrons from this reaction are used by two cytoplasmic protons to form dihydrogen, thus consuming net protons, consequently, counteracting acidification (55). Curli is a major complex extracellular proteinaceous matrix produced by Enterobacteriaceae that helps pathogenic bacteria like Salmonella in adhesion to surfaces, cell aggregation, and biofilm formation (56). Acidic pH strongly enhances biofilm formation in Streptococcus agalactiae (57). We hypothesize that curli fibers might potentially protect bacteria from severe acid stress through the physical barrier and likely by the generation of alkaline compounds, as in oral biofilms (58). PhoP regulates SPI-11 genes, such as envEF, pagCD, and msgA, where the latter three are required by Salmonella to survive low pH within macrophages (59, 60). In Helicobacter pylori, the only organism to colonize in the acidic human stomach, the ExbB/ExbD/TonB complex is required for acid survival and periplasmic buffering (61). Additionally, survival of ΔexbD was diminished compared to the wild type at pH 3 in E. coli (62). The metC gene encoding a key enzyme in methionine biosynthesis, required for the generation of homocysteine, pyruvate, and ammonia, plays a crucial role in bacterial acid stress responses (63). However, there was no overlap between Salmonella enterica serotype Derby genes identified by Gu et al. for growth under acidic conditions due to different experiment design (growth versus survival) and serotypes (64).
(v) CEGs specifically required for starvation stress resistance. Out of 241 Salmonella fitness genes essential for starvation stress, 160 genes were important for resistance against only starvation stress among the five infection-relevant conditions in this study (Fig. S7) (https://version-11-5.string-db.org/cgi/network?networkId=b1V1bvEGNU4v). Major enriched gene pathways were oxidative phosphorylation, pathogenesis, two-component system, and lipopolysaccharide biosynthetic process among others. NADH dehydrogenase, the first component of the respiratory chain, subunit proteins (nuoCEFGHLMN) were required for the fitness of Salmonella during long-term carbon starvation. Salmonella defective in NADH dehydrogenase enzyme exhibits defective energy-dependent proteolysis during carbon starvation (65). Proteolysis of unbound or unemployed proteins helps bacteria to access nutrients as an important survival strategy during carbon starvation (66). SPI-1 (hilACD, iagB, invH, orgAC, prgHIJK, STM14_3500, and STM14_3501) and SPI-2 (ssaMNOPQRSTV, sscB, and sseDEF) encoding type III secretion system (T3SS) and SPI-6 (safABCD, sinR, STM14_0359, and ybeJ) encoding type VI (T6SS) secretion system were required for in vitro survival in long-term starvation stress. Salmonella usually requires SPI-1 genes for the invasion of intestinal epithelial cells (67). HilACD regulates SPI-1 invasion gene expression under multiple environmental conditions, including stationary phase, pH, osmolality, oxygen tension, and short-chain fatty acids (68). SPI-2 genes are expressed under in vitro starvation conditions, indicating the use of nutritional deprivation as a signal (69). T6SS has been hypothesized to confer a growth advantage to bacteria in environmental niches where bacterial competition for a nutrient is critical for survival (70).
Two-component systems (TCs), a basic stimulus-response coupling mechanism, enable microbes to respond to various stimuli, such as pH, osmolality, quorum signals, or nutrient availability, and regulate their cellular functions (71). TCs required for fitness under starvation conditions were envZ/OmpR, cpxA/cpxR, sensory histidine kinase protein (phoQ), and kdpD (Fig. S5B). EnvZ/OmpR regulates the synthesis of porin proteins (ompF and OmpC) that are important for the survival of E. coli in seawater under starvation stress conditions (72). It is believed that carbon starvation causes cell envelope stress. Bacchelor et al. found that cpxA/cpxR in E. coli regulates the expression of porins ompF and ompC, a major component of the outer membrane. However, Kenyon et al. showed that starvation stress of S. Typhimurium does not require cpxR-regulated extracytoplasmic functions (73, 74). Two genes phoQ and kdpD play a role in Mg2+ and K+ homeostasis, respectively, which is critical to the virulence and intracellular survival of S. Typhimurium (71, 75).
The outer membrane of Gram-negative bacteria contains phospholipids and lipopolysaccharides (LPS). LPS molecules act as a permeability barrier to prevent the entry of toxic compounds and allow the entry of nutrient molecules (76). LPS biosynthetic process genes required for fitness under starvation conditions were rfbABCD, rfbUNMKP, galF, udg, wzxE, and wzzB. Starvation of carbon energy sources activates an envelope stress response in S. Typhimurium (77). Additionally, pstSCAB, encoding the Pst ABC transporter, catalyzes the uptake of inorganic phosphate (78). Mutations in the Pst system results in structural modifications of lipid A and an imbalance in unsaturated fatty acids, consequently leading to an increase in outer membrane permeability, making E. coli more vulnerable to environmental stresses, including antimicrobial peptides and low pH (78).
Additional genes required for starvation stress resistance were aroGH, ytfMNP (ytfM [outer membrane protein]), and stcB (putative periplasmic outer chaperone protein). Furthermore, other envelope proteins were outer membrane lipoproteins (stcD and yifL), putative outer membrane proteins (stcC, STM14_0404, and ytfM), and putative inner membrane proteins (STM14_0398, STM14_0402, STM14_2763, STM14_4741, STM14_4742, STM14_4745, STM14_4880, ydiK, and yjeT). Similarly, putative cytoplasmic proteins required for starvation stress were STM14_2759, STM14_4743, STM14_5374, ydiL, and ytfP.
(vi) CEGs required for tolerance to multiple stressors. We found 12 Salmonella genes required for stress resistance in either three or four of the in vitro host stresses in our study, as shown in a STRING protein-protein interaction network (https://version-11-5.string-db.org/cgi/network?networkId=bVnBdnBjt1SH). The enriched gene ontology (GO) biological process/KEGG pathways were noncoding RNA (ncRNA) processing (gidAB and mnmE), DNA metabolic process (dam, uvrD [SOS response], and xerC), and biosynthesis of amino acids (aroB and rpe [microbial metabolism in diverse environments]). Also, other responsive proteins included ATP synthase subunit protein (atpI), putative permease (STM14_4659), inner membrane protein (damX), and flavin mononucleotide phosphatase.
damX, dam, rpe, aroB, uvrD, and yigB were required for fitness under pH 3, starvation, and H2O2. Disruption of damX in S. enterica causes bile sensitivity (79). The DNA adenine methylation gene (dam) plays an important role in bacterial gene expression and virulence (80). A dam mutant of S. enterica is extremely attenuated in mice (81). The gene aroB encodes dehydroquinate synthase, a part of the shikimate pathway, which is essential for bacteria and is absent in mammals (82). In prokaryote species, uvrD is involved in maintaining genomic stability and helps DNA lesion repair, mismatch repair, nucleotide excision repair, and recombinational repair (83). Overproduction of yigB produced higher-level persister cells that exhibit multidrug tolerance in E. coli (84). However, deletion of gidB (glucose-inhibited division gene B) confers high-level antimicrobial resistance in Salmonella and has compromised overall bacterial fitness compared to wild type (85). GidA (together with mnmE) is responsible for the proper biosynthesis of 5-methylaminomethtyl-2-thouridine of tRNAs, and deletion causes attenuation in bacterial pathogenesis (86). In addition, mrp gene involved in thiamine synthesis was shown to be required for survival in both pH3 and starvation (Fig. 3B).
ATP synthase genes are obligatory for Salmonella fitness during in vitro host stressors.
ATP synthase (FoF1-ATPase) is a ubiquitous enzyme largely conserved across all domains of life. All of the eight genes encoding ATP synthase subunit proteins were required for the fitness of S. Typhimurium under all five in vitro conditions of our study (Fig. 2A and 3A). FoF1-ATP synthase complex is required for ATP production from ADP and Pi. ATP synthase also regulates pH homeostasis in bacteria (Listeria monocytogenes and S. Typhimurium) at the expense of ATP (87). In Streptococcal faecalis, upregulation of FoF1-ATPase promotes ATP-dependent H+ extrusion under acidic conditions. However, in E. coli, the expression of ATP synthase is decreased under acidic conditions (88). ATP synthase in Mycobacterium and Staphylococcus has been validated as a promising target for new antimicrobial drugs (87, 89).
Mutant phenotypic assays for growth and survival.
