Abstract
Measuring the organisation of the cellular cytoskeleton and the surrounding extracellular matrix (ECM) is currently of wide interest as changes in both local and global alignment can highlight alterations in cellular functions and material properties of the extracellular environment. Different approaches have been developed to quantify these structures, typically based on fibre segmentation or on matrix representation and transformation of the image, each with its own advantages and disadvantages.
Here we present AFT−Alignment by Fourier Transform, a workflow to quantify the alignment of fibrillar features in microscopy images exploiting 2D Fast Fourier Transforms (FFT). Using pre-existing datasets of cell and ECM images, we demonstrate our approach and compare and contrast this workflow with two other well-known ImageJ algorithms to quantify image feature alignment. These comparisons reveal that AFT has a number of advantages due to its grid-based FFT approach. 1) Flexibility in defining the window and neighbourhood sizes allows for performing a parameter search to determine an optimal length scale to carry out alignment metrics. This approach can thus easily accommodate different image resolutions and biological systems. 2) The length scale of decay in alignment can be extracted by comparing neighbourhood sizes, revealing the overall distance that features remain anisotropic. 3) The approach is ambivalent to the signal source, thus making it applicable for a wide range of imaging modalities and is dependent on fewer input parameters than segmentation methods. 4) Finally, compared to segmentation methods, this algorithm is computationally inexpensive, as high-resolution images can be evaluated in less than a second on a standard desktop computer. This makes it feasible to screen numerous experimental perturbations or examine large images over long length scales.
Implementation is made available in both MATLAB and Python for wider accessibility, with example datasets for single images and batch processing. Additionally, we include an approach to automatically search parameters for optimum window and neighbourhood sizes, as well as to measure the decay in alignment over progressively increasing length scales.
Keywords: alignment, Fast Fourier Transform (FFT), cytoskeleton, extracellular matrix (ECM), fibres, anisotropy
1. INTRODUCTION
Measuring the anisotropy of features in biological images is of increasing interest as the degree of alignment can inform both the underlying cellular behaviours and material properties of the sample. For example, different cell types have unique emergent capacities to align in culture, controlled by cellular packing and geometry (Duclos et al., 2014, 2017). When cells are exposed to external forces such as cyclical stretch, they tend to align their cytoskeleton perpendicular to the axis of stretch (Standley et al., 2002; Yoshigi et al., 2005; Livne et al., 2014). Subcellular cytoskeletal networks can also spontaneously organise in response to the stresses of their environment (Gupta et al., 2015, 2019) or changes in biochemical signalling (Ridley and Hall, 1992). Similar changes in cytoskeletal architecture can be seen in reconstituted protein systems (Falzone et al., 2013; Linsmeier et al., 2016). Additionally, the extracellular matrix (ECM) also has an inherent capacity to align in different tissues or pathologies, such as cancer (Ouellette et al., 2021) or tissue fibrosis (Park et al., 2020; Mascharak et al., 2021), which is thought to alter the ECM network mechanical properties. These examples illustrate the variety of environments where alignment of features reveals important biological properties. As such, it is necessary to develop approaches to efficiently quantify anisotropy of features across a range of length scales, from subcellular organisation to tissue level alignment.
A number of different approaches have been developed to quantify the alignment of image features. These methods can be categorised based on the type of algorithm used to highlight features and subsequently quantify anisotropy. Fibre segmentation tools, such as the Fiji Ridge Detection plugin (Lindeberg, 1998), the curvelet-based CurveAlign/CT-FIRE suite (Bredfeldt et al., 2014a,b; Liu et al., 2017, 2020), and Filament Sensor (Eltzner et al., 2015), provide individual fibre information. Tools based on representation/transformation of the image, such as the Fiji plugins OrientationJ (Rezakhaniha et al., 2012) and FibrilTool (Boudaoud et al., 2014) or the CytoSpectre suite (Kartasalo et al., 2015), supply overall fibre alignment information. Hybrid tools, such as TWOMBLI, which exploit a combination of approaches (i.e., fibre segmentation followed by matrix representation of the image) have also been recently made available (Wershof et al., 2021).
Many image transformation algorithms rely on Fourier transformation of the image and exploit its frequency space representation to obtain alignment information (Chaudhuri et al., 1987; Pourdeyhimi et al., 1997; Marquez, 2006; Sander and Barocas, 2009; Goldyn et al., 2010; Kartasalo et al., 2015; Clemons et al., 2018). These algorithms tend to be computationally efficient (Sander and Barocas, 2009) and insensitive to the image background noise (Liu et al., 2020), which is desirable for rapidly processing large volumes of experimental data (Püspöki et al., 2016). Another desirable trait for an alignment detection tool is its flexibility in terms of size and location of alignment patterns (Püspöki et al., 2016), which is handled well by the only open-source software available in this category (CytoSpectre (Kartasalo et al., 2015)) due to a power spectrum analysis of the image’s discrete Fourier transform.
One feature missing from the available tools in the literature is an analysis of the alignment length scale. Depending on the scientific question of interest, one may want to measure alignment over a small region of the image (e.g., local cytoskeletal features inside cells) or over a broad length scale (e.g., ECM organisation in whole tissues). Additionally, when comparing experimental datasets, it is important to understand the precise length scale over which the observed alignment is significant and determine the spatial decay of the anisotropy, which will allow for better interpretation and analysis of results (e.g., is the alignment of a region of interest a subcellular or supracellular phenomenon). Here we explain the implementation of an open-source alignment quantification algorithm, AFT – Alignment by Fourier Transform, that can be run on a variety of biological images and has a number of advantages over pre-existing approaches. This FFT-based quantification is rapid and computationally efficient, flexible to different types of microscopy images and samples, easy to use with a low number of parameters, and importantly allows for a user-defined length scale over which to measure feature anisotropy.
2. MATERIALS AND METHODS
2.1. Biological mages
Sample datasets of microscopy images were used to illustrate the analysis and are provided as Supplementary Material. In particular, cultured fibroblasts transfected with GFP-actin, fibroblasts fixed and stained for filamentous actin (F-actin, phalloidin), fixed cell-derived matrices immunostained for actin (before decellularisation) and fibronectin (after decellularisation), and second harmonic generation imaging of tissue samples were utilised. Fixed actin images were acquired at different magnifications using either single cells or cells at confluence, to allow for evaluation of the length scale of alignment. Y-27632 treated cells were incubated in the indicated concentration of the drug for 30 minutes and then fixed and stained with phalloidin.
Fixed samples were imaged using a Zeiss LSM 880 equipped with a 40x NA 1.3 Plan-Apochromat oil objective or 63x NA 1.4 Plan-Apochromat oil objective. Decellularised matrices were imaged using a Zeiss LSM 880 equipped with 63x NA 1.4 Plan-Apochromat oil. For second harmonic generation imaging, tissue sections were imaged using a Zeiss LSM 7MP equipped with a 20x NA 1.0 water immersion objective. Y-27632 treated cells were imaged on a 3i Marianas Imaging System consisting of a Zeiss Axio Observer 7 inverted microscope attached to a Yokogawa W1 Confocal Spinning Disk using a 63x NA 1.4 Plan-Apochromat oil objective.
2.2. Synthetic data
Images of fibrillar features were simulated in MATLAB (Mathworks, v2018b) to create sets of synthetic data with specific length scales. To this aim, squared units with a pre-defined size were randomly distributed in the field of view. Each unit was filled with parallel lines with 5 px spacing, and then rotated with a random angle between 0° and 180°. Three 10-image sets were generated, with unit sizes equal to 100, 200, or 400 px.
2.3. Pre-processing
Image pre-processing is not required for this analysis, but can be used to additionally highlight the fibrillar features. In the context of this work, pre-processing was performed only for the cell-derived matrix images, as their surface is often not flat, resulting in uneven signal across the field of view. To account for this, contrast was enhanced in Fiji (0.35% saturated pixels) and subsequently a local contrast adjustment was performed (CLAHE plugin, default parameters). All images shown are maximum projections of Z-stack acquisitions, except for the live cell images which represent a single confocal slice.
2.4. Comparison with other available alignment tools
The performance of the algorithm presented in this work was compared with OrientationJ (Rezakhaniha et al., 2012) and TWOMBLI (Wershof et al., 2021), by using two 10-image samples of fibronectin-stained cell-derived matrices that were either isotropic or anisotropic. To access the alignment vector field for OrientationJ, a Fiji macro was designed to batch run the OrientationJ Vector Field plugin. This operation was either performed on its own or preceded by fibre segmentation by the Ridge Detection plugin (Lindeberg, 1998), this second approach mimicking the TWOMBLI workflow (hybrid method). To obtain a comparable spatial resolution of the vector field, a window of 100 px with 50% overlap was used to run the FFT analysis, and a matching local window σ of 100 px with a grid size of 50 was employed in OrientationJ. The Ridge Detection parameters were set to: line width = 20 px, high contrast = 120, low contrast = 0, sigma = 6.27, lower threshold = 0, upper threshold = 0.17, minimum line length = 10 px. These parameters were selected to match the ones assigned by the TWOMBLI macro when calling the Ridge Detection plugin (see below for the user input parameters that were chosen in TWOMBLI). From the acquired vector fields, an order parameter over neighbourhoods of 5x vectors was calculated and compared for the three approaches.
A second comparison was based on coherency, the metric traditionally used for global alignment output by both OrientationJ and TWOMBLI. Coherency of the FFT was measured on windows of 100 px with 50% overlap and averaged to obtain a median value representing the global alignment score for each image. To obtain the coherency score from OrientationJ, a macro was designed to batch process the images using the OrientationJ Dominant Direction plugin. TWOMBLI was run on the sample data as per the developer instructions. A parameter optimisation step was performed on test images and the obtained parameter file was used for batch processing (line width = 20 px, curvature window = 150 px, branch length = 10).
Computational time was measured on a 3.6 GHz Quad-Core Intel Core i7 machine with 32 GB memory, with suitable functions in MATLAB and ImageJ macro language. Only the effective analysis time for a 10-image sample (1 MB per image) was taken into account, excluding the input and parameter upload. Timings for OrientationJ refer to the designed macro, no manual handling of data was involved.
2.5. Statistical analysis
Statistical analysis was performed in Prism (Graphpad, v9). Mann-Whitney two-tailed tests were used to compare metrics on isotropic vs. anisotropic samples. Significance is reported in figure panels as follows: ‘****’ for p-values lower than 0.0001, ‘***’ lower than 0.001, ‘**’ lower than 0.01, ‘*’ lower than 0.05, ‘ns’ otherwise.
