Abstract
The N-terminal tail domains of the core histones play important roles in gene regulation, but the exact mechanisms through which they act are not known. Recent studies suggest that the tail domains may influence the ability of RNA polymerase to elongate through the nucleosomal DNA and, thus, that posttranslational modification of the tail domains may provide a control point for gene regulation through effects on the elongation rate. We take advantage of an experimental system that uses bacteriophage T7 RNA polymerase as a probe for aspects of nucleosome transcription that are dominated by the properties of nucleosomes themselves. With this system, experiments can analyze the synchronous, real-time, single-passage transcription on the nucleosomal template. Here, we use this system to directly test the hypothesis that the tail domains may influence the “elongatability” of nucleosomal DNA and to identify which of the tail domains may contribute to this. The results show that the tail domains strongly influence the rate of elongation and suggest that the effect is dominated by the N-terminal domains of the (H3-H4)2 tetramer. They further imply that tail-mediated octamer transfer is not essential for elongation through the nucleosome. Acetylation of the tail domains leads to effects on elongation that are similar to those arising from complete removal of the tail domains.
Each of the four core histones of the nucleosome has a ∼15- to 45-amino-acid highly positively charged N-terminal tail domain. These tail domains are of particular significance because they are the sites for posttranslational modifications that are linked to chromosome function. In particular, histone acetylation has been the subject of recent interest because it establishes a link between tail domain function and gene regulation. Each of the core histone proteins can be acetylated in vivo on multiple lysines within the N-terminal domains. Many gene-regulatory proteins have been found to encode histone acetylases or deacetylases, or to act in combination with other proteins that themselves are histone acetylases or deacetylases (14, 18, 39, 40, 45, 52, 54, 55).
Remarkably, despite their evolutionary conservation, individual tail domains can be deleted with little effect on the viability or even the growth rate of yeast (17, 36, 50). In addition, nucleosomes from which the N-terminal tails have been entirely removed are essentially unchanged in overall structure and stability (2, 8, 9, 24). Subtle phenotypes that are observed in yeast mutants lacking one or another of these conserved tail domains are mimicked by point mutations that simulate lysine acetylation (e.g., lysine to glutamine). This suggests that at least some aspects of tail domain function in gene activation (or derepression) can be achieved, equivalently, either by eliminating the distinctive positive charge of lysine residues (by natural acetylation or by lysine to glutamine mutation) or by deleting the tail domains altogether.
The exact mechanisms through which the tail domains contribute to gene regulation are not known. They may be involved in stabilizing higher-order chromatin folding by binding to DNA (or histones) on nucleosomes neighboring in three-dimensional space within the folded chromatin fiber (12, 23, 52). Alternatively, they may recruit non-histone regulatory proteins that establish repressive chromatin regions (for reviews, see references 14, 18, 39, 40, 45, 52, 54, and 55). The tail domains may also play important roles in controlling the dynamics and accessibility of DNA within individual nucleosomes (notwithstanding the absence of significant effects on bulk nucleosome stability when the tail domains are deleted) (13, 16, 19, 46–49). In a recent study we examined this hypothesis quantitatively and found that deletion of the tail domains led to a 1.5- to 14-fold increase in the equilibrium accessibility of DNA target sites buried inside nucleosomes (32).
Other studies have suggested that the histone tail domains may play roles in transcriptional elongation. Earlier studies suggested that histone tail domains may hold the histones to the DNA while the DNA is being traversed by RNA polymerase (RNAP) (10, 29), allowing the polymerase free access to the DNA while maintaining the relationship between particular histone molecules and the underlying DNA. The observation that the histone octamer can apparently step over an elongating RNAP without dissociating from the DNA (41) points to a possible role for the tail domains in this intramolecular transfer. These and other recent studies (4, 6, 44, 45, 53) raise the possibility that the tail domains may contribute to regulating the elongation process.
In earlier work we and others (see references 4, 20, 21, 33, 34, and 41 and references therein) have used bacteriophage RNA polymerases as simple probes for aspects of nucleosome transcription that are dominated by the properties of nucleosomes themselves. The bacteriophage polymerase system offers many advantages for such a study. The polymerase elongation rate can be tuned from much faster to much slower than the natural rate for eukaryotic RNA polymerase II (∼23 nucleotides [nt] s−1 [37]), and experiments can be constructed to analyze synchronous, real-time, single-passage transcription on a nucleosomal template (33, 34). This approach allows effects on the elongation process to be clearly distinguished from alternative effects such as changes in initiation efficiency. Using conditions in which the elongation rate on naked DNA is comparable to that of eukaryotic RNAP II, we found that transcription on nucleosomal templates is slowed relative to that on naked DNA, because the polymerase exhibits longer residence times at pause sites specified by the DNA sequence itself (33).
It seems likely that such slowing down may be a consequence of effects of the tail domains on the dynamic equilibrium accessibility of the nucleosomal DNA discussed above (32). In the present study we take advantage of this experimental system to directly test the hypothesis that the tail domains may influence the “elongatability” of nucleosomal DNA and to identify which of the tail domains may contribute to this.
MATERIALS AND METHODS
Preparation of DNA and histones.
The DNA used in these studies is identical to the 216-bp T7 transcription template described earlier (33, 34) and is synthesized by preparative-scale PCR and purified by anion-exchange high-performance liquid chromatography. For the transcription reactions, the DNA was labeled at both 5′ ends with [γ-32P]ATP and polynucleotide kinase. For the exonuclease III and DNase I experiments, the DNA was singly labeled at the left (T7 promoter) or right (nucleosome positioning sequence) 5′ end during PCR synthesis by inclusion of an appropriately radiolabeled primer and was subsequently gel purified. Tracer quantities of radiolabeled DNA were mixed with optical quantities of unlabeled DNA. The amount of cold DNA used depended on the reconstitution scale and the number of reconstitutes to be prepared; a typical ratio used was 0.5 μg of labeled DNA per 50 μg of cold DNA. The DNA pool was then divided into several aliquots for reconstitution with various sets of histone octamer or kept for use as naked DNA.
Long chicken erythrocyte chromatin (22, 51) and native histone octamer (11) were prepared from chicken erythrocytes as described. Long chicken erythrocyte chromatin was depleted of histone H1 (and H5) by chromatography on a 45-by-5-cm Sephacryl S-200 sizing column in 0.65 M NaCl–1× TE (TE is 10 mM Tris, 1 mM EDTA) with a flow rate of 1 ml min−1. Fractions that contained core histones but were free of detectable H1 and H5 were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then pooled and concentrated with Centriprep-10 concentrators (Amicon). The resulting (H1- and H5-) stripped chromatin was then dialyzed into 25 mM Tris-HCl (pH 7.5)–0.1 mM EDTA and stored at 4°C.
Stripped chromatin was digested with different amounts of clostripain to selectively remove the amino-terminal tails of the (H3-H4)2 tetramer (tet-no-tails) or the amino-terminal tails of all the histones (oct-no-tails) (8, 9). For clostripain digestion, the purified chromatin was dialyzed overnight into diethanolamine-HCl (pH 7.5)–20 mM NaCl–7 mM β-mercaptoethanol at 4°C. A clostripain solution was freshly activated prior to each digestion reaction. Clostripain (Sigma) was weighed out and dissolved in 2.5 mM dithiothreitol (DTT) and 1 mM calcium acetate at 0.02 U μl−1 (the specific activity of clostripain was 100 U mg−1) and was activated at 4°C for 2 h. All digestions were carried out in 20 mM DEA-HCl (pH 7.4)–7 mM β-mercaptoethanol–1 mM CaCl2–2.5 mM DTT–20 mM NaCl. To obtain (H3-H4)2 tetramers lacking their amino-terminal tails, chromatin (1 mg ml−1) was digested with clostripain (0.0002 U μl−1) at 37°C for 60 min. To obtain octamers lacking their amino-terminal tails, chromatin (1 mg ml−1) was digested with clostripain (0.004 U μl−1) at 37°C for 60 min. In both cases, the reaction was quenched with 1 mM TLCK (Nα-p-tosyl-l-lysine chloromethyl ketone). The various histone and chromatin preparations were analyzed by SDS-PAGE as described earlier (33, 34), except that the running buffer contained 0.05% instead of 1% SDS (9).
Stripped chromatin was separately digested with trypsin to yield fully tailless histone octamer donor. The preparation and characterization of the material used in the present study is described elsewhere (32).
HeLa core histones and hyperacetylated HeLa core histones were prepared and purified as described (3) with minor modifications (J. D. Anderson, P. T. Lowary, and J. Widom, unpublished data) using HeLa cells and butyrate-treated HeLa cells produced at the National Cell Culture Center (Minneapolis, Minn.).
Reconstitution of native, tailless, and hyperacetylated nucleosomes.