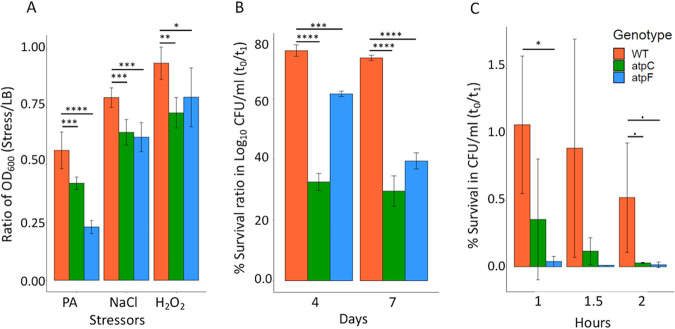
In light of the importance of ATP synthase genes, two single-gene deletion mutants, ΔaptC and ΔatpF, were chosen for phenotypic validation. Both of the mutants had significantly reduced growth compared to wild-type S. Typhimurium in 100 M PA, 3% NaCl, and 1 mM H2O2 after 6 h of growth (Fig. 4A). In fact, growth of the two mutants was impaired in LB broth compared to the wild type. However, the growth defect was even more severe under the stressors as clearly illustrated in Fig. 3A, which allowed for identification of the eight genes in the atp operon as CEGs under all five stress conditions. Similarly, the survival of both mutants was significantly lower than wild-type S. Typhimurium after starvation in 1× phosphate-buffered saline (PBS) for 4 days and 7 days (Fig. 4B). Additionally, the survival fitness of only the ΔatpF mutant was significantly reduced compared to wild-type S. Typhimurium when incubated at pH 3 for 1 h. A trend for a decrease in survival fitness was observed for both mutants compared to the wild type when incubated for 2 h at pH 3 (Fig. 4C). For the survival assays (starvation and pH 3), sampling was performed at various time points to determine survival. However, we picked the time points that can highlight the mutant phenotypes more clearly to present the data in Fig. 4B and C. The mutant phenotypes showed the same trends in reference to the wild type to various degrees over the sampling time points.
FIG 4.
Mutant phenotypic assays for growth and survival. Growth (A) and survival assays (B and C) of wild type (WT), ΔaptC, and ΔatpF. (A) Bacteria were treated with 100 mM propionate, 3% NaCl, and 1 mM H2O2 in LB, and growth (OD600) was measured at 6 h. (B) Bacteria were starved for 4 (t1) days and 7 (t1) days in 1× PBS. Viable bacteria counts were enumerated by plating serial dilutions on LB agar plates. (C) Bacteria were incubated in glycine-HCl buffer (pH 3.0) and incubated for 1, 1.5, and 2 h. Viable cells were enumerated by plating serial dilutions. Bar represents mean ± SE. All of the above-described experiments were performed in ≥3 replicates. Statistical analysis was performed using one-way analysis of variance (ANOVA) with correcting for multiple comparisons using the Holm-Sidak method; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Phenotypic bases of Salmonella in vivo fitness genes required for enteric and systemic infection.
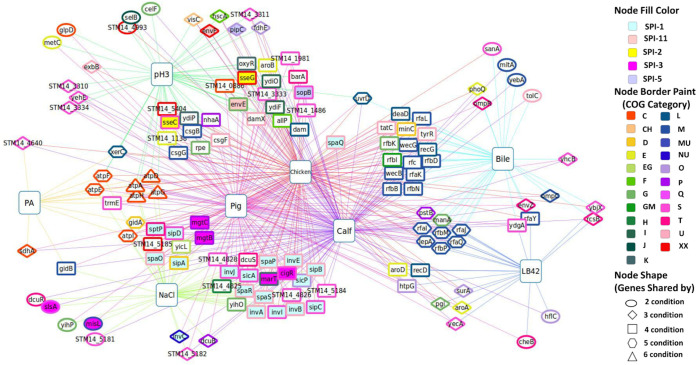
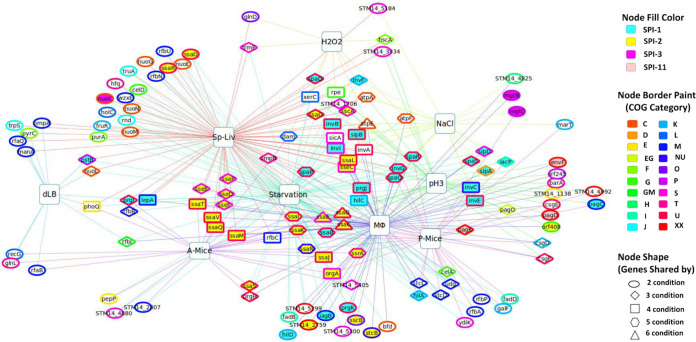
Numerous genes in S. Typhimurium that are required for in vivo fitness during infection in either cell culture or animal infection models have been identified in previous studies, suggesting that they are required by S. Typhimurium to overcome host defenses. However, for a large portion of them, the phenotypic basis by which they are required in particular in vivo niches remain unknown. Therefore, we constructed the genotype-phenotype network diagrams in Fig. 5 (enteric infection) and Fig. 6 (systemic infection) to show all the genes that are important for fitness under at least one of the in vitro conditions (this study and our previous study), which are also important for fitness in at least one of the in vivo infection models (previous studies from other labs). The genes that were important either under the in vitro or in vivo conditions only were excluded from the diagrams. The information on the common requirements of the genes shown in these networks (Fig. 5 and 6) for at least one well-defined in vitro stress and in vivo infection model is valuable because it provides new insights into the nature of the selective pressures S. Typhimurium might be facing during infection in the host and which genetic factors S. Typhimurium uses to overcome each particular stressor.
FIG 5.
Genotype-phenotype network connections illustrating phenotypic basis of S. Typhimurium genetic factors required for enteric infection (in vitro versus in vivo [enteric]). Large square nodes indicate various conditions (studies), and small nodes are fitness genes. Each node (gene) is shared by at least one of the in vitro conditions (i.e., stressors encountered by Salmonella during enteric infection; PA, pH 3, NaCl, bile, and LB42; current study and our previous study) and at least one of the in vivo enteric conditions (pig, calf, and chicken; a previous study). The interactive network through the network data exchange (NDEx) is available at www.ndexbio.org/#/network/027b067d-e209-11e8-aaa6-0ac135e8bacf (118).
FIG 6.
Genotype-phenotype network connections illustrating phenotypic basis of S. Typhimurium genetic factors required for systemic infection (in vitro versus in vivo [systemic]). Large square nodes indicate various conditions (studies), and small nodes are fitness genes. Each node (gene) is shared by at least one of the in vitro conditions (i.e., stressors encountered by Salmonella inside macrophages; NaCl, H2O2, pH 3, starvation, and dLB; current study and our previous study) and at least one of the in vivo systemic conditions (MΦ, Sp-Liv, P-Mice, and A-Mice; previous studies). The interactive network through the network data exchange (NDEx) is available at www.ndexbio.org/#/network/5e78ad70-e209-11e8-aaa6-0ac135e8bacf (118).
(i) Enteric infection. We have identified an overlapping set of 135 CEGs that are commonly required to cause enteric infection in at least one of the hosts (pig, calf, and chicken [10]) and for fitness in one of the in vitro host stressors (LB42, bile [12], pH 3, PA, and NaCl) encountered during enteric infection (Fig. 5; Table S6). Genes in SPI-1 (invABCEIJ, sicAP, sipABCD, spaOPQRS, and sptP) and SPI-3 (cigR, marT, mgtBC, misL, and slsA) were required for fitness in NaCl and all hosts. However, genes encoding SPI-2 (sseCG), SPI-5 (slsA and pipC), and SPI-11 (envEF) were essential for fitness in only one in vitro stressor pH 3 and intestinal colonization in three hosts. Other enriched pathways were lipopolysaccharide biosynthesis (rfaIJKLQY and rfbBDKMNP), oxidative phosphorylation (ATP synthase genes and sdhA), and biosynthesis of amino acids (aroABD, rpe, and metC) including others as shown in the STRING protein-protein interaction against S. enterica LT2 (https://version-11-5.string-db.org/cgi/network?networkId=bHPdi1Ibnskf).
High osmolality, low oxygen, and late log phase induce hilA expression in vitro that in turn regulates the expression of SPI-1 genes (90). Interestingly, we identified SPI-1 genes as fitness genes required for in vitro NaCl stress. Similarly, LPS biosynthetic process genes were enriched in LB42 and bile and in pig, calf, and chicken for fitness during enteric infection. LPS, a critical factor in the virulence of Gram-negative bacterial infection, is required for intestinal colonization, resistance to killing by macrophages, swarming motility, serum resistance, and bile stress (12, 91). A csgBA (curli subunit protein) mutant of S. Typhimurium was attenuated in its ability to elicit fluid accumulation in bovine ligated ileal loops (92) and is required for fitness at pH 3, including csgF and csgG. Additionally, putative proteins STM14_1138, STM14_1486, STM14_1981, and STM14_3333 and STM14_4826, STM14_4828, STM14_5184, and STM14_5185 (hypothetical protein) were required for fitness in vitro under acidic and osmotic stress, respectively, and enteric infection in the three hosts.
