Abstract
The SWI/SNF family of chromatin-remodeling complexes facilitates gene activation by assisting transcription machinery to gain access to targets in chromatin. This family includes BAF (also called hSWI/SNF-A) and PBAF (hSWI/SNF-B) from humans and SWI/SNF and Rsc from Saccharomyces cerevisiae. However, the relationship between the human and yeast complexes is unclear because all human subunits published to date are similar to those of both yeast SWI/SNF and Rsc. Also, the two human complexes have many identical subunits, making it difficult to distinguish their structures or functions. Here we describe the cloning and characterization of BAF250, a subunit present in human BAF but not PBAF. BAF250 contains structural motifs conserved in yeast SWI1 but not in any Rsc components, suggesting that BAF is related to SWI/SNF. BAF250 is also a homolog of the Drosophila melanogaster Osa protein, which has been shown to interact with a SWI/SNF-like complex in flies. BAF250 possesses at least two conserved domains that could be important for its function. First, it has an AT-rich DNA interaction-type DNA-binding domain, which can specifically bind a DNA sequence known to be recognized by a SWI/SNF family-related complex at the β-globin locus. Second, BAF250 stimulates glucocorticoid receptor-dependent transcriptional activation, and the stimulation is sharply reduced when the C-terminal region of BAF250 is deleted. This region of BAF250 is capable of interacting directly with the glucocorticoid receptor in vitro. Our data suggest that BAF250 confers specificity to the human BAF complex and may recruit the complex to its targets through either protein-DNA or protein-protein interactions.
The regulation of gene expression requires the participation of ATP-dependent chromatin-remodeling complexes to destabilize nucleosome structures and allow the binding of transcriptional factors to chromatin (26, 35, 47). All such complexes contain a SWI2/SNF2-type ATPase, which enables each particle to utilize the energy of ATP hydrolysis to alter the structures of nucleosomes. The complexes from multiple species, including Saccharomyces cerevisiae, Drosophila melanogaster, mice, and humans have been discovered and have been implicated in not only transcription but also other cellular processes, such as chromatin assembly and DNA replication.
Chromatin-remodeling complexes can be divided into several families, one of which is exemplified by SWI/SNF, first discovered in the yeast Saccharomyces cerevisiae (5, 11). It is required for optimal expression of several genes, including those involved in the mating-type switching and sucrose fermentation pathways (29, 36, 37). SWI/SNF is closely related to the other chromatin-remodeling complex from yeast, Rsc (7). They have two identical subunits and have at least four other homologous ones (3). Both possess similar ATP-dependent chromatin-remodeling activities in vitro. But despite these similarities, the two yeast complexes have distinct functions. They appear to regulate different sets of genes (3, 8, 16, 41). In addition, Rsc is essential for mitotic growth of yeast, whereas SWI/SNF is not (7).
In Drosophila, one complex containing several components homologous to either SWI/SNF or Rsc has been identified (34). This complex, named BAP or brahma complex, is required for proper expression of developmentally important genes, such as homeotic and segmentation genes (15, 17, 39). The complex also antagonizes the wingless signaling pathway during fly development (40). Flies with BAP inactivated die early during embryogenesis.
In mammals, the complexes of the SWI/SNF family are highly divergent and present in multiple forms (1, 27, 32, 45, 46). We and others have previously purified two different human complexes, BAF (also called hSWI/SNF-A) and PBAF (hSWI/SNF-B) (45; Xue et al., submitted for publication). They possess either BRG1 or hbrm, two different ATPases related to yeast SWI2/SNF2 and STH1 (the ATPases of SWI/SNF and Rsc, respectively) (25, 28). Each complex consists of about 10 subunits (referred to as BAFs for BRG1 or hbrm-associated factors), most of which are orthologous to those of yeast SWI/SNF or Rsc (45, 46, 50). However, the subunits characterized so far are equally similar to components of both yeast complexes, making it impossible to establish which human complex is related to yeast SWI/SNF or Rsc. Here we show that BAF250, a subunit present only in BAF but not PBAF, contains structural motifs conserved in yeast SWI/SNF but not Rsc, suggesting that BAF is more related to SWI/SNF. We provide evidence that BAF250 may act as a targeting subunit that recruits BAF to its chromatin loci through either protein-DNA or protein-protein interactions.
MATERIALS AND METHODS
Complex purification and cloning of BAF250.
The purification of BAF (hSWI/SNF-A) and PBAF (hSWI/SNF-B) complexes has previously been described (45). Briefly, BAF was immunoaffinity purified with a BRG1 antibody from the 0.5 M phosphocellulose fraction. This fraction contains BAF250 but very little BAF180. Most of the BAF180s and PBAFs fractionate in the 0.75 M phophocellulose fraction and were isolated from this fraction using the BRG1 antibody column. The complex on antibody beads was either used directly for functional assays or was eluted with glycine (0.1 M, pH 2.5) for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or immunoblot analysis. For microsequencing, the BAF complex was purified using a cell line that stably expresses hemagglutinin (HA)-tagged BAF57 (44). BAF250 was separated from the other BAFs by preparative SDS-PAGE. The band corresponding to BAF250 was excised and microsequenced by Edman degradation as described previously (44). Nine peptide sequences were obtained and were then used to search the NR and dbEST databases at the National Center for Biotechnology Information with the BLAST algorithm. Six peptide sequences matched those of brain protein B120 and several expressed sequence tags (ESTs). The associated DNAs were used as probes to screen a human cDNA library derived from Jurkat T cells. Several overlapping cDNA clones were obtained; these were completely sequenced and used to assemble the full-length BAF250 cDNA. Sequence alignment was performed using the MacDNASIS 3.1 software (Hitachi Software Engineering Co. Genetic Systems).
A rabbit polyclonal antibody was raised against a fusion protein containing maltose-binding protein (MBP; New England Biolabs) and amino acid residues 968 to 1144 of BAF250. The fusion protein was expressed in Escherichia coli and purified according to the manufacturer's protocol. The antibody was affinity purified and used for immunoblotting and affinity purification as described previously (44).
For immunoprecipitation with GR antibody, whole-cell extracts were prepared from 2305 cells as described previously (18). Extracts were incubated with an anti-GR antibody (BUGR2) or an antibody to E2F1 (Santa Cruz Biotechnology; sc-251) for 1 h and then with protein A-agarose (Santa Cruz Biotechnology) for an additional hour. The immunocomplexes were pelleted, washed three times in phosphate-buffered saline, and boiled for 5 min in 2× SDS loading buffer. Proteins were separated by SDS–8% PAGE, transferred to polyvinylidene difluoride membranes, and immunoblotted with anti-GR and anti-BAF250 antibodies.