Native nucleosomes were prepared either by gradual salt dialysis or by exchange reactions using long, stripped chromatin as the histone donor. Gradual salt dialysis was performed as described (34). Exchange reactions were initiated by mixing DNA pool (0.25 μg μ−1) and long, stripped chromatin (5 mg ml−1) in 1 M NaCl. Following a 30-min incubation at 37°C, the salt concentration was progressively lowered by dilution with addition buffer I (0.5× TE, 5 mM NaCl, 50 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM benzamidine hydrochloride [BZA]) to 0.8, 0.6, 0.4, 0.2, and 0.1 M NaCl. Each dilution step was incubated at 37°C for 30 min. The volume of the solution was then reduced to 50 μl with a Centricon-30 microconcentrator, washed with 1 ml of addition buffer I, and collected in a final volume of approximately 200 μl.
Nucleosomes containing (H3-H4)2 tetramer without amino-terminal tails (tet-no-tails nucleosomes) or histone octamer without amino-terminal tails (oct-no-tails nucleosomes) were prepared by exchange using clostripain-digested stripped chromatin as the histone donor. Exchange reactions were performed exactly as described above for nucleosomes with native histones.
Nucleosomes containing trypsinized octamer were prepared by exchange using trypsinized chromatin as the histone donor. The exchange reaction was initiated by mixing 40 μg of the DNA pool and 1 mg of trypsinized chromatin in 1 M NaCl and soybean pancreatic trypsin inhibitor (0.01 mg ml−1) in a volume of 160 μl. The reaction was incubated at 37°C for 30 min, after which the salt concentration was progressively lowered by dilution with addition buffer II (0.5× TE, 5 mM NaCl, 50 mM PMSF, 1 mM BZA, soybean pancreatic trypsin inhibitor [0.01 mg ml−1]) to 0.8, 0.6, 0.4, 0.2, and 0.1 M NaCl, with each dilution step incubated at 37°C for 30 min. The volume of the solution was then reduced to 25 μl with a Centricon-30 microconcentrator, washed with 1 ml of addition buffer II, and collected in a final volume of 200 μl.
Native or hyperacetylated HeLa nucleosomes were prepared by gradual salt dialysis from native or hyperacetylated HeLa histone octamer as described (34).
Purification of reconstituted nucleosomes and single positioning isomers.
Naked DNA, native reconstitutes, and reconstitutes containing proteolyzed histones were purified by sucrose gradient ultracentrifugation as described (33, 34). Peak nucleosome-containing fractions were pooled and exchanged into 0.5× TE–5 mM NaCl (for naked DNA), 0.5× TE–5 mM NaCl–0.5 mM PMSF–1 mM BZA (for reconstituted nucleosomes containing native or clostripain-digested histones), or 0.5× TE–5 mM NaCl–0.5 mM PMSF–1 mM BZA–SBTI (0.01 mg ml−1) (for reconstituted nucleosomes containing trypsinized histones) using Centricon-30 microconcentrators. Final yields were typically 50 to 100 μl of nucleosomes at 100 to 500 nM.
Individual nucleosome positioning isomers were purified by native acrylamide gel electrophoresis. Native gels (5% acrylamide in 1/3× TBE [0.03 M Tris-borate–0.67 nM EDTA]) were prerun for 3 h at 10 V cm−1 at room temperature before use. The native and proteolyzed reconstitutes were mixed with sucrose loading buffer (10% sucrose in 0.5× TE–5 mM NaCl), loaded, and run at 10 V cm−1 at 4°C. After electrophoresis for 6 h, the wet gel was wrapped in plastic and exposed to X-ray film to locate the desired bands. Desired bands were excised and then eluted overnight in 0.5× TE–5 mM NaCl–bovine serum albumin (BSA) (150 μg ml−1) in silanized tubes using a shaking platform. In all cases the most abundant positioning isomer was purified, and the material that remained was checked to confirm that the proper isomer had in fact been taken. Eluates were separated from gel pieces with brief, low-speed centrifugation and then were concentrated using Centricon-30 microconcentrators. Final volumes were typically 50 μl. Samples then underwent scintillation counting, allowing a determination of the approximate concentration by comparison to the known specific activity of the starting naked DNA pool.
Characterization of reconstituted nucleosomes.
Purified reconstituted nucleosomes were analyzed by native acrylamide gel electrophoresis essentially as described (33, 34), except that the loading buffer contained 10% sucrose instead of 3% Ficoll. After prerunning a 5% native acrylamide–1/3× TBE gel at 10 V cm−1 for 1 h, naked DNA or nucleosomes were loaded onto the gel in 10% sucrose and electrophoresed for 3 to 4 h at room temperature. The gel was then dried and exposed to a phosphorimager plate for quantitative analysis.
For mapping nucleosome positions using exonuclease III, the template DNA was singly labeled on either the left or right 5′ end as described above. The exonuclease III digestions were carried out in 50 mM Tris-HCl (pH 8.0)–5 mM MgCl2, with 50 nM nucleosomes and exonuclease III (0.5 U μl−1) at 37°C. Aliquots were removed after 0.25, 0.5, and 1 min and quenched with an equal volume of formamide mix. The aliquots, together with undigested controls also mixed with formamide, were digested with proteinase K (50 μg ml−1) for 1 h at 37°C. Samples were then electrophoresed on a sequencing-size denaturing 6% polyacrylamide gel. The gel was then dried and exposed to a phosphorimager plate for quantitative analysis.
For DNase I analysis, approximately 50 nM template DNA (as naked DNA or native or trypsinized nucleosomes) was digested with DNase I (0.025 U μl−1) in 1× T7 transcription buffer (40 mM Tris [pH 8.0], 50 μg ml−1 BSA, 5.0 mM DTT) and 10 mM MgCl2 at room temperature. Aliquots were withdrawn after 0.5 and 1.0 min, quenched with an equal volume of formamide mix, and digested with proteinase K (50 μg ml−1) as described above. Samples were separated by electrophoresis and underwent phosphorimager analysis.
Transcription reactions.
A master mixture consisting of transcription buffer, nucleoside triphosphates (NTPs), RNasin, [α-32P]ATP, and MgCl2 was distributed among the different transcription templates (naked DNA and various reconstituted nucleosomes) and kept on ice. T7 RNAP was then separately added to each reaction mixture to initiate transcription. The final concentrations were 50 nM template, 40 mM Tris (pH 8.0), 50-μg ml−1 BSA, 5.0 mM DTT, 5 μM ATP, 25 μM CTP, 200 μM GTP, 1-U μl−1 RNasin (Promega), 0.5 μM [α-32P]ATP (3,000 Ci mmol−1; Amersham), 5 mM MgCl2 and 1-U μl−1 T7 RNA polymerase (U.S. Biochemical). After incubation at 37°C for 5 min, an aliquot was taken for analysis of stalled complexes while the rest of the mixture was cooled to 0°C. The remaining solution was kept at 0°C for 1 min and was then mixed with an equal volume of a prechilled (0°C) solution that contained buffer and all four NTPs to restart elongation. After mixing, the final conditions were 2 mM ATP, 100 μM CTP, 200 μM GTP, and 100 μM UTP. For the reactions on nucleosomes containing clostripain-digested histones, aliquots were taken at 10, 20, and 40 s and 1, 4, and 16 min. For the reactions on nucleosomes containing trypsinized histones, aliquots were taken at 10, 20, and 40 s and 1, 2, 4, 8, and 16 min. Aliquots of stalled complexes or of elongation reaction mixtures at desired time points were immediately mixed with an ice-cold formamide stop solution (94% deionized formamide, 30 mM EDTA, xylene cyanol [125 μg ml−1]), heated to 90°C for 10 min, and run on a sequencing-size denaturing 6% polyacrylamide gel. Gels were dried and analyzed by phosphorimager. The great excess of unlabeled ATP added to the chase solution strongly suppresses but does not suffice to completely prevent the additional incorporation of radiolabel after elongation is restarted. Thus, instead of comparing full-length transcript counts at each time point to the corresponding stalled complex counts that are initially produced from each template, completion efficiencies for the nucleosomal templates are measured relative to naked DNA. For the naked DNA template, transcription is effectively over by 1 min (see Fig. 5A). Completion efficiencies for the nucleosomal templates are therefore measured by quantifying the counts in the full-length transcript at each time point and comparing them to the number of counts in the full-length transcripts produced from the naked DNA template after 1 min of elongation time.