2.6. Implementations
This approach builds upon previous work employed in (Aratyn-Schaus et al., 2011; Cetera et al., 2014; Fernandes et al., 2020) and adds features such as periodic decomposition to improve the accuracy of the FFT angle determination along with the ability to search for the optimal length scale for comparison. The current implementation is available at github.com/OakesLab/AFT-Alignment_by_Fourier_Transform, both in MATLAB and Python languages. The MATLAB version is provided with a simple user interface for inputting parameters, while the Python version is presented as a set of Jupyter notebooks. The code is divided into two separate suites. The first suite can be used to perform the alignment quantification on a sample of images containing fibrillar structures. The second suite can be used to run a search to optimise the analysis parameters, with the aim of maximising differences between two samples, i.e., locating the length scale for which the difference in alignment between two samples is greatest. Further code documentation is provided in the repository.
3. RESULTS
3.1. Measuring alignment
The algorithm is inspired by the general approach of Particle Image Velocimetry (Willert and Gharib, 1991), where the image is broken down into a series of windows that are analysed independently to create a vector field that represents the entire image. Windows are analysed in frequency space to reveal information about both the fibrillar structure and local alignment as illustrated in Fig 1 using filamentous actin images. If the image contains aligned features in the real space, the corresponding FFT in the frequency domain will be asymmetrically skewed, with the direction of skew orthogonal to the original feature orientation (Fig 1AB). This process can then be repeated on each successive window across the image, resulting in a vector field that represents the local alignment in the image. A detailed protocol for determining the local alignment of each window and the order parameter of global alignment follows.
Figure 1. Using FFTs to measure local alignment of biological images.
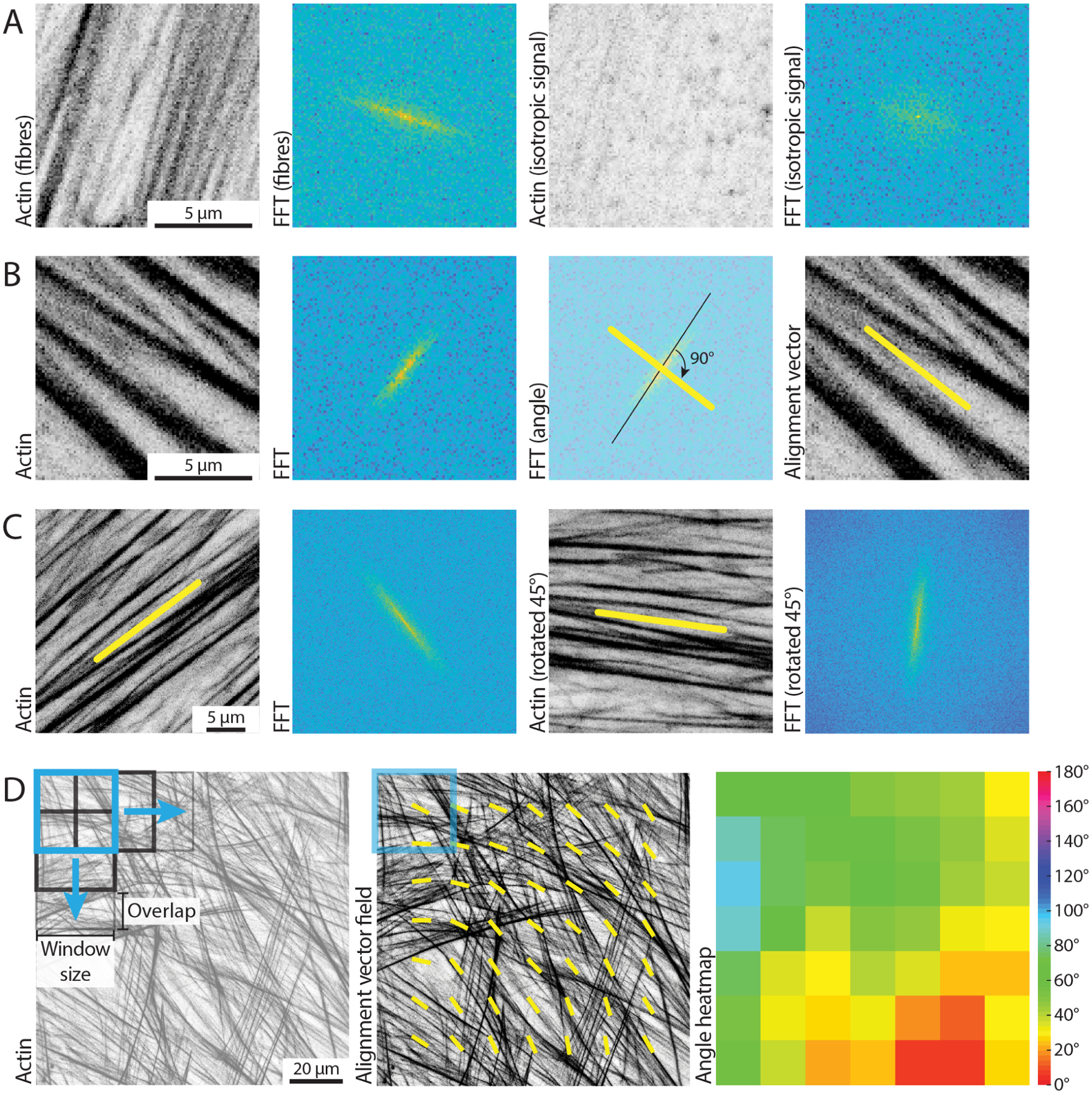
(A) Small windows of a microscopic image are considered, containing either aligned filamentous actin fibres, or isotropic signal. The 2D Fast Fourier Transform (FFT) shows an elongated (i.e., skewed) shape if the image in the real space contains aligned fibrillar features, and displays a more round shape otherwise. Scale bar, 5 μm. (B) A small window of a microscopic image containing aligned filamentous actin fibres is considered. Its FFT shows a predominant skewness (black line) of an angle that is orthogonal to the direction of the fibres in the real space image. The obtained alignment vector (yellow line) can be overlaid to the original image to highlight the measured fibre orientation. Scale bar, 5 μm. (C) The alignment vector is calculated for an image containing aligned actin fibres by exploiting the characteristics of the FFT. The same image, rotated by 45°, is analysed, to demonstrate robustness of the method to rotation. Scale bar, 5 μm. (D) A square representing the window size for analysis (250 px, light blue) is overlaid on a filamentous actin image. The black squares represent subsequent instances of such a window with the defined overlap, as the image is scanned during the analysis to calculate local alignment vectors (yellow). The heatmap shows a different representation of the alignment vector field, with a wrapped colour scale ranging from 0° to 180°. Scale bar, 20 μm.
3.1.1. Local Alignment
Square windows of n × n pixels are chosen from the original real space image and a 2D FFT is performed. Typically, n is chosen as odd to ensure that the zero-order frequency component, which is always the greatest in magnitude, is situated at the centre of the FFT. FFTs of non-periodic signals are often plagued by strong horizontal and vertical components, appearing as a cross, due to the mismatch of intensity in the images at the edges. To avoid this effect, we break down the image into its smooth and periodic components following the approach of Moisan (2011). We use the periodic component of this decomposition to take the FFT and then take its norm to only deal with real numbers. The resulting image is then multiplied by a mask of diameter n/2 to capture all the relevant high frequency components and to ensure the sample is symmetric. The central image moments of the masked FFT are then calculated as
| (1) |
where x and y represent the position and I is the magnitude of the FFT norm. We next construct the covariance matrix of the image as
| (2) |
which has eigenvalues of
| (3) |
The orientation of the FFT can thus be calculated by
| (4) |
and the eccentricity, a measure of how oblong the FFT is, as
| (5) |
The orientation of the features in the real image is orthogonal to the orientation of the FFT, and thus we apply a 90° rotation (Fig 1B). Orientation vectors do not have a polarity, and thus angles of 0° or 180° are considered equivalent. Therefore, all angles are mapped to a range of 0° to 180° (Fig 1D). This methodology is robust to rotational transformations and does not suffer from the inherited bias of FFT for vertical and horizontal components (Fig 1C).
3.1.2. Order Parameter Calculation
Both the size of the window and the degree of overlap between the windows is customisable (Fig 1D). To measure how aligned fibrillar features are within adjacent windows, we define an order parameter by correlating the directionality of vectors within the neighbourhood of customisable size. Specifically, the order parameter can be calculated as
| (6) |
with θij representing the angle between the orientation of a central reference vector and its neighbours (Fig 2A). The order parameter can take values between −1 and 1 (Fig 2A), with 1 representing perfect alignment (i.e., all vectors in the neighbourhood have the same orientation as the reference vector), 0 representing random orientation, and −1 representing opposite alignment (i.e., all vectors in the neighbourhood are pointing in the orthogonal direction compared to the reference vector). While a number of different formulations of the order parameter have been used previously, in the context of the current analysis, the chosen order parameter normally ranges between 0 (random) and 1 (perfectly aligned), as fibre polarity is not taken into account and the neighbourhood area is kept relatively local (Fig 2B). Similar results were obtained for real and simulated data (and Fig 3).
Figure 2. Calculating an image order parameter.

(A) The equation for the order parameter used to assign an alignment score for each neighbourhood is shown, together with examples. The order parameter takes a value of 1 for complete alignment, 0 for random alignment, and −1 for orthogonal alignment. (B) A square representing a neighbourhood of 5x vectors (magenta, neighbourhood radius of 2x vectors around the central reference) is overlaid on an actin image and its corresponding alignment vector field. The black squares represent subsequent instances of such neighbourhood, as the vector field is scanned during the analysis to obtain local alignment scores. The resulting order parameters for each neighbourhood are reported in the heatmap, together with their median value, representing the output of the analysis.
Figure 3. Examples of image neighbourhoods and corresponding order parameter.

(A) Examples of neighbourhoods in simulated data displaying different degrees of alignment are shown together with the calculated order parameter (window size 35 px). Scale bar, 25 px. (B) Examples of neighbourhoods in actin images displaying different degrees of alignment are shown together with the calculated order parameter (window size 100 μm). Scale bar, 50 μm.
To calculate an overall alignment score for each image, we define a neighbourhood size. The previously calculated vector field is then split into all possible overlapping neighbourhoods of such size. The order parameter is subsequently calculated for each neighbourhood, and the overall order parameter for the image is defined as the median value (Fig 2B).
