High-throughput microscopy has outpaced analysis; biomarker-optimized CNNs are a generalizable, fast, and interpretable solution.
Abstract
Cellular events underlying neurodegenerative disease may be captured by longitudinal live microscopy of neurons. While the advent of robot-assisted microscopy has helped scale such efforts to high-throughput regimes with the statistical power to detect transient events, time-intensive human annotation is required. We addressed this fundamental limitation with biomarker-optimized convolutional neural networks (BO-CNNs): interpretable computer vision models trained directly on biosensor activity. We demonstrate the ability of BO-CNNs to detect cell death, which is typically measured by trained annotators. BO-CNNs detected cell death with superhuman accuracy and speed by learning to identify subcellular morphology associated with cell vitality, despite receiving no explicit supervision to rely on these features. These models also revealed an intranuclear morphology signal that is difficult to spot by eye and had not previously been linked to cell death, but that reliably indicates death. BO-CNNs are broadly useful for analyzing live microscopy and essential for interpreting high-throughput experiments.
INTRODUCTION
Observing dynamic biological processes over time with live fluorescence microscopy has played an essential role in understanding fundamental cell biology. Automating such longitudinal live microscopy experiments with robotics has been especially useful to achieve the throughput necessary for performing massive screens of neuronal physiology and detecting rare or transient changes in cells (1–8). Nevertheless, image analysis is often a rate-limiting step for scientific discovery in live microscopy, and this issue is magnified in high-throughput regimes (9–11). In some cases, such as for the detection of death in longitudinal fluorescence imaging, the standard approach for image analysis is to rely on manual curation, which is often slow and labor intensive, and potentially introduces experimental bias. The impact of automated imaging on scientific discovery will be limited without new approaches that can replace manual image analysis.
A popular alternative to manual curation is semiautomated image analysis, available through open-source platforms such as ImageJ/National Institutes of Health (NIH) (12) and CellProfiler (13). This approach relies on feedback loops between manual curation and basic statistical models to incrementally build better computer vision models for rapidly phenotyping samples (14). However, the resulting models rely on visual features that are highly tuned to the specific experimental conditions they were trained on, meaning that new models must be built for every new experiment. These approaches are also ultimately limited by the precision and accuracy with which human curators can score and annotate images. Variability in curator precision, combined with ambiguities present in biological imaging due to technical artifacts, often leads to a noisy ground truth, limiting the success of attempts toward automation.
The recent explosion of computer vision models leveraging deep learning has led to more accurate, versatile, and efficient algorithms for cellular image analysis (15). Convolutional neural networks (CNNs), in particular, have been responsible for massive improvements over classic computer vision for reconstructing cellular morphology captured by light microscopy (16), automated phenotypic biomarker analysis in live-cell microfluidics (17), subcellular protein localization (18), and fluorescent image segmentation (19). For example, CNNs have been found to be able to classify hematoxylin- and eosin-stained histopathology slides with superhuman accuracy (20) and can sometimes generalize between datasets with minimal or no additional labeled data (21). Remarkably, CNNs have shown promise in detecting visual features within images that humans could not, suggesting that they can be harnessed to drive scientific discovery (22). Nevertheless, a major hurdle in achieving such goals with CNNs is their dependency on large labeled datasets, which require massive amounts of human curation and/or annotation (23). Thus, it is critical to develop methods that generate datasets that are sufficiently large to train CNNs but need minimal human curation.
Cell death is a ubiquitous phenomenon during live imaging that requires substantial attention in image analysis. In some cases, cell death is noise that must be filtered from the signal of live cells, while in other cases cell death is the targeted biological phenotype, for instance, in studies of development (24), cancer (25), aging (26), and disease (27).
In cellular models of neurodegenerative disease, neuronal death is frequently used as a disease-relevant phenotype (28, 29) and its analysis as a tool for the discovery of covariates of neurodegeneration (3, 30, 31). Nevertheless, detection of neuronal death can be challenging, especially in live-imaging studies, due to the dearth of acute biosensors capable of detecting death. A plethora of cell death indicators, dyes, and stains have been developed and described, yet these reagents can introduce artificial toxicity into an experiment, they are often difficult to quantify and display substantial batch-to-batch variability, and they have poor control over false-negative detections as they only report very late stages of death after notable decay has already occurred (29). More recently, we established a novel family of genetically encoded death indicators (GEDIs) that acutely mark a stage at which neurons are irreversibly committed to die (32). Our approach uses a robust signal that standardizes the measurement across experiments and is amenable to high-throughput analysis. However, this approach cannot be applied to cells that do not express the GEDI reporter. Moreover, the GEDI construct emits in two fluorescent channels, which restricts its use in cells coexpressing fluorescent biosensors in overlapping emission spectra.
Here, we addressed the extant issues in detecting neuronal death in microscopy by developing a novel quantitative robotic microscopy pipeline that automatically generates GEDI-curated data to train a CNN without human input. The resulting GEDI-CNN is capable of detecting neuronal death from images of morphology alone, alleviating the need for any additional use of GEDI in subsequent experiments. The GEDI-CNN achieves superhuman accuracy and speed in identifying neuronal cell death in high-throughput live-imaging datasets. Through systematic analysis of a trained GEDI-CNN, we find that it learns to detect death in neurons by locating morphology that has classically been linked to death, despite receiving no explicit supervision toward these features. We also show that this model generalizes to images captured with different parameters or displays of neurons and cell types from different species without additional training, including to studies of neuronal death in cells derived from patients with neurodegenerative disease. These data demonstrate that CNNs, directly optimized with a biomarker signal, are powerful tools for analysis and discovery of biology imaged with robotic live microscopy.
RESULTS
An automated pipeline for optimizing a GEDI-based classifier for neuronal death
In prior work, we found that the GEDI biosensor can detect irreversibly elevated intracellular Ca2+ levels to quantitatively and definitively classify neurons as live or dead (Fig. 1, A to C) (32). In contrast to other live/dead indicators, GEDI is constitutively expressed in cells to quantitatively and irreversibly report death only when a cell has reached a level of calcium not found in live cells. Before the GEDI biosensor, human curation of neuronal morphology was often deployed for detection of death in longitudinal imaging datasets and was considered the state of the art (29, 33). It has been long recognized that the transition from life to death closely tracks with changes in a neuron’s morphology that occur as it degenerates (34). However, human curation based on morphology relies on subjective and ill-defined interpretation of features in an image and thus can be both inaccurate and imprecise (29, 32). Furthermore, in cases where human curation is incorrect, it remains unknown whether images of cell morphology convey enough information to indicate an irreversible commitment to death or whether curators incorrectly identify and weigh features within the dying cells. Because the GEDI biosensor gives a readout of a specific physiological marker of a cell’s live/dead state, it provides superior fidelity in differentiating between live and dead cells (32). We hypothesized that the GEDI biosensor signal could be leveraged to recognize consistent morphological features of live and dying cells and help improve classification in datasets that do not include GEDI labeling. In addition, we sought to further understand how closely morphology features track with death and whether early morphological features of dying cells could be precisely identified and defined. To this end, we optimized CNNs with the GEDI biosensor signal to detect death using the morphology signal alone.
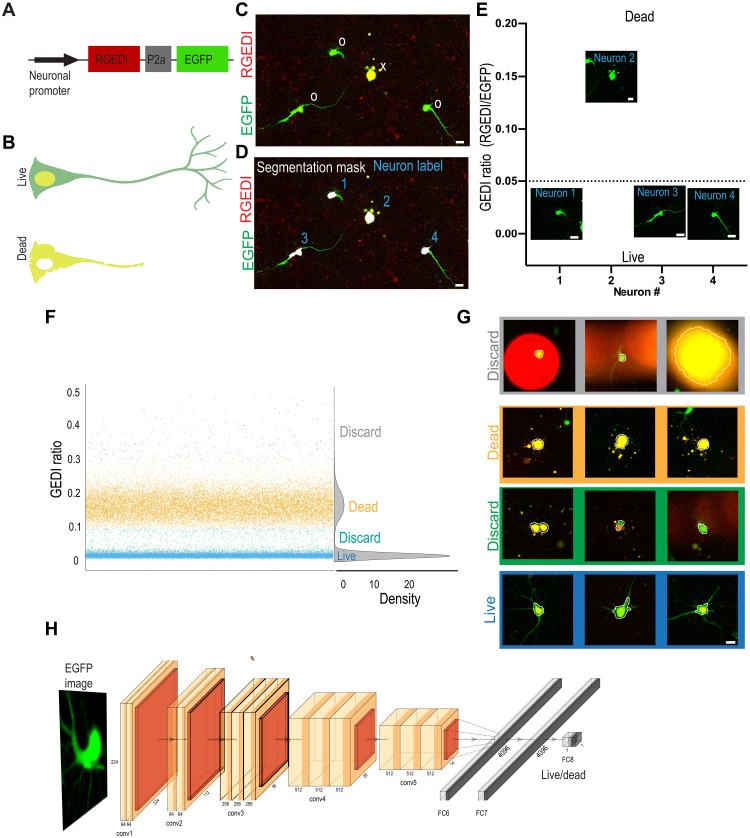
Fig. 1. GEDI signal as a ground truth for training a live/dead classifier CNN from morphology.
(A) GEDI biosensor expression plasmid contains a neuron-specific promoter driving expression of a red fluorescent RGEDI protein, a P2a “cleavable peptide,” and an EGFP protein. Normalizing the RGEDI signal to the EGFP signal (GEDI ratio) at a single-cell level provides a ratiometric measure of a “death” signal that is largely independent of cell-to-cell variation in transfection efficiency and plasmid expression. (B) Schematic overlay of green and red channels illustrating the GEDI sensor’s color change in live neurons (top) and dead neurons (bottom). Live neurons typically contain basal RGEDI signal in the nucleus and in the perinuclear region near intracellular organelles with high Ca2+ (32). (C) Representative red and green channel overlay of neurons expressing GEDI, showing one dead (x) and three live (0) neurons. (D) Segmentation of image in (C) for objects above a specific size and intensity identifies the soma of each neuron (segmentation masks), which is given a unique identifier label (1 to 4). (E) Ratio of RGEDI to EGFP fluorescence (GEDI ratio) in neurons from (D). A ratio above the GEDI threshold (dotted line) indicates an irreversible increase in GEDI signal associated with neuronal death. Cropped EGFP images are plotted at the level of their associated GEDI ratio. (F and G) Generation of GEDI-CNN training datasets from images of individual cells. GEDI ratios from images of each cell (G) were used to create training examples of live and dead cells. Cells with intermediate or extremely high GEDI ratios were discarded to eliminate ambiguity during the training process. Automated cell segmentation boundaries are overlaid in white. (H) Architecture of GEDI-CNN model based off of VGG16 architecture. conv1 to conv5, convolutional layers; FC6 to FC8, fully connected layers (numbers describe the dimensionality of each layer). Scale bar, 20 μm.