For Northern blotting analysis, a BAF250 cDNA fragment from nucleotides 4500 to 5020 was used as a hybridization probe on human multiple-tissue RNA blots (Clontech). Hybridization used ExpressHyb hybridization solution (Clontech) according to the manufacturer's protocols.
Plasmid construction.
BAF250 full-length cDNA was cloned into mammalian expression vector pCI-neo (Promega). The region encoding amino acid residues 975 to 1149 was removed to make a BAF250 mutant with its AT-rich DNA interaction (ARID) domain deleted (BAF250-ΔARID). In C-terminal deletion mutant BAF250-ΔC, the region containing amino acid residues 1637 to 2285 was removed. A construct expressing the fusion protein containing glutathione S-transferase (GST) and the C-terminal region of BAF250, GST-250-C, was constructed by cloning a fragment encoding amino acid residues 1669 to 2171 into bacterial expression vector pGST-His-T1 (Pharmacia).
GST pull-down analysis.
The fusion protein GST-B250-C was expressed in E. coli strain BL21 at 22°C. The protein was induced by addition of IPTG (isopropyl-β-d-thiogalactopyranoside; 0.3 mM) for 4 h. The 35S-labeled GR protein was produced using expression vector pSG5-rGR (kindly provided by K. Yamamoto) and the TNT in vitro transcription and translation system (Promega) according to the manufacturer's protocols. The GR protein obtained was tested for its interaction with the GST-B250-C protein in the presence or absence of dexamethasone (DEX; 10 μM) as described previously (14). Briefly, the GST-B250-C fusion protein or GST itself was loaded onto glutathione-Sepharose beads. The beads were then incubated with 35S-labeled GR. After being washed five times with the binding buffer, the bound products were further eluted with SDS loading buffer and analyzed by SDS-PAGE, followed by autoradiography. The buffer used for the binding reaction contained 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.01% NP-40, 0.1 mM phenylmethylsulfonyl fluoride, 10% glycerol, 1 mM dithiothreitol, 2 mg of bovine serum albumin/ml, and the protease inhibitor cocktail (Sigma).
Cell culture and transfection.
T47D cells were maintained in accordance with instructions from the American Type Culture Collection. Transfection was performed using the DO-TOP Lipofectin reagent (Roche). Briefly, 0.5 μg each of GR expression vector (RSV-GR) and a luciferase reporter [(GRE)3-IL2-LUC] were transfected together with plasmids expressing wild-type or mutant BAF250 (5 μg), as indicated in the figures. The reporter plasmid contained the interleukin-2 minimal promoter fused to three tandem repeats of glucocorticoid response element (GRE). After 24 h, DEX was added to the final concentration of 10−7 M. The cells were harvested 20 h later, and the luciferase assay was performed as described previously (45).
Gel shift analysis.
The DNA fragment containing the pyrimidine-rich element from the δ-globin promoter, δ99, was kindly provided by A. Bank (31). Reactions were carried out as previously described (30), with minor changes. The 10-μl reaction mixture contained 60 mM KCl, 25 mM HEPES (pH 7.6), 1 mM dithiothreitol, 1 mM EDTA, 5% glycerol, and 0.1 ng of 32P-labeled (δ99) fragment. The reaction was at 4°C for 30 min with 0.5 to 1.0 μg of recombinant proteins or 200 to 500 ng of purified complex. Poly(dI-dC), poly(dA-dT), poly (dG-dC), or an unrelated DNA fragment from mouse rhodopsin promoter (bp −551 to −514) was sometimes included in the mixture as a competitor, as indicated in the figures. The mixture was analyzed by PAGE using a gel of either 5% (acrylamide-to-bisacrylamide ratio was 30:1) for recombinant proteins or 3.9% (acrylamide-to-bisacrylamide ratio was 60:1) for the BAF complex.
Chromatin-remodeling assay.
The mononucleosome disruption assay has been previously described (33, 44).
Nucleotide sequence accession number.
The 7,697-bp full-length cDNA obtained in this study has been assigned GenBank accession no. AF231056.
RESULTS
Purification and cloning of BAF250.
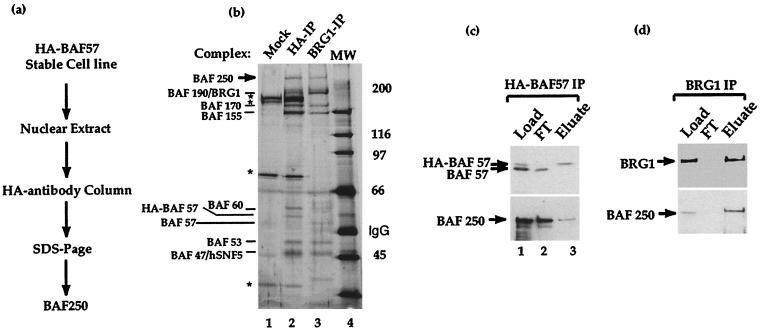
We have previously described the purification of two distinct human chromatin-remodeling complexes, BAF and PBAF, from two different fractions of nuclear extract by using an antibody against BRG1 (45). Both complexes have many common subunits. A 250-kDa protein (referred to as BAF250), however, was observed only in the human BAF, not the PBAF, complex. We have since developed a cell line that stably expresses BAF57 tagged with the HA epitope. It is highly convenient to purify the BAF complex from this cell line using the monoclonal antibody against the HA epitope (44) (Fig. 1a). A 250-kDa protein was similarly detected from the complex purified using this method (Fig. 1b, compare lanes 2 and 3). This protein was separated from the other subunits of the complex by SDS-PAGE and subjected to microsequencing by Edman degradation.
FIG. 1.
Purification of BAF250, the signature subunit of human SWI/SNF family-related BAF complex. (a) Schematic diagram of the purification procedure for BAF250. (b) Silver-stained SDS-polyacrylamide gel of human SWI/SNF family-related BAF complex purified by an antibody against HA-tagged BAF57 (lane 2). The complex purified by anti-BRG1 antibody was used as a control (lane 3). The polypeptides bound to both antibody columns are indicated. Several contaminating polypeptides (∗) were present in the preparation, as shown by a mock purification using the parent cell line that lacks the tagged BAF57 (lane 1). (c and d) Immunoblot of the load, flowthrough (FT), and eluate fractions from the anti-HA-tagged BAF57 (c) or anti-BRG1 antibody (d) column.
