Abstract
Background
Suicide represents a major health concern, especially in developing countries. While many demographic risk factors have been proposed, the underlying molecular pathology of suicide remains poorly understood. A body of evidence suggests that aberrant DNA methylation and expression is involved. In this study, we examined DNA methylation profiles and concordant gene expression changes in the prefrontal cortex of Mexicans who died by suicide.
Methods
In collaboration with the coroner’s office in Mexico City, brain samples of males who died by suicide (n = 35) and age-matched sudden death controls (n = 13) were collected. DNA and RNA were extracted from prefrontal cortex tissue and analyzed with the Infinium Methylation480k and the HumanHT-12 v4 Expression Beadchips, respectively.
Results
We report evidence of altered DNA methylation profiles at 4430 genomic regions together with 622 genes characterized by differential expression in cases vs controls. Seventy genes were found to have concordant methylation and expression changes. Metacore-enriched analysis identified 10 genes with biological relevance to psychiatric phenotypes and suicide (ADCY9, CRH, NFATC4, ABCC8, HMGA1, KAT2A, EPHA2, TRRAP, CD22, and CBLN1) and highlighted the association that ADCY9 has with various pathways, including signal transduction regulated by the cAMP-responsive element modulator, neurophysiological process regulated by the corticotrophin-releasing hormone, and synaptic plasticity. We therefore went on to validate the observed hypomethylation of ADCY9 in cases vs control through targeted bisulfite sequencing.
Conclusion
Our study represents the first, to our knowledge, analysis of DNA methylation and gene expression associated with suicide in a Mexican population using postmortem brain, providing novel insights for convergent molecular alterations associated with suicide.
Keywords: Epigenomics/transcriptomics, Mexico, suicide, postmortem human brain
Significance Statement.
This is, to our knowledge, the first genome-wide study that integrates differential methylation and expression analysis in Latin American suicide cases using postmortem human brain tissue. Seventy genes were both differentially methylated and expressed in cases vs controls. Gene-based functional enrichment analysis, using these 70 genes, identified 10 that were overrepresented in pathways related to synaptic plasticity, signal transduction regulated by the cAMP-responsive element modulator, and neurophysiological processes regulated by the corticotrophin-releasing hormone. ADCY9, which was hypomethylated and overexpressed by suicide, was overrepresented in many of these pathways. In sum, this study reveals novel genomic regions that may be associated with suicide and should be followed-up with functional experiments in future investigations.
Introduction
Suicide represents a major public health concern that results in nearly 800 000 deaths per year, is one of the leading causes of death in young adults aged 15–29 years, and is the fifth leading cause of death in those aged 30–49 years (World Health Organization, 2018). Moreover, 79% of deaths by suicide occur in low-income and middle-income countries (World Health Organization, 2018). In Mexico, suicide rates have been increasing over the past few decades, affecting mostly younger men and older adults (Alicandro et al., 2019).
The underlying etiology of suicide is multifactorial and includes complex interactions between biological predispositions, psychological, and socio-cultural risk factors (World Health Organization, 2014). The socio-cultural risk factors that stand out include financial stress, unemployment, lack of access to health and education systems, poor living conditions, high levels of violence, and malnutrition, among others (World Health Organization, 2014). As Mexico has been placed among the countries with the highest levels of social inequality in the world, a large percentage of the inhabitants reside within the socio-cultural risk context described (Piacentini, 2014). Indeed, the association between poverty and suicide has been previously reported (Vijayakumar et al., 2005; Oyesanya et al., 2015; Iemmi et al., 2016; Lee et al., 2017; Choi et al., 2019). Exposure to impoverished and stressful environments is known to play an important role in perceived stress and can have an impact on stress response pathways and other signaling systems (Barnett Burns et al., 2018). Although the molecular processes underlying these changes are not fully understood, growing evidence has suggested that epigenetic mechanisms, which allow the environment to modulate gene expression, mediate the process by which stressful experiences impact the brain (McEwen and Bulloch, 2019).
Epigenetic modifications alter gene expression without changing the underlying genetic code (Petronis, 2010). This adaptable feature allows an organism to prepare for various environmental conditions such as psychosocial factors and can, therefore, provide insights into the mechanisms underlying gene-environment interactions. DNA methylation is the most investigated and well-characterized epigenetic modification to date (BLUEPRINT Consortium, 2016), which is traditionally defined as the addition of a methyl group to the fifth carbon of a cytosine base and predominantly at those directly followed by guanine (CpG dinucleotide sites). Methylation at promoter regions is generally associated with repression of transcription, either by blocking transcription factor binding or by recruiting other repressive proteins with methyl-binding domains (Moore et al., 2013). However, positive correlations between methylation within gene bodies and expression have also been described (Jones, 2012).
Previous epigenetic studies have identified changes in various biological pathways related to suicide, including the GABAergic, poly-aminergic, neurotrophic, and immune systems as well as the hypothalamic-pituitary-adrenal axis (Poulter et al., 2008; Ernst et al., 2009; Keller et al., 2010; Fiori and Turecki, 2011; Labonte et al., 2012a, 2012b, Maussion et al., 2014; Murphy et al., 2017; Roy and Dwivedi, 2017; Kouter et al., 2019, Policicchio et al., 2020). Importantly, epigenetic changes may be specific to brain regions involved in the pathophysiology of suicide (Keller et al., 2011). Despite these results, few studies have examined genome-wide methylation changes in postmortem brain samples, and none have explored these changes in a Latin American population. Here, we report evidence of altered DNA methylation in postmortem human brain samples from Mexicans who died by suicide and the effects of these alterations on gene expression.
Materials and Methods
Subjects and Samples
Human postmortem brain tissue samples were obtained during autopsy in collaboration with the Mexico City Coroner’s Office, from the Forensic Sciences Institute. All subjects were Mexicans who lived in Mexico City at the time of death, and to reduce ethnic variation and stratification effects, we selected subjects that descended from 2 Mexican generations. This study included male subjects: (1) who died by suicide (cases, n = 35); or (2) who died suddenly without prolonged agonal state (controls, n = 13). The cause and manner of death were determined by the coroner’s office, in accordance with the National Code of Criminal Procedures (2016), after evaluating autopsy results, circumstances of death, data from extensive toxicological testing, police reports, family interviews, and medical records. Groups were matched for age and postmortem interval. The exclusion criteria for both groups included age >65 years old, comorbid medical illness, undetermined cause of death, incomplete forensic records, and poor DNA and RNA quality. Sample characteristics are presented in supplementary Table 1. Information on socio-demographic data (age, years of education, marital status, occupational status), circumstances of the death (method, toxicology reports, suicide note), and clinical information (medical and psychiatric reports from the hospital as well as death certificates) for each donor was obtained through the complete coroner’s records. To ensure comparability of the 2 groups, all individuals were subject to a consensus diagnosis between a pathologist, a psychologist, a criminologist, and a psychiatrist based on the DSM-5 criteria and using proxy-based testimonies as described elsewhere (Romero-Pimentel et al., 2018). Demographic and clinical data were compared between groups and considered for final analyses (supplementary Table 1).
For identification and dissection of Brodmann area 9, which corresponds to the dorsolateral prefrontal cortex, we used well-characterized neuroanatomical maps (Haines, 2000; Nolte, 2002). Briefly, the second prefrontal gyrus and precentral gyrus were identified to landmark the area of interest. In all cases, 3 cm3 of left hemisphere gray matter tissue was carefully dissected and immediately snap-frozen, whereas 1.5 cm3 of the tissue was stored in RNAlater (Qiagen, Singapore) at −80°C until further processing. This study was conducted in accordance with the ethical principles of the last Declaration of Helsinki and was approved by the Bio-Ethical Committee for Human Research at National Institute of Genomic Medicine.
Genome-Wide DNA Methylation Analysis on the Infinium HumanMethylation450 BeadChip
Genomic DNA was isolated from 25 mg of tissue using the manufacturer’s instructions of the QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA) and tested for purity and degradation using the NanoDrop 2000 spectrometer (Thermo Fisher, Wilmington, DE, USA) and agarose gel electrophoresis, respectively. Genomic DNA (200 ng) from each sample was treated with sodium bisulfite using the EZ DNA methylation Kit (Zymo Research, CA, USA) following the manufacture’s standard protocol. DNA methylation was quantified using the Infinium HumanMethylation450 BeadChip Array (Illumina, Inc., San Diego, CA, USA), as previously described (Pidsley et al., 2013). Bisulfite conversion and initial methylation signal detection quality control were performed at the Microarray Core Facility in the National Institute of Genomic Medicine (UMI) located in Mexico City.