Neurons transfected with a GEDI construct coexpress the red fluorescent protein (RGEDI) and enhanced green fluorescent protein (EGFP) (morphology label) in a one-to-one ratio due to their fusion by a porcine teschovirus-1 2a (P2a) “self-cleaving” peptide (Fig. 1, A and B) (32). RGEDI fluorescence increases when the Ca2+ level within a neuron reaches a level indicative of an irreversible commitment to death. We normalize for cell-to-cell variations in the amount of the plasmid transfected by dividing the RGEDI fluorescence by the fluorescence of EGFP, which is constitutively expressed from the same plasmid, to give the GEDI ratio (Fig. 1C and fig. S1). Starting from our previously described time series of GEDI-labeled primary cortical neurons from rats (30), we segmented the EGFP-labeled soma of each neuron with an intensity threshold and minimum and maximum size filter, as previously described (35). Using the intensity weighted centroid from each segmentation mask, images of each neuron were automatically cropped and sorted on the basis of their GEDI ratio as live or dead (Fig. 1, D and E). Plots of the GEDI ratio from the first time point of the imaging dataset showed that the values from most neurons clustered into two groups corresponding to either live or dead cells (Fig. 1F). A minority of “outlier” neurons had GEDI ratios outside the main distribution of live and dead GEDI signals, typically due to segmentation errors (Fig. 1G), and these were discarded from the training set. Cropped images from a total of 53,638 live and 22,533 dead neurons from the first time point of an imaging series, both with and without background subtracted, were sorted to generate a dataset for training computer vision models. We selected a widely used CNN called VGG16, which was initialized with weights from training on the large-scale ImageNet dataset of natural images (36). We refer to this model as GEDI-CNN (Fig. 1H and fig. S1). In validation on held-out data from the first time point of the imaging series, the GEDI-CNN classified dead cells from the EGFP channel images with 96.8% accuracy [0.96 area under the receiver operating characteristic curve (AUC) in fig. S1D]. The GEDI-CNN was also effective at classifying the aforementioned outlier neurons, indicating its robustness for detecting cell death even in the face of experimental noise (fig. S1E). These data indicate that direct utilization of a biomarker signal without human curation can generate a highly accurate biomarker-optimized CNN (BO-CNN) capable of automated classification of microscopy images.
Applying GEDI-CNN to longitudinal imaging of individual neurons
Live-cell imaging over multiple time points exacerbates the problems with classification of dead cells by human curators because each time point multiplies the number of images that must be classified. Therefore, GEDI-CNN curation could be especially advantageous for cell-imaging time series. As the GEDI-CNN model was trained only with the initial time point of our imaging series, we next tested its ability to classify individual neurons at later time points. Across 478,987 neurons tested (fig. S1), the GEDI-CNN was 96.4% accurate (0.92 AUC) in detecting dead neurons, with little deviation across 20 different imaging plate batches (range, 88.4 to 97.4%) and across 46 different days of imaging (range, 87.8 to 98.1%) (Fig. 2A). The GEDI-CNN was slightly better at correctly classifying live neurons than dead neurons but was still able to successfully classify datasets with imbalanced numbers of live versus dead cells (Fig. 2, B and C). GEDI-CNN models trained with different CNN architectures such as VGG19 and ResNet performed with similar accuracy to the VGG16 model (fig. S2). The focal quality of the image (37) did not correlate with the GEDI-CNN accuracy, suggesting that the GEDI-CNN is capable of making quality predictions on out-of-focus images (fig. S3).
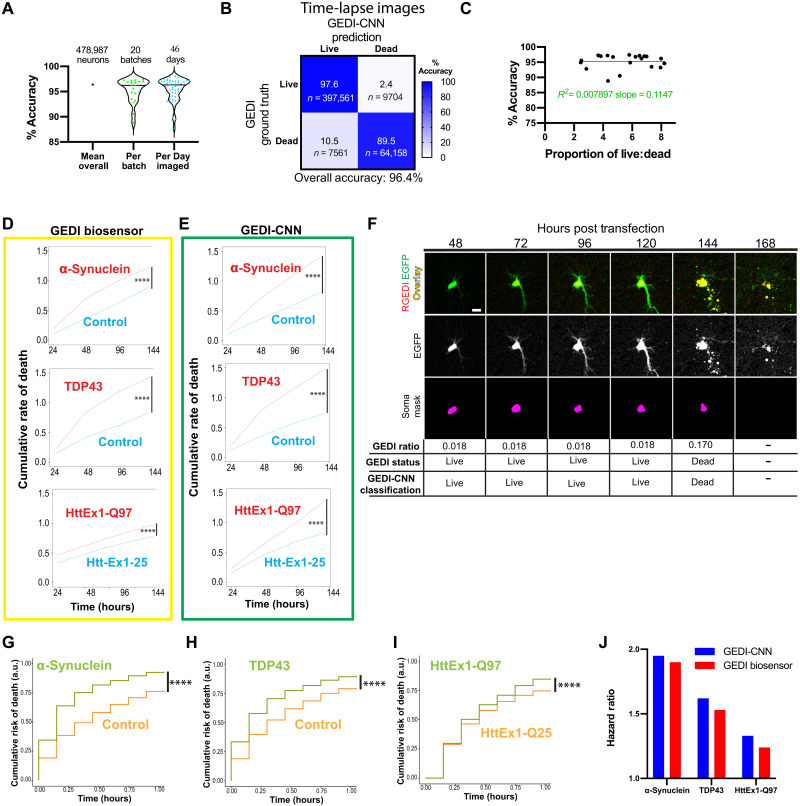
Fig. 2. Application of GEDI-CNN to time-lapse, single-cell imaging of neurons.
(A) Accuracy of GEDI-CNN testing on time-lapse imaging across the entire dataset, per batch (reflecting biological variation between primary neuron preps), and per imaging date (reflecting technical variability of the microscope over time). Black horizonal line represents mean. (B) Confusion matrix comparing GEDI-CNN predictions to ground truth (GEDI biosensor) with percentage (top) and neuron count (bottom). (C) Percentage of accuracy as a function of the proportion of live to dead neurons in dataset. Green line represents linear regression fit to data. (D and E) Cumulative rate of death per well over time for α-synuclein–expressing versus RGEDI-P2a-EGFP (control) neurons (top), TDP43-expressing versus control (middle) neurons, or HttEx1-Q97 (bottom) versus HttEx1-Q25 neurons using GEDI biosensor (D) or GEDI-CNN (E) (n = 48 wells each condition). (F) Representative time-lapse images of a dying single neuron with correct classification showing the overlay (top) (EGFP = green, RGEDI = red), the free EGFP morphology signal (middle) used by GEDI-CNN for classification, and the binary mask of automatically segmented object (bottom) used for quantification of GEDI ratio. By 168 hours after transfection, free EGFP within dead neurons has sufficiently degraded for an object not to be detected. (G) Cumulative risk of death of α-synuclein–expressing versus RGEDI-P2a-control neurons (HR = 1.95, ****P < 2 × 10−16), (H) TDP43 versus RGEDI-P2a-control neurons (HR = 1.62, ****P < 2 × 10−16), and (I) HttEx1-Q97 versus HttEx1-Q25 neurons (HR = 1.33, ***P < 2.11 × 10−13). a.u., arbitrary units. (J) Comparison of the hazard ratio using GEDI-CNN (top) versus GEDI biosensor (bottom) (32). Scale bar, 20 μm.
We next tested whether a GEDI-CNN could detect cell death in longitudinally imaged rat primary cortical neuron models of neurodegenerative disease. Imaging with the GEDI biosensor previously showed that protein overexpression models of Huntington’s disease (HD) (pGW1:HttEx1-Q97) (33), Parkinson’s disease (PD) (pCAGGs:α-synuclein) (38), and amyotrophic lateral sclerosis (ALS) or frontotemporal dementia (FTD) (pGW1:TDP43) (1) have high rates of neuronal death (32). We therefore compared the cumulative rate of death per well (dead neurons/total neurons) as estimated by the GEDI biosensor versus the GEDI-CNN. We began by quantifying cumulative rate of death per well, which does not require tracking each neuron in the sample, simplifying the comparison between the GEDI biosensor and the GEDI-CNN predictions. From the cumulative rate of death, we derived a hazard ratio comparing each disease model with its control using a linear mixed-effects model (Fig. 2, D and E). For each disease model, the GEDI biosensor and the GEDI-CNN detected similar increases in neuronal death in the overexpression lines compared to the control lines, confirming that the GEDI-CNN can be used to automatically and quickly assess the amount of neuronal death in live-imaging data (Fig. 2, D and E). Furthermore, the high accuracy of the GEDI-CNN indicates that there is enough information in the morphological changes of a neuron alone to accurately predict reduced survival within a neuronal disease model with nearly equivalent performance as the GEDI biosensor.
Evaluation of the amount of death in a cross section of cells per well over time can be an effective tool for tracking toxicity within a biological sample, but it cannot resolve dynamic changes within the sample because of changes in the total cell number or heterogeneity within the culture. Changes in cell number are treated as technical variation, which reduces the overall sensitivity of the system and can mask transient changes within the culture. In contrast, single-cell longitudinal analysis has proven advantages over conventional imaging approaches because the live/dead state of each cell and rare or transient changes in cells can be quantitatively linked to their fate and the biological significance of those observations understood (11). In combination with a suite of statistical tools commonly used for clinical trials called survival analysis (39) and Cox proportional hazard (CPH) analysis (40), the analysis of time of death in single cells can provide 100 to 1000 times more sensitivity than the analysis of single snapshots in time (11). In addition, single-cell approaches can facilitate covariate analyses of the factors that predict neuronal death (3, 5, 6, 11, 30). Yet, each of these analyses is limited by an inability to acutely differentiate live and dead cells (29, 32).
We performed automated single-neuron tracking on HD, PD, and ALS/FTD model datasets while quantifying the timing and amount of death detected with GEDI biosensor or GEDI-CNN (Fig. 2F). With accurate single-cell tracking and live/dead classification in place, we applied CPH analysis to calculate the relative risk of death for each model, which closely recapitulated previously reported hazard ratios (Fig. 2, G to J) (32), and mirrored the cumulative rate of death analysis. These data indicate that the GEDI-CNN, trained with GEDI biosensor data from the first time point of live imaging, can accurately predict death in later time points using the morphology signal alone and can therefore be used to identify neurodegenerative phenotypes in longitudinal culture models.