Nine peptide sequences were obtained from microsequencing and used to search the databases for matches. Six of them matched several human ESTs as well as a known protein in the database, brain protein B120 (38). Although B120 was thought to be a cytoplasmic protein of 120 kDa, we noticed that the published B120 cDNA sequence contains an unspliced intron at its 5′ end and a sequencing error, resulting in a frameshift at its 3′ end. In addition, B120 harbors a DNA-binding domain conserved in yeast SWI1 (see below), suggesting that it may be a partial sequence of a nuclear protein. We therefore used BAF120 cDNA as a probe to screen a human cDNA library to recover additional cDNA clones. The new clones obtained were sequenced and used to assemble a full-length cDNA of 7,697 bp. It encodes a predicted product of 2,285 amino acids containing all nine peptides obtained from microsequencing (Fig. 2a). There is an in-frame stop codon before the predicted start codon, suggesting that the cDNA sequence is full length (see Fig. 7b for additional evidence). A polyclonal antibody raised against a region of the putative product recognizes the 250-kDa polypeptide in BAF complexes purified with both the HA antibody and the BRG1 antibody (Fig. 1c and d), indicating that the cDNA encodes BAF250.
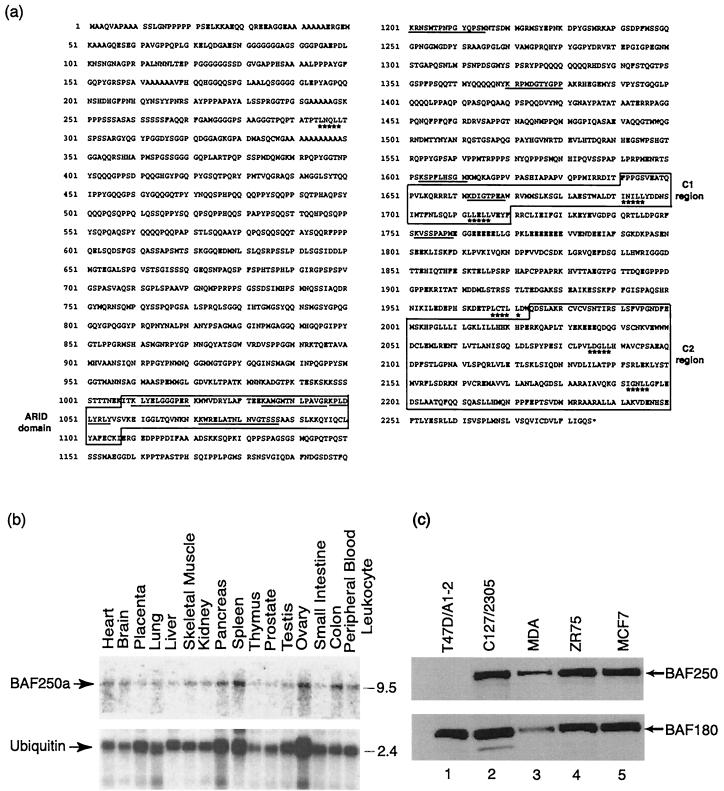
FIG. 2.
BAF250 protein and its expression pattern. (a) Predicted amino acid sequence of BAF250. The underlined sequences indicate the nine peptides obtained from microsequencing the BAF250 protein. Boxed regions, ARID domain, the C1 region, and the C2 region, which are homologous to those of Drosophila Osa. Starred amino acid residues, predicted LXXLL motifs. (b) Northern blot analysis of RNA from different human tissues probed with BAF250 (top) or ubiquitin (bottom). Each tissue is indicated at the top. The molecular size markers (right) are in kilobases. (c) Immunoblot analysis of BAF250 in several human and mouse breast cancer cell lines, as indicated at the top. The analysis of BAF180 is also shown for comparison.
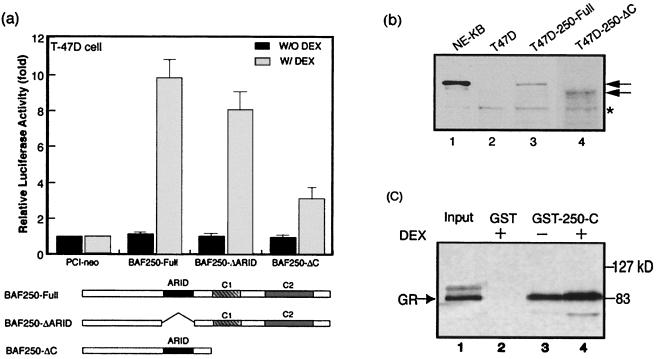
FIG. 7.
The C-terminal region of BAF250 directly interacts with GR. (a) Graph showing that deletion of the C-terminal region of BAF250 decreases its ability to facilitate GR-dependent gene activation. The assays were performed in the presence (shaded bars) or absence (black bars) of DEX (10−7 M). Schematic diagrams of the full-length BAF250 and deletion mutants are shown below. (b) Immunoblot analysis of extract from T47D cells transfected with full-length BAF250 or a C-terminal deletion mutant. The arrows mark each protein. A polypeptide cross-reactive with the antibody (∗) was used as a loading control. The nuclear extract from KB cells was the positive control (lane 1). (c) Autoradiograph showing that the C-terminal region of BAF250 directly interacts with GR in a GST pull-down assay. Either GST alone or GST fused to the C-terminal region of BAF250 (amino acid residues 1670 to 2137) was used. The presence or absence of DEX (10−7 M) is indicated. The 35S-labeled GR protein was produced using an in vitro transcription and translation system.
BAF250 was found widely expressed in different human tissues by Northern blot analysis (Fig. 2b). This is in keeping with the results for other subunits of the human BAF complex (44, 46). Immunoblot analysis with the BAF250 antibody detected a single protein of about 250 kDa in cell extracts of several cell lines (Fig. 2c). Interestingly, BAF250 was missing in breast cancer cell line T47D, which was used for functional studies of BAF250 (see below).
BAF250 belongs to the same family as Drosophila Osa/eyelid and yeast SWI1.
BAF250 was used to search databases of different organisms. In the completed genome of D. melanogaster, the top homolog of the BAF250 gene was identified as Osa (also called eyelid; probability, 10−68), a gene required for proper expression of homeotic and segmentation genes (40, 42). Osa also antagonizes the wingless signaling pathway in the early development of flies (40). BAF250 and Osa share three highly conserved regions. One is an ARID domain, which has also been found in several gene products, including yeast SWI1 (Fig. 3a and b). The other loci are located near the C terminus and were termed C1 and C2 (Fig. 3a, c, and d). The C2 region of Osa was previously noted to be homologous to two human EST sequences (40), one of which (held2) is derived from BAF250. In addition to these conserved regions, the N-terminal regions of BAF250 and Osa also resemble each other in that both are rich in glutamine, proline, and alanine. Osa has previously been shown to interact with the Drosophila SWI/SNF family-related brahma complex (10, 42). Our result is therefore consistent with the observations for flies.
FIG. 3.