(ii) Systemic infection. We compared the CEGs that are shared between at least one of the in vitro host stressors (H2O2, NaCl, pH 3, starvation, and dLB [12]), encountered inside MΦ and in vivo systemic infections (MΦ [9], A-Mice [9], P-Mice [8], and Sp-Liv [11]) and identified an overlapping set of 130 genes (Fig. 6; Table S7) shown in a protein-protein interaction network using STRING (https://version-11-5.string-db.org/cgi/network?networkId=bqRqTdvi8p4u). SPI-1 genes (hilACD, iacP, iagB, invABCEFGI, orgA, prgHIJK, sicA, sipABC, and spaOPQRS) encoding T3SS were essential for fitness in NaCl, starvation, MΦ survival, and systemic infection. Additionally, SPI-2 genes (ssaBCDEGIJKLMNOPQRSTV, orf245, orf408, sscAB, sseCDEF, ssrA, and STM14_1706) encoding T3SS were required for fitness in pH 3, starvation, MΦ survival, and systemic infection. Similarly, SPI-3 genes (marBCT) were required for fitness in NaCl, MΦ survival, and persistent infection in mice (P-Mice). SPI-11 genes (envF and pagCD) were required for fitness in pH 3, MΦ survival, and P-Mice.
Other than SPI genes, the major enriched genes were nucleic acid metabolic process (dam, trpS, MnmE, truA, serc, csgD, ompR, and cra), lipopolysaccharide biosynthetic process (rfbABCNPU, rfaB, udg, and galF), oxidative phosphorylation (ATP synthase genes and NADH dehydrogenase genes), and two-component system (ompR, barA, phoQ, glnDL, and pagKO) among others (Fig. 6). The gene dam was required for fitness in H2O2, NaCl, A-Mice, and Sp-Liv. XerC and rpe were required for H2O2, pH 3, starvation, and Sp-Liv. Interestingly, pagK was not identified as a CEG in A-Mice, P-Mice, and Sp-Liv but in pH 3, starvation, and MΦ. Putative genes essential for either in vitro or in vivo systemic infection were STM14_1138, STM14_4880, STM14_4992, STM14_5184, STM14_2759, STM14_2807, STM14_3334, STM14_4825, STM14_5299, and STM14_5300.
CONCLUSION
A recent study by Kroger et al. presented transcriptomes of S. Typhimurium under 22 distinct infection-relevant environmental conditions in vitro. The study found induction of Salmonella pathogenicity islands under in vitro conditions, such as early stationary phase, anaerobic growth, oxygen shock, nitric oxide shock, and pH 3, NaCl, bile, and peroxide shock among others (93). However, transcription of a gene does not necessarily indicate the requirement for that gene function for fitness under a given particular condition. The transcript can be a leaky expression or required for fitness in the upcoming environment through predictive adaptation, phenomena where bacteria can anticipate and preemptively respond to regular environmental fluctuations (temporally distributed stimuli) that confers a considerable fitness advantage for the survival of an organism (94, 95). Traditionally, it is believed that the “central dogma of life” that information flows from DNA to RNA to proteins is highly concordant. However, there is a modest correlation between levels of transcripts and corresponding proteins (96–98). Thus, functional genomics screening such as Tn-seq is expected to reveal more direct functional aspects of the genes involved in responding to the current stresses.
In this report, we were able to map genotype-to-phenotype links, providing the phenotypic basis of the genetic requirements for fitness for an overlapping set of 221 virulence genes for in vivo fitness (Fig. S8 in the supplemental material). These CEGs were required for fitness for at least one of the in vitro host stressors (PA, NaCl, pH 3, starvation, bile, LB42, and dLB) and enteric infection (calf, chicken, and pig) or systemic infection (mice, including intracellular survival inside macrophages). Forty-four common CEGs were required to cause both systemic and enteric infections (in vivo fitness) and in vitro fitness (Fig. S8; Table 1). Common SPI genes for in vivo and in vitro fitness were SPI-1 (invABCEI, sicA, sipABD, and spaOPQRS), SPI-2 (sseC), SPI-3 (marT and mgtCB), and SPI-11 (envF). Salmonella genes other than SPI essential for fitness under in vitro stresses and in vivo survival were atpAEF, lepA, dam, pstB, xerC, manA, phoQ, rfaQ, rfbBIP, rpe, trmE, rfbIP, ompR, csgF, recG, hscA, barA, and putative genes STM14_1138, STM14_3334, STM14_4825, and STM14_5184 (Table 1).
TABLE 1.
Salmonella genes required for in vitro and in vivo (enteric and systemic) fitness
| Categorya | Genesa,b | Conditions (in vitro, enteric, and systemic)c,d | COGc | Protein name |
|---|---|---|---|---|
| SPI genes | ||||
| SPI-1* | invA | Na, S, C, P, Ch, MΦ, SL | U | Needle complex export protein |
| invB | Na, S, C, P, Ch, MΦ, SL | U | Secretion chaperone | |
| invC | Na, S, C, P, MΦ, PM | NU | ATP synthase SpaL | |
| invE | Na, S, C, P, Ch, MΦ, SL | U | Invasion protein | |
| invI | Na, S, C, P, Ch, MΦ, SL | S | Needle complex assembly protein | |
| sicA | Na, S, C, P, Ch, MΦ, SL | S | Secretion chaperone | |
| sipA | Na, S, C, P, Ch, MΦ | D | Secreted effector protein | |
| sipB | Na, S, C, P, Ch, MΦ, SL | U | Translocation machinery component | |
| sipD | Na, S, C, P, Ch, MΦ | S | Translocation machinery component | |
| spaO | Na, S, C, P, Ch, MΦ, PM, SL | U | Surface presentation of antigens protein SpaO | |
| spaP | Na, S, C, P, Ch, MΦ, AM, SL | U | Surface presentation of antigens protein SpaP | |
| spaQ | Na, S, C, P, Ch, SL | U | Needle complex export protein | |
| spaR | Na, S, C, P, Ch, MΦ, PM, SL | U | Needle complex export protein | |
| spaS | Na, S, C, P, Ch, MΦ | U | Surface presentation of antigens protein SpaS | |
| SPI-2* | sseC | pH, S, C, P, Ch, MΦ, SL | S | Translocation machinery component |
| SPI-3 | marT | Na, C, P, Ch, PM | K | Putative transcriptional regulator |
| SPI-11 | envF | pH, C, Ch, MΦ | XX | Putative envelope lipoprotein |
| Non-SPI genes | ||||
| Two-component system | ompR* | B, S, C, P, MΦ, SL | T | Osmolality response regulator |
| phoQ* | B, S, dLB, C, Ch, AM, SL | E | Sensor protein PhoQ | |
| barA | pH, S, C, P, Ch, MΦ | T | Hybrid sensory histidine kinase BarA | |
| O antigen biosynthetic process | rfbB | B, S, C, P, Ch, AM, SL | M | dTDP-glucose-4,6-dehydratase |
| rfbP | B, S, L4, C, P, Ch, PM | M | Undecaprenol-phosphate galactosephosphotransferase/O-antigen transferase | |
| rfbN | B, S, C, P, Ch, SL | M | Rhamnosyl transferase | |
| ATP synthase genes* | atpA | PA, Na, pH, H2, S, C, P, Ch, SL | C | FoF1-ATP synthase subunit alpha |
| atpE | PA, Na, pH, H2, S, P, Ch, MΦ, SL | C | FoF1-ATP synthase subunit C | |
| atpF | PA, Na, pH, H2, S, C, Ch, MΦ | C | FoF1-ATP synthase subunit B | |
| Mismatch repair | dam* | pH, H2, S, C, P, Ch, AM, SL | L | DNA adenine methylase |
| Chromosome segregation | xerC* | PA, pH, H2, S, C, P, Ch, SL | L | Site-specific tyrosine recombinase XerC |
| Fructose and mannose metabolism | manA* | B, L4, dLB, C, P, Ch, SL | G | Mannose-6-phosphate isomerase |
| Carbon metabolism | rpe* | pH, H2, S, C, P, Ch, SL | G | Ribulose-phosphate 3-epimerase |
| Homologous recombination | recG | B, dLB, C, P, Ch, AM | L | ATP-dependent DNA helicase RecG |
| ABC transporter | pstB* | B, L4, S, dLB, C, P, Ch, SL | P | Phosphate transporter subunit |
| Translational elongation | lepA* | B, L4, S, dLB, C, P, Ch, MΦ, SL | M | GTP-binding protein LepA |
| Iron-sulfur cluster assembly | hscA | pH, H2, C, Ch, MΦ, SL | F | Chaperone protein HscA |
| Others | csgF* | pH, C, P, Ch, MΦ, PM | U | Curli assembly protein CsgF |
| rfaQ | B, L4, dLB, C, P, Ch, SL | M | Lipopolysaccharide core biosynthesis protein | |
| rfbI | B, S, dLB, C, P, Ch, MΦ | GM | CDP-6-deoxy-delta-3,4-glucoseen reductase | |
| trmE* | PA, H2, S, C, P, Ch, SL | S | tRNA modification GTPase TrmE | |
| Putative protein | STM14_1138 | pH, C, P, Ch, MΦ | E | Putative transcriptional regulator |
| STM14_3334 | pH, C, P, SL | S | Putative DNA/RNA helicase | |
| STM14_4825 | Na, C, P, Ch, MΦ | H | Coproporphyrinogen III oxidase | |
| STM_5184 | Na, C, P, Ch, SL | S | Putative inner membrane protein | |
Genes marked with an asterisk (*) have been implicated in vaccine development or drug targeting against a wide range of bacteria.