GEDI-CNN outperforms human curation
The high accuracy with which the GEDI-CNN classifies dead cells in imaging time series suggested that it could also be an improvement over standard human curation. To test this hypothesis, we manually curated 3000 images containing GEDI biosensor that were not included in model training. Four trained human curators classified the same set of neurons as live or dead using the EGFP morphology signature by interacting with a custom-written ImageJ script that displayed each image and kept track of each input and the time each curator took to accomplish the task. The GEDI-CNN was 90.3% correct in detecting death across six balanced batches of 500 images each, significantly higher than each human curator individually (curator range, 79 to 86%) Fig. 3A). In addition to accurately detecting cell death, the GEDI-CNN was two orders of magnitude faster than human curators at rendering its decisions on our hardware; the rate of data analysis was faster than the rate of data acquisition (Fig. 3B). Images correctly classified live by the GEDI-CNN passed classical curation standards such as the presence of clear neurites in live neurons (Fig. 3C). Neurons classified as dead showed patterns of apoptosis such as pyknotic rounding-up of the cell, retraction of neurites, and plasma membrane blebbing (Fig. 3D) (41). These data suggest that the GEDI-CNN learns to detect death using features in neuronal morphology that have been previously recognized to track with cell death.
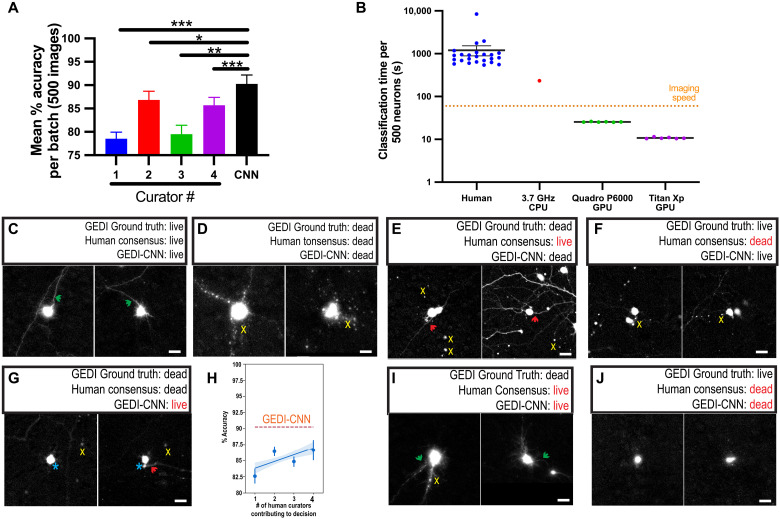
Fig. 3. GEDI-CNN has superhuman accuracy and speed at live/dead classification.
(A) Mean classification accuracy for GEDI-CNN and four human curators across four batches of 500 images using GEDI biosensor data as ground truth [±SEM, analysis of variance (ANOVA) Dunnett’s multiple comparison, ***P < 0.0001, **P < 0.01, and *P < 0.05]. (B) Speed of image curation by human curators versus GEDI-CNN running on a central processing unit (CPU) or graphics processing unit (GPU). Dotted line indicates average imaging speed (±SEM). (C and D) Representative cropped EGFP morphology images in which GEDI biosensor ground truth, a consensus of human curators, and GEDI-CNN classify neurons as live (C) or dead (D). Green arrows point to neurites from central neuron; yellow x’s mark peripheral debris near central neuron. (E to G, I, and J) Examples of neurons that elicited different classifications from the GEDI ground truth, the GEDI-CNN, and/or the human curator consensus. Green arrows point to neurites from central neuron, and red arrows point to neurites that may belong to a neuron other than the central neuron. Yellow x’s indicate peripheral debris near the central neuron. Turquoise asterisk indicates a blurry fluorescence from the central neuron. (H) Number of humans needed for their consensus decisions to reach GEDI-CNN accuracy. Human ensembles are constructed by recording modal decisions over bootstraps, which can reduce decision noise exhibited by individual curators. The blue line represents a linear fit to the human ensembles, and the shaded area depicts a 95% confidence interval.
Images for which the consensus of the human curators differed from that of the GEDI-CNN are of interest in finding examples of morphology related to death that are imperceptible to humans, and instances where the GEDI-CNN but not humans systematically misclassified death. For example, neurites were typically present in images where no human curator detected death, but the GEDI-CNN did (Fig. 3E). This suggests that human curators may be overly reliant on the presence of contiguous neurites to make live/dead classifications. In cases where humans incorrectly labeled a neuron as dead while the GEDI-CNN correctly indicated a live neuron, images were often slightly out of focus, lacked distinguishable neurites, or contained nearby debris and ambiguous morphological features such as a rounded soma that are sometimes, though not always, indicative of death (Fig. 3F). Similarly, in cases where the GEDI-CNN but not humans incorrectly classified dead neurons as alive, the neurons typically lacked distinguished neurites, contained ambiguous morphological features, and were surrounded by debris whose provenance was not clear (Fig. 3G). We recorded no instances of live neurons that were correctly classified by humans but incorrectly classified by the GEDI-CNN. These data suggest that the GEDI-CNN correctly classifies neurons with superhuman accuracy, in part, because it is robust to many imaging nuisances that limit human performance, which would explain how the accuracy of GEDI-CNN exceeded accuracy of curators, taken either individually or as an ensemble of curators (Fig. 3H).
The curator consensus and the GEDI-CNN were still both incorrect in more than 9% of the curated images, suggesting that there are some images in which the live or dead state cannot be determined by EGFP morphology alone. This could be, in part, because the level of Ca2+ within the neuron detected by GEDI is a better indicator of death than EGFP morphology. Close examination of images in which curation and GEDI-CNN were incorrect showed ambiguity in common features used to classify death, such as the combination of intact neurites and debris (Fig. 3I), or lack of neurites and rounded soma (Fig. 3J), suggesting that the morphology of the image was not fully indicative of its viability. These data suggest that GEDI-CNN can exceed accuracy of human curation, but it is also limited by the information present in the EGFP morphology image.
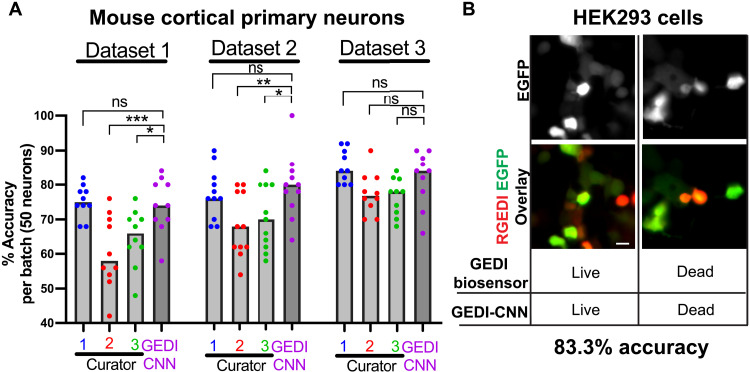
CNNs often fail to translate across different imaging contexts, so we next examined whether GEDI-CNN accurately classifies within datasets collected with different imaging parameters and biology. To probe how well GEDI-CNN translates to neurons from other species, we prepared plates of mouse cortical primary neurons expressing the GEDI biosensor and imaged them with parameters similar though not identical to those used on the rat primary cortical neurons used for initial training and testing. The GEDI-CNN showed human-level accuracy across each mouse primary neuron dataset (Fig. 4A and fig. S4). While the GEDI-CNN significantly outperformed some human curators in some datasets, it did not have a consistent edge in accuracy over all human curators, suggesting that some classification accuracy is lost when translating predictions to new biology or imaging conditions. To probe how far GEDI-CNN could translate, we next transfected immortalized human embryonic kidney (HEK) 293 cells, a cancer cell line, with RGEDI-P2a-EGFP, and exposed the cells to a cocktail of sodium azide and Triton X-100 to induce death in the culture. HEK293 cells lack neurites, and their morphology differs notably from the morphology of primary neurons, yet the GEDI-CNN classification accuracy was still significant—83.3%—in these cells (Fig. 4B and fig. S4). Overall, these data demonstrate that the GEDI-CNN equals or exceeds human-level accuracy across a diverse range of imaging, biological and technical conditions, but with superhuman speed.
Fig. 4. GEDI-CNN classifications translate across different imaging parameters and biology.
(A) Mean classification accuracy of GEDI-CNN in comparison to human curators across three mouse primary cortical neuron datasets. Scale bar, 20 μm (ANOVA and Dunnett’s multiple comparison, ***P < 0.001, **P < 0.01, and *P < 0.05; ns, not significant). (B) Representative cropped EGFP (top) and overlaid RGEDI and EGFP images (bottom) of HEK293 cells. Cell centered on left panels is live by both GEDI biosensor and GEDI-CNN. Cell centered on right panels is dead by both GEDI biosensor and GEDI-CNN.
GEDI-CNN uses membrane and nuclear signal as cues for its superhuman classifications
Our findings indicate that the GEDI-CNN joins a growing number of other examples that use CNNs to achieve superhuman speed and accuracy in biomedical image analysis (20, 42). Nevertheless, a key limitation of these models is the difficulty in interpreting the visual features they rely on for their decisions. One popular technique for identifying the visual features that contribute to CNN decisions is guided gradient-weighted class activation mapping (GradCAM) (43). GradCAM produces a map of the importance of visual features for a given image by deriving a gradient of the CNN’s evidence for a selected class (i.e., dead) that is masked and transformed for improved interpretability (fig. S5A). GradCAM has been found to identify visual features used by leading CNNs trained on object classification that closely align with those used by human observers to classify the same images (43).