3.2. Investigating the optimal alignment length scale between two samples
When comparing two samples displaying different degrees of alignment, it may be of interest to evaluate the length scale for which this difference is greatest. The approach described above allows for this, as it is possible to evaluate the order parameter using a range of window and neighbourhood sizes which correspond to different length scales (i.e., each permutation of window and neighbourhood size define a specific length scale). By comparing the obtained alignment scores between the two experimental conditions for the different parameter permutations, it is possible to investigate the precise length scale at which the difference between the two samples is most pronounced. The comparison can be carried out by looking at the order parameter difference between the two samples or by running statistical tests and analysing the resulting p-values.
We tested this parameter search approach on actin images of cultured fibroblasts with different degree of isotropy (Fig 4A, 10 images for each sample), for window sizes ranging from 25 to 325 px and neighbourhood radii ranging from 1x to 38x vectors (Fig 4BC). We performed the comparison in alignment scores for each window/neighbourhood size permutation by quantifying the difference between the sample order parameters (Fig 4B) or by calculating the p-value of a Mann-Whitney statistical test between the two populations (Fig 4C).
Figure 4. Determining the optimal length scale difference.
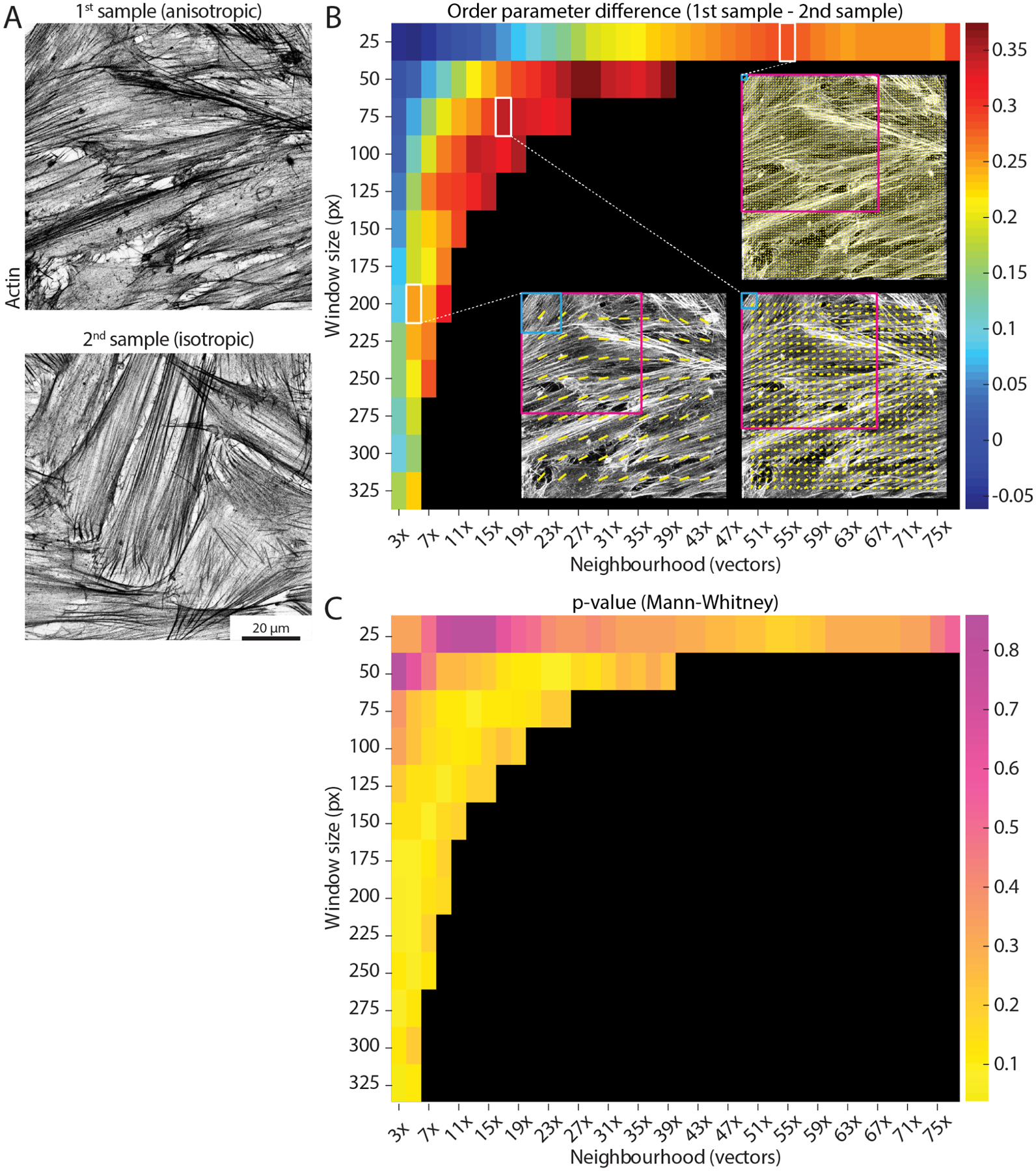
(A) Example actin images taken from two separate samples and displaying different degrees of alignment (higher on the left, more anisotropic; lower on the right, more isotropic). Scale bar, 20 μm. (B) The parameter search runs the analysis for a range of window sizes (y-axis, from 25 px to 325 px) and a range of neighbourhood sizes (x-axis, from 3x to 77x vectors, where the neighbourhood size is defined as 2 * neighbourhood radius + 1). Each permutation of window and neighbourhood represent a length scale. Each coloured square shows the difference in the median order parameter between the anisotropic sample and the isotropic sample (mean values over 10 images for each sample) for a specific pair of window and neighbourhood sizes. Small window sizes paired with small neighbourhoods (upper left corner) display noisy output, due to the limited amount of image information available to process. Three example vector fields are shown for a window size of 25 px and neighbourhood of 55x vectors (corresponding to 92 μm), 75 px and 17x vectors (89 μm), and 200 px and 5x vectors (79 μm), with the light blue and magenta squares showing the size of the window and neighbourhood respectively. Despite relatively similar neighbourhood sizes, the output varies depending on the size of the examined window. Reading the graph horizontally from left to right for each window size up to ~150 px, it is possible to note how an optimal length scale of ~90 μm is detected (i.e., a peak difference value is displayed), likely correlated to relevant biological dimensions (e.g., cell size). (C) A similar graph to the one in (B) is shown for the p-value of Mann-Whitney tests between the two samples for all the combinations of window and neighbourhood sizes. In this case, greater differences are represented by lower values (yellow).
It should be noted that small windows paired with small neighbourhoods display noisy output (Fig 4BC), as the corresponding FFT for such small windows will tend towards low eccentricity values (i.e., increasing the likelihood of spurious vectors, Fig 1A). Interestingly, when comparing two samples with known differences in alignment (Fig 4A), it can be observed how the difference in order parameter between the two samples displays a peak when keeping the window size constant and increasing the neighbourhood (Fig 4B). This suggests a well-defined optimum length scale where a clear difference between the two samples occurs, and it is likely to be correlated with relevant dimensions in the sample (e.g., cell size). It is important to note that the optimal length scale will likely exist for a range of different window/neighbourhood pairs, which together will define a similar local area. In the example shown in Fig 4B, the order parameter difference peaks for a length scale of ~ 90 μm, as calculated by multiplying the window size by the neighbourhood size for pairs displaying high difference values. This suggests that this length scale should be investigated further to evaluate the biological relevance of features of such size.
3.3. Evaluating the length scale decay in alignment
A similar approach to the parameter search can be employed to look at how the alignment decays over increasingly larger length scales, by progressively enlarging the size of the region over which the alignment is measured. To this aim, the window size over which the angle vector field is calculated is kept constant, while progressively increasing the neighbourhood over which the order parameter is evaluated.
To confirm the performance of the algorithm in locating the alignment length scale, we created three 10-image sets of synthetic data. These images show a number of squared units of pre-defined size (i.e., 100, 200, or 400 px), randomly arranged in the field of view. Each unit is filled with parallel lines representing fibres (Fig 5A). We first measured the median order parameter for each image in each set with a window size of 35 px and a neighbourhood of 5x vectors, corresponding to 100 px (Fig 5B). The order parameter increases with the selected unit size, as expected as neighbourhoods of a given size will represent a progressively more local point of view for increasing unit sizes. Moreover, we analysed the decay in the order parameter by keeping a fixed 10 px window size and progressively increasing the neighbourhood size (Fig 5C). The half-life of the fitted decay curve shows values very close to the pre-defined unit size for each set. This result highlights the ability of our approach to correctly identify the alignment length scale within an image.
Figure 5. Evaluating length scale on synthetic data.