The genes listed are required for both in vitro and in vivo fitness (enteric and systemic infection) (i.e., conditions listed in Fig. 5 and 6).
COG, cluster of orthologous groups (same as Fig. 2); SPI, Salmonella pathogenicity island; Na, NaCl; H2, H2O2; S, starvation; C, cattle; P, pig; Ch, chicken; MΦ, macrophage; SL, Sp-Liv; pH, pH 3; B, bile; L4, LB42; AM, A-Mice; PM, P-Mice.
For conditions, italic font indicates in vitro, bold font indicates enteric, and underlining indicates systemic.
Interestingly, most of the common 44 genes required for in vitro and in vivo (enteric and systemic infection) fitness have been implicated in vaccine or drug target development against a broad spectrum of bacteria; for example, ATP synthase genes (87, 89), dam (99, 100), pstB (100), phoQ (101), ompR (102), xerC (103), and rfbBPN (104), manA (105), rpe (106), lepA (107), csgF (108), trmE (109), and SPI-1 and SPI-2 (110) have been used in vaccine development or drug target development (Table 1). Thus, there lies a great potential to explore genes marBCT, envF, barA, hscA, rfaQ, rfbI, and putative proteins STM14_1138, STM14_3334, STM14_4825, and STM_5184 as novel therapeutic and intervention strategies to curb Salmonella infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. Typhimurium 14028s, a spontaneous mutant resistant to nalidixic acid (NA), was grown in LB plates or LB medium (BD Difco, Sparks, MD) on a shaking rack at 225 rpm and incubated at 37°C unless otherwise indicated. The single-gene deletion mutants ΔatpC and ΔatpF in S. Typhimurium 14028s strain background were obtained from BEI Resources, NIAID, NIH, Salmonella enterica subsp. enterica, strain 14028s (serovar Typhimurium) single-gene deletion mutant library, plate SGD_156/157_Kan, NR-42849. NA (ICN Biomedicals Inc., Aurora OH, USA) and kanamycin (Km; Shelton Scientific Inc. CT, USA) were used at 25 μg/mL and 50 μg/mL, respectively. The S. Typhimurium strains were stored in 50% glycerol at −80°C. All procedures involving this bacterial pathogen (biosafety level 2) were conducted according to the protocol approved by the Institutional Biosafety Committee (IBC) at the University of Arkansas.
Construction of transposon mutant library.
To prepare electrocompetent cells, S. Typhimurium 14028s (NAr) was grown overnight in 10 mL of LB medium supplemented with NA, which was subsequently diluted 100-fold in 10 mL of 2× yeast extract tryptone (2xYT) medium (BD Difco, Sparks, MD, USA) containing NA and incubated for 3 h on a shaking rack. Bacterial cells were washed 6 times with wash solution (cold 10% glycerol). Centrifugation was done at 8,000 rpm for 1 min at refrigeration temperature (4°C). The bacterial pellet was resuspended gently in 60 μL of wash solution, preventing aeration. One microliter of the EZ-Tn5 <KAN-2> Tnp transposome complex (Epicentre BioTechnologies, Madison, WI, USA) was added to electrocompetent S. Typhimurium cells and incubated on ice for 10 min. Then, the mixture was gently transferred to an ice-cold cuvette, avoiding the formation of any air bubbles, and electroporated at 2,450 V. Immediately, 500 μL of super optimal broth with catabolite repression (SOC) was added and incubated for 90 min on a shaking rack at 37°C. The reaction was plated on LB plates supplemented with NA and Km to recover the Tn5 mutants. With 3 electroporations, we were able to collect approximately 350,000 Tn5 mutants and stored them in LB medium with 50% glycerol at −80°C (Fig. 1).
In vitro growth-based selections of transposon mutant library.
In vitro selection of the transposon mutant library was done as described previously (111) with some modifications. Briefly, the transposon mutant library was thawed on ice, and an aliquot of 300 μL was added to 60 mL of LB broth with NA and Km (optical density at 600 nm [OD600] = 0.131). The library was incubated at 37°C on a shaking rack for 30 min (OD600 = 0.135) and centrifuged at 5,500 rpm for 8 min at room temperature. The transposon mutant library pellet was resuspended in 50 mL of 1× PBS (OD600 = 0.143), and CFU (4 × 107 CFU/mL) was measured (t1). This step was included to prepare the mutant cells adapted to LB medium at 37°C, shortening the lag phase in the following selective conditions. Ten-milliliter aliquots were saved from t1 as an input pool (IP1). The above procedure was repeated to make a technical replicate of IP1 as input pool 2 (IP2). An aliquot of 0.5 mL from t1 was inoculated in 10 mL of LB, LB containing 100 mM propionate (pH adjusted to pH 7.0; PA), LB with 3% NaCl (NaCl), and LB containing 1 mM H2O2 (H2O2). The initial OD600 of the inoculated medium was 0.009. We then incubated the libraries on a shaking rack (225 rpm) at 37°C with variable incubation times ranging from 3.75 h to 7 h to reach a mid-logarithmic phase (t2). The final OD600 of all output pools was very similar across the selections and around 0.64 at time point t2. Input pool and output pool libraries were centrifuged, and the pellets were stored at −80°C for DNA extraction (Fig. 1) (LB, PA, NaCl, and H2O2).
In vitro survival-based selections of transposon mutant library.
To identify genes negatively selected during starvation, an aliquot of 0.5 mL from t1 was transferred to 10 mL of PBS and incubated at 37°C on a shaking rack for 12 days. On the 12th day, the tube was centrifuged, and the pellet was resuspended in 1 mL of PBS. A 100-μL aliquot was plated and incubated on an LB plate (NA + Km) overnight at 37°C. All colonies were collected in PBS and stored at −80°C for DNA extraction. Whereas for survival at pH 3, 0.5 mL from t1 was exposed to LB medium adjusted to pH 3 for 30 min at 37°C and immediately transferred to 40 mL of PBS. The cells were centrifuged at 8,000 rpm for 8 min, and the pellet was resuspended in 1 mL of PBS. An aliquot of 250 μL was plated and incubated on an LB plate (NA + Km) overnight at 37°C. All colonies were collected in PBS and stored at −80°C for DNA extraction (Fig. 1) (pH 3 and starvation). Under these two conditions (pH 3 and starvation), a subset of the mutants sensitive to the stressors would lose their cell viability to various degrees, while the mutant cells did not multiply in number. Therefore, to capture only those mutant cells that survived the stressors quantitatively by Tn-seq profiles, the output pools were prepared by recovering the mutant colonies on LB agar plates and combining them in sufficient numbers to represent the populations of all surviving mutants.
DNA library preparation for Illumina sequencing.
Genomic DNA (gDNA) was extracted from the bacterial cell pellets of input libraries (IP1 and IP2) and output libraries (LB, PA, NaCl, H2O2, pH 3, and starvation) using a QIAamp DNA minikit (Qiagen, Valencia, CA, USA) following the manufacturer’s protocol and stored at −80°C. Purity and concentration were checked using a Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA) with Qubit assay kits (double-stranded DNA [dsDNA] broad-range [BR] assay) following the manufacturer’s manual.
The sample for Illumina sequencing was prepared as previously described (15, 17, 18, 112). All DNA primers (Table S1 in the supplemental material) used for Tn-seq library construction were custom designed using Primer3 (v. 0.4.0) (113) and ordered from Integrated DNA Technologies (IDT; Coralville, IA). The simplified diagram for the preparation of the Tn-seq amplicon library is shown in Fig. S1A. Briefly, Tn5 junctions at the right end of the transposon were amplified from gDNA extracted from input and output libraries. The single primer linear extension was done with EZ-Tn5 primer3 using Taq DNA polymerase (New England Biolabs, Ipswich, MA, USA). The 50-μL linear PCR extension reaction constituted nuclease-free water (40 μL), ThermoPol buffer (10×, 5 μL), deoxynucleoside triphosphates (dNTPs; 2.5 mM each, 1 μL), EZ-Tn5 primer3 (20 μM, 1 μL), gDNA library (50 ng/μL, 2 μL, ∼100 ng), and Taq DNA polymerase (1 μL added during PCR). The PCR cycle consisted of manual hot start with the initial denaturation at 95°C for 2 min and addition of Taq DNA polymerase followed by 50 cycles of 95°C for 30 s, 62°C for 45 s, and 72°C for 10 s, which was then followed by a hold at 4°C. The linear PCR products were then purified with a MinElute PCR purification kit (Qiagen, Valencia, CA, USA) and eluted in 10 μL of elution buffer (EB) following the manufacturer’s protocol. Then, deoxycytidine homopolymer tail (C-tail) was added to the 3′ end of the linear PCR extension product using terminal transferase (TdT; New England Biolabs, Ipswich, MA, USA) enzyme as previously described (114, 115). The C-tailing reaction consisted of DNA (linear PCR extension product; 10 μL), TdT buffer (10×, 2 μL), CoCl2 (2.5 mM, 2 μL), dCTP (10 mM, 2.4 μL), ddCTP (1 mM, 1 μL), nuclease-free water (1.6 μL), and terminal transferase (1 μL), making a total volume of 20 μL. The reaction mixture was incubated at 37°C for 1 h followed by heat inactivation of the enzyme at 75°C for 20 min on a thermocycler. The C-tailed products were purified using a MinElute PCR purification kit and eluted to 10 μL.