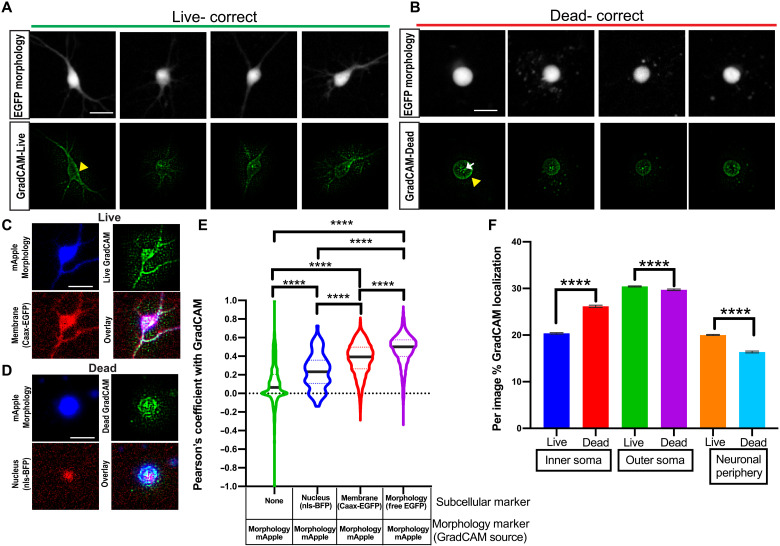
We generated GradCAM feature importance maps for both live and dead decisions for every image (fig. S5, B to D). We found that feature importance maps corresponding to the GEDI-CNN’s ultimate decision placed emphasis on cell body contours and neurites (Fig. 5, A and B). GradCAM importance maps highlighted the central segmented soma rather than peripheral fluorescence signal that might have come from other neurons within the cropped field of view. These importance maps also did not indicate that regions without fluorescence were important for visual decisions (fig. 5, E to H). Thus, the GEDI-CNN learned to base its classification on the neuron in the center of the image rather than background nuisances or other neurons not centered in the image. The backbone architecture of the GEDI-CNN—a VGG16—is uniquely suited for learning this center bias due to its multilayer, fully connected readout, which has distinct synapses for every spatial location and feature in the final convolutional layer of the model. While this bias is often not helpful in natural image classification, where it is important to build invariance to object poses and positions, the same bias is particularly well suited for our imaging pipeline, where neurons always appear in the same position.
Fig. 5. GEDI-CNN learns to use membrane and nuclear signal to classify live/dead cells with superhuman accuracy.
(A and B) Free EGFP fluorescence of neurons (top) and GEDI-CNN GradCAM signal (bottom) associated with correct live (A) and dead (B) classification. Yellow arrowhead indicates GradCAM signal corresponding to the neuron membrane. White arrow indicates GradCAM signal within the soma of the neuron. (C) Cropped image of a neuron classified as live by GEDI-CNN that is coexpressing free mRuby (top left, blue) and CAAX-EGFP (bottom left, red), with the corresponding GradCAM image (top right, green), and the three-color overlay (bottom right). (D) Cropped image of a neuron classified as dead by GEDI-CNN coexpressing free mRuby (top left, blue) and nls-BFP (bottom left, red), with the corresponding GradCAM image (top right, green), and the three-color overlay (bottom right). (E) Violin plot of quantification of the Pearson coefficient after Costes’s automatic thresholding between GradCAM signal and background signal (n = 3083), nls-BFP (n = 86), CAAX-EGFP (n = 3151), and free mRuby (n = 5404, ****P < 0.0001). (F) Per image percentage of GradCAM signal in the inner soma and outer soma across live and dead neurons. Scale bar, 20 μm.
We next looked to identify neuronal morphology that GEDI-CNNs deemed important for detecting dead neurons. The two consensus criteria under which a solitary cell can be regarded as dead according to current expert opinion are either the loss of integrity of the plasma membrane or the disintegration of the nucleus (44). We tested whether the GEDI-CNN had learned to follow the same criteria by performing subcellular colocalization of the GradCAM feature maps. Neurons were cotransfected with free mApple to mark neuronal morphology and either nuclear-targeted blue fluorescent protein (nls-BFP) or membrane-targeted EGFP (Caax-EGFP). Pearson correlation between biomarker-labeled images and GradCAM feature maps was used to measure the overlap between the two (Fig. 5, C to E) (45). GradCAM signal significantly colocalized with Caax-EGFP and, to a lesser extent, nls-BFP fluorescence. To further validate the importance of nuclear and membrane regions for live/dead classification, we devised a heuristic segmentation strategy that further subdivides the neuronal morphology. We subdivided each masked soma into inner and outer regions (fig. S6). Signal outside of the soma was also separated into neuronal periphery (neurites and other neurons within the image crop) signal versus regions with no fluorescent signal. We found that signal within the inner soma was enriched for nls-BFP, whereas signals within the outer soma and neuronal periphery were enriched for membrane-bound EGFP (Caax-EGFP). The importance of the inner soma region to the ultimate decision classification was increased in neurons classified as dead versus those classified as alive, according to GradCAM feature importance analysis. In contrast, GradCAM feature importance decreased in the outer soma and neuronal periphery in live-classified neurons compared to dead-classified neurons (Fig. 5F). These colocalizations indicate that the GEDI-CNN uses opposing weights for features near the nucleus versus the neurites and membrane in generating live/dead classification.
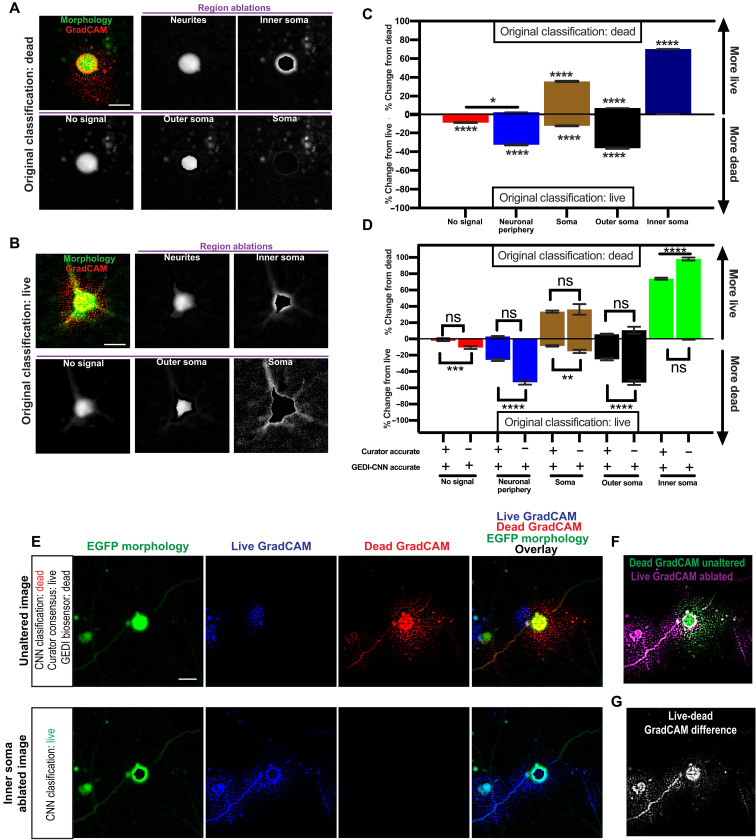
GradCAM signal correlations suggested the nuclear/inner soma region and membrane/outer soma/peripheral neurite regions are important for live/dead classification, but they did not establish causality in the relationship of those features to the final classification. To validate the relationships, we measured GEDI-CNN classification performance as we systematically ablated pixels it deemed important for detecting death. We used heuristic subcellular segmentations to generate a panel of ablation images in which signal within each region of interest (ROI) was replaced with noise, and we used these images to probe the decision making of the GEDI-CNN (Fig. 6, A and B). Ablations of the neurites and outer soma had the greatest effect in changing classifications of live neurons to dead, while ablations of the inner soma had the least effect (Fig. 6C, bottom). In contrast, ablation of the soma, particularly the inner soma, caused the GEDI-CNN to change classifications from “dead” to “live” substantially more often than ablations of no-signal, neurites, and outer soma (Fig. 6C, top). Although the inner soma and outer soma regions were the most sensitive ablations for switching classification to dead or live, respectively, they represented the smallest areas of ablated pixels, suggesting that the correct classification is based on a very specific subcellular morphological change (fig. S7). These data confirm that regions enriched in membrane signal and in nuclear signal are critical for GEDI-CNN live-dead classification, though in opposite ways.
Fig. 6. Inner soma signal important for dead classification and membrane and neurite signals important for live classifications by GEDI-CNN.
(A and B) Top left: Overlay of free EGFP morphology (green) and GradCAM signal (red) from a neuron classified as dead (A) or live (B) by GEDI-CNN and a series of automatically generated ablations of the neurites, inner soma, no-signal area, outer soma, and soma (left to right, top to bottom). (C) Effects of signal ablations on GEDI-CNN classification by region. Panels depict the mean percentage of neurons in each region whose classification incorrectly changed from dead to live after ablation (top, n = 4619) or from live to dead (bottom, n = 5122). Changes significantly different from 0 are marked by * (Tukey’s post hoc with correction for multiple comparisons, ****P < 0.0001 and *P < 0.05). (D) Effects of ablating signal from different subcellular regions on the accuracy of the GEDI-CNN’s classification of neurons that were originally correctly classified by GEDI-CNN but incorrectly classified by at least two human curators (Tukey’s post hoc with correction for multiple comparisons, ****P < 0.0001 and *P < 0.05). (E) Top: Representative image of the EGFP morphology of a dead neuron classified live by a consensus of four human curators and dead by a GEDI-CNN with its associated live and dead GradCAM signals. Bottom: Inner soma–ablated EGFP image that changed GEDI-CNN classification from dead to live and its associated live and dead GradCAM signals. (F) Overlay of the dead GradCAM signal from the dead-classified, unaltered image and the live GradCAM signal from the live-classified inner soma–ablated image. (G) Difference between dead GradCAM signal from the unaltered image and the live GradCAM signal from the inner soma–ablated image. Scale bar, 20 μm.
We next investigated if the source of the superhuman ability of the GEDI-CNN to classify neurons as live or dead was a result of the CNN’s ability to see patterns in images that humans do not easily see within these identified regions. To this end, we analyzed the impact of regional ablations in images that the GEDI-CNN classified correctly but that more than a single curator misclassified (Fig. 6D). Notably, almost all dead neurons that GEDI-CNN correctly classified but human curators incorrectly classified required the inner soma signal, suggesting that the inner soma signal underlies GEDI-CNN’s superhuman classification ability. GradCAM signal showed a shift from within the inner soma to the membrane and neurites after ablation (Fig. 6, E to G). These data show that the GEDI-CNN used the nuclear/inner soma and neurite/membrane morphology to gain superhuman accuracy at live or dead classification of neurons.