Several regions of BAF250 are conserved in Drosophila Osa and yeast SWI1. (a) Schematic representation of BAF250 and its orthologs from several organisms. Each box represents a conserved region. Arrows, predicted LXXLL motifs. Regions rich in glutamines (Q), prolines (P), alanines (A), and asparagines (N) are underlined. Note that the two predicted C. elegans ORFs are next to each other on the chromosome and could be a single gene. (b) Alignment of the ARID domain of BAF250 with related sequences from other proteins. Proteins are listed in order of the similarity of sequences in them to those of the ARID domain of BAF250. Dark shading, residues conserved in different α-helices (H1 to H8) and β-sheets (B1 and B2) according to solution structure of the ARID domain of Dead ringer (24); light shading, residues identical in more than three proteins; stars, amino acid residues interacting with DNA. Abbreviations: D, Drosophila; M, mouse; H, human; C, C. elegans; SP, S. pombe; SC, S. cerevisiae. The ARID domains from Dead ringer and Bright can bind specific DNA sequences. (c and d) Alignment of C1 and C2 regions of BAF250 and its orthologs. Shaded amino acid residues are conserved in two or more proteins. Stars, LXXLL motifs. Note that the conserved regions from yeast genes are shorter than those from other species.
In the genome of S. cerevisiae, the gene encoding the top ortholog of BAF250 was found to be the SWI1 gene. The ARID domain and C1 and C2 regions are all present in SWI1, and their arrangement is as in BAF250 and Osa. The overall BLAST score is low (probability, 10−3), and each homologous region is shorter than those in humans or flies, but there is only one other gene in the entire yeast genome encoding a protein that contains an ARID domain, and this protein completely lacks C1 and C2 regions. Thus, SWI1 is the closest relative of BAF250 in yeast. It is interesting to note that SWI1 is one of few SWI/SNF subunits that have no homologous counterparts in Rsc. None of the Rsc components possesses an ARID domain (B. Cairns, personal communication). Therefore, our data further suggest that the human BAF complex is more related to yeast SWI/SNF than to Rsc.
We have further identified members of the BAF250/Osa/SWI1 family in several other organisms (Fig. 3a). In the fission yeast Schizosaccharomyces pombe (whose genome is 70% sequenced), two orthologs have been identified (probabilities, 10−7 and 10−3). Both contain the ARID domain and the C1 region (Fig. 3a to c), but the C2 region is only present in one of them. In the completed Caenorhabditis elegans genome, no single open reading frame (ORF) was found to encode all three domains present in the BAF250 family of proteins. We have noticed, however, that two adjacent putative ORFs (C01G8.8 and C01G8.7) encode an ARID domain in one and the C1 and C2 regions in the other. Both ORFs are transcribed in the same direction, making it a possibility that these two ORFs are actually the same gene. Alternatively, the gene encoding the BAF250 ortholog may have been split into two genes in the nematode.
BAF250 is an integral component of human BAF.
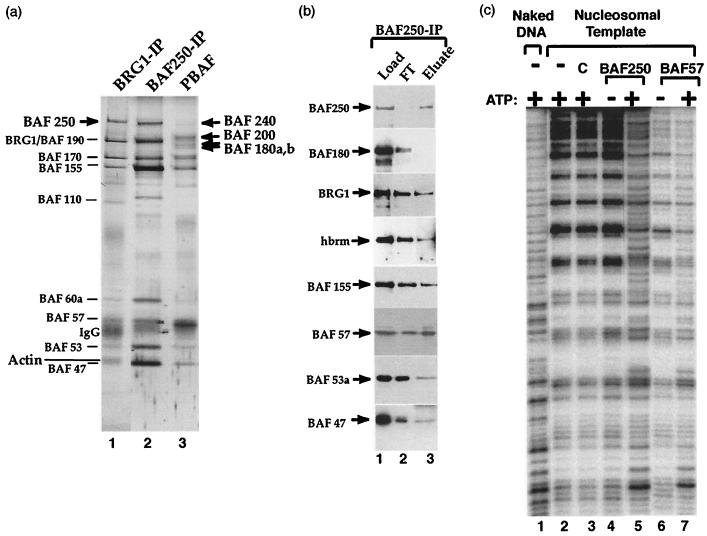
We utilized a BAF250 antibody to immunopurify the associated complex from the fractionated nuclear extract. The complex recovered exhibits a mobility pattern on the SDS-PAGE gel similar to that of the BAF complex purified with BRG1 antibody (Fig. 4a, compare lanes 1 and 2). Immunoblotting confirmed that the BAF250-associated complex contains each subunit from human BAF that was tested (Fig. 4b), indicating that BAF250 is an intrinsic subunit of the BAF complex. We noticed that BAF110, a polypeptide previously observed in complexes isolated by BRG1 and hSNF5 antibodies, is not present in the complex obtained with the BAF250 antibody (Fig. 4a). This polypeptide is similarly absent in the complex isolated with the BAF57 antibody (44). It remains to be determined whether BAF110 is a loosely associated subunit or a contaminant.
FIG. 4.
BAF250 distinguishes human BAF complex from PBAF. (a) Silver-stained SDS-PAGE gel of human BAF complex purified by anti-BRG1 (lane 1) or anti-BAF250 antibody (lane 2). The PBAF complex is shown as a comparison (lane 3). Lines, subunits shared by both complexes; arrows, components unique to each complex. (b) Western blotting of the load, flowthrough (FT), and eluate fractions from the BAF250 antibody column. (c) Autoradiograph showing the mononucleosome disruption activity by human BAF complex purified with a BAF250 antibody (lanes 4 and 5). The results for the BAF complex purified with a BAF57 antibody are also shown for comparison (lanes 6 and 7). The templates and the complexes used in each reaction are shown at the top. The presence (+) or absence (−) of ATP is indicated. C, control in which antibody beads without the complex loaded was tested.
The BAF250-associated complex was also found to possess an ATP-dependent mononucleosome disruption activity similar to that of the BAF complex purified by the BAF57 antibody (Fig. 4c). In this assay, a 176-bp fragment of 5S ribosomal DNA containing a nucleosome-positioning sequence was assembled into a rotationally phased mononucleosome. Digestion of this nucleosome by DNase I produces a characteristic 10-bp ladder on denaturing PAGE gel (Fig. 4c, compare lanes 1 and 2). The BAF complex purified with the BAF250 antibody (lanes 4 and 5) strongly disrupts the 10-bp ladder in the presence but not in the absence of ATP. The disrupted pattern is similar to that generated by the BAF complex purified with BAF57 antibody (lanes 6 and 7). The data provide further evidence that BAF250 is an integral component of the human BAF complex.
BAF250 distinguishes two similar human chromatin-remodeling complexes.