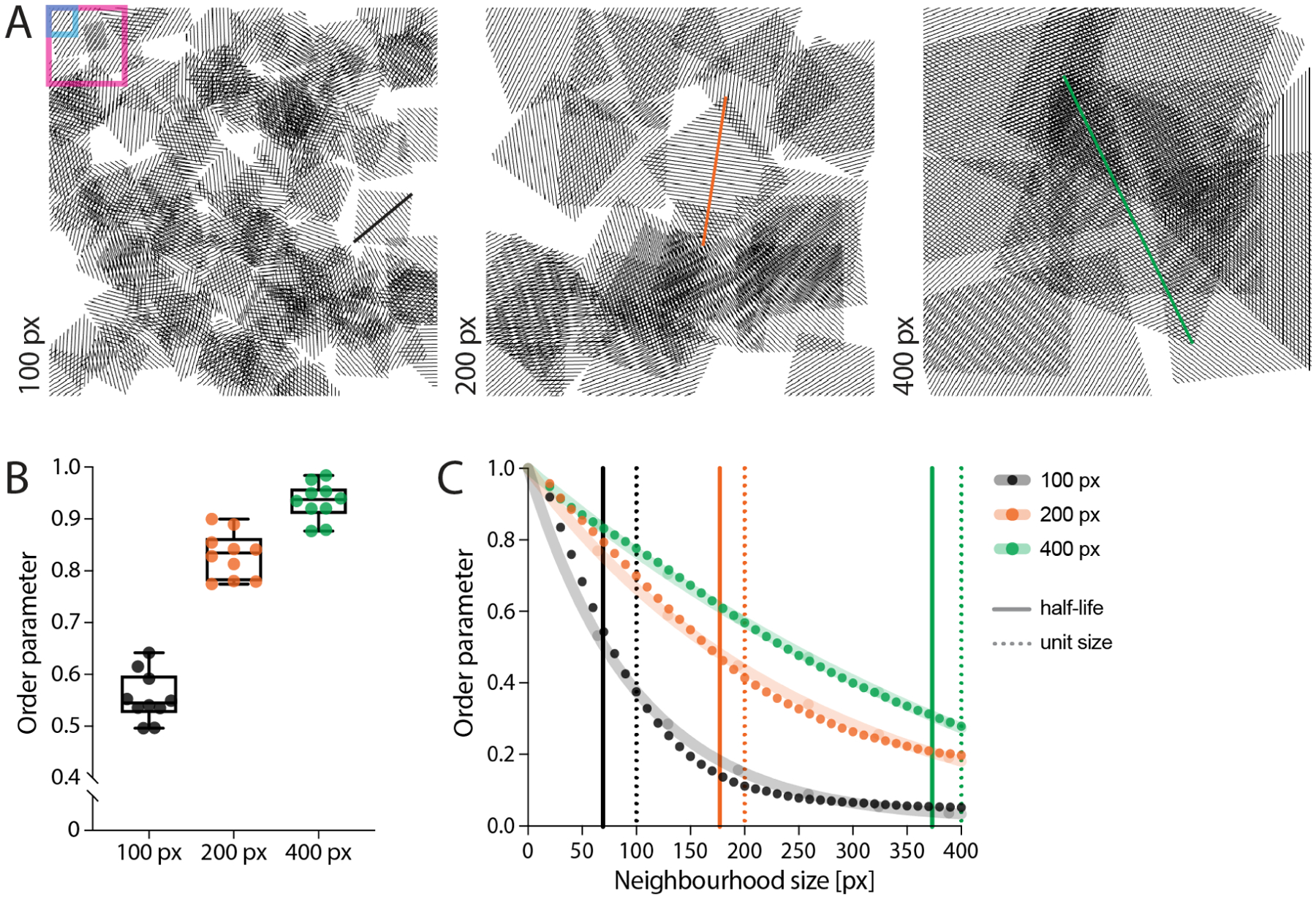
(A) Synthetic images were created by randomly distributing units of parallel lines within the field of view. The size of each unit was pre-defined to be either 100 px (left, black), 200 px (centre, orange) or 400 px (right, green). Each sample is constituted by 10 images. The blue and magenta boxes represent the window and neighbourhood sizes used for the analysis in (B), set to 35 px and 100 px, respectively. (B) Measuring the order parameter with the same parameters for the three set of images show an increase in its median value with the unit size. This can be expected, as the information within this length scale will be more aligned the bigger the unit size. (C) Keeping the window size constant (10 px) and progressively increasing the neighbourhood shows a decay in the order parameter for all three samples (dots). By fitting a one-phase decay to each data set (shaded lines) and measuring its half-life, it is possible to appreciate how the algorithm is able to locate the pre-defined length scale (i.e., unit size).
We then used the alignment decay analysis to quantify the length scale on real biological data, as exemplified by using ~1 mm2 tilescan images of cell-derived matrices (i.e., matrices produced in situ by the cells) stained for F-actin and fibronectin (before and after decellularisation, respectively, Fig 6A). We first calculated the angle vector field for both actin and fibronectin using the FFT approach for a window size of 250 px and overlap of 50% (Fig 6B). Subsequently, we calculated the median order parameter over neighbourhoods of increasing radii, ranging from 1x to 21x vectors, corresponding to 50 × 50 μm to 1050 × 1050 μm in the original image (Fig 6ACD). By plotting the median order parameter over the neighbourhood size, it is possible to observe the alignment decaying from the local to the global length scale (Fig 6D). As expected, actin and fibronectin displayed a similar decay in alignment as cells are responsible for depositing and remodelling the ECM in this experiment.
Figure 6. Measuring the decay in order parameter as a function of length scale.
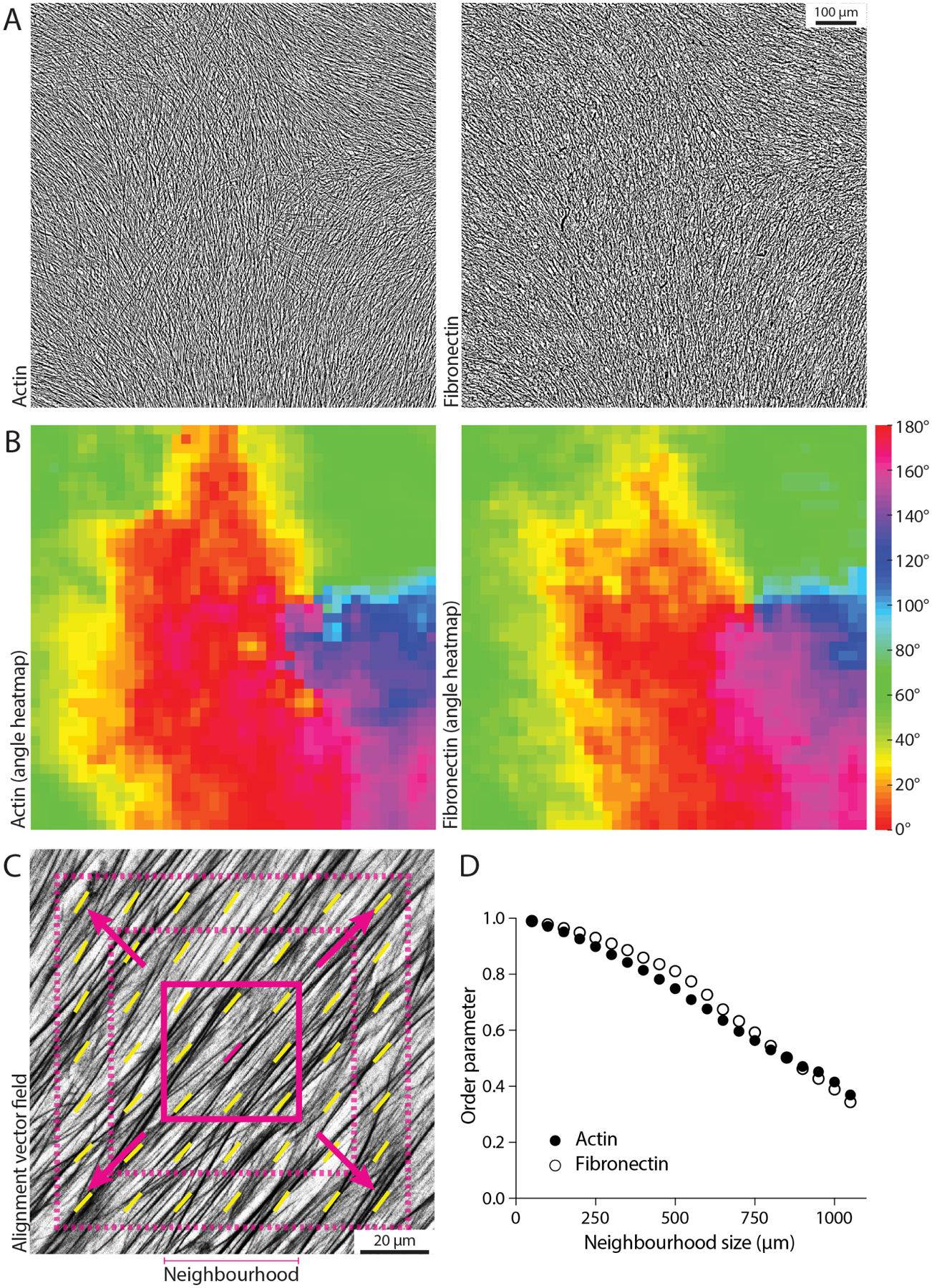
(A) Example tilescan images about 1 mm2 in size for actin and fibronectin on the same sample (fibroblast-derived matrix). Scale bar, 100 μm. (B) Angle heatmap showing the alignment vector field for the images in (A), using a window size of 250 px and 50% overlap. Actin and fibronectin display similar alignment. (C) Schematic of the alignment decay analysis, where the window size is maintained constant while increasing the size of the neighbourhood (i.e., including more and more vectors in the order parameter calculation to evaluate alignment from the local to the global scale). The solid magenta square represents the starting neighbourhood, which then increases in size at each iteration (dashed magenta squares). (D) The calculated order parameter for actin and fibronectin for the images in (A) is graphed for a window size of 250 px, for each neighbourhood ranging from 50 μm (3x vectors, neighbourhood radius of 1x vector) to 1050 μm (43x vectors, neighbourhood radius of 21x vectors). Alignment decays in a similar manner when going from local (small neighbourhood size) to global (large neighbourhood size) length scales.
3.4. Filtering input for alignment analysis
Some biological images might display regions that are either blank (i.e., little to no signal, lack of image features), or isotropic (i.e., no obvious fibrillar features). In certain applications, such regions should be excluded from the alignment analysis, as images containing noisy or blank areas will affect the output order parameter. By exploiting our window-based algorithm, it is possible to implement optional filtering of features to automatically exclude regions with such characteristics from the analysis.
To filter out blank regions, a threshold can be set on the mean intensity for each window, as demonstrated on second harmonic generation imaging of tissue samples (Fig 7A). In this example, windows that are considered background have a mean intensity signal lower than 20 (with intensity values ranging from 0-black to 255-white). This value can be set as a threshold: windows displaying mean intensity values lower than the threshold will be ignored during the calculation of the angle vector field (and subsequent alignment score, Fig 7B).
Figure 7. Filtering vector fields based on intensity and eccentricity.
