Neuropeptide-bound NK1R:G protein complex structures explain mechanisms ranging from insurmountable antagonism to activation.
Abstract
The neurokinin 1 receptor (NK1R) is involved in inflammation and pain transmission. This pathophysiologically important G protein–coupled receptor is predominantly activated by its cognate agonist substance P (SP) but also by the closely related neurokinins A and B. Here, we report cryo–electron microscopy structures of SP-bound NK1R in complex with its primary downstream signal mediators, Gq and Gs. Our structures reveal how a polar network at the extracellular, solvent-exposed receptor surface shapes the orthosteric pocket and that NK1R adopts a noncanonical active-state conformation with an interface for G protein binding, which is distinct from previously reported structures. Detailed comparisons with antagonist-bound NK1R crystal structures reveal that insurmountable antagonists induce a distinct and long-lasting receptor conformation that sterically blocks SP binding. Together, our structures provide important structural insights into ligand and G protein promiscuity, the lack of basal signaling, and agonist- and antagonist-induced conformations in the neurokinin receptor family.
INTRODUCTION
The neurokinin receptor (NKR) family and its endogenous ligands are one of the most thoroughly studied neuropeptide signaling systems (1–4). Initially found in the early 1930s, to date, the human NKR family is known to consist of three members (NK1R, NK2R, and NK3R), which belong to the superfamily of G protein–coupled receptors (GPCRs). NK1R represents the most abundant and widely expressed member of the family, and this receptor has been found in neuronal, muscle, and endothelial cells of diverse tissues, ranging from the central and peripheral nervous system (5, 6) to the digestive (7) and respiratory (8) system, as well as on T cells in the immune synapse (9). Accordingly, NK1R is implicated in major pathophysiological processes, such as inflammation (10), analgesia (11), depression (12), pruritus (13), and possibly asthma (14).
In the clinic, NK1R is today targeted by the two marketed antagonists, aprepitant and netupitant, for the treatment of chemotherapy-induced nausea and vomiting (CINV) (15–17). Nonetheless, because of their widespread role, NK1R ligands may find additional clinical uses, justifying a mechanistic study of this receptor. Recently, we reported crystal structures of the NK1R bound to antagonists from different development stages, which allowed us to propose a molecular mechanistic model for the observed insurmountable antagonism underlying the long-lasting effect of the marketed drugs, which is not shown by other tight-binding tool compounds (18–21). Nevertheless, to verify this hypothesis, a structure of the agonist-bound active state would be needed.
The antagonist-bound structures also provided a mechanistic model for the lack of basal signaling of NK1R: In contrast to other class A GPCRs where an allosteric sodium ion stabilizes the inactive conformation, the NK1R-specific residue E782.50 [number in superscript corresponds to Ballesteros-Weinstein numbering (22)] takes up a position in the receptor core, which stabilizes this state and, hence, prevents basal activity (23). However, in the absence of an agonist-bound structure, these studies could not explain the molecular determinants of receptor activation.
Activation of NK1R is mediated predominantly through substance P (SP), an 11–amino acid–long peptide with the sequence RPKPQQFFGLM, comprising an amidated C terminus. However, two other neurokinins, A (NKA) and B (NKB), are also capable of binding to NK1R, albeit with lower affinity compared to SP (24). NKA and NKB, in turn, are the main endogenous agonists of the two other NKR family members, NK2R and NK3R, respectively. In their C-terminal part, which mediates receptor activation, SP, NKA, and NKB share a high sequence homology with the three terminal amino acids being identical. Numerous pharmacological studies have located the determinants for ligand selectivity to the N-terminal portion of the peptide (25–28). On this basis, a message-address model has been proposed for the endogenous NKR agonists, dividing the ligand in two functionally distinct parts, the C-terminal region inducing receptor activation (message) and the N-terminal region providing receptor subtype selectivity (address) (29). Nonetheless, the precise molecular determinants of ligand binding and receptor activation have remained largely unknown because of the lack of structural information on the agonist-bound receptor in an active, G protein–bound state.
Last, the NK1R has also been shown to couple to different heterotrimeric G protein subtypes, predominantly Gq and Gs, for intracellular signal transduction. Various previous studies suggested a coupling preference (2- to 20-fold difference in median effective concentration, EC50) of the receptor to Gq over Gs (23, 30–32). Here, we elucidated the binding mode of neurokinin peptide ligands exemplified by SP and investigated the mechanism for the reported G protein–coupling preference. We used cryo–electron microscopy (cryo-EM) to determine the structures of SP-bound NK1R complexed with Gq and Gs heterotrimers.
RESULTS
Structure determination
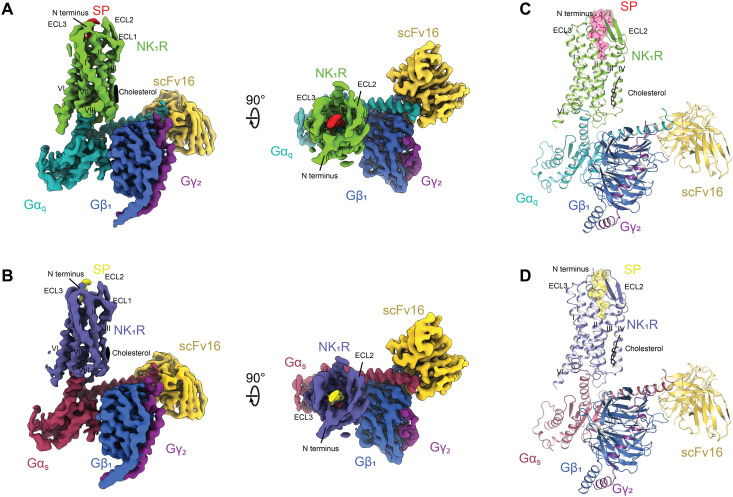
For the preparation of stable, active-state complexes of SP-bound wild-type human NK1R with Gq and Gs, we used the previously engineered G protein versions mini-Gαq and mini-Gαs (33, 34) as heterotrimers with Gβ1γ2. To permit binding of a complex-stabilizing single-chain antibody variable fragment (scFv16) distant from the GPCR/G protein interface (35), the N terminus of mini-Gαq and mini-Gαs was replaced with the N-terminal 18 amino acids of Gαi1. The resulting heterotrimeric G proteins are hereafter, for simplicity, referred to as Gq and Gs. Purified complexes of these G proteins with SP-bound NK1R and scFv16 (fig. S1) were subjected to cryo-EM data acquisition. After iterative rounds of two-dimensional (2D) and 3D classification, the two complexes of NK1R with Gq and Gs were resolved to 2.7 and 2.9 Å, respectively (Fig. 1, Table 1, and figs. S2 to S6).
Fig. 1. Cryo-EM structures of NK1R:SP:Gq:scFv16 and NK1R:SP:Gs:scFv16.
(A) Cryo-EM map of the NK1R:SP:Gq:scFv16 complex resolved to 2.71 Å (FSC 0.143) as viewed from the side and from the top. (B) Cryo-EM map of the NK1R:SP:Gs:scFv16 complex resolved to 2.87 Å (FSC 0.143) as viewed from the side and from the top. (C) Molecular model of the NK1R:SP:Gq:scFv16 complex with SP and cholesterol shown in stick representation. (D) Molecular model of the NK1R:SP:Gs:scFv16 complex with SP and cholesterol shown in stick representation.
Table 1. Cryo-EM processing and modeling statistics.
|
NK1R:SP:Gq:scFv16
(PDB ID: 7P00) |
NK1R:SP:Gs:scFv16
(PDB ID: 7P02) |
|
| Data collection | ||
| Microscope | Titan Krios G3i | Titan Krios G3i |
| Detector | Gatan K3 | Gatan K3 |
| Energy filter slit width (eV) |
20 | 20 |
| Magnification | 130,000 | 130,000 |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e−/Å2) |
62.51 | 63.51 |
| Defocus range (μm) | 0.8–2.4 | 0.8–2.4 |
| Pixel size (Å) | 0.65 | 0.65 |
| Symmetry imposed | C1 | C1 |
| Number of micrographs |
5291 | 6341 |
| Initial particle images (n) |
3,511,990 | 4,227,825 |
| Final particle images (n) |
558,058 | 395,052 |
| Map resolution (Å) | 2.71 | 2.87 |
| FSC threshold | 0.143 | 0.143 |
| Refinement | ||
| Number of atoms | ||
| All | 9110 | 9061 |
| Protein | 8987 | 8957 |
| Ligand | 95 | 76 |
| Cholesterol | 28 | 28 |
| Model validation | ||
| CC map versus model (%) |
79 | 78 |
| RMSD | ||
| Bond lengths (Å) | 0.012 | 0.012 |
| Bond angles (°) | 1.679 | 1.510 |
| Ramachandran statistics |
||
| Favored regions (%) | 96.63 | 95.36 |
| Allowed regions (%) | 3.37 | 4.64 |
| Outliers (%) | 0.00 | 0.00 |
| Rotamer outliers (%) | 0.81 | 1.12 |
| Cβ deviations (%) | 0.74 | 0.00 |
| MolProbity overall score |
4.75 | 6.05 |
In the structures of both complexes, the receptor region of highest resolution was observed in the transmembrane core, whereas the extracellular and intracellular transmembrane helix tips as well as their connecting flexible loops represented regions of lower local resolution (figs. S4 and S5). Intracellular loop 3 (ICL3) was not resolved in either of the complexes. In contrast to the previous crystal structures of antagonist-bound NK1R, both SP-bound NK1R complexes display a resolved receptor N terminus, starting at P22N-term, as well as a resolved extracellular loop 3 (ECL3), due to direct contact of these regions with the ligand (19). ECL2 is packed against ECL1 and adopts the previously observed extended β-hairpin, which is tethered to the extracellular tip of transmembrane helix III through a disulfide between C180ECL2 and C1053.25. ECL3 adopts short α-helical fold between D276ECL3 and L279ECL3. The receptor N terminus, together with all ECLs, establishes the structural framework for extracellular SP binding. The C terminus of the ligand is located within the orthosteric pocket, where it binds deep into the receptor core in an elongated conformation.
On the intracellular portion of NK1R, we detected an additional cholesterol density at the outer interface of helices II and IV in both the Gq-bound and the Gs-bound complexes (fig. S7). The observed binding area overlaps with a previously described cholesterol consensus motif present in the sequence of 21% of all human class A GPCRs (36). The presence of a cholesterol molecule in structures of the active NK1R state may explain the observed cholesterol dependence of NK1R activation (37). Furthermore, a functionally important cholesterol molecule was observed at a similar binding interface in two recently reported active-state complexes of the cholecystokinine A receptor (CCK1R) with Gq and Gs (38).