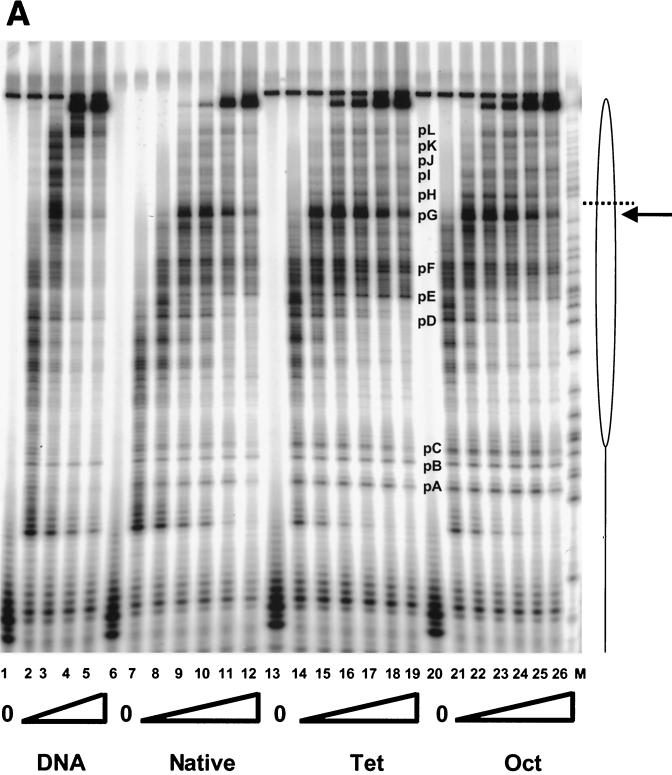
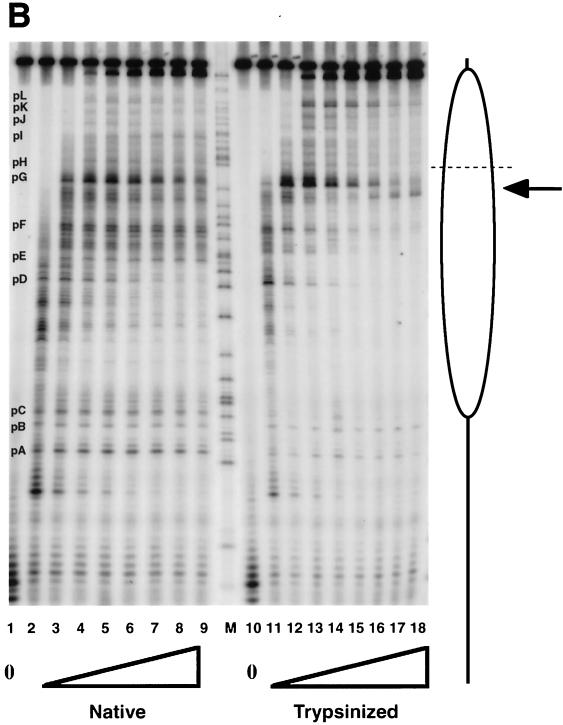
FIG. 5.
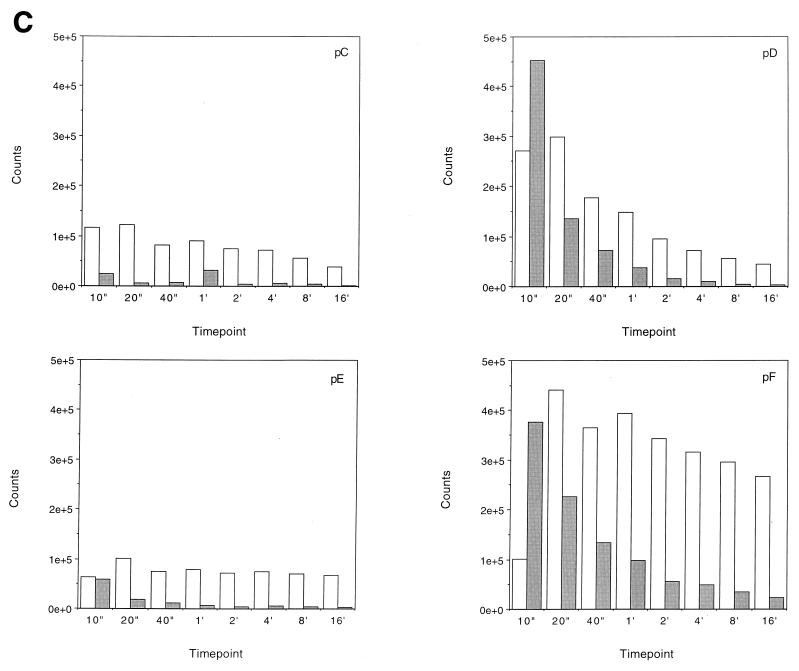
Transcription elongation on the tailless nucleosomes. (A) Clostripain-digested histones. Transcription reactions on naked DNA (lanes 1 to 5), native nucleosomes (lanes 6 to 12), nucleosomes containing tailless tetramers (lanes 13 to 19), and nucleosomes containing tailless octamers (lanes 20 to 26) were run on a 6% denaturing sequencing-size gel and exposed to a phosphorimager plate for analysis. Aliquots were taken after stalled complex formation (lanes 1, 6, 13, and 20) and after 10 s (lanes 2, 7, 14, and 21), 20 s (lanes 3, 8, 15, and 22), 40 s (lanes 4, 9, 16, and 23), 1 min (lanes 5, 10, 17, and 24), 4 min (lanes 11, 18, and 25), and 16 min (lanes 12, 19, and 26) of elongation. The lane labeled M contains RNA markers obtained by transcribing the DNA in the presence of the RNA chain terminator dATP and represents an A ladder. The open oval represents the approximate location of the nucleosome on the template; the dashed line identifies the dyad axis of symmetry. Major polymerase pause sites are labeled (pA to pL). The band corresponding to the major pause site immediately promoter proximal to the dyad axis (pG) is indicated also with an arrow. (B) Trypsin-digested histones. Transcription reactions on native nucleosomes (lanes 1 to 9) and trypsinized nucleosomes (lanes 10 to 18) were run on a 6% denaturing sequencing-size gel and exposed to a phosphorimager plate for analysis. Aliquots were taken after stalled complex formation (lanes 1 and 10) and after 10 s (lanes 2 and 11), 20 s (lanes 3 and 12), 40 s (lanes 4 and 13), 1 min (lanes 5 and 14), 2 min (lanes 6 and 15), 4 min (lanes 7 and 16), 8 min (lanes 8 and 17), and 16 min (lanes 9 and 18) of elongation. (C) Quantitative analysis of pause site residence times for sites pC, pD, pE, and pF from panel B. Open bars, native nucleosomes; shaded bars, fully tailless (trypsinized) histones. Counts present in the bands corresponding to the indicated paused species at each time point are shown (note: all graphs are shown on the same scale for direct comparison). Pause sites pC and pE are essentially unpopulated after removal of the histone tails; sites pD and pF do become well populated but decay (i.e., allow release and continued elongation) much more quickly for the tailless nucleosomes.
RESULTS AND DISCUSSION
Preparation and purification of native and tailless nucleosomes.
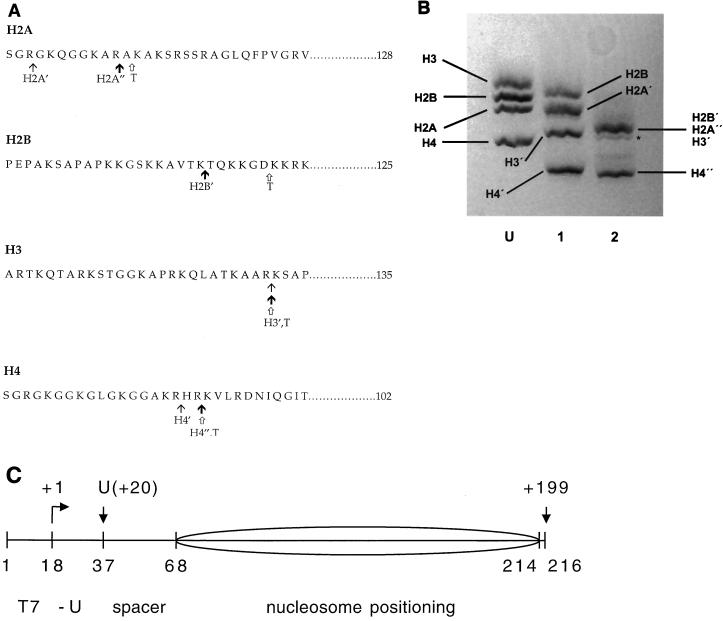
The proteases clostripain and trypsin allow controlled removal of the core histone N-terminal tail domains (5, 8) (Fig. 1A). Limited digestion with clostripain yields histones lacking the tail domains of the (H3-H4)2 tetramer (tet-no-tails), while more extensive digestion with clostripain cleaves off the tail domains from all four histones (oct-no-tails) (Fig. 1B). We wished also to prepare nucleosomes lacking only the tail domains of H2A and H2B. While methods for preparing such species have been reported (2), numerous attempts to implement this procedure in our laboratory have proven unsuccessful; evidently there are critical variables that remain unrecognized. Trypsin digestion yields histone octamers that are similar to histones extensively digested with clostripain, except that slightly greater portions of the tail domains are removed for three of the four histones, referred to subsequently as fully tailless histones. The preparation and characterization of the fully tailless histones used for the present studies are described elsewhere (32).