Cases and controls were randomized among the BeadChip to avoid batch effects. Pre-processing and analysis of raw microarray data were conducted within R (ver 3.4) using the Chip Analysis Methylation Pipeline Bioconductor package (Morris et al., 2014). Sample methylation quality control (QC) was assessed by plotting log median methylated and unmethylated signals. Only samples that passed QC measures (>1% of sites P > .05) were included. Probes were excluded if they showed intensities indistinguishable from the background (detection P > .05) in at least 1% of samples and bead count <3 in 5% of the samples as well as if they were non-specific, showed cross-reactivity, and if they hybridized to single nucleotide polymorphisms.
Sex chromosomes with CpG probes were used to confirm sample sex. Single value decomposition analysis was used to detect technical batches and covariates. For the annotation of probes, the University of California, Santa Cruz (UCSC) RefGene name from Illumina’s annotation file and enhanced annotation to the UCSC Known Gene were used. All annotations used the human February 2009 (GRCh37/hg19) assembly. Beta (β) values were calculated as the ratio of methylated signal to the sum of unmethylated and methylated signals at each CpG site, and log2 transformed β values were used for the remainder of pre-processing steps (Du et al., 2010). Technical batches and covariates were detected and corrected for using single value decomposition analysis via the ComBat method before differential methylation analysis (Johnson et al., 2007).
After these procedures, 474 958 CpG sites were extracted. Differentially methylated regions (DMRs) were identified between cases and controls using the DMRcate method (Peters et al., 2015). DMRcate fits a limma linear model with empirical Bayes adjustment for each individual CpG site. We implemented the default smoothing parameters with bandwidth λ = 1000 bp and scaling factor C = 2. Nominal P < .05 was used to denote significant DMRs between controls and cases and corrected for genome-wide multiple testing using the Benjamini-Hochberg procedure with false discovery rates (FDR) of 0.1. Also, a mean β fold change of ± 0.01 was set as a cutoff value to decrease the number of significant DMRs and to identify sites with more biologically relevant methylation differences.
Genome-Wide mRNA Gene Expression Analysis on the HumanHT-12 v4 Expression BeadChip
Total RNA was extracted from 25 mg of tissue using the RNeasy Kit (Qiagen), according to the manufacturer’s instructions. NanoDrop 2000 spectrometer (Thermo Fisher) and Agilent Bioanalyzer 2100 were used to assess RNA quality across samples, with an RNA integrity number cutoff of 6. RNA samples were then sent to UMI, where an additional RNA QC was conducted. Complementary RNA was prepared using standard Illumina Whole-Genome protocols and was hybridized to Human HT-12 v4 Expression BeadChips (Illumina, USA). Differential gene expression analysis was conducted on the Human HT-12 v4 Expression BeadChip (Illumina), which provides accurate genome-wide expression coverage on up to 47 000 well-known genes, gene candidates, and splice variants. Initial QC of raw probe signals was conducted in GenomeStudio by UMI.
Pre-processing steps and differential gene expression analysis were performed in R using the limma Bioconductor package (Ritchie et al., 2015). Only the subset of samples that appeared in our DNA methylation analysis were included for whole-genome expression analysis (25 cases and 6 controls). Probe signals were detected with the propexp function and normalized with the normalize Between Arrays function. Probe filtering was conducted using a detection P < .05 in at least 20% of samples cut off to denote retained probes, where 27 453 gene probes were preserved for downstream analysis. Differentially expressed genes were identified through linear regression analyses. All analyses were conducted with log2-transformed values. Only probes with a log fold change (LFC) of at least > ±0.3, between cases and controls, were included for further investigation. LFC is the log-ratio of a gene expression value in 2 different conditions, and it is calculated as the ratio of the difference between the final value and the initial value over the initial value. Finally, genes that contained DMRs with ± 0.01 mean β fold change and that appeared in our differential expression analysis were identified using GeneOverlap package within R.
Sensitivity Analyses
We performed sensitivity analyses to determine the effects of toxicology and psychiatric condition on our methylation and expression findings. For each sensitivity analysis, we adjusted the original models to include each additional covariate and then ran linear regressions to compare the original and adjusted LFCs. Altogether, 4 sensitivity analyses were run.
Investigating the Effects of Brain Cell and Genetic Heterogeneity
To address the possibility of confounding effects of brain cell composition, Cell EpigenoType Specific mapper was used to estimate brain cellular heterogeneity in all samples. The mapper was designed for quantification and normalization of differing neuronal proportions in genome-wide DNA methylation data sets (Guintivano et al., 2013). To check for potential genetic heterogeneity, we extracted CpG sites that were population-specific SNPs using the MetyltoSNP R package (LaBarre et al., 2019). The R package extracts CpG sites that are present in 3 discrete levels of methylation: fully methylated, fully unmethylated, and 50% methylation, which correspond to genotypes CC, TT, and CT. Then a principal component analysis was performed in the population-specific SNPs data to identify sample outliers.
Pathway Analysis
The overlapping differentially expressed and methylated gene set was input into pathway analysis package MetaCore version 6.27 (GenoGo, Thomson Reuters, New York, NY, USA) to build top biological networks and list the associated biological processes. P = .05 was used as a cutoff to determine significant pathway enrichment. MetaCore analysis is based on MetaBase (http://metadatabase.org), a 100% manually curated integrated database of mammalian biology that contains over 6 million experimental findings on protein-protein, protein-DNA, protein-RNA, and protein-compound interactions, metabolic, signaling pathways, and others (Bolser et al., 2012).
Targeted Bisulfite Sequencing for Technical Validation
In our selection of the region that we chose to validate, through targeted bisulfite sequencing, we looked at a DMR that was the most overrepresented in our Metacore enrichment analysis. The DMR in ADYC9 not only fit this criterion but was also the most hypomethylated region. Therefore, DNA methylation across the ADYC9-associated DMR (spanning chromosome 16:4102293-4103533) was selected. The region encompasses 3 CpG sites (cg00701890, cg16774375, cg02910037), which overlap with those measured by the 450k array. A single amplicon (222 bp) was amplified with primers designed using the Methyl Primer Express software (ThermoFisher Scientific) and using genome assembly GRCh37/hg19. Specific details for primer design and amplicon library preparation are included in the supplementary Methods. DNA samples of controls (n = 13) and cases (n = 35) were bisulfite-converted using the Epitect 96 Bisulfite kit (Qiagen) as per the manufacturer’s guidelines. All samples were ensured to have optimal molarity of 2 nM before being loaded onto the MiSeq platform with the V3 600 cycle kit (Illumina). On retrieving raw sequencing data, Trimmomatic (v.0.35) was used to trim adaptor sequences (Bolger et al., 2014). Reads with Phred scores <20 were removed and aligned with Bowtie 2 (v 2.1.0) (Langmead et al., 2009). Methylated and non-methylated CpG signals were extracted to calculate the level of methylation at our sites of interest. The average methylation level for the CpG sites was calculated for all CpGs. Results were analyzed using 1-tailed t tests. Correlation of microarray and sequencing methylation values was assessed with Pearson correlation coefficients.
Results
Sample Characterization
As described in Figure 1, 35 cases and 13 controls were collected for the DNA methylation component of this study. For the transcriptomic analysis, 10 cases and 7 controls had to be excluded because of the poor quality of RNA samples (RNA integrity number <6). There were no significant differences in postmortem interval, age, years of education, and occupational status between cases and controls. However, as expected, a significant difference was noted in the presence of a psychiatric disorder (X2 = 18, df = 4, P = .0001) and toxicology (X2 = 6.10, df = 2, P = .04) (Table 1).
Figure 1.
Brief overview of the methodological approach used in this study. Abbreviations: BA, Broadmann area; GW, genome wide; DMRs, differentially methylated regions; Mean β FC, beta fold change; LFC, log fold change.