We recently demonstrated that the human PBAF complex contains idiosyncratic subunit BAF180. The PBAF complex isolated with BAF180 antibody completely lacks BAF250, consistent with the identification of PBAF and BAF as two separate complexes (Xue et al., submitted). Consistent with this notion, the BAF complex purified by BAF250 displayed a pattern on the SDS-polyacrylamide gel different from that displayed by the PBAF complex (Fig. 4a, compare lanes 2 and 3). In particular, it lacked the 180-kDa band. Immunoblotting confirmed that the BAF complex has no detectable level of BAF180 (Fig. 4b).
We have also found that, of the two human SWI/2/SNF2 family-related ATPases, the PBAF complex contains only BRG1 but not hbrm (Xue et al., submitted). In contrast, the BAF complex isolated by BAF250 can include either BRG1 or hbrm (Fig. 4b). These data underscore the distinctness of the two human complexes and suggest that BAF250 is a signature subunit that may confer specificity to the BAF complex.
The ARID domain of BAF250 interacts with the pyrimidine-rich element at the human β-globin locus.
In addition to BAF, two other chromatin-remodeling complexes, ERC-1 and PYR, have been purified from mammalian erythroid cells. ERC-1 was purified based on its activity to stimulate transcription of β-globin in vitro (1). PYR was isolated because of its ability to bind a pyrimidine-rich element termed δ99, which is located upstream of the δ-globin gene and which is required for the fetal-to-adult globin gene switching (32). The polypeptide compositions of all three complexes appear to be similar based on SDS-PAGE analysis, with each complex containing a component of 250 kDa. These complexes also share at least four subunits. In addition, the BAF complex can substitute for ERC-1 in an in vitro assay (1). This leaves open the possibility that some of these complexes could be identical. We therefore investigated whether the BAF complex can bind the pyrimidine-rich element of the δ-globin gene through the ARID domain of BAF250.
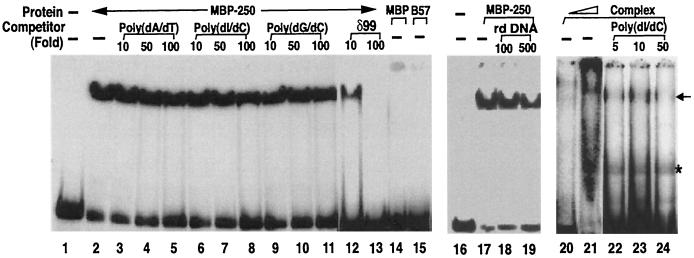
A fusion protein containing MBP and the ARID domain of BAF250 (MBP-250) was found to bind the δ99 fragment containing the pyrimidine-rich element in a gel mobility shift assay (Fig. 5, lanes 1 and 2). This binding should be due to the ARID domain of BAF250 because MBP itself does not bind the same DNA (lane 14). The interaction appears to be specific, because a 100-fold excess of the unlabeled δ99 fragment completely abolished the binding (lanes 12 and 13), whereas the same amount of poly(dA-dT), poly(dI-dC), or poly(dG-dC) has very little effect (lanes 3 to 11). Competition using an unrelated DNA fragment also has very little effect (lanes 16 to 19). The finding that poly(dA-dT) competes with an efficiency similar to those of the other two polymers implies that the BAF250 ARID domain has no preference for an AT-rich sequence.
FIG. 5.
The ARID domain of BAF250 specifically binds the pyrimidine-rich element from the β-globin loci. (a) Autoradiograph showing the results of a gel mobility shift assay for the recombinant BAF250 ARID domain and other proteins as indicated. A DNA fragment containing the pyrimidine-rich element (δ99) located between fetal γ-globin and adult β-globin genes was used as a probe (31). A recombinant protein containing the ARID domain of BAF250 fused to MBP (MBP-250) or MBP alone was tested. B57, HMG domain of BAF57, which can bind the four-way junction DNA (44). Several types of unlabeled DNA were used as competitors (lanes 3 to 13). Rd DNA, 41-bp DNA fragment from the mouse rhodopsin promoter region. The BAF complex purified with HA antibody was also analyzed (lanes 20 to 24). Arrow, specific complex formed between the complex and the probe. A contaminant band (∗) which appears to be derived from preparation of the probe was detected.
The entire BAF complex was also found to bind the δ99 fragment to form a specific complex (lane 20), implying that the ARID domain of BAF250 may be one of the motifs in the BAF complex that could recruit it to a specific DNA sequence. But when the amount of the BAF complex was increased to higher levels, most of the DNA-BAF complex formed aggregates and no longer entered the gel (lane 21). The addition of poly(dI-dC) effectively competed away the aggregates but not the specific complex, suggesting that the aggregates are most likely formed through nonspecific interactions between the BAF complex and the probe. The BAF complex is known to contain at least two additional DNA-binding domains, the HMG domain of BAF57 and the AT-hook motif of BRG1, that can interact with DNA with low or no sequence specificity (2, 44). It is therefore not surprising that BAF can interact with the probe in a sequence-nonspecific way through other DNA-binding domains. We found that the HMG domain of BAF57 does not bind the δ99 fragment (lane 15), hinting that the AT-hook motif of BRG1 might be the one that interacts with the δ99 fragment nonspecifically.
BAF250 may facilitate transcriptional activation by the glucocorticoid receptor through direct protein-protein interaction.
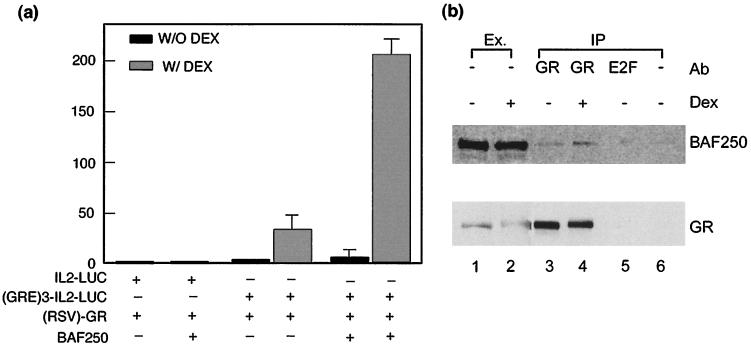
The SWI/SNF family-related chromatin-remodeling complexes have been demonstrated to be required for GR-mediated transcriptional activation in humans as well as yeast (when GR is ectopically expressed) (18, 28, 43, 45, 48). Two subunits of the human BAF complex, hbrm and BRG1, have been shown to stimulate GR-dependent gene activation in transient transfection assays of human cell lines that lack these two proteins. We noted that BAF250 was absent in human breast cancer cell line T47D (Fig. 2c). By using the same transfection assay for this cell line, BAF250 was found to be able to enhance GR-mediated transcriptional activation by about sixfold (Fig. 6a), an effect comparable to that due to stimulation by hbrm and BRG1. This enhancement depends on the presence of GRE, because the promoter that lacks GRE was not stimulated by BAF250. BRG1 and BAF155 can be coimmunoprecipitated with GR in a ligand-dependent manner from nuclear extract (18). Using the same protocol, BAF250 was similarly found to be coimmunoprecipitated by a GR antibody (Fig. 6b). The amount of BAF250 associated with GR was increased in the presence of glucocorticoid (compare lanes 3 and 4). As a negative control, immunoprecipitation with an unrelated antibody or protein A alone yielded very little BAF250 (lanes 5 and 6). These results suggest that BAF250 is involved in GR-mediated gene activation.