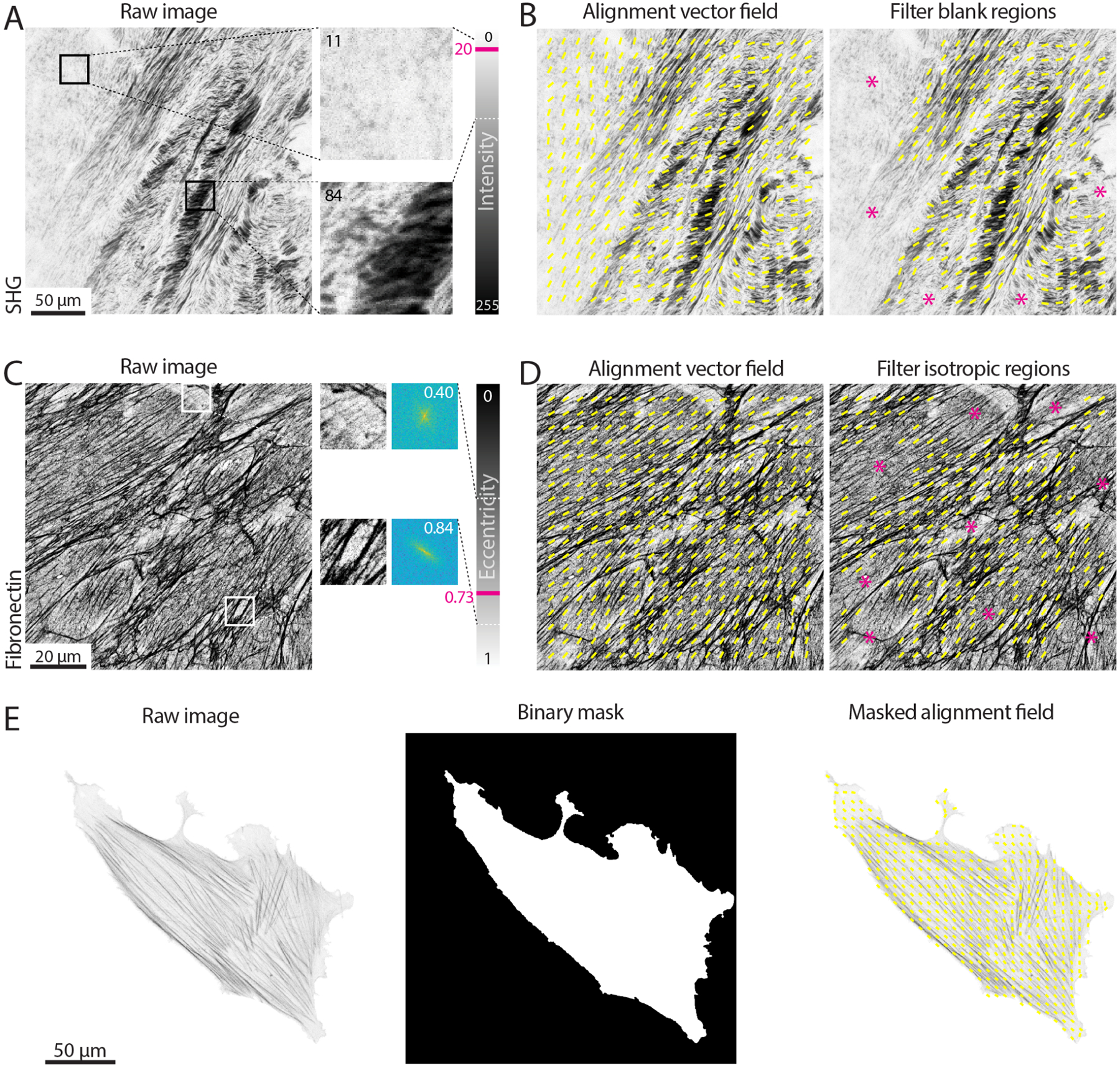
(A) A tissue section second harmonic generation (SHG) image is shown. Two regions with the same size of the window used for the alignment analysis (100 px) are enlarged and their mean intensity value measured. The intensity of an 8-bit image can vary from 0 to 255. The area on the top left contains little to no signal and it is considered background (mean intensity of 11); the area on the right contains many dark pixels, for a mean intensity of 84. A threshold value of 20 is set, meaning that all windows with mean intensity lower than such values will be considered as blank (i.e., background) and excluded from the analysis. Scale bar, 50 μm. (B) The alignment vector field (yellow) is overlaid on the raw SHG image, either without filtering for blank regions (left) or filtering with a threshold on the mean intensity of 20 (right). Magenta asterisks highlight regions excluded from the analysis. (C) An immunostaining for fibronectin in cell-derived matrices is shown. Two regions with the same size of the window used for the alignment analysis (100 px) are enlarged and their 2D Fast Fourier Transform (FFT) displayed. The FFT eccentricity can range from 0 (circular) to 1 (highly elongated). The region on the top lacks clear aligned fibres, resulting in a more uniform FFT (eccentricity of 0.40); the area on the bottom has aligned fibrillar features, leading to a highly skewed FFT (eccentricity of 0.84). A threshold value of 0.73 is set, meaning that all windows with eccentricity lower than such values will be considered as isotropic and excluded from the analysis. Scale bar, 20 μm. (D) The alignment vector field (yellow) is overlaid on the raw fibronectin image, either without filtering for isotropic regions (left) or filtering with a threshold on the eccentricity of 0.73 (right). Magenta asterisks highlight the regions excluded from the analysis. (E) The actin in a single cell is shown. If one wants to exclude the background from the analysis, a binary mask can be provided highlighting the regions of the image to be analysed (window size 50 px with 50% overlap). Scale bar, 50 μm.
Regions in which image features display isotropic organisation can also be excluded by setting a threshold on the eccentricity of the FFT calculated for a given window, as shown for cell derived matrices immunostained for fibronectin in Fig 7C. The FFT eccentricity can be used as a measure of its skewness (Fig 1A): regions containing oriented fibres will display a more elongated FFT, hence higher eccentricity (values closer to 1); regions where no fibres can be detected, or where there are multiple fibres without a clear orientation, will display a more homogeneously shaped (i.e., round) FFT, hence lower eccentricity (values closer to 0). In the example shown in Fig 7C, a threshold on the eccentricity was set to 0.73, allowing for the exclusion of isotropic regions from the angle vector field calculation (Fig 7D).
Finally, it is possible that only a specific region within an image is to be analysed, as in the case of actin in single cells (Fig 7E). A binary image can be used to mask the angle vector field, in order to only consider a given area of the original input.
3.5. Extracting kinetics of organisation from live imaging
Many biological processes involve the evolution of architectures and organisations over time. By measuring an order parameter for each frame of a time series it is possible to extract the kinetics of alignment in response to perturbations. As an example, we transfected an NIH 3T3 fibroblast with GFP-actin and imaged the cell before, during, and after treatment with the Rho kinase inhibitor Y-27632 (Fig 8). Inhibition of Rho kinase leads to a reduction in myosin activity and thus a dissolution of stress fibres (e.g., a loss of fibrillar structures; Fig 8A). As a test case, treatment with Y-27632 is convenient because it acts rapidly and is easily washed out by replacing the media in the sample chamber with fresh media. Upon removal of Y-27632, the actin cytoskeleton begins to reform stress fibres and again takes on an aligned appearance (Fig 8B). As evidenced in the plot of the order parameter, we can see a loss of alignment immediately after addition of Y-27632 and an immediate recovery following washout (Fig 8C).
Figure 8. Measuring kinetics of alignment.
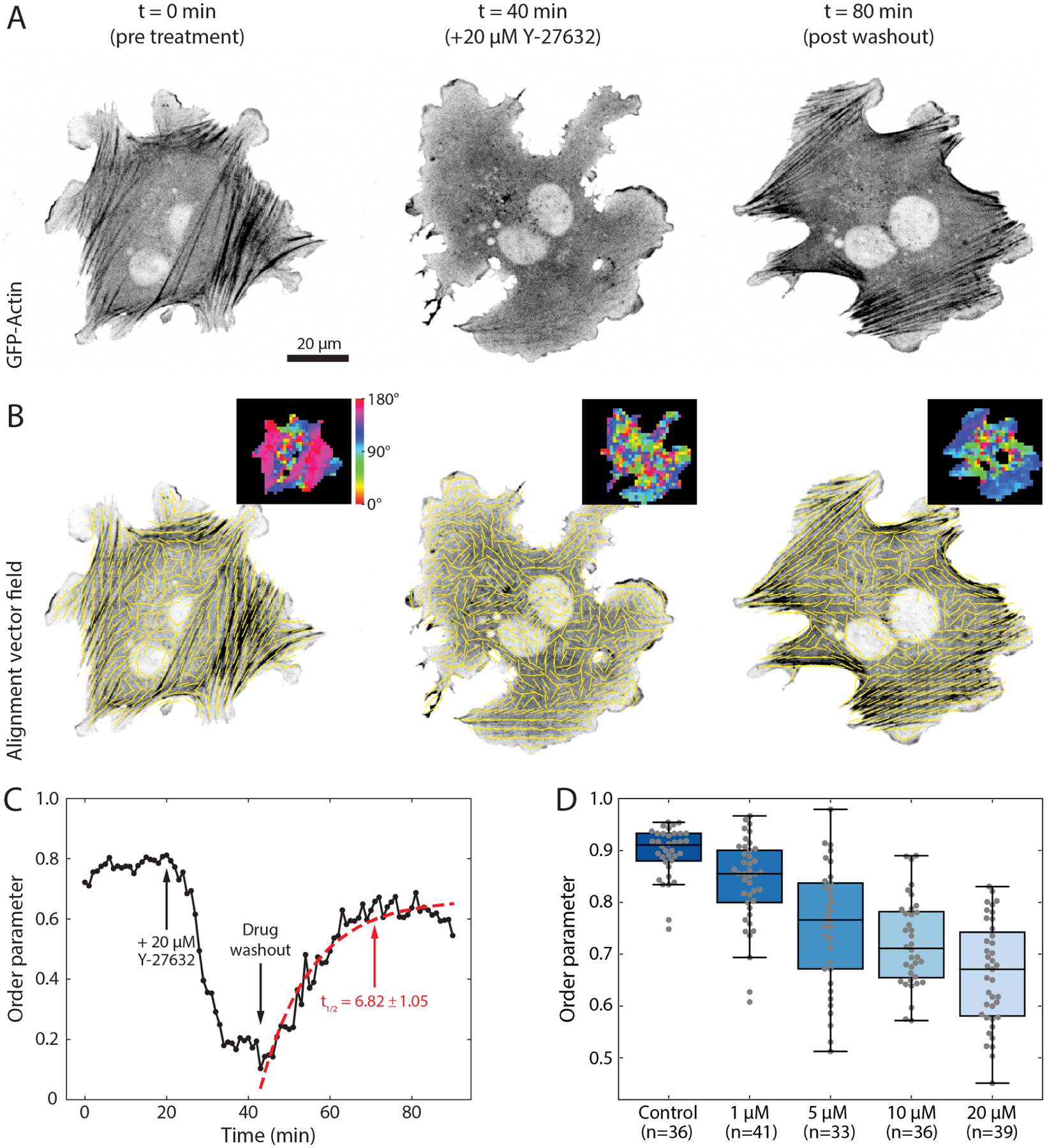
(A) An NIH 3T3 fibroblast transfected with GFP-actin is shown before, during, and after washout of 20 μM Y-27632. (B) Local alignment was calculated for each frame in the time series using a window size of 17 px and an overlap of 50%. Yellow vectors indicate the local direction of alignment in the image while insets show the angle of orientation for each window. (C) Plot of the calculated order parameter over time for a neighbourhood radius of 2x vectors. Arrows indicate the time points of addition of Y-27632 and wash out of the drug. Red dashed line indicates the fit to Eq 7. (D) Boxplots of the average order parameter for fibroblasts treated for 30 min with different concentrations of Y-27632 and then fixed and stained with phalloidin to label actin. Each dot represents the median order parameter of a single cell with a window size of 33 px, overlap of 50%, and neighborhood radius of 2x.