Despite the high resolutions obtained and the monoolein matrix being supplemented with 10% (w/w) cholesterol, cholesterol has not been observed in any of the antagonist-bound NK1R crystal structures (18–20). The presence of this cholesterol molecule may thus be specific to the active state of the receptor, similar to state-dependent lipid interactions of the adenosine A2A receptor (A2AR) (39, 40).
Binding of SP to NK1R
Over the past decades, the binding mode of SP on NK1R has been investigated in numerous mutational and pharmacological studies (41–46). Despite these extensive efforts, the precise molecular details of endogenous agonist interactions have remained unknown. The obtained resolution now allows us to describe the precise interaction mode of SP with NK1R in the fully active receptor state. To also characterize the observed interactions in a quantitative manner, we performed whole-cell ligand binding assays on wild-type human NK1R and single mutants of structurally identified key interaction residues using fluorescently labeled SP (table S1).
Overall, the ligand binding pose of the endogenous agonist SP is highly similar in the Gs and the Gq complexes of NK1R (fig. S8). The interactions of SP with NK1R can broadly be divided into two groups: (i) those of the C terminus (residues F7-M11, subscript indicates SP residue number), which are highly conserved between the endogenous agonists SP, NKA, and NKB, with the transmembrane helical core of the receptor, and (ii) those of the nonconserved N terminus (residues R1-Q6) with the extracellular surface of the receptor (Fig. 2, A and B).
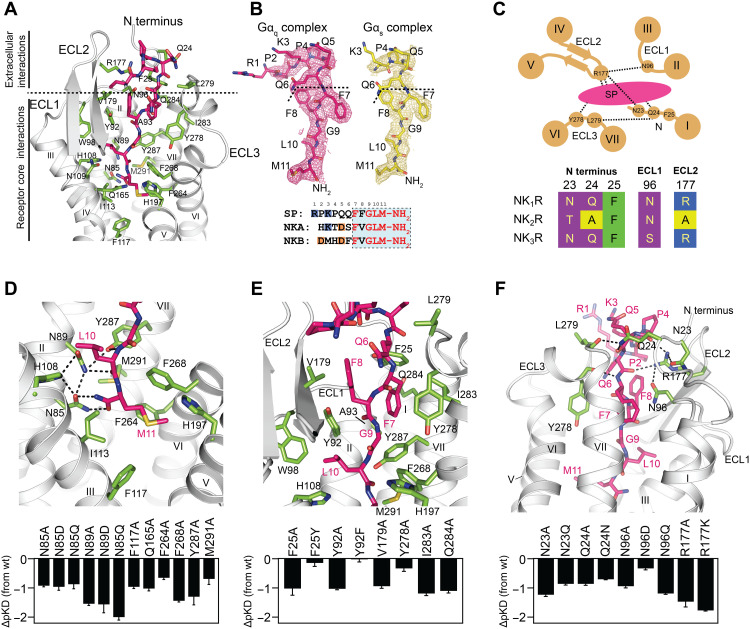
Fig. 2. Interactions of the endogenous agonist SP with NK1R.
(A) SP (stick representation, carbons depicted in pink) establishes a host of interactions with the receptor transmembrane core and the extracellular receptor surface (receptor-interacting residues depicted as sticks with green carbons). (B) Processed cryo-EM maps of SP in the NK1R:Gq complex (colored in pink) and the NK1R:Gs complex (colored in yellow) contoured at 4.5 σ. Bottom: The amino acid sequences (numbers on top correspond to position in SP) of the three neurokinins SP, NKA, and NKB are indicated by residues of the conserved ligand C terminus highlighted in red, and the C-terminal amide is shown. Negatively and positively charged residues are highlighted in orange and blue, respectively. (C) Schematic representation of the polar network shaping the extracellular receptor surface for SP binding, indicating the hydrogen bonds detected in the structure. Bottom: Alignment of residues involved in specific side-chain interactions of the polar network are shown, comparing NK1R with NK2R and NK3R. Framework residues of ECL3 only interact via backbone hydrogen bonding and are therefore not included in the alignment. (D to F) Close-up on the interactions of the amidated SP C terminus at the bottom of the orthosteric binding pocket (D), the interactions of the neurokinin-conserved C-terminal portion of SP with the hydrophobic interaction surface of helices VI and VII of NK1R (E), and the polar network at the extracellular receptor surface (F). Bottom: Agonist affinity profiles of selected mutants compared to wild-type NK1R are shown. pKD values were derived from whole-cell saturation binding experiments (table S1). Data are shown as means ± SEM from three to six independent experiments performed in duplicate.
Interactions of the C terminus of SP
The interactions of the conserved SP C terminus with NK1R are well resolved in both cryo-EM density maps of the Gq and the Gs complexes, allowing to model all side chains of the peptide (Fig. 2B). In contrast, at its N terminus, density can only be assigned to the peptide backbone. Furthermore, in the Gs-bound structure of NK1R, we could not resolve EM densities for the N-terminal residues R1-P2 and have thus modeled only K3-M11 of SP (Fig. 2B). Therefore, because of the higher resolution obtained and the completely resolved ligand, we refer to the Gq-bound structure for the description of the interactions of SP with NK1R.
The pocket to which the C-terminal part of SP binds is lined by transmembrane helices II, III, VI, and VII of NK1R (Fig. 2A). Residues F7-M11 of SP are oriented along a hydrophobic interface established by helices VI and VII in a well-defined conformation, as evidenced by the clear EM density for all ligand side chains in this region (Fig. 2B).
Within the receptor core, the interactions of the agonist can again be divided into two main groups: (i) a hydrogen-bonding network centered at the C-terminal amide of the agonist, spanning helices II, III, and VII (Fig. 2D), and (ii) a hydrophobic contact surface lined mainly by residues from helices VI and VII (Fig. 2E).
C-terminal amidation is a central feature of tachykinins occurring in all vertebrates and invertebrates (2), and its removal has been demonstrated to be detrimental to agonist activity of SP (47, 48). In the structures of SP-bound NK1R, we now find the C-terminal amide of the agonist positioned in close hydrogen-bonding distance (N-O distances of 2.7 and 2.9 Å) to N852.57. Accordingly, we observe a 10-fold decrease in agonist binding affinity upon changing the intermolecular distance of the interacting amides (mutation N852.57Q) and an 8- and 10-fold decrease upon either removal of the side chain or changing its electrostatics (mutations N852.57A and N852.57D, respectively) (Fig. 2D), underlining the importance of this interaction.
The binding pose of the C-terminal portion of SP is further stabilized through hydrogen bonding of the backbone oxygen and nitrogen of the agonist residue L10 with N892.61 and Y2877.35, respectively. The importance of these H bonds is underlined by a 36- to 111-fold decrease in SP binding affinity upon mutation of N892.61 to A, D, or Q, and a 31-fold decrease upon removal of the side chain in Y2877.35A (Fig. 2D). In addition to these hydrophilic contacts, we find His1083.28 in hydrogen-bonding distance to both N852.57 and N892.61, potentially further stabilizing the rotamer orientation of these two residues that bind SP.
On the opposite side of this hydrogen-bonding network at the bottom of the orthosteric pocket, the side chain of M11 is found in a hydrophobic subpocket. This subpocket is lined by residues from helices III to VII, including I1133.33, F1173.37, Q1654.60, H1975.39, F2646.51, F2686.55, and M2917.39, and removal of the hydrophobic moieties from residues F1173.37, Q1654.60, F2646.51, F2686.55, and M2917.39 by mutation to an alanine leads to a 6- to 28-fold reduction in SP binding affinity (Fig. 2D).
From the bottom of the orthosteric ligand binding pocket toward the extracellular opening, the peptide agonist packs with residues F7 and G9 against the hydrophobic surface of helices VI, VII, and ECL3, thereby enforcing an elongated conformation of the SP peptide backbone (Fig. 2E). Thus, F8 and L10 of SP are facing away from helices VI and VII and pack against ECL1 and ECL2 of NK1R. At the extracellular boundary of the orthosteric ligand binding pocket, SP is held in position through hydrophobic interactions with V179ECL2 and Y278ECL3 as well as with Y922.64, A932.65, I2837.31, and Q2847.32 from the helical tips of helices II and VII. In line with the hydrophobic nature of these interactions, SP binding affinity is reduced by a factor of 2 to 16 upon removing interaction surface by mutation of these residues to alanine, while removal of the hydroxy headgroup of Y922.64 by mutation Y922.64F leaves SP binding affinity unchanged (Fig. 2E).
F8 of SP, being the first amino acid in C- to N-terminal direction, which is not conserved between SP, NKA, and NKB, is not involved in a direct interaction with the transmembrane domain of NK1R but rather facing toward the open extracellular space. This orientation explains the relative ease of replacing this residue of the agonist with different amino acids or labeling it with radioactive tracers, as was done in earlier pharmacological studies (49, 50).
Interactions of the N terminus of SP
In the N-terminal portion (residues R1-Q6), the binding of SP to NK1R is governed by several interactions of the receptor with the peptide backbone of the ligand (Fig. 2F and fig. S9). The residue side chains in this region of the agonist are not resolved (except for Q6, which packs against ECL3), likely due to higher flexibility in the receptor:ligand complex (Fig. 2B). Specifically, the backbone carbonyl oxygen of Y278ECL3 engages in a hydrogen bond to the backbone nitrogen of F7 of the ligand. Furthermore, R177ECL2, located at the tip of the β-turn in ECL2, establishes two hydrogen bonds to the backbone carbonyls of Q6 and P4 of SP. R177ECL2 is also within hydrogen-bonding distance of N962.68 at the start of ECL1 and N23N-term. Through its interaction with N23N-term, R177ECL2 helps to position the receptor N terminus from the tip of helix I toward the peptide agonist, enabling the formation of two additional hydrogen bonds of Q24N-term with the backbone carbonyl of Q5 of SP and with the backbone carbonyl of L279ECL3. Hence, R177ECL2 acts as a central residue, which engages a polar network that links all ECLs with the receptor N terminus, thereby shaping the entire extracellular surface for ligand binding (Fig. 2, C and F, and fig. S9). The importance of this polar network for SP binding to NK1R is exemplified by the 7- to 46-fold loss in ligand binding affinity upon mutation of either of the involved residues to alanine, with mutation of the central R177ECL2 showing the most drastic effect. To exclude that the interactions of R177ECL2 solely emerge from its positive charge, we furthermore substituted R177ECL2 with lysine, resulting in a 60-fold reduced binding affinity.