Zero-shot generalization of GEDI-CNN on human iPSC-derived motor neurons
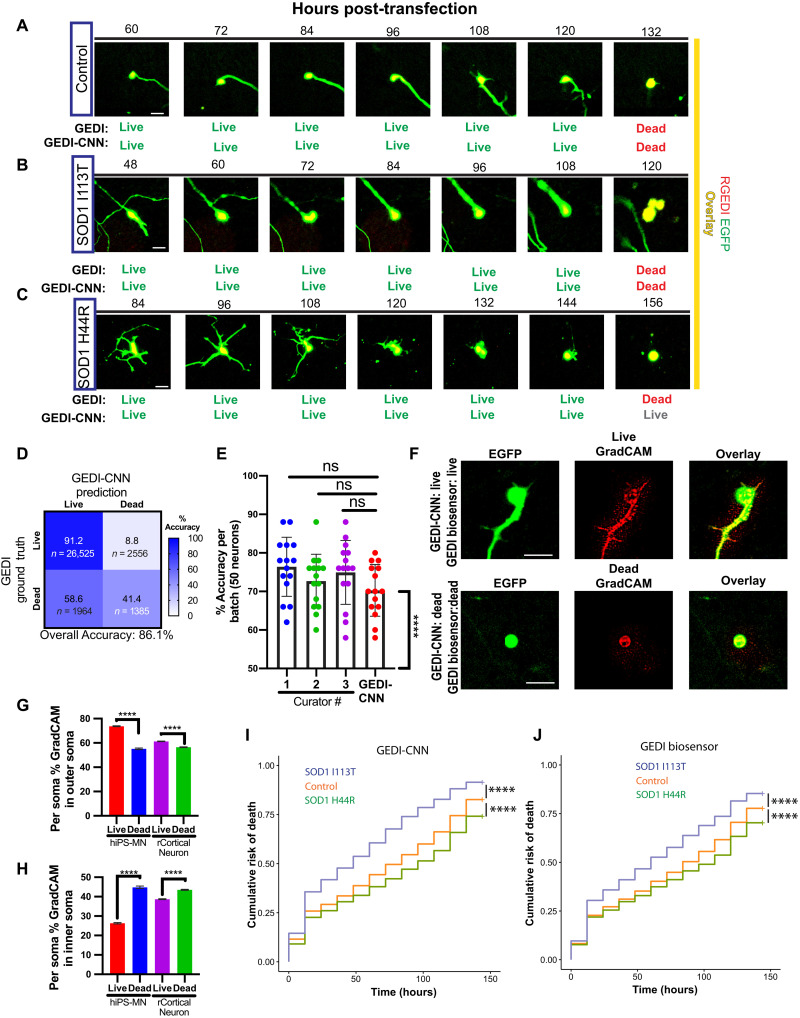
We next asked whether GEDI-CNN could detect death in a neuronal model of ALS derived from human induced pluripotent stem cells (iPSCs). Neurons derived from iPSCs (i-neurons) maintain the genetic information of the patients from whom they are derived, facilitating modeling of neurodegenerative (46), neurological (47), and neurodevelopmental diseases (48) in which cell death can play a critical role. iPSC motor neurons (iMNs) generated from the fibroblasts of a single patient carrying the SOD1 D90A mutation have been previously shown to model key ALS-associated pathologies and display a defect in survival compared to neurons derived from control fibroblasts (32, 49, 50). However, iPSC lines derived from patient fibroblasts with less common SOD1 mutations associated with ALS, I113T (51–53) and H44R (54), have not been previously characterized for survival deficits, so whether reduced survival in culture is a common characteristic of all iMNs with SOD1 mutation remains unknown. To test whether the GEDI-CNN could detect reduced survival in human iPSC-derived neurons, we differentiated iPSCs from SOD1I113T and SOD1H44R ALS patients and healthy volunteers into iMNs, transfected neurons with RGEDI-P2a-EGFP, and imaged them longitudinally (Fig. 7, A to C). Overall classification accuracy based on morphology alone of i-neurons was 86.1% (0.68 AUC), with a markedly higher accuracy for live neurons (91.2%) than dead ones (41.4%) (Fig. 7D). Across balanced randomized batches of data, GEDI-CNN averaged only 70% accuracy and was not significantly different from human curation accuracy, indicating that death is difficult to discern from the EGFP morphology alone in this dataset (Fig. 7E). A potential explanation for this difficulty is that iMNs often underwent a prolonged period of degeneration characterized by neurite retraction before a positive GEDI signal was observed (Fig. 7C), a phenomenon not observed in mouse primary cortical neurons.
Fig. 7. GEDI-CNN classifications translate to studies of neurodegeneration in human iPSC-derived motor neurons.
(A to C) Representative longitudinal time-lapse images overlaying RGEDI (red) and EGFP (yellow) in iPSC-derived motor neurons derived from a healthy control patient (A), an ALS patient with SOD1I113T (B), or an ALS patient with SOD1H44R (C). Comparison of live/dead classifications derived from the GEDI biosensor and GEDI-CNN below each image. (D) Confusion matrix of live/dead classification accuracy of GEDI-CNN on iPSC-derived motor neurons. (E) Mean classification accuracy of GEDI-CNN and human curators on randomized 50% live:dead balanced batches of data (ANOVA with Tukey’s multiple comparisons). Each curator and the GEDI-CNN showed classification accuracy significantly above chance (****P < 0.0001, Wilcoxon signed-rank difference from 50%). (F) Representative images of free EGFP morphology expression (left), GradCAM signal (middle), and overlay of GradCAM and EGFP (right) of iPSC-derived motor neurons classified correctly as live (top) and dead (bottom) by GEDI-CNN. (G and H) GradCAM signal in the soma shifts from outer soma in live neurons (G) to inner soma in dead neurons (H) in both human iPSC-derived motor neurons (hiPS-MN) and rat primary cortical neurons (rCortical Neuron). (I) Cumulative risk of death of SOD1I113T (HR = 1.52, ****P < 0.0001) and SOD1H44R (HR = 0.79, ****P < 0.0001) neurons derived from GEDI-CNN. (J) Cumulative risk of death of SOD1I113T (HR = 1.35, ****P < 0.0001) and SOD1H44R (HR = 0.83, ****P < 0.0001) neurons using GEDI biosensor. Scale bar, 20 μm.
GradCAM signal from correctly classified live and dead neurons defined the contours of iMNs (Fig. 7F) and showed similar signal changes in response to neuronal death as observed in mouse cortical neurons (compare Fig. 7, G and H, to Fig. 4). Single-cell Kaplan-Meier (KM) and Cox proportional hazard (CPH) analysis of SOD1I113T and SOD1H44R neurons yielded similar results with the GEDI-CNN classification and the GEDI biosensor classification. In both cases, SOD1I113T neurons showed reduced survival compared to control neurons, whereas SOD1H44R neurons showed significantly increased survival (Fig. 7, I and J). Human curation using neuronal morphology showed the same trends (fig. S8). The hazard curves and hazard ratio (HR) obtained with GEDI-CNN for SOD1D90A was also similar to previously reported GEDI biosensor data (fig. S9) (32). These data demonstrate that a reduced survival phenotype of iMNs generated from patients with ALS is consistently detected in some cell lines [SOD1D90A, here; (32, 49, 50)] but not detected in others (SOD1H44R), suggesting that higher-powered studies are necessary to overcome inherent study variability and attribute phenotypes found in iMNs to underlying genetic factors. Nevertheless, our data show that the GEDI-CNN can be deployed to make human-level zero-shot predictions of GEDI ground truth on an entirely different cell type than the one on which it was trained.
DISCUSSION
Robotic microscopy is frequently hindered by the rate and accuracy of image analysis. Here, we developed a novel strategy that leverages physiological biomarkers to train computer vision models for image analysis, which can efficiently scale to the high-throughput imaging regimes enabled by robotics. Using signal from GEDI, a sensitive detector of Ca2+ levels presaging cell death (32), we trained a CNN that detects cell death with superhuman accuracy and performs across a variety of tissues and imaging configurations. We demonstrate that the GEDI-CNN learned to detect cell death by locating fundamental morphological features known to be related to cell death, despite receiving no explicit supervision to focus on those features. In addition, we demonstrate that, in combination with automated microscopy, the GEDI-CNN can accelerate the analysis of large datasets to rates faster than the rate of image acquisition, enabling highly sensitive single-cell analyses at large scale. Using this strategy, we identified distinct survival phenotypes in iPSC-derived neurons from ALS patients with different mutations in SOD1. Our results demonstrate that the GEDI-CNN can improve the precision of experimental assays designed to uncover the mechanisms of neurodegeneration, and that the BO-CNN strategy—training CNNs to predict biomarker activity—can accelerate discovery in the field of live-cell microscopy.
GEDI-CNN live/dead classification predictions were significantly above chance across every cell type tested (fig. S4). Given the diversity of cell types tested, this indicates that GEDI-CNN learned to locate morphology features that are fundamental to death across biology. Using a combination of GradCAM and pixel ablation simulations, we show that the predictions of live and dead are sourced from different subcellular compartments such as the plasma membrane and the nucleus. Altered plasma membrane and retracted neurites are prominent features in images of neuronal morphology that are well known to correlate with vitality (55) and are likely easily appreciated by human curators. However, EGFP morphology signal in the region of the nucleus has not been previously appreciated as an indicator of vitality. While rupture of the nuclear envelope is well recognized as an indication of decreasing cell vitality (44), it is not clear how free EGFP signal, which is both inside and outside of the nuclear envelope, could indicate nuclear envelope rupture, or if another cell death–associated phenomenon is responsible for this signal. Either way, EGFP morphology signal in the nuclear region of cells represents a previously unrecognized feature that correlates with cell death and one that helps the GEDI-CNN achieve superhuman classification accuracy. The ability to harness the superhuman pattern recognition abilities of BO-CNN models represents a powerful new direction for the discovery of previously undiscovered biological phenomena and underscores the importance of understanding the visual basis of classification models to interpret this previously unrecognized biology.
Our demonstration that the GEDI-CNN can automate live-imaging studies of cellular models of neurodegenerative disease has tremendous implications for accelerating research in this field. Neuronal death is used as a phenotypic readout in nearly all studies of neurodegeneration (29). We demonstrate the use of the GEDI-CNN for large-scale studies of neurons using accurate cell death as a readout, and its potential for high-throughput screening purposes. Similarly, in large-scale studies or high-throughput screens of neurons in which cell death is not the primary phenotype, the GEDI-CNN can accurately account for cell death or be used to filter out dead objects that could contaminate other analyses. Using the GEDI-CNN to make predictions and map GradCAM on morphology images of neurons is free, open source, and simple to execute and does not require specialized hardware. In addition to facilitating high-throughput screening, which can markedly improve statistical power and sensitivity of analyses (11), generally accounting for cell death in microscopy experiments should help control experimental noise.