Table 1.
Demographics and Clinical Data of Samples Considered in the Epigenomic and Transcriptomic Overlap Analysis
| Control (n = 6) | Cases (n = 25) | Statistical analysis | |
|---|---|---|---|
| Age | 29.50 (18,40) | 30 (16,61) | U = 72.00, P = .89 |
| PMI (h) | 16 (11,19) | 12 (3,16) | U = 36.50, P = .53 |
| RIN | 6.9 (5.2, 7.2) | 5.8 (4.4, 8.1) | U = 62.50, P = .54 |
| Cause of death | Accidental death 100% (n = 6) | Hanging: 92% (n = 32) | |
| Jumping: 4% (n = 1) | |||
| Cutting: 4% (n = 1) | |||
| Years of education | 10.5 (9, 16) | 9 (6, 16) | U = 43, P = .10 |
| Occupational status | |||
| Student | 0% (n = 0) | 8% (n = 2) | X 2 = 1.4, df = 2, P = .48 |
| Employed | 100% (n = 6) | 80% (n = 20) | |
| Unemployed | 0% (n = 0) | 12% (n = 3) | |
| Marital status | |||
| Single | 0% (n = 0) | 52% (n = 13) | X 2 = 5.4, df = 1, P = .02 |
| Married | 100% (n = 6) | 48% (n = 12) | |
| Psych. family history | |||
| Yes | 0% (n = 0) | 0% (n = 0) | |
| No | 100% (n = 6) | 96% (n = 24) | |
| Unknown | 0% (n = 0) | 4% (n = 1) | |
| Previous episodes of suicide attempt | |||
| Yes | 0% (n = 0) | 12% (n = 3) | X 2 = 0.80, df = 1, P = .89 |
| No | 100% (n = 6) | 88% (n = 24) | |
| Toxicology | |||
| Alcohol | 0% (n = 0) | 52% (n = 13) | X 2 = 6.10, df = 2, P = .04 |
| Cocaine | 0% (n = 0) | 4% (n = 1) | |
| Negative | 100% (n = 6) | 44% (n = 11) | |
| Psych. disorder (DSM-5) | |||
| Depressive disorder | 35.29% (n = 12) | 0% (n = 0) | X 2 = 18, df = 4, P = .001 |
| Substance-related disorder | 52.94% (n = 18) | 28.57% (n = 2) | |
| Psychotic disorder | 2.94% (n = 1) | 0% (n = 0) | |
| Personality disorder | 5.88% (n = 2) | 14.29% (n = 1) | |
| None | 2.94% (n = 1) | 57.14% (n = 3) | |
| Postmortem note | |||
| Yes | n/a | 8% (n = 2) | |
| No | n/a | 92% (n = 23) |
Abbreviations: df, degrees of freedom; PMI, postmortem interval; RIN, RNA integrity number; U, Mann–Whitney U test; X2, chi-squared test.
Median (maximum, minimum) are shown.
Differential Methylation Analysis
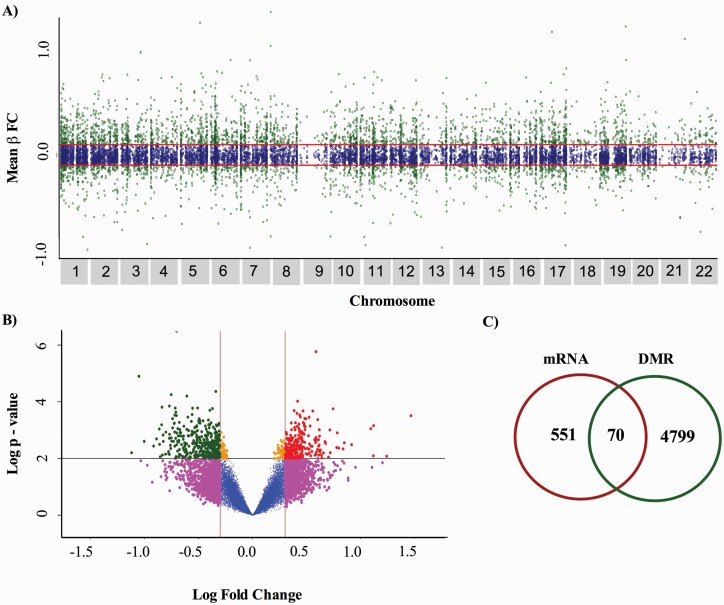
We used DMRcate a limma-based linear regression methods to identify DMRs in suicide cases compared with controls. The regional analysis identified 13 223 significantly methylated regions (P < .05, FDR = 0.10); however, these included DMRs with very low differences in methylation, that is, mean β fold change below ±0.001. Therefore, we applied a mean β fold change cutoff of > ±0.01 to identify 4430 DMRs with methylation changes that are more likely to be biologically relevant (Figure 2A). The top-ranked DMR was found within the Zinc Finger Protein 57 Homolog gene known as ZFP57 located near the transcription start site (chromosome 6:29648161-29649807, in the GRCh37/hg19 assembly, UCSC genome browser, and was consistently hypermethylated across 17 of 22 CpG sites in suicide cases relative to controls (mean β fold change = 0.20, P = 1.22–122). The 100 most significant DMRs (P < 3.16–301) are provided in supplementary Table 2.
Figure 2.
Epigenomic and transcriptomic overlap differences from prefrontal cortex of Mexican individuals who died by suicide. (A) Plot showing suicide-associated methylated regions in human prefrontal cortex. The red line indicated mean β fold change cut-off applied (> ±0.01), 4430 regions were identified as methylated between cases and controls (P < .05, FDR = 0.10). (B) Volcano plot showing raw P values vs fold change values for differential gene expression between cases and controls. The most significant genes with differential transcription are shown in green and red (P < .01, LFC ≥ ±0.3 y FDR = 0.10). (C) Venn diagram showing overlap in the number of DMRs and mRNA expression differences (Exact Hypergeometric Test P < 2.58 × 10−24). Abbreviations: DMR, Differentially methylated regions; Mean β FC, mean beta fold change.
Effects of Altered DNA Methylation on Gene Expression
To understand the possible biological effects of altered methylation, we analyzed gene expression using HumanHT-12 v4 Expression BeadChips. Comparison between cases and controls revealed 621 differentially expressed genes (LFC ≥ ±0.3; Figure 2B). To determine whether DNA methylation differences, between cases and controls, led to a functional impact on transcription, we investigated the overlap of DNA methylation and gene expression data using GeneOverlap package within R. Statistical overlap significance was calculated with the exact hypergeometric test (P < .05) (Shen, 2020). The Venn diagram in Figure 2C shows 70 regions that show altered DNA methylation and altered gene expression, and all overlapped with unique genes (exact hypergeometric test P < 2.58 × 10−24; supplementary Table 3). From the list of differentially methylated regions, 38 (54%) probes indicated hypermethylation and 32 (46%) probes indicated hypomethylation in suicide cases. Noteworthy, B-Cell Receptor CD22 (CD22) located in chromosome 19:35818807-35820181 (GRCh37/hg19) and NLR Family Pyrin Domain Containing 3 (NLRP3) located in chromosome 1: 247578552-247580106 (GRCh37/hg19) were the most significantly hypermethylated regions between suicide cases and controls (P = 5.89E-36, mean β fold change = .056, FDR = 0.1, and P = 8.21E-15, mean β fold change = .045, FDR = 0.1, respectively). Solute Carrier Family 25 Member 34 (SLC25A34) located in chromosome 1:16062361-16063471 (GRCh37/hg19) and Adenylate Cyclase 9 (ADCY9) located in chromosome 16: 4102293-4103533 were, on the other hand, genes with altered expression and the highest hypomethylation after correcting for multiple testing (P = 5.47E-20, mean β fold change = −.034 and P = −8.85E-14, mean β fold change = −.032, q = 0.10, respectively).
Next, to determine the effect of toxicology and psychiatric condition and our methylation and expression findings, we performed a series of sensitivity analyses. Overall, adjusting our models for these variables had minimal effects on our results (supplementary Figure 1).
Brain Cell and Genetic Heterogeneity
The individual proportion of neuronal and non-neuronal cells did not have any specific, significant effects on our primary findings (supplementary Table 4). Our genetic heterogeneity analysis also showed that population-specific SNPs for each sample formed 1 cluster with no outliers (supplementary Figure 2).