The preorientation of the NK1R N terminus through this polar network enables direct interaction of F25N-term with F7 of the ligand (Fig. 2E), a residue conserved in the neurokinin peptide family (Fig. 2B), thereby introducing a pronounced kink in the receptor N terminus (Fig. 2F). The importance of this specific interaction for SP binding is underlined by the 16-fold reduced binding affinity observed for the mutation F25N-termA, removing the hydrophobic moiety, while the mutation F25N-termY retaining the aromatic properties resulted only in a subtle change in binding affinity (Fig. 2E). Together, the participation of N23N-term and Q24N-term in the observed polar network at the extracellular receptor surface and the direct interaction between F25N-term and the conserved F7 of the peptide agonist provide a structural basis for the previously described importance of the receptor N terminus for ligand binding (42).
Implications on ligand subtype selectivity
As early as the late 1970s, a message-address hypothesis was proposed for several neuropeptide receptors (29, 51, 52), which broadly divides the endogenous neurokinin peptide agonists in two functionally distinct domains: a “message” domain responsible for receptor activation and an “address” domain establishing NKR subtype selectivity. The observation of numerous well-resolved interactions of the hydrophobic C terminus of SP with the core of the receptor and the observed effects of mutations are consistent with a major contribution of the neurokinin family–conserved peptide C terminus to the overall binding energy.
Unexpectedly, no defined short-range interactions between side chains of the ligand and NK1R are observed for the charged N terminus of SP, which is divergent in both length and residue composition between the different neurokinin peptides. Instead, it appears that binding interactions are predominantly established through interactions of NK1R with the peptide backbone of SP (Fig. 2F and fig. S9). This finding implies that there are several more subtle components contributing to the preference of SP binding to NK1R over NKA and NKB. The N terminus of SP has a double-positive charge in this region, while NKA is zwitterionic and NKB carries a double-negative charge (Fig. 2B). These differences in charge distribution of the neurokinin peptides favor interactions between the positively charged N terminus of SP and the negatively charged extracellular receptor surface formed by the solvent-exposed residues E172ECL2, E183ECL2, E186ECL2, and D276ECL3 (fig. S10). Notably, mutation of E172ECL2, which is found closest to R1-K3 of SP, to a positively charged lysine, as found in NK3R, increases affinity of NKB, presumably because of the interaction with the NKB N-terminal D1 (corresponding to P2 for SP) (41, 53). Furthermore, the presence of two prolines in SP may alter the preferred conformers of the flexible N terminus, compared to NKA and NKB. Therefore, the subtype specificity does not seem to be achieved through specific short-range side-chain interactions but rather through a mechanism of electrostatic and conformational selection of the N terminus.
Last, the polar network established by residue R177ECL2 points toward an additional layer of complexity to subtype-specific ligand binding in NKRs: The receptor residues involved in establishing this polar network, which structures the entire extracellular receptor surface for ligand binding, are only partially conserved between NK1R, NK2R, and NK3R (Fig. 2C). Because of the geometry of the observed interactions, the exact identity of all of these residues is pivotal to this network, and hence, it can only be established in this form by NK1R, but not by its close relatives NK2R and NK3R.
Activation of NK1R
The comparison of the active, G protein–bound NK1R:SP complexes with the inactive-state structure of NK1R in complex with the antagonist CP-99,994 (19) enables us to identify the molecular changes underlying receptor activation. In the canonical activation mechanism of class A receptors, binding of the agonist to the orthosteric pocket of the receptor induces a cascade of conformational changes in several conserved motifs and culminates in a pronounced outward swing of helix VI to accommodate the C-terminal Cα5 helix of the G protein (54). Unexpectedly, NK1R adopts most but not all of the canonical rearrangements observed for active-state class A GPCRs, and we use the closely related neuropeptide orexin-B receptor (OX2R) to exemplify the specific differences (Fig. 3) (55).
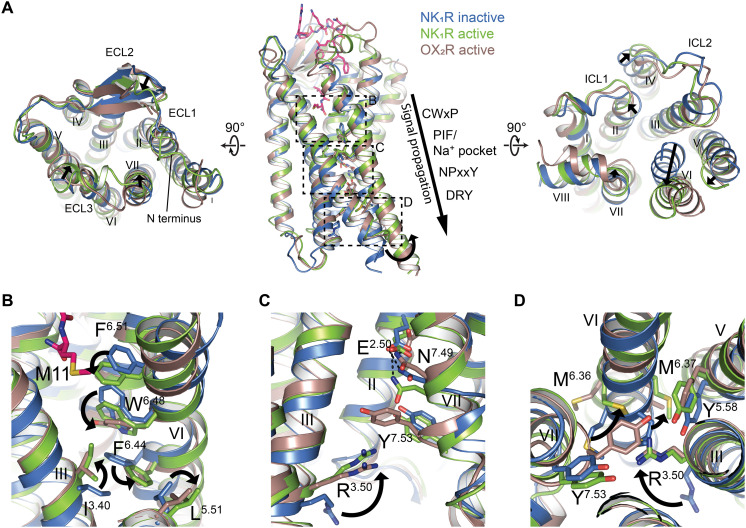
Fig. 3. Molecular characteristics of NK1R activation by SP.
(A) Superposition of the active-state NK1R:SP:Gq (green) with the inactive-state crystal structure of NK1R in complex with the antagonist CP-99,994 (PDB ID: 6HLL) (blue) and the active-state OX2R:Gq complex (PDB ID: 7L1U) (brown). Relative changes from inactive to active NK1R are indicated by black arrows. Left: extracellular view, right: intracellular view, middle: side view with approximate position of conserved motifs involved in receptor activation indicated, and sections of the close-up views in (B) to (D) highlighted by dotted boxes. (B to D) Close-up views on conserved motifs involved in receptor activation. Repositioning of side chains from the inactive to the active state are indicated by arrows.
At the extracellular receptor portion, SP binding causes a contraction of the orthosteric pocket through an inward movement of helices VI and VII, and ECL3 toward the central axis of the receptor (Fig. 3A). These rearrangements reposition F2646.51 at the bottom of the orthosteric pocket to trigger the transmission switch W2616.48, leading to the canonical outward move of F2576.44 of the PIF motif together with L2095.51 and an inward repositioning of I1203.40 (Fig. 3B). On the same horizontal plane through the receptor, an extensive water-mediated hydrogen bonding network, clustered around a central sodium ion that links helices II, III, VI, and VII, was described to stabilize class A GPCRs in the inactive state, involving interactions with the highly (98% of class A GPCRs) conserved D2.50 (56). Upon receptor activation, this sodium ion is expelled, and the conserved D2.50 is repositioned to establish a direct hydrogen bond to N7.49 of the NPxxY motif in helix VII (57). NK1R, however, has a glutamate E782.50 at the position of the conserved D2.50, a sequence variation that has been linked to the unusual lack of constitutive signaling of NK1R in functional studies (23). In the inactive state of the antagonist CP-99,994–bound receptor, the longer side chain of E782.50 enables the establishment of a direct hydrogen bond to N3017.49 without the need of a bridging sodium ion. In the active-state structure, we now find that the entire helix VII is pushed downward and toward the intracellular space owing to the longer side chain of E782.50 (Fig. 3C).
This NK1R-specific translocation restricts the canonical inward shift of helix VII at the intracellular receptor surface upon receptor activation and prevents Y3057.53 of the conserved NPxxY motif from adopting the expected inward-facing orientation toward R1303.50 of the DRY motif (Fig. 3, C and D) (58). As a consequence, the outward swing of helix VI is less pronounced in NK1R, and M2496.36 is rotated further into the receptor core instead of adopting an outward-facing orientation, while the ratchet-like rearrangement of M2506.37 enables the canonical packing against Y2435.58 (Fig. 3D). In summary, NK1R adopts an active conformation with smaller rearrangements of the intracellular tips of helices VI and VII, thus presenting a distinctly shaped intracellular binding surface for the C-terminal Cα5 helix of the Gα protein (Fig. 4, B and C).
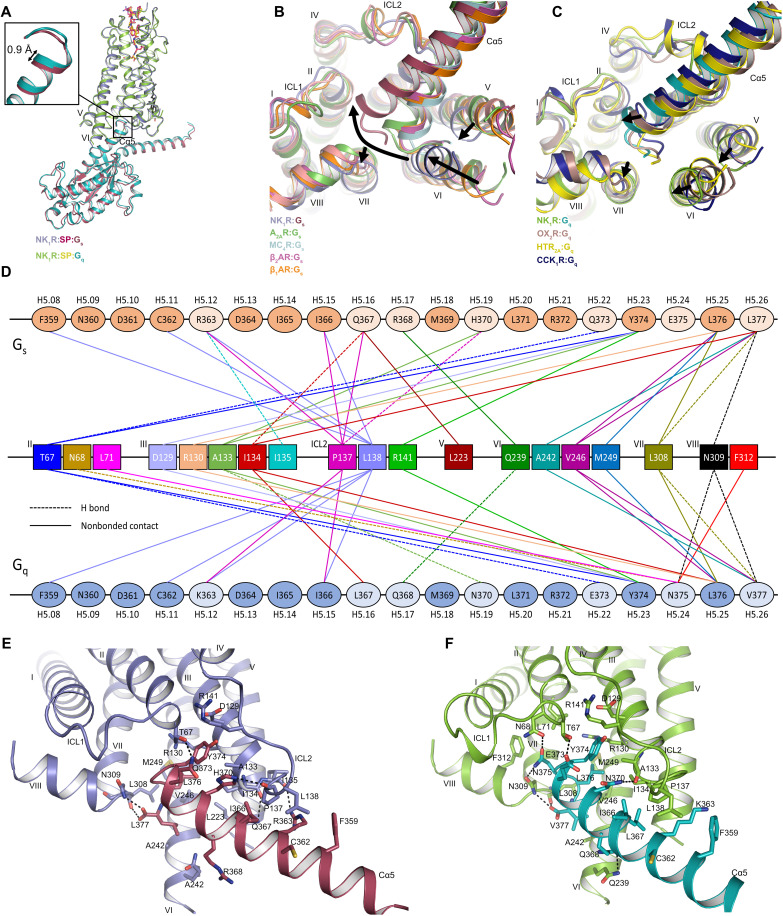
Fig. 4. Interactions of NK1R with Gαs and Gαq.