Subsequently, the C-tailed PCR product was enriched with exponential PCR. PCR constituted nuclease-free water (35 μL), ThermoPol buffer (10×, 5 μL), dNTPs (2.5 mM each, 4 μL), IR2 barcoded (BC) primer (with Illumina adapter and barcode; 10 μM, 2 μL), HTM primer (with Illumina adapter; 20 μM, 1 μL), C-tailed DNA (2 μL), and Taq DNA polymerase (New England Biolabs; 1 μL), making a total volume of 50 μL. The manual hot start PCR cycle was comprised of 95°C for 2 min, followed by 25 cycles of 95°C for 30 s, 58°C for 45 s, and 72°C for 20 s, trailed by a final extension at 72°C for 10 min.
Finally, the exponential PCR products were heated at 65°C for 15 min and run on 1.5% agarose gels. The Tn-seq library showed a smear pattern, whereas S. Typhimurium wild type (negative control) showed almost no amplification (Fig. S1B). The gel was excised ranging from 300 to 500 bp, and DNA was extracted using a QIAquick gel extraction kit (Qiagen, Valencia, CA). The purity and concentration of DNA were measured using a Qubit 2.0 fluorometer. Equal amounts (∼10 ng) of DNA (gel-purified products) from each library were mixed and sent for next-generation sequencing on an Illumina HiSeq 2000 with single-end reads and 100 cycles (Center for Genome Research and Biocomputing, Oregon State University, Corvallis).
Analysis of Tn-seq data.
Raw reads from HiSeq Illumina sequencing were demultiplexed based on the barcodes to their respective libraries using a custom Perl script. The barcode and transposon sequences were trimmed off from the 5′ end. Consequently, the remaining sequence was Tn5 junction sequences with/without poly(C)-tail. Only 20 bp from the Tn5 junction were kept, discarding most of the poly(C)-tails. The reads were then aligned against the S. Typhimurium 14028s complete genome (NC_016856.1) using Bowtie version 0.12.7 (116). The aligned sequence (SAM mapping file) was fed to the ARTIST pipeline to identify conditionally essential genes (CEGs) using Con-ARTIST (27). Briefly, Tn5 insertion frequency was assigned to the S. Typhimurium 14028s genome divided into 100-bp window sizes. Uncorrected raw data (nonnormalized) of input and output libraries were normalized and were compared between the matching input and output pool using a Mann-Whitney U test (MWU). The MWU results were used to train the hidden Markov model (HMM) to predict the likelihood of loci that were not required for growth under either condition, essential under both conditions, and depleted in output library (P < 0.01). Only the insertions in the central 80% of the gene were considered to eliminate any insertions that may not disrupt the gene functions effectively. The cutoff of a >8-fold decrease was applied as an additional filter for identification of the depleted loci to those genes that showed significant changes (P < 0.01).
Comparative analysis of CEGs between in vitro and in vivo stressors.
We compared the in vitro essential genes identified in this study and our previous study (12) with the previously identified in vivo fitness genes. CEGs for acute infection of mice (A-Mice), macrophage survival (MΦ) (9), and persistent infection of mice (P-Mice) (8) were previously identified in S. Typhimurium strain SL1344 background. Additionally, Salmonella genes required for gastrointestinal colonization of pigs, calves, and chickens were identified in S. Typhimurium strain ST4/74 (10), and those for intraperitoneal infection of mice (Sp-Liv) were reported in S. Typhimurium strain 14028s background (11). The CEGs of different strains were searched for the corresponding orthologous genes in S. Typhimurium 14028s background using the prokaryotic genome analysis tool (PGAT) (117). To get insight into the phenotypic basis of CEGs required for in vivo intestinal colonization of pigs, calves, and chickens, these CEGs were compared with CEGs of in vitro host stressors found in the gut (current study: PA, NaCl, and pH 3; reference 12: bile and LB42). Similarly, for the phenotypic basis of CEGs for in vivo systemic infection (A-Mice, MΦ, P-Mice, and Sp-Liv), those CEGs were compared to in vitro macrophage stressors (current study: H2O2, NaCl, starvation, and pH 3; reference 12: dLB). Only the CEGs that were common between at least one of the in vitro host stressors and at least one of in vivo infections were identified and included in the comparative analysis.
Mutant phenotypic testing.
For growth assays, overnight cultures of bacteria were prepared as described above and inoculated in LB medium, LB medium containing 100 mM propionate (pH 7), LB medium containing 3% NaCl, and LB medium containing 1 mM H2O2. The OD600 was monitored every 1 h using a Tecan Infinite M200 plate reader (Tecan Trading AG, Switzerland) during incubation at 37°C with shaking (200 rpm) for 16 h. For survival assays, 0.1 mL of the overnight culture was washed 3 times in PBS (pH 7.0) and inoculated in 10 mL of 1× PBS (pH 7.0) and glycine-HCl buffer (pH 3.0) for starvation survival and pH 3.0 survival, respectively, and the cell suspensions were incubated at 37°C. For the starvation assay, an aliquot of the sample was collected at days 0, 4, and 7, and viable cells were enumerated by plating 10-fold serial dilutions on LB agar plates. For the pH 3 survival assay, an aliquot of the sample was collected at 0, 1.5, and 2 h, and viable cells were enumerated by plating 10-fold serial dilutions on LB agar plates. All of the assays were performed in at least 3 replications.
Data availability.
Sequencing data for Tn-seq analysis in this study are available on the NCBI Sequence Read Archive under BioProject number PRJNA385835.
ACKNOWLEDGMENTS
We thank the Arkansas High-Performance Computing Center (AHPCC), the University of Arkansas, for their computational support.
Footnotes
Supplemental material is available online only.
Contributor Information
Rabindra K. Mandal, Email: rmandal@iu.edu.
Jan Claesen, Lerner Research Institute.
REFERENCES
- 1.Ao TT, Feasey NA, Gordon MA, Keddy KH, Angulo FJ, Crump JA. 2013. Global burden of invasive nontyphoidal Salmonella disease, 2010. Emerg Infect Dis 21:941–949. doi: 10.3201/eid2106.140999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease ‘Burden of Illness’ Studies . 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 3.Carden S, Okoro C, Dougan G, Monack D. 2015. Non-typhoidal Salmonella Typhimurium ST313 isolates that cause bacteremia in humans stimulate less inflammasome activation than ST19 isolates associated with gastroenteritis. Pathog Dis 73:ftu023. doi: 10.1093/femspd/ftu023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crim SM, Griffin PM, Tauxe R, Marder EP, Gilliss D, Cronquist AB, Cartter M, Tobin-D'Angelo M, Blythe D, Smith K, Lathrop S, Zansky S, Cieslak PR, Dunn J, Holt KG, Wolpert B, Henao OL, Centers for Disease Control and Prevention (CDC) . 2015. Preliminary incidence and trends of infection with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 US sites, 2006–2014. MMWR Morb Mortal Wkly Rep 64:495–499. [PMC free article] [PubMed] [Google Scholar]
- 5.Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. 2012. Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 379:2489–2499. doi: 10.1016/S0140-6736(11)61752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 7.Kwon YM, Ricke SC, Mandal RK. 2016. Transposon sequencing: methods and expanding applications. Appl Microbiol Biotechnol 100:31–43. doi: 10.1007/s00253-015-7037-8. [DOI] [PubMed] [Google Scholar]
- 8.Lawley TD, Chan K, Thompson LJ, Kim CC, Govoni GR, Monack DM. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog 2:e11. doi: 10.1371/journal.ppat.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan K, Kim CC, Falkow S. 2005. Microarray-based detection of Salmonella enterica serovar Typhimurium transposon mutants that cannot survive in macrophages and mice. Infect Immun 73:5438–5449. doi: 10.1128/IAI.73.9.5438-5449.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudhuri RR, Morgan E, Peters SE, Pleasance SJ, Hudson DL, Davies HM, Wang J, van Diemen PM, Buckley AM, Bowen AJ, Pullinger GD, Turner DJ, Langridge GC, Turner AK, Parkhill J, Charles IG, Maskell DJ, Stevens MP. 2013. Comprehensive assignment of roles for Salmonella Typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet 9:e1003456. doi: 10.1371/journal.pgen.1003456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silva-Valenzuela CA, Molina-Quiroz RC, Desai P, Valenzuela C, Porwollik S, Zhao M, Hoffman RM, Andrews-Polymenis H, Contreras I, Santiviago CA, McClelland M. 2015. Analysis of two complementary single-gene deletion mutant libraries of Salmonella Typhimurium in intraperitoneal infection of BALB/c mice. Front Microbiol 6:1455. doi: 10.3389/fmicb.2015.01455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khatiwara A, Jiang T, Sung S-S, Dawoud T, Kim JN, Bhattacharya D, Kim H-B, Ricke SC, Kwon YM. 2012. Genome scanning for conditionally essential genes in Salmonella enterica serotype Typhimurium. Appl Environ Microbiol 78:3098–3107. doi: 10.1128/AEM.06865-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Opijnen T, Camilli A. 2013. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol 11:435–442. doi: 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawoud TM, Jiang T, Mandal RK, Ricke SC, Kwon YM. 2014. Improving the efficiency of transposon mutagenesis in Salmonella enteritidis by overcoming host-restriction barriers. Mol Biotechnol 56:1004–1010. doi: 10.1007/s12033-014-9779-4. [DOI] [PubMed] [Google Scholar]