A rapid rate of analysis is particularly necessary where high rates of heterogeneity and biological diversity are present. Studies using human iPSC-derived neurons have begun grappling with the importance of scale for identifying and studying biological phenomena in the incredibly diverse human population (56–58). This is well illustrated in our data applying GEDI and the GEDI-CNN to iMNs from patients with SOD1 mutations. Multiple studies of iMNs from patients with SOD1 mutations linked to ALS, such as A4V and D90A (32, 49), have shown reduced survival phenotypes, and we observed a similar phenotype in cells harboring the I113TE and D90A mutations. However, the current study shows that the H44R mutation improves the survival of iPSC-derived neurons compared to control, indicating that a cell death phenotype in iMNs from ALS patients is not consistent under these conditions. There is high variability across different iPSC differentiation protocols, and it remains to be determined how much variability between iPSC lines contributes to previous underpowered reports of phenotypes associated with different lines. Ultimately, the only way to overcome issues with iPSC line variability is to increase the number of cell lines tested and standardize methods of measurements and analysis compatible with higher-powered studies. As GEDI-CNN significantly reduces the time and resources required to do high-power studies and eliminates human bias and intra-observer variability in live/dead classification, GEDI-CNN live/dead classifications should become a useful tool in future studies attempting to link iMN phenotypes to their genotypes.
While GEDI-CNN model prediction accuracy was always above chance, the accuracy level was reduced in some datasets, suggesting differences in imaging parameters and/or biology that may limit its ability to generalize from one dataset to the next (fig. S4). To offset reduced accuracy, new GEDI-CNN models can be transferred or “primed” with small datasets that better represent the qualities of a particular experiment or microscope setup. These datasets can be generated by imaging the GEDI biosensor on a particular microscope platform and training a model according to the BO-CNN pipeline we outline here. Nevertheless, on some datasets, we observed that both human and GEDI-CNN classification accuracy and GEDI-CNN accuracy were reduced, suggesting that, in these cases, morphology alone may be insufficient to accurately classify a cell as live or dead. Morphology of iMN datasets proved particularly difficult to interpret for both human curators and GEDI-CNN (Fig. 7C). While classification accuracy for live neurons was consistently high across different cell types, accuracy of classification for dead neurons often reduced overall accuracy (Figs. 2B and 6D), suggesting that death classification by morphology is especially difficult. Because of this variability in how different biological systems reflect death, the use of the GEDI biosensor as a ground truth to evaluate GEDI-CNN model predictions as well as for training new models will continue as the state of the art for detecting cell death based on morphology.
While GEDI-CNN represents the first proof of principle in using a live biosensor to train a BO-CNN, we believe that similar strategies could be used to generate new BO-CNNs that could help parse mechanisms underlying other disease-related phenotypes in neurodegenerative disease models. Several features of the GEDI biosensor proved critical for success. First, the GEDI biosensor acts as a nearly binary demarcation of live or dead cell states (32). Other biosensors with similar biological-state demarcation characteristics would make good candidates for this approach. However, other strategies for automatically extracting a classification label, such as using sample age or applied perturbations as the classification label, could also be used as a ground truth in place of the biomarker signal. Second, the GEDI signal is simple and robust enough to extract with currently available image analysis techniques. However, the fluorescent proteins within the GEDI we used here (RGEDI and EGFP) are not the brightest in their respective classes, and many available biosensors could be used that have as much, if not more, signal. Third, the free EGFP morphology signal within GEDI is already well known to indicate live or dead state, as it had been previously used as the state of the art for longitudinal live-imaging studies of neurodegeneration (29, 33). This gave us confidence that the patterned information within the EGFP morphology images would be sufficient to inform the model of the biological state of the neuron. While that information may not be available in all situations, there are now many examples where deep learning has uncovered new unexpected patterns in biological imaging, and the BO-CNN strategy could also be used in a similar way to uncover unexpected relationships between images and known biological classifications. As each of these properties can be found in other biosensors and biological problems, we are confident that other effective BO-CNNs can be developed using the same strategy we used to generate the GEDI-CNN. As the rate of microscopy imaging and scale has continued to accelerate in recent years from the commercialization of automated screening microscopes, we expect the importance of BO-CNN analyses to continue to increase, leading to new discoveries and therapeutic approaches to combat neurodegenerative disease.
MATERIALS AND METHODS
Animals, culturing, and automated time-lapse imaging
All animal experiments complied with University of California, San Francisco (UCSF) regulations. Primary mouse cortical neurons were prepared at embryonic days 20 to 21 as previously described (22). Neurons were plated in a 96-well plate at 0.1 × 106 cells per well and cultured in neurobasal growth medium with 100× GlutaMAX, penicillin/streptomycin, and B27 supplement (neurobasal medium). Automated time-lapse imaging was performed with long time intervals (12 to 24 hours) to minimize phototoxicity while imaging, reduce dataset size, and provide a protracted time window to monitor neurodegenerative processes. Imaging speed was assessed as the average time to image a 4 × 4 montage of EGFP-expressing neurons per well in a 96-well plate (6 s) divided by the average number of neurons transfected per well (49). HEK cells [Research Resource Identifier (RRID): CVCL_0045] were plated in a 96-well plate at 0.01 × 106 cells per well 24 hours before transfection. Sodium azide (0.4%) and Triton X-100 (0.02%) (final concentration) were added to induce cell death. Images were captured 20 min after sodium azide and Triton X-100 application.
Automated imaging and image processing pipeline
Quantification of GEDI ratios was performed as previously described (32). In short, RGEDI and EGFP channel fluorescence images obtained by automated imaging were processed using custom scripts running within a custom-built image processing Galaxy bioinformatics cluster (54, 59). The Galaxy cluster links together workflows of modules including background subtraction of median intensity of each image, montaging of imaging panels, fine-tuned alignment across time points of imaging, segmentation of individual neurons, and cropping image patches where the centroid of individual neurons is positioned at the center. Segmentation was performed by identifying a lower bound intensity threshold and minimum and maximum size filter so that neuronal somas, the brightest part of the image, are separated from debris and other background signal. Each individual object segmented is converted to a segmentation mask by binarizing the image so that each object has a pixel intensity value equal to 1 and the background has a value equal to 0. The resulting black and white image is further processed so that each cell is identified as an independent object with its own numerical identifier. Single-cell tracking was performed using a custom Voronoi tracking algorithm that monitors the movements of regionalized binary objects through space (55). Survival analysis was performed by defining the last time point alive as the time point before the GEDI ratio of a longitudinally imaged neuron exceeds the empirically calculated GEDI threshold for death, the last time point a human curator tracking cells finds a neuron alive, or the time point before the GEDI-CNN classifies a neuron as dead. KM survival curves and CPH were calculated using custom scripts written in R, and survival functions were fit to these curves to derive cumulative survival and risk-of-death curves that describe the instantaneous risk of death for individual neurons as previously described (56). Cumulative risk of death analysis was performed by quantification of cumulative mean of dead cells/total cells per well across time points. Because fluorescent neuronal debris can remain in culture for up to 48 hours past the time point of death (32), time points other than at 48-hour intervals were removed from the analysis to avoid double counting dead cells within the culture. A linear mixed-effects model implemented in R (57) was fit to the cumulative rate of death, using “well” as a random effect and interaction between condition and elapsed hours as a fixed effect to derive hazard ratios. Corrections for multiple comparisons were made using Holm-Bonferroni method.
Training GEDI-CNN
We began with a standard VGG16 (36), implemented in TensorFlow 1.4. The 96,171 images of rodent primary cortical neurons were separated into training and validation folds (79%/21%), normalized to [0, 1] using the maximum intensity observed in our microscopy pipeline, and converted to single precision. We adopted a transfer-learning procedure in which we initialized early layers of the network (i.e., close to the image input) with pretrained weights from natural image categorization (ImageNet) (36) and later layers of the network (i.e., close to the category readout) with Xavier Normal random initializations (58). We empirically selected which layers to put at the end of the network (i.e., from moving from the readout toward the input) by training models with different amounts of these layers randomly initialized and recording their performance on the validation set. Models were trained on Nvidia Titan X GPUs for 20 epochs using the Adam optimizer to minimize class-weighted cross entropy between live/dead prediction and GEDI-derived labels. We ultimately chose to initialize the first three blocks of VGG16 convolutional layers with ImageNet weights and the remaining layers with random weights. Layers initialized with ImageNet weights were trained with a learning rate of 1 × 10−8, and the rest were trained with a learning rate of 3 × 10−4. To improve model robustness, neuron images were augmented with random up/down/left right flips, cropping from 256 × 256 to 224 × 224 pixels using randomly placed bounding boxes, and random rotations of 0°, 90°, 180°, or 270°. These images, which were natively one-channel intensity images, were replicated into three-channel images to match the number of channels expected by the VGG16. We selected the best-performing weights according to validation loss for the experiments reported here. To compare, we trained ResNet50 and VGG19 initialized with ImageNet weights on the same data. For VGG19, we froze the first four convolutional blocks. Both were trained with the Adam optimizer with a learning rate of 3 × 10−4. Experiment code can be found at https://github.com/finkbeiner-lab/GEDI-ORDER.
Testing the GEDI-CNN
To test the trained GEDI-CNN model, microscopy images are passed through the Galaxy image processing pipeline that includes background subtraction of median intensity of each image, montaging of imaging panels, fine-tuned alignment across time points of imaging, segmentation of individual neurons, and cropping image patches where the centroid of individual neurons is positioned at the center, in the same way the training dataset was generated. Cropped images are passed into the GEDI-CNN to generate a classification.
GEDI-CNN GradCAM
Feature importance maps were derived using guided GradCAM—a method for identifying image pixels that contribute to model decisions. Guided GradCAM is designed to control for visual noise that emerges from computing such feature attributions for visual decisions through very deep neural network models. After extracting these feature importance maps from our networks, which were the same height/width/number of channels as the input images, we visualized them by rescaling pixel values into unsigned eight-bit images, converting to grayscale intensity maps, and then concatenating with a grayscale version of the input image to highlight morphology selected by the trained models to make their decisions.