(A) Structural superposition of NK1R:Gs and NK1R:Gq with a close-up of the C-terminal tip of Cα5, highlighting the 0.9 Å deeper binding of Gαq compared to Gαs. Gβ1γ2 and scFv16 are omitted for clarity. (B) Structural superposition of NK1R:Gs with class A GPCR:Gs complexes viewed from the cytoplasmic side, highlighting the distinct Cα5 binding angle in NK1R and differing positions of helices V, VI, and VII. A2A:Gs (PDB ID: 6GDG), MC4R:Gs (PDB ID: 7AUE), β2AR:Gs (PDB ID: 7BZ2), and β1AR:Gs (PDB ID: 7JJO). The arrows indicate the relative helical position from the group of class A GPCR:Gs structures toward NK1R:Gs. (C) Structural superposition of NK1R:Gq with class A GPCR:Gq shown from the cytoplasmic side highlighting deeper insertion position of the Cα5 the NK1R cavity and differing positions of helices V, VI, and VII. OX2R:Gq (PDB ID: 7L1U), HTR2A:Gq (PDB ID: 6WHA), and CCK1R:Gq (PDB ID: 7MBY). The arrows indicate the relative helical position from the group of class A GPCR:Gq structures toward NK1R:Gq. (D) Schematic drawing of direct contacts between NK1R and the Cα5 helix of both Gαs and Gαq subunits with H-bonded and hydrophobic contacts indicated by a dashed and solid line, respectively. Residues differing between Cα5 of Gαs and Gαq are indicated by a lighter color. G protein residues are labeled according to CGN numbering beneath each residue. (E and F) Interactions of Cα5 of Gαs (E) and Gαq (F) with NK1R. Hydrogen bonds are indicated by black, dotted lines.
Coupling of NK1R to different G proteins
Promiscuity for different G proteins has been described for many GPCRs including NK1R, which mainly signals through the Gs- and Gq-coupled pathways (59). Because of the lack of structural information on the signaling-active complexes of NK1R with either of the two G proteins, the molecular details of receptor–G protein interaction and the differences between Gs- and Gq-bound NK1R have remained unknown. Our structures of SP-bound NK1R in complex with both Gs and Gq now reveal detailed information on these interactions.
Overall, in both resolved complexes, Gs and Gq adopt a highly similar conformation relative to NK1R, where the C-terminal Cα5 of Gq is shifted 0.9 Å deeper into the intracellular crevice created by the activated receptor (Fig. 4A). Unexpectedly, in the NK1R:Gs complex, we observe a relative conformation of the two interacting proteins toward each other that is highly distinct from those observed in other class A GPCR:Gs complex structures (Fig. 4B). In the NK1R complex, the tip of Cα5 of Gs is oriented further away from helices V and VI and more toward helix II, when compared to previously reported Gs complexes: melanocortin 4 receptor (MC4R) (60), A2AR (61), β1-adrenergic receptor (β1AR), and β2-adrenergic receptor (β2AR) (62, 63). The distinct orientation of Cα5 of Gs in NK1R can be attributed to the less-pronounced outward swing of helix VI that is stabilized through the reduced inward movement of helix VII and the movement of helix V toward helix VI. Thus, the NK1R:Gs intracellular helical movements more closely resemble those generally observed for Gq complexes, including the NK1R:Gq structure we determined (Fig. 4C). These observations are consistent with the ability of the receptor to couple to both Gq and Gs.
To date, only few structures of class A GPCRs in complex with Gq have been determined, including those of the CCK1R (38), OX2R (55), and 5-hydroxytryptamine receptor 2A (HTR2A) (64). Common to all these complexes is a less-pronounced outward swing of the intracellular tip of helix VI, compared to Gs complexes, along with a Cα5 positioning closer to helix II. Although overall very similar, Cα5 in NK1R:Gq is bound more deeply in the receptor cavity than in the other Gq-bound complex structures. This deeper binding mode is enabled by the NK1R-specific reduced inward shift of helix VII and the shift of helices V and VI toward helix VII (Fig. 4C). In summary, the NK1R-specific intracellular positioning of transmembrane helices V, VI, and VII, resulting from the unexpected inactive-state orientation of Y3057.53 in the conserved NPxxY motif, translates into distinct Cα5 positions in both active-state complexes of NK1R:Gs and NK1R:Gq.
G protein interactions in GPCRs are mainly governed by contacts established through the C-terminal α5 helix of the Gα protein (Fig. 4, D to F, and tables S2 and S3). Owing to the similar overall conformation of NK1R:Gs and NK1R:Gq, the interactions at the contact surface of receptor and G protein are also closely related, whereas the differences are due to the differing residues between Gs and Gq within the respective Cα5 (Fig. 4D). Furthermore, Gq engages in a larger number of hydrogen-bonding interactions (seven in Gq compared to six in Gs) with NK1R due to the deeper binding of Cα5 in the crevice formed by the receptor.
In NK1R:Gs, G protein coupling involves contacts with receptor transmembrane helices II, III, V, VI, and VII; helix VIII; and ICL2, with most contacts in Gs being established by residues L376H5.25, L377H5.26, and Y374H5.23 [common Gα numbering for G proteins indicated in superscript (65)]. Enabled by the distinct orientation of Cα5, NK1R establishes an additional hydrogen-bonding contact between T672.39 at the intracellular tip of transmembrane helix II and Q373H5.22 of the G protein, which had not been observed in previous Gs complexes (Fig. 4E). A similar interaction, also not observed in previous GPCR complexes with Gq, is established between T672.39 and E373H5.22 of Gq (Fig. 4F). In this region of the receptor, we furthermore find two hydrogen bonds of N375H5.24 with N682.40 and N3098.47 and, accordingly, a central role of N375H5.24 in Gq binding, as demonstrated for HTR2A (64). Together, the deeper binding of Cα5 of Gq in NK1R, compared to Gs, and the larger number of interactions may provide a structural rationale for the reported preference of NK1R signaling via Gq over Gs (30).
SP binding is incompatible with the insurmountable antagonist-bound NK1R conformation
The clinically used NK1R antagonists aprepitant and netupitant behave as pseudo-irreversible antagonists, establishing long-lasting inhibition of receptor activation by SP despite their competitive binding to the orthosteric pocket (fig. S11) (21). This so-called insurmountable antagonism and the markedly prolonged in vivo efficacy of these drugs could only in part be explained by slower dissociation kinetics alone.
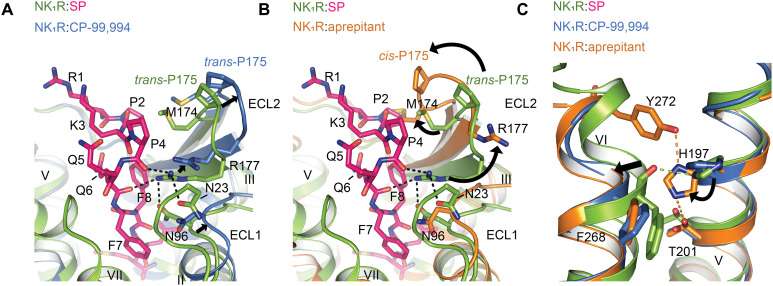
In our previous structural studies of NK1R, we found a distinct conformation of NK1R when in complex with aprepitant and netupitant, compared to the tight-binding, but surmountable progenitor antagonist CP-99,994 (19), and this was backed by clear evidence in the electron density. On the basis of this structural evidence, we were able to pinpoint a hydrogen-bonding network, cross-linking the extracellular tips of transmembrane helices V and VI as a molecular determinant of insurmountable antagonism. Furthermore, in the insurmountable antagonist-bound structures, we observed a cis configuration of the peptide bond preceding P175ECL2 at the tip of the β-hairpin loop of ECL2, which was absent in the CP-99,994–bound state of the surmountable antagonist. Comparison of the active-state NK1R:SP structures with the inactive-state crystal structures of NK1R:aprepitant and NK1R:CP-99,994 now provides evidence for the insurmountable antagonist–induced state to directly interfere with SP binding.
The active-state NK1R adopts a conformation more closely related to the surmountable antagonist complex with CP-99,994 with a root mean square deviation (RMSD) over all atoms of 1.65 Å and 1.77 Å, compared to NK1R:CP-99,994 and NK1R:aprepitant (insurmountable), respectively. Looking more closely at the extracellular portion of the receptor, the most dominant differences between the active state and both of the antagonist-bound structures are rearrangements of ECL2 together with ECL1 and the inward shift of the tip of helices VI and VII due to direct contacts of the agonist SP with residues from these helices (Fig. 5, A and B). However, the rearrangements of ECL2 are different for the two types of antagonists.
Fig. 5. The insurmountable antagonist-bound NK1R state prohibits SP binding.
(A) Superposition of agonist-bound NK1R:SP:Gq (green) with the surmountable antagonist CP-99,994–bound NK1R (PDB ID: 6HLL) (blue) reveals a slightly outward-moved but similar ECL2 conformation for the antagonist conformation. Note the small movement of R177 ECL2. (B) Superposition of agonist-bound NK1R:SP:Gq with the insurmountable antagonist aprepitant-bound NK1R (PDB ID: 6HLO) reveals an inward shifted and twisted ECL2 in the insurmountable conformation that clashes at M174ECL2 with P4 of the endogenous agonist and also poses R177ECL2 away from its central position in the polar network at the extracellular receptor surface. (C) Superposition of agonist-bound NK1R:SP:Gq, CP-99,994- and aprepitant-bound NK1R showing the hydrogen bonds (indicated as orange, dotted lines) that lock helices V and VI in the insurmountable conformation. The rotamer orientation of H1975.39 in the insurmountable conformation prevents the necessary inward movement of helix VI and the formation of a new hydrogen bond between H1975.39 with the backbone carbonyl oxygen of F2686.55 (indicated as green, dotted line) in the active state.
In the surmountable antagonist CP-99,994–bound NK1R, ECL2 adopts a conformation similar to what we now revealed for the active state receptor yet with ECL2 positioned farther away from the central axis of the receptor (Fig. 5A). In contrast, in the insurmountable antagonist-bound NK1R structures, the cis configuration of the peptide bond preceding P175ECL2 causes a twist of the loop toward the central axis of the receptor, where the agonist is located in the SP-bound structures (Fig. 5B). Thus, in addition to an inward shift of the entire ECL2, the cis configuration of P175ECL2 positions M174ECL2 in an orientation where its side chain clashes with P4 of SP and thus directly interferes with agonist binding.
Potentially even more important, the cis configuration of P175ECL2 orients R177ECL2 away from the ligand-binding interface, thereby preventing this residue from establishing its central role in the hydrophilic network connecting ECL1, ECL2, and ECL3 and the receptor N terminus for optimal agonist binding (Fig. 5B). The direct clash of M174ECL2 with SP and the prevention of R177ECL2 to engage in shaping the entire extracellular receptor surface necessary for agonist binding provide an additional structural rationale for the long-lasting effects of insurmountable antagonists on NK1R.