FIG. 6.
BAF250 facilitates glucocorticoid receptor-dependent transcriptional activation. (a) Graph showing that BAF250 enhances glucocorticoid receptor-mediated gene expression in transient cotransfection assays. The assays were performed in the presence (W/ DEX) or absence (W/O DEX) of DEX (10−7 M). The luciferase reporter plasmids contained either interleukin-2 minimal promoter (IL2-LUC) or the same promoter fused with three GR binding sites upstream [(GRE)3-IL2-LUC]. They were cotransfected with expression vectors for GR (RSV-GR) and BAF250 into T47D cells that lacked endogenous BAF250. An empty expression vector was used as a negative control for BAF250 (bars with − for BAF250). The graph represents the averages of luciferase activities from three independent assays. (b) Immunoblot analysis of the immunoprecipitates obtained with an anti-GR antibody (Ab) from lysates of C127/2305 cells. Cells were either untreated (lanes 1 and 3) or treated with DEX for 1 h (lanes 2 and 4). Immunoprecipitation (IP) with an anti-GR antibody (lanes 3 and 4), an anti-E2F1 nonspecific antibody (lane 5), or no antibody (lane 6) indicates that the interaction between GR and BAF250 is specific. The nuclear extract (Ex.; lanes 1 and 2) was used as the control.
We investigated the possibility that BAF250 might interact directly with GR. We first mapped the regions of BAF250 important for GR-mediated gene activation by making several mutants in which the regions of BAF250 conserved in other species were removed. Deletion of the ARID DNA-binding domain reduced GR-dependent activation slightly compared to that produced by full-length BAF250 (Fig. 7a). However, removal of the C-terminal conserved region of BAF250 decreased its activation about 70%. The data thus suggest that the C-terminal region is important for mediating GR-dependent activation. As a control, immunoblot analysis of the extract from DNA-transfected cells showed that the C-terminal deletion mutant is expressed at a level comparable to that for wild-type BAF250 (Fig. 7b, lanes 3 and 4). The same experiment also demonstrated that the BAF250 cDNA clone isolated in this study is full length (compare lanes 1 and 3).
We then analyzed whether the C-terminal region of BAF250 can directly associate with GR in a GST pull-down experiment. Briefly, GR was produced in an in vitro transcription-translation system. A protein resulting from a fusion between GST and the C-terminal region of BAF250 was expressed as a recombinant protein in E. coli. The fusion protein (Fig. 7c, lane 3), but not GST alone (lane 2), can bring down GR, suggesting a direct interaction between GR and the BAF250 C-terminal region. The presence of the glucocorticoid enhances the interaction by about threefold (compare lanes 3 and 4). These results are similar to those observed for interactions between several nuclear receptors and their partners (9, 22). It should be pointed out that GR contains both ligand-dependent and ligand-independent activation domains. It is possible that BAF250 could interact with one or both of these domains. Indeed, the yeast SWI/SNF has recently been shown to interact with the ligand-independent domain of GR (43). Future work will be needed to test for an analogous mechanism for BAF.
DISCUSSION
This study attempts to address three questions regarding the human BAF chromatin-remodeling complex. (i) Is BAF more closely related to SWI/SNF or Rsc? (ii) What are the biochemical markers that distinguish BAF from other related complexes? (iii) Does the BAF250 subunit play a targeting role for the complex? We demonstrated that BAF250, a specific component of the human BAF complex, contains structural motifs conserved in yeast SWI1 but not Rsc, suggesting that BAF is indeed of the SWI/SNF type. In related work, we have found that BAF180, a specific subunit of PBAF, possesses structural domains present only in yeast Rsc, not in SWI/SNF, suggesting that PBAF is of the Rsc type (Xue et al., submitted). Therefore, BAF and SWI/SNF represent one subfamily of evolutionarily conserved complexes, whereas PBAF and Rsc represent the other subfamily. Below, we discuss these findings in more detail.
BAF250 provides specific structural features for human BAF.
The two human complexes BAF and PBAF are highly similar, with as many as eight components in common (Fig. 4) (Xue et al., submitted). The only subunits unique to BAF are hbrm and BAF250. However, the PBAF complex contains hbrm homolog BRG1. These two proteins are 70% identical in amino acid sequence and have the same domain structure. Thus, hbrm is unlikely to be the subunit that provides the most distinctive feature for BAF. Rather, the only apparent subunit that might be a specific marker for BAF is BAF250.
In comparison, yeast SWI/SNF and Rsc have two identical and at least four homologous subunits (3, 7). SWI/SNF has five other components, SWI1, SNF6, SNF11, TFG3, and SWP82, which are not homologous to any Rsc components. Among them, SNF6, SNF11, and SWP82 have no significant matches in the completed databases of Drosophila and C. elegans (data not shown; B. Cairns, personal communication), suggesting that these proteins are not conserved in higher eucaryotes. TFG3 has two different human orthologs, ENL and AF9 (4), but neither of them has been detected in human BAF (unpublished data). Because yeast tfg3 mutants do not display swi or snf phenotypes, TFG3 is probably not a required subunit of SWI/SNF. The data and the analyses support the notion that BAF250 and SWI1 are the only evolutionarily conserved signature subunits in this family of chromatin-remodeling complexes. They provide specific structural markers that discriminate BAF and SWI/SNF from PBAF and Rsc.
The ARID domain of BAF250 may target the BAF complex through a protein-DNA interaction.
If BAF and PBAF have distinct functions, like their yeast counterparts, the subunits unique to each complex may provide specific targeting of the particles to distinct loci. In this regard, BAF250 was noted to possess the ARID DNA-binding domain. This domain has been found in several other proteins and has been shown to be able to bind DNA in a sequence-specific manner (20, 23). In accord with previous results, we found that the ARID domain from BAF250 exhibits a strong affinity to a DNA control element from the β-globin locus (Fig. 5). This DNA element has been shown to be specifically recognized by PYR, a human SWI/SNF family-related complex sharing at least four subunits with BAF (32). Perhaps the ARID domain of BAF250 may be involved in recruiting BAF or PYR to their targets through specific protein-DNA interactions.