- 16.Karash S, Liyanage R, Qassab A, Lay JO, Kwon YM. 2017. A comprehensive assessment of the genetic determinants in Salmonella Typhimurium for resistance to hydrogen peroxide using proteogenomics. Sci Rep 7:17073. doi: 10.1038/s41598-017-17149-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandal RK, Kwon YM. 2017. Global screening of Salmonella enterica serovar Typhimurium genes for desiccation survival. Front Microbiol 8:1723. doi: 10.3389/fmicb.2017.01723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandal RK, Jiang T, Kwon YM. 2017. Essential genome of Campylobacter jejuni. BMC Genomics 18:616. doi: 10.1186/s12864-017-4032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ha SD, Maciorowski KG, Kwon YM, Jones FT, Ricke SC. 1998. Survivability of indigenous microflora and a Salmonella typhimurium marker strain in poultry mash treated with buffered propionic acid. Anim Feed Sci and Technol 75:145–155. doi: 10.1016/S0377-8401(98)00195-3. [DOI] [Google Scholar]
- 20.Nava GM, Bielke LR, Callaway TR, Castaneda MP. 2005. Probiotic alternatives to reduce gastrointestinal infections: the poultry experience. Anim Health Res Rev 6:105–118. doi: 10.1079/ahr2005103. [DOI] [PubMed] [Google Scholar]
- 21.Sleator RD, Hill C. 2002. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol Rev 26:49–71. doi: 10.1111/j.1574-6976.2002.tb00598.x. [DOI] [PubMed] [Google Scholar]
- 22.Smith JL. 2003. The role of gastric acid in preventing foodborne disease and how bacteria overcome acid conditions. J Food Prot 66:1292–1303. doi: 10.4315/0362-028x-66.7.1292. [DOI] [PubMed] [Google Scholar]
- 23.Lee EJ, Choi J, Groisman EA. 2014. Control of a Salmonella virulence operon by proline-charged tRNA(Pro). Proc Natl Acad Sci USA 111:3140–3145. doi: 10.1073/pnas.1316209111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenberger CM, Finlay BB. 2003. Phagocyte sabotage: disruption of macrophage signalling by bacterial pathogens. Nat Rev Mol Cell Biol 4:385–396. doi: 10.1038/nrm1104. [DOI] [PubMed] [Google Scholar]
- 25.van der Heijden J, Bosman ES, Reynolds LA, Finlay BB. 2015. Direct measurement of oxidative and nitrosative stress dynamics in Salmonella inside macrophages. Proc Natl Acad Sci USA 112:560–565. doi: 10.1073/pnas.1414569112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slauch J, Taylor R, Maloy S. 1997. Survival in a cruel world: how Vibrio cholerae and Salmonella respond to an unwilling host. Genes Dev 11:1761–1774. doi: 10.1101/gad.11.14.1761. [DOI] [PubMed] [Google Scholar]
- 27.Pritchard JR, Chao MC, Abel S, Davis BM, Baranowski C, Zhang YJ, Rubin EJ, Waldor MK. 2014. ARTIST: high-resolution genome-wide assessment of fitness using transposon-insertion sequencing. PLoS Genet 10:e1004782. doi: 10.1371/journal.pgen.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, Shales M, Lovett S, Winkler ME, Krogan NJ, Typas A, Gross CA. 2011. Phenotypic landscape of a bacterial cell. Cell 144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Opijnen T, Camilli A. 2012. A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Res 22:2541–2551. doi: 10.1101/gr.137430.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen LJ, Julien P, Kuhn M, von Mering C, Muller J, Doerks T, Bork P. 2008. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res 36:D250–D254. doi: 10.1093/nar/gkm796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barquist L, Langridge GC, Turner DJ, Phan MD, Turner AK, Bateman A, Parkhill J, Wain J, Gardner PP. 2013. A comparison of dense transposon insertion libraries in the Salmonella serovars Typhi and Typhimurium. Nucleic Acids Res 41:4549–4564. doi: 10.1093/nar/gkt148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knuth K, Niesalla H, Hueck CJ, Fuchs TM. 2004. Large‐scale identification of essential Salmonella genes by trapping lethal insertions. Mol Microbiol 51:1729–1744. doi: 10.1046/j.1365-2958.2003.03944.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhang R, Ou HY, Zhang CT. 2004. DEG: a database of essential genes. Nucleic Acids Res 32:D271–D272. doi: 10.1093/nar/gkh024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Q, Wood TK. 2011. Protein acetylation in prokaryotes increases stress resistance. Biochem Biophys Res Commun 410:846–851. doi: 10.1016/j.bbrc.2011.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawhon SD. 2002. Genetic and environmental regulation of virulence genes in Salmonella enterica serovar Typhimurium. PhD dissertation. NC State University, Raleigh, NC. [Google Scholar]
- 36.Hasan CMM, Shimizu K. 2008. Effect of temperature up-shift on fermentation and metabolic characteristics in view of gene expressions in Escherichia coli. Microb Cell Fact 7:35. doi: 10.1186/1475-2859-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt H, Hensel M. 2004. Pathogenicity islands in bacterial pathogenesis. Clin Microbiol Rev 17:14–56. doi: 10.1128/CMR.17.1.14-56.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee EJ, Groisman EA. 2012. Tandem attenuators control expression of the Salmonella mgtCBR virulence operon. Mol Microbiol 86:212–224. doi: 10.1111/j.1365-2958.2012.08188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dorsey CW, Laarakker MC, Humphries AD, Weening EH, Bäumler AJ. 2005. Salmonella enterica serotype Typhimurium MisL is an intestinal colonization factor that binds fibronectin. Mol Microbiol 57:196–211. doi: 10.1111/j.1365-2958.2005.04666.x. [DOI] [PubMed] [Google Scholar]
- 40.Yin J, Xia J, Tao M, Xu L, Li Q, Geng S, Jiao X. 2016. Construction and characterization of a cigR deletion mutant of Salmonella enterica serovar Pullorum. Avian Pathol 45:569–575. doi: 10.1080/03079457.2016.1187708. [DOI] [PubMed] [Google Scholar]
- 41.Figueira R, Watson KG, Holden DW, Helaine S. 2013. Identification of Salmonella pathogenicity island-2 type III secretion system effectors involved in intramacrophage replication of S. enterica serovar typhimurium: implications for rational vaccine design. mBio 4:e00065. doi: 10.1128/mBio.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibson DL, White AP, Snyder SD, Martin S, Heiss C, Azadi P, Surette M, Kay WW. 2006. Salmonella produces an O-antigen capsule regulated by AgfD and important for environmental persistence. J Bacteriol 188:7722–7730. doi: 10.1128/JB.00809-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murinova S, Dercova K. 2014. Response mechanisms of bacterial degraders to environmental contaminants on the level of cell walls and cytoplasmic membrane. Int J Microbiology 2014:873081. doi: 10.1155/2014/873081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weber A, Jung K. 2002. Profiling early osmostress-dependent gene expression in Escherichia coli using DNA macroarrays. J Bacteriol 184:5502–5507. doi: 10.1128/JB.184.19.5502-5507.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imlay JA, Linn S. 1986. Bimodal pattern of killing of DNA-repair-defective or anoxically grown Escherichia coli by hydrogen peroxide. J Bacteriol 166:519–527. doi: 10.1128/jb.166.2.519-527.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wakamatsu T, Kim K, Uemura Y, Nakagawa N, Kuramitsu S, Masui R. 2011. Role of RecJ-like protein with 5'-3' exonuclease activity in oligo(deoxy)nucleotide degradation. J Biol Chem 286:2807–2816. doi: 10.1074/jbc.M110.161596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henderson ML, Kreuzer KN. 2015. Functions that protect Escherichia coli from tightly bound DNA-protein complexes created by mutant EcoRII methyltransferase. PLoS One 10:e0128092. doi: 10.1371/journal.pone.0128092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riordan JT, Tietjen JA, Walsh CW, Gustafson JE, Whittam TS. 2010. Inactivation of alternative sigma factor 54 (RpoN) leads to increased acid resistance, and alters locus of enterocyte effacement (LEE) expression in Escherichia coli O157: H7. Microbiology (Reading) 156:719–730. doi: 10.1099/mic.0.032631-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hartman CE, Samuels DJ, Karls AC. 2016. Modulating Salmonella Typhimurium’s response to a changing environment through bacterial enhancer-binding proteins and the RpoN regulon. Front Mol Biosci 3:41. doi: 10.3389/fmolb.2016.