Furthermore, in the active-state structures, SP establishes contacts to F2646.51 and F2686.55 of transmembrane helix VI. The associated inward shift of helix VI enables the formation of a hydrogen bond between the backbone carbonyl oxygen of F2686.55 and H1975.39, thus further stabilizing the active conformation (Fig. 5C). However, H1975.39 is the central residue of a hydrogen-bonding network cross-linking helices V and VI in the insurmountable antagonist-bound NK1R crystal structures (19). In the structural superposition, we now find that the orientation of H1975.39 in the insurmountable conformation interferes with the inward repositioning of helix VI in the active conformation (C-C distance of H1975.39 and F2686.55 backbone of 2 Å) (Fig. 5C). The formation of the hydrogen-bonding network between Y2726.59, H1975.39, and T2055.43 in the insurmountable antagonist-bound conformation may thus directly prevent the receptor from adopting an active conformation by sterically preventing the necessary translocation of helix VI. In summary, SP binding to NK1R is precluded in the insurmountable antagonist-bound receptor conformation by direct steric incompatibility of ECL2 and helix VI with agonist binding as well as by preventing the formation of a critical polar network established by R177ECL2 of ECL2.
DISCUSSION
The neurokinin family has served as a model system in neuropeptide research since its discovery more than half a century ago, and many of the breakthroughs in this field have been associated with SP, the main endogenous agonist of NK1R (1, 2). Already early studies on NK1R have been hampered by the promiscuity of NK1R toward ligand interaction with NKA and NKB, the cognate agonists of NK2R and NK3R, and the coupling of NK1R to different G proteins for intracellular signal transmission, and these promiscuities have therefore subsequently received much attention (28, 30).
Furthermore, NK1R has been the subject of intense high-throughput screening and medicinal chemistry campaigns over the course of several decades, resulting in marketed drugs against CINV (66). While we recently found the molecular determinants for the efficiency of the marketed drugs aprepitant and netupitant in inactive-state NK1R crystal structures by comparing them with a precursor compound (19), we could not fully describe the mechanism of preventing SP binding in the absence of structural information of the agonist-bound active state of the receptor. Our active-state complexes have allowed us now to advance the understanding of this receptor system in several major aspects: (i) the unexpected mechanism of agonist preference for SP, (ii) the unusual geometry of interaction with Gs and the ensuing Gq preference, (iii) the mechanism of eliminating basal activity, and (iv) the insurmountable antagonism of some clinically used drugs.
Our structures of SP-bound NK1R in complex with Gq and Gs reveal the interactions of the endogenous agonist with its receptor in great detail. While the conserved amidated C terminus binds deep into the pocket in a well-defined orientation to trigger activation, the N terminus engages a polar network, shaping the entire extracellular receptor surface for ligand binding. In the polar N terminus of the ligand, which highly deviates between neurokinin family endogenous agonists SP, NKA, and NKB in both length and residue composition, stable binding is established through defined direct interactions of NK1R with the peptide backbone of SP but not with any of its side chains. Thus, subtype specificity does not seem to be achieved through stable short-range side-chain interactions but electrostatics and conformational fine-tuning of the N terminus. Furthermore, the residues of the extracellular polar network, which establishes the structural framework for binding of the agonist N terminus in NK1R, are not conserved among the three NKR subtypes, implying a distinct ligand interaction mode of NK2R and NK3R. Therefore, for a detailed understanding of the determinants for neurokinin family subtype specificity, further studies including NK2R, NK3R, and their endogenous ligands NKA and NKB are needed.
Only very recently have structures of the same GPCR in complex with several different G proteins become available (61, 67). Similar to CCK1R, we cannot observe notable differences in ligand binding depending on the identity of the coupled G protein. In contrast to all other Gs-bound structures, including CCK1R, NK1R adopts a distinct Cα5 orientation resembling more the conformation that is generally observed in Gq complexes. We find that in the NK1R:Gq complex, Cα5 binds deeper into the intracellular receptor crevice than in any of the previously reported Gq complexes, which is consistent with the preference of Gq over Gs.
Recently, our inactive-state NK1R crystal structures in complex with antagonists of different clinical development stages have allowed us to provide a structural rationale for two peculiarities of NK1R: the reported lack of basal signaling in NK1R and the molecular determinants of insurmountable antagonism (23, 50). The determined active-state structures allowed us to now extend and refine our previous hypotheses.
First, the unusual D782.50E substitution in the otherwise highly conserved allosteric sodium ion binding site enables the establishment of a direct hydrogen bond to N3017.49 in the inactive state. This prevents the canonical rearrangements in the conserved NPxxY motif upon receptor activation, thereby restricting the intracellular outward movement of helix VI, all in all preventing fluctuations, which would allow basal signaling.
Second, we find that the insurmountable antagonist-bound NK1R conformation directly interferes with SP binding through steric clashes of the agonist with ECL2 and the prevention of the inward movement of the extracellular tip of helix VI necessary for receptor activation. A key component is the prevention of the formation of a critical polar network established by R177ECL2 of ECL2. The fact that this conformation of ECL2 contains a cis proline and results in a series of interdependent conformational changes, as described above, makes it plausible that it is not “surmountable” by adding agonist. Quite in contrast, in the surmountable antagonist complex structure, the ECL2 is merely positioned further away from the central axis, but otherwise found in a conformation similar to the one in the active-state receptor. In summary, our structures provide important insights into agonist preferences, antagonist mechanisms, lack of basal activity, and G protein promiscuity in the NKR family.
METHODS
Constructs
The sequences of scFv16 (35) and of wild-type human NK1R (C-terminally truncated after residue 335) were cloned into a modified pFL vector (MultiBac system, Geneva Biotech), each resulting in an expression construct with a melittin signal sequence, followed by a FLAG-tag, a His10-tag, and a human rhinovirus 3C protease cleavage site N-terminal to the gene of interest. To generate the Gαs and Gαq subunits, the N-terminal 20 amino acids of the engineered mini-Gs399 and mini-Gsq70 were replaced by the 18 N-terminal amino acids of Gi1 (34, 35). Each engineered Gα protein sequence was, together with Gβ1 including an N-terminal noncleavable His10-tag and Gγ2 (not containing any tags), cloned into one pFL vector, each gene under the control of its own polyhedrin promoter.
Expression in insect cells
Recombinant baculovirus was generated using the MultiBac expression system. Virus production and protein expression were performed as previously described (19). The expression cassettes were integrated into the DH10EMBacY baculovirus genome, and the resulting bacmids were transfected into Sf9 cells in six-well tissue culture plates (2 ml, density of 4 × 105 cells/ml) using 8 μl of Cellfectin II Reagent (Thermo Fisher Scientific) and Sf-900 II serum-free medium (Thermo Fisher Scientific). Viral P0 stocks were harvested as the supernatant after 5 days and were subsequently amplified to obtain high-titer viral P1 stocks. For expression, 4 to 5 liters of Sf9 insect cells in Sf-900 II SFM medium were infected with P1 virus at a cell density of 3 × 106 cells/ml and a multiplicity of infection of 5. Expression was performed for 72 hours at 27°C under constant shaking. Cells expressing NK1R or G proteins were harvested by centrifugation, washed with phosphate-buffered saline (MilliporeSigma), frozen in liquid nitrogen, and stored at −80°C. Cells secreting scFv16 were separated from the medium by centrifugation immediately before purification.
Purification of human NK1R
Insect cells expressing the human NK1R construct were lysed, and receptor-containing membranes were isolated by repeated Dounce homogenization in hypotonic buffer {10 mM Hepes (pH 7.5), 20 mM KCl, 10 mM MgCl2, and protease inhibitors [Pefabloc SC (50 μg/ml; Carl Roth) and Pepstatin A (1 μg/ml; Carl Roth)]} and hypertonic buffer [10 mM Hepes (pH 7.5), 20 mM KCl, 10 mM MgCl2, 1.0 M NaCl, and protease inhibitors] in the presence of deoxyribonuclease (DNase, MilliporeSigma). Purified membranes were resuspended in hypotonic buffer, frozen in liquid nitrogen, and stored at −80°C until further use.
Frozen membranes were thawed on ice, CP-99,994 (Tocris) was added to a final concentration of 80 μM to maximize solubilization yield, and the suspension was incubated for 30 min while turning on a wheel. Afterward, iodoacetamide (2 mg/ml; MilliporeSigma) was added to the solution followed by another 30 min of incubation. Subsequently, the receptor was solubilized in solubilization buffer [30 mM Hepes (pH 7.5), 500 mM NaCl, 10 mM KCl, 5 mM MgCl2, 40 μM CP-99,994, 1% (w/v) n-dodecyl-β-d-maltopyranoside (DDM, Anatrace), and 0.2% (w/v) cholesteryl hemisuccinate (CHS, MilliporeSigma)] for 3 hours at 4°C. Insoluble material was removed by ultracentrifugation, and the supernatant was incubated with TALON IMAC resin (Cytiva) at 4°C overnight to bind the receptor to the resin.
The receptor-bound resin was washed with 20 column volumes (CVs) of wash buffer I [50 mM Hepes (pH 7.5), 500 mM NaCl, 10 mM MgCl2, 5 mM imidazole, 10% (v/v) glycerol, and 8 mM adenosine triphosphate (ATP, MilliporeSigma)], stepwise reducing the concentration of detergent and CP-99,994 from 1% (w/v) DDM, 0.2% (w/v) CHS, and 20 μM antagonist to 0.5% (w/v) DDM, 0.01% (w/v) CHS, and 10 μM antagonist, respectively. Subsequently, the detergent was exchanged with 16 CVs of wash buffer I supplemented with 1% (w/v) lauryl maltose neopentyl glycol (LMNG, Anatrace), 0.1% (w/v) CHS, and 10 μM CP-99,994, and the antagonist was replaced with SP (WuXi STA) with another 16 CVs of wash buffer I supplemented with 0.5% (w/v) LMNG, 0.05% (w/v) CHS, and 100 μM SP. Agonist-bound NK1R was finally washed with 25 CVs of wash buffer II [50 mM Hepes (pH 7.5), 500 mM NaCl, 15 mM imidazole, 10% (v/v) glycerol, 0.05% (w/v) LMNG, 0.005% (w/v) CHS, and 50 μM SP] and eventually eluted stepwise with 4 CVs of elution buffer [50 mM Hepes (pH 7.5), 500 mM NaCl, 250 mM imidazole, 10% (v/v) glycerol, 0.01% (w/v) LMNG, 0.001% (w/v) CHS, and 50 μM SP].
Protein-containing fractions were concentrated to 0.5 ml using a 50-kDa molecular weight cutoff (MWCO) Vivaspin 2 concentrator (Sartorius Stedim) and added to a PD MiniTrap G-25 column (Cytiva) equilibrated with G25 buffer [50 mM Hepes (pH 7.5), 150 mM NaCl, 10% (v/v) glycerol, 0.01% (w/v) LMNG, 0.001% (w/v) CHS, and 50 μM SP] to remove imidazole. The complex was treated overnight with His-tagged 3C protease and peptide N-glycosidase (PNGase) F (both prepared in-house) to remove the N-terminal affinity tags and to deglycosylate the receptor. After incubation with Ni-nitrilotriacetic acid (Ni-NTA) resin (Cytiva) for 1 hour, cleaved receptor was collected as the flow-through, then concentrated to ~3 to 5 mg/ml with a 50-kDa MWCO Vivaspin 2 concentrator, frozen in liquid nitrogen, and stored at – 80°C until further use. Protein concentrations were determined by absorbance at 280 nm on a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific). Protein purity and monodispersity were assessed by LDS–polyacrylamide gel electrophoresis and analytical size exclusion chromatography (SEC) using a Nanofilm SEC-250 column (Sepax).