FIG. 1.
Protease cleavage of the core histone tail domains. (A) Amino acid sequences of the four chicken erythrocyte core histones showing the locations of the cleavage sites for limited or more-extensive clostripain digestion (see reference 8) and trypsin digestion (see reference 5). The locations of the clostripain cleavage sites shown are those determined in past studies on rat histones (8), which, based on the high degree of amino acid sequence conservation between the two organisms and the close similarity of SDS-PAGE analyses of the time courses of digestion for the two cases, are presumed to be identical. The tet-no-tails sample obtained by limited clostripain digestion (at points indicated by thin arrows, with prime designations on the histones) contains slightly shortened H2A′ (lacking 3 residues from the N terminus), undigested H2B, tailless H3′ (cleaved after residues 26), and tailless H4′ (cleaved after residue 17). The oct-no-tails sample obtained by more-extensive clostripain digestion (at points indicated by bold arrows, with prime or double-prime designations on the histones) contains H2A" (which differs from H2A′ by the loss of an additional 8 residues), fully digested H2B′ (cleaved after residue 20), H3′, and H4" (which differs from H4′ by the loss of an additional two residues). Trypsin digestion (at points indicated by open arrows [histones designated with a T]) yields fully tailless histones. Trypsin also removes short stretches from the C termini of H2A and H3 (5). (B) SDS-PAGE analysis of histone octamer donor chromatin. Lane U, native histone octamer donor (i.e., H1- and H5-stripped but undigested chromatin); lane 1, histone donor for the tet-no-tails sample obtained by limited clostripain digestion; lane 2, histone donor for the oct-no-tails sample obtained by more-extensive clostripain digestion. H2B′, H2A", and H3′ run together as a single, unresolved broad band (8, 9). Trace amounts of an overdigested product are evident (asterisk). For a similar analysis of the fully tailless histone donor obtained by trypsin digestion, see reference 32. (C) Diagram of the nucleosomal template (see reference 34 for details). Numbers below the drawing represent positions (nucleotide number) of key points in the template; numbers above the drawing represent points along the RNA, including the position of the first U (residue +20) and the runoff product (nt 199). The sequence derives from the 5S RNA gene nucleosome positioning sequence (38) with changes to incorporate a promoter for T7 RNAP (nt 1 to 17) and transcription start site and U-less cassette (nt 18 to 37). The predominant nucleosome position is indicated by the ellipse.
The nucleosomal DNA (Fig. 1C) was designed to allow synchronous, real-time, single-passage studies of nucleosome transcription by T7 RNAP and is described in an earlier study (34). It incorporates a natural nucleosome positioning sequence at one end and a promoter recognition sequence for T7 RNAP followed by a U-less cassette (which allows production of stalled complexes and subsequent synchronous elongation) at the other. The U-less cassette is separated from the nucleosome positioning DNA sequence by a short stretch of additional DNA to provide increased access of the polymerase to the promoter despite the presence of the histone octamer nearby.
Nucleosomes were reconstituted with native histone octamer, tet-no-tails histones, oct-no-tails histones, and fully tailless histones (see Materials and Methods). Reconstituted nucleosomes were purified by sucrose gradient ultracentrifugation. Examples of typical separations for each of the three different reconstituted tailless nucleosomes are shown in Fig. 2. The profiles of the different nucleosome preparations reveal small differences in mobility on the gradients. The tet-no-tail nucleosomes sediment with a velocity similar to that of native nucleosomes, while the oct-no-tail and fully-tailess nucleosomes sediment more slowly. These differences in sedimentation velocity correlate with the progressive loss of the tail domains and could reflect either the increasing reduction in overall molecular weight or changes in the time-averaged compactness of the different tailless nucleosomes, or both.
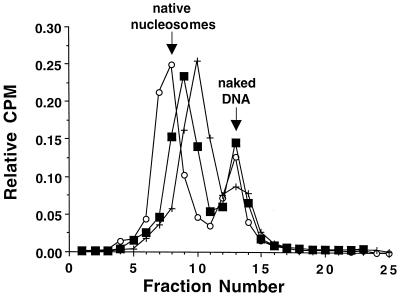
FIG. 2.
Sucrose gradient purification of the various tailless nucleosomes utilized in this study. Tailless nucleosomes were reconstituted by exchange using chromatin digested with trypsin or clostripain as histone donors. After undergoing stepwise dilution from high (1 M NaCl in 1× TE) to low (5 mM NaCl in 0.5× TE) salt conditions, the particles were analyzed on 5 to 30% (wt/vol) linear sucrose gradients having the same salt and buffer conditions. Following fractionation into 0.5-ml fractions, the reconstitutes were located by scintillation counting. Relative counts per minute (CPM) are plotted against fraction number for nucleosomes containing tailless tetramers prepared with clostripain (○), nucleosomes containing tailless octamers prepared with clostripain (■), and nucleosomes containing tailless octamers prepared with trypsin (+). The mobility of nucleosomes reconstituted with native histone octamer (which comigrate with isolated natural nucleosomes) is indicated.
Native gel analysis of the different nucleosome samples following sucrose gradient purification demonstrated the expected mobility shifts relative to naked DNA, but also revealed the presence of multiple positioning isomers as previously noted for nucleosomes reconstituted on this template DNA (data not shown, but see references 33, and 34). We therefore used preparative native gel electrophoresis to enrich for the most predominant species. Following native gel purification, reanalysis on a second native gel shows each sample to now consist of a single predominant band representing a single discretely positioned species (Fig. 3). As found for the sucrose gradients (Fig. 2), the mobilities exhibited by the different nucleosome preparations correlate with the number and extent of tail domains removed by proteolysis (although in the opposite direction, as expected).
FIG. 3.
(A) Native gel analysis of native and tailless nucleosomes. Native histones (N), histones digested with clostripain to generate tailless tetramers (-tet) or tailless octamers (-oct) and histones digested with trypsin to generate fully tailless octamers (tryp) were reconstituted with the T7 transcription template DNA and purified by sucrose gradient ultracentrifugation. Individual positioning isomers were isolated by native gel purification. The resulting samples were analyzed by native gel electrophoresis. Approximately 100 ng of naked DNA or the different nucleosome preparations in sucrose loading buffer (10% sucrose in 5 mM NaCl and 0.5× TE) was run on a 5% acrylamide gel in 1/3× TBE at 10 V cm−1 and exposed to a phosphorimager plate for analysis. Naked DNA (D) was included as a mobility reference. (B) Native gel analysis of native and trypsinized nucleosomes singly end labeled on the left or right 5′ end. Native and trypsinized histones were reconstituted with template DNA labeled on the left or right 5′ end for studies with exonuclease III and DNase I and were analyzed on a 5% polyacrylamide–1/3× TBE gel. Lanes contain naked DNA (D), native nucleosomes (N) and trypsinized nucleosomes (T); the end containing the label is designated below the lane labels.
Nucleosome mapping using exonuclease III and DNase I.
The exact location of native histone octamer on this particular DNA construct has been determined on two separate occasions using two different methods (33, 34). These studies indicated that a single positional isomer predominates. After isolation of this predominant single isomer by preparative native gel electrophoresis, mapping experiments revealed that the histone octamer occupies the nucleosome positioning sequence on the right end of the DNA construct, as desired. In particular, the histone-protected region ends 4 bp from the right end of the template DNA, leaving the left, T7 promoter-containing region free for the polymerase to engage. Removal of the tail domains is not expected to alter this positioning preference (7, 15). Nevertheless, because positioning is important for the present studies, additional experiments were carried out to directly determine the positioning on the purified isomers.