The ARID domain from Drosophila Osa was found to possess little sequence specificity in vitro, a result different from our finding for BAF250 (10). Although these ARID domains are conserved from humans to yeast, their DNA-binding specificities may not be conserved. The solution structures of two different ARID domains have recently been reported (24, 49). They consist of eight α-helices (H1 to H8) and a short two-stranded antiparallel β sheet (Fig. 3b). The middle six helices represent a special helix-turn-helix motif interacting with the major groove of DNA, while the β hairpins and H8 bind the minor groove. The amino acid residues within the DNA recognition helix (H6) for BAF250 and SWI1 are completely different and are only partially conserved between BAF250 and Osa (two out of nine amino acid residues are identical). The first helix (H1) of the ARID domain is present in BAF250 and Osa but is absent in SWI1. Thus, these ARID domains may have adopted different DNA-binding properties during millions of years of evolution.
In addition to the ARID domain of BAF250, both human BAF and PBAF harbor at least two other DNA-binding domains: an HMG domain in BAF57 (44) and an AT-hook motif in hbrm or BRG1 (2). Both of these domains bind to the minor groove of DNA and have low or no sequence specificity. Because they are present in both BAF and PBAF, they are unlikely to serve as the primary targeting domains to recruit each complex to its specific loci. However, they could function to provide secondary interaction sites after the complex is recruited to its target in chromatin.
While this paper was in preparation, Dallas and colleagues published the sequence of p270 (13), a protein originally identified based on its cross-reactivity to an antibody against the transcription cofactor p300 (12). They presented evidence that p270 coimmunoprecipitates with several subunits of human BAF and suggested that p270 could be BAF250. We have found that p270 is a partial sequence of BAF250 (missing 378 amino acids). They also showed that the ARID domain of p270 exhibits no apparent sequence specificity using a PCR-based binding site selection assay. This result is somewhat different from our findings. There could be many possible explanations for the discrepancy. One possibility is that the stringency in their assay may not be high enough to efficiently remove the low-affinity sequences. Alternatively, the high-affinity binding DNA may not be amplified efficiently by PCR because of its unusual sequence (TC rich).
BAF250 may mediate GR-dependent transcriptional activation through direct protein-protein interaction.
Previous studies have suggested that transcriptional activation by GR requires SWI/SNF-like complexes in yeast or humans. However, it remains unclear which subunit(s) in these complexes directly interacts with GR. Our work provides evidence that the human BAF complex can be recruited to GR through direct interaction between BAF250 and GR. First, the cotransfection of BAF250 with GR stimulates GR-dependent transcription. Second, BAF250 coimmunoprecipitates with GR in a ligand-enhanced manner. Because BAF250 is a signature subunit of BAF, the data further suggest that the SWI/SNF-related BAF complex mediates GR activation and interacts with GR. Third, the C-terminal conserved region of BAF250 directly interacts with GR in vitro. Deletion of this region also strongly reduces GR activation in the transfection assay. Careful inspection of the BAF250 sequence revealed the presence of two C-terminal regions conserved in Drosophila Osa and yeast SWI1. The same regions also include several LXXLL motifs, which are known to be able to interact directly with nuclear hormone receptors (21). Future work should further define the minimal region of BAF250 that interacts with GR.
Our working idea that BAF250 recruits BAF to GR is consistent with results from several previous investigations. First, transcriptional activation by GR requires SWI/SNF when GR is ectopically expressed in yeast (6, 48). Interestingly, genetic screens of yeast have identified SWI/SNF but not Rsc, despite the fact that SWI/SNF is present at a level at least 10-fold lower than that of Rsc (7). These data imply that subunits unique to SWI/SNF may target the complex to GR. Second, because GR is able to activate transcription in both yeast and humans, the subunit(s) interacting with GR is probably conserved through evolution. BAF250 is the only subunit in BAF that meets both of these criteria: it is a unique subunit of human SWI/SNF (BAF) and has a conserved yeast SWI/SNF homolog (SWI1). We speculate that BAF250 orthologs in other species may play similar functions in targeting their SWI/SNF-related complexes.
ACKNOWLEDGMENTS
We thank G. Crabtree and R. Tjian for their support of this project. We also thank K. Yamamoto for pSG5-rGR vector, T. Takeuchi for providing B120 cDNA, A. Bank for the pyrimidine-rich fragment, J. Wong for cell lines, A.-J. Kim and C. Fryer for assistance in experiments, and the National Cell Culture Center for providing a large quantity of cells. We are grateful to B. Cairns for communicating results prior to publication. We thank David Schlessinger for the critical reading of the manuscript.