To measure the kinetics of this interaction we can fit the data to any relevant model. For the washout of Y-27632 we fit the post-washout data to a simple exponential recovery curve of the form
| (7) |
We can then define a t1/2 value, or the time it takes to reach half its maximum recovery value, as
| (8) |
This results in a value of t1/2 ≈ 7 min for the Y-27632 washout experiment, consistent with previous reports (Aratyn-Schaus et al., 2011). To further illustrate the robustness of this approach, we imaged numerous cells treated with varying concentrations of Y-27632 (Fig 8D). Cells were treated with concentrations ranging from 0–20 μM for 30 minutes, and then fixed and stained with phalloidin. Analysis of these data sets reveals a steady decrease in order parameter as a function of Y-27632 concentration, as expected. The difference in magnitude of the order parameter between the fixed and live imaging is the product of superior signal to noise and better labelling that is achieved with phalloidin compared to fluorescently expressed actin.
3.6. Comparing with other available algorithms
The performance of the algorithm presented in this work was tested against two other open-source tools widely used by the biology community, OrientationJ (Rezakhaniha et al., 2012) and TWOMBLI (Wershof et al., 2021). OrientationJ is a Fiji plugin based on image representation by structure tensors using real space, and evaluates orientation by the coherency metric, that indicates the degree of feature orientation with values ranging from 0 (random) to 1 (perfectly aligned) (Rezakhaniha et al., 2012; Clemons et al., 2018). Using this approach, it is possible to output either an alignment vector field (OrientationJ Vector Field) or a global coherency score for the image (OrientationJ Dominant Direction). TWOMBLI is a Fiji macro envisioned as a hybrid approach performing fibre segmentation followed by alignment analysis. Fibre segmentation is carried out with the Ridge Detection plugin (Lindeberg, 1998), while the alignment of the binarised image is subsequently measured via OrientationJ. In addition to the coherency metric as a measure of alignment and due to the segmentation step, TWOMBLI also allows the user to obtain further metrics that characterise the fibrillar features, such as number of branch and end points, density, curvature, and fractal dimensions.
We first compared the alignment vector field calculated with the three methods (FFT, structure tensor, and hybrid approach) for cell-derived matrices immunostained for fibronectin displaying different degrees of isotropy (Fig 9). When comparing the alignment vector field for images with strongly anisotropic fibrillar features, similar output was observed for the three methods (Fig 9A). When working with less aligned input, however, the FFT was able to better resolve the feature alignment in the isotropic regions compared to the other two algorithms (Fig 9B).
Figure 9. Comparing approaches to measure organisation in biological images.
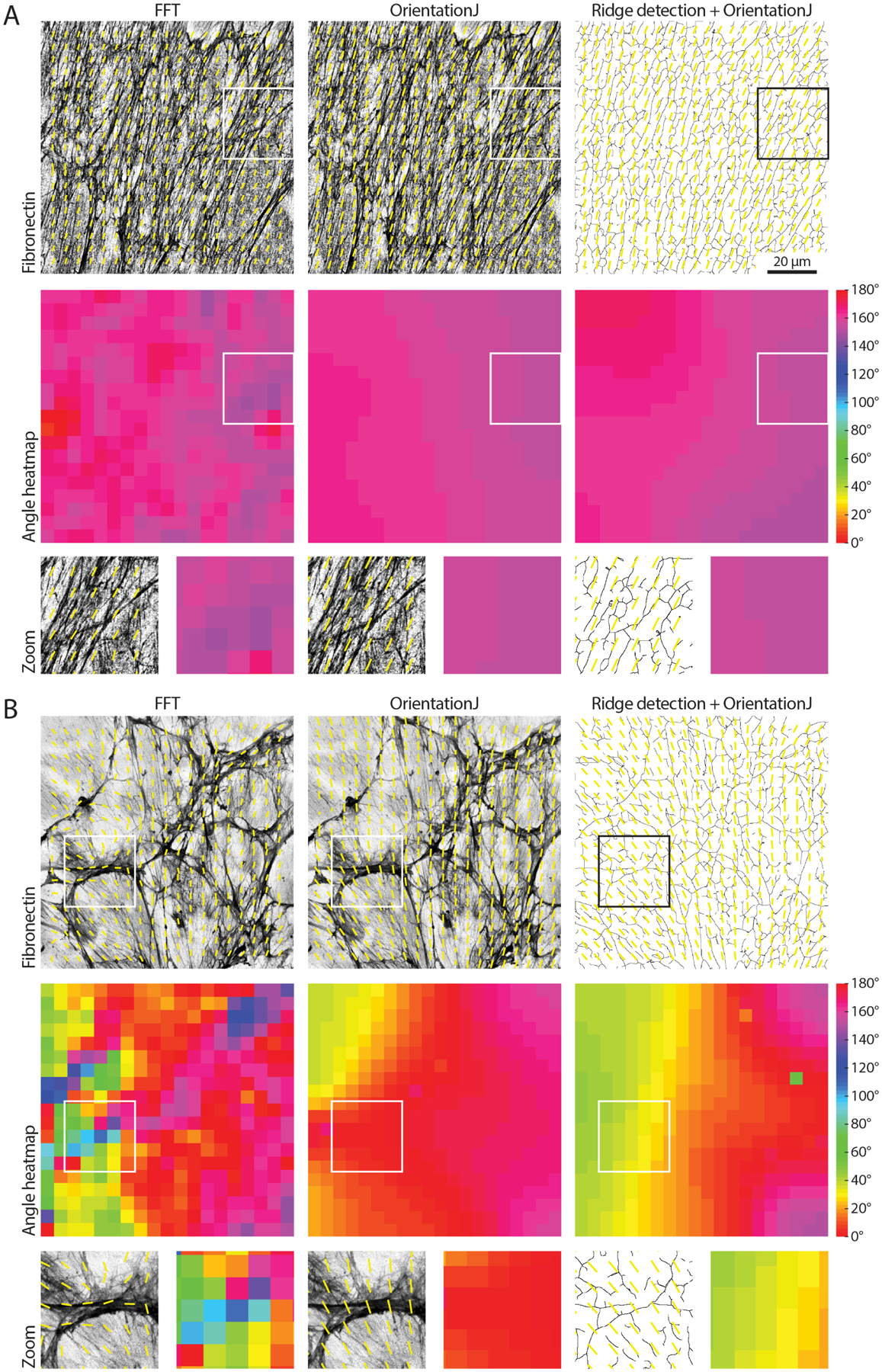
(A) A fibronectin image showing fibrillar features with strong alignment is analysed with the FFT approach presented in this work, with the Fiji OrientationJ Vector Field plugin, and with a combined approach using the Fiji Ridge Detection plugin followed by OrientationJ (mimicking the TWOMBLI hybrid approach, the binary mask calculated by the Ridge Detection is shown). A window size of 100 px with 50% overlap was used, and the order parameter was calculated over neighbourhoods of 5x vectors (neighbourhood radius of 2x vectors). Both the alignment vector field and the angle heatmap are shown for each case; zooms for the areas highlighted by the white boxes are displayed. The three algorithms behave similarly. Scale bar, 20 μm. (B) The analysis of a more randomly organised fibronectin image is compared as in (A). It can be noted how the FFT returns more accurate alignment vectors for the more isotropic regions of the image (insets).
Two alignment metrics were analysed, the order parameter S (Eq 6) and the coherency; the first metric is the one used in this work, while the coherency is the standard output of both OrientationJ and TWOMBLI. The comparison dataset contained two samples of images, with isotropic (Fig 9B) and anisotropic (Fig 9A) features. All the tested algorithms were able to recognise the alignment difference in the data, however, the order parameter scores calculated with the TWOMBLI approach (fibre segmentation via Ridge Detection followed by alignment with OrientationJ) did not reach statistical significance (Fig 10A). Moreover, the order parameter values calculated with both OrientationJ and the hybrid approach were very close to the upper limit of the range (values equal to 1 signify perfect alignment). This suggests that these methods might not be reliable in detecting subtle differences between samples, due to their limited ability to resolve local isotropy (Fig 9). Global coherency obtained with the three methods was also compared, showing that all of them could successfully distinguish the global alignment differences in the two samples (Fig 10B).
Figure 10. Comparing output metrics of different approaches to measure organisation in biological images.