Pathway Analysis
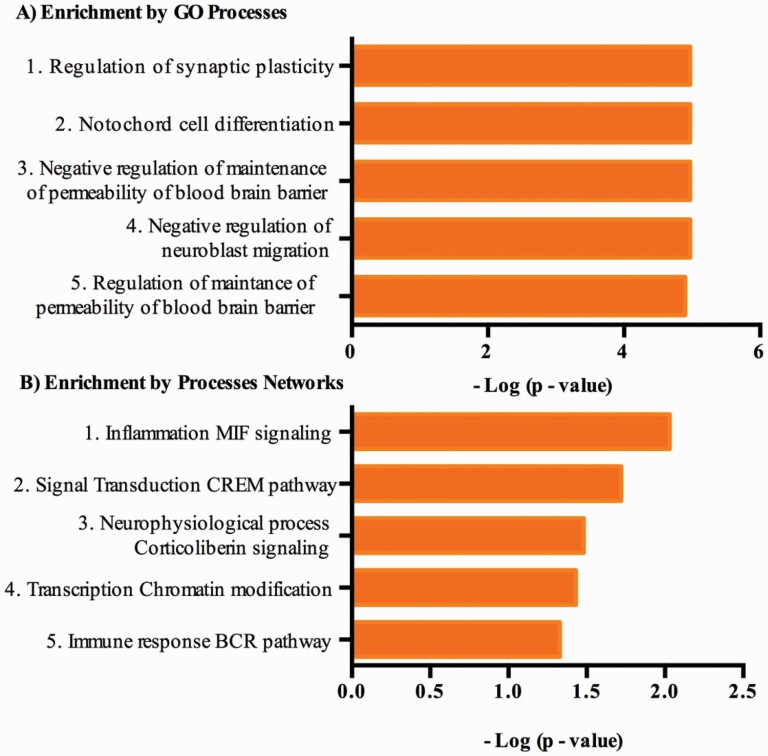
To shed light on the potential biological effects of the 70 genes where expression and methylation was significantly altered in suicide cases vs controls (supplementary Table 3), a pathway analysis was performed based on the Metacore database. The most enriched gene ontology processes, after FDR adjustment (<0.05), included regulation of synaptic plasticity (P = 1.02 × 10−05), notochord cell differentiation (P = 1.06 × 10−05), and the negative regulation of permeability maintenance of brain blood-barrier (P = 1.06–05) (Figure 3A). In addition, Metacore biological processes analysis indicated that these differentially methylated and expressed genes were also highly correlated with (1) inflammation regulated by the macrophage migration inhibitory factor (P = 9.20 × 10−03), (2) signal transduction regulated by the cAMP-responsive element modulator also known as cAMP responsive element modulator (CREM) pathway (P = 1.97 × 10−02), and (3) neurophysiological process regulated by the corticotrophin-releasing hormone (P = 3.35 × 10−02) (Figure 3B). However, these pathways did not survive the FDR adjustment. The main pathways are shown in Table 2.
Figure 3.
Enrichment analysis of epigenomic and transcriptomic overlap differences from prefrontal cortex of Mexican individuals who died by suicide. (A) Top 5 significant process networks from Metacore as sorted by –log P values. (B) Top 5 significant biological processes from GO ontology as sorted by –log P values. Bar length reflects significance and equals to the negative logarithm of enrichment P value. Abbreviations: BCR = B cell receptor; CREM, cAMP-responsive element modulator; GO, Gene Ontology; MIF, macrophage migration inhibitory factor.
Table 2.
Enrichment Analysis of Epigenomic and Transcriptomic Overlap Differences From Prefrontal Cortex of Mexican Individuals Who Died by Suicide
| Term | Counts | P value | FDR | Network objects from active data |
|---|---|---|---|---|
| Gene ontology process | ||||
| Regulation of synaptic plasticity | 8 | 1.00–05 | 4.41–03 | Adenylate cyclase, CRH, Ephrin-A receptors, S100, NFATC4, CBLN1, KAT2A, NF-AT |
| Notochord cell differentiation | 2 | 1.10–05 | 4.41–03 | Ephrin-A receptors, EPHA2 |
| Negative regulation of maintenance of permeability of blood-brain barrier | 2 | 1.10–05 | 4.41–03 | SUR, ABCC8 |
| Negative regulation of neuroblast migration | 2 | 1.10–05 | 4.41–03 | SUR, ABCC8 |
| Regulation of maintenance of permeability of blood-brain barrier | 2 | 1.10–05 | 4.41–03 | SUR, ABCC8 |
| Cell process networks | ||||
| Inflammation_MIF signaling | 4 | .009 | 6.53–01 | Adenylate cyclase, CRH, Adenylate cyclase type IX, HMGA1 |
| Signal transduction_CREM pathway | 3 | .019 | 6.58–01 | Adenylate cyclase, CRH, NFATC4 |
| Neurophysiological process_Corticotrophin signaling | 2 | .033 | 6.58–01 | Adenylate cyclase, CRH |
| Transcription_Chromatin modification | 3 | .037 | 6.58–01 | TRRAP, HMGA1, KAT2A |
| Immune response_BCR pathway | 3 | .046 | 6.58–01 | CD22, NFATC4, NF-AT |
Abbreviations: BCR, B cell receptor; CREM, cAMP-responsive element modulator; FDR, False Discovery Rate; MIF, macrophage migration inhibitory factor.
Top 5 significant pathways from Metacore as sorted by P values.
Technical Validation
We next sought to technically validate our Illumina 450K data using targeted bisulfite sequencing. For this, we overlapped the list of genes identified as network objects from active Metacore data to select a DMR for validation. Ten genes were identified as actively enriched Metacore terms. What was most notable, ADCY9 was enriched in 4 terms and showed the highest negative LFC after correcting for multiple testing (P = −8.85E-14, mean β fold change = −0.032, q = 0.10) (Table 3). Therefore, differential methylation of the ADYC9 region (chromosome 16:4102293-4103533 [GRCh37/hg19]), in our genome-wide analysis was selected for validation. Consistent with the microarray analysis, targeted bisulfite sequencing revealed ADYC9 to be significantly hypomethylated in suicide cases vs controls (P < .001, Student’s t test). Also, we obtained a significant Pearson’s correlation coefficient of r = 0.40 with a P < .009 among the level of CpGs methylation assessed by targeted bisulfite sequencing and microarray methods (Figure 4).
Table 3.
List of Genes that Showed Epigenomic and Transcriptomic Overlap Differences and Enriched Metacore Terms
| DNA methylation | Gene expression | ||||||
|---|---|---|---|---|---|---|---|
| Position (build GRCh37/hg19) | Gene annotated to DMRs | Transcript | P value | LFC | P value | LFC | Number metacore term |
| chr16: 4102293-4103533 | ADCY9 | Adenylate Cyclase 9 | 8.85–14 | −0.032 | .0059 | −0.42 | 4 |
| chr8:67088895-67091580 | CRH | Corticotrophin releasing hormone | 3.59–28 | 0.026 | .0006 | 1.13 | 4 |
| chr14:24834695-24839226 | NFATC4 | Nuclear factor of activated T cells 4 | 6.80–21 | 0.017 | .0007 | 1.12 | 3 |
| chr11:17497693-17498952 | ABCC8 | ATP binding cassette subfamily C member 8 | 9.23–22 | 0.011 | .0057 | −0.55 | 3 |
| chr6:34202568-34204646 | HMGA1 | High mobility group AT-Hook 1 | 2.47–20 | 0.023 | .0088 | 0.33 | 2 |
| chr17:4027290140275359 | KAT2A | Lysine acetyltransferase 2A | 3.80–18 | −0.011 | .0012 | −0.55 | 2 |
| chr1:16481715-16483658 | EPHA2 | EPH receptor 2 | 1.99–27 | 0.016 | .0092 | −0.36 | 2 |
| chr7:98475615-98477438 | TRRAP | Transformation/transcription Domain associated protein | 3.37–47 | −0.013 | .0082 | −0.39 | 1 |
| chr19:35818807-35820181 | CD22 | CD22 molecule | 5.89–36 | 0.056 | .0047 | −0.62 | 1 |
| chr16:49311483-49314257 | CBLN1 | Cerebellin 1 precursor | 9.80–19 | 0.029 | .0058 | 0.33 | 1 |
Abbreviations: Chr, chromosome; DMRs, differentially methylated regions; LFC, log fold change.