Purification of heterotrimeric G proteins
Purification of the engineered heterotrimeric G proteins was similar to receptor purification. All buffers used were supplemented with 10 μM guanosine diphosphate (MilliporeSigma) and 100 μM tris(2-carboxyethyl)phosphine) (Thermo Fisher Scientific). In contrast to the receptor purification, NaCl concentration never exceeded 150 mM, and all buffers were devoid of any receptor ligand and iodoacetamide. Insect cells expressing the heterotrimeric mini-G proteins were lysed by sonication in hypotonic buffer in the presence of DNase I and separated from debris by centrifugation. G protein–containing membranes were washed by Dounce homogenization in physiological buffer [10 mM Hepes (pH 7.5), 150 mM NaCl, 20 mM KCl, 10 mM MgCl2, and protease inhibitors] and directly solubilized in solubilization buffer at 4°C for 2 hours. Insoluble material was removed by ultracentrifugation, and the supernatant was incubated with TALON IMAC resin at 4°C for 1 hour.
The G protein–bound resin was washed with 20 CVs of wash buffer I, stepwise reducing the concentration of DDM and CHS from 1% (w/v) and 0.2% (w/v) to 0.5% (w/v) and 0.01% (w/v), respectively. Subsequently, the detergent was exchanged with 16 CVs of wash buffer I supplemented with 1% (w/v) LMNG and 0.01% (w/v) CHS, and the detergent concentration was reduced with another 16 CVs of wash buffer I supplemented with 0.5% (w/v) LMNG and 0.05% (w/v) CHS. G proteins were finally washed with 25 CVs of wash buffer II and eluted stepwise with 4 CVs of elution buffer containing 750 mM imidazole. Starting from wash buffer II, all buffers contained 1 mM MgCl2.
Protein-containing fractions were concentrated using a 100-kDa MWCO Vivaspin 2 concentrator and further purified using a Superdex 200 10/300 (Cytiva) column equilibrated with SEC buffer [25 mM Hepes (pH 7.5), 150 mM NaCl, 2% (v/v) glycerol, 0.01% (w/v) LMNG, and 0.001% (w/v) CHS]. Fractions containing monodisperse G protein were collected, concentrated to 7 mg/ml with a 100-kDa MWCO Vivaspin 2 concentrator, frozen in liquid nitrogen, and stored at −80°C until further use.
Purification of scFv16
ScFv16-containing supernatant from expression by secretion from Sf9 cells was pH-adjusted by addition of Hepes (pH 7.5). Metal-chelating agents were quenched by incubation with 1 mM CoCl2 and 5 mM CaCl2 for 1 hour at 22°C. Resulting precipitates were removed by centrifugation, and the filtrated supernatant was loaded onto a Co-loaded HiTrap IMAC HP column (Cytiva). The column was washed with 20 CVs of buffer A [20 mM Hepes (pH 7.5), 150 mM NaCl, 1 mM MgCl2, 4 mM ATP, and 5 mM imidazole] followed by 20 CVs of buffer B [20 mM Hepes (pH 7.5), 150 mM NaCl, and 30 mM imidazole]. The protein was eluted with buffer C [20 mM Hepes (pH 7.5), 150 mM NaCl, and 300 mM imidazole]. Monomeric fractions were pooled, concentrated using a 10-kDa MWCO Amicon Ultra concentrator (Merck), and imidazole was removed by applying the concentrate to a PD-10 desalting column (Cytiva) equilibrated with G25 buffer lacking SP. scFv16 was treated for 3 hours with His-tagged 3C protease and PNGase F for removal of affinity tags and deglycosylation. After incubation with Ni-NTA resin for 1 hour, cleaved scFv was collected as the flow-through, concentrated with a 10-kDa MWCO Amicon Ultra concentrator, and further purified using a Superdex 200 10/300 column equilibrated with SEC buffer. Monomeric fractions were pooled, concentrated to ~6 to 7 mg/ml, flash-frozen in liquid nitrogen, and stored at −80°C until further use.
Complex formation
Purified NK1R, engineered heterotrimeric G proteins, and scFv16 were mixed in a molar ratio of 1:1.2:1.4 in complexation buffer [final concentrations: 25 mM Hepes (pH 7.5), 100 mM NaCl, 2 mM MgCl2, 0.01% (w/v) LMNG, 0.001% (w/v) CHS, 100 μM SP, and 100 μm TCEP]. Apyrase (1.25 U/ml; MilliporeSigma) was added to the mixture, and the coupling reaction was allowed to proceed overnight at 4°C. The formed complex was then isolated by SEC on a Superdex 200 10/300 column equilibrated with complexation buffer with reduced detergent concentrations of 0.001% (w/v) LMNG and 0.0001% (w/v) CHS. Monomeric peak fractions were concentrated to 1 mg/ml for EM studies, using a 100-kDa MWCO Vivaspin Turbo PES concentrator.
Sample preparation for cryo-EM and image acquisition
For cryo-EM grid preparation, 3.5 μl of purified NK1R:SP:Gq were applied on glow-discharged holey carbon gold grids (Quantifoil R1.2/1.3, 300 mesh) and subsequently vitrified using a Vitrobot Mark IV (Thermo Fisher Scientific) operated at 100% humidity and 4°C. Cryo-EM imaging was performed on a Titan Krios G3i (Thermo Fisher Scientific), operated at 300 kV, at a nominal magnification of 130,000 using a K3 direct electron detector (Gatan) in superresolution mode, corresponding to a pixel size of 0.325 Å. A BioQuantum energy filter (Gatan) was operated with an energy slit width of 20 eV. A total of 5291 movies were obtained, with an exposure rate of ~14.8 electrons/Å2 per second and a defocus range of −0.8 to −2.4 μm using automatic data acquisition with EPU (Thermo Fisher Scientific). The total exposure time was 1.79 s with an accumulated dose of ~62.51 electrons/Å2 and a total of 67 frames per micrograph.
The NK1R:SP:Gs complex was imaged with the same conditions and parameters. The only difference was that a total of 6341 movies were recorded with an exposure rate of ~15.03 electrons/Å2 per second for an accumulated dose of ~63.51 electrons/Å2.
Cryo-EM data processing
Using MotionCor2 (68), raw movies of NK1R:SP:Gq were binned once to generate a pixel size of 0.65 Å, motion-corrected, and dose-weighted. Contrast transfer function parameters for corrected micrographs were determined by Gctf (69), implemented in cryoSPARC (version 3.0.1) (70). Subsequent cryo-EM processing steps were all performed in cryoSPARC. Initial particle selection was done using the automated blob picker on 100 micrographs, using a 100 Å minimum and 150 Å maximum particle diameter, to extract a total of 36,000 particles. The extracted particles were subjected to one round of 2D classification split into 200 classes, from which 10 classes were selected and used as a template for automatic particle picking using the template picker. A total of 3,511,990 particles were subsequently extracted from all 5291 micrographs, followed by two rounds of 2D classification split into 200 classes each. The particles from the best classes were then subjected to two rounds of ab initio 3D reconstructions split into three classes for the first round and two classes for the second round. A final dataset of 558,058 particles from the best 3D class was subjected to 3D nonuniform refinement, resulting in a 2.71 Å density map after sharpening, determined by gold standard Fourier shell correlation (FSC) using the 0.143 criterion. Local resolution estimation was performed with cryoSPARC.
The NK1R:SP:Gs complex dataset was subjected to the same processing procedure with the following differences. A total of 4,227,825 particles were extracted from 6341 micrographs. After two rounds of 2D classification, the particles from the selected 2D classes were only subjected to one ab initio 3D reconstruction that was split into six 3D classes. A joined total of 395,052 particles from two of the 3D classes were then subjected to nonuniform refinement, resulting in a 2.87-Å density map after sharpening, determined by gold standard FSC using the 0.143 criterion. Local resolution estimation was performed with cryoSPARC.
Model building and refinement
For NK1R:SP:Gq:scFv16, an initial model was assembled by docking of the individual complex components into the cryo-EM map using the “fit in map” routine in UCSF Chimera (71). The following components were used: NK1R [Protein Data Bank (PDB) ID: 6HLO], Gαs (PDB ID: 6GDG), and Gβ1γ2 and scFv16 (PDB ID: 6OIJ). SP was manually built in Coot (72). The initial model was manually rebuilt in Coot, followed by several rounds of manual real-space refinement in Coot and real-space refinement with the software package Phenix.real_space_refine from Phenix (73), using information from both the processed and raw cryo-EM map. For the NK1R:SP:Gs:scFv16 model, the individual components of the refined NK1R:SP:Gq:scFv16 model were docked into the cryo-EM map using the “fit in map” routine in the USCF Chimera, followed by manual model building and iterative real-space refinement in Coot with the Phenix.real_space_refine routine. The quality of both models was assessed using MolProbity (74) before PDB deposition.
Whole-cell ligand binding analysis
Human embryonic kidney (HEK) 293T/17 cells (American Type Culture Collection) were cultivated in Dulbecco’s modified Eagle’s medium (MilliporeSigma) supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml; MilliporeSigma), and 10% (v/v) fetal calf serum (BioConcept). Cells were maintained at 37°C in a humidified atmosphere of 5% CO2, 95% air. Transient transfections were performed with TransIT-293 (Mirus Bio) according to the manufacturer’s instructions.
Ligand-binding experiments were performed on whole HEK293T cells for comparison of affinities for wild-type and receptor mutants using an homogeneous time-resolved fluorescence binding assay. HEK293T cells were seeded and transfected in poly-l-lysine–coated 384-well plates (Greiner) at a cell density of 104 cells per well. Twenty-four hours after transfection, cells were labeled with 50 nM SNAP-Lumi4-Tb (Cisbio) in assay buffer [20 mM Hepes (pH 7.5), 100 mM NaCl, 3 mM MgCl2, and 0.05% (w/v) bovine serum albumin (MilliporeSigma)] for 1.5 hours at 37°C. Cells were washed four times with ice-cold assay buffer and were then incubated for 1 hour at room temperature in assay buffer containing fluorescently labeled peptide SP-HL488 [SP labeled with HiLyte Fluor 488 at Lys-3 (Anaspec)]. Fluorescence intensities were measured on a Spark fluorescence plate reader (Tecan) with an excitation wavelength of 340 nm and emission wavelengths of 620 nm and 510 nm for Tb3+ and the fluorophore HiLyte Fluor 488, respectively. The ratio of fluorescence resonance energy transfer donor and acceptor fluorescence intensities was calculated (F500 nm/F620 nm). Nonspecific binding was determined in the presence of 125-fold excess of unlabeled SP. Data were analyzed by global fitting to a one-site saturation binding equation with the GraphPad Prism software (version 9.1.0, GraphPad Prism).