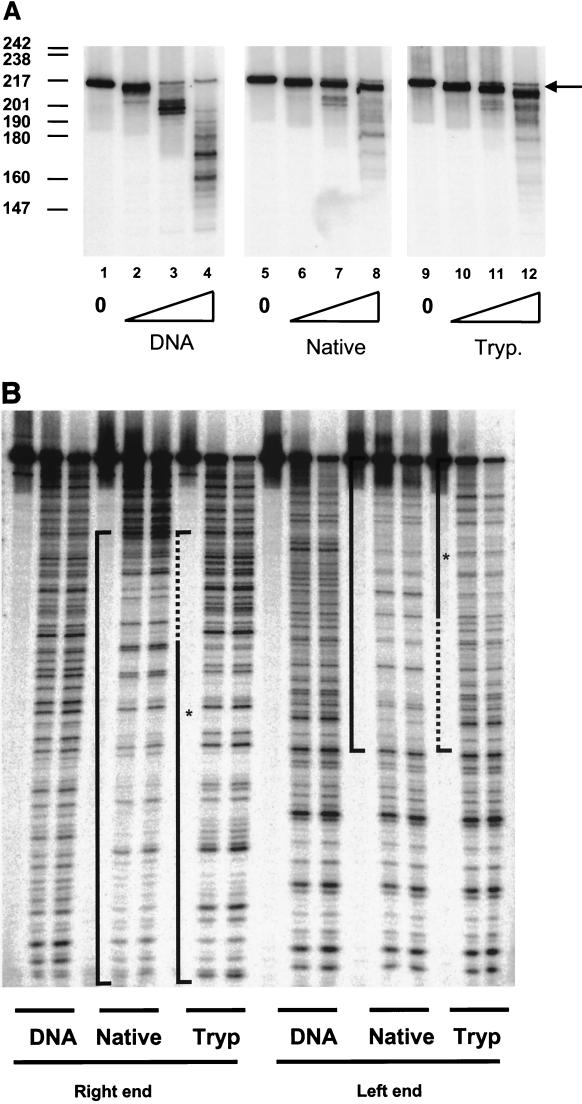
We used exonuclease III (30) and DNase I (27) to map the positioning of the predominant, isolated isomer of native and fully tailless nucleosomes by reconstituting the appropriate histones with DNA labeled on only one end (Fig. 3B). Exonuclease III digestion experiments carried out on nucleosomes labeled on their right end, which analyze the left or promoter-containing end, proved difficult to interpret even for native nucleosomes (data not shown). Presumably, the long distance from the left end to the histone-protected region (which is expected to begin ∼70 bp away) allows the exonuclease molecules to become too desynchronized, blurring the finite protection provided by nucleosomal organization of this modest-affinity DNA sequence (1). Exonuclease III digestion experiments carried out on nucleosomes labeled on their left end, which analyze the right end of the template, were more revealing. Digestion time courses monitoring the right end of naked DNA and of the purified, singly positioned isomers of native and of fully tailless nucleosomes are shown in Fig. 4A. After 30 s of digestion, exonuclease III has digested almost all of the full-length naked DNA substrate to a set of intermediates mainly ∼200 bp in length, and these intermediates are further degraded by the 60-s time point. In contrast, at the 30-s time point, the majority of the native nucleosomal DNA is only slightly shortened to a strong pause site (the right-hand end of the nucleosome core particle) that persists during further digestion. The fully tailless nucleosomes behave similarly to the native ones, implying that the positioning of the two samples is the same.
FIG. 4.
Nucleosome mapping with exonuclease III and DNase I. (A) Exonuclease III. Naked DNA (lanes 1 to 4), native nucleosomes (lanes 5 to 8) and fully tailless (trypsinized) nucleosomes (lanes 9 to 12) labeled at the 5′ T7 promoter (left) end were digested with exonuclease III for 0 min (lanes 1, 5, and 9), 0.25 min (lanes 2, 6, and 10), 0.5 min (lanes 3, 7, and 11), or 1 min (lanes 4, 8, and 12) and analyzed on a 6% denaturing sequencing size gel. The numbers to the left of the figure refer to the mobilities of MspI-digested pBR322 fragments used as markers. The band corresponding to the histone-protected core particle boundary (which runs slightly faster than the initial full-length 216-nt band) is identified by an arrow. Exonuclease III nibbles at the end of the protected core fragment over the time course of digestion illustrated here, slightly increasing the mobility of the protected fragment, whereas naked DNA is extensively digested over the same period. The rate of this nibbling is reproducibly increased in the tailless nucleosomes. With increased digestion time or concentration of exonuclease III even the protected nucleosomal DNA is extensively degraded (32, 33; data not shown). (B) DNase I. Naked DNA (lanes 1 to 3 and 10 to 12), native nucleosomes (lanes 4 to 6 and 13 to 15), and tailless (trypsinized) nucleosomes (lanes 7 to 9 and 16 to 18) labeled on the 5′ phasing sequence (right) end (lanes 1 to 9) or the 5′ T7 promoter (left) end (lanes 10 to 18) were digested with DNase I for 0 min (lanes 1, 4, 7, 10, 13, and 16), 0.5 min (lanes 2, 5, 8, 11, 14, and 17) or 1 min (lanes 3, 6, 9, 12, 15, and 18) and analyzed on a 6% denaturing sequencing size gel. Brackets delineate regions protected by the core particle; dashed lines in the brackets for the tailless nucleosomes indicate regions that are hypersensitive to DNase I relative to the corresponding regions in the native nucleosomes. The asterisks mark the approximate position of the nucleosome's dyad axis of symmetry.
Interestingly, more extensive digestion (Fig. 4A, lanes 8 and 12) reveals that the enzyme can invade the histone-protected region, “nibbling” at the ends of the strongly protected core particle intermediate by an additional few nucleotides, with slightly more nucleotides digested from the tailless nucleosomes. The protection provided by nucleosomal organization against exonuclease invasion is finite, being decreased relative to naked DNA by the equilibrium constant for site exposure (uncoiling) of the DNA (1, 33). In another recent study we find that the equilibrium constants for site exposure are slightly increased by removal of the tail domains (32).
DNase I footprinting was also used to determine whether complete removal of the tail domains alters nucleosome positioning. DNase I footprinting is a particularly sensitive indicator of rotational positioning, or changes in positioning of one or a few base pairs (and possibly additional integral multiples of 10 bp) (28, 38). Native and fully tailless nucleosomes singly labeled at either the left or right ends (Fig. 3B) were digested with DNase I for increasing amounts of time, and the products analyzed by gel electrophoresis. Results from such an experiment are shown in Fig. 4B. For the native nucleosomes, analysis of results from particles labeled on the right and left ends reveals the expected 10-bp ladder of cleavage sites. For the nucleosomes labeled on the right end, the periodic pattern begins from the bottom of the gel and extends approximately 150 bp into the gel. For the nucleosomes labeled on the left end, the converse is true—the periodic pattern of cleavage sites begins from the top of the gel and extends ∼150 bp into the gel. Both sets of data are consistent with the previously determined position of the nucleosome on the template.
For the trypsinized nucleosomes, the periodic pattern characteristic of nucleosomes is also apparent. The maintenance of the cleavage pattern implies that the rotational positioning of the DNA relative to the octamer surface is unaffected by the removal of the tail domains. However, the region where protection is detected is reduced in size. For fully tailless nucleosomes labeled on the right end, the periodic pattern begins from the bottom of the gel and extends only ∼100 bp into the gel. For the fully-tailless nucleosomes labeled on the left end, a corresponding smaller protected area is detected—the periodic pattern begins from the top of the gel and extends only 100 bp into the gel. Thus, both sets of results indicate that the first 40 to 50 bp into the core particle (from the left promoter end) is more accessible to DNase I. This is consistent with a slightly increased equilibrium constant for site exposure caused by removal of the tail domains (see above and reference 32).
Taken together, these mapping studies using exonuclease III and DNase I reveal that the majority of the gel-purified positioned nucleosomes are located over the positioning sequence, toward the right-hand end of the template DNA, as desired. In addition, removal of the tail domains does not alter this preferred positioning, consistent with earlier reports (7, 15).
Transcription reactions.
The different nucleosome preparations were used as templates in transcription reactions with T7 RNAP. We investigated the effects of removing the histone tail domains on the elongation process by taking advantage of our ability to tune the transcription elongation rate (33, 34). We chose a rate that is fast enough to reveal nucleosome-induced rate attenuation characteristic of transcription at the natural in vivo rate (∼23 nt s−1 [37]) but also slow enough to allow convenient sampling of the reaction kinetics by manual pipeting. T7 RNAP is incubated with naked or nucleosomal DNA template under optimal transcription conditions but in the absence of UTP. This allows the polymerase to efficiently initiate and synthesize a 19-nt transcript; the polymerase stalls when the first template A residue is encountered at residue 20. Initiation is carried out in the presence of [α32P]ATP to allow phosphorimager analysis of radiolabeled transcripts after electrophoretic separation on a high-resolution, denaturing gel. When the initiation phase of the reaction has been completed, the samples are shifted to 0°C so that subsequent elongation will be slowed down to ∼3 to 5 nt s−1 (for naked DNA). Synchronous elongation is restarted by addition of a chase solution (also at 0°C) containing all four NTPs and with a great excess of cold ATP such that radiolabeling is largely limited to the stalling phase of the reactions. Thus, comparison of counts in the stalled complex to counts in the full-length transcripts allows the measurement of elongation rates and completion efficiencies on the different templates.
Examples of such transcription reactions on naked DNA, native nucleosomes and nucleosomes containing clostripain-digested histones are shown in Fig. 5A and are summarized quantitatively in Table 1. Initiation, chase out, and completion efficiencies are measured as detailed in our earlier studies (33, 34). Briefly, transcripts are analyzed by phosphorimager analysis, and efficiencies are determined relative to initiation on naked DNA, which was separately determined to be ∼100%.