Figure 4.
Differential methylation between cases and controls at DMR_10219, assessed through 480 k Illumina BeadChip analysis, correlated with values assessed through targeted bisulfite sequencing. (A) Bar graphs show percentage of methylation of cases and controls detected through targeting bisulfite sequencing. (B) Bar graphs show log fold change of gene expression of cases and controls detected through HumanHT-12 v4 Expression BeadChip. (C) Scatterplots show correlation of methylation levels assessed by microarray and targeted bisulfite sequencing platforms. Abbreviations: BS, bisulfite sequencing; R, Pearson correlation coefficient.
Discussion
To our knowledge, this represents the first genome-wide study that integrates differential methylation and expression analyses in postmortem human brain tissue from Latin American individuals who died by suicide. In this study, we identified several differentially methylated regions, with fold changes > ±0.01 between cases and controls. While a cutoff of > ±0.01 may appear relatively low, subtle methylation differences in multiple functional gene networks are often reported in studies of psychiatric disorders. Together, these subtle differences in many genes are thought to reflect the complex etiology of suicidal behavior (Labonte et al., 2013; Haghighi et al., 2014; Nagy et al., 2015; Schneider et al., 2015; Ju et al., 2019; Kouter et al., 2019; Policicchio et al., 2020).
We next combined these DMRs with differential expression analysis and focused only on genes that were also differentially expressed between cases and controls. This allowed for the identification of 70 DMRs that did not share any gene level overlap. Gene set enrichment analysis with these 70 genes revealed that 10 were overrepresented in, and therefore biologically relevant to, processes related to suicide (ADCY9, CRH, NFATC4, ABCC8, HMGA1, KAT2A, EPHA2, TRRAP, CD22, CBLN1). Of these 10 genes, ADCY9 appeared in the greatest number of pathways. We therefore chose to validate the differential methylation we observed in ADCY9 by using a targeted bisulfite sequencing approach.
The product of the ADCY9 gene belongs to a family of transmembrane-bound enzymes that catalyzes the formation of cyclic AMP from ATP and is implicated in intracellular signaling cascades and secondary messenger systems (Dessauer et al., 2017). The dysregulation of ADCY9 has been previously identified in the context of major depressive disorder (Fan et al., 2020). Early studies have shown that adenylate cyclase activity is altered in postmortem human brain samples from individuals with a history of mood disorders as well as those who died by suicide (Cowburn et al., 1994; Reiach et al., 1999). In our investigation, we observed hypomethylation at the second intron of the ADCY9 gene between cases and controls. This hypomethylation corresponded to a decrease in mRNA expression (Table 3). While methylation at a promoter region is typically anticorrelated with expression, our findings of a positive correlation between gene body methylation and expression are consistent with other studies (Ball et al., 2009; Lutz et al., 2018).
As revealed by gene enrichment analysis, genes that were both differentially methylated and expressed by suicide were found to be involved in the regulation of synaptic plasticity (ADCY9, CRH, EPHA2, S100, NFATC4, CBLN1, KAT2A, NF-AT), signal transduction through CREM (ADCY9, CRH, NFATC4), and molecular pathways involved in neurophysiological processes regulated by corticotrophin-releasing hormone (ADCY9, CRH).
We were especially interested in the regulation of synaptic plasticity, since it was found as the most enriched pathway in our list of 70 genes dysregulated by suicide (Table 2). Synaptic plasticity is one of the most fundamental and important functions of the brain. The efficacy of transmission at a synapse depends on modulation of the connectivity between neurons and neuronal circuits during adaptation to the environment (Marsden et al., 2013). Previous reports have highlighted key roles of synaptic plasticity in suicide (Liu et al., 2017). For instance, Nagy et al. (2020) identified cell-type specific differentially expressed genes associated with the regulation of synaptic plasticity in the dorsolateral prefrontal cortex of male suicide cases. Our findings showed that ADCY9 and CRH were both members of the synaptic plasticity pathway and dysregulated in suicide. Accumulating evidence indicates that adenylate cyclase is an essential regulator of synaptic plasticity. For example, it regulates pathways related to long-term potentiation, such as, Ca2+/calmodulin sensitive adenylate cyclase signaling (Wang and Storm, 2003; Abel and Nguyen, 2008; Kim et al., 2011), cAMP-PKA signaling (Wang and Storm, 2003; Banko et al., 2004; Mockett et al., 2004; Valera et al., 2008), and cAMP response element-binding protein (CREB) activity (Benito et al., 2011; Bengtson and Bading, 2012). In addition, dysregulation of both ADCY9 and CREB has been previously observed in the prefrontal cortex of individuals who died by suicide (Dwivedi et al., 2003; Yamada et al., 2003; Pandey et al., 2007). Interestingly, signal transduction through CREM was also enriched in suicide cases. Taken together, the downregulation of ADCY9 may alter CREB signaling and therefore impact the regulation of synaptic plasticity in the prefrontal cortex of individuals who died by suicide. Given that functional studies of adenylate cyclase in the context of suicide are scarce, future investigations are necessary to explain the function of ADCY9 and its role in synaptic plasticity.
Corticotrophin Releasing Hormone or CRH was also identified as belonging to the synaptic plasticity pathway. Previous investigations have consistently found an association between CRH and suicide (Nemeroff et al., 1988; Arato et al., 1989; Hiroi et al., 2001; Austin et al., 2003; Mann et al., 2009; Zhao et al., 2015; Jokinen et al., 2018; Pandey et al., 2019). Stress and pain have been reported to have profound effects on synaptic structure and function (Christoffel et al., 2011; Sandi, 2011; Popoli et al., 2012). Here, we identify increased CRH expression in cases vs controls. This result is consistent with other studies that reported elevated CRH levels in several frontal regions of individuals who died by suicide (Merali et al., 2004; Zhao et al., 2015) as well as with studies in animal models that have shown that stress increases CRH expression in the prefrontal cortex (Meng et al., 2011). Specifically, we identified DNA methylation changes in the promoter region of CRH in suicide cases. Similarly, Jokinen et al. (2018) also reported altered levels of methylation at 2 CpG sites in the same gene region in suicide attempters with a high risk/severe phenotype. Adverse childhood experiences including sexual, physical, and emotional abuse or neglect are also associated with alterations in CRH signaling (Lee et al., 2005; Heim et al., 2008a, 2008b) and suicidal behavior (Read et al., 2001; Bruffaerts et al., 2010; Miller et al., 2013). Several studies have shown how child abuse might epigenetically influence the HPA in subjects who died by suicide (McGowan et al., 2009). Due to the design of our study, child abuse and other exposures to stress were not measured. Therefore, future studies should take into account this important confounder. It has also been shown that glucocorticoid receptor signaling may play a role in synaptic plasticity by increasing glutamate release in prefrontal cortex (Musazzi et al., 2011, Sandi, 2011) and then initializing several adenylate cyclase–related transduction pathways (Wang and Storm, 2003; Banko et al., 2004; Mockett et al., 2004; Abel and Nguyen, 2008; Valera et al., 2008; Kim et al., 2011). The downregulation of AMPAR and NMDAR subunits observed in postmortem brain of suicide cases may, therefore, be explained by the aforementioned mechanism (Cohen et al., 2011; Duric, 2013). We hypothesize that our findings of increased CRH expression in the prefrontal cortex of individuals who died by suicide may relate, at least in part, to the reduced expression of ADCY9 that we observed. We also provide evidence that epigenetic mechanism may be involved in the regulation of CRH expression by identifying hypermethylation in the promoter region of CRH gene (Table 3). Methylation of CpG island promoters is generally associated with the repression of gene expression (Delcuve et al., 2009; Novakovic et al., 2011). Nevertheless, non-typical gene expression and promoter methylation relationships have been described in genes with CpG-poor promoters, of which the CRH promoter falls into (Weber et al., 2007).