Acknowledgments
We thank S. Sorrentino at the Center for Microscopy and Image Analysis at the University of Zurich for helpful discussions in the initial phase of the project, O. Eggenberger for help with protein production and ligand binding assays, and I. Berger, then at the European Molecular Biology Laboratory, for providing us with baculovirus transfer vectors. We also thank the Biomolecular Structure and Mechanism PhD Program of the Life Science Zurich Graduate School.
Funding: This work was supported by the European Research Council (810057-HighResCells to A.P. and O.M.) and Schweizerischer Nationalfonds Grant 31003A_182334 to A.P.
Author contributions: The project was outlined by C.T., J.E., Y.W., and A.P. J.E., Y.W., and J.S. designed the G protein and scFv constructs. C.T., J.E., Y.W., and J.S. established the purification of the G proteins, scFv, and NK1R. C.T., J.E., and Y.W. purified all proteins and prepared the complexes. C.T., J.E., S.V., and Y.W. vitrified the samples on cryo-EM grids. S.V. acquired, processed, and refined cryo-EM density maps. C.T. and J.E. built and refined the structure models. C.T. designed and performed ligand binding experiments. Project management was carried out by C.T., J.E., S.V., Y.W., O.M., and A.P. The manuscript was prepared by C.T., J.E., S.V., Y.W., and A.P. All authors contributed to the final editing and approval of the manuscript.
Competing interests: The authors declare that they have no competing interests:
Data and materials availability: Cryo-EM maps of NK1R:SP:Gq and NK1R:SP:Gs have been deposited in the Electron Microscopy Data Bank under the accession codes EMD-13140 and EMD-13141. The atomic coordinates of the NK1R:SP:Gq and NK1R:SP:Gs models have been deposited in the Protein Data Bank under accession codes 7P00 and 7P02. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Tables S1 to S3
REFERENCES AND NOTES
- 1.Hökfelt T., Pernow B., Wahren J., Substance P: A pioneer amongst neuropeptides. J. Intern. Med. 249, 27–40 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Severini C., Improta G., Falconieri-Erspamer G., Salvadori S., Erspamer V., The tachykinin peptide family. Pharmacol. Rev. 54, 285–322 (2002). [DOI] [PubMed] [Google Scholar]
- 3.von Euler U. S., Gaddum J. H., An unidentified depressor substance in certain tissue extracts. J. Physiol. 72, 74–87 (1931). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinhoff M. S., von Mentzer B., Geppetti P., Pothoulakis C., Bunnett N. W., Tachykinins and their receptors: Contributions to physiological control and the mechanisms of disease. Physiol. Rev. 94, 265–301 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mantyh P., Gates T., Mantyh C., Maggio J., Autoradiographic localization and characterization of tachykinin receptor binding sites in the rat brain and peripheral tissues. J. Neurosci. 9, 258–279 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McLean S., Ganong A., Seeger T., Bryce D., Pratt K., Reynolds L., Siok C., Lowe J., Heym J., Activity and distribution of binding sites in brain of a nonpeptide substance P (NK1) receptor antagonist. Science 251, 437–439 (1991). [DOI] [PubMed] [Google Scholar]
- 7.Buck S. H., Maurin Y., Burks T. F., Yamamura H. I., High-affinity 3H-substance P binding to longitudinal muscle membranes of the guinea pig small intestine. Life Sci. 34, 497–507 (1984). [DOI] [PubMed] [Google Scholar]
- 8.Chu H. W., Kraft M., Krause J. E., Rex M. D., Martin R. J., Substance P and its receptor neurokinin 1 expression in asthmatic airways. J. Allergy Clin. Immunol. 106, 713–722 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Morelli A. E., Sumpter T. L., Rojas-Canales D. M., Bandyopadhyay M., Chen Z., Tkacheva O., Shufesky W. J., Wallace C. T., Watkins S. C., Berger A., Paige C. J., Falo L. D., Larregina A. T., Neurokinin-1 receptor signaling is required for efficient Ca2+ flux in T-cell-receptor-activated T cells. Cell Rep. 30, 3448–3465.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pintér E., Pozsgai G., Hajna Z., Helyes Z., Szolcsányi J., Neuropeptide receptors as potential drug targets in the treatment of inflammatory conditions. Br. J. Clin. Pharmacol. 77, 5–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felipe C. D., Herrero J. F., O'Brien J. A., Palmer J. A., Doyle C. A., Smith A. J. H., Laird J. M. A., Belmonte C., Cervero F., Hunt S. P., Altered nociception, analgesia and aggression in mice lacking the receptor for substance P. Nature 392, 394–397 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Ratti E., Bettica P., Alexander R., Archer G., Carpenter D., Evoniuk G., Gomeni R., Lawson E., Lopez M., Millns H., Rabiner E. A., Trist D., Trower M., Zamuner S., Krishnan R., Fava M., Full central neurokinin-1 receptor blockade is required for efficacy in depression: Evidence from orvepitant clinical studies. J. Psychopharmacol. 27, 424–434 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Ständer S., Siepmann D., Herrgott I., Sunderkötter C., Luger T. A., Targeting the neurokinin receptor 1 with aprepitant: A novel antipruritic strategy. PLOS One 5, e10968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramalho R., Soares R., Couto N., Moreira A., Tachykinin receptors antagonism for asthma: A systematic review. BMC Pulm. Med. 11, 41 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hale J. J., Mills S. G., MacCoss M., Finke P. E., Cascieri M. A., Sadowski S., Ber E., Chicchi G. G., Kurtz M., Metzger J., Eiermann G., Tsou N. N., Tattersall F. D., Rupniak N. M., Williams A. R., Rycroft W., Hargreaves R., MacIntyre D. E., Structural optimization affording 2-(R)-(1-(R)-3, 5-bis(trifluoromethyl)phenylethoxy)-3-(S)-(4-fluoro)phenyl-4- (3-oxo-1,2,4-triazol-5-yl)methylmorpholine, a potent, orally active, long-acting morpholine acetal human NK-1 receptor antagonist. J. Med. Chem. 41, 4607–4614 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann T., Bös M., Stadler H., Schnider P., Hunkeler W., Godel T., Galley G., Ballard T. M., Higgins G. A., Poli S. M., Sleight A. J., Design and synthesis of a novel, achiral class of highly potent and selective, orally active neurokinin-1 receptor antagonists. Bioorg. Med. Chem. Lett. 16, 1362–1365 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Karthaus M., Schiel X., Ruhlmann C. H., Celio L., Neurokinin-1 receptor antagonists: Review of their role for the prevention of chemotherapy-induced nausea and vomiting in adults. Expert Rev. Clin. Pharmacol. 12, 661–680 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Chen S., Lu M., Liu D., Yang L., Yi C., Ma L., Zhang H., Liu Q., Frimurer T. M., Wang M.-W., Schwartz T. W., Stevens R. C., Wu B., Wüthrich K., Zhao Q., Human substance P receptor binding mode of the antagonist drug aprepitant by NMR and crystallography. Nat. Commun. 10, 638 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schöppe J., Ehrenmann J., Klenk C., Rucktooa P., Schütz M., Doré A. S., Plückthun A., Crystal structures of the human neurokinin 1 receptor in complex with clinically used antagonists. Nat. Commun. 10, 17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin J., Chapman K., Clark L. D., Shao Z., Borek D., Xu Q., Wang J., Rosenbaum D. M., Crystal structure of the human NK1 tachykinin receptor. Proc. Natl. Acad. Sci. U.S.A. 115, 13264–13269 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindström E., von Mentzer B., Påhlman I., Ahlstedt I., Uvebrant A., Kristensson E., Martinsson R., Novén A., de Verdier J., Vauquelin G., Neurokinin 1 receptor antagonists: Correlation between in vitro receptor interaction and in vivo efficacy. J. Pharmacol. Exp. Ther. 322, 1286–1293 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Ballesteros J. A., Weinstein H., Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 25, 366–428 (1995). [Google Scholar]
- 23.Valentin-Hansen L., Frimurer T. M., Mokrosinski J., Holliday N. D., Schwartz T. W., Biased Gs versus Gq proteins and β-arrestin signaling in the NK1 receptor determined by interactions in the water hydrogen bond network. J. Biol. Chem. 290, 24495–24508 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakanishi S., Mammalian tachykinin receptors. Annu. Rev. Neurosci. 14, 123–136 (1991). [DOI] [PubMed] [Google Scholar]
- 25.Cascieri M. A., Huang R. R., Fong T. M., Cheung A. H., Sadowski S., Ber E., Strader C. D., Determination of the amino acid residues in substance P conferring selectivity and specificity for the rat neurokinin receptors. Mol. Pharmacol. 41, 1096–1099 (1992). [PubMed] [Google Scholar]
- 26.Otsuka M., Yoshioka K., Neurotransmitter functions of mammalian tachykinins. Physiol. Rev. 73, 229–308 (1993). [DOI] [PubMed] [Google Scholar]
- 27.Severini C., Salvadori S., Guerrini R., Falconieri-Erspamer G., Mignogna G., Erspamer V., Parallel bioassay of 39 tachykinins on 11 smooth muscle preparations. Structure and receptor selectivity/affinity relationship. Peptides 21, 1587–1595 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Yokota Y., Akazawa C., Ohkubo H., Nakanishi S., Delineation of structural domains involved in the subtype specificity of tachykinin receptors through chimeric formation of substance P/substance K receptors. EMBO J. 11, 3585–3591 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwyzer R., Membrane-assisted molecular mechanism of neurokinin receptor subtype selection. EMBO J. 6, 2255–2259 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holst B., Hastrup H., Raffetseder U., Martini L., Schwartz T. W., Two active molecular phenotypes of the tachykinin NK1 receptor revealed by G-protein fusions and mutagenesis. J. Biol. Chem. 276, 19793–19799 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Martini L., Hastrup H., Holst B., Fraile-Ramos A., Marsh M., Schwartz T. W., NK1 receptor fused to β-Arrestin displays a single-component, high-affinity molecular phenotype. Mol. Pharmacol. 62, 30–37 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Sagan S., Chassaing G., Pradier L., Lavielle S., Tachykinin peptides affect differently the second messenger pathways after binding to CHO-expressed human NK-1 receptors. J. Pharmacol. Exp. Ther. 276, 1039–1048 (1996). [PubMed] [Google Scholar]