REFERENCES
- 1.Armstrong J A, Bieker J J, Emerson B M. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- 2.Bourachot B, Yaniv M, Muchardt C. The activity of mammalian brm/SNF2α is dependent on a high-mobility-group protein I/Y-like DNA binding domain. Mol Cell Biol. 1999;19:3931–3939. doi: 10.1128/mcb.19.6.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cairns B R, Erdjument-Bromage H, Tempst P, Winston F, Kornberg R D. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol Cell. 1998;2:639–651. doi: 10.1016/s1097-2765(00)80162-8. [DOI] [PubMed] [Google Scholar]
- 4.Cairns B R, Henry N L, Kornberg R D. TFG/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol Cell Biol. 1996;16:3308–3316. doi: 10.1128/mcb.16.7.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cairns B R, Kim Y J, Sayre M H, Laurent B C, Kornberg R D. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cairns B R, Levinson R S, Yamamoto K R, Kornberg R D. Essential role of Swp73p in the function of yeast Swi/Snf complex. Genes Dev. 1996;10:2131–2144. doi: 10.1101/gad.10.17.2131. [DOI] [PubMed] [Google Scholar]
- 7.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 8.Cao Y, Cairns B R, Kornberg R D, Laurent B C. Sfh1p, a component of a novel chromatin-remodeling complex, is required for cell cycle progression. Mol Cell Biol. 1997;17:3323–3334. doi: 10.1128/mcb.17.6.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavailles V, Dauvois S, L'Horset F, Lopez G, Hoare S, Kushner P J, Parker M G. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins R T, Furukawa T, Tanese N, Treisman J E. Osa associates with the Brahma chromatin remodeling complex and promotes the activation of some target genes. EMBO J. 1999;18:7029–7040. doi: 10.1093/emboj/18.24.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cote J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 12.Dallas P B, Cheney I W, Liao D-W, Bowrin V, Byam W, Pacchione S, Kobayashi R, Yaciuk P, Moran E. p300/CREB binding protein-related protein p270 is a component of mammalian SWI/SNF complexes. Mol Cell Biol. 1998;18:3596–3603. doi: 10.1128/mcb.18.6.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dallas P B, Pacchione S, Wilsker D, Bowrin V, Kobayashi R, Moran E. The human SWI-SNF complex protein p270 is an ARID family member with non-sequence-specific DNA binding activity. Mol Cell Biol. 2000;20:3137–3146. doi: 10.1128/mcb.20.9.3137-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dingwall A K, Beek S J, McCallum C M, Tamkun J W, Kalpana G V, Goff S P, Scott M P. The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol Biol Cell. 1995;6:777–791. doi: 10.1091/mbc.6.7.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du J, Nasir I, Benton B K, Kladde M P, Laurent B C. Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics. 1998;150:987–1005. doi: 10.1093/genetics/150.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elfring L K, Daniel C, Papoulas O, Deuring R, Sarte M, Moseley S, Beek S J, Waldrip W R, Daubresse G, DePace A, Kennison J A, Tamkun J W. Genetic analysis of brahma: the Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics. 1998;148:251–265. doi: 10.1093/genetics/148.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fryer C J, Archer T K. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 19.Gametchu B, Harrison R W. Characterization of a monoclonal antibody to the rat liver glucocorticoid receptor. Endocrinology. 1984;114:274–279. doi: 10.1210/endo-114-1-274. [DOI] [PubMed] [Google Scholar]
- 20.Gregory S L, Kortschak R D, Kalionis B, Saint R. Characterization of the dead ringer gene identifies a novel, highly conserved family of sequence-specific DNA-binding proteins. Mol Cell Biol. 1996;16:792–799. doi: 10.1128/mcb.16.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 22.Heng H H, Chamberlain J W, Shi X M, Spyropoulos B, Tsui L C, Moens P B. Regulation of meiotic chromatin loop size by chromosomal position. Proc Natl Acad Sci USA. 1996;93:2795–2800. doi: 10.1073/pnas.93.7.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herrscher R F, Kaplan M H, Lelsz D L, Das C, Scheuermann R, Tucker P W. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family. Genes Dev. 1995;9:3067–3082. doi: 10.1101/gad.9.24.3067. [DOI] [PubMed] [Google Scholar]
- 24.Iwahara J, Clubb R T. Solution structure of the DNA binding domain from Dead ringer, a sequence-specific AT-rich interaction domain (ARID) EMBO J. 1999;18:6084–6094. doi: 10.1093/emboj/18.21.6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khavari P A, Peterson C L, Tamkun J W, Mendel D B, Crabtree G R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 26.Kornberg R D, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 27.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 28.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nie Z, Chen S, Kumar R, Zack D J. RER, an evolutionarily conserved sequence upstream of the rhodopsin gene, has enhancer activity. J Biol Chem. 1996;271:2667–2675. doi: 10.1074/jbc.271.5.2667. [DOI] [PubMed] [Google Scholar]
- 31.O'Neill D, Bornschlegel K, Flamm M, Castle M, Bank A. A DNA-binding factor in adult hematopoietic cells interacts with a pyrimidine-rich domain upstream from the human delta-globin gene. Proc Natl Acad Sci USA. 1991;88:8953–8957. doi: 10.1073/pnas.88.20.8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Neill D, Yang J, Erdjument-Bromage H, Bornschlegel K, Tempst P, Bank A. Tissue-specific and developmental stage-specific DNA binding by a mammalian SWI/SNF complex associated with human fetal-to-adult globin gene switching. Proc Natl Acad Sci USA. 1999;96:349–354. doi: 10.1073/pnas.96.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Owen-Hughes T, Utley R T, Cote J, Peterson C L, Workman J L. Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science. 1996;273:513–516. doi: 10.1126/science.273.5274.513. [DOI] [PubMed] [Google Scholar]
- 34.Papoulas O, Beek S J, Moseley S L, McCallum C M, Sarte M, Shearn A, Tamkun J W. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development. 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- 35.Pazin M J, Kadonaga J T. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell. 1997;88:737–740. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- 36.Peterson C L, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 37.Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 38.Takeuchi T, Chen B K, Qiu Y, Sonobe H, Ohtsuki Y. Molecular cloning and expression of a novel human cDNA containing CAG repeats. Gene. 1997;204:71–77. doi: 10.1016/s0378-1119(97)00525-8. [DOI] [PubMed] [Google Scholar]
- 39.Tamkun J W, Deuring R, Scott M P, Kissinger M, Pattatucci A M, Kaufman T C, Kennison J A. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 40.Treisman J E, Luk A, Rubin G M, Heberlein U. eyelid antagonizes wingless signaling during Drosophila development and has homology to the Bright family of DNA-binding proteins. Genes Dev. 1997;11:1949–1962. doi: 10.1101/gad.11.15.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsuchiya E, Hosotani T, Miyakawa T. A mutation in NPS1/STH1, an essential gene encoding a component of a novel chromatin-remodeling complex RSC, alters the chromatin structure of Saccharomyces cerevisiae centromeres. Nucleic Acids Res. 1998;26:3286–3292. doi: 10.1093/nar/26.13.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vazquez M, Moore L, Kennison J A. The trithorax group gene osa encodes an ARID-domain protein that genetically interacts with the brahma chromatin-remodeling factor to regulate transcription. Development. 1999;126:733–742. doi: 10.1242/dev.126.4.733. [DOI] [PubMed] [Google Scholar]
- 43.Wallberg A E, Neely K E, Hassan A H, Gustafsson J-Å, Workman J L, Wright A P H. Recruitment of the SWI-SNF chromatin remodeling complex as a mechanism of gene activation by the glucocorticoid receptor τ1 activation domain. Mol Cell Biol. 2000;20:2004–2013. doi: 10.1128/mcb.20.6.2004-2013.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang W, Chi T, Xue Y, Zhou S, Kuo A, Crabtree G R. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc Natl Acad Sci USA. 1998;95:492–498. doi: 10.1073/pnas.95.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang W, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 46.Wang W, Xue Y, Zhou S, Kuo A, Cairns B R, Crabtree G R. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 47.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 48.Yoshinaga S K, Peterson C L, Herskowitz I, Yamamoto K R. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 49.Yuan Y C, Whitson R H, Liu Q, Itakura K, Chen Y. A novel DNA-binding motif shares structural homology to DNA replication and repair nucleases and polymerases. Nat Struct Biol. 1998;5:959–964. doi: 10.1038/2934. [DOI] [PubMed] [Google Scholar]
- 50.Zhao K, Wang W, Rando O J, Xue Y, Swiderek K, Kuo A, Crabtree G R. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]