According to our Metacore gene enrichment analysis, ADCY9 is also enriched in a pathway related to corticotrophin-releasing hormone signaling. The mechanism regulating increased CRH expression remains unclear, although methylation of CpG sites within cyclic-AMP response element (CRE) sensitive regions may be 1 explanation (Rishi et al., 2010; Pan et al., 2015). Molecular studies using CRH promoter-reporter constructs showed that transcription factor complexes bound a CRE located at CRH 200 bp upstream of the major transcription start site mediated the cAMP stimulation (Adler et al., 1990). It has been shown that methylation of the CRH CRE can increase transcription factor binding affinity and enhance CRH gene expression (Pan et al., 2015). Also, the CRH receptor is a G-protein coupled protein receptor and signals through a cAMP-dependent mechanism. Interestingly, as mentioned before, one of the most enriched pathways associated with suicide is CREM. Our observation of CRH and ADCY9 differential methylation and increased expression suggest a positive feedback among these genes in the prefrontal cortex of individuals who died by suicide. However, additional functional studies are needed for a better understanding of the role of ADCY9 in the CRH process in the context of suicide.
The present study represents an important step in identifying differentially methylated regions associated with suicide in postmortem brain of Mexicans who died by suicide. However, our results must be interpreted with caution, as there are limitations to this study. To begin with, the sample size is relatively small; however, we have fully characterized each sample with additional data, such as demographic information and detailed clinical data that were considered in our analyses. Second, a methodological limitation inherent to the methylation microarray platforms is the inability to distinguish hydroxymethylated cytosines from methylated cytosines, and it is reasonable that the differences detected are confounded by other modifications.
Another important point to consider in this study is that the robustness of our findings is limited by several factors. Firstly, due to the lack of independent data set replication, it would be desirable to replicate our results in a different cohort of Mexicans who die by suicide. Additional explanatory data such as genotype were not assessed in our study. Integrative approaches have shown that DNA methylation is best predicted in the combination of genotype information with environmental factors (Czamara et al., 2019). Another relevant limitation for this study is the lack of technical validation in other top candidate gene regions identified as enriched in our pathways analysis; however, we have demonstrated that our findings fit with previously reported pathways, such as synaptic plasticity, which in turn supports the reliability of our results. Still, future studies are needed to replicate our findings in large independent cohorts. Nevertheless, this study represents the first, to our knowledge, to investigate a Mexican population in the context of suicide. As part of an ongoing project, which started in 2018, we expect to collect more samples and to overcome the aforementioned limitations.
In conclusion, we present new evidence that altered DNA methylation is a mechanism that affects these processes in postmortem brain of suicide cases. Overall, the available data suggest that altered methylation and expression of genes involved in synaptic plasticity, signal transduction regulated by the cAMP-responsive element modulator, and neurophysiological processes regulated by the corticotrophin-releasing hormone may serve an important role in the molecular pathology of suicide. In this study, ADCY9 gene was identified to be involved in several pathways and is, therefore, a strong candidate for future functional studies.
Supplementary Material
Acknowledgments
Ana Luisa Romero-Pimentel is a doctoral student from the Programa de Psicología, Universidad Nacional Autónoma de México (UNAM), and was supported by Consejo Nacional de Ciencia y Tecnología (CONACyT) Scholarship #486265.
This study received partial funding from the “Instituto Nacional de Medicina Genomica.”
Statement of Interest
None.
References
- Abel T, Nguyen PV (2008) Regulation of hippocampus-dependent memory by cyclic AMP-dependent protein kinase. Prog Brain Res 169:97–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler GK, Smas CM, Fiandaca M, Frim DM, Majzoub JA (1990) Regulated expression of the human corticotropin releasing hormone gene by cyclic AMP. Mol Cell Endocrinol 70:165–174. [DOI] [PubMed] [Google Scholar]
- Alicandro G, Malvezzi M, Gallus S, La Vecchia C, Negri E, Bertuccio P (2019) Worldwide trends in suicide mortality from 1990 to 2015 with a focus on the global recession time frame. Int J Public Health 64:785–795. [DOI] [PubMed] [Google Scholar]
- Arato M, Bánki CM, Bissette G, Nemeroff CB (1989) Elevated CSF CRF in suicide victims. Biol Psychiatry 25:355–359. [DOI] [PubMed] [Google Scholar]
- Austin MC, Janosky JE, Murphy HA (2003) Increased corticotropin-releasing hormone immunoreactivity in monoamine-containing pontine nuclei of depressed suicide men. Mol Psychiatry 8:324–332. [DOI] [PubMed] [Google Scholar]
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM (2009) Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol 27:361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banko JL, Hou L, Klann E (2004) NMDA receptor activation results in PKA- and ERK-dependent Mnk1 activation and increased eIF4E phosphorylation in hippocampal area CA1. J Neurochem 91:462–470. [DOI] [PubMed] [Google Scholar]
- Barnett Burns S, Almeida D, Turecki G (2018) The epigenetics of early life adversity: current limitations and possible solutions. Prog Mol Biol Transl Sci 157:343–425. [DOI] [PubMed] [Google Scholar]
- Bengtson CP, Bading H (2012) Nuclear calcium signaling. Adv Exp Med Biol 970:377–405. [DOI] [PubMed] [Google Scholar]
- Benito E, Valor LM, Jimenez-Minchan M, Huber W, Barco A (2011) cAMP response element-binding protein is a primary hub of activity-driven neuronal gene expression. J Neurosci 31:18237–18250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BLUEPRINT Consortium (2016) Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat Biotechnol 34:726–737. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolser DM, et al. (2012) MetaBase–the wiki-database of biological databases. Nucleic Acids Res 40:D1250–D1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruffaerts R, et al. (2010) Childhood adversities as risk factors for onset and persistence of suicidal behaviour. Br J Psychiatry 197:20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GG, Gross JA, Lutz PE, Vaillancourt K, Maussion G, Bramoulle A, Théroux JF, Gardini ES, Ehlert U, Bourret G, Masurel A, Lepage P, Mechawar N, Turecki G, Ernst C (2017) Medium throughput bisulfite sequencing for accurate detection of 5-methylcytosine and 5-hydroxymethylcytosine. BMC Genomics 18:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JW, Kim TH, Shin J, Han E (2019) Poverty and suicide risk in older adults: a retrospective longitudinal cohort study. Int J Geriatr Psychiatry 34:1565–1571. [DOI] [PubMed] [Google Scholar]
- Christoffel DJ, Golden SA, Russo SJ (2011) Structural and synaptic plasticity in stress-related disorders. Rev Neurosci 22:535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JW, Louneva N, Han LY, Hodes GE, Wilson RS, Bennett DA, Lucki I, Arnold SE (2011) Chronic corticosterone exposure alters postsynaptic protein levels of PSD-95, NR1, and synaptopodin in the mouse brain. Synapse 65:763–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowburn RF, Marcusson JO, Eriksson A, Wiehager B, O’Neill C (1994) Adenylyl cyclase activity and G-protein subunit levels in postmortem frontal cortex of suicide victims. Brain Res 633:297–304. [DOI] [PubMed] [Google Scholar]
- Czamara D, et al. ; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium (2019) Integrated analysis of environmental and genetic influences on cord blood DNA methylation in new-borns. Nat Commun 10:2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcuve GP, Rastegar M, Davie JR (2009) Epigenetic control. J Cell Physiol 219:243–250. [DOI] [PubMed] [Google Scholar]