- 33.Carpenter B., Tate C. G., Engineering a minimal G protein to facilitate crystallisation of G protein-coupled receptors in their active conformation. Protein Eng. Des. Sel. 29, 583–594 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nehmé R., Carpenter B., Singhal A., Strege A., Edwards P. C., White C. F., Du H., Grisshammer R., Tate C. G., Mini-G proteins: Novel tools for studying GPCRs in their active conformation. PLOS ONE 12, e0175642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maeda S., Koehl A., Matile H., Hu H., Hilger D., Schertler G. F. X., Manglik A., Skiniotis G., Dawson R. J. P., Kobilka B. K., Development of an antibody fragment that stabilizes GPCR/G-protein complexes. Nat. Commun. 9, 3712 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanson M. A., Cherezov V., Griffith M. T., Roth C. B., Jaakola V.-P., Chien E. Y. T., Velasquez J., Kuhn P., Stevens R. C., A specific cholesterol binding site is established by the 2.8 Å structure of the human β2-Adrenergic Receptor. Structure 16, 897–905 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monastyrskaya K., Hostettler A., Buergi S., Draeger A., The NK1 receptor localizes to the plasma membrane microdomains, and its activation is dependent on lipid raft integrity. J. Biol. Chem. 280, 7135–7146 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Mobbs J. I., Belousoff M. J., Harikumar K. G., Piper S. J., Xu X., Furness S. G. B., Venugopal H., Christopoulos A., Danev R., Wootten D., Thal D. M., Miller L. J., Sexton P. M., Structures of the human cholecystokinin 1 (CCK1) receptor bound to Gs and Gq mimetic proteins provide insight into mechanisms of G protein selectivity. PLoS Biol. 19, e3001295 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song W., Yen H.-Y., Robinson C. V., Sansom M. S. P., State-dependent lipid interactions with the A2a receptor revealed by MD simulations using in vivo-mimetic membranes. Structure 27, 392–403.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yen H.-Y., Hoi K. K., Liko I., Hedger G., Horrell M. R., Song W., Wu D., Heine P., Warne T., Lee Y., Carpenter B., Plückthun A., Tate C. G., Sansom M. S. P., Robinson C. V., PtdIns(4,5)P2 stabilizes active states of GPCRs and enhances selectivity of G-protein coupling. Nature 559, 423–427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fong T. M., Huang R. R., Strader C. D., Localization of agonist and antagonist binding domains of the human neurokinin-1 receptor. J. Biol. Chem. 267, 25664–25667 (1992). [PubMed] [Google Scholar]
- 42.Fong T. M., Yu H., Huang R. R. C., Strader C. D., The extracellular domain of the neurokinin-1 receptor is required for high-affinity binding of peptides. Biochemistry 31, 11806–11811 (1992). [DOI] [PubMed] [Google Scholar]
- 43.Huang R. R. C., Yu H., Strader C. D., Fong T. M., Interaction of substance P with the second and seventh transmembrane domains of the neurokinin-1 receptor. Biochemistry 33, 3007–3013 (1994). [DOI] [PubMed] [Google Scholar]
- 44.Pellegrini M., Bremer A. A., Ulfers A. L., Boyd N. D., Mierke D. F., Molecular characterization of the substance P·neurokinin-1 receptor complex. J. Biol. Chem. 276, 22862–22867 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Turcatti G., Zoffmann S., Lowe J. A. III, Drozda S. E., Chassaing G., Schwartz T. W., Chollet A., Characterization of non-peptide antagonist and peptide agonist binding sites of the NK1 receptor with fluorescent ligands. J. Biol. Chem. 272, 21167–21175 (1997). [DOI] [PubMed] [Google Scholar]
- 46.Valentin-Hansen L., Park M., Huber T., Grunbeck A., Naganathan S., Schwartz T. W., Sakmar T. P., Mapping substance P binding sites on the neurokinin-1 receptor using genetic incorporation of a photoreactive amino acid. J. Biol. Chem. 289, 18045–18054 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cascieri M. A., Goldenberg M. M., Liang T., Biological activity of substance P methyl ester. Mol. Pharmacol. 20, 457–459 (1981). [PubMed] [Google Scholar]
- 48.Hanley M. R., Lee C. M., Michell R. H., Jones L. M., Similar effects of substance P and related peptides on salivation and on phosphatidylinositol turnover in rat salivary glands. Mol. Pharmacol. 18, 78–83 (1980). [PubMed] [Google Scholar]
- 49.Boyd N. D., Kage R., Dumas J. J., Krause J. E., Leeman S. E., The peptide binding site of the substance P (NK-1) receptor localized by a photoreactive analogue of substance P: Presence of a disulfide bond. Proc. Natl. Acad. Sci. U.S.A. 93, 433–437 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cascieri M. A., Ber E., Fong T. M., Hale J. J., Tang F., Shiao L. L., Mills S. G., MacCoss M., Sadowski S., Tota M. R., Strader C. D., Characterization of the binding and activity of a high affinity, pseudoirreversible morpholino tachykinin NK1 receptor antagonist. Eur. J. Pharmacol. 325, 253–261 (1997). [DOI] [PubMed] [Google Scholar]
- 51.Schwyzer R., ACTH: A short introductory review. Ann. N.Y. Acad. Sci. 297, 3–26 (1977). [DOI] [PubMed] [Google Scholar]
- 52.Schwyzer R., 100 years lock-and-key concept: Are peptide keys shaped and guided to their receptors by the target cell membrane? Biopolymers 37, 5–16 (1995). [DOI] [PubMed] [Google Scholar]
- 53.Huang R. R., Yu H., Strader C. D., Fong T. M., Localization of the ligand binding site of the neurokinin-1 receptor: Interpretation of chimeric mutations and single-residue substitutions. Mol. Pharmacol. 45, 690–695 (1994). [PubMed] [Google Scholar]
- 54.Wang J., Hua T., Liu Z.-J., Structural features of activated GPCR signaling complexes. Curr. Opin. Struct. Biol. 63, 82–89 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Hong C., Byrne N. J., Zamlynny B., Tummala S., Xiao L., Shipman J. M., Partridge A. T., Minnick C., Breslin M. J., Rudd M. T., Stachel S. J., Rada V. L., Kern J. C., Armacost K. A., Hollingsworth S. A., O’Brien J. A., Hall D. L., McDonald T. P., Strickland C., Brooun A., Soisson S. M., Hollenstein K., Structures of active-state orexin receptor 2 rationalize peptide and small-molecule agonist recognition and receptor activation. Nat. Commun. 12, 815 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mirzadegan T., Benkö G., Filipek S., Palczewski K., Sequence analyses of G-protein-coupled receptors: Similarities to rhodopsin. Biochemistry 42, 2759–2767 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gutiérrez-de-Terán H., Massink A., Rodríguez D., Liu W., Han G. W., Joseph J. S., Katritch I., Heitman L. H., Xia L., Ijzerman A. P., Cherezov V., Katritch V., Stevens R. C., The role of a sodium ion binding site in the allosteric modulation of the A2A adenosine G protein-coupled receptor. Structure 21, 2175–2185 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manglik A., Kruse A. C., Structural basis for G protein-coupled receptor activation. Biochemistry 56, 5628–5634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inoue A., Raimondi F., Kadji F. M. N., Singh G., Kishi T., Uwamizu A., Ono Y., Shinjo Y., Ishida S., Arang N., Kawakami K., Gutkind J. S., Aoki J., Russell R. B., Illuminating G-protein-coupling selectivity of GPCRs. Cell 177, 1933–1947.e25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Israeli H., Degtjarik O., Fierro F., Chunilal V., Gill A. K., Roth N. J., Botta J., Prabahar V., Peleg Y., Chan L. F., Ben-Zvi D., McCormick P. J., Niv M. Y., Shalev-Benami M., Structure reveals the activation mechanism of the MC4 receptor to initiate satiation signaling. Science 372, 808–814 (2021). [DOI] [PubMed] [Google Scholar]
- 61.García-Nafría J., Lee Y., Bai X., Carpenter B., Tate C. G., Cryo-EM structure of the adenosine A2A receptor coupled to an engineered heterotrimeric G protein. eLife 7, e35946 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su M., Zhu L., Zhang Y., Paknejad N., Dey R., Huang J., Lee M.-Y., Williams D., Jordan K. D., Eng E. T., Ernst O. P., Meyerson J. R., Hite R. K., Walz T., Liu W., Huang X.-Y., Structural basis of the activation of heterotrimeric Gs-protein by isoproterenol-bound β1-adrenergic receptor. Mol. Cell 80, 59–71.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y., Yang F., Ling S., Lv P., Zhou Y., Fang W., Sun W., Zhang L., Shi P., Tian C., Single-particle cryo-EM structural studies of the β2AR–Gs complex bound with a full agonist formoterol. Cell Discov. 6, 45 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim K., Che T., Panova O., DiBerto J. F., Lyu J., Krumm B. E., Wacker D., Robertson M. J., Seven A. B., Nichols D. E., Shoichet B. K., Skiniotis G., Roth B. L., Structure of a hallucinogen-activated Gq-coupled 5-HT2A serotonin receptor. Cell 182, 1574–1588.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Flock T., Ravarani C. N. J., Sun D., Venkatakrishnan A. J., Kayikci M., Tate C. G., Veprintsev D. B., Babu M. M., Universal allosteric mechanism for Gα activation by GPCRs. Nature 524, 173–179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hargreaves R., Ferreira J. C. A., Hughes D., Brands J., Hale J., Mattson B., Mills S., Development of aprepitant, the first neurokinin-1 receptor antagonist for the prevention of chemotherapy-induced nausea and vomiting. Ann. N.Y. Acad. Sci. 1222, 40–48 (2011). [DOI] [PubMed] [Google Scholar]
- 67.Qiao A., Han S., Li X., Li Z., Zhao P., Dai A., Chang R., Tai L., Tan Q., Chu X., Ma L., Thorsen T. S., Reedtz-Runge S., Yang D., Wang M.-W., Sexton P. M., Wootten D., Sun F., Zhao Q., Wu B., Structural basis of Gs and Gi recognition by the human glucagon receptor. Science 367, 1346–1352 (2020). [DOI] [PubMed] [Google Scholar]
- 68.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 72.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Afonine P. V., Poon B. K., Read R. J., Sobolev O. V., Terwilliger T. C., Urzhumtsev A., Adams P. D., Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Biol. Crystallogr. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Williams C. J., Headd J. J., Moriarty N. W., Prisant M. G., Videau L. L., Deis L. N., Verma V., Keedy D. A., Hintze B. J., Chen V. B., Jain S., Lewis S. M., Arendall W. B. III, Snoeyink J., Adams P. D., Lovell S. C., Richardson J. S., Richardson D. C., MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Tables S1 to S3