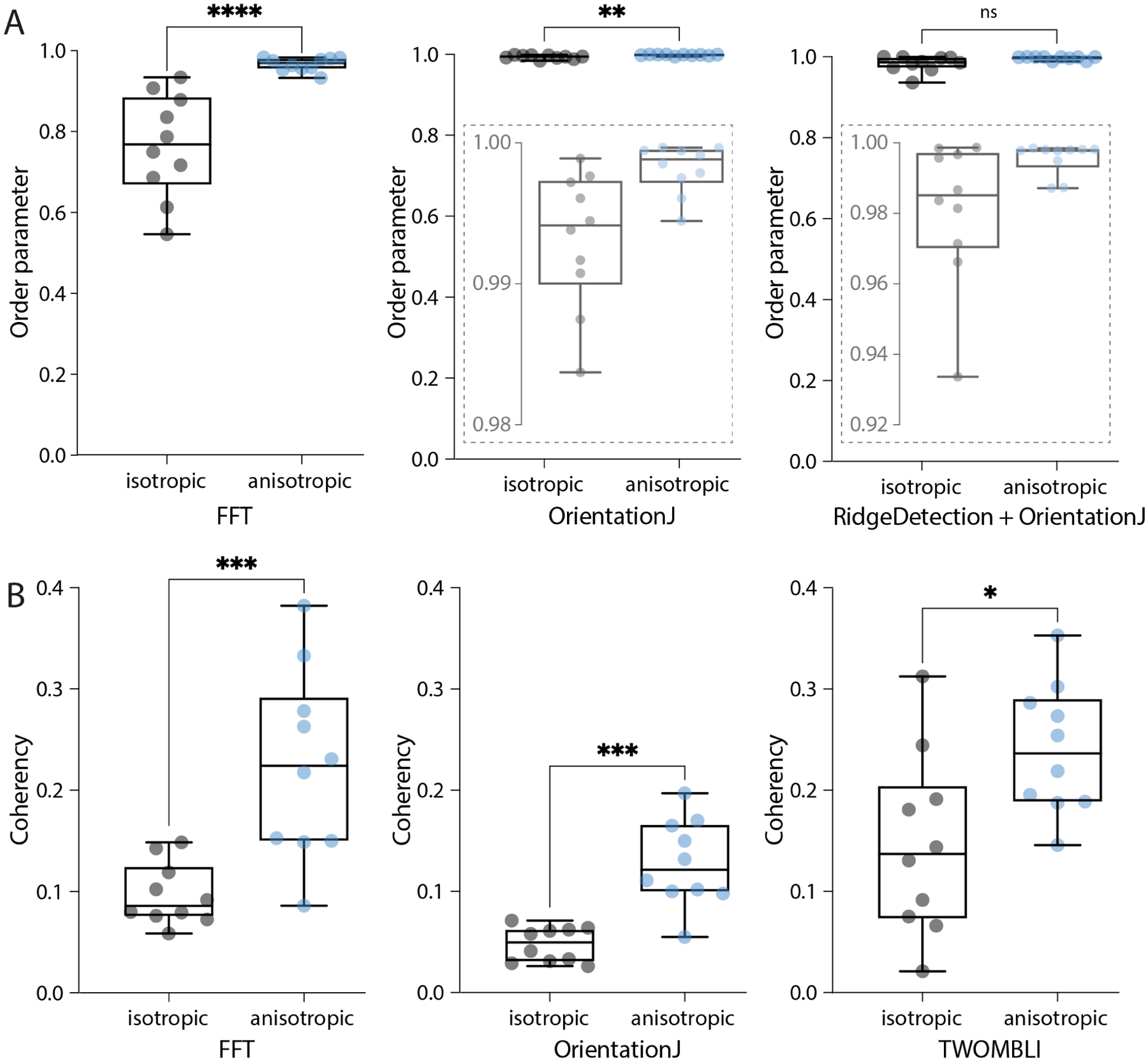
(A) Boxplots of the order parameters for the samples shown in Fig 9 calculated with the three methods, the FFT (as presented here), OrientationJ (Vector Field plugin), and the hybrid approach (Ridge Detection followed by OrientationJ Vector Field). Local windows of 100 px with 50% overlap and 5x vector neighbourhoods were considered. Insets represent zooms of the y-axis to better display distributions. Mann-Whitney two-tailed test (****p < 0.0001, **p = 0.0039, ns = 0.063). (B) Boxplots of the global coherency calculated with the three tools, AFT − Alignment by Fourier Transform (as presented here), OrientationJ (Dominant Direction plugin), and TWOMBLI. Mann-Whitney two-tailed test (***p = 0.0002, ***p = 0.0002, *p = 0.0147).
Computational time is comparable between the FFT and OrientationJ (with a dedicated macro) with each image being evaluated in less than half a second, while it increases for TWOMBLI with about 7 s per image (due to the fibre segmentation step). While this difference might appear insignificant for small samples as the ones used in this context, it might become important should batch processing be required for larger sample sizes or more high-throughput applications. The FFT approach presented here can be easily used to address length scale analysis. While OrientationJ also offers the option of a length scale analysis (Vector Field plugin), this requires ad hoc macros and further post-processing to obtain a unique alignment score for each image for a given window size. TWOMBLI allows the user to obtain a wider range of metrics characterising the fibrillar features, thanks to its fibre segmentation step, but this comes at the cost of setting more parameters to start the analysis. The FFT performs best at resolving local alignment differences (Fig 9), and the available filters on window intensity and eccentricity make it straightforward to automatically exclude regions of the images. This avoids having to manually discard entire images containing unsuitable regions, as would happen with the other two methods. The pros and cons of each methodology should be taken into account depending on the application (Table 1); overall, the AFT approach demonstrated efficient performance and broad flexibility to different types of input images.
Table 1.
A comparison of the performance and characteristics of the three analysis methods.
| AFT | OrientationJ | TWOMBLI | |
|---|---|---|---|
| Computational time | low (1.8–4.8 s*) | low (4.8 s**) | high (72.4 s) |
| Length scale analysis | + | (+) | − |
| Additional metrics | − | (+) | + |
| Automated image filtering | + | − | − |
Computational time was measured on a 3.6 GHz Quad-Core Intel Core i7 machine with 32 GB memory for a 10-image sample. ‘+’: yes, ‘(+)’: yes, but requires further post-processing,
depends on window size;
with a dedicated macro.
3.7. Implementing a streamlined software for measuring alignment
Finally, we have developed a workflow named AFT – Alignment by Fourier Transform to automate image analysis using the approach described above. This includes the pipeline described in Section 3.1, the parameter search in Section 3.2, the analysis of length scale decay in Section 3.3, the filtering options in Section 3.4, and the extraction of organisation kinetics in Section 3.5. Open-source implementation is made available in both MATLAB and Python at github.com/OakesLab/AFT-Alignment_by_Fourier_Transform.
Some considerations regarding the choice of parameters follows. The user is requested to set three mandatory parameters in order to run the alignment analysis: the window size, the overlap, and the neighbourhood size (Section 3.1, Fig 1D and 2B). Each of these parameters will affect the calculated order parameter by changing the length scale of the alignment analysis. While default parameters are suggested, it is important to test different values on individual images to gain some insight on the optimal length scale over which to carry out this analysis. In addition to the mandatory parameters, a number of optional filtering features are available as described in Section 3.4 (Fig 7).
3.7.1. Mandatory parameters
Window size. The size of the local regions in the image to analyse. This value depends on the image resolution, the size of the fibrillar features to be examined, and the length scale of interest (Fig 1D). In practice, within a window one should be able to observe enough information to discern by eye an orientation of features. There is no hard-set lowest limit for the window size, but it should be noted that smaller windows will lead to more spurious vectors as the calculated FFT will be nearly circular (i.e., not skewed, Fig 1A) for regions where fibrillar features are not recognised. A visual check of the output images is always recommended during the parameter optimisation phase.
Overlap. The overlap between adjacent windows (Fig 1D). Increasing the overlap, in conjunction with choosing smaller window sizes, allows for increasing the resolution of the analysis, with more sampled areas (i.e., vectors) within the field of view. A starting value of 50% is recommended.
Neighbourhood radius. The length scale over which to compare neighbouring measures of alignment. This should be set depending on the length scale of interest. It is defined as the number of nearest neighbour vectors to be compared to a central reference in order to obtain an alignment score (Fig 2). Neighbourhood size is calculated as 2 * neighbourhood radius + 1.
3.7.2. Optional parameters
Output images. This parameter enables the ability to save the analysed images, consisting of the original image overlaid with the alignment vector field and the angle heatmap (Fig 1D). The colour scale for the angle heatmap is wrapped, with similar colours for 0° and 180°, as fibres are not considered polar (Section 3.1, Fig 1D).
Filter blank spaces. Windows that contain little or no signal can be excluded from the analysis. When enabled a threshold on the mean window intensity can be set (Fig 7AB). Windows below this mean intensity are not analysed. Threshold values can be estimated by opening a sample image in Fiji, selecting regions considered background with similar size to the window size, and measuring their mean intensity. Expected values range from 0 (black) to 255 (white) for an 8-bit image.
Filter isotropic regions. It is possible to exclude windows from the analysis should the eccentricity of their FFT be lower than a threshold (Fig 7CD). Values for the FFT eccentricity range from 0 (circular, isotropic) to 1 (elongated, anisotropic). To estimate this threshold, the analysis can be run iteratively for increasing values until the desired output is achieved (i.e., until the regions considered isotropic do not contain alignment vectors in the output images).
Masking. By default, the whole image is being analysed. Alternatively, the user can input a folder containing binary masks of selected areas to be analysed for each input image (Fig 7E). A similar output as the one shown in Fig 7E could have also been obtained by filtering out the blank spaces, as the regions of interest have brighter signal than the background. However, if regions devoid of signal would also be present inside the cell outline, they would be disregarded as well. Therefore, a binary masking approach might be preferable in this case.
4. DISCUSSION
Quantifying alignment of fibrillar features in biological images has gained wide interest as a possible biomarker in disease aetiology and progression (Ouellette et al., 2021). While tools are available for this aim (Püspöki et al., 2016; Liu et al., 2017), many are difficult to implement and fairly rigid on the length scale over which anisotropy can be interrogated. The method presented in this work exploits the representation of the image in the frequency domain paired with a custom window-based approach, offering a computationally efficient algorithm that can be easily applied to investigate local to global alignment. Decay in alignment scores with increasing distance can be evaluated to reveal the length scale at which fibrillar features remain anisotropic.
An open-source suite, AFT – Alignment by Fourier Transform, is presented: its flexibility to a wide range of biological images is showcased through the examples, and its performance tested against synthetic data. The AFT – Alignment by Fourier Transform tool is demonstrated to be robust for different imaging modes (fixed, live-cell, SHG, fluorescence), fluorescent probes (F-actin, fibronectin), and image resolutions (from single cells to tissue tilescans). Further applications for which this method could be used that were not shown in the present work could entail the evaluation of alignment across the depth of a sample (measuring alignment for each slice of a Z-stack), or the investigation of cytoskeletal architectures in reconstituted protein systems.
The FFT approach performance was comparable to both OrientationJ and TWOMBLI, with increased accuracy in resolving local isotropic regions. Its computational efficiency makes it a good candidate for analysing high volumes of data. Moreover, the availability of filters and masking options allow for this methodology to be applied to a wide range of microscopy images.
Excitingly, more advanced 3-dimensional imaging modalities are becoming available, with techniques being developed to achieve isotropic resolution. While currently the FFT approach is designed to work with 2D data, the algorithm could be adapted to evaluate alignment of 3D fibrillar features, and its low computational cost could be exploited for these more computationally intensive data.
5. CONCLUSIONS
Here we presented a methodology to measure fibrillar feature alignment in biological images. We created an open-source tool that can be used on a wide range of biological images. Its performance was compared against other common workflows, and it was shown to be computationally efficient without compromising on accuracy. It is easy to implement with a small number of analysis parameters, and allows for interrogating the length scale of fibre anisotropy.