- Dessauer CW, Watts VJ, Ostrom RS, Conti M, Dove S, Seifert R (2017) International union of basic and clinical pharmacology. CI. Structures and small molecule modulators of mammalian adenylyl cyclases. Pharmacol Rev 69:93–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM (2010) Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duric V, Banasr M, Stockmeier CA, Simen AA, Newton SS, Overholser JC, Jurjus GJ, Dieter L, Duman RS (2013) Altered expression of synapse and glutamate related genes in post-mortem hippocampus of depressed subjects. Int J Neuropsychopharmacol 16:69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y, Rao JS, Rizavi HS, Kotowski J, Conley RR, Roberts RC, Tamminga CA, Pandey GN (2003) Abnormal expression and functional characteristics of cyclic adenosine monophosphate response element binding protein in postmortem brain of suicide subjects. Arch Gen Psychiatry 60:273–282. [DOI] [PubMed] [Google Scholar]
- Ernst C, Chen ES, Turecki G (2009) Histone methylation and decreased expression of TrkB.T1 in orbital frontal cortex of suicide completers. Mol Psychiatry 14:830–832. [DOI] [PubMed] [Google Scholar]
- Fan T, Hu Y, Xin J, Zhao M, Wang J (2020) Analyzing the genes and pathways related to major depressive disorder via a systems biology approach. Brain Behav 10:e01502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiori LM, Turecki G (2011) Epigenetic regulation of spermidine/spermine N1-acetyltransferase (SAT1) in suicide. J Psychiatr Res 45:1229–1235. [DOI] [PubMed] [Google Scholar]
- Guintivano J, Aryee MJ, Kaminsky ZA (2013) A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics 8:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighi F, Xin Y, Chanrion B, O’Donnell AH, Ge Y, Dwork AJ, Arango V, Mann JJ (2014) Increased DNA methylation in the suicide brain. Dialogues Clin Neurosci 16:430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines DE (2000) Neuroanatomy: an atlas of structures, sections, and systems. Philadelphia, PA: Lippincott Williams & Wilkins. [Google Scholar]
- Heim C, Mletzko T, Purselle D, Musselman DL, Nemeroff CB (2008a) The dexamethasone/corticotropin-releasing factor test in men with major depression: role of childhood trauma. Biol Psychiatry 63:398–405. [DOI] [PubMed] [Google Scholar]
- Heim C, Newport DJ, Mletzko T, Miller AH, Nemeroff CB (2008b) The link between childhood trauma and depression: insights from HPA axis studies in humans. Psychoneuroendocrinology 33:693–710. [DOI] [PubMed] [Google Scholar]
- Hiroi N, Wong ML, Licinio J, Park C, Young M, Gold PW, Chrousos GP, Bornstein SR (2001) Expression of corticotropin releasing hormone receptors type I and type II mRNA in suicide victims and controls. Mol Psychiatry 6:540–546. [DOI] [PubMed] [Google Scholar]
- Iemmi V, Bantjes J, Coast E, Channer K, Leone T, McDaid D, Palfreyman A, Stephens B, Lund C (2016) Suicide and poverty in low-income and middle-income countries: a systematic review. Lancet Psychiatry 3:774–783. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8:118–127. [DOI] [PubMed] [Google Scholar]
- Jokinen J, Boström AE, Dadfar A, Ciuculete DM, Chatzittofis A, Åsberg M, Schiöth HB (2018) Epigenetic changes in the CRH gene are related to severity of suicide attempt and a general psychiatric risk score in adolescents. Ebiomedicine 27:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13:484–492. [DOI] [PubMed] [Google Scholar]
- Ju C, et al. (2019) Integrated genome-wide methylation and expression analyses reveal functional predictors of response to antidepressants. Transl Psychiatry 9:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller S, et al. (2010) Increased BDNF promoter methylation in the Wernicke area of suicide subjects. Arch Gen Psychiatry 67:258–267. [DOI] [PubMed] [Google Scholar]
- Keller S, Sarchiapone M, Zarrilli F, Tomaiuolo R, Carli V, Angrisano T, Videtic A, Amato F, Pero R, di Giannantonio M, Iosue M, Lembo F, Castaldo G, Chiariotti L (2011) TrkB gene expression and DNA methylation state in Wernicke area does not associate with suicidal behavior. J Affect Disord 135:400–404. [DOI] [PubMed] [Google Scholar]
- Kim M, Park AJ, Havekes R, Chay A, Guercio LA, Oliveira RF, Abel T, Blackwell KT (2011) Colocalization of protein kinase A with adenylyl cyclase enhances protein kinase A activity during induction of long-lasting long-term-potentiation. Plos Comput Biol 7:e1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouter K, Zupanc T, Videtič Paska A (2019) Genome-wide DNA methylation in suicide victims revealing impact on gene expression. J Affect Disord 253:419–425. [DOI] [PubMed] [Google Scholar]
- LaBarre BA, Goncearenco A, Petrykowska HM, Jaratlerdsiri W, Bornman MSR, Hayes VM, Elnitski L (2019) MethylToSNP: identifying SNPs in Illumina DNA methylation array data. Epigenetics Chromatin 12:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonté B, Suderman M, Maussion G, Navaro L, Yerko V, Mahar I, Bureau A, Mechawar N, Szyf M, Meaney MJ, Turecki G (2012a) Genome-wide epigenetic regulation by early-life trauma. Arch Gen Psychiatry 69:722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonte B, Yerko V, Gross J, Mechawar N, Meaney MJ, Szyf M, Turecki G (2012b) Differential glucocorticoid receptor exon 1(B), 1(C), and 1(H) expression and methylation in suicide completers with a history of childhood abuse. Biol Psychiatry 72:41–48. [DOI] [PubMed] [Google Scholar]
- Labonte B, Suderman M, Maussion G, Lopez JP, Navarro-Sánchez L, Yerko V, Mechawar N, Szyf M, Meaney MJ, Turecki G (2013) Genome-wide methylation changes in the brains of suicide completers. Am J Psychiatry 170:511–520. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R, Geracioti TD Jr, Kasckow JW, Coccaro EF (2005) Childhood trauma and personality disorder: positive correlation with adult CSF corticotropin-releasing factor concentrations. Am J Psychiatry 162:995–997. [DOI] [PubMed] [Google Scholar]
- Lee SU, Oh IH, Jeon HJ, Roh S (2017) Suicide rates across income levels: retrospective cohort data on 1 million participants collected between 2003 and 2013 in South Korea. J Epidemiol 27:258–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Ge T, Leng Y, Pan Z, Fan J, Yang W, Cui R (2017) The role of neural plasticity in depression: from hippocampus to prefrontal cortex. Neural Plast 2017:6871089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz PE, Gross JA, Dhir SK, Maussion G, Yang J, Bramoulle A, Meaney MJ, Turecki G (2018) Epigenetic regulation of the kappa opioid receptor by child abuse. Biol Psychiatry 84:751–761. [DOI] [PubMed] [Google Scholar]
- Mann JJ, Arango VA, Avenevoli S, Brent DA, Champagne FA, Clayton P, Currier D, Dougherty DM, Haghighi F, Hodge SE, Kleinman J, Lehner T, McMahon F, Mościcki EK, Oquendo MA, Pandey GN, Pearson J, Stanley B, Terwilliger J, Wenzel A (2009) Candidate endophenotypes for genetic studies of suicidal behavior. Biol Psychiatry 65:556–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden WN (2013) Synaptic plasticity in depression: molecular, cellular and functional correlates. Prog Neuropsychopharmacol Biol Psychiatry 43:168–184. [DOI] [PubMed] [Google Scholar]
- Maussion G, Yang J, Suderman M, Diallo A, Nagy C, Arnovitz M, Mechawar N, Turecki G (2014) Functional DNA methylation in a transcript specific 3’UTR region of TrkB associates with suicide. Epigenetics 9:1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS, Bulloch K (2019) Epigenetic impact of the social and physical environment on brain and body. Metabolism 100S:153941. [DOI] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonté B, Szyf M, Turecki G, Meaney MJ (2009) Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci 12:342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng QY, Chen XN, Tong DL, Zhou JN (2011) Stress and glucocorticoids regulated corticotropin releasing factor in rat prefrontal cortex. Mol Cell Endocrinol 342:54–63. [DOI] [PubMed] [Google Scholar]
- Merali Z, Du L, Hrdina P, Palkovits M, Faludi G, Poulter MO, Anisman H (2004) Dysregulation in the suicide brain: mRNA expression of corticotropin-releasing hormone receptors and GABA(A) receptor subunits in frontal cortical brain region. J Neurosci 24:1478–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AB, Esposito-Smythers C, Weismoore JT, Renshaw KD (2013) The relation between child maltreatment and adolescent suicidal behavior: a systematic review and critical examination of the literature. Clin Child Fam Psychol Rev 16:146–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockett BG, Brooks WM, Tate WP, Abraham WC (2004) Dopamine D1/D5 receptor activation fails to initiate an activity-independent late-phase LTP in rat hippocampus. Brain Res 1021:92–100. [DOI] [PubMed] [Google Scholar]