TABLE 1.
Initiation and elongation on nucleosomes lacking the histone tetramer or octamer tail domains
| Time point | Efficiency (%)a
|
|||
|---|---|---|---|---|
| DNA | Native | Tet-no-tails | Oct-no-tails | |
| Initiation | 100 | 92 | 121 | 92 |
| Chase out | 75 | 80 | 75 | 77 |
| 40 sb | 0.5 | 9 | 12 | |
| 60 sb | 3 | 19 | 24 | |
| 4 minb | 35 | 57 | 68 | |
| 16 minb | 65 | 74 | 99 | |
Efficiencies of stalled transcript synthesis and chase-out for the experiment shown in Fig. 5A. Nucleosomes were prepared with native or with clostripain-digested histones lacking the tail domains of the (H3-H4)2 tetramer (tet-no-tails) or of all core histones (oct-no-tails). Previous studies demonstrate that the absolute efficiency of utilization of the naked DNA template in this system is approximately 100% (33). Thus, the efficiency of initiation for naked DNA is defined here as 100%, and initiation efficiencies on the different nucleosomal templates were determined by measuring the number of counts in the stalled transcripts (19 nt and longer) relative to the counts from naked DNA. Chase out was calculated by comparing the number of counts in the original stalled product to the number of stalled product counts remaining in the restarted reactions. These results are typical for multiple replicates of the experiments. We do not present averages and standard deviations because of run-to-run variability, which is enhanced by the rapid rates and closely spaced sampling times. However, all major conclusions of this paper hold for every instance of each experiment. Importantly, note that the present paper reports (and shows) similar results obtained from four different experimental systems compared to native (chicken erythrocyte or HeLa) nucleosomes: tet-no-tails, oct-no-tails, fully tailless, and hyperacetylated nucleosomes. Thus, all major results are consistent between runs for the same sample type and also for four different types of sample.
Completion efficiencies for the nucleosomal templates are measured by quantifying the counts in the full-length transcript at each time point and comparing them to the number of counts in the full-length transcripts produced from the naked DNA template after 1 min of elongation time (see Materials and Methods).
Quantitation reveals that stalled complexes are formed just as efficiently on the native and tailless nucleosomes. Removal of some or all of the tail domains has no effect on the efficiency of initiation. This is not surprising, since the use of gel-purified positioned nucleosomes in this system ensures that the histone octamers are positioned on the right end of the template, leaving the T7 promoter histone free and accessible to the polymerase. Similarly, chase out efficiencies for each of the four templates are approximately equivalent (75 to 80%). The stalled complex counts that do not elongate after UTP addition likely represent polymerase complexes that initiated but prematurely dissociated from their templates, perhaps due to the incorporation of incorrect NTPs (33). In any case, the constant level of chase out efficiency between the different templates suggests that the processivity of the polymerase molecules at this stage of the elongation process is not affected by the histones located downstream, whether native or lacking some or all of the tail domains.
Effects of the tail domains on transcription elongation.
In contrast to the initiation and chase out steps, the absence of the tail domains has substantial effects on the elongation phase of the reaction. These effects are significant even when the tail domains of H2A and H2B remain present. Under the conditions used for the elongation phase of the present experiments, transcription on naked DNA is essentially complete between 20 and 40 s. In contrast, for native nucleosomal templates, only 0.5% of the complexes have reached full-length after 40 s of elongation time. However, the remaining large majority of the elongation complexes are not permanently stuck. Rather, they are held up at a set of distinct pause sites (a subset of which are highlighted in Fig. 5 and designated as pA-pL), and given sufficient time, they chase out to yield full-length transcripts. We previously showed that these pause sites at which nucleosome transcription is transiently held up are coincident with those found for transcription on naked DNA; the only difference is that the average residence time at these pause sites is greatly increased by the organization of DNA in nucleosomes (33, 34).
Removal of the tail domains from the (H3-H4)2 tetramer (tet-no-tails sample) increases the average rate of transcription elongation and reduces the average residence times at particular pause sites. The increased overall rate of transcription elongation is seen most clearly from the fraction of transcripts that reach full length at earlier times (Table 1). For example, at the 40 s and 1-min time points (for which transcription is complete on naked DNA at both times), transcription has been completed on only ∼0.5 and 3% of native nucleosomal templates, respectively, whereas on the tet-no-tails sample transcription has been completed on ∼9 and 19% of the templates, respectively. The decreased residence times at sequence-specified pause sites are most obvious for pause sites located short distances inside the nucleosome (e.g., site D; see the discussion of fully tailless nucleosomes, below). Events at later pause sites are “blurred out” over time by the increasing asynchrony of the population as more and more pause sites are encountered, making it difficult to judge unequivocally whether the residence times at these sites are similarly affected. Modest decreases in residence times, integrated over multiple pause sites, cause the overall elongation rate to increase. Additional increases in the overall elongation rate may arise from general effects of removing the (H3-H4)2 tail domains that are not attributable to any particular pause sites.
Additional removal of the tail domains of H2A and H2B by clostripain (oct-no-tails sample) leads to only small additional increases in the average velocity of elongation (and correspondingly, in the fraction of templates completing transcription at each time point). The simplest interpretation of this finding is that the (H3-H4)2 tetramer dominates the ability of the tail domains to suppress the rate of transcriptional elongation through the nucleosome. Because we were unable to prepare the reciprocal particles (containing intact H3 and H4 tails but lacking the tails of H2A and H2B), we cannot rule out the alternative possibility that the effects on elongation rate might instead depend only on the number of tail domains remaining intact. However, the recent finding from another group (49) that the tail domains of the (H3-H4)2 tetramer—and not those of H2A and H2B—control the ability of a gene regulatory protein to gain access to a nucleosomal DNA target site lends indirect support for the simpler interpretation of the present study.
The effects seen with removal of the tail domains by clostripain are observed even more strongly in experiments on the fully tailless (trypsin-digested) nucleosomes. Results from such an experiment are illustrated in Fig. 5B and are summarized quantitatively in Table 2. Several aspects of these results are noteworthy. As can be seen quantitatively by analysis of the counts present in particular paused species as a function of time (Fig. 5C), removal of the entirety of the tail domains by trypsin decreases the residence times at particular major pause sites (e.g., sites D and F) or essentially eliminates other pause sites altogether (e.g., sites C and E), leading to substantial increases in the fraction of templates completing transcription at each time point (Table 2), as well as resulting in a greater population of the more distal pause sites (sites G to L).
TABLE 2.
Initiation and elongation on fully tailless nucleosomes
| Time point | Efficiency (%)a
|
||
|---|---|---|---|
| DNA | Native | Fully tailless | |
| Initiation | 100 | 75 | 47 |
| Chase out | 83 | 83 | 72 |
| 40 s | 1.2 | 22 | |
| 60 s | 10 | 51 | |
| 2 min | 21 | 103 | |
| 4 min | 30 | 129 | |
| 8 min | 40 | 101 | |
Efficiencies of stalled transcript synthesis and chase out for the experiment shown in Fig. 5B. Nucleosomes were prepared with native or with trypsin-digested histones (yielding fully tailless nucleosomes). Efficiencies of initiation, chase-out, and relative completion are calculated as described in the footnotes to Table 1.
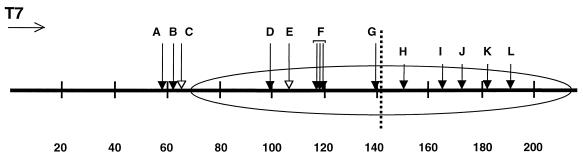
The locations of these sites mapped onto the positioned nucleosome are illustrated in Fig. 6. Of the pause sites exhibiting significant decreases in residence time with removal of the tail domains, sites D, E, and F are located in the region that concomitantly exhibit increased accessibility to DNase I, while site C is not even in the nucleosomal region. Site C is located a few base pairs away from the edge of the nucleosome, suggesting that the tails also interact with linker DNA or constitute steric obstacles for transcription in the linker regions.
FIG. 6.
Locations of T7 RNAP pause sites mapped onto the nucleosomal templates. The open oval represents the region occupied by the nucleosome core particle as determined in the exonuclease III and DNase I mapping experiments. The dashed line indicates the nucleosomal dyad axis of symmetry. Arrows above the oval identify major pause sites of T7 RNAP (here designated A to L), as identified in Fig. 5A and B. All of the identified pause sites are detectable on all the samples, but their lifetimes (residence times) and prominence (which reflects both the residence time at that pause site and the flux of reactant into that pause site) depend strongly on nucleosomal organization (33, 34) and, as shown here, on the histone tail domains. Long arrows with filled heads indicate pause sites that persist but with diminished intensity for the tailless nucleosomes (A and B), or exhibit decreased lifetime for the tailless nucleosomes (D, F, and G). Open arrowheads indicate pause sites (C and E) that are essentially no longer detectable on the tailless nucleosomes. Short arrows indicate the faint pause sites detected on the second half of the nucleosome.