Image ablations and curation tools
Curation was performed using a custom Fiji script that runs a graphical interface with a curator, displaying a blinded batch of cropped EGFP morphology images one at a time while prompting the curator to indicate whether the displayed neuron is live or dead with a keystroke (ImageCurator.ijm). Automated subcellular segmentations were generated and saved as ROIs and measured within a custom Fiji script (measurecell8.ijm). ROIs were used to generate a panel of ablations for each image using another custom Fiji script (cellAblations16bit.ijm). Pearson colocalization was measured using JACoP with Costes background randomization (45) within a custom Fiji script (PearsonColocalization.ijm).
iPSC differentiation to MNs
NeuroLINCS iPSC lines derived from fibroblast tissues from a healthy control individual or ALS patients with SOD1 H44R (CS04iALS-SOD1H44Rnxx) or I114T mutations (SOD1I114Tnxx) were obtained from the Cedars Sinai iPSC Core Repository. Healthy and SOD1 mutant iPSCs were found to be karyotypically normal and were differentiated into MNs using a modified dual-SMAD inhibition protocol (http://neurolincs.org/pdf/diMN-protocol.pdf) (59). iPSC-derived MNs were dissociated using trypsin (Thermo Fisher Scientific), embedded in diluted Matrigel (Corning) to limit cell motility, and plated onto Matrigel-coated 96-well plates. From days 20 to 35, the neurons underwent a medium change every 2 to 3 days. At day 32 of differentiation, neurons were transfected using Lipofectamine and imaged starting on day 33.
Acknowledgments
We thank K. Claiborn and F. Chanut for editorial assistance, K. Nelson and G. Abramova for administrative assistance, and D. Cahill for laboratory neuron curation, laboratory maintenance, and organization. We also thank R. Thomas and the Gladstone Institutes bioinformatics core for assistance with statistical models.
Funding: This work was supported by grants from the NIH (U54 NS191046, R37 NS101996, RF1 AG058476, RF1 AG056151, RF1 AG058447, P01 AG054407, and U01 MH115747), R01 LM013617, as well as support from the Koret Foundation Artificial Intelligence Program for Biomedical Research, the Taube/Koret Center for Neurodegenerative Disease, the Target ALS Foundation, the Amyotrophic Lateral Sclerosis Association Neuro Collaborative, a gift from Mike Frumkin, and the Department of Defense award W81XWH-13-ALSRP-TIA. Gladstone Institutes received support from the National Center for Research Resources (grant RR18928). We acknowledge the support of NVIDIA Corporation with the donation of the Titan Xp and Quadro P6000 GPUs used for this research. T.S. and D.A.L. received support from ONR (N00014-190102029), NSF (IIS-1912280 and EAR-1925481), DARPA (D19AC00015), the Artificial and Natural Intelligence Toulouse Institute (ANR-19-PI3A-0004), and the Carney Brain Science Compute Cluster at Brown.
Author contributions: J.W.L., D.A.L., and S.F. wrote the manuscript. J.W.L., D.A.L., S.F., N.A.C., A.J., and T.S. conceptualized the study. J.W.L., D.A.L., K.S., and N.A.C. performed data analysis and statistics. J.K., J.L., N.A.C., S.W., and V.O. performed robotic microscopy, transfections, iPSC differentiation, and cell culture. D.A.L. developed GEDI-CNN code base. D.A.L., J.L., G.R., and Z.T. performed GEDI-CNN training, GradCAM visualization, and Defocus-CNN evaluations. J.W.L., J.L., and G.R. performed custom Fiji scripts for analysis and curation of imaging experiments. All authors reviewed the manuscript.
Competing interests: The authors declare that they have no competing nonfinancial interests, but the following competing financial interests: S.F. is the inventor of Robotic Microscopy Systems, U.S. patent 7,139,415, and Automated Robotic Microscopy Systems, U.S. patent application 14/737, 325, both assigned to J. David Gladstone Institutes. A provisional U.S. and EPO patent for the GEDI biosensor (inventors J.L., K.S., and S.F.) assigned to The J. David Gladstone Institutes has been placed GL2016-815, May 2019.
Data and materials availability: Stable code for the GEDI-CNN is available at: https://zenodo.org/record/4908177#.YWkcVEbMJhA. A subsample of training, validation, and testing data can be found at https://zenodo.org/record/4908177#.YMGz8S-cbAY. Access to and use of the code are subject to a nonexclusive, revocable, nontransferable, and limited right for the exclusive purpose of undertaking academic, governmental, or not-for-profit research. Use of the code or any part thereof for commercial or clinical purposes is strictly prohibited in the absence of a Commercial License Agreement from the J. David Gladstone Institutes. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The GEDI biosensors can be provided by Gladstone Institutes pending scientific review and a completed material transfer agreement. Requests for the GEDI biosensors should be submitted to S.F.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
REFERENCES AND NOTES
- 1.Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S., Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Barmada S. J., Serio A., Arjun A., Bilican B., Daub A., Ando D. M., Tsvetkov A., Pleiss M., Li X., Peisach D., Shaw C., Chandran S., Finkbeiner S., Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat. Chem. Biol. 10, 677–685 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller J., Arrasate M., Shaby B. A., Mitra S., Masliah E., Finkbeiner S., Quantitative relationships between huntingtin levels, polyglutamine length, inclusion body formation, and neuronal death provide novel insight into Huntington's disease molecular pathogenesis. J. Neurosci. 30, 10541–10550 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsvetkov A. S., Miller J., Arrasate M., Wong J. S., Pleiss M. A., Finkbeiner S., A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. Proc. Natl. Acad. Sci. U.S.A. 107, 16982–16987 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carpenter A. E., Extracting rich information from images. Methods Mol. Biol. 486, 193–211 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Finkbeiner S., Frumkin M., Kassner P. D., Cell-based screening: Extracting meaning from complex data. Neuron 86, 160–174 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rueden C. T., Schindelin J., Hiner M. C., DeZonia B. E., Walter A. E., Arena E. T., Eliceiri K. W., ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamprecht M. R., Sabatini D. M., Carpenter A. E., CellProfiler: Free, versatile software for automated biological image analysis. BioTechniques 42, 71–75 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Jones T. R., Carpenter A. E., Lamprecht M. R., Moffat J., Silver S. J., Grenier J. K., Castoreno A. B., Eggert U. S., Root D. E., Golland P., Sabatini D. M., Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proc. Natl. Acad. Sci. U.S.A. 106, 1826–1831 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LeCun Y., Bengio Y., Hinton G., Deep learning. Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Stringer C., Wang T., Michaelos M., Pachitariu M., Cellpose: A generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Manak M. S., Varsanik J. S., Hogan B. J., Whitfield M. J., Su W. R., Joshi N., Steinke N., Min A., Berger D., Saphirstein R. J., Dixit G., Meyyappan T., Chu H. M., Knopf K. B., Albala D. M., Sant G. R., Chander A. C., Live-cell phenotypic-biomarker microfluidic assay for the risk stratification of cancer patients via machine learning. Nat. Biomed. Eng. 2, 761–772 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ouyang W., Winsnes C. F., Hjelmare M., Cesnik A. J., Åkesson L., Xu H., Sullivan D. P., Dai S., Lan J., Jinmo P., Galib S. M., Henkel C., Hwang K., Poplavskiy D., Tunguz B., Wolfinger R. D., Gu Y., Li C., Xie J., Buslov D., Fironov S., Kiselev A., Panchenko D., Cao X., Wei R., Wu Y., Zhu X., Tseng K. L., Gao Z., Ju C., Yi X., Zheng H., Kappel C., Lundberg E., Analysis of the human protein atlas image classification competition. Nat. Methods 16, 1254–1261 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caicedo J. C., Goodman A., Karhohs K. W., Cimini B. A., Ackerman J., Haghighi M., Heng C. K., Becker T., Doan M., McQuin C., Rohban M., Singh S., Carpenter A. E., Nucleus segmentation across imaging experiments: The 2018 Data Science Bowl. Nat. Methods 16, 1247–1253 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esteva A., Kuprel B., Novoa R. A., Ko J., Swetter S. M., Blau H. M., Thrun S., Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ouyang W., Mueller F., Hjelmare M., Lundberg E., Zimmer C., ImJoy: An open-source computational platform for the deep learning era. Nat. Methods 16, 1199–1200 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Christiansen E. M., Yang S. J., Ando D. M., Javaherian A., Skibinski G., Lipnick S., Mount E., O’Neil A., Shah K., Lee A. K., Goyal P., Fedus W., Poplin R., Esteva A., Berndl M., Rubin L. L., Nelson P., Finkbeiner S., In silico labeling: Predicting fluorescent labels in unlabeled images. Cell 173, 792–803.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sullivan D. P., Winsnes C. F., Åkesson L., Hjelmare M., Wiking M., Schutten R., Campbell L., Leifsson H., Rhodes S., Nordgren A., Smith K., Revaz B., Finnbogason B., Szantner A., Lundberg E., Deep learning is combined with massive-scale citizen science to improve large-scale image classification. Nat. Biotechnol. 36, 820–828 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Fuchs Y., Steller H., Programmed cell death in animal development and disease. Cell 147, 742–758 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labi V., Erlacher M., How cell death shapes cancer. Cell Death Dis. 6, e1675 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tower J., Programmed cell death in aging. Ageing Res. Rev. 23, 90–100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hotchkiss R. S., Strasser A., McDunn J. E., Swanson P. E., Cell death. N. Engl. J. Med. 361, 1570–1583 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serio A., Bilican B., Barmada S. J., Ando D. M., Zhao C., Siller R., Burr K., Haghi G., Story D., Nishimura A. L., Carrasco M. A., Phatnani H. P., Shum C., Wilmut I., Maniatis T., Shaw C. E., Finkbeiner S., Chandran S., Astrocyte pathology and the absence of non-cell autonomy in an induced pluripotent stem cell model of TDP-43 proteinopathy. Proc. Natl. Acad. Sci. U.S.A. 110, 4697–4702 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barmada S. J., Ju S., Arjun A., Batarse A., Archbold H. C., Peisach D., Li X., Zhang Y., Tank E. M. H., Qiu H., Huang E. J., Ringe D., Petsko G. A., Finkbeiner S., Amelioration of toxicity in neuronal models of amyotrophic lateral sclerosis by hUPF1. Proc. Natl. Acad. Sci. U.S.A. 112, 7821–7826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linsley J. W., Reisine T., Finkbeiner S., Cell death assays for neurodegenerative disease drug discovery. Expert Opin. Drug Discov. 14, 901–913 (2019a). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skibinski G., Nakamura K., Cookson M. R., Finkbeiner S., Mutant LRRK2 toxicity in neurons depends on LRRK2 levels and synuclein but not kinase activity or inclusion bodies. J. Neurosci. 34, 418–433 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barmada S. J., Skibinski G., Korb E., Rao E. J., Wu J. Y., Finkbeiner S., Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J. Neurosci. 30, 639–649 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linsley J. W., Shah K., Castello N., Chan M., Haddad D., Doric Z., Wang S., Leks W., Mancini J., Oza V., Javaherian A., Nakamura K., Kokel D., Finkbeiner S., Genetically encoded cell-death indicators (GEDI) to detect an early irreversible commitment to neurodegeneration. Nat. Commun. 12, 5284 (2021).. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.A. Waller, F. R. S. Owen, XX. Experiments on the section of the glossopharyngeal and hypoglossal nerves of the frog, and observations of the alterations produced thereby in the structure of their primitive fibres. Philos. Trans. R. Soc. Lond. 140, 423–429 (1850).