- Moore LD, Le T, Fan G (2013) DNA methylation and its basic function. Neuropsychopharmacology 38:23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris TJ, Butcher LM, Feber A, Teschendorff AE, Chakravarthy AR, Wojdacz TK, Beck S (2014) ChAMP: 450k chip analysis methylation pipeline. Bioinformatics 30:428–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TM, Crawford B, Dempster EL, Hannon E, Burrage J, Turecki G, Kaminsky Z, Mill J (2017) Methylomic profiling of cortex samples from completed suicide cases implicates a role for PSORS1C3 in major depression and suicide. Transl Psychiatry 7:e989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musazzi L, Racagni G, Popoli M (2011) Stress, glucocorticoids and glutamate release: effects of antidepressant drugs. Neurochem Int 59:138–149. [DOI] [PubMed] [Google Scholar]
- Nagy C, Suderman M, Yang J, Szyf M, Mechawar N, Ernst C, Turecki G (2015) Astrocytic abnormalities and global DNA methylation patterns in depression and suicide. Mol Psychiatry 20:320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy C, Maitra M, Tanti A, Suderman M, Théroux JF, Davoli MA, Perlman K, Yerko V, Wang YC, Tripathy SJ, Pavlidis P, Mechawar N, Ragoussis J, Turecki G (2020) Single-nucleus transcriptomics of the prefrontal cortex in major depressive disorder implicates oligodendrocyte precursor cells and excitatory neurons. Nat Neurosci 23:771–781. [DOI] [PubMed] [Google Scholar]
- National Code of Criminal Procedures (2016) Diario Oficial de la Federacion. Mexico: Camara de Diputados de H. Congreso de la Union. [Google Scholar]
- Nemeroff CB, Owens MJ, Bissette G, Andorn AC, Stanley M (1988) Reduced corticotropin releasing factor binding sites in the frontal cortex of suicide victims. Arch Gen Psychiatry 45:577–579. [DOI] [PubMed] [Google Scholar]
- Nolte J (2002) The human brain: an introduction to its functional neuroanatomy. St Louis, MO: Mosby. [Google Scholar]
- Novakovic B, Gordon L, Wong NC, Moffett A, Manuelpillai U, Craig JM, Sharkey A, Saffery R (2011) Wide-ranging DNA methylation differences of primary trophoblast cell populations and derived cell lines: implications and opportunities for understanding trophoblast function. Mol Hum Reprod 17:344–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyesanya M, Lopez-Morinigo J, Dutta R (2015) Systematic review of suicide in economic recession. World J Psychiatry 5:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Bowman M, Scott RJ, Fitter J, Nicholson RC, Smith R, Zakar T (2015) Methylation of the corticotropin releasing hormone gene promoter in BeWo cells: relationship to gene activity. Int J Endocrinol 2015:861302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GN, Dwivedi Y, Ren X, Rizavi HS, Roberts RC, Conley RR (2007) Cyclic AMP response element-binding protein in post-mortem brain of teenage suicide victims: specific decrease in the prefrontal cortex but not the hippocampus. Int J Neuropsychopharmacol 10:621–629. [DOI] [PubMed] [Google Scholar]
- Pandey GN, Rizavi HS, Bhaumik R, Ren X (2019) Increased protein and mRNA expression of corticotropin-releasing factor (CRF), decreased CRF receptors and CRF binding protein in specific postmortem brain areas of teenage suicide subjects. Psychoneuroendocrinology 106:233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, V Lord R, Clark SJ, Molloy PL (2015) De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin 8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronis A (2010) Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature 465:721–727. [DOI] [PubMed] [Google Scholar]
- Piacentini M (2014) Measuring income inequality and poverty at the regional level in OECD countries. OECD Statistics Working Papers. OECD Publishing. https://www.oecd-ilibrary.org/economics/measuring-income-inequality-and-poverty-at-the-regional-level-in-oecd-countries_5jxzf5khtg9t-en [Google Scholar]
- Pidsley R, Y Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC (2013) A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics 14:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Policicchio S, Washer S, Viana J, Iatrou A, Burrage J, Hannon E, Turecki G, Kaminsky Z, Mill J, Dempster EL, Murphy TM (2020) Genome-wide DNA methylation meta-analysis in the brains of suicide completers. Transl Psychiatry 10:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popoli M, Yan Z, McEwen BS, Sanacora G (2012) The stressed synapse: the impact of stress and glucocorticoids on glutamate transmission. Nat Rev Neurosci 13:22–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter MO, Du L, Weaver IC, Palkovits M, Faludi G, Merali Z, Szyf M, Anisman H (2008) GABAA receptor promoter hypermethylation in suicide brain: implications for the involvement of epigenetic processes. Biol Psychiatry 64:645–652. [DOI] [PubMed] [Google Scholar]
- Read J, Agar K, Barker-Collo S, Davies E, Moskowitz A (2001) Assessing suicidality inadults: integrating childhood trauma as a major risk factor. ProfessPsychol Res Pract 32:367–372. [Google Scholar]
- Reiach JS, Li PP, Warsh JJ, Kish SJ, Young LT (1999) Reduced adenylyl cyclase immunolabeling and activity in postmortem temporal cortex of depressed suicide victims. J Affect Disord 56:141–151. [DOI] [PubMed] [Google Scholar]
- Rishi V, Bhattacharya P, Chatterjee R, Rozenberg J, Zhao J, Glass K, Fitzgerald P, Vinson C (2010) CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc Natl Acad Sci U S A 107:20311–20316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Pimentel AL, Mendoza-Morales RC, Fresan A, Garcia-Dolores F, Gonzalez-Saenz EE, Morales-Marin ME, Nicolini H, Borges G (2018) Demographic and clinical characteristics of completed suicides in Mexico City 2014-2015. Front Psychiatry 9:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Dwivedi Y (2017) Understanding epigenetic architecture of suicide neurobiology: a critical perspective. Neurosci Biobehav Rev 72:10–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandi C (2011) Glucocorticoids act on glutamatergic pathways to affect memory processes. Trends Neurosci 34:165–176. [DOI] [PubMed] [Google Scholar]
- Schneider E, El Hajj N, Müller F, Navarro B, Haaf T (2015) Epigenetic dysregulation in the prefrontal cortex of suicide completers. Cytogenet Genome Res 146:19–27. [DOI] [PubMed] [Google Scholar]
- Shen L (2020) GeneOverlap: test and visualize gene overlaps. R package version 1.26.0 (computer program).
- Valera E, Sánchez-Martín FJ, Ferrer-Montiel AV, Messeguer A, Merino JM (2008) NMDA-induced neuroprotection in hippocampal neurons is mediated through the protein kinase A and CREB (cAMP-response element-binding protein) pathway. Neurochem Int 53:148–154. [DOI] [PubMed] [Google Scholar]
- Vijayakumar L, John S, Pirkis J, Whiteford H (2005) Suicide in developing countries (2): risk factors. Crisis 26:112–119. [DOI] [PubMed] [Google Scholar]
- Wang H, Storm DR (2003) Calmodulin-regulated adenylyl cyclases: cross-talk and plasticity in the central nervous system. Mol Pharmacol 63:463–468. [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D (2007) Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39:457–466. [DOI] [PubMed] [Google Scholar]
- World Health Organization (2014) Preventive suicide. A global imperative. https://www.who.int/publications/i/item/preventing-suicide-a-global-imperative. Accessed January 20, 2020.
- World Health Organization (2018) Suicide. https://www.who.int/news-room/fact-sheets/detail/suicide. Accessed January 20, 2020.
- Yamada S, Yamamoto M, Ozawa H, Riederer P, Saito T (2003) Reduced phosphorylation of cyclic AMP-responsive element binding protein in the postmortem orbitofrontal cortex of patients with major depressive disorder. J Neural Transm 110:671–680. [DOI] [PubMed] [Google Scholar]
- Zhao J, Qi XR, Gao SF, Lu J, van Wamelen DJ, Kamphuis W, Bao AM, Swaab DF (2015) Different stress-related gene expression in depression and suicide. J Psychiatr Res 68:176–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.