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hwang S, Jeon B, Yun J, Ryu S. 2011. Roles of RpoN in the resistance of Campylobacter jejuni under various stress conditions. BMC Microbiol 11:207. 207–2180-11–207. doi: 10.1186/1471-2180-11-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Denoncin K, Vertommen D, Arts IS, Goemans CV, Rahuel-Clermont S, Messens J, Collet JF. 2014. A new role for Escherichia coli DsbC protein in protection against oxidative stress. J Biol Chem 289:12356–12364. doi: 10.1074/jbc.M114.554055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou L, Zhao M, Wolf RZ, Graham DE, Georgiou G. 2009. Transcriptional regulation of the Escherichia coli gene rraB, encoding a protein inhibitor of RNase E. J Bacteriol 191:6665–6674. doi: 10.1128/JB.00344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jitprasutwit S, Ong C, Juntawieng N, Ooi WF, Hemsley CM, Vattanaviboon P, Titball RW, Tan P, Korbsrisate S. 2014. Transcriptional profiles of Burkholderia pseudomallei reveal the direct and indirect roles of Sigma E under oxidative stress conditions. BMC Genomics 15:787. doi: 10.1186/1471-2164-15-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gogol EB, Rhodius VA, Papenfort K, Vogel J, Gross CA. 2011. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc Natl Acad Sci USA 108:12875–12880. doi: 10.1073/pnas.1109379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leonhartsberger S, Korsa I, Bock A. 2002. The molecular biology of formate metabolism in enterobacteria. J Mol Microbiol Biotechnol 4:269–276. [PubMed] [Google Scholar]
- 56.Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu Rev Microbiol 60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.D’Urzo N, Martinelli M, Pezzicoli A, De Cesare V, Pinto V, Margarit I, Telford JL, Maione D, Members of the DEVANI Study Group . 2014. Acidic pH strongly enhances in vitro biofilm formation by a subset of hypervirulent ST-17 Streptococcus agalactiae strains. Appl Environ Microbiol 80:2176–2185. doi: 10.1128/AEM.03627-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cotter PD, Hill C. 2003. Surviving the acid test: responses of Gram-positive bacteria to low pH. Microbiol Mol Biol Rev 67:429–453. doi: 10.1128/MMBR.67.3.429-453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gunn JS, Alpuche-Aranda CM, Loomis WP, Belden WJ, Miller SI. 1995. Characterization of the Salmonella Typhimurium pagC/pagD chromosomal region. J Bacteriol 177:5040–5047. doi: 10.1128/jb.177.17.5040-5047.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee YH, Kim S, Helmann JD, Kim B-H, Park YK. 2013. RaoN, a small RNA encoded within Salmonella pathogenicity island-11, confers resistance to macrophage-induced stress. Microbiology (Reading) 159:1366–1378. doi: 10.1099/mic.0.066688-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marcus EA, Sachs G, Scott DR. 2013. The role of ExbD in periplasmic pH homeostasis in Helicobacter pylori. Helicobacter 18:363–372. doi: 10.1111/hel.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahmer BM, Thomas MG, Larsen RA, Postle K. 1995. Characterization of the exbBD operon of Escherichia coli and the role of ExbB and ExbD in TonB function and stability. J Bacteriol 177:4742–4747. doi: 10.1128/jb.177.16.4742-4747.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reid AN, Pandey R, Palyada K, Whitworth L, Doukhanine E, Stintzi A. 2008. Identification of Campylobacter jejuni genes contributing to acid adaptation by transcriptional profiling and genome-wide mutagenesis. Appl Environ Microbiol 74:1598–1612. doi: 10.1128/AEM.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gu D, Xue H, Yuan X, Yu J, Xu X, Huang Y, Li M, Zhai X, Pan Z, Zhang Y, Jiao X. 2021. Genome-wide identification of genes involved in acid stress resistance of Salmonella Derby. Genes 12:476. doi: 10.3390/genes12040476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Archer CD, Wang X, Elliott T. 1993. Mutants defective in the energy-conserving NADH dehydrogenase of Salmonella Typhimurium identified by a decrease in energy-dependent proteolysis after carbon starvation. Proc Natl Acad Sci USA 90:9877–9881. doi: 10.1073/pnas.90.21.9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Michalik S, Liebeke M, Zühlke D, Lalk M, Bernhardt J, Gerth U, Hecker M. 2009. Proteolysis during long‐term glucose starvation in Staphylococcus aureus COL. Proteomics 9:4468–4477. doi: 10.1002/pmic.200900168. [DOI] [PubMed] [Google Scholar]
- 67.Klein G, Pack A, Bonaparte C, Reuter G. 1998. Taxonomy and physiology of probiotic lactic acid bacteria. Int J Food Microbiol 41:103–125. doi: 10.1016/s0168-1605(98)00049-x. [DOI] [PubMed] [Google Scholar]
- 68.Olekhnovich IN, Kadner RJ. 2007. Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J Bacteriol 189:6882–6890. doi: 10.1128/JB.00905-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hensel M. 2000. Salmonella pathogenicity island 2. Mol Microbiol 36:1015–1023. doi: 10.1046/j.1365-2958.2000.01935.x. [DOI] [PubMed] [Google Scholar]
- 70.Brunet YR, Khodr A, Logger L, Aussel L, Mignot T, Rimsky S, Cascales E. 2015. H-NS silencing of the Salmonella pathogenicity island 6-encoded type VI secretion system limits Salmonella enterica serovar Typhimurium interbacterial killing. Infect Immun 83:2738–2750. doi: 10.1128/IAI.00198-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Freeman ZN, Dorus S, Waterfield NR. 2013. The KdpD/KdpE two-component system: integrating K homeostasis and virulence. PLoS Pathog 9:e1003201. doi: 10.1371/journal.ppat.1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Darcan C, Ozkanca R, Idil O, Flint KP. 2009. Viable but non-culturable state (VBNC) of Escherichia coli related to EnvZ under the effect of pH, starvation and osmotic stress in sea water. Pol J Microbiol 58:307–317. [PubMed] [Google Scholar]
- 73.Batchelor E, Walthers D, Kenney LJ, Goulian M. 2005. The Escherichia coli CpxA-CpxR envelope stress response system regulates expression of the porins ompF and ompC. J Bacteriol 187:5723–5731. doi: 10.1128/JB.187.16.5723-5731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kenyon WJ, Sayers DG, Humphreys S, Roberts M, Spector MP. 2002. The starvation-stress response of Salmonella enterica serovar Typhimurium requires σE-, but not CpxR-regulated extracytoplasmic functions. Microbiology (Reading) 148:113–122. doi: 10.1099/00221287-148-1-113. [DOI] [PubMed] [Google Scholar]
- 75.Kato A, Groisman EA. 2008. The PhoQ/PhoP regulatory network of Salmonella enterica, p 7–21. In Utsumi R (ed), Bacterial signal transduction: networks and drug targets. Advances in experimental medicine and biology, vol 631. Springer, New York, NY. [Google Scholar]
- 76.Schakermann M, Langklotz S, Narberhaus F. 2013. FtsH-mediated coordination of lipopolysaccharide biosynthesis in Escherichia coli correlates with the growth rate and the alarmone (p)ppGpp. J Bacteriol 195:1912–1919. doi: 10.1128/JB.02134-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rowley G, Spector M, Kormanec J, Roberts M. 2006. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat Rev Microbiol 4:383–394. doi: 10.1038/nrmicro1394. [DOI] [PubMed] [Google Scholar]
- 78.Lüttmann D, Göpel Y, Görke B. 2012. The phosphotransferase protein EIIANtr modulates the phosphate starvation response through interaction with histidine kinase PhoR in Escherichia coli. Mol Microbiol 86:96–110. doi: 10.1111/j.1365-2958.2012.08176.x. [DOI] [PubMed] [Google Scholar]
- 79.López-Garrido J, Casadesús J. 2010. The DamX protein of Escherichia coli and Salmonella enterica. Gut Microbes 1:285–288. doi: 10.4161/gmic.1.4.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Low DA, Weyand NJ, Mahan MJ. 2001. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect Immun 69:7197–7204. doi: 10.1128/IAI.69.12.7197-7204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jakomin M, Chessa D, Baumler AJ, Casadesus J. 2008. Regulation of the Salmonella enterica std fimbrial operon by DNA adenine methylation, SeqA, and HdfR. J Bacteriol 190:7406–7413. doi: 10.1128/JB.01136-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Mendonca JD, Ely F, Palma MS, Frazzon J, Basso LA, Santos DS. 2007. Functional characterization by genetic complementation of aroB-encoded dehydroquinate synthase from Mycobacterium tuberculosis H37Rv and its heterologous expression and purification. J Bacteriol 189:6246–6252. doi: 10.1128/JB.00425-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kang J, Blaser MJ. 2006. UvrD helicase suppresses recombination and DNA damage-induced deletions. J Bacteriol 188:5450–5459. doi: 10.1128/JB.00275-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hansen S, Lewis K, Vulic M. 2008. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob Agents Chemother 52:2718–2726. doi: 10.1128/AAC.00144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mikheil DM, Shippy DC, Eakley NM, Okwumabua OE, Fadl AA. 2012. Deletion of gene encoding methyltransferase (gidB) confers high-level antimicrobial resistance in Salmonella. J Antibiot (Tokyo) 65:185–192. doi: 10.1038/ja.2012.5. [DOI] [PubMed] [Google Scholar]