Effects of histone hyperacetylation.
The tailless nucleosomes investigated in the reactions in Fig. 5 are widely used as a model system for the possible effects of histone hyperacetylation. But it is important to determine whether the striking effects of the tail domains on transcriptional elongation through nucleosomes are in fact also obtained with hyperacetylated nucleosomes. We therefore reconstituted transcription template DNA into nucleosomes using native or hyperacetylated (butyrate-treated) HeLa histones and carried out single-passage transcription experiments. The results from a typical experiment are illustrated in Fig. 7 and are summarized quantitatively in Table 3. Remarkably, histone hyperacetylation leads to a dramatic increase in the rate of elongation through the nucleosome and a corresponding decrease in pause site residence times. This is again seen most clearly from the fraction of transcripts that reach full length at earlier times (Table 3). For example, at the 40-s and 1-min timepoints, transcription has been completed on only ∼3 and 11% of native HeLa nucleosomal templates, respectively, whereas on the hyperacetylated HeLa nucleosomes transcription has been completed on ∼36 and 67% of the templates, respectively. The decreased residence times at sequence-specified pause sites are particularly apparent for pause sites located a short distance inside the nucleosome (Fig. 7).
FIG. 7.
Transcription elongation on native and hyperacetylated nucleosomes. Transcription reactions on naked DNA (lanes 3 to 8), nucleosomes prepared with native HeLa histones (lanes 9 to 14), and nucleosomes prepared with hyperacetylated HeLa histones (lanes 15 to 20) were run on a 6% denaturing sequencing-size gel and exposed to a phosphorimager plate for analysis. Aliquots were taken after stalled complex formation (lanes 3, 9, and 15) and after 10 s (lanes 4, 10, and 16), 20 s (lanes 5, 11, and 17), 40 s (lanes 6, 12, and 18), 1 min (lanes 7, 13, and 19), and 4 min (lanes 8, 14, and 20). Lanes 1 and 2 contain size standards (lane 1, 10-bp ladder; lane 2, A ladder [see Fig. 5 legend]). The open oval represents the approximate location of the nucleosome on the template; the dashed line identifies the dyad axis of symmetry. The band corresponding to the major pause site immediately promoter proximal to the dyad axis is indicated with a long arrow. Pause sites present near the promoter-proximal end of the nucleosome, whose residence times are decreased by histone hyperacetylation, are indicated by short arrows.
TABLE 3.
Initiation and elongation with native and hyperacetylated HeLa histones
| Time point | Efficiency (%)a
|
||
|---|---|---|---|
| DNA | Native HeLa | Hyperacetylated HeLa | |
| Initiation | 100 | 107 | 83 |
| Chase out | 87 | 86 | 92 |
| 10 s | 0 | 0 | |
| 20 s | 0 | 1 | |
| 40 s | 3 | 36 | |
| 60 s | 11 | 66 | |
| 4 min | 104 | 188 | |
Looping during transcription.
A striking aspect of the data in Fig. 5A and B and 7 is the presence of a strong pause site immediately proximal to the nucleosome dyad, together with greatly diminished pausing in the promoter-distal half of the nucleosome, even at later time points when most of the templates have reached completion. These properties have been noticed previously (4, 41–43) and have been attributed to the ability of DNA behind the polymerase to loop around and get captured by the histone octamer. Loop formation then allows the histone octamer to step around the elongating polymerase, producing a nucleosome at a new location that is displaced backwards toward the promoter. This behavior is essentially unchanged with the deletion of the tail domains of the (H3-H4)2 tetramer or the entire octamer by clostripain (Fig. 5A), in the fully tailless nucleosomes produced with trypsin (Fig. 5B and 6) and in the hyperacetylated nucleosomes (Fig. 7). Assuming that this distinctive pattern of pause site intensities is indeed attributable to the histone octamer stepping around the elongating polymerase (4, 41–43), these data imply that the tail domains are not responsible for this ability of the histone octamer to undergo such a translocation, suggesting instead that such a translocation occurs via interactions between naked DNA behind the polymerase and unoccupied DNA-binding surface of the histone octamer's globular domains.
Conclusions.
The present study points to three chief conclusions. First, histone acetylation, or partial or complete removal of the tail domains, significantly increases the average rate of transcription elongation through the nucleosome. Evidently, the core histone tail domains contribute significantly to a nucleosome-dependent reduction of the average transcription elongation rate, and posttranslational modification of the tail domains provides the possibility for regulation of the elongation rate in vivo. We showed earlier that native (and unacetylated) nucleosomes decrease the rate of transcriptional elongation compared to the rate on naked DNA by increasing the residence times spent by the polymerase at pause sites that are specified by the DNA sequence itself (33, 34). We find in the present study that acetylation or removal of the tail domains reduces this nucleosome-dependent pausing at particular pause sites. These findings extend the results of a previous study (6), which showed that the tail domains contribute to a decreased transcription rate on oligonucleosomal templates but did not investigate how this was accomplished, which tail domains contributed to the effect, or the effects of histone acetylation. Our new findings also complement our recent observation that removal of the tail domains leads to 1.5- to 14-fold increases in the equilibrium accessibility of DNA target sites that (in the time average) are buried within the nucleosome (32).
Second, the effects observed here appear primarily attributable to the tail domains of the (H3-H4)2 tetramer. While we cannot exclude any role for H2A-H2B heterodimers, we find that given nucleosomes already lacking the tail domains of the (H3-H4)2 tetramer, additional removal of the tail domains of H2A-H2B heterodimers leads to only small additional decreases in pausing (i.e., to only small additional increases in average elongation rate).
Finally, assuming that the distinctive pattern of pause site intensities (strong pausing just proximal to the dyad, relatively weaker pausing at distal sites) reflects—and is attributable to—the histone octamer stepping around the elongating polymerase, the present results establish that this ability of the octamer to step around the polymerase is inherent to the globular histone fold domains of the octamer. This finding is significant in part because it implies that eukaryotes and histone-containing archaebacteria (which conserve many aspects of the eukaryotic transcription apparatus but have histones lacking tail domains [25, 26, 31, 35]) may utilize similar mechanisms for transcriptional elongation through their natural chromatin substrate.
ACKNOWLEDGMENTS
This work was supported by a grant from the NIH. We acknowledge with gratitude the use of instruments in the Keck Biophysics Facility, which was established with a grant from the W. M. Keck Foundation.
We also thank the National Cell Culture Center for the production of HeLa cells and butyrate-treated HeLa cells.