Supplementary Material
FUNDING
This project has been funded from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 681808 - SM, BS), the Wellcome Trust (grant no. 107859/Z/15/Z - FK, BS), the London Interdisciplinary Doctoral Programme (BB/J014567/1 - DBDF), the National Institutes of Health (NIH) National Institute of Allergy and Infectious Disease (NIAID) (Award # P01 AI02851 - LDT, PWO), and the National Science Foundation (NSF) (CAREER award # 2000554 - PWO). For the purpose of open access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
DATA AVAILABILITY STATEMENT
All the experimental datasets used as examples are included in the Supplementary Material (https://doi.org/10.6084/m9.figshare.15326472.v1), further inquiries can be directed to the corresponding authors. The software implementations can be found at github.com/OakesLab/AFT-Alignment_by_Fourier_Transform together with further documentation on how to run the code.
REFERENCES
- Aratyn-Schaus Y, Oakes PW, and Gardel ML (2011). Dynamic and structural signatures of lamellar actomyosin force generation. Molecular Biology of the Cell 22, 1121–1419. doi: 10.1091/mbc.e10-11-0891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudaoud A, Burian A, Borowska-Wykret D, Uyttewaal M, Wrzalik R, Kwiatkowska D, et al. (2014). FibrilTool, an ImageJ plug-in to quantify fibrillar structures in raw microscopy images. Nature Protocols 9, 457–463. doi: 10.1038/nprot.2014.024 [DOI] [PubMed] [Google Scholar]
- Bredfeldt JS, Liu Y, Conklin MW, Keely PJ, Mackie TR, and Eliceiri KW (2014a). Automated quantification of aligned collagen for human breast carcinoma prognosis. Journal of Pathology Informatics 5, 28. doi: 10.4103/2153-3539.139707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredfeldt JS, Liu Y, Pehlke CA, Conklin MW, Szulczewski JM, Inman DR, et al. (2014b). Computational segmentation of collagen fibers from second-harmonic generation images of breast cancer. Journal of Biomedical Optics 19, 016007. doi: 10.1117/1.jbo.19.1.016007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetera M, Ramirez-San Juan GR, Oakes PW, Lewellyn L, Fairchild MJ, Tanentzapf G, et al. (2014). Epithelial rotation promotes the global alignment of contractile actin bundles during Drosophila egg chamber elongation. Nature Communications 5. doi: 10.1038/ncomms6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S, Nguyen H, Rangayyan RM, Walsh S, and Frank CB (1987). A Fourier Domain Directional Filtering Method for Analysis of Collagen Alignment in Ligaments. IEEE Transactions on Biomedical Engineering BME-34, 509–518. doi: 10.1109/TBME.1987.325980 [DOI] [PubMed] [Google Scholar]
- Clemons TD, Bradshaw M, Toshniwal P, Chaudhari N, Stevenson AW, Lynch J, et al. (2018). Coherency image analysis to quantify collagen architecture: Implications in scar assessment. RSC Advances 8, 9661–9669. doi: 10.1039/c7ra12693j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclos G, Erlenkämper C, Joanny JF, and Silberzan P (2017). Topological defects in confined populations of spindle-shaped cells. Nature Physics 13, 58–62. doi: 10.1038/nphys3876 [DOI] [Google Scholar]
- Duclos G, Garcia S, Yevick HG, and Silberzan P (2014). Perfect nematic order in confined monolayers of spindle-shaped cells. Soft Matter 10, 2346–2353. doi: 10.1039/c3sm52323c [DOI] [PubMed] [Google Scholar]
- Eltzner B, Wollnik C, Gottschlich C, Huckemann S, and Rehfeldt F (2015). The filament sensor for near real-time detection of cytoskeletal fiber structures. PLoS ONE 10, 1–28. doi: 10.1371/journal.pone.0126346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzone TT, Oakes PW, Sees J, Kovar DR, and Gardel ML (2013). Actin assembly factors regulate the gelation kinetics and architecture of f-actin networks. Biophysical Journal 104, 1709–1719. doi: 10.1016/j.bpj.2013.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes NRJ, Reilly NS, Schrock DC, Hocking DC, Oakes PW, and Fowell DJ (2020). CD4+ T Cell Interstitial Migration Controlled by Fibronectin in the Inflamed Skin. Frontiers in Immunology 11, 1–14. doi: 10.3389/fimmu.2020.01501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldyn AM, Kaiser P, Spatz JP, Ballestrem C, and Kemkemer R (2010). The kinetics of force-induced cell reorganization depend on microtubules and actin. Cytoskeleton 67, 241–250. doi: 10.1002/cm.20439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M, Doss BL, Kocgozlu L, Pan M, Mège RM, Callan-Jones A, et al. (2019). Cell shape and substrate stiffness drive actin-based cell polarity. Physical Review E 99, 1–16. doi: 10.1103/PhysRevE.99.012412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M, Sarangi BR, Deschamps J, Nematbakhsh Y, Callan-Jones A, Margadant F, et al. (2015). Adaptive rheology and ordering of cell cytoskeleton govern matrix rigidity sensing. Nature Communications 6. doi: 10.1038/ncomms8525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartasalo K, Pölönen R, Ojala M, Rasku J, Lekkala J, Aalto-Setälä K, et al. (2015). CytoSpectre: A tool for spectral analysis of oriented structures on cellular and subcellular levels. BMC Bioinformatics 16, 1–23. doi: 10.1186/s12859-015-0782-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg T (1998). Edge Detection and Ridge Detection with Automatic Scale Selection. International Journal of Computer Vision 30, 117–156. doi: 10.1023/a:1008097225773 [DOI] [Google Scholar]
- Linsmeier I, Banerjee S, Oakes PW, Jung W, Kim T, and P MM (2016). Disordered actomyosin networks are sufficient to produce cooperative and telescopic contractility. Nature Communications 7, 12615. doi: 10.1038/ncomms12615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Keikhosravi A, Mehta GS, Drifka CR, and Kevin W (2017). Methods for Quantifying Fibrillar Collagen Alignment. Methods Mol Biol 1627, 429–451. doi: 10.1007/978-1-4939-7113-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Keikhosravi A, Pehlke CA, Bredfeldt JA, Dutson M, Liu H, et al. (2020). Fibrillar Collagen Quantification With Curvelet Transform Based Computational Methods. Frontiers in Bioengineering and Biotechnology 8, 1–14. doi: 10.3389/fbioe.2020.00198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livne A, Bouchbinder E, and Geiger B (2014). Cell reorientation under cyclic stretching. Nature Communications 5. doi: 10.1038/ncomms4938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez JP (2006). Fourier analysis and automated measurement of cell and fiber angular orientation distributions. International Journal of Solids and Structures 43, 6413–6423. doi: 10.1016/j.ijsolstr.2005.11.003 [DOI] [Google Scholar]
- Mascharak S, des Jardins-Park HE, Davitt MF, Griffin M, Borrelli MR, Moore AL, et al. (2021). Preventing Engrailed-1 activation in fibroblasts yields wound regeneration without scarring. Science 372. doi: 10.1126/science.aba2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisan L (2011). Periodic plus smooth image decomposition. Journal of Mathematical Imaging and Vision 39, 161–179. doi: 10.1007/s10851-010-0227-1 [DOI] [Google Scholar]
- Ouellette JN, Drifka CR, Pointer KB, Liu Y, Lieberthal TJ, Kao WJ, et al. (2021). Navigating the collagen jungle: The biomedical potential of fiber organization in cancer. Bioengineering 8, 1–19. doi: 10.3390/bioengineering8020017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Wershof E, Boeing S, Labernadie A, Jenkins RP, George S, et al. (2020). Extracellular matrix anisotropy is determined by TFAP2C-dependent regulation of cell collisions. Nature Materials 19, 227–238. doi: 10.1038/s41563-019-0504-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourdeyhimi B, Dent R, and Davis H (1997). Measuring Fiber Orientation in Nonwovens Part III: Fourier Transform. Textile Research Journal 67, 143–151. doi: 10.1177/004051759706700211 [DOI] [Google Scholar]
- Püspöki Z, Storath M, Sage D, and Unser M (2016). Transforms and operators for directional bioimage analysis: A survey. In Focus on Bio-Image Informatics. Advances in Anatomy Embryology and Cell Biology, eds. De Vos W, Munck S, and Timmermans J (Springer, Cham; ), vol. 219. 69–93. doi: 10.1007/978-3-319-28549-8 [DOI] [PubMed] [Google Scholar]
- Rezakhaniha R, Agianniotis A, Schrauwen JT, Griffa A, Sage D, Bouten CVC, et al. (2012). Experimental investigation of collagen waviness and orientation in the arterial adventitia using confocal laser scanning microscopy. Biomechanics and Modeling in Mechanobiology 11, 461–473. doi: 10.1007/s10237-011-0325-z [DOI] [PubMed] [Google Scholar]
- Ridley AJ and Hall A (1992). The small gtp-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70, 389–399. doi: 10.1016/0092-8674(92)90163-7 [DOI] [PubMed] [Google Scholar]
- Sander EA and Barocas VH (2009). Comparison of 2D fiber network orientation measurement methods. Journal of Biomedical Materials Research - Part A 88, 322–331. doi: 10.1002/jbm.a.31847 [DOI] [PubMed] [Google Scholar]
- Standley PR, Camaratta A, Nolan BP, Purgason CT, and Stanley MA (2002). Cyclic stretch induces vascular smooth muscle cell alignment via NO signaling. American Journal of Physiology -Heart and Circulatory Physiology 283, 1907–1914. doi: 10.1152/ajpheart.01043.2001 [DOI] [PubMed] [Google Scholar]
- Wershof E, Park D, Barry DJ, Jenkins RP, Rullan A, Wilkins A, et al. (2021). A FIJI macro for quantifying pattern in extracellular matrix. Life Science Alliance 4, e202000880. doi: 10.26508/lsa.202000880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willert CE and Gharib M (1991). Digital particle image velocimetry. Experiments in Fluids 10, 181–193. doi: 10.1007/BF00190388 [DOI] [Google Scholar]
- Yoshigi M, Hoffman LM, Jensen CC, Yost HJ, and Beckerle MC (2005). Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. Journal of Cell Biology 171, 209–215. doi: 10.1083/jcb.200505018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the experimental datasets used as examples are included in the Supplementary Material (https://doi.org/10.6084/m9.figshare.15326472.v1), further inquiries can be directed to the corresponding authors. The software implementations can be found at github.com/OakesLab/AFT-Alignment_by_Fourier_Transform together with further documentation on how to run the code.