- 86.Shippy DC, Fadl AA. 2014. tRNA modification enzymes GidA and MnmE: potential role in virulence of bacterial pathogens. Int J Mol Sci 15:18267–18280. doi: 10.3390/ijms151018267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Balemans W, Vranckx L, Lounis N, Pop O, Guillemont J, Vergauwen K, Mol S, Gilissen R, Motte M, Lancois D, De Bolle M, Bonroy K, Lill H, Andries K, Bald D, Koul A. 2012. Novel antibiotics targeting respiratory ATP synthesis in Gram-positive pathogenic bacteria. Antimicrob Agents Chemother 56:4131–4139. doi: 10.1128/AAC.00273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Krulwich TA, Sachs G, Padan E. 2011. Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol 9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lu P, Lill H, Bald D. 2014. ATP synthase in mycobacteria: special features and implications for a function as drug target. Biochim Biophys Acta 1837:1208–1218. doi: 10.1016/j.bbabio.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 90.Lostroh CP, Lee CA. 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect 3:1281–1291. doi: 10.1016/S1286-4579(01)01488-5. [DOI] [PubMed] [Google Scholar]
- 91.Kong Q, Yang J, Liu Q, Alamuri P, Roland KL, Curtiss R III.. 2011. Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar Typhimurium. Infect Immun 79:4227–4239. doi: 10.1128/IAI.05398-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tükel C, Raffatellu M, Humphries AD, Wilson RP, Andrews-Polymenis HL, Gull T, Figueiredo JF, Wong MH, Michelsen KS, Akçelik M, Adams LG, Bäumler AJ. 2005. CsgA is a pathogen‐associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll‐like receptor 2. Mol Microbiol 58:289–304. doi: 10.1111/j.1365-2958.2005.04825.x. [DOI] [PubMed] [Google Scholar]
- 93.Kröger C, Colgan A, Srikumar S, Händler K, Sivasankaran SK, Hammarlöf DL, Canals R, Grissom JE, Conway T, Hokamp K, Hinton JCD. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 94.Mitchell A, Romano GH, Groisman B, Yona A, Dekel E, Kupiec M, Dahan O, Pilpel Y. 2009. Adaptive prediction of environmental changes by microorganisms. Nature 460:220–224. doi: 10.1038/nature08112. [DOI] [PubMed] [Google Scholar]
- 95.Tagkopoulos I, Liu YC, Tavazoie S. 2008. Predictive behavior within microbial genetic networks. Science 320:1313–1317. doi: 10.1126/science.1154456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Foss EJ, Radulovic D, Shaffer SA, Ruderfer DM, Bedalov A, Goodlett DR, Kruglyak L. 2007. Genetic basis of proteome variation in yeast. Nat Genet 39:1369–1375. doi: 10.1038/ng.2007.22. [DOI] [PubMed] [Google Scholar]
- 97.Fu J, Keurentjes JJB, Bouwmeester H, America T, Verstappen FWA, Ward JL, Beale MH, de Vos RCH, Dijkstra M, Scheltema RA, Johannes F, Koornneef M, Vreugdenhil D, Breitling R, Jansen RC. 2009. System-wide molecular evidence for phenotypic buffering in Arabidopsis. Nat Genet 41:166–167. doi: 10.1038/ng.308. [DOI] [PubMed] [Google Scholar]
- 98.Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN, Farber CR, Sinsheimer J, Kang HM, Furlotte N, Park CC, Wen P-Z, Brewer H, Weitz K, Camp DG, Pan C, Yordanova R, Neuhaus I, Tilford C, Siemers N, Gargalovic P, Eskin E, Kirchgessner T, Smith DJ, Smith RD, Lusis AJ. 2011. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet 7:e1001393. doi: 10.1371/journal.pgen.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Garcia-Del Portillo F, Pucciarelli MG, Casadesus J. 1999. DNA adenine methylase mutants of Salmonella Typhimurium show defects in protein secretion, cell invasion, and M cell cytotoxicity. Proc Natl Acad Sci USA 96:11578–11583. doi: 10.1073/pnas.96.20.11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Garmory HS, Titball RW. 2004. ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect Immun 72:6757–6763. doi: 10.1128/IAI.72.12.6757-6763.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Miller SI, Kukral AM, Mekalanos JJ. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A 86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dougan G, Chatfield SN, Higgins CF, Dorman CJ. 1996. Vaccines comprising attenuated salmonella having mutations in the ompr genes. US patent US5527529A.
- 103.Hur J, Xiang Z, Feldman EL, He Y. 2011. Ontology-based Brucella vaccine literature indexing and systematic analysis of gene-vaccine association network. BMC Immunol 12:49. doi: 10.1186/1471-2172-12-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sturm S, Timmis KN. 1986. Cloning of the rfb gene region of Shigella dysenteriae 1 and construction of an rfb-rfp gene cassette for the development of lipopolysaccharide-based live anti-dysentery vaccines. Microb Pathog 1:289–297. doi: 10.1016/0882-4010(86)90054-9. [DOI] [PubMed] [Google Scholar]
- 105.Amineni U, Pradhan D, Marisetty H. 2010. In silico identification of common putative drug targets in Leptospira interrogans. J Chem Biol 3:165–173. doi: 10.1007/s12154-010-0039-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Edwards A, Dharamsi A, Vedadi M, Domagala M, Pinder B, Alam MZ, Nethery K, Houston S, Kimber M, Clarke T. 2004. Crystal structures of bacterial ribulose-phosphate 3-epimerases. Worldwide patent application WO2003CA01672.
- 107.Patton BS. 2007. Applications and mechanisms of colicin E1. PhD dissertation. Iowa State University, Ames, IA. [Google Scholar]
- 108.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. 2008. The biology and future prospects of antivirulence therapies. Nat Rev Microbiol 6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shippy DC, Fadl AA. 2014. Bacterial tRNA modification enzymes: potential role in biology and virulence. Microb Pathog 89:100–107. doi: 10.1016/j.micpath.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 110.Matulová M, Havlickova H, Sisak F, Rychlik I. 2012. Vaccination of chickens with Salmonella pathogenicity island (SPI) 1 and SPI2 defective mutants of Salmonella enterica serovar Enteritidis. Vaccine 30:2090–2097. doi: 10.1016/j.vaccine.2012.01.050. [DOI] [PubMed] [Google Scholar]
- 111.van Opijnen T, Lazinski DW, Camilli A. 2014. Genome‐wide fitness and genetic interactions determined by Tn‐seq, a high‐throughput massively parallel sequencing method for microorganisms. Curr Protoc Microbiol 36:1E.3.1–1E.3.24. doi: 10.1002/9780471729259.mc01e03s36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mandal RK. 2016. Genetic determinants of Salmonella and Campylobacter required for in vitro fitness. PhD dissertation. University of Arkansas, Fayetteville, AR. [Google Scholar]
- 113.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. 2012. Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lazinski DW, Camilli A. 2013. Homopolymer tail-mediated ligation PCR: a streamlined and highly efficient method for DNA cloning and library construction. Biotechniques 54:25–34. doi: 10.2144/000113981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Karash S, Jiang T, Samarth D, Chandrashekar R, Kwon YM. 2019. Preparation of transposon library and Tn-seq amplicon library for Salmonella Typhimurium. Methods Mol Biol 2016:3–15. doi: 10.1007/978-1-4939-9570-7_1. [DOI] [PubMed] [Google Scholar]
- 116.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brittnacher MJ, Fong C, Hayden HS, Jacobs MA, Radey M, Rohmer L. 2011. PGAT: a multistrain analysis resource for microbial genomes. Bioinformatics 27:2429–2430. doi: 10.1093/bioinformatics/btr418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pratt D, Chen J, Welker D, Rivas R, Pillich R, Rynkov V, Ono K, Miello C, Hicks L, Szalma S, Stojmirovic A, Dobrin R, Braxenthaler M, Kuentzer J, Demchak B, Ideker T. 2015. NDEx, the network data exchange. Cell Syst 1:302–305. doi: 10.1016/j.cels.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download Spectrum.00155-21-s0001.xlsx, XLSX file, 0.1 MB (105KB, xlsx)
Supplemental material. Download Spectrum.00155-21-s0002.pdf, PDF file, 1.7 MB (1.7MB, pdf)
Data Availability Statement
Sequencing data for Tn-seq analysis in this study are available on the NCBI Sequence Read Archive under BioProject number PRJNA385835.