REFERENCES
- 1.Anderson J D, Widom J. Sequence- and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J Mol Biol. 2000;296:979–987. doi: 10.1006/jmbi.2000.3531. [DOI] [PubMed] [Google Scholar]
- 2.Ausio J, Dong F, van Holde K E. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J Mol Biol. 1989;206:451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- 3.Ausio J, van Holde K E. Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry. 1986;25:1421–1428. doi: 10.1021/bi00354a035. [DOI] [PubMed] [Google Scholar]
- 4.Bednar J, Studitsky V M, Grigoryev S A, Felsenfeld G, Woodcock C L. The nature of the nucleosomal barrier to transcription: direct observation of paused intermediates by electron cryomicroscopy. Mol Cell. 1999;4:377–386. doi: 10.1016/s1097-2765(00)80339-1. [DOI] [PubMed] [Google Scholar]
- 5.Böhm L, Crane-Robinson C. Proteases as structural probes for chromatin: the domain structure of histones. Biosci Rep. 1984;4:365–386. doi: 10.1007/BF01122502. [DOI] [PubMed] [Google Scholar]
- 6.Chirinos M, Hernández F, Palacián E. Repressive effect on oligonucleosome transcription of the core histone tail domains. Biochemistry. 1998;37:7251–7259. doi: 10.1021/bi9729817. [DOI] [PubMed] [Google Scholar]
- 7.Dong F, Hansen J C, van Holde K E. DNA and protein determinants of nucleosome positioning on sea urchin 5S rRNA gene sequences in vitro. Proc Natl Acad Sci USA. 1990;87:5724–5728. doi: 10.1073/pnas.87.15.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dumuis-Kervabon A, Encontre I, Etienne G, Jauregui-Adell J, Mery J, Mesnier D, Parello J. A chromatin core particle obtained by selective cleavage of histones by clostripain. EMBO J. 1986;5:1735–1742. doi: 10.1002/j.1460-2075.1986.tb04418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Encontre I, Parello J. Chromatin core particle obtained by selective cleavage of histones H3 and H4 by clostripain. J Mol Biol. 1988;202:673–676. doi: 10.1016/0022-2836(88)90296-3. [DOI] [PubMed] [Google Scholar]
- 10.Ericsson C, Grossbach U, Bjorkroth B, Daneholt B. Presence of histone H1 on an active balbiani ring gene. Cell. 1990;60:73–83. doi: 10.1016/0092-8674(90)90717-s. [DOI] [PubMed] [Google Scholar]
- 11.Feng H-P, Scherl D S, Widom J. Lifetime of the histone octamer studied by continuous-flow quasielastic light scattering: test of a model for nucleosome transcription. Biochemistry. 1993;32:7824–7831. doi: 10.1021/bi00081a030. [DOI] [PubMed] [Google Scholar]
- 12.Fletcher T M, Hansen J C. Core histone tail domains mediate oligonucleosome folding and nucleosomal DNA organization through distinct molecular mechanisms. J Biol Chem. 1995;270:25359–25362. doi: 10.1074/jbc.270.43.25359. [DOI] [PubMed] [Google Scholar]
- 13.Godde J S, Nakatani Y, Wolffe A P. The amino-terminal tails of the core histones and the translational position of the TATA box determine TBP/TFIIA association with nucleosomal DNA. Nucleic Acids Res. 1995;23:4557–4564. doi: 10.1093/nar/23.22.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 15.Hayes J J, Clark D J, Wolffe A P. Histone contributions to the structure of DNA in the nucleosome. Proc Natl Acad Sci USA. 1991;88:6829–6833. doi: 10.1073/pnas.88.15.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howe L, Ranalli T A, Allis C D, Ausio J. Transcriptionally active Xenopus laevis somatic 5S ribosomal RNA genes are packaged with hyperacetylated histone H4, whereas transcriptionally silent oocyte genes are not. J Biol Chem. 1998;273:20693–20696. doi: 10.1074/jbc.273.33.20693. [DOI] [PubMed] [Google Scholar]
- 17.Kayne P S, Kim U-J, Han M, Mullen J R, Yoshizaki F, Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988;55:27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- 18.Kuo M H, Allis C D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 19.Lee D Y, Hayes J J, Pruss D, Wolffe A P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 20.Lorch Y, LaPointe J W, Kornberg R D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- 21.Losa R, Brown D D. A bacteriophage RNA polymerase transcribes in vitro through a nucleosome core without displacing it. Cell. 1987;50:801–808. doi: 10.1016/0092-8674(87)90338-2. [DOI] [PubMed] [Google Scholar]
- 22.Lowary P T, Widom J. Higher order structure of Saccharomyces cerevisiae chromatin. Proc Natl Acad Sci USA. 1989;86:8266–8270. doi: 10.1073/pnas.86.21.8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Structure of the nucleosome core particle at 2.8Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 24.Luger K, Rechsteiner T J, Flaus A J, Waye M M Y, Richmond T J. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 25.Luger K, Richmond T J. DNA binding within the nucleosome core. Curr Opin Struct Biol. 1998;8:33–40. doi: 10.1016/s0959-440x(98)80007-9. [DOI] [PubMed] [Google Scholar]
- 26.Luger K, Richmond T J. The histone tails of the nucleosome. Curr Opin Genet Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 27.Lutter L C. Digestion of nucleosomes with deoxyribonucleases I and II. Methods Enzymol. 1989;170:264–269. doi: 10.1016/0076-6879(89)70051-3. [DOI] [PubMed] [Google Scholar]
- 28.Lutter L C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978;124:391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- 29.Nacheva G A, Guschin D Y, Preobrazhenskaya O V, Karpov V L, Ebralidse K K, Mirzabekov A D. Change in the pattern of histone binding to DNA upon transcriptional activation. Cell. 1989;58:27–36. doi: 10.1016/0092-8674(89)90399-1. [DOI] [PubMed] [Google Scholar]
- 30.Neubauer B, Horz W. Analysis of nucleosome positioning by in vitro reconstitution. Methods Enzymol. 1989;170:630–644. doi: 10.1016/0076-6879(89)70069-0. [DOI] [PubMed] [Google Scholar]
- 31.Pereira S L, Reeve J N. Histones and nucleosomes in Archaea and Eukarya: a comparative analysis. Extremophiles. 1998;2:141–148. doi: 10.1007/s007920050053. [DOI] [PubMed] [Google Scholar]
- 32.Polach K J, Lowary P T, Widom J. Effects of core histone tail domains on the equilibrium constants for dynamic DNA site accessibility in nucleosomes. J Mol Biol. 2000;298:211–223. doi: 10.1006/jmbi.2000.3644. [DOI] [PubMed] [Google Scholar]
- 33.Protacio R U, Polach K J, Widom J. Coupled enzymatic assays for the rate and mechanism of DNA site-exposure in a nucleosome. J Mol Biol. 1997;274:708–721. doi: 10.1006/jmbi.1997.1440. [DOI] [PubMed] [Google Scholar]
- 34.Protacio R U, Widom J. Nucleosome transcription studied in a real-time synchronous system: test of the lexosome model and direct determination of effects due to histone octamer. J Mol Biol. 1996;256:458–472. doi: 10.1006/jmbi.1996.0101. [DOI] [PubMed] [Google Scholar]
- 35.Reeve J N, Sandman K, Daniels C J. Archaeal histones, nucleosomes, and transcription initiation. Cell. 1997;89:999–1002. doi: 10.1016/s0092-8674(00)80286-x. [DOI] [PubMed] [Google Scholar]
- 36.Schuster T, Han M, Grunstein M. Yeast histone H2A and H2B amino termini have interchangeable functions. Cell. 1986;45:445–451. doi: 10.1016/0092-8674(86)90330-2. [DOI] [PubMed] [Google Scholar]
- 37.Shermoen A W, O'Farrell P H. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 1992;67:303–310. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simpson R T, Stafford D W. Structural features of a phased nucleosome core particle. Proc Natl Acad Sci USA. 1983;80:51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strahl B D, Allis C D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 40.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 41.Studitsky V M, Clark D J, Felsenfeld G. A histone octamer can step around a transcribing polymerase without leaving the template. Cell. 1994;76:371–382. doi: 10.1016/0092-8674(94)90343-3. [DOI] [PubMed] [Google Scholar]
- 42.Studitsky V M, Clark D J, Felsenfeld G. Overcoming a nucleosomal barrier to transcription. Cell. 1995;83:19–27. doi: 10.1016/0092-8674(95)90230-9. [DOI] [PubMed] [Google Scholar]
- 43.Studitsky V M, Kassavetis G A, Geiduschek E P, Felsenfeld G. Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science. 1997;278:1960–1963. doi: 10.1126/science.278.5345.1960. [DOI] [PubMed] [Google Scholar]
- 44.Tse C, Sera T, Wolffe A, Hansen J. Disruption of higher order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turner B M. Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell Mol Life Sci. 1998;54:21–31. doi: 10.1007/s000180050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ura K, Kurumizaka H, Dimitrov S, Almouzni G, Wolffe A P. Histone acetylation: influence on transcription, nucleosome mobility and positioning, and linker histone-dependent transcriptional repression. EMBO J. 1997;16:2096–2107. doi: 10.1093/emboj/16.8.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vettese-Dadey M, Grant P A, Hebbes T R, Crane-Robinson C, Allis C D, Workman J L. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 48.Vettese-Dadey M, Walter P, Chen H, Juan L-J, Workman J L. Role of the histone amino termini in facilitated binding of a transcription factor, GAL4-AH, to nucleosome cores. Mol Cell Biol. 1994;14:970–981. doi: 10.1128/mcb.14.2.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vitolo J M, Thiriet C, Hayes J J. The H3–H4 N-terminal tail domains are the primary mediators of transcription factor IIIA access to 5S DNA within a nucleosome. Mol Cell Biol. 2000;20:2167–2175. doi: 10.1128/mcb.20.6.2167-2175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallis J W, Rykowski M, Grunstein M. Yeast histone H2B containing large amino terminus deletions can function in vivo. Cell. 1983;35:711–719. doi: 10.1016/0092-8674(83)90104-6. [DOI] [PubMed] [Google Scholar]
- 51.Widom J. Physicochemical studies of the folding of the 100Å nucleosome filament into the 300Å filament. J Mol Biol. 1986;190:411–424. doi: 10.1016/0022-2836(86)90012-4. [DOI] [PubMed] [Google Scholar]
- 52.Widom J. Structure, dynamics, and function of chromatin in vitro. Annu Rev Biophys Biomol Struct. 1998;27:285–327. doi: 10.1146/annurev.biophys.27.1.285. [DOI] [PubMed] [Google Scholar]
- 53.Wolffe A. Chromatin structure and function. 3rd ed. San Diego, Calif: Academic Press; 1998. [Google Scholar]
- 54.Wolffe A P, Hayes J J. Chromatin disruption and modification. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]