- 30.Sharma P., Ando D. M., Daub A., Kaye J. A., Finkbeiner S., High-throughput screening in primary neurons. Methods Enzymol. 506, 331–360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M., Berg A. C., Fei-Fei L., ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 115, 211–252 (2015). [Google Scholar]
- 32.Yang S. J., Berndl M., Michael Ando D., Barch M., Narayanaswamy A., Christiansen E., Hoyer S., Roat C., Hung J., Rueden C. T., Shankar A., Finkbeiner S., Nelson P., Assessing microscope image focus quality with deep learning. BMC Bioinformatics 19, 77 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamura K., Nemani V. M., Azarbal F., Skibinski G., Levy J. M., Egami K., Munishkina L., Zhang J., Gardner B., Wakabayashi J., Sesaki H., Cheng Y., Finkbeiner S., Nussbaum R. L., Masliah E., Edwards R. H., Direct membrane association drives mitochondrial fission by the Parkinson disease-associated protein α-synuclein. J. Biol. Chem. 286, 20710–20726 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dekker F. W., de Mutsert R., van Dijk P. C., Zoccali C., Jager K. J., Survival analysis: Time-dependent effects and time-varying risk factors. Kidney Int. 74, 994–997 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Wolfe R. A., Strawderman R. L., Logical and statistical fallacies in the use of Cox regression models. Am. J. Kidney Dis. 27, 124–129 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Shaby B. A., Skibinski G., Ando M., LaDow E. S., Finkbeiner S., A three-groups model for high-throughput survival screens. Biometrics 72, 936–944 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kroemer G., Galluzzi L., Vandenabeele P., Abrams J., Alnemri E. S., Baehrecke E. H., Blagosklonny M. V., el-Deiry W. S., Golstein P., Green D. R., Hengartner M., Knight R. A., Kumar S., Lipton S. A., Malorni W., Nuñez G., Peter M. E., Tschopp J., Yuan J., Piacentini M., Zhivotovsky B., Melino G.; Nomenclature Committee on Cell Death 2009 , Classification of cell death: Recommendations of the nomenclature committee on cell death 2009. Cell Death Differ. 16, 3–11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ting D. S. W., Cheung C. Y. L., Lim G., Tan G. S. W., Quang N. D., Gan A., Hamzah H., Garcia-Franco R., San Yeo I. Y., Lee S. Y., Wong E. Y. M., Sabanayagam C., Baskaran M., Ibrahim F., Tan N. C., Finkelstein E. A., Lamoureux E. L., Wong I. Y., Bressler N. M., Sivaprasad S., Varma R., Jonas J. B., He M. G., Cheng C. Y., Cheung G. C. M., Aung T., Hsu W., Lee M. L., Wong T. Y., Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA 318, 2211–2223 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selvaraju R. R., Das A., Vedantam R., Cogswell M., Parikh D., Batra D., Grad-CAM: Visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vis. 128, 336–359 (2019). [Google Scholar]
- 40.Galluzzi L., Aaronson S. A., Abrams J., Alnemri E. S., Andrews D. W., Baehrecke E. H., Bazan N. G., Blagosklonny M. V., Blomgren K., Borner C., Bredesen D. E., Brenner C., Castedo M., Cidlowski J. A., Ciechanover A., Cohen G. M., de Laurenzi V., de Maria R., Deshmukh M., Dynlacht B. D., el-Deiry W. S., Flavell R. A., Fulda S., Garrido C., Golstein P., Gougeon M. L., Green D. R., Gronemeyer H., Hajnóczky G., Hardwick J. M., Hengartner M. O., Ichijo H., Jäättelä M., Kepp O., Kimchi A., Klionsky D. J., Knight R. A., Kornbluth S., Kumar S., Levine B., Lipton S. A., Lugli E., Madeo F., Malorni W., Marine J. C. W., Martin S. J., Medema J. P., Mehlen P., Melino G., Moll U. M., Morselli E., Nagata S., Nicholson D. W., Nicotera P., Nuñez G., Oren M., Penninger J., Pervaiz S., Peter M. E., Piacentini M., Prehn J. H. M., Puthalakath H., Rabinovich G. A., Rizzuto R., Rodrigues C. M. P., Rubinsztein D. C., Rudel T., Scorrano L., Simon H. U., Steller H., Tschopp J., Tsujimoto Y., Vandenabeele P., Vitale I., Vousden K. H., Youle R. J., Yuan J., Zhivotovsky B., Kroemer G., Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ. 16, 1093–1107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolte S., Cordelières F. P., A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Haston K. M., Finkbeiner S., Clinical trials in a dish: The potential of pluripotent stem cells to develop therapies for neurodegenerative diseases. Annu. Rev. Pharmacol. Toxicol. 56, 489–510 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L., Chao J., Shi Y., Modeling neurological diseases using iPSC-derived neural cells: iPSC modeling of neurological diseases. Cell Tissue Res. 371, 143–151 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu C., Oikonomopoulos A., Sayed N., Wu J. C., Modeling human diseases with induced pluripotent stem cells: From 2D to 3D and beyond. Development 145, dev156166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen H., Qian K., du Z., Cao J., Petersen A., Liu H., Blackbourn L. W. IV, Huang C. T. L., Errigo A., Yin Y., Lu J., Ayala M., Zhang S. C., Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons. Cell Stem Cell 14, 796–809 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luigetti M., Conte A., Madia F., Marangi G., Zollino M., Mancuso I., Dileone M., del Grande A., di Lazzaro V., Tonali P. A., Sabatelli M., Heterozygous SOD1 D90A mutation presenting as slowly progressive predominant upper motor neuron amyotrophic lateral sclerosis. Neurol. Sci. 30, 517–520 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Lopate G., Baloh R. H., al-Lozi M. T., Miller T. M., Fernandes Filho J. A., Ni O., Leston A., Florence J., Schierbecker J., Allred P., Familial ALS with extreme phenotypic variability due to the I113T SOD1 mutation. Amyotroph. Lateral Scler. 11, 232–236 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Enayat Z. E., Orrell R. W., Claus A., Ludolph A., Bachus R., Brockmuller J., Ray-Chaudhuri K., Radunovic A., Shaw C., Wilkinson J., king A., Swash M., Leigh P. N., deBelleroche J., Powell J., Two novel mutations in the gene for copper zinc superoxide dismutase in UK families with amyotrophic lateral sclerosis. Hum. Mol. Genet. 4, 1239–1240 (1995). [DOI] [PubMed] [Google Scholar]
- 49.Orrell R. W., Habgood J. J., Gardiner I., King A. W., Bowe F. A., Hallewell R. A., Marklund S. L., Greenwood J., Lane R. J. M., deBelleroche J., Clinical and functional investigation of 10 missense mutations and a novel frameshift insertion mutation of the gene for copper-zinc superoxide dismutase in UK families with amyotrophic lateral sclerosis. Neurology 48, 746–751 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Wei Q., Zhou Q. Q., Chen Y. P., Ou R. W., Cao B., Xu Y. Q., Yang J., Shang H. F., Analysis of SOD1 mutations in a Chinese population with amyotrophic lateral sclerosis: A case-control study and literature review. Sci. Rep. 7, 44606 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rubin L. L., Neuronal cell death: When, why and how. Br. Med. Bull. 53, 617–631 (1997). [DOI] [PubMed] [Google Scholar]
- 52.Kilpinen H., Goncalves A., Leha A., Afzal V., Alasoo K., Ashford S., Bala S., Bensaddek D., Casale F. P., Culley O. J., Danecek P., Faulconbridge A., Harrison P. W., Kathuria A., McCarthy D., McCarthy S. A., Meleckyte R., Memari Y., Moens N., Soares F., Mann A., Streeter I., Agu C. A., Alderton A., Nelson R., Harper S., Patel M., White A., Patel S. R., Clarke L., Halai R., Kirton C. M., Kolb-Kokocinski A., Beales P., Birney E., Danovi D., Lamond A. I., Ouwehand W. H., Vallier L., Watt F. M., Durbin R., Stegle O., Gaffney D. J., Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 546, 370–375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Linsley J. W., Tripathi A., Epstein I., Schmunk G., Mount E., Campioni M., Oza V., Barch M., Javaherian A., Nowakowski T. J., Samsi S., Finkbeiner S., Automated four-dimensional long term imaging enables single cell tracking within organotypic brain slices to study neurodevelopment and degeneration. Commun. Biol. 2, 155 (2019c). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvier D., Čech M., Chilton J., Clements D., Coraor N., Eberhard C., Grüning B., Guerler A., Hillman-Jackson J., von Kuster G., Rasche E., Soranzo N., Turaga N., Taylor J., Nekrutenko A., Goecks J., The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44, W3–W10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Y. H. Chang, J. Linsley, J. Lamstein, J. Kalra, I. Epstein, M. Barch, K. Daily, P. Synder, L. Omberg, L. Heiser, S. Finkbeiner, Single cell tracking based on Voronoi partition via stable matching, in 2020 59th IEEE Conference on Decision and Control (CDC) (IEEE, 2020). [Google Scholar]
- 56.Skibinski G., Hwang V., Ando D. M., Daub A., Lee A. K., Ravisankar A., Modan S., Finucane M. M., Shaby B. A., Finkbeiner S., Nrf2 mitigates LRRK2- and α-synuclein–induced neurodegeneration by modulating proteostasis. Proc. Natl. Acad. Sci. U.S.A. 114, 1165–1170 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bates D., Mächler M., Bolker B., Walker S., Fitting linear mixed-effects models Usinglme4. J. Stat. Softw. 67, (2015). [Google Scholar]
- 58.X. Glorot, Y. Bengio, in AISTATS, Y. W. Teh, D. M. Titterington, Eds. (JMLR.org, 2010), vol. 9, pp. 249–256.
- 59.Chambers S. M., Fasano C. A., Papapetrou E. P., Tomishima M., Sadelain M., Studer L., Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9